Abstract
A detailed protocol for preparing small molecule samples for microcrystal electron diffraction (MicroED) experiments is described. MicroED has been developed to solve structures of proteins and small molecules using standard electron cryo-microscopy (cryo-EM) equipment. In this way, small molecules, peptides, soluble proteins, and membrane proteins have recently been determined to high resolutions. Protocols are presented here for preparing grids of small-molecule pharmaceuticals using the drug carbamazepine as an example. Protocols for screening and collecting data are presented. Additional steps in the overall process, such as data integration, structure determination, and refinement are presented elsewhere. The time required to prepare the small-molecule grids is estimated to be less than 30 min.
Introduction
Microcrystal electron diffraction (MicroED) is an electron cryo-microscopy (cryo-EM) method for determining atomic resolution structures from sub-micrometer sized crystals1,2. Crystals are applied to standard transmission electron microscope (TEM) grids and frozen by either plunging into liquid ethane or liquid nitrogen. Grids are then loaded into a TEM operating at cryogenic temperatures. Crystals are located on the grid and screened for initial diffraction quality. Continuous rotation MicroED data are collected from a subset of the screened crystals, where the data are saved using a fast camera as a movie3. These movies are converted to a standard crystallographic format and processed almost identically as an X-ray crystallography experiment4.
MicroED was originally developed to investigate protein microcrystals1,2. A bottleneck in protein crystallography is growing large, well-ordered crystals for traditional synchrotron X-ray diffraction experiments. As electrons interact with matter orders of magnitude stronger than X-rays, the limitations of the crystal size needed to produce detectable diffraction is considerably smaller5. Additionally, the ratio of elastic to inelastic scattering events is more favorable for electrons, suggesting that more useful data can be collected with a smaller overall exposure5. Constant developments have allowed for MicroED data to be collected from the most challenging microcrystals6,7,8,9.
Recently, MicroED has been shown to be a powerful tool for determining the structures of small molecule pharmaceuticals from apparently amorphous materials10,11,12,13. These powders can come straight from a bottle of purchased reagent, a purification column, or even from crushing a pill into a fine powder10. These powders appear amorphous by eye, but may be either entirely composed of nanocrystals or merely contain trace amounts of nanocrystalline deposits in a greater non-crystalline, amorphous fraction. Application of the material to the grid is facile, and the subsequent steps of crystal identification, screening, and data collection might even be automated in the near future14. While others may use different methods for sample preparation and data collection, here the protocols developed and used in the Gonen laboratory for preparing samples of small molecules for MicroED and for data collection are detailed.
Protocol
1. Preparing small molecule samples
Transfer a small amount (0.01 - 1 mg) of powder, liquid, or solids into a small vial or tube.
-
For samples already in powder form, seal the tube using the cap until the sample is needed. Dry the liquid samples into powders prior to attempts at method 1 (step 3) or 2 (step 4).
NOTE: Samples dissolved in liquid may use method 3 (5.X) below
2. Preparing TEM grids
NOTE: Some TEMs with autoloader systems require that the grids be clipped and placed into a cassette prior to loading into the TEM column. Clipping involves physically securing the 3 mm TEM grid into a metal ring that the autoloader can manipulate. This step and subsequent steps can be performed using either normal TEM grids, or TEM grids that have been clipped. For these experiments, it is often easier to manipulate the grids if they have been clipped ahead of time.
Wrap plastic film around one end of a glass cover slide.
Place the TEM grids onto the film on the top of the cover slide with the carbon side facing up. Identify the two sides of the grid under a light. The copper side shines and appears metallic, whereas the carbon side appears a drab, brown color (Figure 1C,D). For clipped grids, the carbon side should face the flat face of the clip ring.
Place the slide with grids into the glow discharge chamber. Glow discharge the coverside for approximately 30 s using the negative setting at 15 pA. Store the grids on the cover slide inside of a glass Petri dish lined with filter paper prior to adding sample to the grids.
Figure 1: Preparation of a pre-clipped TEM grid for small molecule investigation.
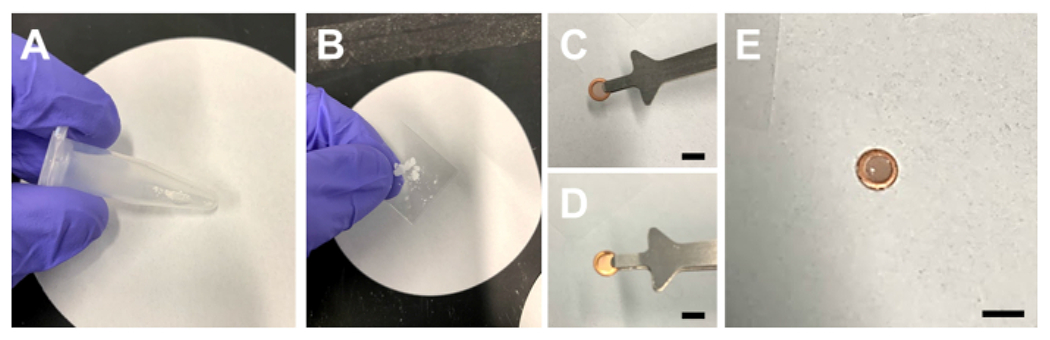
(A) A tube with a small portion of sample for investigation. (B) Crushed sample between two microscope slides. (C) The carbon side of the pre-clipped grid, and (D) the copper side of the pre-clipped grid. (E) A pre-clipped TEM grid after the crushed powder has been dropped onto it. Scale bars 3mm in (C), (D), and (E). Please click here to view a larger version of this figure.
3. Applying sample to grids by creating a homogenous fine powder (Method 1)
Remove a glow discharged TEM grid from the covered Petri dish using tweezers. Place the grid onto a circular filter paper with the carbon side facing up.
Using a small spatula, remove a very small scoop of powder (approximately 0.1 mg) and place it onto a small, square, glass coverslip just next to the TEM grid on the filter paper. Place another small square glass slide or coverslip on top of the powder.
With fingers, gently rub the two glass slides together to make a fine powder.
Angle the coverslips and position them just above the TEM grid on the filter paper and continue to rub the coverslips together, just a few cm above the glow discharged TEM grid (Figure 1).
Observe to see if the powder is falling towards the grid. Uncover the finely ground powder by removing one of the two glass coverslips. Gently brush the fine powder off of the coverslip using a piece of filter paper onto the TEM grid (Figure 1).
4. Applying sample to grids by applying the “shaking” method (Method 2)
Grab a grid using a pair of tweezers and drop it into the vial or tube of powder sample. Close the vial using the plastic cap to ensure no material will escape when shaken.
Grabbing the vial in the hand, shake the vial such that the powder and grid are both moving for approximately 10 - 30 s.
-
Empty the vial contents onto a circular filter paper. Grabbing the grid on an edge, gently tap the grid edge on the filter paper to remove any excess material from the grid.
NOTE: Depending on the type of vial and size, one may also use tweezers to remove the TEM grid without emptying the contents.
5. Applying sample to grids using the evaporation method (Method 3)
Place a grid (clipped or not) onto the center of a circular piece of filter paper with the carbon side facing up and the copper side facing down.
Using a 10 μL gas-tight syringe, apply a small drop (approximately 1 - 3 μL) of dissolved compound onto the carbon side of the grid.
Gently move the filter paper with grids into a vacuum desiccation chamber. Cover the chamber and turn on the vacuum. Leave the grids under vacuum to dry for up to one day.
Turn off the vacuum and allow the chamber to vent for 5 min. Venting the chamber avoids the grids moving or flying away when it is uncovered.
6. Freezing and loading grids into the TEM
Grab the edge of the TEM grid using a set of tweezers, assuring that the tips do not puncture any of the grid squares. Lift the grid 1 - 2 cm above the filter paper and angle the grid at 90° to the paper below. Gently tap the tweezers while keeping the grid firmly tweezed to remove any loose powder.
Freeze the grid by moving the tip of the tweezers with the grid directly into a liquid nitrogen container by hand. This container is typically the grid loading station for the TEM, but can be any safe container, such as a liquid nitrogen safe thermos, is acceptable for transferring or storage. Liquid nitrogen is −196 °C.
Wait until the grid and tweezers stop boiling before further manipulations.
Under liquid nitrogen or in nitrogen vapors, place the grid in the sample holder with the carbon side oriented such that the sample will be hit by the beam prior to the carbon support film.
-
Load the sample holder into the TEM assuring that the grid is kept at liquid nitrogen temperatures at all times.
For autoloader systems, place the clipped grids into a cassette in a liquid nitrogen cooled container. This cassette transferred into a shuttling container than allows the autoloader robotics to accept the cassette while keeping the samples safe for shuttling between the autoloader and the column.
For side-entry TEM setups, secure the grid to a commercial side-entry TEM holder. These holders have a sample preparation container that is filled with liquid nitrogen to allow transfer of the grid to the holder without warming the sample. The side-entry holder is inserted into the TEM directly with the sample secured at the end.
7. Collecting MicroED data
-
Locating and screening nanocrystals
Open the TEM column valves. Adjust the magnification using the hand panels to the lowest magnification possible. Find the beam by adjusting the intensity knob on the hand panels such that a round, bright area is visible on the fluorescent screen.
Take an all-grid atlas at a low magnification (50 - 300x) using appropriate software14,15,16 (Figure 2). Ensure that the microscope is well aligned for both low and high magnification imaging prior to collecting high-resolution MicroED data.
Identify grid squares without broken carbon and have a visible black or dark material/grains on the film (Figure 2). Navigate around the grid, either physically using the joystick on the hand panels, or virtually on the collected Atlas, in order to search for grid squares that are not broken and contain microcrystals.
Add the center of each of these squares to a list of grid locations for investigation. These locations can be added to a notebook, in the microscope user interface, or in microscope automation software.
Increase the magnification to 500-1,300x and adjust the eucentric height at each stored grid location and update the saved Z value to the positions noted in 7.1.4.
Search either on the fluorescent screen or on a fast camera at this higher magnification for small black spots/grains on the grid. A good sample with often have sharp edges at high magnification, suggesting crystalline order.
-
Move a located potential crystal to the center of the screen and increase the magnification such that the TEM enters high magnification mode.
NOTE: This is referred to as either high-magnification, Zoom2, or SA mode on different instruments and typically corresponds to magnifications of 3,000x or higher.
Insert a selected area aperture. Change the aperture to a larger or smaller size to assure that the selected area is just larger than the crystal (Figure 3).
-
Switch to diffraction mode by pressing the diffraction button on the TEM hand panels, assuring that the fluorescent screen is inserted. Adjust the camera length using the magnification knob such that the edge is at least 1 Å resolution.
NOTE: Calibration of the diffraction lengths should be performed by a service engineer using a gold waffle grating or known specimen prior to attempting MicroED experiments.
Adjust the diffraction focus such that the central spot is as sharp and small as possible. Using the diffraction shift knobs, move the central beam to the center of the fluorescent screen
Insert the beam stop and make sure the beam is behind it. Lift the fluorescent screen. Take a short (approximately 1 s) exposure on the camera (Figure 4).
Inspect the corresponding diffraction pattern. A good candidate for collecting a full dataset will have sharp spots that are regularly arranged in columns and rows, and the diffraction will extent to beyond 2 Å, preferably at least to 1 Å (Figure 4). Save the crystal coordinates in either the TEM user interface or by writing them down.
Repeat diffraction screening for all the potential crystals of interest on the current grid square.
-
Data collection
Center a screened crystal on the screen at a high magnification (> 1,000x). Adjust the eucentric height of the crystal using either an automatic routine or by hand.
Insert the selected area aperture that best fits the crystal size and shape as determined in 7.1.8 (Figure 3). Tilt the stage in the negative and positive directions until the image is occluded by the grid bars (Figure 3). Note these angles for data collection purposes.
Determine the number of frames that will be required to span the total angular wedge of the dataset. It is typical to use 0.5 - 1.0° wedges for each frame, with frames being read out every 1 to 5s depending on the sensitivity of the camera. For example, a wedge spanning from −30° to +30°, rotating at a constant rate of 1° s−1 with a frame read out every 1s, would require 60 total frames.
-
Considering the maximum total tilt range of −72° to +72°, begin rotating the stage at a constant rate of 1° s−1 and then begin reading the data out every 1s on a modern camera with a rolling shutter readout mode (Movie 1). This process can be performed manually or by using TEM specific software.
-
Set the rotation rate by specifying the % of the maximum tilt speed and when to stop in the microscope user interface or dedicated software. In this investigation, this was measured to be 0.3° s−1 for each % of maximum speed but will need to be independently verified for each microscope.
NOTE: The tilting and camera data collection are set up independently here, but software programs14 can coordinate this to simplify the process.
Discard approximately 1° on each side of the specified wedge in most cases as dead time, where the stage is either ramping up or slowing down to the desired rotation speed.
-
Save data in a variety of formats as either individual frames or as a stack of images (Movie 1).
Convert these diffraction frames to a typical crystallographic format using MicroED tools - available here (https://cryoem.ucla.edu). Diffraction images saved in SMV format are readily processed using documented crystallographic software17,18.
Figure 2: Identification of small molecule crystals in the TEM.
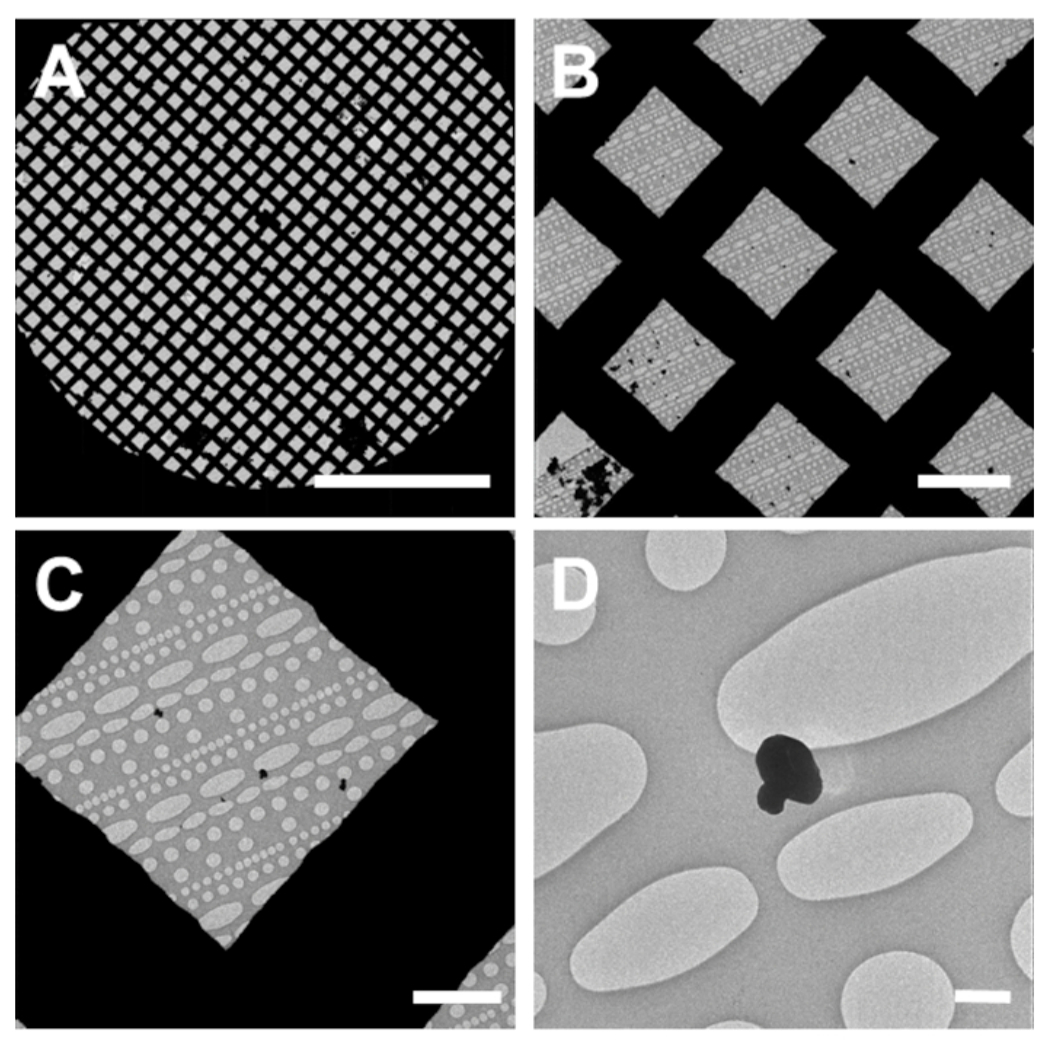
(A) An all-grid atlas or montage at low magnification. (B) A single low magnification image used for screening. (C) Higher magnification image used to identify smaller grains. (D) High magnification micrograph of a clumped small molecule crystal. Scale bar 750 μm in (A), 50 μm in (B), 10 μm in (C), 1 μm in (D). Please click here to view a larger version of this figure.
Figure 3: Screening and aligning microcrystals for MicroED data collection.
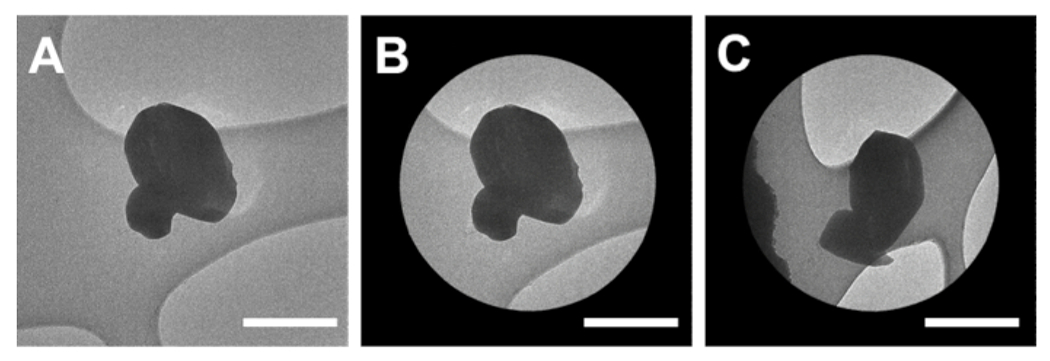
(A) High magnification micrograph of a microcrystal. (B) Micrograph of the isolated crystal within the selected area aperture. (C) The same microcrystal in the aperture with the stage tilted to −69 °. Scale bars all 1 μm. Please click here to view a larger version of this figure.
Figure 4: Examples of MicroED data.
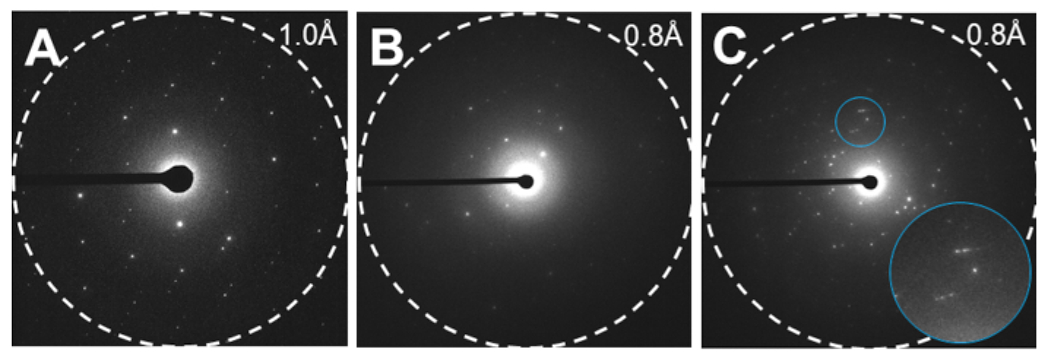
(A) high quality MicroED data with clear, sharp spots suitable for high-resolution structure determination. (B) Weak, smeared MicroED data with poor lattice definition. In this example the alignment is also off so the diffraction is only apparent on one side of the image (C) Poor MicroED data showing multiple lattices and split and/or smeared spots. Inset of blue area enhanced in bottom right showing smeared, split spots. Please click here to view a larger version of this figure.
Representative Results
MicroED is a cryoEM method that leverages the strong interactions between electrons and matter, which allows for the investigation of vanishingly small crystals12,13. After these steps, it is expected to have a diffraction movie in crystallographic format collected from microcrystals (Movie 1). Here, the technique is demonstrated using carbamazepine12. The results show a continuous rotation MicroED dataset from a carbamazepine microcrystal identified on a TEM grid (Movie 1). A good dataset has strong, clear spots that are not smeared or split, and has only a single lattice on each frame that can easily be followed by stepping through the movie19. These data are easily indexed, integrated, and scaled using standard X-ray crystallography software4. Split spots can be seen from a crystal that has been cracked, and two orientations of the same crystal are closely aligned, but not quite coincident19. Multiple lattices can also occur, particularly for these small-molecule crystals, where multiple single crystals have stuck together in a clump on the grid. Another common scenario occurs where the crystals have been frozen incorrectly or treated too harshly during fragmentation, and no diffraction is observed9.
After data collection, integration, and structure solution, it is expected that a high-resolution structure is determined (Figure 5). Obtaining a clear structure solution will ultimately depend on the quality and completeness of the data.
Figure 5: MicroED structure of Carbamazepine.
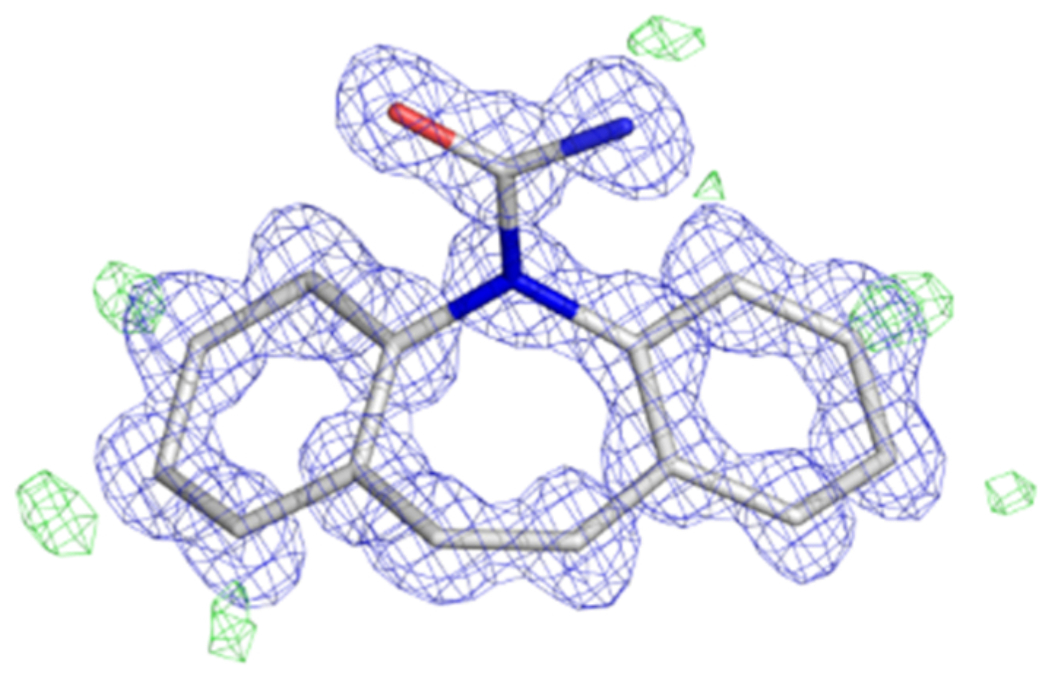
Atomic model shown as sticks with carbon atoms colored white, oxygen red, and nitrogen blue. The 2Fo-FC map is contoured at the 1.5 σ level and colored blue. The Fo-Fc map showing hydrogens is contoured at the 3.0 σ level and colored green. This figure was adapted from the deposited maps of EMDB-928410. Please click here to view a larger version of this figure.
Discussion
Sample preparation is typically an iterative process, where optimizations are made after sessions of screening and data collection. For small-molecule samples, it is often prudent to first attempt grid preparation without glow-discharging the grids, since many pharmaceuticals tend to be hydrophobic10,11. If the grids have too few nanocrystalline deposits, it is a good idea to try again after first glow-discharging the grids. It may be the case that the crystals from lyophilized powders are too large and thick to collect good data. In these cases, it may be possible to collect data from an edge or thinner part of a larger crystal. If this proves difficult, grinding the powder down to a finer consistency using a rougher surface, such as a mortar and pestle may be necessary.
MicroED data are typically collected with the TEM operating in microprobe mode4,20. Here, the size of the TEM beam that corresponds to an exposure rate of 0.01 e− Å−2 s−1 is typically around 10 μm in diameter, which is much larger than the typical microcrystals1,21. The signal is then isolated from the crystals of interest using a selected area aperture (Figure 3)2,20. Various aperture sizes allow for quick adjustment of the setup to varying sizes of crystals. Alternatively, it is possible to collect data with the TEM operating in nano probe mode. This reduces the size of the beam by approximately a factor of 5. A smaller beam corresponds to a commensurately higher exposure rate in the beam footprint. Since many TEMs are two condenser lens systems, the parallel condition will dictate that the beam be a single size in either microprobe or nano probe mode. Reaching an exposure rate of 0.01 e− Å−2 s−1 in nano probe without adjusting the gun lens is challenging. The choice between the two is up to the user. An advantage of nano probe is that there is less of a need to insert and retract the selected area aperture between screening in imaging and diffraction modes of operation. However, with modern microscopes insertion and retraction of the SA aperture is automatic and accurate. Microprobe offers larger flexibility in isolating diffraction by having access to multiple sizes of selected area apertures. The larger beam in microprobe may also expose nearby crystals, whereas nano probe can more precisely target individual crystals.
The presented protocol is the standard approach to MicroED data collection for small molecules used in our laboratory10,11,12,13. There are many adaptations and modifications that could be implemented. The best approach to making grids with high crystal density is most dependent on the familiarity of the user with a given approach. There are many cases where drugs are present as large crystals that are too fragile to physically fragment without losing diffracting power19. In these cases, the recently adapted method of focused ion-beam milling to thin the crystals to make them more accessible to MicroED6,7,8,9,22.
Supplementary Material
Acknowledgments
The Gonen lab is supported by funds from the Howard Hughes Medical Institute. This study was supported by the National Institutes of Health P41GM136508.
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/62313.
Disclosures
The authors have nothing to disclose.
References
- 1.Shi D, Nannenga BL, ladanza MG, Gonen T Three-dimensional electron crystallography of protein microcrystals. eLife. 2, e01345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nannenga BL, Shi D, Leslie AGW, Gonen T High-resolution structure determination by continuous-rotation data collection in MicroED. Nature Methods. 11 (9), 927–930 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hattne J, Martynowycz MW, Penczek PA, Gonen T MicroED with the Falcon III direct electron detector. IUCrJ. 6 (5), 921–926 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hattne J et al. MicroED data collection and processing. Acta Crystallographica Section A Foundations and Advances. 71 (4), 353–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderson R The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Quarterly Reviews of Biophysics. 28 (2), 171–193 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Martynowycz MW et al. MicroED structure of the human adenosine receptor determined from a single nanocrystal in LCP. BioRxiv. 316109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martynowycz MW, Zhao W, Hattne J, Jensen GJ, Gonen T Collection of continuous rotation MicroED data from ion beam-milled crystals of any size. Structure. 27 (3), 545–548.e2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martynowycz MW, Gonen T Ligand incorporation into protein microcrystals for MicroED by on-grid soaking. Structure. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martynowycz MW, Khan F, Hattne J, Abramson J, Gonen T MicroED structure of lipid-embedded mammalian mitochondrial voltage-dependent anion channel. Proceedings of the National Academy of Sciences. 117 (51), 32380–32385 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones CG et al. The CryoEM method MicroED as a powerful tool for small molecule structure determination. ACS Central Science. 4 (11), 1587–1592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dick M, Sarai NS, Martynowycz MW, Gonen T, Arnold FH Tailoring tryptophan synthase TrpB for selective quaternary carbon bond formation. Journal of the American Chemical Society. 141 (50), 19817–19822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher-Jones M et al. Sub-ångström cryo-EM structure of a prion protofibril reveals a polar clasp. Nature Structural & Molecular Biology. 25 (2), 131–134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ting CP et al. Use of a scaffold peptide in the biosynthesis of amino acid-derived natural products. Science. 365 (6450), 280–284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Cruz MJ, Martynowycz MW, Hattne J, Gonen T MicroED data collection with SerialEM. Ultramicroscopy. 201, 77–80 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. Journal of Structural Biology. 152 (1), 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Schorb M, Haberbosch I, Hagen WJH, Schwab Y, Mastronarde DN Software tools for automated transmission electron microscopy. Nature Methods. 16 (6), 471–477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabsch W XDS. Acta Crystallographica Section D Biological Crystallography. 66 (2), 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winter G et al. DIALS: Implementation and evaluation of a new integration package. Acta Crystallographica Section D. 74 (2), 85–97 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de la Cruz MJ et al. Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED. Nature Methods. 14 (4), 399–402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi D et al. The collection of MicroED data for macromolecular crystallography. Nature Protocols. 11 (5), 895–904 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nannenga BL, Shi D, Hattne J, Reyes FE, Gonen T Structure of catalase determined by MicroED. eLife. 3, e03600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martynowycz MW, Zhao W, Hattne J, Jensen GJ, Gonen T Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED. Structure. 27 (10), 1594–1600.e2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


