CD161 characterizes an inflamed subset of cytotoxic T lymphocytes that have high expression of immune checkpoints while retaining high cytotoxic capability. These cells can be reinvigorated by immune checkpoint blockade and are a biomarker for clinical benefit.
Abstract
Human papillomavirus (HPV)–driven oropharyngeal carcinoma (OPSCC) is distinct from tobacco- or alcohol-associated OPSCC and has a unique immune landscape. Studies have supported the heterogeneity of T cells, accompanied by a broad repertoire of T-cell responses, within tumors driven by HPV infection. However, the phenotype and function of these HPV-related T cells remain unclear. Using a combination of single-cell RNA sequencing, flow cytometry, pharmacologic inhibition, and immunofluorescence staining, we explored the prognostic implication of HPV-related T cells and further validated our findings in two independent cohorts. Cytotoxic T lymphocytes (CTL) within OPSCC displayed a spectrum of transcriptional signatures. Among which, we identified CD161 receptor, encoded by KLRB1, as a potential marker to distinguish the CTL subsets in HPV-positive OPSCC with a divergent evolutionary trajectory. In-depth analysis revealed that CD161+ CTLs exhibited a more robust immune response over the CD161− counterparts and a T cell–inflamed phenotype that could be further reinvigorated by immune-checkpoint blockade. Despite the high expression of exhaustion markers, reinforcement of CD161+ CTL reactivity was expected to boost immune responses, considering their functional reversibility. We further confirmed that the high level of intratumoral CD161+ CTLs associated with a favorable treatment response and prolonged overall survival. Therefore, our research not only provides an insight into the immune landscape of HPV-driven OPSCC but also sheds light on a special subset of CTLs with prognostic and therapeutic significance.
Introduction
Persistent infection of high-risk human papillomavirus (HPV) is a major etiologic factor that contributes to the malignant transformation in head and neck squamous cell carcinoma (HNSCC; ref. 1). The incidence of oropharyngeal squamous cell carcinoma (OPSCC) has increased rapidly over the past decade, and it has become the most prevalent HPV-driven HNSCC among the Western countries (2). Despite the fact that it arises in a similar anatomic location, virus-driven OPSCC is distinct from tobacco- or alcohol-associated OPSCC and has unique biological behavior and clinical presentation (3–5). Consistent with the prognostic stratification driven by HPV, the sensitivity to radio-/chemotherapy observed in HPV-positive patients reveals a generally vigorous reactivity to conventional antitumor strategies. Our current treatments do not yet differ based upon tumor HPV status. However, accumulating evidence from completed or ongoing clinical trials has emphasized the potential for treatment deintensification as a means of improving quality of life while maintaining survival benefits (1, 6, 7). Whether HPV status predicts the response to immunotherapy remains ambiguous so far but many studies have indicated a trend toward higher sensitivity in HPV-positive patients. In the phase Ib KEYNOTE-012 trial, recurrent or metastatic HPV-positive HNSCCs show a higher objective response rate (ORR) to pembrolizumab alone as a first-line or subsequent-line treatment compared with HPV-negative counterparts (25% vs. 14%; ref. 8). Ensuing data from an expansion cohort of KEYNOTE-012 also show a larger ORR benefit (32% vs. 14%), which results in better 6-month progression-free survival (PFS; 37% vs. 20%) and overall survival (OS; 70% vs. 56%) in HPV-positive patients (9). Similar increases of ORR are reported with nivolumab (15.9% vs. 8.0%; ref. 10) and the anti–PD-L1 agent durvalumab (30% vs. 10.8%; ref. 11) in recurrent or metastatic HNSCCs. In the CheckMate 358 trial, nivolumab yields much higher pathologic regressions in HPV-positive patients than in negative patients (23.5% vs. 5.9%) when applied in a neoadjuvant setting (prior to surgery; ref. 12). Moreover, evidence from the implementation of dual immune-checkpoint blockade (ICB) also supports patients with HPV infection having the greatest benefit from additional ICB (13). The increased ORR and improved OS in HPV-positive patients were further validated in several pooled analyses (14, 15). These data collectively underline a higher immunotherapeutic sensitivity associated with HPV infection, which might be attributed to the preexisting and vigorous immune microenvironment triggered by exogenous viral antigens.
In contrast with the intrinsic immunosuppression of nonviral-induced HNSCC, HPV is known to reshape immune infiltration and enhance innate/adaptive immunity (16, 17). A high abundance of lymphocytes, natural killer (NK) cells, dendritic cells (DC), and M1 macrophages (18, 19), concomitant with the excessive release of type I cytokines and specific antibodies against HPV viral antigens, can evoke an effective local/systemic immune response (20). However, enrichment of regulatory T cells (Treg) and the upregulation of T-cell exhaustion markers, including PD-L1, CTLA-4, LAG3, Tim-3, and TIGIT, are also observed and can modulate immune responses. Despite this, antitumor immune responses ultimately lead to prolonged survival instead of poor outcomes (18, 21, 22). This paradox is known as the T-cell inflamed phenotype and reflects the double-edged sword of HPV infection whereby it induces immune reactivity but not its overactivation (18). Conventional methods of assessing immune activity based on single or several parameters are inadequate, given the inconsistency between phenotype and function. Notwithstanding numerous explorations on a broad repertoire of T-cell responses within tumors driven by HPV infection, the scarcity of deep phenotypic and functional interrogation of subpopulation heterogeneity still highlights an urgent need to comprehensively understand the immune landscape of HPV-driven OPSCC, which will aid in prioritizing which molecules or cell types to target.
Here, we systematically analyzed the immune profiles of HPV-positive versus-negative OPSCC in treatment-naïve patients by single-cell RNA sequencing. According to the transcriptional states and differentiation trajectories of these immune cells, we found KLRB1, which encodes a C-type lectin like receptor, CD161, enriched in HPV-positive tumors and additionally characterized a unique subset of cytotoxic T lymphocytes (CTL) exhibiting robust antitumor immunity and had a T-cell inflamed phenotype compared with their CD161− counterparts. We focused on the phenotypic characteristics and immunologic functions of CD161 and further verified the reversible T-cell state of CD161+ CTLs, rather than terminal dysfunction. In contrast to completely exhausted T cells that presented irreversible dysfunction, overexpression of inhibitory molecules did not compromise the cytotoxic capability of CD161+ CTLs that could be reinvigorated with immunotherapy to potentiate their antitumor effects. The therapeutic and prognostic implications of infiltrating CD161+ CTLs were then significantly validated in the cohorts of OPSCC.
Materials and Methods
Study design and patient cohorts
Two independent cohorts of patients with OPSCC were included in this study. The test cohort consisted of 55 patients with prospectively collected fresh blood samples and 28 matched fresh tumor tissues prior to treatment in Fudan University Shanghai Cancer Center (FUSCC) from March 2019 to September 2022. Lymphocytes were isolated from these fresh blood or tumor samples within 2 hours after collection to detect the fractions of CD161-expressing CTLs or the immunologic functions of corresponding subsets as described below. A pair of HPV-positive and -negative tumor tissue was subjected to single-cell RNA sequencing to explore the immune landscape at the transcriptomic level as indicated. Patients in the test cohort received definitive radiotherapy following induction chemotherapy, and their responses to treatment were recorded in detail for further analyses. Another 83 patients from our retrospective study with archived formalin-fixed paraffin-embedded (FFPE) tumor and cervical lymph node tissues between January 2007 and July 2019 (23) were enrolled as the validation cohort to evaluate immune cell infiltration, and among these patients, 78 cases with the completed follow-up information (medium 32.5 months) were included for survival analyses. All OPSCC patients were staged according to the 8th edition of the UICC/AJCC staging system. FFPE tissue sections were collected and stored at room temperature in the dark. HPV-DNA genotyping and p16INK4A IHC staining was performed to identify the HPV status as previously described (23). In brief, a total of 18 high-risk (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, 83, and 82) and 5 low-risk HPV subtypes (HPV6, 11, 42, 43, and 81) were tested by ISH. In our study, all samples were HPV16 DNA-positive. Only the high-risk HPV-DNA-positive cases with diffuse staining of p16INK4A in >70% of tumor cells were finally considered as HPV-positive. Assessments of response rates were performed by physicians using the RECIST (Response Evaluation Criteria in Solid tumors) v1.1 criterion. Informed written consent was obtained from all patients, and the study was approved by the Ethics Committee of FUSCC in accordance with the ethical standards prescribed by the Declaration of Helsinki.
Processing of samples
Peripheral blood mononuclear cells (PBMC) were isolated from the blood samples of OPSCC patients after using gradient centrifugation (centrifuge 400 × g for 30 minutes at 18°C) over Ficoll-Paque PLUS (GE, cat. #17-1440-03) and incubated with red cell lysis buffer (BioLegend, cat. #420301) for 10 minutes at room temperature. Freshly isolated PBMCs were washed with PBS (HyClone, cat. #SH30256) and then divided into two sections, one was stored with serum-free cell freezing medium (Amsbio, cat. #11914) at −80°C for further checkpoint blockade experiment as described below, and the other was used to detect the fractions of CD161-expressing CTLs and their immunologic function immediately. In the lymphocyte stimulation test, PBMCs were cultured in DMEM (HyClone, cat. #SH30022) for adhesion purification.
Adherent monocytes were roughly separated from the suspended lymphocytes and stained with anti-CD14 (BioLegend, cat. #301803) at 4°C for 15 minutes and analyzed by flow cytometry (BD FACSAria II, purity>80%), and then cultured in DMEM containing 10% FBS supplemented with 20 ng/mL rhGM-CSF for 3 days to prepare autologous antigen-presenting cells (APC). Suspended lymphocytes were activated with 5 μg/mL anti-CD3/CD28 (Novoprotein, cat. #GMP-A018, cat. #GMP-A063) for 1 day and then expanded in DMEM containing 10% FBS and supplemented with 2 U/mL IL7/IL15 (Novoprotein, TSCM Expander, cat. #GMP-1647) for 4 days to obtain proliferative T lymphocytes.
Fresh tumor tissues were minced on ice to small pieces and digested with 1 mg/mL type IV collagenase (Sigma, cat. #C5138), 1 mg/mL hyaluronidase (Sigma, cat. #H3506), and 0.2 mg/mL DNase I (Sigma, cat. #DN25) for 0.5 hours at 37°C and then were gently dissociated through a 70-mesh stainless sieve to prepare the single-cell suspensions at 4°C. Cells from tumor samples were washed after red cell lysis, and then they were divided into two sections as mentioned for blood. A pair of HPV-positive and -negative cell suspensions (viability > 95%) were processed and stored at 4°C for single-cell RNA sequencing within 1 hour after enzyme digestion. Tumor-infiltrating lymphocytes (TIL) were activated and expanded with the same cell culture condition for PBMCs.
Single-cell RNA sequencing
Fresh single-cell suspensions from tumor samples were used at a concentration of 1 × 106 cells/mL, and suspension from the HPV-negative patient served as a control. Sample processing and data acquisition were performed by OE Biotech. Briefly, single-cell gel beads in emulsions were generated by loading single-cell suspensions onto a Chromium Single-Cell Controller Instrument (10× Genomics) using Chromium Next GEM Single-Cell 3′ Kit v3.1 (10× Genomics, cat. #PN-1000268). After that, reverse transcription, cDNA PCR amplification and library preparation were performed according to the 10× Genomics’ gene-expression protocol. Finally, these libraries were sequenced using the Illumina NovaSeq 6000 with 2×150 bp paired-end reads.
The Cell Ranger software pipeline (version 3.1.0) provided by 10× Genomics was used to demultiplex cellular barcodes, map reads to the genome (GRCh38) and transcriptome using the STAR aligner, and downsample reads as required to generate normalized aggregate data across samples, producing a matrix of gene counts versus cells. Cell Ranger also returned Q30 Bases related sequencing quality targets. All Q30-related targets [e.g., Q30 Bases in Barcode, Q30 Bases in RNA Read, Q30 Bases in unique molecular identifier (UMI)] were greater than 89.9%. We processed the UMI count matrix using the R package Seurat (version 3.1.1). To remove the batch effects in single-cell RNA-sequencing data, the mutual nearest neighbors was performed with the R package batchelor. To remove low-quality cells and likely multiplet captures, which is a major concern in microdroplet-based experiments, we applied a criterion to filter out cells with UMI/gene numbers out of the limit of mean value ± 2-fold of standard deviations, assuming a Gaussian distribution of each cells’ UMI/gene numbers. Following a visual inspection of the distribution of cells by the fraction of mitochondrial genes expressed, we further removed the cells with more than 20% mitochondrial-derived UMI counts. Library size normalization was performed in Seurat on the filtered matrix to obtain the normalized count. The average expression and dispersion were calculated for each gene, and genes were subsequently placed into several bins based on the expression. Cells were clustered based on a graph-based clustering approach and were visualized in two-dimension using t-distributed stochastic neighbor embedding (t-SNE). The FindMarkers function in Seurat was run to analyze marker genes of each cluster and differentially expressed genes (DEG).
Heat map and the GO enrichment analysis of DEGs were performed using DoHeatmap and enrichGO in R, respectively. The developmental pseudotime was determined with the Monocle2 package (version 2.9.0; ref. 24). The raw count was first converted from the Seurat object into CellDataSet object with the importCDS function in Monocle. The differentialGeneTest function of the Monocle2 package was used to select ordering genes (q < 0.01) that were likely to be informative in the ordering of cells along the pseudotime trajectory. The dimensional reduction clustering analysis was performed with the reduceDimension function, followed by trajectory inference with the orderCells function using default parameters. Gene expression was plotted with the plot_genes_in_pseudotime function to track changes over pseudotime. To evaluate putative interactions between T cells and cancer cells, the CellPhoneDB (version 2.0; ref. 25) was used to identify biologically relevant ligand–receptor interactions from single-cell transcriptomics data. A ligand or a receptor was defined as “expressed” in a particular cell type if 10% of the cells of that type had nonzero read counts for the ligand/receptor encoding gene. Statistical significance was then assessed by randomly shuffling the cluster labels of all cells and repeating the above steps, which generated a null distribution for each ligand/receptor pair in each pairwise comparison between the two cell types. After running 1,000 permutations, P values were calculated with the normal distribution curve generated from the permuted ligand/receptor pair interaction scores. To define networks of cell–cell communication, we linked any two cell types where the ligand was expressed in the former cell type and the receptor in the latter. R packages Igraph and Circlize were used to display the cell–cell communication networks. We used the AddModuleScore function in Seurat to calculate the average expression of each program on a single-cell level. All analyzed features described in a previous report (26) were binned based on averaged expression, and the control features were randomly selected from each bin. In order to determine if any of the subsets of T cells identified were tissue-resident memory T cells, the related signature (CA10, ITGA1, ITGAE, IL2, IL10, CXCR6, CXCL13, KCNK5, RGS1, CRTAM, DUSP6, and PDCD1) was calculated as well.
Cell treatments
For the lymphocyte stimulation test, clinical-grade HPV16 E7 peptides (22-mers with 12 amino acids overlap; Supplementary Table S1 “HPV E7 Peptide Pool”), as well as recombinant E7 protein, were synthesized (purity > 95%) to assess the reactivity of lymphocytes. Freshly dispersed lymphocytes from blood or tumor tissues were seeded in a 48-well plate (5 ×105 cells/well) and stimulated with the HPV16 E7 peptide pools (10 μg/mL) or recombinant E7 protein (10 μg/mL)-pulsed autologous monocytes in triplicate wells for 5 days, and lymphocytes with unloaded monocytes served as the negative controls (27). The expression of surface markers was conducted by flow cytometry as described below. For the checkpoint blockade experiment, cryopreservated TILs or PBMCs were thawed, expanded, and seeded (5 × 105 cells/well) as described, and treated with PD-1 blocking monoclonal antibody (mAb; BioLegend, clone EH12.2H7, cat. #329926, 10 μg/mL), CD39 blocking mAb (Bio-Rad, clone A1, cat. #MCA1268EL, 10 μg/mL), or Tim-3 blocking mAb (BioLegend, clone F28-2E2, cat. #345004, 10 μg/mL) for 7 days. The expression of aforementioned cytotoxic molecules, including TNFα, IFNγ, and granzyme B, was assessed and compared with respective IgG isotype controls (BioLegend, cat. #400165, 10 μg/mL; Bio-Rad, cat. #MCA928, 10 μg/mL; BioLegend, cat. #400124, 10 μg/mL) within CD161+ or CD161− CTL subsets by flow cytometry as indicated. In the experiments with CD161 blocking mAb, lymphocytes from HPV-positive samples were cultured in the presence of 10 mg/mL CD161 mAb (BioLegend, clone HP-3G10, cat. #339902, 10 μg/mL) or its IgG isotype control (BioLegend, cat. #401402, 10 μg/mL) for 72 hours and labeled with a secondary fluorescent antibody rat anti-mouse IgG1 PE-Cyanine7 (Invitrogen, cat. #25-4015-82) before detection. Expression of relevant cell markers was evaluated by flow cytometry as stated below.
Flow cytometry
The expression of cell-surface makers CD8, CD161, PD-1, Tim-3, CD39, and the intracellular cytokines TNFα, IFNγ, and granzyme B in PBMCs, TILs, or treated cell suspensions was determined and analyzed by flow cytometry as indicated. In brief, cell populations could be prestimulated by a cell activation cocktail with Brefeldin A (BioLegend, cat. #423303) at 37°C for 4 hours if needed to inhibit protein transport for intracellular staining. Dead cells were excluded with a zombie dye fixable viability kit (BioLegend, cat. #423105) for 15 minutes at room temperature, and the remaining living cells were protected from light and preincubated with an Fc receptor blocking solution (BioLegend, cat. #422301) for 5 minutes at room temperature. Cell suspensions were labeled with anti-CD8 (cat. #301006) and anti-CD161 (cat. #339917) and incubated at 4°C for 15 minutes. For the phenotypic analysis, cell-surface antigens were additionally stained with anti–PD-1 (cat. #329906), anti–Tim-3 (cat. #345004), and anti-CD39 (cat. #328209). For functional analysis, intracellular antigens could be subsequently stained with anti-TNFα (cat. #502913), anti-IFNγ (cat. #502531), and anti-granzyme B (cat. #372203) after fixation and permeabilization (BD Biosciences, Fixation/Permeabilization Kit, cat. #554714). Cells were washed two times and resuspended with staining buffer included in the kit. These samples were acquired using a CytoFLEX (Beckman Coulter) and analyzed using FlowJo (version 10). All antibodies were purchased from BioLegend and labeled according to the manufacturer's protocols.
Immunofluorescence staining of tissue sections
The expression of CD8 and CD161 was assessed in the cohort of patients with archival specimens from biopsies or resections prior to treatments. Paraffin-embedded, 4-μm-thick tumor sections were deparaffinized, rehydrated, retrieved, and incubated with the following primary antibodies: anti-CD8 (clone C8/144B, CST, cat. #70306, primary antibody concentration 25 μg/mL) and anti-CD161 (Abcam, cat. #ab197979, primary antibody concentration 1 mg/mL) at a 1:200 dilution for 30 minutes. Isotype-specific secondary fluorescent antibodies goat anti-mouse IgG Alexa 488 (Servicebio, cat. #GB25301, 1:400) or goat anti-rabbit IgG Cy3 (Servicebio, cat. #GB21303, 1:600) were added after washing and then incubated for 10 minutes. Nuclei were visualized with DAPI (Invitrogen, cat. #D1306). Tumor sections from HPV-positive patients were treated with PBS buffer and served as the negative controls, whereas sections from cervical lymph nodes were tested as the positive controls. Because of the low amount of cells within biopsy specimens, we scanned the slides at low magnification (100×) to reduce unnecessary loss of staining intensity, and the images were excluded if they contained either <20% tumor epithelium or <20% stroma. Three representative images of each sample were captured randomly using a confocal scanning microscope (Zeiss; ref. 28). The compartmental location of immune cells expressing CD8 and/or CD161 within the epithelial intratumoral nests or in the stroma adjacent to tumor nests was independently measured by two independent observers blinded to clinical outcomes using the LSM 5 Image Examiner software. No disagreements arose between the two observers. Finally, the average number of staining cells for each slide was calculated and represented as the number of cells per mm2.
Data set analyses
Four public data sets were used at the different steps of our study. We first queried TCGA database in order to retrieve RSEM-normalized log2 bulk mRNA-seq expression data, as well as clinical data of 523 patients with HNSCC. We then filtered 72 HPV-positive and 415 -negative patients who underwent testing for HPV status via p16INK4A IHC. A similar approach was used to retrieve normalized microarray-based gene-expression data of 31 HPV-positive and 64 -negative patients from GSE113282, normalized transcripts per million data of 262 HPV-positive and 49 HPV-negative patients from GSE171898, and normalized counts per million of 11 HPV-positive and 49 HPV-negative patients from GSE159067. From these resources, we utilized gene-expression data available from TCGA database, GSE171898, or GSE113282 to assess the relationship between expression of the given gene (e.g., KLRB1, CLEC2D) and HPV status, and exhibited results with the boxplot function in R. In addition, we utilized survival data from TCGA database to assess the clinical significance of the LLT1/CD161 axis and immune phenotype score data from GSE159067 to analyze the prognostic power of KLRB1 expression in a clinical trial testing PD-1/PD-L1 inhibitors.
Statistical analysis
All values are presented as mean ± standard deviation (SD). The number of samples used (n) is indicated in the figure legends. Data of patients in different groups were compared using a paired or an unpaired Student t test. The Fisher exact test and Pearson chi-square test were used to analyze categorical data in a contingency table. Survival curves were constructed using the Kaplan–Meier method, and the log-rank test was used to compare the groups. The Cox proportional hazards model was applied to evaluate hazard ratios (HR) with a 95% confidence interval (CI). P < 0.05 was considered statistically significant, with P < 0.01 and P < 0.001 as highly significant. All statistical analyses were conducted using SPSS software (version 26.0) or GraphPad Prism (version 8.3).
Data availability
The single-cell RNA sequencing data generated in this study have been deposited in the NCBI database with BioProject (PRJNA843317) or available within the article and its supplementary data files. Expression profile data analyzed in this study are publicly available in Gene-Expression Omnibus at GSE113282, GSE159067, GSE171898, or TCGA database.
Results
Patient characteristics
OPSCC patients treated with induction chemotherapy and definitive radiotherapy with or without concurrent chemotherapy were analyzed in the test and validation cohorts, with p16INK4A + cases accounting for 67.3% (37/55) and 46.2% (36/78), respectively. Baseline patient characteristics from both cohorts are shown in Table 1.
Table 1.
Patient characteristics.
| Fresh samples | Paraffin samples | |||||||
|---|---|---|---|---|---|---|---|---|
| Total | HPV+ | HPV− | P | Total | HPV+ | HPV− | P | |
| Variables | n = 55 (100.0) | n = 37 (67.3) | n = 18 (32.7) | value | n = 78 (100.0) | n = 36 (46.2) | n = 42 (53.8) | value |
| Age medium (range) | 56 (22–77) | 55 (22–77) | 54 (48–77) | 0.002a | 57 (22–80) | 51 (22–74) | 58 (36–80) | 0.026a |
| Gender, N (%) | ||||||||
| Male | 46 (83.6) | 29 (52.7) | 17 (30.9) | 0.262 | 69 (88.5) | 29 (37.2) | 40 (51.3) | 0.095 |
| Female | 9 (16.4) | 8 (14.5) | 1 (1.8) | 9 (11.5) | 7 (9.0) | 2 (2.6) | ||
| Smoking, N (%) | ||||||||
| No | 21 (57.1) | 18 (32.7) | 3 (5.5) | 0.022 | 38 (48.7) | 21 (26.9) | 17 (21.8) | 0.116 |
| Yes | 34 (42.9) | 19 (34.5) | 15 (27.3) | 40 (51.3) | 15 (19.2) | 25 (32.1) | ||
| Anatomic site, N (%) | ||||||||
| Tonsil | 46 (83.6) | 34 (61.8) | 12 (21.8) | 37 (47.4) | 23 (29.5) | 14 (17.9) | ||
| Base of tongue | 4 (7.3) | 2 (3.6) | 2 (3.6) | 0.037b | 30 (38.5) | 11 (14.1) | 19 (24.4) | 0.032b |
| Soft palate | 4 (7.3) | 1 (1.8) | 3 (5.5) | 4 (5.1) | 1 (1.3) | 3 (3.8) | ||
| Other | 1 (1.8) | 0 (0) | 1 (1.8) | 7 (9.0) | 1 (1.3) | 6 (7.7) | ||
| T Stage, N (%) | ||||||||
| T1 | 5 (9.1) | 3 (5.5) | 2 (3.6) | 17 (21.8) | 9 (11.5) | 8 (10.3) | ||
| T2 | 27 (49.1) | 20 (36.4) | 7 (12.7) | 0.115b | 31 (39.7) | 16 (20.5) | 15 (19.2) | 0.110b |
| T3 | 18 (32.7) | 13 (23.6) | 5 (9.1) | 24 (30.8) | 11 (14.1) | 13 (16.7) | ||
| T4 | 5 (9.1) | 1 (1.8) | 4 (7.3) | 6 (7.7) | 0 (0) | 6 (7.7) | ||
| N Stage, N (%) | ||||||||
| N0 | 1 (1.8) | 1 (1.8) | 0 (0) | 4 (5.1) | 1 (1.3) | 3 (3.8) | ||
| N1 | 27 (49.1) | 25 (45.5) | 2 (3.6) | 0.000b | 24 (30.8) | 21 (26.9) | 3 (3.8) | 0.000b |
| N2 | 12 (21.8) | 3 (5.5) | 9 (16.4) | 17 (21.8) | 12 (15.4) | 5 (6.4) | ||
| N3 | 15 (27.3) | 8 (14.5) | 7 (12.7) | 33 (42.3) | 2 (2.6) | 31 (39.7) | ||
| Tumor Stage, N (%) | ||||||||
| I | 18 (32.7) | 18 (32.7) | 0 (0) | 23 (29.5) | 17 (21.8) | 6 (7.7) | ||
| II | 10 (18.2) | 10 (18.2) | 0 (0) | 0.000b | 20 (25.6) | 17 (21.8) | 3 (3.8) | 0.000 |
| III | 11 (20.0) | 9 (16.4) | 2 (3.6) | 10 (12.8) | 2 (2.6) | 8 (10.3) | ||
| IV | 16 (29.1) | 0 (0) | 16 (29.1) | 25 (32.1) | 0 (0) | 25 (32.1) | ||
at test.
bFisher exact test.
Single-cell landscape of HPV-positive OPSCC
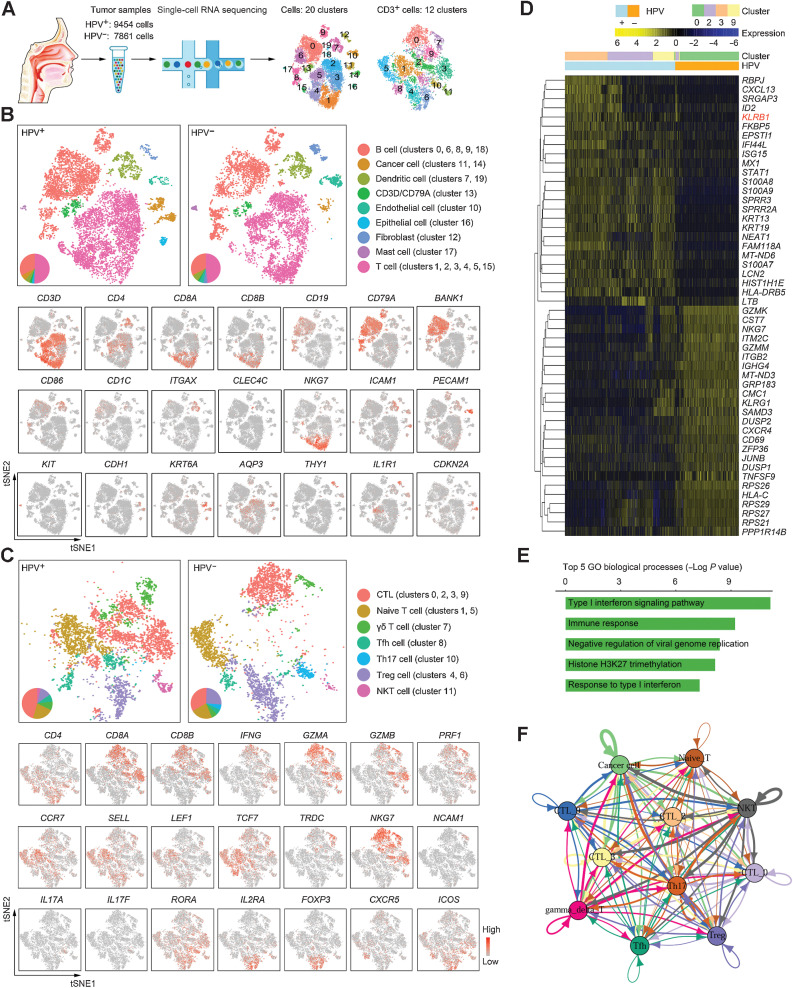
We prepared single-cell suspensions from fresh OPSCC tumors, and then performed single-cell transcriptomics on viable cells that passed quality control filtering. Profiles of 9454 and 7861 cells were yielded in HPV-positive and HPV-negative OPSCC biopsies, respectively, which were categorized, irrespective of HPV status, into 20 clusters (cluster 0–19) among all cell types and 12 clusters (cluster 0–11) among T cells (CD3-positive; Fig. 1A). Analysis of marker genes was conducted to identify cell types, including nonimmune and immune cell fractions (Fig. 1B; Supplementary Fig. S1A). Within the dynamic and heterogeneous microenvironment, nonimmune cells primarily consisted of endothelial cells (marked by PECAM1 and ICAM1; cluster 10), fibroblasts (marked by THY1 and IL1R1; cluster 12), epithelial cells (marked by CDH1, KRT6A, and AQP3; cluster 16), and cancer cells (part of epithelial cells marked by CDKN2A; clusters 11 and 14). Immune cells comprised T cells (marked by CD3D, CD4, and CD8A/CD8B; clusters 1, 2, 3, 4, 5, and 15), B cells (marked by CD19, CD79A, and BANK1; clusters 0, 6, 8, 9, and 18), mast cells (marked by KIT; cluster 17), DCs (marked by CD86, CD1C, and ITGAX; clusters 7 and 19), and a small group of double-positive cells (marked by CD3D and CD79A; cluster 13) that was excluded from the subsequent analysis for coexpression of T-cell and B-cell marker genes. All these cellular compositions were shared in both HPV-positive and HPV-negative OPSCCs with different proportions.
Figure 1.
The immune landscape of OPSCC. A, Graphical overview of the experimental setting. Single cells were prepared from a pair of HPV-positive and HPV-negative OPSCC tumors and processed for single-cell RNA sequencing. Clusters in all cells and within CD3+ cells were determined. B, Top: t-SNE plot of all single-cell suspensions (HPV+, 9454 cells; HPV−, 7861 cells) from tumors partitioned into 20 clusters. Cell subsets were annotated and marked by color code. Bottom: representative genes are shown. C, Top: t-SNE plot of all CD3-expressing cells from tumors partitioned into 12 clusters. Bottom: representative genes are shown. D, Heat map displaying scaled expression values of discriminative genes from all CD8+ T cells between HPV− and HPV+ samples. Top bars show the mapping of each cell to clusters and the HPV status. E, The top 5 enriched GO biological processes of DEGs as defined in D. Individual bars represent the P value after the Benjamini–Hochberg correction for enrichment of GO biological processes. F, The cross-talk between CTLs (clusters 0, 2, 3, and 9), cancer cells, and other immune cells.
In agreement with the intratumoral immune abundance of OPSCCs (29–31), T lymphocytes made up a considerable part of the infiltrate, which could be subdivided into CTLs (marked by CD8A/CD8B, IFNG, GZMA/GZMB, and PRF1; clusters 0, 2, 3, and 9), naïve T cells (marked by CCR7, SELL, and TCF7; clusters 1 and 5), γδ T cells (marked by TRDC; cluster 7), helper follicular T cells (Tfh, marked by CD4, CXCR5, and ICOS; cluster 8), Th17 cells (marked by CD4, IL17A/IL17F, and RORA; cluster 10), Tregs (marked by CD4, IL2RA, and FOXP3; clusters 4 and 6), natural killer T cells (NKT, marked by NCAM1 and NKG7; cluster 11; Fig. 1C; Supplementary Fig. S1B). Despite a little nuance in cell infiltrates, GO enrichment analysis of the DEGs in intratumoral CD4+ T-cell subsets revealed major functions in immune regulation (Supplementary Fig. S1C–S1D), in terms of the similar transition trajectory shared in HPV-positive and HPV-negative samples (Supplementary Fig. S1E). This was not observed for intratumoral CD8+ T cells. Differing from the homogeneous cluster 0 of HPV-negative patients, CTLs from HPV-positive OPSCC displayed increased heterogeneity and could be further subdivided into clusters 2, 3, and 9 (Fig. 1A and C). A total of 50 downregulated and 124 upregulated DEGs were found in CTLs from HPV-positive patients (the top 25 up-/downregulated genes shown in Fig. 1D; Supplementary Table S1 “Gene List for Fig. 1D”), and the top 5 GO biological processes of those DEGs indicated an upregulated interferon-related immune response within HPV-driven OPSCC (Fig. 1E). Cell–cell communication network analysis also suggested that CTLs derived from clusters 2, 3, and 9 interacted more tightly with the cancer cell cluster and other T-cell clusters (Fig. 1F). Collectively, we characterized the immune landscape between HPV-positive and HPV-negative OPSCCs using gene profiles from single-cell RNA sequencing and propose that the persistent infection of HPV affects cell infiltration and transcription features, which subsequently results in the divergent immune responses.
HPV infection reshapes T-cell infiltration into the tumor microenvironment
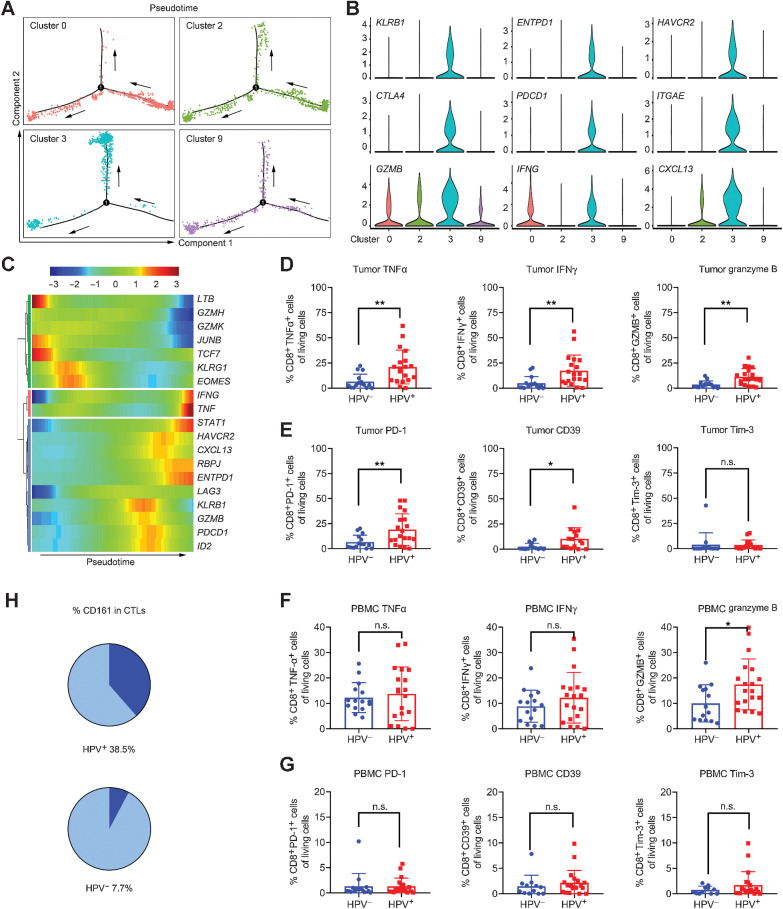
Given the heterogeneity of CTLs in HPV-positive OPSCC, we conducted transcriptional comparisons among clusters 2, 3, and 9 to infer predominant functions. Cluster 3 was most enriched for CTLs with functions mainly involved in the regulation of chemotaxis, apoptosis, and immune response (Supplementary Fig. S2A–S2C). We bioinformatically separated 4 transcriptionally distinct clusters (clusters 0, 2, 3, and 9; Fig. 1C) from TILs and observed that cells belonging to cluster 3 exhibited a distinctive differentiation trajectory (Fig. 2A), and their overexpression of T-cell exhaustion markers [e.g., HAVCR2 (gene for Tim-3), PDCD1 (gene for PD-1), ENTPD1 (gene for CD39), CTLA4], as well as stimulatory cytokines [e.g., GZMB (gene for granzyme B), IFNG (gene for IFNγ)], associated with cytotoxicity (Fig. 2B). We further investigated the transcriptional features of representative genes and observed that the stemlike TCF7 and cytotoxic GZMH and GZMK were expressed only in early effector state, whereas cytotoxic GZMB, IFNG, and TNF were expressed nearer to the terminal exhaustion stage. The checkpoint marker LAG3 was modestly upregulated during this transition, whereas inhibitory PDCD1, HAVCR2, and ENTPD1 were gradually increased before exhaustion (Fig. 2C). These findings demonstrate a delay of gene expression among effector molecules and immunosuppressive checkpoints, which leads to the heterogeneity of CTLs during the transition from early effector to terminal exhaustion. As shown in Fig. 2D and E, elevated cytotoxic molecules TNFα, IFNγ, and granzyme B, as well as exhaustion markers PD-1, CD39, and Tim-3, were confirmed via flow cytometry (Supplementary Fig. S2D) in HPV-positive tumors, consistent with the molecular signatures reported in previous studies whereby HPV-driven OPSCC usually display a mixture of immune activation and dysfunction (22, 29). We failed to duplicate the trend in PBMC samples (Fig. 2F and G), probably due to the relative absence of pathogens (i.e., HPV) in circulation.
Figure 2.
The heterogeneity and cytotoxicity of CTLs in HPV-positive OPSCC. A, Trajectory analysis for the CD8+ T-cell clusters (clusters 0, 2, 3, and 9). B, Expression of marker genes in CD8+ T cells annotated and marked by color code. C, Pseudotime analysis of marker genes in CD8+ T cells. D, Expression of cytotoxic cytokines in OPSCC biopsies (HPV−, n = 14–15; HPV+, n = 19) via flow cytometry, unpaired Student t test. E, Expression of inhibitory receptors in OPSCC biopsies (HPV−, n = 13–15; HPV+, n = 15–19) via flow cytometry, unpaired Student t test. F, Expression of cytotoxic cytokines in PBMCs (HPV−, n = 12–15; HPV+, n = 19–20) via flow cytometry, unpaired Student t test. G, Expression of inhibitory receptors in PBMCs (HPV−, n = 14–15; HPV+, n = 19) via flow cytometry, unpaired Student t test. H, The proportion of CD161+ CTLs (dark blue) based on the data from single-cell RNA sequencing. *, P < 0.05 and **, P < 0.01. n.s.: no statistical significance.
Several studies have proposed the prognostic role of immune cells expressing PD-1/PD-L1 with desirable clinical outcomes in HPV-positive patients (18, 21). This contradiction prompted us to hypothesize that a subset of phenotypically exhausted T cells may be able to exert potent antitumor effects. Related to this, we additionally found that KLRB1, which was widely expressed in CTLs (Fig. 2H; 38.5% of CTLs in HPV+ vs. 7.7% in HPV−) and significantly enriched in cluster 3, and its corresponding product CD161 distinguished a subset exhibiting similarly contradictory characteristics as described above. Despite acting as a coinhibitory receptor in NK cells, CD161 is associated with HPV infection. Some studies find that CD161 expression on T cells represents enhanced cytotoxic potential, long-term memory, and tissue-homing property that mediates protection at pathogen sites of entry (32).
CD161+ CTLs are highly abundant in HPV-driven OPSCC patients
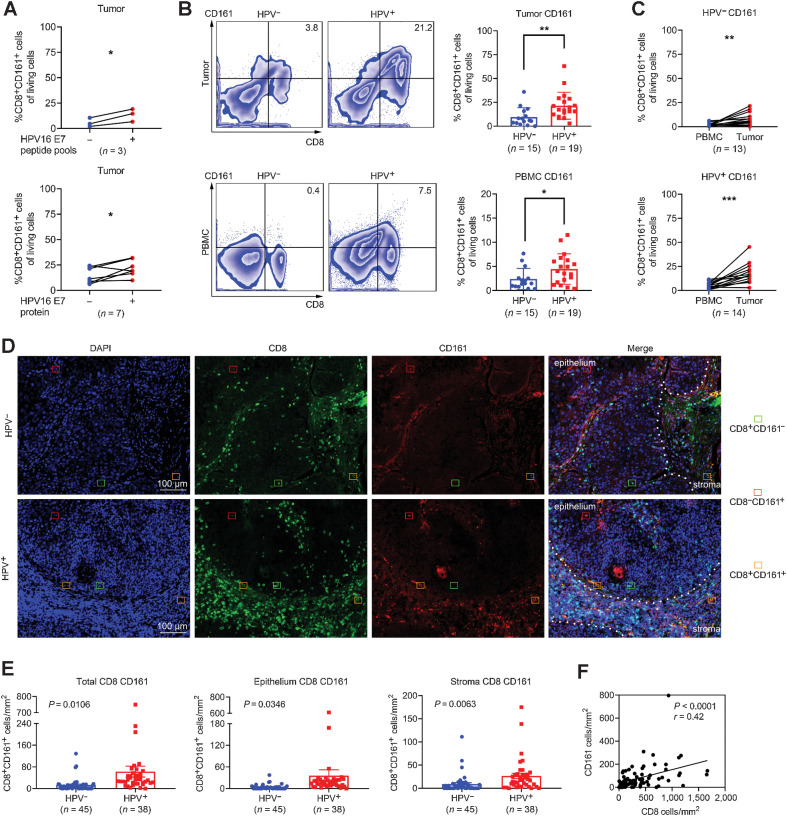
The positive correlation between KLRB1 expression and checkpoint gene expression, as well as the association with tumor mutational burden and MSI status, also provides a rationale to speculate the prognostic implication of CD161 (33). First, we compared the transcript expression of KLRB1 in 3 independent public cohorts with known HPV status (TCGA database, GSE171898, and GSE113282) and validated the prevalent upregulation of KLRB1 in HPV-positive samples (Supplementary Fig. S3A). To explore the association between HPV infection and CTL phenotype, we coincubated TILs or PBMCs derived from HPV-positive patients of the test cohort with their autologous monocytes pulsed with HPV16 E7 peptide pools or recombinant E7 protein for 5 days (Fig. 3A; Supplementary Fig. S3B). The phenotypic reactivity to antigen stimulation was examined directly in a total of 10 fresh tumor samples and 10 unpaired blood samples. An increased proportion of CD161 was found after exposure to viral antigens, indicating the impact of HPV16 infection on T-cell phenotype. Next, we observed a similar enrichment of CD8+CD161+ T cells in HPV-positive samples (Fig. 3B), and the frequency of this subset was higher in tumors than in peripheral blood (Fig. 3C), which implies HPV-dependent expression of CD161. Conversely, a decline of the same subset was synchronously found in peripheral blood after receiving induction chemotherapy (Supplementary Fig. S3C), which made it feasible to use CD161 as a surrogate biomarker on account of its predictive effect on residual antigen load.
Figure 3.
The abundance and distribution of CD161 in HPV16-positive OPSCC. A, Frequencies of CD161 on CTL subsets from HPV16-positive tumors after response to HPV16 E7 peptide pools (n = 3) or recombinant oncoprotein E7 (n = 7) via flow cytometry, paired Student t test. B, Flow cytometry for expression of CD161 on CTLs in tumors and PBMCs (HPV−, n = 15; HPV+, n = 19), unpaired Student t test. C, Pairwise proportion of CD161+ CTLs in tumors and matched PBMCs (HPV−, n = 13; HPV+, n = 14), paired Student t test. D, Representative images of epithelial and stromal sites. The densities of CD8+ T cells and CD161+ cells were counted in HPV− (n = 45) and HPV+ (n = 38) OPSCC biopsies by immunofluorescence. E, The density of total CD161+ CTLs in the epithelium and stroma for each patient. F, Correlation analysis between the number of total CD8+ T cells and total CD161+ cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To depict the spatial localization pattern of CD161, we verified the abundance and composition of CTLs in the validation cohort of 83 patients using multispectral fluorescence IHC (Fig. 3D). The distribution of TILs within a given specimen was very focal. Except for some single cells scattered in the stroma or tumor nest, the majority of CD161+ CTLs were found around the periphery of the tumor mass or in adjacent stromal bands dissecting the mass. In addition, tumor samples displayed one of four patterns: (i) little or no CTL infiltration in epithelium or stroma; (ii) presence of CTL infiltrates that resided solely outside the edge of tumor mass; (iii) presence of intratumoral CTL infiltrates with minimal to no expression of CD161; and (iv) presence of intratumoral CTL infiltrates with intermediate to the maximal expression of CD161. Single-positive cells (CD8+CD161− or CD8−CD161+) and double-positive cells (CD8+CD161+) were evaluated in epithelium or stroma, respectively. Substantial differences were observed in the amount and type of infiltrating T cells across individuals. As expected, HPV-positive OPSCCs were more densely infiltrated with CD161+ CTLs over HPV-negative samples (Supplementary Fig. S3D; Fig. 3E), and the expression of CD161 significantly correlated with that of CD8 (Fig. 3F).
CD161 characterizes a phenotypically and functionally distinct subset of CTLs
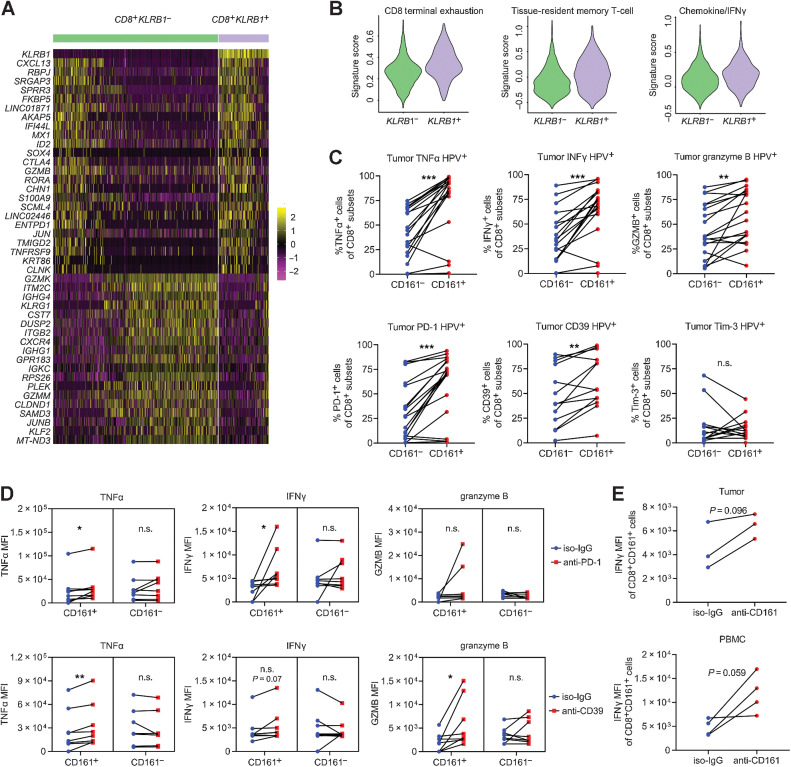
Among T cells, CD161 had been found on memory/effector CD4+ and CD8+ αβ T cells, Th1/17 cells, FoxP3+ Tregs, γδ T cells, NKT cells, as well as mucosal-associated invariant T (MAIT) cells (34). CD161-expressing T cells are thus extremely heterogeneous in spite of their robust cytotoxicity that has been linked with the memory/effector phenotype and rapid response upon antigen encounter (34). The transcriptomic profiles between KLRB1+ CTLs and KLRB1− counterparts displayed increased expression of CXCL13, CTLA4, GZMB, ENTPD1, and AKAP5 accompanied by repression of GZMK, KLRG1, and JUNB expression (Fig. 4A; Supplementary Table S1 “Gene List for Fig. 4A”). The similarity of expression signatures in KLRB1+ CTL subset and cluster 3 (Supplementary Fig. S2B) suggested that CD161 could potentially delineate an enriched subset with coexpression of stimulatory and inhibitory molecules. We calculated relatively higher scores for the exhaustion signature, tissue-resident memory T-cell signature, and chemokine/IFNγ signature in the KLRB1+ T-cell cluster (Fig. 4B). Elevated TNFα, IFNγ, and granzyme B were confirmed in local or peripheral CD161+ CTLs, as likewise for checkpoints PD-1 and CD39 (Fig. 4C; Supplementary Fig. S4A). Although the mechanism behind this remains unclear, to some extent, CD161 did distinguish an inflamed T-cell subset with vigorous reactivity compared with CD161− counterparts.
Figure 4.
The phenotype and function of CD161+ CTLs. A, Heat map displaying expression values of discriminative genes between KLRB1− and KLRB1+ CTLs (from clusters 0, 2, 3, and 9) based on the data from single-cell RNA sequencing. B, Scores for the terminal exhaustion signature, tissue-resident memory T-cell signature, and chemokine/IFNγ signature in KLRB1− and KLRB1+ CTLs based on the data from single-cell RNA sequencing. C, Coexpression of cytotoxic cytokines and inhibitory receptors on CD161− or CD161+ CTLs in OPSCC biopsies (n = 13–19) via flow cytometry, paired Student t test. D, MFI of cytotoxic cytokines coexpressed on CD161− or CD161+ CTLs treated with PD-1 or CD39 blocking antibodies in OPSCC biopsies (n = 7–8), paired Student t test. E, MFI of IFNγ coexpressed on CD161+ CTLs treated with CD161-blocking antibodies for 72 hours in HPV+ biopsies (n = 3) and blood samples (n = 4), paired Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s.: no statistical significance.
It is reported that HNSCC patients with high transcript levels of KLRB1 retain active tumor immunity (33), partially accounting for why the phenotypically exhausted immune cells infiltrating HPV-positive cancer respond better to treatment. Owing to the interplay between the CD161 receptor and its ligand LLT1 lectin-like transcript 1 (LLT1), the transcripts of the LLT1-coding gene CLEC2D were reanalyzed using the public data of TCGA database and another cohort GSE171898. Of note, the synchronous expression of CLEC2D and KLRB1 (Supplementary Figs. S3A and S4B) within a given sample correlated with an active LLT1/CD161 interaction, which led to a considerable survival benefit (Supplementary Fig. S4C; Supplementary Table S1 “TCGA for Supplementary Fig. S4C”). One explanation for the prolonged OS was the positive correlation between KLRB1 expression and the HOT phenotype score (Supplementary Fig. S4D; Supplementary Table S1 “GSE159067 for Supplementary Fig. S4D”), a scoring system developed to identify immunologically active tumors; thus, patients with a higher score are more likely to respond to PD-1/PD-L1 inhibitors (35). Receiver operating characteristic (ROC) analysis was also performed using the public data of 60 HNSCCs with known HPV status from GSE159067, and we validated its power to predict immune phenotype (hot or cold) based on KLRB1 expression (Supplementary Fig. S4D; AUC of ROC = 0.85).
We showed that CTLs exhibited a continuous progression from an early effector state toward a dysfunctional T-cell state. Instead of being at the terminal phase of exhaustion, CD161+ CTLs are more like a transitional subgroup because of their plasticity in responses. The enrichment of immune checkpoints in CD161+ CTLs implies that they would be promising targets for checkpoint inhibitors. Therefore, we attempted to block PD-1, CD39, and Tim-3 and evaluated CTL responses to treatment. According to the median fluorescence intensity (MFI) of TNFα, IFNγ, and granzyme B, inhibition of PD-1 and CD39 increased cytotoxic capability of CD161+ CTLs (Fig. 4D) and the increased MFI was not observed with Tim-3 blockade, which might be due to its negligible difference at gene expression. A trending, but nonsignificant, elevation in IFNγ was found in CD161+ CTLs after a 72-hour blockade with anti-CD161 (Fig. 4E), indicating the therapeutic potential of blocking the LLT1/CD161 axis to fully unleash CTL antitumor effects. This again reflected an ongoing protective immune response with checkpoints representing activation rather than exhaustion; thus, patients with high infiltration of CD161+ CTLs might exhibit a hotter tumor microenvironment and could preferentially benefit from checkpoint inhibitors.
Infiltration of CD161+ CTLs correlates with better treatment response and prolonged survival
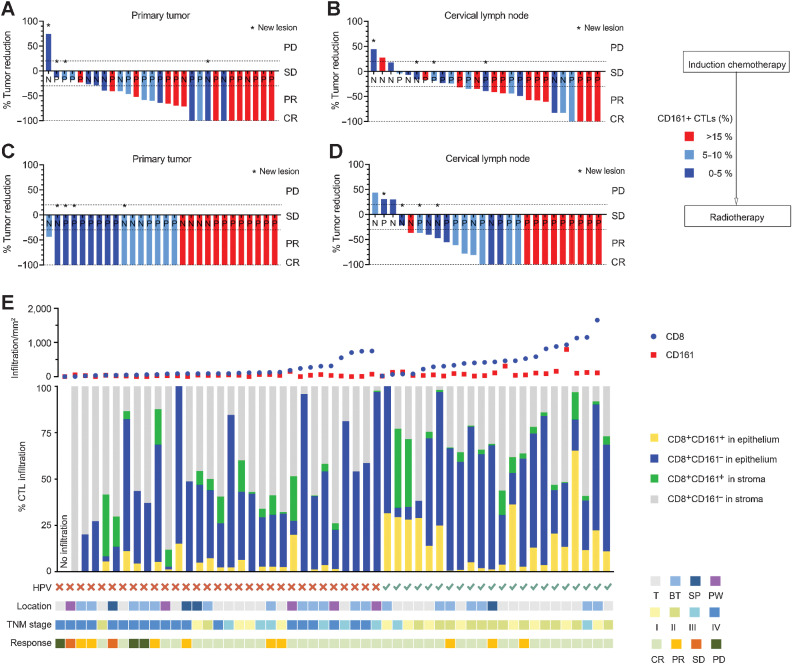
It is well established that HPV infection and no smoking exposure correlate with the improved prognosis in OPSCC patients and is consistent with our analysis (Supplementary Fig. S5A–S5D). However, the impact of CD161+ CTLs on clinical outcomes has not yet been determined. We speculated that the function of CD8+CD161+ T cells might be parallel to that of CD4+CD161+ T cells, in which CD161 defines an activated effector/memory phenotype associated with survival benefits in HPV16-related tumors (19, 36). In the test cohort, patients received definitive radiotherapy following induction chemotherapy, so tumor responses to treatment were assessed after the induction treatment and the second after radiotherapy (Fig. 5A–D). The infiltrating density of CD161+ CTLs prior to treatments was evaluated in tumor specimens and then patients were grouped based on the degree of cell infiltration (low, <5%; medium, 5–10%; high, >15%). Patients with higher proportions of CD161+ CTLs tended to show more objective responses to induction chemotherapy in both primary tumors and metastatic lymph nodes (Fig. 5A and B). Although no obvious differences in disease control were observed in primary sites after radiotherapy, owing to the general sensitivity to radiation in HPV+ OPSCCs (Fig. 5C), we found cases where patients with higher immune infiltration tended to achieve completed response (CR) in metastatic lymph nodes, supporting the potent cytotoxicity of CD161+ T cells (Fig. 5D). No patient developed new lesions among patients with high CD161 density in the following 2-year observation. These effects were further reconfirmed in 53 patients from the validation cohort with available radiologic response data. Patients were grouped according to HPV status and then ordered by the infiltrating density of CTLs (Fig. 5E, top). Combining the composition, spatial distribution (middle), and clinical features (bottom) of each individual, we confirmed that patients with a higher density of CD161+ CTLs, especially the functional infiltration into the epithelium, achieved better posttreatment responses. Whereas patients with no to a few CTLs or those with abundant CTLs confined to areas just around the adjacent stroma were poorly controlled as a result of an ineffective immune response. Tumor regression was also a high probability event in HPV-negative patients with dense infiltration of CD161+ CTLs, suggesting that the percentage of intratumoral CD161+ CTLs prior to treatment could serve as a therapeutic predictor, irrespective of HPV status, and conducive to select HPV-negative OPSCC patients who would respond well.
Figure 5.
The relationship between CD161+ CTLs and treatment response. A and B, Waterfall plot showing response of patients from the test cohort (n = 27–29) to induction chemotherapy according to the proportion of CD161+ CTLs in the tumor microenvironment. X axis represents each patient annotated with their HPV status; Y axis represents the percentage of maximum tumor reduction. Asterisks represent new lesions after the treatment. Dashed lines mark tumor progression of 20% or reduction of 30%, 100%, which defined PD or PR and CR, respectively. C and D, Waterfall plot showing the response of patients (n = 26–28) to the radiotherapy following the induction chemotherapy according to the proportion of CD161+ CTLs in the tumor microenvironment. E, Patients (n = 53) from the validation cohort were divided by HPV status and ordered by the density of CD8+ T cells. Top: number of CD8+ cells and CD161+ cells in the tumor microenvironment; middle: composition and spatial distribution within CD8+ T cells; bottom: HPV status, location, stage, and response to treatment of each patient. N, HPV-negative; P, HPV-positive; T, tonsil; BT, base of tongue; SP, soft palate; PW, posterior wall; CR, complete response; PD, progressive disease; PR, partial response; SD, stable disease.
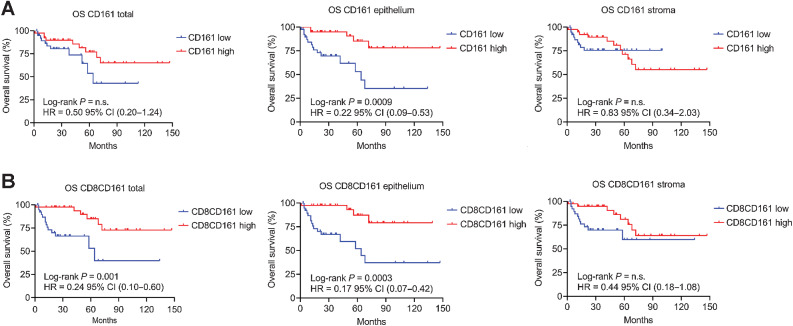
Finally, we analyzed the clinical outcomes in the validation cohort of 78 patients with OPSCC based on the expression and distribution of the CD161 receptor. As previously reported (24), patients with higher infiltration of CD8+ T cells had higher PFS and OS (Supplementary Fig. S5E–S5G), which was not found for CD161 alone (Fig. 6A). However, CTLs with the coexpression of CD161 yielded a significant segregation in PFS and OS, especially when in the epithelium (Fig. 6B; Supplementary Fig. S5H), highlighting the importance of effective infiltration into the epithelium for disease control. OS based on the density of CD161+ CTLs was similar to that based on HPV status, both of which showed a reduction of 83% in the risk of death (Fig. 6B; Supplementary Fig. S5B). All these findings corroborate the conclusion that patients with immunologically “hotter” tumors can be identified based on the expression of CD161 on CTLs. As a quantifiable parameter, the prognostic stratification of CD161 might distinguish patients who would benefit more from treatment, independent of HPV status.
Figure 6.
The prognostic significance of CD161+ CTLs in OPSCC. A, Kaplan–Meier survival plots showing the OS of the validation cohort between CD161 low (n = 39) and CD161 high (n = 39) according to the median cell density of CD161 (total 47 cells/mm2, intraepithelial 19 cells/mm2, stromal 23 cells/mm2) or (B) Kaplan–Meier survival plots between CD8+CD161+ low (n = 39) and CD8+CD161+ high (n = 39) according to the median cell density of CTLs with coexpression of CD8 and CD161 (total 16 cells/mm2, intraepithelial 5.5 cells/mm2 or stromal 8 cells/mm2). The HR with 95% CI and log-rank P value are given. n.s.: no statistical significance.
Discussion
The survival benefit for HPV-positive OPSCC has been associated with a dense T-cell infiltration triggered by HPV infection, but the role of immune responses against HPV is still not completely understood. To elucidate the clinicopathologic impact of HPV on OPSCC, transcriptional profiles of the tumor microenvironment were mapped at the single-cell level. Our work demonstrated that viral infection contributes to divergent CD8+ T-cell subsets and affected the expression of CD161, which distinguished a subset of CTLs enriched in primary tumors and detectable in peripheral blood. Deep analysis into CD161+ CTLs revealed a mixture of cells with an exhausted phenotype but also exhibited robust responses, which enhanced antitumor effects and facilitated a clinically favorable microenvironment in HPV-driven OPSCC.
Chronic HPV infection induces alterations in the tumor microenvironment, with distinct local inflammation and systematic effects. A variety of immunologic parameters have been compared between HPV-positive and HPV-negative HNSCC and concluded that HPV initiates more responsive immunity. Among tumor-infiltrating cells, the predominant T cells and their functional status can directly influence the progression of the disease (37, 38). An earlier study describes that HPV-specific T cells can be stimulated by autologous APCs presenting cognate HPV16-antigen and can acquire a T cell–inflamed phenotype that is proportional to the viral antigen load (22). The presence of inflamed T cells is often accompanied by increased expression of multiple immune-checkpoint molecules that then can lead to T-cell anergy. However, this was not observed for immune responses in HPV-associated tumors.
Our findings offer a possible explanation for the paradox of an exhausted phenotype and robust response in HPV-driven OPSCC. Canonically, CD161 is known as a coinhibitory receptor in NK cells, but its role on T cells remains controversial in the context of tumor immunology (26). Molecular profiles of a pan-cancer resource indicate the favorable prognostic gene KLRB1 from ∼18,000 human tumors across 39 malignancies (39). In agreement with this, high expression of CD161 in tumor-resident immune infiltrates is associated with improved outcomes (26, 40) due to rapid and vigorous responses (32, 36, 41, 42). A preferential expression of CD161 is reported in effector and memory T cells, Th17 cells, MAIT cells, and Tregs at mucosal sites, with the highest amounts of type I cytokines upon activation (36). Apart from the CD8+CD161+ T cells, survival benefit is found in OPSCC patients with high infiltration of active CD4+CD161+ T cells (36, 41). The upregulation of C–C motif chemokine receptors CCR6, CCR5, and CCR2 also suggests altered homing characteristics of cells expressing CD161, which may contribute to tissue pathogenesis during chronic viral infection because of their tissue-resident properties (43). It is becoming increasingly clear that CD161 characterizes a subset of heterogeneous T cells sharing the mixture feature of stemlike and terminal differentiation (44). The engagement of CD161 by the ligand LLT1 favors adaptive immunity, based on the capability to costimulate T-cell proliferation and IFNγ/IL17 production (40). Although studies have tried to explain the underlying mechanism of LLT1/CD161 interaction involved in downstream immune responses, how the LLT1/CD161 axis works and affects the phenotype or function of T cells remains unclear and warrants further investigation.
The PD-1 antibodies nivolumab and pembrolizumab were approved for patients with metastatic, refractory HNSCC based on evidence from the phase III trials CheckMate-141 and KEYNOTE-040, respectively. The latter was also approved as the first-line monotherapy for patients with PD-L1+ (combined-positive score ≥1%) metastatic or unresectable HNSCC based on data from the phase III trial KEYNOTE-048 (45). Although immune-checkpoint inhibitors have emerged as novel and effective therapeutic options, long-lasting responses occur in only ∼20% of patients (10). One challenge is how to select patients most likely to benefit from immunotherapy. Studies indicate that dysfunctional CD8+ T cells can be subdivided into “progenitor” or “stemlike” exhausted and terminally exhausted CD8+ T cells (44, 46–48), meaning T cells with abundant immune checkpoints, but not functionally terminally exhausted, would be promising targets (26, 46). Our research demonstrates that CD161 marked a subgroup of CTLs that retain excellent immune reactivity, albeit concomitant with the overexpression of inhibitory checkpoints. The transitional state of CD161+ CTLs makes it feasible to reinvigorate the subset by checkpoint blockade to boost antitumor responses. It is estimated that around 20% of HPV-driven OPSCC patients have an inferior prognosis, meanwhile, a portion of HPV-negative patients miss the opportunity to benefit from immunotherapy. Our work provides preliminary evidence for the prognostic value of CD161+ CTLs, which can be used as a quantifiable biomarker conducive to screen patients, as well as to predict the clinical outcome irrespective of HPV status.
There are certain limitations to our study. Further validation experiments are required to identify the immunologic role of CD161 within the tumor microenvironment and how cells respond to tumor-associated antigens in terms of the functional variability of the CD161+ T-cell subset. The study does not clearly demonstrate how HPV infection affects the activity of the CD161 receptor, nor what role receptor ligation might play in the downstream regulation. A need continues to exist for further explorations to elucidate the molecular signatures and underlying mechanisms of T-cell exhaustion during chronic viral infection, as well as tumorigenesis. Lastly, the study lacked compelling evidence from clinical data to support the prognostic value of CD161+ CTLs in patients treated with ICB regarding their abundant therapeutic targets.
In conclusion, we demonstrate the unique immunologic features in patients with HPV-positive OPSCC, that CD161 characterizes a subpopulation of CTLs with vigorous immune response, and that an inflamed phenotype is associated with better clinical outcomes and can aid in the selection of eligible patients for immunotherapy.
Supplementary Material
Supplementary Table 1
Supplementary figures and legends
Acknowledgments
This study was supported by grants from the Fudan University Shanghai Cancer Center Foundation Project (YJRC1903 to X. Lu), the National Natural Science Foundation of China (82172666 to F. Su) and the Western Talent Program of the Chinese Academy of Sciences (E023222Y to F. Su). We gratefully thank all the patients who participated in this research. We also thank Yongbing Ba from OE Biotech Co., Ltd (Shanghai, China) for assistance with bioinformatics analysis.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
X. Ou reports grants from the Chinese Society of Clinical Oncology and the National Natural Science Foundation of China outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
Y. Wei: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. T. Xu: Resources, investigation. C. Li: Software, formal analysis, investigation, writing–review and editing. X. Zhou: Resources, investigation, writing–review and editing. W. Qian: Resources. C. Shen: Resources. Q. Wang: Resources, validation. X. Xing: Resources. X. Ou: Resources. X. He: Resources. H. Ying: Resources. C. Hu: Resources. Y. Wang: Resources. Q. Ji: Resources. F. Su: Supervision, funding acquisition, investigation, project administration, writing–review and editing. X. Lu: Conceptualization, resources, supervision, funding acquisition, investigation, writing–review and editing.
References
- 1. Chow LQM. Head and neck cancer. N Engl J Med 2020;382:60–72. [DOI] [PubMed] [Google Scholar]
- 2. de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer 2017;141:664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koneva LA, Zhang Y, Virani S, Hall PB, McHugh JB, Chepeha DB, et al. HPV integration in HNSCC correlates with survival outcomes, immune response signatures, and candidate drivers. Mol Cancer Res 2018;16:90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer 2018;18:269–82. [DOI] [PubMed] [Google Scholar]
- 5. Chen YP, Wang YQ, Lv JW, Li YQ, Chua MLK, Le QT, et al. Identification and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: implications for immunotherapy. Ann Oncol 2019;30:68–75. [DOI] [PubMed] [Google Scholar]
- 6. Ozcan-Wahlbrink M, Schifflers C, Riemer AB. Enhanced radiation sensitivity of human papillomavirus-driven head and neck cancer: focus on immunological aspects. Front Immunol 2019;10:2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen AM, Felix C, Wang PC, Hsu S, Basehart V, Garst J, et al. Reduced-dose radiotherapy for human papillomavirus-associated squamous-cell carcinoma of the oropharynx: a single-arm, phase 2 study. Lancet Oncol 2017;18:803–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol 2016;17:956–65. [DOI] [PubMed] [Google Scholar]
- 9. Chow LQM, Haddad R, Gupta S, Mahipal A, Mehra R, Tahara M, et al. Antitumor activity of pembrolizumab in biomarker-unselected patients with recurrent and/or metastatic head and neck squamous cell carcinoma: results from the phase Ib KEYNOTE-012 expansion cohort. J Clin Oncol 2016;34:3838–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferris RL, Blumenschein G Jr, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med 2016;375:1856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zandberg DP, Algazi AP, Jimeno A, Good JS, Fayette J, Bouganim N, et al. Durvalumab for recurrent or metastatic head and neck squamous cell carcinoma: Results from a single-arm, phase II study in patients with ≥25% tumour cell PD-L1 expression who have progressed on platinum-based chemotherapy. Eur J Cancer 2019;107:142–52. [DOI] [PubMed] [Google Scholar]
- 12. Ferris RL, Spanos WC, Leidner R, Goncalves A, Martens UM, Kyi C, et al. Neoadjuvant nivolumab for patients with resectable HPV-positive and HPV-negative squamous cell carcinomas of the head and neck in the CheckMate 358 trial. J Immunother Cancer 2021;9:e002568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Semrau S, Gostian AO, Traxdorf M, Eckstein M, Rutzner S, von der Grun J, et al. Implementation of double immune checkpoint blockade increases response rate to induction chemotherapy in head and neck cancer. Cancers (Basel) 2021;13:1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu Y, Zhu G, Maroun CA, Wu IXY, Huang D, Seiwert TY, et al. Programmed death-1/programmed death-ligand 1-axis blockade in recurrent or metastatic head and neck squamous cell carcinoma stratified by human papillomavirus status: a systematic review and meta-analysis. Front Immunol 2021;12:645170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galvis MM, Borges GA, Oliveira TB, Toledo IP, Castilho RM, Guerra ENS, et al. Immunotherapy improves efficacy and safety of patients with HPV positive and negative head and neck cancer: a systematic review and meta-analysis. Crit Rev Oncol Hematol 2020;150:102966. [DOI] [PubMed] [Google Scholar]
- 16. Wang HF, Wang SS, Tang YJ, Chen Y, Zheng M, Tang YL, et al. The double-edged sword-how human papillomaviruses interact with immunity in head and neck cancer. Front Immunol 2019;10:653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Subbarayan RS, Arnold L, Gomez JP, Thomas SM. The role of the innate and adaptive immune response in HPV-associated oropharyngeal squamous cell carcinoma. Laryngoscope Investig Otolaryngol 2019;4:508–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gameiro SF, Ghasemi F, Barrett JW, Koropatnick J, Nichols AC, Mymryk JS, et al. Treatment-naive HPV+ head and neck cancers display a T-cell-inflamed phenotype distinct from their HPV- counterparts that has implications for immunotherapy. OncoImmunology 2018;7:e1498439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Welters MJP, Ma W, Santegoets S, Goedemans R, Ehsan I, Jordanova ES, et al. Intratumoral HPV16-specific T cells constitute a Type I-oriented tumor microenvironment to improve survival in HPV16-driven oropharyngeal cancer. Clin Cancer Res 2018;24:634–47. [DOI] [PubMed] [Google Scholar]
- 20. Mytilineos D, Ezic J, von Witzleben A, Mytilineos J, Lotfi R, Furst D, et al. Peripheral cytokine levels differ by HPV status and change treatment-dependently in patients with head and neck squamous cell carcinoma. Int J Mol Sci 2020;21:5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Solomon B, Young RJ, Bressel M, Urban D, Hendry S, Thai A, et al. Prognostic significance of PD-L1(+) and CD8(+) immune cells in HPV(+) oropharyngeal squamous cell carcinoma. Cancer Immunol Res 2018;6:295–304. [DOI] [PubMed] [Google Scholar]
- 22. Krishna S, Ulrich P, Wilson E, Parikh F, Narang P, Yang S, et al. Human papilloma virus specific immunogenicity and dysfunction of CD8(+) T cells in head and neck cancer. Cancer Res 2018;78:6159–70. [DOI] [PubMed] [Google Scholar]
- 23. Xu T, Shen C, Wei Y, Hu C, Wang Y, Xiang J, et al. Human papillomavirus (HPV) in Chinese oropharyngeal squamous cell carcinoma (OPSCC): a strong predilection for the tonsil. Cancer Med 2020;9:6556–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32:381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020;15:1484–506. [DOI] [PubMed] [Google Scholar]
- 26. Mathewson ND, Ashenberg O, Tirosh I, Gritsch S, Perez EM, Marx S, et al. Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell 2021;184:1281–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Poelgeest MI, Visconti VV, Aghai Z, van Ham VJ, Heusinkveld M, Zandvliet ML, et al. Potential use of lymph node-derived HPV-specific T cells for adoptive cell therapy of cervical cancer. Cancer Immunol Immunother 2016;65:1451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Komdeur FL, Prins TM, van de Wall S, Plat A, Wisman GBA, Hollema H, et al. CD103+ tumor-infiltrating lymphocytes are tumor-reactive intraepithelial CD8+ T cells associated with prognostic benefit and therapy response in cervical cancer. OncoImmunology 2017;6:e1338230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cillo AR, Kurten CHL, Tabib T, Qi Z, Onkar S, Wang T, et al. Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity 2020;52:183–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chakravarthy A, Henderson S, Thirdborough SM, Ottensmeier CH, Su X, Lechner M, et al. Human papillomavirus drives tumor development throughout the head and neck: improved prognosis is associated with an immune response largely restricted to the oropharynx. J Clin Oncol 2016;34:4132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen X, Yan B, Lou H, Shen Z, Tong F, Zhai A, et al. Immunological network analysis in HPV associated head and neck squamous cancer and implications for disease prognosis. Mol Immunol 2018;96:28–36. [DOI] [PubMed] [Google Scholar]
- 32. Konduri V, Oyewole-Said D, Vazquez-Perez J, Weldon SA, Halpert MM, Levitt JM, et al. CD8(+)CD161(+) T-cells: cytotoxic memory cells with high therapeutic potential. Front Immunol 2020;11:613204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cheng X, Cao Y, Wang X, Cheng L, Liu Y, Lei J, et al. Systematic pan-cancer analysis of KLRB1 with prognostic value and immunological activity across human tumors. J Immunol Res 2022;2022:5254911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Braud VM, Meghraoui-Kheddar A, Elaldi R, Petti L, Germain C, Anjuere F. LLT1-CD161 interaction in cancer: promises and challenges. Front Immunol 2022;13:847576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Foy JP, Karabajakian A, Ortiz-Cuaran S, Boussageon M, Michon L, Bouaoud J, et al. Immunologically active phenotype by gene expression profiling is associated with clinical benefit from PD-1/PD-L1 inhibitors in real-world head and neck and lung cancer patients. Eur J Cancer 2022;174:287–98. [DOI] [PubMed] [Google Scholar]
- 36. Duurland CL, Santegoets SJ, Abdulrahman Z, Loof NM, Sturm G, Wesselink TH, et al. CD161 expression and regulation defines rapidly responding effector CD4+ T cells associated with improved survival in HPV16-associated tumors. J Immunother Cancer 2022;10:e003995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colbert LE, El MB, Lynn EJ, Bronk J, Karpinets TV, Wu X, et al. Expansion of candidate HPV-specific T cells in the tumor microenvironment during chemoradiotherapy is prognostic in HPV16(+) cancers. Cancer Immunol Res 2022;10:259–71. [DOI] [PubMed] [Google Scholar]
- 38. Shamseddine AA, Burman B, Lee NY, Zamarin D, Riaz N. Tumor immunity and immunotherapy for HPV-related cancers. Cancer Discov 2021;11:1896–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015;21:938–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Braud VM, Biton J, Becht E, Knockaert S, Mansuet-Lupo A, Cosson E, et al. Expression of LLT1 and its receptor CD161 in lung cancer is associated with better clinical outcome. OncoImmunology 2018;7:e1423184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Santegoets SJ, van Ham VJ, Ehsan I, Charoentong P, Duurland CL, van Unen V, et al. The anatomical location shapes the immune infiltrate in tumors of same etiology and affects survival. Clin Cancer Res 2019;25:240–52. [DOI] [PubMed] [Google Scholar]
- 42. Li Z, Zheng B, Qiu X, Wu R, Wu T, Yang S, et al. The identification and functional analysis of CD8+PD-1+CD161+ T cells in hepatocellular carcinoma. NPJ Precis Oncol 2020;4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Konduri V, Joseph SK, Byrd TT, Nawas Z, Vazquez-Perez J, Hofferek CJ, et al. A subset of cytotoxic effector memory T cells enhances CAR T cell efficacy in a model of pancreatic ductal adenocarcinoma. Sci Transl Med 2021;13:eabc3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eberhardt CS, Kissick HT, Patel MR, Cardenas MA, Prokhnevska N, Obeng RC, et al. Functional HPV-specific PD-1(+) stem-like CD8 T cells in head and neck cancer. Nature 2021;597:279–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lechner M, Liu J, Masterson L, Fenton TR. HPV-associated oropharyngeal cancer: epidemiology, molecular biology and clinical management. Nat Rev Clin Oncol 2022;19:306–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 2019;50:195–211. [DOI] [PubMed] [Google Scholar]
- 47. Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, et al. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity 2016;45:415–27. [DOI] [PubMed] [Google Scholar]
- 48. Fergusson JR, Huhn MH, Swadling L, Walker LJ, Kurioka A, Llibre A, et al. CD161(int)CD8+ T cells: a novel population of highly functional, memory CD8+ T cells enriched within the gut. Mucosal Immunol 2016;9:401–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Supplementary figures and legends
Data Availability Statement
The single-cell RNA sequencing data generated in this study have been deposited in the NCBI database with BioProject (PRJNA843317) or available within the article and its supplementary data files. Expression profile data analyzed in this study are publicly available in Gene-Expression Omnibus at GSE113282, GSE159067, GSE171898, or TCGA database.