Abstract
In order to assess the microbial water quality in canal waters throughout the Florida Keys, a survey was conducted to determine the concentration of microbial fecal indicators and the presence of human pathogenic microorganisms. A total of 19 sites, including 17 canal sites and 2 nearshore water sites, were assayed for total coliforms, fecal coliforms, Escherichia coli, Clostridium perfringens, enterococci, coliphages, F-specific (F+) RNA coliphages, Giardia lamblia, Cryptosporidium parvum, and human enteric viruses (polioviruses, coxsackie A and B viruses, echoviruses, hepatitis A viruses, Norwalk viruses, and small round-structured viruses). Numbers of coliforms ranged from <1 to 1,410, E. coli organisms from <1 to 130, Clostridium spp. from <1 to 520, and enterococci from <1 to 800 CFU/100 ml of sample. Two sites were positive for coliphages, but no F+ phages were identified. The sites were ranked according to microbial water quality and compared to various water quality standards and guidelines. Seventy-nine percent of the sites were positive for the presence of enteroviruses by reverse transcriptase PCR (polioviruses, coxsackie A and B viruses, and echoviruses). Sixty-three percent of the sites were positive for the presence of hepatitis A viruses. Ten percent of the sites were positive for the presence of Norwalk viruses. Ninety-five percent of the sites were positive for at least one of the virus groups. These results indicate that the canals and nearshore waters throughout the Florida Keys are being impacted by human fecal material carrying human enteric viruses through current wastewater treatment strategies such as septic tanks. Exposure to canal waters through recreation and work may be contributing to human health risks.
The islands that form the Florida Keys are adjacent to the only living coral reef system within the territorial borders of the continental United States. The island chain extending from Key Largo to Key West supports a significant population of human residents and vacationers and is well known as a premier recreational site. With the exception of a few of the smaller islands within this region, the development of residential communities and vacation resorts has been continuous over the last 20 years. The community on Key West is the only population served by a full-scale sewage treatment plant and outfall disposal. Two other communities, Key Colony Beach on Marathon and Ocean Reef Resort on Key Largo, are served by full-scale treatment plants and injection well disposal. All other island communities rely on septic tanks, cesspools and package treatment plants combined with injection wells for disposal of sewage. Currently, there are approximately 30,000 septic tanks and 600 injection wells utilized in the Keys (20, 33). The depths of the injection wells vary from ∼1 to 30 m, with current regulations requiring a drill depth of 27.4 m with casing to 18.3 m (33). Due to the porous nature of limestone (which makes up the strata of the islands), coupled with natural physical dynamics of the region such as flushing from precipitation and tidal pumping (33), these types of sewage disposal practices may be inadequate for the protection of water quality from microbial pollutants. Several studies conducted in the Florida Keys have demonstrated the movement of contaminants from septic tanks and injection wells to the surrounding marine environment. One study conducted in the lower Keys demonstrated elevated nutrient levels in areas where septic tanks were being utilized, compared to levels in areas outside the influence of septic tanks (20). Studies in the upper Keys demonstrated the presence of fecal indicator bacteria in surface waters and the subsurface aquifer, with levels of microbes decreasing with distance from areas under the influence of septic tanks (26). In the same area, viral tracers moved within 11 h from a seeded septic tank out into the surrounding environment (25). Similar tracer studies conducted in the middle Keys demonstrated an 8-h migration of viral tracers from a seeded injection well into the surrounding groundwater and a 53-h migration to surface waters located on the opposite side of the island (24).
One of the concerns of recreational water use is the risk of illness resulting from exposure to waters contaminated by human sewage. The risk to human health associated with contaminated marine water has been well documented. In a study conducted along beaches in Hong Kong, swimmers were at higher risk of illness than nonswimmers, and there was a greater risk at beaches affected by pollution than at beaches considered unpolluted (7). Children swimming in contaminated seawater are more likely to develop illnesses than those who do not (2). A dose-response relationship between the level of indicator organisms in recreational marine waters and the risk of illness has been reported (11).
The U.S. Environmental Protection Agency (USEPA) has suggested an ambient water quality goal for Escherichia coli of 126 CFU/100 ml for freshwater environments. The USEPA guidance level for marine waters is a geometric mean of 35 enterococcal CFU/100 ml for five samples spaced over a 30-day period (9). The single-sample guidance level of 104 enterococcal CFU/100 ml of sample has been suggested. Fujioka and Shizumura (12) have suggested that Clostridium spp. (<50 CFU/100 ml) are a better indicator for tropical waters. The State of Florida Department of Health (DOH) ambient water quality standards for total coliforms are a geometric mean of 1,000 CFU/100 ml, a maximum single-sample level of 2,400 CFU/100 ml, and a level of <1,000 CFU/100 ml for 80% of samples. The DOH ambient water quality standards for fecal coliforms are a geometric mean of 200 CFU/100 ml, a maximum single-sample level of 800 CFU/100 ml, and a level of <400 CFU/100 ml for 90% of samples.
Total and fecal coliform bacterial indicators often do not indicate the persistence of pathogens, especially viruses in surface waters. Human viral and protozoal pathogens are more persistent in these waters than are coliform bacteria and are not removed as efficiently by treatment processes such as chlorination (17). In addition, total and fecal coliforms can readily be isolated in tropical waters from areas far removed from human activity and thus are not adequate indicators of fecal contamination and human health risks (29). Simultaneous monitoring of water samples for alternate indicators, F-specific (F+) RNA coliphages, enterococci, and Clostridium perfringens, as well as direct pathogen monitoring for enteroviruses and enteric protozoa (Cryptosporidium and Giardia spp.), enables better assessment of fecal contamination and public health risks. Protocols are now available for the detection of F+ RNA coliphages (18) and enteroviruses, enabling the determination of the fecal source as human or animal. A number of researchers have used enteroviruses to assess water quality (1, 8, 13, 19, 22, 34).
In order to make improvements to wastewater management in the Florida Keys, a better understanding is needed of the sources of microbial contaminants (human versus animal); their transport, prevalence, and fate in the marine environments; and the resulting public health risks. This study was specifically designed to assess microbial water quality in the canal systems of the residential communities of the Florida Keys.
MATERIALS AND METHODS
Sites.
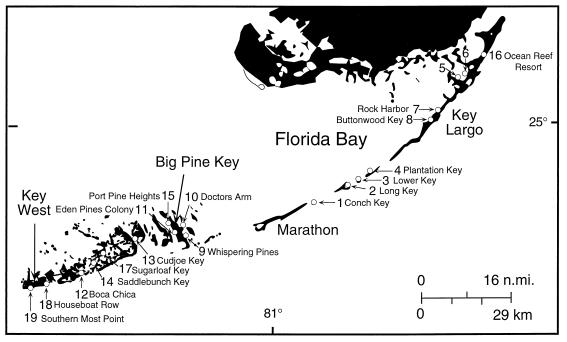
Nineteen sites from the northern portion of Key Largo (Ocean Reef Resort) to Southern Most Point in Key West (Table 1 and Fig. 1) were sampled. Seventeen of the sites were residential canals identified by the USEPA as sites of suspected poor water quality. Two of the sites were nearshore water sites. Sites 1 through 8 were sampled once between 29 September 1997 and 4 October 1997, and sites 9 through 19 were sampled once between 16 August 1998 and 22 August 1998. Table 1 lists each sample site and its location.
TABLE 1.
Sample sites
| Sample identification | Site location | Mile marker |
|---|---|---|
| EPA-Keys 1 | Conch Key, Seaview Ave., canal, Gulf side at bridge | 63 |
| EPA-Keys 2 | Long Key, Layton Dr., canal across the street from the Florida Keys Marine Laboratory house | 69 |
| EPA-Keys 3 | Lower Matecumbe Key, past Sandy Cove Ave., Captains Cove | 75 |
| EPA-Keys 4 | Plantation Key, Venetian Shores, Palo-De Oro Dr., canal at empty lot | 86 |
| EPA-Keys 5 | Key Largo, Tropical Ln., mobile home park off a boat ramp, innermost part of the canal | 103 |
| EPA-Keys 6 | Key Largo, Sexton Cove Estates, corner of Sexton Cove Rd. and Grassy Rd., off end of canal and mobile home park | 106 |
| EPA-Keys 7 | Rock Harbor, Jolly Roger Dr., Buccaneer Point, private boat ramp | 99 |
| EPA-Keys 8 | Buttonwood Bay, bayside boat ramp just past mile marker 97 | 97 |
| EPA-Keys 9 | Big Pine Key, Whispering Pines subdivision, at the very end of the canal off of Gordon Dr. | 30 |
| EPA-Keys 10 | Big Pine Key, Doctors Arm subdivision, first canal off of W. Ortega Ln., corner lot | 30 |
| EPA-Keys 11 | Big Pine Key, Eden Pines Colony, canal off of W. Shore Dr., corner lot | 30 |
| EPA-Keys 12 | Boca Chica Key, Boca Chica Ocean Shores, at the end of the canal off of Scopio Ln. | 10 |
| EPA-Keys 13 | Cudjoe Key, Cudjoe Gardens, canal behind the sheriff’s station | 21 |
| EPA-Keys 14 | Saddlebunch Keys, Bay Point subdivision, E. Circle Dr., first canal on the right | 15 |
| EPA-Keys 15 | Big Pine Key, Port Pine Heights, the canal at Kyle Blvd. and Driftwood St. | 30 |
| EPA-Keys 16 | Key Largo, Ocean Reef Resort, the canal behind the chapel, near the bridge | NAa |
| EPA-Keys 17 | Sugarloaf Key, Sugarloaf Shores, the canal at W. Bonita Ln. and Sugarloaf Blvd. | 17 |
| EPA-Keys 18 | Key West, Houseboat Row | 0 |
| EPA-Keys 19 | Key West, near Southern Most Point, at the seawall located on the end of Simonton St. | 0 |
NA, not applicable.
FIG. 1.
Florida Keys sample site locations.
Detection protocols.
Grab samples were collected by using sterile 2-liter bottles or were concentrated by cartridge filtration (for large-volume analysis of protozoa and viruses) and vortex flow filtration (collected with sterile 20-liter carboys) concentrates (27).
Total coliforms.
Volumes of 50.0, 5.0 (grab samples), and 1.0 ml (vortex flow filtration retentate) of each water sample were filtered through membrane filters (0.45 μm [pore size] and 47 mm; Gelman Sciences). Each volume was assayed in duplicate. The filters were placed on mENDO medium and incubated for 24 h at 37°C. The colonies that produced a metallic sheen were enumerated as total coliforms according to the Standard Methods for the Examination of Water and Wastewater (2a).
Fecal coliforms and E. coli.
Water samples were filtered as described above. The filters were placed on M-FC medium and sealed in plastic bags within 30 min of filtration. The plates were incubated for 24 h in a water bath at 44.5°C. The bacterial colonies with various shades of blue were counted as fecal coliform bacteria (34). The filters with blue colonies were then transferred to EC-MUG medium and incubated at 35.5°C for 24 h. At 24 h the filters were exposed to UV light, and the fluorescing colonies (E. coli) were enumerated.
Enterococci.
Water samples were filtered as described above. The filters were placed on MEI medium and incubated at 41.0°C. After 24 h of incubation, enterococci showed pink or red colonies on the membrane filters. The colonies, which develop a black or reddish-brown precipitate on the underside of the filter, were counted as enterococci (28).
C. perfringens.
Water samples were filtered as described above. The filters were placed on the M-CP plates and sealed with anaerobic gas packs (BBL GasPak; Becton Dickinson). After 24 h of incubation at 45.0°C, the yellow colonies were exposed to ammonium hydroxide fumes; colonies that turned red or dark pink were enumerated as C. perfringens (5).
Protozoan analysis (Cryptosporidium spp. and Giardia spp.).
Samples were processed and assayed for enteric protozoa by filtration and immunofluorescence microscopy techniques (10). Between 50.0 and 220.0 liters were collected from each site by filtration through filter cartridges. Volumes were monitored by attached flow meters. After collection, the filters were placed on ice for transport to the University of South Florida, where they were processed by cutting the filter and washing the collected material from the filter to recover the protozoan cysts and oocysts. The eluent was centrifuged to a concentrated pellet representing the initial volume of water collected. An aliquot of concentrated pellet was then clarified by using Percoll-sucrose gradient centrifugation. The final concentrates were examined by using an indirect antibody-fluorescein isothiocyanate epifluorescence assay. Equivalent concentrations of cysts and oocysts per 100 liters were then calculated.
PCR detection of enteroviruses.
Approximately 110 liters of water was filtered at each site by using Filterite filters (DFN 0.45-10UN; Filterite/MEMTEC A. Corp., Timonium, Md.) as outlined in Standard Methods for the Examination of Water and Wastewater (2a). Viruses were eluted with beef extract (pH 9.5) and concentrated by using organic flocculation. The concentrates were stored at −20°C until analysis. Portions (100 μl) of each sample were purified and concentrated to 60.0 μl by using spin-column chromatography (RNEasy Mini Kit; Qiagen, Santa Clarita, Calif.). Then, 10 μl of this sample was utilized for reverse transcriptase PCR (RT-PCR) for each group of viruses assayed. The primer sets and biotinylated oligonucleotide probes (Table 2) used for viral detection included a set for the detection of 25 different enteroviruses, a set for the detection of hepatitis A viruses, a set for the detection of Norwalk viruses (31), and a set for the detection of small round-structured viruses (SRSVs) (4). Biotinylated probes were synthesized at the University of Florida DNA Synthesis Laboratory (Gainesville). RT-PCR profiles and master mixes were used as previously published per respective primer groups and sets, i.e., separate profiles and master mixes for the Schwab et al. (31) and Ando et al. (4) assays. Detection of PCR product included gel electrophoresis (gels are stained with 0.5 μg of ethidium bromide per ml and visualized by using UV light) and chemiluminescent dot blotting (Southern Star Chemiluminescent Detection System for Biotin-Labeled Probes, version A.2; Tropix, Inc., Bedford, Mass.).
TABLE 2.
Human virus primer sets and probe sequences
| Virus | Primer and probe sequences | Amplicon and target |
|---|---|---|
| Panenterovirus | Upstream, 5′-CCTCCGGCCCTGAATG-3′ | 197-bp highly conserved 5′ untranslated region |
| Downstream, 5′-ACCGGATGGCCAATC-3′ | ||
| Probe, 5′-TACTTTGGGTGTCCGTGTTTC-3′ | ||
| Hepatitis A virus | Upstream, 5′-CAGCACATCAGAAAGGTGAG-3′ | 192-bp VP1 and VP2 capsid protein interphase |
| Downstream, 5′-CTCCAGAATCATCTCCAAC-3′ | ||
| Probe, 5′-TGCTCCTCTTTATCATGCTATG-3′ | ||
| Norwalk | Upstream, 5′-CAAATTATGACAGAATCCTTC-3′ | 260-bp viral polymerase |
| Downstream, 5′-GAGAAATATGACATGGATTGC-3′ | ||
| Probe, 5′-ATGTCATCAGGGTCAAAGAGG-3′ | ||
| SRSV (Ando) | Upstream, 5′-TGTCACGATCTCATCATCACC-3′ | 123-bp RNA polymerase region |
| Downstream, 5′-TGGAATTCCATCGCCCACTGG-3′ | ||
| Probes, 5′-ATGTCAGGGGACAGGTTTGT-3′, 5′-ATGTCGGGGCCTAGTCCTGT-3′, 5′-ACATCGGGTGATAGGCCTGT-3′ |
Nonspecific coliphage assay.
Aliquots (1.0 ml) of grab samples and sample concentrates were assayed with a lawn of E. coli ATCC 15597 by using a standard overlay technique. Our experience with E. coli ATCC 15597 has demonstrated that this strain is a sensitive host for the detection of DNA and RNA coliphages. The plates were allowed to solidify and were then incubated at 35.0°C for 24 h. Plaques were then enumerated as total coliphage.
Genotyping F+ RNA coliphages by using nucleic probes.
We used a modified assay adapted from that of Hsu et al. (18). Aliquots (1.0 ml) of grab samples and sample concentrates were assayed with a lawn of E. coli Famp by using a standard overlay technique. After an overnight incubation at 35.0°C, plaques were picked with a Pasteur pipette and suspended in 1.0 ml of 0.5 M Tris (pH ∼8.0). Plaques were then verified by spotting 10.0 μl of the Tris-plaque suspension onto a fresh lawn of E. coli Famp by using a standard medium and 10.0 μl onto a lawn of E. coli Famp by using medium containing RNase (to differentiate RNA from DNA coliphages). Isolates that resulted in plaques on the standard-medium plate and did not produce plaques on the plates containing RNase were picked again by using a Pasteur pipette. A total of 10 agar plugs were picked from the cleared zone and suspended in a buffer containing 537.0 μl of 20× SSC (1 liter of 20× SSC is 175.3 g of NaCl plus 88.2 g of sodium citrate [pH 7.0]), 495.0 μl of 37% (wt/wt) formaldehyde (Fisher Scientific F79-500), and 400.0 μl of filter-sterilized water which had been irradiated with UV light. These suspensions were then incubated at 65.0°C for 30.0 min. Aliquots of 335.0 μl (four samples, one for each virus probe group) were then applied to 0.45-μm-pore-size nylon filter paper by using a dot blot apparatus. Viral RNA was fixed to the filters by using UV light and hybridized with the appropriate probe. The probes (Table 3) for the detection of group II and group III F+ RNA coliphages (found predominantly in human feces) and for the detection of group I and group IV F+ RNA coliphages (found predominantly in animal feces) were used for hybridization and detection. As with the enterovirus protocol, chemiluminescence was used for target and probe detection.
TABLE 3.
F+ RNA coliphage probe data
| Virus | Group | Probe sequence | Target region |
|---|---|---|---|
| MS2 | I | 5′-CTAAGGTATGGACCATCGAGAAAGGA-3′ | Maturation protein |
| GA | IIa | 5′-CATGTTATCCCCCAAGTGCTGGCTAT-3′ | Maturation protein |
| IIb | 5′-GTTTTCCTTATGTTTTGCTTTCAGACCCA-3′ | ||
| QB | III | 5′-ATACTCAGTGAA(A/G)TACTGCTGTGT-3′ | 5′ Nontranslated region |
| SP/FI | IV | 5′-GGCATAGATTCTCCTCTGTAGTGCG-3′ | 5′ Nontranslated region |
Canal type and number and age of homes.
Table 4 lists the sample sites and canal descriptions (ages and numbers of homes on each canal screened; sites 1 through 17). Sites 18 and 19, which were both nearshore sites located on Key West, were not included in Table 4. There were four types of canals: (i) flowthrough canals, (ii) long and short single dead-end canals, (iii) feeder and/or side canals in a multicanal network, and (iv) main canals in a multicanal network.
TABLE 4.
Site descriptions and prokaryote ranks
| Site | Prokaryote rank | No. of homes on canal/no. of lots on canal | No. of condo and/or apt. units on canal | No. of homes built by yr (1930s to present by decade)
|
Avg age of homes | Canal typea | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 30 | 40 | 50 | 60 | 70 | 80 | 90 | ||||||
| 16 | 1 | 66/75 | 40 | 28 | 21 | 7 | 10 | 23 | FT | |||
| 6 | 17 | 17/24 | 4 | 9 | 3 | 1 | 23 | SDE | ||||
| 5 | 8 | 22/23 | 1 | 18 | 3 | 0 | 23 | SDE* | ||||
| 7 | 12 | 15/25 | 2 | 4 | 9 | 30 | SDE | |||||
| 8 | 6.5 | 260 | FT | |||||||||
| 4 | 5 | 24/32 | 1 | 9 | 11 | 3 | 18 | SDE | ||||
| 3 | 19 | 27/33 | 5 | 17 | 5 | 15 | MCN | |||||
| 2 | 9 | 7/17 | 2 | 4 | 1 | 17 | FCN | |||||
| 1 | 14 | 15/19 | 2 | 2 | 6 | 3 | 1 | 46 | SDE* | |||
| 9 | 11 | 8/16 | 4 | 2 | 1 | 1 | 26 | SDE | ||||
| 10 | 10 | 15/45 | 12 | 3 | 11 | FCN | ||||||
| 11 | 16 | 16/24 | 2 | 2 | 7 | 5 | 14 | MCN | ||||
| 15 | 13 | 3/16 | 1 | 2 | 6 | FCN | ||||||
| 13 | 3 | 10/34 | 2 | 8 | 5 | MCN | ||||||
| 17 | 2 | 9/16 | 1 | 1 | 6 | 1 | 16 | SDE* | ||||
| 14 | 6.5 | 58/85 | 12 | 14 | 15 | 13 | 4 | 26 | FT | |||
| 12 | 7 | 27/27 | 3 | 18 | 5 | 1 | 22 | FCN* | ||||
Abbreviations: FCN, feeder canal in a multicanal network; FT, flowthrough canal; MCN, main canal in a multicanal network; SDE, single dead-end canal. An asterisk indicates a canal or multicanal network of ≤100 m.
RESULTS
Indicator water quality.
Six indicators (bacteria and phages) of fecal pollution were assessed at each site (Table 5). Total coliform levels averaged 212 CFU/100 ml (the geometric mean value was 54 CFU/100 ml). None of the sites were in violation of the State of Florida ambient water quality standard. Average fecal coliform levels were 124 CFU/100 ml (geometric mean, 24 CFU/100 ml), and no single site was in violation of the State of Florida ambient water quality standard. Average E. coli concentrations were 45 CFU/100 ml, and the geometric mean was 13 CFU/100 ml. C. perfringens levels averaged 31 CFU/100 ml, and the geometric mean value was 3 CFU/100 ml. A C. perfringens level of 520 CFU/100 ml was detected at site 19, which was the only site with a level greater than the Hawaiian guidance level of <50 CFU/100 ml. Enterococcus levels averaged 120 CFU/100 ml, and the geometric mean value was 23 CFU/100 ml. Three sites (site 3, 800 CFU/100 ml; site 11, 240 CFU/100 ml; and site 19, 680 CFU/100 ml) had levels above a single-sample guidance level of 107 CFU/100 ml. Nonspecific coliphages were detected at low levels at sites 1 and 4 (10 PFU/100 ml in both cases). F+ RNA coliphages were not detected at any of the other sites.
TABLE 5.
EPA-Keys microbial grab sample data
| Site | No. of CFU or coliphages/100 mla
|
|||||
|---|---|---|---|---|---|---|
| Total coliforms | Fecal coliforms | E. coli | Clostridium spp. | Enterococcus spp. | Nonspecific coliphage | |
| 1 | 328 | 56 | 15 | 5 | 78 | 10 |
| 2 | 15 | 9 | 8 | 5 | 12 | 0 |
| 3 | 480b | 770 | 120 | 26 | 800 | 0 |
| 4 | 1b | 6 | 6 | 0 | 0 | 10 |
| 5 | 36 | 19 | 5 | 1 | 1 | 0 |
| 6 | 390 | 294 | 180 | 6 | 80 | 0 |
| 7 | 30 | 26 | 21 | 22 | 30 | 0 |
| 8 | 11 | 10 | 8 | 1 | 18 | 0 |
| 9 | 40 | 20 | 13 | 4 | 51 | 0 |
| 10 | 60 | 44 | 40 | 0 | 27 | 0 |
| 11 | 330 | 220 | 170 | 1 | 240 | 0 |
| 12 | 14 | 1 | 0 | 0 | 80 | 0 |
| 13 | 20 | 5 | 2 | 0 | 0 | 0 |
| 14 | 11 | 6 | 1 | 3 | 23 | 0 |
| 15 | 168 | 150 | 70 | 0 | 94 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 30 | 0 | 0 | 0 | 0 | 0 |
| 18 | 710 | 130 | 130 | 2 | 63 | 0 |
| 19 | 1,410 | 600 | 80 | 520 | 680 | 0 |
| Avg | 212 | 124 | 46 | 32 | 120 | 1 |
| Geometric mean | 54 | 24 | 13 | 3 | 23 | 1 |
The coliphage data had a detection limit of <10/100 ml, as only 10 ml of sample was assayed at each site (see the definition of the detection limit in the text).
Accompanied by overgrowth which prevented accurate enumeration. These values were not included in calculating the averages.
Table 6 lists the sites in order from the north end of Key Largo to Southern Most Point on Key West. The indicator (bacterium and phage) concentrations were given rankings of 1 (the lowest levels of bacteria and coliphages within each group) to 19 (the highest levels of bacteria and coliphages within each group) in order to compare the sites to each other. After each category was ranked, a total score was calculated and an overall ranking was assigned per site. Site 16, which was located within the Ocean Reef Resort (Key Largo), had the lowest ranking (best microbial water quality). Site 3, which was located on the Lower Matecumbe Key, had the highest ranking (worst microbial water quality). Site 19, which was located at Southern Most Point on Key West, had the second highest rating (rank 18).
TABLE 6.
Site rankings based on the prokaryote data (sites are listed in order from north to south)a
| Location | Site | Total coliforms | Fecal coliforms | E. coli | Clostridium spp. | Enterococcus spp. | Coliphage | Total | Rank |
|---|---|---|---|---|---|---|---|---|---|
| Ocean Reef Resort | 16 | 1 | 1.5 | 2 | 3.5 | 2 | 8.5 | 18.5 | 1 |
| Key Largo | 6 | 16 | 17 | 19 | 16 | 14 | 8.5 | 90.5 | 17 |
| Key Largo | 5 | 10 | 9 | 6 | 9.5 | 5 | 8.5 | 48 | 8 |
| Rock Harbor | 7 | 8.5 | 11 | 12 | 17 | 10 | 8.5 | 67 | 12 |
| Buttonwood Bay | 8 | 3 | 8 | 8 | 8 | 7 | 8.5 | 42.5 | 6.5 |
| Plantation Key | 4 | 2 | 5 | 8 | 3.5 | 2 | 18.5 | 39 | 5 |
| Lower Matecumbe | 3 | 18 | 19 | 16 | 18 | 19* | 8.5 | 98.5 | 19* |
| Long Key | 2 | 6 | 7 | 8 | 14.5 | 6 | 8.5 | 50 | 9 |
| Conch Key | 1 | 14 | 13 | 11 | 14.5 | 13 | 18.2 | 74 | 14 |
| Big Pine | 9 | 11 | 10 | 10 | 13 | 11 | 8.5 | 63.5 | 11 |
| Big Pine | 10 | 12 | 12 | 13 | 3.5 | 9 | 8.5 | 58 | 10 |
| Big Pine | 11 | 15 | 16 | 18 | 9.5 | 17* | 8.5 | 84 | 16* |
| Big Pine | 15 | 13 | 15 | 14 | 3.5 | 16 | 8.5 | 70 | 13 |
| Cudjoe | 13 | 7 | 4 | 5 | 3.5 | 2 | 8.5 | 30 | 3 |
| Sugarloaf | 17 | 8.5 | 1.5 | 2 | 3.5 | 2 | 8.5 | 26 | 2 |
| Saddlebunch | 14 | 4 | 6 | 4 | 12 | 8 | 8.5 | 42.5 | 6.5 |
| Boca Chica | 12 | 5 | 3 | 2 | 3.5 | 15 | 8.5 | 37 | 4 |
| Key West, Houseboat Row 1 | 18 | 17 | 14 | 17 | 11 | 12 | 8.5 | 79.5 | 15 |
| Key West, Southern Most Point | 19 | 19 | 18 | 15 | 19 | 18* | 8.5 | 97.5 | 18* |
An asterisk indicates a value that exceeds USEPA guidance levels for single sampling (enterococci). The sites are ranked according to the number of organisms found at each site. A ranking of 19 equals the highest number of pathogens. After each category was ranked (total coliforms, fecal coliforms, etc.), the rankings were totaled, and an overall ranking was assigned to each site.
Protozoa.
No protozoa were detected in any of the samples. The assay detection limits ranged from <2 cysts or oocysts/100 liters to <23 cysts or oocysts/100 liters.
Human enteric viruses.
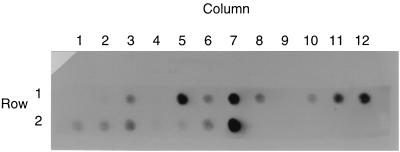
Table 7 lists the sites from north to south and includes the prokaryote and coliphage site rankings in addition to the RT-PCR human virus data. The RT-PCR virus data was reported as presence or absence. All viral data was confirmed by dot blotting. Figure 2 is a photograph of the panenteroviral dot blot. Seventy-nine percent of the sites were positive when assayed with the panenterovirus primer set. Sixty-three percent of the sites were positive for hepatitis A viruses. Ten percent of the sites were positive for Norwalk viruses. No site was positive for SRSVs.
TABLE 7.
RT-PCR virus results by site (sites are listed in order from north to south)a
| Location | Sample site | Prokaryote and coliphage ranking | Enteroviruses | Hepatitis A viruses | Norwalk viruses | SRSVs |
|---|---|---|---|---|---|---|
| Ocean Reef Resort | 16 | 1 | + | − | − | − |
| Key Largo | 6 | 17 | + | − | + | − |
| Key Largo | 5 | 8 | + | + | − | − |
| Rock Harbor | 7 | 12 | + | + | − | − |
| Buttonwood Bay | 8 | 6.5 | + | + | − | − |
| Plantation Key | 4 | 5 | − | + | − | − |
| Lower Matecumbe | 3 | 19* | + | + | + | − |
| Long Key | 2 | 9 | + | + | − | − |
| Conch Key | 1 | 14 | − | + | − | − |
| Big Pine | 9 | 11 | − | + | − | − |
| Big Pine | 10 | 10 | + | − | − | − |
| Big Pine | 11 | 16* | + | + | − | − |
| Big Pine | 15 | 13 | + | − | − | − |
| Cudjoe | 13 | 3 | + | − | − | − |
| Sugarloaf | 17 | 2 | − | − | − | − |
| Saddlebunch | 14 | 6.5 | + | + | − | − |
| Boca Chica | 12 | 7 | + | + | − | − |
| Key West, Houseboat Row | 18 | 15 | + | + | − | − |
| Key West, Southern Most Point | 19 | 18* | + | − | − | − |
An asterisk indicates a value that exceeds USEPA guidance levels for single sampling (enterococci). −, none detected; +, detected. The percent positive values (not including site 17) for enteroviruses, hepatitis A viruses, Norwalk viruses, and SRSVs were 79, 63, 10, and 0%, respectively.
FIG. 2.
Panenteroviral dot blot results by sample site. Row 1, columns 1 to 12: sample sites 1 to 12 (Conch Key, Long Key, Lower Matecumbe, Plantation Key, Key Largo [Tropical Lane], Key Largo [Sexton Cove Estates], Rock Harbor, Buttonwood Bay, Big Pine Key [Whispering Pines], Big Pine Key [Doctors Arm], Eden Pines Colony, and Boca Chica Key, respectively). Row 2, columns 1 to 4: sample sites 13 to 16 (Cudjoe Key, Saddlebunch Key, Big Pine Key Port Pine Heights, and Key Largo Ocean Reef Resort, respectively). Row 2, columns 5 and 6: sample sites 18 and 19 (Key West Houseboat Row and Southern Most Point, respectively). Row 2, column 7: positive control. Row 2, column 8: negative control. Site 17 was completed at a later date and was negative.
DISCUSSION
Previous research has demonstrated that coliforms are not good indicators in tropical waters (15, 16, 29). We have identified numerous E. coli isolates in the Marquesas Keys, which is a chain of islands approximately 25 miles due west of Key West, where there are no human residences and there is minimal human activity. It is believed that these isolates are the result of the animal and bird presence and that, once deposited in these types of warm shallow marine environments, they are able to multiply. Research conducted in our laboratory has suggested that Florida waters should be considered tropical waters and that coliforms are not adequate predictors of fecal contamination and public health risks. Others have suggested that alternate indicators are more reflective of pollution; for example, the USEPA has promoted the use of enterococcus levels in marine waters as a better indicator of health risks. Cabelli (6) showed that levels greater than 35 CFU/100 ml were associated with an increased risk of illness. The average level (23 CFU/100 ml) found in this study was just below that level. Three sites (site 3, site 11, and site 19) with the highest single-sample levels were among the highest-ranked sites for fecal pollution.
Canal type (Table 4; flowthrough, multicanal network, etc.) and the age and number of homes on the canals within each type appeared to influence prokaryote and/or indicator prevalence. The general trend was that canals that could flush easily, such as the flowthrough canals and the short single dead-end canals, had the lower rankings. Exceptions were related to the ages of the homes. Most of the canals that were part of a multicanal network had the higher rankings. Several of the sites and noted characteristics listed in Table 4 are described below in order from north (Key Largo) to southwest (Key West). The cleanest site sample (site 16, rank 1) was taken from a flowthrough canal within the Ocean Reef Resort on the north end of Key Largo. This site differs from the other canal sites in that the homes are connected to a central sewer system and thus do not utilize septic tanks for sewage disposal. Sites 6 and 5 were located on Key Largo and both were located in single dead-end canals (ranks of 17 and 8, respectively). The sites had 17 and 22 homes per canal (sites 6 and 5, respectively) with an average age of 23 years. The difference between these sites is that site 5 was a short canal with greater tidal flushing. Site 8 was located in Buttonwood Bay, in a flowthrough canal (rank 6.5). This site had 260 condominium units around it. Site 4 was located on Plantation Key, in a long single dead-end canal (rank 5). The canal had 24 homes on it, with an average age of 18 years. Site 3 was located on Lower Matecumbe, in Captains Cove, which is a small protected boat basin that is linked to the Gulf by a main residential canal of a multicanal network; it had the highest ranking of all the sites (rank 19). Site 1 was located on Conch Key, in a short single dead-end canal. This site had the highest ranking (rank 14) of all the short dead-end canals and was unique in that it had the homes (15 homes) with the highest average age (46 years). Site 13 was located on Cudjoe Key, in the main canal of a multicanal network (rank 3). This site had the lowest ranking of all the multicanal network sites and was unique in that it had the homes (10 homes) with the lowest average age (5 years), of all the sites. Site 14 was located on Saddlebunch Key, in a flowthrough canal (rank 6.5). This site, which was one of the cleanest, had 58 homes on it with an average age of 26 years.
The second-highest-ranked site (rank 18) was located off of Southern Most Point on Key West. The island of Key West utilizes full-scale sewage treatment. However, sewer lines in this section of the City of Key West are in need of replacement, as the city experiences a salt-water intrusion level of 65% at this site (65% of the wastewater coming from this section of the city is marine in origin). Site 18, which was taken near Houseboat Row on Key West, had a rank of 15. Numerous live-aboard houseboats (many of these have been destroyed in a hurricane since this site was sampled) were located at this site, and the houseboats were hooked up to the City of Key West’s sewer system.
The low numbers of coliphages isolated in this study may indicate rapid die-off of phages, which may have been a result of salinity and the high water temperatures noted during the sampling dates. Salinity values ranged from 22 to 37 ppt (average, 27 ppt) during the first sampling dates and 32 to 36 ppt (average, 34 ppt) during the second sampling dates. Water temperatures averaged 29.0 and 33.0°C, respectively. The only phage isolation occurred during the first sampling dates, when both the salinity and water temperature were lower.
The numbers of sites positive for enteroviruses (79%, panenterovirus primer set), hepatitis A viruses (63% of sites positive), and Norwalk viruses (10% positive) suggest that wastewater is impacting the canals and nearshore waters of the Florida Keys. It should be noted that there was a marked difference in RT-PCR positive control signal (amplicon) between the four primer sets used. The panenterovirus and the hepatitis A virus primer sets produced a positive control signal, which could be detected by gel electrophoresis. The Norwalk and SRSV primer sets required dot blot analysis with overnight X-ray film exposure to detect a positive control signal. The published sensitivity levels of the primer sets used in this study demonstrated that the detection limits vary. The sensitivities of the panenterovirus and hepatitis A virus primer sets varied from 103 to 0.01 poliovirus and hepatitis A virus PFU as determined with various virus recovery techniques (30, 32). The Norwalk primer required at least 105 amplifiable units (approximate number of virions) before amplicon was detected (19). The inefficiency of the SRSV primer sets targeting short regions of polymerase gene has also been noted (3). A new RT-PCR protocol, which was published after the start of this study, has demonstrated a detection limit of <21 SRSVs by nested RT-PCR (14). That study demonstrated the presence of SRSVs in samples (contaminated shellfish) previously determined to be negative by single-round RT-PCR. It is also interesting that preliminary recovery assays conducted in our laboratory resulted in enhanced RT-PCR detection of Norwalk and SRSV seeded samples when magnetic poly(T) capture of viral RNA was used, compared to the assay method utilized in this study (data not shown).
The panenterovirus RT-PCR data in this study mirrors the results obtained from a similar study conducted in Sarasota County, Florida. The water quality study conducted in Sarasota County (21) on the impact of septic tank effluents on microbial water quality reported 12 of 15 samples in violation of Florida State Standards for Safe Swimming (200 fecal coliforms/100 ml), with averages of 152 to 2,780 fecal coliforms/100 ml. Enterococci, Clostridium spp., and coliphages were also found in concentrations indicating significant fecal contamination. Enteroviruses were detected by cell culture in 88% of the samples and by RT-PCR in 91% of the samples. In the current study an additional three viral RT-PCR primer sets and assays were utilized, and 95% of the sites were positive for at least one of the groups of viruses. Site 3, which was ranked highest for indicator prevalence, was positive for enteroviruses, hepatitis A viruses, and Norwalk viruses. RT-PCR detection of these virus groups does not address the question of viability. Salinity and water temperature, as the phage data suggests, may significantly impact viral viability. Research has demonstrated that both hepatitis A viruses and polio virus 1 viruses can survive for a period of time in marine and estuarine environments (a 3-log reduction of seeded viruses over a period of 5 days as determined by cell culture [23]). In contrast to the Sarasota County study (which had lower salinity and water temperatures for both fresh and tidally influenced samples), where a high percentage of the samples positive by RT-PCR were also positive by cell culture, many of the RT-PCR positive results in this study may represent inactivated viruses.
The waste disposal studies conducted in the Florida Keys to date have directly demonstrated microbial and nutrient loading in the nearshore water column throughout the region. The objectives of this survey were to broadly assess throughout the Florida Keys the level of fecal input and the possible presence of human pathogens in canals. This study presents a preliminary assessment of water quality, but it does not address the temporal variability at each site. Seasonal differences will likely influence the variation in water quality reflected by environmental conditions such as water temperature, percent UV irradiation, precipitation, and occupancy of homes. These results have prompted further studies to examine the seasonal differences and the presence of cultivatable viruses in the waters of the Florida Keys.
ACKNOWLEDGMENTS
This work was funded by a grant from the U.S. Environmental Protection Agency, Region 4 (984295-97-0).
We thank the following individuals for their assistance on this project: Gus Rios of the Florida Department of Environmental Protection, Marathon, for providing assistance in sample site selection; David Coleman of Lindahl Browning Ferrari and Hellstrom, Inc., Consulting Engineers, Palm City, Fla., for the GIS data; and Robert C. Hudson and Doug Wilder of the Florida Department of Environmental Protection, St. Petersburg, for assistance in analyzing the GIS data.
REFERENCES
- 1.Abbaszadegan M, Stewart P, LeChevallier M, Yates M, Gerba C. American Water Works Association, Washington, D.C. 1995. Occurrence of enteroviruses in groundwater and correlation with water quality parameters. Water Quality Technology Conference, New Orleans, La. [Google Scholar]
- 2.Alexander L M, Heaven A, Tennant A, Morris R. Symptomatology of children in contact with sea water contaminated with sewage. J Epidemiol Community Health. 1992;46:340–344. doi: 10.1136/jech.46.4.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.American Public Health Association. Standard methods for the examination of water and wastewater. 20th ed. Washington, D.C: American Public Health Association; 1998. [Google Scholar]
- 3.Ando T, Monroe S S, Noel J S, Glass R I. A one-tube method of reverse transcription-PCR to efficiently amplify a 3-kilobase region from the RNA polymerase gene to the poly(A) tail of small round-structured viruses (Norwalk-like viruses) J Clin Microbiol. 1997;35:570–577. doi: 10.1128/jcm.35.3.570-577.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ando T, Monroe S S, Gentsch J R, Jin Q, Lewis D C, Glass R I. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and Southern hybridization. J Clin Microbiol. 1995;33:64–71. doi: 10.1128/jcm.33.1.64-71.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armon R, Payment P. A modified m-CP medium for enumerating Clostridium perfringens from water samples. Can J Microbiol. 1988;34:78–79. doi: 10.1139/m88-014. [DOI] [PubMed] [Google Scholar]
- 6.Cabelli V J. Health effects for marine recreation waters. USEPA 600/1-80-031. Research Triangle Park, N.C: Health Effects Research Laboratory; 1983. [Google Scholar]
- 7.Cheung W H S, Chang K C K, Hung R P S. Health effects of beach water pollution in Hong Kong. Epidemiol Infect. 1990;105:139–162. doi: 10.1017/s0950268800047737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLeon R, Shieh C, Baric S, Sobsey M D. Detection of enteroviruses and hepatitis A virus in environmental samples by gene probes and polymerase chain reaction. Water Quality Technology Conference Proceedings. Washington, D.C: American Water Works Association; 1990. [Google Scholar]
- 9.Dufour A P, Ericksen T H, Ballentine R K, Cabelli V J, Goldberg M, Fox W E. Bacteriological ambient water quality criteria for marine and fresh recreational waters. Ambient water quality criteria for bacteria. EPA44075-84-002. U.S. Washington, D.C: Environmental Protection Agency; 1986. [Google Scholar]
- 10.Federal Register. Appendix C to subpart M: proposed ICR protozoan method for detecting Giardia cysts and Cryptosporidium oocysts in water by fluorescent antibody technique. Fed. Regist. Vol. 59 1994. [Google Scholar]
- 11.Fliesher J M, Kay D, Wyer M, Merrett H. The enterovirus test in the assessment of recreational water-associated gastroenteritis. Water Res. 1996;30:2341–2346. [Google Scholar]
- 12.Fujioka R S, Shizumura L K. Clostridium perfringens a reliable indicator of stream water quality. J Water Pollut Control Fed. 1985;57:986–992. [Google Scholar]
- 13.Gantzer C, Maul A, Audic J M, Schwartzbrod L. Detection of infectious enteroviruses, enterovirus genomes, somatic coliphages, and Bacteroides fragilis phages in treated wastewater. Appl Environ Microbiol. 1998;64:4307–4312. doi: 10.1128/aem.64.11.4307-4312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green J, Henshilwood K, Gallimore C I, Brown D W G, Lees D N. A nested reverse transcriptase PCR assay for detection of small round-structured viruses in environmentally contaminated molluscan shellfish. Appl Environ Microbiol. 1998;64:858–863. doi: 10.1128/aem.64.3.858-863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagler A N, Rosa C A, Morris P B, Mendonca-Hagler L C, Franco G M O, Araujo F V, Soares C A G. Yeasts and coliform bacteria of water accumulated in bromeliads of mangrove and sand dune ecosystems of southeast Brazil. Can J Microbiol. 1993;39:973–976. doi: 10.1139/m93-146. [DOI] [PubMed] [Google Scholar]
- 16.Hardina C M, Fujioka R S. Soil: the environmental source of Escherichia coli and enterococci in Hawaii’s streams. Environ Toxicol Water Qual Int J. 1991;6:185–195. [Google Scholar]
- 17.Hejkal T W, Wellings F M, Lewis A L, LaRock P A. Distribution of viruses associated with particles in wastewater. Appl Environ Microbiol. 1981;41:628–634. doi: 10.1128/aem.41.3.628-634.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu F-C, Shieh Y-S C, van Duin J, Beekwilder M J, Sobsey M D. Genotyping male-specific RNA coliphages by hybridization with oligonucleotide probes. Appl Environ Microbiol. 1995;61:3960–3966. doi: 10.1128/aem.61.11.3960-3966.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaykus L-A, DeLeon R, Sobsey M D. A virion concentration method for detection of human enteric viruses in oysters by PCR and oligoprobe hybridization. Appl Environ Microbiol. 1996;62:2074–2080. doi: 10.1128/aem.62.6.2074-2080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapointe B E, O’Connell J D, Garrett G S. Nutrient couplings between on-site waste disposal systems, groundwater, and nearshore surface waters of the Florida Keys. Biogeochemistry. 1990;10:289–307. [Google Scholar]
- 21.Lipp E, Farrah S, Rose J. A study on the presence of human viruses in surface waters of Sarasota County. Sarasota. Sarasota, Fla: Florida Department of Health Water Quality; 1997. [Google Scholar]
- 22.Pallin R, Wyn-Jones A P, Place B M, Lightfoot N F. The detection of enteroviruses in large volume concentrates of recreational waters by the polymerase chain reaction. J Virol Methods. 1997;67:57–67. doi: 10.1016/s0166-0934(97)00076-1. [DOI] [PubMed] [Google Scholar]
- 23.Patti A M, Santi A L, Gabrieli R, Fiamma S, Cauletti M, Pana A. Hepatitis A virus and poliovirus 1 inactivation in estuarine water. Water Res. 1987;21:1335–1338. [Google Scholar]
- 24.Paul J H, Rose J B, Jiang S C, Zhou X, Cochran P, Kellogg C, Kang J B, Griffin D, Farrah S, Lukasik J. Evidence for groundwater and surface marine water contamination by waste disposal wells in the Florida Keys. Water Res. 1997;31:1448–1454. [Google Scholar]
- 25.Paul J H, Rose J B, Jiang S, Kellogg C, Shinn E. Occurrence of fecal indicator bacteria in surface waters and the subsurface aquifer in Key Largo, Florida. Appl Environ Microbiol. 1995;61:2235–2241. doi: 10.1128/aem.61.6.2235-2241.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul J H, Rose J B, Brown J, Shinn E A, Miller S, Farrah S R. Viral tracer studies indicate contamination of marine waters by sewage disposal practices in Key Largo, Florida. Appl Environ Microbiol. 1995;61:2230–2234. doi: 10.1128/aem.61.6.2230-2234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul J H, III, Jiang S C, Rose J B. Concentration of viruses and dissolved DNA from aquatic environments by vortex flow filtration. Appl Environ Microbiol. 1991;57:2197–2204. doi: 10.1128/aem.57.8.2197-2204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhodes M W, Kator J. Enumeration of Enterococcus sp. using a modified mE method. J Appl Microbiol. 1998;83:120–126. doi: 10.1046/j.1365-2672.1997.00200.x. [DOI] [PubMed] [Google Scholar]
- 29.Santiago-Mercado J, Hazen T C. Comparison of four membrane filter methods for fecal coliform enumeration in tropical waters. Appl Environ Microbiol. 1987;53:2922–2928. doi: 10.1128/aem.53.12.2922-2928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwab K J, De Leon R, Sobsey M D. Immunoaffinity concentration and purification of waterborne enteric viruses for detection by reverse transcriptase PCR. Appl Environ Microbiol. 1996;62:2086–2094. doi: 10.1128/aem.62.6.2086-2094.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwab K J, DeLeon R, Sobsey M D. Concentration and purification of beef extract mock eluates from water samples for the detection of enteroviruses, hepatitis A virus, and Norwalk virus by reverse transcription-PCR. Appl Environ Microbiol. 1995;61:531–537. doi: 10.1128/aem.61.2.531-537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shieh Y-S C, Baric R S, Sobsey M D. Detection of low levels of enteric viruses in metropolitan and airplane sewage. Appl Environ Microbiol. 1997;63:4401–4407. doi: 10.1128/aem.63.11.4401-4407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinn E A, Reese R S, Reich C D. Fate and pathways of injection-well effluent in the Florida Keys. Washington, D.C: Department of the Interior/U.S. Geological Survey; 1994. [Google Scholar]
- 34.Vantarakis A C, Papapetropoulou M. Detection of enteroviruses and adenoviruses in coastal waters of SW Greece by nested polymerase chain reaction. Water Res. 1998;32:2365–2372. [Google Scholar]