Summary
The goal of oncology is to provide the longest possible survival outcomes with the therapeutics that are currently available without sacrificing patients’ quality of life. In lung cancer, several data points over a patient’s diagnostic and treatment course are relevant to optimizing outcomes in the form of precision medicine, and artificial intelligence (AI) provides the opportunity to use available data from molecular information to radiomics, in combination with patient and tumor characteristics, to help clinicians provide individualized care. In doing so, AI can help create models to identify cancer early in diagnosis and deliver tailored therapy on the basis of available information, both at the time of diagnosis and in real time as they are undergoing treatment. The purpose of this review is to summarize the current literature in AI specific to lung cancer and how it applies to the multidisciplinary team taking care of these complex patients.
Keywords: lung cancer, artificial intelligence, machine learning, deep learning, natural language processing, big data, neural network, radiomics, computer vision
Ladbury et al. review the current landscape of artificial intelligence (AI) research in lung cancer. Although much AI research is limited to single-institution datasets without external validation, there is promising foundational work in imaging, pathology, and patient treatment management. Future work should emphasize robust data sources, accuracy, reproducibility, and collaboration.
Introduction
Medicine continues to strive for efficiency in the era of increasing data available to the clinician to interpret and make appropriate treatment decisions. With these increasing demands and the large volumes of data generated from imaging and molecular testing, streamlining the vast amount of clinical data is important to help guide oncologists in the management of their patients. Over the past decade, the applications of artificial intelligence (AI) have grown dramatically, due in part to the advent of improved algorithms, increased computational power, and expansion and organizational streamlining of available data. Big data has provided a unique opportunity for data science and AI research to flourish in the field of oncology.1 Oncology in general, and specifically the management of lung cancer, has progressed significantly over the past several decades, including improved imaging, staging techniques, and incorporation of patient-centered treatment using molecular markers to guide therapy, all of which yield large quantities of information that might guide management of future patients.2,3 The large amount of data now gathered at the time of diagnosis and during treatment can be vast and often difficult to filter and interpret, which can represent a barrier to routine incorporation into the thoracic oncology clinic. AI presents a possible avenue for addressing this issue by offering improved means to distill and interpret available data through overlapping methodologies including machine learning (ML), neural networks (NN), deep learning (DL), computer vision (CV), and natural language processing (NLP).4,5
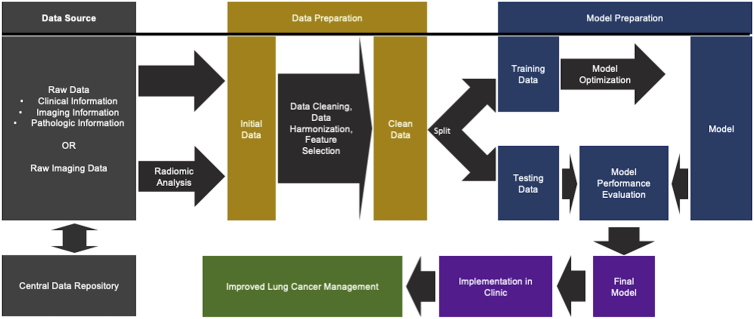
Lung cancer remains the leading cause of cancer-related mortality worldwide.6 The complexity of personalized lung cancer management, including screening, diagnosis, treatment, and follow up, can be extensive, complicated by a range of relevant data inclusive of clinical presentation, tumor stage, pathology, radiologic features, tumor genomics, liquid biopsies, treatment options, treatment response assessment, and overall outcomes. Because of these complexities, the incorporation of AI has become both necessary and exciting. There are currently several U.S. Food and Drug Administration (FDA) approvals for AI applications in clinical oncology, including lung cancer. In this review, we summarize applications of AI to the overall management of lung cancer. A general framework of how AI workflows might operate in the lung cancer clinic is visualized in Figure 1. This included data sources, data preparation (inclusive of data cleaning, data harmonization, and feature selection, whereby data are made suitable for entry into a model), model preparation, and finally, implementation. Critically, to date external validation and clinical implementation of lung cancer AI research is limited, and the studies included in this review are no exception. In cases in which only internal validation is performed, reported performance metrics are likely overly optimistic and may not validate to other datasets, which limits their clinical utility. Thus, the potential for many of the studies included in this review to actually influence clinical care is limited. Therefore, this review primarily illustrates how small-scale research has been done in lung cancer AI, which will optimally provide a framework for future expanded research, further validation, and actual incorporation into clinical workflows.
Figure 1.
Clinical AI workflow schema
Simplified schema of workflow for implementation of AI in lung cancer clinic on the basis of artificial intelligence best practices.
AI data sources
The common denominator among all AI research is a requirement for suitable quantities of training, testing and validation data. The electronic medical record (EMR) offers a potential source of data to feed into AI, as it contains information from every aspect of patient care, including radiology, pathology, and the treatment team. For modern research, the EMR almost invariably is used for patient identification and initial source of data. The difficulty lies in harmonization of data extraction and types, and maintaining patient privacy, particularly when the data are not stored in a discrete form that can be easily queried. Nevertheless, several studies have provided approaches to pulling data from the EMR to improve management of lung cancer patients.
In one such study, Wang et al.7 developed a model identifying patients at risk for new lung cancers using EMR data from the Main Health Information Exchange network. They extracted data from 873,598 patients in a retrospective cohort for model training and 836,659 patients in a prospective cohort for validation. These data were fed into an extreme gradient boosting (XGBoost) ML algorithm to predict the risk for developing lung cancer within a year time frame. The model yielded an area under the receiver operating characteristic curve (AUC) of 0.88 in the test set. The goal of this model is to identify at-risk individuals to facilitate more intensive screening and intervention, if needed. In another study, Kehl et al.8 trained a NLP model using more than 300,000 imaging reports from 16,780 patients to predict cancer progression. They successfully created a model that could predict patient prognosis and treatment change, with concordance index of 0.76 and an AUC of 0.77. The authors suggested that this application could be used to potentially identify candidates for targeted clinical trials in real time. These studies demonstrate the feasibility of automated data extraction from the EMR toward AI applications, being able to generate large quantities of data that would otherwise be prohibitive to collect manually. However, it will still be critical to validate these approaches in clinical practice as these models deal with imbalanced data, low event rates, and contamination of data by patients with pre-existing lung cancers. The next question is how available data can be used by AI to improve lung cancer management.
Diagnostic modalities
Imaging
The use of AI to augment imaging technology has found success in several disciplines, including computer-aided detection and diagnosis (CAD), convolutional neural networks (CNNs), and radiomics. CAD systems are typically standalone with a unified goal of detection or diagnosis of disease.9 At its core, it is simply trying to aid practitioners with identification of disease, with primary focus on that binary outcome. The field of radiomics seeks to use medical imaging to generate high-dimensional quantitative data, which can in turn be used for analysis that seeks to better understand the underlying characteristics of disease.10 Radiomics is inherently meant to support the overall diagnosis and management of patients at any point in the imaging workflow and can be combined with other patient characteristics to produce powerful support tools, and therefore can be considered a natural extension of CAD. As opposed to radiomics, where features are extracted and subsequently selected from images and can then be used to predict outcomes when input into tradition regression or ML models, CNNs are a subclass of DL in which the images are input into the predictive algorithm directly.11
In lung cancer, multiple imaging modalities are now used, including magnetic resonance imaging (MRI), computed tomography (CT), and positron emission tomography (PET), providing valuable sources of data that can be analyzed using radiomics, CAD, or CNNs.12 These data can be combined with other factors such as gene expressions/mutations and molecular signatures to refine prognostication. AI has been used to augment image analysis at diagnosis and to assess/monitor treatment response.
Screening
Lung cancer screening has expanded on the basis of data demonstrating improvement in disease-specific mortality. The National Lung Screening Trial (NLST) demonstrated that low-dose computed tomography (LDCT) was associated with a 20% reduction in overall mortality in current and high-risk former smokers. Currently, the U.S. Preventive Task Force supports annual screening for lung cancer with LDCT for adults aged 50–80 years who have a 20 pack-year smoking history and currently smoke or have quit within the past 15 years. Although implementation of screening is key to improving lung cancer mortality, it is not without its issues.
In particular, challenges arise because of issues with low specificity, a known problem for cancer screening modalities, particularly when relying on single-time-point imaging.13 Lung cancer screening is non-specific and often identifies indeterminate nodules, few of which are ultimately found to be malignant. In NLST, of the nodules identified during screening, the majority (>90%) were not malignant.13 There are currently no guidelines to assist radiologists and pulmonologists in classifying small indeterminate nodules as benign or malignant, and therefore these nodules are monitored closely with serial CT scans and biopsies if indicated. These additional CT scans and biopsies can be anxiety provoking, while biopsies are also a non-trivial invasive procedure with significant opportunity for morbidity.14 When the decision is made to monitor nodules with serial imaging, there is limited guidance on the appropriate time for intervention. The American College of Radiology Lung CT Screening Reporting and Data System (Lung-RADS) provides recommendations for workup and follow up of incidental lung nodules, but these include both quantitative and qualitative measures, with potential for variance in interpretation. In such scenarios, AI may be able to better identify suspicious nodules worthy of intervention on either screening or follow-up imaging.
AI has been applied to classify early cancers on imaging. When radiologists are performing such a classification, they use imaging features including density, shape, size, and surrounding changes. Application of AI can potentially better use these characteristics to create risk models. Some tools are designed to augment radiologists’ classification abilities. CAD systems have been applied to aid in finding nodules on CT imaging, including the ability to identify nodules as small as 3 mm while distinguishing nodules from normal pulmonary vascular anatomy.15,16,17 More specifically, CAD systems are classified into two groups: computer-aided detection (CADe) systems and computer-aided diagnosis (CADx) systems. CADe is used primarily to aid in detecting the presence and location of lesions, while the aim of CADx is to characterize lesions, including identification as malignant.18 Several of these systems have been approved by the FDA.19 These systems use various ML algorithms to allow automatic identification and segmentation of lung nodules and normal anatomy on chest CT, thereby offering both a “second opinion” to the radiologist’s judgment and a gathering of quantitative metrics useful for monitoring patients. Notably, these systems generally do not actually classify nodules as benign or malignant but draw attention to potential regions of interest.
Other approaches aim to improve classification of identified nodules. One of the most critical AI (and, more specifically, DL) algorithms for image analysis and associated classification is CNNs.20 CNNs are inspired by biological neural networks, with a goal of recognizing visual patterns by passing raw pixel data through a series of transformations and filters. CNNs have been used to classify nodules on screening LDCTs. In a study by Paul et al.,21 CNNs were used on select participants from NLST, achieving an accuracy of 89.45% and an AUC of 0.96. In another study, Ardila et al.22 also analyzed LDCT scans from NLST, achieving an AUC of 0.94. Importantly, their models’ predictions were compared with those of six radiologists. When prior CT imaging was not available, the model outperformed all radiologists and led to absolute reductions of 11% in false positives and 5% in false negatives. When prior imaging was available, performance was comparable to that of the radiologists. Image analyses aren’t limited to identification of malignant versus benign either. Chen et al.23 used radiomics to differentiate between non-small-cell lung cancer (NSCLC) and peripherally located small-cell lung cancer (SCLC) with an AUC of 0.93, which further supports use of radiomics as a non-invasive approach for early diagnosis and treatment of lung cancer. Overall, AI offers an opportunity to improve accuracy of nodule classification, either on initial screening or follow-up, and may even provide information on tumor histology. Future work will need to clarify which models/approaches yield the best performance and should be incorporated into standard clinical practice, with a critical consideration being avoiding false positives and overdiagnosis.
Outcome prediction
In addition to assisting with identifying lung cancers, AI can also help predict oncologic outcomes overall and who will respond to therapy. Predicting outcomes including locoregional and distant recurrence, progression-free survival, and overall survival (OS) can be challenging, given that factors that influence these outcomes are multivariate. Imaging features are no doubt highly relevant, but these must be combined with patient and tumor characteristics to produce accurate radiomic models.
Presently, primary tumor staging is based on the American Joint Committee on Cancer (AJCC) staging system, which largely uses tumor size in its staging algorithm. Although tumor size and response generally correlate with overall survival outcomes, there is some level of subjectivity, and additional information is critical. There are efforts to incorporate radiologic findings with additional radiomic features to better assess tumor characteristics. The Computer Aided Nodule Assessment and Risk Yield (CANARY) tool is one such approach where radiomic features were incorporated with imaging findings to predict a subset of lung adenocarcinoma patients, diagnosed as part of NLST, that have more aggressive disease.24
There are a number of studies that have attempted to combine patient characteristics, genomic features, and radiomic changes to predict for overall outcomes with some success.25,26 In a study by D’Antonoli et al.,25 the authors extracted radiomic features from patients with resected NSCLC in order to predict risk for recurrence. This study demonstrated how radiomics and clinical information can synergize to improve models; when using tumor-node-metastasis (TNM) stage or radiomics alone to predict local recurrence, the model achieved AUCs of 0.58 and 0.73, respectively. When combined, the AUC was improved to 0.75. Lee et al.26 used radiomic features to predict OS in patients with stage I NSCLC. In this study, combining radiomic and genomic features led to a concordance index of 0.70, compared with 0.62 using molecular features alone. Other similar studies in patients with brain metastases have been performed using MRI radiomics to predict survival and mutational status.27,28
Use of radiomics has not been limited to pre- or post-treatment scans. In a study conducted by Buizza et al.,29 the authors demonstrated that PET/CT radiomic features could help distinguish early responders from non-responders in patients undergoing definitive chemoradiation for lung cancer by obtaining PET/CT prior to initiating treatment and during the first 3 weeks of treatment. They entered available features into a support vector machine (SVM) ML model intended to identify early responders, and achieved AUC as high as 0.98 and 0.93 on their test data for patients treated with sequential and concurrent chemoradiotherapy, respectively.
More advanced pipelines have also been used, which can combine image analysis and classification into a single analysis framework, rather than feeding extracted features into a predictive model. In one such example, Xu et al.30 used CNNs and other DL algorithms on CT imaging obtained prior to treatment and over the course of follow-up to predict mortality risk in locally advanced NSCLC. Each additional scan that was included in the model increased model performance, with an AUC as high as 0.74. This represents one of many approaches in which DL can incorporate multi-time-point imaging to improve clinical outcome predication, with minimal invasiveness and need for human input.
Other radiomics studies seek to identify suitable candidates for available treatments. By itself, PD-L1 (programmed cell death ligand 1) expression can predict for outcomes in lung cancer, yet this is determined from a biopsy and may not represent the heterogeneous microenvironment of lung cancer. Using AI to identify radiomic signatures as surrogates for responsiveness to PD-L1 directed therapies is an alternative. There is already evidence suggesting certain radiomic features may predict for those who will respond to immunotherapy.31,32
Response assessment
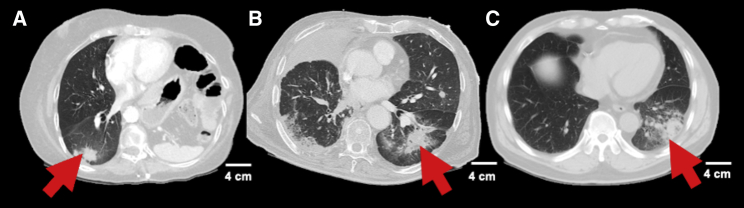
Although significant advances have been made in the management of NSCLC, between 10% and 20% of patients with early-stage NSCLC33 and approximately 66% of patients with advanced NSCLC34 have disease progression or die within five years of treatment. Given that certain progressive patients may still be curable, there is a significant need for accurate assessment of treatment response and identification of progression. Traditionally and currently on most prospective clinical trials, evaluation of tumor response relies upon RECIST (Response Evaluation Criteria in Solid Tumors) and World Health Organization (WHO) criteria,35 though in the era of immunotherapy immune-related RECIST (irRECIST) is increasingly being used.36 These criteria, although useful, often times do not correlate with treatment response and may lead to the interpretation of tumor progression when in fact it is merely post-treatment changes.37,38 This issue is amplified in patients treated with immunotherapy and radiation; both are known to cause treatment changes, inflammation, and pneumonitis, which can sometimes be difficult to distinguish from local progression.39,40 Examples of changes related to radiation, immunotherapy, and progression, to illustrate their similarity, are visualized in Figure 2. Distinguishing treatment changes from progression is challenging but critical, as it can lead to unnecessary tests including invasive biopsies, early changes in therapy when it is not indicated, and potentially disqualifying patients from an enrolled clinical trial where they have limited options. AI and radiomics in particular, have been demonstrated to refine response assessment to improve patient management.
Figure 2.
Examples of radiographic changes that may benefit from being distinguished using artificial intelligence
Representative non-contrast computed tomography slices showing patients who experienced (A) radiation pneumonitis, (B) immunotherapy pneumonitis, or (C) disease progression following cancer treatment are shown. The similarity of these images exemplify an area in which artificial intelligence might help providers distinguish subtle differences in imaging to make the correct diagnosis.
In a study by Mattonen et al.,37 the authors used CT texture changes following stereotactic body radiation therapy (SBRT) to predict recurrence and distinguish it from radiation-induced lung injury (RILI). Their dataset included scans from 13 lesions with moderate to severe RILI and 11 with recurrence. In this sample, RECIST achieved an accuracy of 65.2%, with a false-negative rate of 45.5% and a false-positive rate of 27.3%. In comparison, using radiomics, accuracy was increased to 77%, with an AUC as high as 0.81. Therefore, AI may be able to improve ability to distinguish treatment-related changes from progression, which is key for counseling of patients.
Pathology
Pathology plays a key role in the diagnosis and treatment of lung cancer. Presently, pathology in lung cancer is more complex than just grade and histology as it provides information on tumor microenvironment, biomarker expression that help select treatment and predict response (PD-L1), and genomic profiling to identify potential targets that can be used to direct therapy (epidermal growth factor receptor [EGFR], anaplastic lymphoma kinase [ALK], Kirsten rat sarcoma viral oncogene [KRAS], and others). AI/DL can potentially assist in interpreting pathologic tumor characteristics.41
Histologic classification
Currently, CNNs are the most frequent model being applied for tumor characterization, largely because of lending themselves nicely to image analysis.42 At the most fundamental histologic level, DL has been used to help in lung cancer diagnosis, differentiating between non-malignant, adenocarcinoma and squamous cell carcinoma. In a study by Coudray et al.,43 CNNs were used to classify histology sample, achieving an AUC of 0.97. This study took classification a step further and sought to predict for gene mutations. This classification is more nuanced, so unsurprisingly the AUCs were lower at 0.73–0.86.
Immunohistochemistry is another field that is critical for subtyping of NSCLC via the use of molecular markers. These analyses often require many stains, which can be difficult to perform when there is limited tissue available. To address this problem, Koh et al. used decision tree and SVM ML classifiers to aid with subtyping, specifically in cases with equivocal findings.44 The accuracy varied from 72.2% to 91.7% depending on the marker pattern, but overall provided a framework for subtyping using just a three marker panel if only small NSCLC biopsies are available.
In a similar study by Wang et al., CNNs were used to classify cell types in tumor microenvironment.45 The model, which they named ConvPath, achieved an overall accuracy of 90.1%. The authors postulate that understanding of the tumor microenvironment may help better understand tumor progression and metastasis. Although this information may be useful, it is potentially prohibitive for pathologists to manually perform the required classification, so having viable AI options is a necessity for expanding research into the microenvironment.
Analysis of liquid biopsies
In addition to tissue biomarker analysis, liquid biopsies have become more common. Liquid biopsy is analysis of any tumor-derived product in the blood or serum. Compared with tissue biopsies, which can assess the spatial heterogeneity of the tumor on the basis of the location of the biopsy, it can capture temporal heterogeneity by obtaining biopsies in the blood stream at different times, which may be able to identify progression.46 This modality is useful in different settings. In early disease, plasma next-generation sequencing (NGS) with large panels can provide high sensitivity and specificity and may help distinguish benign from malignant nodules, guide neoadjuvant therapy, and guide adjuvant therapy for surgically resectable patients. Zhang et al.47 used synthetic minority oversampling (SMOTE), a technique used to aid AI when the classification of interest is in the minority and therefore liable to have its defining feature that must be learned lost among the volume of the majority class, combined with random forests to identify lung cancer using circulating microRNA (miRNA), achieving an AUC as high as 0.99. Critically, this study used a case-control design with samples not limited to early-stage disease, which likely inflated performance. Nevertheless, research on the use of AI/ML in analysis of liquid biopsies is needed; human analysis of such multi-modal high-dimensional data is simply not possible, so AI will play a key role in finding ways to improve sensitivity and specificity of liquid biopsies, which will benefit patients by being less invasive and catching cancers earlier.
Genetic mutations and gene expression
Although most of the current application of AI in lung cancer has focused on immunohistochemistry there has been some incorporation of ML techniques in gene expression profile analysis. For example, adenocarcinomas which present with mutations that can be targeted with therapies is one area in which ML has been applied to better understand genomic pathways and potentially identify other actionable biomarkers downstream.48 With major improvements in genomic sequencing through commercial platforms such as NGS, the presence of more data points and pathways will improve ML analyses to aid clinicians in selecting appropriate treatments and gaining better understanding of the tumor biology and potential outcomes including risk for metastatic potential.49
In work by Cook et al.,50 a novel ML algorithm was used to sub-classify lung adenocarcinoma and lung squamous cell carcinoma. This facilitated identification of novel mutations, such as PIGX (an oncogenic driver in breast cancer) worthy of future investigation. This research specifically harnessed a sub-type of ML algorithms called unsupervised learning, that searches for commonalities and patterns without assigning specific labels as part of a classification problem, which is what enabled identification of potentially novel mutations.
Treatment
AI has also been applied to treatment decision making. A clinical decision support system (CDSS) is a tool to assist physicians in making clinical decisions on the basis of analyses of multiple data points on a particular patient. Watson for Oncology (WFO) is one example of a CDSS that has been applied to the treatment management of lung cancer. A study comparing decisions made by WFO to a multidisciplinary team found relatively high concordance in recommendations for early stage and metastatic disease (92.4%–100%) but lower rates of concordance in stage II or III (80.8%–84.6%).51 Therefore, although there is room for improvement for decision support, these tools will be critical for standardizing lung cancer treatment across available treatment options and disciplines, thereby enhancing outcomes.
Medical oncology
Drug discovery
A primary area in which AI can benefit medical oncology is drug development. A major challenge for the oncologic pharmaceutical industry is optimization of drug development and in silico drug screening. Drug development can be timely and costly, not to mention the possibility that the required investment ultimately proves futile because of lack of efficacy in human trials. AI offers the potential to support drug development in two ways: it can be used to screen previously developed drugs for new uses,52 and it can help identify potential drug candidates worthy of further investigation before undergoing years of research requiring millions of dollars.53 In work by Li et al.,54 transcriptomic and chemical structures were used as inputs into a DL algorithm intended for drug repurposing. Using this algorithm, pimozide, an anti-dyskinesia agent previously used for Tourette’s disorder, was identified as a strong candidate for treatment of NSCLC. The authors further validated these finding in in vitro experiments, demonstrating efficacy against certain NSCLC cell lines.
Response prediction
Work is ongoing to use AI tools to predict response to immune checkpoint inhibitors and targeted therapies. For immunotherapy, Charoentong et al.55 performed a pan-cancer, inclusive of lung cancer, immunogenomic ML analysis to predict response to checkpoint inhibitors. They used this approach to generate an “immunophenoscore,” which outperformed PD-L1 expression for predicting response to immunotherapies in certain histologies. ML applications in predicting failure have also been applied to patients undergoing targeted therapies. Kureshi et al.56 applied ML, specifically SVM and decision tree classifiers, to evaluate multiple factors in predicting tumor response in EGFR positive NSCLC receiving erlotinib or gefitinib and found a predictive accuracy of the data-driven decision support model of 76% and an AUC of 0.76. Overall, these data support use of AI to optimize systemic therapy selection in lung cancer, where the range of options, particularly in the metastatic setting, is quite broad.
Radiation oncology
Treatment planning
In addition to aiding with interventions that improve outcomes, AI might also be used to streamline treatment workflows. Radiation oncology in particular is an example of a field that is resource intensive. Treatment of patients requires, at minimum, CT simulation, delineation of organs at risk (OARs), definition of target volumes, treatment plan optimization, plan evaluation, and quality assurance. Many of these tasks are repetitive, and their burden could be reduced by AI, leading to shorter durations between consultation and delivery of the first fraction.
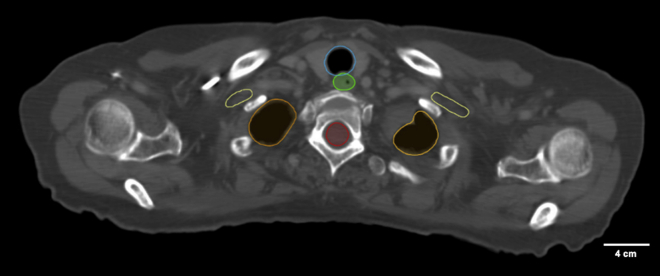
Wu et al.57 reported on AAR-RT, a system designed to automatically contour OARS on CT images, a key and often time-consuming step in the radiation treatment process. The authors specifically focused on treatment of patients with head and neck and thoracic malignancies, which require careful definition of normal organs to permit optimization that avoids excessive toxicity. The framework enabled improved structure recognition and subsequent delineation. Example auto-contoured OARs are visualized in Figure 3. Other work has aimed to automate the planning process once OARs and targets have been drawn. Planning is inherently an optimization problem, which lends itself nicely to automation. Zhang et al.58 reported on an automatic planning algorithm for lung intensity-modulated radiation therapy. When auto-generated plans were compared with plans generated by experienced medical dosimetrists, they consistently have equivalent or improved performance with regard to target coverage, OAR sparing, and overall quality. Last, Wall et al.59 reported on the use of ML models for predicting quality assurance outcomes. They evaluated multiple different types of ML algorithms, ultimately leading to selection of a SVM, with a mean absolute error of 3.85%. The authors asserted that such work could help guide the plan optimization process to avoid issues that would decrease quality assurance pass rates. In total, these are all steps in the automation of radiotherapy planning which will reduce time required to optimally plan a case from about a week to a day or less.
Figure 3.
Examples of auto-contoured organs at risk for stage III lung cancer
Normal organ image segmentation outlines (lung [orange], spinal cord [red], brachial plexus [yellow], esophagus [green], and trachea [blue]) on a computed tomographic scan generated using an in-house deep learning algorithm designed to streamline radiation treatment workflows.
Treatment selection
Another scenario in which AI has been shown to improve lung cancer radiotherapy is helping with treatment selection. Although there are certain cases in which radiation is clearly indicated, other cases are borderline. For example, two recently published randomized controlled trials have called into question routine postoperative radiotherapy (PORT) in patients with completely resected N2 NSCLC.60,61 However, certain populations may still benefit from treatment. In work by Zarinshenas et al.,62 an XGBoost ML model was used to identify patients who may still benefit from PORT on the basis of nodal burden. The model identified positive nodal count thresholds of ≥3 and positive nodal ratios of ≥0.34 as predictive of benefit from PORT, achieving a concordance index of 0.65, outperforming Cox regression.
Surgery
Decision making
Surgical resection is standard of care for management of localized lung cancer.12 Extent of surgery depends on several factors, including disease progression and patient eligibility. When possible, lobectomy has been established as standard, with improved disease control and/or survival compared with smaller wedge resections63 and larger pneumonectomies.64 Furthermore, the mortality rate of lobectomies is 2.3% compared with 6.8% with pneumonectomies.65 However, not every patient will be a candidate for lobectomy, because of factors such as medical history, smoking history, and lung function. AI offers an opportunity to better risk-stratify patients to come up with an optimal treatment plan, which might also include no surgery at all if risk is too high.
Santos-Garcia et al.66 used a NN to predict postoperative cardio-respiratory in patients with NSCLC. Their model achieved an AUC of 0.98, thereby proving to be a valuable tool for avoiding unnecessary toxicity. Similarly, in a study by Esteva et al.,67 NNs were used to estimate postoperative prognosis and major postoperative complications using clinical and laboratory variables from a training set of 113 patients. The model had 100% testing set sensitivity and specificity at correctly classifying risk for mortality and postoperative morbidity in a test set of 28 patients.
Specific data critical to assessing candidacy for surgery is pulmonary function tests (PFTs), as patients with poor lung function at baseline may be unable to sustain sufficient function with a portion of their lung removed. In a study by Topalovic et al.,68 PFTs were interpreted by a decision tree ML framework and pulmonologists, and the resulting interpretations were compared with guideline gold standards. A total of 120 pulmonologists each evaluated 50 cases. The pulmonologist interpretation matched the guideline interpretation 74.4% of the time (interrater variability 0.67), leading to diagnosis of the correct underlying pathology 45% of the time (interrater variability 0.35). In contrast, the ML model, developed and validated on the 1,500 historical patient cases, matched guidelines 100% of the time, yielding the correct diagnosis in 82% of the time. Thus, by being able to predict underlying patient factors, AI can help predict morbidity and mortality, thereby leading to improvements in treatment selection.
Robotic surgery
Modern thoracic surgery regularly makes use of robotic surgical systems, which can facilitate minimally invasive procedures with decreased surgical trauma.69 Robotic-assisted surgical systems are by definition not AI, as they are not programmed but serve as extensions of the surgeon, thereby improving precision. However, there may be a role for robotic systems to gain autonomy to perform certain repetitive tasks. In one such example, Shademan et al.70 reported on a “smart tissue autonomous robot” (STAR) that uses AI to optimize and automate complex surgery planning. The authors compared anastomosis metrics between the STAR system, manual laparoscopic surgery, and standard robotic-assisted surgery. STAR outperformed both alternative techniques with respect to suture consistency, leakage, number of mistakes, completion time, and lumen reduction. Therefore, incorporation of AI into actual surgical is potentially feasible and may improve quality, though this testing was performed in pigs and is not part of routine clinical care. There are still significant unknowns with regard to legal, regulatory, and ethical considerations, and there are currently no FDA approvals for autonomous surgery.71 Therefore the impact of AI in this field of lung cancer is limited but might prove helpful in the future when surgeons seek no operate around complicated thoracic anatomy.
Network meta-analysis
A rapidly emerging area in lung cancer research, with implications for all treatment modalities, are Bayesian network meta-analyses. Traditional systematic meta-analysis aims to pool clinical trial data to compare two interventions. However, as is commonly the case when managing lung cancer in the modern era, there might be more than two options, in which case traditional meta-analysis is insufficient. Network analysis helps address this problem through one of two approaches: strictly indirect treatment comparison through a shared comparator or a mixed approach where both indirect comparisons and direct comparisons from available trial are used.72 A downside of some network meta-analysis approaches is that they do not produce forest plots, which practitioners are accustomed to interpreting, and therefore results may be difficult to understand.73 The Bayesian network approach helps address this issue by producing novel outputs such as treatment rankings, odds ratios, and probability distributions.74 Several such studies have been performed in lung cancer, including evaluating safety and efficacy of bevacizumab biosimilars,75 determining optimal platinum-based chemotherapy for early-stage resected NSCLC,76 how smoking status influences effect of targeted therapy,77 and choice of first-line treatment for patients on the basis of PD-L1 expression.78,79,80
Current challenges to applying AI to lung cancer
Although AI is clearly an invaluable tool to the multidisciplinary lung cancer care team, several barriers remain to its widespread implementation and availability. First, AI relies heavily on data, and data acquisition and organization continue to be a challenge that AI will need to overcome. Efforts will optimally focus on ways of efficiently extracting EMR data to create large databases for AI research. Sample size is important in AI research, as it must be sufficiently large to train, test and validate models. Presently, most outcomes-based research studies include relatively small numbers of patients (between tens and hundreds of patients) that are somewhat heterogeneous as far as patient demographics, genomics, and imaging features are concerned. Though it is sometimes possible to perform AI analyses on datasets of that size, sample sizes in the thousands might be required for many applications.81 Otherwise, models may be inaccurate, poorly generalizable, and not applicable or reproducible to clinical outcomes. Additionally, although the EMR system has provided the opportunity to extract data into models for AI-based research, a number of variables are recorded as free text, which cannot directly be extracted for data analysis.
Another challenging aspect of AI research is reproducibility. Although many institutions are performing research in AI, methodology and reproducibility vary and are somewhat at the discretion of the researcher. It has therefore become critical that publications of AI-based research include detailed descriptions of methodology and data curation in order to verify the accuracy and interpretation of the data being presented. An approach to improve reproducibility is to have reporting standards, which has been published for radiomics.82 This challenge is exemplified by the fact that the studies in this review almost exclusively lack external validation or prospective evaluation, which may explain their excellent (and potentially overly optimistic) performance. Thus, although these studies are hypothesis generating and lay a solid foundation, significant future work is required before widespread clinical implementation is feasible. Moreover, much of AI-based research has focused on retrospectively analyzing patterns and outcomes, but there are few studies that have actually applied AI-based interventions to patient care and compared them with the gold standard of patient care. Future studies are needed to compare the effects of AI-based interventions to standard of care on patient outcomes.
AI has the potential to incorporate thousands of variables and features, including patient demographics, tumor characteristics, genomic and radiomic features, treatment details such systemic therapy agents and radiation dose to tumor and organs at risk, to predict outcomes for lung cancer patients and potentially guide treatment decisions. However, these datasets currently are often stored in institutional repositories. In order to ensure reproducibility and permit validation of results, datasets will need to be more widely available. Centralized repositories will also be required for multimodality integration, essential for unifying available sources of information to generate the best possible models.83
A related but distinct challenge will be ensuring appropriate study designs. One aspect of this is related to sample size; included in the study design, there must be a means of model testing and validation, whether through a hold-out dataset, an external dataset, or at the very least via statistical resampling. Ignoring such considerations will lead to the models that overfit the training data and/or introduce bias, and would not generalize to clinical practice.84 This is emphasized in Figure 1.
It is also critical to acknowledge ethical concerns of implementing AI/ML in the clinic.85 How should the risks and benefits of implementing AI in the clinic be balanced? Although AI may improve health care delivery efficiency and efficiency there are concerns for patient confidentiality and autonomy as their data are used to create or evaluate models that make decisions. Do patients need to consent to AI tools being used to guide their care? This is especially relevant when one considers that models are certainly fallible, with major ramifications for patient outcomes if they are incorrect. Furthermore, where does liability lie when such shortcomings adversely affect patient care? Answers to these questions are still in their infancy, but will not doubt need to be explored as the role of AI/ML expands in the lung cancer clinic.
Last, adoption of AI must also overcome resistance to its inherent complexity. For example, the output of widely used Cox proportional hazards models are ubiquitous in oncology research and relatively simple to understand. In contrast, radiomic features or the DL models in general are not nearly as intuitive. This represents a barrier to implementation of AI in the clinic, because of lack of understandability or trust, regardless of demonstrated efficacy. There are solutions to this. First, there are frameworks in a discipline designated “explainable AI” (XAI), which seek to visualize the inner working of the “black box” that is AI.86,87 Popular frameworks include locally interpretable model agnostic explanations (LIME) and shapley additive explanation (SHAP), which provide estimates of how model features lead to ultimate model predictions, similar to how regression coefficients are used in standard regression models.88 Alternatively, the field can seek to use inherently high-interpretability methods when possible without sacrificing predictive performance. These include “simple” decision trees, regression models such as least absolute shrinkage and selection operator (LASSO), and Bayesian probabilistic causal networks, though these are not applicable to all clinical problems/models.89,90,91,92,93,94,95
Future directions
The above limitations need to be addressed prior to general incorporation of AI in the lung cancer clinic. We must work as a field to build robust, accessible, and validated models designed to improve patient care. With that accomplished, AI provides a major avenue for personalized medicine in lung cancer. Logical next steps include efforts to improve sample sizes and data availability. Examples of such initiatives are the Cancer Imaging Archive and The Cancer Genome Atlas.96,97 If researchers have access to large, centralized sources of data, many of the challenges associated with adoption of AI will be addressed; sample sizes will increase, enabling appropriately sized training and testing datasets. The importance of this is emphasized by inclusion in Figure 1. Furthermore, working with a single dataset also allows researchers to ensure reproducibility, increase collaboration, and overall maximize patient benefit. Obstacles to this effort include data sharing difficulties, challenges in pooling diverse data, and significant effort requirements. In addition to structural improvements, there will be algorithmic improvements as well. Computational power is always increasing, and as research in AI for lung cancer continues to grow, so too will performance metrics and efficiency, both of which will stimulate applicability to the clinic. However, at the same time it is also critical that we not make AI out to be something it is not; it is not a silver bullet, with many challenges to translation to the lung cancer clinic.98 Neither it is an absolutely autonomous, encapsulated system, and nor should it be. We seek to develop systems informed by all stakeholders, including patients, physicians, and administrators, to ensure our models are useable, applicable, and valid.
Conclusions
The present is an exciting time for lung cancer treatment, as the available treatment options, and the precision with which we can select them, have improved dramatically in recent years. However, these increasingly tailored treatment options are accompanied by a need for data to inform clinical decisions, and therefore a need to be able to make sense of large volumes of data throughout a hypothetical patients’ treatment course. The overarching field of AI, inclusive of ML, NNs, DL, NLP, XAI, and other domains and methodologies, offers a promising avenue for improving all aspects of lung cancer management with data-driven approaches. Advances in radiomics allow us to derive additional value from existing diagnostic imaging, while ML algorithms help with optimizing treatment selection. Although there are limitations to AI and challenges as discussed, with large databases and suitable platforms AI research will continue to grow and become more reproducible, accurate, and applicable. With the rise in AI-based research over the past decade and increasing interest toward AI in the oncology community, including young trainees, AI-based interventions in lung cancer management will play a key role in the future.
Acknowledgments
A.R. is supported by the National Institutes of Health under grants U01CA232216 and R01LM013138. Additional support by the Dr. Susumu Ohno Chair in Theoretical Biology (held by A.R.) is kindly acknowledged.
Author contributions
Conceptualization, A.A., A.G., I.M., and R.S.; methodology, C.L., A.A., A.G., I.M., and R.S.; investigation, C.L., A.A., and A.G.; data curation, C.L. and A.A.; writing – original draft, C.L. and A.A.; writing – review & editing, C.L., A.A., A.G., I.M., D.R., E.M., T.W., A.R., and R.S.; visualization, C.L.; supervision, A.A. and R.S.; project administration, C.L., A.A., and R.S.
Declaration of interests
The authors declare no competing interests.
References
- 1.Dlamini Z., Francies F.Z., Hull R., Marima R. Artificial intelligence (AI) and big data in cancer and precision oncology. Comput. Struct. Biotechnol. J. 2020;18:2300–2311. doi: 10.1016/j.csbj.2020.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones G.S., Baldwin D.R. Recent advances in the management of lung cancer. Clin. Med. 2018;18(Suppl 2):s41–s46. doi: 10.7861/clinmedicine.18-2s-s41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roosan M.R., Mambetsariev I., Pharaon R., Fricke J., Baroz A.R., Chao J., Chen C., Nasser M.W., Chirravuri-Venkata R., Jain M., et al. Evaluation of somatic mutations in solid metastatic pan-cancer patients. Cancers. 2021;13:2776. doi: 10.3390/cancers13112776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chua I.S., Gaziel-Yablowitz M., Korach Z.T., Kehl K.L., Levitan N.A., Arriaga Y.E., Jackson G.P., Bates D.W., Hassett M. Artificial intelligence in oncology: path to implementation. Cancer Med. 2021;10:4138–4149. doi: 10.1002/cam4.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Achuthan S., Chatterjee R., Kotnala S., Mohanty A., Bhattacharya S., Salgia R., Kulkarni P. Leveraging deep learning algorithms for synthetic data generation to design and analyze biological networks. J. Biosci. 2022;47:43. [PubMed] [Google Scholar]
- 6.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA. Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 7.Wang X., Zhang Y., Hao S., Zheng L., Liao J., Ye C., Xia M., Wang O., Liu M., Weng C.H., et al. Prediction of the 1-year risk of incident lung cancer: prospective study using electronic health records from the state of Maine. J. Med. Internet Res. 2019;21:e13260. doi: 10.2196/13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kehl K.L., Groha S., Lepisto E.M., Elmarakeby H., Lindsay J., Gusev A., Van Allen E.M., Hassett M.J., Schrag D. Clinical inflection point detection on the basis of EHR data to identify clinical trial-ready patients with cancer. JCO Clin. Cancer Inform. 2021;5:622–630. doi: 10.1200/CCI.20.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Omenn G.S., Nass S.J., Micheel C.M. Evolution of Translational Omics: Lessons Learned and the Path Forward. National Academic Press; 2012. [DOI] [PubMed] [Google Scholar]
- 10.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita R., Nishio M., Do R.K.G., Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9:611–629. doi: 10.1007/s13244-018-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ettinger D.S., Wood D.E., Aisner D.L., Akerley W., Bauman J.R., Bharat A., Bruno D.S., Chang J.Y., Chirieac L.R., D'Amico T.A., et al. Non–small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc. Netw. 2022;20:497–530. doi: 10.6004/jnccn.2022.0025. [DOI] [PubMed] [Google Scholar]
- 13.Coughlin J.M., Zang Y., Terranella S., Alex G., Karush J., Geissen N., Chmielewski G.W., Arndt A.T., Liptay M.J., Zimmermann L.J., et al. Understanding barriers to lung cancer screening in primary care. J. Thorac. Dis. 2020;12:2536–2544. doi: 10.21037/jtd.2020.03.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruano-Ravina A., Pérez-Ríos M., Casàn-Clará P., Provencio-Pulla M. Low-dose CT for lung cancer screening. Lancet Oncol. 2018;19:e131–e132. doi: 10.1016/S1470-2045(18)30121-9. [DOI] [PubMed] [Google Scholar]
- 15.Fraioli F., Serra G., Passariello R. CAD (computed-aided detection) and CADx (computer aided diagnosis) systems in identifying and characterising lung nodules on chest CT: overview of research, developments and new prospects. Radiol. Med. 2010;115:385–402. doi: 10.1007/s11547-010-0507-2. [DOI] [PubMed] [Google Scholar]
- 16.Keshani M., Azimifar Z., Tajeripour F., Boostani R. Lung nodule segmentation and recognition using SVM classifier and active contour modeling: a complete intelligent system. Comput. Biol. Med. 2013;43:287–300. doi: 10.1016/j.compbiomed.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Lo S.B., Freedman M.T., Gillis L.B., White C.S., Mun S.K. JOURNAL CLUB: Computer-aided detection of lung nodules on CT with a computerized pulmonary vessel suppressed function. AJR Am. J. Roentgenol. 2018;210:480–488. doi: 10.2214/AJR.17.18718. [DOI] [PubMed] [Google Scholar]
- 18.Firmino M., Angelo G., Morais H., Dantas M.R., Valentim R. Computer-aided detection (CADe) and diagnosis (CADx) system for lung cancer with likelihood of malignancy. Biomed. Eng. Online. 2016;15:2. doi: 10.1186/s12938-015-0120-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kann B.H., Hosny A., Aerts H.J.W.L. Artificial intelligence for clinical oncology. Cancer Cell. 2021;39:916–927. doi: 10.1016/j.ccell.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anwar S.M., Majid M., Qayyum A., Awais M., Alnowami M., Khan M.K. Medical image analysis using convolutional neural networks: a review. J. Med. Syst. 2018;42:226–313. doi: 10.1007/s10916-018-1088-1. [DOI] [PubMed] [Google Scholar]
- 21.Paul R., Hall L., Goldgof D., Schabath M., Gillies R. International Joint Conference on Neural Networks. IJCNN; 2018. Predicting nodule malignancy using a CNN ensemble approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ardila D., Kiraly A.P., Bharadwaj S., Choi B., Reicher J.J., Peng L., Tse D., Etemadi M., Ye W., Corrado G., et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019;25:954–961. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- 23.Chen B.T., Chen Z., Ye N., Mambetsariev I., Fricke J., Daniel E., Wang G., Wong C.W., Rockne R.C., Colen R.R., et al. Differentiating peripherally-located small cell lung cancer from non-small cell lung cancer using a CT radiomic approach. Front. Oncol. 2020;10:593. doi: 10.3389/fonc.2020.00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maldonado F., Duan F., Raghunath S.M., Rajagopalan S., Karwoski R.A., Garg K., Greco E., Nath H., Robb R.A., Bartholmai B.J., Peikert T. Noninvasive computed tomography-based risk stratification of lung adenocarcinomas in the national lung screening trial. Am. J. Respir. Crit. Care Med. 2015;192:737–744. doi: 10.1164/rccm.201503-0443OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akinci D'Antonoli T., Farchione A., Lenkowicz J., Chiappetta M., Cicchetti G., Martino A., Ottavianelli A., Manfredi R., Margaritora S., Bonomo L., et al. CT radiomics signature of tumor and peritumoral lung parenchyma to predict nonsmall cell lung cancer postsurgical recurrence risk. Acad. Radiol. 2020;27:497–507. doi: 10.1016/j.acra.2019.05.019. [DOI] [PubMed] [Google Scholar]
- 26.Lee J., Li B., Cui Y., Sun X., Wu J., Zhu H., Yu J., Gensheimer M.F., Loo B.W., Jr., Diehn M., Li R. A quantitative CT imaging signature predicts survival and complements established prognosticators in stage I non-small cell lung cancer. Int. J. Radiat. Oncol. Biol. Phys. 2018;102:1098–1106. doi: 10.1016/j.ijrobp.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Chen B.T., Jin T., Ye N., Mambetsariev I., Wang T., Wong C.W., Chen Z., Rockne R.C., Colen R.R., Holodny A.I., et al. Predicting survival duration with MRI radiomics of brain metastases from non-small cell lung cancer. Front. Oncol. 2021;11:621088. doi: 10.3389/fonc.2021.621088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen B.T., Jin T., Ye N., Mambetsariev I., Daniel E., Wang T., Wong C.W., Rockne R.C., Colen R., Holodny A.I., et al. Radiomic prediction of mutation status based on MR imaging of lung cancer brain metastases. Magn. Reson. Imaging. 2020;69:49–56. doi: 10.1016/j.mri.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buizza G., Toma-Dasu I., Lazzeroni M., Paganelli C., Riboldi M., Chang Y., Smedby Ö., Wang C. Early tumor response prediction for lung cancer patients using novel longitudinal pattern features from sequential PET/CT image scans. Phys. Med. 2018;54:21–29. doi: 10.1016/j.ejmp.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y., Hosny A., Zeleznik R., Parmar C., Coroller T., Franco I., Mak R.H., Aerts H.J.W.L. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin. Cancer Res. 2019;25:3266–3275. doi: 10.1158/1078-0432.CCR-18-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dercle L., Fronheiser M., Lu L., Du S., Hayes W., Leung D.K., Roy A., Wilkerson J., Guo P., Fojo A.T., et al. Identification of non-small cell lung cancer sensitive to systemic cancer therapies using radiomics. Clin. Cancer Res. 2020;26:2151–2162. doi: 10.1158/1078-0432.CCR-19-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trebeschi S., Drago S.G., Birkbak N.J., Kurilova I., Cǎlin A.M., Delli Pizzi A., Lalezari F., Lambregts D.M.J., Rohaan M.W., Parmar C., et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann. Oncol. 2019;30:998–1004. doi: 10.1093/annonc/mdz108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang J.Y., Mehran R.J., Feng L., Verma V., Liao Z., Welsh J.W., Lin S.H., O'Reilly M.S., Jeter M.D., Balter P.A., et al. Stereotactic ablative radiotherapy for operable stage I non-small-cell lung cancer (revised STARS): long-term results of a single-arm, prospective trial with prespecified comparison to surgery. Lancet Oncol. 2021;22:1448–1457. doi: 10.1016/S1470-2045(21)00401-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spigel D.R., Faivre-Finn C., Gray J.E., Vicente D., Planchard D., Paz-Ares L., Vansteenkiste J.F., Garassino M.C., Hui R., Quantin X., et al. Five-year survival outcomes from the PACIFIC trial: durvalumab after chemoradiotherapy in stage III non–small-cell lung cancer. J. Clin. Oncol. 2022;40:1301–1311. doi: 10.1200/JCO.21.01308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisenhauer E.A., Therasse P., Bogaerts J., Schwartz L., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) European journal of cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 36.Bohnsack O., Hoos A., Ludajic K. Adaptation of the immune related response criteria: irRECIST. Ann. Oncol. 2014;25:iv369. [Google Scholar]
- 37.Mattonen S.A., Ward A.D., Palma D.A. Pulmonary imaging after stereotactic radiotherapy—does RECIST still apply? Br. J. Radiol. 2016;89:20160113. doi: 10.1259/bjr.20160113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayes S.A., Pietanza M.C., O’Driscoll D., Zheng J., Moskowitz C.S., Kris M.G., Ginsberg M.S. Comparison of CT volumetric measurement with RECIST response in patients with lung cancer. Eur. J. Radiol. 2016;85:524–533. doi: 10.1016/j.ejrad.2015.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schoenfeld J.D., Nishino M., Severgnini M., Manos M., Mak R.H., Hodi F.S. Pneumonitis resulting from radiation and immune checkpoint blockade illustrates characteristic clinical, radiologic and circulating biomarker features. J. Immunother. Cancer. 2019;7:112–117. doi: 10.1186/s40425-019-0583-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrara R., Mezquita L., Texier M., Lahmar J., Audigier-Valette C., Tessonnier L., Mazieres J., Zalcman G., Brosseau S., Le Moulec S., et al. Hyperprogressive disease in patients with advanced non-small cell lung cancer treated with PD-1/PD-L1 inhibitors or with single-agent chemotherapy. JAMA Oncol. 2018;4:1543–1552. doi: 10.1001/jamaoncol.2018.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Echle A., Rindtorff N.T., Brinker T.J., Luedde T., Pearson A.T., Kather J.N. Deep learning in cancer pathology: a new generation of clinical biomarkers. Br. J. Cancer. 2021;124:686–696. doi: 10.1038/s41416-020-01122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S., Yang D.M., Rong R., Zhan X., Xiao G. Pathology image analysis using segmentation deep learning algorithms. Am. J. Pathol. 2019;189:1686–1698. doi: 10.1016/j.ajpath.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coudray N., Ocampo P.S., Sakellaropoulos T., Narula N., Snuderl M., Fenyö D., Moreira A.L., Razavian N., Tsirigos A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koh J., Go H., Kim M.Y., Jeon Y.K., Chung J.H., Chung D.H. A comprehensive immunohistochemistry algorithm for the histological subtyping of small biopsies obtained from non-small cell lung cancers. Histopathology. 2014;65:868–878. doi: 10.1111/his.12507. [DOI] [PubMed] [Google Scholar]
- 45.Wang S., Wang T., Yang L., Yang D.M., Fujimoto J., Yi F., Luo X., Yang Y., Yao B., Lin S., et al. ConvPath: a software tool for lung adenocarcinoma digital pathological image analysis aided by a convolutional neural network. EBioMedicine. 2019;50:103–110. doi: 10.1016/j.ebiom.2019.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roosan M.R., Mambetsariev I., Pharaon R., Fricke J., Husain H., Reckamp K.L., Koczywas M., Massarelli E., Bild A.H., Salgia R. Usefulness of circulating tumor DNA in identifying somatic mutations and tracking tumor evolution in patients with non-small cell lung cancer. Chest. 2021;160:1095–1107. doi: 10.1016/j.chest.2021.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y.-H., Jin M., Li J., Kong X. Identifying circulating miRNA biomarkers for early diagnosis and monitoring of lung cancer. Biochim. Biophys. Acta, Mol. Basis Dis. 2020;1866:165847. doi: 10.1016/j.bbadis.2020.165847. [DOI] [PubMed] [Google Scholar]
- 48.Kikuchi T., Daigo Y., Katagiri T., Tsunoda T., Okada K., Kakiuchi S., Zembutsu H., Furukawa Y., Kawamura M., Kobayashi K., et al. Expression profiles of non-small cell lung cancers on cDNA microarrays: identification of genes for prediction of lymph-node metastasis and sensitivity to anti-cancer drugs. Oncogene. 2003;22:2192–2205. doi: 10.1038/sj.onc.1206288. [DOI] [PubMed] [Google Scholar]
- 49.Rabbani M., Kanevsky J., Kafi K., Chandelier F., Giles F.J. Role of artificial intelligence in the care of patients with nonsmall cell lung cancer. Eur. J. Clin. Invest. 2018;48:e12901. doi: 10.1111/eci.12901. [DOI] [PubMed] [Google Scholar]
- 50.Cook M.P., Qorri B., Baskar A., et al. Small patient datasets reveal genetic drivers of non-small cell lung cancer subtypes using a novel machine learning approach. 2021. Preprint at medRxiv. [DOI] [Google Scholar]
- 51.Kim M.S., Park H.Y., Kho B.G., Park C.K., Oh I.J., Kim Y.C., Kim S., Yun J.S., Song S.Y., Na K.J., et al. Artificial intelligence and lung cancer treatment decision: agreement with recommendation of multidisciplinary tumor board. Transl. Lung Cancer Res. 2020;9:507–514. doi: 10.21037/tlcr.2020.04.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gottlieb A., Stein G.Y., Ruppin E., Sharan R. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol. Syst. Biol. 2011;7:496. doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang G., Fan W., Luo H., Zhu X. The emerging roles of artificial intelligence in cancer drug development and precision therapy. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2020;128:110255. doi: 10.1016/j.biopha.2020.110255. [DOI] [PubMed] [Google Scholar]
- 54.Li B., Dai C., Wang L., Deng H., Li Y., Guan Z., Ni H. A novel drug repurposing approach for non-small cell lung cancer using deep learning. PLoS One. 2020;15:e0233112. doi: 10.1371/journal.pone.0233112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 56.Kureshi N., Abidi S.S.R., Blouin C. A predictive model for personalized therapeutic interventions in non-small cell lung cancer. IEEE J. Biomed. Health Inform. 2016;20:424–431. doi: 10.1109/JBHI.2014.2377517. [DOI] [PubMed] [Google Scholar]
- 57.Wu X., Udupa J.K., Tong Y., Odhner D., Pednekar G.V., Simone C.B., McLaughlin D., Apinorasethkul C., Apinorasethkul O., Lukens J., et al. AAR-RT–a system for auto-contouring organs at risk on CT images for radiation therapy planning: principles, design, and large-scale evaluation on head-and-neck and thoracic cancer cases. Med. Image Anal. 2019;54:45–62. doi: 10.1016/j.media.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X., Li X., Quan E.M., Pan X., Li Y. A methodology for automatic intensity-modulated radiation treatment planning for lung cancer. Phys. Med. Biol. 2011;56:3873–3893. doi: 10.1088/0031-9155/56/13/009. [DOI] [PubMed] [Google Scholar]
- 59.Wall P.D., Fontenot J.D. Application and comparison of machine learning models for predicting quality assurance outcomes in radiation therapy treatment planning. Inform. Med. Unlocked. 2020;18:100292. [Google Scholar]
- 60.Le Pechoux C., Pourel N., Barlesi F., Lerouge D., Antoni D., Lamezec B., Nestle U., Boisselier P., Dansin E., Paumier A., et al. Postoperative radiotherapy versus no postoperative radiotherapy in patients with completely resected non-small-cell lung cancer and proven mediastinal N2 involvement (Lung ART): an open-label, randomised, phase 3 trial. Lancet Oncol. 2021;23:104–114. doi: 10.1016/S1470-2045(21)00606-9. [DOI] [PubMed] [Google Scholar]
- 61.Hui Z., Men Y., Hu C., Kang J., Sun X., Bi N., Zhou Z., Liang J., Lv J., Feng Q., et al. Effect of postoperative radiotherapy for patients with pIIIA-N2 non–small cell lung cancer after complete resection and adjuvant chemotherapy. JAMA Oncol. 2021;7:1178–1185. doi: 10.1001/jamaoncol.2021.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zarinshenas R., Ladbury C., McGee H., Raz D., Erhunmwunsee L., Pathak R., Glaser S., Salgia R., Williams T., Amini A. Machine learning to refine prognostic and predictive nodal burden thresholds for post-operative radiotherapy in completely resected stage III-N2 non-small cell lung cancer. Radiother. Oncol. 2022;173:10–18. doi: 10.1016/j.radonc.2022.05.019. [DOI] [PubMed] [Google Scholar]
- 63.Ginsberg R.J., Rubinstein L.V. Randomized trial of lobectomy versus limited resection for T1 N0 non-small cell lung cancer. Lung Cancer Study Group. Ann. Thorac. Surg. 1995;60:615–622. doi: 10.1016/0003-4975(95)00537-u. [DOI] [PubMed] [Google Scholar]
- 64.Albain K.S., Swann R.S., Rusch V.W., Turrisi A.T., 3rd, Shepherd F.A., Smith C., Chen Y., Livingston R.B., Feins R.H., Gandara D.R., et al. Radiotherapy plus chemotherapy with or without surgical resection for stage III non-small-cell lung cancer: a phase III randomised controlled trial. Lancet. 2009;374:379–386. doi: 10.1016/S0140-6736(09)60737-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brunelli A., Salati M., Rocco G., Varela G., Van Raemdonck D., Decaluwe H., Falcoz P.E., ESTS Database Committee European risk models for morbidity (EuroLung1) and mortality (EuroLung2) to predict outcome following anatomic lung resections: an analysis from the European Society of Thoracic Surgeons database. Eur. J. Cardio. Thorac. Surg. 2017;51:490–497. doi: 10.1093/ejcts/ezw319. [DOI] [PubMed] [Google Scholar]
- 66.Santos-García G., Varela G., Novoa N., Jiménez M.F. Prediction of postoperative morbidity after lung resection using an artificial neural network ensemble. Artif. Intell. Med. 2004;30:61–69. doi: 10.1016/s0933-3657(03)00059-9. [DOI] [PubMed] [Google Scholar]
- 67.Esteva H., Marchevsky A., Núñez T., Luna C., Esteva M. Neural networks as a prognostic tool of surgical risk in lung resections. Ann. Thorac. Surg. 2002;73:1576–1581. doi: 10.1016/s0003-4975(02)03418-5. [DOI] [PubMed] [Google Scholar]
- 68.Topalovic M., Das N., Burgel P.-R., Daenen M., Derom E., Haenebalcke C., Janssen R., Kerstjens H.A.M., Liistro G., Louis R., et al. Artificial intelligence outperforms pulmonologists in the interpretation of pulmonary function tests. Eur. Respir. J. 2019;53:1801660. doi: 10.1183/13993003.01660-2018. [DOI] [PubMed] [Google Scholar]
- 69.Kumar A., Asaf B.B. Robotic thoracic surgery: the state of the art. J. Minim. Access Surg. 2015;11:60–67. doi: 10.4103/0972-9941.147693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shademan A., Decker R.S., Opfermann J.D., Leonard S., Krieger A., Kim P.C.W. Supervised autonomous robotic soft tissue surgery. Sci. Transl. Med. 2016;8:337ra64. doi: 10.1126/scitranslmed.aad9398. [DOI] [PubMed] [Google Scholar]
- 71.O'Sullivan S., Nevejans N., Allen C., Blyth A., Leonard S., Pagallo U., Holzinger K., Holzinger A., Sajid M.I., Ashrafian H. Legal, regulatory, and ethical frameworks for development of standards in artificial intelligence (AI) and autonomous robotic surgery. Int. J. Med. Robot. 2019;15:e1968. doi: 10.1002/rcs.1968. [DOI] [PubMed] [Google Scholar]
- 72.Lumley T. Network meta-analysis for indirect treatment comparisons. Stat. Med. 2002;21:2313–2324. doi: 10.1002/sim.1201. [DOI] [PubMed] [Google Scholar]
- 73.Hu D., O'Connor A.M., Winder C.B., Sargeant J.M., Wang C. How to read and interpret the results of a Bayesian network meta-analysis: a short tutorial. Anim. Health Res. Rev. 2019;20:106–115. doi: 10.1017/S1466252319000343. [DOI] [PubMed] [Google Scholar]
- 74.Dias S., Welton N.J., Caldwell D.M., Ades A.E. Checking consistency in mixed treatment comparison meta-analysis. Stat. Med. 2010;29:932–944. doi: 10.1002/sim.3767. [DOI] [PubMed] [Google Scholar]
- 75.Xu X., Zhang S., Xu T., Zhan M., Chen C., Zhang C. Efficacy and safety of bevacizumab biosimilars compared with reference biologics in advanced non-small cell lung cancer or metastatic colorectal cancer patients: a network meta-analysis. Front. Pharmacol. 2022;13:880090. doi: 10.3389/fphar.2022.880090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pang L.L., Gan J.D., Huang Y.H., Liao J., Lv Y., Ali W.A.S., Zhang L., Fang W.F. Investigation of the optimal platinum-based regimen in the postoperative adjuvant chemotherapy setting for early-stage resected non-small lung cancer: a Bayesian network meta-analysis. BMJ Open. 2022;12:e057098. doi: 10.1136/bmjopen-2021-057098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin K., Lin J., Huang Z., Fu J., Yi Q., Cai J., Khan M., Yuan Y., Bu J. Impact of smoking on response to the first-line treatment of advanced ALK-positive non-small cell lung cancer: a bayesian network meta-analysis. Front. Pharmacol. 2022;13:881493. doi: 10.3389/fphar.2022.881493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim J.H., Jeong S.Y., Lee J.J., Park S.T., Kim H.S. A bayesian network meta-analysis of first-line treatments for non-small cell lung cancer with high programmed Death ligand-1 expression. J. Clin. Med. 2022;11:1492. doi: 10.3390/jcm11061492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhai J., Lu J., Zhang Z., Wang Y., Li X., Zhang S., Mu S., Zhi X., Ge X., Lu D., et al. First-line PD-1/PD-L1 inhibitors plus chemotherapy versus bevacizumab plus chemotherapy for advanced non-squamous non-small cell lung cancer: a Bayesian network meta-analysis of randomized controlled trials. Cancer Med. 2022;11:2043–2055. doi: 10.1002/cam4.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mao Z., Jiang P., Zhang Y., Li Y., Jia X., Wang Q., Jiao M., Jiang L., Shen Y., Guo H. First-line immune-based combination therapies for advanced non-small cell lung cancer: a Bayesian network meta-analysis. Cancer Med. 2021;10:9139–9155. doi: 10.1002/cam4.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vabalas A., Gowen E., Poliakoff E., Casson A.J. Machine learning algorithm validation with a limited sample size. PLoS One. 2019;14:e0224365. doi: 10.1371/journal.pone.0224365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zwanenburg A., Abdalah M., Ashrafinia S., et al. Results from the image biomarker standardisation initiative. Radiother. Oncol. 2018;127:S543–S544. [Google Scholar]
- 83.Lambin P., Leijenaar R.T.H., Deist T.M., Peerlings J., de Jong E.E.C., van Timmeren J., Sanduleanu S., Larue R.T.H.M., Even A.J.G., Jochems A., et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 84.Ying X. An overview of overfitting and its solutions. J. Phys, Conf. Ser. 2019;1168:022022. [Google Scholar]
- 85.Rigby M.J. Ethical dimensions of using artificial intelligence in health care. AMA Journal of Ethics. 2019;21:121–124. [Google Scholar]
- 86.Lundberg S.M., Lee S.-I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017;30 [Google Scholar]
- 87.Ribeiro MT, Singh S, Guestrin C. " Why should i trust you?" Explaining the predictions of any classifier. Paper presented at: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining2016.
- 88.Ladbury C., Zarinshenas R., Semwal H., Tam A., Vaidehi N., Rodin A.S., Liu A., Glaser S., Salgia R., Amini A. Utilization of model-agnostic explainable artificial intelligence frameworks in oncology: a narrative review. Transl Cancer Res. 2022;11:3853–3868. doi: 10.21037/tcr-22-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kalet A.M., Doctor J.N., Gennari J.H., Phillips M.H. Developing Bayesian networks from a dependency-layered ontology: a proof-of-concept in radiation oncology. Med. Phys. 2017;44:4350–4359. doi: 10.1002/mp.12340. [DOI] [PubMed] [Google Scholar]
- 90.Trilla-Fuertes L., Gámez-Pozo A., Arevalillo J.M., López-Vacas R., López-Camacho E., Prado-Vázquez G., Zapater-Moros A., Díaz-Almirón M., Ferrer-Gómez M., Navarro H., et al. Bayesian networks established functional differences between breast cancer subtypes. PLoS One. 2020;15:e0234752. doi: 10.1371/journal.pone.0234752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Park S.B., Hwang K.-T., Chung C.K., Roy D., Yoo C. Causal Bayesian gene networks associated with bone, brain and lung metastasis of breast cancer. Clin. Exp. Metastasis. 2020;37:657–674. doi: 10.1007/s10585-020-10060-0. [DOI] [PubMed] [Google Scholar]
- 92.Wang X., Branciamore S., Gogoshin G., Ding S., Rodin A.S. New analysis framework incorporating mixed mutual information and scalable bayesian networks for multimodal high dimensional genomic and epigenomic cancer data. Front. Genet. 2020;11:648. doi: 10.3389/fgene.2020.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Djulbegovic B., Hozo I., Dale W. Transforming clinical practice guidelines and clinical pathways into fast-and-frugal decision trees to improve clinical care strategies. J. Eval. Clin. Pract. 2018;24:1247–1254. doi: 10.1111/jep.12895. [DOI] [PubMed] [Google Scholar]
- 94.Gupta S., Vundavilli H., Osorio R.S.A., Itoh M.N., Mohsen A., Datta A., Mizuguchi K., Tripathi L.P. Integrative network modeling highlights the crucial roles of rho-GDI signaling pathway in the progression of non-small cell lung cancer. IEEE J. Biomed. Health Inform. 2022;26:4785–4793. doi: 10.1109/JBHI.2022.3190038. [DOI] [PubMed] [Google Scholar]
- 95.Hinton T., Karnak D., Tang M., Jiang R., Luo Y., Boonstra P., Sun Y., Nancarrow D.J., Sandford E., Ray P., et al. Improved prediction of radiation pneumonitis by combining biological and radiobiological parameters using a data-driven Bayesian network analysis. Transl. Oncol. 2022;21:101428. doi: 10.1016/j.tranon.2022.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Clark K., Vendt B., Smith K., Freymann J., Kirby J., Koppel P., Moore S., Phillips S., Maffitt D., Pringle M., et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tomczak K., Czerwińska P., Wiznerowicz M. Review the cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 2015;19:68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kann B.H., Hosny A., Aerts H.J.W.L. Artificial intelligence for clinical oncology. Cancer Cell. 2021;39:916–927. doi: 10.1016/j.ccell.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]