Summary
This study develops a method combining a convolutional neural network model, INSIGHT, with a self-attention model, WiseMSI, to predict microsatellite instability (MSI) based on the tiles in colorectal cancer patients from a multicenter Chinese cohort. After INSIGHT differentiates tumor tiles from normal tissue tiles in a whole slide image, features of tumor tiles are extracted with a ResNet model pre-trained on ImageNet. Attention-based pooling is adopted to aggregate tile-level features into slide-level representation. INSIGHT has an area under the curve (AUC) of 0.985 for tumor patch classification. The Spearman correlation coefficient of tumor cell fraction given by expert pathologist and INSIGHT is 0.7909. WiseMSI achieves a specificity of 94.7% (95% confidence interval [CI] 93.7%–95.7%), a sensitivity of 84.7% (95% CI 82.6%–86.9%), and an AUC of 0.954 (95% CI 0.948–0.960). Comparative analysis shows that this method has better performance than the other five classic deep learning methods.
Keywords: colorectal cancer, microsatellite instability, whole slide images, machine learning, convoluted neural network, self-attention, tumor purity
Graphical abstract
Highlights
-
•
INSIGHT achieves better tumor purity prediction performance
-
•
The self-attention mechanism assigns weight for different whole slide image (WSI) pixels
-
•
INSIGHT + WiseMSI capture Asian-ancestry-specific colorectal cancer histopathological features
-
•
CRC patients predicted by WiseMSI to be MSI-H present better disease-free survival
Chang et al. develop a method combining a CNN model, INSIGHT, with a self-attention model, WiseMSI, to predict microsatellite instability (MSI) based on the tiles in colorectal cancer (CRC) patients from a multicenter Chinese cohort. Comparative analysis shows that this method has better performance than five classic deep learning methods.
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide and the fourth most common malignancy in China.1,2 Mismatch repair (MMR) deficiency, which causes microsatellite instability (MSI), is recognized as a distinct mechanism promoting tumorigenesis in 15% of CRC.3 MSI and MMR defect are associated with a poor response to chemotherapy in intermediate stage CRC.4 Increased neoantigen load is a result of defective MMR,5 so MSI has been used to predict immune checkpoint blockade (ICB) therapy response, which is a major early breakthrough for precision oncology.6,7,8,9,10 ICB alone and in combination with other therapy targets have shown great promise; as a result, MSI has been increasingly used to guide chemotherapy and immunotherapy of CRC.11,12,13,14
Conventionally, multiplex polymerase chain reaction (PCR) assays or multiplex immunohistochemistry (IHC) panels for evaluating the expression of the most common gene products associated with MMR (MLH1, MSH2, MSH6, and PMS2) are used to examine MSI of CRC patients.15,16,17 Meanwhile, MSI PCR assays have a sensitivity of 100% and specificity of 61.1% or a specificity of 92.5% and sensitivity of 66.7%.18,19 IHC is more commonly used clinically compared with PCR assays, and its sensitivity and specificity range from 80.8% to 100% and 80.5% to 91.9%, respectively.20,21 Despite a high sensitivity and specificity of PCR assays and IHC for MSI in CRC, these approaches could be labor intensive and require pathologists with years of experience to achieve high accuracy and consistency.
Artificial intelligence has revolutionized pathological diagnosis. Distinct pathomorphological features of whole slide images (WSIs) of CRC, such as tumor-infiltrating lymphocytes and mucinous differentiation, are indicative of underlying molecular events such as defective MMR and could be explored by a deep learning approach for supervised feature extraction of MSI status in cancer patients.22,23,24,25,26,27,28 Deep learning models using WSIs for prediction of MSI status in CRC26,27,28,29,30,31 and other cancer patients have emerged.22,32 The AUROC values of MSI predictions in CRC patients by deep learning are mostly between 0.7 and 0.9 with training and validation datasets varying from about 300 to about 1,500 WSIs (Table S1). The performance increased to 0.96 for AUROC value when a training and testing dataset increased to 8,836 patients.24
The methods in these studies generally adopted two types of strategies: classical weakly supervised approach and multiple instance learning (MIL) approach.33 The first step was image preprocessing, which tessellates the digitized WSIs into small image tiles, typically of 512 × 512 pixels, followed by quality control and color normalization. Different WSIs produce different numbers of tiles, varying from hundreds to tens of thousands of tiles. The classical weakly supervised approach presumes each tile inherits the slide’s MSI status and randomly selects a fixed number of tiles. Convolutional neural network (CNN) models were then trained to make tile-level prediction of MSI status. Patient-level predictions were made by aggregating the tile-level predictions, except Cao’s study, which adopted an ensemble-learning approach to optimize the prediction accuracy.
Instead of sub-sampling a fixed number of tiles, the MIL approaches use all tiles from a patient as a “bag” without assuming every single tile in the bag reflects the MSI status of the patient in order to address the issue of heterogeneous tiles from different regions of the WSI. However, the benchmark analysis of these two approaches by Laleh et al.33 showed that classical weakly supervised methods unexpectedly outperform the more sophisticated MIL approach. One likely reason is that MIL-based methods assigned the highest prediction scores to image tiles with tissue edge and other artifacts. A few studies, including Yamashita et al.26 and Bilal et al.,31 combined two CNN models: one for tissue classification and one using only tumor tiles as input to improve MSI prediction accuracy (Table S1).
Self-attention models are a class of neural network that can learn features from supervised data. The major advance of the self-attention mechanism lies in its capability to enable the decoder to access the whole of the encoded information, assigning attention weights over the input data, which captures the importance of each token and prioritizes them for generating output tokens at each step. The self-attention mechanism, whose receptive field is the whole image, employs self-captured features for prediction and enables the interpretation of the output.34 Self-attention networks, which are widely used for natural language processing, have been shown to be superior to other deep learning approaches in analyzing pathology reports.35
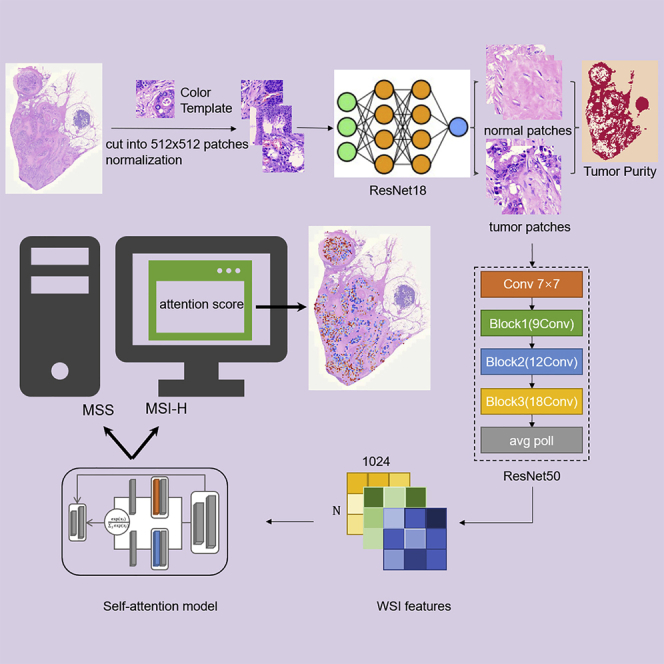
In this study, we constructed a hybrid method WiseMSI by combining a CNN model with a self-attention model to predict MSI based on the tiles in CRC patients from cohorts from multiple medical centers across China. After a trained CNN model differentiated tumor tiles from normal tissue tiles from a WSI, features of tumor tiles were extracted with a ResNet model trained on ImageNet, with bags of tiles labeled following patient’s MSI status, like the MIL-based methods. Attention-based pooling was adopted to aggregate tile-level features into slide-level representation like CLAM36 but with dot product attention replaced by cosine similarity, which works better for image summaries.
Results
Construction and performance of a CNN tumor detector model, INSIGHT
In the current study, we developed a self-attention-based CNN for prediction of MSI in colorectal adenocarcinoma. To construct a highly accurate model for tumor detection, we first trained the tumor detector model INSIGHT with ResNet-18 on 254 WSIs including 117 MSS and 137 MSI-H WSIs from the TongShu MSI Colorectal Cancer (TSMCC) Cohort (Figure 1), with a total of 25,349 normal patches and 25,235 tumor patches labeled by experienced pathologists. The overall architecture is shown in Figure 2. INSIGHT was then tested on the test set consisting of 51 WSIs with a total of 2,549 normal patches and 2,373 tumor patches. The tumor detector achieved an area under the curve (AUC) of 0.985.
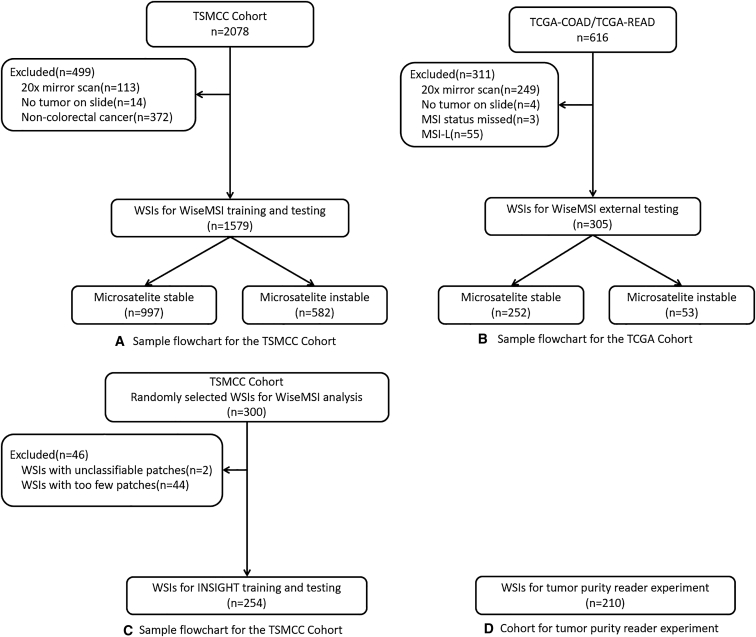
Figure 1.
The flowchart for the disposition of patient WSI
(A) A total of 2,708 patient WSIs in the TSMCC cohort were included. Among them, 499 WSIs were excluded for quality issues. The remaining 1,579 WSIs were used for WiseMSI training and testing, including 997 MSSs and 582 MSIs.
(B) A total of 616 WSIs of the COAD and READ cohorts were downloaded from TCGA. 311 WSIs were excluded for quality reasons. The remaining 305 WSIs were used for external testing of WiseMSI.
(C) In total 300 WSIs were randomly selected from the TSMCC cohort, and 46 of them were excluded in the following quality check step. The remaining 254 WSIs were used for INSIGHT training and testing.
(D) The WSIs of 210 extra patients were involved in the reader experiment. TongShu MSI Colorectal Cancer; WSI: whole slide image.
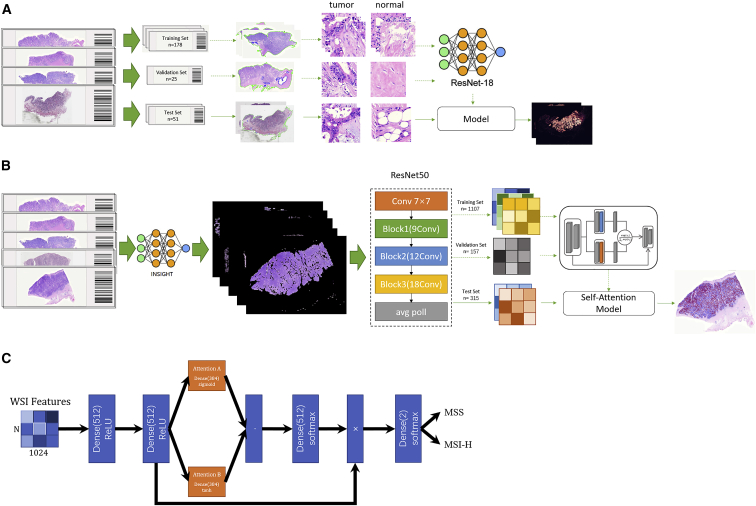
Figure 2.
Schemas showing the data acquisition workflow and training and testing of neural networks INSIGHT and WiseMSI
(A) In the randomly selected 254 WSIs, 178 were used for training, 25 for validation, and 51 for testing. These WSIs were cut into square patches of 512-pixel length and were color normalized with staintools. A pathologist labeled tumor and normal patches that were fed into the ResNet18 tumor detection model.
(B) Patches from 1,579 WSIs were passed to INSIGHT, and only the tumor patches predicted by INSIGHT were further fed into the ImageNet pre-trained ResNet 50 model for feature extraction. The extracted feature matrix for each patch was passed to the self-attention model. The 1,579 WSIs were split into a 7:1:2 ratio for training, validation, and testing of the self-attention model.
(C) Plot illustrates the architecture of the self-attention model in WiseMSI. MSI, microsatellite instability; MSI-H, high MSI; MSS, microsatellite stability.
Construction and performance of a self-attention-based CNN MSI prediction model
The TSMCC cohort included 1,579 WSIs, with 997 MSS and 582 MSI-H WSIs. These 1,579 WSIs tessellated into patches and were color normalized with staintools. Through random sampling, patches from 1,107 randomly selected WSIs, including 699 MSS WSIs and 408 MSI-H WSIs, were assigned as training set. Patches from 157 WSIs including 99 MSS WSIs and 58 MSI-H WSIs worked as the validation set. And patches from 315 WSIs were randomly assigned to the internal test set, including 199 MSS and 116 MSI-H WSIs (Data S1). All the patches from the training set, the validation set, and the test set were fed into the trained ResNet18 tumor patch detector INSIGHT. The patches categorized by INSIGHT as tumor patches were then passed onto the ResNet50 pre-trained on the ImageNet. The feature matrix derived from tumor patches predicted by INSIGHT in the training set and validation set was fed to the self-attention model with standard cross-entropy loss function. The model with the minimum loss in validation set was selected as the optimized MSI classifier WiseMSI. WiseMSI is the combination of the ResNet50 CNN model for feature vector encoding and the self-attention model that processes the feature vectors and output attention score and MSI score. WiseMSI’s performance was then evaluated with the internal test set.
To evaluate the variability of our dataset, and the reliability of WiseMSI, 10-fold cross-validation was conducted by randomly dividing the 1,579 WSIs into 10 subsets. The entire training and testing process of WiseMSI was repeated 10 times with each time using eight different subsets as training and validation sets, and two different subsets as internal test set. WiseMSI achieved with the internal test set a mean specificity of 94.7% (95% confidence interval [CI] 93.7% to 95.7%) and a mean sensitivity of 84.7% (95% CI 82.6% to 86.9%), with an accuracy rate of 91.1% and an AUC of 0.954 (95% CI 0.948 to 0.960) (Table 1 and Figure 3A I). If no pre-selection of tumor region patches was applied, and all the normalized patches were passed onto the ResNet50 CNN model for feature extraction and the self-attention model for MSI prediction, there was an average of AUC 0.93 (95% CI 0.91 to 0.94), specificity 95.7% (95% CI 93.2% to 98.2%), and sensitivity 77.4% (95% CI 72.5% to 82.3%). The pre-selection of tumor region tiles improved MSI-H prediction sensitivity from 77.4% to 84.7% and AUC value from 0.93 to 0.954.
Table 1.
Diagnostic performance of the self-attention-based CNN for prediction of microsatellite instability in colorectal adenocarcinoma with 10-fold cross-validation with the TSMCC cohort
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| MSS | 185/199 | 192/199 | 189/199 | 191/199 | 189/199 | 185/199 | 191/199 | 190/199 | 188/199 | 184/199 |
| MSI-H | 97/116 | 93/116 | 100/116 | 97/116 | 93/116 | 102/116 | 97/116 | 101/116 | 103/116 | 100/116 |
| Specificity | 0.9296 | 0.9648 | 0.9497 | 0.9598 | 0.9497 | 0.9296 | 0.9598 | 0.9548 | 0.9447 | 0.9246 |
| Sensitivity | 0.8362 | 0.8017 | 0.8621 | 0.8362 | 0.8017 | 0.8793 | 0.8362 | 0.8707 | 0.8879 | 0.8621 |
| AUC | 0.9466 | 0.9504 | 0.9504 | 0.9656 | 0.9346 | 0.9604 | 0.9594 | 0.9606 | 0.9542 | 0.9562 |
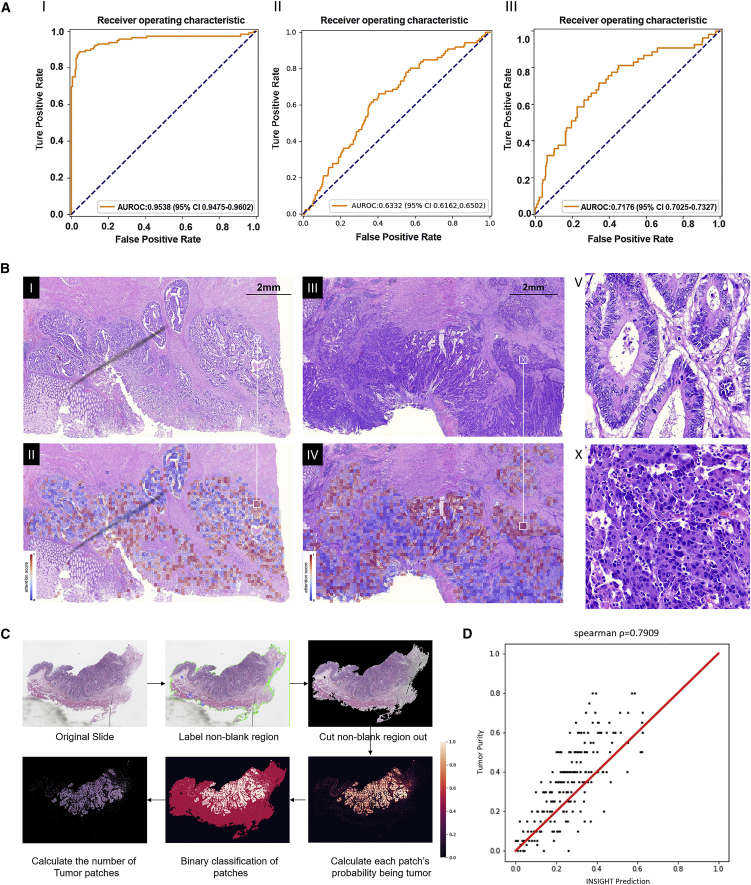
Figure 3.
The performance and visualization of WiseMSI and INSIGHT
(A) The ROC curve of the self-attention-based CNN for MSI prediction in the internal test set from the TSMCC cohort (Ⅰ). The ROC curve of the self-attention-based CNN for MSI prediction in the external test set from the TCGA-COAD and TCGA-READ cohorts (Ⅱ). The ROC curve of the self-attention-based CNN for MSI prediction in the external test set from the 305 WSIs scanned with a 40x objective lens (Ⅲ).
(B) Attention score map for tumor regions in two WSIs. Ⅰ is the original tumor region for a MSS patient, and Ⅱ is the corresponding attention score map. Red dots are pixels with high attention score, and blue dots are pixels with low attention scores. Similarly, Ⅲ is the original tumor region for an MSI-H patient, and Ⅳ is the corresponding attention score map. V and X are enlarged regions in Ⅱ and Ⅳ, as shown by white squares and lines.
(C) The workflow of INSIGHT’s tumor purity prediction (Ⅰ) and the scatterplot (Ⅱ) of the tumor purities manually estimated by an expert pathologist (y axis) vs. the tumor purities predicted by INSIGHT (x axis). In the prediction of tumor purity, first the model detects the patches with sample tissue present; then the probability of containing tumor cells is evaluated by INSIGHT for each patch. Patches with probability value larger than 0.5 are grouped into tumor patches and otherwise as non-tumor patches. Tumor purity is calculated as the percent of tumor patches among non-blank patches.
(D) Spearman correlation coefficient between manual estimation and INSIGHT prediction is given on the top of the scatterplot. And the red line is the diagonal.
We also examined 609 WSIs, including 523 MSS and 86 MSI-H WSIs, from TCGA-COAD and TCGA-READ as the external test set for our WiseMSI model trained based on the TSMCC cohort, including 12 Asian individuals, 68 Black individuals, 283 White individuals, and 245 individuals of unknown ethnic group. A mean specificity of 35.3% (95% CI 27.5% to 43.1%) and a mean sensitivity 71.5% (95% CI 53.1% to 89.9%) was achieved, and the mean AUC was 0.632 (95% CI 0.703 to 0.733) (Figure 3AⅡ). When only the 305 WSIs scanned with 40x objective lens, including 252 MSS and 53 MSI-H WSIs, was used as the external test set, the mean AUC increased to 0.718 (95% CI 0.703 to 0.733), specificity to 46.5% (95% CI 33.3% to 59.8%), and sensitivity to 76.6% (95% CI 65.3% to 87.9%) (Table 2 and Figure 3A Ⅲ). Among different ethnic groups, the Asian group had sensitivity and specificity, both being 100% in nine out of ten testing rounds. The White group had a specificity of 86% but sensitivity of only 50.2%. The Black and not reported populations have a similar pattern to the White population (Table S3).
Table 2.
Diagnostic performance of the self-attention-based CNN for prediction of microsatellite instability in colorectal adenocarcinoma with 10-fold cross-validation with the TCGA-COAD and TCGA-READ cohort
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| MSS | 89/252 | 132/252 | 227/252 | 129/252 | 86/252 | 50/252 | 124/252 | 130/252 | 114/252 | 91/252 |
| Specificity | 0.3532 | 0.5238 | 0.9008 | 0.5119 | 0.3413 | 0.1984 | 0.4921 | 0.5159 | 0.4524 | 0.3611 |
| MSI-H | 46/53 | 38/53 | 18/53 | 42/53 | 45/53 | 48/53 | 43/53 | 42/53 | 42/53 | 42/53 |
| Sensitivity | 0.8679 | 0.7170 | 0.3396 | 0.7925 | 0.8491 | 0.9057 | 0.8113 | 0.7925 | 0.7925 | 0.7925 |
| AUC | 0.7504 | 0.6816 | 0.7206 | 0.7378 | 0.6898 | 0.7096 | 0.7100 | 0.7162 | 0.7315 | 0.7285 |
We further trained the self-attention model with TCGA-STAD cohort and tested its performance by cross-validation (Table S4). The AUC values of the self-attention model are 0.750 (95% CI 0.699 to 0.802) in 10 rounds of testing. The specificity of MSI-H prediction is high, with an average value of 96.3% and 95% CI from 93.6% to 99.1%. The sensitivity is poor with an average value of 18.5% and 95% CI from 6.2% to 30.7%. This cohort was used to train and test the ViT methods, which have the top performance in Laleh’s benchmark analysis33 for CRC MSI prediction. Similar results were obtained with an average AUC value of only 0.682 (95% CI 0.635 to 0.729), average specificity 0.972 (95% CI 0.948 to 0.997), and average sensitivity 13.1% (95% CI 3.01% to 23.1%).
An attention score map of tumor region reflects the differences in tumor region between MSI-H and MSS WSIs captured by WiseMSI (Figure 3B). The red dots with high attention score contributed more to the predictions of MSI status than the blue dots with low attention scores. The morphologies of cells in the red dots from MSI-H WSIs have poor differentiation, and the ones from MSS WSIs have high differentiation. This is similar to previous findings by Greenson37 and Kather.22 This finding further confirmed that histopathological features could work as predictors of MSI status and illustrated the rationale of detection of MSI from hematoxylin and eosin (H&E) stained slides by deep learning methods. Therefore, the area highlighted by attention score provides a way to visualize and explain the prediction of the deep learning model.
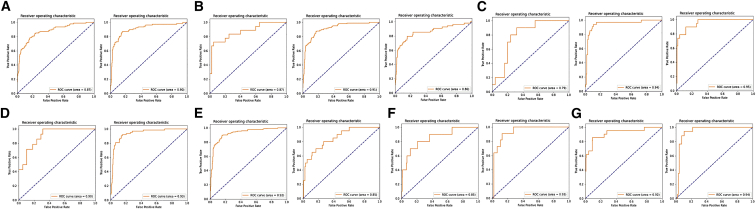
We further studied the robustness of WiseMSI’s prediction performance across heterogenous clinicopathologic subgroups of CRC patients. The CRC patients in the TSMCC cohort comprise seven demographically, anatomically, and/or pathologically distinct subgroups, including colon cancer, rectum cancer, right- and left-sided colon cancer, low and median pathological differentiation degree, having or not having Lynch syndrome, and different cancer stage, age, and gender. There are some variations of WiseMSI’s performance regarding disease stage and patient age: the AUC value was 0.79 for stage 1 cancer (n = 35 WSIs), 0.94 for stage 2 (n = 164 WSIs), 0.95 for stage 3 (n = 133 WSIs), 0.87 for patients’ age ≤ 40 years old (n = 58 WSIs), 0.90 for patients with 40 < age ≤ 70 (n = 823 WSIs), and 0.80 for patients older than 70 years (n = 216 WSIs). Variations in WiseMSI’s classification performance related to anatomic features are relatively small: AUC value of 0.92 for colon cancer (n = 753 WSIs), 0.85 for rectum cancer (n = 247 WSIs), 0.85 for left-sided cancer (n = 228 WSIs), and 0.93 for right-sided cancer (n = 58 WSIs). WiseMSI’s classification performance was very stable (AUC value around 0.90) across subgroups related to pathological differentiation degree, Lynch syndrome, and gender. Overall, the WiseMSI model had a good predictive performance of MSI across all subgroups examined (Figure 4).
Figure 4.
Performance of WiseMSI on MSI prediction across heterogenous clinicopathologic subgroups of CRC patients
(A–G) The ROC curves for male (left panel) or female patients (right panel) (A); patients aged no more than 40 years (left panel), between 41 and 64 years (mid panel), or at least 65 years (right panel) (B); patients with stage I (left panel), II (mid panel), or III CRC (right panel) (C); patients with poorly (left panel) or moderately differentiated CRC (right panel) (D); patients with colon (left panel) and rectum cancer (right panel) (E); patients with left (left panel) or right CRC (right panel) (F); and patients with (left panel) or without Lynch syndrome (right panel) (G).
Comparative analysis of WiseMSI with four other computational pathology MSI classifiers
We systematically estimated the performance difference between WiseMSI; two representative classical weakly supervised methods, EfficientNet and ViT; two MIL methods, MIL and AttMIL, which have the top performances in Laleh’s benchmark analysis; and VarMIL in DeepSMILE. Cross-validation within TSMCC cohort was carried out, and the performance metrics are given in Table 3. The performances of ViT and EfficientNet are slightly better than their performance in Laleh’s study (ViT 0.939 vs. 0.885, EfficientNet 0.905 vs. 0.883), and ViT remains the method with the best performance among the four methods being compared. But the performance difference between classical weakly supervised approach and MIL approach is not as obvious as in Laleh’s study as the AUC values of MIL and AttMIL are all close to 0.90 as well. The specificities of these five methods are all very high (ViT 97.8%, EfficientNet 97.3%, MIL 97.0%, AttMIL 99.5%, VarMIL 99.2%), but their sensitivities are relatively poor, with AttMIL at 63.2%, MIL at 66.8%, ViT at 74.3%, VarMIL at 62.4%, and EfficientNet only at 36.0%. The low sensitivity is due to MSI score threshold and could be adjusted. The AUC value of WiseMSI, 0.954, is higher than these five methods. With the same MSI score threshold of 0.5, WiseMSI achieved similar specificity and much better sensitivity than the other five methods. In general, the hybrid approach WiseMSI has better performance than both classical weakly supervised and MIL approaches, as demonstrated in Table 3 and Figure S1.
Table 3.
Performance statistics of the five MSI classification models and two feature extractors for comparative analysis and WiseMSI on TSMCC cohort
| Model | Specificity (95% CI) | Sensitivity (95% CI) | AUC (95% CI) |
|---|---|---|---|
| EfficientNet | 0.973 (0.920, 1.026) | 0.3603 (0.071, 0,650) | 0.9050 (0.865, 0.946) |
| ViT | 0.978 (0.966, 0.990) | 0.7431 (0.681, 0.806) | 0.9385 (0.928, 0.949) |
| MIL | 0.9698 (0.950, 0.990) | 0.6680 (0.635, 0.701) | 0.9073 (0.884, 0.931) |
| AttMIL | 0.9950 (0.989, 1.001) | 0.6318 (0.579, 0.685) | 0.8993 (0.872, 0.927) |
| VarMIL | 0.992 (0.983, 1.000) | 0.624 (0.577, 0.671) | 0.903 (0.890, 0.915) |
| WiseMSI (Moco V2 version) | 0.926 (0.871, 0.980) | 0.793 (0.723, 0.863) | 0.937 (0.920, 0.95) |
| WiseMSI (ImageNet version) | 0.947 (0.937, 0.957) | 0.847 (0.826, 0.869) | 0.954 (0.948, 0.968) |
Performance is reported on WSI level and 95% confidence level (CI) is calculated based on 5-fold cross-validation for EfficientNet, Vit, MIL, and AttMIL, and 10-fold cross-validation of WiseMSI.
We also benchmarked the Moco V2 feature extractor with the ImageNet pre-trained feature extractor model by re-training Moco V2 with 622 WSIs from TCGA-COAD + READ cohorts and 400 WSIs from our in-house TSMCC data. Replacing the feature extraction model with the in-house trained Moco V2 model did not significantly improve the performance, with an AUC value of 0.937 for Moco V2 version of WiseMSI and AUC value of 0.954 for ImageNet version of WiseMSI, as demonstrated in Table 3 and Figure S1.
The running time for these models was also evaluated by using these 1,579 WSIs for validation. As shown in Table S2, the average running time of WiseMSI from tumor tile detection, feature extraction, and MSI classification is 807.8 s per WSI, slightly shorter than MIL and AttMIL on Intel I9 10900K with 3,090 GPU. A more powerful GPU server could be used to shorten the running time to be less 10 minutes, an acceptable time for clinical application, though longer than EfficientNet and ViT. The shorter time of WiseMSI even with an extra step is due to the smaller number of tumor tiles passed onto feature extraction. Classical weakly supervised approaches only used a fraction of tiles from WSIs and need a much shorter time to give MSI prediction, with ViT being around 3.3 seconds and EfficientNet at 4.0 seconds.
Survival of CRC patients according to MSI status by molecular typing and by WiseMSI prediction model
We additionally investigated the diagnostic performance of WiseMSI model for survival of CRC patients. 293 patients from the TSMCC cohort had demographic and clinical information. We excluded 63 CRC patients who had no survival data, and 226 CRC patients were included in the survival analysis. Their demographic and baseline variables are shown in Table S5. 21 patients had MSI-H and 205 patients had MSS as measured by PCR + capillary electrophoresis assays (see STAR Methods). Thirty-three disease-free survival (DFS) events occurred in the MSS patients and none in the MSI-H patients. MSI-H patients had a higher 60-month DFS rate than MSS patients (MSI-H: 100.0% vs. MSS: 83.9%) (Figure S2A). The WiseMSI model had 14 MSI-H patients and 212 MSS patients in the cohort. Patients with predicted MSI-H also had a higher 60-month DFS rate than patients with predicted MSS (MSI-H: 100.0% vs. MSS: 84.4%) (Figure S2B). Furthermore, MSS patients had a DFS comparable to patients with predicted MSS (MSS: 58.3 ± 1.5 months, 95% CI 55.3 to 61.3 months vs. predicted MSS: 58.6 ± 1.5 months, 95% CI 55.7 to 61.5 months) (Figure S2C).
The effect of MSI status on prognosis is associated with tumor stage. MSI is a positive prognostic factor in stage II–III CRC. Subgroup survival analysis was performed to control the stage confounder and investigate the survival benefit of MSI status detected by PCR and AI. In stage Ⅱ and stage Ⅲ subgroups, both the predicted MSI-H and labeled MSI-H subgroups had higher 60-month DFS rate than patients with predicted or labeled MSS (Figure S3). In the stage Ⅳ subgroup, there are no predicted or labeled MSI-H patients.
Tumor purity prediction by INSIGHT is highly correlated with pathologist estimation
Our model training procedures produced two models, the ResNet18 model INSIGHT and WiseMSI, consisting of the ResNet50 CNN model and the self-attention model. INSIGHT differentiated tumor tissues from normal tissues and classified a patch as tumor patch or normal patch. The patches predicted to be tumor patches by INSIGHT were passed to WiseMSI model to predict their MSI status. Based on INSIGHT’s classification of patches as tumor patch or normal patch, the tumor purity of a WSI could be estimated by calculating the percentage of tumor patches among the patches from a CRC WSI. Therefore, we compared tumor purity prediction function by INSIGHT with the tumor purity estimation done by human pathologists. The final estimation was confirmed and scored by an independent, seasoned pathologist.
The cohort for this purpose was another cohort with 208 filtered WSIs. Pathologist tumor purity estimations were recorded in the format of 0 to 5 on unit place, while INSIGHT provided estimation with more refined decibels but at the same total range.
The spearman correlation coefficient was 0.7909 (Figure 3D). Manual inspection by the independent certified pathologist inspector with more than 15 years’ experience was conducted for the 41 slides with either estimated tumor purity difference between pathologist and INSIGHT larger than 0.20 or non-zero tumor purity by INSIGHT but tumor-free designation by the pathologist. For 16 slides, the tumor purity predicted by INSIGHT was judged to be more accurate than the pathologist’s manual estimation. These slides usually have scattered tissues on them, which makes it difficult for a pathologist to estimate the overall tumor purity, or they contain infiltrating normal cells in the tumor, leading to a pathologist’s overestimate, as illustrated by the example case in Figure 3C. In this example, the pathologist gave 50% tumor purity, but there are many infiltrating normal cells in the darkly stained tumor regions, and 50% was an overestimation. INSIGHT’s binary classification of small-size patches into tumor and non-tumor helps to overcome this problem and give a more accurate prediction, which was determined to be 21% tumor purity.
However, in another example, INSIGHT gave 20% tumor purity for one slide that was eventually judged as tumor free. Manual re-inspection by the independent pathologist inspector found that this slide was a rare sample that contained a lot of adenoma cells. Adenoma cells are morphologically similar to cancer cells but are treated as normal cells by pathologists. This type of cell is underrepresented in our training dataset and therefore leads to the misjudgment by INSIGHT. In addition, INSIGHT gave tumor purity values between 0 and 0.05 for the other six slides that are tumor free. The non-zero predictions of INSIGHT are the result of a small fraction of glandular tube cells and lymphocytes being treated as tumor cells by the tumor detector model.
The values predicted by INSIGHT were smaller than the manual ones because of the omission of mucinous adenocarcinoma cells for 14 slides. The true tumor purities for two slides should lie between the values given by pathologist and INSIGHT. And for three slides, INSIGHT’s prediction values were higher than the manual ones as a result of miscounting small fractions of glandular tube cells and lymphocytes as tumor cells by the tumor detector model.
Discussion
In this study, we built a MSI prediction model that combines a self-attention model with a CNN model. This multicenter study demonstrated that WiseMSI has a good diagnostic performance in MSI prediction and achieved prediction levels similar to those of PCR assays and IHC methods currently used for detection of MSI in CRC.18,19 Notably, MSI predicted by WiseMSI, similar to MSI by PCR assays and IHC methods, could stratify the clinical outcome of colorectal adenocarcinoma patients38,39,40,41,42,43 WiseMSI is a hybrid method combining a CNN model with a self-attention model to predict MSI. The CNN model INSIGHT differentiates tumor tiles from normal tissue tiles. The self-attention model enables an improved feature extraction and aggregate tile-level features into slide-level representation similar to CLAM,36 but with dot product attention replaced by cosine similarity, which works better for image summaries. Comparative analysis of WiseMSI with five other representative methods of classical weakly supervised methods and MIL methods showed that WiseMSI has better performance. The internal test set performance of WiseMSI achieved a specificity of 94.7% and a sensitivity of 84.7%, which is comparable to PCR assays and IHC methods currently used for detection of MSI in CRC patients.20,21 In particular, the proposed model generally had a high accuracy rate (91.1%), illustrating the model’s predictive reliability. We also tried to make MSI prediction by directly feeding the entire WSIs including both tumor and normal tissues into the self-attention model and got an AUC of the model of 0.84, which is comparable to the AUC of 0.85 in an Asian ancestry cohort in the study by Cao et al. that combined patch-level MSI prediction and WSI-level prediction.28 The AUC value increased to 0.954 by exclusion of normal tissue and only using detected tumor tissue by INSIGHT for MSI prediction. Utilization of INSIGHT to exclude non-tumor regions not only helps to improve the performance metrics, but it also helps to reduce the computing time as only tumor tiles are passed onto the feature extraction step.
The quality and the sample size of the training data are critical for the success of deep learning models. In this study, we utilized data sets from different medical centers and open databases to secure sufficient disparities in our training and reflect heterogeneous patient features.44 The MSI prediction model was also tested in multiple cohorts from different medical centers, showing consistent performance of the model across different cohorts. In addition, we examined the performance of the WiseMSI prediction using WSIs from TCGA-COAD and TCGA-READ in which WSIs were prepared from formalin-fixed and paraffin-embedded tissues of mainly White CRC patients. Our MSI prediction model using the TCGA-COAD and TCGA-READ cohort as the external test set showed an AUC of 0.718 and a sensitivity of 76.6%. Training and cross-validation of the self-attention model of WiseMSI and VIT displayed similar performance on the TCGA-STAD cohort. But the sensitivity and specificity on the 12 Asian ancestry WSIs in TCGA-COAD and TCGA-READ cohorts are both 100%. This external Asian ancestry cohort is very small in size, but it reflects the reality that many so-claimed diversified cohorts only contain very limited Asian ancestry samples for their model training. In Laleh’s benchmark study, ViT and other methods were trained with European descendant cohorts. The AUC value of ViT decreased slightly from 0.906 to 0.885 in external validation since both the training cohort and validation cohort were mostly European descendants. But when we trained these methods with our Chinese cohorts in this study, ViT showed significantly decreased performance, with the AUC value only being 0.682 (Table S4). This is an interesting ethnicity-related observation and emphasized the importance of WiseMSI for the Asian population, which has large absolute number but is still underrepresented in the research literature. Cao et al. found that their deep learning model ensemble patch likelihood aggregation (EPLA) that was trained on TCGA-COAD showed reduced performance in Asian individuals with CRC in the absence of transfer learning.28 A deep learning MSI model that had been trained on TCGA achieved an AUC less than 0.70 in a Japanese cohort.22 These findings highlight the need for a high-performance prediction model that can be universally applied across different ethnicities and diverse healthcare settings.
MSI is becoming a theragnostic marker to guide the therapy of CRC.45 Our study further showed that MSI predicted by the self-attention-based CNN model could stratify the DFS of CRC patients, suggesting that predicted MSI could be potentially useful as a theragnostic marker to guide CRC management. In addition, WiseMSI has a high specificity, and its performance is comparable to that of an MSI assay based on PCR-capillary electrophoresis (Tongshu Biotechnology Co.). The current study indicates that the self-attention-based CNN model, WiseMSI, could be employed for early screening of MSI status in CRC patients as a substitute or supplement for PCR-capillary electrophoresis assays.
The high correlation between the tumor detection model INSIGHT and expert pathologists on percent tumor purity estimation provides the possibility to support pathologists and lighten their load of routine tedious and time-consuming work by giving an AI’s evaluation first followed by the pathologist’s re-inspection and confirmation. INSIGHT’s performance could be further improved if some detailed, cell-level labeling data could be provided for tumor detection model training. For samples with scattered tissues on the slides, or samples with infiltrating normal cells in tumor, INSIGHT’s estimation tends to be more accurate than that of pathologists. Inter- and intra-pathologist variation can also be minimized by an AI’s prompt and high reproducibility. However, INSIGHT’s ability to differentiate tumor tissues from normal tissues depends on the training WSIs. It could not handle rare cases like cancerous adenoma cells, which are not included in the training set. Cells like glandular tube cells and lymphocytes may be mis-identified as cancer cells because of nonnormal H&E staining. More detailed tumor and normal region-based annotation rather than patch-based annotation by a pathologist would help overcome this problem.
In summary, we have constructed a powerful and explainable MSI prediction model, WiseMSI, that demonstrates excellent diagnostic performance across multiple cohorts of CRC patients including both Chinese and White individuals. In the future, we plan to evaluate the model by examining clinicopathologic variables and clinical outcomes so that an optimized MSI prediction model can be constructed.
Limitations of the study
The histological and molecular features of WSIs from the TSMCC have both been validated by an expert pathologist and NMPA-approved gold-standard PCR test set. The hybrid approach of WiseMSI, using tumor patches predicted by INSIGHT, helps to reduce the impact of within-slide tissue heterogeneity. However, current molecular testing is on the per-patient level. Therefore, labeling of MSI status is at the per-slide level, and the unknown within-slide intratumor heterogeneity may lead to noise. The self-attention mechanism enables WiseMSI to learn well-annotated histopathological features and assign attention scores to different regions of the slide. However, we have not used these learned features to generate captions for the users for full transparency. WiseMSI has excellent performance on Asian ancestry samples, including the Asian ancestry samples from TCGA cohorts, and is a complement to the research literature for the underrepresented Asian population. However, a larger, multi-ethnicity training dataset will help to boost the prediction performance of WiseMSI on European or African descendants’ samples.
STAR★Methods
Key resources table
Resource availability
Lead contact
Any further information and requests for resources and codes should be directed to and will be fulfilled by the lead contact, Xueping Quan (quanxueping@tongshugene.com).
Materials availability
This study did not generate new unique reagents.
Experimental models and subject details
The study protocol was approved by the Medical Ethics Committee of each participating institution. No patient consent was required given the retrospective nature of the study. The H&E− WSIs of 2078 deidentified patients [the TongShu MSI Colorectal Cancer (TSMCC) Cohort] who underwent primary colorectal adenocarcinoma resection were obtained. TSMCC cohort consist of paraffin section, and H&E staining, and slide scanning data from multiple medical centers across China ( Acknowledgments). For quality control, we excluded 372 non-CRC WSIs. Fourteen tumor-free WSIs and 113 tumor WSIs were evaluated by two experienced pathologists under 20× objective lens as a central pathological review. In total, 1579 WSIs were included in this study. The level of MSI at microsatellite loci BAT-25, BAT-26, D5S346, D2S123 and D17S250 of each sample was determined using commercially available PCR + capillary electrophoresis assays (Tongshu Biotechnology Co., Changzhou, Jiangsu China, CFDSM 20213400070). Samples show ≥2 positive loci were deemed to be high MSI (MSI-H) per ESMO recommendations,7 otherwise the sample was categorized to be MSS. The WSIs were tessellated into non-overlapping square patches of 512-pixel edge length and saved at a resolution of ∼0.5 mm per pixel. All image patches were color normalized using staintools (https://staintools.readthedocs.io/en/latest/index.html;https://github.com/Peter554/StainTools). These patches were manually annotated by experienced pathologists as tumor patches or normal patches.
The flowchart for patient disposition is shown in Figure 1. First, we randomly chose 300 WSIs from the TSMCC Cohort. We excluded 2 WSIs that were not classifiable, and 44 WSIs with insufficient numbers of patch. Two hundred and fifty-four WSIs were used for the tumor detector model INSIGHT. And 1579 WSIs from the TSMCC Cohort were also used for MSI prediction model WiseMSI.
Additionally, 616 WSIs of colorectal adenocarcinoma were acquired from TCGA-COAD and TCGA-READ (https://portal.gdc.cancer.gov/projects/TCGA-READ; https://portal.gdc.cancer.gov/projects/TCGA-COAD). For quality control, 3 WSIs were excluded due to lack of MSI analysis conclusion, and additionally 55 WSIs were excluded as they were deemed to be MSI-L. Finally, 4 WSIs were excluded for missing tumor tissue in the slides. Both WSIs scanned under 20x objective lens and 40x lens were included. In total, 609 WSIs, 523 labeled as MSS and 86 labeled as MSI-H, were used as the external test set for the self-attention model.
An extra 210 WSIs were obtained from multi-centers for the tumor purity reader experiment to check the consistency between our trained tumor detection model INSIGHT and expert pathologist.
Method details
Neural network training and testing
Figure 2 shows the workflow of the data, training, and testing of the neural networks. A residual convolutional neural network INSIGHT (ResNet-18,38 the tumor detector model) was trained and tested on the image patches coming from 254 WSIs. Each patch was categorized as normal patch or tumor patch by experienced pathologists. These patches were randomly assigned in a 7:1:2 ratio to the training set, the validation set and the test set. Then patches of the training set and the validation set was fed into the ResNet18 tumor patch detector. Maximally 20 iterations of training were undertaken, and the learning rate was 2e-5. Binary cross-entropy loss was calculated upon completion of each iteration and the minimal loss was updated. Training was terminated when no further decreasing on binary cross-entropy loss in 5 uninterrupted iterations with at least 10 training iterations. The network architecture was optimized in the validation set and the model weights were preserved when minimal loss was achieved and this model with minimal loss was used as the tumor detector model named as INSIGHT. The precision rate, recall rate or sensitivity, and area under the curve (AUC) of the network were obtained using the test set. Precision predicted the correct value in the case of predicting positive samples while recall predicted the correct value in instances with positive labels.
Furthermore, 1579 WSIs from the TSMCC Cohort were tessellated into patches and normalized as described above, and then the patches were randomly assigned in a 7:1:2 ratio to the training set, the validation set and the test set and fed into the trained ResNet18 tumor patch detector INSIGHT. A SoftMax layer was used as the output layer to obtain the probabilities of each patch belonging to a tumor class; an input patch whose p value was ≥0.5 was classified as a tumor patch. The tumor patches were then entered into Residual Network with 50 layers (Resnet50) that had been pre-trained on the ImageNet.39 We chose to encode each tissue patch with a 1024-dimensional feature vector, yielding a feature matrix with a size of N × 1024, where N represented the number of tumor patches.
The feature matrix derived from the 1579 WSIs was fed into the Self-Attention model. The Self-Attention model was trained on the training set and verified on the validation set. Standard cross-entropy loss was calculated and the minimal loss was updated upon completion of each iteration. Training was terminated when no further decreasing on cross-entropy loss in 40 uninterrupted iterations with at least 100 training iterations. The precision rate, recall rate and AUC of the Self-Attention model were then obtained using the test set.
The Rectified Linear Unit (ReLU), as the activation function of the Self-Attention neural network, was used to define the nonlinear output of the neuron after linear transformation. The 1024-dimensional feature vector was reduced to 512-dimenional vector after two linear transformations using the equation:
where W and b represent the summed learnable weight matrix and bias of fully connected layers, respectively, and Zk represents the 1024-dimensional feature vector and hk the 512-dimenional vector after two linear transformations with activation of ReLU. Then, the attention score for the corresponding patch was obtained by attention pooling using the formula:
where V and U represent the summed learnable weight, and c and d represent the bias, respectively, and ak represents the attention score of patch k. Then, the 512-dimenional vector of a WSI was obtained by calculating the weighted sum of ak of all the patches of a WSI using the equation:
where hslide represents the 512-dimenional vector of a WSI. The MSI score was obtained by projecting the vector of a WSI using the classification layer followed by the softmax activation:
where Wcis and bcis represent the summed learnable weight, and bias of linear layers, respectively. SoftMax function was applied with default threshold 0.5 of MSI score used to classify a WSI into MSS or MSI-H.
The self-attention model of WiseMSI was separately tested as well using all the patches of the 1579 WSIs without the tumor pre-selection step.
Comparative analysis of WiseMSI with other models
For direct comparison of the performance of WiseMSI with other published methods, we select two classical weakly supervised methods EfficientNet40 and ViT41 and two MIL methods MIL and AttMIL42,43 which have been proved to have the top performance in Laleh’ benchmark analysis.33 DeepSMILE30 uses two-stage multiple instance learning approach, with a self-trained CNN model for feature extraction and VarMIL model for MSI classification. Only the untrained code of DeepSMILE was available to the public. The VarMIL model was included in our comparative analysis as well. Benchmark testing of these five methods vs. WiseMSI was carried on TSMCC cohort for end-to-end prediction of MSI status. The source codes of these methods were downloaded and re-trained and tested using TSMCC cohort data randomly assigned in a 7:1:2 ratio to the training set, the validation set and the test set. The training-validation-testing process was repeated for five iterations for each method. Similar to WiseMSI, fixed threshold 0.5 of MSI score were used in these four methods to classify a WSI into MSS or MSI-H.
Saillard C et al.44 used a self-trained RestNet50 model Moco V2 for feature extraction to improve dMMR/MSI detection, and open sourced the untrained Moco V2 model to public. Saillard trained Moco V2 using the same parameters and data augmentation scheme as the description in Dehaene et al.45 but a bigger ResNet backbone. We downloaded the untrained Moco V2 model and self-trained it following Dehaene’s description using 622 WSIs from TCGA COAD + READ cohorts and 400 WSIs from TSMCC. Standard ResNet 50 backbone was used in our training. Each WSI was divided into 20 tiles. Each model was trained for 600 epochs. The obtained features have a dimension of 1024. This self-trained Moco V2 feature extractor was benchmarked with the ImageNet pre-trained ResNet 50 model in WiseMSI.
WiseMSI on other cancer type
Performance of the self-attention model in WiseMSI and the classical weakly supervised method ViT on other cancer type were estimated by the TCGA-STAD cohort. 442 WSIs of stomach adenocarcinoma FFPE samples were acquired. 35 WSIs were excluded due to poor image quality. Of the remaining 407 WSIs, 342 were labeled as MSS and 65 as MSI-H by TCGA. 286 WSIs were randomly selected into training set, 40 into validation set, and 81 into testing set. These WSIs were tessellated into patches at 512 × 512 pixels, normalized, and passed into the Resnet50 pre-trained on the ImageNet for feature extraction. MSI prediction were made by the self-attention model of WiseMSI. Similarly, these patches were passed to ViT for MSI prediction.
Tumor purity reader experiment
Tumor purity is the fraction of cancer cells in tumor tissue. Tumor purity estimation by expert pathologist is a critical step of sample selection and result interpretation in cancer molecular test. To compare the performance of our tumor detection model INSIGHT on tumor purity estimation with that of pathologists, we carried a reader experiment in which the tumor purity of an additional external cohort with 210 WSIs were estimated by a board-certified, experienced pathologist, and by the tumor detect model INSIGHT independently. Two WSIs were excluded from performance comparison due to slide damage and wrong tumor type. The pathologist estimated tumor purity by reading the H&E stained hispathological slides. Tumor purity from INSIGHT was the percentage of patches with detected tumor among the patches from a CRC WSI. Spearman correlation coefficient was used for performance evaluation. The slides with larger than 20% difference in tumor purity estimations between the pathologist and INSIGHT were further inspected by another independent experienced pathologist.
Quantification and statistical analysis
For performance evaluation, in both the internal and external validation of WiseMSI, and in the comparative analysis of different models, ten-fold cross-validation was conducted by randomly dividing the input WSIs into 10 subsets. The entire training and testing process of WiseMSI were repeated 10 times with each time using 8 different subsets as training and validation sets, and 2 different subsets as internal test set. The 95% CI for the mean specificity, sensitivity, and AUC value were calculated.
Statistical analysis was undertaken using İBM SPSS Statistics 22. Disease-free survival (DFS) was calculated as the duration from the date of surgery to the date of recurrence, second cancer, or death from any cause, whichever occurred earlier. The data cutoff date was July 31, 2021. Kaplan-Meier survival analysis was used to determine whether there were differences in the survival distribution for the different group.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81773022). The institutions that participated in this project are Union Hospital, Tongji Medical College, Huazhong University of Science and Technology; Fujian Medical University Cancer Hospital, Fujian Cancer Hospital; The First Affiliated Hospital of Xi’an Jiaotong University; Shanxi Provincial Cancer Hospital; and Tianjin’s Clinical Research Center for Cancer, Tianjin Medical University Cancer Institute and Hospital.
Author contributions
X.Q., X.M., and X.N. contributed to the conception and design of the research. X.M., X.N., X.C., J.W., G.Z., M.Y., Y.X., and G.C. contributed to the sample and data collection and sample preprocessing and labeling. X.Q. and C.X. contributed to the AI model development and statistical analysis. X.Q. drafted the manuscript. X.Q., X.M., and X.N. contributed to the revision of the manuscript for important intellectual content. All authors contributed to final approval of the submitted version.
Declaration of interests
Patent is pending, and the patent application does not affect reproduction of this study’s results for research purposes.
Published: January 30, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100914.
Contributor Information
Xiu Nie, Email: niexiuyishi@126.com.
Bin Meng, Email: mbincn@163.com.
Xueping Quan, Email: quanxueping@tongshugene.com.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. The original codes have been deposited on GitHub: https://github.com/woshihang01/WiseMSI and are publicly available. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Siegel R.L., Miller K.D., Goding Sauer A., Fedewa S.A., Butterly L.F., Anderson J.C., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2020. CA A Cancer J. Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.Feng R.M., Zong Y.N., Cao S.M., Xu R.H. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun. 2019;39:22. doi: 10.1186/s40880-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vilar E., Gruber S.B. Microsatellite instability in colorectal cancer-the stable evidence. Nat. Rev. Clin. Oncol. 2010;7:153–162. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gavin P.G., Colangelo L.H., Fumagalli D., Tanaka N., Remillard M.Y., Yothers G., Kim C., Taniyama Y., Kim S.I., Choi H.J., et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. Clin. Cancer Res. 2012;18:6531–6541. doi: 10.1158/1078-0432.CCR-12-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Germano G., Lamba S., Rospo G., Barault L., Magrì A., Maione F., Russo M., Crisafulli G., Bartolini A., Lerda G., et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature. 2017;552:116–120. doi: 10.1038/nature24673. [DOI] [PubMed] [Google Scholar]
- 6.Diao Z., Han Y., Chen Y., Zhang R., Li J. The clinical utility of microsatellite instability in colorectal cancer. Crit. Rev. Oncol. Hematol. 2021;157 doi: 10.1016/j.critrevonc.2020.103171. [DOI] [PubMed] [Google Scholar]
- 7.Luchini C., Bibeau F., Ligtenberg M.J.L., Singh N., Nottegar A., Bosse T., Miller R., Riaz N., Douillard J.Y., Andre F., Scarpa A. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann. Oncol. 2019;30:1232–1243. doi: 10.1093/annonc/mdz116. [DOI] [PubMed] [Google Scholar]
- 8.Kather J.N., Halama N., Jaeger D. Genomics and emerging biomarkers for immunotherapy of colorectal cancer. Semin. Cancer Biol. 2018;52:189–197. doi: 10.1016/j.semcancer.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Boland C.R., Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087.e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binnewies M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M., Coussens L.M., Gabrilovich D.I., Ostrand-Rosenberg S., Hedrick C.C., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morse M.A., Hochster H., Benson A. Perspectives on treatment of metastatic colorectal cancer with immune checkpoint inhibitor therapy. Oncol. 2020;25:33–45. doi: 10.1634/theoncologist.2019-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lizardo D.Y., Kuang C., Hao S., Yu J., Huang Y., Zhang L. Immunotherapy efficacy on mismatch repair-deficient colorectal cancer: from bench to bedside. Biochim. Biophys. Acta, Rev. Cancer. 2020;1874 doi: 10.1016/j.bbcan.2020.188447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganesh K., Stadler Z.K., Cercek A., Mendelsohn R.B., Shia J., Segal N.H., Diaz L.A., Jr. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019;16:361–375. doi: 10.1038/s41575-019-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franke A.J., Skelton W.P., Starr J.S., Parekh H., Lee J.J., Overman M.J., Allegra C., George T.J. Immunotherapy for colorectal cancer: a review of current and novel therapeutic approaches. J. Natl. Cancer Inst. 2019;111:1131–1141. doi: 10.1093/jnci/djz093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Funkhouser W.K., Jr., Lubin I.M., Monzon F.A., Zehnbauer B.A., Evans J.P., Ogino S., Nowak J.A. Relevance, pathogenesis, and testing algorithm for mismatch repair-defective colorectal carcinomas: a report of the association for molecular pathology. J. Mol. Diagn. 2012;14:91–103. doi: 10.1016/j.jmoldx.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J. Mol. Diagn. 2008;10:293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapusot C., Martin L., Puig P.L., Ponnelle T., Cheynel N., Bouvier A.M., Rageot D., Roignot P., Rat P., Faivre J., Piard F. What is the best way to assess microsatellite instability status in colorectal cancer? Am. J. Surg. Pathol. 2004;28:1553–1559. doi: 10.1097/00000478-200412000-00002. [DOI] [PubMed] [Google Scholar]
- 18.Poynter J.N., Siegmund K.D., Weisenberger D.J., Long T.I., Thibodeau S.N., Lindor N., Young J., Jenkins M.A., Hopper J.L., Baron J.A., et al. Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol. Biomarkers Prev. 2008;17:3208–3215. doi: 10.1158/1055-9965.EPI-08-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnetson R.A., Tenesa A., Farrington S.M., Nicholl I.D., Cetnarskyj R., Porteous M.E., Campbell H., Dunlop M.G., Roseanne Cetnarskyj P.D., Mary E., et al. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N. Engl. J. Med. 2006;354:2751–2763. doi: 10.1056/NEJMoa053493. [DOI] [PubMed] [Google Scholar]
- 20.Stjepanovic N., Moreira L., Carneiro F., Balaguer F., Cervantes A., Balmaña J., Martinelli E., ESMO Guidelines Committee. Electronic address: clinicalguidelines@esmo.org Hereditary gastrointestinal cancers: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-updagger. Ann. Oncol. 2019;30:1558–1571. doi: 10.1093/annonc/mdz233. [DOI] [PubMed] [Google Scholar]
- 21.Sepulveda A.R., Hamilton S.R., Allegra C.J., Grody W., Cushman-Vokoun A.M., Funkhouser W.K., Kopetz S.E., Lieu C., Lindor N.M., Minsky B.D., et al. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American society for clinical pathology, College of American pathologists, association for molecular pathology, and American society of clinical oncology. Arch. Pathol. Lab Med. 2017;141:625–657. doi: 10.5858/arpa.2016-0554-CP. [DOI] [PubMed] [Google Scholar]
- 22.Kather J.N., Pearson A.T., Halama N., Jäger D., Krause J., Loosen S.H., Marx A., Boor P., Tacke F., Neumann U.P., et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019;25:1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niazi M.K.K., Parwani A.V., Gurcan M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20:e253–e261. doi: 10.1016/S1470-2045(19)30154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saltz J., Gupta R., Hou L., Kurc T., Singh P., Nguyen V., Samaras D., Shroyer K.R., Zhao T., Batiste R., et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181–193.e7. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhargava R., Madabhushi A. Emerging themes in image informatics and molecular analysis for digital pathology. Annu. Rev. Biomed. Eng. 2016;18:387–412. doi: 10.1146/annurev-bioeng-112415-114722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita R., Long J., Longacre T., Peng L., Berry G., Martin B., Higgins J., Rubin D.L., Shen J. Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet Oncol. 2021;22:132–141. doi: 10.1016/S1470-2045(20)30535-0. [DOI] [PubMed] [Google Scholar]
- 27.Echle A., Grabsch H.I., Quirke P., van den Brandt P.A., West N.P., Hutchins G.G.A., Heij L.R., Tan X., Richman S.D., Krause J., et al. Clinical-Grade detection of microsatellite instability in colorectal tumors by deep learning. Gastroenterology. 2020;159:1406–1416.e11. doi: 10.1053/j.gastro.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao R., Yang F., Ma S.C., Liu L., Zhao Y., Li Y., Wu D.H., Wang T., Lu W.J., Cai W.J., et al. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics. 2020;10:11080–11091. doi: 10.7150/thno.49864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Echle A., Ghaffari Laleh N., Quirke P., Grabsch H.I., Muti H.S., Saldanha O.L., Brockmoeller S.F., van den Brandt P.A., Hutchins G.G.A., Richman S.D., et al. Artificial intelligence for detection of microsatellite instability in colorectal cancer-a multicentric analysis of a pre-screening tool for clinical application. ESMO Open. 2022;7 doi: 10.1016/j.esmoop.2022.100400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schirris Y., Gavves E., Nederlof I., Horlings H.M., Teuwen J. DeepSMILE: contrastive self-supervised pre-training benefits MSI and HRD classification directly from H&E whole-slide images in colorectal and breast cancer. Med. Image Anal. 2022;79 doi: 10.1016/j.media.2022.102464. [DOI] [PubMed] [Google Scholar]
- 31.Bilal M., Raza S.E.A., Azam A., Graham S., Ilyas M., Cree I.A., Snead D., Minhas F., Rajpoot N.M. Development and validation of a weakly supervised deep learning framework to predict the status of molecular pathways and key mutations in colorectal cancer from routine histology images: a retrospective study. Lancet. Digit. Health. 2021;3:e763–e772. doi: 10.1016/S2589-7500(21)00180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong R., Liu W., DeLair D., Razavian N., Fenyö D. Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghaffari Laleh N., Muti H.S., Loeffler C.M.L., Echle A., Saldanha O.L., Mahmood F., Lu M.Y., Trautwein C., Langer R., Dislich B., et al. Benchmarking weakly-supervised deep learning pipelines for whole slide classification in computational pathology. Med. Image Anal. 2022;82 doi: 10.1016/j.media.2022.102474. [DOI] [PubMed] [Google Scholar]
- 34.Ashish Vaswani N.S., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser Ł., Polosukhin I. Attention is all you need. arXiv. 2017 doi: 10.48550/arXiv.1706.03762. Preprint at. [DOI] [Google Scholar]
- 35.Gao S., Qiu J.X., Alawad M., Hinkle J.D., Schaefferkoetter N., Yoon H.J., Christian B., Fearn P.A., Penberthy L., Wu X.C., et al. Classifying cancer pathology reports with hierarchical self-attention networks. Artif. Intell. Med. 2019;101 doi: 10.1016/j.artmed.2019.101726. [DOI] [PubMed] [Google Scholar]
- 36.Lu M.Y., Williamson D.F.K., Chen T.Y., Chen R.J., Barbieri M., Mahmood F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 2021;5:555–570. doi: 10.1038/s41551-020-00682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenson J.K., Bonner J.D., Ben-Yzhak O., Cohen H.I., Miselevich I., Resnick M.B., Trougouboff P., Tomsho L.D., Kim E., Low M., et al. Phenotype of microsatellite unstable colorectal carcinomas: well-differentiated and focally mucinous tumors and the absence of dirty necrosis correlate with microsatellite instability. Am. J. Surg. Pathol. 2003;27:563–570. doi: 10.1097/00000478-200305000-00001. [DOI] [PubMed] [Google Scholar]
- 38.Wang F., Wang Z.X., Chen G., Luo H.Y., Zhang D.S., Qiu M.Z., Wang D.S., Pan Z.Z., Shen L., Li J., et al. Expert opinions on immunotherapy for patients with colorectal cancer. Cancer Commun. 2020;40:467–472. doi: 10.1002/cac2.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stelloo E., Jansen A.M.L., Osse E.M., Nout R.A., Creutzberg C.L., Ruano D., Church D.N., Morreau H., Smit V.T.H.B.M., van Wezel T., Bosse T. Practical guidance for mismatch repair-deficiency testing in endometrial cancer. Ann. Oncol. 2017;28:96–102. doi: 10.1093/annonc/mdw542. [DOI] [PubMed] [Google Scholar]
- 40.Lupinacci R.M., Goloudina A., Buhard O., Bachet J.B., Maréchal R., Demetter P., Cros J., Bardier-Dupas A., Collura A., Cervera P., et al. Prevalence of microsatellite instability in intraductal papillary mucinous neoplasms of the pancreas. Gastroenterology. 2018;154:1061–1065. doi: 10.1053/j.gastro.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Heather Hampel M.S., Wendy Frankel M.D.L., Edward Martin M.D., Mark Arnold M.D., Karamjit Khanduja M.D., Philip Kuebler M.D., P.D., Hidewaki Nakagawa M.D., P.D., Kaisa Sotamaa M.D., et al. Screening for the lynch syndrome (hereditary nonpolyposis colorectal cancer) N. Engl. J. Med. 2005;352:10. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 42.Buhard O., Cattaneo F., Wong Y.F., Yim S.F., Friedman E., Flejou J.F., Duval A., Hamelin R. Multipopulation analysis of polymorphisms in five mononucleotide repeats used to determine the microsatellite instability status of human tumors. J. Clin. Oncol. 2006;24:241–251. doi: 10.1200/JCO.2005.02.7227. [DOI] [PubMed] [Google Scholar]
- 43.Southey M.C., Jenkins M.A., Mead L., Whitty J., Trivett M., Tesoriero A.A., Smith L.D., Jennings K., Grubb G., Royce S.G., et al. Use of molecular tumor characteristics to prioritize mismatch repair gene testing in early-onset colorectal cancer. J. Clin. Oncol. 2005;23:6524–6532. doi: 10.1200/JCO.2005.04.671. [DOI] [PubMed] [Google Scholar]
- 44.Xu J., Xue K., Zhang K. Current status and future trends of clinical diagnoses via image-based deep learning. Theranostics. 2019;9:7556–7565. doi: 10.7150/thno.38065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tieng F.Y.F., Abu N., Lee L.H., Ab Mutalib N.S. Microsatellite instability in colorectal cancer liquid biopsy-current updates on its potential in non-invasive detection, prognosis and as a predictive marker. Diagnostics. 2021;11:544. doi: 10.3390/diagnostics11030544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng J., Dong W., Socher R., Li L.J., Li K., Fei-Fei L. ImageNet: a large-scale hierarchical image database. IEEE Conference on Computer Vision and Pattern Recognition. 2009;2009:248–255. [Google Scholar]
- 47.Olivier Dehaene A.C., Moindrot O., Axel de Lavergne Self-supervision closes the gap between weak and strong supervision in histology. arXiv. 2012 doi: 10.48550/arXiv.2012.03583. Preprint at. [DOI] [Google Scholar]
- 48.Tan M.X.V.L.Q. EfficientNetV2: smaller models and faster training. arXiv. 2021 doi: 10.48550/arXiv.2104.00298. Preprint at. [DOI] [Google Scholar]
- 49.Ilse M.T.J., Welling M. Attention-based deep multiple instance learning. arXiv. 2018 doi: 10.48550/arXiv.1802.04712. Preprint at. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. The original codes have been deposited on GitHub: https://github.com/woshihang01/WiseMSI and are publicly available. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.