A multiplex-edited mouse model of Richter syndrome transformation from CLL reveals dysregulated signaling pathways and potential therapeutic targets.
Abstract
Transformation to aggressive disease histologies generates formidable clinical challenges across cancers, but biological insights remain few. We modeled the genetic heterogeneity of chronic lymphocytic leukemia (CLL) through multiplexed in vivo CRISPR-Cas9 B-cell editing of recurrent CLL loss-of-function drivers in mice and recapitulated the process of transformation from indolent CLL into large cell lymphoma [i.e., Richter syndrome (RS)]. Evolutionary trajectories of 64 mice carrying diverse combinatorial gene assortments revealed coselection of mutations in Trp53, Mga, and Chd2 and the dual impact of clonal Mga/Chd2 mutations on E2F/MYC and interferon signaling dysregulation. Comparative human and murine RS analyses demonstrated tonic PI3K signaling as a key feature of transformed disease, with constitutive activation of the AKT and S6 kinases, downmodulation of the PTEN phosphatase, and convergent activation of MYC/PI3K transcriptional programs underlying enhanced sensitivity to MYC/mTOR/PI3K inhibition. This robust experimental system presents a unique framework to study lymphoid biology and therapy.
Significance:
Mouse models reflective of the genetic complexity and heterogeneity of human tumors remain few, including those able to recapitulate transformation to aggressive disease histologies. Herein, we model CLL transformation into RS through multiplexed in vivo gene editing, providing key insight into the pathophysiology and therapeutic vulnerabilities of transformed disease.
This article is highlighted in the In This Issue feature, p. 101
INTRODUCTION
Despite notable discoveries of cancer-related genes inferred by large-scale genomic analyses over recent years, how these diverse putative drivers functionally interact to contribute to oncogenesis remains incompletely explored. Furthermore, transformation from indolent to aggressive histology is a recognized problem across cancers, yet is still poorly understood, due to the lack of suitable cellular and mouse models. Recent advances in genetic engineering have increasingly facilitated the ability to assess the in vivo effects of candidate disease drivers (1). For chronic lymphocytic leukemia (CLL), these approaches have enabled improved modeling of genetic lesions introduced into the B-cell lineage [e.g., del(13q), Ikzf3, Sf3b1/Atm; refs. 2–4]. However, although these models have all demonstrated the development of CLL with indolent kinetics, consistent with the phenotype common to human disease, they do not reflect the expression of combinations of multiple alterations typical of most tumors. Moreover, they do not shed light on the histologic transformation of CLL to Richter syndrome (RS), which represents a formidable clinical challenge with limited current therapeutic outcomes and dismal prognosis (5).
Given the propensity of CLL to clonally evolve, even in the absence of therapeutic pressure (6), we asked whether combinatorial gene editing by CRISPR-Cas9 could recapitulate patterns of clonal evolution typical of human disease and thereby permit the modeling of coevolution of cooccurring traits in vivo in mice. Here, we develop a robust approach for the multiplexed introduction of driver mutations into B cells, consisting of up to 6 loss-of-function (LOF) lesions typical of CLL (7). We report not only the feasibility of generating a genetically heterogeneous cohort of indolent leukemias carrying combinatorial drivers that bear striking phenotypic, transcriptional, genetic, and epigenetic similarities with human disease, but remarkably, also clonal evolution into aggressive lymphoma. Our model yields critical new understanding on predisposing genetic lesions for transformation, demonstrates coevolution of combinatorial traits in vivo, reveals underlying signaling pathways and functional dependencies, and provides new information on actionable therapeutic vulnerabilities.
RESULTS
In Vivo Modeling of Multiplexed LOF Lesions Leads to CLL Development and Transformation into RS
To model genetic heterogeneity typical of human CLL disease, we developed an in vivo system combining multiple LOF alterations [i.e., the DNA damage response genes Atm (mutated or deleted in 18.9% CLL), Trp53 (10%), Samhd1 (1.4%), the negative regulator of MYC signaling Mga (4.2%), the NF-κB signaling modulator Birc3 (3.2%), and the chromatin remodeler Chd2 (7.5%); ref. 7] into del(13q)-B cells, as this is the most common genomic aberration of human CLL (48.5% cases; ref. 7). Expression of del(13q) alone can induce leukemia at low penetrance in elderly mice (2) and cooccurs with the 6 LOF mutations in patients (cooccurrence with mut-ATM: 10.1%; mut-TP53: 4.8%; mut-CHD2: 2.8%; mut-MGA: 2.3%; mut-BIRC3: 1.7%; mut-SAMHD1: 0.4%), as demonstrated in a recent large-scale characterization of CLL (7), thus representing a reliable CLL-predisposing background for our models.
We chose to model LOF lesions, as these can be most easily introduced via Cas9-based gene editing and have shown cooperative leukemogenic potential in human studies (7, 8). We established a transplant model that can rapidly introduce combinatorial lesions in B cells via engineering of Lin−Sca-1+c-Kit+ (LSK) cells from donor mice expressing homozygous del(13q) (2) and B cell–restricted Cas9-GFP (ref. 9; Fig. 1A). We chose LSKs because of their high transducibility and long-term repopulating potential. We lentivirally transduced LSKs (sorting strategy in Supplementary Fig. S1A) with mCherry+–single-guide RNA (sgRNA) targeting either all 6 (“6-plex”) or 5 (“5-plex”) LOF mutations (“targeting pool”); the Trp53 sgRNA was removed from the “5-plex” in order to assess Trp53-dependent and independent mechanisms of disease progression (Fig. 1A). LSKs were then transplanted into either sublethally irradiated immune-competent CD45.1 or immune-deficient NSG recipients (n = 30–35/group). Parallel control cohorts of equal size were generated with pools of 5 or 6 nontargeting sgRNAs (“nontargeting pool”).
Figure 1.
Multiplexed CRISPR editing of CLL-LOF lesions generates CLL disease models. A, Schema for the generation of transplant lines. BM donor lines containing homozygous del(13q) and B cell–restricted Cas9-GFP [i.e., del(13q)-Cd19Cas9-GFP] were obtained by intercrossing del(13q)-Cd19cre/cre and del(13q)-Cas9/Cas9 mice. LSKs isolated from 8- to 12-week-old del(13q)-Cd19Cas9 animals were transduced in vitro with a pool of lentivirus-expressing sgRNAs against the 6 or 5 LOF mutations of interest (Atm, Mga, Samhd1, Chd2, Birc3, Trp53 in “6-plex”; Trp53 absent in “5-plex”) or corresponding nontargeting pools of scrambled sgRNAs and transplanted into 8- to 12-week-old immune-competent (CD45.1) or immune-deficient (NSG) recipients (n = 30–35/group). B, Single-cell qPCR-based analysis of sgRNA expression in LSK from the 3 targeting and the 3 nontargeting cohorts, as assessed at 72 hours after transduction and in vitro culture. The number of sgRNAs quantified in individual single cells is displayed. C, Percent (%) animals with circulating CLL in targeting and nontargeting cohorts. P, Fisher exact test. D, Longitudinal tumor burden assessments by flow-cytometric analysis of peripheral bleeds. A logistic mixed-effects model was used to estimate the average trajectories of peripheral longitudinal tumor burden (%GFP+mCh+B220+CD5+Igκ+ cells) in individual mice from the targeting (6-plex: dark brown; 5-plex: salmon) and nontargeting cohorts (gray). Disease onset and survival (median and range) for each cohort are indicated.
We verified our ability to efficiently introduce combinatorial sgRNAs into individual LSKs by qPCR analysis on 76 to 96 single LSKs per transplant group, assayed 72 hours following transduction and in vitro culture. We detected 5 to 6 sgRNAs in most LSKs from both the 5- or 6-plex targeting and nontargeting cohorts, confirming the successful combinatorial introduction of pooled sgRNAs in individual cells and recapitulating diverse sgRNA assortments (Fig. 1B; Supplementary Fig. S1B). We then monitored the transplanted mice through peripheral blood flow-cytometric analysis for the development of CLL, as defined by GFP+mCherry+ (i.e., Cas9/sgRNA-expressing) B220+CD5+Igκ+ small lymphocytes, at least every 4 months starting at 4 months of age. Remarkably, we detected CLL development, as defined by these surface markers, in 28 of 35 (80%) 6-plex CD45.1, 17 of 30 (56%) NSG 6-plex, and 19 of 30 (63%) NSG 5-plex mice from the targeting cohorts. In contrast, we detected only the expected incidence, consistent with known penetrance of the del(13q) background strain (2), in mice that received nontargeting sgRNA pools [i.e., 5 of 35 (14%) CD45.1, 3 of 30 (10%) NSG 6-plex, 3 of 30 (10%) NSG 5-plex; P < 0.0001; Fig. 1C and D]. Disease onset was faster and overall survival was lower in NSG 6-plex compared with either CD45.1 or NSG 5-plex cohorts (P ≤ 0.003; Fig. 1D; Supplementary Table S1).
At euthanasia, we identified three distinct disease entities in mice that had been identified as having circulating CLL in the targeting cohorts based on flow cytometry and IHC (see Methods). All observed patterns were preceded by evidence of circulating CLL (Fig. 1D). We observed cases consistent with CLL (defined both by the presence of the markers B220+CD5+Igκ+ by flow cytometry and PAX5+CD5high by IHC), transformed lymphoma [RS, defined as lymphocytes with increased forward side scatter (FSC), expressing GFP+mCherry+B220medCD5med/low Igκ+ and morphologically large lymphocytes PAX5+CD5med/low], or with coexistence of the two (CLL/RS; Fig. 2A and B; Supplementary Fig. S2A and S2B). In contrast, the nontargeting control mice that developed disease generated primarily CLL-like disease, with only one instance of de novo diffuse large B-cell lymphoma (DLBCL). Similarly, in a parallel cohort of 40 del(13q)-Cd19Cas9 mice on a B6 background, we also detected development of CLL-like disease in 20% mice [del(13q)-CLL; Supplementary Fig. S2C], consistent with the expected penetrance of the transgenic strain (2). These results confirmed that RS transformation arises because of the combinatorial introduction of CLL-associated LOF lesions together with del(13q). Moreover, RS predominantly developed in CD45.1 hosts, rather than in the NSG 6-plex group (P = 0.06; Fig. 2C). No RS cases were observed to arise from the NSG 5-plex cohort, implicating mutation in Trp53 as necessary to drive full-blown lymphoma transformation in our models (Fig. 2C).
Figure 2.
Transformation into RS shows phenotypes consistent with human disease. A, Representative flow-cytometric plots highlighting the presence of small (CLL), small and large (CLL/RS), or large (RS) cells in the SP of one representative animal per disease pattern, at euthanasia. B, Representative H&E and CD5/PAX5 images from SP and BM of one case per group. Arrows indicate the presence of both small (blue) and large (red) cells in the representative CLL/RS case. Images captured at 10× magnification; black bar,100 μm; white bar, 20 μm. C, Pie charts displaying number of total cases of CLL (blue), CLL/RS (purple), and RS (red) in CD45.1, NSG 6-plex, NSG 5-plex and all nontargeting cohorts. Overall penetrance for each cohort is indicated. P, Fisher exact test. D, Disease burden as represented by %GFP+mCherry+B220+CD5+ Igκ+ cells in PB, SP, and BM of 11 NT-CLL, 21 CLL, 18 CLL/RS, and 25 RS cases at euthanasia. NT-CLL, nontargeting CLL controls. Horizontal lines, group median values. P values, ANOVA with Tukey correction for multiple comparisons. E, Number of cases displaying visible nodal infiltrations. LN, Lymph nodes. F, Representative IHC staining from one murine (left) and one human RS case (right) stained for CD5/PAX5, MUM1, BCL6, CD30, MYC, and Ki67. Images were taken at a 40× magnification; scale bar, 50 μm. G, Number of animals per pattern, and IGHV homology to germline, as assessed by BCR Immuno-Seq. H, Percent (%) clonotype frequency of shared BCR rearrangements as analyzed at time of CLL and upon RS transformation (euthanasia) in 2 CLL/RS and 3 RS cases sampled serially. The dominant RS clonotype is shown in purple (CLL/RS) and red (RS).
Consistent with an aggressive disease phenotype, RS-bearing animals had an overall increased tumor burden in the peripheral blood (PB), spleen (SP), and bone marrow (BM), increased SP size, and lymph nodes (LN; Fig. 2D and E; Supplementary Fig. S2D and S2E). Across SP sections from 10 cases identified as RS based on the criteria above [assessed in parallel with sections from CLL (n = 3), CLL/RS (n = 3), and del(13q)-CLL (n = 3)], we detected positive MUM1 expression and lack of BCL6 and CD30 expression (10, 11). Ki-67 (median: 92% positive cells; range, 83%–99%) and MYC expression (median: 90%; range, 64%–99%) were also uniformly elevated across these 10 RS cases. All of the above features were consistent with human RS histology (Fig. 2F; Supplementary Fig. S2F–S2H; Supplementary Table S2). IGHV as assessed by next-generation sequencing (NGS) was unmutated (≥98% homology to germline) in all 54 analyzed cases across patterns (Fig. 2G; Supplementary Table S3). In 2 CLL/RS and 3 RS cases for which serial PB samples were available, we identified shared B-cell receptor (BCR) clonotypes between RS and antecedent CLL (Fig. 2H), consistent with clonal relatedness of the two disease entities, also a common feature of human RS (ref. 5; Supplementary Table S4). One RS case showed instead clonally unrelated transformation (Supplementary Fig. S2I; Supplementary Table S4). Altogether, our analyses established that our multiplex gene-edited models generated a spectrum of phenotypes recapitulating the bona fide transformation of indolent CLL into RS.
Clonal Evolutionary Analyses Reveal Coselection of Cooccurring Trp53, Mga, and Chd2 Lesions in RS
To verify the robust introduction of the targeted lesions into the engrafted B cells, we performed targeted PCR-based NGS (i.e., CRISPR-seq) on flow-sorted nonleukemic B cells (i.e., CD5−B220+GFP+mCherry+) from PB, collected 4 months after transplant. We detected the presence (defined as ≥0.1% edited sequences per gene, by CRISPResso (12), see Methods and Supplementary Table S5) of a median of 5 LOF mutations in 5-plex and 6 LOF mutations in 6-plex derived cohorts (range, 2–6 in 6-plex NSG; Fig. 3A). At euthanasia, we again identified these lesions as mostly enriched in flow-sorted CLL, CLL/RS, and RS tissue, with a high prevalence of Trp53 mutations [50% tumors harboring clonal Trp53 (defined as >85% gene indels)], Mga (27%), and Chd2 (23%; Fig. 3A], consistent with the abundant detection of these lesions observed in human RS (61%, 29%, and 5.2% cases, respectively; ref. 13).
Figure 3.
Coselection of combinatorial traits is associated with RS transformation. A, Heat map showing CRISPR-edited lesions (in preleukemic normal B cells and at euthanasia) in relationship with disease pattern, recipient strain, and the number of introduced LOFs. Cases for which longitudinal CRISPR-seq, single-cell DNA-seq, WGS, and BCR-IGHV analyses were performed are indicated in green. Percent cases carrying clonal mutations (>85% indels) are shown in the bar diagram (blue, CLL; purple, CLL/RS; red, RS). B, Total number of clonal drivers in all cases with CLL (n = 21), CLL/RS (n = 18), and RS (n = 25). P value, ANOVA with Tukey correction for multiple comparisons. C, The proportion in which a recurrent driver is found as clonal or subclonal across the CLL, CLL/RS, and RS samples is provided (top), along with the individual % indels values (and group medians, horizontal lines) for each sample. P, Fisher exact test comparing the proportions of clonal and subclonal events in CLL versus CLL/RS or CLL versus RS, with Bonferroni correction for multiple comparisons. D, Number of mice carrying individual or selected combinations of clonal drivers, as analyzed by bulk CRISPR-seq at euthanasia. E, Longitudinal clonal trajectories of gene edits as analyzed by CRISPR-seq in 4 RS cases. Pre-leuk, preleukemic B cells. Clonality thresholds of 85% presence of indels are indicated by the horizontal dotted black line. Filled lines, clonal drivers; dotted lines, subclonal. F, Modification rates across single cells for the six on-target loci (columns) using single-cell amplicon sequencing. Splenocytes from 2 CLL/RS (n = 2,499 and 829) and 2 RS cases (n = 4,018 and 505) were sampled at euthanasia. A scale bar ranging from 0% (blue color) to 100% (red color) modifications is shown. G, Total number of chromosomal amplifications and deletions (right bars) in all cases with CLL (n = 5), CLL/RS (n = 4), and RS (n = 8) analyzed by WGS. P value, ANOVA with Tukey correction for multiple comparisons. H, Overlay of copy-number profiles from 5 del(13q)-CLLs, 2 CLL/RS 5-plex, 2 CLL/RS 6-plex, and 8 RS samples, as analyzed by WGS. The y axis shows the mean copy-number variant (CNV) ratio within a running window of 1-Mb pairs in each chromosome within the different cohorts. Recurrent human and murine RS drivers are indicated in the figure.
A high degree of genetic heterogeneity was evident across the three disease patterns, with enrichment of driver gene edits at euthanasia when compared with preleukemia (P ≤ 0.01 for Trp53, Mga, Chd2, Birc3, with Trp53 as the most enriched lesion; Supplementary Fig. S3A) and evidence of clonal selection of distinct combinatorial assortments, particularly in RS (P = 0.001; Supplementary Fig. S3B). The total number of clonal LOF drivers increased upon transformation to RS [P (CLL vs. CLL/RS) = 0.06; P (CLL vs. RS) = 0.0025; Fig. 3B], and clonal Mga showed increased abundance in CLL/RS (P = 0.006) and RS (P = 0.01) compared with CLL. Chd2 was also enriched in RS compared with CLL (P = 0.05; Fig. 3C), with 4 of 21 instances (19%) in CLL, 12 of 18 (66%) in CLL/RS, and 14 of 25 (56%) in RS cases carrying these drivers (alone or combined with Trp53) at euthanasia (Fig. 3D). Similar trends of enrichment in Mga and Chd2 lesions were observed when the 5-plex and 6-plex experiments were analyzed separately (Supplementary Fig. S3C). Trp53 was predominantly clonal across the three disease patterns, whereas Samhd1, Birc3, and Atm were mostly subclonal (Supplementary Fig. S3C), and the total number of cases harboring clonal mutations in Mga, Chd2, Atm, Samhd1, and Birc3 was lower in the 5-plex experiments compared with the 6-plex cohort (P = 0.007, Fisher exact test; Supplementary Fig. S3D), highlighting the role of Trp53 in favoring clonal evolution of cooccurring lesions. Of note, in our 5-plex cohort, we observed transforming disease (i.e., CLL/RS) carrying clonal Mga-only (2 cases), Chd2-only (3 cases), or combined Mga-Chd2 (1 case; Fig. 3D), emphasizing the role of these LOF mutations as RS mutational drivers rather than passengers.
Further support for the coselection of Trp53, Mga, and Chd2 clonal lesions upon RS transformation was gained through longitudinal CRISPR-seq analyses of sorted cells from PB in 8 cases (4 CLL/RS, 4 RS) at preleukemia (i.e., normal B cells), at CLL onset, and at time of RS (Fig. 3E; Supplementary Fig. S3E and S3F). Of these cases, the cooccurrence of combinatorial traits was definitively confirmed by the single-cell DNA sequencing (DNA-seq) analysis of CRISPR-introduced gene edits in splenocytes from 4 animals (2 CLL/RS, 2 RS cases; n = 505–4,018 single cells; Fig. 3F; Supplementary Fig. S4A; ref. 14).
Consistent with the high degree of genomic instability observed in human disease, RS cases were characterized by a higher number of chromosome amplifications or deletions (median = 12.5; range, 4–17) compared with CLL (1;1–2, P = 0.0001) or CLL/RS (3; 1–4, P = 0.001; Fig. 3G; Supplementary Table S6), as detected by whole-genome sequencing (WGS) analysis of a subset of cases [5 del(13q)-CLL, 2 CLL/RS 5-plex, 2 CLL/RS 6-plex, and 8 RS cases]. Common to most samples, we observed amplification of chromosomes (chr)15 (16 of 17 cases) and 17 (11 of 17), similar to previously reported genetically engineered CLL models (2–4). Loss of chr12 and chr16 was commonly observed in RS (6 of 8 cases, and 4 of 8 cases, respectively) and in CLL/RS 6-plex (2 of 2), whereas deletion of chr8 (5 of 8 cases) and amplification of chr10 (4 of 8) were unique to RS (Fig. 3H; Supplementary Fig. S4B). These regions contain frequently amplified putative human disease drivers, including Myc (chr15), Ccnd3 (chr17), and Cdk family genes (chr10), whereas the frequent human LOF lesions Dnmt3a (chr12) and Irf2bp2 (chr8) were part of recurrently deleted regions (Fig. 3H, right). We also identified a single-nucleotide variant (SNV) in the histone modifier Setd2 in 1 RS case (Supplementary Fig. S4C; Supplementary Table S7). All these represent recurrent putative drivers in human RS genomic data sets (13), confirming the high level of faithfulness of these mouse models to human disease.
Key RS Pathways Revealed by Murine RS Transcriptomes Show Similarity with Human RS
To understand the molecular events underlying lymphoma transformation, we evaluated the transcriptional profiles of flow-sorted del(13q)-CLL (n = 5), CLL/RS (n = 4), RS cases (n = 11), and normal B-cell control splenocytes (n = 4) from age-matched wild-type animals. We prioritized the analysis of CD45.1 mice, to be able to assess changes resulting from transformation occurring in an immune-competent environment. Using ANOVA, we identified 1,662 genes that were differentially expressed among the 4 different histologic groups (FDR < 0.1; Supplementary Table S8); this analysis revealed RS cells to have a distinct expression profile (Fig. 4A). Notably, RS cases showed upregulation of pathways involved in cell survival and proliferation (i.e., E2F/MYC targets, G2–M checkpoint) and downregulation of immune response genes (i.e., IFNγ response, IL6/JAK/STAT3, and IL2/STAT5 signaling; Fig. 4B; Supplementary Fig. S5A; Supplementary Table S9). Consistently, pairwise comparisons revealed distinct expression profiles between RS and CLL (682 upregulated and 999 downregulated genes, FC > 2, FDR < 0.05), whereas CLL/RS cases showed similarity to both RS and CLL (43 upregulated and 82 downregulated; 133 upregulated and 280 downregulated genes, respectively; Supplementary Fig. S5B; Supplementary Table S10), indicative of ongoing transcriptional reprogramming associated with disease transformation.
Figure 4.
Murine transcriptomes show strong similarity with human disease. A, Heat map of differentially expressed genes among the four different groups of mice (4 normal B, 5 del(13q)-CLL, 4 CLL/RS, and 11 RS). ANOVA FDR < 0.1 was used as a cutoff. B, Signaling pathways enriched for the differentially expressed genes in A. C, Concordantly modulated genes in human and murine RS and respective functional categories. P, Pearson correlation. D, Gene set enrichment plots showing the correlation of human RS versus CLL gene sets (upregulated, top; downregulated, bottom plot) in murine RS versus CLL data. NES, normalized enrichment score; FDR, false discovery rate. E, Representative IGV tracks of MYC ChIP-seq and RNA-seq data of two MYCMGA top targets in Mga-mutant RS and del(13q)-CLL. F, Representative IGV tracks of CHD2 ChIP-seq and RNA-seq data of two CHD2 top targets involved in the interferon γ pathway in Chd2-mutant RS and del(13q)-CLL. G, Log2FC [RS vs. del(13q)-CLL] of MGA targets included in MYC targets and IFNγ response categories. The top 500 MGA cistromeGO targets were used in the analysis. H, Log2FC [RS vs. del(13q)-CLL] of CHD2 targets included in E2F targets and IFNγ response categories. The top 500 CHD2 CistromeGO targets were used in the analysis. I, Immunoblot analysis of phospho-STAT1 and STAT3 (and respective totals), after 5- and 15-minute stimulation with soluble IFNα and IFNγ. One representative normal B-cell, del(13q)-CLL and RS sample is shown. GAPDH was probed, as control. J, Relative abundance of P-STAT1 and P-STAT3 levels across the three replicates of normal B cells, CLL and RS samples. Mean with SEM is displayed. P, ANOVA. K, Bar plot showing HLA class I and II MFI ratios (compared with isotype controls), as assessed by flow cytometry on normal B, del(13q)-CLL and RS cases (n = 5/group). Mean with SEM is displayed. P, ANOVA. L, Mean expression levels (in transcripts per million) of class I (HLA-A, -B, -C) and class II family (HLA-DR, -DQ) on RNA-seq data from 5 human CLL and matched RS cases. P, paired t test.
To further confirm the consistency of our mouse models to human disease, we assessed transcriptional similarity of the murine RS tumors to 5 primary TP53-deleted human RS samples for which matched antecedent CLL RNA sequencing (RNA-seq) was available (Supplementary Table S2). As for the murine tumors, human RS cases were highly enriched for upregulated E2F targets and cell cycle–related pathways, whereas IL2/STAT5 signaling and DNA damage response were predominantly downregulated (Supplementary Fig. S5C; Supplementary Tables S9 and S11). Of note, when mapping murine RS signatures (per Fig. 4A) to human differentially expressed genes in RS versus paired CLL (n = 1,413 upregulated and n = 534 downregulated genes per Supplementary Table S11), we identified 157 signature genes (97 upregulated and 60 downregulated) that were concordantly dysregulated between murine and human lymphoma, including upregulated cell cycle regulators (i.e., CDK1, CCNA2, AURKB), and downmodulated immune signaling adapters (i.e., CD74, ITGB7; r = 0.80, P < 0.0001; Fig. 4C; Supplementary Table S12). E2F/MYC and mTORC1 signaling were consistently observed as top upregulated pathways in RS bulk transcriptomes compared with paired antecedent CLL in a recent report (15), which we further confirmed through comparative transcriptomic analysis of 107 CLL (4) and 36 independent RS cases (by Enrichr, FC > 2, FDR < 0.001; Supplementary Table S13; ref. 13). Gene set enrichment analysis (GSEA) revealed the enrichment of upregulated human RS signatures in upregulated genes of murine RS versus del(13q)-CLL (Fig. 4D, top; Supplementary Table S10), whereas downregulated human RS signatures were significantly enriched in downregulated genes of murine RS, suggesting global transcriptome similarity (Fig. 4D, bottom). Conversely, comparing the murine signatures to human data, we identified positive enrichment of the murine signatures in human RS versus CLL (Supplementary Fig. S5D), but not in human RS versus de novo DLBCL [36 clonally related RS vs. 28 de novo DLBCLs from Broseus et al. (16); Supplementary Fig. S5E], confirming clonally related human and murine RS as representing distinct entities from de novo DLBCL. Altogether, these analyses support the existence of remarkably similar evolutionary paths for histologic transformation between the two species.
Given the unique enrichment of Mga and Chd2 clonal lesions in RS cases, we asked whether these driver genes were implicated in the transcriptional reprogramming occurring in CLL cells on their path to RS transformation. To this end, we reanalyzed existing Chromatin Immunoprecipitation followed by sequencing (ChIP-seq) data of MYC (which shares transcriptional targets with Mga; refs. 17–19) and of CHD2, generated from the mouse CD5+ B1a-derived B-cell lymphoma cell line CH12 (20), as well as RNA-seq data originating from our murine RS cases carrying clonal Mga or Chd2 mutations (n = 4/group) compared with del(13q)-CLL. Using the tool CistromeGO (21), which integrates ChIP-seq and RNA-seq data sets to provide functional enrichment analysis of ChIP-seq peaks, we identified targets most likely to be directly affected by Mga or Chd2 loss (Fig. 4E and F; Supplementary Tables S14 and S15). As expected, the top 500 targets affected by Mga loss were mostly enriched for upregulated MYC targets, but also for downregulated IFNγ response genes (Fig. 4G; Supplementary Fig. S5F; Supplementary Tables S14 and S16). In contrast, the top 500 targets affected by Chd2 loss were highly enriched for upregulated E2F targets, and downregulated genes were involved in IFNγ and IFNα response and IL2/STAT5 signaling (Fig. 4H; Supplementary Fig. S5G, Supplementary Tables S15 and S16).
We confirmed diminished interferon responsiveness of RS through immunoblotting for STAT1 and STAT3 phosphorylation, after in vitro IFNα and IFNγ stimulation compared with normal B and CLL cells of 3 independent animals per group (P ≤ 0.04; Fig. 4I and J; Supplementary Fig. S5H–S5I). Consistent with diminished in vivo interferon signaling, we detected diminished baseline HLA class I and II expression on the surface of murine RS cells compared with CLL or normal B cells by flow cytometry (P ≤ 0.003; Fig. 4K). Of note, class I (HLA-A, -B, -C) and the most common class II family genes (HLA-DR, -DQ) were downmodulated in human RS RNA-seq data, compared with antecedent CLL (P ≤ 0.03, Fig. 4L; and recently confirmed by paired bulk transcriptomes from Nadeu and colleagues, ref. 15), again confirming cross-species evolutionary similarities.
Tonic PI3K Signaling Is a Characteristic Feature of RS
Given the strong MYC/mTOR transcriptional activity typical of human and murine RS, we reasoned that constitutive activation of the PI3K/mTOR signaling pathway could be a characteristic feature of RS. We therefore examined baseline expression and phosphorylation of AKT and S6, alongside c-MYC and the PTEN phosphatase by immunoblotting in 5 del(13q)-CLL, 6 CLL/RS, 12 RS primary murine splenocyte samples and 14 human primary or patient-derived xenograft (PDX)-derived RS (characterized by FISH and/or TP53 targeted sequencing; Supplementary Table S2; refs. 22, 23; Fig. 5A; Supplementary Fig. S6A). PTEN is a well-known negative regulator of AKT activity, and its protein levels can be diminished in the presence of aberrant MYC activity (24, 25). As anticipated, we observed constitutive baseline phosphorylation of AKT and/or S6 kinases in most murine and human RS specimens (P ≤ 0.02; Fig. 5B) and an inverse correlation between c-MYC and PTEN expression levels in murine and human RS (P = 0.05; Fig. 5C). Through our murine analyses, we additionally identified the Ptpn6 gene (encoding for the protein tyrosine phosphatase SHP1, a negative PI3K signaling regulator; ref. 26) as a putative target of CHD2, downmodulated in the presence of Chd2 mutation, as inferred by CistromeGO (Fig. 5D; Supplementary Table S15). Indeed, Ptpn6 and SHP1 expression appeared lower at both the RNA and protein levels, respectively, in clonal Chd2-mutant murine RS, when compared with cases carrying subclonal or absent mutation (P ≤ 0.01; Fig. 5E and F; Supplementary Fig. S6B and S6C). In line with these results, we observed increased baseline accessibility (i.e., deviation z score) of the transcription factor 3 (TCF3), which may contribute to enhanced PI3K signaling by downregulating SHP1 and upregulating BCR signaling components (27), in single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) analysis of murine RS, compared with del(13q)-CLL and to normal BM and SP from a wild-type B6 mouse (P < 0.0001, Fig. 5G and H; Supplementary Fig. S6D; ref. 28). Similarly, an increased baseline accessibility of TCF3 transcription factor motifs was detected in LN and peripheral blood mononuclear cells (PBMC) from a TP53-deleted RS patient (29), when compared with matched antecedent CLL (P < 0.0001; Fig. 5I–J). Consistent with aberrant PI3K signaling, the mean expression of 114 PI3K signature genes (i.e., PI3K score; ref. 30) was increased upon RS transformation (P ≤ 0.02; Fig. 5K), in murine and paired human RNA-seq data sets. In support of cross-talk between the MYC and PI3K signaling axes in transforming disease, we identified a consistent correlation of the PI3K score with the mean expression of 192 MYC signaling genes (i.e., MYC score; ref. 31) in both murine and human RS (Fig. 5L, P < 0.0001, including 37 primary cases with known TP53 status). All these lines of evidence point to the synergistic activity of MYC/PI3K signaling as a feature of human and murine RS.
Figure 5.
Tonic PI3K signaling is a characteristic feature of RS. A, Immunoblot analysis of 12 murine RS and 14 human RS cases, assessed for baseline expression of c-MYC, PTEN, and phosphorylation and total expression of AKT and S6 kinases. GAPDH is shown as a loading control. Separate gels, run concomitantly, were used for the analysis. B, Heat map of relative phosphorylation intensities in AKT and S6, as analyzed by Western blot in del(13q)-CLL (n = 5), CLL/RS (n = 6), RS (n = 12), and human RS (n = 14). P, ANOVA. C, Linear regression of c-MYC and PTEN protein expression levels (normalized to GAPDH) in 12 murine and 14 human RS cases. P, Pearson correlation. D, Representative IGV tracks of Chd2 ChIP-seq and RNA-seq data of the Ptpn6 gene region from one representative normal B, one del(13q)-CLL, one Chd2-wt RS, and one clonal Chd2-mut RS case. E, Western blot analysis of SHP1 levels in primary murine RS carrying either wild-type (WT) or clonal Chd2 mutation (MUT). GAPDH is displayed, as control. F, Relative SHP1 expression (normalized to GAPDH) in 5 murine RS carrying subclonal/wt Chd2, and 5 cases carrying clonal Chd2 lesions. Mean with SEM is displayed. P, Mann–Whitney. G, Annotation of Uniform Manifold Approximation and Projection (UMAP) representation of single-cell chromatin accessibility profiles by cell type of murine data (left). B cells (black), CLL (blue), and RS (red) cells, T cells (brown), myeloid (gray), and erythroid (beige) compartment and deviation scores of TCF3 transcription factor motifs in murine data sets (right). H, Distribution of deviation z-scores of murine TCF3 binding motif across T cells, B cells, CLL, and RS. Median and interquartile ranges are displayed. P, ANOVA with Tukey correction for multiple comparisons. I, Annotation of UMAP representation of human single-cell chromatin accessibility profiles by cell type (left). B cells (black), CLL (blue), RS (lymph node, red; BM and PB, dark pink) cells, T cells (brown), and deviation scores of TCF3 transcription factor motifs (right). J, Distribution of deviation z-scores of human TCF3 binding motif across T cells, B cells, CLL, and RS. Median and interquartile ranges are displayed. P, paired t test. K, PI3K signature gene expression in the 4 normal B, 5 del(13q)-CLL, 4 CLL/RS, and 11 RS murine cases (mean with SEM) and in the 5 paired CLL/RS samples, analyzed by RNA-seq. P (murine), ANOVA; P (human), paired t test. L, Correlation between PI3K scores and MYC scores in the 4 CLL/RS and 11 RS murine cases and 37 human RS analyzed by RNA-seq. P value, Pearson correlation.
MYC/mTOR/PI3K Signaling Is a Key Vulnerability of RS
To interrogate therapeutic vulnerabilities of RS, we exposed 7 CLL, 3 CLL/RS, 12 RS primary splenocyte samples, and 5 human primary RS cases to a panel of 9 compounds targeting key prosurvival pathways of CLL and RS in vitro for 24 hours followed by the CellTiter-Glo assessment of cellular viability (Fig. 6A; see Methods for drug concentrations). We observed strong sensitivity of human RS and murine RS and CLL/RS samples to the BRD4 inhibitor JQ1, the mTOR inhibitor everolimus, the PI3Kγδ inhibitor duvelisib, the CDK4/6 inhibitor palbociclib, and the Aurora kinase inhibitor alisertib, compared with del(13q)-CLL (P ≤ 0.04; Fig. 6B). In contrast, murine and human RS exhibited reduced response to the chemotherapeutic agent fludarabine (P = 0.04; Supplementary Fig. S7A). Overall, modest sensitivity to BTK kinase inhibition by ibrutinib and acalabrutinib and similar sensitivity to BCL2 inhibition by venetoclax were detected in murine and human RS samples (Supplementary Fig. S7B). Of note, the levels of baseline AKT and S6 phosphorylation (detected by immunoblotting, including 10 additional human primary RS cases from Fig. 5A) correlated with increased response to duvelisib and everolimus, respectively, indicating the increased efficacy of these compounds in samples carrying higher PI3K/mTOR baseline pathway activity (P = 0.004; Fig. 6C and D). The efficacy of single-agent palbociclib and alisertib was modest in secondary NSG-based transplant models (P = 0.002; Supplementary Fig. S7C). Combinatorial treatments with duvelisib-based combinations (with either everolimus or JQ1) reduced MYC expression in vitro (P = 0.003; Fig. 6E; Supplementary Fig. S7D and S7E) and diminished RS cell survival both in vitro (P ≤ 0.0004; Fig. 6F) and in vivo, in transplantable models (P < 0.001 across all comparisons, Fig. 6G), without impacting body weight (Supplementary Fig. S7F). These studies confirm MYC/mTOR/PI3K as key vulnerabilities and actionable targets of RS.
Figure 6.
MYC/mTOR/PI3K signaling are targetable vulnerabilities of RS. A, Summary heat map of drug treatment studies of murine primary splenocytes [7 del(13q)-CLL, 3 CLL/RS, and 12 RS] and 5 human RS cases. Percent (%) viability compared with the DMSO control is displayed. B, Bar plots showing percent (%) viability after JQ1, everolimus, duvelisib, palbociclib, and alisertib treatment. Mean with SEM is displayed. P, ANOVA. C, Correlation between response to duvelisib (% viability) and phospho-AKT levels, as detected by immunoblot in 5 del(13q)-CLL, 2 CLL/RS, 8 murine RS, and 14 human RS cases. P, Pearson correlation. D, Correlation between response to everolimus (% viability) and phospho-S6 levels on the same set of samples per C, as detected by immunoblot. P, Pearson correlation. E, Immunoblot assessment of c-MYC expression levels following a 12-hour treatment with single-agent duvelisib, everolimus, JQ1 or respective duvelisib-based combinations. One representative mouse and human samples are shown and GAPDH is used as loading control. F, Percent (%) viability normalized to DMSO control of 4 murine and 3 human RS primary specimens treated for 40 hours with single-agent duvelisib, everolimus, JQ1, or respective duvelisib-based combinations. Mean with SEM is displayed. P value, ANOVA with Tukey correction for multiple comparisons. G, Transplant schema for combination treatment studies and survival curve of NSG mice transplanted with RS splenocytes and then treated with 50 mg/kg duvelisib, 10 mg/kg everolimus, or combination (left), or 50 mg/kg duvelisib, 50 mg/kg JQ1, or combination (right) for 2 weeks followed by observation until survival endpoints. P values, log-rank test with Bonferroni correction for multiple comparisons. The pink shaded area indicates the treatment period.
DISCUSSION
In B lymphoid tumors, notable well-acknowledged difficulties in genetic manipulation of B cells have thus far largely challenged functional genomic analyses and complex disease modeling. We herein report the generation of a unique system that reliably recapitulates cooccurrence and clonal evolution of combinatorial lesions in B cells in vivo, thus broadening the applicability of multiplexed CRISPR-Cas9 gene editing beyond epithelial cell cancers (32–34) and acute myeloid and erythroid malignancies (35–37). Through our platform, we were able to model clonal evolution and histologic switch in lymphoid malignancies, gaining unique insights into the molecular mechanisms and therapeutic vulnerabilities associated with the evolution of indolent CLL into RS. Molecular mechanisms underlying transformation into RS have been thus far largely unknown because of the lack of cell lines, paucity of primary patient material for functional analyses, and limited availability of genetically engineered mouse models faithful to the step-wise evolution of human indolent malignancy. Indeed, RS PDXs (22) do not allow studies of disease transformation from antecedent CLL. Similarly, the few cases of RS transformation reported in Eμ-TCL1–based crosses (i.e., the most commonly used CLL model; ref. 38) in the context of expression of high-risk alterations (i.e., Trp53, Akt, and Cdkn2a/b; refs. 39–41) cannot faithfully recapitulate evolution from indolent CLL, because TCL1 is inherently not a CLL-specific genetic lesion, and can alone induce aggressive disease.
By analyzing combinatorially edited CLL and RS mice, generated through the CRISPR-Cas9 gene editing of del(13q)-expressing hematopoietic stem cells with B cell–restricted CLL LOF lesions, we demonstrated coselection of Trp53, Mga, and Chd2 mutations on the path to transformation, with a key role for Trp53 in facilitating the clonal evolution of cooccurring traits, and that Mga/Chd2 mutations were associated with dysregulated MYC/E2F and interferon signaling. Our system uniquely demonstrates that when a diverse pool of gene edits is introduced in maturing B cells, only a limited set of assortments of combinations of lesions (primarily Trp53, Mga, and Chd2) undergo clonal selection upon transformation; these data strongly support the notion that only a restricted set of coselected drivers are enriched on the path to RS. These findings align with the strong prevalence of TP53 mutation in human RS (13, 15, 42–44), and with the notion that MGA functions as an MYC suppressor and is frequently lost in RS (13, 15, 45). In an independent human study characterizing 97 human RS cases, the prevalence of TP53-mutated RS was 61% [combining TP53 mutation and del(17p) frequency]; total MGA inactivation was ∼29% [18% focal deletion (15q.15.1), 11.3% frameshift mutation], thus more prevalent than MYC amplifications (∼14%); and CHD2 frameshift mutations were captured in 5.2% cases (13). Importantly, the modeled combinatorial assortments showed significant copresence in human RS, with 19 of 97 (19.6%) cases with cooccurring TP53/MGA, 3 of 97 (3.09%) with TP53/CHD2, 2 of 97 (2.06%) with TP53/MGA/CHD2, and 1 case with MGA/CHD2 lesions in the absence of TP53 loss (13). Another recent study also reported chromothriptic events targeting (among others) TP53 and/or MGA and CHD2 lesions in 2 of 19 (10.5%) analyzed cases, and diminished expression levels of CHD2 in transformed disease, again confirming the relevance of loss of these genes in RS transformation (15). Aside from TP53, one of the most frequent human RS drivers is NOTCH1 (mutated in ∼39% cases; ref. 13); we note that mut-NOTCH1 occurs primarily as a 2-bp deletion, leading to truncation of a small C-term domain of the protein; thus, it cannot be efficiently modeled through S. piogenes Cas9-based methods (i.e., those used in the current study), which generally introduce random indels up to 15 base pairs downstream or upstream of the sgRNA cut site. Modeling of NOTCH1 mutations is a priority for future studies, which will require incorporating base editing approaches to more feasibly introduce this type of alteration. A limitation of our study is the inability to control the timing of mutational acquisition, because all lesions are introduced upon Cd19-driven Cas9 expression during early and throughout mature B-cell development; this aspect will be overcome in future studies by using inducible (i.e., Tet-based) and/or alternative conditional approaches (i.e., Dre-lox).
In contrast to GC-derived B-cell lymphomas (46), and consistent with previously published CLL models (2–4, 38), our RS tumors invariably expressed unmutated immunoglobulins, similar to the majority of human RS (11, 13, 42–44), and indicative of a lack of somatic hypermutation, and therefore of a cell of origin that has not yet transited the GC reaction. We, therefore, propose a model whereby strong MYC-driven signals occurring in tumor cells prior to GC entry facilitate the acquisition of transcriptional and functional programs similar to GC-derived B-cell lymphomas, including diminished PTEN expression and TCF3 activation, which ultimately facilitate tonic PI3K/mTOR signaling (47, 48). One of the known mechanisms underlying PTEN downregulation is upregulated expression of the mir-17/92 cluster (25), resulting from aberrant MYC signaling. The strong MYC activity typical of human and murine RS and the focal amplification of the mir-17/92 cluster observed in ∼9% human RS (13) support this explanation. We here also propose an association between the presence of Chd2 mutations and diminished SHP1 phosphatase expression, another key regulator of PI3K activity (26), in line with the notion that Chd2 mutation interferes with chromatin remodeling in human CLL cells (49). Besides downregulated phosphatase activity, tonic PI3K signaling could also stem from autonomous BCR signaling activity, as previously proposed for CLL (50), albeit this aspect remains to be functionally validated. The predominant role of MYC in our model, the frequent mutual exclusivity of NOTCH1 and MYC mutations in human RS (51), and the functional interdependence of the NOTCH–MYC signaling axis (52) may explain the absence of a predominant NOTCH signature in these mice. Consistent with the strong synergy between MYC and PI3K signaling activity, our drug treatment studies point to the strong efficacy of combination therapy of PI3K-targeted agents with MYC/mTOR inhibitors, which jointly reduce MYC expression. The stronger efficacy of PI3K/mTOR inhibitors in cells carrying higher baseline signaling activity supports opportunities for combinatorial treatments in the context of mutation and/or biomarker-based selection of RS cases, using novel targeted agents. This aligns with ongoing clinical trials exploring duvelisib and copanlisib (pan-PI3K inhibitor)-based combination treatments in RS (NCT03534323 and NCT03892044), with promising results despite short follow-up (53).
The predominant incidence of RS disease in immunocompetent models may be at least partly explained by the lack of CD4+ T cells and of fully functional monocyte/macrophages in NSG mice, given their known relevance in CLL development and progression in mouse models (54, 55). The generally increased lifespan of CD45.1 recipients (up to ∼24 months compared with ∼15 months for NSG) also allows for a prolonged time frame for both genetic and microenvironmental factors to contribute to disease transformation. While our findings shed light on tumor-intrinsic changes on the path to transformation, the immune-competent nature of the CD45.1-based models opens future opportunities to dissect immune-microenvironmental composition and function at different disease stages, and to analyze responses to novel immunotherapies.
Overall, this study demonstrates the feasibility of modeling complex disease phenotypes by multiplexing relevant disease drivers in a lymphoid setting and advances the ability to dissect mechanistic biology and therapeutic vulnerabilities underlying disease transformation in a B-cell context. The remarkable similarity with human disease renders this platform unique for the study of lymphoma biology and therapy. The success of our approach now creates a road map applicable across B-cell malignancies, opening new opportunities for in vivo modeling of complex cooccurring traits more broadly.
METHODS
Generation of Multiplexed Mouse Models
All animal protocols were approved by the Institutional Care and Use Committee of the Dana-Farber Cancer Institute (DFCI). We generated del(13q)-Cd19cre/cre mice (deposited as JAX ID: 037070) by interbreeding MDR mice [JAX ID: B6.129S1-ls(14)2Rdf/J; MDR represents the minimally deleted region of murine chr14, syntenic to human del(13q); ref. 2] with Cd19cre/cre mice (JAX ID: B6.129P2(C)-Cd19tm1(cre)Cgn/J); similarly, we generated del(13q)-Cas9/Cas9 (deposited as JAX ID: 037071) by interbreeding MDR mice with homozygotes Cas9/Cas9 mice (JAX ID: B6J.129(Cg)-Gt(ROSA)26Sortm1(CAG-cas9*,-EGFP)Fezh/J). We then interbred del(13q)-Cd19cre/cre with del(13q)-Cas9/Cas9 to obtain del(13q)-Cd19Cas9 [i.e., expressing B cell–restricted del(13q) and Cas9-GFP] BM donors for all transplant cohorts. Forty del(13q)-Cd19Cas9 mice were also longitudinally observed and monitored for disease development. BM cells were extracted from 8- to 12-week-old donors, and hematopoietic progenitors were enriched (EasySep Mouse Hematopoietic Progenitor Cell Enrichment kit, STEMCELL Technologies). Lineage− Sca-1+ c-Kit+ (LSK) cells were then isolated by cell sorting, with Pacific-Blue anti-Lineage (RRID: AB_11150779), PE anti-mouse Sca-1 (clone D7, RRID: AB_313345), and APC anti-mouse c-Kit (clone 2B9, RRID: AB_313221) antibody staining performed in the presence of FcR blocking antibodies (clone 93, all from BioLegend). Isolated LSKs were precultured at 20,000 cells/mL in vitro at 37°C for 1 hour in StemSpan (STEMCELL Technologies) supplemented with 50 ng/mL mTPO and 50 ng/mL mSCF (PeproTech), prior to transduction with pooled high-titer lentivirus expressing sgRNAs against the target genes of interest (Atm: 5′-TGGATCACGGAGTACATCCA-3′; Trp53: 5′-GAGCGCTGCTCCGATGGTGA-3′; Chd2: 5′-AAGCAACCTAAGATTCAGCG-3′; Samhd1: 5′-CCTCCGAACCTGGGAACCGG-3′; Birc3: 5′-AAAGTCATACTTCAACTCAA-3′; Mga: 5′-AAGTAAAAGTATGAACACCG-3′) or nontargeting scramble controls (nt-1: 5′-AACTCGCGTGGGAAGTCCGG-3′; nt-2: 5′-CAACCTGCCTAGCGACCCGC-3′; nt-3: 5′-GTGGTTACGTTAACGACTAC-3′; nt-4: 5′-TTGGCGGCTCGCTCCTGCGT-3′; nt-5: 5′-GCTTACCTACTCCGCCCCGC-3′; nt-6: 5′-GTCTTCACAGGGTGCAACGA-3′). After overnight incubation, cells were harvested, washed twice in PBS-10% FBS, then resuspended in PBS, and injected at 20,000 LSK cells/recipient in 100 μL PBS. Each animal also received 200,000 BM “helper” cells (i.e., unsorted hematopoietic progenitors). Female 8- to 12-week-old CD45.1 (JAX ID: B6.SJL-PtprcaPepcb/BoyJ) recipient mice were preconditioned with 400 rads split dose, whereas female 8- to 12-week-old NSGs (JAX ID: NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) received 125 rads split dose irradiation, 1 day prior to LSK transfer. The presence of sgRNAs in individual LSKs was evaluated at 72 hours after transduction, by plate-sorting of mCherry+ LSKs followed by single-cell qPCR-based analysis on the Fluidigm Biomark platform, as previously described (14).
Human CLL and RS Samples
CLL and RS samples were collected through sample collection protocols from DFCI, Weill Cornell Medicine, and the University of Ulm in Germany. RS-PDX–derived specimens (either subcutaneous or intraperitoneal tumor) were generously gifted by Drs. Silvia Deaglio and Tiziana Vaisitti from the University of Torino, Italy, and previously reported (22, 23). All biospecimen collection protocols were conducted after obtaining written informed consent from the patients in accordance with the principles of the Declaration of Helsinki and with the approval of the Institutional Review Boards (IRB) of the respective institutions. Detailed patient and sample characteristics are provided in Supplementary Table S2. Cryopreserved primary RS (splenocytes, nodal preparations, BM aspirate, or circulating lymphocytes; RS7, RS12-21), or PDX-passaged RS cells (RS 8–11) were thawed in prewarmed complete RPMI and incubated at 37°C for 30 minutes prior to use for downstream analysis. The viability of all utilized preparations was >85%.
CRISPR-seq of Murine Gene Edits
To assess the presence of gene editing, B cells were isolated from the PB of animals by cell sorting at 4 months after transplant and at euthanasia. DNA was isolated via the Qiagen DNAeasy MicroKit and amplified by multiplexed PCR, as previously described (14). Ten nanograms of genomic DNA was amplified by PCR, as follows: 1 cycle of 1 minute at 98°C, 35 cycles of 98°C for 15 seconds, 65°C for 30 seconds, 72°C for 30 seconds, and 1 cycle of 5 minutes at 72°C. The primers targeted a 200-bp region flanking the sgRNA cut site (14). The PCR product was then purified and submitted for DNA-seq though the Massachusetts General Hospital (MGH) DNA core. FASTQ files were processed by CRISPResso2 (12) software analysis to quantitatively and qualitatively assess the presence of gene edits in individual samples. The presence of CRISPR-induced mutations was defined as ≥ 0.1% indels (lower limit of detection of CRISPResso2) and clonal lesions defined as those with >85% indels, consistent with the cancer cell fraction (CCF) cutoff (i.e., CCF > 85%) we previously reported (8). Comparisons between the proportion of cases in which the LOF-mutated driver was found as clonal or subclonal across the 3 disease entities were performed using the Fisher exact test with Bonferroni correction for multiple comparisons. Change between preleukemia and euthanasia samples was assessed for each gene with a two-sided Wilcoxon signed-rank test. Differences between paired samples were categorized as a decrease (≤10%), an increase (≥10%), or stable otherwise, consistent with the previously reported CCF ≥ 10% threshold (8). For each gene, a one-sided exact binomial test was performed to assess whether the proportion of each category (decrease, increase, and stable) significantly exceeded 0.5; false discovery rate was controlled using the Benjamini–Hochberg method with a significance threshold of 0.1. Mission Bio Tapestri single-cell DNA-seq detection of CRISPR edits on murine splenocyte preparations was performed according to previously published methodology (14).
Disease Blood Monitoring in Mouse Cohorts and Secondary Engraftments
Mice were monitored at least every 4 months starting at 4 months after transplant by submandibular blood draws followed by flow cytometry. One-hundred microliters of blood was collected from mice into EDTA-containing tubes. Erythrocyte lysis was carried out by incubating the blood in 1 mL of ammonium–chloride–potassium (ACK) lysis buffer for 5 minutes and then washing with PBS supplemented with 2% FCS and 2 mmol/L EDTA. Cells were then stained with the following antibodies (all from BioLegend): BV711 anti-mouse CD45.2 (clone 104, RRID: AB_2616859), Pacific-Blue anti-mouse B220 (clone RA3-6B2 RRID: AB_492876), APC anti-mouse Cd11b (clone M1/70, RRID: AB_312795), APC-Cy7 anti-mouse CD3 (clone 17A2, RRID: AB_2242784), PE-Cy7 anti-mouse CD5 (clone 100622, RRID: AB_2562773), and Alexa700 anti-mouse Igκ (clone RMK-45, RRID: AB_2563583) for 15 minutes at 4°C. Next, cells were washed and analyzed using flow cytometry. GFP (Cas9) and Cherry (sgRNAs) were used as intracellular markers for edited B cells. CLL was defined by the presence of at least 2% GFP+mCherry+B220+CD5+Igκ+ circulating disease with most (55 of 64 animals) showing the presence of greater of equal 5% circulating cells at disease onset. Experimental endpoints were defined as the presence of >70% tumor cells in the PB and/or hunched posture, visible hepatosplenomegaly or lymphadenopathy, labored breathing, body weight loss of 15%. Secondary grafts were generated by injecting 2 million viably cryopreserved splenocytes from primary RS tumors into 8- to 12-week-old immunodeficient recipient NSG mice. Animals were randomized for treatment 1 week after engraftment by submandibular blood draw and flow-cytometric analysis of engrafted GFP+mCherry+B220+CD5+Igκ+ cells, detected as ∼10–20% circulating cells.
B-cell Purification, In Vitro Stimulation, and Flow Cytometry
B cells were immunomagnetically selected from splenic single-cell suspension using the MACS mouse B-cell Isolation Kit (Miltenyi Biotec). For IFNα and IFNγ in vitro stimulation, normal B, del(13q)-CLL, and RS cells were resuspended at 5 × 106/mL in complete RPMI, then stimulated for 5 and 15 minutes with 1,000 IU/mL of ligand. For flow-cytometric analysis of murine HLA class I and II expression, the following antibodies were used: BV 510 anti-mouse H-2Kb (clone AF6-88.5, RRID: AB_2800584), PerCP-Cy5.5 anti-mouse I-A/I-E (clone M5/114.15.2, RRID: AB_2191071), BV510 mouse IgG2a, κ isotype control (clone MOPC-173, RRID: AB_2734131), and PerCP/Cy5.5 Rat IgG2b, κ isotype control (clone RTK4530, RRID:AB_893677), all from BioLegend. MFI ratios were calculated after normalization with isotype control staining.
qPCR Analysis and Immunoblotting
cDNA was generated from murine RS RNA through PrimeScript RT Master Mix (Takara). Ptpn6 gene expression was quantified by TaqMan probe Mm00469153_m1 (Thermo Scientific), and Gapdh through mouse Gapdh Mix 20X (Applied Biosystems), with a 40-cycle qPCR at 60°C annealing temperature. Two −Ct values were used for analysis, and Ptpn6/Gapdh ratios were reported. Western blotting was performed as previously described (3). Twenty micrograms of protein per sample was loaded. Membranes were probed with antibodies (all from Cell Signaling Technology, rabbit mAbs unless otherwise stated) against phosphorylated (phospho) STAT1 (Y701, clone 58D6, RRID: AB_561284), phospho-STAT3 (Y705, clone D3A7, RRID:AB_2491009), phospho-AKT (S473, clone D9E, RRID: AB_2315049), phospho-S6 (T389, clone D5U10, RRID: AB_2800283), and respective totals (STAT1, rabbit polyclonal; STAT3, clone D3Z2G, RRID: AB_2629499; AKT, clone 40D4, RRID: AB_329827; S6, clone 49D7, RRID: AB_390722). SHP1, c-MYC, and PTEN total levels were evaluated by probing membranes with an anti-SHP1 mAb (clone C14H6, RRID: AB_2173694), c-MYC (D84C12, RRID: AB_1903938), or PTEN (D4.3, RRID: AB_2253290). A GAPDH-HRP (clone D16H11, RRID: AB_11129865) antibody was used as a control. All antibodies were used at a dilution of 1:1,000 overnight (except GAPDH-HRP incubated for 2 hours followed by detection), followed by anti-rabbit secondary antibody (1:10,000; Cell Signaling Technology) and detection after a 5-minute incubation with SuperSignal West Pico Chemiluminescent substrate (Thermo Fisher). Total proteins were probed after a short stripping (5–10 minutes) of phospho-protein signals with Restore Western Blot Stripping Buffer (Thermo Fisher). For CLL, CLL/RS, and RS baseline PI3K signaling analysis, balanced proportions of the three sample groups were included in every gel. Image acquisition was performed on a Bio-Rad Chemidoc with a 1-minute exposure time; when multiple gels were run concomitantly, exposure was also concomitant. Image analysis was performed by ImageJ and reported as the ratio between phosphorylated and total protein expression. C-MYC and PTEN expression levels were instead normalized to GAPDH. In the P-STAT1 and P-STAT3 evaluations, the 5- and 15-minute stimulation values were averaged within each sample.
IHC and Pathology
Murine tissues were fixed in 10% buffered neutral formalin overnight, and BM sections underwent decalcification in RapidCal Immuno Buffer (Statlab) for 2 hours. Tissues were then paraffin-embedded and cut into 4 μm–thick sections for IHC staining. Hematoxylin and eosin staining, CD5/PAX5 (ref. 56; CD5, 1:300 from Sino Biological; PAX5, RRID: AB_2798001 1:100 from Cell Signaling Technology), Ki67 (1:800, RRID: AB_2620142; Cell Signaling Technology), MUM1 (1:500, RRID: AB_2877647; Cell Signaling Technology), BCL6 (1:1,000; Sigma), MYC (1:1,000; RRID: AB_731658, Abcam), and CD30 (1:100, RRID: AB_2799462, Cell Signaling Technology) stainings were performed as described in the section using standard procedures (3). SP, BM, lung, kidney, liver, and LN from each diseased mouse were evaluated by a board-certified pathologist, and analysis and classification of leukemias/lymphomas were performed following published criteria (57, 58). CLL cases were characterized by the expansion of small PAX5/CD5 double-positive cells, with a relatively low proliferation rate as assessed by Ki-67, predominantly adjacent to the edges of T-cell areas in the SP and small focal infiltrates in other organs, resembling human CLL. RS cases revealed the effacement of normal SP architecture by diffuse proliferation of large PAX5+ CD5med/low MUM1+ BCL6− CD30− cells with a high proliferation rate resembling human RS. Large atypical cells were also abundantly detected within other organs, including BM, liver, lung, and kidney. Pleomorphism, large rather than intermediate size, absence of a “starry sky” pattern, and presence of antecedent circulating small lymphocytic CD5+B220+ disease ruled out both Burkitt and de novo DLBCL diagnosis. CLL/RS cases were characterized by a concomitant presence of large and small lymphocytes, with at least 10% residual CLLs in 2 or more of the analyzed organs. Cases with <10% residual CLLs by combined flow and IHC analysis were classified in the “RS” category. For the Ki-67 and MYC quantification, slides from 10 RS samples were scanned using Vectra 2 Intelligent Slide Analysis System (Caliper LifeSciences) and analyzed with HALO Image Analysis Software (Indica Labs). The percentage of positive cells was assessed in three representative regions of interest, each ∼300,000 μm2, per case. Human tissue samples were processed according to CLIA-approved protocols from the hematopathology service at BWH, Pathology Department.
BCR-IGHV Sequencing
Genomic DNA was isolated using the QIAGEN DNeasy Blood and Tissue Kit (Qiagen) from the PBMCs of diseased mice; 500 ng of genomic DNA per sample was used for sequencing. Sample data were generated using the mouse-IGH immunoSEQ Assay (Adaptive Biotechnologies). BCR repertoire analyses were performed on individual samples using the immunoSEQ Analyzer 3.0, and sequences were subjected to analysis using IMGT/Vquest software to define all V, D, and J genes as well as CDR3 sequences. Mutational status was defined as unmutated when ≥98% homology to germline was measured (59).
Drug Treatment Studies
For in vitro single-agent drug treatment studies, primary murine CLL, CLL/RS, and RS samples and human RS samples were resuspended at 2 to 3 million/mL in RPMI 1640 supplemented with 10% FBS. After incubation for 24 hours at 37°C with 5% CO2 in the presence of either 5 μmol/L JQ1, 10 μmol/L everolimus, 10 μmol/L palbociclib, 10 nmol/L venetoclax, 1 μmol/L ibrutinib, 5 μmol/L acalabrutinib, 5 μmol/L duvelisib, 10 μmol/L alisertib, or 10 μmol/L fludarabine, cell viability was measured by the CellTiter-Glo Luminescent Assay in technical duplicates, and the percentage of viability obtained by normalizing the value from treated samples to the DMSO vehicle control. For in vitro combination treatments, we used suboptimal drug doses for each of the tested compounds: 1 μmol/L duvelisib, 0.5 μmol/L everolimus, and 1 μmol/L JQ1. Viability analyses were performed after a 40-hour incubation, whereas Western blotting for c-MYC protein detection was performed after a 12-hour treatment to limit cellular apoptosis. For in vivo studies, everolimus was administered at 10 mg/kg (60) orally in 30% propylene glycol, 5% Tween-80 in water; JQ1 at 50 mg/kg by intraperitoneal injection (i.p.; ref. 61) in 2% DMSO, 30% PEG300, 5% Tween-80 in water; alisertib at 30 mg/kg orally (62) in 10% 2-hydroxypropyl-β-cyclodextrin in saline; palbociclib at 50 mg/kg orally (63) in 10% DMSO and 20% 2-SBE-β-CD in saline; duvelisib at 50 mg/kg orally (64) in 10% DMSO and 20% 2-SBE-β-CD in saline. All drugs were purchased from Selleckchem and administered for 2 weeks on a 5 days on–2 days off schedule either as single agents or combined (duvelisib + everolimus; duvelisib + JQ1).
RNA-seq
Human RNA-seq was generated from 5 CLL/RS paired samples; murine RNA-seq was performed on viably cryopreserved and FACS-sorted GFP+mCherry+ B220+CD5High/Med cells from 5 del(13q)-CLL, 4 CLL/RS, or 11 RS samples. Four normal B-cell samples were also assayed, isolated from age-matched del(13q)-Cd19Cas9 wild-type mice. Cells were washed in cold PBS and total RNA was extracted using a Qiagen RNAeasy kit from at least 1 × 106 cells. Twenty nanograms of total RNA (RIN quality >9.5) was utilized for library construction. For cDNA library construction, total RNA was quantified using the Quant-iT RiboGreen RNA Assay Kit and normalized to 5 ng/μL. The cDNA libraries were prepared using NEBnext single-cell/low input RNA library prep kit for Illumina (NEB, E6420), followed by cDNA fragmentation and adaptor ligation. Dual indexed PCR was performed using an adapted method, as previously reported (65). After Ampure beads cleaning, final sequencing libraries were quantified using a High-Sensitivity DNA Kit on Bioanalyzer (Agilent). RNA-seq libraries were normalized to 4 nmol/L concentration and pooled before loading onto Illumina sequencer. Human RNA-seq reads were aligned to the human reference genome hg38 and murine RNA-seq reads were aligned to the mouse reference genome mm10 using STAR (v2.4.0.1; ref. 66) and transcripts per kilobase million (TPM) value was calculated. Differentially expressed genes between two groups were assessed using DESeq2 (67). Those with FC > 2 and FDR < 0.05 were considered statistically significant. ANOVA was used to determine differentially expressed genes among multiple groups (i.e., normal B, del(13q)-CLL, CLL/RS, and RS) in murine data, and FDR < 0.1 was considered statistically significant. The murine RS signature genes identified through ANOVA analysis were further compared with human signatures (RS vs. paired CLL, FC > 2, FDR < 0.05) and commonly dysregulated genes were plotted based on Log2FC (RS vs. CLL) values. Pathway analyses were performed using the Enrichr database and the MsigDB hallmark gene sets. Differentially expressed genes in murine RS versus del(13q)-CLL, human RS versus paired CLL, and human RS versus de novo DLBCL (16) were utilized for GSEA v4.3.2 (68) to evaluate interspecies similarities. For these analyses, RNA-seq from 36 additional human RS and 28 de novo DLBCLs was utilized, as previously described (16). The following parameters were utilized across all runs: (i) gene name remapping platforms: Human_Gene_Symbol_with_Remapping_SigDB.v2022.1 and Mouse_Gene_Symbol_with_Remapping_Human_Orthologs_SigDB.v2022.1; (ii) number of permutations: 1,000; (iii) permutation type: phenotype if n > 7 in each group, otherwise gene_set; (iv) gene collapsing: aggregate by gene symbol (median); (v) scoring scheme: weighted. Integrative analysis of RNA-seq and ChIP-seq data was performed using Cistrome-GO (21). A differential expression (DE) rank RDE was calculated based on the DESeq2-FDR of differential gene expression between Mga-mutant or Chd2-mutant RS and del(13q)-CLL. A regulatory potential rank RRP was defined based on the weighted sum of Myc (Encode SRX140369) or Chd2 (Encode SRX140389) CHIP-seq peaks. The two ranks were integrated by rank product (RDE × RRP) to generate the final rank of target genes. The top 500 targets were used for analysis. Class I and II gene expression in human CLL and RS samples was computed based on mean expression (based on TPM values) of HLA-A, -B, -C (class I) and HLA-DR and -DQ family (class II) in paired human RNA-seq data. The 114 PI3K signature genes and MYC “scores” were computed utilizing previously published gene sets (30, 31); “scores” were computed as mean expression of all genes within the signature in murine and human RNA-seq data sets and log2 values comparatively reported. Comparative analyses between 107 CLLs and an external independent set of 36 RS primary cases (13) were performed with FC > 2 and FDR < 0.001 cutoffs for differential gene expression followed by the Enrichr pathway analysis using MsigDB hallmark gene sets.
scATAC-seq and Analysis
scATAC-seq was performed as previously described (29). After fixation with 1% formaldehyde and quenching with 0.125 mol/L glycine, cells were permeabilized and washed with modified lysis (10 mmol/L Tris-HCl pH 7.4, 10 mmol/L NaCl, 3 mmol/L MgCl2 0.1% Nonidet P40 substitute, 1% BSA, nuclease-free H2O) and wash buffers (10 mmol/L Tris-HCl pH 7.4, 10 mmol/L NaCl, 3 mmol/L MgCl2, 1% bovine serum albumin (BSA), nuclease-free H2O). After resuspension in nuclei buffer (10X Genomics) at 5,000 cells per μL, cells were loaded onto a Chromium Chip E (10X Genomics) with targeted recovery of 7,000 cells. Library preparation with the Chromium Single-Cell ATAC Library and Gel Bead Kit was followed by quality control using a 2100 High-Sensitivity DNA Kit on Bioanalyzer (Agilent) and pooling of libraries. Sequencing was performed on an Illumina NovaSeq 6000 S2 platform with 50-bp paired-end reads, 8 bp for index 1 and 16 bp for index 2. Raw sequencing reads were aligned against the mm10 reference genome and preprocessed using Cell Ranger ATAC version 1.2.0. followed by downstream analyses with the ArchR package (69). Reference chromatin profiles of BM and SP were downloaded as bam files from the Mouse sci-ATAC-seq Atlas (28) and realigned against mm10 using GATK (70), Bowtie2 (71), and Samtools (72). Following the ArchR manual, after the exclusion of low-quality cells (transcription start site enrichment <4, fragments per cell <1,000) and doublets, standard preprocessing was performed including annotation of cells based on canonical gene activity scores. Differential gene activity scores between B cells, CLL, and RS were detected using FDR < 0.01 and Log2FC ≥ 1.25. Differential accessibility of transcription factor motifs between CLL and RS was detected based on the cisbp motif set (73). Statistical testing was performed using a Wilcoxon rank sum test. The human scATAC-seq data were generated and analyzed as previously described (29).
WGS Data Generation and Analysis
Genome sequencing was performed on flow cytometry–sorted tumor cells from the SP of 5 del(13q)-CLL, 2 CLL/RS “5-plex,” 2 CLL/RS “6-plex,” and 8 RS cases and matched Cd11b+ cells from the BM, as germline control. One microgram genomic DNA (∼1–2 × 106 cells) was used for library preparation. Shearing was performed acoustically using a Covaris focused ultrasonicator, targeting 385-bp fragments. Following fragmentation, additional size selection was performed using an SPRI cleanup. DNA libraries were constructed using the KAPA Library Prep Kits (KAPA Biosystems) using palindromic forked adapters with unique 8 base index sequences embedded within the adapter (IDT). Following sample preparation, libraries were quantified using quantitative PCR (KAPA Biosystems), with probes specific to the ends of the adapters. On the basis of qPCR quantification, libraries were normalized to 2.2 nmol/L and pooled into 24 plexes. Sample pools were combined with NovaSeq Cluster Amp Reagents DPX1, DPX2, and DPX3 and loaded into single lanes of a NovaSeq 6000 S4 flowcell cell using the Hamilton Starlet Liquid Handling system. Cluster amplification and sequencing were performed on NovaSeq 6000 Instruments utilizing sequencing-by-synthesis kits to produce 151-bp paired-end reads. Sequencing data from the Illumina software were aligned to the GRCm38 reference sequence with bwa version 0.7.15. Aligned reads were then deduplicated, and base quality scores were recalibrated with GATK version 4.1.8.1. Somatic SNVs and InDels were primarily detected by the intersection of two callers: Mutect2 (bioRxiv: 2019.10.1101/861054), GATK and Strelka2 version 2.9.3 (74). Additional variants called by either Mutect2 or Strelka2 were also included if the variants were related to putative RS driver mutations. Candidate InDels were first called by Manta version 1.5.0 before being passed to Strelka2. Copy ratios over chromosomal segments were first computed with a GATK-based pipeline and somatic copy-number alterations (sCNA) were subsequently called with GISTIC version 2.0.23 (75). Recurrent arm-level aberrations were identified from these calls by requiring that the G-score of the primary aberration > 0.25, the G-score of the secondary aberration < 0.1, and the q-value of primary aberration < 0.35, and reported as amplifications (2), deletions (0), or copy neutral (−1). These filters respectively verify that the primary event is recurrent across samples and cells, appears more often than the secondary event, and is statistically significant. CNV plots were generated using average sample/normal B-cell copy ratio within a running window of 0.1 M base pairs.
Statistical Analyses
Statistical analyses were performed using GraphPad Prism 6, unless otherwise specified in Methods. Three- or four-group data sets were analyzed by ANOVA with Tukey correction for the multiplicity of testing; two-group comparisons were performed with two-sided Mann–Whitney (unpaired comparisons) or paired t test (paired comparisons). Pearson correlation was used to assess the correlation coefficient between two variables. Survival analyses were conducted using the log-rank test, with Bonferroni correction for multiple comparisons. For the longitudinal tumor burden assessments of peripheral bleeds, a logistic mixed-effects model (lme4 R package) was used to estimate average trajectories of peripheral longitudinal tumor burden in individual mice. For all the above, the significance threshold was set at P ≤ 0.05. CRISPR-seq, RNA-seq, WGS, and scATAC-seq statistical analyses are described within related paragraphs.
Data Availability
Murine RNA-seq and scATAC-seq data have been deposited at GEO with accession number GSE186137. Human scATAC-seq data have been deposited at GEO with accession number GSE163579. Human RNA-seq data have been deposited in dbGAP (accession number phs002458.v2.p1, 5 paired CLL and RS from ref. 13) and in EGA under accession number EGAD00001007922 (36 clonally related RS and 28 de novo DLBCLs from ref. 16), and will be available at the time of publication. All other data will be available from the corresponding author upon reasonable request.
Supplementary Material
Supplementary file contains all supplementary tables S1-S16 in Excel format in the order in which they appear in the text.
Supplementary file contains a PDF version of supplementary figures S1-S7 and associated figure legends.
Acknowledgments
The authors thank the current and former members of the Wu lab, in particular Cynthia Hahn, Satyen Gohil, and Jacqueline Ho, for valuable discussions and contributions. The authors are also thankful to Anthony Letai and Salma Parvin for valuable input on the drug treatment studies. The authors are also grateful to the Dana-Farber Cancer Institute Animal Research Facility technical team for excellent technical assistance with in vivo treatment studies, Mei Zheng from the BWH Pathology core for assistance with mouse IHC, and the DFCI Flow Cytometry Core for assistance with flow sorting. This study was supported by grants from the NIH/NCI (P01 CA206978 and R01CA216273). E. ten Hacken is a scholar of the American Society of Hematology. S. Yin was supported by a research fellowship from the Lauri Strauss Leukemia Foundation and is currently supported by NIH/NCI R21CA267527-01. K. Clement is supported by NHGRI K99HG011658. L. Penter was supported by a research fellowship from the German Research Foundation (DFG, PE 3127/1-1) and is a current scholar of the American Society of Hematology. M. Gruber was supported by a EU Marie Curie IOF (PIOF-2013-624924). E.M. Parry is supported by a Doris Duke Physician-Scientist Fellowship, a DFCI Flames Flair Grant and an ASCO Conquer Cancer Young Investigator Award. M. Hernandez-Sanchez is supported by a Sara Borrell postdoctoral contract (CD19/00222) from Instituto de Salud Carlos III (ISCIII), cofunded by Fondo Social Europeo (FSE) “El Fondo Social Europeo invierte en tu futuro.” B.A. Knisbacher was supported by a long-term EMBO fellowship (ALTF 14-2018). S. Li is supported by the NCI Research Specialist Award (R50CA251956). S.S. and E.T. were supported by a grant from the DFG (SFB1074, B1). M.S. Davids is a scholar in Clinical Research of the Leukemia and Lymphoma Society.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
E. ten Hacken reports grants from the American Society of Hematology. S. Yin reports grants from the Lauri Strauss Leukemia Foundation and the NIH/NCI. M. Gruber reports grants from the European Union, Marie Curie IOF during the conduct of the study. K. Clement reports personal fees from Edilytics, Inc. outside the submitted work; in addition, K. Clement has a patent for US20220028491A1 pending, licensed, and with royalties paid from General Hospital Corp. N. Ruthen reports grants from NIH during the conduct of the study. E.M. Parry reports grants from ASCO Conquer Cancer Foundation, DFCI Flames Flair Grant and a DDCF physician scientist fellowship during the conduct of the study; in addition, E.M. Parry has a patent for U.S. provisional patent application serial number 63/244,625, filed on September 15, 2021, and U.S. provisional patent application serial number 63/291,213, filed on December 17, 2021, both of which are entitled “Diagnosis and Prognosis of Richter's Syndrome” pending (to E. Parry, G. Getz and C.J. Wu). J. Broseus reports personal fees and nonfinancial support from AstraZeneca and nonfinancial support from AbbVie outside the submitted work. R. Guieze reports other support from AbbVie, Janssen, AstraZeneca, Amgen, Roche, and other support from BeiGene outside the submitted work. B.A. Knisbacher reports a patent for compositions, panels, and methods for characterizing chronic lymphocytic leukemia (US 63/063,798) pending to Broad Institute and a patent for methods for treatment selection for chronic lymphocytic leukemia (US 63/408,452) pending to Broad Institute. S. Li reports grants from NIH/NCI during the conduct of the study. P. Feugier reports grants, personal fees, and nonfinancial support from Janssen and AbbVie personal fees and nonfinancial support from Gilead, Astra, BeiGene, and personal fees and nonfinancial support from Loxo outside the submitted work. E. Tausch reports grants, personal fees, and nonfinancial support from Roche and AbbVie; personal fees and nonfinancial support from BeiGene and Janssen-Cilag, personal fees from AstraZeneca outside the submitted work. M.S. Davids reports personal fees from AbbVie and BeiGene; grants and personal fees from Ascentage Pharma, AstraZeneca, Secura Bio, TG Therapeutics, MEI Pharma, Verastem and Genentech, BMS, Eli Lilly, Janssen, Merck, Novartis, Ono Pharmaceuticals, and Takeda; and grants from Surface Oncology outside the submitted work. G. Getz reports personal fees from Scorpion Therapeutics and grants from IBM and Pharmacyclics outside the submitted work; in addition, G. Getz is an inventor on patent applications related to MSMuTect, MSMutSig, MSIDetect, Polysolver, and SignatureAnalyzer-GPU. K.J. Livak reports grants from NIH/NCI, DoD/USAMRAA, NIH/NIAID, and FNIH during the conduct of the study; and other support from Standard BioTools Inc. outside the submitted work. I. Bozic reports grants from Johnson & Johnson outside the submitted work. C.J. Wu reports grants from NIH/NCI (P01 CA206978 and R01CA216273) during the conduct of the study; and other support from BioNTech and Pharmacyclics outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
E. ten Hacken: Conceptualization, data curation, formal analysis, funding acquisition, investigation, visualization, methodology, writing–original draft, writing–review and editing. T. Sewastianik: Formal analysis. S. Yin: Formal analysis. G. Brunsting Hoffmann: Data curation. M. Gruber: Conceptualization, data curation. K. Clement: Formal analysis. L. Penter: Formal analysis. R.A. Redd: Formal analysis. N. Ruthen: Formal analysis. S. Hergalant: Resources, formal analysis. A. Sholokhova: Formal analysis. G. Fell: Formal analysis. E.M. Parry: Resources. J. Broséus: Resources. R. Guieze: Resources. F. Lucas: Resources. M. Hernández-Sánchez: Methodology. K. Baranowski: Data curation. J. Southard: Data curation. H. Joyal: Data curation. L. Billington: Data curation. F.F.D. Regis: Data curation. E. Witten: Data curation. M. Uduman: Formal analysis. B.A. Knisbacher: Formal analysis. S. Li: Data curation. H. Lyu: Data curation. T. Vaisitti: Resources. S. Deaglio: Resources. G. Inghirami: Resources. P. Feugier: Resources. S. Stilgenbauer: Resources. E. Tausch: Resources. M.S. Davids: Resources. G. Getz: Methodology. K.J. Livak: Methodology. I. Bozic: Formal analysis. D.S. Neuberg: Formal analysis, methodology. R.D. Carrasco: Data curation. C.J. Wu: Conceptualization, supervision, funding acquisition, writing–original draft, writing–review and editing.
References
- 1. Ten Hacken E, Wu CJ. Understanding CLL biology through mouse models of human genetics. Blood 2021;138:2621–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 2010;17:28–40. [DOI] [PubMed] [Google Scholar]
- 3. Lazarian G, Yin S, Ten Hacken E, Sewastianik T, Uduman M, Font-Tello A, et al. A hotspot mutation in transcription factor IKZF3 drives B cell neoplasia via transcriptional dysregulation. Cancer Cell 2021;39:380–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yin S, Gambe RG, Sun J, Martinez AZ, Cartun ZJ, Regis FFD, et al. A murine model of chronic lymphocytic leukemia based on B Cell-restricted expression of Sf3b1 mutation and Atm deletion. Cancer Cell 2019;35:283–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Allan JN, Furman RR. Current trends in the management of Richter's syndrome. Int J Hematol Oncol 2018;7:IJH09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gruber M, Bozic I, Leshchiner I, Livitz D, Stevenson K, Rassenti L, et al. Growth dynamics in naturally progressing chronic lymphocytic leukaemia. Nature 2019;570:474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Knisbacher BA, Lin Z, Hahn CK, Nadeu F, Duran-Ferrer M, Stevenson KE, et al. Molecular map of chronic lymphocytic leukemia and its impact on outcome. Nat Genet 2022;54:1664–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations driving CLL and their evolution in progression and relapse. Nature 2015;526:525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014;159:440–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tandon B, Peterson L, Gao J, Nelson B, Ma S, Rosen S, et al. Nuclear overexpression of lymphoid-enhancer-binding factor 1 identifies chronic lymphocytic leukemia/small lymphocytic lymphoma in small B-cell lymphomas. Mod Pathol 2011;24:1433–43. [DOI] [PubMed] [Google Scholar]
- 11. Rossi D, Spina V, Deambrogi C, Rasi S, Laurenti L, Stamatopoulos K, et al. The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood 2011;117:3391–401. [DOI] [PubMed] [Google Scholar]
- 12. Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 2019;37:224–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parry E, Leshchiner I, Guieze R, Johnson C, Tausch E, Parikh S, et al. Evolutionary history of transformation from chronic lymphocytic leukemia to Richter syndrome. Nat Med 2023;29:158–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ten Hacken E, Clement K, Li S, Hernandez-Sanchez M, Redd R, Wang S, et al. High throughput single-cell detection of multiplex CRISPR-edited gene modifications. Genome Biol 2020;21:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nadeu F, Royo R, Massoni-Badosa R, Playa-Albinyana H, Garcia-Torre B, Duran-Ferrer M, et al. Detection of early seeding of Richter transformation in chronic lymphocytic leukemia. Nat Med 2022;28:1662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Broseus J, Hergalant S, Vogt J, Tausch E, Kreuz M, Mottok A, et al. Molecular characterization of Richter syndrome identifies de novo diffuse large B-cell lymphomas with poor prognosis. Nat Commun 2023;14:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Llabata P, Mitsuishi Y, Choi PS, Cai D, Francis JM, Torres-Diz M, et al. Multi-omics analysis identifies MGA as a negative regulator of the MYC pathway in lung adenocarcinoma. Mol Cancer Res 2020;18:574–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mathsyaraja H, Catchpole J, Freie B, Eastwood E, Babaeva E, Geuenich M, et al. Loss of MGA repression mediated by an atypical polycomb complex promotes tumor progression and invasiveness. eLife 2021;10:e64212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holmes AG, Parker JB, Sagar V, Truica MI, Soni PN, Han H, et al. A MYC inhibitor selectively alters the MYC and MAX cistromes and modulates the epigenomic landscape to regulate target gene expression. Sci Adv 2022;8:eabh3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bishop GA, Haughton G. Induced differentiation of a transformed clone of Ly-1+ B cells by clonal T cells and antigen. Proc Natl Acad Sci U S A 1986;83:7410–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li S, Wan C, Zheng R, Fan J, Dong X, Meyer CA, et al. Cistrome-GO: a web server for functional enrichment analysis of transcription factor ChIP-seq peaks. Nucleic Acids Res 2019;47:W206–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vaisitti T, Braggio E, Allan JN, Arruga F, Serra S, Zamo A, et al. Novel richter syndrome xenograft models to study genetic architecture, biology, and therapy responses. Cancer Res 2018;78:3413–20. [DOI] [PubMed] [Google Scholar]
- 23. Vaisitti T, Arruga F, Vitale N, Lee TT, Ko M, Chadburn A, et al. ROR1 targeting with the antibody drug-conjugate VLS-101 is effective in Richter syndrome patient-derived xenograft mouse models. Blood 2021;137:3365–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Olive V, Sabio E, Bennett MJ, De Jong CS, Biton A, McGann JC, et al. A component of the mir-17-92 polycistronic oncomir promotes oncogene-dependent apoptosis. eLife 2013;2:e00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 2008;9:405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Getahun A, Beavers NA, Larson SR, Shlomchik MJ, Cambier JC. Continuous inhibitory signaling by both SHP-1 and SHIP-1 pathways is required to maintain unresponsiveness of anergic B cells. J Exp Med 2016;213:751–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 2012;490:116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, et al. A Single-cell atlas of in vivo mammalian chromatin accessibility. Cell 2018;174:1309–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Penter L, Gohil SH, Lareau C, Ludwig LS, Parry EM, Huang T, et al. Longitudinal single-cell dynamics of chromatin accessibility and mitochondrial mutations in chronic lymphocytic leukemia mirror disease history. Cancer Discov 2021;11:3048–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med 2010;2:26ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006;439:353–7. [DOI] [PubMed] [Google Scholar]
- 32. Sanchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 2014;516:428–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 2014;514:380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dow LE, Fisher J, O'Rourke KP, Muley A, Kastenhuber ER, Livshits G, et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol 2015;33:390–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol 2014;32:941–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tothova Z, Krill-Burger JM, Popova KD, Landers CC, Sievers QL, Yudovich D, et al. Multiplex CRISPR/Cas9-based genome editing in human hematopoietic stem cells models clonal hematopoiesis and myeloid neoplasia. Cell Stem Cell 2017;21:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iacobucci I, Qu C, Varotto E, Janke LJ, Yang X, Seth A, et al. Modeling and targeting of erythroleukemia by hematopoietic genome editing. Blood 2021;137:1628–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci U S A 2002;99:6955–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Knittel G, Rehkamper T, Korovkina D, Liedgens P, Fritz C, Torgovnick A, et al. Two mouse models reveal an actionable PARP1 dependence in aggressive chronic lymphocytic leukemia. Nat Commun 2017;8:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kohlhaas V, Blakemore SJ, Al-Maarri M, Nickel N, Pal M, Roth A, et al. Active Akt signaling triggers CLL toward Richter transformation via overactivation of Notch1. Blood 2021;137:646–60. [DOI] [PubMed] [Google Scholar]
- 41. Chakraborty S, Martines C, Porro F, Fortunati I, Bonato A, Dimishkovska M, et al. B Cell Receptor signaling and genetic lesions in TP53 and CDKN2A/CDKN2B cooperate in Richter transformation. Blood 2021;138:1053–66. [DOI] [PubMed] [Google Scholar]
- 42. Chigrinova E, Rinaldi A, Kwee I, Rossi D, Rancoita PM, Strefford JC, et al. Two main genetic pathways lead to the transformation of chronic lymphocytic leukemia to Richter syndrome. Blood 2013;122:2673–82. [DOI] [PubMed] [Google Scholar]
- 43. Klintman J, Appleby N, Stamatopoulos B, Ridout K, Eyre TA, Robbe P, et al. Genomic and transcriptomic correlates of Richter's transformation in chronic lymphocytic leukemia. Blood 2020;137:2800–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fabbri G, Khiabanian H, Holmes AB, Wang J, Messina M, Mullighan CG, et al. Genetic lesions associated with chronic lymphocytic leukemia transformation to Richter syndrome. J Exp Med 2013;210:2273–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hurlin PJ, Steingrimsson E, Copeland NG, Jenkins NA, Eisenman RN. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J 1999;18:7019–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol 2015;15:137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sander S, Calado DP, Srinivasan L, Kochert K, Zhang B, Rosolowski M, et al. Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell 2012;22:167–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wilke AC, Doebele C, Zindel A, Lee KS, Rieke SA, Ceribelli M, et al. SHMT2 inhibition disrupts the TCF3 transcriptional survival program in Burkitt lymphoma. Blood 2022;139:538–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rodriguez D, Bretones G, Quesada V, Villamor N, Arango JR, Lopez-Guillermo A, et al. Mutations in CHD2 cause defective association with active chromatin in chronic lymphocytic leukemia. Blood 2015;126:195–202. [DOI] [PubMed] [Google Scholar]
- 50. Duhren-von Minden M, Ubelhart R, Schneider D, Wossning T, Bach MP, Buchner M, et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature 2012;489:309–12. [DOI] [PubMed] [Google Scholar]
- 51. Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med 2011;208:1389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ryan RJH, Petrovic J, Rausch DM, Zhou Y, Lareau CA, Kluk MJ, et al. A B cell regulome links notch to downstream oncogenic pathways in small B cell lymphomas. Cell Rep 2017;21:784–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Crombie J, Tyekucheva S, Wang Z, Savell A, Brennan L, Lowney J, et al. Updated results from a phase I/II study of duvelisib and venetoclax in patients with relapsed and refractory CLL/SLL or Richter's syndrome. Blood 2020;136:46–7. [Google Scholar]
- 54. Hanna BS, McClanahan F, Yazdanparast H, Zaborsky N, Kalter V, Rossner PM, et al. Depletion of CLL-associated patrolling monocytes and macrophages controls disease development and repairs immune dysfunction in vivo. Leukemia 2016;30:570–9. [DOI] [PubMed] [Google Scholar]
- 55. Grioni M, Brevi A, Cattaneo E, Rovida A, Bordini J, Bertilaccio MTS, et al. CD4+ T cells sustain aggressive chronic lymphocytic leukemia in Emu-TCL1 mice through a CD40L-independent mechanism. Blood Adv 2021;5:2817–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ten Hacken E, Valentin R, Regis FFD, Sun J, Yin S, Werner L, et al. Splicing modulation sensitizes chronic lymphocytic leukemia cells to venetoclax by remodeling mitochondrial apoptotic dependencies. JCI Insight 2018;3:e121438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rossi D, Spina V, Gaidano G. Biology and treatment of Richter syndrome. Blood 2018;131:2761–72. [DOI] [PubMed] [Google Scholar]
- 58. Campo E, Swerdlow SH, Harris NL, Pileri S, Stein H, Jaffe ES. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood 2011;117:5019–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ghia P, Stamatopoulos K, Belessi C, Moreno C, Stilgenbauer S, Stevenson F, et al. ERIC recommendations on IGHV gene mutational status analysis in chronic lymphocytic leukemia. Leukemia 2007;21:1–3. [DOI] [PubMed] [Google Scholar]
- 60. Lane HA, Wood JM, McSheehy PM, Allegrini PR, Boulay A, Brueggen J, et al. mTOR inhibitor RAD001 (everolimus) has antiangiogenic/vascular properties distinct from a VEGFR tyrosine kinase inhibitor. Clin Cancer Res 2009;15:1612–22. [DOI] [PubMed] [Google Scholar]
- 61. Ott CJ, Federation AJ, Schwartz LS, Kasar S, Klitgaard JL, Lenci R, et al. Enhancer architecture and essential core regulatory circuitry of chronic lymphocytic leukemia. Cancer Cell 2018;34:982–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kurokawa C, Geekiyanage H, Allen C, Iankov I, Schroeder M, Carlson B, et al. Alisertib demonstrates significant antitumor activity in bevacizumab resistant, patient derived orthotopic models of glioblastoma. J Neurooncol 2017;131:41–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vijayaraghavan S, Karakas C, Doostan I, Chen X, Bui T, Yi M, et al. CDK4/6 and autophagy inhibitors synergistically induce senescence in Rb positive cytoplasmic cyclin E negative cancers. Nat Commun 2017;8:15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Horwitz SM, Koch R, Porcu P, Oki Y, Moskowitz A, Perez M, et al. Activity of the PI3K-delta, gamma inhibitor duvelisib in a phase 1 trial and preclinical models of T-cell lymphoma. Blood 2018;131:888–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li S, Sun J, Allesoe R, Datta K, Bao Y, Oliveira G, et al. RNase H-dependent PCR-enabled T-cell receptor sequencing for highly specific and efficient targeted sequencing of T-cell receptor mRNA for single-cell and repertoire analysis. Nat Protoc 2019;14:2571–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Granja JM, Corces MR, Pierce SE, Bagdatli ST, Choudhry H, Chang HY, et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat Genet 2021;53:403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 2017;14:975–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kim S, Scheffler K, Halpern AL, Bekritsky MA, Noh E, Kallberg M, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods 2018;15:591–4. [DOI] [PubMed] [Google Scholar]
- 75. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file contains all supplementary tables S1-S16 in Excel format in the order in which they appear in the text.
Supplementary file contains a PDF version of supplementary figures S1-S7 and associated figure legends.
Data Availability Statement
Murine RNA-seq and scATAC-seq data have been deposited at GEO with accession number GSE186137. Human scATAC-seq data have been deposited at GEO with accession number GSE163579. Human RNA-seq data have been deposited in dbGAP (accession number phs002458.v2.p1, 5 paired CLL and RS from ref. 13) and in EGA under accession number EGAD00001007922 (36 clonally related RS and 28 de novo DLBCLs from ref. 16), and will be available at the time of publication. All other data will be available from the corresponding author upon reasonable request.



![Figure 1. Multiplexed CRISPR editing of CLL-LOF lesions generates chronic lymphocytic leukemia disease models. A, Schema for the generation of transplant lines. BM donor lines containing homozygous del(13q) and B-cell–restricted Cas9-GFP [i.e., del(13q)-Cd19Cas9-GFP] were obtained by intercrossing del(13q)-Cd19cre/cre and del(13q)-Cas9/Cas9 mice. LSKs isolated from 8- to 12-week-old del(13q)-Cd19Cas9 animals were transduced in vitro with a pool of lentivirus expressing sgRNAs against the 6 or 5 LOF mutations of interest (Atm, Mga, Samhd1, Chd2, Birc3, Trp53 in “6-plex”; Trp53 absent in “5-plex”) or corresponding nontargeting pools of scrambled sgRNAs and transplanted into 8- to 12-week-old immune-competent (CD45.1) or immune-deficient (NSG) recipients (n = 30–35/group). B, Single-cell qPCR-based analysis of sgRNA expression in LSK from the 3 targeting and the 3 nontargeting cohorts, as assessed at 72 hours after transduction and in vitro culture. The number of sgRNAs quantified in individual single cells is displayed. C, Percent (%) animals with circulating CLL in targeting and nontargeting cohorts. P, Fisher exact test. D, Longitudinal tumor burden assessments by flow-cytometric analysis of peripheral bleeds. A logistic mixed-effects model was used to estimate the average trajectories of peripheral longitudinal tumor burden (%GFP+mCh+B220+CD5+Igκ+ cells) in individual mice from the targeting (6-plex: dark brown; 5-plex: salmon) and nontargeting cohorts (gray). Disease onset and survival (median and range) for each cohort are indicated.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/b1d4/9975769/aeaaca96d749/150fig1.jpg)
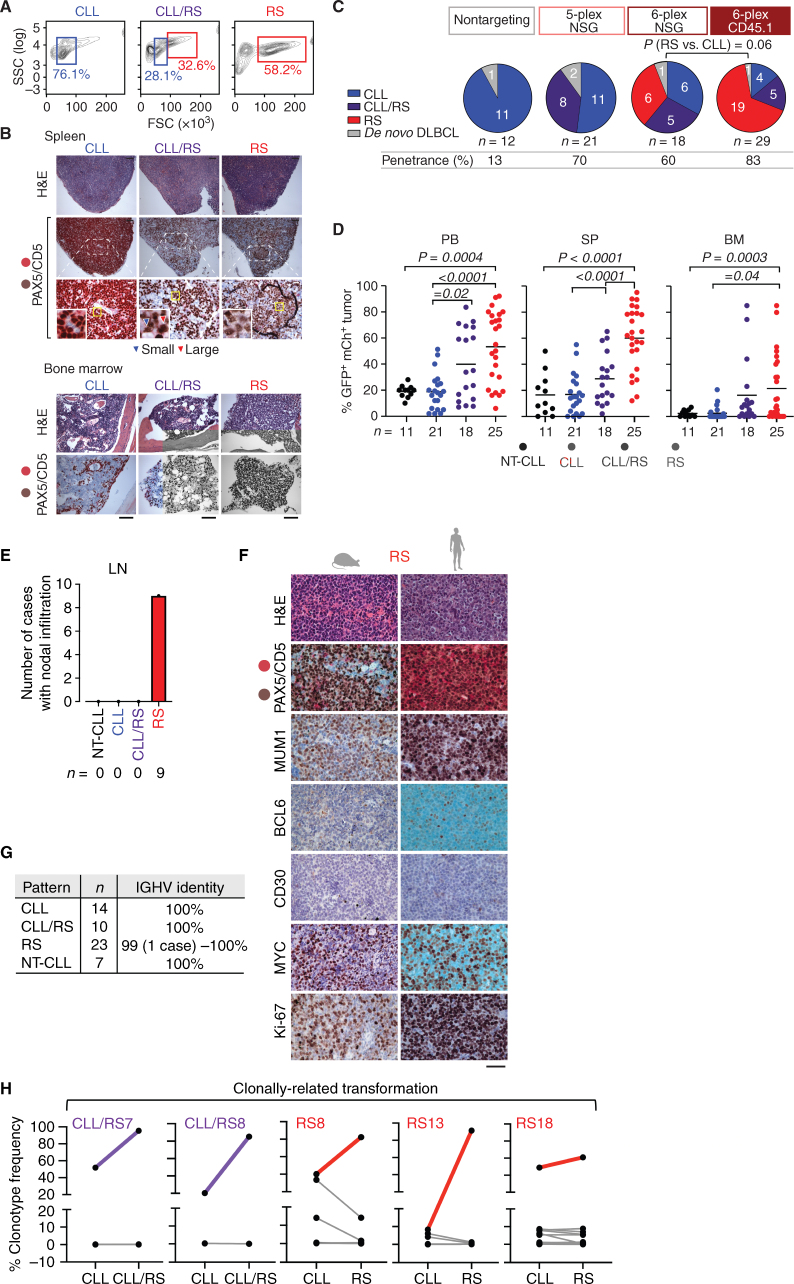
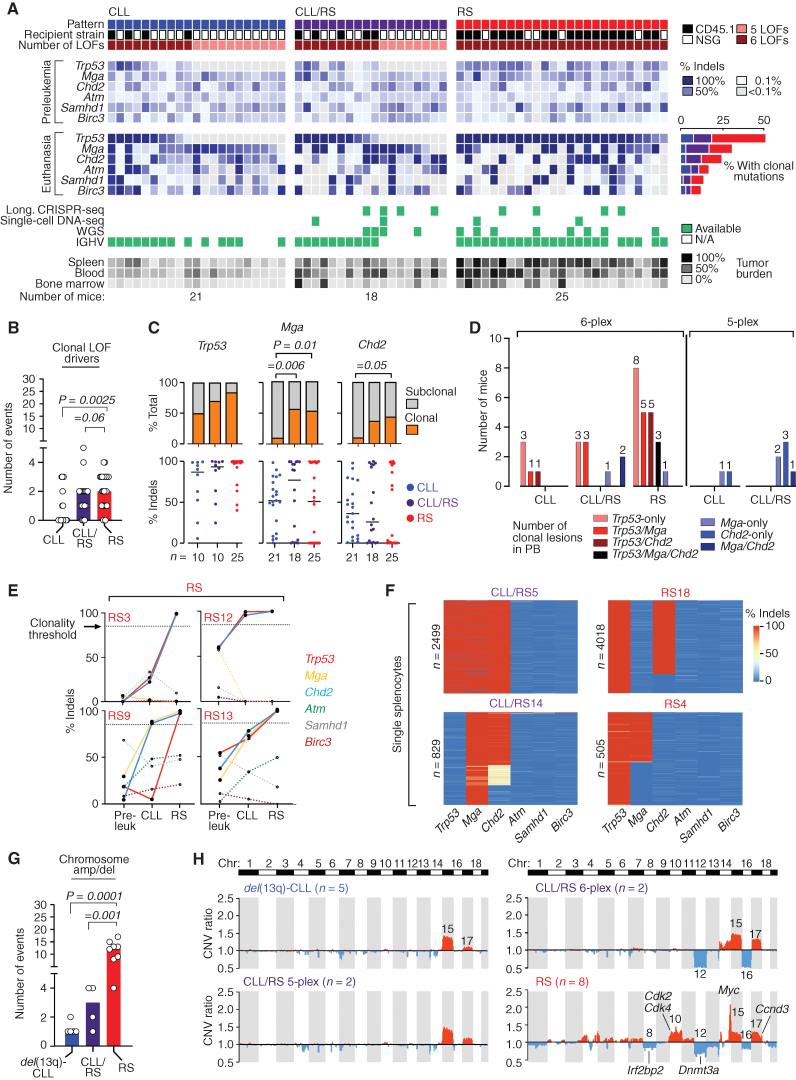
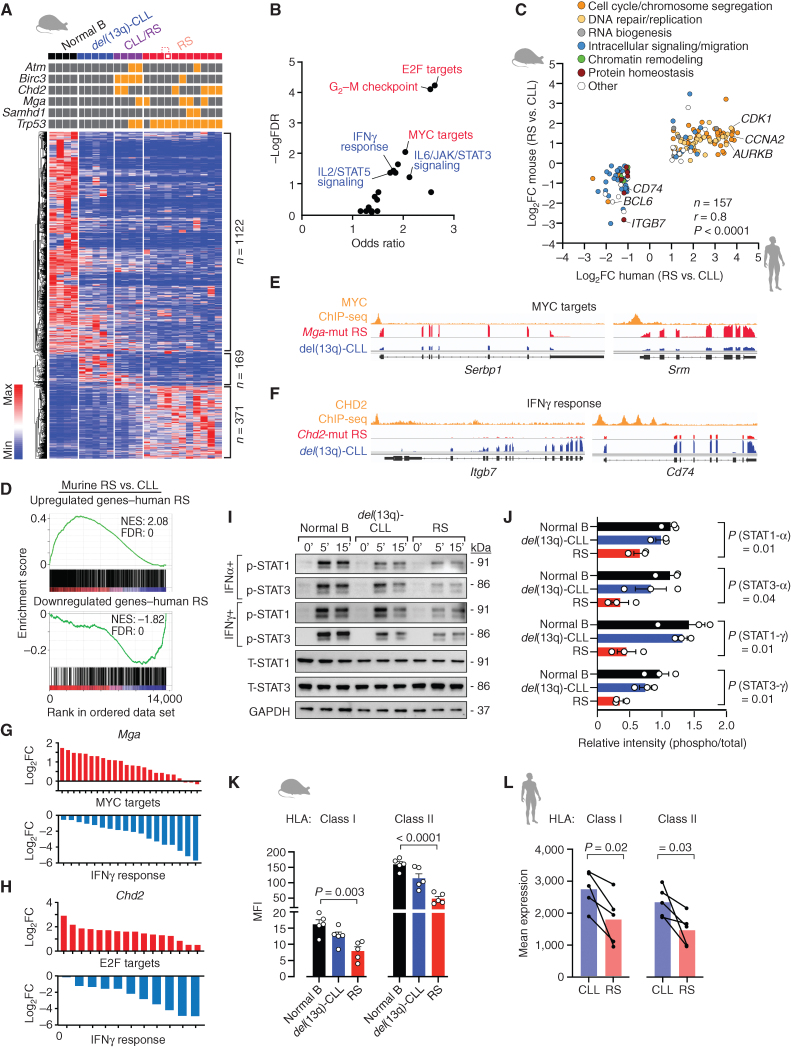
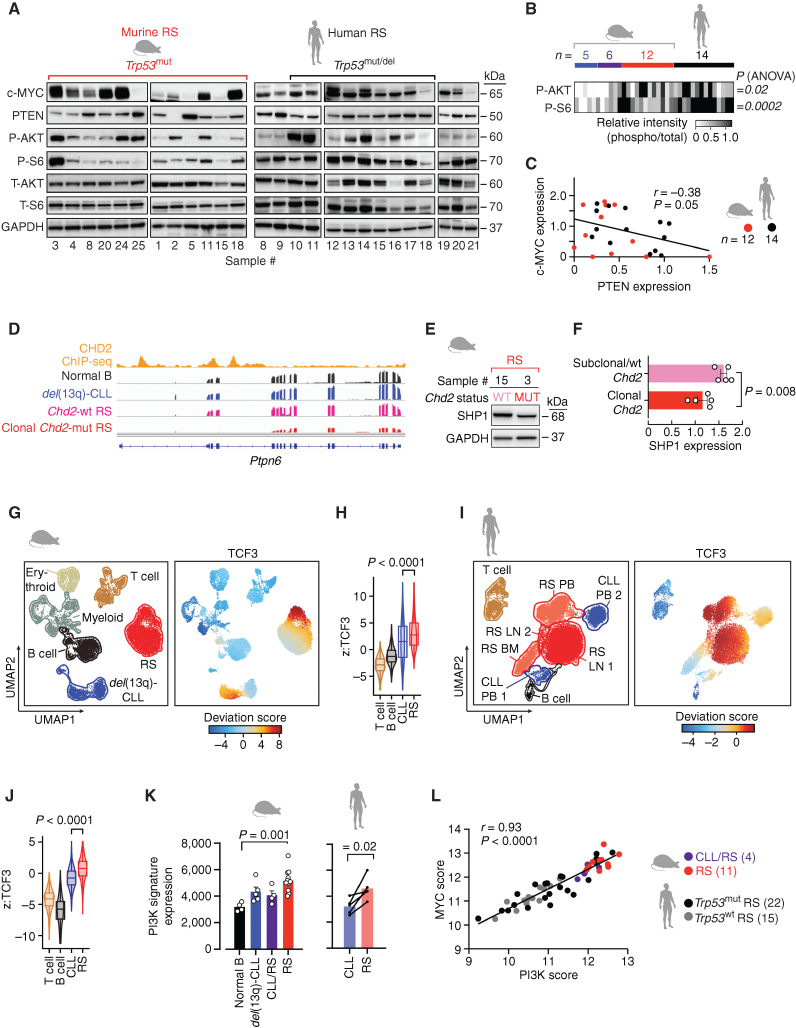
![Figure 6. MYC/mTOR/PI3K signaling are targetable vulnerabilities of RS. A, Summary heat map of drug treatment studies of murine primary splenocytes [7 del(13q)-CLL, 3 CLL/RS, and 12 RS] and 5 human RS cases. Percent (%) viability compared with the DMSO control is displayed. B, Bar plots showing percent (%) viability after JQ1, everolimus, duvelisib, palbociclib, and alisertib treatment. Mean with SEM is displayed. P, ANOVA. C, Correlation between response to duvelisib (% viability) and phospho-AKT levels, as detected by immunoblot in 5 del(13q)-CLL, 2 CLL/RS, 8 murine RS, and 14 human RS cases. P, Pearson correlation. D, Correlation between response to everolimus (% viability) and phospho-S6 levels on the same set of samples per C, as detected by immunoblot. P, Pearson correlation. E, Immunoblot assessment of c-MYC expression levels following a 12-hour treatment with single-agent duvelisib, everolimus, JQ1 or respective duvelisib-based combinations. One representative mouse and human samples are shown and GAPDH is used as loading control. F, Percent (%) viability normalized to DMSO control of 4 murine and 3 human RS primary specimens treated for 40 hours with single-agent duvelisib, everolimus, JQ1, or respective duvelisib-based combinations. Mean with SEM is displayed. P value, ANOVA with Tukey correction for multiple comparisons. G, Transplant schema for combination treatment studies and survival curve of NSG mice transplanted with RS splenocytes and then treated with 50 mg/kg duvelisib, 10 mg/kg everolimus, or combination (left), or 50 mg/kg duvelisib, 50 mg/kg JQ1, or combination (right) for 2 weeks followed by observation until survival endpoints. P values, log-rank test with Bonferroni correction for multiple comparisons. The pink shaded area indicates the treatment period.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/b1d4/9975769/27ef79f3802c/150fig6.jpg)