Abstract
Immunoblot analyses performed with three monoclonal antibodies (MAbs) that recognized the nitrite oxidoreductase (NOR) of the genus Nitrobacter were used for taxonomic investigations of nitrite oxidizers. We found that these MAbs were able to detect the nitrite-oxidizing systems (NOS) of the genera Nitrospira, Nitrococcus, and Nitrospina. The MAb designated Hyb 153-2, which recognized the α subunit of the NOR (α-NOR), was specific for species belonging to the genus Nitrobacter. In contrast, Hyb 153-3, which recognized the β-NOR, reacted with nitrite oxidizers of the four genera. Hyb 153-1, which also recognized the β-NOR, bound to members of the genera Nitrobacter and Nitrococcus. The molecular masses of the β-NOR of the genus Nitrobacter and the β subunit of the NOS (β-NOS) of the genus Nitrococcus were identical (65 kDa). In contrast, the molecular masses of the β-NOS of the genera Nitrospina and Nitrospira were different (48 and 46 kDa). When the genus-specific reactions of the MAbs were correlated with 16S rRNA sequences, they reflected the phylogenetic relationships among the nitrite oxidizers. The specific reactions of the MAbs allowed us to classify novel isolates and nitrite oxidizers in enrichment cultures at the genus level. In ecological studies the immunoblot analyses demonstrated that Nitrobacter or Nitrospira cells could be enriched from activated sludge by using various substrate concentrations. Fluorescence in situ hybridization and electron microscopic analyses confirmed these results. Permeated cells of pure cultures of members of the four genera were suitable for immunofluorescence labeling; these cells exhibited fluorescence signals that were consistent with the location of the NOS.
Nitrification, the microbial oxidation of ammonia to nitrate, is an integral part of the nitrogen cycle. Chemolithoautotrophic ammonia oxidizers convert ammonia to nitrite, and subsequently nitrite is oxidized to nitrate by chemolithoautotrophic nitrite oxidizers. The two groups of organisms occur together and have been isolated from diverse aerobic environments (reviewed in references 5 and 19).
In natural samples nitrifiers have commonly been analyzed by the most-probable-number technique (23), which is often criticized because the culture conditions are not optimal (3). Antibodies or rRNA-targeted oligonucleotide probes are used for in situ analyses in order to avoid the limitations of the most-probable-number technique. Immunological detection of nitrifiers is limited by the serological diversity of cells originating from the same ecosystem (4, 16, 33). Furthermore, the organisms need to be isolated prior to antibody development. Thus, unknown and possibly unculturable nitrifiers are not detectable. Nitrobacter species have commonly been isolated by standard procedures and therefore are considered the dominant nitrite oxidizers in freshwater and terrestrial ecosystems (5). Therefore, mainly antibodies that recognize Nitrobacter species are known so far. However, in situ analyses performed with rRNA-targeted oligonucleotide probes recently revealed that Nitrospira species and not Nitrobacter species are the dominant nitrite oxidizers in sewage sludge, aquaria, and bioreactors (10, 15, 17, 25).
Genus-specific monoclonal antibodies (MAbs) that recognize the nitrite oxidoreductase (NOR) of Nitrobacter species may be used to overcome the problem of serological diversity. The NOR is ubiquitous in Nitrobacter species, and the MAbs react similarly with members of the species Nitrobacter hamburgensis, Nitrobacter winogradskyi, and Nitrobacter vulgaris (1). The MAbs designated Hyb 153-1 and Hyb 153-3 bind to the β subunit of the NOR (β-NOR), whereas the MAbs designated Hyb 153-2 recognize an epitope of the α-NOR (1). Immunological analyses revealed recently that Hyb 153-3 also detects the nitrite-oxidizing system (NOS) of Nitrospira species (29, 30).
In this study, immunoblot analyses provided evidence that the MAbs recognized the key enzymes of all genera of nitrite oxidizers that have been described so far. Since the immunoreactions were specific for each genus of nitrite oxidizers, the MAbs were also used to identify undescribed isolates and enrichment cultures. Immunoblot analyses of enrichment cultures obtained from activated sludge allowed us to identify Nitrobacter and Nitrospira strains which were cultivated on media containing different substrate concentrations. In addition, immunofluorescence (IF) labeling could be used to visualize whole cells from pure cultures and was therefore used to examine enrichment cultures obtained from activated sludge.
(This paper is based on the doctoral study of S. Bartosch at the University of Hamburg).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Nitrobacter hamburgensis X14 and Nitrobacter winogradskyi Engel (6, 7) were isolated from soil from the old Botanical Garden in Hamburg, Germany. Nitrobacter vulgaris K55 was obtained from sandstone of Cologne Cathedral (7). Nitrospira moscoviensis M-1 originated from an iron pipe in a heating system in Moscow, Russia (11), and Nitrospira marina 295 was isolated from seawater from the Gulf of Maine (38). The marine organisms Nitrospina gracilis 3/211 and Nitrococcus mobilis 231 have been described by Watson and Waterbury (37). Nitrobacter alkalicus AN 1 and AN 4 were isolated from a soda lake in Siberia and a soda soil in Kenya, respectively (27). Strains BS 5/6 and BS 5/13, which originated from the sulfidic ore mine in Baia Sprie, Romania, have not been described previously. Other previously undescribed nitrite-oxidizing bacteria, designated Ns (42°C) and Ns (47°C), were enriched from steel pipes in a heating system in Moscow. All of the strains have been deposited in the culture collection of the Institut für Allgemeine Botanik, Abteilung Mikrobiologie, Universität Hamburg.
Nitrobacter hamburgensis X14, Nitrobacter winogradskyi Engel, and Nitrobacter vulgaris K55 were grown mixotrophically, and Nitrobacter alkalicus AN 1 and AN 4 were grown lithoautotrophically in the presence of 2 g of NaNO2 liter−1 (7). Nitrospira moscoviensis M-1 and strains BS 5/6, BS 5/13, Ns (42°C), and Ns (47°C) were cultivated in lithoautotrophic medium supplemented with 0.2 g of NaNO2 liter−1 (11). Nitrospira marina 295 was grown in a seawater medium containing 0.4 g of NaNO2 liter−1 (38). Nitrospina gracilis 3/211 and Nitrococcus mobilis 231 were cultivated in seawater media as described by Watson and Waterbury (37). Most cultures were incubated at 28°C; the only exceptions were the Nitrospira moscoviensis M-1, Ns (42°C), and Ns (47°C) cultures, which were incubated at 37, 42, and 47°C, respectively.
Escherichia coli K-12 strain ATCC 23716, Pseudomonas putida ATCC 12633, and Micrococcus denitrificans NCIP 8944 were grown under anaerobic conditions in the presence of 2 g of NaNO3 liter−1. Nitrate and nitrite concentrations were monitored regularly by high-performance liquid chromatography (HPLC) (11).
Bradyrhizobium japonicum DSM 30131 and Rhodopseudomonas palustris DSM 126 were obtained from the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany. These organisms were cultivated as recommended by the German Collection of Microorganisms and Cell Cultures.
Activated sludge samples.
The activated sludge samples used originated from the aeration stage of the sewage treatment plant in Dradenau near Hamburg, Germany. Nitrite-oxidizing bacteria were enriched in mixotrophic medium containing 55 mg of sodium pyruvate liter−1, 150 mg of yeast extract liter−1, 150 mg of peptone liter−1, and 2 g of NaNO2 liter−1 or 0.2 g of NaNO2 liter−1.
MAbs.
MAbs were produced by Aamand et al. (1), who used purified NOR of Nitrobacter hamburgensis X14 as the antigenic peptide. The MAbs designated Hyb 153-1 and Hyb 153-3 recognize the β-NOR, while the MAbs designated Hyb 153-2 bind to the α-NOR.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting.
Cells were harvested by centrifugation, washed with 0.9% NaCl, and sonicated on ice for 15 to 30 min by using a Biorupter apparatus. The protein concentrations of the crude extracts were determined colorimetrically by using the method of Bradford (9), as modified by Spector (28). The protein concentrations of crude extracts of pure cultures were adjusted to 0.5 mg ml−1, and the protein concentrations of crude extracts of enrichment cultures were adjusted to 1 to 5 mg ml−1. Samples were diluted (1:1) with 10 mM Tris-HCl buffer (pH 6.8) containing 2% sodium dodecyl sulfate, 20% glycerol, 1% 2-mercaptoethanol, and 0.001% bromophenol blue and boiled for 8 min. Samples (10 μl) were loaded onto lanes of 1-mm-thick polyacrylamide gels prepared as described by Laemmli (22). The stacking and separating gels contained 4.5 and 10% polyacrylamide, respectively. Electrophoresis was performed at 30 mA by using a Minigel Twin apparatus (Biometra). The separated proteins were electroblotted (Pegasus, PHASE) onto a cellulose nitrate membrane (pore size, 0.2 μm; Schleicher & Schuell) by using a discontinuous buffer system (21). The proteins were transferred for 2 h at 0.8 mA per cm2 of cellulose nitrate membrane. The membrane was then blocked overnight in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA). The proteins on the cellulose nitrate membrane were incubated with MAbs (diluted 1:1,000 in PBS containing 0.05% BSA and 0.025% Tween 20) for 1 h at room temperature. Then they were incubated with alkaline phosphatase-conjugated secondary antibodies (Sigma) diluted 1:1,000 in PBS containing 0.05% BSA, 0.025% Tween 20, and 5% goat serum for 1 h at room temperature. After incubation with the antibodies, the cellulose nitrate membrane was washed twice with 10 mM Tris-HCl (pH 8.6) containing 0.02% BSA and 0.05% Tween 20. The membrane was then incubated with a substrate solution containing 0.005% 5-bromo-4-chloro-3-indolylphosphate (BCIP), 0.001% 4-nitroblue tetrazolium, 0.1 M NaHCO3, 0.05 M Na2CO3, and 0.004 M MgCl2. The enzymatic reaction was stopped by adding distilled water. A dense blue color indicated that a reaction was positive.
Cell fixation, IF labeling, and FISH.
Three different modified fixation procedures were used, as described by Beimfohr et al. (2). Cells were fixed in 3% formaldehyde for 1 h on ice and stored in PBS-ethanol (1:1) at −20°C; cells were fixed in 0.3% formaldehyde in ethanol for 1 h on ice and stored in PBS-ethanol at −20°C; or cells were not fixed with formaldehyde and were stored in PBS-ethanol at −20°C. The samples were placed on gelatin-coated slides and dehydrated by using 50, 80, and 96% ethanol (3 min each) (13). In the case of Nitrobacter cells an additional lysozyme treatment enhanced permeation of the MAbs (12). All of the samples were blocked with PBS containing 3% BSA for 30 min at room temperature. The samples were then incubated with the MAbs diluted 1:10 in PBS containing 0.05% BSA and 0.025% Tween 20 for 1 h at room temperature and with Cy3-labeled secondary antibodies (Biotrend) diluted 1:100 in PBS containing 0.05% BSA, 0.025% Tween 20, and 5% goat serum for 1 h at room temperature. The reactions were stopped by washing the slides in PBS. Control preparations without MAbs were included in every experiment.
Fluorescence in situ hybridization (FISH) was performed with oligonucleotide probes S-*-Ntspa-1026-a-A-18, specific for Nitrospira moscoviensis (17), and NIT3, specific for the genus Nitrobacter (36). To detect total cells, samples were stained with 4′,6-diamidino-2-phenylindole (DAPI) (10 μg ml−1) for 5 min.
Fluorescence microscopy and confocal laser scanning microscopy.
DAPI staining results were visualized by using Leica filter set A (BP 340-380 exc.; RKP 400; LP 425 em.). IF labeling and FISH results were visualized with a confocal laser scanning microscope (CLSM) (model TCS 4D; Leica); excitation was supplied by an argon-krypton laser (568 exc.; LP 590 em.). Image processing was performed with the standard software (Scanware 5.1; Leica).
Electron microscopy.
The methods used for cell fixation, embedding, ultrathin sectioning, and shadow casting were the methods described by Ehrich et al. (11). Electron microscopy was performed with a Philips model 420 transmission electron microscope.
RESULTS
Immunoblot analyses.
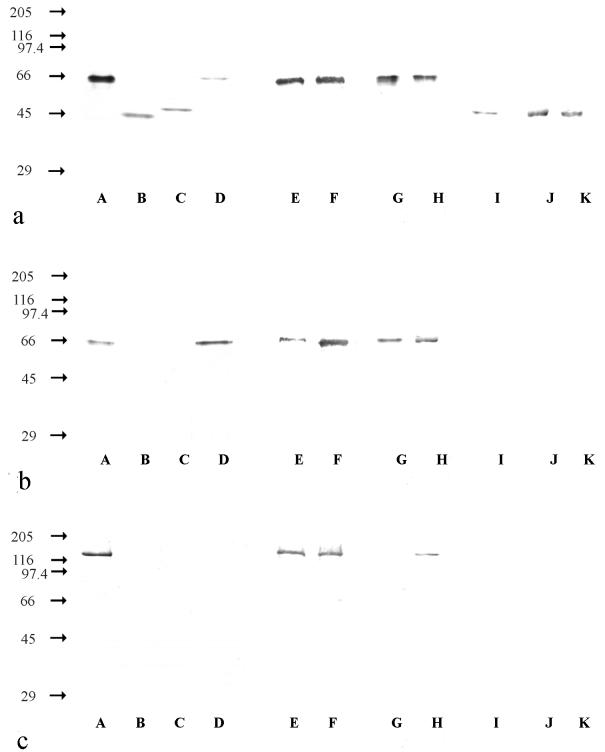
Figure 1a shows that MAb Hyb 153-3 was suitable for detecting all described genera of nitrite oxidizers. The MAb recognized proteins with molecular masses of 65 kDa in Nitrobacter hamburgensis X14, Nitrobacter alkalicus AN 1 and AN 4, and Nitrococcus mobilis 231. In Nitrospira moscoviensis M-1 and Nitrospira marina 295 Hyb 153-3 detected 46-kDa proteins. In addition, this MAb bound to a 48-kDa protein in Nitrospina gracilis 3/211. The proteins recognized by the MAbs in Nitrococcus, Nitrospira, and Nitrospina strains were considered the β-subunits of the NOS (β-NOS), analogous to the β-NOR of Nitrobacter strains. Hyb 153-1, which recognized the β-NOR of Nitrobacter strains, like Hyb 153-3, bound to proteins in Nitrobacter hamburgensis X14, Nitrobacter alkalicus AN 1 and AN 4, and Nitrococcus mobilis 231 that had molecular masses of 65 kDa (Fig. 1b). Unlike Hyb 153-3, Hyb 153-1 did not react with the β-NOS of Nitrospina gracilis 3/211, Nitrospira moscoviensis M-1, or Nitrospira marina 295. Hyb 153-2, which bound to the α-NOR of Nitrobacter strains, were specific for this genus. These MAbs recognized a protein with a molecular mass of 130 kDa in Nitrobacter hamburgensis X14 and Nitrobacter alkalicus AN 1 and AN 4 (Fig. 1c), but no cross-reactions with the members of the other genera anlayzed were observed. The immunoblot results are summarized in Table 1, which shows that the MAbs reacted specifically with the members of the different genera of nitrite oxidizers.
FIG. 1.
Immunoblots of different nitrite-oxidizing bacteria. (a) Hyb 153-3, which recognized the β-NOR. (b) Hyb 153-1, which recognized the β-NOR. (c) Hyb 153-2, which recognized the α-NOR. The values on the left are molecular masses (in kilodaltons). Lane A, Nitrobacter hamburgensis X14; lane B, Nitrospira moscoviensis M-1; lane C, Nitrospina gracilis 3/211; lane D, Nitrococcus mobilis 231; lane E, Nitrobacter alkalicus AN 1; lane F, Nitrobacter alkalicus AN 4; lane G, strain BS 5/6; lane H, strain BS 5/13; lane I, Nitrospira marina 295; lane J, strain Ns (42°C); lane K, strain Ns (47°C). All cells were disrupted by sonication and were added to the gel at protein concentrations of 0.25 to 1 mg ml−1.
TABLE 1.
Reactions of different MAbs with the nitrite oxidizers analyzed and phylogenetic relationships based on 16S rRNA sequences
| Taxon(s) | Phylogenetic position | MAb(s) | Size of NOS (kDa) |
|---|---|---|---|
| Nitrobacter hamburgensis, Nitrobacter vulgaris, Nitrobacter winogradskyi, Nitrobacter alkalicus | α-Proteobacteria | Hyb 153-1 | 65 |
| Hyb 153-2 | 130 | ||
| Hyb 153-3 | 65 | ||
| Nitrococcus mobilis | γ-Proteobacteria | Hyb 153-1 | 65 |
| Hyb 153-3 | 65 | ||
| Nitrospina gracilis | δ-Proteobacteria | Hyb 153-3 | 48 |
| Nitrospira moscoviensis, Nitrospira marina | Nitrospira phylum | Hyb 153-3 | 46 |
Based on the genus-specific reactions, the MAbs were used to determine the taxonomic affiliations of four unknown nitrite oxidizers. Two strains, designated BS 5/6 and BS 5/13, were isolated from the sulfidic ore mine in Baia Sprie, Romania (17a). Hyb 153-1 and Hyb 153-3 detected 65-kDa proteins in both of these strains (Fig. 1a and b). Hyb 153-2 reacted with a 130-kDa protein of strain BS 5/13 but not with strain BS 5/6 (Fig. 1c). Thus, strain BS 5/13 was identified as a member of the genus Nitrobacter, but BS 5/6 could not be classified yet.
Two additional strains, designated Ns (42°C) and Ns (47°C), were enriched from a heating system in Moscow (22a). Hyb 153-3 detected 46-kDa proteins (Fig. 1a) in crude extracts of these organisms, whereas no signals were obtained with Hyb 153-1 and Hyb 153-2 (Fig. 1b and c). These results indicated that Nitrospira cells were present in both cultures.
In addition, immunoblot analyses were performed with natural samples collected from activated sludge from the sewage treatment plant in Dradenau. In order to obtain detectable quantities of the nitrite-oxidizing enzymes, the nitrite oxidizers in the samples had to be enriched. This process was carried out by using mixotrophic media containing 2 or 0.2 g of NaNO2 liter−1. In the enrichment culture containing the high substrate concentration (2 g of NaNO2 liter−1), Hyb 153-3 recognized a protein with a molecular mass of 65 kDa (Fig. 2a), whereas Hyb 153-2 detected a 130-kDa protein (Fig. 2b). These results indicated that Nitrobacter cells were present. However, in the culture containing the lower substrate concentration (0.2 g of NaNO2 liter−1), Hyb 153-3 detected a protein with a molecular mass of 46 kDa (Fig. 2a). Since no signals were obtained with the Nitrobacter-specific MAbs Hyb 153-2 (Fig. 2b), the presence of Nitrospira cells was confirmed.
FIG. 2.

Immunoblots of enrichment cultures from activated sludge from the sewage treatment plant in Dradenau. (a) Hyb 153-3, which recognized the β-NOR. (b) Hyb 153-2, which recognized the α-NOR. The values on the left are molecular masses (in kilodaltons). Lane A, Nitrobacter hamburgensis X14; lane B, enrichment culture containing 2 g of NaNO2 liter−1; lane C, enrichment culture containing 0.2 g of NaNO2 liter−1; lane D, Nitrospira moscoviensis M-1. All cells were disrupted by sonication and were added to the gel at protein concentrations of 0.25 to 2.5 mg ml−1.
In order to prove that the MAbs were specific, additional control experiments had to be performed. In addition to the control experiments of Aamand et al. (1), nitrate-reducing pure cultures of E. coli, P. putida, and M. denitrificans were analyzed by the immunoblot procedure in order to examine possible serological similarities between the genetically closely related nitrate reductases NRZ and NRA of E. coli and NOR of Nitrobacter hamburgensis X14 (18). In the crude extracts of each strain, no signals were obtained with any of the MAbs (data not shown). In addition, immunoblot analyses were carried out with B. japonicum DSM 30131 and R. palustris DSM 126 because these strains are phylogenetically closely related to the genus Nitrobacter (35). Again, the MAbs did not react with crude extracts of these organisms.
IF labeling.
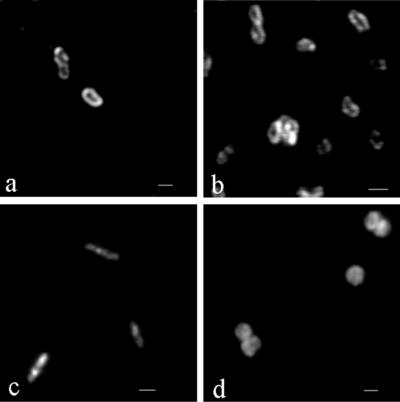
For microscopic in situ detection of nitrite oxidizers with the MAbs, IF labeling was performed with whole cells from pure cultures of members of the different genera. To obtain successful antibody penetration into the bacteria, various permeation procedures had to be used. For IF labeling of members of the genus Nitrobacter (Nitrobacter hamburgensis X14, Nitrobacter winogradskyi Engel, and Nitrobacter vulgaris K55) cells were stored directly in PBS-ethanol at −20°C and then treated with lysozyme. IF labeling was successful with Hyb 153-1, Hyb 153-2, and Hyb 153-3. After IF labeling, the cells exhibited bright signals at the cell periphery (Fig. 3a). Cells of Nitrospira moscoviensis M-1, Nitrospina gracilis 3/211, and Nitrococcus mobilis 231 could be labeled by the MAbs after treatment with formaldehyde. In the case of Nitrospira moscoviensis M-1 IF labeling occurred only with Hyb 153-3; fluorescence signals were observed at the cell periphery (Fig. 3b). After IF labeling of Nitrospina gracilis 3/211 with Hyb 153-3, the signals appeared to be spread over the whole cell (Fig. 3c). IF labeling of Nitrococcus mobilis 231 occurred with Hyb 153-3 and Hyb 153-1. In these cases, fluorescence signals were present at the cell periphery as well as in the cytoplasm (Fig. 3d). In control experiments without MAbs the cells exhibited no fluorescence signals. In addition, no IF labeling occurred with nitrate-reducing cultures of E. coli, P. putida, and M. denitrificans.
FIG. 3.
IF labeling with Hyb 153-3. (a) Nitrobacter vulgaris K55 (zoom step 7.4). (b) Nitrospira moscoviensis M-1 (zoom step 9.9). (c) Nitrospina gracilis 3/211 (zoom step 7.3). (d) Nitrococcus mobilis 231 (zoom step 7.3). Bars = 1 μm. The objective used was a Neoflutar objective (100×/1.4oil). The images were obtained with a CLSM by using different zoom steps (model TCS 4D microscope; Leica); excitation was provided by an argon krypton laser (568 exc.; LP 590 em.).
Based on the results described above, enrichment cultures obtained from activated sludge from the sewage treatment plant in Dradenau were analyzed by IF labelling. As determined by the immunoblot analysis, Nitrobacter cells were found in the enrichment culture containing 2 g of NaNO2 liter−1. Accordingly, the cells had to be permeated by storage in PBS-ethanol at −20°C and an additional lysozyme treatment, and IF labeling occurred with all of the MAbs (Fig. 4). In the enrichment culture containing 0.2 g of NaNO2 liter−1 Nitrospira cells were present according to the immunoblot results. In this case IF labeling failed the use of different permeation procedures use of 13, 14).
FIG. 4.
Epifluorescence micrographs of the enrichment culture from activated sludge from the sewage treatment plant in Dradenau containing 2 g of NaNO2 liter−1. (a) DAPI staining. (b) IF labeling with the Nitrobacter-specific Hyb 153-2. Bars = 5 μm. The objective used was a Neoflutar objective (100×/1.4oil). DAPI was visualized with Leica filter set A (BP 340-380 exc.; RKP 400; LP 425 em.), and IF labeling was visualized with Leica filter set I3 (BP 450-490 exc.; RKP 510; LP 515 em.).
FISH.
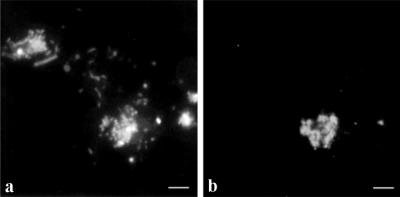
The immunoblot analysis indicated that Nitrobacter cells were present in the activated sludge enrichment culture containing 2 g of NaNO2 liter−1. This result was confirmed by FISH. The dominant cells were stained with the Nitrobacter-specific oligonucleotide probe NIT3 (36) (Fig. 5a and b). In contrast, FISH of the activated sludge enrichment culture containing the low substrate concentration (0.2 g of NaNO2 liter−1) succeeded with probe S-*-Ntspa-1026-a-A-18, which was specific for Nitrospira moscoviensis (17). Microcolonies of Nitrospira-like organisms occurring in tetrads were detected (Fig. 5c and d). This finding is consistent with the results of the immunoblot analysis, in which Nitrospira cells were detected as the dominant nitrite oxidizers. Nitrospira-like organisms were also found in the activated sludge from the sewage treatment plant in Dradenau when it was analyzed by FISH. Consistent with the results of the FISH analysis of the enrichment culture containing 0.2 g of NaNO2 liter−1, the organisms occurred in microcolonies (Fig. 5e and f). However, oligonucleotide probe NIT3 (36) did not detect any Nitrobacter cells in the activated sludge.
FIG. 5.
FISH analyses of activated sludge from the sewage treatment plant in Dradenau and subsequent enrichment of nitrite oxidizers. (a) Epifluorescence micrograph of DAPI-stained enrichment culture containing 2 g of NaNO2 liter−1. (b) CLSM image after FISH of panel a with oligonucleotide probe NIT3, which recognizes Nitrobacter species (36). (c) Epifluorescence micrograph of DAPI-stained enrichment culture containing 0.2 g of NaNO2 liter−1. (d) CLSM image after FISH of panel c with oligonucleotide probe S-∗-Ntspa-1026-a-A-18, which is specific for Nitrospira moscoviensis (17). (e) Epifluorescence micrograph of DAPI-stained activated sludge from the sewage treatment plant in Dradenau. (f) CLSM image after FISH of panel e with oligonucleotide probe S-∗-Ntspa-1026-a-A-18, which is specific for Nitrospira moscoviensis (17). Bars = 5 μm. The objective used was a Neoflutar objective (100×/1.4oil). DAPI was visualized with Leica filter set A (BP 340-380 exc.; RKP 400; LP 425 em.), and FISH was visualized with a CLSM (Leica model TCS 4D); excitation was provided by an argon krypton laser (568 exc.; LP 590 em.)].
Electron microscopic analyses.
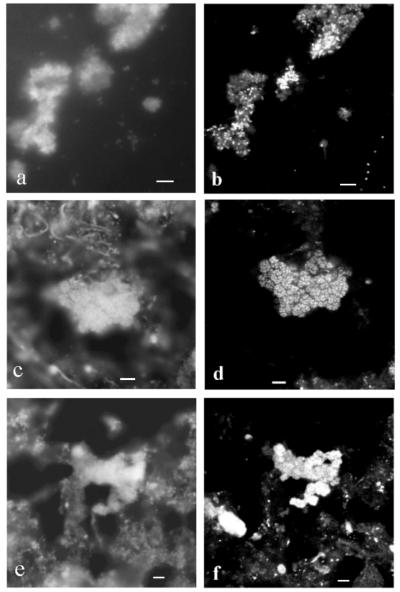
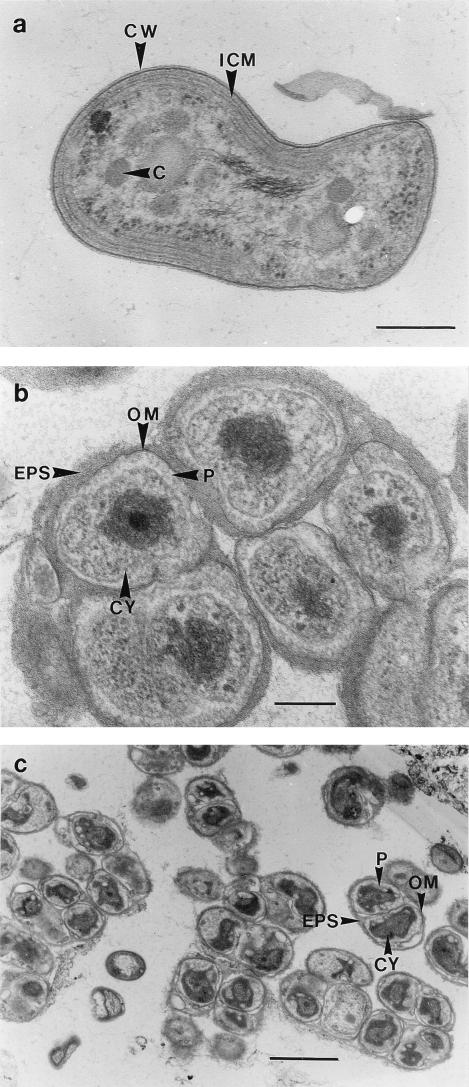
Electron microscopic investigations were used to visualize the dominant organisms in the enrichment cultures obtained from activated sludge. The dominant cells growing with 2 g of NaNO2 liter−1 had a morphology similar to the morphology of Nitrobacter cells (5, 7). They had a characteristic asymmetric cell wall with an electron-dense inner layer and a polar cap consisting of intracytoplasmic membranes (Fig. 6a). These findings are consistent with the results of the immunoblot analysis of this enrichment culture, in which Nitrobacter cells were identified (Fig. 2). In the enrichment culture growing in the presence of 0.2 g of NaNO2 liter−1, the dominant cells lacked intracytoplasmic membranes and carboxysomes and had characteristic enlarged periplasmic spaces (Fig. 6b). These properties are typical of both Nitrospira species (11, 38). Correspondingly, a 46-kDa protein was identified in the immunoblot analysis, which indicated that Nitrospira cells were present (Fig. 2a). Unlike cells of Nitrospira moscoviensis and Nitrospira marina, these cells were surrounded by a layer of extracellular polymeric substances (EPS). Furthermore, ultrathin sections of the activated sludge revealed microcolonies of Nitrospira-like cells, which were also surrounded by EPS (Fig. 6c).
FIG. 6.
Electron micrographs of ultrathin sections of activated sludge and enrichment cultures containing nitrite oxidizers. (a) Pleomorphic short rods were the dominant cells in the nitrite-oxidizing enrichment culture containing 2 g of NaNO2 liter−1. The cells each had an asymmetric cell wall and cytomembranes with an electron-dense layer on the inner side, a polar cap consisting of intracytoplasmic membranes, and carboxysomes like Nitrobacter. Bar = 0.25 μm. (b) Nitrospira-like cells were the dominant cells in the nitrite-oxidizing enrichment culture containing 0.2 g of NaNO2 liter−1. The cells had no intracytoplasmic membranes and carboxysomes but had enlarged periplasmic spaces. Bar = 0.25 μm. (c) Activated sludge from the sewage treatment plant in Dradenau contained microcolonies of Nitrospira-like cells. Bar = 1 μm. C, carboxysome; CW, cell wall; CY, cytoplasm; ICM, intracytoplasmic membranes; OM, outer membrane; P, periplasm.
DISCUSSION
We found that the MAbs that recognize the NOR of Nitrobacter species (1) allowed us to detect the NOS of Nitrococcus, Nitrospina, and Nitrospira species (Fig. 1). Table 1 shows that the immunoreactions were specific for each genus of nitrite oxidizers. The MAbs reacted identically with the four known Nitrobacter species (1; this study). Nitrobacter alkalicus was recently described by Sorokin et al. (27). As demonstrated in this study, the NOR of this species (Fig. 1) was serologically similar to the NOR of Nitrobacter winogradskyi, Nitrobacter hamburgensis, and Nitrobacter vulgaris (1). Nitrospira-specific reactions were observed with both of the Nitrospira species that have been described, Nitrospira moscoviensis and Nitrospira marina. The genera Nitrococcus and Nitrospina each consist of only one species, (37), which exhibited a genus-specific immunoreaction as well.
The reactions of the three MAbs were correlated with the phylogeny of nitrite oxidizers based on 16S rRNA sequences (Table 1). This indicated that the levels of similarity of the NOS of the genera Nitrococcus, Nitrospina, and Nitrospira to the NOR of the genus Nitrobacter were higher the more closely the organisms are related to the genus Nitrobacter. Originally, the three MAbs were developed for Nitrobacter hamburgensis X14 (a member of the alpha subclass of the class Proteobacteria [alpha-Proteobacteria] [35]). Nitrococcus mobilis, which belongs to the gamma-Proteobacteria (35), reacted with two of the MAbs (Fig. 1a and b). The molecular masses of the proteins detected were identical to the molecular mass of the β-NOR of Nitrobacter species (Fig. 1a and b). Accordingly, the locations of the nitrite-oxidizing enzymes of Nitrobacter and Nitrococcus species are similar. These enzymes are associated with the inner sides of the cytoplasmic and intracytoplasmic membranes (29, 31, 34). The genus Nitrospina (a member of the delta-Proteobacteria [35]) and the genus Nitrospira (which belongs to its own phylum [11]) are not as closely related to the genus Nitrobacter as the genus Nitrococcus is. Members of these genera reacted only with Hyb 153-3, and the molecular masses of their β-NOS differed from the molecular mass of the β-NOR of Nitrobacter species (Fig. 1a). Furthermore, the cellular locations of the NOS of Nitrospira and Nitrospina species are different than the cellular location of the NOR of Nitrobacter species. They have been found to be associated with the periplasmic sides of the cytoplasmic membranes (30, 31).
The genus-specific reactions of the MAbs were used to taxonomically classify three undescribed nitrite oxidizers. Immunoblot analyses revealed that strain BS 5/13 belonged to the genus Nitrobacter, whereas Ns (42°C) and Ns (47°C) were identified as members of the genus Nitrospira (Fig. 1a). Microscopic analyses performed by Kirstein (17a) and by Lebedeva (22a) confirmed these conclusions.
However, classification of strain BS 5/6 was not possible. This strain had morphological and ultrastructural similarities to Nitrobacter species (28a), and the 16S rRNA sequence revealed greater than 98% similarity with this genus (22b). In contrast, the protein profile (20) and G+C content (63%) (17a) of strain BS 5/6 differed from the protein profiles and G+C contents of Nitrobacter winogradskyi, Nitrobacter hamburgensis, and Nitrobacter vulgaris. Thus, strain BS 5/6 might belong to a new Nitrobacter species. Surprisingly, the Nitrobacter-specific MAbs Hyb 153-2 did not react with strain BS 5/6 (Fig. 1c). This result indicated that the NOS of strain BS 5/6 and the NOR of Nitrobacter species developed in different ways, although the 16S rRNA sequences of these organisms remained very similar. Since the genus Nitrobacter belongs to a phylogenetically young group (24, 26), few modifications of the 16S rRNA sequence are found in this genus. The Nitrobacter cluster is also closely associated with B. japonicum, R. palustris, Afipia clevelandensis, and Blastobacter denitrificans (24, 26, 35). Thus, 16S rRNA sequences alone could not be used to determine the taxonomic affiliations of such closely related organisms. According to Stackebrandt and Goebel (32), DNA-DNA reassociation studies are necessary to determine clear taxonomic affiliations for bacteria whose levels of 16S rRNA sequence similarity are greater than 97%. Additional studies to characterize strain BS 5/6 are in progress.
Moreover, the specific immunoreactions were useful for ecological studies. Different nitrite oxidizers originating from the activated sludge from the sewage treatment plant in Dradenau were identified by the MAbs after enrichment in particular media. Nitrobacter cells were enriched by using a high substrate concentration (2 g of NaNO2 liter−1) and were detected by the MAbs that bound to the 65-kDa β-NOR and the 130-kDa α-NOR (Fig. 2). This result was confirmed by a FISH analysis performed with Nitrobacter-specific oligonucleotide probe NIT3 (Fig. 5b). In addition, the electron microscopic analysis revealed pleomorphic rods with an ultrastructure like that of Nitrobacter cells (Fig. 6a). In the enrichment culture containing a low substrate concentration (0.2 g of NaNO2 liter−1), Nitrospira cells were identified in the immunoblot analysis by Hyb153-3, which reacted with 46-kDa β-NOS. A FISH analysis performed with oligonucleotide probe S-*-Ntspa-1026-a-A-18 (Fig. 5d) specific for Nitrospira moscoviensis (17) confirmed this result. Furthermore, investigations of ultrathin sections (Fig. 6b) revealed mainly cells with a morphology and ultrastructure like the morphology and ultrastructure of Nitrospira cells (11, 38). These results demonstrated that Nitrobacter and Nitrospira cells were present in the activated sludge. The different genera of nitrite-oxidizing bacteria could be selectively enriched by varying the substrate concentration. Whereas Nitrobacter strains tolerate high concentrations of nitrite (5, 7), Nitrospira strains are inhibited by 1 g of NaNO2 liter−1 (11, 38). Combined FISH analyses of nitrite ixodizers and in situ measurements of nitrite with microelectrodes in nitrifying biofilms (24a, 25) confirmed that Nitrobacter cells prefer microenvironments with higher nitrite concentrations (>0.5 mM), whereas Nitrospira cells dominate in microenvironments with lower nitrite concentrations (0 to 0.5 mM). However, in the activated sludge from the sewage treatment plant in Dradenau, the genus Nitrospira seemed to be the dominant genus of nitrite oxidizers. In this case FISH performed with oligonucleotide probe S-*-Ntspa-1026-a-A-18 (17) (Fig. 5f) and electron microscopic investigations (Fig. 6c) revealed characteristic microcolonies containing Nitrospira-like cells. Immunoblot analyses did not detect Nitrospira cells in the activated sludge. We supposed that the NOS concentration remained below the detection limit of this technique. In the activated sludge Nitrobacter cells were detected neither by FISH nor by the electron microscopic analysss. Obviously, the number of Nitrobacter cells remained below the detection limit of microscopic analyses. Accordingly, the concentration of Nitrobacter NOR was too low for detection with immunoblot analyses. These findings agreed with the results of recent studies (10, 15, 17, 25) in which Nitrospira species and not Nitrobacter species were described as the most abundant nitrite oxidizers in freshwater aquaria and sewage sludge. If Nitrospira species had been the dominant nitrite oxidizers in the activated sludge from the sewage treatment plant in Dradenau, Nitrobacter cells might have outcompeted Nitrospira cells during enrichment with 2 g of NaNO2 liter−1. Isolation of Nitrospira cells from this activated sludge is in progress.
IF labeling with the MAbs was possible when carried out with whole cells from pure cultures of nitrite oxidizers (Fig. 3). Unlike antibodies that recognize strain-specific epitopes outside the cell wall, the MAbs needed to penetrate the cells to reach the membrane-bound enzymes involved in nitrite oxidation. We accomplished this by using different permeation procedures for Nitrobacter, Nitrococcus, Nitrospina, and Nitrospira species. Although Nitrobacter species are gram negative, these organisms possess an additional layer on the inner side of the cell wall (8). Therefore, we had to use permeation procedures that are known to improve the permeation of gram-positive bacteria (2). Formaldehyde treatment enabled antibody penetration in Nitrococcus, Nitrospina, and Nitrospira species. IF labeling was successful with the same MAbs that reacted in the immunoblot analyses with the different genera of nitrite oxidizers. The IF signals in cells corresponded to the locations of the key enzymes at the cytomembranes (Fig. 3). In situ detection of Nitrobacter cells in the enrichment culture was possible with IF labeling (Fig. 4) with all of the MAb. In situ detection of Nitrospira-like cells has not been successful when we used a permeation procedure that permeated Nitrobacter cells in pure cultures. FISH was more successful. As revealed by ultrathin sections, Nitrospira-like cells were surrounded by slime coats consisting of EPS (Fig. 6b and c). Since the oligonucleotide probe molecules were smaller than the antibody molecules, these EPS might have hindered antibody penetration.
ACKNOWLEDGMENTS
We thank Jens Aamand of the Geological Survey of Denmark and Greenland for providing the MAbs, Christian Noah for helpful assistance, Helen Lebedeva and Dimitry Sorokin of the Academy of Science in Moscow for contributing enrichment cultures Ns (42°C) and Ns (47°C), respectively, and Nitrobacter alkalicus AN 1 and AN 4, and Michael Wagner of the Technical University in Munich for providing the oligonucleotide probes.
REFERENCES
- 1.Aamand J, Ahl T, Spieck E. Monoclonal antibodies recognizing nitrite oxidoreductase of Nitrobacter hamburgensis, N. winogradskyi, and N. vulgaris. Appl Environ Microbiol. 1996;62:2352–2355. doi: 10.1128/aem.62.7.2352-2355.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beimfohr C, Krause A, Amann R, Ludwig W, Schleifer K-H. In situ identification of lactococci, enterococci and streptococci. Syst Appl Microbiol. 1993;16:450–456. [Google Scholar]
- 3.Belser L W. Population ecology of nitrifying bacteria. Annu Rev Microbiol. 1979;33:309–333. doi: 10.1146/annurev.mi.33.100179.001521. [DOI] [PubMed] [Google Scholar]
- 4.Belser L W, Schmidt E L. Serological diversity within a terrestrial ammonia-oxidizing population. Appl Environ Microbiol. 1978;36:589–593. doi: 10.1128/aem.36.4.589-593.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bock E, Koops H-P. The genus Nitrobacter and related genera. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 2302–2309. [Google Scholar]
- 6.Bock E, Sundermeyer-Klinger H, Stackebrandt E. New facultative lithoautotrophic nitrite-oxidizing bacteria. Arch Microbiol. 1983;136:281–284. [Google Scholar]
- 7.Bock E, Koops H-P, Möller U C, Rudert M. A new facultatively nitrite oxidizing bacterium, Nitrobacter vulgaris sp. nov. Arch Microbiol. 1990;153:105–110. [Google Scholar]
- 8.Bock E, Koops H-P, Harms H, Ahlers B. The biochemistry of nitrifying organisms. In: Shively J M, Barton L L, editors. Variations in autotrophic life. London, United Kingdom: Academic Press; 1991. pp. 171–200. [Google Scholar]
- 9.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Burrell P C, Keller J, Blackall L L. Microbiology of a nitrite-oxidizing bioreactor. Appl Environ Microbiol. 1998;64:1878–1883. doi: 10.1128/aem.64.5.1878-1883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol. 1995;164:16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- 12.Hahn D, Amann R, Ludwig W, Akkermans A D, Schleifer K-H. Detection of micro-organisms in soil after in situ hybridisation with rRNA-targeted, fluorescently labelled oligonucleotides. J Gen Microbiol. 1992;138:879–887. doi: 10.1099/00221287-138-5-879. [DOI] [PubMed] [Google Scholar]
- 13.Hahn D, Amann R, Zeyer J. Detection of mRNA in Streptomyces cells by whole-cell hybridization with digoxigenin-labeled probes. Appl Environ Microbiol. 1993;59:2753–2757. doi: 10.1128/aem.59.8.2753-2757.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn D, Amann R, Zeyer J. Whole-cell hybridization of Frankia strains with fluorescence- or digoxigenin-labeled, 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1993;59:1709–1716. doi: 10.1128/aem.59.6.1709-1716.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hovanec T A, Taylor L T, Blakis A, Delong E F. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl Environ Microbiol. 1998;64:258–264. doi: 10.1128/aem.64.1.258-264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Josserand A, Cleyet-Marel J C. Isolation from soils of Nitrobacter and evidence for novel serotypes using immunofluorescence. Microb Ecol. 1979;5:197–205. doi: 10.1007/BF02013526. [DOI] [PubMed] [Google Scholar]
- 17.Juretschko S, Timmermann G, Schmid M, Schleifer K-H, Pommerening-Röser A, Koops H-P, Wagner M. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Kirstein, K. Personal communication.
- 18.Kirstein K, Bock E. Close genetic relationship between Nitrobacter hamburgensis nitrite oxidoreductase and Escherichia coli nitrate reductase. Arch Microbiol. 1993;160:447–453. doi: 10.1007/BF00245305. [DOI] [PubMed] [Google Scholar]
- 19.Koops H-P, Möller U C. The lithotrophic ammonia-oxidizing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 2625–2637. [Google Scholar]
- 20.Krause-Kupsch T. Entwicklung einer Schnellmethode zur Identifizierung und Klassifizierung nitritoxidierender Bakterien. Ph.D. thesis. Hamburg, Germany: University of Hamburg; 1993. [Google Scholar]
- 21.Kyse-Anderson J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22a.Lebedeva, H. Personal communication.
- 22b.Ludwig, W. Personal communication.
- 23.Matulewich V A, Strom P F, Finstein M S. Length of incubation for enumerating nitrifying bacteria present in various environments. Appl Microbiol. 1975;29:265–268. doi: 10.1128/am.29.2.265-268.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orso S, Gouy M, Navarro E, Normand P. Molecular phylogenetic analysis of Nitrobacter spp. Int J Syst Bacteriol. 1994;44:83–86. doi: 10.1099/00207713-44-1-83. [DOI] [PubMed] [Google Scholar]
- 24a.Schramm, A. Personal communication.
- 25.Schramm A, De Beer D, Wagner M, Amann R. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol. 1998;64:3480–3485. doi: 10.1128/aem.64.9.3480-3485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seewaldt E, Schleifer K-H, Bock E, Stackebrandt E. The close phylogenetic relationship of Nitrobacter and Rhodopseudomonas palustris. Arch Microbiol. 1982;131:287–290. [Google Scholar]
- 27.Sorokin D Y, Muyzer G, Brinkhoff T, Kuenen J G, Jetten M S M. Isolation and characterization of a novel facultatively alkaliphilic Nitrobacter species, N. alkalicus sp. nov. Arch Microbiol. 1998;170:345–352. doi: 10.1007/s002030050652. [DOI] [PubMed] [Google Scholar]
- 28.Spector T. Refinement of Coomassie-blue method of protein quantification. Ann Biochem. 1978;86:142–146. doi: 10.1016/0003-2697(78)90327-5. [DOI] [PubMed] [Google Scholar]
- 28a.Spieck, E. Unpublished data.
- 29.Spieck E, Aamand J, Bartosch S, Bock E. Immunocytochemical detection and location of the membrane-bound nitrite oxidoreductase in cells of Nitrobacter and Nitrospira. FEMS Microbiol Lett. 1996;139:71–76. [Google Scholar]
- 30.Spieck E, Ehrich S, Aamand J, Bock E. Isolation and immunocytochemical location of the nitrite oxidizing system in Nitrospira moscoviensis. Arch Microbiol. 1998;169:225–230. doi: 10.1007/s002030050565. [DOI] [PubMed] [Google Scholar]
- 31.Spieck, E., and E. Bock. The nitrite-oxidizing bacteria. In G. M. Garrity, T. W. Stanley, J. T. Staley, D. J. Brenner, J. G. Holt, D. R. Boone, R. W. Castenholz, N. R. Krieg, and H.-H. Schleifer (ed.), Bergey’s manual of systematic bacteriology, 2nd ed., in press. The Williams & Wilkins Co., Baltimore, Md.
- 32.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 33.Stanley P M, Schmidt E L. Serological diversity of Nitrobacter spp. from soil and aquatic habitats. Appl Environ Microbiol. 1981;41:1069–1071. doi: 10.1128/aem.41.4.1069-1071.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundermeyer H, Bock E. Characterization of the nitrite-oxidizing system in Nitrobacter. In: Bothe H, Trebst A, editors. Biology of inorganic nitrogen and sulfur. Berlin, Germany: Springer-Verlag; 1981. pp. 317–324. [Google Scholar]
- 35.Teske A, Alm E, Regan J M, Toze S, Rittmann B E, Stahl D A. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J Bacteriol. 1994;176:6623–6630. doi: 10.1128/jb.176.21.6623-6630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner M, Rath G, Koops H-P, Flood J, Amann R. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci Technol. 1996;34:237–244. [Google Scholar]
- 37.Watson S W, Waterbury J B. Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch Microbiol. 1971;77:203–230. [Google Scholar]
- 38.Watson S W, Bock E, Valois F W, Waterbury J B, Schlosser U. Nitrospira marina gen. nov. sp. nov: a chemolithotrophic nitrite-oxidizing bacterium. Arch Microbiol. 1986;144:1–7. [Google Scholar]