Abstract
The isoamylase gene from Flavobacterium odoratum KU was cloned into and expressed in Escherichia coli JM109. The promoter of the gene was successful in E. coli, and the enzyme produced was excreted into the culture medium, depending on the amount of the enzyme expressed. The enzyme found in the culture medium showed almost the same Mr, heat-inactivating constant, and N-terminal sequence as those of the enzyme accumulated in the periplasmic space. This result indicated that the enzyme accumulated in an active form at the periplasm was transported out of the cell. The primary sequence of the enzyme, which was deduced from its nucleotide sequence, showed that the mature enzyme consisted of 741 amino acid residues. By changing five possible residues to Ala independently, it was found that Asp-374, Glu-422, and Asp-497 were essential. The sequences around those residues were highly conserved in isoamylases of different origins and the glycogen operon protein X, GlgX. The comparison of the distance between these essential residues with those of various amylases suggested that the bacterial and plant isoamylase but not GlgX had a longer fourth loop than the other amylases. This longer fourth loop had a possible role in accommodating the long branched chains of native glycogens and starches.
Isoamylase (glycogen 6-glucanohydrolase; EC 3.2.1.68) completely hydrolyzes an α-1,6-glucosidic linkage inside starches and glycogens (48). Thus, isoamylase is useful not only for the structural analysis of these polysaccharides and derived oligosaccharides but also for the starch industry in producing glucose, maltose, and higher oligosaccharides from starch with the concomitant action of exo-type hydrolases. Isoamylase is also used to introduce branches into cyclodextrins (1) to improve their solubility and hemolytic properties (27) through the reversed action of the enzyme.
The enzyme was first reported to be present in yeast (22), and then Harada and his coworkers (11, 47, 48) found isoamylase in a bacterium, Pseudomonas amyloderamosa, isolated from soil. The latter group examined the nature of the enzyme, its action, and the differences in properties between isoamylase and bacterial pullulanase, which debranches pullulan and starches, but not glycogen. Since then, several bacteria and one yeast have been reported to produce isoamylase (8, 9, 15, 33, 36), although the enzyme from P. amyloderamosa is the only enzyme available commercially. The genes of isoamylase from Pseudomonas have been cloned (3, 6, 44) and expressed (6).
We have identified a strain, Flavobacterium odoratum KU, isolated from soil, that produces extracellular isoamylase (37). The enzymatic properties are similar to those of Pseudomonas isoamylase for practical purposes except for the optimum pH for action (12). The enzyme from Flavobacterium shows the highest activity at pH 6.0, while the optimum activity of Pseudomonas occurs at pH 3.5. Because many amylolytic enzymes prefer mild-acidic pH for their optimal activity, the enzyme of Flavobacterium is much more suitable for the concomitant action with exo-acting amylases, such as soybean β-amylase or Pseudomonas stutzeri maltotetraose-forming amylase (34), to produce maltooligosaccharides from starch.
As our final aim is to understand the relationship between the function and structure of isoamylase, we first sequenced the entire isolated DNA fragment and deduced the primary sequence of isoamylase of F. odoratum. In this report, we describe the expression of the enzyme in Escherichia coli and the catalytic residues of isoamylase clarified by site-directed mutagenesis and then discuss the difference in the architecture of the enzyme between isoamylase and α-amylase. We also discuss the significance of the long loop 4 in isoamylases for their debranching action.
MATERIALS AND METHODS
Bacterial strains and DNAs.
F. odoratum KU is maintained in our laboratory (37). E. coli JM109 (Δ[lac-pro] endA1 gyrA96 hsdR17 λ− relA1 supE44 thi [F′ lacIq lacZΔM15 proAB traD36]) was used throughout this study. Multicopy plasmids pHSG398 (Takara Shuzo, Kyoto, Japan), having camr, and pUC18 and pUC19, having Ampr, were used as vectors. A low-copy plasmid, pMW119, was obtained from Nippon Gene Co. Ltd. (Toyama, Japan). Nucleotide primers for PCR were synthesized by Nippon Gene Co. Ltd.
DNA manipulation.
General DNA manipulation was done according to the method of Sambrook et al. (31). DNA fragments separated by agarose gel electrophoresis were purified with Gene Pure (Nippon Gene Co.). Plasmids were introduced into E. coli by electroporation with an 0.1-cm cell (1,250 V, 100 Ω, 50 μF) on The Electroporator II (Invitrogen). DNA sequencing was carried out on both strands on the A.L.F. DNA sequencer (Pharmacia Biotech) with an AutoRead sequencing kit (Pharmacia Biotech) after creating nested deletions with exonuclease III and S1 nuclease. DNA sequence was analyzed by Genetyx-Win version 3 (Software Development Co. Ltd., Tokyo, Japan).
Sequence alignment.
The primary sequences of isoamylases were aligned by ClustalX (43) and GeneDoc (26). The sequences of isoamylases of P. amyloderamosa SB-15, Pseudomonas sp. strain SMP-1, Flavobacterium sp., and Zea mays were obtained from the DDBJ/EMBL/GenBank database. These have accession no. J03871, M25247, U90120, and U18908, respectively. The sequences of GlgX from Arabidopsis thaliana, E. coli, Chlamydia trachomatis, Haemophilus influenzae, Mycobacterium tuberculosis, Synechocystis sp. (two sequences), and Sulfolobus solfataricus have accession no. AC005278 (PID:g3850573), AE000419 (PID:g2367229), AE001278 (PID:g3328433), U32815 (PID:g1574821), Z74020 (PID:g1403478), D90900 (PID:g1651771) and D90908 (PID:g1652733), and Y08256 (PID:g1707700), respectively.
Enzyme assay.
Isoamylase activity was measured by the iodine method (37). When necessary, the enzyme solution was diluted with 50 mM sodium acetate (pH 6.0) containing 0.005% bovine serum albumin and 1 mM CaCl2, not exceeding an ΔA580 of 0.2. The value is the limit of the linearity (amount of enzyme versus activity) in this method. One unit of isoamylase activity is defined as the amount of enzyme that increases 0.1 A580 per h.
Cloning of the isoamylase gene.
Chromosomal DNA of F. odoratum KU was partially hydrolyzed with Sau3AI, and the fragments of 4 to 7 kbp in size were isolated through electrophoresis on 0.8% agarose. Then, the fragments were ligated into the dephosphorylated BamHI site of pUC19. The resulting plasmids were introduced into E. coli JM109 by electroporation, and the recombinant having the iam gene was selected as a colony surrounded by a blue zone after flooding 0.2% I2–2% KI onto Luria-Bertani agar medium containing 0.5% waxy rice starch.
Cell fractionation.
The cells of E. coli JM109 harboring constructed plasmids were grown overnight on Luria-Bertani or M9 medium (50 ml) containing suitable antibiotics. The latter medium was supplemented with thiamine and glucose as a carbon source. The cells were fractionated as reported previously (2).
ELISA.
Polyclonal antibody against Flavobacterium isoamylase was raised in New Zealand White rabbits by using TiterMax R-1 (Vaxcel, Inc.) as the first-time adjuvant. The enzyme-linked immunosorbent assay (ELISA) was done according to the work of Yagi et al. (45) with anti-rabbit immunoglobulin G-alkaline phosphatase complex from goat (EY Laboratories, Inc.) and an ELISA plate reader (Bio-Rad Japan, Tokyo, Japan).
N-terminal sequencing.
The N-terminal sequence of the purified enzyme was determined on an Applied Biosystems 477A protein sequencer.
Site-directed mutagenesis.
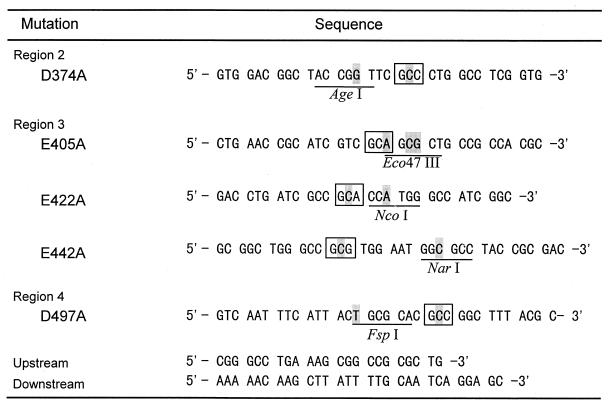
Site-directed mutagenesis was done by the megaprimer-PCR method (7) with primers (Fig. 1) and Ultma DNA polymerase (Perkin-Elmer Applied Biosystems, Chiba, Japan). A new restriction site was created on each primer as a silent mutation.
FIG. 1.
Primers for site-directed mutagenesis. Shading indicates substituted nucleotides, and boxes indicate the positions of mutations where Asp or Glu is replaced with Ala. New restriction enzyme sites are underlined.
Briefly, a fragment obtained by the first PCR with an upstream primer and one of the mutant primers was purified by agarose gel electrophoresis and recovered with Gene Pure (Nippon Gene). Then the second PCR was performed with this fragment as a megaprimer and a downstream primer. A product was isolated and cut by NotI and PpuMI. Then the fragment was ligated with a large fragment of pHIF2 cut with the same enzymes. The fragments were confirmed to be free from any error due to misincorporation of PCR by the sequencing of both strands.
Nucleotide sequence accession number.
The DDBJ accession no. for the sequence reported in this paper is D88029.
RESULTS
Cloning of isoamylase gene and its expression.
We obtained one clone, which was positive by the starch-iodine test, from 3,000 transformants. The clone had a 3.4-kbp insert on the vector (pIF1). The small ApaI fragment (1.1 kbp) of this inserted DNA was isolated and labeled with digoxigenin according to the directions of the manufacturer. The hybridization of this labeled fragment with either BamHI-cut or PstI-cut chromosomal DNA of F. odoratum KU gave a single band, but not with E. coli chromosomal DNA or plasmid vector (data not shown). This proved that the isolated gene originated from the genomic DNA of F. odoratum.
The PstI-XbaI fragment (3.1 kbp) of pIF1 was subcloned into pUC19 and pUC18, giving pIF12 and pIF13, in which the fragment was integrated in the same direction as and the opposite direction from pIF1 in respect to the lac promoter on the vector, respectively. E. coli JM109(pIF12) and E. coli JM109(pIF13) produced isoamylase without addition of IPTG (isopropyl-β-d-thiogalactopyranoside), and the specific activities of the enzymes were 85.9 and 85.5 U/A600, respectively. The productivity of 91 U/ml of culture was attained in both the transformants, and this value was eight times as high as that for the original bacterium, F. odoratum KU.
pHIF2 and pMIF3 were constructed by inserting the above PstI-XbaI fragments into pHSG398 and pMW119, respectively.
Location and properties of the enzyme.
Fifty-two percent of the isoamylase activity was found in the culture broth, and the remainder was found in the periplasmic (46%) and intracellular (2%) spaces of E. coli JM109(pIF12) grown in M9 minimal medium (Table 1). Another periplasmic protein, β-lactamase, was located in the extracellular fluid and periplasmic and intracellular spaces in the ratio of 56:42:1, while 93% of malate dehydrogenase, a cytoplasmic protein, was detected in the intracellular fraction. These values were almost the same when E. coli JM109(pIF13) was fractionated.
TABLE 1.
Distribution of activities of isoamylase, β-lactamase, and malate dehydrogenase in transformantsa
| Introduced vector | Fraction | Isoamylase (%) | β-Lactamase (%) | Malate dehydrogenase (%) |
|---|---|---|---|---|
| pIF13 | Ex | 56 | 50 | 3 |
| Pe | 42 | 50 | 5 | |
| In | 1 | 0 | 92 | |
| pIF12 | Ex | 52 | 56 | 4 |
| Pe | 46 | 42 | 6 | |
| In | 2 | 1 | 90 | |
| pUC19 | Ex | 0 | 6 | 0 |
| Pe | 0 | 90 | 6 | |
| In | 0 | 4 | 94 | |
| pMF13 | Ex | 6 | ND | ND |
| Pe | 93 | ND | ND | |
| In | 1 | ND | ND |
The values are the averages of three experiments. Ex, extracellular; Pe, periplasmic; In, intracellular; ND, not determined.
This transportation was independent of the incubation temperature since cultivation at both 30 and 37°C gave similar distribution ratios. The enzyme activity was not needed for the translocation, since the inactive, mutant enzyme, E422A (described below), was found to be distributed in the extracellular and periplasmic fractions in the ratio of 55:45 by ELISA. When pMIF3 was introduced into JM109, the total amount of enzyme produced decreased to 9.0 U/ml of culture, and this value was 9.8% of the total activity produced by E. coli JM109(pIF13). Ninety-three percent of isoamylase activity produced by E. coli JM109(pMIF3) was located in the periplasmic space after cultivation for 12 h (Table 1).
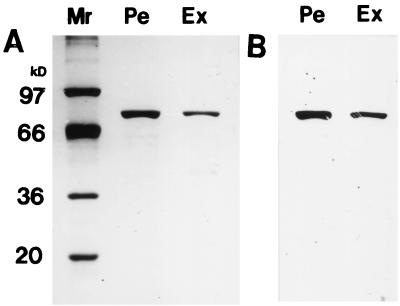
Both the enzymes from the culture fluid of E. coli JM109(pIF13) and the periplasmic fraction of the cells were purified by starch adsorption (Fig. 2). The purity of both enzymes was more than 97% by densitometry with the purification factor of 350-fold and the yield of 50% in both cases. They were found to have almost identical properties, such as Mr, the constant for heat inactivation, N-terminal sequence, etc., as shown in Table 2.
FIG. 2.
SDS-PAGE of the purified extracellular (Ex) and periplasmic (Pe) isoamylase produced by E. coli JM109(pIF13). The purified enzymes were subjected to SDS-PAGE and detected with Coomassie Brilliant Blue R-250 (A) and by Western blotting (B). Mr markers: phosphorylase a (97,000), bovine serum albumin (66,000), glyceraldehyde 3-phosphate dehydrogenase (36,000), and soybean trypsin inhibitor (20,100).
TABLE 2.
Characterization of extracellular and periplasmic isoamylases
| Characteristic | Value for fractiona:
|
|
|---|---|---|
| Ex | Pe | |
| Mr | 80,000 | 80,000 |
| Sp act (U/mg) | 6.2 × 104 | 6.3 × 104 |
| Thermal inactivation rateb (min−1) | −5.1 × 10−3 | −5.2 × 10−3 |
| N-terminal sequence | Identicalc | Identicalc |
| Antigenicity | Identicald | Identicald |
Ex, extracellular; Pe, periplasmic.
The purified enzymes were incubated in 50 mM sodium acetate (pH 6.0) containing 1 mM CaCl2 at 50°C.
AINPNKLGAA.
Fused on Ouchterlony test.
Sequence of the iam gene.
The entire sequence of the insert on pIF13 was determined on both strands. The insert was 3,056 bp in size, and two overlapping open reading frames were found, namely, nucleotides (nt) 276 to 2600 (2,325 bp and 774 amino acid residues) and nt 312 to 2600 (2,289 bp and 762 residues). The fragment sizes were large enough to encode a protein of 80,000 in Mr (37). There were several promoter-like sequences that were effective in E. coli. The nt 166 to 171, 207 to 212, 210 to 215, and 263 to 268 for the −35 region and nt 189 to 194, 233 to 238, and 284 to 292 for the −10 region, respectively, were found as the candidate for the promoter of E. coli by Genetyx-Win promoter search. The program is based on the results of Mulligan et al. (24a). As for the Shine-Dalgarno sequence, we could find GGAG at nt 302.
Identification of essential residues for isoamylase activity.
In this study, we replaced all five possible candidates (three Glu residues and two Asp residues [see Discussion]) with Ala. All the mutant plasmids were sequenced on both strands, and we confirmed that no error other than the intended mutation was introduced. The mutant plasmid was electroporated into E. coli JM109, and the enzyme produced in the periplasmic space was purified by raw-starch adsorption. The purified enzymes showed a single band by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) after silver staining and immunological detection (data not shown). Every mutation of D374, E422, or D497 to Ala abolished enzyme activity, while E405A and E442A enzymes had 94 and 97%, respectively, of the specific activity of the wild-type isoamylase (Table 3).
TABLE 3.
Specific activities of mutant and wild-type isoamylases
| Mutant | Sp act (U/mg) |
|---|---|
| D374A | <10 |
| E405A | 6.0 × 104 |
| E422A | <10 |
| E442A | 5.8 × 104 |
| D497A | <10 |
| Wild type | 6.2 × 104 |
Comparison of the primary structures.
In order to find the sequence specific to debranching enzymes, the deduced primary sequence of mature F. odoratum isoamylase (FODIAM) was compared with those of isoamylases and GlgX proteins (Fig. 3). GlgX protein from E. coli is reported elsewhere as a debranching enzyme (46).
FIG. 3.
Sequence alignment of isoamylases. FSPIAM, Flavobacterium sp. isoamylase; PSPIAM, Pseudomonas sp. strain JD210 isoamylase (6); ZMAIAM, Z. mays sugary-1 protein (13); ECOGLG, E. coli GlgX; CTRGLG, C. trachomatis GlgX. SSOGLG, S. solfataricus GlgX; SSPGLG, Synechocystis sp. GlgX. The first 93 residues of ZMAIAM were omitted. Black shading indicates 100%, dark gray shading indicates 80%, and light gray shading indicates 60% conservation, according to the Blosum 62 matrix table.
The primary sequence of FODIAM and that of Flavobacterium sp. isoamylase (FSPIAM) were 75% identical; however, the number and location of Cys residues differed (Fig. 3). In the latter enzyme, four Cys residues were found, whereas the former had six Cys residues. Two Cys residues (Cys-387 and Cys-391) of FSPIAM corresponded to Cys-383 and Cys-387 of FODIAM, but the rest (Cys-136 and Cys-156) were unique. The locations of six Cys residues found in FODIAM were almost conserved in Pseudomonas isoamylases (Fig. 3).
We then aligned the primary sequences of Pseudomonas enzyme (data not shown) and found that the sequences of P. amyloderamosa JD210 isoamylase (PSPIAM) and Pseudomonas sp. strain SMP-1 isoamylase were identical, with a mismatch of one residue, and that the enzyme from P. amyloderamosa SB-15 had a different stretch of amino acid residues at positions 427 to 457. Finally, the sequences of FODIAM, FSPIAM, and PSPIAM as a representative of Pseudomonas enzymes, Z. mays sugary-1 protein (ZMAIAM) and GlgXs were compared (Fig. 3). The sequences of FODIAM and PSPIAM resembled each other, although deletion and insertion in FODIAM at positions 386 to 388 and 623 to 630, respectively, were noted.
DISCUSSION
We have cloned the isoamylase gene of F. odoratum. During the assay of gene expression, we found that the promoter of the isoamylase gene of F. odoratum could be effective in E. coli, since the enzyme was produced regardless of its orientation with respect to the lac promoter of the vector in the absence of IPTG. The signal sequence of Flavobacterium isoamylase was effective in E. coli; furthermore, the mature enzyme was transported out of the cell, since isoamylase activity was detected in both the periplasmic and the extracellular fractions. This enzyme accumulation in culture fluid was not due to cell lysis, since malate dehydrogenase was always recovered as the intracellular fraction. The small amount of isoamylase found in the intracellular fraction may due to the incomplete conversion of the cells into protoplasts.
Pseudomonas isoamylase expressed in E. coli HB101 is secreted almost exclusively into the culture fluid (6), as found by measuring the activity in the medium and cell extract obtained by sonication. We were afraid that cytoplasmic α-amylase (29) in E. coli might interfere with the determination of isoamylase activity located inside the cell by the iodine method. We, therefore, measured separately the activity in both the periplasmic and the intracellular fractions, since isoamylase was expected in the periplasmic space. The activity was distributed almost equally in both the extracellular and the periplasmic fractions, and their properties resembled each other (Table 2). From these results, we concluded that the enzyme was first accumulated in the periplasmic space of E. coli in the active form and then transported to the medium. This transport did not depend on the cultivation time and its activity but on the amount of enzyme produced in the periplasm. Therefore, this is the leakage of the enzyme protein. Further study is needed to clarify how a folded, large molecule can penetrate the outer membrane of E. coli.
The sequencing of the entire DNA fragment in pIF13 gave two overlapping open reading frames; however, the open reading frame from nt 312 to 2600 was likely the one, since the Shine-Dalgarno region (nt 302 to 306) was found only for this frame. From the comparison of the deduced primary sequences and the result of N-terminal sequencing of the mature enzyme from F. odoratum, we found that 21 amino acid residues corresponded to the leader peptide. This signal peptide was very different from that of Flavobacterium sp. isoamylase (18) (24 residues long). Several extracellular proteins of Flavobacterium have been cloned, and those genes have been expressed in E. coli thus far. Those signal peptides were reported to be 45 (41, 42), 42 (21), 40 (5), 39 (41), and 21 (32) residues long, showing the diversity of Flavobacterium signal peptides in length.
The enzymes, which belong to the amylase family, share four conserved regions, Regions 1 to Region 4, and the catalytic residues of the enzyme are located at Region 2, Region 3, and Region 4 (25). We could find Region 1, Region 2, and Region 4 in FODIAM, although Region 3 was difficult to discern. The sequence surrounding 454EWSV of P. amyloderamosa SB-15, the region that was reported by Nakajima et al. (25) as Region 3, was not conserved among the bacterial enzymes (data not shown). The analogy suggested that D374 and D497 of F. odoratum isoamylase corresponded to D206 and D297 of α-amylase from Aspergillus oryzae (24) (Taka amylase) based on its characteristic sequence near the residues (Region 2 and Region 4). As to the glutamic residue, which corresponds to E230 (Region 3) of Taka amylase, we found three candidates, i.e., E405, E422, and E442. E405 and E442 of the Flavobacterium enzyme corresponded to E416 and E454 of Pseudomonas strain SB-15 isoamylase, respectively. E416 of Pseudomonas isoamylase was rationally predicted as one of the essential residues by analogy to the structure of α-amylases (14), and the latter was suggested to be essential by Nakajima et al. (25). We mutated three Glu residues (E405, E422, and E442) in addition to two Asp residues (D374 and D497) to Ala.
The results of the activity determination of the mutant enzymes clearly indicated that D374, E422, and D497 were essential for Flavobacterium isoamylase (Table 3), and those residues were well conserved in all the sequences listed in Fig. 3. Recently, the three-dimensional structure of isoamylase from P. amyloderamosa has been solved by Katsuya et al. (16). According to their model, D375, E435, and D510 of Pseudomonas isoamylase are located at the bottom of a deep cleft, and their three-dimensional location resembles those of the catalytic residues of Taka amylase. These residues corresponded to D374, E422, and D497 of F. odoratum isoamylase, respectively (Fig. 3). This also supported our conclusion. Since all the mutant enzymes adsorbed to raw starch granules, the enzyme activity was not essential to the binding of the isoamylase to the granules.
It is interesting that Region 3 of isoamylases and GlgX proteins share a Glu-(Pro/Ala)-Trp-(Ala/Asp) stretch (Fig. 3). The third residues in this region of all the proteins were noted to be Trp, which is suggested to be located at the P1 site in Pseudomonas isoamylase (16). Branching enzymes have different sequences at Region 3 of Glu-Asp-(Ser/Val)-(Thr/Ser) (20, 40), suggesting that isoamylase and branching enzymes have different structures in the vicinity of one of the catalytic residues and different substrate-binding modes.
Although FODIAM, FSPIAM, and PSPIAM resembled each other, particularly at their central areas, they showed differences in properties such as optimum pH and temperature, resistance to heat inactivation, and stabilization by Ca2+. The study to find the components responsible for those differences is in progress. The central areas of ZMAIAM and GlgX proteins were also similar to those of FODIAM and PSPIAM, suggesting that all the enzymes had a common architecture in their main domain.
Table 4 shows the comparison of distances between the essential residues of various amylases. Every amylase has three essential residues, Asp, Glu, and Asp, and X-ray analysis of some amylases shows those three residues located at the end of the fourth, fifth, and seventh β-strands of the (α/β)8 barrel structure. Isoamylases from bacteria, sugary-1 protein, and branching enzymes for starch or glycogen were shown to have clearly longer distances between the first Asp and Glu than α-amylases and cyclodextrin glucanotransferase, which act solely on α-1,4-glucosidic linkage, and GlgX proteins.
TABLE 4.
Span lengths between essential residues of the enzymes in the amylase family
| Enzyme | Source and reference | Span length (no. of residues)
|
|
|---|---|---|---|
| Region 2-D ∼ 3-Ea | Region 3-E ∼ 4-Db | ||
| α-Amylase | Bacillus subtilis (38) | 31 | 60 |
| Aspergillus oryzae (24) | 23 | 66 | |
| Barley (isozyme 1) (35) | 24 | 85 | |
| Porcine pancrease (28) | 35 | 66 | |
| Cyclodextrin glycosyltransferase | B. circulans (17) | 27 | 70 |
| Neopullulanase | Bacillus stearothermophilus (19) | 28 | 66 |
| Amylopullulanase | Thermoanaerobacter ethanolicus (23) | 28 | 76 |
| Pullulanase | Klebsiella aerogenes (14) | 28 | 127 |
| Isoamylase | F. odoratum (this study) | 47 | 74 |
| P. amyloderamosa (JD210) (predicted in this study) | 59 | 74 | |
| Maize (sugary-1) (predicted in this study) | 53 | 72 | |
| Chlamydia (GlgX) (predicted in this study) | 36 | 70 | |
| E. coli (GlgX) (predicted in this study) | 34 | 71 | |
| Branching enzyme | B. stearothermophilus (40) | 42 | 68 |
| Maize (isozyme II) (20) | 55 | 68 | |
Span length between Asp in Region 2 and Glu in Region 3.
Span length between Glu in Region 3 and Asp in Region 4.
Isoamylase has been shown elsewhere to have an (α/β)8 architecture similar to that of α-amylase (16). However, the enzyme must have a different shape of active site, because isoamylase has to accommodate two chains, main and side chains, of a substrate to hydrolyze its branch point, while one chain is enough for α-amylase to act. Most of the longer region of isoamylases could correspond to the fourth loop of the (α/β)8 structure. According to the three-dimensional structure, the loop consists of the side wall of the active-site cleft of the enzyme. The role of this unique loop has not been clarified, and any similarity in their sequences has not been found yet. However, the loop, particularly its conformation, may play an important role in the action of isoamylases on native polysaccharides as described below.
ZMAIAM has been expressed in E. coli, and the action properties of the protein have been found to be similar to those of bacterial isoamylases (30), although its primary sequence is rather similar to that of GlgX proteins (Fig. 3). One debranching enzyme has been isolated from E. coli (15), and it was reported that phosphorylase-limit dextrins of glycogens, which have a branch degree of polymerization of 4, are good substrates, but native starches or glycogens are inert. Yang et al. (46) reported that this enzyme corresponds to GlgX. Maize sugary-1 protein and E. coli GlgX resemble each other but differ in the length of the fourth loop (Fig. 3 and Table 4). The latter enzyme, which has the shorter fourth loop, prefers the substrate of short branches as mentioned above, suggesting that loop 4 has a possible role in accommodating a long branched chain in α-glucans. From these comparisons, it was suggested that the isoamylases of Flavobacterium and Pseudomonas and maize sugary-1 protein belong to one of the subfamilies of isoamylases, while GlgX proteins belong to another subfamily.
Pullulanase is an enzyme that cleaves the pullulan and starch, and its essential residues have not been elucidated yet. However, this enzyme seemed to have rather short and very long distances between the first Asp residue and Glu and between Glu and the second Asp residue, respectively, from the predicted catalytic residues (14) as shown in Table 4. Neopullulanase acts on both α-1,4- and α-1,6-glucosidic bonds (39) but has the short first Asp-Glu distance like α-amylases. Thus, these enzymes may have architectures rather different from that of isoamylase.
ACKNOWLEDGMENTS
We thank Professor Yagi (Kagoshima University) for N-terminal sequencing of the enzymes.
This study was performed as a part of the project entitled “High and Ecological Utilization of Regional Carbohydrates,” through Special Coordination Funds for Promoting Science and Technology (Leading Research Utilizing Potential of Regional Science and Technology) of the Science and Technology Agency of the Japanese Government, 1997.
REFERENCES
- 1.Abe J, Mizowaki N, Hizukuri S, Koizumi K, Utamura T. Synthesis of branched cyclomalto-oligosaccharides using Pseudomonas isoamylase. Carbohydr Res. 1986;154:81–92. doi: 10.1016/s0008-6215(00)90024-7. [DOI] [PubMed] [Google Scholar]
- 2.Abe J, Shibata Y, Fujisue M, Hizukuri S. Expression of periplasmic α-amylase of Xanthomonas campestris K-11151 in Escherichia coli and its action on maltose. Microbiology. 1996;142:1505–1512. doi: 10.1099/13500872-142-6-1505. [DOI] [PubMed] [Google Scholar]
- 3.Amemura A, Chakraborty R, Fujita M, Noumi T, Futai M. Cloning and nucleotide sequence of the isoamylase gene from Pseudomonas amyloderamosa SB-15. J Biol Chem. 1988;263:9271–9275. [PubMed] [Google Scholar]
- 4.Ara K, Saeki K, Ito S. Purification and characterization of an alkaline isoamylase from an alkalophilic strain of Bacillus. J Gen Microbiol. 1993;139:781–786. [Google Scholar]
- 5.Barsomian G D, Johnson T L, Borowski M, Denman J, Ollington J, Hirani S, McNeilly D S, Rasmussen J R. Cloning and expression of peptide-N4-(N-acetyl-beta-D-glucosaminyl)asparagine amidase F in Escherichia coli. J Biol Chem. 1990;265:6967–6972. [PubMed] [Google Scholar]
- 6.Chen J H, Chen Z Y, Chow T Y, Chen J C, Tan S T, Hsu W H. Nucleotide sequence and expression of the isoamylase gene from an isoamylase-hyperproducing mutant, Pseudomonas amyloderamosa JD210. Biochim Biophys Acta. 1990;1087:309–315. doi: 10.1016/0167-4781(90)90004-l. [DOI] [PubMed] [Google Scholar]
- 7.Datta A K. Efficient amplification using ’megaprimer’ by asymmetric polymerase chain reaction. Nucleic Acids Res. 1995;23:4530–4531. doi: 10.1093/nar/23.21.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gastro G R, Garcia G F, Siñeriz F. Extracellular isoamylase produced by Bacillus circulans MIR-137. J Appl Bacteriol. 1992;73:520–523. [Google Scholar]
- 9.Gunja-Smith Z, Marshall J J, Smith E E, Whelan W J. A glycogen-debranching enzyme from Cytophaga. FEBS Lett. 1970;12:96–100. doi: 10.1016/0014-5793(70)80572-5. [DOI] [PubMed] [Google Scholar]
- 10.Guo Z, Arfman N, Ong E, Gilkes N R, Kilburn D G, Warren R A J, Miller R C., Jr Leakage of Cellulomonas fimi cellulases from Escherichia coli. FEMS Microbiol Lett. 1988;49:279–283. [Google Scholar]
- 11.Harada T, Misaki A, Akai H, Yokobayashi K, Sugimoto K. Characterization of Pseudomonas isoamylase by its actions on amylopectin and glycogen: comparison with Aerobacter pullulanase. Biochim Biophys Acta. 1972;268:497–505. doi: 10.1016/0005-2744(72)90345-2. [DOI] [PubMed] [Google Scholar]
- 12.Hizukuri S, Kozuma T, Yoshida H, Abe J, Takahashi K, Yamamoto M, Nakamura N. Properties of Flavobacterium odoratum KU isoamylase. Starch/Stärke. 1996;48:295–300. [Google Scholar]
- 13.James M G, Robertson D S, Myers A. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell. 1995;7:417–429. doi: 10.1105/tpc.7.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaspersen H M, MacGregor E A, Henrissat B, Sierks M R, Svensson B. Starch- and glycogen-debranching and branching enzymes: prediction of structural features of the catalytic (β/α)8-barrel domain and evolutionary relationship to other amylolytic enzymes. J Protein Chem. 1993;12:791–805. doi: 10.1007/BF01024938. [DOI] [PubMed] [Google Scholar]
- 15.Jeanningros R, Creuzet-Sigal N, Frixon C, Cattaneo J. Purification and properties of a debranching enzyme from Escherichia coli. Biochim Biophys Acta. 1976;438:186–199. doi: 10.1016/0005-2744(76)90235-7. [DOI] [PubMed] [Google Scholar]
- 16.Katsuya Y, Mezaki Y, Kubota M, Matsuura Y. Three-dimensional structure of Pseudomonas isoamylase at 2.2 Å resolution. J Mol Biol. 1998;281:885–897. doi: 10.1006/jmbi.1998.1992. [DOI] [PubMed] [Google Scholar]
- 17.Klein C, Hollender J, Bender H, Schulz G E. Catalytic center of cyclodextrin glycosyltransferase derived from X-ray structure analysis combined with site-directed mutagenesis. Biochemistry. 1992;31:8740–8746. doi: 10.1021/bi00152a009. [DOI] [PubMed] [Google Scholar]
- 18.Krohn B M, Barry G F, Kishore G M. An isoamylase with neutral pH optimum from a Flavobacterium species: cloning, characterization and expression of the iam gene. Mol Gen Genet. 1997;254:469–478. doi: 10.1007/s004380050441. [DOI] [PubMed] [Google Scholar]
- 19.Kuriki T, Takata H, Okada S, Imanaka T. Analysis of the active center of Bacillus stearothermophilus neopullulanase. J Bacteriol. 1991;173:6147–6152. doi: 10.1128/jb.173.19.6147-6152.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuriki T, Guan H, Sivak M, Preiss J. Analysis of the active center of branching enzyme II from maize endosperm. J Protein Chem. 1996;15:305–313. doi: 10.1007/BF01887119. [DOI] [PubMed] [Google Scholar]
- 21.Lemp D, Haselbeck A, Klebl F. Molecular cloning and heterologous expression of N-glycosidase F from Flavobacterium meningosepticum. J Biol Chem. 1990;265:15606–15610. [PubMed] [Google Scholar]
- 22.Maruo B, Kobayashi T. Enzymic scission of the branch links in amylopectin. Nature. 1951;167:606–607. doi: 10.1038/167606a0. [DOI] [PubMed] [Google Scholar]
- 23.Mathupala S P, Lowe S E, Podkovyrov S M, Zeikus J G. Sequencing of the amylopullulanase (apu) gene of Thermoanaerobacter ethanolicus 39E, and identification of the active site by site-directed mutagenesis. J Biol Chem. 1993;268:16332–16344. [PubMed] [Google Scholar]
- 24.Matsuura Y, Kusunoki M, Harada W, Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem (Tokyo) 1984;95:697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- 24a.Mulligan M E, Hawley D K, Entriken R, McClue W R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984;12:789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakajima R, Imanaka T, Aiba S. Comparison of amino acid sequences of eleven different α-amylases. Appl Microbiol Biotechnol. 1986;23:355–360. [Google Scholar]
- 26.Nicholas K B, Nicholas H B, Jr, Deerfield D W., II . GeneDoc: analysis and visualization of genetic variation. [Online] 1997. http://www.cris.com/ http://www.cris.com/K̃etchup/genedoc.shtml. [3 August 1999, last date accessed.] etchup/genedoc.shtml. [3 August 1999, last date accessed.] [Google Scholar]
- 27.Okada Y, Kubota Y, Koizumi K, Hizukuri S, Ohfuji T, Ogata K. Some properties and the inclusion behavior of branched cyclodextrins. Chem Pharm Bull. 1988;36:2176–2185. doi: 10.1248/cpb.36.2176. [DOI] [PubMed] [Google Scholar]
- 28.Qian M, Haser R, Buisson G, Duée E, Payan F. The active center of a mammalian α-amylase. Structure of the complex of a pancreatic α-amylase with a carbohydrate inhibitor refined to 2.2 Å resolution. Biochemistry. 1994;33:6284–6294. doi: 10.1021/bi00186a031. [DOI] [PubMed] [Google Scholar]
- 29.Raha M, Kawagishi I, Müller V, Kihara M, Macnab R M. Escherichia coli produces a cytoplasmic α-amylase, amyA. J Bacteriol. 1992;174:6644–6652. doi: 10.1128/jb.174.20.6644-6652.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman A, Wong K, Jane J T, Myers A M, James M G. Characterization of SU1 isoamylase, a determinant of storage starch structure in maize. Plant Physiol. 1998;117:425–435. doi: 10.1104/pp.117.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Sasisekharan R, Biulmer M, Moremen K W, Cooney C L, Langler R. Cloning and expression of heparinase I gene from Flavobacterium heparinum. Proc Natl Acad Sci USA. 1993;90:3660–3664. doi: 10.1073/pnas.90.8.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato H H, Park Y K. Purification and characterization of extracellular isoamylase from Flavobacterium sp. Starch/Stärke. 1980;32:132–136. [Google Scholar]
- 34.Schmidt J, John M. Starch metabolism in Pseudomonas stutzeri. I. Studies on maltotetraose-forming amylase. Biochim Biophys Acta. 1979;566:88–99. doi: 10.1016/0005-2744(79)90252-3. [DOI] [PubMed] [Google Scholar]
- 35.Søgaard M, Kadziola A, Haser R, Svensson B. Site-directed mutagenesis of histidine 93, aspartic acid 180, glutamic acid 205, histidine 290, and aspartic acid 291 at the active site and tryptophan 279 at the raw starch binding site in barley amylase. J Biol Chem. 1993;268:22480–22484. [PubMed] [Google Scholar]
- 36.Spencer-Martins I. Extracellular isoamylase produced by the yeast Lipomyces kononenkoae. Appl Environ Microbiol. 1982;44:1253–1257. doi: 10.1128/aem.44.6.1253-1257.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi K, Abe J, Kozuma T, Yoshida M, Nakamura N, Hizukuri S. Production and application of an isoamylase from Flavobacterium odoratum. Enzyme Microb Technol. 1996;19:456–461. [Google Scholar]
- 38.Takase K. Interaction of catalytic-site mutants of Bacillus subtilis alpha-amylase with substrates and acarbose. Biochim Biophys Acta. 1992;1122:278–282. doi: 10.1016/0167-4838(92)90405-3. [DOI] [PubMed] [Google Scholar]
- 39.Takata H, Kuriki T, Okada S, Takesada Y, Iizuka M, Minamiura N, Imanaka T. Action of neopullulanase. Neopullulanase catalyzes both hydrolysis and transglycosylation at α-(1→4)- and α-(1→6)-glucosidic linkages. J Biol Chem. 1992;267:18447–18452. [PubMed] [Google Scholar]
- 40.Takata H, Takaha T, Kuriki T, Okada S, Takagi M, Imanaka T. Properties and active center of the thermostable branching enzyme from Bacillus stearothermophilus. Appl Environ Microbiol. 1994;60:3096–3104. doi: 10.1128/aem.60.9.3096-3104.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tarentino A L, Quinones G, Changchien L, Plummer T H., Jr Multiple endoglycosidase F activities expressed by Flavobacterium meningosepticum endoglycosidases F2 and F3. J Biol Chem. 1993;208:9702–9708. [PubMed] [Google Scholar]
- 42.Tarentino A L, Quinones G, Hauer C R, Changchien L M, Plummer T H., Jr Molecular cloning and sequence analysis of Flavobacterium meningosepticum glycosylasparaginase: a single gene encodes the alpha and beta subunits. Arch Biochem Biophys. 1995;316:399–406. doi: 10.1006/abbi.1995.1053. [DOI] [PubMed] [Google Scholar]
- 43.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tognoni A, Carrera P, Galli G, Lucchese G, Camerini B, Grandi G. Cloning and nucleotide sequence of the isoamylase gene from a strain of Pseudomonas sp. J Gen Microbiol. 1989;135:37–45. doi: 10.1099/00221287-135-1-37. [DOI] [PubMed] [Google Scholar]
- 45.Yagi F, Sawada R, Imada T, Toyonaga S, Tadera K, Ishihata K. Two isolectins from leaves of winged bean, Psophocarpus tetragonolobus (L.) DC. Plant Cell Physiol. 1994;35:1087–1095. [PubMed] [Google Scholar]
- 46.Yang H, Liu M Y, Romeo T. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the csrA gene product. J Bacteriol. 1996;178:1012–1017. doi: 10.1128/jb.178.4.1012-1017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yokobayashi K, Akai H, Sugimoto T, Hirao M, Sugimoto K, Harada T. Comparison of the kinetic parameters of Pseudomonas isoamylase and Aerobacter pullulanase. Biochim Biophys Acta. 1973;293:197–202. doi: 10.1016/0005-2744(73)90391-4. [DOI] [PubMed] [Google Scholar]
- 48.Yokobayashi K, Misaki A, Harada T. Purification and properties of Pseudomonas isoamylase. Biochim Biophys Acta. 1970;212:458–469. doi: 10.1016/0005-2744(70)90252-4. [DOI] [PubMed] [Google Scholar]