Abstract
Recently Bacillus spp. has gained much attention as potential probiotics due to the production of resistant cells. So, this research is purposeful for evaluation of probiotic characteristics of Bacillus isolates from camel milk as a suitable source for growth and isolation of microorganisms that can be candidate to be used as probiotic. First, forty-eight colonies were screened by using morphological and biochemical analysis. Among the isolates, two of them were recognized as Bacillus subtilis CM1 and CM2 by partial 16SrRNA sequencing that, probiotic potentials of them were evaluated. Both of them, in the preliminary safety screening, were found negative for hemolysis and lecithinase activity. Also, in vitro characteristics such as acid, bile salts and artificial gastric juice resistant, cell surface hydrophobicity, auto-aggregation, antioxidant characteristics, and adherent capability to HT-29 cells were determined for them approximately in the range of other probiotic strains. Two strains were susceptible to various antibiotics and enterotoxigenic activities were not detected by PCR which means isolated Bacillus strains could be classified as safe. Altogether, results demonstrate that Bacillus CM1 and CM2 strains could have the potential of consideration as probiotics, however more extensive in vitro/vivo studies are needed.
Subject terms: Microbiology, Applied microbiology
Introduction
Live microorganisms that administration of sufficient quantities of them could have beneficial healthy effects are defined as probiotics1. Today, owing to recognition of health benefits of probiotics as food supplements that include inhibition of intestinal pathogens by promoting the growth of healthy microflora in gastrointestinal tract, reduction in cholesterol level, control of diarrhea, immune response enhancement, anti-mutagenic and anti-carcinogenic activity, alleviation of lactose intolerance and etc., the market for probiotics has grown too2,3. Although two main genera Lactobacillus and Bifidobacteria are largely represented on the market as convential probiotics that mostly isolated from sources such as parts of GIT, feces, milk and fermented food products, but different species from Streptococcus, Propionibacterium, Bacillus, Enterococcus and Saccharomyces from various sources are claimed as probiotics too3–6. Also, however plenty of accessible probiotic strains are belonged to the lactic acid bacteria (LAB) as a group of non-sporulating bacteria but it is important to know that, in comparable of vegetative cells, spore forming bacteria such as Bacillus species due to their interesting properties have gained much attention1,7. A probiotic strain must fulfill some essential standards and must tolerate manufacturing, storage, transportation, application steps and so on. The extremely resistance properties of spores to heat, UV irradiation, pH conditions, desiccation and solvents offers capability of long time periods of storage at low or room temperature, higher stability in heat processing and better acid tolerance which they are important traits for overcoming to some difficulties in term of LAB usage as probiotics4,8,9. So, the possibility of incorporation of them in food products can be raised and could be dominant microorganisms in pasteurized milk-based products5,10. Although several Bacillus strains with probiotic potential have been evaluated in various in vitro and in vivo studies9 but some of them such as B. subtilis, B. polyfermenticus, B. clausii, B. coagulans, B. licheniformis and B. pumillus have been approved for commercial use as dietary supplements or growth promoters in aquaculture and in animals respectively, and much effort has been devoted to research on the Bacillus isolation from various sources for probiotic products development5,11,12. Bacteria of Bacillus genus specially B. subtilis are dominant in soil, but they have been identified in water, air, human and animal gut, vegetables, fermented foods, raw and pasteurized milk and dairy products4. Thus, owing to their ubiquitous in different environment, they could easily find their way in food products and are often present in milk microflora5.
In many countries of dry land and desert ecosystems, camels due to high adaptation to the hostile climatic conditions, have significant role in life of these types of communities by providing meat, milk and transportation13. In addition, camels have medical importance through their milk and urine14. Camel milk has good nutritional and medicinal properties and as a medicinal drink in Middle Eastern, Asian and African cultures has been used. Camel milk is reputed as an anti-diabetic, anti-cancerous and anti-infectious food and the therapeutic effects of camel milk have been investigated in case reports, in vitro or in vivo studies and clinical trials15. Camel milk, in some aspects is different from other ruminant milk. It contains all the essential nutritious needed for humans and its biochemical composition is close to human milk thus it can be served as alternative of cow milk16. It has many groups of water and fat-soluble vitamin which the vitamins and iron content of it, is 3times and 10folds higher than in cow milk respectively15,17 and because of high vitamin C content, camel milk has powerful antioxidant activity18. It contains a large amounts of various proteins like albumin, immunoglobulins and lactoferrin19,20 that are apparently more heat resistant than those of cowʼs milk17. It is also rich in amount and type of amino acids such as valine, methionine, lysine, arginine and phenylalanine19. The milk has low sugar, low fat content, high minerals especially zinc, calcium and kalium and large concentration of insulin15,21. Another advantage of this milk, is its low allergenicity18. Aside from the high nutritional value and physicochemical characteristics of milk, a rich bacterial diversity exists in the milk and fermented products that have been received low attention, also few researches have been reported about isolation of strains with probiotic potential from them22. so, this research was aimed for assessment of probiotic properties of B. subtilis strains from camel milk.
Results:
Strain isolation and molecular identification
Initially, 48 bacterial colonies were isolated from milk samples and primarily identification of them was performed by morphological and biochemical tests (Fig. 1, Table 1). Selected isolates were Gram positive, catalase, citrate, nitrate reduction and motility positive and had a shape of bacilli containing oval spores in the center or subterminal and were able to hydrolyze mannose, glucose, maltose and starch were known as Bacillus genus. Then among spore- forming isolates, two isolates designated as CM1 and CM2 were selected for molecular identification. 16SrRNA gene sequence analysis of these isolates revealed that these bacteria belonged to the B. subtilis and sequences of the 16SrRNA gene from the two isolates of B. subtilis were deposited in the NCBI database (national center of Biotechnology Information) with the accession numbers MK559537 and MK611084 (Fig. 2, Table 2). https://www.ncbi.nlm.nih.gov/nuccore/MK611084.1https://www.ncbi.nlm.nih.gov/nuccore/MK559537.1.
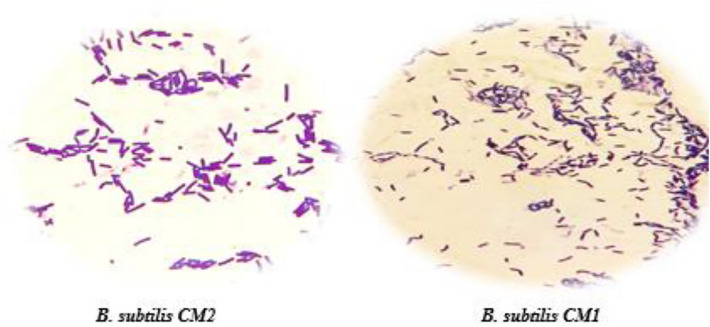
Figure 1.
Gram staining reaction of bacteria isolates.
Table 1.
Biochemical characterization of Bacillus isolates from camel milk.
| Tests | CM1 | CM2 | Tests | CM1 | CM2 |
|---|---|---|---|---|---|
| Gram staining | + | + | Glucose | + | + |
| Spore formation | + | + | Maltose | + | + |
| Starch hydrolysis | + | + | Mannose | + | + |
| Simon ʼs citrate | + | + | Nitrate reduction | + | + |
| Methyl red | − | − | VP | + | + |
| Catalase | + | + | Growth at 50 °C | + | + |
| Urease | − | − | SIM | −/−/+ | −/−/+ |
Plus (+) and minus sign (−) indicate the positive and negative results of reaction/test, respectively.
Figure 2.
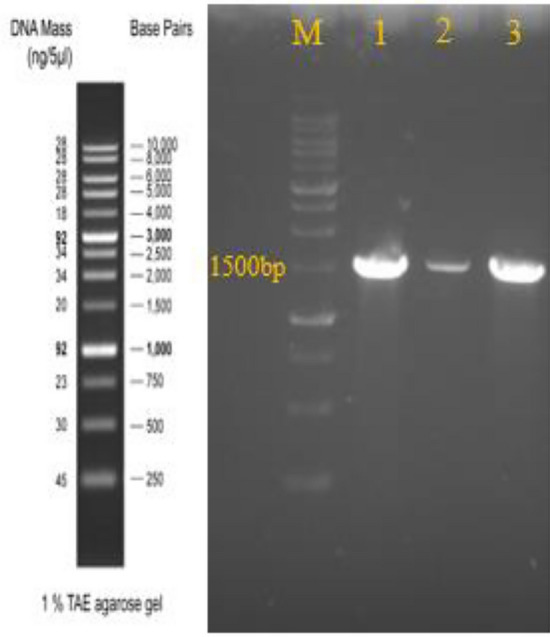
PCR products of 16SrRNA from Bacillus isolates. M: 1 Kb DNA ladder. Lane 1: Positive control. Lane 2 and 3: CM1 and CM2 strains.
Table 2.
Accession numbers and similarity of Bacillus species isolated from camel milk.
| Bacteria | Accession number | Query coverage | percent identity |
|---|---|---|---|
| Bacillus subtilis strain CM1 | MK559537 | 99% | 99.65% |
| Bacillus subtilis strain CM2 | MK611084.1 | 100% | 99.30% |
Hemolytic and Lecithinase activity of isolates
Both of the selected bacterial isolates showed α-hemolysis pattern. Also, these two bacteria were lecithinase negative while B. cereus was lecithinase positive (Table 3).
Table 3.
The culture results for selected isolates from camel milk in different conditions.
| Bacteria | Growth at | |||||
|---|---|---|---|---|---|---|
| pH4 | Gastric juice tolerance | pH2 | Bile 0.3% | Lecithinase activity | Hemolytic activity | |
| B. subtilis Strain CM1 | 87.62 ± 0.86a | 73.58 ± 0.68a | 88.58 ± 2.14a | 0.35 ± 0.02a | – | α |
| B. subtilis strain CM2 | 95.22 ± 1.21b | 74.47 ± 0.63a | 86.64 ± 0.78a | 0.37 ± 0.03a | – | α |
Data for three replications are presented as mean ± SD. Mean within the same column followed by different supercript letters differ significantly (P < 0.05).
Tolerance of isolates to acid and bile
Tolerance of two obtained B. subtilis strains to low pH (pH 2 and pH 4) and 0.3% bile salts was evaluated. Approximately more than 80% of bacterial populations of two strains survived at acidic conditions, so tolerance of these isolates was high. Also, both of them showed resistance to bile salts (Table 3).
Gastric juice tolerance assay
The isolates were investigated for artificial gastric juice tolerance by determination of total viable cell count at 0 and 4 h after exposure to gastric conditions. The viability count of more than 70% indicated that these two strains could be survived after 4 h and have the ability to pass through stomach conditions (Table 3).
Determination of cell surface properties
Surface characteristics were evaluated based on auto-aggregation, adhesion capacity to HT-29 cells and hydrophobic traits. In order to determine colonization quality, the bacterial adhesion ability to hydrocarbons (Chloroform, Ethyl acetate and Toluene) was assessed that results are reported in Table 4. Regarding to auto-aggregation, this attribute for two strains varied from 39% (CM2) to 49% (CM1) but in comparison, about 62% cells aggregation were showed for isolates after 24 h. Also, the isolates showed adhesion characteristics to colonic adenocarcinoma cells and B. subtilis CM1 and B. subtilis CM2 strains were adhered 49.66% and 47.35% to the HT-29 cells respectively (Table4).
Table 4.
Percentage of auto-aggregation, cell attachment and hydrophobicity traits of isolates.
| Strain | Auto-aggregation (%) | Hydrophobicity(%) | ||||
|---|---|---|---|---|---|---|
| 4 h | 24 h | Chloroform | Toluene | Ethyl acetate | HT-29 attachment | |
| B. subtilis strain CM1 | 48.78 ± 5.55a | 62.07 ± 2.07a | 55.00 ± 1.80a | 56.24 ± 2.58a | 42.44 ± 2.94a | 49.66 ± 0.82a |
| B. subtilis strain CM2 | 39.10 ± 1.74b | 62.48 ± 0.31a | 48.96 ± 1.18b | 60.93 ± 1.33b | 60.73 ± 0.67b | 47.35 ± 0.02b |
Cell surface characteristics are presented as mean ± SD of three replications. Isolates having different superscript letters differ significantly (P < 0.05).
Antibiotic susceptibility
The inhibition zone of selected antibiotics was revealed the antibiotic sensitivity of B. subtilis CM1 and CM2 to several antibiotics which are presented in Table 5.
Table 5.
Antibiotic susceptibility and antioxidant activity of the Bacillus isolates.
| Strain | Chloramphenicol | Tetracycline | Erythromycin | Streptomycin | Vancomycin | Gentamycin | Clindamycin | Penicillin |
|---|---|---|---|---|---|---|---|---|
| B. subtilis CM1 | 34.00 ± 3.61S | 30.33 ± 1.53S | 27.33 ± 2.08S | 17.00 ± 1.00S | 21.33 ± 1.53S | 27.33 ± 1.53S | 18.33 ± 0.57S | 30.66 ± 3.05S |
| B. subtilis CM2 | 37.00 ± 2.08S | 34.66 ± 3.05S | 24.33 ± 2.51S | 20.33 ± 1.53S | 23.67 ± 1.53S | 30.33 ± 1.53S | 22.67 ± 1.15S | 32.67 ± 2.08S |
Mean ± SD expressing data of inhibition diameter (mm) in three replications. Sensitive (s), Resistance (r).
Antioxidant activity
DPPH scavenging activity was done for determination of the antioxidant nature of Bacillus strains and antioxidant activity recorded 33.8 ± 1.37% and 42.39 ± 2.59% for cell- free supernatant of CM2 and CM1 respectively.
Detection of enterotoxin genes
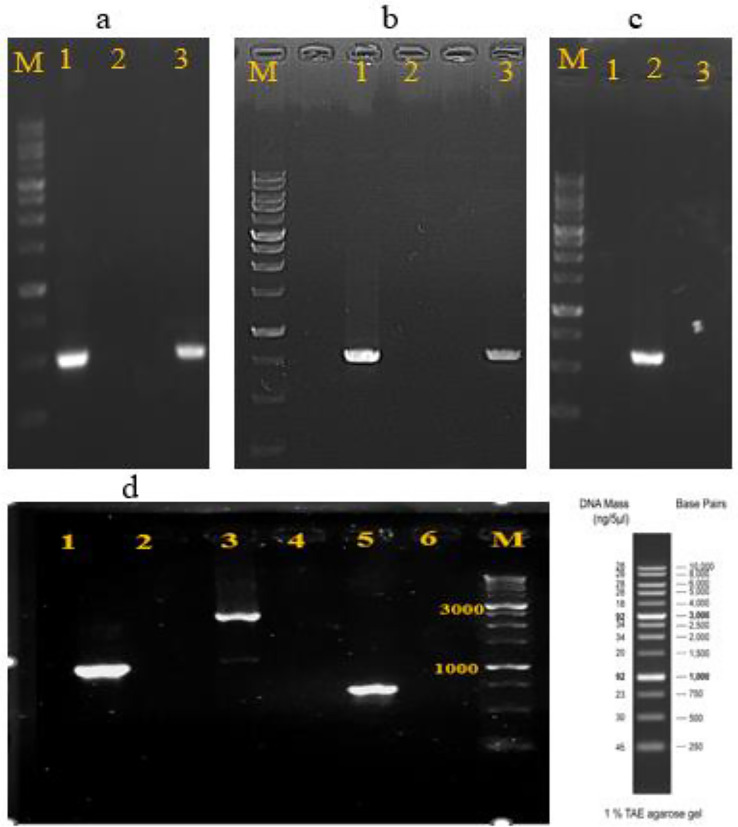
This test in order to evaluation of safety of strains was done based on PCR. B. subtilis CM1 did not carry non-hemolytic enterotoxin (nhe) and hemolysin (hbl) genes but in contrast, B. subtilis CM2 was found to carry nheA and nheB genes. Also, we could detect all six enterotoxin genes in positive control (Fig. 3).
Figure 3.
PCR products of enterotoxin genes from the isolated Bacillus subtilis strains and Bacillus cereus. Lane M: 1 kb DNA ladder. (a) nhe A gene 1: positive control, 2: CM1, 3: CM2. (b) nheB gene 1: positive control, 2: CM1, 3: CM2. (c) nheC gene 1: CM1, 2: positive control, 3: CM2. (d) 1, 3 and 5: hblA, hblB and hblC genes from positive control, 2,4 and 6: negative results of hbl genes for CM1 (negative results observed for both of CM1 and CM2 strains).
Discussion
Camel milk and its products have been given much attention in the world owing to their beneficial effects such therapeutic and nutritional values13,15. Recent scientific reports have indicated the camel milk as a rich source for probiotics19 that aside from nutritional composition as well as the therapeutic and physicochemical properties of milk, information about its microbiota is limited23. Results of microflora diversity in camel milk satisfy to high diversity across countries24 that lactic acid bacteria be as a one of the predominant bacteria which isolated from it25. Therefore, according to the latest studies and available reports, there are no data on the isolation of Bacillus probiotics from camel milk So, here camel milk samples were prepared for isolation Bacillus strains with probiotic potentials as a new source. The isolated strains based on results of biochemical and molecular tests were related to B. subtilis and further used for probiotic evaluation.
It is noticeable to know that probiotic characteristics be strain specific that its own ability is mainly dependent on strain isolation sources and its target, thus for consideration of one microorganism as a probiotic, in vitro/vivo probiotic properties must be evaluated26–28. Hemolysis and Lecithinase activity are usually considered for destroying host cells and tissues29, so screening of bacteria for these products is important for ensuring safety of one isolate27. Some Bacillus species produced hemolysis which this capability is considered a disadvantage for probiotic strain and could be a health risk for the host8. In this study, both of the strains showed no lecithinase activity. Similar observations have been reported for probiotic candidates like B. clausii UBBC07 and Bacillus strains BS3 and BS3130,31. Among screened isolates, B. subtilis CM1 and CM2 strains display α-hemolytic activity results. Similar results were shown by Keubutornye et al.32. Also, Naeem et al. had worked on probiotic potential of Bacillus strains and their results showed no hemolysis for isolated strains8. Although γ-hemolytic and α-hemolytic strains are remarkable as safe that means the Bacillus species did not show any risk to host, but γ-hemolytic isolates are ideal for consideration and usage as probiotics29,32.
Assessment of the antibiotic susceptibility of bacterial cells was conducted to ensure inability of strains for transferring of antibiotic resistance determinants that is other essential aspects for investigation of probiotic safety33. Antibiotic resistance pattern indicated the susceptibility of B. subtilis CM1 and CM2 strains to antibiotics, that ensures their inability to possess antibiotic resistance34. These results were similar to previous studies about B. subtilis NC11, B. subtilis TPS4, Bacillus velezensis TPS3N and Bacillus amyloliquefaciens TPS17 which were found to be sensitive to antibiotics32,35.
Since, acid and bile salts in the stomach and intestine respectively, are the first biological barriers that a probiotic strain must be overcome after ingestion to reach its place of action28, acid and gastric juice tolerance as well as bile resistance are as a most essential factors for viability and growth of probiotic strains during their transit to the gastrointestinal tract36. Two isolates could tolerate the acidic pH and artificial gastric juice condition and both strains showed bile salt resistance. So, present findings show similarity to previous results about Bacillus strains with probiotic potential37–40.
Other factors that should be considered for probiotic potential are cell surface hydrophobicity, auto-aggregation and epithelial cell adherence that be required for adhesion to the target sites of gastrointestinal tract41. The capability of bacteria to bind to themselves in addition to binding to the extracellular matrix of host tissues or host cells known as auto-aggregation33. Auto-aggregation of probiotics prevents their elimination from the body and could give them a superiority trait and capability for interaction with other bacteria27. Moreover, to determination of cell surface properties, microbial adhesion to hydrocarbons was performed as another important property of bacteria which aids attachment of microorganisms to the intestinal epithelium. Bacterial adhesion characteristic considered as a complex process which needed bacterial cell membranes contact with interacting surfaces, so this ability offers a competitive advantage for probiotic bacteria12,41,42. In addition to mechanisms of interaction between the strain and the superficial components of intestinal cells, the types of cell lines (HT29 or Caco2) can also be affected the adhesion capacities of bacteria to epithelial cells43. In present study, two isolates exhibited auto-aggregation that increasing over time. Similar observations were obtained by Ragul et al. for Bacillus species and Dial et al. for Lactobacillus plantarum40,44. Also, the results of MATH in this research are comparatively similar to results of Lee et al. and Thirabunyamon et al. for Bacillus isolates35,45. In contrast, in a study by Kuebutornye et al., B. subtilis TPS4, B. velezensis TPS3N and B. amyloliquefaciens TPS17 were reported as Bacillus isolates from the gut of Nile tilapia that exhibited approximately 85 to 97% hydrophobicity measured with chloroform and 74 to 90% hydrophobicity with ethyl acetate. The higher hydrophobicity results of them in comparable of Bacillus isolates in our report, indicating higher electron donation (chloroform) and acceptability (ethyl acetate) of isolates, therefore better adhesion to epithelial cells comparable with Bacillus strains in our study32. The HT-29 cells attachment percentage of B. subtilis CM1 and CM2 strains were higher than Bacillus isolates reported by Talebi et al. on Caco2 cell line37. The results are in line with Mahmoudi et al. that introduced several levels of attachment for Lactobacillus isolates from camel milk on Caco2 and HT-29 MXT as various groups of cell lines46.
The antioxidant potential of probiotic microorganisms is another beneficial and therapeutic value of probiotics47. Generation of free radicals in the body cause damage to macromolecules like lipids, proteins and DNA, therefore, probiotics could neutralize free radicals with their antioxidant potential that would be beneficial for the host48. In this study, two selected isolates exhibited antioxidant activity relatively similar to those reported by Talebi et al. for Bacillus atrophaeus and Bacillus safensis37.
Also, two major complexes, the hemolysin BL (Hbl) and the non-hemolytic enterotoxin (Nhe), that cause diarrhea are noticeable as a reason for food poisoning which included B. cereus49. So, one of the important criteria to ensuring from safety of Bacillus isolates is investigation of enterotoxins production. Hbl and Nhe consist of the protein parts B, L1 and L2 codification by hbl A, hbl C and hbl D as well as Nhe A, Nhe B and Nhe C encoded by nhe A, nhe B and nhe C respectively50. When PCR was carried out, only B. subtilis CM2 was positive for nhe A and nhe B genes. It should be noted that combination of all three parts of the Hbl and Nhe enterotoxin complexes is required to show the enterotoxigenic traits29. Therefore, current results could be considered negative for two isolates and our findings could be similar to most of the studied Bacillus strains, they could not express all genes of enterotoxin complex together and were safe29,49.
Finally, it can be concluded that B. Subtilis CM1 and B. subtilis CM2 could be notable as probiotic candidates. This selection regarded based on analyzing the results of all tests that showed the strains had desirable probiotic potential, however other in vitro and in vivo evaluations including enzymatic activity, co-aggregation, antimicrobial activity, biofilm formation, cholesterol reduction and animal models must be performed in the future for the final decision about their application as probiotic strains.
Methods
Camelʼs milk samples preparation
The raw milk samples were gathered from Varamin (Tehran province, Iran) under aseptic condition and in accordance with the ethical principles and standards guidelines for conducting Veterinary Research in Iran, stored at 4 °C and serial dilutions were prepared in saline buffer, heated at 80 °C, 15 min. Afterward 0.1 ml of any sample was streaked on nutrient agar plates (Merck, Germany), incubated for 24 h, 37 °C and different morphological colonies were purified, checked for Gram staining and catalase activity. Finally, biochemical tests and sequencing of 16SrRNA genes was done for further identification of obtained isolates17,37. The animal experiments have acquired approval of Research Ethics Committees of University of Isfahan (Approval ID: IR.UI.REC.1400.024) and performed under the ARRIVE guidelines.
Identification of B. subtilis isolates
First biochemical tests were used for identification of isolates as described elsewhere according to Bergey manual of Systematic Bacteriology. Then for identifying the bacteria with PCR, after 18 h incubation of the selected isolates, boiling method was used for DNA extraction from them. Respectively, 27F (5′-AGAGTTTGATCCTGGCTCAC-3′) and 1492R (5′CGGTTACCTTGTTACGACTT-3′) were used as forward and reverse primers with an expected product size of 1500 bp. Finally, the PCR products were sequenced and analyzing of the sequence was done by BLAST algorithm (NCBI)51,52.
Screening of B. subtilis isolates for probiotic properties
Hemolysis activity
The selected isolates were streaked on blood agar plates (Merck, Germany) and incubated for 24 h, 37 °C. Then hemolysis pattern was classified as α, β or γ-hemolysis. S. aureus ATCC25923 was used as the control53.
Lecithinase activity
A loopfull of each strain and B. cereus ATCC14579 as positive control were streaked as a straight line on the egg yolk agar (Biomark, India) and incubated (24–48 h, 37 °C) for detection of lecithinase production30.
Acid tolerance
For acid tolerance determination, several pH grades were prepared by hydrochloric acid solution (Merck, Germany) 5N to pH 2.0 and 4.0 in Phosphate-Buffered Saline (PBS). The isolates were incubated in nutrient broth (Biolife, Italy) for 18 h at 37 °C, then cell pellet was harvested and washed in PBS, resuspended in both pH solutions, including 2 and 4, and incubation was done for 4 h, 37 °C. Plate counts on nutrient agar at 0 h and 4 h were used for assessment of survivability according to this equation:
where N1 and N0 represent (log cfu/ml) count of selected species after and before treatment respectively37.
Gastric juice tolerance
For gastric juice test, pepsin (Sigma-Aldrich, USA) (0.3 w/v) and NaCl (Merck, Germany) (0.5% w/v) was added to nutrient broth and adjusted to pH 2.5. First, strains were incubated (nutrient broth, 18 h, 37 °C), and pellet washed twice in PBS, then cell suspension was diluted in gastric juice pH 2.5 and incubated. Viable cells count was investigated at 0 and 4 h for samples.
where N0 and Nt are initial and final viable cells (cfu/ml)53.
Bile salts resistance
For assessment of bile salts resistance, 100 μl bacterial suspensions were inoculated into nutrient broth containing bile salts (Sigma-Aldrich, USA) at concentration of 0.3% and nutrient broth without salts, followed by incubation (37 °C, 8 h). Inhibition rate was calculated by recording the absorbance at 600 nm.
where T8 and T0 represent the OD at time 0 h and after 8 h incubation. Cinh (inhibitory) of less than 0.4 is acceptable for probiotic candidate38.
Assessment of cell surface hydrophobicity
Isolates were cultured in nutrient broth for 18 h and harvested pellets by centrifugation 3 min at 9400g, were washed and resuspended in 2 ml of PBS. To determine percentage of hydrophobicity, its optical density (OD) was measured and recorded as A0. After adding equal volume of Chloroform (Merck, Germany), Ethyl acetate (Merck, Germany) and Toluene (Merck, Germany) to bacterial suspension, blended them by vortexing for 5 min, and OD600 of aqueous phase was recorded as A1 after 30 min incubation at room temperature. The isolate adhering to solvents was estimated as below:
where A0 and A1 initial and final OD at 600 nm45.
Auto-aggregation
For auto-aggregation test, after centrifugation of bacterial cells from overnight culture (nutrient broth, 37 °C), the pellet was washed and resuspended in buffer till absorbance of suspension reach to 0.3 ± 0.05 at 600 nm. Bacterial suspensions were vortexed for 10 s and OD600 of samples was recorded after incubation for 4 h and 24 h in 37 °C. Auto-aggregation was presented using the equation below:
where At represented absorbance of samples in different times (4 or 24 h), A0 represented the absorbance at the beginning of the assay54.
Bacterial adhesion assay
Adherence potential of candidate probiotic isolates was carried out with colonic adenocarcinoma cells that were obtained from Iranian Biological Resource Center (HT-29 IBRC C10097). First, 1 × 105 cells/ml were seeded and incubated to obtained 80–90% confluency. Then cells in a 24-well plate were washed and medium was changed to antibiotic-free Dulbeccoʼs Modified Eagle Medium (DMEM) (Bioidea, Iran). Subsequently, 108 cells/ml of test isolates were inoculated to cells in each well and incubated for 3 h (37 °C, 5% CO2). After incubated, three times washing of cells with PBS (Bioidea, Iran) was used for removing the non-adherent bacteria. Trypsinization of HT-29 monolayers by trypsin–EDTA solution (Bioidea, Iran) was done and finally, the cell attachment capacity was determined by Counting of adherent cells at time 0 (N0) and after 3 h (Nt) in triplicate on nutrient agar55.
Antibiotic resistance
Assessment of antibiotic resistance of B. subtilis isolates based on the recommendation of CLSI (Clinical and laboratory standards institute) was done by disc diffusion test. Briefly, approximately 108 cfu/ml of overnight cultures were swabbed on the Mueller–Hinton Agar plates and antibiotic discs (Padtan Teb Co, Iran) containing tetracycline (30 μg), chloramphenicol (30 μg), erythromycin (15 μg), streptomycin (10 μg), vancomycin (30 μg), gentamycin (10 μg), clindamycin (2 μg) and penicillin (10 μg) were placed on the agar plates, and after 24 h incubation at 37 °C, sensitivity of bacteria was determined by measuring the diameter of inhibition zone56.
DPPH scavenging assay
DPPH scavenging effect of cell free supernatants of two isolates was evaluated by mixing a volume of 100 μl filtrate culture with equal volume of DPPH solution (Merck, Germany) (0.2 mM) and left at 30 °C in darkness for 30 min. Deionized water was used as a control. Determination of absorbance at 517 nm using Synergy HTX multimode reader was done for measuring DPPH scavenging potency as below:
Enterotoxin genes detection
DNA from B. subtilis isolates and B. cereus ATCC14579 as a positive control were extracted via the boiling method. In the next stage, PCR analyses were carried out to detect 6 enterotoxigenic genes. Table 6 show primer sequencers and PCR conditions58.
Table 6.
Primer names and sequences, target size and PCR conditions for enterotoxin genes detection58.
| Primers | Target size (bp) | Sequences (5′–3′) | Reaction conditions |
|---|---|---|---|
|
hblA F hblA R |
1154 |
AAGCAATGGAATACAATGGG AGAATCTAAATCATGCCACTGC |
94 °C,2 min/(94 °C,60 s*56 °C,60 s*72 °C,120 s)35cycles /72 °C,5 min |
|
hblC F hblC R |
740 |
GATACCAATGTGGCAACTGC TTGAGACTGCTCGCTAGTTG |
94 °C,2 min/(94 °C,60 s*58 °C,60 s*72 °C,120 s)35cycles /72 °C,5 min |
|
hblD F hblD R |
829 |
ACCGGTAACACTATTCATGC GAGTCCATATGCTTAGATGC |
94 °C,2 min/(94 °C,60 s*58 °C,60 s*72 °C,120 s)35cycles /72 °C,5 min |
|
nheA F nheA R |
499 |
TACGCTAAGGAGGGGCA GTTTTTATTGCTTCATCGGCT |
94 °C,2 min /(94 °C,60 s*56 °C,60 s*72 °C,120 s)35cycles /72 °C,5 min |
|
nheB F nheB R |
769 |
CTATCAGCACTTATGGCAG ACTCCTAGCGGTGTTCC |
94 °C,2 min/(94 °C,60 s*54 °C,60 s*72 °C,120 s)35cycles /72 °C,5 min |
|
nheC F nheC R |
581 |
CGGTAGTGATTGCTGGG CAGCATTCGTACTTGCCAA |
94 °C,2 min/(94 °C,60 s*58 °C,60 s*72 °C,120 s)35cycles /72 °C,5 min |
Statistical analysis
Analyzing results of three repetitions of experiments was done by IBM SPSS Statistics Software (version 26.0, SPSS Inc., USA) and presented as mean ± SD. Also, for finding significant difference (p ˂ 0.05) across means ANOVA analysis (One-way analysis of variance) followed by Post Hoc method (Duncan) was used.
Acknowledgements
The authors thankful to Dr. Esmaeili for his assistance in camel milk collection.
Author contributions
All authors listed have made substantial or indirect contributions to the work and in the design of experiments. R.D. performed the experiments, collected the samples and data. M.R., R.D. analyzed the data and R.D. wrote the manuscript. M.R., A.S.H.A. and J.H.K. read the paper and revised the manuscript. All authors approved the final manuscript.
Data availability
All data generated or analyzed during this study are included in this published article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lefevre M, et al. Safety assessment of Bacillis subtilis CU1 for use as a probiotic in humans. Regul. Toxicol. Pharmacol. 2017;83:54–65. doi: 10.1016/j.yrtph.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 2.Kim BJ, Hong JH, Jeong YS, Jung HK. Evaluation of two Bacillus subtilis strains isolated from Korean fermented food as probiotics against loperamide-induced constipation in mice. J. Korean. Soc. Appl. Biol. Chem. 2014;57(6):797–806. doi: 10.1007/s13765-014-4106-0. [DOI] [Google Scholar]
- 3.Hameed A, et al. Isolation and characterization of a cholesterol-lowering bacteria from Bubalus bubalis raw milk. Fermentation. 2022;8:163. doi: 10.3390/fermentation8040163. [DOI] [Google Scholar]
- 4.Olmos J, Paniagua-Michel J. Bacillus subtilis a potential probiotic bacterium to formulate functional feeds for aquaculture. J. Microb. Biochem. Technol. 2014;6(7):361–365. doi: 10.4172/1948-5948.1000169. [DOI] [Google Scholar]
- 5.Sorokulova I. Modern status and perspectives of Bacillus bacteria as probiotics. J. Prob. Health. 2013;1:e106. doi: 10.4172/2329-8901.1000e106. [DOI] [Google Scholar]
- 6.Sanders ME, Merenstein D, Merrifield CA, Hutkins R. Probiotics for human use. Nutr. Bull. 2018;43:212–225. doi: 10.1111/nbu.12334. [DOI] [Google Scholar]
- 7.Nawaz Khan A, et al. Antagonistic, anrioxidant, anti-inflammatory and anti-diabetic probiotic potential of Lactobacillus agilis isolated from the rhizosphere of the medicinal plants. Saudi J. Biol. Sci. 2021;28:6069–6076. doi: 10.1016/j.sjbs.2021.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naeem M, Ahmed I, Ahmed S, Ahmed Z, Riaz MN, Ghazanfar S. Screening of cattle gut associated Bacillus strains for their potential use as animal probiotic. Indian J. Anim. Res. 2018 doi: 10.18805/ijar.B-948. [DOI] [Google Scholar]
- 9.Elshaghabee FMF, Rokana N, Gulhane RD, Sharma C, Panwar H. Bacillus as potential probiotics: Status, concerns and future perspectives. Front. Microbiol. 2017;8:1490. doi: 10.3389/fmicb.2017.01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Permpoonpattana P, Hong HA, Khaneja R, Cutting SM. Evaluation of Bacillus subtilis strains as probiotics and their potential as a food ingredient. Benef. Microbes. 2017;3(2):127–135. doi: 10.3920/BM2012.0002. [DOI] [PubMed] [Google Scholar]
- 11.Lee NK, Kim WS, Paik HD. Bacillus strains as human probiotics: Characterization, safety, microbiome and probiotic carrier. Food. Sci. Biotechnol. 2019;28:1297–1305. doi: 10.1007/s10068-019-00691-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nwagu TN, Ugwoudu C, Onwosi OC, Inyiama O, Uchendu O, Akpuru C. Evaluation of the probiotic attributes of Bacillus strains isolated from traditional fermented African locust beanseeds (Parkia biglobosa), "daddawa". Ann. Microbiol. 2020 doi: 10.1186/s13213-020-01564-x. [DOI] [Google Scholar]
- 13.Berhe T, Ipsen R, Seifu E, Kurtu MY, Fugl A, Hansen EB. Metagenomic analysis of bacterial community composition in Dhanaan: Ethiopian traditional fermented camel milk. FEMS. Microbiol. Lett. 2019;366(11):127–132. doi: 10.1093/femsle/fnz128. [DOI] [PubMed] [Google Scholar]
- 14.Noor SO, Alenini MS. Effects of oral administration of camel milk and urine on gut microbiota: Biochemical and microbiological profiling in rats. Am. J. Mol. Biol. 2018;8:1–12. doi: 10.4236/ajmb.2018.81001. [DOI] [Google Scholar]
- 15.Zibaee S, Hosseini SM, Yousefi M, Taghipour A, Kiani MA, Noras MR. Nutritional and therapeutic characteristics of camel milk in children: A systematic review. Electron. Physician. 2015;7(7):1523–1528. doi: 10.19082/1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalesi M, Salami M, Moslehishad M, Winterburn J, Moosavi-Movahedi AA. Biomolecular content of camel milk: A traditional superfood towards future healthcare industry. Trends. Food. Sci. Technol. 2017;62:49–58. doi: 10.1016/j.tifs.2017.02.004. [DOI] [Google Scholar]
- 17.Ziane M, Couvert O, Le-Chevalier P, Moussa-Boudjemaa B, Leguerinel I. Identification and characterization of aerobic spore forming bacteria isolated from commercial camel̕ s milk in south of Algeria. Small Rumin. Res. 2016;137:59–64. doi: 10.1016/j.smallrumres.2016.03.004. [DOI] [Google Scholar]
- 18.Sisay F, Awoke K. Review on production, quality and use of Camel milk in Ethiopia. J. Fish. Livest. Prod. 2015;3(3):145. [Google Scholar]
- 19.Sikarchi A, Fozouni L. Inhibitory effect of probiotic bacteria isolated from camel milk on clinical strains of drug-resistant Helicobacter pylori. Med. Lab. J. 2018;12(2):20–26. doi: 10.29252/mlj.12.2.20. [DOI] [Google Scholar]
- 20.Swelum AA, et al. Nutritional, antimicrobial and medicinal properties of Camel̕ s milk: A review. Saudi. J. Biol. Sci. 2021;28:3126–3136. doi: 10.1016/j.sjbs.2021.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abrhaley A, Leta S. Medicinal value of camel milk and meat. J. Appl. Anim. Res. 2018;46(1):552–558. doi: 10.1080/09712119.2017.1357562. [DOI] [Google Scholar]
- 22.Shori AB. Camel milk and its fermented products as a source of potential probiotic strains and novel food cultures: A mini review. Pharma. Nutr. 2017;5:84–88. [Google Scholar]
- 23.Zhao J, et al. Analyses of physicochemical properties, bacterial microbiota, and lactic acid bacteria of fresh camel milk collected in Inner Mongolia. J. Dairy. Sci. 2020;103(1):106–116. doi: 10.3168/jds.2019-17023. [DOI] [PubMed] [Google Scholar]
- 24.Konuspayeva G, Faye B. Recent advances in camel milk processing. Animals. 2021;11(4):1045. doi: 10.3390/ani11041045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahmeh, R., Alomirah, H., Akbar, A. & Sidhu, J. Composition and properties of camel milk. In MilkProduction,ProcessingandMarketing. (IntechOpen, 2019) 10.5772/intechopen.82592.
- 26.Fijan S. Microorganisms with claimed probiotic properties: An overview of recent literature. Int. J. Environ. Res. Public health. 2014;11:4745–4767. doi: 10.3390/ijerph110504745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.AlGburi A, et al. Safety properties and probiotic potential of Bacillus subtilis KATMIRA 1933 and Bacillus amyloliquefaciens B-1895. Adv. Microbiol. 2016;6:432–452. doi: 10.4236/aim.2016.66043. [DOI] [Google Scholar]
- 28.Shakira G, Qubtia M, Ahmed I, Hasan F, Anjum MI, Imran M. Effect of indigenously isolated Saccharomyces cerevisiae probiotics on milk production, nutrient digestibility, blood chemistry and fecal microbiota in lactating dairy cows. J. Anim. Plant Sci. 2018;28(2):407–420. [Google Scholar]
- 29.Mohkam M, et al. Multifaceted toxin profile of Bacillus probiotic in newly isolated Bacillus spp. from soil rhizosphere. Biologia. 2020;75:309–315. doi: 10.2478/s11756-019-00357-1. [DOI] [Google Scholar]
- 30.Sorokulova IB, et al. The safety of two Bacillus probiotic strains for human use. Dig. Dis. Sci. 2008;53:954–963. doi: 10.1007/s10620-007-9959-1. [DOI] [PubMed] [Google Scholar]
- 31.Lakshmi SG, Jayanthi N, Saravanan M, Ratna MS. Safety assessment of Bacillus clausii UBBC07, a spore forming probiotic. Toxicol. Rep. 2017;4:62–71. doi: 10.1016/j.toxrep.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuebutornye FK, Lu Y, Abarike ED, Wang Z, Li Y, Sakyi ME. In vitro assessment of the probiotic characteristics of three Bacillus species from the gut of nile tilapia, Oreochromis niloticus. Probiot. Antimicro. Prot. 2017;12:412–424. doi: 10.1007/s12602-019-09562-5. [DOI] [PubMed] [Google Scholar]
- 33.Fayyaz Khan F, et al. Recent innovations in non-dairy prebiotics and probiotics: Physiological potential, applications, and characterization. Probiot. Antimicro. Prot. 2022 doi: 10.1007/s12602-022-09983-9. [DOI] [PubMed] [Google Scholar]
- 34.Manhar AK, et al. Cellulolytic potential of probiotic Bacillus subtilis AMS6 isolated from traditional fermented soybean (Churpi): An in-vitro study with regards to application as an animal feed additive. Microbiol. Res. 2016;186–187:62–70. doi: 10.1016/j.micres.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 35.Thirabunyanon M, Thongwittaya N. Protection activity of a novel probiotic strain of Bacillus subtilis against Salmonella Enteritidis infection. Res. Vet. Sci. 2012;93(1):74–81. doi: 10.1016/j.rvsc.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Nair AS, Dubhashi AV. In-vitro transit tolerance of probiotic Bacillus species in human gastrointestinal tract. Int. J. Sci. Res. 2015;5(6):1899–1902. [Google Scholar]
- 37.Talebi S, Makhdoumi A, Bahreini M, Matin MM, Moradi HS. Three novel Bacillus strains from a traditional lacto-fermented pickle as potential probiotics. J. Appl. Microbiol. 2018;125(3):888–896. doi: 10.1111/jam.13901. [DOI] [PubMed] [Google Scholar]
- 38.Ebnetorab SMA, Ahari H, Kakoolaki S. Isolation, biochemical and molecular detection of Bacillus subtilis and Bacillus licheniformis from the digestive system of rainbow trout (Oncorhynchus mykiss) and its inhibitory effect on Aeromonas hydrophila. Iran. J. Fish. Sci. 2020;19(6):2824–2845. [Google Scholar]
- 39.Lee A, Cheng KC, Liu JR. Isolation and characterization of a Bacillus amyloliquefaciens strain with zearalenone removal ability and its probiotic potential. PLoS ONE. 2017;12(8):e0182220. doi: 10.1371/journal.pone.0182220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ragul K, Syiem I, Sunder K, Shetty PH. Characterization of probiotic potential of Bacillus species isolated from a traditional brine pickle. J. Food Sci. Technol. 2017;54(13):4473–4483. doi: 10.1007/s13197-017-2928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duary RK, Rajput YS, Batish VK, Grover S. Assessing the adhesion of putative indigenous probiotic lactobacilli to human colonic epithelial cells. Indian. J. Med. Res. 2011;134(5):664–671. doi: 10.4103/0971-5916.90992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shivangi S, Devi PB, Ragul K, Shetty PH. Probiotic potential of Bacillus strains isolated from an acidic fermented food Idli. Probiot. Antimicro. Prot. 2020;12(4):1502–1513. doi: 10.1007/s12602-020-09650-x. [DOI] [PubMed] [Google Scholar]
- 43.Fonseca HC, De Sousa Melo D, Ramos CL, Dias DR, Schwan RF. Probiotic properties of Lactobacilli and their ability to inhibit the adhesion of enteropathogenic bacteria to Caco-2 and HT-29 cells. Probiot. Antimicro. Prot. 2021;13:102–112. doi: 10.1007/s12602-020-09659-2. [DOI] [PubMed] [Google Scholar]
- 44.Dias FS, Duarte WF, Schwan RF. Evaluation of adhesive properties of presumptive probiotic Lactobacillus plantarum strains. Biosci. J. 2013;29:1678–1686. [Google Scholar]
- 45.Lee S, et al. Probiotic characteristics of Bacillus strains isolated from Korean traditional soy sauce. Food Sci. Technol. 2017;79:518–524. [Google Scholar]
- 46.Mahmoudi I, et al. Functional in vitro screening of Lactobacillus strains isolated from Tunisian camel raw milk toward their selection as probiotic. Small Rum. Res. 2016;137:91–98. doi: 10.1016/j.smallrumres.2016.03.016. [DOI] [Google Scholar]
- 47.Wang Y, et al. Antioxidant properties of probiotic bacteria. Nutrients. 2017;9(5):521. doi: 10.3390/nu9050521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mishra V, Shah Ch, Mokashe N, Chavan R, Yadav H, Prajapati J. Probiotics as potential antioxidants: A systematic review. J. Agric. Food. Chem. 2015;63(14):3615–3626. doi: 10.1021/jf506326t. [DOI] [PubMed] [Google Scholar]
- 49.Abdulmawjood A, Herrmann J, Riede S, Jimenez G, Becker A, Breves G. Evaluation of enterotoxin gene expression and enterotoxin production capacity of the probiotic strain Bacillus toyonensis BCT-7112. PLoS ONE. 2019;14(4):e0214536. doi: 10.1371/journal.pone.0214536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Senesi S, Ghelardi E. Production, secretion and biological activity of Bacillus cereus enterotoxins. Toxins. 2010;2(7):1690–1703. doi: 10.3390/toxins2071690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edalati, E., Sanaeei, B., Alizadeh, M., Hosseini, S.S., Zahedi Bialvaei, A. & Taheri, K. Isolation of probiotic bacteria from raw camel ʼ s milk and their antagonistic effects on two bacteria causing food poisoning. NewMicrobes.NewInfect. 27, 64–68 (2019). [DOI] [PMC free article] [PubMed]
- 52.Zulkhairi Amin FA, et al. Probiotic properties of Bacillus strains isolated from stingless bee (Heterotrigona itama) honey collected across Malaysia. Int. J. Environ. Res. Public Health. 2020;17:278–293. doi: 10.3390/ijerph17010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kavitha M, Raja M, Perumal P. Evaluation of probiotic potential of Bacillus spp. isolated from the digestive tract of freshwater fish Labeo calbasu (Hamilton 1822) Aquac. Rep. 2018;11:59–69. doi: 10.1016/j.aqrep.2018.07.001. [DOI] [Google Scholar]
- 54.Jeon HL, Lee NK, Yang SJ, Kim WS, Paik HD. Probiotic characterization of Bacillus subtilis P223 isolated from kimchi. Food. Sci. Biotechnol. 2017;26(6):1641–1648. doi: 10.1007/s10068-017-0148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rokana N, Singh BP, Thakur N, Sharma Ch, Gulhane RD, Panwar H. Screening of cell surface properties of potential probiotic lactobacilli isolated from human milk. J. Dairy. Res. 2018;85(3):347–354. doi: 10.1017/S0022029918000432. [DOI] [PubMed] [Google Scholar]
- 56.Nithya V, Halami PM. Antibacterial peptides, probiotic properties and biopreservative efficacy of native Bacillus species isolated from different food sources. Probiot. Antimicro. Prot. 2012;4:279–290. doi: 10.1007/s12602-012-9115-x. [DOI] [PubMed] [Google Scholar]
- 57.Xing J, et al. Determination antioxidant activities of Lactobacilli cell-free supernatants by cellular antioxidant assay: A comparison with traditional methods. PLoS ONE. 2015;10(3):e0119058. doi: 10.1371/journal.pone.0119058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim JB, Kim JM, Cho SH, Oh HS, Choi NJ, Oh DH. Toxin genes profiles and toxin production ability of Bacillus cereus isolated from clinical and food samples. J. Food. Sci. 2011;76(1):25–29. doi: 10.1111/j.1750-3841.2010.01958.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.