Abstract
Common sensitizing mutations in epidermal growth factor receptor (cEGFR), including exon 19 deletions (19-Del) and exon 21 L858R substitution, are associated with high sensitivity to EGFR-TKIs in NSCLC patients. The treatment for NSCLC patients with uncommon EGFR (uEGFR) mutations remains a subject of debate due to heterogeneity in treatment responses. In this manuscript, the targeted next-generation sequencing (NGS) data of a large cohort of EGFR-mutated NSCLC patients was assessed to elucidate genomic profiles of tumors carrying cEGFR or uEGFR mutations. The results showed that NSCLC patients with uEGFR mutations were more likely to harbor co-occurring genetic alterations in the Hippo pathway and a higher TMB compared with cEGFR-positive patients. Smoking-related mutations were found to significantly enriched in uEGFR-positive patients. Subgroup analyses were performed to identify potential prognostic biomarkers in patients harboring various EGFR subtype mutations. L858R-positive patients with co-existing ARID2 mutations had shorter progression-free survival (PFS) than those who were L858R- or 19-Del-positive but ARID2-negative (median: 2.3 vs. 12.0 vs. 8.0 months, P = 0.038). Furthermore, mutational profiles, such as top frequently mutated genes and mutational signatures of patients with various EGFR subtype mutations were significantly different. Our study analyzed the mutational landscape of NSCLC patients harboring cEGFR and uEGFR mutations, revealing specific genomic characteristics associated with uEGFR mutations that might explain the poor prognosis of first-generation EGFR-TKIs.
Keywords: Common EGFR, Uncommon EGFR, NSCLC, EGFR-TKI
Abbreviations: EGFR, epidermal growth factor receptor; cEGFR, common EGFR; uEGFR, uncommon EGFR; 19-Del, EGFR exon 19 deletion; 20-Ins, EGFR exon 20 insertions; TKI, tyrosine kinase inhibitor; NSCLC, non-small-cell lung cancer; NGS, next-generation sequencing; TMB, tumor mutational burden; FFPE, formalin-fixed paraffin-embedded; SNVs, single-nucleotide variations; MAF, mutant allele frequency; COSMIC, Catalog of Somatic Mutations in Cancer; PFS, progression-free survival; OS, overall survival
Introduction
Lung cancer is the most common cause of cancer-related mortality worldwide, with non-small cell lung cancer (NSCLC) accounting for ∼80% of newly diagnosed lung cancer cases annually [1], [2], [3]. The epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) have demonstrated profound clinical efficacies in NSCLC patients harboring EGFR exon 19 deletion (19-Del) or L858R mutations, showing superior survival benefit over platinum-based chemotherapy and fewer side effects [4,5].
Activation of EGFR can promote cell proliferation, angiogenesis, tumor invasion, and metastasis, therefore playing an essential role in the development and progression of NSCLC [6], [7], [8]. EGFR mutations have been reported to occur almost exclusively in exons 18 to 21 of the gene, which encodes the EGFR kinase domain [9,10]. About 80-90% of such mutations occur as 19-Del (codons 746 to 754 in exon 19) and L858R (exon 21), termed common EGFR (cEGFR) mutations [11], [12], [13]. With extensive analysis of the molecular features of patients harboring cEGFR mutations, accumulating evidence has demonstrated distinguishing clinical characteristics between 19-Del and L858R [4,14]. Moreover, patients with 19-Del or L858R mutations showed differential sensitivity to various chemotherapy or targeted therapies, although some conclusions are still controversial [4,5,[14], [15], [16], [17]].
Despite the high prevalence of cEGFR mutations in NSCLC, approximately 10-20% of cases involve other mutation sites within exons 18 to 21 of EGFR, termed uncommon EGFR (uEGFR) mutations. G719X (exon 18), L861Q (exon 21), S768I (exon 20), and exon 20 insertions (20-Ins) are the most frequent subtypes for uEGFR mutations [18], [19], [20]. NSCLC patients with these uEGFR mutations benefit from first-generation (G) EGFR-TKIs such as erlotinib and gefitinib [18], [19], [20], [21], [22], [23]. Increasing evidence demonstrated improved outcomes for patients with uEGFR mutations such as G719X, L861Q, and S768I mutations upon treatment with second-generation EGFR-TKIs [23]. On the other hand, EGFR 20-Ins are traditionally believed insensitive to anti-EGFR therapies [24], [25], [26]. Overall, data from most retrospective studies of uEGFR mutations is highly heterogeneous. There is no consensus to determine the therapeutic intervention for populations with uEGFR mutations.
This study aims to systemically review the targeted sequencing results of a large cohort of EGFR-mutated NSCLC patients to investigate the molecular landscape of patients with different EGFR mutation subtypes associated with clinical response to first-generation EGFR-TKIs. The genetic-clinical association would provide valuable information to support treatment selection in heterogeneous subgroups of NSCLC patients with rare EGFR mutations.
Materials and methods
Patient enrollment and sample collection
Tumor specimens were collected from EGFR-mutated NSCLC patients as a routine diagnosis at all participating hospitals between June 2015 and May 2020. This study was approved by the institutional research ethics committee of the Nanjing Drum Tower Hospital. Written consent was acquired from each patient before sample collection. Qualified samples were subjected to targeted next-generation sequencing by a CLIA-certified and CAP-accredited clinical testing laboratory (Nanjing Geneseeq Technology Inc., Nanjing, China) using pan-cancer gene panels (GENESEEQPRIME™ and RADIOTRONTM, Geneseeq Technology Inc.). Formalin-fixed paraffin-embedded (FFPE) tissue samples were confirmed by pathologists from the centralized clinical testing center before genetic testing. Clinical characteristics and treatment history were extracted from medical records. The mutational and corresponding clinical data of the validation cohort comprising 156 EGFR-mutated lung adenocarcinoma patients were downloaded from the cBioPortal for Cancer Genomics (https://www.cbioportal.org/) [27,28].
Targeted next-generation sequencing
DNA extraction, library construction, and targeted capture enrichment were carried out following standard protocols as previously described with modifications [29,30]. FFPE samples were de-paraffinized first with xylene before genomic DNA extraction using QIAamp DNA FFPE Tissue Kit (Qiagen Cat. No. 56404) according to the manufacturer's instructions. Genomic DNA extracted from tumor samples was qualified using Nanodrop2000 (Thermo Fisher Scientific, Waltham, MA), then quantified using the dsDNA HS assay kit on a Qubit 3.0 fluorometer (Life Technology, US) according to the manufacturer's recommendations. Targeted NGS libraries were prepared using the KAPA Hyper Prep kit (KAPA Biosystems) with an optimized manufacturer's protocol for different sample types. Targeted capture enrichment was performed as previously described [31]. According to the manufacturer's instructions, the target-enriched library was then sequenced on HiSeq4000 and HiSeq4000 NGS platforms (Illumina).
Mutation calling
Sequencing data was first demultiplexed and subjected to FASTQ file quality control using Trimmomatic [32]. Only data without extra nucleotide bases and passed quality control (QC above 15) were subjected to the following analyses. Raw reads were mapped to the reference Human Genome (hg19) using Burrows-Wheeler Aligner (BWA-mem, v0.7.12; https://github.com/lh3/bwa/tree/master/bwakit). Genome Analysis Toolkit (GETK 3.4.0; https://software.broadinstitute.org/gatk/) was employed to perform local realignment around the insertions/deletions (INDELs) and base quality score recalibration. Picard was used to remove PCR duplicates. VarScan2 was applied to detect single-nucleotide variations (SNVs) and INDELs. SNVs were filtered out if the mutant allele frequency (MAF) was less than 1% for tumor tissue and 0.3% for plasma samples. Eight out of ten oncogenic signaling pathway alterations were compared among patients with EGFR mutations [33]. The Nrf2 pathway was excluded from the pathway-level analyses of gene alterations co-mutated in EGFR-mutated NSCLC patients, given that KEAP1, NFE2L2, and CUL3 were not included in the 416-cancer-related gene panel. The RTK/RAS pathway was also excluded, given that all patients in the study cohort harbored EGFR mutations.
Identifying mutation signatures
Information on mutational signatures prevalent in EGFR-mutated NSCLC patients was downloaded from version 3.0 of the Catalog of Somatic Mutations in Cancer (COSMIC) database (https://cancer.sanger.ac.uk/cosmic/signatures/) [34,35]. The mutation signature was determined by matching observed mutations to the most frequent nucleotide base changes and the trinucleotide context in each signature. If multiple signatures were compatible with the mutation in question, the most active signature in a specific biological context referring to the scientific literature was used.
Statistical methods
Plots in this study were generated using the R Project for Statistical Computing (version 3.4.0). Statistical analyses were performed using R. Fisher's exact tests were used to test the categorical variables between groups. Kaplan-Meier curves were used to analyze progression-free survival (PFS) and overall survival (OS) of various patient groups, and the statistical difference was analyzed using the log-rank test. A two-sided P value of less than 0.05 was considered significant for all tests unless indicated otherwise (*P < 0.05, 0.01<**P < 0.05, ***P < 0.001).
Results
Patient overview
2,280 NSCLC patients with known EGFR mutations who underwent targeted NGS of 416 cancer-related genes were included in the following analyses (Supplementary Figs. 1, 2). Patients were further divided into two subgroups depending on whether the patient harbored common or uncommon EGFR mutations at diagnosis. In particular, the cEGFR-positive cohort comprised 1,022 patients with L858R (44.8%) and 973 patients with 19-Del (42.7%). There was no significant difference in the clinical characteristics between these two cohorts (Supplementary Table S1). On the other hand, four uEGFR mutation subtypes were analyzed, including 20-Ins, G719X, L861Q, and S768I. Notably, since many patients with L861Q or S768I harbored co-existing G719X, patients with double EGFR mutations were separated from patients with single mutations (Supplementary Fig. 2). In brief, the uEGFR-positive cohort contained 139 patients with 20-Ins (6.1%), 59 patients with G719X (2.6%), 45 patients with L861Q (2.0%), 6 patients with L861Q and G719X compound mutations (0.3%), 8 patients with S768I (0.4%), and 28 patients with S768I and G719X compound mutations (1.2%). The demographic and clinical characteristics of patients with varying types of EGFR mutations were summarized in Table 1. A cohort combined using two datasets [27,28] comprising 156 EGFR-mutated patients with NSCLC was used as an external validation set to test the robustness of our mutational profiling results. Corresponding clinical characteristics of these 159 NSCLC patients were summarized in Supplementary Table. S2.
Table 1.
Clinical characteristics among patients with different EGFR mutation subtypes (N = 2280).
| Characteristics | Common N (%) |
Uncommon N (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| L858R | 19-Del | 20-Ins | G719X | L861Q | S768I | L861Q+G719X | S768I+G719X | P value | |
| Gender | |||||||||
| Male | 412 (40.3) | 417 (42.9) | 58 (41.7) | 34 (57.6) | 15 (33.3) | 6 (75.0) | 4 (66.7) | 16 (57.1) | 0.018 |
| Female | 610 (59.7) | 556 (57.1) | 81 (58.3) | 25 (42.4) | 30 (66.7) | 2 (25.0) | 2 (33.3) | 12 (42.9) | |
| Age | |||||||||
| ≥ 60 years | 602 (58.9) | 453 (46.6) | 69 (49.6) | 29 (49.2) | 30 (66.7) | 3 (37.5) | 1 (16.7) | 17 (60.7) | <0.001 |
| < 60 years | 412 (40.3) | 510 (52.4) | 70 (50.4) | 30 (50.8) | 15 (33.3) | 4 (50.0) | 5 (83.3) | 10 (35.7) | |
| NA | 8 (0.8) | 10 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (12.5) | 0 (0.0) | 1 (3.6) | |
| Histology | |||||||||
| ADC | 868 (84.9) | 806 (82.8) | 120 (86.3) | 52 (88.1) | 37 (82.2) | 5 (62.5) | 3 (50.0) | 23 (82.1) | 0.022 |
| SCC | 11 (1.1) | 23 (2.4) | 5 (3.6) | 1 (1.7) | 1 (2.2) | 1 (12.5) | 1 (16.7) | 1 (3.6) | |
| ASC | 14 (1.4) | 14 (1.4) | 2 (1.4) | 1 (1.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| NOS | 129 (12.6) | 130 (13.4) | 12 (8.6) | 5 (8.5) | 7 (15.6) | 2 (25.0) | 2 (33.3) | 4 (14.3) | |
| Stage at diagnosis | |||||||||
| Ⅰ | 79 (7.7) | 64 (6.6) | 11 (7.9) | 4 (6.8) | 2 (4.4) | 1 (12.5) | 0 (0.0) | 0 (0.0) | 0.071 |
| Ⅱ | 29 (2.8) | 26 (2.7) | 4 (2.9) | 2 (3.4) | 3 (6.7) | 0 (0.0) | 0 (0.0) | 1 (3.6) | |
| Ⅲ | 41 (4.0) | 49 (5.0) | 12 (8.6) | 6 (10.2) | 2 (4.4) | 0 (0.0) | 0 (0.0) | 1 (3.6) | |
| Ⅳ | 382 (37.4) | 414 (42.6) | 57 (41.0) | 30 (50.9) | 10 (22.2) | 3 (37.5) | 3 (50.0) | 10 (35.7) | |
| NA | 491 (48.0) | 420 (43.2) | 55 (39.6) | 17 (28.8) | 28 (62.3) | 4 (50.0) | 3 (50.0) | 16 (57.1) | |
P values are based on the Fisher's exact test; ADC, adenocarcinoma; SCC, squamous cell carcinoma; ASC, adenosquamous cell carcinoma; NA: not available; NOS: not otherwise specified; Bold represents significant P values.
Somatic alterations associated with different EGFR mutation subtypes
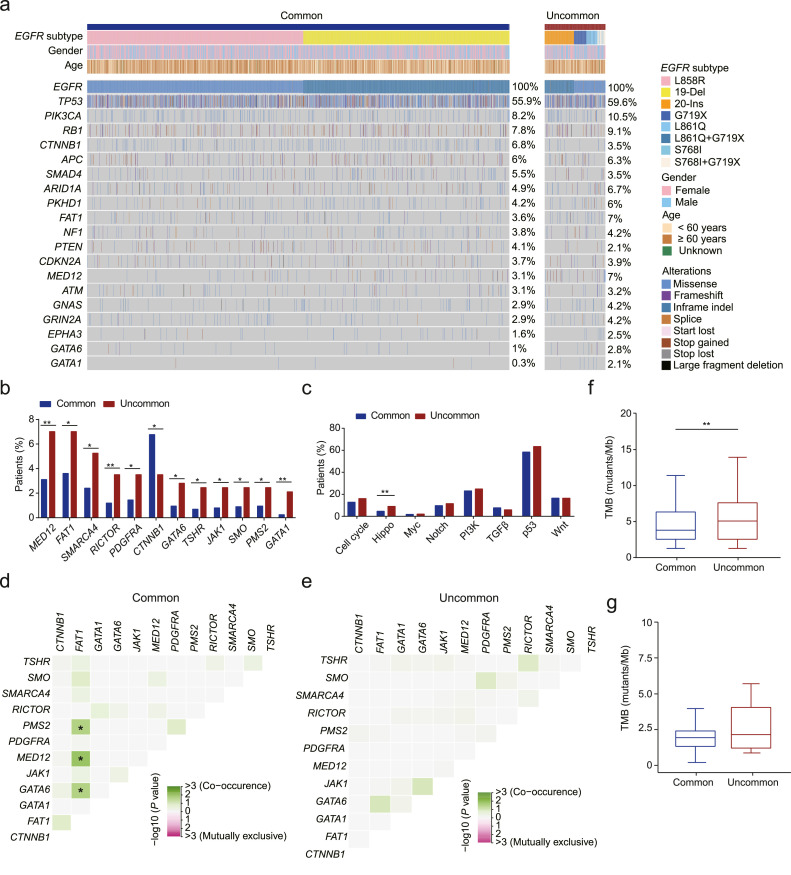
Since there has been accumulating evidence showing that EGFR mutation subtypes confer different responses to EGFR-TKIs, we analyzed the NGS data of 2,250 EGFR-mutated NSCLC patients and characterized the genomic landscape of these patients to explore the genetic-clinical associations (Fig. 1a). The top frequently co-mutated genes in the cEGFR and uEGFR cohorts include TP53 (55.9% vs. 59.6%), PIK3CA (8.2% vs. 10.5%), RB1 (7.8% vs. 9.1%), CTNNB1 (6.8% vs. 3.5%), APC (6% vs. 6.3%), SMAD4 (5.5% vs. 3.5%) and ARID1A (4.9% vs. 6.7%). Gene-level analyses showed that CTNNB1, which encodes β-catenin protein previously implicated in the pathogenesis of several cancers, was significantly more enriched in patients with cEGFR than those with uEGFR (P < 0.05, Fig.1b). In great contrast, genes such as MED12, RICTOR, and GATA1 showed a higher mutation frequency in patients with uEGFR mutations (P<0.01). Pathway-level analyses revealed that hippo pathway gene alterations were more frequently observed in patients harboring uEGFR mutations than those with cEGFR mutations (Fig. 1c). In addition, FAT1 showed a strong correlation with PMS2, MED12, and GATA6 in cEGFR-positive patients (P < 0.05, Fig. 1d). In contrast, no significant correlation was observed in the uEGFR cohort (Fig. 1e).
Fig. 1.
Somatic alterations associated with common and uncommon EGFR mutations. a. The genomic landscape of patients with common or uncommon EGFR mutations. Individual gene mutations in baseline tumor samples of EGFR-mutated NSCLC patients (N = 2280) were assessed by targeted NGS. Each column represents one patient. Clinical characteristics of patients are shown at the top. The frequency of each gene alteration is listed on the right. b. The bar plot shows the different distribution of somatic mutations in the cEGFR and uEGFR patient cohorts. c. The correlation between signaling pathways in which the concurrent mutations occur and EGFR subtypes. d-e. The heatmap demonstrates how frequently two somatic mutations occur in patients with cEGFR (d) or uEGFR mutations (e). f-g. Higher TMB was more likely associated with patients harboring uEGFR mutations than those carrying cEGFR mutations assessed by using the discovery study cohort (f) or the external validation cohort (g).
We next assessed the tumor mutational burden (TMB) in the cEGFR and uEGFR cohorts. TMB associated with uEGFR mutations was significantly higher than with cEGFR mutations (5.07 vs. 3.80, P < 0.01, Fig. 1f). Consistent with this finding, a higher TMB was observed in uEGFR-positive patients in the validation cohort (P = 0.1746, Fig. 1g). We reasoned that the insignificant difference could be due to the limited sample size of validation patients harboring uEGFR mutations (Supplementary Table. S2). Overall, our findings indicate that although patients with either cEGFR or uEGFR mutations showed some similarities in their genomic landscapes, for example, high mutational frequency in TP53, PI3KCA, and RB1. Gene- and pathway-level analyses implicated unique molecular features of patients with uEGFR mutations that might be associated with differential responses to targeted therapy.
Mutational signatures in patients with common or uncommon EGFR mutations
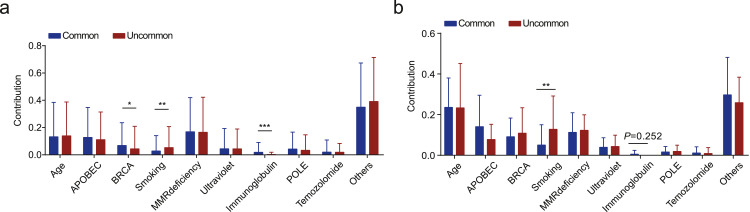
We then analyzed our data set of somatic mutational signatures associated with common and uncommon EGFR mutations. BRCA1/2-related signature (signature 3) and immunoglobulin-related signature (signature 9) showed a higher prevalence in patients with cEGFR mutations (Fig. 2a). In great contrast, smoking-induced mutations (signature 4) showed enrichment in the uEGFR subgroup, consistent with results obtained by assessing the external validation cohort (Fig. 2b). Unlike findings obtained using the discovery patient cohort, no significant difference in the contribution of mutations in neither the BRCA- nor the immunoglobulin-related signatures was observed, presumably due to the relatively small sample size in cEGFR-positive patients in the validation cohort (1,995 discovery cEGFR vs. 138 validation cEGFR, P<0.001, Supplementary Table S2). Overall, these results suggest that NSCLC patients carrying common or uncommon EGFR mutations may have undergone distinct mutational processes, resulting in unique mutational patterns and specific activities in the genome during tumorigenesis and cancer development.
Fig. 2.
The smoking-related mutational signature was enriched in uEGFR patients. a. Stacked bar plot illustrates the contribution of mutational signatures in cEGFR and cEGFR cohorts using the discovery cohort (N = 2,280). b. Stacked bar plot demonstrates the contribution of mutational signatures in cEGFR and cEGFR cohorts using the external validation cohort (N = 156).
Subgroup analysis reveals unique molecular features of EGFR mutation subtypes
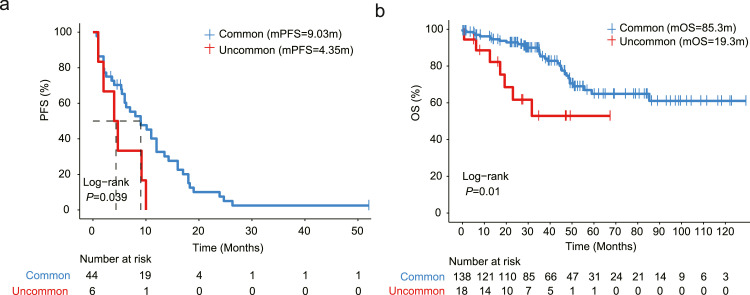
Fifty patients in our discovery cohort underwent first-generation EGFR-TKI treatment (Supplementary Table S3). The progression-free survival (PFS) of patients with uEGFR mutations was significantly shorter than those harboring cEGFR mutations (median: 9.03 vs. 4.35 months, P = 0.039, Fig. 3a), suggesting that NSCLC patients with uEGFR mutations are associated with a poorer prognosis than those with cEGFR mutations on first-generation EGFR-TKI treatment. Consistent with this notion, the overall survival (OS) greatly benefited from first-generation EGFR-TKIs when patients harbored cEGFR mutations rather than uEGFR mutations (median: 85.3 vs. 19.3 months, P = 0.01, Fig. 3b). Further analyses indicated that none of the clinical characteristics, specifically the patients’ gender, age or line of treatment, contributed to the efficacy of EGFR-TKIs (Table 2, Supplementary Fig. 3a). However, the PFS of patients with PI3K (median: 5.3 vs. 10.0 months, P=0.047) or TGFβ pathway gene alterations (median: 7.0 vs. 24.7 months, P=0.028) was significantly shorter than their wild-type counterparts (Supplementary Fig. 3b, 3c), implicating that there are other non-negligible factors associated with the differential responses to first-generation EGFR-TKIs in EGFR-mutated NSCLC patients.
Fig. 3.
uEGFR mutations confer a poor prognosis on first-generation EGFR-TKI treatment. a. Kaplan-Meier estimates of progression-free survival (PFS) in NSCLC patients with cEGFR or uEGFR mutations who had received first-generation EGFR-TKI treatment (N = 50). b. Kaplan-Meier estimates of overall survival (OS) using the external validation cohort patients (N = 156).
Table 2.
Univariate analysis of factors associated with first-generation EGFR-TKI clinical responses.
| Factor | N (%) | HR for PFS | P value |
|---|---|---|---|
| Gender (female) | 27 (54.0) | 1.08 (0.60∼1.93) | 0.805 |
| Age (≥60) | 21 (42.0) | 1.25 (0.69∼2.26) | 0.464 |
| EGFR mutation subtype (uEGFR) | 6 (12.0%) | 2.51 (1.01∼6.21) | 0.039 |
| Signaling pathways | |||
| Cell cycle | 9 (18.0) | 0.58 (0.28∼1.24) | 0.157 |
| Hippo | 2 (4.0) | 2.66 (0.61∼11.52) | 0.175 |
| Myc | 3 (6.0) | 0.74 (0.22∼2.51) | 0.627 |
| Notch | 4 (8.0) | 0.97 (0.30∼3.16) | 0.961 |
| PI3K | 11 (22.0) | 2.21 (1.00∼4.93) | 0.047 |
| TGFβ | 3 (6.0) | 0.22 (0.05∼0.94) | 0.028 |
| p53 | 38 (76.0) | 1.50 (0.75∼3.00) | 0.244 |
| Wnt | 9 (18.0) | 1.32 (0.60∼2.88) | 0.484 |
Bold represents statistically significant P values based on the log-rank test.
Analysis of molecular features of patients with mutation of 19-Del or L858R
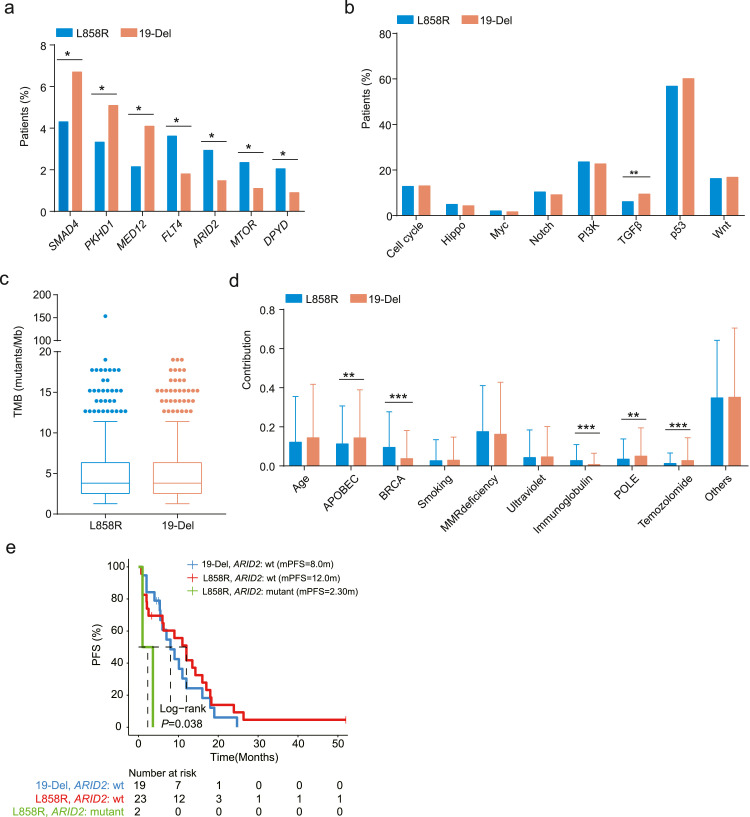
Since previous studies have demonstrated a prolonged survival of NSCLC patients with EGFR 19-Del than those with L858R [4,15,16], we evaluated the molecular features of patients with either 19-Del or L858R to explain the different clinical responses to EGFR-TKIs. Interestingly, mutations in FLT4, ARID2, MTOR, and DPYD showed a higher prevalence in patients with L858R, whereas mutations in SMAD4, PKHD1, and MED12 were more commonly identified in patients with 19-Del (Fig. 4a). Pathway-level analysis results demonstrated that gene alterations related to the TGFβ signaling pathway were markedly enriched in patients with 19-Del rather than L858R (Fig. 4b). Although no significant difference in TMB was observed in the 19-Del and L858R cohorts, somatic mutations belonging to the BRCA- and immunoglobulin-related signatures showed a higher prevalence in patients with L858R than those with 19-Del (Fig. 4c, d). Of the four significantly enriched genes in patients with L858R, only ARID2 was associated with patients’ survival after first-generation EGFR-TKI treatment. Specifically, the PFS of patients with L858R-positive and ARID2-positive tumors was significantly shorter than patients with cEGFR-positive and ARID2-negative tumors (Fig. 4e, Supplementary Fig. 3d). Although TGFβ gene alterations showed a higher prevalence in patients with 19-Del, no significant difference was observed in the PFS of 19-Del-positive patients with or without mutated TGFβ pathway (median: 16.4 vs. 7.0 months, P=0.17, Supplementary Fig. 3e). In addition, the PFS of uEGFR-positive patients carrying TGFβ gene alterations showed no significant difference compared to uEGFR-positive patients with unaltered TGFβ pathway (P=0.13, Supplementary Fig. 3f).
Fig. 4.
Comparing the molecular features of NSCLC patients with L858R or 19-Del mutations. a. Somatic mutations showed differential distribution in patients with L858R and 19-Del mutations. b. Mutations in the TGFβ signaling pathway were enriched in patients with 19-Del. c. No significant difference was observed in TMB between the two cohorts. d. Mutational signatures attributed by L858R and 19-Del mutations. e. Kaplan-Meier estimates of PFS in L858R-positive or 19-Del-positive NSCLC patients with or without ARID2 mutations.
Analysis of gene-level and pathway-level changes among EGFR mutation subtypes
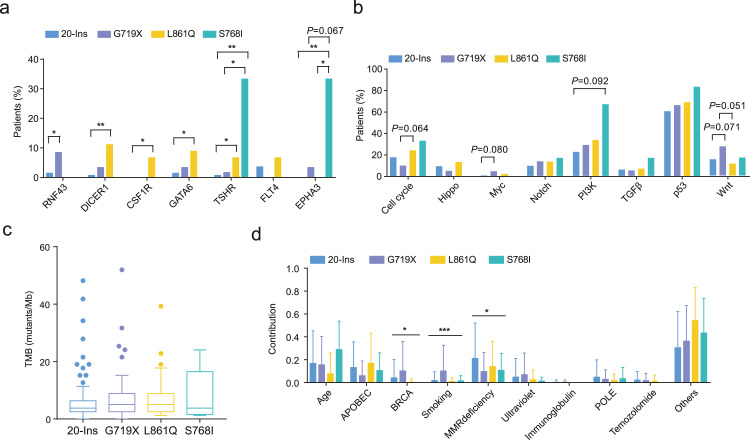
Given that uEGFR mutations in NSCLC are heterogeneous, EGFR-TKIs can have different efficacy in this specific population. Therefore, further assessment is required to decipher the genetic-clinical associations among these patients. Similarly, we assessed gene-level and pathway-level changes among EGFR mutation subtypes, specifically 20-Ins, G719X, L861Q and S768I. Notably, RNF43 mutation was more frequently observed in the G719X cohort, while DICER1, CSF1R, and GATA6 mutations were more likely associated with L861Q (Fig. 5a). Patients with S768I, on the other hand, tended to carry TSHR and EPHA3 mutations compared to other EGFR subtypes. We also showed that mutational signatures were significantly different among EGFR mutation subtypes (Fig. 5b). No significant difference regarding the pathway-level changes or TMB was observed in our analysis (Fig. 5c, d). Lastly, we compared the top frequently mutated genes in patients with single and compound EGFR mutations, and no significantly differentially mutated genes were identified (Supplementary Fig. 4). Indeed, previous studies suggest that G719X is frequently observed in patients with L861Q or S768I as a co-existing mutation [36], though the mechanism behind this co-mutation remains unclear.
Fig. 5.
Subgroup analysis of the molecular features of NSCLC patients with uEGFR mutations associated with different clinical responses. a. Somatic mutations showed differential distribution among patients with different EGFR mutation subtypes. b. No significant difference was observed in the pathway-level analysis of somatic mutations identified among EGFR mutation subtypes. c. TMB associated with each type of uEGFR mutation showed no significant difference. d. Mutational signatures attributed by uEGFR mutations.
Discussion
In this study, we retrospectively analyzed targeted next-generation sequencing results of baseline tumor specimens collected from 2,280 EGFR-mutated patients with NSCLC. We used an external validation cohort comprising 156 patients from two public-available patient databases to cross-validate our findings. This study indicates that patients with common or uncommon EGFR mutations have a unique molecular landscape, which probably underlies their differential responses to targeted therapies such as first-generation EGFR-TKIs.
Previous studies have demonstrated the efficacy of EGFR-TKIs on NSCLC patients harboring EGFR 19-Del and L858R mutations. However, very little was known about why the median progression-free survival time of patients harboring 19-Del was significantly improved compared with patients carrying L858R mutation following first-line EGFR-TKI treatment. The current study found that the presence of somatic mutations in ARID2, a gene that encodes a protein that functions in the chromatin remodelling complex to promote gene transcription, might confer a significantly shorter PFS in patients harboring L858R compared to patients with 19-Del (Fig. 4a, e, Supplementary Fig. 3d). We also demonstrated that the enrichment of BRCA1/2- and immunoglobulin-related mutational signatures in the L858R cohort might explain the differential sensitivity to EGFR-TKIs (Fig. 4d).
Approximately 10-20% of EGFR mutation sites occur within or, even more rarely, outside the kinase domain of the receptor (i.e., uEGFR mutations). Prior data derived from retrospective series of uEGFR mutations are highly heterogeneous in which the efficacy data are not distinguished between single and compound mutations. An initial objective of this study was to provide explanations for different sensitivity to EGFR-TKIs among patients with various EGFR mutation subtypes by characterizing somatic alterations, TMB and mutational signatures of these patients. This study confirms that uEGFR mutations are associated with a poorer prognosis after first-generation EGFR-TKI treatment (Fig. 3). One possible explanation for this was that patients harboring uEGFR mutations tend to have more Hippo pathway-related gene alterations and higher TMB than those with cEGFR mutations (Fig. 1c, f). Surprisingly, S768I was associated with somatic mutations in TSHR and EPHA3, whereas L861Q was more frequently observed in patients with concurrent DICER, CSF1R, and GATA6 mutations (Fig. 5a). Unfortunately, this study was unable to identify differential genetic alterations associated with single or compound mutations. One possible explanation for this is that S768I or L861Q shows a lower penetrance in the genome, thereby being more likely to form compound mutations with G719X than other uEGFR mutations.
Despite the above-mentioned promising results, some limitations were associated with the study. For example, due to the limited number of patients with available clinical data, we could not assess the efficacy of EGFR-TKIs in patients harboring EGFR 20-Ins. Previous studies have demonstrated that EGFR 20-Ins-positive patients responded poorly to targeted EGFR inhibitors, including the third-generation EGFR-TKI, osimertinib [37], [38], [39]. Future research should be undertaken to investigate why patients with this mutation adopt poor clinical responses and how to improve the survival benefit of these patients.
In conclusion, we systematically elucidated molecular profiles of tumors collected from NSCLC patients harboring different EGFR mutation subtypes, which showed specific genomic characteristics that might explain differential treatment responses to first-generation EGFR-TKIs.
Ethics approval and consent to participate
The study was approved by the Ethical Committee of the Nanjing Drum Tower Hospital (2020-158). Written consent was obtained from each patient.
Consent for publication
Written informed consent form was obtained from each patient.
Author's contributions
Conception and design: Yongkang Bai, Xiang Liu, Yuan Mao, Louqian Zhang
Collection and assembly of data: Shi Xiong, Pengfei Zhang, Zichen Jiao, Gefei Zhao, Chu Zhou, Junli Zhang, Jiaohui Pang
Data analysis: Yongkang Bai, Xiang Liu, Limin Zheng, Shi Xiong, Pengfei Zhang, Zichen Jiao, Gefei Zhao, Chu Zhou, Song Wang, Junli Zhang, Jiaohui Pang, Yang Xu
Manuscript writing: Yongkang Bai, Xiang Liu, Song Wang, Yang Xu
Study supervision: Yuan Mao, Louqian Zhang
Final approval of manuscript: all authors
Funding
This research is supported by the grants from the National Natural Science Foundation (no. 82073389) and Jiangsu Province “333 Project” Research Projects (2022)3-1-327).
Declaration of Competing Interest
The authors de-clare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank the patients and their families. We are grateful for generous help from investigators and researchers involved in the current study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2023.100888.
Contributor Information
Yuan Mao, Email: ymaoent@njmu.edu.cn.
Louqian Zhang, Email: zhanglouqian@126.com.
Appendix. Supplementary materials
Availability of data and materials
The datasets generated and/or analysed during this current study are available from the corresponding author on reasonable request.
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics. CA Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Ettinger D.S., Akerley W., Bepler G., Blum M.G., Chang A., Cheney R.T., Chirieac L.R., D'Amico T.A., Demmy T.L., Ganti A.K., et al. Non-small cell lung cancer. J. Natl. Compr. Cancer Netw. 2010;8:740–801. doi: 10.6004/jnccn.2010.0056. [DOI] [PubMed] [Google Scholar]
- 3.Molina J.R., Yang P., Cassivi S.D., Schild S.E., Adjei A.A. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee C.K., Wu Y.L., Ding P.N., Lord S.J., Inoue A., Zhou C., Mitsudomi T., Rosell R., Pavlakis N., Links M., et al. Impact of Specific Epidermal Growth Factor Receptor (EGFR) mutations and clinical characteristics on outcomes after treatment with EGFR tyrosine kinase inhibitors versus chemotherapy in egfr-mutant lung cancer: a meta-analysis. J. Clin. Oncol. 2015;33:1958–1965. doi: 10.1200/JCO.2014.58.1736. [DOI] [PubMed] [Google Scholar]
- 5.Maemondo M., I A., Kobayashi K., Sugawara S., Oizumi S., Isobe H., Gemma A., Harada M., Yoshizawa H., Kinoshita I., Fujita Y., Okinaga S., Hirano H., Yoshimori K., Harada T., Ogura T., Ando M., Miyazawa H., Tanaka T., Saijo Y., Hagiwara K., Morita S., Nukiwa T., for the North-East Japan Study Group* Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010 doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 6.Cooper W.A., Lam D.C., O'Toole S.A., Minna J.D. Molecular biology of lung cancer. J. Thorac. Dis. 2013;5(Suppl 5):S479–S490. doi: 10.3978/j.issn.2072-1439.2013.08.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciardiello F., Tortora G. EGFR antagonists in cancer treatment. N. Engl. J. Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 8.Pao W., Miller V.A., Politi K.A., Riely G.J., Somwar R., Zakowski M.F., Kris M.G., Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji H., Li D., Chen L., Shimamura T., Kobayashi S., McNamara K., Mahmood U., Mitchell A., Sun Y., Al-Hashem R., et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9:485–495. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 10.Sordella R., Bell D.W., Haber D.A., Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 11.Kosaka T., Yatabe Y., Endoh H., Kuwano H., Takahashi T., Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 12.Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. U. S. A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paez J.G., Janne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y., He D., Fang W., Kang S., Chen G., Hong S., Sheng J., Zhan J., Chen N., Hu Z., et al. The difference of clinical characteristics between patients with Exon 19 deletion and those with L858R mutation in nonsmall cell lung cancer. Medicine. 2015;94:e1949. doi: 10.1097/MD.0000000000001949. (Baltimore) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong W., Wu Q., Zhang J., Zhou Y. Prognostic value of EGFR 19-del and 21-L858R mutations in patients with non-small cell lung cancer. Oncol. Lett. 2019;18:3887–3895. doi: 10.3892/ol.2019.10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackman D.M., Yeap B.Y., Sequist L.V., Lindeman N., Holmes A.J., Joshi V.A., Bell D.W., Huberman M.S., Halmos B., Rabin M.S., et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin. Cancer Res. 2006;12:3908–3914. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- 17.Riely G.J., Pao W., Pham D., Li A.R., Rizvi N., Venkatraman E.S., Zakowski M.F., Kris M.G., Ladanyi M., Miller V.A. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin. Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 18.Shi J., Yang H., Jiang T., Li X., Zhao C., Zhang L., Zhao S., Liu X., Jia Y., Wang Y., et al. Uncommon EGFR mutations in a cohort of Chinese NSCLC patients and outcomes of first-line EGFR-TKIs and platinum-based chemotherapy. Chin. J. Cancer Res. 2017;29:543–552. doi: 10.21147/j.issn.1000-9604.2017.06.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Wang Z., Hao X., Hu X., Wang H., Wang Y., Ying J. Clinical characteristics and response to tyrosine kinase inhibitors of patients with non-small cell lung cancer harboring uncommon epidermal growth factor receptor mutations. Chin. J. Cancer Res. 2017;29:18–24. doi: 10.21147/j.issn.1000-9604.2017.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J., Jin B., Chu T., Dong X., Yang H., Zhang Y., Wu D., Lou Y., Zhang X., Wang H., et al. EGFR tyrosine kinase inhibitor (TKI) in patients with advanced non-small cell lung cancer (NSCLC) harboring uncommon EGFR mutations: a real-world study in China. Lung Cancer. 2016;96:87–92. doi: 10.1016/j.lungcan.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe S., Minegishi Y., Yoshizawa H., Maemondo M., Inoue A., Sugawara S., Isobe H., Harada M., Ishii Y., Gemma A., et al. Effectiveness of gefitinib against non-small-cell lung cancer with the uncommon EGFR mutations G719X and L861Q. J. Thorac. Oncol. 2014;9:189–194. doi: 10.1097/JTO.0000000000000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krawczyk P., Kowalski D.M., Ramlau R., Kalinka-Warzocha E., Winiarczyk K., Stencel K., Powrozek T., Reszka K., Wojas-Krawczyk K., Bryl M., et al. Comparison of the effectiveness of erlotinib, gefitinib, and afatinib for treatment of non-small cell lung cancer in patients with common and rare EGFR gene mutations. Oncol. Lett. 2017;13:4433–4444. doi: 10.3892/ol.2017.5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y.L., Hirsh V., Sequist L.V., Hu C.P., Feng J., Lu S., Huang Y., Schuler M., Mok T., Yamamoto N., et al. Does EGFR mutation type influence patient-reported outcomes in patients with advanced EGFR mutation-positive non-small-cell lung cancer? Analysis of two large, Phase III studies comparing afatinib with chemotherapy (LUX-Lung 3 and LUX-Lung 6) Patient. 2018;11:131–141. doi: 10.1007/s40271-017-0287-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oxnard G.R., Lo P.C., Nishino M., Dahlberg S.E., Lindeman N.I., Butaney M., Jackman D.M., Johnson B.E., Janne P.A. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J. Thorac. Oncol. 2013;8:179–184. doi: 10.1097/JTO.0b013e3182779d18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greulich H., Chen T.H., Feng W., Janne P.A., Alvarez J.V., Zappaterra M., Bulmer S.E., Frank D.A., Hahn W.C., Sellers W.R., et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J.Y., Wu S.G., Yang C.H., Gow C.H., Chang Y.L., Yu C.J., Shih J.Y., Yang P.C. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin. Cancer Res. 2008;14:4877–4882. doi: 10.1158/1078-0432.CCR-07-5123. [DOI] [PubMed] [Google Scholar]
- 27.Chen J., Yang H., Teo A.S.M., Amer L.B., Sherbaf F.G., Tan C.Q., Alvarez J.J.S., Lu B., Lim J.Q., Takano A., et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat. Genet. 2020;52:177–186. doi: 10.1038/s41588-019-0569-6. [DOI] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research N Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z., Yang N., Ou Q., Xiang Y., Jiang T., Wu X., Bao H., Tong X., Wang X., Shao Y.W., et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin. Cancer Res. 2018;24:3097–3107. doi: 10.1158/1078-0432.CCR-17-2310. [DOI] [PubMed] [Google Scholar]
- 30.Shu Y., Wu X., Tong X., Wang X., Chang Z., Mao Y., Chen X., Sun J., Wang Z., Hong Z., et al. Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci. Rep. 2017;7:583. doi: 10.1038/s41598-017-00520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang W., Ma Y., Yin J.C., Hong S., Zhou H., Wang A., Wang F., Bao H., Wu X., Yang Y., et al. Comprehensive Genomic profiling identifies novel genetic predictors of response to Anti-PD-(L)1 therapies in non-small cell lung cancer. Clin. Cancer Res. 2019;25:5015–5026. doi: 10.1158/1078-0432.CCR-19-0585. [DOI] [PubMed] [Google Scholar]
- 32.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S., et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337. doi: 10.1016/j.cell.2018.03.035. e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E., et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefebvre C., Bachelot T., Filleron T., Pedrero M., Campone M., Soria J.C., Massard C., Levy C., Arnedos M., Lacroix-Triki M., et al. Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med. 2016;13 doi: 10.1371/journal.pmed.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho J.H., Lim S.H., An H.J., Kim K.H., Park K.U., Kang E.J., Choi Y.H., Ahn M.S., Lee M.H., Sun J.M., et al. Osimertinib for patients with non-small-cell lung cancer harboring uncommon EGFR mutations: a multicenter, open-label, Phase II Trial (KCSG-LU15-09) J. Clin. Oncol. 2020;38:488–495. doi: 10.1200/JCO.19.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi Y., Mitsudomi T. Not all epidermal growth factor receptor mutations in lung cancer are created equal: perspectives for individualized treatment strategy. Cancer Sci. 2016;107:1179–1186. doi: 10.1111/cas.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Veggel B., Van Der Wekken A., Hashemi S., Cornelissen R., Monkhorst K., Heideman D., Radonic T., Schuuring E., Smit E., De Langen J. P2.13-42 osimertinib treatment for patients with EGFR exon 20 insertion positive non-small-cell lung cancer. J. Thorac. Oncol. 2018;13:S815. [Google Scholar]
- 39.Vyse S., Huang P.H. Targeting EGFR exon 20 insertion mutations in non-small cell lung cancer. Signal Transduct. Target. Ther. 2019;4:5. doi: 10.1038/s41392-019-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during this current study are available from the corresponding author on reasonable request.