Triple-vaccinated myeloma patients lack CD8 responses and display suboptimal antibody responses, particularly against newer SARS-CoV-2 variants. Live-virus neutralization activity, rather than antibody titers, correlate with breakthrough infections.
Abstract
Patients with multiple myeloma (MM) mount suboptimal neutralizing antibodies (nAb) following 2 doses of SARS-CoV-2 mRNA vaccines. Currently, circulating SARS-CoV-2 variants of concern (VOC) carry the risk of breakthrough infections. We evaluated immune recognition of current VOC including BA.1, BA.2, and BA.5 in 331 racially representative patients with MM following 2 or 3 doses of mRNA vaccines. The third dose increased nAbs against WA1 in 82%, but against BA variants in only 33% to 44% of patients. Vaccine-induced nAbs correlated with receptor-binding domain (RBD)–specific class-switched memory B cells. Vaccine-induced spike-specific T cells were detected in patients without seroconversion and cross-recognized variant-specific peptides but were predominantly CD4+ T cells. Detailed clinical/immunophenotypic analysis identified features correlating with nAb/B/T-cell responses. Patients who developed breakthrough infections following 3 vaccine doses had lower live-virus nAbs, including against VOC. Patients with MM remain susceptible to SARS-CoV-2 variants following 3 vaccine doses and should be prioritized for emerging approaches to elicit variant-nAb and CD8+ T cells.
Significance:
Three doses of SARS-CoV-2 mRNA vaccines fail to yield detectable VOC nAbs in nearly 60% and spike-specific CD8+ T cells in >80% of myeloma patients. Patients who develop breakthrough infections following vaccination have low levels of live-virus nAb.
This article is highlighted in the In This Issue feature, p. 101
INTRODUCTION
Preclinical studies have demonstrated an important role for neutralizing antibodies (nAb) and CD8+ T cells in protective immunity against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2; refs. 1, 2). Patients with hematologic malignancies, including multiple myeloma (MM), exhibit increased mortality following SARS-CoV-2 infection (3, 4). Although initial studies described >80% rates of spike/receptor-binding domain (RBD)–specific antibodies following mRNA vaccination in patients with MM (3, 5), studies evaluating the presence of nAbs, either using live virus (6) or surrogate assays (7–10), demonstrated suboptimal induction of nAbs following the first 2 doses of mRNA vaccines. The dominant circulating strains in the SARS-CoV-2 pandemic have continued to evolve, with recent Omicron (BA) variants of concern (VOC) acquiring several RBD mutations (11). A third dose of the mRNA vaccines yields higher nAb titers, including against several VOC (12, 13), as well as cellular responses in healthy individuals (14–17). Booster vaccines also led to enhanced induction of nAbs in MM, although vaccine-mediated protection against current circulating SARS-CoV-2 variants (such as BA.5) in patients with MM is not known (18, 19). Due to the small numbers of patients studied to date for cellular responses, clinical or immunologic variables that affect such responses in MM also remain underexplored (18, 19). Black populations carry an increased risk of developing MM, but prior studies also have an underrepresentation of Black patients (8, 18, 19). In this study, we have analyzed blood specimens from a diverse cohort of patients with MM after second or third vaccine dose, to evaluate virus/VOC-specific nAb and B/T cellular responses combined with high-dimensional immunophenotyping (Supplementary Fig. S1).
RESULTS
Dose 3 led to higher titers of RBD-specific antibodies (Supplementary Fig. S2), as well as nAb detected by pseudovirus neutralization (Fig. 1A). Environmental exposure of SARS-CoV-2 infection in this cohort was monitored by measuring nucleocapsid (NC) antibodies (Supplementary Fig. S3), and NC Ab+ patients were analyzed separately to distinguish between vaccine-induced and hybrid immunity. Dose 3 led to higher RBD-specific antibodies (Supplementary Fig. S4) as well as nAbs (Fig. 1B) in both NCAb+ and NCAb− cohorts. Subanalysis of patients with paired samples is shown in Supplementary Figs. S5 and S6. These antibodies were detectable up to 4 months following each dose of the vaccines (Supplementary Fig. S7A and S7B).
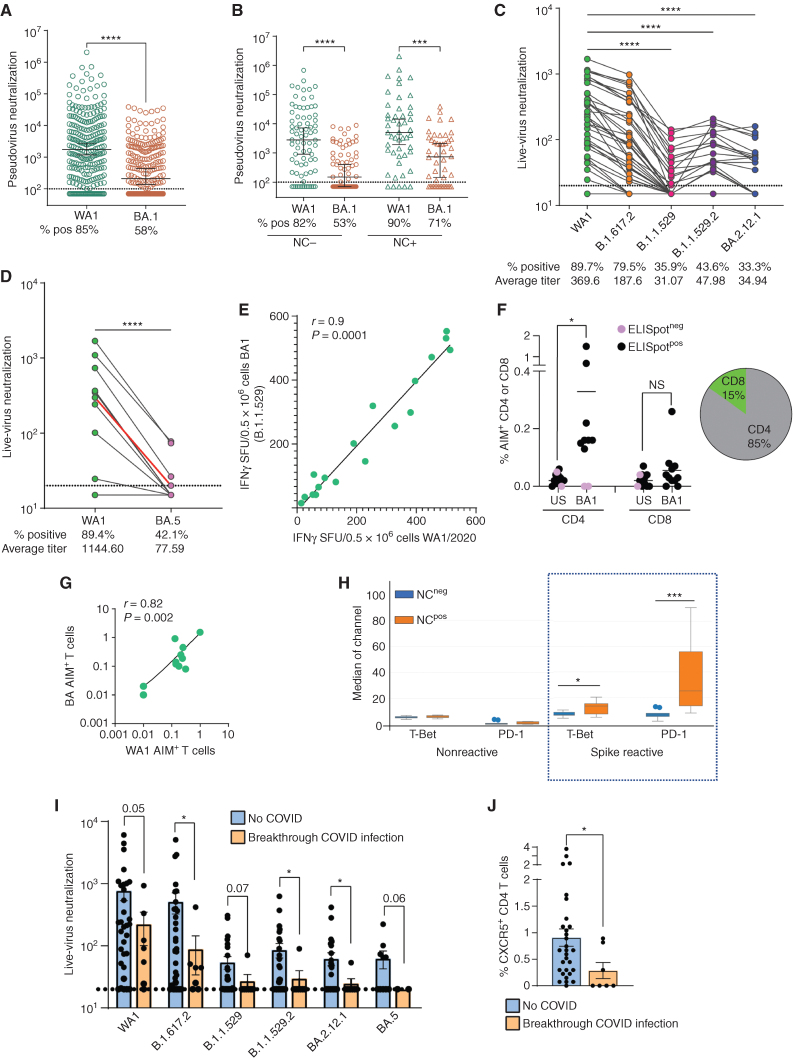
Figure 1.
Vaccine-induced nAb, B- and T-cell responses to WA1 SARS-CoV-2. Blood was collected from myeloma patients 1 week to 3 months following dose 2 and dose 3 of the SARS-CoV-2 mRNA vaccine. Plasma was analyzed to determine serologic responses to WA1 SARS-CoV-2 using RBD-specific endpoint titer and pseudovirus neutralization assay. Peripheral blood mononuclear cells were examined for the presence of RBD-specific B cells using flow cytometry and single-cell mass cytometry. T-cell response directed against WA1 spike protein was detected using the IFNγ-ELISpot assay as well as antigen-induced marker expression (AIM) assay using cytometry by time of flight (CyTOF). In some patients, immunosequencing of the CDR3 regions of human TCRβ chains was performed using the ImmunoSEQ T-Detect Assay (Adaptive Biotechnologies) to identify SARS-CoV-2–specific T cells. A, Pseudovirus neutralization IC50 following 2 (D2, n = 342) or 3 doses (D3, n = 253). Data, median with a 95% confidence interval (****, P < 0.0001, Mann–Whitney test). B, Pseudovirus neutralization IC50 following 2 (D2) or 3 doses (D3), based on nucleocapsid (NC) Ab reactivity. NC− (D2: n = 269, D3: n = 78), NC+ (D2: n = 73, D3: n = 49). Data, median with a 95% confidence interval (**, P < 0.01; ****, P < 0.0001, Kruskal–Wallis). C, RBD-specific B cells as % of all B cells following dose 2 (D2: n = 107) or dose 3 (D3: n = 60). Figures show median with 95% confidence interval (*, P < 0.05, Mann–Whitney test). D and E, Correlation between RBD-specific B cells and RBD-specific endpoint titer (last dilution for positive assay; D) and pseudovirus neutralization (E). F, RBD-specific IgG+ B cells between dose 2 and dose 3 in patients with detectable RBD-specific B cells. Figures show median with 95% confidence interval (*, P < 0.05; Mann–Whitney test). G, CyTOF was performed to examine RBD-specific B-cell response. Hierarchical consensus clustering was performed on RBD-specific B cells from healthy control (HC, n = 7) as well as patients following 2 or 3 doses of the SARS-CoV-2 vaccine (n = 26 and n = 19, respectively). The figure shows FlowSOM map for all samples, as well as a heat map of markers expressed by the four B-cell metaclusters (MC; MC1, MC2, MC3, and MC4). The bar graph shows the proportion of RBD+ cells in individual metaclusters. H–J, WA1 spike-specific T cells detected by interferon-γ ELISpot assay. H, IFNγ ELISPOT assay in the entire cohort (dose 2: n = 130, dose 3: n = 60) by dose. I, ELISpot assay split by nucleocapsid reactivity [NC-D2 (n = 100), NC-D3 (n = 35), NC + D2 (n = 22), NC + D3 (n = 25)]. J, ELISpot assay by serum RBD reactivity (seropositive (RBD+ n = 150) and seronegative (RBD− n = 32) nucleocapsid antibody-negative patients. K, Detection of WA1 spike-specific T cells. Graph shows AIM+ CD4 and CD8 T cells in patients with detectable spike-specific IFNγ-specific T cells by the ELISpot assay (n = 9) as well as patients who did not have detectable spike-specific IFNγ-specific T cells by ELISPOT assay (n = 2). Pie chart shows the mean proportions of spike-specific CD4+ and CD8+ T cells for 9 patients with detectable spike-reactive T cells. US = unstimulated control. *, P < 0.05; #, P = 0.05, paired t test. L, Detection of SARS-CoV-2–specific T cells by Adaptive Biotechnologies T-Detect COVID assay. Note that increases in surface glycoprotein-reactive TCRs as assessed by COVID T-cell breadth are mostly seen for CD4+ TCRs.
The presence of RBD-specific B cells was analyzed by flow and mass cytometry (Supplementary Fig. S8) and correlated with each other (Supplementary Fig. S9). The proportion of RBD-specific B cells was higher following dose 3 compared with dose 2 (Fig. 1C). Subanalysis of RBD-specific B cells in patients with paired Dose2/Dose3 samples is shown in Supplementary Fig. S10. RBD-specific B cells correlated with levels of anti-RBD antibodies (Fig. 1D), as well as nAbs (Fig. 1E). Dose 3 also led to a higher proportion of class-switched (IgG+) RBD-specific B cells (Fig. 1F). FlowSOM analysis revealed that RBD-specific B cells following dose 2 exhibited more IgM+ phenotypes, whereas those following dose 3 were similar to healthy donors with a dominance of class-switched IgM−CD27+ memory B cells (metacluster 4; Fig. 1G). Of note, RBD-specific B cells in patients with MM also had a higher proportion of cells with T-bet+CD11c+ phenotype (Fig. 1G), implicated in aging and extrafollicular responses (20). These phenotypic differences are specific for RBD+ B cells, as the distribution of B cell subsets in these cohorts was similar (Supplementary Fig. S11A–S11C).
Vaccine-induced T-cell responses were measured with 3 orthogonal methods [interferon-γ ELISpot, activation-induced marker (AIM) assay and T-cell receptor (TCR) sequencing], which yielded corroborative data. Spike-specific T cells were detected by ELISpot following both dose 2 and dose 3 (Fig. 1H), with the highest levels following dose 3 in NC+ patients, consistent with hybrid immunity (Fig. 1I). T-cell responses were higher in seropositive patients but were also detected in seronegative patients (Fig. 1J). There was no correlation between ELISpot and nAb titers (Supplementary Fig. S12). AIM expression assay demonstrated that most of the spike-specific T cells were predominantly CD4+ T cells and correlated with ELISpot reactivity (Fig. 1K). Vaccine-induced spike-specific CD8+ T cells were not detectable above background in patients with MM, although such responses are detectable with these assays in healthy donors, consistent with prior studies (ref. 21; Supplementary Fig. S13). Analysis of T-cell responses by Adaptive Biotechnologies T-Detect COVID assay revealed a selective increase in TCRs against surface glycoprotein relative to other viral proteins, consistent with the vaccine effect (Supplementary Fig. S14). As with the AIM assay, T-Detect analysis also suggested the dominance of CD4+ T-cell responses (Fig. 1L). The phenotype of AIM+ CD4+ T cells was consistent with effector T cells, similar to that seen in other studies (Supplementary Fig. S15; ref. 21).
In order to better understand the efficacy of the booster vaccines to mediate protection against current circulating variants, we next analyzed the induction of nAbs and T-cell responses against VOC. Vaccine-induced nAbs exhibited a significantly lower capacity to neutralize BA1 (Omicron) variant in pseudovirus neutralization assay, irrespective of nucleocapsid (NC) reactivity (Fig. 2A and B). Live-virus neutralization was tested to compare the neutralizing capacity of booster-induced antibodies against several Omicron subvariants. Although the third dose of the vaccine led to higher nAbs against the ancestral strain (WA1) and B.1.617.2 variant (89% and 79%, respectively), the capacity to neutralize BA variants (BA.1 and BA.2.12.1) was markedly lower and detected in only 33% to 43% of patients (Fig. 2C). Similarly, nAb titers against BA.5 were 14-fold lower than against WA1, even following booster vaccination (Fig. 2D). In contrast to viral neutralization, spike-specific T cells against BA1-spike by ELISpot were highly correlated with those against WA1, indicating that the T cells were capable of recognizing BA spike-derived peptides (Fig. 2E). Extension of AIM assay to BA1 peptides again demonstrated that the majority of this response consists of CD4+ and not CD8+ T cells (Fig. 2F). As with ELISpot, detection of BA spike-specific T cells by the AIM assay correlated with WA spike-specific T cells (Fig. 2G). Patients with NC reactivity (reflecting hybrid immunity) not only had a higher proportion of spike-reactive T cells following dose 3 (Fig. 1I), but these cells also had a distinct phenotype with higher expression of PD-1 and T-bet (Fig. 2H).
Figure 2.
Variant-specific nAb/T cells and immunophenotypic correlates of immunity following 3 doses of SARS-CoV-2 vaccine. A, Pseudovirus neutralization IC50 against WA1 (n = 253) and Omicron (BA1: n = 249) variant. The figure shows the median with a 95% confidence interval (****, P < 0.0001, Mann–Whitney). Dotted lines show the threshold for detection at 100. B, Pseudovirus neutralization IC50 against WA1 and BA1, split by nucleocapsid reactivity (NC-WA: n = 78, NC-BA: n = 75, NC + WA: n = 49 and NC + BA: n = 48). Data, median with a 95% confidence interval (***, P < 0.001; ****, P < 0.0001 Kruskall Wallis). Dotted lines show the threshold for detection at 100. C, Live-virus neutralization (FRNT50) against ancestral strain (WA1), B.1.617.2, and Omicron variants [BA1 (B.1.1.529), BA2 (B.1.1.529.2), and BA2.12.1; n = 39]. Lines connect data points from the same sample. Dotted line represents the lower limit of positive live-virus neutralization (<20; ****, P < 0.0001, Kruskal–Wallis). D, Live-virus neutralization (FRNT50) against ancestral strain (WA1) and BA.5 variant. The red line represents the median of all measurements. Lines connect datapoints from the same sample. The dotted line signifies a lower limit of positive neutralization (<20; ****, P < 0.0001 Mann–Whitney). E, Correlation between WA1 spike and BA1 spike reactivity as evaluated by interferon-gamma ELISpot. F, BA1-spike-reactive CD4+ and CD8+ T cells as assayed by AIM assay. Pie chart shows the mean proportions of BA1 spike-specific CD4+ and CD8+ T cells for all patients. US = unstimulated control. *, P < 0.05, paired t test. G, Correlation between WA spike- and BA spike-reactive AIM+ T cells. H, Phenotype of spike-reactive CD4+ T cells in NC+ or NC− patients, as analyzed by mass cytometry. Data, median expression of markers. ****, P < 0.0001, t test. I, Live-virus neutralization (FRNT50) against ancestral strain (WA1), and Omicron variants in patients with MM who developed breakthrough COVID infection, or did not, following booster mRNA vaccines. Data, mean + SEM. *, P < 0.05, t test with Welch correction. Values >0.05 are noted in the figure. J, Differences in the CXCR5+ T-cell cluster in patients with MM who developed breakthrough COVID infection, or did not, following booster mRNA vaccines. Figure shows mean + SEM. *, P < 0.05, t test with Welch correction.
In view of marked interindividual variation in vaccine responses, immunophenotypic analysis was performed utilizing mass cytometry to identify the impact of underlying immune status on vaccine-induced SARS-CoV-2–specific nAb/B/T cells (Supplementary Fig. S16). As expected, seronegativity was associated with a lower proportion of B cells (Supplementary Fig. S16). Interestingly, seronegativity also correlated with altered proportions of CD4+ T and myeloid cells (Supplementary Fig. S16). Focused FlowSOM analysis of CD4 T cells identified 10 metaclusters (MC; Supplementary Fig. S17A and S17B). Of these, seronegativity was associated with a decline in naïve CD4+ (MC6) and an increase in CD27− MCs (MC2, 3, 7; Supplementary Fig. S17A and S17B), whereas an increased proportion of MC9 (CXCR5+ subset) correlated with higher ELISpot reactivity (Supplementary Fig. S18). We also performed FlowSOM analysis focused on CD8+ T cells. Seronegative patients had a lower proportion of naïve (MC2) and higher CD27−CD8+ T (MC5) cells (Supplementary Fig. S19A and S19B). Interestingly, the proportion of CD16+CD11c+CD14−DR+ myeloid cells, consistent with activated myeloid or dendritic cell phenotype correlated with higher seropositivity as well as T-cell responses (Supplementary Fig. S20A–S20C).
Another distinct feature of this data set is the inclusion of a significant proportion of Black patients with MM (38% of the cohort). Consistent with prior analysis, Black patients had higher levels of nAbs after dose 2, but these differences were not observed after dose 3 (Supplementary Fig. S21). Interestingly, the phenotype of RBD-specific B cells in Black patients revealed a higher proportion of T-bet+ B cells (Supplementary Fig. S22 and MC3 in heat map in Fig. 1G), previously linked to extrafollicular responses and aging (20). In contrast to dose 2 (6), the vaccine type (Moderna or Pfizer) did not affect the induction of nAbs following dose 3 (Supplementary Fig. S23). Clinical correlates of nAb responses (Table 1) were analyzed separately from those for cellular responses (Table 2). nAb responses were lower in males and in patients with hypogammaglobulinemia. From a therapy perspective, anti–B cell maturation antigen (BCMA) therapy had an adverse effect on nAbs, whereas patients on lenalidomide maintenance had higher nAbs. The adverse effect of anti-CD38 antibodies on nAb, which was previously detected in samples after the first two doses of the vaccine (6), was not detected following booster (dose 3) vaccination. Correlates of RBD-specific B cells were similar to those for nAbs and were affected adversely by hypogammaglobulinemia and anti-BCMA therapy. T-cell responses were higher in patients with prior SARS-CoV-2 exposure. Interestingly, patients with hypogammaglobulinemia also trended toward lower ELISpots. nAb titers were lower in patients with hypogammaglobulinemia (Supplementary Fig. S24A and S24B). The finding that patients with hypogammaglobulinemia had a deficiency of both nAbs and T-cell responses led us to evaluate immunophenotypic features in these patients. In addition to an expected decline in B cells, these patients also had lower proportions of CXCR5+CD4+ cells consistent with T follicular helper (TFH) cells (Supplementary Fig. S25A and S25B).
Table 1.
Correlates of WA1- and BA-neutralizing Ab response following booster vaccination.
| WA1 neutralization | BA (Omicron) neutralization | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All patients (N = 187) | NC negative (N = 65) | All patients (N = 187) | NC negative (N = 65) | ||||||||
| Covariate | Level | N | Odds ratio (95% CI) | P value | N | Odds ratio (95% CI) | P value | Odds ratio (95% CI) | P value | Odds ratio (95% CI) | P value |
| Sex | Female | 92 | 1.73 (0.72–4.18) | 0.223 | 31 | 6.04 (1.21–30.28) | 0.029 | 1.00 (0.55–1.81) | 0.991 | 1.78 (0.66–4.78) | 0.252 |
| Male | 95 | — | — | 34 | — | — | — | — | — | — | |
| Racea | Black | 68 | 1.53 (0.60–3.90) | 0.376 | 24 | 0.86 (0.24–3.09) | 0.815 | 1.07 (0.57–2.00) | 0.834 | 0.47 (0.16–1.32) | 0.150 |
| Other | 114 | — | — | 38 | — | — | — | — | — | — | |
| Age ≤65 | Yes | 108 | 0.65 (0.26–1.60) | 0.347 | 34 | 1.69 (0.48–6.02) | 0.417 | 1.07 (0.58–1.96) | 0.830 | 0.93 (0.35–2.46) | 0.878 |
| No | 79 | — | — | 31 | — | — | — | — | — | — | |
| Prior LOT (>2) | Yes | 46 | 0.46 (0.18–1.15) | 0.081 | 21 | 0.36 (0.09–1.35) | 0.130 | 0.42 (0.21–0.84) | 0.013 | 0.27 (0.09–0.82) | 0.021 |
| No | 135 | — | — | 40 | — | — | — | — | — | — | |
| Prior SARS-CoV-2 exposure | Positive | 40 | 2.79 (0.74–10.59) | 0.131 | 2.00 (0.87–4.60) | 0.103 | |||||
| Negative | 65 | — | — | — | — | ||||||
| IgG ≤400 | Yes | 48 | 0.35 (0.14–0.84) | 0.019 | 21 | 0.26 (0.07–0.94) | 0.040 | 0.81 (0.41–1.58) | 0.53 | 0.69 (0.24–1.96) | 0.487 |
| No | 139 | — | — | 44 | — | — | — | — | — | — | |
| Vaccine type | Moderna | 54 | 1.85 (0.57–5.97) | 0.306 | 19 | 1.57 (0.27–9.04) | 0.611 | 1.43 (0.71–2.87) | 0.319 | 1.71 (0.54–5.48) | 0.363 |
| Pfizer | 101 | — | — | 32 | — | — | — | — | — | — | |
| Anti-CD38 | Yes | 60 | 0.76 (0.31–1.85) | 0.599 | 21 | 1.54 (0.37–6.41) | 0.551 | 0.85 (0.45–1.61) | 0.624 | 1.22 (0.43–3.47) | 0.713 |
| No | 127 | — | — | 44 | — | — | — | — | — | — | |
| Len maintenance | Yes | 44 | 8.24 (1.08–62.82) | 0.042 | 14 | 3.57 (0.42–30.40) | 0.243 | 1.93 (0.90–4.13) | 0.107 | 4.12 (1.03–16.56) | 0.046 |
| No | 143 | — | — | 51 | — | — | — | — | — | — | |
| Anti-BCMAb,c | Yes | 6 | 0.06 (0.01–0.36) | 0.002 | 4 | 0.06 (0.01–0.62) | 0.018 | 0.10 (0.01–0.91) | 0.041 | ||
| No | 181 | — | — | 61 | — | — | — | — | |||
NOTE: Anti-BCMA could not be evaluated for Omicron neutralization (NC negative cohort) due to inestimable ORs.
Abbreviations: LOT, lines of therapy; IgG, immunoglobulin G.
aAmong the n = 114 with other race, 110 were White, 2 Asian, 1 Native American, and 1 Native Hawaiian or Pacific Islander.
b P < 0.05 on MVA for WT neutralization (all).
c P < 0.05 on MVA for WT neutralization (NC negative).
Table 2.
Correlates of antigen-specific T- and B-cell response following booster vaccination.
| Spike-specific T cells (ELISpot; N = 190) | RBD-specific B cells (n = 136) | ||||||
|---|---|---|---|---|---|---|---|
| Covariate | Level | N | Odds ratio (95% CI) | P value | N | Odds ratio (95% CI) | P value |
| Sex | Female | 87 | 1.50 (0.80–2.81) | 0.209 | 63 | 0.99 (0.48–2.03) | 0.980 |
| Male | 103 | — | — | 73 | — | — | |
| Race | Black | 79 | 1.57 (0.81–3.03) | 0.182 | 62 | 1.09 (0.52–2.31) | 0.818 |
| Othera | 105 | — | — | 67 | — | — | |
| Age ≤65 | Yes | 91 | 0.66 (0.35–1.25) | 0.201 | 64 | 1.45 (0.70–2.99) | 0.321 |
| No | 99 | — | — | 72 | — | — | |
| Prior LOT (>2) | Yes | 57 | 0.70 (0.34–1.43) | 0.325 | 42 | 0.43 (0.19–0.96) | 0.041 |
| No | 124 | — | — | 89 | — | — | |
| Prior SARS-CoV-2 exposureb,c | Positive | 47 | 2.18 (1.14–4.18) | 0.019 | 31 | 5.39 (1.82–15.97) | 0.002 |
| Negative | 135 | — | — | 103 | — | — | |
| IgG ≤400 | Yes | 53 | 0.49 (0.25–0.96) | 0.039 | 40 | 0.30 (0.13–0.70) | 0.005 |
| No | 137 | — | — | 96 | — | — | |
| Vaccine type | No booster | 31 | 1.05 (0.47–2.34) | 0.908 | 30 | 1.22 (0.48–3.13) | 0.672 |
| Moderna | 45 | 1.39 (0.63–3.08) | 0.420 | 32 | 1.24 (0.49–3.15) | 0.657 | |
| Pfizer | 88 | — | — | 59 | — | — | |
| Anti-CD38 | Yes | 56 | 0.70 (0.35–1.42) | 0.323 | 40 | 0.75 (0.34–1.67) | 0.477 |
| No | 134 | — | — | 96 | — | — | |
| Lenalidomide maintenance | Yes | 38 | 2.03 (0.94–4.37) | 0.071 | 24 | 1.67 (0.68–4.07) | 0.261 |
| No | 152 | — | — | 112 | — | — | |
| Anti-BCMA | Yes | 5 | 0.91 (0.52–1.62) | 0.761 | 3 | 0.46 (0.32–0.66) | <0.001 |
| No | 185 | — | — | 133 | — | — | |
| Dose type | Boost | 60 | 1.59 (0.96–2.62) | 0.069 | 36 | 1.30 (0.64–2.62) | 0.465 |
| Dose 2 | 130 | — | — | 100 | — | — | |
NOTE: Odds of ELISpot ≥ 33 and odds of RBD B cells ≥ 0.024 are reported. Sample size N reflects the number of observations, rather than the number of patients.
Abbreviations: LOT, lines of therapy; IgG, immunoglobulin G.
aAmong the 105 Other race in ELISpot data, 103 were White, 1 Asian, and 1 Native American. Among the 67 Other race in B cell data, 65 were White, 1 Asian, and 1 Native American.
b P < 0.05 on MVA for ELISpot.
c P < 0.05 on MVA for RBD B cells.
We identified a cohort of patients who developed symptomatic SARS-CoV-2 breakthrough infection (BTI) requiring antiviral therapy following booster vaccines. Patients who developed BTI had significantly lower live-virus nAbs, including against BA variants (Fig. 2I). Clinical features of this cohort were similar to other boosted patients as a control (Supplementary Table S1). nAbs detected by pseudovirus neutralization against BA1 in the BTI cohort trended lower, although those against WA1 were comparable with controls (Supplementary Table S1). Detailed immunophenotypic analysis in patients with available mass cytometry data identified a lower proportion of a distinct subset of CXCR5+ CD4 T cells consistent with TFH phenotype (metacluster9) in the BTI patients (Fig. 2J). No other significant differences in CD8+ T cells, B cells, or myeloid subsets were identified.
DISCUSSION
These data provide a detailed landscape of immunophenotypic and clinical correlates that affect the induction of vaccine-induced nAbs, as well as antigen-specific B- and T-cell responses in MM and inform the risk of SARS-CoV-2 infection and future vaccination strategies in these patients. Booster vaccines led to enhanced induction of nAbs; however, these nAbs fail to neutralize Omicron variants in nearly 60% of patients. Booster dose also led to an increase in RBD-specific class-switched memory B cells, whereas B cells elicited with earlier doses revealed more transitional memory phenotypes. Repeat dosing, therefore, seems to affect both quantitative and qualitative aspects of B/antibody response and may also be essential for the emerging application of variant-specific vaccines in these patients. In a recent randomized trial, tandem influenza vaccination was needed to achieve high seroconversion rates in patients with MM (22). Therefore, repeat vaccinations may be particularly important for protective immunity against several pathogens in patients with MM.
In addition to nAbs, CD8+ T cells have also been shown to mediate protection against SARS-CoV-2 in preclinical models (2). Our finding that vaccine-induced spike-specific T cells were predominantly CD4+ T cells is consistent with another study testing T-cell responses in hematologic malignancies, although functional responses were not analyzed (23). It is already recognized that spike-specific T cells induced by current SARS-CoV-2 mRNA vaccines in healthy donors are biased toward CD4 responses (21). It is notable that the reagents/methods utilized in the current paper are identical to those utilized in some of these studies (21). It has been proposed that the paucity of CD8+ T-cell responses following current SARS-CoV-2 mRNA vaccines may be related to vaccine design being limited to spike regions, which are not the dominant source of CD8+ T-cell epitopes in natural infection (15). Therefore, the lack of detectable spike-specific CD8 T cells in vaccinated patients with MM may represent a quantitative rather than a qualitative difference. New strategies to induce SARS-CoV-2–specific CD8+ T cells are needed, and such vaccines should be prioritized in patients with hematologic malignancies, particularly as new variants are expected to continue to emerge.
Another distinct aspect of this paper is the detailed immunophenotypic analysis to understand immune correlates of vaccine response. Variance in seroconversion following vaccines correlated with not just changes in B cells but also distinct changes in T and myeloid cells. Specific changes in immune phenotypes such as a decline in naïve T cells or an increase in CD27- T cells may also be relevant for the emerging application of immune therapies in MM (24). Immune response to vaccines is also affected by disease/therapy-related factors. Booster vaccines blunted the adverse effect of prior anti-CD38 antibody therapy, but not prior BCMA-directed therapies on nAbs as observed following the first 2 doses (6). In this regard, patients with MM with hypogammaglobulinemia may represent a particularly high-risk cohort, and our data suggest that immune paresis in these patients may extend beyond B cells/antibodies as initially thought, to also include changes in T-cell phenotypes. Along these lines, it is notable that monoclonal gammopathy of undetermined significance patients with hypogammaglobulinemia were recently shown to be at increased risk of COVID (25).
Patients who developed BTI following boosters had lower titers of live-virus nAbs, particularly against BA variants. These data, therefore, illustrate the importance of measuring live-virus nAbs (as opposed to binding assays) to assess vaccine-mediated protection. This issue may be particularly relevant in immune-compromised hosts such as MM, where vaccine-induced CD8+ T cells seem to be deficient. Patients experiencing BTI also had a reduction in CXCR5+ CD4 T cells, consistent with the role of TFH cells in humoral immunity (26).
Strengths of this paper include a racially diverse cohort, with concurrent analyses of serologic and cellular responses utilizing several platforms, including live-virus neutralization against current variants and detailed immunophenotypic analyses. Although this is one of the largest analyses of SARS-CoV-2 vaccine-induced cellular responses in MM to date, further studies are needed to link these changes to impact on COVID-related mortality.
In summary, booster (third dose) SARS-CoV-2 mRNA vaccines lead to increased nAbs in patients with MM, and these patients seem to require the third dose to generate class-switched memory B cells. However, most patients with MM may remain susceptible to current circulating SARS-CoV-2 Omicron subvariants even following booster vaccination, as these vaccines do not elicit detectable variant-nAbs or CD8+ T cells. These data illustrate ongoing challenges to achieve protection from SARS-CoV-2 infection in MM. These patients should therefore be prioritized for novel approaches to elicit (or administer) variant nAbs and may require multiple doses of emerging variant-specific vaccines. They may also benefit from novel strategies to induce SARS-CoV-2–specific CD8+ T cells, including strategies targeting immunogenic epitopes outside the spike region (27, 28). High-dimensional analysis of antigen-specific cellular responses described here will also inform emerging vaccines/immune therapies against cancer and other pathogens in these patients (24).
METHODS
Patients and Patient Selection
The study was approved by the Institutional Review Board of Emory University. Per protocol, patients were approached in myeloma clinics of Winship Cancer Institute without any selection bias, and all ethnicities and racial backgrounds (self-reported) were considered. Patients who provided written informed consent were eligible for a research blood draw. Studies were conducted in accordance with the declaration of Helsinki. Blood samples were collected from eligible patients 1 week to 3 months after the second or third dose of the vaccine, concurrent with their routine clinic visit. Samples were processed to isolate plasma and mononuclear cells.
RBD and Nucleocapsid Binding Assay
Detection of SARS-CoV-2 spike RBD binding antibodies as well as nucleocapsid antibodies was performed as previously described (29).
Viruses and Cells
VeroE6-TMPRSS2 cells were generated and cultured as previously described (30). nCoV/USA_WA1/2020 (WA1), closely resembling the original Wuhan strain, was propagated from an infectious SARS-CoV-2 clone as previously described (31). icSARS-CoV-2 was passaged once to generate a working stock. The BA.1 variant was isolated and propagated as previously described (10, 30). The BA.5 isolate was kindly provided by Dr. Richard Webby (St Jude Children's Research Hospital, Memphis, TN) and was plaque purified and propagated once in VeroE6-TMPRSS2 cells to generate a working stock. All viruses used in this study were deep-sequenced and confirmed as previously described (30).
Pseudovirus Neutralization Assay
Neutralization activity against SARS-2-CoV was measured in a single-round-of-infection assay with pseudotyped virus particles (pseudoviruses), as previously described (6, 32). In addition to pseudoviruses expressing full-length spikes from WA1 parental strain, pseudoviruses expressing spikes from Omicron (BA1) were also utilized. Pseudoviruses were mixed with serial dilutions of plasma or antibodies and then added to monolayers of ACE-2-overexpressing 293T cells. Twenty-four hours after infection, cells were lysed, luciferase was activated with the Luciferase Assay System (Promega), and relative light units (RLU) were measured on a synergy Biotek reader. After subtraction of background RLU (uninfected cells), % neutralization was calculated as 100 × [(virus only control) − (virus plus antibody)]/(virus only control). Dose–response curves were generated with a 4-parameter nonlinear function, and titers were reported as the serum dilution or antibody concentration required to achieve 50% (50% inhibitory dilution [ID50]) neutralization. The input dilution of serum is 1:60, and 100 is set as the lower limit of quantification.
Focus Reduction Neutralization Test
FRNT assays were performed as described previously (28, 30, 33, 34). Briefly, plasma samples were serially diluted and incubated with 100 to 200 FFU of WA1, B.1.617.2, B.1.1.529, B.1.1.529.2, or BA.5 at 37°C for 1 hour. The virus–antibody mixture was then added to VeroE6-TMPRSS2 cells and incubated at 37°C for 1 hour. After incubation, the virus–antibody mixture was removed and replaced by a 0.85% methylcellulose (Sigma-Aldrich, #M0512-250G) overlay. Cells were incubated at 37°C for 18 to 40 hours, after which the methylcellulose overlay was removed. The cells were washed, fixed with 2% paraformaldehyde, permeabilized, and incubated with Alexa Fluor-647–conjugated anti-SARS-CoV-2 spike antibody (CR3022-AF647) for up to 4 hours at room temperature. Cells were then washed and visualized on an ELISpot reader (CTL Analyzer). Antibody neutralization was quantified by counting the number of foci for each sample using Virodot (35). Neutralization titers were calculated as follows: 1 − (ratio of the mean number of foci in the presence of plasma and foci at the highest dilution of the respective sample). Specimens were tested in duplicate. Samples that do not neutralize at the limit of detection at 50% are plotted at 20 and were used for geometric mean calculations.
Immunophenotyping
Immunophenotypic was performed utilizing a 36-marker mass cytometry panel and complemented with flow cytometry. Both platforms were also utilized for the detection of RBD-specific B cells, utilizing fluorochrome- or metal-conjugated RBD protein. Briefly, thawed bone marrow mononuclear cells were stained with custom panels of metal-conjugated antibodies at manufacturer-suggested concentrations (Fluidigm; antibodies as noted in Supplementary Table S2). Cells were fixed, permeabilized, and washed in accordance with the manufacturer's cell-surface and nuclear staining protocol as described (36–38). After antibody staining, cells with incubated with intercalation solution, mixed with EQ Four Element Calibration Beads (cat. #201708) and acquired using a Helios mass cytometer (all from Fluidigm). Gating and data analysis were performed using Cytobank (https://www.cytobank.org). Viable cells and doublets were excluded using cisplatin intercalator and DNA content with iridium intercalator. For the flow cytometry assay, peripheral blood mononuclear cells were stained with a panel of antibodies (Supplementary Table S3), to detect the presence of RBD-specific B cells, as described (29).
Detection of Spike-Specific T Cells with Interferon-γ ELISpot and AIM Expression Assay
The presence of spike-reactive T cells was analyzed by interferon-γ ELISpot following stimulation with an overlapping 15-mer peptide library derived from WA1 or BA1 spike, as described previously (21). The same peptide libraries were utilized for the AIM assay, which measures activation-induced marker expression following antigenic stimulation, as described earlier (21, 27), with the exception that T cells were analyzed by mass cytometry instead of flow cytometry, after staining with a 33-marker panel (antibodies used are shown in Supplementary Table S4). AIM+ CD4+ T cells were identified as OX40+CD137+, whereas AIM+ CD8+ T cells were identified as CD69+CD137+ T cells.
T-cell Receptor Variable Beta Chain Sequencing
Immunosequencing of the CDR3 regions of human TCRβ chains was performed using the ImmunoSEQ Assay (Adaptive Biotechnologies). Extracted genomic DNA was amplified in a bias-controlled multiplex PCR, followed by high-throughput sequencing. Sequences were collapsed and filtered in order to identify and quantitate the absolute abundance of each unique TCRβ CDR3 region for further analysis as previously described (39–41). The fraction of T cells was calculated by normalizing TCRβ template count to the total number of cells sequenced on the assay using a panel of reference genes found in all nucleated cells.
T-DETECT
All samples were classified as positive or negative for the detection and enrichment of SARS-CoV-2–specific T cells using Adaptive's T-DETECT COVID classifier. The classifier was trained by comparing peripheral repertoires from COVID+ and convalescent subjects with control samples collected prepandemic. T-cell responses are categorized as negative, positive, and “No Call” (representing samples with an insufficient number of T-cell rearrangements to make a definitive negative call).
Mapping of SARS-CoV-2 TCRβ Sequences
Peripheral repertoires were mapped against a set of TCR sequences that are known to react to SARS-CoV-2. Briefly, these sequences were first identified by Multiplex Identification of T-cell Receptor Antigen Specificity (MIRA; ref. 42). SARS-CoV-2–specific TCR sequences identified via MIRA correspond to antigen-specific addresses, which detail MHC class presentation and were used to correlate the response with viral ORF and cellular subsets (CD8 vs. CD4). Reactive TCRs were further screened for enrichment in a previously COVID-19–infected repertoire compared with COVID-19–negative repertoires collected as part of ImmuneCODE, a publicly available open database (adaptivebiotech.com/Immunecode), to remove TCRs that may be very frequent or cross-reactive to common antigens. The filtered list represents a set of TCRs that are both experimentally observed expanding to SARS-CoV-2 antigens and enriched in COVID-19 subjects (43, 44). Responses were quantified by the number and/or frequency of SARS-CoV-2–specific TCRs. Specifically, clonal breadth (defined as the proportion of distinct TCRs that are COVID-19 specific divided by the number of unique TCRs sequenced in the sample) and clonal depth (defined as the sum frequency of the COVID-19–specific TCRs in the repertoire) were determined for each sample. These two metrics have been shown to have a good separation of cases (defined as either COVID-19 infected or vaccinated) and controls previously (43, 44).
Statistical Analysis
The Mann–Whitney test was used to calculate the difference between groups. Correlation between the assays was performed by the Pearson r correlation method and linear regression analysis. Statistical analysis was performed using GraphPad Prism version 9, SPSS package version 27.0, and SAS 9.4 (SAS Institute Inc.). Antibody neutralization was quantified by counting the number of foci for each sample using the Viridot program (35). The neutralization titers were calculated as follows: 1 − (ratio of the mean number of foci in the presence of sera and foci at the highest dilution of the respective sera sample). Each specimen was tested in duplicate. The FRNT-50 titers were interpolated using a 4-parameter nonlinear regression in GraphPad Prism 9.4.1. Samples that do not neutralize at the limit of detection at 50% are plotted at 20 and were used for geometric mean calculations. Patient data were obtained from the myeloma database, which captures patient demographics and outcomes data and is continuously updated with periodic quality checks. Chi-square tests and Fisher exact tests were used while comparing differences between categorical variables, and the nonparametric Mann–Whitney test was used for continuous variables. Binomial logistic regression was used to identify the predictors for nAb responses and RBD responses, for both WT neutralization and Omicron neutralization. Observations were selected based on peak Omicron neutralization for each patient among boost doses. Generalized linear models with a binary response variable were fit for ELISpot and RBD B cells using a generalized estimating equations approach for within-subject correlation. An exchangeable working correlation structure was selected. ELISpot and RBD B cells were dichotomized using median values. All observations, both dose 2 and boost, with an ELISpot value or an RBD B-cell value were included for the analysis, respectively. Among variables statistically significant on univariate analysis, backward elimination was utilized to identify relevant characteristics for inclusion in the final multivariable models, with an alpha value of 0.20. All statistical tests were two-sided unless otherwise noted, and statistical significance was assessed at the 0.05 level.
Data Availability
Data described in this paper are available upon request from the corresponding author. T-cell repertoire profiles and antigen annotation data are available through the ImmuneCODE resource and can be downloaded from the Adaptive Biotechnologies immuneACCESS site at clients.adaptivebiotech.com/pub/azeem-2023-bcd.
Supplementary Material
Suppl Table 1. Correlative analysis of breakthrough SARS CoV-2 infection following booster vaccination.
Supplementary Table 2: Antibodies used for immune-phenotyping (CyTOF analysis). Supplementary Table 3: Antibodies used for flow cytometry. Supplementary Table 4: Antibodies used for AIM Assay (CyTOF analysis).
This file contains supplementary figures 1-25
Acknowledgments
This work is supported in part by NCI U54 Seronet award CA260563. M.V. Dhodapkar is also supported in part by funds from the NIH R35CA197603 and SCOR award from the Leukemia and Lymphoma Society (LLS). K.M. Dhodapkar is supported in part by funds from NIH CA238471 and AR077926 and LLS. The authors acknowledge the support of Cancer Tissue and Pathology, Immune Monitoring, and Data and Technology Applications, and Biostatistics shared resource of the Winship Cancer Institute of Emory University and NIH/NCI under award number P30CA138292. The authors acknowledge the support of M. Johns and H. Von Hollen and their team for help with specimen collection; and Barb Banbury at Adaptive Biotechnologies for help with T-Detect assay. M.S. Suthar is partly supported by grants NIH P51 OD011132, HHSN272201400004C, and U19AI090023, by the Emory Executive Vice President for Health Affairs Synergy Fund award, the Pediatric Research Alliance Center for Childhood Infections and Vaccines and Children's Healthcare of Atlanta, COVID-Catalyst-I3 Funds from the Woodruff Health Sciences Center and Emory School of Medicine, Woodruff Health Sciences Center 2020 COVID-19 CURE Award, and the Emory–UGA Center of Excellence for Influenza Research and Surveillance.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
N. Cheerdarla reports grants from NIH/NCI during the conduct of the study. A. Moreno reports grants from NIH during the conduct of the study. B. Wali reports grants from NIH during the conduct of the study. J.L. Kaufman reports grants from NIH during the conduct of the study; personal fees from AbbVie, BMS, Janssen, and Incyte outside the submitted work. S. Lonial reports personal fees from Amgen, grants and personal fees from Takeda, Janssen, and BMS, personal fees from Pfizer, Genentech, AbbVie, and Celgene outside the submitted work; and Board of Directors TG Therapeutics with stock. J.D. Roback reports grants from NIH during the conduct of the study; other support from Cambium and personal fees and other support from Secure Transfusion Solutions outside the submitted work. M.S. Suthar reports grants and personal fees from Moderna and Ocugen outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
M.I. Azeem: Writing–review and editing, data interpretation and analysis, performed immune assays. A.K. Nooka: Writing–review and editing. U. Shanmugasundaram: Writing–review and editing. N. Cheedarla: Writing–review and editing. S. Potdar: Writing–review and editing. R.J. Manalo: Writing–review and editing. A. Moreno: Writing–review and editing. J.M. Switchenko: Writing–review and editing. S. Cheedarla: Writing–review and editing. D.B. Doxie: Writing–review and editing. R. Radzievski: Writing–review and editing. M.L. Ellis: Writing–review and editing. K.E. Manning: Writing–review and editing. B. Wali: Writing–review and editing. R.M. Valanparambil: Writing–review and editing. K.T. Maples: Writing–review and editing, data interpretation and analysis, helped with collection of breakthrough infection data. E. Baymon: Writing–review and editing. J.L. Kaufman: Writing–review and editing. C.C. Hofmeister: Writing–review and editing. N.S. Joseph: Writing–review and editing. S. Lonial: Writing–review and editing. J.D. Roback: Writing–review and editing. A. Sette: Writing–review and editing. R. Ahmed: Writing–review and editing. M.S. Suthar: Writing–review and editing. A.S. Neish: Writing–review and editing. M.V. Dhodapkar: Conceptualization, resources, formal analysis, supervision, funding acquisition, writing–original draft, project administration. K.M. Dhodapkar: Conceptualization, resources, formal analysis, supervision, funding acquisition, writing–original draft, project administration.
References
- 1. Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 2021;27:1205–11. [DOI] [PubMed] [Google Scholar]
- 2. Liu J, Yu J, McMahan K, Jacob-Dolan C, He X, Giffin V, et al. CD8 T cells contribute to vaccine protection against SARS-CoV-2 in Macaques. Sci Immunol 2022:eabq7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ribas A, Dhodapkar MV, Campbell KM, Davies FE, Gore SD, Levy R, et al. How to provide the needed protection from COVID-19 to patients with hematologic malignancies. Blood Cancer Discov 2021;2:562–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dhodapkar MV, Dhodapkar KM, Ahmed R. Viral immunity and vaccines in hematologic malignancies: implications for COVID-19. Blood Cancer Discov 2021;2:9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Van Oekelen O, Gleason CR, Agte S, Srivastava K, Beach KF, Aleman A, et al. Highly variable SARS-CoV-2 spike antibody responses to two doses of COVID-19 RNA vaccination in patients with multiple myeloma. Cancer Cell 2021;39:1028–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nooka AK, Shanmugasundaram U, Cheedarla N, Verkerke H, Edara VV, Valanparambil R, et al. Determinants of neutralizing antibody response after SARS CoV-2 vaccination in patients with myeloma. J Clin Oncol 2022;40:JCO2102257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chung DJ, Shah GL, Devlin SM, Ramanathan L, Doddi S, Pessin M, et al. Disease and therapy-specific impact on humoral immune responses to COVID-19 vaccination in hematologic malignancies. Blood Cancer Discov 2021;2( Sep 13):577–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Terpos E, Gavriatopoulou M, Ntanasis-Stathopoulos I, Briasoulis A, Gumeni S, Malandrakis P, et al. The neutralizing antibody response post COVID-19 vaccination in patients with myeloma is highly dependent on the type of anti-myeloma treatment. Blood Cancer J 2021;11:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pajon R, Doria-Rose NA, Shen X, Schmidt SD, O'Dell S, McDanal C, et al. SARS-CoV-2 omicron variant neutralization after mRNA-1273 booster vaccination. N Engl J Med 2022;386:1088–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edara VV, Manning KE, Ellis M, Lai L, Moore KM, Foster SL, et al. mRNA-1273 and BNT162b2 mRNA vaccines have reduced neutralizing activity against the SARS-CoV-2 omicron variant. Cell Rep Med 2022;3:100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jacobs JL, Haidar G, Mellors JW. COVID-19: challenges of viral variants. Annu Rev Med 2022Jul 18 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 12. Moreira ED Jr, Kitchin N, Xu X, Dychter SS, Lockhart S, Gurtman A, et al. Safety and efficacy of a third dose of BNT162b2 covid-19 vaccine. N Engl J Med 2022;386:1910–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu J, Collier AY, Rowe M, Mardas F, Ventura JD, Wan H, et al. Neutralization of the SARS-CoV-2 omicron BA.1 and BA.2 variants. N Engl J Med 2022;386:1579–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Z, Mateus J, Coelho CH, Dan JM, Moderbacher CR, Galvez RI, et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell 2022;185:2434–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grifoni A, Sidney J, Vita R, Peters B, Crotty S, Weiskopf D, et al. SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe 2021;29:1076–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Muik A, Lui BG, Wallisch AK, Bacher M, Muhl J, Reinholz J, et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science 2022;375:678–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sahin U, Muik A, Vogler I, Derhovanessian E, Kranz LM, Vormehr M, et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature 2021;595:572–7. [DOI] [PubMed] [Google Scholar]
- 18. Enssle JC, Campe J, Buchel S, Moter A, See F, Griessbaum K, et al. Enhanced but variant-dependent serological and cellular immune responses to third-dose BNT162b2 vaccination in patients with multiple myeloma. Cancer Cell 2022;40:587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aleman A, Van Oekelen O, Upadhyaya B, Beach K, Kogan Zajdman A, Alshammary H, et al. Augmentation of humoral and cellular immune responses after third-dose SARS-CoV-2 vaccination and viral neutralization in myeloma patients. Cancer Cell 2022;40:441–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Myles A, Sanz I, Cancro MP. T-bet(+) B cells: A common denominator in protective and autoreactive antibody responses? Curr Opin Immunol 2019;57:40–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tarke A, Coelho CH, Zhang Z, Dan JM, Yu ED, Methot N, et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 2022;185:847–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Branagan AR, Duffy E, Gan G, Li F, Foster C, Verma R, et al. Tandem high-dose influenza vaccination is associated with more durable serologic immunity in patients with plasma cell dyscrasias. Blood Adv 2021;5:1535–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Greenberger LM, Saltzman LA, Gruenbaum LM, Xu J, Reddy ST, Senefeld JW, et al. Anti-spike T cell and antibody responses to SARS-CoV-2 mRNA vaccines in patients with hematologic malignancies. Blood Cancer Discov 2022;3:481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dhodapkar MV. The immune system in multiple myeloma and precursor states: lessons and implications for immunotherapy and interception. Am J Hematol 2022Oct 4 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ho M, Zanwar S, Buadi FK, Ailawadhi S, Larsen J, Bergsagel L, et al. Risk factors for severe infection and mortality in COVID-19 and monoclonal gammopathy of undetermined significance. Blood 2022;140:1997–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lederer K, Castano D, Gomez Atria D, Oguin TH 3rd, Wang S, Manzoni TB, et al. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity 2020;53:1281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cohen KW, Linderman SL, Moodie Z, Czartoski J, Lai L, Mantus G, et al. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep Med 2021;2:100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suthar MS, Zimmerman MG, Kauffman RC, Mantus G, Linderman SL, Hudson WH, et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep Med 2020;1:100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Edara VV, Pinsky BA, Suthar MS, Lai L, Davis-Gardner ME, Floyd K, et al. Infection and vaccine-induced neutralizing-antibody responses to the SARS-CoV-2 B.1.617 variants. N Engl J Med 2021;385:664–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 2020;27:841–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anderson EJ, Rouphael NG, Widge AT, Jackson LA, Roberts PC, Makhene M, et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N Engl J Med 2020;383:2427–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vanderheiden A, Edara VV, Floyd K, Kauffman RC, Mantus G, Anderson E, et al. Development of a rapid focus reduction neutralization test assay for measuring SARS-CoV-2 neutralizing antibodies. Curr Protoc Immunol 2020;131:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chang A, Akhtar A, Linderman SL, Lai L, Orellana-Noia VM, Valanparambil R, et al. Humoral responses against SARS-CoV-2 and variants of concern after mRNA vaccines in patients with non-Hodgkin lymphoma and chronic lymphocytic leukemia. J Clin Oncol 2022;40:JCO2200088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Katzelnick LC, Coello Escoto A, McElvany BD, Chavez C, Salje H, Luo W, et al. Viridot: an automated virus plaque (immunofocus) counter for the measurement of serological neutralizing responses with application to dengue virus. PLoS Negl Trop Dis 2018;12:e0006862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dhodapkar KM, Cohen AD, Kaushal A, Garfall AL, Manalo RJ, Carr AR, et al. Changes in bone marrow tumor and immune cells correlate with durability of remissions following BCMA CAR T therapy in myeloma. Blood Cancer Discov 2022;3:490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kaushal A, Nooka AK, Carr AR, Pendleton KE, Barwick BG, Manalo J, et al. Aberrant extrafollicular B cells, immune dysfunction, myeloid inflammation, and MyD88-mutant progenitors precede Waldenstrom macroglobulinemia. Blood Cancer Discov 2021;2:600–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bailur JK, McCachren SS, Pendleton K, Vasquez JC, Lim HS, Duffy A, et al. Risk-associated alterations in marrow T cells in pediatric leukemia. JCI Insight 2020;5:e140179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Robins H, Desmarais C, Matthis J, Livingston R, Andriesen J, Reijonen H, et al. Ultra-sensitive detection of rare T cell clones. J Immunol Methods 2012;375:14–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 2009;114:4099–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun 2013;4:2680. [DOI] [PubMed] [Google Scholar]
- 42. Klinger M, Pepin F, Wilkins J, Asbury T, Wittkop T, Zheng J, et al. Multiplex identification of antigen-specific T cell receptors using a combination of immune assays and immune receptor sequencing. PLoS One 2015;10:e0141561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swanson PA 2nd, Padilla M, Hoyland W, McGlinchey K, Fields PA, Bibi S, et al. AZD1222/ChAdOx1 nCoV-19 vaccination induces a polyfunctional spike protein-specific TH1 response with a diverse TCR repertoire. Sci Transl Med 2021;13:eabj7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alter G, Yu J, Liu J, Chandrashekar A, Borducchi EN, Tostanoski LH, et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature 2021;596:268–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl Table 1. Correlative analysis of breakthrough SARS CoV-2 infection following booster vaccination.
Supplementary Table 2: Antibodies used for immune-phenotyping (CyTOF analysis). Supplementary Table 3: Antibodies used for flow cytometry. Supplementary Table 4: Antibodies used for AIM Assay (CyTOF analysis).
This file contains supplementary figures 1-25
Data Availability Statement
Data described in this paper are available upon request from the corresponding author. T-cell repertoire profiles and antigen annotation data are available through the ImmuneCODE resource and can be downloaded from the Adaptive Biotechnologies immuneACCESS site at clients.adaptivebiotech.com/pub/azeem-2023-bcd.



![Figure 1. Vaccine-induced nAb, B- and T-cell responses to WA1 SARS-CoV-2. Blood was collected from myeloma patients 1 week–3 months following dose 2 and dose 3 of the SARS-CoV-2 mRNA vaccine. Plasma was analyzed to determine serologic responses to WA1 SARS-CoV-2 using RBD-specific endpoint titer and pseudovirus neutralization assay. PBMCs were examined for the presence of RBD-specific B cells using flow cytometry and single-cell mass cytometry. T-cell response directed against WA1 spike protein was detected using the IFNγ-ELISpot assay as well as antigen-induced marker expression (AIM) assay using CyTOF. In some patients, immunosequencing of the CDR3 regions of human TCRβ chains was performed using the ImmunoSEQ T-Detect Assay (Adaptive Biotechnologies) to identify SARS-CoV-2–specific T cells. A, Pseudovirus neutralization IC50 following 2 (D2, n = 342) or 3 doses (D3, n = 253). Data, median with a 95% confidence interval (****, P < 0.0001, Mann–Whitney test). B, Pseudovirus neutralization IC50 following 2 (D2) or 3 doses (D3), based on nucleocapsid (NC) Ab reactivity. NC− (D2: n = 269, D3: n = 78), NC+ (D2: n = 73, D3: n = 49). Data, median with a 95% confidence interval (**, P < 0.01; ****, P < 0.0001, Kruskal–Wallis). C, RBD-specific B cells as % of all B cells following dose 2 (D2: n = 107) or dose 3 (D3: n = 60). Figures show median with 95% confidence interval (*, P < 0.05, Mann–Whitney test). D and E, Correlation between RBD-specific B cells and RBD-specific endpoint titer (last dilution for positive assay; 1D) and pseudovirus neutralization (1E). F, RBD-specific IgG+ B cells between dose 2 and dose 3 in patients with detectable RBD-specific B cells. Figures show median with 95% confidence interval (*, P < 0.05; Mann–Whitney test). G, CyTOF was performed to examine RBD-specific B-cell response. Hierarchical consensus clustering was performed on RBD-specific B cells from healthy control (HC, n = 7) as well as patients following 2 or 3 doses of the SARS-CoV-2 vaccine (n = 26 and n = 19, respectively). The figure shows FlowSOM map for all samples, as well as a heat map of markers expressed by the four B-cell metaclusters (MC; MC1, MC2, MC3, and MC4). The bar graph shows the proportion of RBD+ cells in individual metaclusters. H–J, WA1 spike-specific T cells detected by interferon-γ ELISpot assay. H, IFNγ ELISPOT assay in the entire cohort (dose 2: n = 130, dose 3: n = 60) by dose. I, ELISpot assay split by nucleocapsid reactivity [NC-D2 (n = 100), NC-D3 (n = 35), NC + D2 (n = 22), NC + D3 (n = 25)]. J, ELISpot assay by serum RBD reactivity (seropositive (RBD+ n = 150) and seronegative (RBD− n = 32) nucleocapsid antibody-negative patients. K, Detection of WA1 spike-specific T cells. Graph shows AIM+ CD4 and CD8 T cells in patients with detectable spike-specific IFNγ-specific T cells by the ELISpot assay (n = 9) as well as patients who did not have detectable spike-specific IFNγ-specific T cells by ELISPOT assay (n = 2). Pie chart shows the mean proportions of spike-specific CD4+ and CD8+ T cells for 9 patients with detectable spike-reactive T cells. US = unstimulated control (*, P < 0.05; #, P = 0.05, paired t test). L, Detection of SARS-CoV-2–specific T cells by Adaptive Biotechnologies T-Detect COVID assay. Note that increases in surface glycoprotein-reactive TCRs as assessed by COVID T-cell breadth are mostly seen for CD4+ TCRs.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/9706/9975771/d0296719535e/106fig1.jpg)