Comprehensive genomics reveal high prevalence of low hypodiploidy in older patients with B-ALL and track its origin to TP53-deficient age-associated clonal hematopoiesis.
Abstract
Low hypodiploidy defines a rare subtype of B-cell acute lymphoblastic leukemia (B-ALL) with a dismal outcome. To investigate the genomic basis of low-hypodiploid ALL (LH-ALL) in adults, we analyzed copy-number aberrations, loss of heterozygosity, mutations, and cytogenetics data in a prospective cohort of Philadelphia (Ph)-negative B-ALL patients (n = 591, ages 18–84 years), allowing us to identify 80 LH-ALL cases (14%). Genomic analysis was critical for evidencing low hypodiploidy in many cases missed by cytogenetics. The proportion of LH-ALL within Ph-negative B-ALL dramatically increased with age, from 3% in the youngest patients (under 40 years old) to 32% in the oldest (over 55 years old). Somatic TP53 biallelic inactivation was the hallmark of adult LH-ALL, present in virtually all cases (98%). Strikingly, we detected TP53 mutations in posttreatment remission samples in 34% of patients. Single-cell proteogenomics of diagnosis and remission bone marrow samples evidenced a preleukemic, multilineage, TP53-mutant clone, reminiscent of age-related clonal hematopoiesis.
Significance:
We show that low-hypodiploid ALL is a frequent entity within B-ALL in older adults, relying on somatic TP53 biallelic alteration. Our study unveils a link between aging and low-hypodiploid ALL, with TP53-mutant clonal hematopoiesis representing a preleukemic reservoir that can give rise to aneuploidy and B-ALL.
See related commentary by Saiki and Ogawa, p. 102.
This article is highlighted in the In This Issue feature, p. 101
INTRODUCTION
B-cell precursor acute lymphoblastic leukemia (B-ALL) represents a rare malignancy in adults associated with a poor prognosis, especially in the older range of patients (1–3). Despite significant improvements provided by pediatric-inspired treatment regimens (4), there is still an important discrepancy between B-ALL outcomes in children and adults, likely owing to two major factors. First, treatment-related toxicity increases with age, which prevents the use of intensive chemotherapy (1, 5). Second, adult B-ALL have decreased sensitivity to treatments when compared with children due to different leukemia genomic backgrounds. Hence, large genomic studies have highlighted the age-related prevalence of distinct genetic aberrations, with a drop in low-risk abnormalities (i.e., high hyperdiploidy and ETV6::RUNX1) beyond the age of 10, together with the progressive increase in high-risk subtypes, i.e., BCR::ABL1, BCR::ABL1-like, KMT2A-rearranged and low hypodiploidy (6–9). Therefore, a better understanding of the pathogenesis of high-risk adult B-ALL is needed to improve clinical management.
B-ALL with low hypodiploidy (LH-ALL) is a distinct entity primarily defined by karyotype harboring 32 to 39 chromosomes with a nonrandom pattern of chromosome losses (10). Afterward, this entity was renamed “low hypodiploidy/near triploidy” to include cases with duplicated low-hypodiploid genome sharing the same poor prognosis (11). Within childhood B-ALL, LH-ALL represents a very small subset associated with a dismal outcome despite being treated with the most intensive procedures (12). A previous study in pediatric ALL showed that most LH-ALL harbored TP53 mutations, with half of them being of germline origin, connecting LH-ALL to the spectrum of the Li–Fraumeni cancer-predisposing syndrome (13). In adults, LH-ALL is also associated with poor outcomes (8) and frequent TP53 mutations (14–16), but few data addressed the somatic origin of TP53 mutations.
Clonal hematopoiesis is a condition characterized by the clonal expansion of hematopoietic stem/progenitor cells (HSPC) carrying somatic mutations at detectable levels. Sequencing of large cohorts of healthy subjects has shown that the prevalence of clonal hematopoiesis increased continuously with age, defining age-related clonal hematopoiesis (ARCH; refs. 17–19). Importantly, TP53 is one of the most frequent genes involved in ARCH. In addition, TP53-mutant clonal hematopoiesis is associated with an increased risk of developing myeloid malignancies, including secondary and treatment-related myeloid neoplasms (20–22).
Here, we identified and characterized a large cohort of adults with LH-ALL. Then, considering the dramatically increased proportion of LH-ALL with age, we sought to investigate the possible role of TP53-mutant clonal hematopoiesis in the pathogenesis of adult LH-ALL. Using paired diagnosis/remission samples and a single-cell multiomics platform, we demonstrate that LH-ALL can arise from a preleukemic TP53-mutant HSPC clone.
RESULTS
DNA Sequencing–Based Assessment of Copy-Number Aberrations and Loss-of-Heterozygosity Is Critical for Identifying LH-ALL
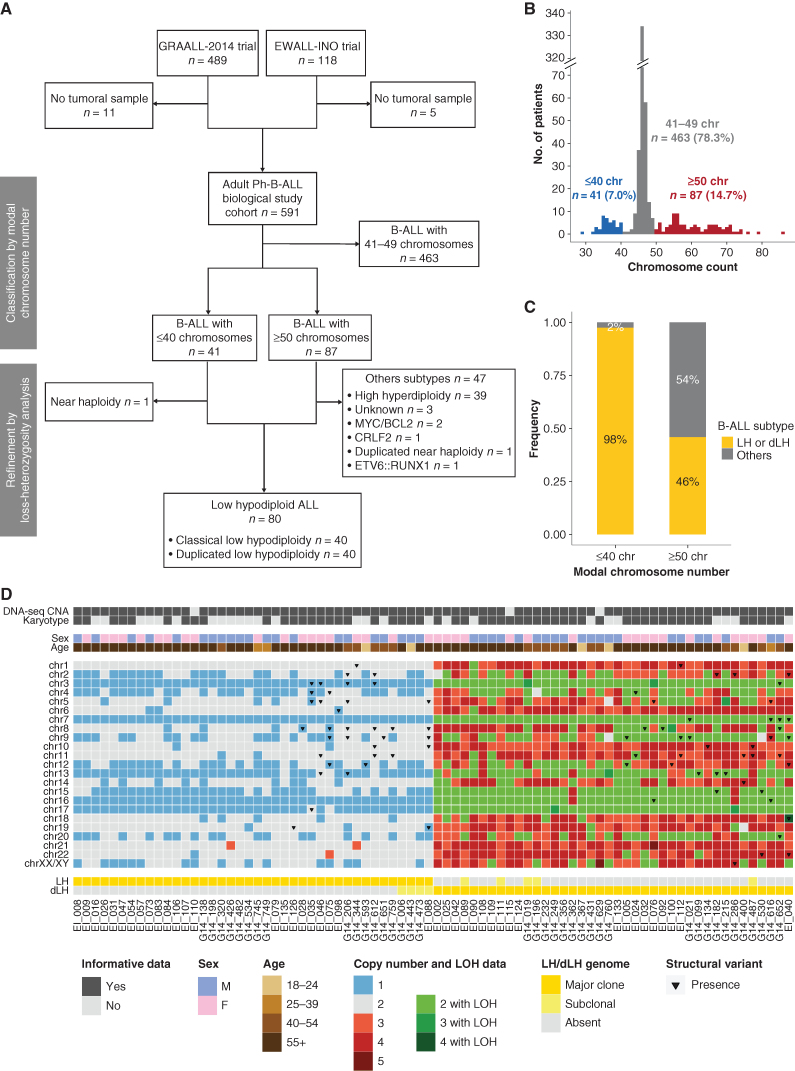
We studied a prospective cohort of adults with newly diagnosed Philadelphia (Ph)-negative B-ALL with available diagnostic sample (n = 591; Fig. 1A). We used targeted-capture sequencing to carry out copy-number aberrations (CNA) analysis and classified cases by a modal chromosomal number based on both karyotype and CNA data (Fig. 1B; Supplementary Fig. S1A and Supplementary Table S1). Noticeably, hypodiploid leukemic cells can sometimes undergo endoreplication, leading to a high number of chromosomes. Therefore, we examined all the cases presenting with either ≤40 (n = 41) or ≥50 chromosomes (n = 87). All cases with ≤40 chromosomes corresponded to LH-ALL, except one having near haploidy, confirming that the latter corresponds to a subtype virtually not found in adult ALL. Within cases with ≥50 chromosomes, loss-of-heterozygosity (LOH) analysis enabled to identify cases with a duplicated low-hypodiploid genome, as harboring multiple uniparental disomies corresponding to the same pattern as that of chromosome losses in classic low hypodiploidy (Supplementary Fig. S1B). Importantly, LOH analysis unequivocally distinguished duplicated low hypodiploidy from high hyperdiploidy, the latter corresponding to a distinct good-risk B-ALL subtype.
Figure 1.
Identification of a cohort of 80 LH-ALL cases by karyotyping and sequencing-based assessment of CNA and LOH. A, Flow diagram describing the study cohort of Ph-negative B-ALL adult patients. B, Distribution of B-ALL cases according to their modal chromosome number assessed by karyotyping and/or sequencing-based CNA analysis. C, Proportion of LH-ALL within cases with ≤40 chromosomes and cases with ≥50 chromosomes after integration of sequencing-based LOH analysis. D, Heat map of chromosome anomalies as determined by karyotyping or CNA/LOH analysis in the 80 LH-ALL cases.
Nearly half (40/87, 46%) of all cases with ≥50 chromosomes turned out to have a duplicated low-hypodiploid genome (Fig. 1C). The ancestral low-hypodiploid clone was detectable by karyotype in only a few patients (5/40, 13%; Fig. 1D), suggesting that they could have been misclassified as high hyperdiploidy based on cytogenetics only. Moreover, in 26 of 80 cases (33%) eventually classified as LH-ALL, conventional cytogenetics concluded to culture failure or normal karyotype, suggesting that leukemic cells failed to grow in vitro. We also identified two LH-ALL cases as having the canonical pattern of chromosome losses, albeit with a modal chromosome number >39 due to trisomy 21. Therefore, our data illustrate that CNA and LOH analyses are critical to allocate correctly B-ALL to distinct aneuploidies representing clinically relevant B-ALL subtypes.
LH-ALL Represents a Major B-ALL Subtype in Older Adults
Overall, combined cytogenetic and molecular analyses of 591 Ph-negative B-ALL adult patients identified 80 patients with LH-ALL (13.5%), either harboring classic low hypodiploidy (n = 40) or duplicated low hypodiploidy (n = 40). We analyzed the baseline characteristics of LH-ALL patients with regard to all the other Ph-negative B-ALL from adult patients (Table 1). LH-ALL patients were significantly older than other B-ALL patients (median 59 vs. 39 years, P < 0.0001) and there was a dramatic increase in LH-ALL prevalence with age, ranging from 3% (8/267) in patients below the age of 40 to 32% (55/171) in patients over 55 years. Other distinct features of LH-ALL included a lower white blood cell count (3.2 vs. 7.5 G/L, P < 0.001) and a lower marrow blast infiltration (79 vs. 92%, P < 0.001). Overall, LH-ALL represents a large fraction of adult Ph-negative B-ALL and has an age-related distribution, from being rare in young adults to become very common in older patients.
Table 1.
Demographic and clinical characteristics of patients at baseline.
| LH-ALL | Non LH-ALL | P | |
|---|---|---|---|
| Patients, n | 80 | 511 | |
| Sex, n (%) | |||
| Male | 42 (53) | 269 (53) | 1 |
| Female | 38 (47) | 242 (47) | |
| Age, year | |||
| Median | 59 | 39 | < 0.0001 |
| Range | 18–84 | 18–84 | |
| Age group, n (%) | |||
| 18–24 y | 5 (6) | 123 (24) | <0.0001 |
| 25–39 y | 3 (4) | 136 (27) | |
| 40–54 y | 17 (21) | 136 (27) | |
| ≥ 55 y | 55 (69) | 116 (23) | |
| White blood cell counta, Giga per liter | |||
| Median | 3.2 | 7.5 | <0.0001 |
| Range | 0.4–87.1 | 0.4–712.0 | |
| Marrow blast infiltrationb, % | |||
| Median | 79 | 92 | <0.0001 |
| Range | 21–99 | 12–100 | |
| CD10 expressionc, n (%) | |||
| Positive | 62 (79) | 382 (76) | 0.67 |
| Negative | 16 (21) | 118 (24) | |
aWhite blood cell counts were available for 582 patients.
bMarrow blast cell counts were assessed for 569 patients.
cCD10 expression was assessed by flow cytometry for 578 patients. Two-tailed independent t tests and two-tailed Fisher tests were used for continuous and categorical variables, respectively, and statistical significance was defined as a P < 0.05.
Adult LH-ALL Exhibits TP53 Biallelic Alteration and a Distinct Pattern of Chromosomal and Gene Abnormalities
We performed targeted DNA sequencing (DNA-seq) of a panel of genes recurrently altered in B-ALL and integrated karyotype, CNA, LOH, and mutation data to characterize the full spectrum of genomic alterations in adult LH-ALL. The pattern of chromosome losses was highly recurrent, including chromosomes 3, 7, 16, and 17 in virtually all cases and chromosomes 13 and 15 in over 80% of cases, whereas chromosomes 2, 4, 9, 12, and 20 were more variably lost, in half of cases each (Fig. 1D; Supplementary Fig. S1C). Of note, chromosome 21 was retained in all cases. In addition, structural abnormalities were present in 31 of 80 (39%) cases.
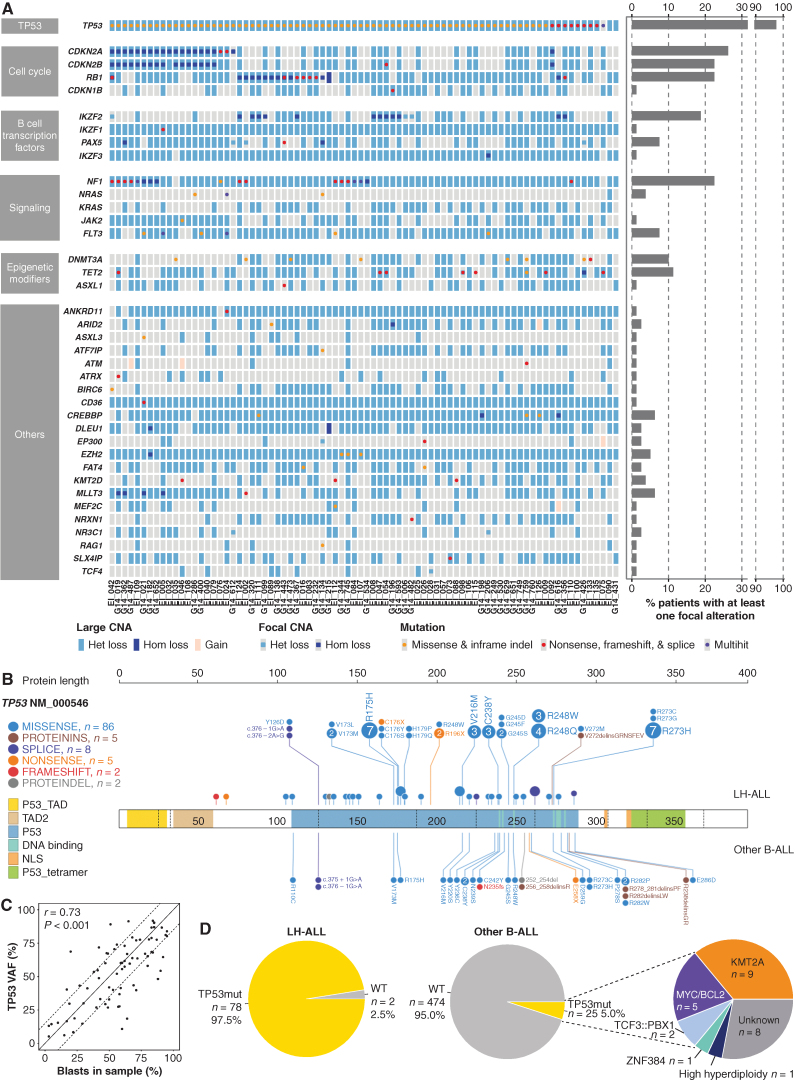
We found TP53 mutations in 78 of 80 (98%) cases (Fig. 2A; Supplementary Table S2). They included missense, nonsense, splice-site mutations, and short insertions/deletions, all predicted to be pathogenic (Fig. 2B). Monosomy 17 or uniparental disomy led to the loss of the second TP53 allele in all cases but one having biallelic TP53 mutations. TP53 mutations were present at variant allele frequencies (VAF) correlated with leukemic cell infiltration (Fig. 2C), suggesting that alterations of both alleles were present in the major clone. Noticeably, considering the whole cohort of adult Ph-negative B-ALL, 76% (78/102) of patients with TP53 mutation had LH-ALL, whereas TP53 mutations were rarely detected in other B-ALL (5% vs. 98%, P < 0.001, two-tailed Fisher test; Fig. 2D). Therefore, biallelic alteration of TP53 is a hallmark of LH-ALL within adult B-ALL.
Figure 2.
Landscape of recurrent genomic alterations in adult LH-ALL. A, Heat map of recurrent CNA and mutations in the 80 LH-ALL cases. Focal alterations refer to CNA not involving whole chromosome arms and mutations. Cases with several mutations are referred to as multihit. B, Lollipop plot depicting TP53 mutations detected in both LH-ALL and other Ph-negative B-ALL. C, Correlation between TP53 mutation VAF and blast percentage, as assessed by flow cytometry after ficoll or morphology on marrow smear, using Pearson correlation coefficient. D, Proportion of TP53 mutations in LH-ALL and other Ph-negative B-ALL with available data from the study cohort (n = 499). Het, heterozygous; Hom: homozygous; indel: insertion/deletion.
Alterations of genes involved in cell-cycle regulation defined the second most frequently altered pathway. Specifically, focal deletions and loss-of-function mutations in the CDKN2A and RB1 genes (Fig. 2A; Supplementary Fig. S2A), associated with monosomies 9 and 13, led to biallelic inactivation in 21 (26%) and 18 (23%) cases, respectively, in a mutually exclusive pattern. The lymphoid transcription factor gene IKZF2 was recurrently affected by focal deletions, observed in 15 cases (19%). By contrast, no intragenic deletion or mutation of IKZF1, one of the most frequently altered genes in adult B-ALL, was found, although hemizygosity was observed in all cases as a result of monosomy 7. Similarly, PAX5 was rarely targeted by focal deletions or mutations (8%).
Aberrations involving cell signaling genes were present in 25 (31%) LH-ALL cases, including loss-of-function mutations and focal deletions of NF1 in 18 (23%) cases (Supplementary Fig. S2B). We also identified FLT3 mutations in 6 cases (8%; Supplementary Fig. S2C), NRAS mutations in 3 cases (4%), and remarkably, JAK2 p.V617F mutation in one case (EI_046). Finally, we detected TET2 and DNMT3A mutations corresponding to classic mutations reported in ARCH and myeloid malignancies in 11% and 10% of cases, respectively (Supplementary Fig. S2D–S2E).
Altogether, adult LH-ALL is characterized by a distinct pattern of aneuploidy, consistent biallelic alteration of TP53, and a constellation of additional gene alterations, the most frequent involving CDKN2A, RB1, NF1, and IKZF2.
TP53 Mutations Are Detected at Remission in a Substantial Proportion of Adults with LH-ALL
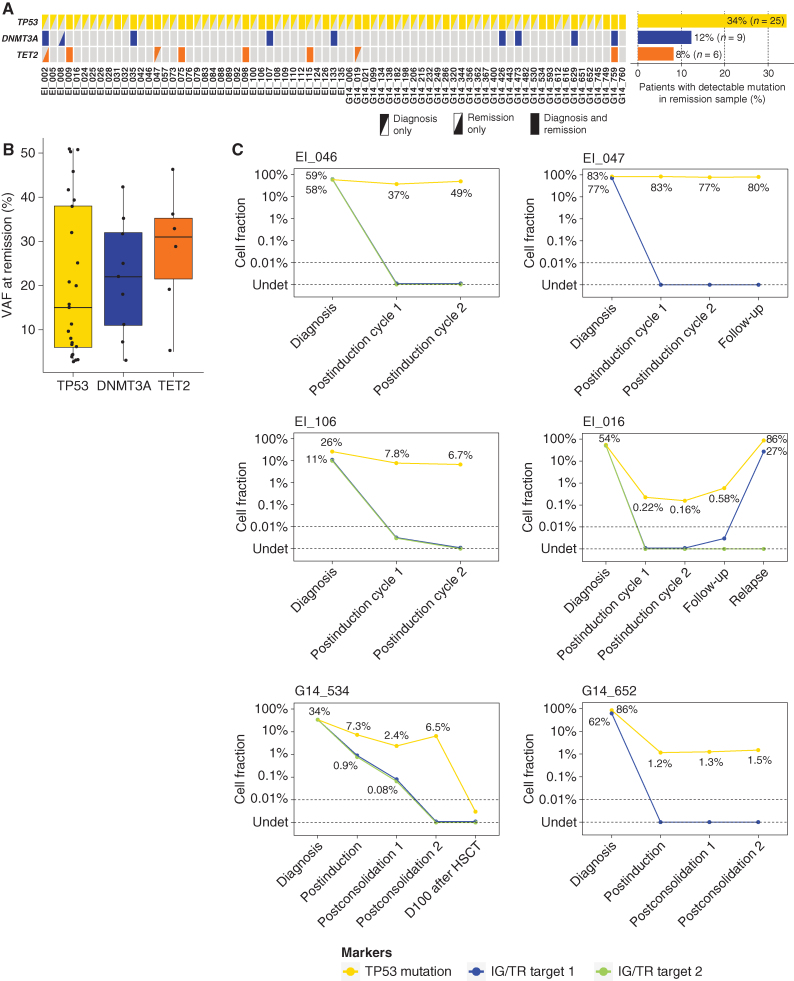
Given the age-related distribution of LH-ALL and the role of TP53 mutations in ARCH, we hypothesized that LH-ALL may be secondary to TP53-mutant ARCH. To test this hypothesis, we looked for TP53 mutations in posttreatment remission bone marrow samples assessed for minimal residual disease (MRD). Among the 73 patients with available samples, we observed persistence of the TP53 mutation identified at diagnosis in 25 patients (34%), at VAF ranging from 2.6% to 51.2% (cutoff value for positivity set at 2%; Fig. 3A and B; Supplementary Table S3). DNMT3A and TET2 mutations were also detected in remission samples from 9 and 6 patients, respectively. MRD levels based on the quantification of clonal rearrangements of immunoglobulin or T-cell receptor genes (IG/TR) were either undetectable or measured at much lower levels (Supplementary Table S4), indicating that those mutations were not related to residual B-ALL leukemic cells.
Figure 3.
Tracking of ARCH-related mutations in remission samples of LH-ALL patients. A, Heat map of mutations in paired diagnosis and remission samples for 71 patients. Data are based on sequencing data with a threshold for positivity at 2% VAF. B, Box plot for VAF of mutations detected by sequencing at remission. C, Longitudinal assessment of TP53-mutant cell fraction as determined by ddPCR (0.05–0.1% sensitivity), along with clonal IG/TR-based MRD quantification (0.01%–0.001% sensitivity), for six patients with TP53 mutation detected in remission samples, including two below the sequencing detection threshold.
It should be noticed that 5 patients had a TP53 mutation with a VAF >40%, raising the possibility of a germline origin, as previously observed in pediatric LH-ALL. This could be ruled out in two patients for whom TP53 mutation was not detected in nonleukemic cells (Supplementary Table S3). In the three remaining patients, including one with detectable TP53 mutation in sorted T cells and two without available nonleukemic material, a germline origin could not be ruled out.
Next, to evaluate the dynamics of the TP53-mutant cell fraction, we used digital droplet PCR (ddPCR) to quantify TP53 mutations in 9 patients with available longitudinal postremission bone marrow samples (Fig. 3C; Supplementary Fig. S3). In two patients with confirmed somatic TP53 mutation, the TP53-mutant clone remained in a steady state at high rates at all time points. Four patients displayed a reduction of the TP53-mutant fraction over time, yet uncoupled from lower or undetectable IG/TR MRD levels. Noticeably, in two of these patients, the higher sensitivity of ddPCR allowed to detect a low fraction of persistent TP53-mutant cells not detected by sequencing. In three other patients, no TP53-mutant cells were detected. Overall, these results reveal that a substantial proportion of adults with LH-ALL carry somatic TP53 mutations in nonleukemic cells repopulating bone marrow after treatment.
Integrated Single-Cell Genotyping and Immunophenotyping Reveals Multilineage TP53-Mutant Clonal Population at Remission Stage
We sought to investigate the contribution of the TP53-mutant clone to the different hematopoietic lineages and infer the oncogenic route toward LH-ALL. Using antibody-derived tag (ADT) sequencing and a custom single-cell DNA-seq (scDNA-seq) panel (23–25), we performed simultaneous single-cell immunophenotyping and genotyping on three bone marrow specimens obtained after induction. The three patients were treated in the EWALL-INO trial and had a TP53 mutation detected at postinduction remission, whereas MRD measured on the same sample was either low or undetectable.
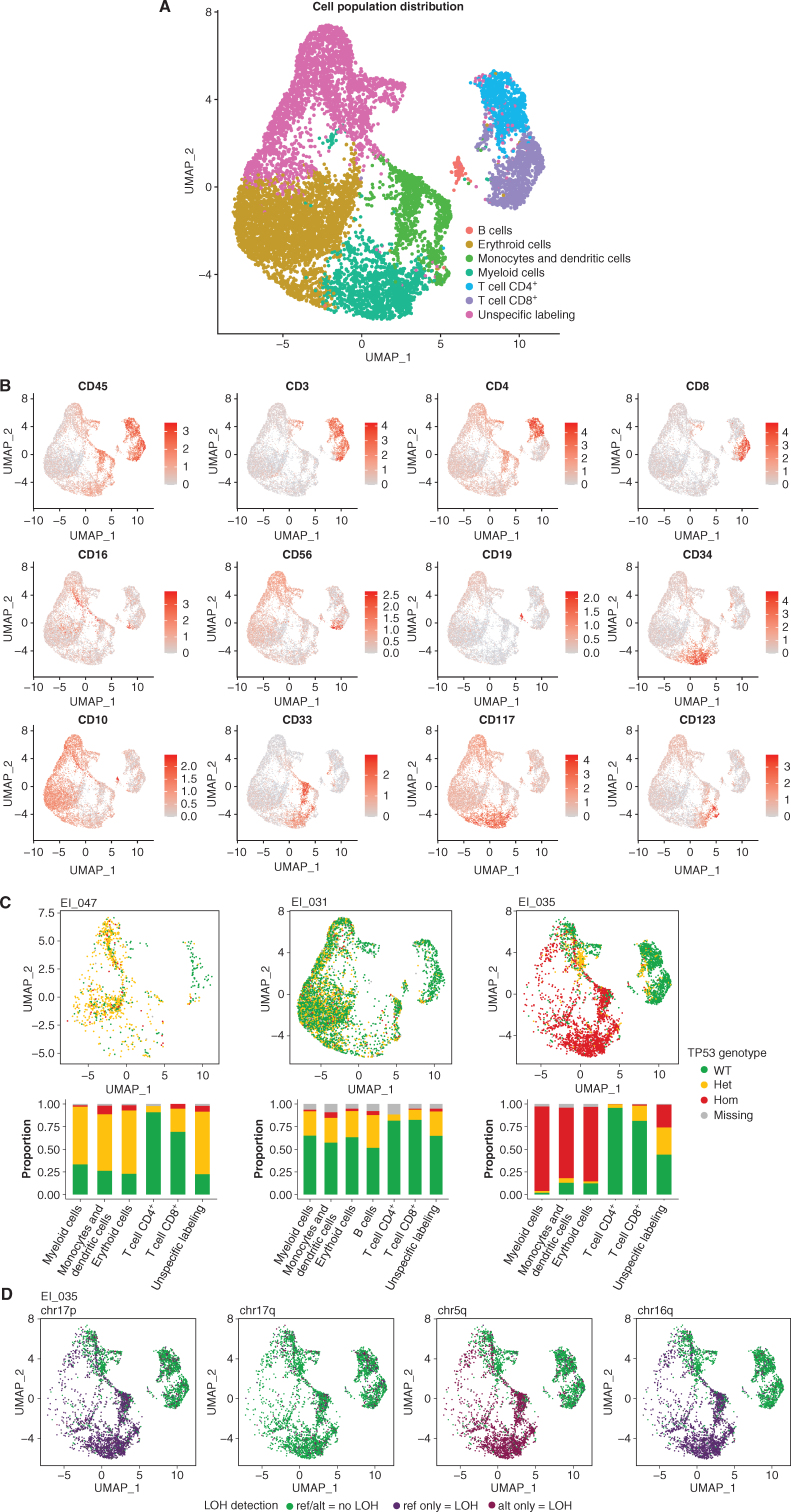
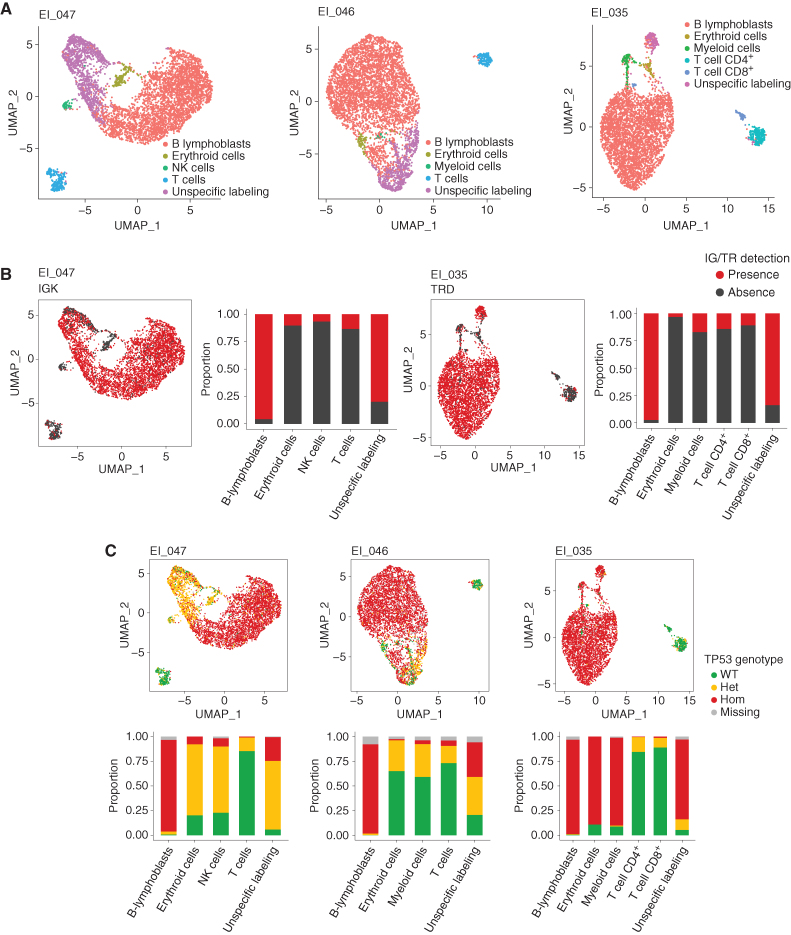
Protein expression data allowed discrimination of 7 distinct clusters (Fig. 4A). Based on lineage-specific markers, we were able to identify confidently T cells, B cells, monocytes, dendritic cells (DC), and myeloid and erythroid cell clusters (Fig. 4B; Supplementary Fig. S4A and S4B). Cell identity could not be assigned to the remaining cluster owing to unspecific and weak labeling of frail cells (thereafter named “unspecific labeling”). Of note, few B cells were detected, which may be related to therapy including B cell–directed antibodies. Cell type composition was comparable across patients, but analyses on each sample individually allowed better identification of discrete cell populations not detected in all patients, such as natural killer (NK) cells and plasmacytoid dendritic cells (pDC; Supplementary Fig. S5).
Figure 4.
Integrated single-cell genotyping and immunophenotyping reveal a multilineage TP53-mutant clonal population at the remission stage in LH-ALL patients. A, Uniform Manifold Approximation and Projection (UMAP) plot of 10,598 cells from remission samples of three patients (EI_047, EI_031, and EI_035 having 1,017, 5,450, and 4,131 cells available for analysis, respectively). Cells are clustered by expression of cell-surface markers and colored according to assigned clusters. B, Same UMAP plots with cells colored by expression of specific cell-surface markers used to assign cell clusters. C, Same UMAP plots (top) for individual patient's remission samples, with cells colored according to TP53 genotype. Histograms (bottom) show the proportion of each TP53 genotype within each cell cluster. D, UMAP plots for the EI_035 remission sample, with cells colored according to genotype for several heterozygous SNPs allowing LOH assessment. ref, reference allele; alt, alternative allele.
Projection of the TP53 genotype on the protein uniform manifold approximation and projection (UMAP) representation allowed us to describe the cell architecture of the TP53-mutant clone (Fig. 4C; Supplementary Fig. S6). Patients EI_031 and EI_047 had similar patterns with TP53-heterozygous mutant cells detected in substantial fractions of myeloid, erythroid, and monocyte/DC clusters (hereafter collectively named myeloid clusters). The “unspecific labeling” cluster was likely related to myeloid populations according to the similar proportion of mutant cells. T-cell populations also contained mutant cells, yet at lower rates. For patient EI_031, NK cells, pDC, and B-cell clusters could be identified and also harbored mutant cells. Of note, the observation of rare cells with a homozygous genotype was likely related to allelic dropout and biologically not relevant, in agreement with similar proportions observed for constitutional heterozygous single-nucleotide polymorphisms (SNP; Supplementary Fig. S7).
Patient EI_035 harbored a large fraction of cells in myeloid clusters carrying TP53-homozygous mutation, pinpointing a concomitant CNA or copy-neutral LOH in those cells. By contrast, T cells were mainly wild-type, with a minority being heterozygous. Including amplicons covering common SNPs in the scDNA-seq panel allowed us to investigate allelic imbalance throughout the genome at the single-cell level. Thus, we evidenced LOH at 17p (but not at 17q) in myeloid clusters, suggesting a cytogenetic aberration at 17p resulting in the loss of the wild-type allele (Fig. 4D; Supplementary Fig. S8). Those cells also had LOH at 5q and 16q, whereas no other LOH possibly related to the LH-ALL clone was observed. Thus, the presence of those aberrations restricted to the myeloid compartments may be related to a distinct, myelodysplastic clone, only sharing the TP53 alteration with the LH-ALL clone. Altogether, these results show that patients with LH-ALL carry TP53-mutant HSPCs able to repopulate bone marrow after intensive treatment.
LH-ALL Arises from Preexisting TP53-Mutant Clonal Hematopoiesis
We next aimed to address whether the TP53-mutant multilineage cell population arose from clonal selection under the stress of cytotoxic therapy for B-ALL or preexisted at diagnosis. We profiled 14,518 cells from three diagnostic samples, including two paired with remission samples already presented (EI_047 and EI_035). ADT-sequencing and IG/TR clono-specific sequences enabled to recognize B-ALL leukemic cells and to identify minor nonleukemic cell populations (Fig. 5A and B; Supplementary Fig. S9). In B-ALL cells from the 3 patients, TP53 and SNP single-cell genotyping showed homozygous (hemizygous) TP53 mutation and LOH at multiple loci (Fig. 5C; Supplementary Fig. S10), in agreement with bulk data (Fig. 1D; Supplementary Table S2). In EI_046 and EI_047, substantial fractions of TP53-heterozygous mutant cells were observed within myeloid and NK cell populations, as observed in the paired-remission sample of EI_047. Similarly, in EI_035, the myeloid population was predominantly TP53-homozygous mutant. By contrast, the small subset of TP53-heterozygous mutant T cells was likely related to the cell doublet artifact, in agreement with a similar rate of detection of clonal IG/TR rearrangements (Supplementary Table S5). Consistent results were obtained through analyses of the fluorescence-activated cell sorting (FACS) of cell fractions (Supplementary Table S6).
Figure 5.
Single-cell analysis at LH-ALL diagnosis identifies a preexisting multilineage TP53-mutant clonal population. A, UMAP plots for individual diagnostic samples from three patients (EI_47, EI_46, and EI_035), having 5,228, 4,947, and 4,343 cells available for analysis, respectively. Cells are clustered by expression of cell-surface markers and colored according to assigned clusters. B, Same UMAP plots for patients EI_047 and EI_035, with cells colored according to the presence of specific clonal IG/TR sequences allowing to identify B-ALL cells. Histograms show the proportion of positive cells within each cell cluster. C, Same UMAP plots (top) with cells colored according to TP53 genotype. Histograms (bottom) show the proportion of each TP53 genotype within each cell cluster.
Overall, these results confirm that the TP53-mutant myeloid population preexisted at B-ALL diagnosis and indicate that TP53-mutant ARCH likely preceded the onset of B-ALL.
Other ARCH-Related Genes Can Be Involved in the Pathogenesis of LH-ALL
Because other ARCH-related mutations were identified in bulk diagnostic samples from several patients with LH-ALL, we aimed to investigate their relation with TP53-mutant clonal hematopoiesis and LH-ALL and decipher the sequence of mutation acquisition.
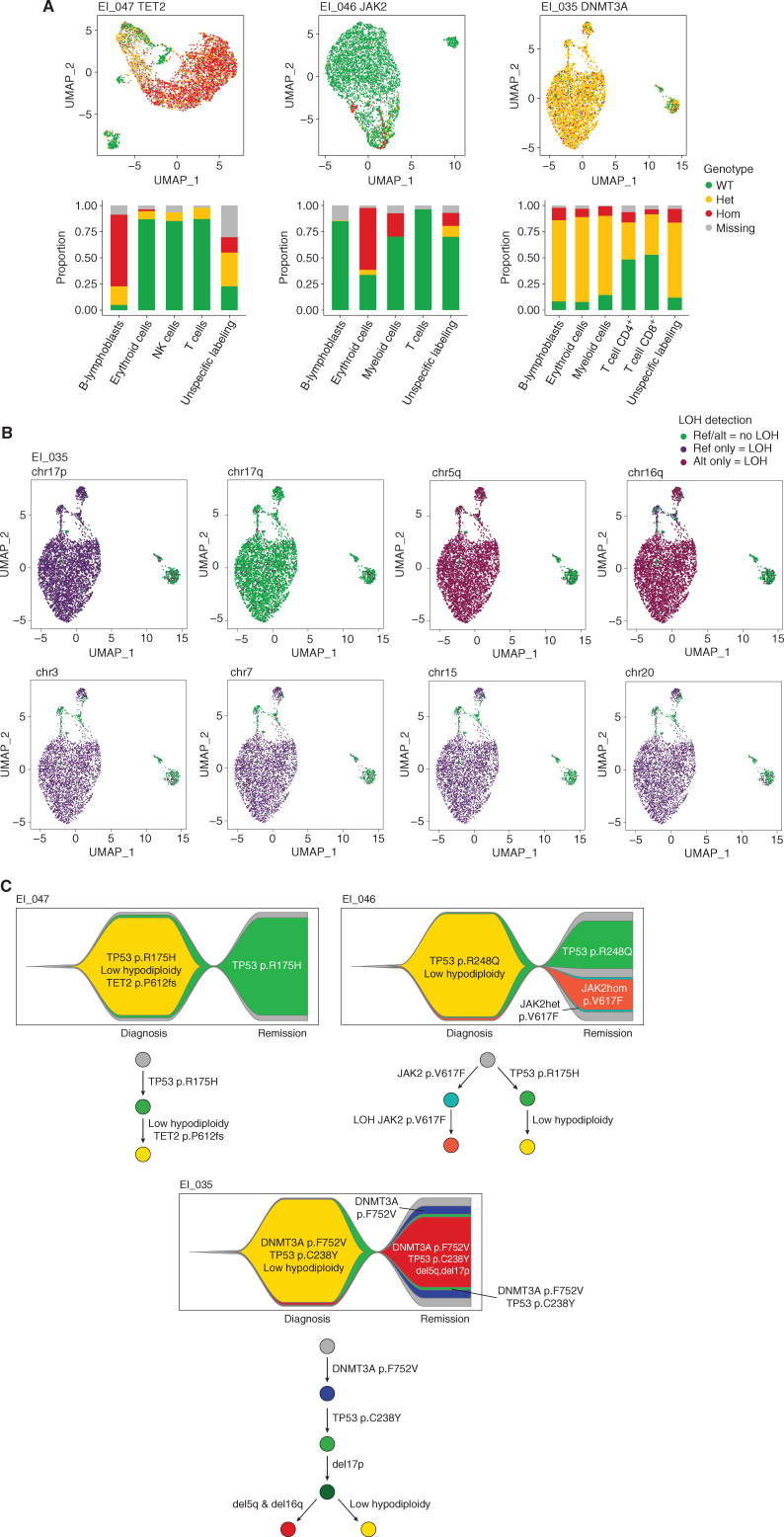
EI_047 had a TET2 mutation detected in bulk sequencing at diagnosis at 48% VAF. Unexpectedly, single-cell analysis of the diagnosis sample revealed that this mutation was restricted to B-ALL cells and was not carried by myeloid cells (Fig. 6A). Accordingly, it was not detected in the bulk remission sample.
Figure 6.
Integration of ARCH mutations into the clonal architecture of LH-ALL. A, UMAP plots for individual diagnostic samples from three patients with cells clustered by expression of cell-surface markers and colored according to genotypes of TET2, JAK2, and DNMT3A mutations in EI_047, EI_046, and EI_035, respectively. Histograms show the proportion of each genotype within each cell cluster. B, UMAP plots for the EI_035 diagnostic sample, with cells colored according to genotype for several heterozygous SNPs allowing LOH assessment. C, Fish plots and phylogenetic trajectories depicting the clonal architecture and the sequential acquisition of genetic abnormalities.
EI_046 harbored a JAK2 V617F detected at 7% VAF in the bulk diagnosis sample. Single-cell analysis revealed that it was restricted to myeloid and erythroid cells, in a homozygous state, reminiscent of myeloproliferative neoplasms (MPN). Moreover, the TP53 and JAK2 mutations appeared to be in independent clones, as no cell carried both mutations (Supplementary Fig. S11). Therefore, single-cell analysis uniquely evidenced a JAK2-mutant ARCH or MPN-like clone, concomitant but clonally unrelated to B-ALL.
EI_035 had a DNMT3A mutation at diagnosis (VAF 45%), still detected at remission (VAF 42%). Single-cell analysis of the diagnosis sample showed that almost all B-ALL and nonleukemic cells carried heterozygous DNMT3A mutation. The presence of TP53-wild-type DNMT3A-mutant cells in the remission sample (Supplementary Fig. S11) indicated that DNMT3A mutation occurred earlier than TP53 mutation. In addition, and as observed at remission, myeloid cells had LOH at 5q, 16q, and 17p, suggesting a premalignant, concurrent clone at B-ALL diagnosis (Fig. 6B). All B-ALL cells had LOH at these loci and at additional loci in relation with low hypodiploidy. However, the discordant homozygous genotype at 16q between B-ALL and myeloid cells indicated a branched rather than linear relation between the myeloid and B-ALL clones.
Overall, these data allow to reconstruct the sequence of mutation acquisition (Fig. 6C) and show that adult LH-ALL can develop on a background of clonal hematopoiesis resembling myelodysplastic syndrome (MDS) or MPN.
DISCUSSION
Our study establishes a comprehensive genomic characterization of the largest series of LH-ALL reported so far. By combining sequencing-based CNA and LOH analyses to cytogenetics data in a prospective cohort of 591 Ph-negative adult ALL, we identified a high proportion of LH-ALL, reaching 32% in the older range of patients. This increased prevalence in older adults is in agreement with, but higher than, that observed in previous studies, including the recent UKALL study of older adult ALL (9). Although the diagnostic pitfall of duplicated hypodiploidy/near triploidy is recognized, our data suggest that a number of LH-ALL cases are still being missed or misclassified owing to misleading normal or hyperdiploid karyotypes. Hence, CNA/LOH analyses in our study allowed to rescue more than a quarter of LH-ALL cases with normal/failed karyotype, whereas duplicated low hypodiploidy represented up to half of all LH-ALL. Of note, we also showed that LH-ALL patients present with lower blast counts in bone marrow and peripheral blood at diagnosis, which may explain underrepresentation in retrospective molecular studies relying on frozen samples. These findings warrant the implementation of CNA/LOH analysis in diagnostic laboratory practices, because genetic classification is critical for risk-adapted treatment stratification in most modern pediatric and adult clinical trials. The improved genetic assignment will also allow the refinement of future clinical correlation studies.
Our single-cell sequencing data on diagnostic and remission samples indicate that TP53 mutation is a preleukemic event preceding aneuploidy with monosomy 17 leading to the loss of the TP53 wild-type allele. Although a dominant-negative effect of TP53 missense mutations has been proposed to mediate the nonmutational inactivation of wild-type TP53 (26), it is noteworthy that genomic alteration of the second allele seems to be mandatory in the pathogenesis of LH-ALL. The specific pattern of chromosomal losses in adult LH-ALL, including the core association of 3, 7, 16, and 17 monosomies, is consistent with that observed in childhood LH-ALL and may inform about key genes involved in LH-ALL oncogenesis. Loss of chromosome 7 results in haploinsufficiency for IKZF1, a critical gene for normal B-cell differentiation, the recurrent loss of which in B-ALL drives a pejorative impact (27). Chromosome 16 contains CREBBP, which is also targeted by focal alterations in B-ALL, especially in relapsed cases (28). Noteworthy, as the CD19 gene is also located on chromosome 16, LH-ALL having only one allele may be prone to CD19 expression loss in the context of selective pressure with CD19-directed therapeutic agents (29). Chromosome 3 contains several tumor suppressor genes previously shown to be involved in B-ALL, i.e., CD200/BTLA locus and SETD2 (27, 30, 31). Thus, by occurring at once, all these monosomies resulting in the loss of several key tumor suppressor genes may drive transformation and shape the tumor biology of LH-ALL. Additionally, this combinatorial pattern could also be the result of the counter-selection of other aneuploidies possibly lethal to the target cell.
The genomic profiling of LH-ALL evidenced additional recurrent alterations in LH-ALL. Frequent and mutually exclusive alterations of the cell-cycle genes CDKN2A and RB1 suggest their functional redundancy and an oncogenic cooperation with a TP53-deficient background. We identified NF1 as a frequent target gene in adult LH-ALL, whereas it was not observed in their pediatric counterpart (13).
DNMT3A and TET2 were among the top mutated genes, found in 10% of patients. As these genes are the most frequently mutated in ARCH (17), these mutations may simply reflect the older age of LH-ALL patients. Single-cell analysis revealed different possible clonal involvement of those mutations not inferred by bulk sequencing. In one patient, DNMT3A mutation was the earliest somatic event, found in an ancestral multilineage hematopoietic clone, as previously shown in acute myeloid leukemia (AML; ref. 32). By contrast, in another patient, TET2 mutation was found as a secondary genetic event restricted to leukemic cells, suggesting an unanticipated oncogenic role of TET2 in B-ALL. In addition, in one patient with JAK2 V617F mutation detected in bulk diagnosis, the single-cell analysis demonstrated that it was clonally unrelated to TP53-mutant ALL cells, which is reminiscent of post-MPN TP53-mutant AML lacking the MPN driver mutation (33).
The major finding of our study is that TP53 mutations are not only frequently associated with LH-ALL, but they are indispensable genetic events preceding aneuploidy. Hence, we detected TP53 mutations in virtually all LH-ALL cases and demonstrated that TP53-mutant heterozygous cells constituted a preleukemic compartment. This oncogenic route relying on initiating TP53 mutation is thus distinct from that observed in other malignancies, including ALL (34), where TP53 mutations often occur during tumor progression as a late event. In LH-ALL, TP53 mutation likely acts as the triggering event for genome instability, allowing the emergence of the low-hypodiploid clone responsible for leukemia onset. The further step of whole-genome duplication, whereas otherwise rare in ALL, may be also related to TP53 deficiency, as reported in other tumors (35).
Moreover, we show that somatic TP53 mutations in LH-ALL affect preleukemic HSPCs that retain their ability to generate mature blood cells, defining clonal hematopoiesis. This connects LH-ALL to the spectrum of hematologic malignancies related to TP53-mutant clonal hematopoiesis, namely, secondary and therapy-related AML and MDS having TP53 biallelic alteration and complex karyotype (20–22). We also show that, at least in some cases, a myeloid premalignant clone may evolve concurrently to LH-ALL, which may support treatments aiming at eradicating not only B-ALL but TP53-mutant clonal hematopoiesis. Altogether, our study sheds light on an unsuspected link between LH-ALL, TP53-mutant clonal hematopoiesis, and related myeloid malignancies and paves the way for future investigations regarding its clinical and therapeutic relevance.
METHODS
Patients and Samples
The study cohort included patients with Ph-negative B-ALL ages 18 to 59 years enrolled in the GRAALL-2014 trial between 2016 and 2020 and French patients ages ≥55 years enrolled in the EWALL-INO trial between 2017 and 2022 (ClinicalTrials.gov number NCT02617004 and NCT03249870, respectively). The GRAALL-2014 protocol was an intensive pediatric-inspired treatment similar to the GRAALL-2005 (5), but with age adaptation of doses for patients 45 to 59 years old. The standard induction comprised prednisone, daunorubicin, vincristine, cyclophosphamide, L-asparaginase, and intrathecal prophylaxis. The EWALL-INO protocol was a treatment based on a low-intensity chemotherapy backbone and intrathecal prophylaxis with the addition of inotuzumab ozogamicin (INO) during the two-phase induction. The first induction phase comprised 3 doses of INO (D1: 0.8 mg/m2, D8 and D15: 0.5 mg/m2) in combination with weekly vincristine 2 mg i.v. and dexamethasone 40 mg for 4 weeks, and the second phase comprised 2 doses of INO at 0.5 mg/m2 in combination with dexamethasone 20 mg (D1 and D8) and cyclophosphamide 300 mg/m2/day i.v. D1–3. All the patients provided written informed consent for sample banking and analyses. The study was in accordance with the Declaration of Helsinki. Cytogenetic analyses at diagnosis were performed by local laboratories using standard procedures. Molecular analyses were performed centrally as previously described (36). Briefly, mononuclear cells from pretreatment bone marrow or peripheral blood samples were isolated by Ficoll centrifugation, and blast percentage was assessed by flow cytometry in most cases before nucleic acid extraction. MRD was assessed by the quantification of clonal IG/TR rearrangements, according to the EuroMRD guidelines.
Targeted DNA Sequencing
A custom panel of genes previously known to be targeted by recurrent mutations or CNA in B-ALL was analyzed by pan-exon capture-based target enrichment (SureSelectXT Low Input Target Enrichment System, Agilent) followed by library sequencing on the Illumina NextSeq500 platform (Illumina) as previously described (gene list in Supplementary Table S7; ref. 36). Data were analyzed for variant calling using Varscan (37) and Pindel (38), and CNA analysis was conducted using Viscap (39) and Facets (40). Lollipop plots were drawn with ProteinPaint (41).
B-ALL Classification Based on Cytogenetic and Molecular Data
B-ALL cases were first classified by the modal chromosome number of leukemia cells into three categories (≤40, 41–49, and ≥50 chromosomes), referring to the major abnormal clone at karyotype analysis when it was informative (i.e., presence of abnormal mitosis) and DNA sequencing-based CNA analysis. Cases with ≤40 or ≥50 chromosomes were further examined as possibly being LH-ALL. By definition, all cases with 32 to 39 chromosomes (corresponding to 7–14 chromosome losses) were considered LH-ALL. Two additional cases having 40 chromosomes with a similar pattern of 7 chromosome losses and trisomy 21 were considered LH-ALL. Near haploidy was defined as 24 to 31 chromosomes. Within cases with ≥50 chromosomes, LOH analysis allowed us to identify duplicated LH-ALL cases as cases with LOH affecting at 7 to 14 chromosomes. One case with LOH affecting 20 chromosomes was considered as duplicated near haploidy. High hyperdiploidy was defined as 51 to 65 chromosomes in the absence of other subtype-defining alteration.
TP53 Mutation Quantitation by Digital Droplet PCR
We performed ddPCR using the QX200 Droplet Digital PCR System (Bio-Rad) to track TP53 mutations detected in bulk sequencing of diagnosis samples, in serial bone marrow posttreatment remission samples. Commercial assays for TP53 p.R175H (dHsaMDV2010105), p.H214R (dHsaMDS2510824), and p.R248Q (dHsaMDV2010127) were used on the QX200 Droplet Digital PCR System (Bio-Rad). Reaction mix was prepared using 2× ddPCR TM Supermix for Probes (no dUTP), primers and probe mix (20×), restriction enzyme MseI 2U and 100 ng of genomic DNA as a template for ddPCR assay in a 96-well plate according to the manufacturer's protocol. Patients’ diagnosis DNAs were used as positive control, peripheral blood mononuclear cells from healthy donors were used as negative control, and water was used instead of DNA for the nontemplate control reaction. Data were analyzed using QX Manager Software Standard edition, version 1.2 (Bio-Rad).
Combined Single-Cell Immunophenotyping and Genotyping
A custom scDNAseq panel was designed to cover relevant somatic mutations and common SNPs allowing the detection of LOH (Supplementary Table S8). Additional primers for amplicons of clone-specific IG/TR rearrangements were also included (Supplementary Table S9). Frozen bone marrow mononuclear cells were thawed, stained using the ADT panel TotalSeq-D Human Heme Oncology Cocktail, V1.0 (BioLegend; antibody list in Supplementary Table S10), and loaded into the Tapestri single-cell DNA genotyping platform (Mission Bio) to perform microfluidic encapsulation, lysis, and barcoding according to the manufacturer's protocol (Chemistry V2, PN_3360A). To improve erythroid cells identification, TotalSeq-D0574 anti-human CD235a (Glycophorin A) Antibody (BioLegend) was spiked at 1 μg during the resuspension procedure (available only for EI_046 diagnostic sample). DNA and ADT libraries were prepared and underwent 2 × 150 bp paired-end sequencing on a NextSeq 500 platform (Illumina).
Bioinformatics Analysis
Raw data were processed using Mission Bio's Tapestri Pipeline (DNA pipeline version 2.0.2 and Protein pipeline version 2.0.1) for preprocessing, alignment (reference genome hg19), cell barcode correction, cell identification, variant calling of DNA amplicons, and ADT-seq reads counting. Multiomics h5 files were then analyzed on R version 4.1.2 (R Foundation for Statistical Computing). TapestriR package was used to annotate variants: a genotype was considered informative if the single-cell sequencing depth was ≥5 reads. Variants were categorized as wild-type (WT) if VAF <10%, heterozygous (het) if VAF comprised between 10% and 90%, and homozygous (hom) if VAF ≥90%. Variants covered by <5 reads were considered noninformative and labeled as “missing genotype.” No genotype filtering was performed before integration with ADT-seq data, in order to avoid data attrition, as all genotypes (i.e., WT, heterozygous and hemizygous/homozygous mutations) are expected. The rate of the allelic dropout was evaluated based on heterozygous SNPs. False homozygous genotypes related to allelic dropout were observed in 1% to 18% of cells (Supplementary Fig. S7). Patient-specific amplicons targeting clone-specific IG/TR rearrangements were considered informative if detected in the diagnostic sample. The IG/TR amplicons were considered positive if covered by ≥5 reads.
ADT read counts and cell barcodes were extracted from h5 files and were analyzed using Seurat package V4.1.0 (42). Remission samples were merged before proceeding to protein expression analysis as similar cell compositions were expected. Diagnostic samples were analyzed separately. Cells with total ADT-seq read counts ≥100k were excluded to minimize the presence of doublet cells in further analyses (Supplementary Table S11). Centered log-ratio transformation was performed to normalize reads across cells before scaling, and then principal component analysis was performed on the normalized and scaled data. Dimensional reduction of protein expression data was performed using UMAP embedding and K-nearest neighbors and Louvain clustering algorithms to identify cell populations based on ADT sequencing before undergoing manual cell population labeling based on cell-surface marker expression. Genotype data were then merged, and relative mutated proportions were calculated for each cluster having at least 10 cells for interpretation.
FACS of Cell Populations
As a cross-validation experiment, diagnostic samples from two patients were subjected to FACS in order to characterize genetic alterations in distinct cell populations. Briefly, frozen cells were thawed, stained with lineage-specific antibodies, and sorted on FACSAria III (BD Biosciences). Cell fractions underwent DNA extraction using QIAamp DNA Microkit (Qiagen) followed by targeted sequencing for somatic mutations and ddPCR quantitation for IG/TR clonal rearrangements.
Data Availability
Single-cell and bulk targeted sequencing data are accessible through the EGA database (https://www.ega-archive.org) under accession numbers EGAS00001006784 and EGAS00001006901, respectively. Other data are available upon reasonable request to the principal investigator.
Supplementary Material
Supplementary Table 1. Karyotypes of LH-ALL patients at diagnosis. Supplementary Table 2. Somatic variants detected in LH-ALL patients at diagnosis. Supplementary Table 3. ARCH related variants detected in LH-ALL patients at remission. Supplementary Table 4. Minimal residual disease values of remission samples used for mutation analysis. Supplementary Table 5. Somatic alterations in cell populations from diagnostic samples (BMMC) based on single-cell analyses. Supplementary Table 6. Somatic alterations in FACS-sorted cell populations from diagnostic samples. Supplementary Table 7. Panel of genes for targeted sequencing. Supplementary Table 8. Single cell DNA amplicons for genotyping and LOH analyses. Supplementary Table 9. Single cell DNA amplicons for B-ALL clono-specific IG/TR detection. Supplementary Table 10. ADT-seq panel and spike-in antibodies. Supplementary Table 11. Single cell sequencing metrics.
Supplementary figure 1. Sequencing-based analysis of CNA and LOH in adult LH-ALL. Supplementary figure 2. Representation of mutations in the frequent targeted genes in adult LH-ALL. Supplementary figure 3. Longitudinal assessment of TP53-mutant cell fraction as determined by ddPCR along with clonal IG/TR-based MRD quantification in three patients with undetectable TP53 mutation at remission. Supplementary figure 4. Merged single-cell analysis of cell-surface markers by ADT-sequencing of remission samples from three LH-ALL patients. Supplementary figure 5. Individual single-cell analyses of cell-surface markers by ADT-sequencing of remission samples from three LH-ALL patients. Supplementary figure 6. Individual analyses of single-cell immunophenotyping and genotyping of remission samples from three LH-ALL patients. Supplementary figure 7. Evaluation of allelic dropout rate on heterozygous single nucleotide polymorphism (SNPs) in remission samples. Supplementary figure 8. Individual single-cell analysis of cell-surface markers by ADT-sequencing of diagnosis samples from three LH-ALL patients. Supplementary figure 9. Single-cell genotyping of heterozygous SNPs allowing LOH assessment in diagnosis samples from three LH-ALL patients. Supplementary Figure 10. Distribution of single-cell genotypes of two LH-ALL patients with persistent TP53 and JAK2 (EI_046) or DNMT3A (EI_035) mutations in remission samples.
Acknowledgments
R. Kim was a recipient of a PhD grant with financial support from ITMO Cancer of Aviesan on funds administered by INSERM. The study was supported by a grant from Force Hémato (2020). The authors thank the patients and all the GRAALL investigators, physicians, and biologists who contributed samples and data for this study. The authors thank the Saint-Louis Molecular Hematology lab for technical support, especially Hélène Boyer, Emilie Gaudas, Léna Yousfi, and Océanne Richard. The authors also thank Stéphanie Mathis, Pierre Lemaire, and Clémentine Chauvel for the flow cytometry evaluation of ALL diagnostic samples. This work benefited from the genomic platform facility of Institut de Recherche Saint-Louis (IRSL), supported by Association Saint-Louis. This work was supported by THEMA, the national center for precision medicine in leukemia.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
S. Gachet reports grants from Fondation de France during the conduct of the study. V. Asnafi reports personal fees from Servier outside the submitted work. L. Ades reports grants and personal fees from BMS, personal fees from Novartis, AbbVie, and JAZZ pharma outside the submitted work. Y. Chalandon reports other support from MSD, Novartis, Incyte, BMS, Pfizer, AbbVie, Roche, Jazz, Gilead, Amgen, Astra-Zeneca, Servier, MSD, Roche, Gilead, Amgen, Incyte, AbbVie, Janssen, Jazz, and Astra-Zeneca outside the submitted work. F. Huguet reports personal fees from Novartis, Incyte, Amgen, Pfizer, and Servier outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
R. Kim: Conceptualization, formal analysis, investigation, writing–original draft, writing–review and editing. H. Bergugnat: Data curation, investigation. L. Larcher: Data curation, investigation. M. Duchmann: Methodology, writing–review and editing. M. Passet: Data curation, investigation, writing–review and editing. S. Gachet: Methodology. W. Cuccuini: Data curation. M. Lafage-Pochitaloff: Data curation, writing–review and editing. C. Pastoret: Resources. N. Grardel: Resources. V. Asnafi: Resources. B.W. Schafer: Resources. E. Delabesse: Resources. R. Itzykson: Resources. L. Adès: Resources. Y. Hicheri: Resources. Y. Chalandon: Resources. C. Graux: Resources. P. Chevallier: Resources. M. Hunault: Resources. T. Leguay: Resources. F. Huguet: Resources. V. Lhéritier: Data curation, project administration. H. Dombret: Project administration. J. Soulier: Supervision, writing–review and editing. P. Rousselot: Project administration. N. Boissel: Project administration. E. Clappier: Conceptualization, supervision, funding acquisition, writing–original draft, writing–review and editing.
References
- 1. Sive JI, Buck G, Fielding A, Lazarus HM, Litzow MR, Luger S, et al. Outcomes in older adults with acute lymphoblastic leukaemia (ALL): results from the international MRC UKALL XII/ECOG2993 trial. Br J Haematol 2012;157:463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hunault-Berger M, Leguay T, Huguet F, Leprêtre S, Deconinck E, Ojeda-Uribe M, et al. A phase 2 study of L-asparaginase encapsulated in erythrocytes in elderly patients with Philadelphia chromosome negative acute lymphoblastic leukemia: The GRASPALL/GRAALL-SA2-2008 study. Am J Hematol 2015;90:811–8. [DOI] [PubMed] [Google Scholar]
- 3. Gökbuget N, Dombret H, Ribera J-M, Fielding AK, Advani A, Bassan R, et al. International reference analysis of outcomes in adults with B-precursor Ph-negative relapsed/refractory acute lymphoblastic leukemia. Haematologica 2016;101:1524–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huguet F, Leguay T, Raffoux E, Thomas X, Beldjord K, Delabesse E, et al. Pediatric-inspired therapy in adults with Philadelphia chromosome-negative acute lymphoblastic leukemia: the GRAALL-2003 study. J Clin Oncol 2009;27:911–8. [DOI] [PubMed] [Google Scholar]
- 5. Huguet F, Chevret S, Leguay T, Thomas X, Boissel N, Escoffre-Barbe M, et al. Intensified therapy of acute lymphoblastic leukemia in adults: report of the randomized GRAALL-2005 clinical trial. J Clin Oncol 2018;36:2514–23. [DOI] [PubMed] [Google Scholar]
- 6. Chiaretti S, Vitale A, Cazzaniga G, Orlando SM, Silvestri D, Fazi P, et al. Clinico-biological features of 5202 patients with acute lymphoblastic leukemia enrolled in the Italian AIEOP and GIMEMA protocols and stratified in age cohorts. Haematologica 2013;98:1702–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roberts KG, Gu Z, Payne-Turner D, McCastlain K, Harvey RC, Chen I-M, et al. High frequency and poor outcome of Philadelphia chromosome-like acute lymphoblastic leukemia in adults. J Clin Oncol 2017;35:394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lafage-Pochitaloff M, Baranger L, Hunault M, Cuccuini W, Lefebvre C, Bidet A, et al. Impact of cytogenetic abnormalities in adults with Ph-negative B-cell precursor acute lymphoblastic leukemia. Blood 2017;130:1832–44. [DOI] [PubMed] [Google Scholar]
- 9. Creasey T, Barretta E, Ryan SL, Butler E, Kirkwood AA, Leongamornlert D, et al. Genetic and genomic analysis of acute lymphoblastic leukemia in older adults reveals a distinct profile of abnormalities: analysis of 210 patients from the UKALL14 and UKALL60+ clinical trials. Haematologica 2022;107:2051–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harrison CJ, Moorman AV, Broadfield ZJ, Cheung KL, Harris RL, Reza Jalali G, et al. Three distinct subgroups of hypodiploidy in acute lymphoblastic leukaemia. Br J Haematol 2004;125:552–9. [DOI] [PubMed] [Google Scholar]
- 11. Charrin C, Thomas X, Ffrench M, Le Q-H, Andrieux J, Mozziconacci M-J, et al. A report from the LALA-94 and LALA-SA groups on hypodiploidy with 30 to 39 chromosomes and near-triploidy: 2 possible expressions of a sole entity conferring poor prognosis in adult acute lymphoblastic leukemia (ALL). Blood 2004;104:2444–51. [DOI] [PubMed] [Google Scholar]
- 12. Nachman JB, Heerema NA, Sather H, Camitta B, Forestier E, Harrison CJ, et al. Outcome of treatment in children with hypodiploid acute lymphoblastic leukemia. Blood 2007;110:1112–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet 2013;45:242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mühlbacher V, Zenger M, Schnittger S, Weissmann S, Kunze F, Kohlmann A, et al. Acute lymphoblastic leukemia with low hypodiploid/near triploid karyotype is a specific clinical entity and exhibits a very high TP53 mutation frequency of 93%. Genes Chromosomes Cancer 2014;53:524–36. [DOI] [PubMed] [Google Scholar]
- 15. Stengel A, Schnittger S, Weissmann S, Kuznia S, Kern W, Kohlmann A, et al. TP53 mutations occur in 15.7% of ALL and are associated with MYC-rearrangement, low hypodiploidy, and a poor prognosis. Blood 2014;124:251–8. [DOI] [PubMed] [Google Scholar]
- 16. Kanagal-Shamanna R, Jain P, Takahashi K, Short NJ, Tang G, Issa GC, et al. TP53 mutation does not confer a poor outcome in adult patients with acute lymphoblastic leukemia who are treated with frontline hyper-CVAD-based regimens. Cancer 2017;123:3717–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014;371:2477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014;20:1472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015;518:552–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gillis NK, Ball M, Zhang Q, Ma Z, Zhao Y, Yoder SJ, et al. Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: a proof-of-concept, case-control study. Lancet Oncol 2017;18:112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abelson S, Collord G, Ng SWK, Weissbrod O, Mendelson Cohen N, Niemeyer E, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 2018;559:400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy R, et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 2020;587:477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morita K, Wang F, Jahn K, Hu T, Tanaka T, Sasaki Y, et al. Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nat Commun 2020;11:5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Demaree B, Delley CL, Vasudevan HN, Peretz CAC, Ruff D, Smith CC, et al. Joint profiling of DNA and proteins in single cells to dissect genotype-phenotype associations in leukemia. Nat Commun 2021;12:1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boettcher S, Miller PG, Sharma R, McConkey M, Leventhal M, Krivtsov AV, et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 2019;365:599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mullighan CG, Su X, Zhang J, Radtke I, Phillips LAA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 2009;360:470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 2011;471:235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao Y, Aldoss I, Qu C, Crawford JC, Gu Z, Allen EK, et al. Tumor-intrinsic and -extrinsic determinants of response to blinatumomab in adults with B-ALL. Blood 2021;137:471–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mar BG, Bullinger LB, McLean KM, Grauman PV, Harris MH, Stevenson K, et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun 2014;5:3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mar BG, Chu SH, Kahn JD, Krivtsov AV, Koche R, Castellano CA, et al. SETD2 alterations impair DNA damage recognition and lead to resistance to chemotherapy in leukemia. Blood 2017;130:2631–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014;506:328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beer PA, Delhommeau F, LeCouédic J-P, Dawson MA, Chen E, Bareford D, et al. Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood 2010;115:2891–900. [DOI] [PubMed] [Google Scholar]
- 34. Hof J, Krentz S, van Schewick C, Körner G, Shalapour S, Rhein P, et al. Mutations and deletions of the TP53 gene predict nonresponse to treatment and poor outcome in first relapse of childhood acute lymphoblastic leukemia. J Clin Oncol 2011;29:3185–93. [DOI] [PubMed] [Google Scholar]
- 35. Bielski CM, Zehir A, Penson AV, Donoghue MTA, Chatila W, Armenia J, et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat Genet 2018;50:1189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Passet M, Boissel N, Sigaux F, Saillard C, Bargetzi M, Ba I, et al. PAX5 P80R mutation identifies a novel subtype of B-cell precursor acute lymphoblastic leukemia with favorable outcome. Blood 2019;133:280–4. [DOI] [PubMed] [Google Scholar]
- 37. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009;25:2865–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pugh TJ, Amr SS, Bowser MJ, Gowrisankar S, Hynes E, Mahanta LM, et al. VisCap: inference and visualization of germ-line copy-number variants from targeted clinical sequencing data. Genet Med 2016;18:712–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou X, Edmonson MN, Wilkinson MR, Patel A, Wu G, Liu Y, et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat Genet 2016;48:4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Karyotypes of LH-ALL patients at diagnosis. Supplementary Table 2. Somatic variants detected in LH-ALL patients at diagnosis. Supplementary Table 3. ARCH related variants detected in LH-ALL patients at remission. Supplementary Table 4. Minimal residual disease values of remission samples used for mutation analysis. Supplementary Table 5. Somatic alterations in cell populations from diagnostic samples (BMMC) based on single-cell analyses. Supplementary Table 6. Somatic alterations in FACS-sorted cell populations from diagnostic samples. Supplementary Table 7. Panel of genes for targeted sequencing. Supplementary Table 8. Single cell DNA amplicons for genotyping and LOH analyses. Supplementary Table 9. Single cell DNA amplicons for B-ALL clono-specific IG/TR detection. Supplementary Table 10. ADT-seq panel and spike-in antibodies. Supplementary Table 11. Single cell sequencing metrics.
Supplementary figure 1. Sequencing-based analysis of CNA and LOH in adult LH-ALL. Supplementary figure 2. Representation of mutations in the frequent targeted genes in adult LH-ALL. Supplementary figure 3. Longitudinal assessment of TP53-mutant cell fraction as determined by ddPCR along with clonal IG/TR-based MRD quantification in three patients with undetectable TP53 mutation at remission. Supplementary figure 4. Merged single-cell analysis of cell-surface markers by ADT-sequencing of remission samples from three LH-ALL patients. Supplementary figure 5. Individual single-cell analyses of cell-surface markers by ADT-sequencing of remission samples from three LH-ALL patients. Supplementary figure 6. Individual analyses of single-cell immunophenotyping and genotyping of remission samples from three LH-ALL patients. Supplementary figure 7. Evaluation of allelic dropout rate on heterozygous single nucleotide polymorphism (SNPs) in remission samples. Supplementary figure 8. Individual single-cell analysis of cell-surface markers by ADT-sequencing of diagnosis samples from three LH-ALL patients. Supplementary figure 9. Single-cell genotyping of heterozygous SNPs allowing LOH assessment in diagnosis samples from three LH-ALL patients. Supplementary Figure 10. Distribution of single-cell genotypes of two LH-ALL patients with persistent TP53 and JAK2 (EI_046) or DNMT3A (EI_035) mutations in remission samples.
Data Availability Statement
Single-cell and bulk targeted sequencing data are accessible through the EGA database (https://www.ega-archive.org) under accession numbers EGAS00001006784 and EGAS00001006901, respectively. Other data are available upon reasonable request to the principal investigator.