Abstract
This article aims to explore hub genes related to different clinical types of cases with COVID-19 and predict the therapeutic drugs related to severe cases. The expression profile of GSE166424 was divided into four data sets according to different clinical types of COVID-19 and then calculated the differential expression genes (DEGs). The specific genes of four clinical types of COVID-19 were obtained by Venn diagram and conducted enrichment analysis, protein–protein interaction (PPI) networks analysis, screening hub genes, and ROC curve analysis. The hub genes related to severe cases were verified in GSE171110, their RNA-specific expression tissues were obtained from the HPA database, and potential therapeutic drugs were predicted through the DGIdb database. There were 536, 266, 944, and 506 specific genes related to asymptomatic infections, mild, moderate, and severe cases, respectively. The hub genes of severe specific genes were AURKB, BRCA1, BUB1, CCNB1, CCNB2, CDC20, CDC6, KIF11, TOP2A, UBE2C, and RPL11, and also differentially expressed in GSE171110 (P < 0.05), and their AUC values were greater than 0.955. The RNA tissue specificity of AURKB, CDC6, KIF11, UBE2C, CCNB2, CDC20, TOP2A, BUB1, and CCNB1 specifically enhanced on lymphoid tissue; CCNB2, CDC20, TOP2A, and BUB1 specifically expressed on the testis. Finally, 55 drugs related to severe COVID-19 were obtained from the DGIdb database. Summary, AURKB, BRCA1, BUB1, CCNB1, CCNB2, CDC20, CDC6, KIF11, TOP2A, UBE2C, and RPL11 may be potential diagnostic biomarkers for severe COVID-19, which may affect immune and male reproductive systems. 55 drugs may be potential therapeutic drugs for severe COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10142-023-00998-1.
Keywords: COVID-19, Bioinformatic Analysis, Hub Genes, Diagnostic Biomarker, Therapeutic Drug Prediction
Introduction
Coronavirus Disease 2019 (COVID-19) is a newly emerged infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The SARS-CoV-2 using spike glycoprotein attaches to the ACE2 receptor protein on the surface of human respiratory cells and is primed for TMPRSS2, invading the body by binding to it and inducing infection (Hoffmann et al. 2020). As of CEST, 24 April 2022, there have been just over 500 million confirmed cases of COVID-19 worldwide and over 6 million cumulative deaths (WHO 2022), which seriously threaten global public health.
COVID-19 was divided into five types: asymptomatic infections, mild, moderate, severe, and critical cases (Gao et al. 2021). Asymptomatic infections refer to those who test positive for nucleic acid but do not have typical clinical symptoms or chest imaging findings, but it could spread pathogens, which is a potential source of infection. A meta-analysis showed that 20% of asymptomatic infections included 5829 pediatric patients from 48 studies as of 30 April 2020 (Cui et al. 2021). By analyzing more than 350 studies as of April 2, 2021, Pratha Sah et al. (2021) found that the percentage of asymptomatic infections was 35.1%. Studies have shown that vaccination is effective in reducing the severity of symptoms associated with COVID-19 (Dagan et al. 2021; Sadoff et al. 2021; Lumley et al. 2022). Therefore, the proportion of asymptomatic infections or mild patients with SARS-CoV-2 infections will increase with the popularization of vaccines. For asymptomatic infections, the most important thing is to be able to identify them early and effectively so that they can be isolated in time and control the spread of COVID-19. As of 19 March 2020, there were 24% severe cases of 12,960 cases in Italy (Ortenzi et al. 2020). By 10 May 2020, about 20% of confirmed patients required hospitalization, and 25% of them required intensive care (Uddin et al. 2020). The in-hospital mortality rate of 28.7% among 326,993 COVID-19 patients from Kaiser Permanente Southern California in the year since February 2021 (Huang et al. 2022). Hence, there is an urgent need to find specific biomarkers to identify or predict severe cases, which will be beneficial to achieving secondary prevention and improving the prognosis, while providing a reference for predicting targeted drugs.
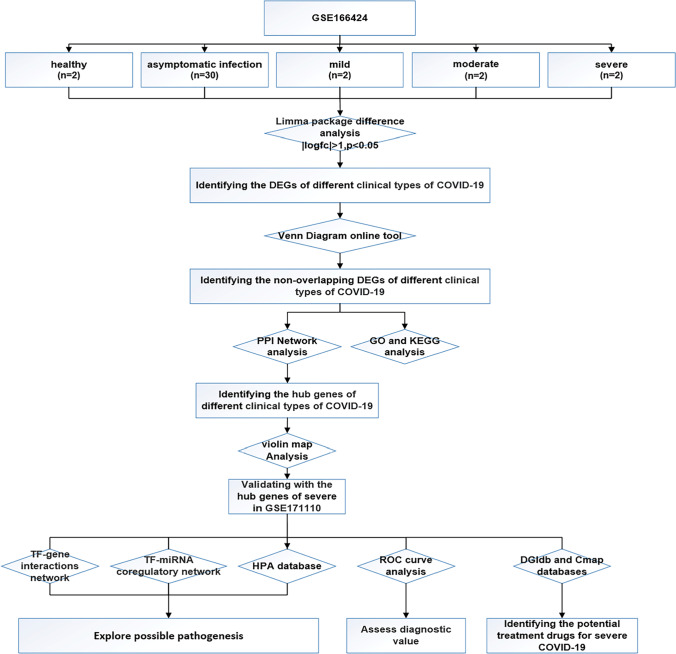
In the study, the expression profile of GSE166424 was used for differential analysis to identify hub genes of four clinical types of COVID-19. And then the hub genes of severe specific genes were verified by evaluating their diagnostic value for severe cases by the ROC curve in GSE171110. Finally, the potential targeted drugs for severe cases were predicted based on the hub genes. The flow chart of the integrated bioinformatics approach was shown in Fig. 1.
Fig. 1.
Flow chart of integrated bioinformatics approach
Methods
The data source
GSE166424 and GSE171110 were expression profiles by high throughput sequencing from Gene Expression Omnibus (GEO) database. GSE166424 is based on the GPL20301 platform and included 38 samples, containing 2 healthy controls, 2 mild cases, 2 moderate cases, 2 severe cases, and 30 asymptomatic infections of COVID-19. GSE166424 was divided into four data sets based on the clinical types of COVID-19: 2 healthy controls and 2 mild cases, 2 healthy controls and 2 moderate cases, 2 healthy controls and 2 severe cases, 2 healthy controls and 30 asymptomatic infections. GSE171110 is based on the GPL16791 platform and included 54 samples, containing 10 healthy controls and 44 severe COVID-19. Merged the Asymptomatic sub-dataset in GSE166424 with GSE17110, and used the Combat function of the sva package to remove the batch effect. The RNA sequencing data set information was shown in Table 1.
Table 1.
Details of gene expression profiling
Data standardization and DEGs screening
Based on R (Version 3.6.3), the expression profiling was converted to TPM format for standardization, and the limma package was used to screen the differentially expressed genes (DEGs) between healthy and infected cases. The screening conditions were P < 0.05 and |log2FC|> 1. To better display the DEGs, the volcano map and heat map were analyzed. The heat map was made with a heatmap package. Furthermore, the non-overlapping up-regulated DEGs and non-overlapping down-regulated DEGs among mild, moderate, severe cases and asymptomatic infections (hereinafter referred to as mild, moderate, severe, and asymptomatic specific gene) were produced by the Venn Diagram online tool.
Functional analysis of DEGs
DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/) was used for Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of specific genes among mild, moderate, severe cases and asymptomatic infections, and then visualization was performed using R. The screening condition was P < 0.05.
Construct the PPI network and screen hub genes
Protein–Protein Interaction (PPI) networks were constructed for specific genes through the STRING (v11.5) (https://string-db.org/), and the filtering condition was a comprehensive score > 0.4. Then, the interactive information was downloaded and visualized by Cytoscape (v3.8.0). Hub genes were screened by CytoHubba in Cytoscape, using five algorithms (MCC, DMNC, MNC, Degree, EPC) to calculate the top 15 hub genes. Finally, the intersection of the results of the five algorithms was taken, and the genes that appeared more than or equal to three times was selected as the final hub genes.
Construct TF-gene interactions and TF-miRNA coregulatory network
NetworkAnalyst (https://www.networkanalyst.ca) is a database for comprehensive gene expression profiling & network visual analytics. The TF-gene interactions network and TF-miRNA coregulatory network of hub genes of severe COVID-19 were constructed through the ENCODE database and RegNetwork repository included in the NetworkAnalyst database, respectively.
Predict potential therapeutic drugs for severe cases
Potential therapeutic drugs for severe cases were predicted based on the 11 hub genes of severe specific genes by the Drug Gene Interaction Database (DGIdb 4.0) (https://dgidb.org/) databases. And the preset filter was approved by the Food and Drug Administration (FDA). Finally, we constructed a gene-drug network through Cytoscape.
Statistic analysis
The 15 hub genes between asymptomatic infections and healthy controls were analyzed for the ROC curve in GSE166424-171110. And the 11 hub genes selected between severe cases and healthy controls were verified in GSE171110. The limma package was used for screening the DEGs, and the ggplot2 package was used to draw the violin map. Subsequently, the pROC package was used to draw the ROC curve for genes. Statistical analysis using t-test and P < 0.05 was considered statistically significant.
Results
DEGs
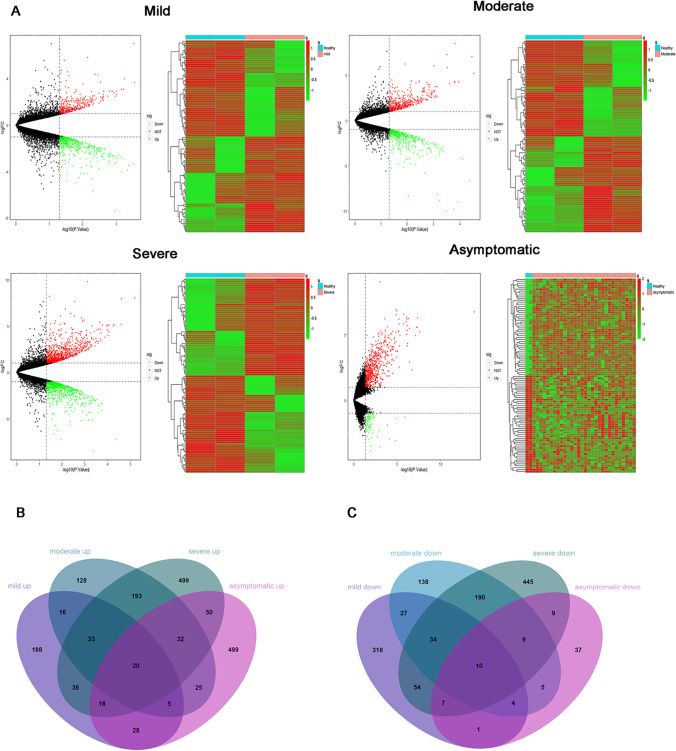
The limma package was used to screen the DEGs between healthy controls and four clinical types of COVID-19 in GSE166424 based on R (Version 3.6.3), with P < 0.05 and |log2FC|> 1. And 799 DEGs were detected in the mild cases, of which 344 genes were up-regulated, and 455 genes were down-regulated. In the moderate cases, 869 DEGs were detected, containing 452 genes up-regulated, and 417 genes down-regulated. And 1639 DEGs were detected in the severe cases, of which 881 genes were up-regulated and 758 genes were down-regulated. The asymptomatic infections revealed 753 DEGs, of which 671 genes were up-regulated and 82 genes were down-regulated. These DEGs were visualized using volcano maps and heatmaps (Fig. 2A). Afterward, 188 up-regulated mild specific genes and 318 down-regulated mild specific genes, 128 up-regulated moderate specific genes and 138 down-regulated moderate specific genes, 499 up-regulated severe specific genes and 445 down-regulated severe specific genes, and 499 up-regulated asymptomatic specific genes and 37 down-regulated asymptomatic specific genes were screened by the Venn Diagram online tool (Fig. 2B, C).
Fig. 2.
Identification of the specific genes of COVID-19. A Volcano map and heat map of DEGs in mild, moderate, severe cases and asymptomatic infections. B Venn plot of up-regulated specific genes in mild, moderate, severe cases and asymptomatic infections. C Venn plot of down-regulated specific genes in mild, moderate, severe cases and asymptomatic infections
GO functional enrichment analysis
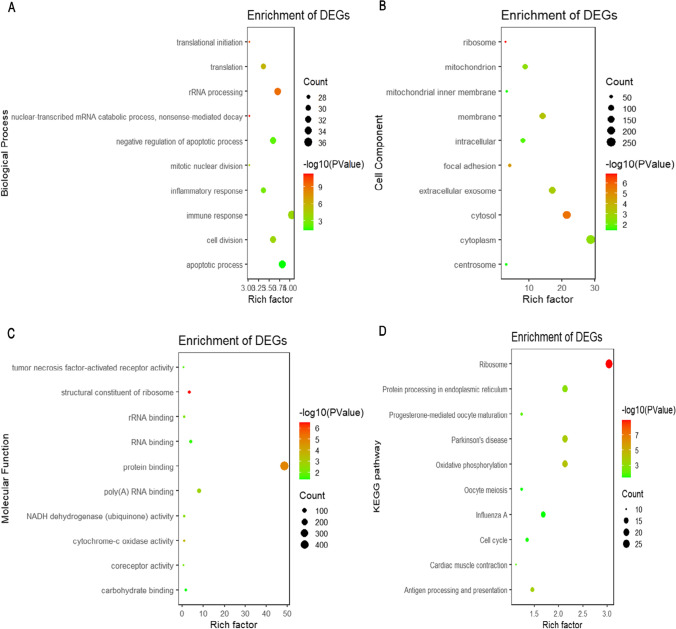
In terms of biological processes, the mild specific genes were significantly enriched in regulating cell division, mitotic nuclear division, and Moderate/M transition of the mitotic cell cycle (Fig S1A); And the moderate specific genes were mainly enriched in the steroid hormone-mediated signaling pathway (Fig S2A); Meanwhile, the severe specific genes were mainly enriched in the nuclear-transcribed mRNA catabolic process, rRNA processing, and translational initiation (Fig. 3A); The asymptomatic specific genes were significantly enriched in the regulation of cell adhesion, extracellular matrix organization, and Moderate/M transition of the mitotic cell cycle (Fig S3A). In terms of cell composition, the mild specific genes mainly mediated nucleoplasm, cytosol, intracellular, and membrane (Fig S1B), while the moderate specific genes were not mediated cellular components; and the severe specific genes mainly mediated ribosome, cytosol, and focal adhesion (Fig. 3B), with the asymptomatic specific genes, a mediated integral component of the plasma membrane, extracellular exosome (Fig S3B). In terms of molecular function, the mild specific genes were mainly significantly enriched in nucleic acid binding, metal ion binding, and epidermal growth factor receptor binding (Fig S1C); As well, the moderate specific genes were mainly enriched in steroid hormone receptor activity (Fig S2C). The structural constituent of Ribosome, protein binding and cytochrome-c oxidase were significantly enriched in the severe specific genes (Fig. 3C). And the asymptomatic specific genes were mainly enriched in mannosyltransferase activity, and carbohydrate binding (Fig S3C).
Fig. 3.
GO enrichment and KEGG pathway analysis of severe specific genes. A Biological process. B Cell composition. C Molecular function. D KEGG pathway
KEGG pathway analysis
KEGG pathway analysis showed that the mild specific genes were mainly concentrated in the Cell cycle, Progesterone-mediated oocyte maturation, N-Glycan biosynthesis, and Transcriptional misregulation in cancer (Fig S1D); Rheumatoid arthritis and Hedgehog signaling pathway were the main sources of the moderate specific genes (Fig S2C). And the severe specific genes were mainly enriched in Ribosome, Oxidative phosphorylation, Parkinson's disease, Antigen processing and presentation, Protein processing in the endoplasmic reticulum, Progesterone-mediated oocyte maturation, Cardiac muscle contraction, Hematopoietic cell lineage, Cell cycle, Oocyte meiosis, Influenza A, p53 signaling pathway (Fig. 3D); ECM-receptor interaction, Proteoglycans in cancer, Focal adhesion, Protein digestion and absorption, PI3K-Akt signaling pathway, Bladder cancer and Pathways in cancer were mainly enriched in the asymptomatic specific genes (Fig S3D).
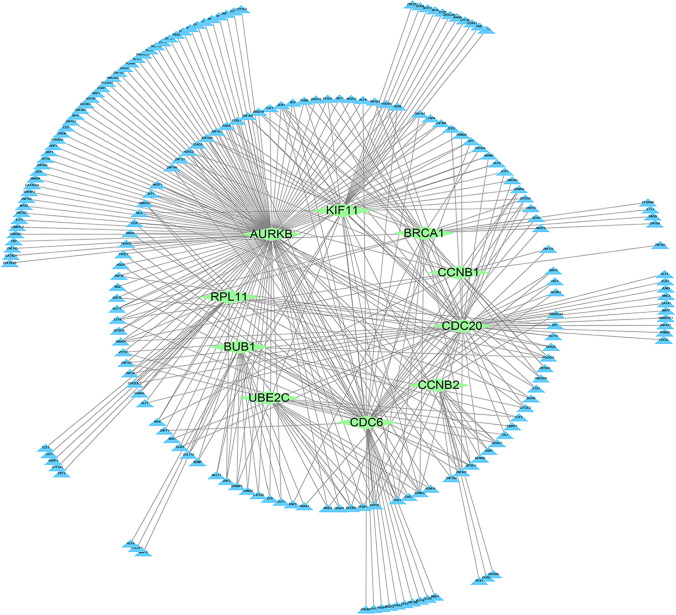
PPI interactive network and hub genes selection
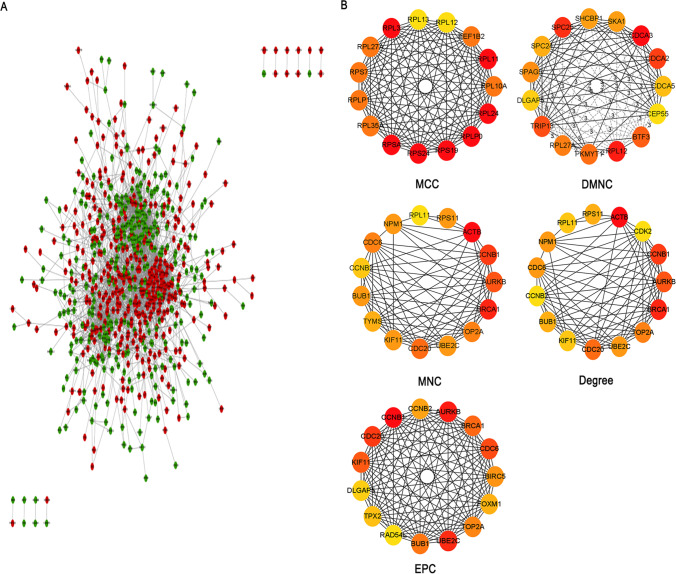
The PPI interaction network of mild specific genes consisted of 444 nodes and 869 edges, with the average local clustering coefficient being 0.348 (Fig S4A). And the PPI network of moderate specific genes was composed of 222 nodes and 109 edges, and the average local clustering coefficient was 0.268 (Fig S5A). As well, the PPI network of severe specific genes consisted of 3,911 edges and 849 nodes, and the average local clustering coefficient was 0.381(Fig. 4A). Finally, the PPI interaction network of asymptomatic specific genes consisted of 421 edges and 419 nodes, and the average local clustering coefficient was 0.360(Fig S6A), and then visualized by Cytoscape (V3.8.0). By CytoHubba calculated the top 15 hub genes of five algorithms (MCC, DMNC, MNC, Degree, EPC), and the genes that appeared more than or equal to 3 times were selected as the final hub genes. The hub genes of mild specific genes were 17 down-regulated genes (FBXO5, KIF20A, MCM6, TPX2, CCNB1, DLGAP5, HJURP, KIF11, MELK, PLK1, TRIP13, CDCA8, CHEK1, CHEK2, CKS1B, ECT2, H4C3) (Figure S4B); There were 9 up-regulated genes (BRCA2, CXCL5, H2AX, RECQL4, ATP5F1D, HBEGF, XIST, RARA, ZBTB16), and 6 down-regulated genes (RAD52, ATP6V1D, NDUFS5, ATP6V1E2, CCR4, TOP1MT) in the hub genes of moderate specific genes(Figure S5B); The hub genes of severe specific genes were 10 up-regulated genes (AURKB, BRCA1, BUB1, CCNB1, CCNB2, CDC20, CDC6, KIF11, TOP2A, UBE2C), and 1 down-regulated gene (RPL11) (Fig. 4B, Table S1). At the last one, the hub genes of asymptomatic specific genes were 14 up-regulated genes (CCL3, CD9, IL4, IL7, NCAM1, PDGFRB, TBX21, CDH1, IL10, IL6, ITGB3, THBS1, COL6A2, ITGB4), and 1 down-regulated gene (CD27) (Fig S6B).
Fig. 4.
The PPI interaction network of specific genes and top 15 genes of five algorithms in severe COVID-19. A The PPI interaction network of severe specific genes. The red nodes represent up-regulated DEGs and the green nodes represent down-regulated DEGs. B The top 15 genes of five algorithms in severe cases
ROC curve analysis of hub genes
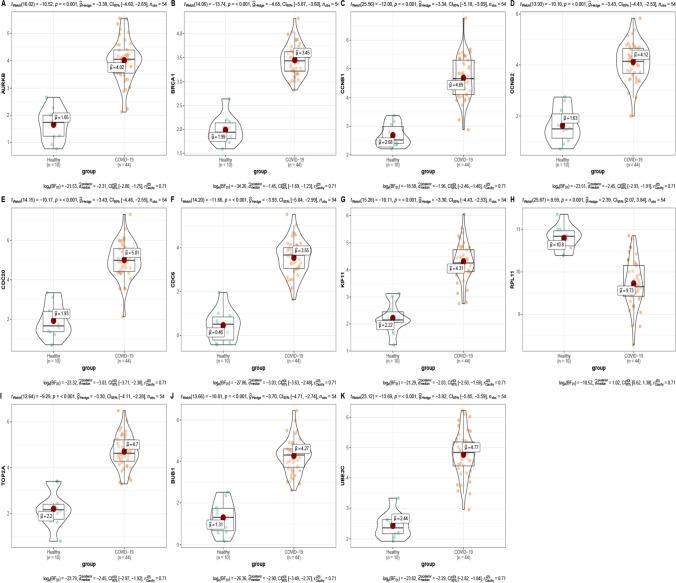
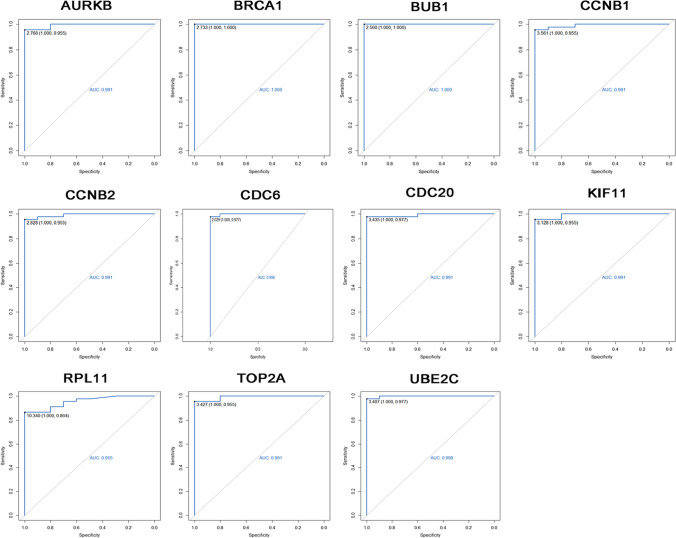
The expression and diagnostic value evaluation of 15 hub genes of asymptomatic infection were verified in the GSE166424-171110 dataset. The expression of CD27, ITGB3, THBS1, IL10, and COL6A2 was statistically different between healthy and asymptomatic infections, but the expression trend of COL6A2 was the opposite of GSE166424 (Fig S7). The AUC values of CD27, ITGB3, THBS1, and IL10 in GSE166424-171110 were greater than 0.830 (Fig S8). To verify the expression of 11 hub genes of severe specific genes, the expression profile of GSE171110 was used to verify. The expression of AURKB, BRCA1, BUB1, CCNB1, CCNB2, CDC20, CDC6, KIF11, TOP2A, UBE2C, and RPL11 were statistically significant between severe cases and healthy controls (P < 0.05) in GSE171110 (Fig. 5), which were the same expression trend of 11 hub genes in GSE166424 (Table S2). ROC curve analysis was used to predict the diagnostic level of 11 hub genes for severe specific genes. The AUC values of 11 hub genes of severe specific genes in GSE171110 were greater than 0.955 (Fig. 6).
Fig. 5.
The violin map of 11 hub genes of severe specific genes in GSE171110. A-K is violin diagram of AURKB, BRCA1, CCNB1, CCNB2, CDC20, CDC6, KIF11, RPL11, TOP2A, BUB1, and UBE2C, respectively
Fig. 6.
The ROC curve of 11 hub genes of severe COVID-19 in GSE171110
RNA tissue expression of hub genes
The Human Protein Atlas (HPA) database (https://www.proteinatlas.org/) detected the RNA tissue expression of hub genes of severe specific genes as shown in Table 2. In the HPA database, RNA tissue expression of the 11 hub genes was exhibited in Fig S8. The RNA tissue specificity of AURKB, CDC6, KIF11, UBE2C, CCNB2, CDC20, TOP2A, BUB1, and CCNB1 specifically enhanced on lymphoid tissue; CCNB2, CDC20, TOP2A, and BUB1 specifically expressed in testis. AURKB was also specifically expressed in the bone marrow.
Table 2.
RNA tissue specificity expression of hub genes of severe specific genes
| Tissue | Gene |
|---|---|
| Lymphoid tissue | AURKB, CDC6, KIF11, UBE2C, CCNB2, CDC20, TOP2A, BUB1, CCNB1 |
| Testis | CCNB2, CDC20, TOP2A, BUB1 |
| Bone marrow | AURKB |
TF-gene interactions
The TF-gene interactions network was constructed through the NetworkAnalyst database. The TF-gene interactions of 11 hub genes of severe COVID-19 contain 213 nodes and 369 edges. This network contains 203 TF-genes and 10 hub genes. Except TOP2A was not found to be regulated by TF-genes, these 203 TF-genes regulated more than one hub gene (Fig. 7, Table S3). Among them, AURKB had the highest degree score and was targeted by 131 TF- genes. Furthermore, SAP30 (degree score = 6), PHF8 (degree score = 6), and KDM5B (degree score = 6) were the top three interactive TF-genes regulating the most hub genes.
Fig. 7.
The TF-gene interactions network. The green nodes represent hub genes of severe COVID-19. The blue nodes represent TF-genes
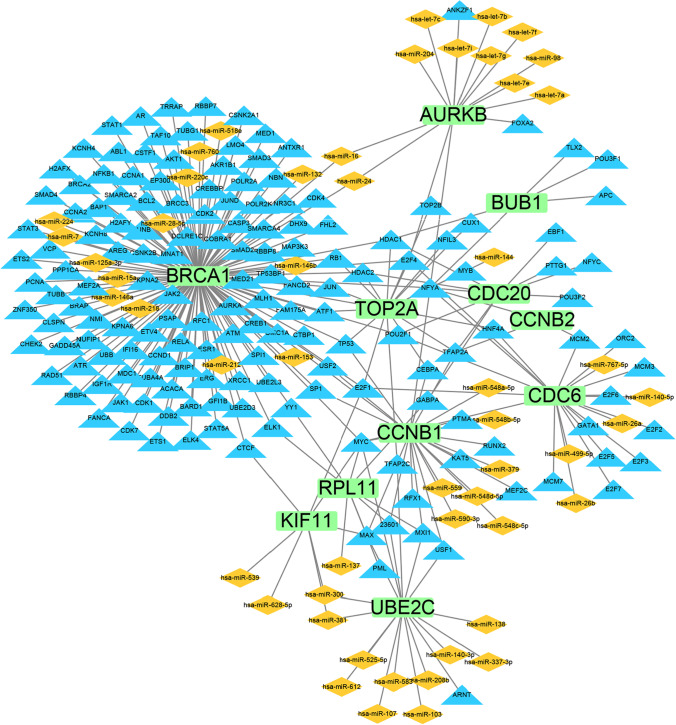
TF-miRNA coregulatory network
The TF-miRNA coregulatory network was constructed using the NetworkAnalyst database. This network consists of 226 nodes and 271 edges, containing 11 hub genes, and 52 miRNAs, 163 TF-genes (Fig. 8, Table S4). BRCA1 (degree score = 142) was the top interactive gene that was regulated by 16 miRNAs and 126 TF-genes.
Fig. 8.
The TF-miRNA coregulatory network. The green nodes represent hub genes of severe COVID-19. The orange nodes represent miRNA. The blue nodes represent TF-genes
Predict potential therapeutic drugs of hub genes of severe specific genes
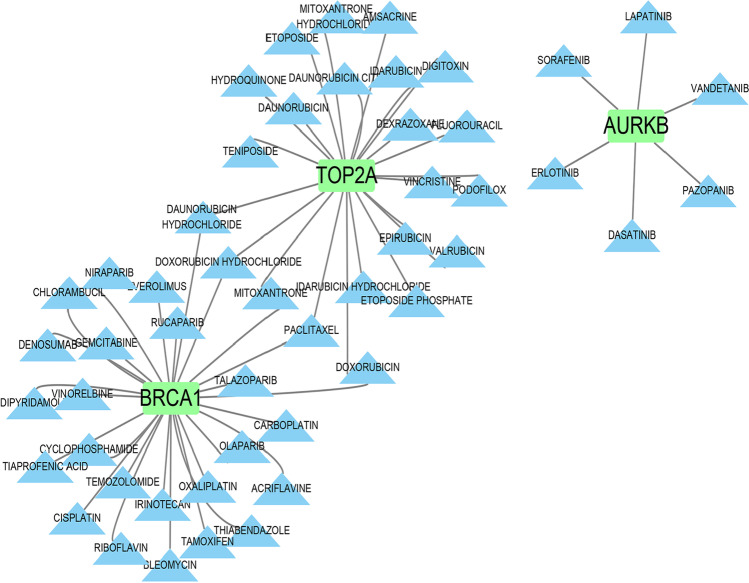
The DGIdb database was used to predict the drugs related to 11 hub genes of severe specific genes, and 55 drugs were obtained (Table S5). And the gene-drug interaction network was shown in Fig. 9. The BRCA1 (27/55) interacts with most potential therapeutic drugs, followed by TOP2A (22/55).
Fig. 9.
Genes and drugs interaction network of hub genes of severe COVID-19 from DGIdb database
Discussion
COVID-19 is a newly emerged infectious disease of the global pandemic caused by SARS-CoV-2, which is a serious threat to global public health. In this study, we found that AURKB, BRCA1, BUB1, CCNB1, CCNB2, CDC20, CDC6, KIF11, TOP2A, UBE2C, and RPL11 were the hub genes of severe specific genes and had high diagnostic value (AUC > 0.955). In the HPA database, we found the hub genes of severe COVID-19 were mainly specifically expressed in lymphoid tissue, followed by the testis. Furthermore, we identified 55 potential therapeutic drugs for severe cases from the DGIdb database.
In terms of the KEGG pathway, the severe specific genes were mainly enriched in ribosomes, Influenza A, Parkinson's disease, Cardiac muscle contraction, and so on. Thoms, M. et al. (Thoms et al. 2020) discovered that nonstructural protein 1 of SARS-Cov-2 binding to 40S ribosomal subunit, prevented the translation of mRNA and accelerated degradation of cellular mRNA, which resulted in host protein translation shut down. Both COVID-19 and influenza are respiratory infections caused by viral infections with similar symptoms (Manzanares-Meza and Medina-Contreras 2020). As previously mentioned, regarding SARS-Cov-2 entry to the host by ACE2, which was enriched in the heart, and central nervous system (Barrantes 2020; Gkogkou et al. 2020). Pavel A, et al. (Pavel et al. 2020) proposed that COVID-19 may be related to Parkinson's susceptibility, which indicated that clinically, attention should be paid to the long-term effects of COVID-19 survivors in neurodegenerative diseases such as Parkinson's. Yang J, et al. (Yang et al. 2021) discovered that ACE2 prioritize enriched in cardiomyocytes, and is mainly enriched in the processes of cardiac muscle contraction.
This study found the hub genes of asymptomatic specific genes were 14 up-regulated genes (CCL3, CD9, IL4, IL7, NCAM1, PDGFRB, TBX21, CDH1, IL10, IL6, ITGB3, THBS1, COL6A2, ITGB4), and 1 down-regulated gene (CD27). CCL3, CD9, IL4, IL7, IL10, IL6, and CD27 are all cytokines that participate in the immune response to COVID-19, especially IL6, which is related to the severity of COVID-19. Also, the anti-IL6 treatment drug Tocilizumab is an effective treatment for severe cases (Ye et al. 2020). TBX21, a human homologous gene of the mouse Tbx21/Tbet gene, encothe ded Tbx21 protein as a Th1 cell-specific transcription factor, which controls the expression of the iconic Th1 cytokine IFNG (Leng et al. 2016).
Meanwhile, the hub genes of severe specific genes were verified by GSE171110, and the hub genes were 10 up-regulated genes (AURKB, BRCA1, BUB1, CCNB1, CCNB2, CDC20, CDC6, KIF11, TOP2A, UBE2C), and 1 down-regulated gene (RPL11). AURKB is a member of the aurora kinase, and the abnormal expression of Aurora kinase has been confirmed to be related to the occurrence and development of various cancers, such as breast cancer, Lung cancer (Tang et al. 2017). Wang B, et al. (Wang and Huang 2020) identified that lung and colorectal cancer patients had higher susceptibility to SARS-CoV-2 compared to other types of cancer. Bertran-Alamillo J, et al. (Bertran-Alamillo et al. 2019) found that AURKB is a potential therapeutic target in NSCLC patients progressing on EGFR TKIs and not harboring resistance mutations. Meanwhile, Cheng J, et al. (Cheng et al. 2021) discovered that TMPRSS2 was expressed at the highest level in the small intestine, followed by the prostate, moreover, the TMPRSS2 gene was significantly increased in prostate cancer tissue, indicating susceptibility to SARS-CoV-2 and correlation with the severity of COVID-19. In this article, AURKB was an up-regulated gene in severe cases of COVID-19, and the result was consistent with those of the above studies. BUB1 (Davidson et al. 2014) encodes a serine/threonine protein kinase and is strongly associated with AURKA and AURKB mRNA levels. TOP2A (Lee and Berger 2019) plays an important role in DNA replication and mitosis. Because the AUC values of the 11 hub genes were greater than 0.955 and had higher sensitivity and specificity, these genes were potential biomarkers of severe cases.
Our results showed the RNA tissue specificity expression of hub genes of severe cases concentrated on lymphoid tissue, testis tissue, and bone marrow, indicating that severe cases of COVID-19 may affect immunity and male reproductive systems. Moderate COVID-19 had lower levels of inflammatory markers, enriched tissue repair genes, and increased inflammatory markers with disease severity, leading to cytokine storms (Lucas et al. 2020). The severity of COVID-19 in adults is inversely correlated with the number of innate lymphoid cells (ILCs) (Silverstein et al. 2022). Jana Ihlow et al. (Ihlow et al. 2021) observed massive B cell reduction in 64% of 11 COVID-19 decedents. The severity of COVID-19 is closely related to immune function, and elderly patients with underlying diseases are more likely to develop severe diseases or even die (Zhavoronkov 2020).
In this study, CCNB2, CDC20, TOP2A, and BUB1 specificity expression on the testis. Docherty AB et al. (Docherty et al. 2020) discovered that 60% of 20,133 UK COVID-19 hospitalizations were male. A meta-analysis including 3,111,714 global COVID-19 cases, found similar numbers of male and female infections, however, compared with women, men have higher disease severity and mortality (Peckham et al. 2020). These results suggest gender disparities in severe COVID-19, and growing evidence suggests that expression of ACE2 and TMPRSS2 mediates sex differences in viral entry mechanisms (Nassau et al. 2022). Zhang, J et al. (Zhang et al. 2021)found that ACE2 has a higher expression level in the testis compared to other tissues. Besides, androgen regulates TMPRSS2 gene expression, which was elevated by androgen (Gkogkou et al. 2020). In postmortem testicular tissue from 12 patients with COVID-19, Yang et al. (Yang et al. 2020) found that no SARS-CoV-2 virus RNA was found in the testes in most (90%) cases, however, pathological examination revealed damage to testicular tissue. Therefore, attention should be paid to the reproductive prognosis of male COVID-19 patients and should be determined to receive timely treatment. At present, there are more and more reports on the infection of the new coronavirus and the extrapulmonary organs. Mousavi et al. found that the infection of the SARS-CoV-2 changed the gene expression of the choroid plexus cells, and upregulated of inflammation and immune response-related pathways involved in key functions of hepatocytes (Mousavi et al. 2022).
The TF-gene interactions network contained 203 TF-genes and 10 hub genes, and SAP30, PHF8, and KDM5B were the top three interactive TF-genes. SAP30 is a member of the msin3-histone deacetylase (HDAC) co-repressor complex and is the binding protein of the human herpesvirus 8 (HHV-8) latency-associated nuclear antigen (LANA) (Krithivas et al. 2000). The interaction between Rift Valley fever virus non-institutional protein NSs and SAP30 is necessary for the establishment of interaction between NSs and host DNA (Mansuroglu et al. 2010). PHF8 is a histone demethylase involved in tumor development and malignant progression in multiple types of cancer (Li et al. 2017). KDM5B is an epigenetic regulator of chromatin that exerts demethylation (Zhao et al. 2022). Regulates ACE2 expression by modulating KDM5B demethylation activity to influence SARS-CoV-2 virus entry into host cells in an epigenetically altered manner (Jit et al. 2021).
The TF-miRNA coregulatory network contained 11 hub genes, 52 miRNAs, and 163 TF-genes. We found that has-miR-16 and has-miR-24 have a higher degree value of 2. Using data from single-cell RNA sequencing, Li, C et al. (Li et al. 2022) identified potential SARS-CoV-2 virus-targeting miRNAs, has-miR-302c-5p and has-miR-16-5p. Wicik, Z et al. (2020) has-miR-16-5p regulate the ACE2 network and SARS-CoV-2 virus-associated proteins. Wang, Y et al. (Wang et al. 2021) found that exosome circulating miR-24-3p reduced SARS-CoV-2 replication and spike glycoprotein expression, moreover, long-term exercise can increase the antiviral function of miR-24-3p.
This study obtained 55 drugs related to severe COVID-19 hub genes from the DGIdb database. Acriflavine (ACF) is a specific inhibitor of SARS-CoV-2 papain-like protease. It has been confirmed by in vitro and in vivo experiments that ACF has the activity of inhibiting the replication of SARS-CoV-2 (Napolitano et al. 2022). Everolimus, an FDA-approved mTOR inhibitor, has been developed for the treatment of cancer (Hua et al. 2019), however, everolimus exerts immunostimulatory effects at low doses. Everolimus is currently considered a geriatric protector, which can improve antiviral immunity in the elderly, although evidence from large clinical studies is still lacking. As we all know, the elderly are susceptible to SARS-CoV-2, so the efficacy of the elderly protective agent in the treatment of elderly patients with a new crown can be further studied (Zhavoronkov 2020). About 20% of COVID-19 patients develop acute respiratory distress syndrome (ARDS), and about 90% of non-survivors have ARDS. The study found that a combination of doxorubicin (DOX) and BSA nanoparticles can significantly reduce sepsis-related lung damage in mice (Qiao et al. 2021). COVID-19 patients may trigger blood clots by triggering the production of autoantibodies (Hampton 2021). Dipyridamole is an antiplatelet drug that inhibited the replication of SARS-CoV-2 (Hampton 2021). Moreover, an observational study found that antiplatelet drugs significantly reduce COVID-19 in-hospital mortality (Chow et al. 2021). Tamoxifen (TAM) is an anti-estrogen drug, and in vitro and in vivo experiments have shown that TAM can inhibit SARS-CoV-2 infection and have anti-inflammatory effects (Zu et al. 2021). Most drugs are anti-tumor drugs, the main mechanism is to inhibit virus entry, virus replication, and inhibition of viral enzyme activity.
The limitations of this study are the lack of data to verify the hub genes of other subtypes except for severe COVID-19, and further verification is needed; furthermore, further experiments are needed to verify the diagnostic value of the hub genes of severe COVID-19.
In conclusion, AURKB, BRCA1, BUB1, CCNB1, CCNB2, CDC20, CDC6, KIF11, TOP2A, UBE2C, and RPL11 may be potential diagnostic biomarkers for severe cases with COVID-19. Severe COVID-19 may affect the immune and male reproductive systems. 55 drugs targeted to AURKB, BRCA1, or TOP2A, may be potential therapeutic drugs for severe COVID-19.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Professor Li Su and Lifang Zhou for their detailed reading and their helpful suggestions for this manuscript. Our sincere thanks to the GEO data providers. We also thank each of the authors who participated in this study.
Authors’ contributions
Li Su, Lifang Zhou, and Jianxiong Long were mainly responsible for the control of analysis ideas; Shengying Liu was responsible for writing and data analysis; Tian Liang was responsible for writing R code for data cleaning; Miao Lva, Xiaolan Huang, and Xueying Liang were responsible for using of databases.
Data availability
The data analyzed by the Institute comes from the NCBI GEO website that can be obtained for free (https://www.ncbi.nlm.nih.gov/geo/).
Declarations
Ethical approval and Consent to participate
Not applicable.
Human and animal ethics
Not applicable.
Consent for publication
The results/data/figures in this manuscript have not been published elsewhere, nor are they under consideration (from you or one of your Contributing Authors) by another publisher.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Shengying Liu, Jianxiong Long and Tian Liang are the first co-authors.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Li Su, Email: suli2018@hotmail.com.
Lifang Zhou, Email: zlf20210623@126.com.
References
- Barrantes FJ. Central Nervous System Targets and Routes for SARS-CoV-2: Current Views and New Hypotheses. ACS Chem Neurosci. 2020;11(18):2793–2803. doi: 10.1021/acschemneuro.0c00434. [DOI] [PubMed] [Google Scholar]
- Bertran-Alamillo J, Cattan V, Schoumacher M, et al. AURKB as a target in non-small cell lung cancer with acquired resistance to anti-EGFR therapy. Nat Commun. 2019;10(1):1812. doi: 10.1038/s41467-019-09734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Zhou J, Fu S, et al. Prostate adenocarcinoma and COVID-19: The possible impacts of TMPRSS2 expressions in susceptibility to SARS-CoV-2. J Cell Mol Med. 2021;25(8):4157–4165. doi: 10.1111/jcmm.16385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JH, Yin Y, Yamane DP, et al. Association of prehospital antiplatelet therapy with survival in patients hospitalized with COVID-19: A propensity score-matched analysis. J Thromb Haemost JTH. 2021;19(11):2814–2824. doi: 10.1111/jth.15517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Zhao Z, Zhang T, et al. A systematic review and meta-analysis of children with coronavirus disease 2019 (COVID-19) J Med Virol. 2021;93(2):1057–1069. doi: 10.1002/jmv.26398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan N, Barda N, Kepten E, et al. BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N Engl J Med. 2021;384(15):1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B, Nymoen DA, Elgaaen BV, et al. BUB1 mRNA is significantly co-expressed with AURKA and AURKB mRNA in advanced-stage ovarian serous carcinoma. Virchows Archiv. 2014;464(6):701–707. doi: 10.1007/s00428-014-1577-7. [DOI] [PubMed] [Google Scholar]
- Docherty AB, Harrison EM, Green CA, et al. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ (clinical Research Ed) 2020;369:m1985. doi: 10.1136/bmj.m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Xu Y, Sun C, et al. A systematic review of asymptomatic infections with COVID-19. J Microbiol Immunol Infect. 2021;54(1):12–16. doi: 10.1016/j.jmii.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkogkou E, Barnasas G, Vougas K, et al. Expression profiling meta-analysis of ACE2 and TMPRSS2, the putative anti-inflammatory receptor and priming protease of SARS-CoV-2 in human cells, and identification of putative modulators. Redox Biol. 2020;36:101615. doi: 10.1016/j.redox.2020.101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton T. Autoantibodies May Drive COVID-19 Blood Clots. JAMA. 2021;325(5):425. doi: 10.1001/jama.2020.25699. [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua H, Kong Q, Zhang H, et al. Targeting mTOR for cancer therapy. J Hematol Oncol. 2019;12(1):71. doi: 10.1186/s13045-019-0754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang BZ, Sidell MA, Wu BU, et al. Pre-Existing Pancreatitis and Elevated Risks of COVID-19 Severity and Mortality. Gastroenterology. 2022;162(6):1758–1760.e3. doi: 10.1053/j.gastro.2022.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihlow J, Michaelis E, Greuel S, et al. B cell depletion and signs of sepsis-acquired immunodeficiency in bone marrow and spleen of COVID-19 deceased. Int J Infect Dis. 2021;103:628–635. doi: 10.1016/j.ijid.2020.12.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jit BP, Qazi S, Arya R, et al. An immune epigenetic insight to COVID-19 infection. Epigenomics. 2021;13(6):465–480. doi: 10.2217/epi-2020-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krithivas A, Young DB, Liao G, et al. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J Virol. 2000;74(20):9637–9645. doi: 10.1128/jvi.74.20.9637-9645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Berger JM (2019) Cell Cycle-Dependent Control and Roles of DNA Topoisomerase II. Genes 10 (11):859 [DOI] [PMC free article] [PubMed]
- Leng RX, Pan HF, Liu J, et al. Evidence for genetic association of TBX21 and IFNG with systemic lupus erythematosus in a Chinese Han population. Sci Rep. 2016;6:22081. doi: 10.1038/srep22081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Sun A, Liang X, et al. Histone demethylase PHF8 promotes progression and metastasis of gastric cancer. Am J Cancer Res. 2017;7(3):448–461. [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang R, Wu A, et al. SARS-COV-2 as potential microRNA sponge in COVID-19 patients. BMC Med Genomics. 2022;15(Suppl 2):94. doi: 10.1186/s12920-022-01243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C, Wong P, Klein J, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumley SF, Rodger G, Constantinides B, et al. An Observational Cohort Study on the Incidence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection and B.1.1.7 Variant Infection in Healthcare Workers by Antibody and Vaccination Status. Clin Infect Dis. 2022;74(7):1208–1219. doi: 10.1093/cid/ciab608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansuroglu Z, Josse T, Gilleron J, et al. Nonstructural NSs protein of rift valley fever virus interacts with pericentromeric DNA sequences of the host cell, inducing chromosome cohesion and segregation defects. J Virol. 2010;84(2):928–939. doi: 10.1128/JVI.01165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanares-Meza LD, Medina-Contreras O. SARS-CoV-2 and influenza: a comparative overview and treatment implications. Boletin Medico Del Hospital Infantil De Mexico. 2020;77(5):262–273. doi: 10.24875/BMHIM.20000183. [DOI] [PubMed] [Google Scholar]
- Mousavi SZ, Rahmanian M, Sami A. Organ-specific or personalized treatment for COVID-19: rationale, evidence, and potential candidates. Funct Integr Genomics. 2022;22(3):429–433. doi: 10.1007/s10142-022-00841-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano V, Dabrowska A, Schorpp K et al (2022) Acriflavine, a clinically approved drug, inhibits SARS-CoV-2 and other betacoronaviruses. Cell Chem Biol 29(5):774–784.e8 [DOI] [PMC free article] [PubMed]
- Nassau DE, Best JC, Kresch E, et al. Impact of the SARS-CoV-2 virus on male reproductive health. BJU Int. 2022;129(2):143–150. doi: 10.1111/bju.15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortenzi F, Albanese E, Fadda M (2020) A Transdisciplinary Analysis of COVID-19 in Italy: The Most Affected Country in Europe. Int J Environ Res Public Health 17(24):9488 [DOI] [PMC free article] [PubMed]
- Pavel A, Murray DK, Stoessl AJ. COVID-19 and selective vulnerability to Parkinson's disease. Lancet Neurol. 2020;19(9):719. doi: 10.1016/S1474-4422(20)30269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham H, de Gruijter NM, Raine C, et al. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat Commun. 2020;11(1):6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, Liu X, Yang T, et al. Nanomedicine for acute respiratory distress syndrome: The latest application, targeting strategy, and rational design. Acta Pharmaceutica Sinica B. 2021;11(10):3060–3091. doi: 10.1016/j.apsb.2021.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff J, Gray G, Vandebosch A, et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. New Engl J Med. 2021;384(23):2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah P, Fitzpatrick MC, Zimmer CF et al (2021) Asymptomatic SARS-CoV-2 infection: A systematic review and meta-analysis. Proc Natl Acad Sci USA 118 (34):e2109229118 [DOI] [PMC free article] [PubMed]
- Silverstein NJ, Wang Y, Manickas-Hill Z et al (2022) Innate lymphoid cells and COVID-19 severity in SARS-CoV-2 infection. eLife 11: e74681 [DOI] [PMC free article] [PubMed]
- Tang A, Gao K, Chu L, et al. Aurora kinases: novel therapy targets in cancers. Oncotarget. 2017;8(14):23937–23954. doi: 10.18632/oncotarget.14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M, Buschauer R, Ameismeier M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science (New York, N.Y.) 2020;369(6508):1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Mustafa F, Rizvi TA et al (2020) SARS-CoV-2/COVID-19: Viral Genomics, Epidemiology, Vaccines, and Therapeutic Interventions. Viruses 12(5):526 [DOI] [PMC free article] [PubMed]
- Wang B, Huang Y. Which type of cancer patients are more susceptible to the SARS-COX-2: Evidence from a meta-analysis and bioinformatics analysis. Crit Rev Oncol Hematol. 2020;153:103032. doi: 10.1016/j.critrevonc.2020.103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhu X, Jiang XM, et al. Decreased inhibition of exosomal miRNAs on SARS-CoV-2 replication underlies poor outcomes in elderly people and diabetic patients. Signal Transduct Target Ther. 2021;6(1):300. doi: 10.1038/s41392-021-00716-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2022) Weekly epidemiological update on COVID-19 - 27 April 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---27-april-2022. Accessed 29 Apr 2022
- Wicik Z, Eyileten C, Jakubik D et al (2020) ACE2 Interaction Networks in COVID-19: A Physiological Framework for Prediction of Outcome in Patients with Cardiovascular Risk Factors. J Clin Med 9(11):3743 [DOI] [PMC free article] [PubMed]
- Yang M, Chen S, Huang B, et al. Pathological Findings in the Testes of COVID-19 Patients: Clinical Implications. Eur Urol Focus. 2020;6(5):1124–1129. doi: 10.1016/j.euf.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Chen T, Zhou Y. Mediators of SARS-CoV-2 entry are preferentially enriched in cardiomyocytes. Hereditas. 2021;158(1):4. doi: 10.1186/s41065-020-00168-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Wang B, Mao J. The pathogenesis and treatment of the `Cytokine Storm' in COVID-19. J Infect. 2020;80(6):607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wu Y, Li S, et al. Bioinformatic and mouse model reveal the potential high vulnerability of Leydig cells on SARS-CoV-2. Ann Transl Med. 2021;9(8):678. doi: 10.21037/atm-21-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LF, Qi FY, Zhang JG, et al. Identification of the upstream regulators of KDM5B in gastric cancer. Life Sci. 2022;298:120458. doi: 10.1016/j.lfs.2022.120458. [DOI] [PubMed] [Google Scholar]
- Zhavoronkov A. Geroprotective and senoremediative strategies to reduce the comorbidity, infection rates, severity, and lethality in gerophilic and gerolavic infections. Aging. 2020;12(8):6492–6510. doi: 10.18632/aging.102988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu S, Luo D, Li L, et al. Tamoxifen and clomiphene inhibit SARS-CoV-2 infection by suppressing viral entry. Signal Transduct Target Ther. 2021;6(1):435. doi: 10.1038/s41392-021-00853-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed by the Institute comes from the NCBI GEO website that can be obtained for free (https://www.ncbi.nlm.nih.gov/geo/).