Summary
While the association between colorectal cancer (CRC) features and Fusobacterium has been extensively studied, less is known of other intratumoral bacteria. Here, we leverage whole transcriptomes from 807 CRC samples to dually characterize tumor gene expression and 74 intratumoral bacteria. Seventeen of these species, including 4 Fusobacterium spp., are classified as orally derived and are enriched among right-sided, microsatellite instability-high (MSI-H), and BRAF-mutant tumors. Across consensus molecular subtypes (CMSs), integration of Fusobacterium animalis (Fa) presence and tumor expression reveals that Fa has the most significant associations in mesenchymal CMS4 tumors despite a lower prevalence than in immune CMS1. Within CMS4, the prevalence of Fa is uniquely associated with collagen- and immune-related pathways. Additional Fa pangenome analysis reveals that stress response genes and the adhesion FadA are commonly expressed intratumorally. Overall, this study identifies oral-derived bacteria as enriched in inflamed tumors, and the associations of bacteria and tumor expression are context and species specific.
Keywords: cancer, colorectal cancer, microbiome, microbiota, intratumoral bacteria, oral microbiome, gut microbiome, Fusobacterium, TCGA, pangenome
Graphical abstract

Highlights
-
•
Oral bacteria are enriched among right-sided, MSI-H, and BRAF-mutant tumors
-
•
Mesenchymal tumors with Fusobacterium animalis have greater collagen and immune genes
-
•
Intratumorally, F. animalis expresses stress response genes and the adhesion FadA
Younginger et al. dually characterize tumor gene expression and 74 bacteria across 807 CRC tumors. They show that oral bacteria are enriched in right-sided, microsatellite-unstable, and BRAF-mutant tumors and that Fusobacterium animalis is associated with increased expression of collagen- and immune-related genes in mesenchymal tumors.
Introduction
Among solid tumors, colorectal cancer (CRC) is predicted to be the third leading cause of death, with rising incidence in younger individuals.1,2 CRC is a heterogeneous disease with tumors varying based on topography, i.e., location along the gastrointestinal (GI) tract, and mutation status, e.g., alterations in the BRAF oncogene gene, as well as molecular subtype, as determined by transcriptional profiles.3 In prior analyses, each of these variables was identified as a prognostic and/or predictive biomarker of CRC. For example, right-sided tumors respond less well to anti-epidermal growth factor receptor (EGFR) therapy,4 and tumors classified as the mesenchymal subtype have lower survival.3
Beyond traditional biomarkers, the microbiota, including both bacteria in the GI tract and within the tumor, is a growing area of interest in regard to CRC. More specifically, recent meta-analyses identified conserved differences in the gut microbiomes of patients with CRC compared with healthy controls.5,6,7 Several bacterial species, most notably Fusobacterium nucleatum, have elevated abundance in CRC tumors compared with adjacent normal or GI tissue from non-cancerous patients.8,9,10 This tumor/normal discrepancy and preclinical data suggest that Fusobacterium may play a causative role in CRC pathogenesis, progression, and response.11,12,13,14,15,16 Notably, Fusobacterium is present in only a subset of tumors and often coexists with other bacterial species including Bacteroides fragilis, Gemella morbillorum, Peptostreptococcus stomatis, and Selenomonas sputigena.8,17,18 The nature of these coexisting species remains poorly understood.
In this study, we generated whole-transcriptome RNA sequencing of 807 tumor tissues from patients with CRC to simultaneously characterize gene expression of both the tumor and microbes residing therein. To contextualize these microbial profiles, we compared our results with those of two meta-analyses that examined microbes in 208 saliva and 852 stool samples from patients with CRC and healthy controls.6,19 Additionally, we benchmarked our findings against 587 samples from The Cancer Genome Atlas (TCGA), which has previously been utilized for tumor/microbial associations.11,18,20,21 Using our bacterial profiles, we detailed links between community composition of oral bacteria and tumor location, mutation status, and molecular subtype. Notably, multiple species, including four F. nucleatum subspecies, were associated with clinical variables at a scale of hundreds of patients, exceeding prior studies (Figure S1A; Table S1A). We revealed that a microbe’s association with tumor gene expression varies depending on the CRC subtype. Finally, we identified which Fusobacterium animalis (Fa) genes are actively expressed in a pangenome analysis. Overall, this increased understanding of tumor microbes could lead to improved subtype stratification and novel prognostic biomarkers.
Results
To investigate both host and microbial transcriptomes that are poorly captured by widely used poly-(A) enrichment or exome capture approaches, we utilized whole-transcriptome RNA sequencing with rRNA depletion to detect microbial species that are metabolically active in tumor biopsies. Patient samples for this study were collected from AVANT,22 a randomized phase 3 trial that enrolled patients with resected stage III or high-risk stage II colon carcinomas and aimed to compare oxaliplatin-based chemotherapy with or without the anti-angiogenesis agent bevacizumab as adjuvant treatment post-surgery. Clinical characteristics of the 807 patients in the biomarker evaluable population were similar to the overall intent-to-treat population (Table S1B). Consistent with these non-metastatic patients being putatively cured by surgery, the number of recurrence events was low, and the AVANT trial did not meet its primary endpoint of improved disease-free survival (Figure S1B). Therefore, we deprioritized response-based analyses and focused on associating diverse microbes with tumor characteristics, including gene expression.
Gut and oral bacterial species are prevalent in the tumor
To identify the microbial content of the 807 whole transcriptomes, we mapped non-human reads against a bacterial and archaeal Genome Taxonomy Database (GTDB) with Kraken and Bracken (see STAR Methods, Figure S1C, and Table S1C).23,24,25 Across all samples, 882 M reads mapped to 2,043 unique species, with a median of 0.47 M (interquartile range [IQR], 0.27–0.99 M) per sample (Figures S1D and S1E). After implementing strict quality-control filters, 75 species remained (see STAR Methods, Figure S1C, and Table S1D). These species spanned common phyla including Bacteriodota, Firmicutes, and Fusobacteriota. Of the 75, all but the Cyanobacterium Aliterella sp000332075 were identified in at least one of 1,060 previously published oral and/or gut microbiome samples from healthy donors and patients with CRC6,19 reanalyzed with the same methods (Tables S1E–S1G). While the majority of these 74 species were highly prevalent in gut microbiome samples, 20 species were more prevalent in oral samples than stool (Figures 1A and 1B). Henceforward, those 20 species are designated as “oral” and the remaining 54 are “gut.” The oral taxa included several Fusobacterium species such as Fa, Fusobacterium polymorphum, Fusobacterium vincentii, and F. nucleatum (Fn). Prior to the GTDB, these four Fusobacterium species were grouped as Fn and delineated as separate subspecies, e.g., Fn subsp. animalis.
Figure 1.
74 bacteria, 54 gut and 20 oral species, were detected in the AVANT CRC samples
(A) Percentage of prevalence of 74 bacteria in AVANT. Bars colored by phyla.
(B) Dark gray bars (left) indicate prevalence across 852 stool samples from healthy and CRC donors6,19; light gray bars indicate prevalence in 208 saliva samples.19 Blue dots indicate the prevalence in control samples (stool, n = 568; saliva, n = 184), while red dots indicate CRC (stool, n = 284; saliva, n = 24).
(C) Each dot corresponds to 43 species overlapping between Kraken and Pathseq (Table S1H). Kraken prevalences are based on Bracken cutoff of 250 reads plus coverage cutoff of 0.5%. Pathseq prevalences are based on a score exceeding 250 plus coverage cutoff of 0.5%.
(D) For species in (A), the mean (excluding zeros) Kraken-assigned reads (prior to any Bracken reassignment) is shown. Point size reflects the percentage of samples that exceeded 250 Kraken-assigned reads for a given species.
(E) For 43 species in (C), the mean Pathseq score (excluding zeros) is shown. Point size reflects the percentage of samples that exceeded a score of 250 for a given species.
(C–E) Color of the point corresponds to the phyla and the shape to the gut or oral designation.
(F) Tile color reflects the log(OR), and shades of red indicate a taxa was more prevalent in BRAF-mutant versus wild type, MSI high (MSI-H) versus MSS, or right versus left, while blue shades indicate the opposite. Fisher’s exact test significance, ∗FDR < 0.05, ∗∗FDR < 0.01, ∗∗∗FDR < 0.001.
See also Figures S1–S3 and Tables S1D–S1Q.
Strikingly, prevalences of the gut species were consistently higher in stool samples from patients with CRC (n = 284) compared with healthy donors (n = 568; Figure 1B; Table S1E).6 Similarly, the oral taxa were more prevalent in saliva samples from patients with CRC (n = 24) compared with healthy controls (n = 184; Table S1F).19 While the presence of Fusobacterium in CRC tumors is widely appreciated,9,10 we detected an additional 15 oral bacterial species. Several of these oral taxa, including G. morbillorum, Parvimonas micra, and P. stomatis, were previously identified as predictive of CRC in samples of the gut microbiome.5,6,7,26 Overall, this indicates that the presence of a colorectal tumor may increase the overall permissiveness of the GI tract to oral bacteria.27,28,29,30,31
To verify our findings, we utilized Pathseq,32 an alternative program. Of our 74 taxa, 43 had an equivalent in the Pathseq database (Table S1H). Promisingly, prevalences for all 43 were strongly concordant across Kraken and Pathseq (Spearman’s Rho = 0.86, p = 2.2e−13; Figures 1C and S2A). As an additional benchmark, we queried 587 colon and rectal TCGA samples generated with poly-(A) enrichment (Table S1I). When applying the 250 read cutoff used for AVANT, only 20 species were detected in TCGA (Figures 1D, S2B, and S2C; Table S1J). Lowering the cutoff to 5 reads recovered all 74 species; however, at this threshold, an additional 1,030 species, including many known contaminants, were identified across the samples (Table S1K). This is a consequence of Kraken mapping 1,289 ± 19,659 (mean ± SD) reads per species in AVANT and only 15 ± 183 in TCGA, thus making it challenging to reliably differentiate signal from noise. Notably, we also observed this discrepancy with Pathseq32 (Figures 1E, S2D, and S2E; Table S1L). Altogether, this comparison highlights the utility of the AVANT dataset and the advantage of using rRNA depletion for identification of a high diversity of species, including taxa at lower abundances, at levels distinguishable from noise.
Right-sided, MSI-H, and BRAF-mutant tumors have greater prevalences of oral bacterial species
Having identified 74 diverse intratumoral bacteria across individuals, we investigated the association between their presence and 21 metadata variables (Table S1B), including demographic factors, such as sex, age, and race, as well as tumor characteristics, e.g., disease stage, tumor classification, location, and mutation status. Of these, 7 variables were associated with at least one intratumoral species (chi-squared test, false discovery rate [FDR] < 0.05; Figure S3A; Table S1M). The top three included microsatellite instability (MSI) status, tumor location, and BRAF-mutation status with 23, 21, and 8 species associations, respectively. MSI status indicates whether an individual had a mismatch repair deficiency in their tumor.33 Here, tumor location was based on the annotated surgical procedure performed (Figure S3B). Notably, these three traits were highly correlated such that 66% of MSI-high and 62% of BRAF-mutant tumors were right-sided, while 44% of BRAF-mutant tumors were MSI high (Figures S3C and S3D).
Across those 3 variables, 31 bacterial species, including 13 oral species, were consistently more prevalent in right- versus left-sided, MSI-high versus microsatellite stable (MSS), and/or BRAF-mutant versus wild-type (WT) tumors (Fisher’s exact test, FDR < 0.05, mean odds ratio [OR] 3.94 [3.48, 4.4]; Figure 1F; Tables S1N and S1O). This is consistent with these tumors having more reads mapping to microbes, despite equivalent starting reads, and a greater portion having any detectable microbial signal (Figures S3E–S3G). It is also consistent with previous studies that identified right-sided tumors as having a greater incidence of biofilms and hence greater microbial burden.8,17 These multitaxa results expand on the prior finding that Fusobacterium is consistently enriched in these tumors20,34,35,36,37,38 (Figure S1A). To account for intercorrelation between these variables (Figures S3C and S3D), we performed multivariate analysis and found that the majority of associations remained significant (Table S1P; Figure S3H).
The impact of F. animalis on tumor gene expression varies by consensus molecular subtype
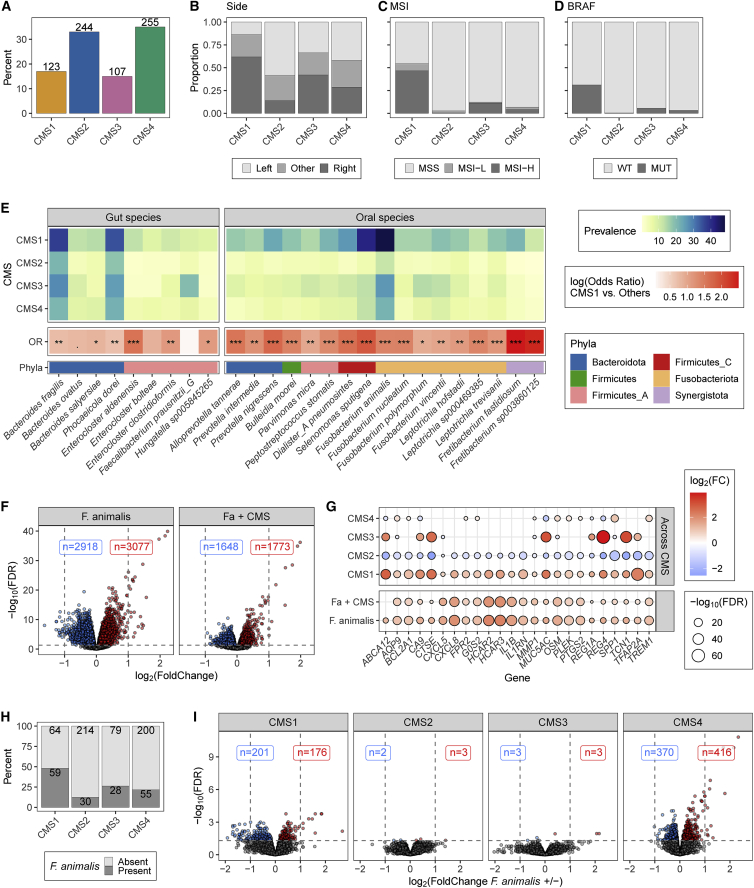
As the aforementioned variables are hallmarks of CRC subtypes, we binned samples into consensus molecular subtypes based on tumor gene expression (Figure 2A).39 As expected, consensus molecular subtype 1 (CMS1) tumors, known as immune high, were more likely to be right-sided, MSI-high, and BRAF-mutant compared with CMS2-4 tumors (Figures 2B–2D).3 Consistent with this and our meta-data results, 25 of the species, including 17 oral species, were most prevalent in CMS1 tumors (chi-squared test, FDR < 0.05; Figure 2E; Table S1Q). Overall, these associations highlight that many oral species beyond Fusobacterium display strong specificity for the inflamed CRC subtype.
Figure 2.
The association of Fa and tumor gene expression varies by CMS
(A) Bars indication percentage of AVANT samples in each CMS. Values indicate the number of samples.
(B) Bars indicate the proportion of samples by location.
(C) Bars indicate the proportion of samples by MSI status.
(D) Bars indicate the proportion of samples that were BRAF-wild type (WT) and mutant (MUT).
(E) For species differentially present between CMSs (chi-squared FDR < 0.05), top heatmap shows the species prevalence by subtype. In the middle, tile color reflects the log(OR) of CMS1 versus the other CMSs. Fisher’s exact test significance, ∗FDR < 0.05, ∗∗FDR < 0.01, ∗∗∗FDR < 0.001. The bottom row indicates the species’ phyla.
(F) Volcano plot of differentially abundant genes between samples with or without Fa based on Voom-Limma. CMS was included as a covariate for the results on the right. Blue dots indicate genes with FDR <0.05 and log2(fold change) <0; red dots are genes with FDR <0.05 and log2(fold change) >0. Labels indicate how many genes were statistically significantly up or down.
(G) Balloon plot shows 24 genes with FDR <0.05 and log2(fold change) >1 in (F). Circle size corresponds to the −log10(FDR), while color is the log2(fold change). The bottom row represents the differential expression values for all samples with or without Fa. The next row includes CMS as a covariate. The CMS1 column represents the differential expression values for all CMS1 samples versus the CMS2, -3, and -4 samples. Similarly, the CMS2 column represents the CMS2 samples versus the CMS1, -3, -4, etc., samples.
(H) Bars indicate percentage of Fa positivity by subtype. Values indicate the number of samples.
(I) Volcano plot of differentially abundant genes between samples with or without Fa stratified by CMS. Colors and labels are the same as (F).
See also Figures S4 and S5 and Tables S1M, S1N, and S1Q–S1S.
Next, we identified host genes associated with intratumoral bacteria and the effect of CMS status on those associations. To address this, we initially focused on the most prevalent Fusobacterium species, Fa. Based on differential expression analysis, Fa presence was associated with changes in 5,995 genes (FDR < 0.05), including 24 with greater than a 2-fold increase in expression (Figure 2F). These included several immune-related genes, e.g., CXCL8 (IL8), IL1B, IL6, OSM, CXCL5, MMP1, and PTGS2 (COX-2) (Table S1R), many of which were previously identified as increased in TCGA samples with detectable Fusobacterium.11,20 To understand if these associations were artifacts of Fa’s strong association with CMS1 (Figure 2E), we included CMS as a covariate in the statistical model. While doing so lowered the number of differentially expressed genes to 3,421, all but 1 of the top 24 genes retained statistical significance, indicating that at least some of these associations were independent of Fa’s enrichment in CMS1 (Figures 2F and 2G; Table S1R).
To determine whether these genes were specific to Fa or a more broadly observed response to intratumoral microbes, we conducted identical analysis, controlling for CMS, on the three gut taxa with prevalences greater than 20% (B. fragilis, Phocaeicola dorei [formerly Bacteroides dorei], and Ruminococcus_C sp000433635) and the second most prevalent oral taxon (S. sputigena) (Figure 1A; Table S1S). Even after false discovery correction, all species were associated with more than 2,000 differentially expressed genes (FDR < 0.05; Figure S4A). Among the top overexpressed genes (log fold change [logFC] > 0.75; Figure S4B), there was strong overlap between the oral species Fa and S. sputigena and, to a lesser extent, R. sp000433635. Interestingly, hydroxycarboxylic acid receptors 2 and 3 (HCAR2 and HCAR3) were the only genes strongly changed across all five species. Altogether, these results reflect that some tumor gene associations are conserved, while others are taxa specific.
To understand if the effect of Fa was variable across CRC subtypes, we analyzed each CMS independently (see STAR Methods, Figures 2H and 2I, and Table S1R). Notably, in the CMS2 and CMS3 tumors, Fa presence was associated with only 5 and 6 differentially expressed genes, respectively. By contrast, 377 genes were differentially expressed in CMS1 tumors, which had the greatest Fa prevalence at 48%. Despite a prevalence of only 22% in CMS4 tumors, Fa presence was associated with 786 differentially expressed genes, of which 116 overlapped with CMS1. The heightened alteration of gene expression by Fa in CMS4 CRC tumors is of particular interest as CMS4 mesenchymal tumors are associated with worse prognosis than other CMS subtypes in this (Figure S4C) and other cohorts.3,20 Additionally, because high Fusobacterium levels have been associated with further reduced survival in patients with CMS4 tumors,20 we were compelled to understand how Fa uniquely impacts this subtype.
Fa is associated with upregulation of collagen and immune pathways in CMS4 tumors
Gene set enrichment analysis (GSEA) revealed 69 REACTOME40 pathways that were differentially expressed based on Fa presence in CMS4 tumors (Table S1T). Of the top pathways, five related to collagen degradation and formation as well as many immune-related pathways, including signaling of the cytokines interleukin-6 (IL-6), IL-12, IL-1, IL-4, and IL-13, were increased (Figures 3A–3C). Within these pathways, IL1B, IL1RN, CXCL8, matrix metallopeptidase 1 (MMP1), and oncostatin M (OSM) were the most highly upregulated in CMS4 tumors (Figure 3D). Notably, increased levels of IL-8, encoded by the gene CXCL8, have been associated with CRC growth, progression, and recurrence in patients.41 Additionally, OSM, a member of the IL-6 cytokine family, IL-1, and MMP1 have each been associated with decreased survival in patients with CRC.42,43,44,45 Altogether, each of these alterations represent multiple means by which tumor-resident Fa may associate with worse CRC outcomes.20
Figure 3.
Fa is associated with upregulation of collagen- and immune-related pathways in CMS4 tumors
(A) For each pathway, a gray square denotes a gene’s presence. Only genes differentially expressed based on Fa presence in at least one of the CMS strata are shown.
(B) For genes differentially expressed in a particular CMS, the log2(fold change) is shown. Genes upregulated in the presence of Fa are shown in red shades, while downregulated genes are in blue.
(C) The circle color indicates in which CMS the REACTOME pathway was statistically significantly enriched based on GSEA in a given species, while size indicates the −log10(FDR) value.
(D) Volcano plots from Figure 2I with addition of red dots highlighting genes of interest in (B). Labeled genes had log2(fold change) <−1 or >1.
(E) Relative abundance plots indicate average cell-type composition in patients with or without Fa in all samples and across the different CMSs.
(F) Percentage of neutrophils across all samples and by CMS and Fa presence/absence. Wilcoxon-test FDR values are shown.
See also Table S1T.
To verify whether these pathways were specific to Fa, we conducted GSEA on the same four taxa as before (Figure S4A; Table S1T). When stratified by CMS status, only P. dorei was associated with any collagen pathways. Importantly, these P. dorei associations were significant only in canonical CMS2 tumors, characterized by WNT and MYC activation, not CMS4. (Figure 3C). As for the immune pathways, IL-10, IL-4, IL-13, and IL-6 signaling were increased in CMS2 and CMS3 tumors, while IL-4 and IL-13 signaling were increased in CMS4 tumors with S. sputigena present (Figure 3C). As such, these results highlight that the association of the intratumoral microbiome with host gene expression is both species- and tumor-context-specific.
To further understand how intratumoral bacteria influence the tumor microenvironment, we used immune deconvolution to infer cell composition from the tumor gene expression (Figure 3E).46 Consistent with the neutrophil chemotaxis induced by CXCL8, across all tumors, the average proportion of neutrophils was increased in Fa-positive (Fa+) samples (34.8%, 95% confidence interval [CI] [33.1%, 36.5%] versus 31% [30.1%, 31.8%]; Figure 3F). Across CMSs, Fa was associated with the greatest neutrophil frequency, 37.2% [33.5%, 40.8%], in CMS4 tumors, mirroring the high expression of CXCL8 observed in this setting (Figure 3F). Importantly, these results are consistent with increased neutrophils in ApcMin/+ mice exposed to Fn.11 Overall, these data highlight how further experiments are necessary to resolve whether Fa can shape the immune milieu as well as the collagen architecture of the tumor microenvironment.
Pangenome analysis reveals highly expressed Fa genes
To explore how Fa may be impacting the tumor, we investigated which Fa genes are actively expressed intratumorally. This analysis builds on prior studies of the Fa transcriptome in vitro.47,48,49 To start, we built a Fa pangenome composed of 4,355 non-redundant gene clusters (see STAR Methods). There were 1,228 genes that were core, i.e., present in all 26 strains used to build the pangenome, while 31 novel genes were added per each additional genome (Figures 4A and 4B; Tables S1U and S1V).
Figure 4.
Fa gene expression in pangenome analysis
(A) Gene accumulation curves for pangenome (blue) and core genome (green) as a function of genome sequences (N). Both are fit by a power law regression. Points are means of n for 200 simulations. Error bars indicate the SDs for the 200 simulations.
(B) Accumulation of new genes (n) discovered with the addition of new genome sequences (N) fits a power law regression.
(C) Boxplots indicate the number of Fa genes identified in the Fa+ and Fa− samples. Wilcoxon p value is shown.
(D) Density plot shows the distribution of prevalence of the pangenome genes. 1,546 genes were identified in 0 of the Fa+ tumors. 948 were identified in at least 10% of the Fa+ tumors.
(E) Bars indicate proportion of genes annotated as ribosomal in the whole pangenome (gray), in the part expressed in at least 10% of the Fa+ samples (light blue), in at least 20% (dark blue), and 50% (green). Fisher’s exact test significance, ∗FDR < 0.05, ∗∗FDR < 0.01, ∗∗∗FDR < 0.001, between the entire pangenome (cutoff 0) and the three prevalence cutoffs (10, 20, and 50).
(F) Same as (E) but non-ribosomal KEGG pathways.
(G) Same as (E) but annotations to CARD and VFDB.
(H) For the 35 non-ribosomal genes expressed in more than half of the Fa+ samples, the far left columns indicate whether the gene was annotated as part of the pangenome core or mapped to something in the CARD or VFDB. Middle column indicates KEGG pathway annotation. On the right, gray dots indicate the prevalence in Fa− samples, while blue dots indicate Fa+ prevalence.
See also Tables S1U and S1V.
Next, we mapped reads to this pangenome database (see STAR Methods) and found 297 ± 397 (mean ± SD) genes in Fa+ tumors and only 31 ± 61 in the negative ones (Figure 4C). Across the pangenome, 948 of the genes were expressed in at least 10% of the Fa+ samples, 451 in ≥20%, and 79 in ≥50% (Figure 4D). Using KEGG annotations, we found that ribosomal genes were overexpressed compared with the pangenome, i.e., genes annotated as ribosomal made up 56% of the genes expressed in at least half of Fa+ samples but only 1.2% of the pangenome (Figure 4E). The quorum-sensing pathway was also overexpressed, suggesting that bacterial loads in the tumor were sufficiently high to induce a signaling cascade. Other overexpressed pathways include ABC transporters, glycolysis gluconeogenesis, RNA degradation, and RNA polymerase (Figure 4F). Further, we annotated the clusters with the Comprehensive Antibiotic Resistance Database (CARD)50 and Virulence Factor Database (VFDB)51 and identified that genes in both were overexpressed compared with the pangenome (Figure 4G).
We then interrogated the 35 most commonly expressed, non-ribosomal genes and found that 32 were in the core of the pangenome and that all were expressed at greater prevalences in Fa+ versus Fa− tumors (Fisher’s exact test, FDR < 0.001; Figure 4H), implying they were Fa specific. Among them were several genes associated with bacterial response to stress, e.g., elongation factor Tu, thiroredoxin, and GroEL enable adaptation to oxidative stress, DnaK provides resistance to thermosensitivity, and the Clp proteases, e.g., ClpB, ClpP, and ClpX, have been identified in many pathogens, including Staphylococcus aureus, as central to stress survival and virulence.52,53,54,55,56 These expression patterns could be reflective of Fa attempting to survive intracellularly against a barrage of host defenses including antimicrobial peptides, e.g., S100A8 and S100A9, which were increased in Fa+ tumors (Table S1R). A handful of other proteases, including the serine protease fusolisin, were also identified as expressed (Table S1V). These results combined with the observation that fusolisin can degrade the extracellular matrix protein collagen57,58 provide a probable mechanism of why many collagen-related genes were upregulated in Fa+ tumors (Figures 3A–3C). Additionally, the well-studied adhesin FadA,16,59,60 which interacts with E-cadherin to mediate Fusobacterium attachment and invasion into epithelial cells, was identified in 56% of Fa+ tumors. In summary, the prevalent tumor expression of FadA and the previous observation that blocking Fad-A/E-cadherin binding can abolish Fad-A-induced CRC cell growth in vivo means that blocking this interaction in patients could be a promising therapeutic opportunity.16
Discussion
CRC tumors harbor diverse microbes that are heterogeneous across individuals. Here, we demonstrate that the prevalences of bacterial species are strongly associated with tumor subtypes as characterized by location, mutation status, and gene expression (Figures 1F and 2E). Notably, oral-derived bacteria displayed a strong association with CMS1, immune-high tumors. Evolutionarily, this preference could imply that physiological features of CMS1 tumors, e.g., oxygen concentration or pH, were similar to that of dental plaques, where these oral species often aggregate into biofilms,61 meaning that oral species are well poised to survive in the CMS1 environment. Consistent with this hypothesis, expression of HIF1A, indicative of tissue hypoxia, was highest in CMS1 tumors and lowest in CMS2 tumors (Figure S5A), which had the lowest prevalence of the oral microbes (Figure 2E). Similarly, H1F1A expression was consistently higher in tumors with any of the 17 oral species that were enriched in CMS1 (Figure S5B). Since the majority of these species are anaerobic, it is reasonable that oxygen concentration would be an important factor underlying the differential bacterial colonization across subtypes.
While strains of Fusobacterium identified in the mouth have been matched to strains in the tumor,62 the method of translocation in patients is unclear. Intriguingly, in preclinical models, Fn was shown to colonize rectal tumors more efficiently when injected intravenously compared with oral gavage.63 At the same time, in the ApcMin/+ mouse model, oral gavage of Fn was sufficient to promote colorectal tumor development, suggesting that transmission via the GI tract was also possible.11 In both scenarios, Fn may initially attach to the CRC tumors via the adhesion Fap2, which binds Gal-GalNAc sugar residues, overexpressed on CRC tumors.12 Notably, Fap2 was identified as being expressed in 9% of Fa+ tumors. Once in the tumor, Fn may act as a keystone species to retain fellow oral bacteria in the tumor, similar to oral biofilms, where Fn connects early and late colonizing bacteria.14 In support of this, the adhesin RadD, known to promote oral biofilms,64 was expressed in 10% of Fa+ tumors.
From the perspective of host gene expression, integrating bacterial presence and tumor gene expression revealed that the impact on expression was variable and dependent on CMS and the bacterial species present. For example, the oral species S. sputigena was associated with upregulated IL-6 and IL-10 signaling pathways exclusively in canonical CMS2 and metabolic CMS3 tumors. In contrast, CMS2 and CMS3 tumors had only 5 and 6 genes, respectively, that were differentially expressed based on Fa presence, while there were 377 and 786 genes in CMS1 and CMS4 tumors, respectively. Of the top genes, many, including CXCL8 (IL8), IL1B, IL6, MMP3, and PTGS2 (COX-2) (Figure 2G), were previously identified as increased in ApcMin/+ mice after being administered Fn.11 Additionally, in vitro, Fusobacterium has been shown to induce expression of CXCL8 in colon and CXCL8 and MMP1 in oral cancer cell lines.65,66 Notably, many of these upregulated genes have been previously associated with metastasis and/or poor prognosis in CRC,41,42,43,44,45 so while Fa was not associated with response in CMS4 patients in this adjuvant cohort (Figures S5C–S5E), these pathways represent possible mechanisms by which Fa could negatively impact response in patients with more advanced, including metastatic, disease.20 Therefore, from a clinical perspective, earlier detection and resection of Fa+ tumors could be particularly prudent for patients, as those tumors are more likely to progress rapidly.
In conclusion, with whole-transcriptome sequencing, we discovered that oral-derived bacteria, in addition to Fusobacterium, are prevalent in inflamed, CMS1 CRC tumors. Within the tumors, the association of bacteria and tumor gene expression, however, is highly context and species specific. Beyond the findings in this paper, the rich metadata and 800-plus deeply sequenced samples provided here will be a valuable resource for future hypothesis generation and testing around diverse bacteria in CRC.
Limitations of the study
Our study has a number of limitations. First, the tumor samples were collected and sequenced without considering the microbiome. In the absence of recommended positive and negative controls,67 we implemented strict computational filters in an attempt to differentiate true signal from artifact. There is a risk that those filters may have been too strict and removed true signals or were too lenient and allowed contaminants to persist. Second, all findings presented here are purely associations. Future experiments to determine whether Fa and other bacterial species are eliciting the observed alterations in tumor gene expression are required.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Raw tumor and microbial RNA-sequencing data and clinical metadata variables | This paper | EGA: EGAS00001006757 |
| Stool metagenomic data from controls and patients with CRC | Yu et al.68 | SRA: PRJEB10878 |
| Stool metagenomic data from controls and patients with CRC | Vogtmann et al.69 | SRA: PRJEB12449 |
| Stool metagenomic data from controls and patients with CRC | Wirbel et al.6 | SRA: PRJEB27928 |
| Saliva metagenomic data from controls and patients with CRC | Belstrøm et al.70 | SRA: PRJEB28422 |
| Stool metagenomic data from controls and patients with CRC | Zeller et al.71 | SRA: PRJEB6070 |
| Stool metagenomic data from controls and patients with CRC | Feng et al.72 | SRA: PRJEB7774 |
| Stool and saliva metagenomic data from controls | Zhang et al.73 | SRA: PRJEB6997 |
| Stool and saliva metagenomic data from controls | Brito et al.74 | SRA: PRJNA217052 |
| Stool and saliva metagenomic data from controls | Heintz-Buschart et al.75 | SRA: PRJNA289586 |
| COAD and READ RNA-seq, Release 7.0 | TCGA | Portal.gdc.cancer.gov; RRID:SCR_003193 |
| Genome Taxonomy Database: GTDB v95 | Parks et al.23 | https://data.ace.uq.edu.au/public/gtdb/data/releases/release95/95.0/; RRID:SCR_019136 |
| Human reference genome NCBI build 38, GRCh38 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| KEGG 2013.10.14 | Kanehisa et al.76 | https://www.genome.jp/kegg/pathway.html; RRID:SCR_012773 |
| CARD and VFDB (mid-2017) from ShortBRED | Kaminski et al.77 | https://github.com/biobakery/shortbred |
| Software and algorithms | ||
| Kraken2; v2.1.1 | Wood et al.24 | https://github.com/DerrickWood/kraken2 |
| Bracken v2.5 | Lu et al.25 | https://github.com/jenniferlu717/Bracken |
| HTSeqGenie; V4.2.2 | Pau et al.78 | https://bioconductor.org/packages/release/bioc/html/HTSeqGenie.html |
| gSNAP; V2013-10-10-v2 | Wu et al.79,80 | https://bioinformaticshome.com/tools/rna-seq/descriptions/GSNAP.html#gsc.tab=0; RRID:SCR_005483 |
| Gencode genes database (GENCODE 27) | GENCODE | http://www.gencodegenes.org/human/release_27.html; RRID:SCR_014966 |
| edgeR, v3.38.2 | Chen at al.81 | https://bioconductor.org/packages/release/bioc/html/edgeR.html; RRID:SCR_012802 |
| Limma v3.52.2 | Liu et al.82 Smyth et al.83 |
https://bioconductor.org/packages/release/bioc/html/limma.html; RRID:SCR_010943 |
| REACTOME | Gillespie et al.40 | https://reactome.org/; RRID:SCR_003485 |
| Immunedeconv, v2.0.4 | Sturm et al.,84 Finotello et al.46 |
https://github.com/omnideconv/immunedeconv |
| Trimmomatic v0.39 | Bolger et al.85 | https://github.com/usadellab/Trimmomatic; RRID:SCR_011848 |
| prinseq-lite v0.20.4 | Schmieder et al.86 | https://github.com/uwb-linux/prinseq; RRID: SCR_005454 |
| Bowtie2 v2.4.1 | Langmead et al.87 | https://github.com/BenLangmead/bowtie2; RRID:SCR_016368 |
| bedtools v2.30.0 | Quinlan et al.88 | https://github.com/arq5x/bedtools2; RRID: SCR_006646 |
| Cutadapt v3.7 | Martin et al.89 | https://cutadapt.readthedocs.io/en/stable/; RRID:SCR_011841 |
| Metaphlan3, v3.0.7 | Beghini et al.90 | https://huttenhower.sph.harvard.edu/metaphlan/; RRID:SCR_004915 |
| mOTUs, v3.0.2 | Milanese et al.91 | https://github.com/motu-tool/mOTUs |
| PathSeq via GATK v4.1.4.0 | Kostic et al.32 | https://software.broadinstitute.org/pathseq/; RRID:SCR_005203 |
| CMScaller v0.99.2 | Eide et al.39 | https://github.com/peterawe/CMScaller |
| USEARCH v11.0.667_i86linux32 | Edgar et al.92 | https://www.drive5.com/usearch/ |
| BLASTp v2.6.0 | Camacho et al.93 | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastDocs&DOC_TYPE=Download; RRID:SCR_001010 |
| DIAMOND v2.0.11 | Buchfink et al.94 | https://github.com/bbuchfink/diamond; RRID:SCR_016071 |
| Samtools v1.11 | Li et al.95 | https://github.com/samtools/samtools; RRID:SCR_002105 |
| R v4.1.1 | 96 | http://www.r-project.org/; RRID: SCR_001905 |
| rmarkdown v2.11 | Allaire et al.97 | https://cran.r-project.org/web/packages/rmarkdown/index.html |
| knitr v1.37 | Xie et al.98 | https://cran.r-project.org/web/packages/knitr/index.html; RRID:SCR_018533 |
| dplyr v1.0.7 | Wickham et al.99 | https://cran.r-project.org/web/packages/dplyr/index.html; RRID:SCR_016708 |
| reshape2 v1.4.4 | Wickham et al.100 | https://cran.r-project.org/web/packages/reshape2/index.html; RRID:SCR_018983 |
| plotrix v3.8-2 | Lemon et al.101 | https://cran.r-project.org/web/packages/plotrix/index.html |
| gtools v3.9.2 | Warnes et al.102 | https://cran.r-project.org/web/packages/gtools/index.html |
| ggplot2 v3.3.5 | Wickham et al.103 | https://cran.r-project.org/web/packages/ggplot2/index.html; RRID:SCR_014601 |
| ggeasy v0.1.3 | Carroll et al.104 | https://cran.r-project.org/web/packages/ggeasy/index.html |
| scales v1.1.1 | Wickham et al.105 | https://cran.r-project.org/web/packages/scales/index.html |
| cowplot v1.1.1 | Wilke et al.106 | https://cran.r-project.org/web/packages/cowplot/index.html; RRID:SCR_018081 |
| ggrepel v0.9.1 | Slowikowski et al.107 | https://cran.r-project.org/web/packages/ggrepel/index.html; RRID:SCR_017393 |
| RColorBrewer v1.1-2 | Neuwirth et al.108 | https://cran.r-project.org/web/packages/RColorBrewer/index.html; RRID:SCR_016697 |
| viridis v0.6.2 | Garnier et al.109 | https://cran.r-project.org/web/packages/viridis/index.html; RRID:SCR_016696 |
| Openxlsx v4.2.5 | Schauberger et al.110 | https://cran.r-project.org/web/packages/openxlsx/index.html; RRID:SCR_019185 |
| Survival v3.2-13 | Therneau et al.111 | https://cran.r-project.org/web/packages/survival/index.html; RRID:SCR_021137 |
| Survminer v0.4.9 | Kassambara et al.112 | https://cran.r-project.org/web/packages/survminer/index.html; RRID:SCR_021094 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Allyson Byrd (byrd.allyson@gene.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Clinical trial information
835 samples were obtained from 821 patients enrolled in the AVANT clinical trial.22 All patients provided informed consent for the AVANT study. The AVANT protocol was approved by the Ethics Review Committee or Institutional Review Board at participating sites. There were 14 patients that had 2 samples each, all of which were excluded for downstream analyses leaving 807 in the biomarker-evaluable population. The trial examined differences in disease-free survival of patients with stage III or high-risk stage II colon carcinomas in response to bevacizumab when combined with oxaliplatin-based chemotherapy in an adjuvant setting. Analysis of patient demographics, including age and sex, from the biomarker evaluable population in relation to the intent to treat population are included in Table S1B. Following surgical resection of tumor tissue, samples were formalin-fixed paraffin-embedded (FFPE) prior to RNA isolation.
Method details
RNA extraction, library prep, and sequencing
Quality control (QC) of RNA samples, library construction and sequencing data generation was performed by an external service provider, Q2 Solutions. QC of RNA samples was done prior to their processing by RNA-seq. The concentration of RNA was determined with Qubit (Thermo Fisher Scientific) and RNA integrity was measured using DV200 on a 2100 Bioanalyzer (Agilent Technologies). Sequencing libraries were generated with the TruSeq Stranded Total RNA kit (Illumina) following rRNA depletion with the Ribo-Zero Gold kit (Illumina). Briefly, starting with total RNA, rRNA was removed using biotinylated probes that selectively bind rRNA species. This process preserves mRNA and other non-coding RNA species including lincRNA, snRNA and snoRNAs. The resulting rRNA-depleted RNA was fragmented using heat in the presence of divalent cations, with fragmentation times varying based on input RNA degradation. Fragmented RNA was converted into double-stranded cDNA with dUTP used in place of dTTP in the second strand master mix. This facilitates the preservation of strand information as PCR amplification will stall when it encounters uracil, rendering the first strand as the only viable amplification template. The double stranded cDNA was used for library generation according to the manufacturer’s protocol (Illumina). The final libraries were PCR amplified, quantified, normalized and pooled in preparation for sequencing. The libraries were sequenced on the HiSeq4000 (Illumina) with a sequencing protocol of 75 bp paired-end sequencing and target total read depth of 60M reads per sample.
Quantification and statistical analysis
Tumor gene expression analysis
Following sequencing, 73.3 billion reads were obtained with 90.8 ± 13.2 (mean ± SD) million reads per sample. All post-sequencing QC steps were performed with the HTSeqGenie package (V4.2.2)78 in BioConductor.113 Reads were quality filtered with a minimum phred quality set to 23 over 70% of the read. Any reads shorter than 18 bp were discarded. Illumina adapters were removed and any reads detected as rRNA against the GRCh38_rRNA database were discarded. Reads remaining after quality control steps were then aligned to the GRCh38.p10 genome with the gSNAP aligner79,80 (version: V2013-10-10-v2), allowing a maximum of two mismatches per 75 base sequence (parameters: ‘-M 2 -n 10 -B 2 -i 1 -N 1 -w 200000 -E 1 --pairmax-rna = 200,000 --clip-overlap'). Transcript annotation was based on the Gencode genes database (GENCODE 27). To quantify gene expression levels, the number of reads mapping unambiguously to the exons of each gene was calculated.
The initial quality control and alignment steps resulted in a total of 56.3 billion reads mapping across all samples, averaging 69.8 ± 18 million reads per sample. From these alignments, reads that mapped successfully to the human genome were converted into a count matrix with ENSEMBL gene IDs and further analyzed in the host expression pipeline, while reads that did not properly align (i.e. nomapping bam files) were processed in the microbial pipeline.
Genes within the gene count matrix were retained with edgeR (v3.38.2)’s81 default filtering strategy, which only retains genes with coverage above a counts per million (CPM) equivalent to a count of 10 (adjusted for differences in library size) in at least n samples, where n is determined from the smallest group in the experimental design (parameters: ‘large.n = 0’). The retained genes were then transformed into log CPM with trimmed mean of M-values normalization (TMM). Genes with a mean of less than 1 log CPM were further removed from the matrix. Differential expression was performed with the voomWithQualityWeights function implemented in Limma (v3.52.2)82,83 with quality weighting and robust empirical Bayes shrinkage to control outlier samples and genes (parameters: lmFit() = default, eBayes() = ‘robust = TRUE’). Models were generated with clinical and microbiome data, explained below. To examine significant groups of genes, gene set enrichment analysis (GSEA) was run with the hypergeometric test and the REACTOME database.40 Significant gene sets had FDR corrected p values less than 0.05.
The immune cell landscape within tumors was estimated with immune deconvolution implemented in the immunedeconv package (v2.0.4) with the method set to quantiseq46,84 (parameters: method = ‘quantiseq’, indications = NULL, tumor = TRUE, arrays = FALSE, column = ‘gene_symbol’, rmgenes = NULL, scale_mRNA = TRUE, expected_cell_types - NULL).
Microbial quantification
To further process the non-human reads for microbiome analysis, any remaining Illumina TruSeq adapters were trimmed with Trimmomatic85 (v0.39, parameters: ‘PE ILLUMINACLIP:TruSeq.fa:3:30:10 MINLEN:50’), low-quality and low-complexity reads were removed with prinseq-lite86 (v0.20.4, parameters: ‘-min_len 50 -min_qual_mean 28 -derep 4 -derep_min 50 -lc_method dust -lc_threshold 40’), and Bowtie287 (v2.4.1, parameters: ‘–very-sensitive –un-conc’) was used to remove reads mapping to PhiX or the PacBio human genome. Reads were then mapped to a custom database for microbial quantification.
We built the custom Kraken-GTDB microbial database as previously described.114 First the following files were downloaded:
ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/bacteria/assembly_summary.txt, ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/bacteria/assembly_summary.txt, ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/archaea/assembly_summary.txt, ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/archaea/assembly_summary.txt, https://data.ace.uq.edu.au/public/gtdb/data/releases/release95/95.0/bac120_taxonomy_r95.tsv, https://data.ace.uq.edu.au/public/gtdb/data/releases/release95/95.0/ar122_taxonomy_r95.tsv23 on January 22, 2021. These files were merged based on accession number, and only those genomes present in both databases were considered, i.e. RefSeq and GenBank genomes with a GTDB taxonomy. To avoid biasing the database toward those species with large numbers of genomes, while balancing the added information provided by additional isolates per species, we selected up to five genomes per GTDB species to include in our database. Genomes were first ordered by their assembly quality, i.e., reference genome, representative genome, complete genome, chromosome, contig, and scaffold, and then randomly selected. Based on these criteria, 52,052 genomes representing 29,634 unique bacterial species and 2,633 genomes representing 1,646 archaea were downloaded and formatted into a Kraken2 database.24 To incorporate the GTDB taxonomy into the Kraken2 database, files mimicking the NCBI-like taxonomy files from ftp://ftp.ncbi.nih.gov/pub/taxonomy/new_taxdump/new_taxdump.zip were created for names.dmp, complete_names.dmp, nodes.dmp, and accession2taxid. A matching Bracken database was then generated with bracken-build (parameters: ‘-k 35 -L 76’).25
Using our custom GTDB-Kraken database, Kraken224 (v2.0.8, parameters: ‘–threads 9 –use-names –confidence 0.2’) and Bracken25 (v2.5, parameters: ‘-r 76 -L S -t 250’) were run on the 807 quality-controlled samples to generate a species bacterial count matrix. With the Bracken cutoff of 250 reads, 8 samples had 0 species which passed. In total, 2,043 species were identified across 799 samples. The median per-sample fraction of total (including human) reads mapped to microbial species was 0.66% (IQR, 0.354-1.43%) (Figures S1D and S1E).
Filtering taxonomic profiles
To account for false positives in the original list of 2,043 taxa, we first prioritized bacterial species present in greater than 5% of samples (n = 41/807) which resulted in 118 highly prevalent species. Next, to reduce false positives due to mismapping reads, we detected horizontal coverage of a genome in a given sample. To do this, taxonomy IDs were parsed from the Bracken output and linked to microbial genomes from the GTDB database. The scaffolds from these microbial genomes were concatenated and a Bowtie287 database was generated. Finally, non-human reads were aligned to the bowtie2 database (parameters: ‘-p 8 --very-sensitive -k 10’) and the coverage of each genome was estimated with the genomecov program of bedtools88 (v2.30.0). When less than 0.5% of the genome was covered by reads for a given sample, a zero was inputted into the matrix for that taxon by sample combination. Ultimately, this step removed 15 species which did not have coverage greater than 0.5% in any sample.
Finally, acknowledging that these samples were not collected or processed with microbiome sterility in mind and that contamination issues are prevalent in low biomass samples,67 we next proceeded to remove putative contaminant organisms. Due to a lack of recommended positive and negative controls, we could not utilize many of the standard contamination removal procedures previously recommended.67 Therefore, we used a correlation-based approach to differentiate true microbial signal from artifacts. To accomplish this, we clustered the remaining 103 species based on spearman correlations of abundance revealing two distinct groupings (Table S1W). One of the clusters included several species previously identified as members of the reagentome, e.g. Ralstonia and Burkholderia species.67,115 Therefore, out of an abundance of caution, we removed the 28 species with a positive correlation (Spearman’s Rho >0) to Ralstonia sp001078575 from further analysis (Table S1D). Lastly, in the resultant filtered matrix, one taxon, Aliterella sp000332075 (Cyanobacteria), was identified that did not occur in two large meta-analyses of gut and oral bacteria,6,19 nor in an analysis of stool microbiota from a 1000-person healthy cohort.114 Therefore, this one taxon was removed, resulting in 74, high-confidence bacterial species that were considered in downstream analyzes (Figure S1C).
Analysis of gut and oral metagenomic samples
To complement results from AVANT, the same microbial quantification pipeline described above was run on 852 stool and 208 oral microbiome samples from patients with CRC and controls in two meta-analyses (Figure 1B, Tables S1E–S1G).6,19 Metadata for the CRC and control samples were pulled from the supplemental tables.6,19
Paired-end samples were downloaded from PRJEB10878,68 PRJEB12449,69 PRJEB27928,6 PRJEB28422,70 PRJEB6070,71 PRJEB7774,72 PRJEB6997,73 PRJNA217052,74 and PRJNA289586.75 When multiple samples were available per donor, the baseline sample was utilized and the others were excluded. A complete list and summary table of the samples utilized is in Table S1F. When a SampleID was associated with more than 1 SRA accession those samples were merged (Table S1G). Cleaned, merged samples were then run through the same Kraken|Bracken pipeline as above and prevalences of the 74 taxa were reported. Notably, samples from PRJEB6070 and PRJEB28422 required additionally cleaning with Cutadapt v3.7 (default parameters) to remove lingering Illumina universal adapter sequences.89
Microbial analysis of TCGA samples
We obtained bulk RNA-seq data, prepared with poly-(A) enrichment, from the TCGA COAD and READ indications (Release 7.0) from portal.gdc.cancer.gov in order to compare our results from the AVANT clinical trial with the microbiome profiles and host expression patterns in a widely-analyzed dataset. A total of 587 files consisting of non-human reads were obtained from the two indications and processed with the same microbial quantification pipeline described above (Table S1I). A subset of samples from both indications was sequenced with single-end Illumina technology; therefore our QC and Kraken|Bracken pipelines were updated to accommodate both the single-end, 70 base-pair and the paired-end, 2 x 50 base-pair reads.
PathSeq, Metaphlan, and mOTUs analysis
The PathSeq functions within the Genome Analysis Toolkit (GATK v4.1.4.0)32,116 were leveraged for comparison with previous intratumoral analyses of TCGA data20 and the kmer-based read mapping with Kraken2 described herein. Reference files were obtained from the Broad’s gcp public data site for hg38 v0 (https://storage.googleapis.com/gcp-public-data--broad-references/hg38/v0/). The PathSeqPipelineSpark function was run on bam files constructed from reads that did not map to GRCh38 from TCGA and AVANT (parameters: ‘--min-clipped-read-length 40 --min-score-identity 0.90 --identity-margin 0.02’). Normalized scores and reads were compiled from separate output files per sample. To compare the Pathseq results, based on NCBI taxonomy, to our Kraken|Bracken ones, based on the GTDB, we mapped each of the 74 GTDB species to their closest match in the Pathseq reference databases using the GTDB to NCBI tool (https://gtdb.ecogenomic.org/taxon-history). When the GTDB split a species compared to NCBI, we rolled up the GTDB results to be comparable to those of NCBI. Notably, this rollup process collapsed the original 74 species down to 68. For example, for these comparative analyzes, within a sample the Bracken read counts from F. animalis, F. vincentii, F. polymorphum, and F. nucleatum were summed prior to calculating prevalence for the rolluped F. nucleatum. With this strategy, 43 rolled up species were identified as overlapping between the programs (Table S1H). Notably, 18 of the Kraken species we were unable to map had “sp” in their names.
As further benchmarking, we also processed the AVANT samples with Metaphlan390 (v3.0.7) and mOTUs91 (v3.0.2) with default parameters. Using the same strategy as described above for Pathseq, we identified 44 of the 68 rolluped species in the Metaphlan database and 42 in that of mOTUs (Table S1X). As expected, given both of these methods align reads only to marker genes, which themselves were not selected due to their being expressed but rather due to their uniqueness (Metaphlan) or level of SNPs (mOTUs), prevalences with mOTUs and Metaphlan were greatly reduced across all species compared to Kraken|Bracken (Table S1X, Figures S6A–S6C). This is reflective of only the samples with the highest number of reads per species (as measured by the Bracken mapping) being identified as positive based on these approaches. Because these methods utilize only a small proportion of the available reads and are thus more prone to false negatives in RNA-seq samples, we prioritized the Kraken|Bracken and Pathseq results.
Analysis with AVANT clinical covariates
Twenty-one categorical variables were obtained from the clinical trial case report form and included in the microbiome and host expression analysis (Table S1B). Tumor location was split into 3 categories based on the annotated surgical procedure performed, with right and transverse colectomies as “right”, left colectomies, sigmoidectomies, and lower anterior resections as “left”, and total colectomies as “other” (Figure S3B). BRAF and MSI status were obtained from calls made on overlapping samples from a project analyzing the AVANT clinical trial with nanostring technology.117 Samples that did not overlap between Daemen et al. 2021 and the project described herein (n = 112) were recorded as NAs and dropped from respective models. CMS calls were made on the host expression matrix with ENSEMBL IDs using CMScaller39 (v0.99.2, Figure 2A).
As we chose to rely upon microbial prevalence due to the extensive filtering of putative contaminant reads in our dataset, we conducted Chi-squared analyses to test for significant associations with clinical covariates. Contingency tables were constructed with the prevalence of each microbial taxon within a clinical subgroup versus those not found in the remaining subgroups. Chi-squared analysis was then tested on each taxon by clinical covariate combination and multiple p values were corrected with the false discovery rate (FDR).118 Odds ratios were calculated on contingency tables constructed for the presence and absence of each of the 74 species in right- versus left-sided, MSI-H versus MSS, BRAF mutated versus wild-type tumors, and CMS1 versus CMS2,3,4 with Fisher exact tests.119 Odds ratios were similarly calculated for the 43 Pathseq-identified species (Table S1O). To account for the co-occurrence between Right-sided, MSI-H, and BRAF mutant tumors (Figures S3C and S3D), in R, we generated generalized linear models (glm) for each species, with the formula species ∼ MSI + SIDE + BRAF, and family = "binomial".
Testing a range of Bracken read cutoffs
To stress test our results, we also tested a range of Bracken cutoffs, including 50, 150, and 250 reads, prior to the coverage-based filter. As expected, prevalences were on average higher at the lower Bracken cutoffs (Table S1Y, Figure S6D). To determine whether these more permissive prevalences changed our downstream conclusions, we repeated the metadata associations with the prevalences determined at the 50 read cutoff. Reassuringly, at the 50 read cutoff, the broad trends held true (Table S1Z, Figure S6E). In other words, oral species were still enriched in right-sided, MSI-H, BRAF-mutant, and CMS1 tumors. Because of this, we ultimately selected the more conservative 250 cutoff in the manuscript.
F. animalis pangenome
To generate the F. animalis pangenome, we first compiled all protein sequences for the 26 genomes classified as F. animalis in the v207 version of the GTDB from NCBI (Table S1U). Notably, to avoid incomplete genomes skewing the results, we did exclude 3 F animalis metagenomic assemblies (MAGs) from the analysis. The 57,220 amino acid protein sequences were then clustered into 4,355 non-redundant orthologs with usearch92 (v11.0.667_i86linu x32, -id 0.50). Gene accumulation curves for these clusters were generated as in Conlan et al.120 The curves showed that new genes discovered with additional genomes, the core genome, and the pangenome all followed a power law curve (Figures 4A and 4B). These gene clusters were then annotated by BLASTp in BLAST+93 (v2.6.0, parameters: -evalue 1e-10) against the KEGG database,76 as well as CARD50 and VFDB51 mid-2017 databases from ShortBRED.77 To identify the F. animalis genes actively expressed in the AVANT samples, reads were mapped to gene cluster/pangenome database using DIAMOND94 (v2.0.11, parameters: diamond blastx --evalue 0.001000 --id 0.600000 -k 25 --verbose --outfmt 101 --max-hsps 10). Coverage of the individual genes was then determined with the samtools95 (v1.11) coverage function. A gene was subsequently considered present only when 50% of its length was covered with reads.
Figures and table generation
All correlations and statistical tests were performed in R96 (v4.1.1), documented via rmarkdown documents97 (v2.11), and compiled with knitr98 (v1.37). Within R, tables were manipulated with functions of the dplyr99 (v1.0.7), reshape2100 (v1.4.4), plotrix101 (v3.8-2), and gtools102 (v3.9.2) packages. The majority of figures were rendered with ggplot2103 (v3.3.5), adjusted with ggeasy104 (v0.1.3) and scales105 (v1.1.1), and arranged with cowplot106 (v1.1.1). Volcano plot gene labels were added with ggrepel107 (v0.9.1). Colors were selected with the help of RColorBrewer108 (v1.1-2) and viridis109 (v0.6.2). Supplemental tables were generated with Openxlsx110 (v4.2.5).
Kaplan-Meier curves were constructed with patient censoring, disease-free and overall survival metrics with treatment arm, CMS, and F. animalis presence as the grouping variable. Curves were generated with the survfit function implemented in Survival111 (v3.2-13) and visualized with Survminer112 (v0.4.9). Throughout, 95% Confidence Intervals (CI) of the mean were calculated in R using the t-distribution. The graphical abstract and Figure S3B were created with BioRender.com.
Acknowledgments
We thank all of the patients for their participation in the study. We thank F. Tamburini, J. Mar, K. Storek, D. Wolan, I. Mellman, and O. Harrison for critical reading of the manuscript and helpful comments. We also thank L. Abrey, J. Carroll, and Z. Khan for help with clinical annotations; T. Behrens and H. Chen for help with data generation; L. Chu, L. Polischuk, and S. Robson for help with data upload; R. Yang for infrastructure support; and M. Grimmer, M. Das Thakur, and M. Tan for their general advice. We also kindly acknowledge TCGA, as some of the results shown here are in whole or in part based upon data generated by the TCGA Research Network (https://www.cancer.gov/tcga).
Author contributions
Conceptualization, M.L.A. and A.L.B.; methodology, B.S.Y., O.M., and A.L.B.; software, B.S.Y. and A.L.B.; formal analysis, B.S.Y., O.M., and A.L.B.; resources, J.R. and Z.M.; data curation, J.R.; writing – original draft, B.S.Y. and A.L.B.; writing – review & editing, B.S.Y., O.M., D.R.N., Z.M., M.L.A., and A.L.B.; visualization, A.L.B.; supervision, M.L.A. and A.L.B.; project administration, D.R.N. and Z.M.
Declaration of interests
O.M., J.R., D.R.N., Z.M., and A.L.B. are Genentech/Roche employees. B.S.Y. and M.L.A. were Genentech/Roche employees.
Published: January 26, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.100920.
Supplemental information
Data and code availability
-
•
Raw RNA-sequencing data, including host and microbial reads, have been deposited at European Genome-Phenome Archive along with the clinical metadata. To request access, contact the Genentech Data Access committee at devsci-dac-d@gene.com. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. CA. Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Goding Sauer A., Fedewa S.A., Butterly L.F., Anderson J.C., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2020. CA. Cancer J. Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 3.Guinney J., Dienstmann R., Wang X., de Reyniès A., Schlicker A., Soneson C., Marisa L., Roepman P., Nyamundanda G., Angelino P., et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee K.-H., Chen W.-S., Jiang J.-K., Yang S.-H., Wang H.-S., Chang S.-C., Lan Y.-T., Lin C.-C., Lin H.-H., Huang S.-C., et al. The efficacy of anti-EGFR therapy in treating metastatic colorectal cancer differs between the middle/low rectum and the left-sided colon. Br. J. Cancer. 2021;125:816–825. doi: 10.1038/s41416-021-01470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas A.M., Manghi P., Asnicar F., Pasolli E., Armanini F., Zolfo M., Beghini F., Manara S., Karcher N., Pozzi C., et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019;25:667–678. doi: 10.1038/s41591-019-0405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wirbel J., Pyl P.T., Kartal E., Zych K., Kashani A., Milanese A., Fleck J.S., Voigt A.Y., Palleja A., Ponnudurai R., et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019;25:679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu N.-N., Jiao N., Tan J.-C., Wang Z., Wu D., Wang A.-J., Chen J., Tao L., Zhou C., Fang W., et al. Multi-kingdom microbiota analyses identify bacterial–fungal interactions and biomarkers of colorectal cancer across cohorts. Nat. Microbiol. 2022;7:238–250. doi: 10.1038/s41564-021-01030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drewes J.L., White J.R., Dejea C.M., Fathi P., Iyadorai T., Vadivelu J., Roslani A.C., Wick E.C., Mongodin E.F., Loke M.F., et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. Npj Biofilms Microbiomes. 2017;3:34. doi: 10.1038/s41522-017-0040-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kostic A.D., Gevers D., Pedamallu C.S., Michaud M., Duke F., Earl A.M., Ojesina A.I., Jung J., Bass A.J., Tabernero J., et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castellarin M., Warren R.L., Freeman J.D., Dreolini L., Krzywinski M., Strauss J., Barnes R., Watson P., Allen-Vercoe E., Moore R.A., Holt R.A. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kostic A.D., Chun E., Robertson L., Glickman J.N., Gallini C.A., Michaud M., Clancy T.E., Chung D.C., Lochhead P., Hold G.L., et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abed J., Emgård J.E.M., Zamir G., Faroja M., Almogy G., Grenov A., Sol A., Naor R., Pikarsky E., Atlan K.A., et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gur C., Ibrahim Y., Isaacson B., Yamin R., Abed J., Gamliel M., Enk J., Bar-On Y., Stanietsky-Kaynan N., Coppenhagen-Glazer S., et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan C.A., Garrett W.S. Fusobacterium nucleatum — symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019;17:156–166. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu T., Guo F., Yu Y., Sun T., Ma D., Han J., Qian Y., Kryczek I., Sun D., Nagarsheth N., et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170:548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubinstein M.R., Wang X., Liu W., Hao Y., Cai G., Han Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dejea C.M., Wick E.C., Hechenbleikner E.M., White J.R., Mark Welch J.L., Rossetti B.J., Peterson S.N., Snesrud E.C., Borisy G.G., Lazarev M., et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. USA. 2014;111:18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullman S., Pedamallu C.S., Sicinska E., Clancy T.E., Zhang X., Cai D., Neuberg D., Huang K., Guevara F., Nelson T., et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt T.S., Hayward M.R., Coelho L.P., Li S.S., Costea P.I., Voigt A.Y., Wirbel J., Maistrenko O.M., Alves R.J., Bergsten E., et al. Extensive transmission of microbes along the gastrointestinal tract. Elife. 2019;8:e42693. doi: 10.7554/elife.42693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salvucci M., Crawford N., Stott K., Bullman S., Longley D.B., Prehn J.H.M. Patients with mesenchymal tumours and high Fusobacteriales prevalence have worse prognosis in colorectal cancer (CRC) Gut. 2021;71:1600–1612. doi: 10.1136/gutjnl-2021-325193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poore G.D., Kopylova E., Zhu Q., Carpenter C., Fraraccio S., Wandro S., Kosciolek T., Janssen S., Metcalf J., Song S.J., et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579:567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.de Gramont A., Van Cutsem E., Schmoll H.-J., Tabernero J., Clarke S., Moore M.J., Cunningham D., Cartwright T.H., Hecht J.R., Rivera F., et al. Bevacizumab plus oxaliplatin-based chemotherapy as adjuvant treatment for colon cancer (AVANT): a phase 3 randomised controlled trial. Lancet Oncol. 2012;13:1225–1233. doi: 10.1016/s1470-2045(12)70509-0. [DOI] [PubMed] [Google Scholar]
- 23.Parks D.H., Chuvochina M., Waite D.W., Rinke C., Skarshewski A., Chaumeil P.-A., Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 24.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu J., Breitwieser F.P., Thielen P., Salzberg S.L. Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci. 2017;3:e104. doi: 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- 26.Baxter N.T., Ruffin M.T., Rogers M.A.M., Schloss P.D. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 2016;8:37. doi: 10.1186/s13073-016-0290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flynn K.J., Baxter N.T., Schloss P.D. Metabolic and community synergy of oral bacteria in colorectal cancer. mSphere. 2016;1:e00102-16. doi: 10.1128/msphere.00102-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flemer B., Warren R.D., Barrett M.P., Cisek K., Das A., Jeffery I.B., Hurley E., O‘Riordain M., Shanahan F., O‘Toole P.W. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67:1454–1463. doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flemer B., Lynch D.B., Brown J.M.R., Jeffery I.B., Ryan F.J., Claesson M.J., O’Riordain M., Shanahan F., O’Toole P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66:633–643. doi: 10.1136/gutjnl-2015-309595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warren R.L., Freeman D.J., Pleasance S., Watson P., Moore R.A., Cochrane K., Allen-Vercoe E., Holt R.A. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1:16. doi: 10.1186/2049-2618-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakatsu G., Li X., Zhou H., Sheng J., Wong S.H., Wu W.K.K., Ng S.C., Tsoi H., Dong Y., Zhang N., et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat. Commun. 2015;6:8727. doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kostic A.D., Ojesina A.I., Pedamallu C.S., Jung J., Verhaak R.G.W., Getz G., Meyerson M. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat. Biotechnol. 2011;29:393–396. doi: 10.1038/nbt.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boland C.R., Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087.e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mima K., Nishihara R., Qian Z.R., Cao Y., Sukawa Y., Nowak J.A., Yang J., Dou R., Masugi Y., Song M., et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamada T., Zhang X., Mima K., Bullman S., Sukawa Y., Nowak J.A., Kosumi K., Masugi Y., Twombly T.S., Cao Y., et al. Fusobacterium nucleatum in colorectal cancer relates to immune response differentially by tumor microsatellite instability status. Cancer Immunol. Res. 2018;6:1327–1336. doi: 10.1158/2326-6066.cir-18-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tahara T., Yamamoto E., Suzuki H., Maruyama R., Chung W., Garriga J., Jelinek J., Yamano H.o., Sugai T., An B., et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311–1318. doi: 10.1158/0008-5472.can-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mima K., Sukawa Y., Nishihara R., Qian Z.R., Yamauchi M., Inamura K., Kim S.A., Masuda A., Nowak J.A., Nosho K., et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 2015;1:653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nosho K., Sukawa Y., Adachi Y., Ito M., Mitsuhashi K., Kurihara H., Kanno S., Yamamoto I., Ishigami K., Igarashi H., et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J. Gastroenterol. 2016;22:557–566. doi: 10.3748/wjg.v22.i2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eide P.W., Bruun J., Lothe R.A., Sveen A. CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci. Rep. 2017;7:16618. doi: 10.1038/s41598-017-16747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillespie M., Jassal B., Stephan R., Milacic M., Rothfels K., Senff-Ribeiro A., Griss J., Sevilla C., Matthews L., Gong C., et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50:D687–D692. doi: 10.1093/nar/gkab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee Y.S., Choi I., Ning Y., Kim N.Y., Khatchadourian V., Yang D., Chung H.K., Choi D., LaBonte M.J., Ladner R.D., et al. Interleukin-8 and its receptor CXCR2 in the tumour microenvironment promote colon cancer growth, progression and metastasis. Br. J. Cancer. 2012;106:1833–1841. doi: 10.1038/bjc.2012.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voronov E., Apte R.N. IL-1 in colon inflammation, colon carcinogenesis and invasiveness of colon cancer. Cancer Microenviron. 2015;8:187–200. doi: 10.1007/s12307-015-0177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waldner M.J., Foersch S., Neurath M.F. Interleukin-6 - a key regulator of colorectal cancer development. Int. J. Biol. Sci. 2012;8:1248–1253. doi: 10.7150/ijbs.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Said A.H., Raufman J.-P., Xie G. The role of matrix metalloproteinases in colorectal cancer. Cancers. 2014;6:366–375. doi: 10.3390/cancers6010366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sunami E., Tsuno N., Osada T., Saito S., Kitayama J., Tomozawa S., Tsuruo T., Shibata Y., Muto T., Nagawa H. MMP-1 is a prognostic marker for hematogenous metastasis of colorectal cancer. Oncol. 2000;5:108–114. doi: 10.1634/theoncologist.5-2-108. [DOI] [PubMed] [Google Scholar]
- 46.Finotello F., Mayer C., Plattner C., Laschober G., Rieder D., Hackl H., Krogsdam A., Loncova Z., Posch W., Wilflingseder D., et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11:34. doi: 10.1186/s13073-019-0638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ponath F., Tawk C., Zhu Y., Barquist L., Faber F., Vogel J. RNA landscape of the emerging cancer-associated microbe Fusobacterium nucleatum. Nat. Microbiol. 2021;6:1007–1020. doi: 10.1038/s41564-021-00927-7. [DOI] [PubMed] [Google Scholar]
- 48.Zhao T., Chen J., Liu S., Yang J., Wu J., Miao L., Sun W. Transcriptome analysis of Fusobacterium nucleatum reveals differential gene expression patterns in the biofilm versus planktonic cells. Biochem. Biophys. Res. Commun. 2022;593:151–157. doi: 10.1016/j.bbrc.2021.11.075. [DOI] [PubMed] [Google Scholar]
- 49.Cochrane K., Robinson A.V., Holt R.A., Allen-Vercoe E. A survey of Fusobacterium nucleatum genes modulated by host cell infection. Microb. Genom. 2020;6:e000300. doi: 10.1099/mgen.0.000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alcock B.P., Raphenya A.R., Lau T.T.Y., Tsang K.K., Bouchard M., Edalatmand A., Huynh W., Nguyen A.-L.V., Cheng A.A., Liu S., et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., Jin Q. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frees D., Gerth U., Ingmer H. Clp chaperones and proteases are central in stress survival, virulence and antibiotic resistance of Staphylococcus aureus. Int. J. Med. Microbiol. 2014;304:142–149. doi: 10.1016/j.ijmm.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 53.Michel A., Agerer F., Hauck C.R., Herrmann M., Ullrich J., Hacker J., Ohlsen K. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 2006;188:5783–5796. doi: 10.1128/jb.00074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balsera M., Buchanan B.B. Evolution of the thioredoxin system as a step enabling adaptation to oxidative stress. Free Radic. Biol. Med. 2019;140:28–35. doi: 10.1016/j.freeradbiomed.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 55.Susin M.F., Baldini R.L., Gueiros-Filho F., Gomes S.L. GroES/GroEL and DnaK/DnaJ have distinct roles in stress responses and during cell cycle progression in Caulobacter crescentus. J. Bacteriol. 2006;188:8044–8053. doi: 10.1128/jb.00824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harvey K.L., Jarocki V.M., Charles I.G., Djordjevic S.P. The diverse functional roles of elongation factor Tu (EF-Tu) in microbial pathogenesis. Front. Microbiol. 2019;10:2351. doi: 10.3389/fmicb.2019.02351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bachrach G., Rosen G., Bellalou M., Naor R., Sela M.N. Identification of a Fusobacterium nucleatum 65 kDa serine protease. Oral Microbiol. Immunol. 2004;19:155–159. doi: 10.1111/j.0902-0055.2004.00132.x. [DOI] [PubMed] [Google Scholar]
- 58.Doron L., Coppenhagen-Glazer S., Ibrahim Y., Eini A., Naor R., Rosen G., Bachrach G. Identification and characterization of fusolisin, the Fusobacterium nucleatum autotransporter serine protease. PLoS One. 2014;9:e111329. doi: 10.1371/journal.pone.0111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han Y.W., Ikegami A., Rajanna C., Kawsar H.I., Zhou Y., Li M., Sojar H.T., Genco R.J., Kuramitsu H.K., Deng C.X. Identification and characterization of a novel adhesin unique to oral fusobacteria. J. Bacteriol. 2005;187:5330–5340. doi: 10.1128/jb.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu M., Yamada M., Li M., Liu H., Chen S.G., Han Y.W. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J. Biol. Chem. 2007;282:25000–25009. doi: 10.1074/jbc.m611567200. [DOI] [PubMed] [Google Scholar]
- 61.Mark Welch J.L., Ramírez-Puebla S.T., Borisy G.G. Oral microbiome geography: micron-scale habitat and niche. Cell Host Microbe. 2020;28:160–168. doi: 10.1016/j.chom.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Komiya Y., Shimomura Y., Higurashi T., Sugi Y., Arimoto J., Umezawa S., Uchiyama S., Matsumoto M., Nakajima A. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. 2019;68:1335–1337. doi: 10.1136/gutjnl-2018-316661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abed J., Maalouf N., Manson A.L., Earl A.M., Parhi L., Emgård J.E.M., Klutstein M., Tayeb S., Almogy G., Atlan K.A., et al. Colon cancer-associated Fusobacterium nucleatum may originate from the oral cavity and reach colon tumors via the circulatory system. Front. Cell. Infect. Microbiol. 2020;10:400. doi: 10.3389/fcimb.2020.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo S., Li L., Xu B., Li M., Zeng Q., Xiao H., Xue Y., Wu Y., Wang Y., Liu W., Zhang G. A simple and novel fecal biomarker for colorectal cancer: ratio of Fusobacterium nucleatum to probiotics populations, based on their antagonistic effect. Clin. Chem. 2018;64:1327–1337. doi: 10.1373/clinchem.2018.289728. [DOI] [PubMed] [Google Scholar]
- 65.Casasanta M.A., Yoo C.C., Udayasuryan B., Sanders B.E., Umaña A., Zhang Y., Peng H., Duncan A.J., Wang Y., Li L., et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci. Signal. 2020;13:eaba9157. doi: 10.1126/scisignal.aba9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harrandah A.M., Chukkapalli S.S., Bhattacharyya I., Progulske-Fox A., Chan E.K.L. Fusobacteria modulate oral carcinogenesis and promote cancer progression. J. Oral Microbiol. 2020;13:1849493. doi: 10.1080/20002297.2020.1849493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eisenhofer R., Minich J.J., Marotz C., Cooper A., Knight R., Weyrich L.S. Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 2019;27:105–117. doi: 10.1016/j.tim.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 68.Yu J., Feng Q., Wong S.H., Zhang D., Liang Q.Y., Qin Y., Tang L., Zhao H., Stenvang J., Li Y., et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66:70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 69.Vogtmann E., Hua X., Zeller G., Sunagawa S., Voigt A.Y., Hercog R., Goedert J.J., Shi J., Bork P., Sinha R. Colorectal cancer and the human gut microbiome: reproducibility with whole-genome shotgun sequencing. PLoS One. 2016;11:e0155362. doi: 10.1371/journal.pone.0155362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Belstrøm D. The salivary microbiota in health and disease. J. Oral Microbiol. 2020;12:1723975. doi: 10.1080/20002297.2020.1723975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zeller G., Tap J., Voigt A.Y., Sunagawa S., Kultima J.R., Costea P.I., Amiot A., Böhm J., Brunetti F., Habermann N., et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feng Q., Liang S., Jia H., Stadlmayr A., Tang L., Lan Z., Zhang D., Xia H., Xu X., Jie Z., et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat. Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 73.Zhang X., Zhang D., Jia H., Feng Q., Wang D., Liang D., Wu X., Li J., Tang L., Li Y., et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015;21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 74.Brito I.L., Yilmaz S., Huang K., Xu L., Jupiter S.D., Jenkins A.P., Naisilisili W., Tamminen M., Smillie C.S., Wortman J.R., et al. Mobile genes in the human microbiome are structured from global to individual scales. Nature. 2016;535:435–439. doi: 10.1038/nature18927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heintz-Buschart A., May P., Laczny C.C., Lebrun L.A., Bellora C., Krishna A., Wampach L., Schneider J.G., Hogan A., de Beaufort C., Wilmes P. Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes. Nat. Microbiol. 2016;2:16180. doi: 10.1038/nmicrobiol.2016.180. [DOI] [PubMed] [Google Scholar]
- 76.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaminski J., Gibson M.K., Franzosa E.A., Segata N., Dantas G., Huttenhower C. High-specificity targeted functional profiling in microbial communities with ShortBRED. PLoS Comput. Biol. 2015;11:e1004557. doi: 10.1371/journal.pcbi.1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pau G., Barr C., Reeder J., Lawrence M., Degenhardt J., Wu T., Huntley M., Brauer M. 2021. HTSeqGenie: A Software Package to Analyse High-Throughput Sequencing Experiments. [Google Scholar]
- 79.Wu T.D., Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu T.D., Reeder J., Lawrence M., Becker G., Brauer M.J. Statistical genomics, methods and protocols. Methods Mol. Biol. 2016;1418:283–334. doi: 10.1007/978-1-4939-3578-9_15. [DOI] [PubMed] [Google Scholar]
- 81.Chen Y., Lun A.T.L., Smyth G.K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 2016;5:1438. doi: 10.12688/f1000research.8987.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu R., Holik A.Z., Su S., Jansz N., Chen K., Leong H.S., Blewitt M.E., Asselin-Labat M.-L., Smyth G.K., Ritchie M.E. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015;43:e97. doi: 10.1093/nar/gkv412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smyth G.K. 2005. pp. 397–420. (Limma: Linear Models for Microarray Data). [DOI] [Google Scholar]
- 84.Sturm G., Finotello F., Petitprez F., Zhang J.D., Baumbach J., Fridman W.H., List M., Aneichyk T. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35:i436–i445. doi: 10.1093/bioinformatics/btz363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. j. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 90.Beghini F., McIver L.J., Blanco-Míguez A., Dubois L., Asnicar F., Maharjan S., Mailyan A., Manghi P., Scholz M., Thomas A.M., et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife. 2021;10:e65088. doi: 10.7554/elife.65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Milanese A., Mende D.R., Paoli L., Salazar G., Ruscheweyh H.-J., Cuenca M., Hingamp P., Alves R., Costea P.I., Coelho L.P., et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat. Commun. 2019;10:1014. doi: 10.1038/s41467-019-08844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 93.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Buchfink B., Reuter K., Drost H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods. 2021;18:366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Team R.C. 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 97.Allaire J., Xie Y., McPherson J., Luraschi J., Ushey K., Atkins A., Wickham H., Cheng J., Chang W., Iannone R. 2021. Rmarkdown: Dynamic Documents for R. [Google Scholar]
- 98.Xie Y. 2021. Knitr: A General-Purpose Package for Dynamic Report Generation in R. [Google Scholar]
- 99.Wickham H., François R., Henry L., Müller K. 2021. Dplyr: A Grammar of Data Manipulation. [Google Scholar]
- 100.Wickham H. 2020. reshape2: Flexibly Reshape Data: A Reboot of the Reshape Package. [Google Scholar]
- 101.Lemon J., Bolker B., Oom S., Klein E., Rowlingson B., Wickham H., Tyagi A., Eterradossi O., Grothendieck G., Toews M., et al. 2021. Plotrix: Various Plotting Functions. [Google Scholar]
- 102.Warnes G.R., Bolker B., Lumley T. 2021. Gtools: Various R Programming Tools. [Google Scholar]
- 103.Wickham H., Chang W., Henry L., Pedersen T.L., Takahashi K., Wilke C., Woo K., Yutani H., Dunnington D. 2021. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. [Google Scholar]
- 104.Carroll J., Schep A., Sidi J. 2021. Ggeasy: Easy Access to Ggplot2 Commands. [Google Scholar]
- 105.Wickham H., Seidel D. 2020. Scales: Scale Functions for Visualization. [Google Scholar]
- 106.Wilke C.O. 2020. Cowplot: Streamlined Plot Theme and Plot Annotations for Ggplot2. [Google Scholar]
- 107.Slowikowski K. 2021. Ggrepel: Automatically Position Non-overlapping Text Labels with Ggplot2. [Google Scholar]
- 108.Neuwirth E. 2014. RColorBrewer: ColorBrewer Palettes. [Google Scholar]
- 109.Garnier S. 2021. Viridis: Colorblind-Friendly Color Maps for R. [Google Scholar]
- 110.Schauberger P., Walker A. 2021. Openxlsx: Read, Write and Edit Xlsx Files. [Google Scholar]
- 111.Therneau T., Lumley T. R Core Team; 2013. R Survival Package. [Google Scholar]
- 112.Kassambara A., Kosinski M., Biecek P., Fabian S. 2017. Package ‘survminer.’ Drawing Survival Curves Using ‘ggplot2’(R Package Version 03 1) [Google Scholar]
- 113.Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T., et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Byrd A.L., Liu M., Fujimura K.E., Lyalina S., Nagarkar D.R., Charbit B., Bergstedt J., Patin E., Harrison O.J., Quintana-Murci L., et al. Gut microbiome stability and dynamics in healthy donors and patients with non-gastrointestinal cancers. J. Exp. Med. 2021;218:e20200606. doi: 10.1084/jem.20200606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Salter S.J., Cox M.J., Turek E.M., Calus S.T., Cookson W.O., Moffatt M.F., Turner P., Parkhill J., Loman N.J., Walker A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Daemen A., Udyavar A.R., Sandmann T., Li C., Bosch L.J.W., O’Gorman W., Li Y., Au-Yeung A., Takahashi C., Kabbarah O., et al. Transcriptomic profiling of adjuvant colorectal cancer identifies three key prognostic biological processes and a disease specific role for granzyme B. PLoS One. 2021;16:e0262198. doi: 10.1371/journal.pone.0262198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1994 [Google Scholar]
- 119.Fisher S.R.A. Confidence limits for a cross-product ratio. Aust. J. Stat. 1962;4:41. doi: 10.1111/j.1467-842X.1962.tb00285.x. [DOI] [Google Scholar]
- 120.Conlan S., Mijares L.A., NISC Comparative Sequencing Program. Becker J., Blakesley R.W., Bouffard G.G., Brooks S., Coleman H., Gupta J., Gurson N., et al. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 2012;13:R64. doi: 10.1186/gb-2012-13-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw RNA-sequencing data, including host and microbial reads, have been deposited at European Genome-Phenome Archive along with the clinical metadata. To request access, contact the Genentech Data Access committee at devsci-dac-d@gene.com. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.