Abstract
Background and Aims:
Obesity predisposes to type 2 diabetes (T2D) and non-alcoholic fatty liver disease (NAFLD), but underlying mechanisms are incompletely understood. Potassium channel tetramerization domain-containing protein 17 (Kctd17) levels are increased in livers from obese mice and humans. In this study, we investigated mechanism of increased Kctd17 and whether it is causal to obesity-induced metabolic complications.
Methods:
We transduced Rosa26-LSL-Cas9 knock-in mice with AAV8-TBG-Cre (Control), AAV8-U6-Kctd17 sgRNA-TBG-Cre (L-Kctd17), AAV8-U6-Oga sgRNA-TBG-Cre (L-Oga), or AAV8-U6-Kctd17/Oga sgRNA-TBG-Cre (DKO). We fed mice high-fat diet (HFD) and assessed for hepatic glucose and lipid homeostasis. We generated Kctd17, O-GlcNAcase (Oga), or Kctd17/Oga-knockout hepatoma cells by CRISPR/Cas9, and Kctd17-directed antisense oligonucleotide (ASO) to test therapeutic potential in vivo. We analyzed transcriptomic data from patients with NAFLD.
Results:
Hepatocyte Kctd17 expression was increased in HFD-fed mice due to increased Srebp1c activity. HFD-fed L-Kctd17 or Kctd17 ASO-treated mice show improved glucose tolerance and hepatic steatosis, while forced Kctd17 expression caused glucose intolerance and hepatic steatosis even in lean mice. Kctd17 induced Oga degradation, resulting in increasing Chrebp protein, so concomitant Oga knockout negated metabolic benefits of hepatocyte Kctd17 deletion. In patients with NAFLD, KCTD17 mRNA was positively correlated with expression of Chrebp target and other lipogenic genes.
Conclusions:
Srebp1c-induced hepatocyte Kctd17 expression in obesity disrupted glucose and lipid metabolism by stabilizing Chrebp, and may represent a novel therapeutic target for obesity-induced T2D and NAFLD.
Keywords: Kctd17, Chrebp, Type 2 Diabetes, NAFLD
Lay Summary
Hepatocyte Kctd17 expression is increased in obesity, which predisposes to insulin resistance and liver fat accumulation. Blocking excess Kctd17 by either genetic or pharmacologic means reverses these pathologic processes, by decreasing Chrebp protein stability, a key factor that regulates both glucose and lipid metabolism.
Introduction
Non-alcoholic fatty liver disease (NAFLD), characterized by excessive hepatic fat accumulation, affects up to 30% of the overall adult population and 70-80% of individuals with obesity and diabetes 1, 2. NAFLD exists across a spectrum of severity 3, 4, but has quickly become a leading cause of liver transplantation 5-7, due to lack of effective therapeutic agents 8-10. A better understanding of mechanisms underlying NAFLD pathogenesis is necessary to develop novel targeted therapies.
Kctd17 (Potassium Channel Tetramerization Domain Containing 17), is a member of the Kctd family 11, soluble non-channel proteins involved in a wide variety of cell functions, including regulation of cellular proliferation, gene regulation, and cytoskeleton organization 12, 13. Kctd17 acts as an adaptor for the Cul3-RING E3 ubiquitin ligase, and has known roles in ciliogenesis and synaptogenesis 14-16, but Kctd17 functions outside the central nervous system are largely unknown, despite wide tissue distribution. For instance, we identified that Kctd17 expression is increased in livers from obese mice and patients with NAFLD/NASH (non-alcoholic steatohepatitis), where it acts to degrade Pleckstrin homology domain leucine-rich repeat protein phosphatase 2 (PHLPP2) to prolong insulin signaling by dephosphorylating Akt 17. But this study left many important questions, in particular, putative upstream regulators and other downstream effects of increased Kctd17.
Here, we show that hepatocyte Kctd17 is a sensitive Srebp1c transcriptional target in the obese liver. Further, forced hepatocyte Kctd17 expression induced glucose intolerance and hepatic lipid accumulation even in normal chow diet (NCD)-fed mice. Conversely, hepatocyte-specific Kctd17 deletion or treatment with Kctd17-directed antisense oligonucleotide (ASO) in high-fat diet (HFD)-fed mice improved glucose intolerance and fatty liver, by increasing carbohydrate response element-binding protein (Chrebp) protein stability via degradation of O-GlcNAcase (Oga). Correspondingly, hepatic KCTD17 expression correlates with expression of Pyruvate Kinase UR (PKLR) and other glycolytic and lipogenic genes in patients with NAFLD. These findings reveal Kctd17 as a key regulatory node in multiple liver metabolic processes, and a novel therapeutic target for obesity-induced insulin resistance and NAFLD.
Materials and Methods
Constructs
20 nucleotide single-guide RNA (sgRNA) sequences for Kctd17 or Oga were designed using a CRISPR design tool, cloned into LentiCRISPRv2 or pAAV-sgRNA-TBG-Cre. pAAV-TBG-HA-Kctd17 were subcloned from pcDNA3/HA/Kctd17 17. AAV8-sgRNA-TBG-Cre, AAV8-TBG-Cre, or AAV8-TBG-HA-Kctd17 were generated by Vigene (Rockville, MD). AAV8-U6-shScramble or AAV8-U6-shKctd17 was first cloned into the pENN-AAV-U6 vector, which was co-transfected with pAAV-RC2/8 and pAAV-Helper vector into AAVpro 293T cells (Takara) to generate recombinant AAV.
Animals
C57BL/6J (#664) and Rosa26-LSL-Cas9 knockin (#026175) 18 mice were purchased from Jackson Labs. Eight-week-old Cre- control or L-Kctd17 mice fed standard chow diet (Purina Mills 5053) were created by transduction of male Rosa26-LSL-Cas9 knockin with either AAV8-TBG-Cre or AAV8-U6-Kctd17 sgRNA-TBG-Cre. AAVs were used at a dose of 1.5 × 1011 genome copies per mouse. Mice were fed either standard chow diet or high-fat diet (HFD; Envigo, TD.06414). Mice were fasted for 18 h and then refed either standard chow or HFD for 8 h prior to sacrifice. Mice were housed 3-5 animals per cage, with a 12 h light/dark cycle, in a temperature-controlled environment. All animal experiments were approved by the Columbia University and Inha University Institutional Animal Care and Utilization Committee.
CRISPR generation of knockout (KO) cells and cell cultures studies
Lentivirus encoding Kctd17 or Oga sgRNAs was generated by co-transfection of 293T cells with the lentiviral vectors psPAX2 and pMD2.G. Viral supernatant was applied to Hepa1c1c7 cells with polybrene (Sigma), which were selected with puromycin (ThermoFisher). Knockout was confirmed by western blot. Primary hepatocytes were transfected with plasmid expressing Kctd17 or Oga using Lipofectamine 3000.
Cell isolation and glucose production
Primary hepatocytes and nonparenchymal cells (NPCs) were isolated as previously described 19-22. Isolated primary hepatocytes from C57BL/6J mice were plated on collagen-coated six-well dishes, and then transduced 2 h after plating with Ad-shControl or Ad-shKctd17 17 adenoviruses at MOI = 10. At 28 h after plating, cells were incubated for 4 h in glucose-free Krebs-Ringer buffer containing 20 mM sodium lactate and 2 mM sodium pyruvate, with or without dexamethasone and forskolin. Glucose released was measured using Glucose (HK) assay kit (Sigma) and normalized to cellular protein concentration (BCA assay, Pierce).
Antibodies, western blots and immunoprecipitation
Immunoblots were conducted on 3-7 samples randomly chosen within each experimental cohort with antibodies against DYKDDDDK-tag (#14793), HA-tag (#3724 or #2367), Myc-tag (#2276), Ogt (#24083), GFP (#2956), Lamin A/C (#4777) and β-actin (#4970) from Cell Signaling; Oga (ab124807) from Abcam; and Chrebp (NB400-135) from Novus, Inc; α-tubulin (T5158) from Sigma. Antibodies directed to Kctd17 were generated by immunizing rabbits with recombinant proteins, verified by ELISA and affinity purified (Lifetein, NJ).
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed by using SimpleChIP Enzymatic Chromatin IP kit following manufacturer’s instructions (Cell Signaling). Anti-SREBP1 (ab3259) from Abcam was used. Kctd17 promoter-specific primers or the negative control primers were as follows: forward (P1; Primer flanking the negative control region): 5’-CTA TTC TAA AAC TCC CCT TCC CC-3’; reverse (P1; Primer flanking the negative control region): 5’-TCC ATA TGT GCT TGA CCT GAA G-3’; forward (P2; Primer flanking the SRE binding site): 5’-TGT CCG AGC AAC ACC AGG ACA AG-3’; reverse (P2; Primer flanking the SRE binding site): 5’-CGC GTT GTC TGC ATC CTT CCC T-3’.
Dual luciferase reporter assay
Hepa1c1c7 cells seeded in 24-well culture plates (8 x 104 cells/well) were co-transfected with different luciferase plasmids (200 ng) and pRL-tk (Renillar luciferase, 10 ng) together with pcDNA3/SREBP1a, pcDNA3/SREBP1c plasmid 23 or empty vector control (200 ng). 24 h after transfection, cells were lysed and subjected to a luciferase activity assay using the Dual-Glo Luciferase Assay System (E1910, Promega). All experiments were performed in at least triplicate.
Metabolic analyses
Blood tail vein glucose was measured using a glucose meter (Bayer). Glucose tolerance tests, pyruvate tolerance tests, or insulin tolerance tests were performed by intraperitoneal injection after a 16 h or 5 h fast as per 24. Hepatic lipids were extracted by the Folch method 25, and plasma and hepatic triglyceride, Cholesterol (ThermoFisher), or NEFA (Wako) measured using colorimetric assays according to the manufacturer’s protocol. Plasma insulin was measured using a mouse insulin ELISA kit (Mercodia).
RNA/quantitative PCR
RNA was isolated by TRIzol (Invitrogen) or NucleoSpin RNA (Clontech), and cDNA synthesized with the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems), followed by quantitative RT-PCR with Power SYBR Green PCR master mix (Applied Biosystems) in a CFX96 Real-Time PCR detection system (Bio-Rad).
Human transcriptomic analysis
All raw transcriptomic data related to human NAFLD samples are publicly available on Gene Expression Omnibus under accession number GSE135251 26. Detailed phenotypic descriptions and demographics were reported previously 26. The cohort was stratified according to NAFLD activity score (NAS) and fibrosis score. The expression value of each gene in the two disease states was compared as previously reported 27, 28. A Wilcoxon one-sided test was employed to test statistical significance and the results were visualized in boxplots (median ± quartiles). The Kruskal Wallis test was used to evaluate the expression of KCTD17 at the parameter level, and results presented as a p-value. Correlation matrices were generated based on Spearman’s correlation coefficient rho. All computations were conducted using the R program (RStudio 1.4).
ASOs
Non-specific control ASO, Kctd17 ASO-1 (5’-AGG TAA TGA TTG TAG C-3’), and Kctd17 ASO-2 (5’- AAG GTA ATG ATT GTA G-3’) were synthetized by Ionis Pharmaceuticals, diluted in saline before injection and administered by intraperitoneal (IP) injection to male C57BL/6J mice at a dose of 25 mg/kg body weight (BW) once weekly for 6 weeks prior to sacrifice.
Statistical analysis
All data shown as mean ± s.d. Differences between two groups were calculated using a two-tailed Student’s t test or one-tailed Wilcoxon rank sum test. Analyses involving multiple groups were performed using one-way ANOVA or Kruskal-Wallis test. P < 0.05 was considered statistically significant.
Results
Hepatocyte-specific knockout of Kctd17 ameliorates HFD-induced liver steatosis and glucose homeostasis
To determine the source of increased Kctd17 in livers from obese mice, we isolated parenchymal cells (PCs) and non-parenchymal cells (NPCs) from livers of C57BL/6 wild-type (WT) mice fed either normal chow diet (NCD) or high-fat diet (HFD). While NPC Kctd17 expression did not change, hepatocyte Kctd17 was specifically induced in HFD-fed mice (Figure 1A).
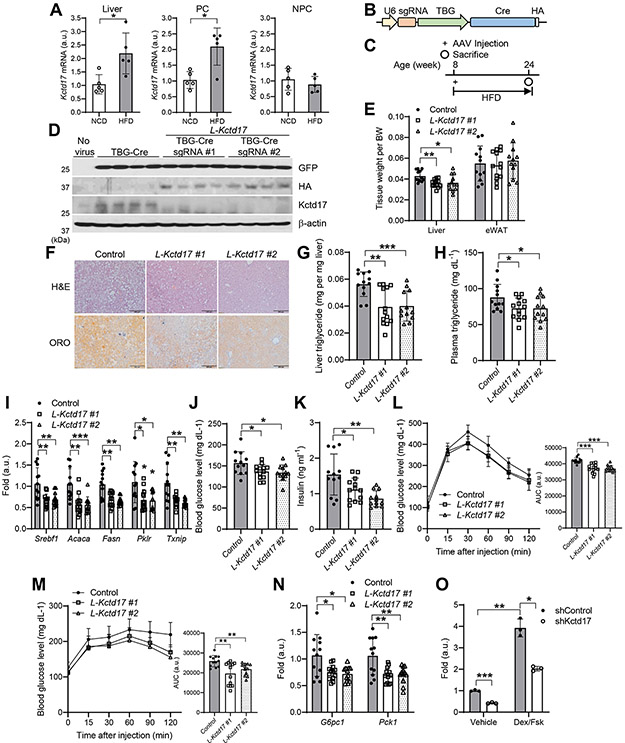
Figure 1. Hepatocyte-specific knockout of Kctd17 ameliorates HFD-induced liver steatosis and glucose homeostasis.
(A) Kctd17 expression in whole liver, or isolated parenchymal cells (PC) and nonparenchymal cells (NPC) in C57BL/6 WT mice fed normal chow (NCD) or high-fat diet (HFD) for 4 weeks (n=5 per group). (B, C) Cas9 knockin (KI) mice (B) were transduced with AAV8 expressing U6-driven Kctd17 sgRNA or TBG-Cre and then fed HFD diet for 16 weeks prior to sacrifice (C). (D) Western blots in livers from NCD-fed control (no virus) or HFD-fed Cas9 KI mice transduced with AAV8 expressing TBG-Cre alone or TBG-Cre with different Kctd17 sgRNAs (L-Kctd17 #1 and #2). (E) Tissue weight, (F) H&E and Oil Red O (ORO) staining, (G and H) liver and plasma triglyceride, and (I) liver gene expression in control and L-Kctd17 mice fed HFD for 16 weeks (n=12 to 13 per group). (J and K) Fasted blood glucose and insulin, (L and M) glucose and pyruvate tolerance tests (left), AUC of GTT and PTT (right), and (N) liver gluconeogenic gene expression in control and L-Kctd17 mice fed HFD for 16 weeks (n=12 to 13 per group). (O) Glucose production in primary hepatocytes transduced with adenovirus expressing shControl or shKctd17, normalized to total protein (n=3 per group). *P < 0.05, **P < 0.01 as compared to the indicated control by two-way ANOVA. All data are shown as the means ± s.e.m.
In order to investigate repercussion of increased hepatocyte Kctd17 in obesity, we generated hepatocyte-specific Kctd17 knockout (L-Kctd17) mice using CRISPR/Cas9 (Figure 1B) . Cre-dependent Cas9 KI mice were transduced with AAV8 expressing U6-sgRNA targeting Kctd17 and Cre recombinase controlled by the hepatocyte-specific TBG promoter, or AAV8-TBG-Cre as a control, then fed HFD for 16 weeks (Figure 1C). To ensure reproducibility, we used two different sgRNA (hereafter referred to as L-Kctd17 #1 or L-Kctd17 #2), both of which showed strong liver-specific, especially hepatocyte-specific, deletion (Figure 1D, Supplementary Figure 1A and 1B). Despite unchanged body weight (Supplementary Figure 1C) , both HFD-fed L-Kctd17 strains showed lower liver weight, hepatic lipid droplet accumulation and hepatic and plasma TG levels as compared to control littermates (Figure 1E-1H), without differences in adiposity, hepatic and plasma cholesterol (Figure 1E and Supplementary Figure 1D, 1E). Hepatic expression of genes involved in lipogenesis were decreased in livers of HFD-fed L-Kctd17 mice (Figure 1I), while the expression of genes involved in fatty acid oxidation (FAO) (Supplementary Figure 1F), fatty acid uptake and VLDL secretion (data not shown) were similar to that of control littermates.
These data largely match that of mice overexpressing hepatic PHLPP2, which is targeted by Kctd17 for degradation 17, 21. But not seen with PHLPP2 manipulation, expression of Pklr (Pyruvate kinase l/r) and Txnip (Thioredoxin interacting protein), direct targets of Chrebp 29, 30, was markedly lower in livers of HFD-fed L-Kctd17 mice (Figure 1I). As Pklr is one of the rate-limiting enzymes of glycolysis, we evaluated whether Kctd17 affected glucose homeostasis. Kctd17 deletion appeared to increase insulin sensitivity (Supplementary Figure 1G), with lower fasted glucose (Figure 1J) and insulin levels (Figure 1K). Consistent with improved hepatic insulin sensitivity, L-Kctd17 mice showed improved glucose and pyruvate tolerance and lower gluconeogenic gene expression (Figure 1L-1N). Primary hepatocytes transduced with adenovirus encoding Kctd17 shRNA produced less glucose (Figure 1O), implying a cell-autonomous mechanism.
As a second method to test these effects, we generated AAV8 expressing shRNA directed against either scramble or Kctd17 (Supplementary Figure 2A). Similar to L-Kctd17 mice, shRNA-mediated knockdown of Kctd17 lowered liver and plasma triglyceride, as well as lipogenic and glycolytic gene expression, with commensurately improved glucose and pyruvate tolerance as compared to shScramble-transduced mice, without change in other metabolic parameters (Supplementary Figure 2B-2I). In sum, these data suggest that hepatocyte Kctd17 regulates hepatic insulin sensitivity and downstream effects on lipogenesis and glucose production.
Forced hepatocyte Kctd17 expression provokes liver steatosis
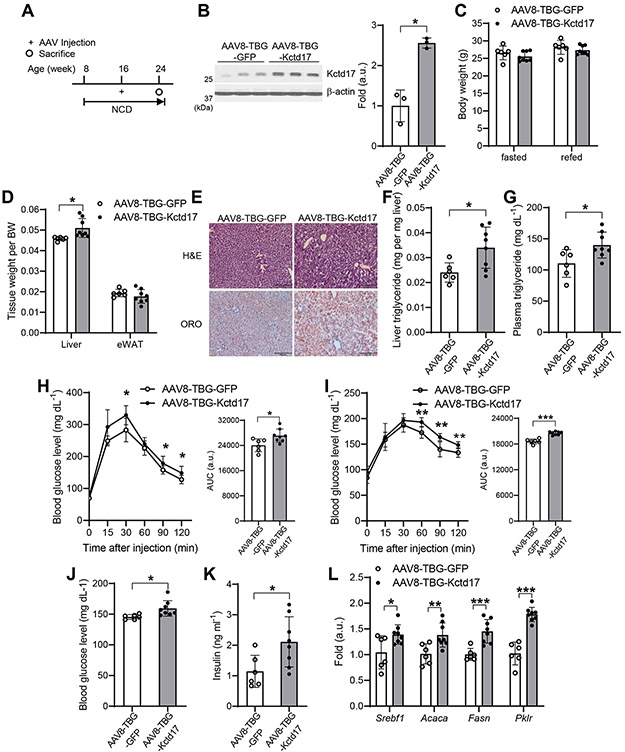
To study whether Kctd17 is sufficient to induce obesity-related phenotypes, we generated AAV8 expressing Kctd17 under the control of the TBG promoter (Figure 2A, 2B and Supplementary Figure 3A, 3B). Despite unchanged body weight and food intake (Figure 2C and data not shown), but forced hepatocyte Kctd17 expression increased liver weight, lipid droplet accumulation, hepatic TG content, plasma TG and phosphorylation of Akt (Figure 2D-2G and Supplementary Figure 3C), without change in cholesterol levels (Supplementary Figure 3D, 3E). In parallel to disrupted lipid metabolism, increased hepatocyte Kctd17 induced glucose and pyruvate intolerance as well as impaired insulin sensitivity, leading to increased glucose and insulin levels (Figure 2H-2K and Supplementary Figure 3F). Upon sacrifice, we observed increased lipogenic, Pklr and Pnpla3 (Patatin-like phospholipase domain-containing protein 3) gene expression in Kctd17 transgenic animals, while genes associated with FAO, (Ppara, Acox, Cpt1a), cholesterol synthesis (Hmgcr, Hmgcs1, Ldlr), VLDL secretion (Mttp) and fatty acid uptake (Cd36) were unchanged as compared to control littermates (Figure 2L and Supplementary Figure 3G-3J). These data suggest increased Srebp1c and Chrebp, but not Srebp2, activity. Consistent with these data, a similar increase of Kctd17 expression in vitro resulted in a similar induction of lipogenic and glycolytic gene expression, compared with no further changes seen at higher Kctd17 (Supplementary Figure 3K), suggesting that even modest increase in hepatocyte Kctd17 is sufficient to disrupt hepatic lipid and glucose metabolism, likely through effects on lipogenic and glycolytic pathways.
Figure 2. Forced hepatocyte Kctd17 expression provokes liver steatosis.
(A) NCD-fed C57BL/6 WT mice were transduced with AAV8-TBG-GFP or AAV8-TBG-Kctd17 and sacrificed 8 weeks after transduction. (B) Kctd17 protein levels, (C) body weight, (D) tissue weight, (E) liver H&E and ORO staining, (F and G) liver and plasma triglyceride levels, (H and I) glucose and pyruvate tolerance tests (left), AUC of GTT and PTT (right), (J and K) blood glucose and insulin, and (L) liver gene expression (n=6 to 8 per group). *P < 0.05, **P < 0.01, ***P < 0.001 as compared to the indicated control by two-way ANOVA. All data are shown as the means ± s.e.m.
Oga-Chrebp pathway mediates the lipogenic action of Kctd17
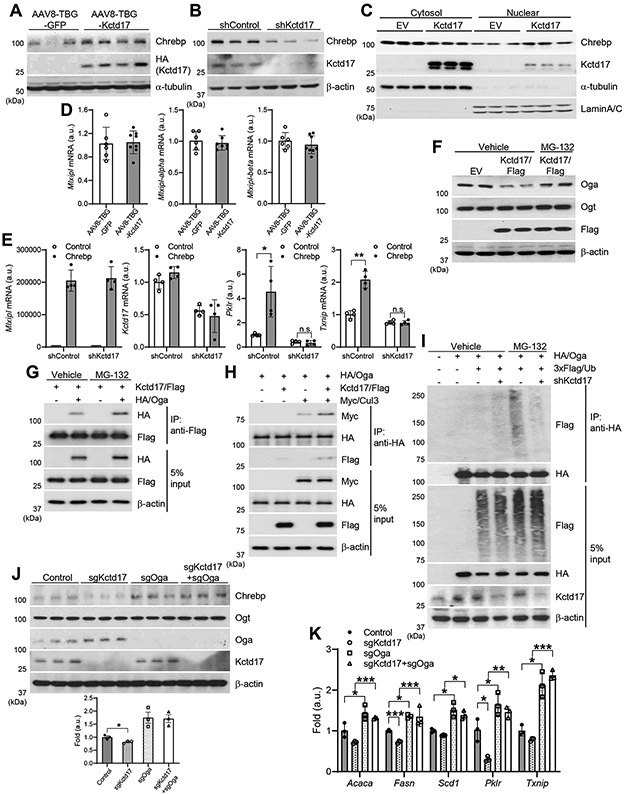
We next investigated mechanism by which Kctd17 affected hepatic metabolic pathways in obese mice. We focused on Chrebp due to its known roles in glycolysis and lipogenesis 29, as well as our preliminary data on Pklr. Indeed, Chrebp protein levels, especially in the nuclear fraction, were markedly affected by Kctd17 – higher in gain- and lower in Kctd17 loss-of-function, respectively (Figure 3A-3C) – but surprisingly, with unchanged expression of total Mlxipl (Chrebp), including both alpha and beta isoforms (Figure 3D). These data suggested a post-transcriptional impact of Kctd17 to regulate Chrebp protein. Consistent with this hypothesis, forced expression of Chrebp induced Pklr and Txnip, but not in cells with simultaneous Kctd17 knockdown (Figure 3E).
Figure 3. Kctd17 prevents Oga-mediated Chrebp degradation.
(A) Western blots in livers from AAV8-TBG-GFP or AAV8-TBG-Kctd17-transduced mice. (B) Western blots from primary hepatocytes transduced with adenovirus expressing shControl or shKctd17. (C) Western blots in cytosol and nuclear fractions isolated from control (empty vector) or Kctd17-overexpressed cells. (D) Chrebp expression in livers from AAV8-TBG-GFP and AAV8-TBG-Kctd17-transduced mice. (E) Gene expression in primary hepatocytes transduced with adenovirus expressing shControl or shKctd17 with or without Chrebp. (F) Western blots in primary hepatocytes transfected with empty vector or Flag-tagged Kctd17 with or without MG-132. (G and H) Co-immunoprecipitation of transfected Oga, Kctd17 and Cul3 in primary hepatocytes with or without MG-132. (I) Oga ubiquitination in 3x Flag/Ub-transfected primary hepatocytes. (J and K) Western blots and gene expression in control, Kctd17 KO, Oga KO, or Kctd17/Oga double KO Hepa1c1c7 cells. *P < 0.05, **P < 0.01, ***P < 0.001 as compared to the indicated control by two-way ANOVA. All data are shown as the means ± s.e.m.
Chrebp is modified by O-linked β-N-acetylglucosamine (O-GlcNAc) 31. O-GlcNAc modifications are highly dynamic, mediated by two opposing enzymes: O-GlcNAc transferase (Ogt) and O-GlcNAcase (Oga). Oga catalyzes the hydrolytic cleavage of O-GlcNAc from protein substrates 32, 33. Of the two, we observed a specific reduction in Oga protein when Kctd17 levels are high, without change in Oga mRNA (Figure 3F and Supplementary Figure 4A, 4B). Kctd17-mediated Oga reduction was fully prevented by concomitant MG132 treatment to inhibit proteasomal degradation (Figure 3F), which also increased the Kctd17-Oga interaction (Figure 3G). As Kctd17 can act as an adaptor that links substrates with the Cul3-RING ubiquitin ligase to promote proteasomal degradation 14, 17, we tested and observed that Kctd17 increased Oga-Cul3 interaction (Figure 3H), and that Kctd17 knockdown diminished Oga ubiquitination (Figure 3I).
We next tested consequence of Kctd17-mediated Oga degradation on Chrebp protein stability. We transduced hepatoma cells with gRNA to reduce Oga levels, which completely reversed effects of Kctd17 deletion on Chrebp protein stability (Figure 3J), as well as on Chrebp transcriptional activity (Figure 3K). In sum, these data show that increased Kctd17 in the obese liver leads to Oga proteasomal degradation, which provokes Chrebp activity, leading to increased expression of lipogenic and glycolytic target genes.
Oga deletion negates Kctd17 effects on glucose and lipid metabolism
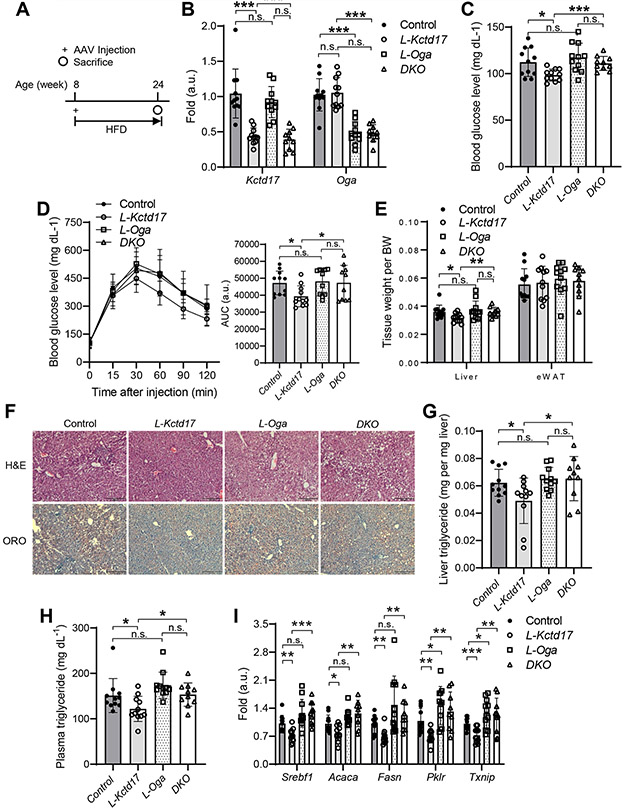
To determine the functional consequences of Kctd17-mediated Oga degradation on lipid accumulation and glucose homeostasis in vivo, we transduced HFD-fed Cas9 KI mice with sgRNA to Kctd17 as before, but simultaneously deleted Oga (Figure 4A, 4B and Supplementary Figure 5A). Despite similar body weight (Supplementary Figure 5B), we again found that mice lacking hepatocyte Kctd17 showed improved glucose homeostasis, with reduced fasting glucose and improved glucose intolerance as compared with control littermates (Figure 4C, 4D). Concomitant deletion of Oga, however, fully reversed this protection, as double-knockout (DKO) mice showed similar glucose intolerance as control animals (Figure 4C, 4D). As in prior cohorts, L-Kctd17 mice had smaller livers and decreased hepatic steatosis, but these phenotypes too were prevented by absence of Oga (Figure 4E-4H and Supplementary Figure 5C, 5D). These data parallel increased lipogenic and Chrebp target gene expression (Figure 4I), suggesting that Kctd17 affects glucose and lipid homeostasis by regulation of Oga, and consequently Chrebp.
Figure 4. Oga deletion negates Kctd17 effects on glucose and lipid metabolism.
(A) Cas9 KI mice were transduced with AAV8 expressing TBG-Cre (Control) or U6-driven Kctd17 (L-Kctd17), Oga (L-Oga), or both Kctd17/Oga (DKO) sgRNA prior to HFD diet-feeding for 16 weeks before sacrifice. (B) Liver gene expression, (C) blood glucose and (D) glucose tolerance test (left), AUC of GTT (right), (E) tissue weight, (F) liver H&E/ORO staining, (G) liver and (H) plasma triglyceride, and (I) liver lipogenic or Chrebp target gene expression (n=10 to 11 per group). *P < 0.05, **P < 0.01 as compared to the indicated control by two-way ANOVA. All data are shown as the means ± s.e.m.
Kctd17 expression is dependent on hepatocyte Srebp1c activity
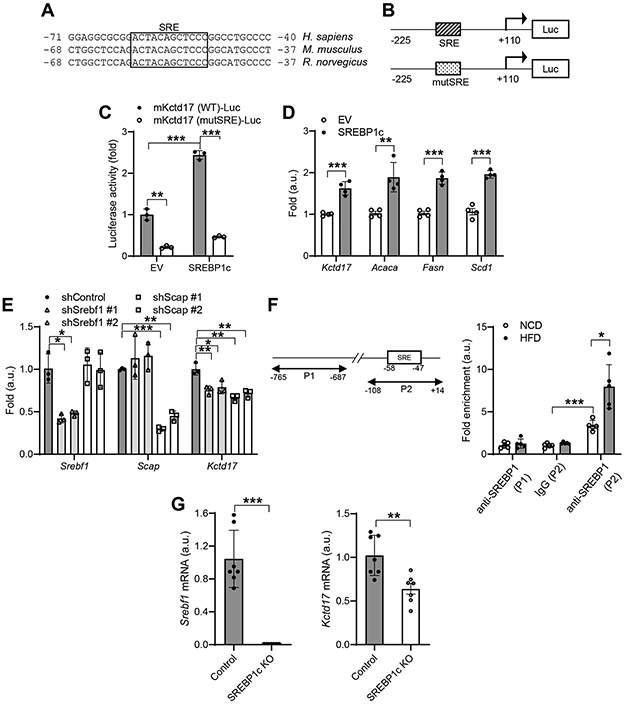
We next investigated mechanism of increased hepatocyte Kctd17 expression in obesity. We noted a consensus, evolutionarily conserved Sterol regulatory element-binding protein (SREBP)-response element (SRE) in the Kctd17 promoter (Figure 5A). As SREBP1c activity is increased in the obese liver, to test whether this was a functional promoter element, we generated Kctd17-luciferase constructs with wildtype or mutated SRE (Figure 5B). Using these tools, we found that Kctd17 promoter activity was significantly increased by SREBP1c expression in Hepa1c1c7 cells, but not when the SRE was mutated (Figure 5C). Commensurately, forced SREBP1c expression increased endogenous Kctd17 gene expression (Figure 5D), while shRNA-mediated knockdown of Srebpf1 suppressed Kctd17 gene expression (Figure 5E). Importantly, we saw the same reduction in Kctd17 with knockdown of SREBP cleavage-activating protein (Scap) (Figure 5E), which is necessary for endogenous, insulin-stimulated SREBP1c activity 34, 35. These data parallel results from chromatin immunoprecipitation (ChIP) assays showing both basal, but in particular, HFD-stimulated SREBP1 binding of the SRE site in the Kctd17 promoter (Figure 5F). To test this regulation in vivo, we analyzed livers from HFD-fed control or SREBP1c KO mice 36. Consistent with previous results, liver Kctd17 was increased in HFD-fed control mice, but this increase was significantly blunted in HFD-fed SREBP1c KO mice (Figure 5G). Effects of Kctd17 knockdown or overexpression on Plkr and Txnip gene expression were preserved in in Srebpf1 or Scap-deficient cells, supporting the conclusion that Srebp1c is an upstream regulator of Kctd17 (Supplementary Figure 6A, 6B). Overall, these data show that increased SREBP1c activity induces hepatocyte Kctd17 expression in the obese liver. When combined with previous results, these data suggest a feed-forward Srebp1c-Kctd17-Chrebp loop to augment hepatic lipid accumulation and glucose production.
Figure 5. Kctd17 is activated by hepatocyte Srebp1c.
(A) Evolutionarily-conserved SREBP-response element (SRE) in the Kctd17 promoter. (B) Schematic of WT SRE and mutated (mut) luciferase constructs of the Kctd17 promoter and (C) Kctd17 promoter luciferase activity, or (D) expression of Kctd17 mRNA in Hepa1c1c7 cells transfected with empty vector (EV) or SREBP1c (n=3 per group). (E) Kctd17 expression in Hepa1c1c7 cells after shRNA-mediated knockdown of Srebp1 or Scap. (F) Primers for the ChIP assay flanking a negative control region (P1) or SRE binding site (P2) in the Kctd17 promoter (left). ChIP assay for SREBP occupancy at the Kctd17 promoter in livers from mice fed NCD or HFD for 16 weeks (n=5 per group) (right). (G) Liver Srebp1c and Kctd17 in control or SREBP1c knockout mice fed HFD for 12 weeks (n=7 to 8 per group). *P < 0.05, **P < 0.01, ***P < 0.001 as compared to the indicated control by two-way ANOVA. All data are shown as the means ± s.e.m.
Liver KCTD17 expression tracks with ChREBP activity in patients with NAFLD
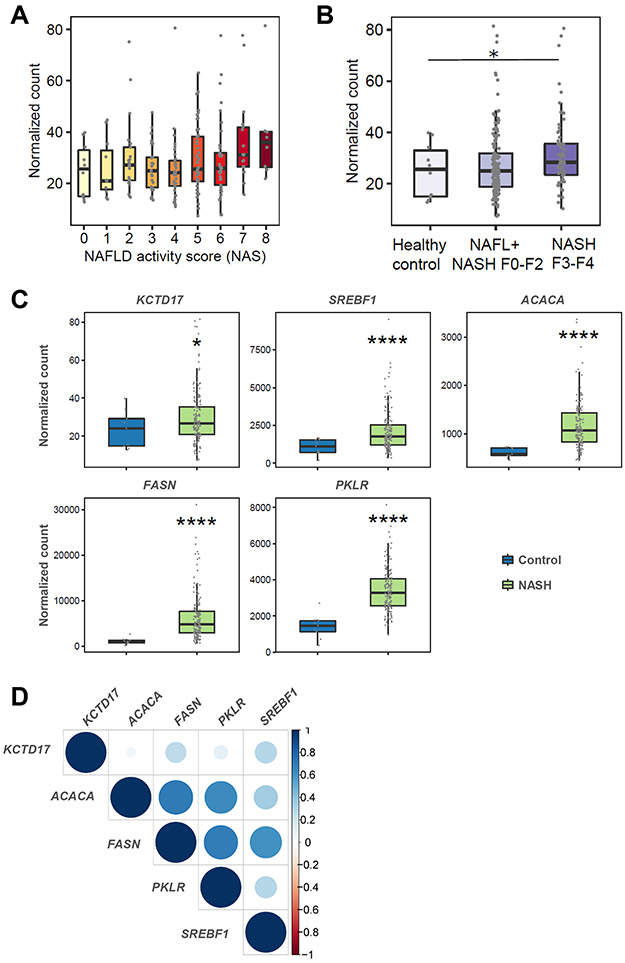
We next investigated potential associations between liver KCTD17 gene expression and markers of NAFLD severity, leveraging transcriptomic data collected from patients with NAFLD. Consistent with data from an independent patient cohort 17, we found a positive association between KCTD17 and NAFLD activity score (NAS), but now additionally show a significant association with NAFLD-induced liver fibrosis across the groups (Figure 6A, 6B). In paired analysis, KCTD17 expression was significantly higher in 206 patients with NASH as compared with healthy controls (Figure 6C), which paralleled increased lipogenic and glycolytic gene expression in this cohort (Figure 6C, 6D). These data suggest that the functional Srebp1c-Kctd17-Chrebp loop identified in mice likely translates to patients with NAFLD.
Figure 6. KCTD17 expression tracks with hepatic ChREBP activity and NAFLD severity in patients.
(A and B) KCTD17 expression in patients as related to NAS (p=0.004) (A) or NAFLD-induced liver fibrosis (B). Each dot represents hepatic KCTD17 expression in one subject. (C) KCTD17, lipogenic and glycolytic gene expression in patients with NAFLD/NASH as compared with healthy controls. (D) Correlation between KCTD17 and PKLR, as well as key lipogenic genes. Correlogram represents the value of Spearman's correlation coefficient rho. The intensity of color and the size of circle indicates Spearman's rho. *P < 0.05, **P < 0.01, ****P < 0.0001; p-values calculated using Kruskal–Wallis test or one-tailed Wilcoxon rank sum test.
Kctd17 ASO protects from diet-induced glucose intolerance and liver steatosis
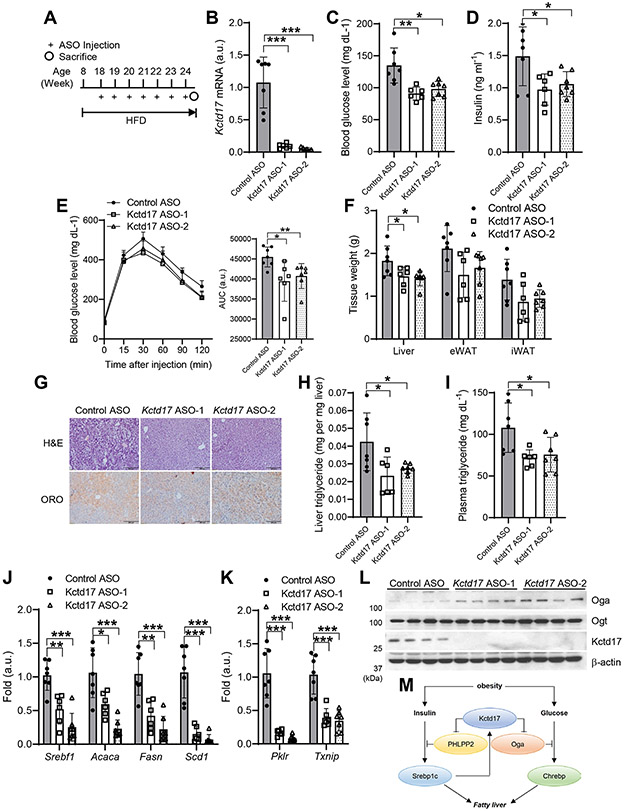
These results led us to evaluate whether liver-selective Kctd17 antagonism may provide a novel therapeutic strategy for obesity-induced metabolic dysfunction. To this end, we screened 384 ASOs for in vitro ability to reduce Kctd17 mRNA expression (not shown) and moved the two most potent ASOs forward for in vivo testing in WT mice. We administered these ASO (Kctd17 ASO-1 and −2) as well as a control ASO of the same chemistry and oligonucleotide length to HFD-fed WT mice (Figure 7A). Kctd17-directed ASO treatment reduced liver Kctd17 mRNA (Figure 7B), fasting glucose and insulin, and improved glucose tolerance (Figure 7C-7E), without significant change in body weight (Supplementary Figure 7A). Also consistent with results from L-Kctd17 mice, application of Kctd17 ASOs reduced liver weight, lipid droplet accumulation, hepatic and plasma triglyceride (Figure 7F-7I), and decreased lipogenic and glycolytic gene expression (Figure 7J, 7K) without significant change in fatty acid oxidation-related gene expression (Supplementary Figure 7B), likely due to increased Oga (Figure 7L). These data suggest that Kctd17 inhibition can ameliorate obesity-related glucose and lipid metabolic derangements.
Figure 7. Kctd17 ASO protects from diet-induced glucose intolerance and liver steatosis.
(A) HFD-fed C57BL/6 WT mice were administered control or Kctd17-directed antisense oligonucleotides (ASOs) by weekly intraperitoneal injection. (B) Kctd17 expression in livers, (C) blood glucose and (D) insulin, (E) glucose tolerance test, (F) tissue weight, (G) liver H&E/ORO staining, (H) liver and (I) plasma triglycerides, (J) liver lipogenic and (K) Chrebp target gene expression, and (L) Western blots in control or Kctd17 ASO-treated HFD mice (n=6 to 7 per group). (M) Model representing the parallel effects of Kctd17 to synergistically induce Srebp1c- and Chrebp-mediated hepatic lipid accumulation. *P < 0.05, **P < 0.01, ***P < 0.001 as compared to the indicated control by two-way ANOVA. All data are shown as the means ± s.e.m.
Discussion
Obesity is a risk factor for T2D and NAFLD, which combined provoke substantial liver-related morbidity and all-cause mortality 37. New pharmacologic targets are necessary for an increasingly obese population, but of perhaps equal importance is understanding how liver metabolic processes are disrupted by obesity. Our work reveals Kctd17 as a novel mediator of the crosstalk between glucose and insulin signaling. We find that hepatocyte Kctd17 expression is increased by Srebp1c activity in obesity. As we have previously shown that Kctd17 mediates PHLPP2 degradation, resulting in prolonged Akt phosphorylation and Srebp1c activity 17, this suggests a rare, positive feedback loop. But as a parallel path, Kctd17 also increases Oga degradation, leading to increased Chrebp activity, which leads to synergistic increase in lipogenic gene expression, but also promotes glucose intolerance. These synergistic effects (Figure 7M) raise the possibility of therapeutic silencing of hepatocyte Kctd17 – by ASO or other means – for potential benefits in obesity-induced insulin resistance and fatty liver.
We found that Kctd17 controls hepatic lipid accumulation and glucose production through Oga-Chrebp, but further studies might be needed for a more detailed explanation of this regulation. Chrebp is required for induction of lipogenic genes and glycolytic enzymes in response to glucose and insulin 3, 38-40. Thus, liver-specific inhibition of Chrebp improves hepatic steatosis and insulin resistance in obese ob/ob mice 41, 42, while liver Chrebp overexpression protein causes hepatic steatosis 43. Chrebp stabilization by O-GlcNAc can increase its transcriptional activity on downstream targets 44, 45, so understanding the opposing forces of Oga and Ogt 33 will be important to understand Chrebp-mediated effects. By corollary, mechanisms by which Ogt and Oga are regulated are largely unknown. A recent study identified histone demethylase (LSD2) as an E3 ubiquitin ligase and promotes the ubiquitination and proteasomal degradation of Ogt to inhibit cancer cell growth and proliferation 46, an interesting parallel to Kctd17/Cul3-mediated Oga degradation. Whether there is coordinate regulation of these processes in pathophysiology is unknown, as are other potential repercussions of altered O-GlcNAc modifications with Kctd17 manipulation. In this vein, recent studies have demonstrated that Cullin-Ring ubiquitin ligases (CRLs) recognize substrates containing serine-rich domains that can be phosphorylated 17, 47, 48. Along these lines, it would be great interest to test whether this is also true for Oga.
Our results may provide rationale for the development of Kctd17 inhibitors for simultaneous treatment of patients with type 2 diabetes and NAFLD. Along these lines, we observed that Kctd17 ASO treatment generally recapitulated phenotypes of hepatocyte-specific Kctd17 knockout genetic models, it appeared to have stronger effects, which may be due to effects on both parenchymal and nonparenchymal cells, as well as potentially on adipose tissue (Supplementary Figure 7C, 7D). Kctd17 actions in nonparenchymal cells adipose require further study.
In sum, our study suggests a novel means of regulation of Chrebp, by Srebp1c-mediated increase in Kctd17. We anticipate that interrupting this feed-forward lipogenic loop by targeting Kctd17 may prove effective for the twin glucose and lipid abnormalities that occur in the obese liver.
Supplementary Material
What You Need to Know.
Background and Context
Obesity predisposes to type 2 diabetes (T2D) and non-alcoholic fatty liver disease (NAFLD), but underlying mechanisms are incompletely understood.
New Findings
Hepatocyte Kctd17 expression is increased in obesity. Hepatic Kctd17 deficiency or inhibition reverses these pathologic processes by blocking Oga degradation, resulting in lower Chrebp protein stability, which improves both glucose and lipid metabolism.
Limitations
This study was performed primarily using mouse models of obesity. Future studies are necessary to assess efficacy of KCTD17 inhibitors in patients with T2D and NAFLD.
Impact
Discovery of a novel regulatory node in liver metabolism that may be therapeutically targeted treats obesity-induced T2D and NAFLD.
Acknowledgements
We thank A. Flete and T. Kolar (Columbia University) for excellent technical support, F. Zhang for the LentiCRISPRv2 plasmid, as well as Luca Valenti, Paola Dongiovanni and members of the Pajvani and Kim laboratories for insightful discussion.
Grant Support:
This work was supported by NIH DK103818 and DK119767 (U.B.P), an INHA UNIVERSITY Research Grant (K.K), and National Research Foundation of Korea (NRF) grants funded by the Korea government (MSIT) [No. 2020R1C1C1014281 and 2021R1A5A8029876 (S.B.L), 2020R1C1C1004015, 2021R1A5A2031612, and 2022S1A5A2A03052617 (K.K)].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: The authors disclose no conflicts.
Data Transparency Statement: Data, analytic methods, and study materials will not be made available to other researchers.
References
- 1.Araujo AR, Rosso N, Bedogni G, et al. Global epidemiology of non-alcoholic fatty liver disease/non-alcoholic steatohepatitis: What we need in the future. Liver International 2018;38:47–51. [DOI] [PubMed] [Google Scholar]
- 2.Hossain P, Kawar B, Nahas ME. Obesity and diabetes in the developing world - A growing challenge. New England Journal of Medicine 2007;356:213–215. [DOI] [PubMed] [Google Scholar]
- 3.Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol 2013;10:656–65. [DOI] [PubMed] [Google Scholar]
- 4.Ray K NAFLD-the next global epidemic. Nature Reviews Gastroenterology & Hepatology 2013;10:621–621. [DOI] [PubMed] [Google Scholar]
- 5.Younossi Z, Anstee QM, Marietti M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 2018;15:11–20. [DOI] [PubMed] [Google Scholar]
- 6.Pais R, Barritt ASt, Calmus Y, et al. NAFLD and liver transplantation: Current burden and expected challenges. J Hepatol 2016;65:1245–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doycheva I, Issa D, Watt KD, et al. Nonalcoholic Steatohepatitis is the Most Rapidly Increasing Indication for Liver Transplantation in Young Adults in the United States. Journal of Clinical Gastroenterology 2018;52:339–346. [DOI] [PubMed] [Google Scholar]
- 8.Oh H, Jun DW, Saeed WK, et al. Non-alcoholic fatty liver diseases: update on the challenge of diagnosis and treatment. Clin Mol Hepatol 2016;22:327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan S, Kirk EP, Mittendorfer B, et al. Randomized trial of exercise effect on intrahepatic triglyceride content and lipid kinetics in nonalcoholic fatty liver disease. Hepatology 2012;55:1738–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalasani N, Younossi Z, Lavine JE, et al. The Diagnosis and Management of Nonalcoholic Fatty Liver Disease: Practice Guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology 2012;142:1592–1609. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z, Xiang Y, Sun G. The KCTD family of proteins: structure, function, disease relevance. Cell Biosci 2013;3:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skoblov M, Marakhonov A, Marakasova E, et al. Protein partners of KCTD proteins provide insights about their functional roles in cell differentiation and vertebrate development. Bioessays 2013;35:586–596. [DOI] [PubMed] [Google Scholar]
- 13.Teng X, Aouacheria A, Lionnard L, et al. KCTD: A new gene family involved in neurodevelopmental and neuropsychiatric disorders. CNS Neurosci Ther 2019;25:887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasahara K, Kawakami Y, Kiyono T, et al. Ubiquitin-proteasome system controls ciliogenesis at the initial step of axoneme extension. Nat Commun 2014;5:5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petroski MD, Deshaies RJ. Function and regulation of Cullin-RING ubiquitin ligases. Nature Reviews Molecular Cell Biology 2005;6:9–20. [DOI] [PubMed] [Google Scholar]
- 16.Inaba H, Goto H, Kasahara K, et al. Ndel1 suppresses ciliogenesis in proliferating cells by regulating the trichoplein-Aurora A pathway. J Cell Biol 2016;212:409–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K, Ryu D, Dongiovanni P, et al. Degradation of PHLPP2 by KCTD17, via a Glucagon-Dependent Pathway, Promotes Hepatic Steatosis. Gastroenterology 2017;153:1568–1580 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Platt RJ, Chen S, Zhou Y, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014;159:440–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Zhu C, Wang X, et al. Hepatocyte TLR4 triggers inter-hepatocyte Jagged1/Notch signaling to determine NASH-induced fibrosis. Sci Transl Med 2021;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pajvani UB, Qiang L, Kangsamaksin T, et al. Inhibition of Notch uncouples Akt activation from hepatic lipid accumulation by decreasing mTorc1 stability. Nat Med 2013;19:1054–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim K, Qiang L, Hayden MS, et al. mTORC1-independent Raptor prevents hepatic steatosis by stabilizing PHLPP2. Nat Commun 2016;7:10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim K, Goldberg IJ, Graham MJ, et al. gamma-Secretase Inhibition Lowers Plasma Triglyceride-Rich Lipoproteins by Stabilizing the LDL Receptor. Cell Metab 2018;27:816–827 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim KH, Shin HJ, Kim K, et al. Hepatitis B virus X protein induces hepatic steatosis via transcriptional activation of SREBP1 and PPARgamma. Gastroenterology 2007;132:1955–67. [DOI] [PubMed] [Google Scholar]
- 24.Pajvani UB, Shawber CJ, Samuel VT, et al. Inhibition of Notch signaling ameliorates insulin resistance in a FoxO1-dependent manner. Nat Med 2011;17:961–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957;226:497–509. [PubMed] [Google Scholar]
- 26.Govaere O, Cockell S, Tiniakos D, et al. Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci Transl Med 2020;12. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y, Li MC, Konate MM, et al. TPM, FPKM, or Normalized Counts? A Comparative Study of Quantification Measures for the Analysis of RNA-seq Data from the NCI Patient-Derived Models Repository. J Transl Med 2021;19:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao S, Ye Z, Stanton R. Misuse of RPKM or TPM normalization when comparing across samples and sequencing protocols. RNA 2020;26:903–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Decaux JF, Antoine B, Kahn A. Regulation of the expression of the L-type pyruvate kinase gene in adult rat hepatocytes in primary culture. J Biol Chem 1989;264:11584–90. [PubMed] [Google Scholar]
- 30.Denechaud PD, Bossard P, Lobaccaro JM, et al. ChREBP, but not LXRs, is required for the induction of glucose-regulated genes in mouse liver. J Clin Invest 2008;118:956–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guinez C, Filhoulaud G, Rayah-Benhamed F, et al. O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes 2011;60:1399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeidan Q, Hart GW. The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. Journal of Cell Science 2010;123:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ong Q, Han W, Yang X. O-GlcNAc as an Integrator of Signaling Pathways. Front Endocrinol (Lausanne) 2018;9:599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997;89:331–40. [DOI] [PubMed] [Google Scholar]
- 35.Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci U S A 1999;96:11041–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen TTP, Kim DY, Lee YG, et al. SREBP-1c impairs ULK1 sulfhydration-mediated autophagic flux to promote hepatic steatosis in high-fat-diet-fed mice. Mol Cell 2021;81:3820–3832 e7. [DOI] [PubMed] [Google Scholar]
- 37.Valenti L, Bugianesi E, Pajvani U, et al. Nonalcoholic fatty liver disease: cause or consequence of type 2 diabetes? Liver Int 2016;36:1563–1579. [DOI] [PubMed] [Google Scholar]
- 38.Dentin R, Pegorier JP, Benhamed F, et al. Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. J Biol Chem 2004;279:20314–26. [DOI] [PubMed] [Google Scholar]
- 39.Ishii S, Iizuka K, Miller BC, et al. Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc Natl Acad Sci U S A 2004;101:15597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma L, Tsatsos NG, Towle HC. Direct role of ChREBP.MIx in regulating hepatic glucose-responsive genes. J Biol Chem 2005;280:12019–27. [DOI] [PubMed] [Google Scholar]
- 41.Dentin R, Benhamed F, Hainault I, et al. Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes 2006;55:2159–70. [DOI] [PubMed] [Google Scholar]
- 42.Iizuka K, Miller B, Uyeda K. Deficiency of carbohydrate-activated transcription factor ChREBP prevents obesity and improves plasma glucose control in leptin-deficient (ob/ob) mice. Am J Physiol Endocrinol Metab 2006;291:E358–64. [DOI] [PubMed] [Google Scholar]
- 43.Benhamed F, Denechaud PD, Lemoine M, et al. The lipogenic transcription factor ChREBP dissociates hepatic steatosis from insulin resistance in mice and humans. J Clin Invest 2012;122:2176–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsatsos NG, Towle HC. Glucose activation of ChREBP in hepatocytes occurs via a two-step mechanism. Biochem Biophys Res Commun 2006;340:449–56. [DOI] [PubMed] [Google Scholar]
- 45.Bricambert J, Miranda J, Benhamed F, et al. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J Clin Invest 2010;120:4316–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, Yin X, Yang H, et al. Histone demethylase LSD2 acts as an E3 ubiquitin ligase and inhibits cancer cell growth through promoting proteasomal degradation of OGT. Mol Cell 2015;58:47–59. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Q, Shi Q, Chen Y, et al. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc Natl Acad Sci U S A 2009;106:21191–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xing H, Hong Y, Sarge KD. PEST sequences mediate heat shock factor 2 turnover by interacting with the Cul3 subunit of the Cul3-RING ubiquitin ligase. Cell Stress Chaperones 2010;15:301–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.