Abstract
Purpose:
Circulating tumor DNA (ctDNA) has the potential to guide therapy selection and monitor treatment response in patients with metastatic cancer. However, germline and clonal hematopoiesis–associated alterations can confound identification of tumor-specific mutations in cell-free DNA (cfDNA), often requiring additional sequencing of tumor tissue. The current study assessed whether ctDNA-based treatment response monitoring could be performed in a tumor tissue–independent manner by combining ultra-deep targeted sequencing analyses of cfDNA with patient-matched white blood cell (WBC)-derived DNA.
Experimental Design:
In total, 183 cfDNA and 49 WBC samples, along with 28 tissue samples, from 52 patients with metastatic colorectal cancer participating in the prospective phase III CAIRO5 clinical trial were analyzed using an ultra-deep targeted sequencing liquid biopsy assay.
Results:
The combined cfDNA and WBC analysis prevented false-positives due to germline or hematopoietic variants in 40% of patients. Patient-matched tumor tissue sequencing did not provide additional information. Longitudinal analyses of ctDNA were more predictive of overall survival than standard-of-care radiological response evaluation. ctDNA mutations related to primary or acquired resistance to panitumumab were identified in 42% of patients.
Conclusions:
Accurate calling of ctDNA mutations for treatment response monitoring is feasible in a tumor tissue–independent manner by combined cfDNA and patient-matched WBC genomic DNA analysis. This tissue biopsy-independent approach simplifies sample logistics and facilitates the application of liquid biopsy ctDNA testing for evaluation of emerging therapy resistance, opening new avenues for early adaptation of treatment regimens.
Translational Relevance.
Treatment response monitoring of patients with metastatic colorectal cancer is currently performed by CT imaging, which assesses tumor volume. Liquid biopsy circulating tumor DNA testing has the potential to replace or complement CT imaging by assessing the presence and abundance of tumor-specific mutations, allowing for personalized treatment and early adaptation of treatment regimens based on the emergence of therapy resistance mutations. We here demonstrate that a combined cell-free DNA and patient-matched white blood cell genomic DNA analysis from a single blood draw is sufficient to eliminate the confounding germline and clonal hematopoiesis–associated alterations and results in the accurate calling of tumor-specific circulating tumor DNA mutations in a tumor tissue–independent manner in patients with metastatic colorectal cancer. In this way, liquid biopsy circulating tumor DNA testing for treatment response monitoring can be offered to patients with cancer in a widely accessible manner.
Introduction
Monitoring cancer treatment to identify response or progression provides physicians with the opportunity to adapt a patients' treatment regimen. As one-size-fits-all systemic therapy makes room for more personalized treatment approaches, and ineffective treatment continuation has serious side effects for the patient, there is a clinical need for biomarkers that can guide the treatment course (1). Currently, clinical treatment response evaluation is performed by standard CT imaging following RECIST (2), which is focused on tumor burden (3). Clinical imaging has limitations for treatment response monitoring because tumor mass does not always correlate with the clinical outcome. For example, clinical imaging does not provide information about the viability of the tumor tissue nor does it constitute the genomic changes of the tumor, that is, the development and outgrowth of subclones following treatment-induced selection (4). As a consequence, determining treatment effectiveness by radiological CT imaging soon after the start of therapy is challenging. Circulating tumor DNA (ctDNA) derived from minimally invasive liquid biopsies is a biomarker indicative of the presence of tumor cells (5, 6). Liquid biopsies allow for longitudinal follow-up and provide the possibility to track intratumoral heterogeneity caused by different subclones without a repeated tumor biopsy and can therefore be a helpful disease monitoring tool (7–10). However, ctDNA analyses are impeded by germline variants and white blood cell (WBC) variants related to clonal hematopoiesis of indeterminate potential (11, 12), which cloud the detection of tumor-specific mutations (13–16). Therefore, ctDNA analyses are often performed in a tumor tissue–informed manner, for which a tumor biopsy is needed to avoid false-positive calls from germline and hematopoietic variants. This requires additional logistic steps, is more complex to perform in daily clinical practice, and is sometimes not feasible. A liquid biopsy-only approach would therefore be an attractive alternative. Here, we investigated whether liquid biopsy cell-free DNA (cfDNA) analyses filtered by liquid biopsy WBC-derived genomic DNA for germline and hematopoietic variants can improve detection of tumor-derived alterations in cfDNA compared with the tissue-informed approach using patient-matched tumor tissue DNA (Fig. 1A). We assessed the applicability of this approach in a cohort of patients with metastatic colorectal cancer (mCRC), with both RAS/BRAF wildtype and left-sided primary tumors, who received doublet chemotherapy and were eligible for anti-EGFR mAb therapy (17–20). These patients are suited to evaluate treatment response monitoring as not all patients respond (21), while those who initially respond are prone to develop acquired resistance over time (22–24).
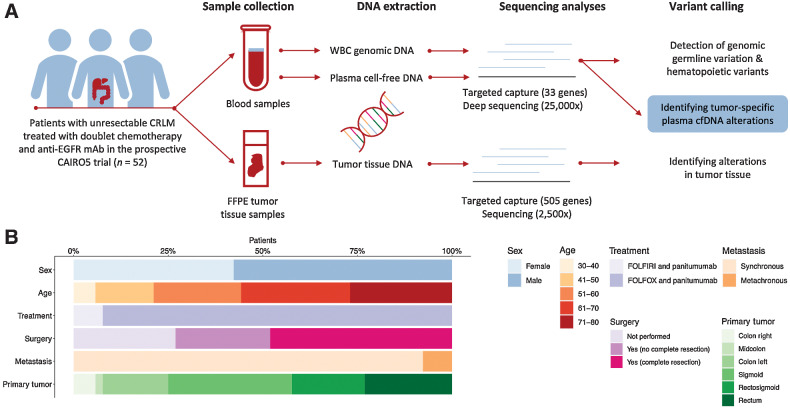
Figure 1.
Analyses of cfDNA in patients with CRLM. A, Schematic representation of the identification of tumor-specific somatic mutations. Blood and tumor tissue samples were collected from 52 patients with CRLM. WBC genomic DNA was isolated from the blood samples and used to remove germline and hematopoietic variants. Tumor tissue DNA was isolated and tumor alterations were identified. Tumor-specific somatic mutations were identified by subtracting the identified WBC hematopoietic and germline variants from the identified plasma cfDNA alterations. B, Baseline characteristics of the 52 patients with CRLM included. CRLM; colorectal liver metastases; EGFR, epidermal growth factor receptor; FFPE, formalin-fixed paraffin-embedded; FOLFIRI, folinic acid, fluorouracil, and irinotecan; FOLFOX, folinic acid, fluorouracil, and oxaliplatin; WBC, white blood cell.
Materials and Methods
Study design
This translational research is a retrospective analysis of liquid biopsies collected from patients with histologically proven colorectal cancer with isolated, previously untreated, initially unresectable liver metastases collected in the prospective multicenter CAIRO5 clinical trial (NCT02162563) of the Dutch Colorectal Cancer Group (25). The ongoing phase-III CAIRO5 trial investigates the optimal first-line systemic therapy for patients with initially unresectable colorectal cancer liver metastases. Patients were eligible for the current study when initially KRAS, NRAS, and BRAF wildtype and randomized for treatment with panitumumab and doublet chemotherapy consisting of 5-fluorouracil, leucovorin, and oxaliplatin or irinotecan (FOLFOX or FOLFIRI), and when at least two liquid biopsy samples were collected. A total of 52 patients, enrolled between November 2014 and April 2019, were included in this study. The study was performed in accordance with the Declaration of Helsinki and a medical ethical committee approved the trial and all patients signed written informed consent for study participation as well as liquid biopsy and tumor tissue collection for translational research. The objective of this research was to identify the best strategy to determine tumor-specific somatic mutations in liquid biopsy ctDNA. Therefore, we analyzed serial ctDNA samples with patient-matched tumor tissue DNA and WBC-derived genomic DNA. As a second endpoint, we assessed the prognostic and predictive clinical power of our genomic profiling approach to evaluate treatment response.
Patient characteristics
Of the 52 patients with initially unresectable colorectal cancer liver metastases included in this study, 42% were female, 92% had synchronous metastases, and patients had a mean age of 61 years. All patients had an adenocarcinoma, were mismatch repair (MMR) proficient, and had tested negative for KRAS and NRAS mutations on tumor tissue samples. However, plasma deep sequencing did reveal baseline KRAS alterations. Non-hotspot mutations in combination with the used tumor tissue mutation assays, or tumor heterogeneity, might explain these discordant results, as discussed previously (26). From February 2017 onward, sidedness and BRAF V600 mutation status were included in the selection criteria and subsequently, only patients with a left-sided primary tumor without a KRAS, NRAS, or BRAF mutation were eligible. No significant differences were found in baseline mutant allele frequency (MAF) levels among patients with left- and right-sided primary tumors (independent sample t test; P = 0.135), among patients with metachronous or synchronous metastases (independent sample t test; P = 0.339), and among males and females (Mann–Whitney test; P = 0.155). All patients were treated with a combination of panitumumab and chemotherapy, consisting of FOLFIRI in 4 patients (8%), and FOLFOX in 48 patients (92%). Fourteen patients remained permanently unresectable, whereas complete radical resection of the primary tumor and liver metastases was achieved in 25 patients. Additional radiotherapy was given in follow-up after systemic treatment to metastases of 18 patients, and 3 patients received a radioembolization procedure. Radiological complete response was observed after treatment in 3 patients, partial response was seen in 30 patients, stable disease in 7 patients, and 12 patients had progressive disease. In addition, 3 patients switched to cetuximab, 20 patients received bevacizumab, 5 patients received CAPOX, and 4 patients received capecitabine monotherapy. In addition, 7 patients were treated with trifluridine and tipiracil, and 4 patients with tegafur, gimeracil, and oteracil. Extrahepatic disease, mainly in the lung and peritoneum, was observed in 17 patients (33%; Supplementary Table S1). An overview of ctDNA dynamics, treatments, and radiological response measurements is depicted per patient in Supplementary Data S1.
Sample characteristics
Liquid biopsies were collected prior to study treatment and every 3 months during follow-up until progression or end of treatment. In total, 186 liquid biopsies were analyzed before treatment (baseline), during treatment, after treatment, and included the latest timepoint closest to progression when available. Six patients missed a baseline blood withdrawal, and no WBCs were stored for 2 patients, resulting in 45 patients with a sample available for the pretreatment analysis. Blood was collected using 10 mL cell-free DNA BCT tubes (Streck) and shipped to the Netherlands Cancer Institute (Amsterdam, the Netherlands). Here, plasma and cell pellet were obtained after a two-step centrifugation process (10 minutes at 1,700 × g followed by 10 minutes at 20,000 × g) and stored at −80°C until further processing. Furthermore, patient-matched formalin-fixed paraffin-embedded (FFPE) tissue blocks from surgical resection or biopsies of the primary tumors were available for 30 patients, and DNA was isolated using the Qiagen AllPrep DNA/RNA/miRNA Universal Kit (Qiagen). The available plasma samples, matched blood-derived WBCs, and FFPE tumor tissue DNA from the 52 patients were sent to Personal Genome Diagnostics (PGDx). Five plasma samples were already part of an earlier analysis using a 58-gene panel (27).
Next-generation sequencing of tumor tissue DNA, plasma cfDNA, and WBC-derived genomic DNA
At PGDx, cfDNA was isolated using the Circulating Nucleic Acid Purification QIAamp kit (Qiagen), with an elution volume of 55 μL. Concentrations of cfDNA were assessed using the Qubit dsDNA High-Sensitivity Assay (Thermo Fisher Scientific). For three plasma cfDNA, one WBC genomic DNA, and two tumor tissue DNA samples, the quality of the material was insufficient for library construction and sequencing analysis. Next, library preparation, hybrid capture, and sequencing of the plasma cfDNA, WBC-derived DNA, and FFPE tumor tissue DNA were performed. For the FFPE tumor tissue DNA, genomic libraries were prepared from 100 ng of DNA by shearing, end-repair, A-tailing, and adapter ligation. Afterward, the libraries were PCR amplified. Target enrichment was performed by hybridized DNA library capture using the PGDx elio tissue complete kit (Research Use Only, RUO) covering single-nucleotide variants (SNV), amplifications, translocations, and microsatellite instability (MSI-H) using a panel consisting of 505 genes (Supplementary Table S2). Samples were pooled and sequenced with 150 bp paired-end reads on the Illumina NextSeq instrument (Illumina) targeting 2,500× depth across the targeted regions, taking along a verified control and a no template control. Two tissue DNA samples did not pass the quality control process and were excluded (Supplementary Table S3).
For the contrived cfDNA, plasma cfDNA, and WBC-derived DNA samples, genomic libraries were prepared from 40 ng of DNA, following normalization, end-repair, A-tailing, adapter ligation, and PCR amplification. For 17 plasma samples, cfDNA yield was limited and genomic libraries were prepared from DNA input ranging from 25 to 40 ng. Targeted capture was performed by hybridization using the custom PGDx elio plasma resolve kit (RUO, version 4.27) covering SNVs, amplifications, translocations, and MSI-H using a panel consisting of 33 genes (Supplementary Table S4), covering over 237,000 bp. After pooling, the captured libraries were sequenced using a targeted deep-sequencing approach with 150 bp paired-end reads on the Illumina NextSeq instrument targeting 25,000× depth across the targeted regions, with an average distinct coverage of 2,900×, taking along a verified control and a no template control. Two plasma samples (Supplementary Table S5) and one WBC genomic DNA sample (Supplementary Table S6) did not pass the quality control process and were excluded.
Somatic mutation calling
Somatic variant identification of tumor tissue DNA, plasma cfDNA, and WBC genomic DNA was performed using machine learning software developed by PGDx, which has shown high accuracy for somatic mutation calling via characterization of germline mutations and sequencing artifacts (28). In the case of less than 500 bp distinct coverage in plasma cfDNA, assessment at specific regions of interest for coverage requirement was done manually, using a 100 bp cutoff to pass the positive threshold and a 475 bp cutoff to pass the negative threshold. Results of the plasma cfDNA samples were scanned for mutations found in other samples of the same patients but not recorded by the PGDx pipeline. Alterations were considered germline when present in all plasma cfDNA timepoints with a MAF over 35%, and when not considered a hotspot (>20 cases reported) in the COSMIC database.
Tumor-specific somatic mutations were identified for all plasma samples by subtracting the identified WBC mutations at baseline from the identified plasma cfDNA mutations, as described previously (14). In short, WBC genomic DNA samples were analyzed for mutations found in one of the plasma samples of the same patient. When present in all samples with at least three duplicate reads of the same mutant, the mutation variant was considered as a hematopoietic variant, except when the MAF in the WBC genomic DNA was more than 100× lower than the MAF in plasma cfDNA, which might indicate circulating tumor cells in the WBC genomic DNA instead of a hematopoietic variant. For mutations identified by cfDNA sequencing but not identified by WBC sequencing, we computed the posterior probability that the mutation was tumor derived using the Bayesian model described in Leal and colleagues (14).
Response evaluation
After removing germline and hematopoietic variants, molecular response evaluation using ctDNA was calculated on the basis of the percentage difference of the most abundant alteration after treatment compared with baseline. Patients without a baseline cfDNA sample were excluded. The molecular response was compared with the radiological response evaluated after systemic treatment. A molecular responder was defined as a patient with elimination of more than 98% of ctDNA after treatment compared with the measurement before treatment initiation. On the basis of our previous studies, we used the elimination of 98% of ctDNA as the threshold for molecular response (8). For comparison, we also evaluated cut-offs of absolute levels of ctDNA at baseline.
After treatment completion, clinical follow-up was performed according to the standard of care. In the case of resectable liver metastases, this included a clinical review every 3 months as well as serum carcinoembryonic antigen (CEA) and CT imaging every 6 months. In the case of unresectable liver metastases, patients continued chemotherapy and were continuously evaluated until disease progression by serum CEA and CT imaging every 2 months. CEA levels were compared between two patient groups, that is, patients with serial CEA levels below 5 ng/mL upon treatment and patients with serial CEA levels above 5 ng/mL. The follow-up was recorded until February 1, 2021. The clinical response evaluation was performed according to standard of care, making use of RECIST version 1.1.
Statistical analyses
Assessment of analytic performance was determined using standardized methods guided by the Clinical and Laboratory Standards Institute (29), including positive (PPA) and negative percent agreement (NPA) for limit of blank, limit of detection, within-run repeatability, between-operator precision, between-day precision, between-instrument precision, and accuracy. Survival analyses were performed using Mantel–Cox log-rank tests. A Bonferroni-corrected threshold of P < 0.02 for significance was set for comparison of three groups. A one-way ANOVA with Tukey multiple comparisons test was used to compare the difference in the number of mutations among the RECIST clinical response criteria. Kolmogorov–Smirnov testing was performed to evaluate the fragment distribution. Lead time between CT imaging and ctDNA, and the number of mutations detected in plasma before and after treatment, was analyzed using Wilcoxon matched-pairs signed-rank testing. Pearson correlation tests were used to calculate the correlation between the number of WBC variants and age, as well as the correlation between MAFs in ctDNA and matched WBC samples. Fisher exact tests were used to compare frequencies. Analyses were performed with Prism version 8 (GraphPad Software, Inc.) or R Statistical Software (version 3.6.1 Foundation for Statistical Computing). Unless otherwise noted, hypothesis tests were two sided with a type 1 error of 5% for determining statistical significance.
Data availability
All data associated with this study are presented in the main text or Supplementary Materials and Methods. Raw sequencing data are available via EGA (EGAS00001006695).
Results
Design and analytic performance of an ultra-deep targeted sequencing liquid biopsy assay
To perform noninvasive tumor profiling and assessment of ctDNA alterations for patients with colorectal cancer, we first designed a targeted gene panel containing common driver genes in colorectal cancer and other tumor types. The FDA-recognized OncoKb database (ref. 30; levels 1, 2, and R1) was utilized to ensure that genes relevant for the management of approved therapies in colorectal cancer were included, which identified BRAF, ERBB2, KRAS, NRAS, and MSI-H. We then expanded this to 33 genes in total to encompass relevant genes and alterations across other common solid tumor types, including non–small cell lung cancer and melanoma. First, we investigated the in silico performance of these gene regions across 353 colorectal cancers previously profiled through The Cancer Genome Atlas project (31), and established that 99.2% of tumors harbored at least one somatic mutation covered by the panel, while 76.8% contained three or more covered alterations. To assess the analytic performance of this ultra-deep targeted sequencing liquid biopsy assay, currently known and further referred to as the PGDx elio plasma resolve assay, a comprehensive set of verification and validation studies were conducted to assess the limit of blank (specificity), limit of detection (sensitivity), within-run repeatability, between-operator precision, between-day precision, between-instrument precision, and accuracy (concordance). In summary, across 39 noncancerous donors, 62 patients with cancer (including 29 patients with colorectal cancer across a total of 13 tumor types), and 18 contrived samples, a total of >450 sample replicates were evaluated, which exhibited exceptional performance for detection of SNVs, insertions/deletions, amplifications, translocations, and MSI-H (Supplementary Tables S7 and S8). Overall, these data demonstrated >99% specificity, >95% sensitivity for detection of alterations 0.25%–1.0% variant allele fraction, >95% repeatability and precision across different laboratory conditions, as well as >95% PPA and NPA compared with orthogonal methods across the majority of alteration types assessed (Supplementary Tables S7 and S8).
Comparison of patient-matched tumor tissue DNA, plasma cfDNA, and WBC DNA
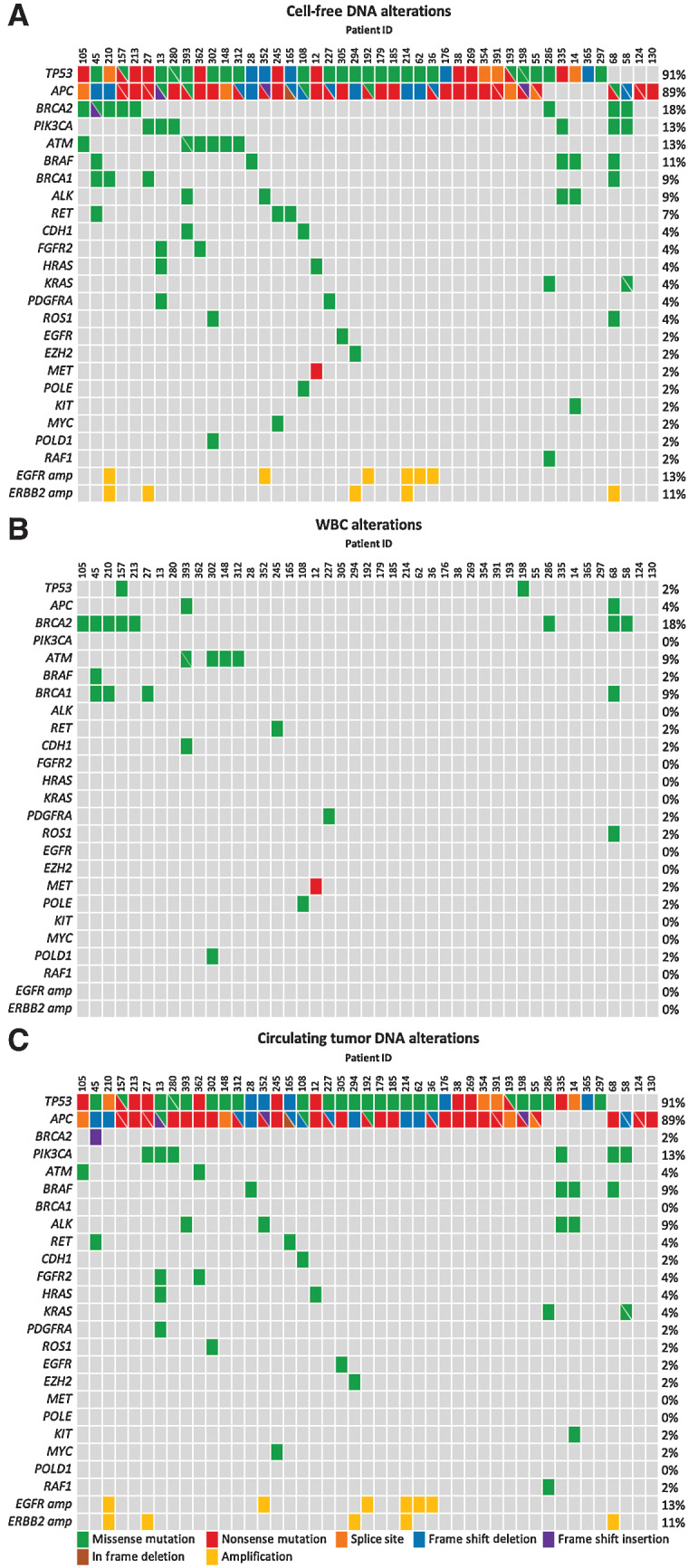
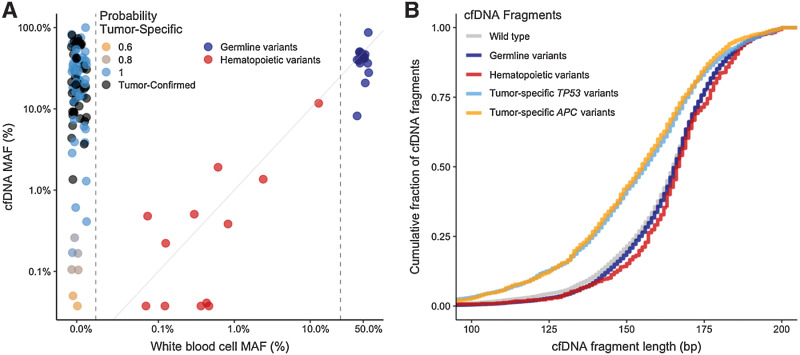
To investigate whether variants in cfDNA were tumor specific or instead were germline or hematopoietic in origin, we performed targeted sequencing of patient-matched cfDNA (Supplementary Table S5), WBC-derived genomic DNA (Supplementary Table S6), and tumor tissue DNA material (Supplementary Table S3) obtained from 52 patients with initially unresectable colorectal liver metastases (CRLM) who were included in the arm of the CAIRO5 clinical trial (NCT02162563) receiving treatment with doublet chemotherapy (FOLFOX or FOLFIRI) and panitumumab (ref. 25; Fig. 1B). Patients were enrolled in this arm of the CAIRO5 study when they had no somatic mutations in KRAS, NRAS, or BRAF, based on tumor tissue analysis. For the current study, DNA alterations were analyzed in both cfDNA and patient-matched WBC-derived genomic DNA from liquid biopsies that could be obtained at baseline, that is, before treatment, which was feasible in 45 of the 52 patients (Supplementary Fig. S1). Ultra-deep targeted sequencing analyses were performed using the PGDx elio plasma resolve 33-gene panel at approximately 25,000× coverage (Supplementary Table S4; refs. 28, 32). A total of 157 alterations were identified in the plasma cfDNA samples (Fig. 2A). All 45 patients (100%) had at least one detectable plasma cfDNA mutation before treatment initiation. Parallel ultra-deep sequence analyses of the patient-matched WBC genomic DNA revealed 39 alterations in total (Supplementary Table S9), of which 29 alterations were also present in the baseline cfDNA (Fig. 2B). These variants comprised rare changes not excluded on the basis of variant databases, as well as potentially pathogenic germline alterations in BRCA1 and BRCA2 genes specifically, which were not filtered because of their clinical utility. By filtering cfDNA alterations by the WBC-derived germline and hematopoietic somatic alterations, the plasma ctDNA alterations were obtained (Fig. 2C). This combined sequencing analysis of plasma cfDNA together with WBC genomic DNA prevented the calling of false-positive ctDNA mutations in 18 patients (40%). After filtering, 45 patients (100%) had at least one detectable plasma ctDNA mutation before treatment initiation, with an average of three tumor-derived mutations per patient with a median MAF of 23% (Supplementary Fig. S2). There was a strong correlation between MAFs of variants observed in both cfDNA and matched WBC samples (Pearson correlation; r = 0.85; P < 0.0001; Fig. 3A). Variants with a WBC MAF of approximately 50% and similarly high cfDNA MAFs most likely represent germline variants, whereas the variants with substantially lower WBC MAFs and similarly low cfDNA MAFs are more likely hematopoietic variants. The number of WBC variants was not significantly correlated with age (Pearson correlation; r = 0.14, P = 0.33; Supplementary Fig. S3). As cfDNA fragments derived from tumor cells are on average slightly smaller than cfDNA fragments derived from WBCs (33), we used this cfDNA characteristic to verify whether cfDNA fragments containing germline variants differed in size from those with tumor-specific variants in cfDNA molecules containing TP53, BRCA, and APC alterations. As expected, we observed that fragments carrying tumor-derived mutations were shorter than wildtype, germline, and WBC-derived hematopoietic fragments (Kolmogorov–Smirnov test, P < 0.001; Fig. 3B; Supplementary Fig. S4A).
Figure 2.
Identification of WBC and ctDNA variants in cfDNA. A, cfDNA alterations identified using a targeted 33-gene panel in 45 baseline plasma samples. B, Germline and hematopoietic alterations were identified on the basis of targeted sequencing of patient-matched WBC-derived genomic DNA. C, ctDNA alterations identified after correction for germline and hematopoietic variants detected in WBC-derived genomic DNA. cfDNA, cell-free DNA; WBC, white blood cell.
Figure 3.
cfDNA and WBC variant frequencies and fragment length distributions. A, WBC-derived MAF (x-axis) and corresponding plasma cfDNA MAF levels (y-axis; Pearson correlation coefficient = 0.85, P < 0.001). The different colors represent the probability of a variant being tumor derived when not observed in WBC genomic DNA. Variants with a WBC MAF of approximately 50% (right vertical dotted line) and high cfDNA MAFs likely represent germline variants. The variants with both low WBC and cfDNA MAFs likely represent hematopoietic variants. B, Cumulative distribution of cfDNA fragment length (bp) for tumor-specific TP53 (light blue) and APC (orange) variants revealed shorter fragment lengths than wild-type (gray), germline (dark blue), and hematopoietic variants (red; Kolmogorov–Smirnov test, P < 0.001). cfDNA, cell-free DNA; MAF, mutant allele frequency; WBC, white blood cell.
Next, we investigated whether the sequencing of patient-matched tumor tissue DNA, which was available for 28 patients (54%; Supplementary Fig. S1), provided additional information to the WBC-informed cfDNA calling. Targeted sequencing analysis was performed using the PGDx elio tissue complete 505-gene panel for detecting alterations in tumor tissue DNA (Supplementary Table S2; ref. 34). This tumor tissue analysis revealed all but one of the germline variants in the samples analyzed but did not identify the majority of hematopoietic variants as identified by the WBC analyses (Supplementary Fig. S5, Supplementary Tables S10 and S11). Interestingly, 1 patient had a somatic TP53 V173G alteration with a tumor tissue MAF of 5% and a plasma cfDNA MAF of 12%, which might be interpreted as a tumor tissue–confirmed plasma ctDNA mutation. However, WBC analyses also reported a MAF of 14%, indicating that this TP53 alteration is a hematopoietic variant. This patient also harbored another TP53 (R248W) variant that was not found in the WBCs but only detected in cfDNA and tumor tissue. Comparing within this patient the tumor-specific TP53 R248W to the hematopoietic TP53 V173G variant by fragment length distributions revealed that TP53 R248W harbored a shorter fragment length than wildtype TP53 and the hematopoietic TP53 V173G, providing additional evidence that the TP53 V173G variant is indeed hematopoietic and not tumor derived (Supplementary Fig. S4B). This demonstrates that tumor tissue analyses can result in false-positive calls due to the presence of hematopoietic variants in WBC.
Treatment response monitoring
Next, we explored the applicability of serial WBC-filtered cfDNA analyses for treatment response monitoring. A total of 183 longitudinal liquid biopsy cfDNA samples were analyzed from 52 patients with CRLM. All patients had an adenocarcinoma, were MMR proficient, and were treated with a combination of panitumumab and chemotherapy (Fig. 1B). In these 183 liquid biopsy samples, 474 cfDNA alterations were observed (Supplementary Table S12). Sequencing the patient-matched WBC genomic DNA samples revealed 39 alterations, of which 16 germline and 23 hematopoietic variants, resulting in filtering 106 non–tumor-specific somatic alterations from the plasma cfDNA (Supplementary Fig. S6; Supplementary Table S13).
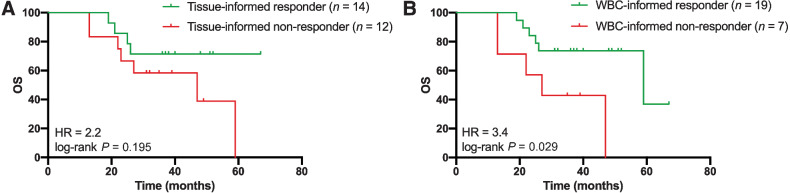
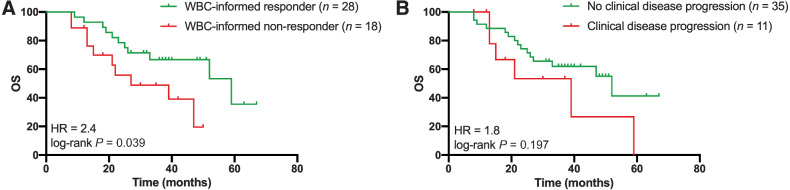
The prognostic value of liquid biopsy ctDNA monitoring was assessed by evaluating the overall survival for patients with or without a molecular response to treatment. On the basis of previous analyses (8), and additional cut-off evaluations (Supplementary Fig. S7), we defined a molecular response as elimination of more than 98% of ctDNA upon treatment based on the highest MAF. Molecular nonresponse was defined as patients with an increase of ctDNA or elimination of less than 98% of ctDNA upon treatment. Supplementary Figure S8A shows an example of a patient classified as a molecular responder, illustrated by clearance of all ctDNA upon treatment. Supplementary Figure S8B and S8C depicts patients classified as molecular nonresponders, presented by MAF levels that do not drop below 20% at any time (Supplementary Fig. S8B) or emerging EGFR ectodomain mutations (Supplementary Fig. S8C). Of the 46 patients whose ctDNA levels could be determined at baseline, 28 (61%) were classified as molecular responders, whereas 18 (39%) patients were classified as molecular nonresponders to treatment. We evaluated the molecular response to treatment using the tissue-informed (Fig. 4A) and WBC-informed approach (Fig. 4B) for all 26 patients with tumor tissue DNA, cfDNA, and WBC genomic DNA analyses. The molecular response to treatment was less prognostic for overall survival when ctDNA was not filtered for germline and hematopoietic variants [not filtered: log-rank P = 0.195; HR = 2.2; 95% confidence interval (CI) = 0.7–7.2 vs. filtered: log-rank P = 0.029; HR = 3.4; 95% CI = 0.8–15.0], highlighting the importance of the combined cfDNA and WBC analyses over the tissue-informed approach. Next, the WBC-informed molecular response evaluation was compared with the clinical response evaluation for all 46 patients. Using the WBC-informed approach, molecular responders showed a significantly longer overall survival compared with molecular nonresponders (median 59 vs. 27 months; log-rank P = 0.039; HR = 2.4; 95% CI = 0.9–6.1; Fig. 5A). In contrast, the overall survival prediction between patients with and without progressive disease based on RECIST evaluation of CT imaging was less strong, with a median overall survival of 39 months and 52 months, respectively (log-rank P = 0.197; HR = 1.8; 95% CI = 0.6–5.6; Fig. 5B; Supplementary Fig. S9). In addition, patients with a radiological complete and partial response had a significant decline in the number of mutations detected in plasma after treatment compared with baseline than patients with progressive disease (Supplementary Fig. S10).
Figure 4.
Comparison of tissue-informed and WBC-informed approaches. Tissue-informed and WBC-informed approaches are compared for the 26 patients with plasma cfDNA, tumor tissue DNA, and WBC genomic DNA available prior to treatment. The molecular response was defined as ctDNA clearance over 98% after treatment compared with the initial baseline ctDNA measurement. A, Assessment of overall survival based on tissue-informed ctDNA analyses (HR = 2.2; 95% CI = 0.7–7.2; log-rank P = 0.195). B, Assessment of overall survival based on WBC-informed cfDNA analyses (HR = 3.4; 95% CI = 0.8–15.0; log-rank P = 0.029). cfDNA, cell-free DNA; ctDNA, circulating tumor DNA; HR, hazard ratio; WBC, white blood cell.
Figure 5.
Overall survival based on molecular response assessment and radiological response evaluation after treatment. Overall survival was compared for the 46 patients with cfDNA assessment prior to treatment. A, Overall survival and molecular response evaluation to treatment based on ctDNA (HR = 2.4; log-rank P = 0.039). The molecular response was defined as ctDNA clearance over 98% after treatment compared with the initial baseline ctDNA measurement. B, Overall survival and radiological response evaluation of CT images (HR = 1.8; log-rank P = 0.197). The radiological response was based on the RECIST evaluation after treatment. cfDNA, cell-free DNA; ctDNA, circulating tumor DNA; CT, computed tomography; HR, hazard ratio; WBC, white blood cell.
In addition to evaluating ctDNA dynamics, that is, changes in ctDNA upon treatment, we examined the prognostic value of ctDNA levels at baseline. Absolute levels of ctDNA at baseline were not prognostic for overall survival (Supplementary Fig. S11). Whereas ctDNA was able to make a valid response prediction after treatment (Fig. 5A), potentially allowing for early switching of therapies, serum CEA levels after treatment were not prognostic for overall survival (HR = 1.1; 95% CI = 0.3–5.1; log-rank P = 0.867; Supplementary Fig. S12A). CEA levels could only discriminate clinical response based on overall survival when taking all longitudinal measurements after treatment into account (Supplementary Fig. S12B).
Detection of disease recurrence and residual disease
After a molecular response to treatment, that is, elimination of >98% of ctDNA, lead time until disease recurrence could be evaluated for 30 patients when restricting the analyses to patients with a blood sample at least 6 months before clinical disease progression (Supplementary Fig. S13A). We observed significantly earlier detection of disease progression based on ctDNA analyses compared with conventional CT imaging, with a median difference of 3.2 months (Wilcoxon matched-pairs signed-rank test; P < 0.001; Supplementary Fig. S13B). Progression was missed using ctDNA analyses of 4 patients (13%; patients 12, 108, 130, 294), and 2 patients (7%) had a complete clinical response and therefore showed no progression of disease on CT imaging nor on ctDNA.
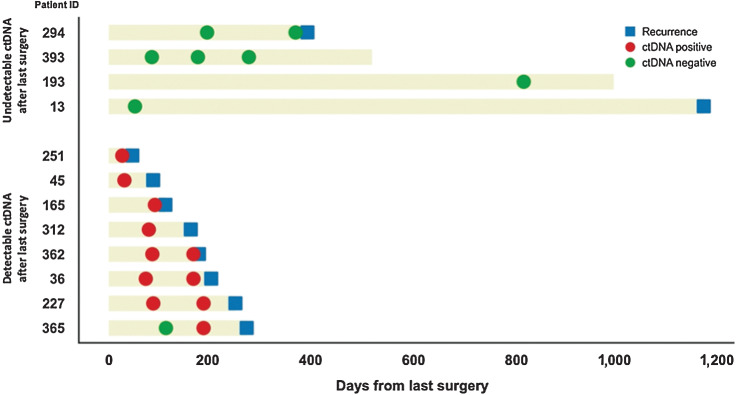
A subset of the patients in the CAIRO5 study was ultimately deemed resectable and had surgical resection of their solitary liver metastasis (n = 38). For the 12 patients among these with a postoperative liquid biopsy sample (range, 31–818 days; Fig. 6), the event-free survival (EFS) calculated as the time from last surgery to disease recurrence or death, whichever came first, was significantly lower for 8 patients with detectable ctDNA postoperatively (median EFS of 5.7 months) compared with 4 patients with undetectable postoperative ctDNA (median EFS of 39.1 months; HR = 6.2; log-rank P = 0.003; Supplementary Fig. S14A). All patients with postoperatively positive ctDNA results developed a recurrence (100% specificity). Postoperative ctDNA measurement missed disease recurrence in 2 patients, one of whom developed the recurrence more than 3 years after surgery (80% sensitivity; Fisher exact test P < 0.0001; Supplementary Fig. S14B).
Figure 6.
Evaluation of disease progression based on postoperative ctDNA in patients with CRLM eligible for surgery. Detectable ctDNA after the resection was indicative of disease recurrence, whereas patients with undetectable levels of ctDNA had less risk of disease recurrence. ctDNA, circulating tumor DNA; CRLM, colorectal liver metastases.
Primary and acquired anti-EGFR mAb treatment resistance
Systemic treatment with therapeutic mAb directed against EGFR can be offered to patients with mCRC, provided that no mutation in either KRAS, NRAS, or BRAF is present and the location of the primary tumor is left sided (17, 18), but it has shown to be effective in only a limited percentage (10%–20%) of these patients (35). Somatic mutations in other genes like PIK3CA, EGFR, PDGFRA, FGFR, as well as amplifications of EGFR, ERBB2, and MET, are known to be involved in the primary and acquired anti-EGFR mAb treatment resistance in patients with mCRC (22, 36–39). Because all patients in our study received panitumumab as an anti-EGFR agent, we evaluated the presence of somatic DNA alterations in KRAS, NRAS, BRAF, EGFR, PIK3CA, PDGFRA, FGFR, and amplifications in EGFR, MET, and ERBB2 as a mechanism of primary and acquired treatment resistance. In addition to the evaluation of somatic alterations in ctDNA, we assessed amplifications in tumor tissue (Supplementary Table S14) and in plasma ctDNA (Supplementary Table S15). Gene mutations associated with primary resistance, which were detected at baseline before the start of treatment, occurred in 17 patients (33%). Mutations associated with acquired resistance, which were detected after the start of treatment, occurred in 5 patients (10%) who were mutually exclusive from the patients with primary resistance (Supplementary Fig. S15). These data imply there is selection pressure to escape the anti-EGFR mAb treatment, yet only in patients lacking these resistance mechanisms at baseline. In addition, for the patients who show acquired resistance on ctDNA analyses, the resistance mechanisms were not detected in the available tumor tissue, highlighting the potential of ctDNA analyses for the detection of anti-EGFR mAb treatment resistance.
Discussion
Liquid biopsy testing for ctDNA is a promising new diagnostic entity. We here demonstrate that correcting for germline and WBC variants is both essential and sufficient for identifying tumor-specific mutations in patients with mCRC. Our analyses showed that combined deep sequencing of plasma cfDNA with patient-matched WBC-derived genomic DNA is able to distinguish tumor-specific mutations from germline variants and WBC alterations associated with clonal hematopoiesis.
The novelty of our approach was the possibility of comparing plasma, WBC-derived genomic DNA, and tumor tissue DNA on an individual patient level and using this information to follow patients during the course of treatment. Other studies investigating patients with mCRC have utilized tissue-informed plasma sequencing without analyses of WBCs (40–46), or did not monitor ctDNA longitudinally (47–49). WBC-informed serial liquid biopsies have been evaluated in a study with patients with stage III colorectal cancer, but only using a pooled set of healthy controls and did not include matched patient-specific filtering (50). In the current study, sequencing matched WBC DNA prevented misclassification of germline and clonal hematopoietic variants in 40% of patients, whereas sequencing of patient-matched tumor tissue DNA for the tissue-informed cfDNA analyses had no additional benefit to the WBC-informed approach. These results indicate that liquid biopsy analyses can be performed for patients with mCRC without the requirement of sequencing tumor tissue. Because of the fact that tumor tissue is often not readily available, liquid biopsy-only testing without the need for patient-matched tissue sequencing has important advantages, including reduced logistical complexity and faster turnaround times, thereby facilitating clinical implementation.
The clinical relevance of cfDNA sequencing was assessed by applying a genomic profiling approach on serial liquid biopsies to evaluate the molecular response to treatment in patients with CRLM receiving chemotherapy and anti-EGFR mAb treatment in a well-controlled clinical trial setting. Longitudinal liquid biopsy ctDNA measurements yield both quantitative (mutant allele fractions) and qualitative (gene mutation) information, allowing simultaneous tracking of different tumor clones. Detecting resistance mechanisms to anti-EGFR treatment in potentially actionable treatment targets, like EGFR and PIK3CA mutations or ERBB2 amplifications (22) could provide opportunities for dynamic adaptation of patients’ treatment regimens or enrollment into new clinical trials. Although panitumumab was not given as monotherapy in the current study, we observed primary resistance in 33% and acquired resistance in another 10% of patients, indicating that a substantial proportion of these patients might benefit from such analyses. For example, 1 patient had a pretreatment EGFR S464 L mutation, a known resistance mechanism to anti-EGFR treatment. This patient showed an initial response, which presumably must have been due to chemotherapy rather than anti-EGFR therapy. Furthermore, all 5 patients with acquired resistance showed clinical disease progression at the blood evaluation timepoint or developed disease progression soon afterward.
The current study also showed that ctDNA dynamics after treatment are informative for patient outcome and can discriminate molecular responders from nonresponders at an early phase of a line of therapy. When investigating all patients with no detectable ctDNA after treatment, who had a complete resection of both the primary tumor and the liver metastases, 11 of 12 patients were still alive at the last moment of follow-up. Molecular response evaluation was more indicative of overall survival than clinical response evaluation (i.e., RECIST) or serum CEA, although it could be useful to evaluate the combination of ctDNA and CEA in the future. As for predicting recurrence of disease after liver resection, despite the limited number of patients analyzed, detection of postoperative ctDNA was indicative of earlier disease recurrence with high sensitivity and specificity and a lead time in favor of ctDNA over CT imaging. These observations suggest that patients with postoperative detectable ctDNA are at high risk for recurrence and might benefit from intensified postoperative disease management.
It is important to note that this study included patients with colorectal cancer liver–only metastases, which have among the highest levels of ctDNA compared with other colorectal cancer metastatic sites (51) and other types of cancer (52). The extent to which the results of the current study using patients with CRLM can be transferred to patients with other sites of metastases, other types of cancer, or earlier stages of colorectal cancer will need to be evaluated.
In conclusion, this study shows that WBC-informed plasma ctDNA testing provides a minimally invasive approach that allows for tissue-independent longitudinal treatment response monitoring that may facilitate clinical intervention in patients with mCRC.
Supplementary Material
The Supplementary Figures file consists of Supplementary Figures 1 to 15 belonging to the manuscript: "Metastatic colorectal cancer treatment response evaluation by ultra-deep sequencing of cell-free DNA and matched white blood cells"
The Supplementary Tables file consists of Supplementary Tables 1 to 15 belonging to the manuscript "Metastatic colorectal cancer treatment response evaluation by ultra-deep sequencing of cell-free DNA and matched white blood cells"
Supplementary Data 1 shows an overview of ctDNA dynamics, treatments, and radiological response measurements per patient.
Acknowledgments
This work was funded in part by the Dutch Cancer Society grant 10438, Stand Up to Cancer–Dutch Cancer Society International Translational Cancer Research Dream Team Grant SU2C-AACR-DT1415, The Mark Foundation for Cancer Research, Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, The Gray Foundation, The Commonwealth Foundation, Cole Foundation, and U.S. NIH grants CA121113, CA006973, and CA233259.
We thank all participating hospitals and their research teams involved in the CAIRO5 study. We thank Pien Delis-van Diemen, Margriet Lemmens, Anne Bolijn, and Marianne Tijssen for laboratory assistance with the FFPE DNA isolations. We would like to acknowledge the NKI-AVL Core Facility Molecular Pathology & Biobanking (CFMPB) for supplying NKI-AVL Biobank material.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 831
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
A. Leal reports other support from Delfi Diagnostics, Inc. outside the submitted work; in addition, A. Leal is an inventor on patent applications submitted by Johns Hopkins University related to cell-free DNA for cancer detection and is a co-founder of Delfi Diagnostics. Under a license agreement between Delfi Diagnostics and Johns Hopkins University, A. Leal and Johns Hopkins University are entitled to royalty distributions related to technology developed at Johns Hopkins University. In addition, A. Leal owns stock in Delfi Diagnostics. J. Phallen reports other support from Delfi Diagnostics during the conduct of the study; in addition, J. Phallen has a patent for cell-free DNA for assessing and/or treating cancer pending, licensed, and with royalties paid from Delfi Diagnostics. V. Adleff reports personal fees from Delfi Diagnostics Inc. outside the submitted work; in addition, V. Adleff has a patent for cell-free DNA for assessing and/or treating cancer and related patents pending, issued, licensed, and with royalties paid from Delfi Diagnostics Inc. J.K. Simmons reports other support from Personal Genome Diagnostics and Natera during the conduct of the study. M. Sausen reports other support from Labcorp outside the submitted work. R.B. Scharpf reports grants and personal fees from Delfi Diagnostics outside the submitted work; in addition, R.B. Scharpf has a patent for US2022-0325343 licensed to Delfi Diagnostics. R.B. Scharpf is a founder of and holds equity in Delfi Diagnostics and also serves as the head of data science; this arrangement has been reviewed and approved by Johns Hopkins University in accordance with its conflict of interest policies. G.A. Meijer reports grants from SU2C/Dutch Cancer Society during the conduct of the study as well as other support from Hartwig Medical Foundation, Sysmex, and Exact Sciences and grants from CZ Health Insurance outside the submitted work; in addition, G.A. Meijer has several patents pending for protein biomarkers for detection of colorectal cancer issued and licensed to CRCbioscreen BV and is co-founder and board member (CSO) of CRCbioscreen BV. G.A. Meijer has research collaborations with Exact Sciences, Sysmex, Sentinel Ch. SpA, Personal Genome Diagnostics (PGDX), Delfi, and Hartwig Medical Foundation; these companies provide materials, equipment, and/or sample/genomic analyses. V.E. Velculescu reports grants, personal fees, and other support from Delfi Diagnostics during the conduct of the study as well as other support from Viron Therapeutics and Epitope outside the submitted work; in addition, V.E. Velculescu has patents and patent applications related to noninvasive detection of cancer pending, issued, licensed, and with royalties paid from PGDx and Delfi Diagnostics. V.E. Velculescu is a founder of Delfi Diagnostics, serves on the board of directors and as a consultant for this organization, and owns Delfi Diagnostics stock, which is subject to certain restrictions under university policy. In addition, Johns Hopkins University owns equity in Delfi Diagnostics. V.E. Velculescu divested his equity in Personal Genome Diagnostics (PGDx) to LabCorp in February 2022. V.E. Velculescu is an inventor on patent applications submitted by Johns Hopkins University related to cancer genomic analyses and cell-free DNA for cancer detection that have been licensed to one or more entities, including Delfi Diagnostics, LabCorp, Qiagen, Sysmex, Agios, Genzyme, Esoterix, Ventana, and ManaT Bio. Under the terms of these license agreements, the university and inventors are entitled to fees and royalty distributions. V.E. Velculescu is an advisor to Viron Therapeutics and Epitope; these arrangements have been reviewed and approved by Johns Hopkins University in accordance with its conflict-of-interest policies. R.J.A. Fijneman reports grants from Dutch Cancer Society, Stand Up to Cancer–Dutch Cancer Society International Translational Cancer Research Dream Team, and other support from Personal Genome Diagnostics during the conduct of the study as well as grants and other support from Personal Genome Diagnostics, Delfi Diagnostics, and Cergentis BV; grants from Merck BV; and other support from Pacific Biosciences outside the submitted work; in addition, R.J. Fijneman has a patent for P127799EP00 pending, a patent for WO2021015619A1 pending, and a patent for EP3631453A1 pending. No disclosures were reported by the other authors.
Authors' Contributions
I. van 't Erve: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J.E. Medina: Conceptualization, data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. A. Leal: Conceptualization, investigation, writing–review and editing. E. Papp: Conceptualization, data curation, validation, investigation, writing–review and editing. J. Phallen: Conceptualization. V. Adleff: Resources. E.J. Chiao: Resources. A.S. Arun: Resources. K. Bolhuis: Resources. J.K. Simmons: Conceptualization, resources, funding acquisition. A. Karandikar: Resources. K.C. Valkenburg: Resources. M. Sausen: Conceptualization, resources, data curation, formal analysis, validation, writing–review and editing. S.V. Angiuoli: Funding acquisition. R.B. Scharpf: Supervision, funding acquisition, methodology, writing–review and editing. C.J.A. Punt: Conceptualization, supervision, funding acquisition, writing–review and editing. G.A. Meijer: Supervision, funding acquisition, writing–review and editing. V.E. Velculescu: Conceptualization, supervision, funding acquisition, investigation, writing–original draft, writing–review and editing. R.J.A. Fijneman: Conceptualization, supervision, funding acquisition, writing–original draft, writing–review and editing.
References
- 1. Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA 2021;325:669–85. [DOI] [PubMed] [Google Scholar]
- 2. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 3. Van Cutsem E, Verheul HM, Flamen P, Rougier P, Beets-Tan R, Glynne-Jones R, et al. Imaging in colorectal cancer: progress and challenges for the clinicians. Cancers 2016;8:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bai HX, Lee AM, Yang L, Zhang P, Davatzikos C, Maris JM, et al. Imaging genomics in cancer research: limitations and promises. Br J Radiol 2016;89:20151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med 2018;379:1754–65. [DOI] [PubMed] [Google Scholar]
- 6. Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov 2014;4:650–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anagnostou V, Forde PM, White JR, Niknafs N, Hruban C, Naidoo J, et al. Dynamics of tumor and immune responses during immune checkpoint blockade in non-small cell lung cancer. Cancer Res 2019;79:1214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Phallen J, Leal A, Woodward BD, Forde PM, Naidoo J, Marrone KA, et al. Early noninvasive detection of response to targeted therapy in non-small cell lung cancer. Cancer Res 2019;79:1204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mauri G, Vitiello PP, Sogari A, Crisafulli G, Sartore-Bianchi A, Marsoni S, et al. Liquid biopsies to monitor and direct cancer treatment in colorectal cancer. Br J Cancer 2022;127:394–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sartore-Bianchi A, Pietrantonio F, Lonardi S, Mussolin B, Rua F, Crisafulli G, et al. Circulating tumor DNA to guide rechallenge with panitumumab in metastatic colorectal cancer: the phase 2 CHRONOS trial. Nat Med 2022;28:1612–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014;20:1472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abbosh C, Swanton C, Birkbak NJ. Clonal haematopoiesis: a source of biological noise in cell-free DNA analyses. Ann Oncol 2019;30:358–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leal A, van Grieken NCT, Palsgrove DN, Phallen J, Medina JE, Hruban C, et al. White blood cell and cell-free DNA analyses for detection of residual disease in gastric cancer. Nat Commun 2020;11:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Razavi P, Li BT, Brown DN, Jung B, Hubbell E, Shen R, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med 2019;25:1928–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weber S, van der Leest P, Donker HC, Schlange T, Timens W, Tamminga M, et al. Dynamic changes of circulating tumor DNA predict clinical outcome in patients with advanced non-small-cell lung cancer treated with immune checkpoint inhibitors. JCO Precis Oncol 2021;5:1540–53. [DOI] [PubMed] [Google Scholar]
- 17. Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023–34. [DOI] [PubMed] [Google Scholar]
- 18. Tejpar S, Stintzing S, Ciardiello F, Tabernero J, Van Cutsem E, Beier F, et al. Prognostic and predictive relevance of primary tumor location in patients with RAS wild-type metastatic colorectal cancer: retrospective analyses of the CRYSTAL and FIRE-3 trials. JAMA Oncol 2017;3:194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Van Cutsem E, Cervantes A, Nordlinger B, Arnold D, Group EGW. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2014;25:iii1–9. [DOI] [PubMed] [Google Scholar]
- 20. Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:1626–34. [DOI] [PubMed] [Google Scholar]
- 21. Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov 2014;4:1269–80. [DOI] [PubMed] [Google Scholar]
- 22. Bertotti A, Papp E, Jones S, Adleff V, Anagnostou V, Lupo B, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature 2015;526:263–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012;486:532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Strickler JH, Loree JM, Ahronian LG, Parikh AR, Niedzwiecki D, Pereira AAL, et al. Genomic landscape of cell-free DNA in patients with colorectal cancer. Cancer Discov 2018;8:164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huiskens J, van Gulik TM, van Lienden KP, Engelbrecht MR, Meijer GA, van Grieken NC, et al. Treatment strategies in colorectal cancer patients with initially unresectable liver-only metastases, a study protocol of the randomised phase 3 CAIRO5 study of the Dutch Colorectal Cancer Group (DCCG). BMC Cancer 2015;15:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van 't Erve I, Greuter MJE, Bolhuis K, Vessies DCL, Leal A, Vink GR, et al. Diagnostic strategies toward clinical implementation of liquid biopsy RAS/BRAF circulating tumor DNA analyses in patients with metastatic colorectal cancer. J Mol Diagn 2020;22:1430–7. [DOI] [PubMed] [Google Scholar]
- 27. Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wood DE, White JR, Georgiadis A, Van Emburgh B, Parpart-Li S, Mitchell J, et al. A machine learning approach for somatic mutation discovery. Sci Transl Med 2018;10:eaar7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clinical Laboratory and Standards Institute. Available from: http://clsi.org.
- 30. Chakravarty D, Gao J, Phillips S, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017;2017:PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 2016;22:1342–50. [DOI] [PubMed] [Google Scholar]
- 32. Al Zoughbi W, Fox J, Beg S, Papp E, Hissong E, Ohara K, et al. Validation of a circulating tumor DNA-based next-generation sequencing assay in a cohort of patients with solid tumors: a proposed solution for decentralized plasma testing. Oncologist 2021;26:e1971–e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019;570:385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deak KL, Jackson JB, Valkenburg KC, Keefer LA, Gerding KMR, Angiuoli SV, et al. Next-generation sequencing concordance analysis of comprehensive solid tumor profiling between a centralized specialty laboratory and the decentralized personal genome diagnostics elio tissue complete kitted solution. J Mol Diagn 2021;23:1324–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bertotti A, Sassi F. Molecular pathways: sensitivity and resistance to anti-EGFR antibodies. Clin Cancer Res 2015;21:3377–83. [DOI] [PubMed] [Google Scholar]
- 36. Sforza V, Martinelli E, Ciardiello F, Gambardella V, Napolitano S, Martini G, et al. Mechanisms of resistance to anti-epidermal growth factor receptor inhibitors in metastatic colorectal cancer. World J Gastroenterol 2016;22:6345–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol 2011;12:594–603. [DOI] [PubMed] [Google Scholar]
- 38. Leto SM, Trusolino L. Primary and acquired resistance to EGFR-targeted therapies in colorectal cancer: impact on future treatment strategies. J Mol Med 2014;92:709–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parseghian CM, Napolitano S, Loree JM, Kopetz S. Mechanisms of innate and acquired resistance to anti-EGFR therapy: a review of current knowledge with a focus on rechallenge therapies. Clin Cancer Res 2019;25:6899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamauchi M, Urabe Y, Ono A, Miki D, Ochi H, Chayama K. Serial profiling of circulating tumor DNA for optimization of anti-VEGF chemotherapy in metastatic colorectal cancer patients. Int J Cancer 2018;142:1418–26. [DOI] [PubMed] [Google Scholar]
- 41. Hsu HC, Lapke N, Wang CW, Lin PY, You JF, Yeh CY, et al. Targeted sequencing of circulating tumor DNA to monitor genetic variants and therapeutic response in metastatic colorectal cancer. Mol Cancer Ther 2018;17:2238–47. [DOI] [PubMed] [Google Scholar]
- 42. Osumi H, Shinozaki E, Yamaguchi K, Zembutsu H. Early change in circulating tumor DNA as a potential predictor of response to chemotherapy in patients with metastatic colorectal cancer. Sci Rep 2019;9:17358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scholer LV, Reinert T, Orntoft MW, Kassentoft CG, Arnadottir SS, Vang S, et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res 2017;23:5437–45. [DOI] [PubMed] [Google Scholar]
- 44. Lyskjaer I, Kronborg CS, Rasmussen MH, Sorensen BS, Demuth C, Rosenkilde M, et al. Correlation between early dynamics in circulating tumour DNA and outcome from FOLFIRI treatment in metastatic colorectal cancer. Sci Rep 2019;9:11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tie J, Wang Y, Cohen J, Li L, Hong W, Christie M, et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: a prospective cohort study. PLoS Med 2021;18:e1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Max Ma X, Bendell JC, Hurwitz HI, Ju C, Lee JJ, Lovejoy A, et al. Disease monitoring using post-induction circulating tumor DNA analysis following first-line therapy in patients with metastatic colorectal cancer. Clin Cancer Res 2020;26:4010–7. [DOI] [PubMed] [Google Scholar]
- 47. Pellini B, Pejovic N, Feng W, Earland N, Harris PK, Usmani A, et al. ctDNA MRD detection and personalized oncogenomic analysis in oligometastatic colorectal cancer from plasma and urine. JCO Precis Oncol 2021;5:PO.20.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang F, Yang Y, Chen X, Jiang H, Wang H, Shen M, et al. Chemotherapy-associated clonal hematopoiesis mutations should be taken seriously in plasma cell-free DNA KRAS/NRAS/BRAF genotyping for metastatic colorectal cancer. Clin Biochem 2021;92:46–53. [DOI] [PubMed] [Google Scholar]
- 49. Parikh AR, Van Seventer EE, Siravegna G, Hartwig AV, Jaimovich A, He Y, et al. Minimal residual disease detection using a plasma-only circulating tumor DNA assay in colorectal cancer patients. Clin Cancer Res 2021;27:5586–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tie J, Cohen JD, Wang Y, Christie M, Simons K, Lee M, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol 2019;5:1710–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. van 't Erve I, Rovers KP, Constantinides A, Bolhuis K, Wassenaar EC, Lurvink RJ, et al. Detection of tumor-derived cell-free DNA from colorectal cancer peritoneal metastases in plasma and peritoneal fluid. J Pathol Clin Res 2021;7:203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplementary Figures file consists of Supplementary Figures 1 to 15 belonging to the manuscript: "Metastatic colorectal cancer treatment response evaluation by ultra-deep sequencing of cell-free DNA and matched white blood cells"
The Supplementary Tables file consists of Supplementary Tables 1 to 15 belonging to the manuscript "Metastatic colorectal cancer treatment response evaluation by ultra-deep sequencing of cell-free DNA and matched white blood cells"
Supplementary Data 1 shows an overview of ctDNA dynamics, treatments, and radiological response measurements per patient.
Data Availability Statement
All data associated with this study are presented in the main text or Supplementary Materials and Methods. Raw sequencing data are available via EGA (EGAS00001006695).