Abstract
This study examined the effects of NH4NO3 fertilizer on the size and activity of nitrifying, autotrophic, ammonia-oxidizing populations of the β subdivision of the class Proteobacteria in arable soils. Plots under different long-term fertilizer regimes were sampled before and after NH4NO3 additions, and the rates of nitrification were determined by 15N isotopic pool dilution assays. Ammonia-oxidizing populations in the plots were quantified by competitive PCR assays based on the amoA and ribosomal 16S genes. Prior to fertilizer addition, ammonium concentrations and nitrification rates in the plots were comparatively low; ammonia-oxidizing populations were present at 104 to 105 gene copies g of soil−1. Three days after the application of fertilizer, nitrification rates had risen considerably but the size of the ammonia-oxidizing population was unchanged. Six weeks after fertilizer treatment, ammonium concentrations and nitrification rates had fallen while the ammonia-oxidizing populations in plots receiving fertilizer had increased. The rapidity of the rise in nitrification rates observed after 3 days suggests that it results from phenotypic changes in the ammonia-oxidizing bacterial population. Associated increases in population sizes were only observed after 6 weeks and did not correlate directly with nitrifying activity. Phylogenetic analyses of PCR products from one of the plots revealed a population dominated by Nitrosospira-type organisms, similar to those prevalent in other soils.
The process of nitrification, defined as the oxidation of ammonia to nitrate via nitrite, is of considerable importance in the terrestrial nitrogen cycle (28). Soil nitrogen (N) in the form of ammonium is bound by the soil matrix and so is retained by the soil. When converted to nitrate, which is mobile within soils, the N becomes susceptible to leaching and denitrification, both of which result in wasteful and potentially polluting N loss from the soil. The initial and rate-limiting step in the process of nitrification is the oxidation of ammonia to nitrite. In arable soils, the majority of this activity is carried out by autotrophic ammonia-oxidizing bacteria (AOB) of the β subdivision of the class Proteobacteria (4, 21). These organisms are able to utilize the redox potential associated with the oxidation of ammonia and to fix CO2 via the Calvin cycle. The initial enzymatic step in autotrophic ammonia oxidation is the conversion of ammonia to hydroxylamine by ammonia monooxygenase. Three genes, amoA, amoB, and amoC, encode this enzyme, which is unique to AOB (5, 22, 29). The amoA gene product contains the enzyme’s active site.
Autotrophic ammonia oxidizers are difficult to grow in culture. Those that have been grown in vitro have been shown to be unrepresentative of soil populations (6). As a result, little is known about the in situ population dynamics of these ubiquitous soil organisms. Molecular analysis of ribosomal 16S and amoA genes has shown that β-subdivision AOB form a monophyletic group with two divisions, the genera Nitrosospira and Nitrosomonas (18, 36). Studies involving the culturing of environmental AOB isolates, as well as those based on the PCR amplification of environmental AOB sequences, suggest that the majority of ammonia oxidizers from a variety of soils belong to the genus Nitrosospira (6, 17, 20, 37, 40). Members of the genus Nitrosomonas are more prevalent in ammonia-rich environments such as sewage sludge (30, 37).
The aim of this work was to examine the responses of AOB in arable soils to NH4NO3 fertilizer addition. Samples were taken from arable plots under different long-term fertilizer regimes before and after NH4NO3 addition. Rates of nitrification were estimated by 15N isotopic pool dilution assays and related to AOB population sizes as determined by competitive PCR (cPCR).
15N isotopic pool dilution is a method suitable for measuring gross rates of nitrification in soil. Isotopically labelled nitrate, the product of nitrification, is added to soil, and the 15NO3−/14NO3− ratio of the soil nitrate pool is measured over time. The rate at which the 15NO3− is diluted by endogenous 14NO3− production can then be deduced, and the rate of nitrification can be determined (3). This method does not require the addition of ammonium, the substrate of nitrification, to the soil. Any such ammonia addition is likely to stimulate nitrifying activity, which would result in an overestimation of the in situ nitrification rates (12).
cPCR assays are robust and convenient methods of quantifying small amounts of DNA. These assays involve PCR amplification of DNA in reaction mixtures containing known quantities of competitor molecules. The competitor molecules have the same primer sites as the target sequence and so coamplify, but they generate a differently sized product which can be separated from the target product on an agarose gel. By comparing target and competitor product band intensities, it is possible to estimate the initial number of target sequences in the assay. Such methods have been used to enumerate both culturable and nonculturable organisms, including AOB, directly from soil (2, 16, 23, 26, 35, 39). In this study, assays based on the amoA and 16S genes were developed and used to enumerate the AOB in the soil. The phylogenetic makeup of the population from one of the soils was determined by sequence analysis of the amoA and 16S PCR products.
MATERIALS AND METHODS
Soil properties and experimental design.
Samples were taken from part of the Broadbalk continuous wheat experiment (1) at Rothamsted Experimental Station, in which plots of approximately 6 by 23 m have been under continuous fertilizer regimes since 1852. The soils are clayey loams of the Batcombe series whose physical properties vary due to long-term differences in management (Table 1). Three plots were sampled: plot NO, which receives no applied nitrogen; plot N144, which in spring receives 144 kg of N ha−1 year−1 as NH4NO3; and plot FYM/N, which receives the equivalent of 48 kg of N ha−1 year−1 as NH4NO3 in spring and 35 metric tons of farmyard manure, containing 248 kg of N, ha−1 year−1 in autumn (1). All plots were under winter wheat as part of a 5-year crop rotation. Soil samples were taken from each of the plots on three separate occasions: immediately before spring fertilizer application, 3 days after spring fertilizer application, and 6 weeks after spring fertilizer application. At each timepoint, 10 independent samples from the top 0 to 10 cm of the soil were removed, mixed, and passed through a 4-mm sieve to give a homogeneous composite sample from which subsamples were taken for analyses. 15N isotopic pool dilution assays were begun immediately, and soil from which DNA was to be extracted was stored at −70°C.
TABLE 1.
The ammonium inputs and selected soil properties of plots NO, N144, and FYM/N
| Plot | Fertilizer addition (ha−1 yr−1)a
|
Total Cb (μg of C g of soil−1) | Total Nb (μg of N g of soil−1) | pHbc | |
|---|---|---|---|---|---|
| Inorganic | Organic | ||||
| NO | None | None | 0.75 | 0.087 | 8.2 |
| N144 | 144 kg of N as NH4NO3 in spring | None | 0.94 | 0.10 | 8.0 |
| FYM/N | 48 kg of N as NH4NO3 in spring | 35 metric tons of manure from bullocks in autumn (equivalent to 248 kg of N) | 2.8 | 0.28 | 7.9 |
15N isotopic pool dilution assays.
For the 15N isotopic pool dilution assays, a modified version of the method described by Barraclough and coworkers (4, 42) was used. Three 200-g subsamples from each of the composite soil samples were placed in individual jars, and 16.28 ml of K15NO3 (10.1 atom %) was added at a rate of 5 μg of N g of soil−1. To stimulate field conditions, the jars were incubated in the dark at 15°C for 7 days. Samples of 40 g were taken from each jar at days 1, 3, and 7. These were shaken for 1 h with 80 ml of 2 M KCl and filtered through Whatman no. 42 paper to extract the soil ammonium and nitrate. Ammonium and nitrate concentrations in the extracts were determined colorimetrically by using a Skalar continuous-flow analyzer, as described previously (19, 25). The samples were diffused onto acidified glass wool discs (7), and the 15N atom percent excess was determined by using an Integra-CN mass spectrometer (Europa Scientific, Crewe, United Kingdom). Rates of nitrification were determined between days 1 and 7 of the incubation period as described by Barraclough (3).
Primer design, PCR conditions, cloning, and sequencing.
All sequence analyses were carried out with the GCG8 (Genetics Computer Group, Madison, Wis.) and PHYLIP V3.57 (13) suites of programs. The program FASTA was used to search the GenBank/EMBL database. Universal amoA primers were designed by aligning all publicly available amoA sequences and selecting areas of homology. The primers chosen were 5′ATYATGTAYTACYTGTGGGT and 5′ACCACCAGTARAAWCCCCAG, which were designed to amplify the region corresponding to nucleotides 486 to 597 of the amoA gene of Nitrosomonas europaea (accession no. L08050); database searches revealed no significant homology to non-amoA sequences, including that of the methane monooxygenase gene pmoA. Primers with published sequences that were specific for AOB 16S genes were assessed by comparison to the database. Primers 5′AGAAAAGCAGGGGATCG (30) and 5′CCTTGTAGTTTCAAACGC (9) were selected because they exhibited maximal homology to AOB 16S genes without having significant homology to non-AOB sequences. PCR mixtures contained 10 mM Tris-HCl (pH 8.3), 100 mM KCl, 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphate mix, 0.8 μM primers, 0.04 U of Taq polymerase (Roche Diagnostics, Lewes, United Kingdom) μl−1, and DNA (equivalent to 50 μg of fresh soil) extracted by the method described by Cullen and Hirsch (10). Conditions for both PCR primer sets were as follows: 45 cycles of 94°C for 1 min, 57.5°C for 1 min, and 72°C for 1 min, followed by 72°C for 5 min. Gel-purified PCR products from plot N144 were cloned into pGEM-T (Promega, Southampton, United Kingdom). Plasmids were prepared with Qiagen Mini Kits (Qiagen, Crawley, United Kingdom) and sequenced by using ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kits (Perkin-Elmer, Warrington, United Kingdom). Phylogenetic trees were constructed by using the programs PROTDIST (Kimura two-parameter method), DNADIST (Kimura two-parameter method), and NEIGHBOUR (unweighted-pair group method using average linkages [UPGMA]) and viewed with Treeview 1.4 (32).
Competitor construction.
To construct a competitor for the amoA primers, soil PCR clone amo8 was digested with NarI and the enzyme was heat inactivated. This digest was ligated to a phenol-chloroform-extracted TaqI digest of pBR322. A competitor for the 16S primers was constructed by digesting soil PCR clone 16S-7 with SmaI and heat inactivating the enzyme. This preparation was further digested with SnaBI (5 U μg of DNA−1), phenol-chloroform extracted, and religated. Both ligation products were transformed into SURE competent cells (Stratagene, Amsterdam, The Netherlands). Resulting colonies were screened by PCR with amoA and 16S primers for products of suitable sizes. The quality of plasmids for cPCR assays, prepared by using Qiagen Midi Kits, was assessed by gel electrophoresis, and three independent dilutions of each plasmid were quantified by determining their optical densities at 260 nm.
cPCRs.
Each cPCR assay consisted of six reaction mixtures containing serial dilutions of 106 to 101 competitor molecules and aliquots of sample DNA. Control reaction mixtures contained either no template, sample DNA alone, or competitor alone. Products were separated on 2.75% agarose gels, which were digitized by using an EagleEye II system (Stratagene). Band intensities were quantified by using GelDoc1000 software (Bio-Rad, Hemel Hempstead, United Kingdom). Standard curves were generated for both primer sets and used both to calibrate the systems and to demonstrate that they were quantitative. Assays for the standard curves were carried out in triplicate. Each contained the following: either 101, 102, 103, or 104 target molecules of cloned soil PCR product; competitor molecules; and an aliquot of soil DNA previously incubated with DNase I at 37°C for 1 h prior to heat inactivation of the enzyme. The log of the number of competitor molecules per reaction was plotted against the log of the ratio of the competitor and target product band intensities, taking into account differences in product length. The number of target molecules in each assay was estimated as the value at which a straight line through points from the three replicates crossed the x axis (i.e., log [target band intensity/competitor band intensity ratio] = 0), and standard errors were determined (Fig. 1). The numbers of target molecules, as estimated by the cPCR assays, were plotted against the actual numbers of molecules to give standard curves (Fig. 2). Soil DNA for cPCR assays was extracted by the method of Cullen and Hirsch (10) from three 1-g subsamples taken from each composite soil sample. Each extraction product was assayed independently by the protocol described for the standard curves, with the modification that only soil DNA and competitor molecules were added to the reaction mixtures. Values and standard errors were determined as described above after sample dilutions, soil moisture contents, and the standard curves were taken into account.
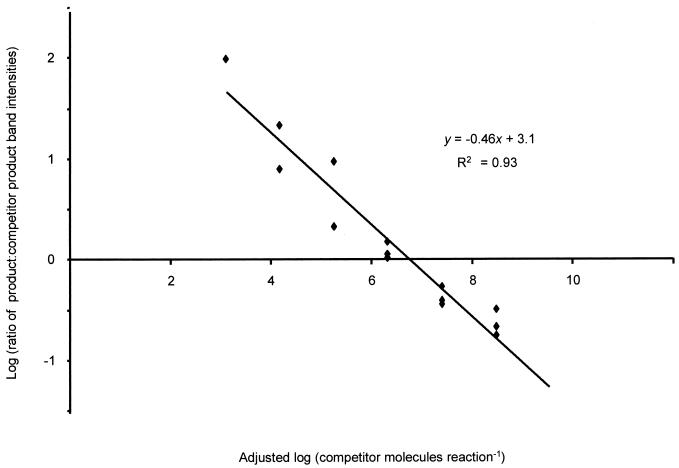
FIG. 1.
Plot of the logs of the ratios of target and competitor product intensities from three replicate cPCR assays using the amoA primers versus the logs of the numbers of competitor molecules in the reactions. Sample dilutions, soil moisture contents, and standard curves have been accounted for. Each point represents a single PCR. The estimated number of target molecules in the assay (the value at which the fitted line crosses the x axis) is 106.6; the standard error of this estimate is 100.22.
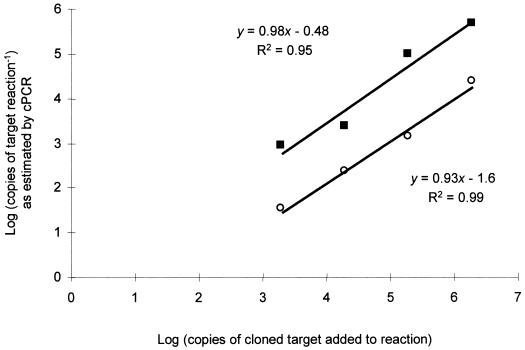
FIG. 2.
The standard curves for cPCRs using the amoA (○) and 16S (■) primers, relating the number of target sequences, as estimated by cPCR, to the actual number of target sequences present in each assay. Each point is derived from a plot similar to Fig. 1.
Nucleotide sequence accession numbers.
The environmental amoA sequences have been deposited in the GenBank and EMBL databases under accession no. Aj238189 to Aj238197. The environmental 16S sequences have been deposited under accession no. Aj238198 to Aj238203 and Aj238205.
RESULTS
15N isotopic pool dilution assays.
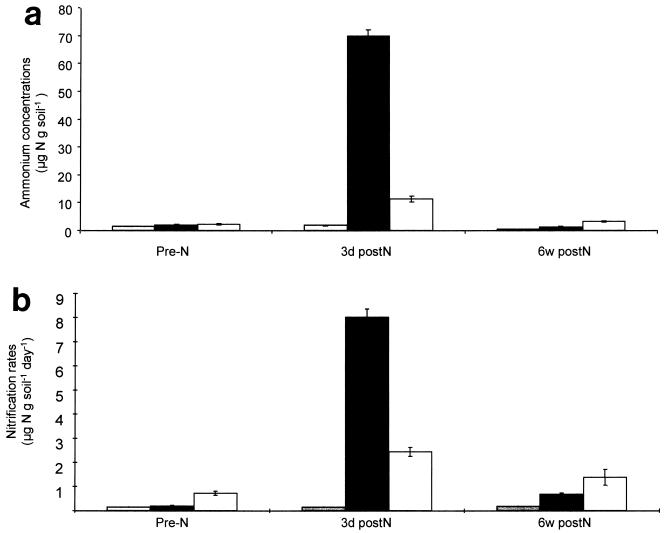
Ammonium concentrations in the three plots prior to fertilizer addition were between 1.5 and 2.3 μg of N g of soil−1 (Fig. 3a). Three days after NH4NO3 fertilizer addition, the ammonium concentrations in plots N144 and FYM/N had risen to 70 and 11 μg of N g of soil−1, respectively, but remained low (1.9 μg of N g of soil−1) in plot NO. Six weeks later, the concentrations in all three plots had fallen back to between 0.46 and 3.3 μg of N g of soil−1.
FIG. 3.
Ammonium concentrations (a) and nitrification rates (b) as estimated by 15N isotopic pool dilution assays of samples from plots NO ( ), N144 (■), and FYM/N (□) (Table 1) taken immediately before fertilizer addition (Pre-N), 3 days after fertilizer addition (3d postN), and 6 weeks after fertilizer addition (6w postN). Error bars represent the standard errors.
), N144 (■), and FYM/N (□) (Table 1) taken immediately before fertilizer addition (Pre-N), 3 days after fertilizer addition (3d postN), and 6 weeks after fertilizer addition (6w postN). Error bars represent the standard errors.
Nitrification rates in the plots correlated with the soil ammonium concentrations. Prior to fertilizer addition, rates of nitrification were low, 0.16 and 0.21 μg of N g of soil−1 day−1, in plots NO and N144, respectively, and 0.76 μg of N g of soil−1 day−1 in plot FYM/N (Fig. 3b). These rates are consistent with those recorded previously (43). Three days after fertilizer addition, the rates had arisen to 8.0 μg of N g of soil−1 day−1 in plot N144 and 2.4 μg of N g of soil−1 day−1 in plot FYM/N. Six weeks after fertilizer addition, nitrification rates had fallen to 0.69 and 1.4 μg of N g of soil−1 day−1 in plots N144 and FYM/N, respectively. The nitrification rate in plot NO did not change significantly, indicating that the observed changes in plots N144 and FYM/N were a result of fertilizer additions. It should be noted that these assays do not measure field ammonia oxidation rates directly. The assays were ex situ, although conditions were designed to reflect those in the field, and determined rates of nitrate, rather than nitrite, production. Nitrite is the product of autotrophic ammonia oxidation, but in the soil it is rapidly converted to nitrate (8), the oxidation of ammonia being the rate-limiting step in nitrification (28).
cPCR.
Individual cPCR assays gave linear relationships between the log of product ratios (target/competitor) and the log of the number of competitor molecules over more than 5 orders of magnitude (Fig. 1). To calibrate the cPCR assays, it was necessary to construct standard curves that to be quantitative must fit the theoretical equation log (Prodtar/Prodcomp) = log (Temptar/Tempcomp) + [n × log (efftar/effcomp)], where Prodtar/Prodcomp is the final ratio of target and competitor products, Temptar/Tempcomp is the initial ratio of target and competitor templates, n is the number of cycles, and eff is the efficiency of amplification (38). This equation states that the ratio of products must equal the initial ratio of the templates, taking into account the number of cycles and the efficiencies of amplification. The amplification efficiency of a given template depends on several factors, most notably length, to which it is inversely proportional (27). However, so long as the ratio of amplification efficiencies remains constant in all reactions, the standard curve can account for this and other reaction variables, e.g., inhibition by coextracted humic substances in soil extracts. Theoretically, a standard curve for a quantitative cPCR assay will be linear with a gradient of 1 (34). The linear regressions for the standard curves in this study were as follows: y = 0.93x − 1.6, R2 = 0.99 for the amoA assays; and y = 0.98x − 0.48, R2 = 0.95 for the 16S sequence-based assays (Fig. 2). These curves indicate that although the amplification efficiencies of target and competitor were not equal in either assay, the ratio between them was constant, and so the assays were quantitative (Fig. 2). Thus, the assays were able to enumerate the number of gene copies in soil DNA extracts and so measure the relative sizes of soil AOB populations.
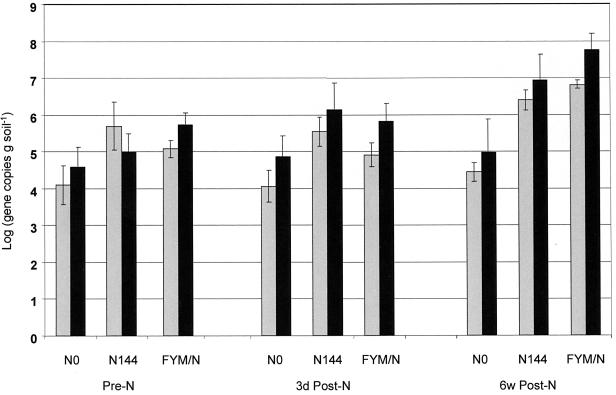
The amoA and 16S primers amplified products of the predicted sizes, 112 and 458 bp, respectively, directly from soil DNA extracts. cPCR assays using the amoA primers showed that prior to NH4NO3 addition, plots FYM/N and N144 contained similarly sized populations of 1.2 × 105 and 5 × 105 gene copies g of soil−1, respectively, and that the population in plot NO was significantly lower, i.e., 1.3 × 104 gene copies g of soil−1 (Fig. 4). Three days after fertilizer addition, the population sizes had not changed significantly. Six weeks after fertilizer addition, populations in plots receiving NH4NO3 had risen; those in plots FYM/N and N144 were 6.5 × 106 and 2.5 × 106 gene copies g of soil−1, respectively. The population in plot NO had not changed significantly, indicating that the increased populations in plots FYM/N and N144 were due to fertilizer addition. The rate of population growth in plot FYM/N was higher than that in plot N144. Assays using the 16S primers gave results similar to those of the amoA-based assays. The results were, however, more variable, and standard errors were greater. In all but one case the 16S-based cPCR assays gave higher estimates of the AOB population size than amoA-based assays (Fig. 4).
FIG. 4.
The number of amoA ( ) and 16S rRNA (■) gene copies in DNA extracts from samples taken from plots NO, N144, and FYM/N (Table 1) immediately before fertilizer addition (Pre-N), 3 days after fertilizer addition (3d Post-N), and 6 weeks after fertilizer addition (6w Post-N). Error bars represent the standard errors of the means.
) and 16S rRNA (■) gene copies in DNA extracts from samples taken from plots NO, N144, and FYM/N (Table 1) immediately before fertilizer addition (Pre-N), 3 days after fertilizer addition (3d Post-N), and 6 weeks after fertilizer addition (6w Post-N). Error bars represent the standard errors of the means.
Phylogenetic analyses.
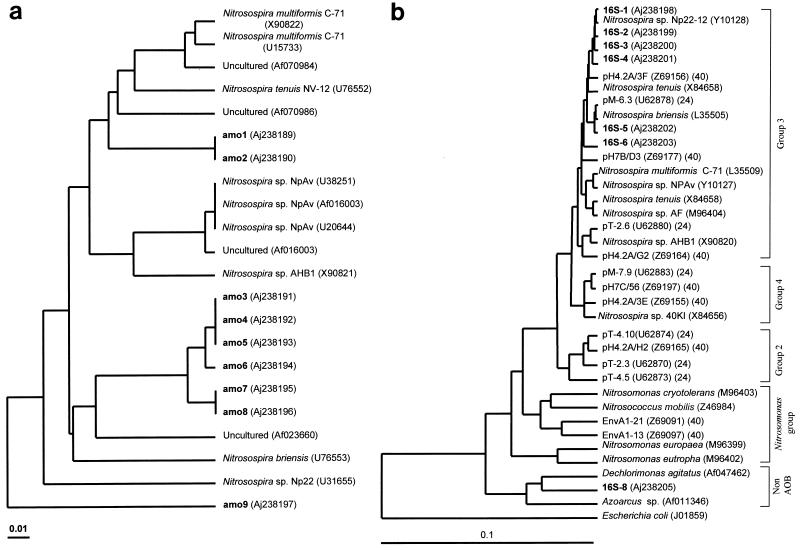
Initial phylogenetic analyses of the nine amoA gene fragments used inferred peptide sequences because these are less biased by multiple or silent nucleotide substitutions and so probably are more phylogenetically accurate (37, 44). All of the amoA peptide sequences grouped with Nitrosospira-type organisms (data not shown). Subsequent phylogenetic analyses of the nucleotides, which are potentially more accurate for closely related organisms (37, 44), placed the sequences in three groups (Fig. 5a). The majority of sequences, amo3 to amo8, formed a unique and tight group with affiliation to Nitrosospira briensis and an environmental sequence from a Norwegian pine forest; amo1 and amo 2 were most closely related to Nitrosospira tenuis NV-12 and Nitrosospira multiformis C-71 (Fig. 5a), while amo9 was related to Nitrosospira sp. strain Np22. The level of homology of the sequences to pmoA, the equivalent methane monooxygenase gene, was significantly lower than that to amoA. The eight 16S sequences obtained from the N144 plot were also dominated by sequences with strong homology to the Nitrosospira group (Fig. 5b). Most of the 16S sequences, 16S-1 to 16S-6, were similar to those defined as group 3 by Stephen et al. (40) (Fig. 5b). Sequence 16S-7 was identified as chimeric and so was not included in the rest of the study or submitted to the public databases. Sequence 16S-8 did not fall into the AOB clade, being most closely related to Dechlorimonas agitatus.
FIG. 5.
Phylogenetic tree constructed with partial amoA and Nitrosospira-type sequences from the GenBank/EMBL database (a) and partial 16S sequences from that database (b). All sequences from this study are derived from plot N144 soil and are shown in bold. The nomenclature of the 16S sequences, from the study by Stephen et al. (40), refers to the pH of the soil from which the sequence was obtained and the isolate number. Groupings are as suggested by Stephen et al. (40). The 16S gene of Escherichia coli has been used as an outgroup. Accession numbers are given in parentheses and are sometimes followed by reference numbers.
DISCUSSION
amoA and 16S genes.
Throughout this study, the amoA primers performed better than the 16S primers; the cloned amoA PCR product always contained target sequence, and the amoA cPCRs were more reproducible. This relatively high degree of primer specificity is due to the uniqueness of the amoA gene, which to date has only been found in organisms capable of oxidizing ammonia. As the ammonia monooxygenase enzyme is involved directly in ammonia oxidation, we can surmise that the amoA sequences amplified are derived from AOB. By contrast, although the ubiquity of 16S genes allows AOB to be placed on the same phylogenetic tree as other prokaryotes, it also results in the amplification of products from nontarget organisms by 16S primers (24, 40). The metabolic capabilities of the organisms characterized by such environmental 16S sequences can only be inferred.
cPCR assays.
The cPCR assays were able to enumerate the number of gene copies in soil DNA extracts and so measure the relative sizes of soil AOB populations. However, the number of gene copies in a soil DNA extract cannot be assumed to be directly equivalent to the number of AOB in the soil. Variations in gene copy number (31), DNA from moribund organisms, and losses during DNA extraction, estimated at between 52 and 78% (10), all may prejudice results. Variability among extractions has been accounted for by the standard errors given. Differences between amoA- and 16S-based cPCR estimates may be explained by differences in primer specificities, as demonstrated by phylogenetic analysis of the products. As would be expected, the less-specific 16S primers tend to give higher estimates of AOB population size.
Ammonia oxidizer population dynamics.
Prior to fertilizer addition, plots N144 and FYM/N had AOB populations corresponding to approximately 105 gene copies g of soil−1; this is similar to estimates for other soils made by most-probable-number culture methods (6) and cPCR (39). The population of plot NO still corresponded to 1.3 × 104 gene copies g of soil−1, despite having received no agricultural N inputs since 1852. Assuming that after such a long period of unchanging fertilizer management the AOB populations are stable year after year, the considerable AOB population of plot NO must survive solely on ammonium inputs from atmospheric NH4+ deposition (6.7 kg of NH4+ N ha−1 year−1 [15]) and from crop residues. The considerable quantities of N added to plots N144 and FYM/N have not resulted in large changes in population sizes when compared to that of the NO plot; however, they may have substantially altered the population structures in these plots.
The addition of fertilizer to plots N144 and FYM/N caused rapid rises in ammonium concentrations and a corresponding rise in nitrification rates, suggesting that the activity of these AOB populations was limited by the ammonia supply. In previous studies, nitrification rates in samples from plot NO have also risen quickly following addition of ammonium (11). Compared to soil ammonium concentrations, nitrification rates in plot FYM/N were greater than those in plot N144. This probably reflects the more rapid turnover of the soil ammonium pool that results from the higher rates of mineralization and immobilization associated with soils rich in organic matter (12, 14). The increases in nitrification rates are too rapid to be explained entirely by population increases and so must reflect phenotypic changes in the AOB. Indeed, data from cPCRs show that after 3 days, despite a large increase in nitrification rates, the AOB populations had not grown significantly (Fig. 4). The speed with which the AOB in all of the plots can respond to ammonium addition implies that despite minimal activity during the winter months, the populations maintain considerable metabolic potential and remain in a state of readiness. Six weeks after fertilizer addition, the AOB populations in plots N144 and FYM/N, which had received NH4NO3, had grown at rates equivalent to approximately 0.5 and 1 cellular division week−1, respectively. These increases were not reflected in the rates of nitrification, which had fallen toward the levels observed prior to fertilizer application. It is not possible to determine whether the AOB population had grown uniformly or whether we were observing changes caused by a faster-growing subpopulation within the AOB. The AOB populations, increased by the temporary increases in substrate concentrations that resulted from fertilizer addition, cannot be sustainable. Assuming that the AOB populations are stable year after year, the populations must decline during the succeeding months to reach prefertilization levels by the spring.
Phylogenetic trees.
As has been observed previously (24, 40), Nitrosospira-type sequences dominated the PCR products amplified from the soil. Direct comparisons of the amoA sequences from this and previous studies which have examined environmental amoA sequences (37, 39) are not possible, since the amplified regions do not overlap. However, comparison of amoA and 16S phylogenies suggests that all amoA sequences from this study represent organisms similar to those defined as group 3 by their 16S sequences (40). Such comparisons are possible since both genes describe similar phylogenetic structures for the AOB (37), although there are to date no amoA sequences from AOB characterized by their 16S sequence as being members of groups 2 and 4. All of the AOB 16S sequences amplified in this study also fall into group 3, which is dominated by sequences from agricultural soils at close to pH 7 (41).
The small number of sequences from this study and the restricted number of amoA sequences from characterized AOB isolates make a more detailed consideration of the population structure in this soil difficult. The amoA and 16S primers, which are likely to exhibit different primer biases, describe organisms that occupy similar phylogenetic positions, suggesting that the amplified sequences do represent the AOB present in the soil. However, the possibility of groups of AOB remaining undetected cannot be overlooked. Other studies have recorded phylogenetically similar AOB sequences (24, 41), suggesting that common groups of AOB dominate in diverse and geographically separate soils. It would be interesting to compare population structures in the three plots to determine whether the differing long-term fertilizer regimes have altered the AOB populations.
Summary.
By developing new cPCR assays specific for AOB and combining these with 15N isotopic pool dilution assays, this study has related the physiological activity of a defined group of uncultured AOB to their abundance under different fertilizer regimes. The addition of ammonium to the soil caused rapid increases in nitrifying activity which can only be explained by phenotypic changes in the AOB population. Over the longer term, this activity resulted in growth of the AOB populations to levels that are not sustainable over the year. Phylogenetic analysis of the amplified products indicates that the AOB population is similar to those described previously (24, 40, 41), suggesting that such organisms dominate in a range of widely distributed soils.
This work has furthered our understanding of the dynamics of the terrestrial nitrogen cycle, which ultimately relates to how N is supplied to crops and is lost from soil. The methods used are applicable to a wide range of organism, and are particularly suited to the investigation of soil functions known to be directly associated with a single gene product. Such a gene provides a suitable target with which to detect and quantify the organisms of interest.
ACKNOWLEDGMENTS
We thank Toby Willison and Dan Murphy for advice and support with the 15N isotopic pool dilution assays, Wendy Wilmer for ammonium and nitrate analyses of soil extracts, Rik Dunn for performing mass spectrometry analyses, and Darren Murray for statistical advice.
IACR—Rothamsted receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom.
REFERENCES
- 1.Anonymous. Rothamsted—the classical experiments. Harpenden, United Kingdom: Lawes Agricultural Trust; 1991. pp. 8–18. [Google Scholar]
- 2.Baek J M, Kenerley C M. Detection and enumeration of a genetically modified fungus in soil environments by quantitative competitive polymerase chain reaction. FEMS Microbiol Ecol. 1998;25:419–428. [Google Scholar]
- 3.Barraclough D. The use of mean abundances to interpret nitrogen-15 tracer experiments. Plant Soil. 1991;131:17–22. [Google Scholar]
- 4.Barraclough D, Puri G. The use of 15N pool dilution and enrichment to separate the heterotrophic and autotrophic pathways of nitrification. Soil Biol Biochem. 1995;27:17–22. [Google Scholar]
- 5.Bédard C, Knowles R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol Rev. 1989;53:68–84. doi: 10.1128/mr.53.1.68-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belser L W, Schmidt E L. Diversity in the ammonia-oxidizing nitrifier population of a soil. Appl Environ Microbiol. 1978;36:584–588. doi: 10.1128/aem.36.4.584-588.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks P D, Stark J M, McInteer B B, Preston T. Diffusion method to prepare soil extracts for automated nitrogen-15 analysis. Soil Sci Soc Am J. 1989;53:1701–1711. [Google Scholar]
- 8.Burns L C, Stevens R J, Laughlin R J. Determination of the simultaneous production and consumption of soil nitrite using 15N. Soil Biol Biochem. 1995;27:839–844. [Google Scholar]
- 9.Chandler D P, Schreckhise R W, Smith J L, Bolton J H. Electroelution to remove humic compounds from soil DNA and RNA extracts. J Microbiol Methods. 1997;28:11–18. [Google Scholar]
- 10.Cullen D W, Hirsch P R. Simple and rapid method for direct extraction of microbial DNA from soil for PCR. Soil Biol Biochem. 1998;30:983–993. [Google Scholar]
- 11.Dendooven, L. Unpublished data.
- 12.Donaldson J M, Henderson G S. Nitrification potential of secondary succession upland oak forests: mineralization and nitrification during laboratory incubations. Soil Sci Soc Am J. 1990;54:335–349. [Google Scholar]
- 13.Felstein J. PHYLIP V3.57 (phylogeny inference package). Seattle: Department of Genetics, University of Washington; 1994. [Google Scholar]
- 14.Fisk M C, Schmidt S K. Microbial responses to nitrogen additions in alpine tundra soil. Soil Biol Biochem. 1996;28:751–755. [Google Scholar]
- 15.Goulding K W T, Bailey N J, Bradbury N J, Hargreaves P, Howe M, Murphy D V, Poulton P R, Willison T W. Nitrogen deposition and its contribution to nitrogen cycling and associated soil processes. New Phytol. 1998;139:49–58. [Google Scholar]
- 16.Hallier-Soulier S, Ducrocq V, Mazure N, Truffaut N. Detection and quantification of degradative genes in soils contaminated by toluene. FEMS Microbiol Ecol. 1996;20:121–133. [Google Scholar]
- 17.Hastings R C, Ceccherini M T, Miclaus N, Saunders J R, Bazzicalupo M, McCarthy A J. Direct molecular analysis of ammonia-oxidising bacterial populations in cultivated soil plots treated with swine manure. FEMS Microbiol Ecol. 1997;23:45–54. [Google Scholar]
- 18.Head I M, Hiorns W D, Embley T M, McCarthy A J, Saunders J R. The phylogeny of autotrophic ammonia-oxidising bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J Gen Microbiol. 1993;139:1147–1153. doi: 10.1099/00221287-139-6-1147. [DOI] [PubMed] [Google Scholar]
- 19.Henriksen A, Selmer-Olsen A R. Automatic methods for determining nitrate and nitrite in water and soil extracts. Analyst. 1970;95:514–518. [Google Scholar]
- 20.Hiorns W D, Hastings R C, Head I M, McCarthy A J, Saunders J R, Pickup R W, Hall G H. Amplification of 16S ribosomal-RNA genes of autotrophic ammonia oxidising bacteria demonstrates the ubiquity of Nitrosospiras in the environment. Microbiology (Reading) 1995;141:2793–2800. doi: 10.1099/13500872-141-11-2793. [DOI] [PubMed] [Google Scholar]
- 21.Killham K. Heterotrophic nitrification. In: Prosser J I, editor. Nitrification. Oxford, United Kingdom: IRL Press; 1986. pp. 117–126. [Google Scholar]
- 22.Klotz M G, Alzerreca J J, Norton J M. A gene encoding a membrane protein exists upstream of the amoA/amoB genes in ammonia-oxidising bacteria; a third member of the amo operon. FEMS Microbiol Lett. 1997;150:65–73. doi: 10.1111/j.1574-6968.1997.tb10351.x. [DOI] [PubMed] [Google Scholar]
- 23.Kowalchuk G A, Naoumenko Z S, Derikx P J L, Felske A, Stephen J R, Arkhipchenko I A. Molecular analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in compost and composted materials. Appl Environ Microbiol. 1999;65:396–403. doi: 10.1128/aem.65.2.396-403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kowalchuk G A, Stephen J R, De Boer W, Prosser J I, Embley T M, Woldendorp J W. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krom M. Spectrophotometric determination of ammonia; a study of modified Berthelot reaction using salicylate and dichloroisocyanurate. Analyst. 1980;105:305–316. [Google Scholar]
- 26.Lee S-Y, Bollinger J, Bezdicek D, Ogram A. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl Environ Microbiol. 1996;62:3787–3793. doi: 10.1128/aem.62.10.3787-3793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCulloch R K, Choong C S, Hurley D M. An evaluation of competitor type and size for use in the determination of mRNA by competitive PCR. PCR Methods Appl. 1995;4:219–226. doi: 10.1101/gr.4.4.219. [DOI] [PubMed] [Google Scholar]
- 28.McDonald R M L. Nitrification in soil: an introductory history. In: Prosser J I, editor. Nitrification. Oxford, United Kingdom: IRL Press; 1979. pp. 1–16. [Google Scholar]
- 29.McTavish H, Fuchs J A, Hooper A B. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J Bacteriol. 1993;175:2436–2444. doi: 10.1128/jb.175.8.2436-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mobarry B K, Wagner M, Urbain V, Rittmann B E, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Norton J M, Low J M, Klotz M G. The gene encoding ammonia monooxygenase subunit A exists in three nearly identical copies in Nitrosospira sp. NpAV FEMS Microbiol Lett. 1996;139:181–188. doi: 10.1111/j.1574-6968.1996.tb08200.x. [DOI] [PubMed] [Google Scholar]
- 32.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 33.Poulton, P. R. Unpublished data.
- 34.Raeymaekers L. Quantitative PCR: theoretical considerations with practical implications. Anal Biochem. 1993;214:582–585. doi: 10.1006/abio.1993.1542. [DOI] [PubMed] [Google Scholar]
- 35.Reilly K, Attwood G T. Detection of Clostridium proteoclasticum and closely related strains in the rumen by competitive PCR. Appl Environ Microbiol. 1998;64:907–913. doi: 10.1128/aem.64.3.907-913.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rotthauwe J-H, de Boer W, Liesack W. Comparative analysis of gene sequences encoding ammonia mono-oxygenase of Nitrosospira sp. AHB1 and Nitrosolobus multiformis C-71. FEMS Microbiol Lett. 1995;133:131–135. doi: 10.1111/j.1574-6968.1995.tb07873.x. [DOI] [PubMed] [Google Scholar]
- 37.Rotthauwe J-H, Witzel K-P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneeberger C P, Speiser F K, Zellinger R. Quantitative detection of reverse transcriptase-PCR products by means of a novel and sensitive DNA stain. PCR Methods Appl. 1995;4:234–238. doi: 10.1101/gr.4.4.234. [DOI] [PubMed] [Google Scholar]
- 39.Stephen J R, Chang Y-J, MacNaughton S J, Kowalchuk G A, Leung K T, Flemming C A, White D C. Effect of toxic metals on indigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl Environ Microbiol. 1999;65:95–101. doi: 10.1128/aem.65.1.95-101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephen J R, McCaig A E, Smith Z, Prosser J I, Embley T M. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephen J R, Kowalchuk G A, Bruns M-A V, McCaig A E, Phillips C J, Embley T M, Prosser J I. Analysis of β-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl Environ Microbiol. 1998;64:2958–2965. doi: 10.1128/aem.64.8.2958-2965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watkins N, Barraclough D. Gross rates of N mineralization associated with the decomposition of plant residues. Soil Biol Biochem. 1996;28:169–175. [Google Scholar]
- 43.Willison T W, Baker J C, Murphy D V, Goulding K W T. Comparison of a wet and dry 15N isotopic dilution technique as a short-term nitrification assay. Soil Biol Biochem. 1998;30:661–663. [Google Scholar]
- 44.Yamamoto S, Harayama S. Phylogenetic analysis of Acinetobacter strains based on the nucleotide sequences of gyrB genes and on the amino acid sequences of their products. Int J Syst Bacteriol. 1996;46:506–511. doi: 10.1099/00207713-46-2-506. [DOI] [PubMed] [Google Scholar]