Summary
Myelodysplastic syndromes (MDSs) are a heterogeneous group of clonal hematopoietic stem cell disorders characterized by myeloid dysplasia, peripheral blood cytopenias, and increased risk of progression to acute myeloid leukemia (AML). The standard of care for patients with MDS is hypomethylating agent (HMA)-based therapy; however, nearly 50% of patients have no response to the treatment. Patients with MDS in whom HMA therapy has failed have a dismal prognosis and no approved second-line therapy options, so enrollment in clinical trials of experimental agents represents these patients’ only chance for improved outcomes. A better understanding of the molecular and biological mechanisms underpinning MDS pathogenesis has enabled the development of new agents that target molecular alterations, cell death regulators, signaling pathways, and immune regulatory proteins in MDS. Here, we review novel therapies for patients with MDS in whom HMA therapy has failed, with an emphasis on the biological rationale for these therapies’ development.
Keywords: myelodysplastic syndromes, hypomethylating agents, secondary acute myeloid leukemia, novel therapeutic agents
Graphical abstract

Highlights
-
•
Novel therapies for patients with MDS after HMA failure are urgently needed
-
•
Exploratory clinical trials hold promise in MDS after HMA failure
-
•
Combination therapies targeting specific stem cell populations are needed
In this review, Rodriguez-Sevilla et al. discuss novel therapies for patients with myelodysplastic syndromes whose disease failed hypomethylating agent therapy, with an emphasis on the biological rationale of current explorative clinical trials of these therapies.
Introduction
Myelodysplastic syndromes (MDSs) arise from a small population of disease-initiating hematopoietic stem cells (HSCs) that persist and expand during conventional therapy and are major contributors to disease progression and relapse.1
Except for allogeneic transplantation, no new curative treatment for MDS has been developed in the last 10 years.2,3 The current standard of care for patients with MDS remains therapy with hypomethylating agents (HMAs), such as 5-azacytidine (5-aza) and its analog 5-aza-2′-deoxycytidine (decitabine), which results in clinical improvements in over 50% of patients.4,5 According to the International Working Group,6 primary HMA failure is defined by the absence of response after at least 4–6 cycles of therapy,7,8,9 when MDSs progress to a higher-risk disease or transform to secondary acute myeloid leukemia (sAML) or when therapy is discontinued as a result of side effects, such as hypoplastic bone marrow (BM) or pancytopenia.10 Secondary response failure, or resistance, is defined by a loss of response or disease progression following an initial response.11
Despite the high frequency of patients with MDS whose disease does not respond to frontline treatment with HMAs or fails HMA therapy after an initial response, there are no approved second-line therapies for these patients, possibly because of the highly heterogeneous mutational and biological landscape driving HMA therapy failure.7
In the last decade, genomic technologies12,13,14,15 coupled with mouse genetic studies16,17,18,19 have greatly improved our understanding of the genetic elements driving MDS initiation and progression and the ways in which such elements functionally contribute to specific aspects of the disease’s pathobiology. These studies have revealed that MDSs are driven by the multistep acquisition of genetic alterations that affect a recurrent set of genes, which promotes the self-renewal of mutant HSCs and leads to their clonal expansion over their normal counterparts.20,21,22
The most common alterations at the onset of MDSs include recurrent somatic mutations that affect the function of the splicing factors SF3B1, SRSF2, U2AF1, and ZRSR223 and the DNA methylation and chromatin modifiers DNMT3A, TET2, and ASXL1.24 Other frequent alterations include cytogenetic aberrations such as the partial deletion of chromosomes 5 (5q) and 20 (20q) or the complete loss of chromosome 7 (monosomy 7) or chromosome 7’s partial deletion involving its long arm.25 Other mutations in MDSs, such as those affecting IDH1/2, RUNX1, GATA2, and CUX1 function, can occur at different times during the course of the disease.26
The progression of MDS to sAML is mostly associated with the expansion of HSC clones carrying preexisting or newly acquired recurrent mutations that affect hematopoietic transcription factor genes that abrogate normal differentiation or HSC clones carrying activating mutations in signaling pathways that control cellular proliferation.21,27,28 The most frequent secondary mutations in MDSs affect the functions of TP53 and RUNX1, the signaling pathway regulators RAS and FLT3, and the cohesion complex components STAG2, SMC1, and SMC329,30,31,32,33 (Figure 1).
Figure 1.
Somatic mutational landscape in MDS
Schematic of the most frequent somatic mutations driving the onset (founder mutations) or progression of MDS to sAML (secondary mutations).
MDS onset can be accelerated by cytotoxic chemotherapy or transplantation after other cancers34, the presence of germline predisposition mutations, such as those affecting telomere maintenance genes,35 the DEAD box protein member DDX41,36 the GATA2 transcriptional factor,37 or any type of stress (e.g., inflammation) that confers a fitness advantage to the mutant clone.38
New agents targeting altered signaling pathways that induce mutant HSCs’ clonal advantage dependently (i.e., in a cell-intrinsic matter) or independently (i.e., in a cell-extrinsic matter) of specific genetic alterations hold promise for overcoming the dismal outcomes patients with MDS experience after HMA therapy failure.
Here, we review novel therapeutic approaches for patients with MDS in whom HMA therapy has failed (Figure 2; Table 1) and discuss the biological and molecular mechanisms supporting these therapies.
Figure 2.
Novel emerging therapies for patients with MDS after HMA failure
Schematic of the experimental agents in clinical trials for patients with MDS whose disease failed HMA therapy. IDH, isocitrate dehydrogenase; TCA, tricarboxylic acid cycle; α-KG, alpha-ketoglutarate; 2-HG, 2-hydroxyglutarate; CTLA-4, cytotoxic T lymphocyte-associated protein 4; PD-1, programmed cell death protein 1; PD-L1/2, programmed death-ligand 1/2; PAMP, pathogen-associated molecular patterns; TLR, Toll-like receptor; MyD88, myeloid differentiation primary response 88; TRIF, TIR-domain-containing adaptor-inducing interferon-β; NF-κB, nuclear factor kappa B; IRFs, interferon regulatory factors; BCL2, B cell lymphoma 2; MCL1, myeloid cell leukemia 1; NEDD8, neuronal precursor cell expressed developmentally downregulated protein-8. IRAK4, interleukin-1 receptor-associated kinase.
Table 1.
Summary of the ongoing therapeutic approaches for patients with MDS whose disease failed HMA therapy
| Therapy | Target | Clinical trial phase | Number of patients enrolled | Patient characteristics | Type of response | NCT number/reference |
|---|---|---|---|---|---|---|
| Therapies targeting anti-apoptotic proteins | ||||||
| Venetoclax + azacitidine | BCL2 | I/II, R | 12 | IPSS-R: - INT: 16% - high: 42% - very high: 42% |
ORR: 75% CR (n = 1) mCR (n = 7) mOS: 8.5 months |
NCT04550442 Desikan et al.39 |
| Venetoclax + azacitidine | BCL2 | 1b ANR |
44 | IPSS-R: - low: 9% - INT: 18% - high: 37% - very high: 36% |
ORR: 39.6% CR: 6.8% mCR: 31.8% mPFS: 8.6 months mOS: 12.6 months |
NCT02966782 Zeidan et al.40 |
| Venetoclax + azacitidine | BCL2 | I/II R |
2 | IPSS: - INT-2: 75% - high: 25% |
ORR: 75% mCR: 75% SD: 25% |
NCT04160052 Morita et al.41 |
| NEDDylation inhibitors | ||||||
| Pevonedistat + azacitidine | NEDD8-activating enzyme | II ANR |
21 | IPSS: - INT-2/high: 65% |
ORR: 42.9% CR (n = 1) mCR (n = 4) DoR: 8.7 months |
NCT03238248 Moyo et al.42 |
| Signal transduction inhibitors | ||||||
| Rigosertib ± low-dose cytarabine | RAS PI3K PLK | III C |
199 | IPSS-R: - low: 1% - INT: 7% - high: 34% - very high: 47% - unknown: 11% |
mOS: - global: 8.2 months - primary failure: 8.6 months - IPSS-R very high: 7.6 months |
NCT01241500 Garcia-Manero et al.43 |
| Rigosertib | RAS PI3K PLK | III ANR |
240 | NA | ITT mOS: 6.4 months |
NCT02562443 Onconova-Therapeutics et al.44 |
| Rigosertib + azacitidine | RAS PI3K PLK | II C |
17 | IPSS-R: - low: 4% - INT: 19% - high: 31% - very high: 45% - unknown: 1% |
ORR: 59% |
NCT01926587 Navada et al.45 |
| Isocitrate dehydrogenase inhibitors | ||||||
| Ivosidenib | IDH1 | I R |
16 | NS | ORR: 81% CR: 44% mCR: 31% DoR at 12 months: 60% |
NCT02074839 Sallman et al.46 |
| Ivosidenib | IDH1 | II R |
13 | NA | ORR: 54% CR: 23% mOS: 7.7 months |
NCT03503409 Sebert et al.47 |
| Enasidenib | IDH2 | I/II ANR |
13 | IPSS: - INT-2/high: 53% IPSS-R: - high/very high: 53% |
ORR: 46% | NCT01915498Stein et al.48 |
| Enasidenib | IDH2 | II ANR |
23 | IPPS-R: - INT: 35% - low: 13% - high: 30% - very high: 22% |
ORR: 35% CR: 22% |
NCT03383575 DiNardo et al.49 |
| Enasidenib | IDH2 | II R |
11 | NA | ORR: 27% CR: 18% 1 year OS: 55.4% |
NCT03744390 Ades et al.50 |
| Olutasidenib FT-2102 + azacitidine | IDH2 | I/II ANR |
8 | NA | ORR: 44% CR: 11% mCR: 33% |
NCT02719574 Jorge et al.51 |
| Inflammation pathway inhibitors | ||||||
| Tomaralimab | TLR2 | I/II C |
51 | NA | ORR: 50% major responders: 27% minor responders: 23% |
NCT02363491 Garcia-Manero et al.52 |
| Emavusertib | IRAK4 | I/II R |
7 | NA | ORR: 57% mCR: 57% |
NCT04278768 Garcia-Manero et al.53 |
| Immune checkpoint inhibitors | ||||||
| Pembrolizumab + azacitidine | PD-1 | II ANR |
20 | IPSS: - INT-1: 65% - INT-2: 20% - high: 15% |
ORR: 25% CR: 5% mCR: 10% HI: 10% |
NCT03094637Chien et al.54 |
| Pembrolizumab | PD-1 | 1b C |
28 | IPSS-R: - low: 11% - INT: 25% - high: 14% - very high: 43% - unknown: 7% |
ORR: 0% mCR: 19% SD: 44% mOS: 6 months |
NCT01953692Garcia-Manero et al.55 |
| Nivolumab and/or ipilimumab ± azacitidine | PD-1 CTLA-4 |
II R |
35 | NA | ORR: - nivolumab: 13% - ipilimumab: 35% CR: - ipilimumab: 15% mEFS: 7.1 months mOS: 8 months |
NCT02530463Garcia-Manero et al.56 |
| Nivolumab and/or ipilimumab ± azacitidine | PD-1 CTLA-4 |
basket exploratory phase II | 11 | IPSS: - INT-1: 46% - INT-2: 18% - high: 36% |
ORR: 36% CR: 9% CRi: 9% HI: 18% mPFS: 7.1 months mOS: 11.4 months |
NCT02530463Morita et al.57 |
| Atezolizumab ± azacitidine | PD-L1 | 1b C |
25 cohort A: 11cohort B: 14 |
IPSS-R: - low: 4% - INT: 12% - NA: 84% |
cohort A: - ORR: 0% - mPFS: 4.5 months - mOS: 8.7 months cohort B: - ORR: 14.3% - mPFS: 7.9 months - mOS: 11.9 months |
NCT02508870Gerds et al.58 |
ANR, active not recruiting; BCL2, B cell lymphoma 2; C, completed; CR, complete response; CRi, complete remission with incomplete count recovery; CTLA-4, cytotoxic T lymphocyte-associated protein 4; DoR, duration of response; HI, hematologic improvement; HMA, hypomethylating agent; IDH, isocitrate dehydrogenase; INT, intermediate; IPSS, International Prognostic Scoring System; IPSS-R, Revised International Prognostic Scoring System; IRAK4, interleukin-1 receptor-associated kinase 4; ITT, intention-to-treat; mCR, marrow complete response; MDS, myelodysplastic syndrome; mEFS, median event-free survival; mOS, median overall survival; mPFS, median progression-free survival; NA, not available; NCT, national clinical trial; NEDD8, neuronal precursor cell-expressed developmentally downregulated-8; NS, not specified; ORR, overall response rate; OS, overall survival; PD-1, programmed cell death 1; PD-L1, programmed cell death ligand-1; PI3K, phosphatidylinositol 3-kinase; PLK, Polo-like kinase 1; R, recruiting; SD, stable disease; TLR2, Toll-like receptor 2.
Therapies targeting anti-apoptotic proteins
Apoptosis is a form of programmed cell death that eliminates unwanted or damaged cells during embryonic development and tissue homeostasis. Apoptosis occurs through either a death receptor-mediated extrinsic pathway or a mitochondria-dependent intrinsic pathway. The intrinsic pathway is activated by several exogenous and endogenous stimuli, which include DNA damage, UV and gamma radiation, hypoxia, oxidative stress, and hormone or cytokine deprivation. The effector components of the intrinsic pathway are the BCL2 family of proteins. BCL2 family members are divided into 3 groups based on their function and number of BCL2 homology (BH) domains: (1) anti-apoptotic members with 4 BH domains (BCL2, BCLxL, BCLw, BCL2A1, BCL2L10, and MCL1); (2) pro-apoptotic members with 4 BH domains (BAX, BAK, and BOK); and (3) pro-apoptotic members with 3 BH domains (BAD, BID, BIK, BIM, BMF, HRK, NOXA, and PUMA).59,60 The activation of pro-apoptotic BCL2 proteins induces the permeabilization of the mitochondrial outer membrane, which elicits the release of the cytochrome complex in the cytoplasm and the activation of caspase, the final regulators of apoptotic cell dismantling. In contrast, the activation of anti-apoptotic BCL2 proteins inhibits cytochrome complex release, thereby preventing apoptosis.61
Venetoclax (ABT199) is a highly selective BCL2 inhibitor that promotes the oligomerization of BAX and BAK, thus inducing apoptosis62,63,64 (Figure 3). Since its approval by the US Food and Drug Administration (FDA) in 2019, venetoclax has substantially improved the treatment outcomes of patients with AML. In older patients with AML who were not otherwise candidates for more aggressive treatments, such as intensive induction chemotherapy, venetoclax combined with low-dose cytarabine yielded high response rates and long remission durations.65 Moreover, among previously untreated patients with AML who were ineligible for intensive chemotherapy, those who received 5-aza plus venetoclax had a longer overall survival duration and a higher remission rate than those who received 5-aza alone.66
Figure 3.
Mechanisms of action of venetoclax and AMG 176
The anti-apoptotic proteins BCL2 and MCL1 bind and sequester the apoptotic effectors BAX/BAK to prevent these proteins’ oligomerization and subsequent induction of apoptosis. Venetoclax or AMG 176 selectively binds to the BH3 domain of BCL2 and MCL1, respectively, allowing for the release of pro-apoptotic proteins, which results in the permeabilization of the mitochondrial outer membrane, cytochrome c release, and caspase cascade activation.
In a phase 1b multicenter clinical trial (ClinicalTrials.gov: NCT02966782) evaluating the safety and efficacy of venetoclax alone or in combination with 5-aza in patients with relapsed/refractory (R/R) MDS, the combination cohort had an overall response rate (ORR) of 39%.40 Moreover, results emerging from a single-center phase I/II clinical trial (ClinicalTrials.gov: NCT04160052) confirmed the potential benefit of combining venetoclax with 5-aza in the treatment of patients with R/R MDS.67 Although these clinical trials were designed to identify the optimal dose of the combination therapy and evaluate its clinical activity and safety, they also offered an unparalleled opportunity to gain insight into the biological mechanisms of venetoclax response and resistance in MDS.
Indeed, the findings of our previous study provide a rationale for using different therapeutic approaches depending on the architecture of MDS HSCs to overcome MDS progression. We performed integrative molecular profiling of hematopoietic stem and progenitor cells (HSPCs) from more than 400 samples from patients with MDS and found that MDS HSCs in 2 differentiation states—long-term HSCs or lymphoid-primed multipotent progenitors (LMMPs)—give rise to 2 distinct patterns of progenitor differentiation, either a “common myeloid progenitor (CMP) pattern” or a “granulocytic-monocytic progenitor (GMP) pattern.”68 These 2 HSPC architectures were maintained throughout HMA therapy and expanded at progression after HMA therapy failure, depending on the recurrent activation of BCL2-or nuclear factor κB (NF-κB)-mediated survival pathways, respectively. Pharmacologically inhibiting these pathways depleted MDS HSCs and reduced tumor burden in experimental systems. Overall, we found that the HSC architectures in MDSs are potential predictive biomarkers for guiding the design or choice of specific therapeutic approaches targeting these cells.68 We then performed a preliminary validation of our preclinical studies in the setting of clinical trials of venetoclax-based therapy, hypothesizing that targeting BCL2 with venetoclax could elicit a durable response in patients with “CMP pattern” MDS. Consistent with our hypothesis, among patients with MDS whose disease progressed after the failure of frontline HMA-based therapy, those with “CMP pattern” HSPC architecture who were treated with venetoclax had a shorter cumulative time to complete remission (p = 0.018) and a longer relapse-free survival duration than those with “GMP pattern” HSC architecture (16.3 vs. 5.2 months). Together, these results suggest that the cellular architecture of MDS should be considered a biomarker for predicting the intrinsic vulnerabilities of the cells that expand at relapse and thus for guiding the design or choice of specific therapeutic approaches targeting these cells, particularly in the setting of venetoclax-based therapy.68
We also observed that patients with the “GMP pattern” HSC architecture overexpressed the downstream NF-κB pathway effector and anti-apoptotic regulator MCL1, which explains why this group of patients is resistant to venetoclax treatment. These data suggest that targeting MCL1, but not BCL2, activity can overcome HMA therapy resistance in these patients. Consistent with this hypothesis, recent preclinical studies showed that drug synergy from combining BCL2 and MCL1 inhibitors was achieved across all subtypes and mutational profiles of MDS, even in the presence of RAS family mutations that conferred resistance to venetoclax monotherapy in AML and chronic myelomonocytic leukemia (CMML).69 Based on these results, we recently opened a phase I clinical trial of the MCL1 inhibitor AMG 176 in combination with 5-aza in patients with MDS with R/R disease after HMA therapy failure (ClinicalTrials.gov: NCT05209152) (Figure 3). After the first phase of this trial establishes the optimal biological dose/minimum safe biologically effective dose, the study will be extended to venetoclax-naive and venetoclax-exposed patients with R/R MDS.
Whereas the molecular and biological mechanisms underpinning AML resistance to venetoclax have recently been recently elucidated,70,71,72 we still do not know why patients with MDS whose disease failed HMA therapy acquire secondary resistance to venetoclax after an initial response. Preliminary data from our group showed that “CMP pattern” MDS that initially responded to therapy underwent clonal evolution and acquired previously undetectable mutations (e.g., STAG2) that changed the cellular architecture of the disease and induced expansion of LMPPs depending on NF-κB pathway activation to maintain survival (S.C., unpublished data). These data suggest that combination therapies targeting specific HSPC populations are needed to overcome venetoclax-resistance in MDS.
NEDDylation inhibitors
Intracellular protein production, degradation, and clearance are controlled by a programmed turnover that maintains cellular homeostasis. NEDDylation is a multistep enzymatic process initiated by the NEDD8-activating enzyme (NAE), which conjugates a ubiquitin-like molecule, neuronal precursor cell-expressed developmentally downregulated-8 (NEDD8), to a conserved lysine residue and promotes protein degradation.73 The most characterized target of NEDDylation is the cullin-RING E3 ubiquitin ligase (CRL).74 The conjugation of NEDD8 and CRL is catalyzed by NAE, which activates CRL’s ligase activity and facilitates the ubiquitination of multiple substrates involved in cycle progression (e.g., p21, p27, cyclin E, c-Myc), DNA damage, tumor suppression (e.g., TP53), and stress responses. Different types of cancer, including AML and MDS,75 have increased levels of NEDD8 and NAE, which promote the growth of tumor cells and their evasion of programmed cell death.76
Agents targeting NEDDylation include pevonedistat (MLN4924), a small molecule that targets NAE’s adenylation active site, thus triggering multiple cellular responses, including cell-cycle arrest, apoptosis, senescence, and autophagy. Previous studies showed that pevonedistat decreases AML cells’ colony-forming ability and induces their apoptosis in vitro77 and demonstrated that the combination of pevonedistat and 5-aza synergistically induces AML cells’ apoptosis in xenograft models.78 A phase 1b clinical trial (ClinicalTrials.gov: NCT01814826) showed that the combination of pevonedistat and 5-aza was well tolerated in older patients with AML who were ineligible for high-dose induction therapy, and the timing and frequency of these patients’ responses suggested that the addition of pevonedistat to a standard regimen of single-agent 5-aza has a greater therapeutic benefit than 5-aza alone.79 The combination therapy is now being evaluated in a phase II clinical trial (ClinicalTrials.gov: NCT03238248) in patients with MDS in whom HMA therapy has failed.42 Preliminary results from 21 patients with R/R MDS demonstrated an ORR of 43%. Several clinical trials of pevonedistat in combination with other agents (e.g., belinostat, fludarabine, cytarabine) in patients with R/R MDS are also ongoing (ClinicalTrials.gov: NCT03772925, NCT03813147, and NCT03459859).
Signal transduction inhibitors
Signaling transduction pathways are disrupted in cancer cells owing to the activation of oncogenes or the inactivation of tumor-suppressor genes.80 The hyperactivation of some specific signal transduction pathways can cause aberrant cell-cycle progression and promote tumor cells’ proliferation, differentiation, and survival. Therefore, agents that target signaling transduction regulators could provide new treatments for patients with a broad range of cancers, including MDS.
Among transduction regulators, receptor tyrosine kinases (RTKs)81 are cytoplasmatic membrane receptors with an intracellular catalytic domain that is involved in signal transduction. The interaction of signaling molecules (typically growth factors) with RTKs’ extracellular region induces intracellular dimerization and activates tyrosine kinase domains (TKDs), which bind to specific phosphotyrosine residues within the receptor and engage downstream effectors that propagate critical cellular signaling pathways.82 The most frequently altered oncogenic signaling pathway in cancer cells is the RTK-RAS cascade.83,84 Once activated, RAS initiates different signaling responses, including the mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) pathways, which promote cancer development by activating oncogenic transcription, cell-cycle progression, cellular survival, cell growth and metabolism, and cell motility and migration.85 RAS’s active state depends on its intrinsic ability to bind and hydrolyze GTP via the conserved G-domain. RAS GTPase activity is mediated by the guanine nucleotide exchange factor/GTPase-activating protein system, which regulates the shift between inactive GDP-RAS and active GTP-RAS. Gain-of-function mutations in HRAS, KRAS, or NRAS, which are common in several types of cancer, constitutively maintain RAS in its active form, thus enhancing the proliferation and survival of mutant cells over those of their normal counterparts.86
RAS pathway-activating mutations are rare in patients with newly diagnosed MDS but occur in about 20% of patients with MDS whose disease has progressed to sAML.31 Therefore, therapeutic approaches that target RAS signaling by inhibiting RAS’s enzymatic activity or its effectors and regulators are under investigation in patients with MDS who have a higher risk of disease progression. For example, the small molecule rigosertib,87 a synthetic benzyl styryl sulfone, blocks the interaction between RAS and the RAS-binding domain of RAF, one of its effectors, thus inhibiting the activation of the downstream MAPK and PI3K/AKT signaling cascades and inducing cell-cycle arrest and apoptosis.88 However, in a phase III clinical trial (ClinicalTrials.gov: NCT01241500) in patients with MDS with excess blasts after HMA therapy failure, the addition of rigosertib to best supportive care did not significantly improve overall survival compared with best supportive care alone.43 Although these results were disappointing, other inhibitors directly targeting RAS mutations, such as the newly approved KRAS inhibitor MRTX1133,89 hold promise for patients with KRAS-mutant cancer, including those with MDS in whom HMA therapy has failed.
Another RTK that plays a key role in controlling the survival, proliferation, and differentiation of hematopoietic cells is FMS-like tyrosine kinase 3 (FLT3). FLT3 has 5 immunoglobulin-like domains in the extracellular region, a transmembrane domain, a juxtamembrane domain, and 2 intracellular TKDs linked by a kinase insert.90 In physiological conditions, FLT3 is in a monomeric inactive form. The binding of FLT3 with its ligand, FLT3L, induces FLT3’s dimerization, which promotes the phosphorylation of TKDs and leads to the activation of downstream signal transduction networks through the PI3K and RAS signaling cascades.91
FLT3 gene alterations that cause the constitutive phosphorylation of the TKD are recurrent genetic abnormalities in AML.92 They include FLT3 internal tandem duplications (FLT3-ITDs) within the juxtamembrane domain as well as point mutations that mainly involve the TKD (FLT3-TKD mutations).93 In AML, FLT3-ITDs are predictors of poor prognosis, particularly in terms of relapse-free and overall survival, whereas FLT3-TKD mutations are not associated with a prognostic impact.94 FLT3-ITDs and FLT3-TKD mutations at MDS onset are infrequent, occurring in less than 1% of patients, but they are present in up to 10% of patients at disease progression to sAML. FLT3 alterations predict a shorter duration to sAML transformation and very poor outcomes.95
FLT3 inhibitors are divided into first-generation multikinase inhibitors (e.g., sorafenib, lestaurtinib, midostaurin) and next-generation inhibitors (e.g., quizartinib, crenolanib, gilteritinib).93 However, given the limited number of ongoing clinical trials of FLT3 inhibitors in patients with R/R MDS with FLT3 genetic alterations (ClinicalTrials.gov: NCT03661307, NCT04493138, and NCT01892371), we do not yet understand whether these patients would benefit from these agents.
Isocitrate dehydrogenase inhibitors
Isocitrate dehydrogenase (IDH), a key enzyme in the tricarboxylic acid cycle, reversibly catalyzes the conversion of isocitrate to α-ketoglutarate (α-KG). Up to 20% of patients with newly diagnosed AML96 and 4%–12% of patients with newly diagnosed MDS97 carry IDH1/2 heterozygous gain-of-function mutations affecting the amino acids R132, R172, and R140.97,98 These mutations inhibit the conversion of isocitrate to α-KG and lead to the acquisition of neomorphic IDH1/2, whose enzymatic activity instead reduces α-KG to 2-hydroxyglutarate (2-HG),99,100 an oncogenic protein that competitively inhibits α-KG-dependent dioxygenases, including TET2 and histone demethylases.101 High levels of 2-HG induce the hypermethylation of histones and DNA, thus changing epigenetic patterns and significantly impairing hematopoietic differentiation.102
IDH1/2 mutations mainly occur in patients with MDS in whom previous therapies have failed, which suggests that these mutations drive MDS progression.103 These patients’ survival outcomes have improved greatly with the recent development of therapies targeting mutant IDH1/2 proteins. The oral small molecules enasidenib (AG-221) and ivosidenib (AG-120) inhibit the production of 2-HG in IDH2- and IDH1-mutant cells, respectively, and lead to the differentiation of mutant cells without inducing BM aplasia. Enasidenib was approved by the FDA in August 2017 after a phase I/II trial in 239 patients with R/R IDH2-mutated AML (ClinicalTrials.gov: NCT01915498), including 30 patients whose disease progressed from MDS.104 Therapy with enasidenib was associated with an ORR of 40%, and the ORR of patients with IDH2R172 mutations (53.3%) was higher than that of patients with IDH2R140 mutations (35.4%). More recently, a phase II trial (ClinicalTrials.gov: NCT03383575) in patients with MDS and IDH2 mutations showed that enasidenib in combination with 5-aza is an effective option for treatment-naive patients with high-risk MDS and that enasidenib alone is an effective option for patients with MDS in whom HMA therapy has failed.49 Ivosidenib received FDA approval in July 2018 after a phase I clinical trial (ClinicalTrials.gov: NCT02074839) demonstrated that in patients with advanced R/R IDH1-mutated AML (n = 179), ivosidenib had few treatment-related adverse events, seldom resulted in transfusion independence, and elicited durable remissions, including molecular remissions in some patients who had complete remission.105
Several new IDH inhibitors for the treatment of patients with MDS with IDH1/2 mutations are under investigation. For example, an ongoing phase I/II clinical trial (ClinicalTrials.gov: NCT02719574) is evaluating the safety and efficacy of olutasidenib (FT-2102),106 an orally active, highly potent, selective inhibitor of mutant IDH1, alone or in combination with 5-aza or cytarabine in patients with R/R MDS or AML.
Although IDH1/2 inhibitors improved the outcomes of patients with AML and MDS with IDH1/2 mutations, these agents cannot cure the disease, probably because IDH1/2 mutations are frequently subclonal and cooperate with other mutations (e.g., SRSF2 mutations) to induce the leukemic phenotype.107 Thus, to improve the overall survival of these patients, future studies should be designed to identify combination therapies of IDH1/2 inhibitors with agents targeting these mutations’ cooperative pathways.
Inflammation pathway inhibitors
The dysregulation of innate immune and inflammatory signaling is a hallmark of MDS.108,109 To escape immune recognition, malignant cells employ several mechanisms, including losing their antigenicity and/or immunogenicity and inducing an immunosuppressive tumor microenvironment.110 Aberrant activation of enhanced Toll-like receptor (TLR) signaling contributes to ineffective hematopoiesis111 and has emerged as a potential therapeutic opportunity in the treatment of MDS.112,113
TLRs are a family of transmembrane pattern-recognition receptors that initiate innate immune responses.114 TLRs are activated by recognizing non-self-danger signals from invading pathogens (pathogen-associated molecular patterns) or self-danger signals from damage-associated molecular patterns (endogenous damage-associated molecular patterns). The 10 different TLRs in humans are expressed on different immune cell types (neutrophils, macrophages, dendritic cells, natural killer cells, and B and T cells), non-immune cell types (fibroblasts and endothelial and epithelial cells), and HSPCs. The interaction between TLRs and their ligands induces the dimerization of the Toll/interleukin-1 receptor’s cytosolic domain, which leads to the recruitment of adaptor proteins.115 These adaptor proteins include myeloid differentiation primary response 88 (MyD88), which activates the innate immune signaling cascade mediated by the IRAK family kinases, and the Toll/interleukin-1 receptor domain-containing adaptor-inducing interferon-β, which activates interferon signaling and the production of inflammatory cytokines.116
TLRs are often overexpressed in cancer cells and can either promote or inhibit tumor progression depending on the cellular context. MDS HSPCs overexpress TLR2, TLR4, TLR6, and MyD88. TLR2 agonists activate the histone demethylase JMJD3, which enhances NF-κB activity and significantly decreases erythropoiesis.113 These results led to a phase I/II clinical trial (ClinicalTrials.gov: NCT02363491) of tomaralimab (OPN-305), a fully humanized immunoglobulin G4 (IgG4) monoclonal antibody against TLR2, in transfusion-dependent patients with low- or intermediate-risk MDS in whom HMA therapy had failed. These heavily pretreated patients had an ORR of 50%, which suggests that targeting TLR2 can improve erythropoiesis in this cohort.52
Interleukin-1 receptor-associated kinase 4 (IRAK4) is a key mediator of TLR and interleukin-1 receptor-induced NF-κB signaling pathway activation and triggers inflammatory responses and survival mechanisms in many types of cancer cells.117 Preclinical studies in MDS and AML showed that U2AF1 and SF3B1 mutations lead to aberrant splicing of IRAK4, which results in a longer isoform retaining exon 4 (IRAK4-L). Compared with the shorter IRAK4 isoform, IRAK4-L more significantly actives the NF-κB pathway and NF-κB’s downstream innate immune signaling. The genetic inhibition of IRAK4-L expression induces differentiation of AML cells and decreases tumoral burden in in vivo experiments.118,119 Therefore, IRAK4 inhibitors are promising therapeutic options to target the innate immune system.120 Further preclinical studies also revealed that IRAK signaling is a mechanism of adaptive resistance in the setting of FLT3-mutant AML.121 These results led to a phase I/II trial (ClinicalTrials.gov: NCT04278768) of Emavusertib (CA-4948), a novel oral IRAK4 inhibitor with dual targeting of IRAK4 and FLT3 in patients with R/R MDS/AML. Preliminary results from this trial showed that CA-4948 as a single agent or in combination with 5-aza or venetoclax has efficacy in patients with SF3B1, U2AF1, or FLT3 mutations. Importantly, in the 7 patients with MDS with spliceosome mutations, 57% reached marrow complete remission, including 1 with red blood cell transfusion independence.53
Immune checkpoint inhibitors
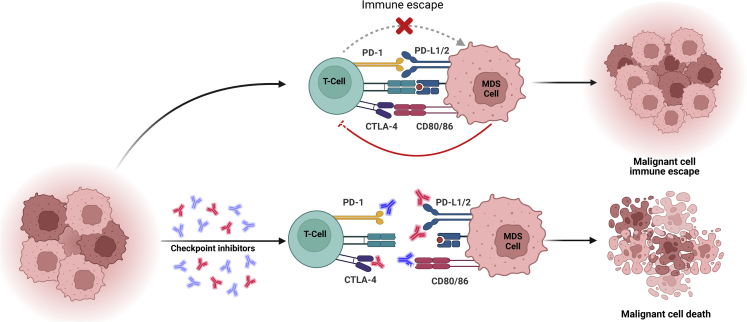
The maintenance of immune homeostasis along the T cell lifespan is crucial to preventing autoimmunity.122 Checkpoint proteins, which are based on ligand-receptor pairs and are expressed on immune cells, tumor cells, and other types of cells, may have inhibitory or stimulatory effects on immune responses123(Figure 4). Among the various immune checkpoints that are abnormally expressed in tumor cells, cytotoxic T lymphocyte-associated protein 4 (CTLA-4) and programmed cell death 1 (PD-1), which are negative regulators of T cell activity, are the most studied.124 Immune checkpoint inhibitors that block the function of CTLA-4 or PD-1—and thus enhance T cell responses against tumor cells—have revolutionized cancer therapy over the last 15 years.125
Figure 4.
Mechanisms of action of immune checkpoints
The expression of checkpoint proteins, such as PD-1, PD-L1, and CTLA-4, is upregulated in MDS cells and leads to T cell functional exhaustion and abrogation of the anti-tumoral response. Treatment with antibodies targeting PD-1/PD-L1 or CTLA-4 overcomes immune response inhibition and enhances antitumor activity.
In CD34+ MDS cells, CTLA-4, PD-1, and PD-ligand-L1/2 (PD-L1/2) are upregulated and induce immune surveillance evasion.108 In addition, HMA therapy upregulates PD-1 and PD-L1/2, which suggests that they may have a role in therapy resistance.126 Several clinical trials are investigating the effects of PD-1 or CTLA-4 blockade in patients with MDS in whom HMA therapy has failed. A phase 1b clinical trial (ClinicalTrials.gov: NCT01953692) of pembrolizumab (MK3475), a humanized IgG4 monoclonal antibody that blocks PD-1’s interaction with its ligand, PD-L1, showed that the antibody did not significantly improve the ORR of patients in whom HMA therapy had failed. However, among the overall cohort of patients, those with an intermediate risk had an improved 2 year ORR of 46%.55 A similar phase II clinical trial confirmed that pembrolizumab in combination with 5-aza (ClinicalTrials.gov: NCT03094637) elicited a modest ORR overall but improved the survival of patients with intermediate-risk disease,54 which suggests that although targeting PD-1 cannot prevent the poor outcomes of patients with MDS after HMA failure, it may have some clinical activity in certain subgroups of patients. For example, an exploratory phase II basket trial (ClinicalTrials.gov: NCT02530463) showed that the combination of the checkpoint inhibitors ipilimumab and nivolumab with or without 5-aza had some clinical activity with tolerable safety profiles in patients with R/R MDS.57 However, a larger study with longer follow up is needed to clarify the efficacy of this regimen and identify biomarkers of response or resistance to these agents.
Conclusions
The development of more effective therapies for patients with MDS whose disease failed HMA therapy requires an improved understanding of how MDS HSCs contribute to therapy failure and disease progression. Studies employing advanced sequencing technologies have shown that aberrant MDS cells that reside in the immunophenotypically defined HSC compartment drive the progression of MDS to sAML,127 which suggests that targeting these cells is the only way to improve the outcomes of patients whose MDS has progressed to higher-risk disease.
Emerging therapies aimed at improving the overall survival of patients with R/R MDS have shown promising clinical efficacy, but none has been shown to overcome these patients’ dismal prognosis, likely because almost all HSCs are mutated at the onset of MDS.68 Thus, future efforts should focus on developing therapeutic strategies that prevent MDS progression or target early-stage MDS, such as clonal cytopenia of undetermined significance, when the mutational burden is low and symptoms are minimal.
Acknowledgments
This work was supported by philanthropic contributions to The University of Texas MD Anderson Cancer Center’s AML/MDS Moon Shot and by the Edward P. Evans Foundation. S.C. is a scholar of the Leukemia and Lymphoma Society. The authors thank Joe Munch at MD Anderson’s Research Medical Library and Kelly Soltysiak for editing the manuscript and Dr. Irene Ganan-Gomez for helpful suggestions. Figures were made using Biorender.com.
Author contributions
J.J.R.-S., V.A., and S.C. wrote the manuscript. G.G.-M. made critical intellectual contributions throughout the manuscript. S.C. designed and finalized the study.
Declaration of interests
G.G.-M. declares research support and an advisory role at Bristol Myers Squibb, Astex, and Helsinn and research support from Amphivena, Novartis, AbbVie, H3 Biomedicine, Onconova, and Merck.
References
- 1.Shastri A., Will B., Steidl U., Verma A. Stem and progenitor cell alterations in myelodysplastic syndromes. Blood. 2017;129:1586–1594. doi: 10.1182/blood-2016-10-696062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Platzbecker U. Treatment of MDS. Blood. Blood. 2019;133:1096–1107. doi: 10.1182/blood-2018-10-844696. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Manero G., Chien K.S., Montalban-Bravo G. Myelodysplastic syndromes: 2021 update on diagnosis, risk stratification and management. Am. J. Hematol. 2020;95:1399–1420. doi: 10.1002/ajh.25950. [DOI] [PubMed] [Google Scholar]
- 4.Fenaux P., Mufti G.J., Hellstrom-Lindberg E., Santini V., Finelli C., Giagounidis A., Schoch R., Gattermann N., Sanz G., List A., et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–232. doi: 10.1016/s1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kantarjian H., Issa J.P.J., Rosenfeld C.S., Bennett J.M., Albitar M., DiPersio J., Klimek V., Slack J., de Castro C., Ravandi F., et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 6.Komrokji R.S., Al Ali N.H., Sallman D., Padron E., DeZern A.E., Barnard J., Roboz G.J., Garcia-Manero G., List A., Steensma D.P., Sekeres M.A. Validation of International Working Group response criteria in higher-risk myelodysplastic syndromes: a report on behalf of the MDS Clinical Research Consortium. Cancer Med. 2021;10:447–453. doi: 10.1002/cam4.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheson B.D., Greenberg P.L., Bennett J.M., Lowenberg B., Wijermans P.W., Nimer S.D., Pinto A., Beran M., de Witte T.M., Stone R.M., et al. Clinical application and proposal for modification of the International Working Group (IWG) response criteria in myelodysplasia. Blood. 2006;108:419–425. doi: 10.1182/blood-2005-10-4149. [DOI] [PubMed] [Google Scholar]
- 8.Jabbour E., Garcia-Manero G., Batty N., Shan J., O'Brien S., Cortes J., Ravandi F., Issa J.P., Kantarjian H. Outcome of patients with myelodysplastic syndrome after failure of decitabine therapy. Cancer. 2010;116:3830–3834. doi: 10.1002/cncr.25247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prébet T., Gore S.D., Esterni B., Gardin C., Itzykson R., Thepot S., Dreyfus F., Rauzy O.B., Recher C., Adès L., et al. Outcome of high-risk myelodysplastic syndrome after azacitidine treatment failure. J. Clin. Oncol. 2011;29:3322–3327. doi: 10.1200/JCO.2011.35.8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jabbour E.J., Garcia-Manero G., Strati P., Mishra A., Al Ali N.H., Padron E., Lancet J., Kadia T., Daver N., O'Brien S., et al. Outcome of patients with low-risk and intermediate-1-risk myelodysplastic syndrome after hypomethylating agent failure: a report on behalf of the MDS Clinical Research Consortium. Cancer. 2015;121:876–882. doi: 10.1002/cncr.29145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santini V. How I treat MDS after hypomethylating agent failure. Blood. 2019;133:521–529. doi: 10.1182/blood-2018-03-785915. [DOI] [PubMed] [Google Scholar]
- 12.Bejar R., Stevenson K., Abdel-Wahab O., Galili N., Nilsson B., Garcia-Manero G., Kantarjian H., Raza A., Levine R.L., Neuberg D., Ebert B.L. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haferlach T., Nagata Y., Grossmann V., Okuno Y., Bacher U., Nagae G., Schnittger S., Sanada M., Kon A., Alpermann T., et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernard E., Tuechler H., Greenberg P.L., Hasserjian R.P., Arango Ossa J.E., Nannya Y., Devlin S.M., Creignou M., Pinel P., Monnier L., et al. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evidence. 2022;1 doi: 10.1056/EVIDoa2200008. [DOI] [PubMed] [Google Scholar]
- 15.Ogawa S. Genetic basis of myelodysplastic syndromes. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2020;96:107–121. doi: 10.2183/pjab.96.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim E., Ilagan J.O., Liang Y., Daubner G.M., Lee S.C.-W., Ramakrishnan A., Li Y., Chung Y.R., Micol J.-B., Murphy M.E., et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27:617–630. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obeng E.A., Chappell R.J., Seiler M., Chen M.C., Campagna D.R., Schmidt P.J., Schneider R.K., Lord A.M., Wang L., Gambe R.G., et al. Physiologic expression of Sf3b1K700E causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. 2016;30:404–417. doi: 10.1016/j.ccell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayle A., Yang L., Rodriguez B., Zhou T., Chang E., Curry C.V., Challen G.A., Li W., Wheeler D., Rebel V.I., Goodell M.A. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125:629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z., Cai X., Cai C.-L., Wang J., Zhang W., Petersen B.E., Yang F.-C., Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sperling A.S., Gibson C.J., Ebert B.L. The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat. Rev. Cancer. 2017;17:5–19. doi: 10.1038/nrc.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makishima H., Yoshizato T., Yoshida K., Sekeres M.A., Radivoyevitch T., Suzuki H., Przychodzen B., Nagata Y., Meggendorfer M., Sanada M., et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat. Genet. 2017;49:204–212. doi: 10.1038/ng.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guess T., Potts C.R., Bhat P., Cartailler J.A., Brooks A., Holt C., Yenamandra A., Wheeler F.C., Savona M.R., Cartailler J.-P., Ferrell P.B., Jr. Distinct patterns of clonal evolution drive myelodysplastic syndrome progression to secondary acute myeloid leukemia. Blood Cancer Discov. 2022;3:316–329. doi: 10.1158/2643-3230.Bcd-21-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M., et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy J.A., Ebert B.L. Clinical implications of genetic mutations in myelodysplastic syndrome. J. Clin. Oncol. 2017;35:968–974. doi: 10.1200/jco.2016.71.0806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haase D. Cytogenetic features in myelodysplastic syndromes. Ann. Hematol. 2008;87:515–526. doi: 10.1007/s00277-008-0483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogawa S. Genetics of MDS. Blood. 2019;133:1049–1059. doi: 10.1182/blood-2018-10-844621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corces-Zimmerman M.R., Hong W.J., Weissman I.L., Medeiros B.C., Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. USA. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.da Silva-Coelho P., Kroeze L.I., Yoshida K., Koorenhof-Scheele T.N., Knops R., van de Locht L.T., de Graaf A.O., Massop M., Sandmann S., Dugas M., et al. Clonal evolution in myelodysplastic syndromes. Nat. Commun. 2017;8 doi: 10.1038/ncomms15099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernard E., Nannya Y., Hasserjian R.P., Devlin S.M., Tuechler H., Medina-Martinez J.S., Yoshizato T., Shiozawa Y., Saiki R., Malcovati L., et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat. Med. 2020;26:1549–1556. doi: 10.1038/s41591-020-1008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cumbo C., Tota G., Anelli L., Zagaria A., Specchia G., Albano F. TP53 in myelodysplastic syndromes: recent biological and clinical findings. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21103432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K., Jabbour E., Wang X., Luthra R., Bueso-Ramos C., Patel K., Pierce S., Yang H., Wei Y., Daver N., et al. Dynamic acquisition of FLT3 or RAS alterations drive a subset of patients with lower risk MDS to secondary AML. Leukemia. 2013;27:2081–2083. doi: 10.1038/leu.2013.165. [DOI] [PubMed] [Google Scholar]
- 32.Tothova Z., Valton A.L., Gorelov R.A., Vallurupalli M., Krill-Burger J.M., Holmes A., Landers C.C., Haydu J.E., Malolepsza E., Hartigan C., et al. Cohesin mutations alter DNA damage repair and chromatin structure and create therapeutic vulnerabilities in MDS/AML. JCI Insight. 2021;6 doi: 10.1172/jci.insight.142149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kon A., Shih L.-Y., Minamino M., Sanada M., Shiraishi Y., Nagata Y., Yoshida K., Okuno Y., Bando M., Nakato R., et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat. Genet. 2013;45:1232–1237. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 34.Leone G., Pagano L., Ben-Yehuda D., Voso M.T. Therapy-related leukemia and myelodysplasia: susceptibility and incidence. Haematologica. 2007;92:1389–1398. doi: 10.3324/haematol.11034. [DOI] [PubMed] [Google Scholar]
- 35.Colla S., Ong D.S.T., Ogoti Y., Marchesini M., Mistry N.A., Clise-Dwyer K., Ang S.A., Storti P., Viale A., Giuliani N., et al. Telomere dysfunction drives aberrant hematopoietic differentiation and myelodysplastic syndrome. Cancer Cell. 2015;27:644–657. doi: 10.1016/j.ccell.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sébert M., Passet M., Raimbault A., Rahmé R., Raffoux E., Sicre de Fontbrune F., Cerrano M., Quentin S., Vasquez N., Da Costa M., et al. Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood. 2019;134:1441–1444. doi: 10.1182/blood.2019000909. [DOI] [PubMed] [Google Scholar]
- 37.Hahn C.N., Chong C.E., Carmichael C.L., Wilkins E.J., Brautigan P.J., Li X.C., Babic M., Lin M., Carmagnac A., Lee Y.K., et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat. Genet. 2011;43:1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindsley R.C. Uncoding the genetic heterogeneity of myelodysplastic syndrome. Hematology. Am. Soc. Hematol. Educ. Program. 2017;2017:447–452. doi: 10.1182/asheducation-2017.1.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desikan S.P., Montalban-Bravo G., Ohanian M., Daver N.G., Kadia T.M., Venugopal S., Chien K.S., Kanagal-Shamanna R., Kantarjian H.M., Garcia-Manero G. Results of a phase 1 trial of azacitidine with venetoclax in relapsed/refractory higher-risk myelodysplastic syndrome (MDS) J. Clin. Oncol. 2022;40:e19068. doi: 10.1200/JCO.2022.40.16_suppl.e19068. [DOI] [Google Scholar]
- 40.Zeidan A.M., Borate U., Pollyea D.A., Brunner A.M., Roncolato F., Garcia J.S., Filshie R., Odenike O., Watson A.M., Krishnadasan R., et al. A phase 1b study of venetoclax and azacitidine combination in patients with relapsed or refractory myelodysplastic syndromes. Am. J. Hematol. 2023;98:272–281. doi: 10.1002/ajh.26771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morita K., Naqvi K., Montalban Bravo G., Thompson P.A., Takahashi K., Alvarado Y., Jabbour E., Kantarjian H.M., Garcia-Manero G. Initial results of a phase I/II study of venetoclax in combination with azacitidine in treatment-naive and relapsed/refractory high-risk myelodysplastic syndrome (MDS) or chronic myelomonocytic leukemia (CMML) Blood. 2020;136:39–40. doi: 10.1182/blood-2020-140644. [DOI] [Google Scholar]
- 42.Moyo T.K., Watts J.M., Skikne B.S., Mendler J.H., Klimek V.M., Chen S.-C., Fan R., Anderson I.A., Sochacki A., Strickland S.A., et al. Preliminary results from a phase II study of the combination of pevonedistat and azacitidine in the treatment of MDS and MDS/MPN after failure of DNA methyltransferase inhibition. Blood. 2019;134:4236. doi: 10.1182/blood-2019-130003. [DOI] [Google Scholar]
- 43.Garcia-Manero G., Fenaux P., Al-Kali A., Baer M.R., Sekeres M.A., Roboz G.J., Gaidano G., Scott B.L., Greenberg P., Platzbecker U., et al. Rigosertib versus best supportive care for patients with high-risk myelodysplastic syndromes after failure of hypomethylating drugs (ONTIME): a randomised, controlled, phase 3 trial. Lancet Oncol. 2016;17:496–508. doi: 10.1016/s1470-2045(16)00009-7. [DOI] [PubMed] [Google Scholar]
- 44.Onconova . Oncova Therapeutics Announces Topline Results from the Pivotal Phase 3 INSPIRE Trial. 2020. https://investor.onconova.com/news-releases/news-release-details/onconova-therapeutics-announces-topline-results-pivotal-phase-3 [Google Scholar]
- 45.Navada S.C., Garcia-Manero G., Atallah E.L., Rajeh M.N., Shammo J.M., Griffiths E.A., Khaled S.K., Dakhil S.R., Young D.E., Odchimar-Reissig R., et al. Phase 2 expansion study of oral rigosertib combined with azacitidine (AZA) in patients (Pts) with higher-risk (HR) myelodysplastic syndromes (MDS): efficacy and safety results in HMA treatment naïve & relapsed (Rel)/refractory (Ref) patients. Blood. 2018;132:230. [Google Scholar]
- 46.Sallman D.A., Foran J.M., Watts J.M., Stein E., De Botton S., Fathi A.T., Prince G.T., Stone R.M., Patel P.A., Roboz G.J., et al. Ivosidenib in patients with IDH1-mutant relapsed/refractory myelodysplastic syndrome (R/R MDS): updated enrollment and results of a phase 1 dose-escalation and expansion substudy. J. Clin. Oncol. 2022;40:7053. doi: 10.1200/JCO.2022.40.16_suppl.7053. [DOI] [Google Scholar]
- 47.Sebert M., Cluzeau T., Beyne Rauzy O., Stamatoulas Bastard A., Dimicoli-Salazar S., Thepot S., Peterlin P., Park S., Gourin M.-P., Brehar O., et al. Ivosidenib monotherapy is effective in patients with IDH1 mutated myelodysplastic syndrome (MDS): the idiome phase 2 study by the GFM group. Blood. 2021;138:62. doi: 10.1182/blood-2021-146932. [DOI] [Google Scholar]
- 48.Stein E.M., Fathi A.T., DiNardo C.D., Pollyea D.A., Roboz G.J., Collins R., Sekeres M.A., Stone R.M., Attar E.C., Frattini M.G., et al. Enasidenib in patients with mutant IDH2 myelodysplastic syndromes: a phase 1 subgroup analysis of the multicentre, AG221-C-001 trial. Lancet. Haematol. 2020;7:e309–e319. doi: 10.1016/s2352-3026(19)30284-4. [DOI] [PubMed] [Google Scholar]
- 49.DiNardo C.D., Venugopal S., Lachowiez C.A., Takahashi K., Loghavi S., Montalban-Bravo G., Wang X., Carraway H.E., Sekeres M.A., Sukkur A., et al. Targeted therapy with the mutant IDH2 inhibitor enasidenib for high-risk IDH2-mutant myelodysplastic syndrome. Blood Adv. 2022 doi: 10.1182/bloodadvances.2022008378. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ades L., Dimicoli-Salazar S., Sebert M., Cluzeau T., Stamatoulas Bastard A., Laribi K., Fossard G., Itzykson R., Beyne Rauzy O., Garnier A., et al. Enasidenib (ENA) is effective in patients with IDH2 mutated myelodysplastic syndrome (MDS) : the ideal phase 2 study by the GFM group. Blood. 2021;138:63. doi: 10.1182/blood-2021-147898. [DOI] [Google Scholar]
- 51.Cortes J.E., Esteve J., Bajel A., Yee K., Braun T., De Botton S., Peterlin P., Recher C., Thomas X., Watts J., et al. Olutasidenib (FT-2102) in combination with azacitidine induces durable complete remissions in patients with mIDH1 acute myeloid leukemia. Blood. 2021;138:698. [Google Scholar]
- 52.Garcia-Manero G., Jabbour E.J., Konopleva M.Y., Daver N.G., Borthakur G., DiNardo C.D., Bose P., Patel P., Komrokji R.S., Shastri A., et al. A clinical study of tomaralimab (OPN-305), a toll-like receptor 2 (TLR-2) antibody, in heavily pre-treated transfusion dependent patients with lower risk myelodysplastic syndromes (MDS) that have received and failed on prior hypomethylating agent (HMA) therapy. Blood. 2018;132:798. [Google Scholar]
- 53.Garcia-Manero G., Winer E.S., DeAngelo D.J., Tarantolo S.R., Sallman D.A., Dugan J., Groepper S., Giagounidis A., Gotze K.S., Metzeler K., et al. Phase 1/2a study of the IRAK4 inhibitor CA-4948 as monotherapy or in combination with azacitidine or venetoclax in patients with relapsed/refractory (R/R) acute myeloid leukemia or lyelodysplastic syndrome. J. Clin. Oncol. 2022;40:7016. doi: 10.1200/JCO.2022.40.16_suppl.7016. [DOI] [Google Scholar]
- 54.Chien K.S., Kim K., Nogueras-Gonzalez G.M., Borthakur G., Naqvi K., Daver N.G., Montalban-Bravo G., Cortes J.E., DiNardo C.D., Jabbour E., et al. Phase II study of azacitidine with pembrolizumab in patients with intermediate-1 or higher-risk myelodysplastic syndrome. Br. J. Haematol. 2021;195:378–387. doi: 10.1111/bjh.17689. [DOI] [PubMed] [Google Scholar]
- 55.Garcia-Manero G., Ribrag V., Zhang Y., Farooqui M., Marinello P., Smith B.D. Pembrolizumab for myelodysplastic syndromes after failure of hypomethylating agents in the phase 1b KEYNOTE-013 study. Leuk. Lymphoma. 2022;63:1660–1668. doi: 10.1080/10428194.2022.2034155. [DOI] [PubMed] [Google Scholar]
- 56.Garcia-Manero G., Sasaki K., Montalban-Bravo G., Daver N.G., Jabbour E.J., Alvarado Y., DiNardo C.D., Ravandi F., Borthakur G., Bose P., et al. A phase II study of nivolumab or ipilimumab with or without azacitidine for patients with myelodysplastic syndrome (MDS) Blood. 2018;132:465. doi: 10.1182/blood-2018-99-119424. [DOI] [Google Scholar]
- 57.Morita K., Kantarjian H.M., Montalban Bravo G., Sasaki K., Daver N., Jabbour E., Alvarado Y., Chien K.S., DiNardo C.D., Ravandi F., et al. A phase II study of double immune checkpoint inhibitor blockade with nivolumab and ipilimumab with or without azacitidine in patients with myelodysplastic syndrome (MDS) Blood. 2020;136:7–9. doi: 10.1182/blood-2020-142003. [DOI] [Google Scholar]
- 58.Gerds A.T., Scott B.L., Greenberg P., Lin T.L., Pollyea D.A., Verma A., Dail M., Feng Y., Green C., Ma C., et al. Atezolizumab alone or in combination did not demonstrate a favorable risk-benefit profile in myelodysplastic syndrome. Blood Adv. 2022;6:1152–1161. doi: 10.1182/bloodadvances.2021005240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Letai A., Bassik M.C., Walensky L.D., Sorcinelli M.D., Weiler S., Korsmeyer S.J. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 60.Bhola P.D., Letai A. Mitochondria-judges and executioners of cell death sentences. Mol. Cell. 2016;61:695–704. doi: 10.1016/j.molcel.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Czabotar P.E., Lessene G., Strasser A., Adams J.M. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 62.Fischer M., Song Y., Gbyli R., Arrate M., Villaume M., Childress M., Smith B., Stricker T., Halene S., Savona M. MCL1 dependence across MDS subtypes and dual inhibition of MCL1 and BCL2 in MISTRG6 mice. bioRxiv. 2020 doi: 10.1101/2020.06.05.133090. Preprint at. [DOI] [Google Scholar]
- 63.Jilg S., Reidel V., Müller-Thomas C., König J., Schauwecker J., Höckendorf U., Huberle C., Gorka O., Schmidt B., Burgkart R., et al. Blockade of BCL-2 proteins efficiently induces apoptosis in progenitor cells of high-risk myelodysplastic syndromes patients. Leukemia. 2016;30:112–123. doi: 10.1038/leu.2015.179. [DOI] [PubMed] [Google Scholar]
- 64.Reidel V., Kauschinger J., Hauch R.T., Müller-Thomas C., Nadarajah N., Burgkart R., Schmidt B., Hempel D., Jacob A., Slotta-Huspenina J., et al. Selective inhibition of BCL-2 is a promising target in patients with high-risk myelodysplastic syndromes and adverse mutational profile. Oncotarget. 2018;9:17270–17281. doi: 10.18632/oncotarget.24775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei A.H., Montesinos P., Ivanov V., DiNardo C.D., Novak J., Laribi K., Kim I., Stevens D.A., Fiedler W., Pagoni M., et al. Venetoclax plus LDAC for newly diagnosed AML ineligible for intensive chemotherapy: a phase 3 randomized placebo-controlled trial. Blood. 2020;135:2137–2145. doi: 10.1182/blood.2020004856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DiNardo C.D., Jonas B.A., Pullarkat V., Thirman M.J., Garcia J.S., Wei A.H., Konopleva M., Döhner H., Letai A., Fenaux P., et al. Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N. Engl. J. Med. 2020;383:617–629. doi: 10.1056/NEJMoa2012971. [DOI] [PubMed] [Google Scholar]
- 67.Bazinet A., Darbaniyan F., Jabbour E., Montalban-Bravo G., Ohanian M., Chien K., Kadia T., Takahashi K., Masarova L., Short N., et al. Azacitidine plus venetoclax in patients with high-risk myelodysplastic syndromes or chronic myelomonocytic leukaemia: phase 1 results of a single-centre, dose-escalation, dose-expansion, phase 1-2 study. Lancet. Haematol. 2022;9:e756–e765. doi: 10.1016/S2352-3026(22)00216-2. [DOI] [PubMed] [Google Scholar]
- 68.Ganan-Gomez I., Yang H., Ma F., Montalban-Bravo G., Thongon N., Marchica V., Richard-Carpentier G., Chien K., Manyam G., Wang F., et al. Stem cell architecture drives myelodysplastic syndrome progression and predicts response to venetoclax-based therapy. Nat. Med. 2022;28:557–567. doi: 10.1038/s41591-022-01696-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fischer M.A., Song Y., Arrate M.P., Gbyli R., Villaume M.T., Smith B.N., Childress M.A., Stricker T.P., Halene S., Savona M.R. Selective inhibition of MCL1 overcomes venetoclax-resistance in a murine model of myelodysplastic syndromes. Haematologica. 2022 doi: 10.3324/haematol.2022.280631. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pei S., Pollyea D.A., Gustafson A., Stevens B.M., Minhajuddin M., Fu R., Riemondy K.A., Gillen A.E., Sheridan R.M., Kim J., et al. Monocytic subclones confer resistance to venetoclax-based therapy in patients with acute myeloid leukemia. Cancer Discov. 2020;10:536–551. doi: 10.1158/2159-8290.Cd-19-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones C.L., Stevens B.M., Pollyea D.A., Culp-Hill R., Reisz J.A., Nemkov T., Gehrke S., Gamboni F., Krug A., Winters A., et al. Nicotinamide metabolism mediates resistance to venetoclax in relapsed acute myeloid leukemia stem cells. Cell Stem Cell. 2020;27:748–764.e4. doi: 10.1016/j.stem.2020.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stevens B.M., Jones C.L., Pollyea D.A., Culp-Hill R., D'Alessandro A., Winters A., Krug A., Abbott D., Goosman M., Pei S., et al. Fatty acid metabolism underlies venetoclax resistance in acute myeloid leukemia stem cells. Nat. Can. (Que.) 2020;1:1176–1187. doi: 10.1038/s43018-020-00126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rabut G., Peter M. Function and regulation of protein neddylation. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duda D.M., Borg L.A., Scott D.C., Hunt H.W., Hammel M., Schulman B.A. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Majidi F., Neukirchen J., Brille S., Fey I., Neumann F., Berger A., Cadeddu R.-P., Rudelius M., Germing U., Haas R., Gattermann N. Altered expression of neddylation pathway proteins in myelodysplastic syndromes. Blood. 2017;130:5298. doi: 10.1182/blood.V130.Suppl_1.5298.5298. [DOI] [Google Scholar]
- 76.Hua S., Feng T., Yin L., Wang Q., Shao X. NEDD9 overexpression: prognostic and guidance value in acute myeloid leukaemia. J. Cell Mol. Med. 2021;25:9331–9339. doi: 10.1111/jcmm.16870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swords R.T., Kelly K.R., Smith P.G., Garnsey J.J., Mahalingam D., Medina E., Oberheu K., Padmanabhan S., O'Dwyer M., Nawrocki S.T., et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 78.Smith P.G., Traore T., Grossman S., Narayanan U., Carew J.S., Lublinksky A., Kuranda M., Milhollen M. Azacitidine/decitabine synergism with the NEDD8-activating enzyme inhibitor MLN4924 in pre-clinical AML models. Blood. 2011;118:578. doi: 10.1182/blood.V118.21.578.578. [DOI] [Google Scholar]
- 79.Swords R.T., Coutre S., Maris M.B., Zeidner J.F., Foran J.M., Cruz J., Erba H.P., Berdeja J.G., Tam W., Vardhanabhuti S., et al. Pevonedistat, a first-in-class NEDD8-activating enzyme inhibitor, combined with azacitidine in patients with AML. Blood. 2018;131:1415–1424. doi: 10.1182/blood-2017-09-805895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kolch W., Halasz M., Granovskaya M., Kholodenko B.N. The dynamic control of signal transduction networks in cancer cells. Nat. Rev. Cancer. 2015;15:515–527. doi: 10.1038/nrc3983. [DOI] [PubMed] [Google Scholar]
- 81.Du Z., Lovly C.M. Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer. 2018;17 doi: 10.1186/s12943-018-0782-4. 58-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S., et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Simanshu D.K., Nissley D.V., McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gimple R.C., Wang X. RAS: striking at the core of the oncogenic circuitry. Front. Oncol. 2019;9:965. doi: 10.3389/fonc.2019.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Prior I.A., Hood F.E., Hartley J.L. The frequency of ras mutations in cancer. Cancer Res. 2020;80:2969–2974. doi: 10.1158/0008-5472.Can-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Athuluri-Divakar S.K., Vasquez-Del Carpio R., Dutta K., Baker S.J., Cosenza S.C., Basu I., Gupta Y.K., Reddy M.V.R., Ueno L., Hart J.R., et al. A small molecule RAS-mimetic disrupts RAS association with effector proteins to block signaling. Cell. 2016;165:643–655. doi: 10.1016/j.cell.2016.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chapman C.M., Sun X., Roschewski M., Aue G., Farooqui M., Stennett L., Gibellini F., Arthur D., Pérez-Galán P., Wiestner A. ON 01910 Na is selectively cytotoxic for chronic lymphocytic leukemia cells through a dual mechanism of action involving PI3K/AKT inhibition and induction of oxidative stress. Clin. Cancer Res. 2012;18:1979–1991. doi: 10.1158/1078-0432.Ccr-11-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hallin J., Bowcut V., Calinisan A., Briere D.M., Hargis L., Engstrom L.D., Laguer J., Medwid J., Vanderpool D., Lifset E., et al. Anti-tumor efficacy of a potent and selective non-covalent KRAS(G12D) inhibitor. Nat. Med. 2022;28:2171–2182. doi: 10.1038/s41591-022-02007-7. [DOI] [PubMed] [Google Scholar]
- 90.Kazi J.U., Rönnstrand L. FMS-Like tyrosine kinase 3/FLT3: from basic science to clinical implications. Physiol. Rev. 2019;99:1433–1466. doi: 10.1152/physrev.00029.2018. [DOI] [PubMed] [Google Scholar]
- 91.Grafone T., Palmisano M., Nicci C., Storti S. An overview on the role of FLT3-tyrosine kinase receptor in acute myeloid leukemia: biology and treatment. Onco Rev. 2012;6:e8. doi: 10.4081/oncol.2012.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Levis M., Small D. FLT3: ITDoes matter in leukemia. Leukemia. 2003;17:1738–1752. doi: 10.1038/sj.leu.2403099. [DOI] [PubMed] [Google Scholar]
- 93.Antar A.I., Otrock Z.K., Jabbour E., Mohty M., Bazarbachi A. FLT3 inhibitors in acute myeloid leukemia: ten frequently asked questions. Leukemia. 2020;34:682–696. doi: 10.1038/s41375-019-0694-3. [DOI] [PubMed] [Google Scholar]
- 94.Daver N., Venugopal S., Ravandi F. FLT3 mutated acute myeloid leukemia: 2021 treatment algorithm. Blood Cancer J. 2021;11:104. doi: 10.1038/s41408-021-00495-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Badar T., Patel K.P., Thompson P.A., DiNardo C., Takahashi K., Cabrero M., Borthakur G., Cortes J., Konopleva M., Kadia T., et al. Detectable FLT3-ITD or RAS mutation at the time of transformation from MDS to AML predicts for very poor outcomes. Leuk. Res. 2015;39:1367–1374. doi: 10.1016/j.leukres.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A.G., Hoadley K., Triche T.J., Jr., Laird P.W., Baty J.D., et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.DiNardo C.D., Jabbour E., Ravandi F., Takahashi K., Daver N., Routbort M., Patel K.P., Brandt M., Pierce S., Kantarjian H., Garcia-Manero G. IDH1 and IDH2 mutations in myelodysplastic syndromes and role in disease progression. Leukemia. 2016;30:980–984. doi: 10.1038/leu.2015.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mondesir J., Willekens C., Touat M., de Botton S. IDH1 and IDH2 mutations as novel therapeutic targets: Current perspectives. J. Blood Med. 2016;7:171–180. doi: 10.2147/jbm.S70716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dang L., White D.W., Gross S., Bennett B.D., Bittinger M.A., Driggers E.M., Fantin V.R., Jang H.G., Jin S., Keenan M.C., et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ward P.S., Patel J., Wise D.R., Abdel-Wahab O., Bennett B.D., Coller H.A., Cross J.R., Fantin V.R., Hedvat C.V., Perl A.E., et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S.-H., Ito S., Yang C., Wang P., Xiao M.-T., et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Figueroa M.E., Abdel-Wahab O., Lu C., Ward P.S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H.F., et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Testa U., Castelli G., Pelosi E. Isocitrate dehydrogenase mutations in myelodysplastic syndromes and in acute myeloid leukemias. Cancers. 2020;12 doi: 10.3390/cancers12092427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stein E.M., DiNardo C.D., Pollyea D.A., Fathi A.T., Roboz G.J., Altman J.K., Stone R.M., DeAngelo D.J., Levine R.L., Flinn I.W., et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130:722–731. doi: 10.1182/blood-2017-04-779405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.DiNardo C.D., Stein E.M., de Botton S., Roboz G.J., Altman J.K., Mims A.S., Swords R., Collins R.H., Mannis G.N., Pollyea D.A., et al. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N. Engl. J. Med. 2018;378:2386–2398. doi: 10.1056/NEJMoa1716984. [DOI] [PubMed] [Google Scholar]
- 106.De Botton S., Yee K.W.L., Recher C., Wei A., Montesinos P., Taussig D., Pigneux A., Braun T., Curti A., Esteve J., et al. Effect of olutasidenib (FT-2102) on complete remissions in patients with relapsed/refractory (R/R) mIDH1 acute myeloid leukemia (AML): results from a planned interim analysis of a phase 2 clinical trial. J. Clin. Oncol. 2021;39:7006. doi: 10.1200/JCO.2021.39.15_suppl.7006. [DOI] [Google Scholar]
- 107.Papaemmanuil E., Gerstung M., Malcovati L., Tauro S., Gundem G., Van Loo P., Yoon C.J., Ellis P., Wedge D.C., Pellagatti A., et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barreyro L., Chlon T.M., Starczynowski D.T. Chronic immune response dysregulation in MDS pathogenesis. Blood. 2018;132:1553–1560. doi: 10.1182/blood-2018-03-784116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sallman D.A., List A. The central role of inflammatory signaling in the pathogenesis of myelodysplastic syndromes. Blood. 2019;133:1039–1048. doi: 10.1182/blood-2018-10-844654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Greten F.R., Grivennikov S.I. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Esplin B.L., Shimazu T., Welner R.S., Garrett K.P., Nie L., Zhang Q., Humphrey M.B., Yang Q., Borghesi L.A., Kincade P.W. Chronic exposure to a TLR ligand injures hematopoietic stem cells. J. Immunol. 2011;186:5367–5375. doi: 10.4049/jimmunol.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nagai Y., Garrett K.P., Ohta S., Bahrun U., Kouro T., Akira S., Takatsu K., Kincade P.W. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wei Y., Dimicoli S., Bueso-Ramos C., Chen R., Yang H., Neuberg D., Pierce S., Jia Y., Zheng H., Wang H., et al. Toll-like receptor alterations in myelodysplastic syndrome. Leukemia. 2013;27:1832–1840. doi: 10.1038/leu.2013.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Farooq M., Batool M., Kim M.S., Choi S. Toll-like receptors as a therapeutic target in the era of immunotherapies. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.756315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.El-Zayat S.R., Sibaii H., Mannaa F.A. Toll-like receptors activation, signaling, and targeting: an overview. Bull. Natl. Res. Cent. 2019;43:187–212. [Google Scholar]
- 116.Akira S., Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 117.Rhyasen G.W., Starczynowski D.T. IRAK signalling in cancer. Br. J. Cancer. 2015;112:232–237. doi: 10.1038/bjc.2014.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Smith M.A., Choudhary G.S., Pellagatti A., Choi K., Bolanos L.C., Bhagat T.D., Gordon-Mitchell S., Von Ahrens D., Pradhan K., Steeples V., et al. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol. 2019;21:640–650. doi: 10.1038/s41556-019-0314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Choudhary G.S., Pellagatti A., Agianian B., Smith M.A., Bhagat T.D., Gordon-Mitchell S., Sahu S., Pandey S., Shah N., Aluri S., et al. Activation of targetable inflammatory immune signaling is seen in myelodysplastic syndromes with SF3B1 mutations. Elife. 2022;11 doi: 10.7554/eLife.78136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bennett J., Starczynowski D.T. IRAK1 and IRAK4 as emerging therapeutic targets in hematologic malignancies. Curr. Opin. Hematol. 2022;29:8–19. doi: 10.1097/MOH.0000000000000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Melgar K., Walker M.M., Jones L.M., Bolanos L.C., Hueneman K., Wunderlich M., Jiang J.K., Wilson K.M., Zhang X., Sutter P., et al. Overcoming adaptive therapy resistance in AML by targeting immune response pathways. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aaw8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Smigiel K.S., Srivastava S., Stolley J.M., Campbell D.J. Regulatory T-cell homeostasis: steady-state maintenance and modulation during inflammation. Immunol. Rev. 2014;259:40–59. doi: 10.1111/imr.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Grywalska E., Pasiarski M., Góźdź S., Roliński J. Immune-checkpoint inhibitors for combating T-cell dysfunction in cancer. OncoTargets Ther. 2018;11:6505–6524. doi: 10.2147/ott.S150817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sharma P., Siddiqui B.A., Anandhan S., Yadav S.S., Subudhi S.K., Gao J., Goswami S., Allison J.P. The next decade of immune checkpoint therapy. Cancer Discov. 2021;11:838–857. doi: 10.1158/2159-8290.Cd-20-1680. [DOI] [PubMed] [Google Scholar]
- 126.Yang H., Bueso-Ramos C., DiNardo C., Estecio M.R., Davanlou M., Geng Q.R., Fang Z., Nguyen M., Pierce S., Wei Y., et al. Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic syndromes is enhanced by treatment with hypomethylating agents. Leukemia. 2014;28:1280–1288. doi: 10.1038/leu.2013.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen J., Kao Y.R., Sun D., Todorova T.I., Reynolds D., Narayanagari S.R., Montagna C., Will B., Verma A., Steidl U. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat. Med. 2019;25:103–110. doi: 10.1038/s41591-018-0267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]