Abstract
Tubulin posttranslational modifications have been predicted to control cytoskeletal functions by coordinating the molecular interactions between microtubules and their associating proteins. A prominent tubulin modification in neurons is polyglutamylation, the deregulation of which causes neurodegeneration. Yet, the underlying molecular mechanisms have remained elusive. Here, using in‐vitro reconstitution, we determine how polyglutamylation generated by the two predominant neuronal polyglutamylases, TTLL1 and TTLL7, specifically modulates the activities of three major microtubule interactors: the microtubule‐associated protein Tau, the microtubule‐severing enzyme katanin and the molecular motor kinesin‐1. We demonstrate that the unique modification patterns generated by TTLL1 and TTLL7 differentially impact those three effector proteins, thus allowing for their selective regulation. Given that our experiments were performed with brain tubulin from mouse models in which physiological levels and patterns of polyglutamylation were altered by the genetic knockout of the main modifying enzymes, our quantitative measurements provide direct mechanistic insight into how polyglutamylation could selectively control microtubule interactions in neurons.
Keywords: katanin, kinesin‐1, microtubule‐associated Tau, microtubules, polyglutamylation, tubulin posttranslational modifications
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Post-translational Modifications & Proteolysis
In vitro assays with purified tubulin from mice lacking tubulin‐modifying enzymes show that physiologically relevant tubulin modification patterns selectively impact microtubule‐protein interactions.
Introduction
Spatio‐temporal control of cytoskeletal functions is one of the keys to understand how morphologically complex and functionally specialised cells maintain homeostasis over long periods of time, adapting to changing physiological conditions and potential pathogenic insults. The recent discovery that perturbation of a tubulin posttranslational modification (PTM), polyglutamylation, causes neurodegeneration (Magiera et al, 2018; Shashi et al, 2018) has provided direct evidence for this hypothesis. Polyglutamylation adds secondary peptide chains of glutamate to its target proteins. In the brain, the most prominent substrates of polyglutamylation are α‐ and β‐tubulin (Eddé et al, 1990; Alexander et al, 1991; Rüdiger et al, 1992; Audebert et al, 1993). The possibility to generate glutamate chains of different lengths on both subunits of the tubulin dimer inspired the “tubulin code” hypothesis, which suggests that specific PTM patterns “encode” specific functions into microtubules (Verhey & Gaertig, 2007; Janke, 2014; Janke & Magiera, 2020; Roll‐Mecak, 2020). The discovery of the tubulin tyrosine ligase‐like (TTLL) enzymes that catalyse polyglutamylation revealed that different enzymes modify either α‐ or β‐tubulin, and also generate glutamate chains of different lengths (Janke et al, 2005; van Dijk et al, 2007; Bodakuntla et al, 2021). This demonstrated that a selective control of microtubule functions by different polyglutamylation patterns, which are generated and controlled by the choice of specific enzymes is theoretically possible. To what extent this is the case under physiological conditions, and which are the mechanisms that read specific polyglutamylation patterns has so far barely been explored.
A direct way to test the impact of polyglutamylation on microtubule functions is in vitro interaction assays between microtubules and interacting or associated proteins. Such assays have, however, been limited by the restricted availability of tubulin with controlled PTMs. The role of polyglutamylation in controlling spastin‐mediated microtubule severing has been characterised with tubulin purified from cell lines in which different TTLL enzymes were overexpressed (Lacroix et al, 2010), or with tubulin modified in vitro with purified TTLL7 (Valenstein & Roll‐Mecak, 2016). Only very recently, using a similar approach another severing enzyme – katanin – was shown to be also controlled by tubulin polyglutamylation (Szczesna et al, 2022). The impact of microtubule polyglutamylation on molecular motors was first probed using recombinant yeast‐human tubulin hybrids on which glutamate chains were coupled chemically (Sirajuddin et al, 2014). While these studies demonstrated that different patterns of tubulin polyglutamylation can have selective roles in controlling microtubule severing or motor protein motility, those experimental approaches have in common the use of artificially bolstered tubulin polyglutamylation, thus generating modification levels and patterns that might not be found under physiological conditions. Finally, a more recent study reported higher kinesin‐3 landing rates on brain microtubules compared with microtubules from HeLa cells (Lessard et al, 2019). HeLa tubulin, however, differs from brain tubulin in many PTMs as well as in tubulin isotype composition, which made it difficult to attribute these findings to a unique tubulin PTM.
Here we determine how physiological levels of polyglutamylation affect microtubule interactions that occur in cells using tubulin purified from the native tissue. Following the discovery that TTLL1 and TTLL7 are the main polyglutamylases responsible for the polyglutamylation observed in neuronal tissue (Janke et al, 2005; Ikegami et al, 2006), and that they differ in their specificity towards α‐ and β‐tubulin (Bodakuntla et al, 2021), we purified tubulin from mouse brains lacking either TTLL1, TTLL7, or both enzymes. This allowed us to determine the selective impact of polyglutamylation catalysed by these two enzymes on the behaviour of three types of neuronal microtubule interactors through in vitro reconstitution assays: the structural MAP Tau (Wang & Mandelkow, 2016), the microtubule‐severing enzyme katanin (McNally & Roll‐Mecak, 2018), and the molecular motor kinesin‐1, which is involved in the axonal transport of several cargoes that have recently been shown to be sensitive to polyglutamylation levels in neurons (Magiera et al, 2018; Gilmore‐Hall et al, 2019; Bodakuntla et al, 2020b, 2021). To determine the specificity of polyglutamylation as a regulator of interactions taking place on the microtubule surface, we further tested the impact of tubulin acetylation. Acetylation is a tubulin PTM occurring in the microtubule lumen (Eshun‐Wilson et al, 2019), and might, thus, not affect the association of proteins at the outer microtubule wall. For this, we purified acetylation‐free tubulin from the brains of mice in which the α‐tubulin N‐acetyltransferase 1 (ATAT1) was genetically knocked out (Kalebic et al, 2013). We demonstrate that the distinct polyglutamylation patterns generated by TTLL1 and TTLL7 at physiological levels specifically influence the binding and activity of those microtubule interactors, thus providing a mechanistic rationale for the tubulin code hypothesis under physiologically relevant conditions.
Results
Purification and characterisation of tubulin with controlled posttranslational modifications from murine brains
Tubulin for in vitro reconstitution experiments is classically purified by repeated cycles of polymerisation and depolymerisation, and co‐purified microtubule‐associated proteins (MAPs) are removed by chromatography (Vallee, 1986). In a refined version of this widely used method, microtubules are polymerised in high‐molarity buffer that prevents the association of MAPs (Castoldi & Popov, 2003). We have recently demonstrated that this protocol, which was designed for the purification of large quantities of tubulin from kilograms of bovine or porcine brains, can be downscaled to a single mouse brain that merely weights 400 mg (Souphron et al, 2019). Combined with the availability of mouse models lacking a variety of tubulin‐modifying enzymes, this novel purification protocol opens the possibility of systematically studying the role of tubulin PTMs in vitro using native brain tubulin.
Following the recent discovery that two major glutamylases in the brain, TTLL1 and TTLL7, catalyse distinct subtypes of polyglutamylation (Fig 1A), which differentially affect cellular and physiological functions of this PTM (Bodakuntla et al, 2021), we aimed at determining the molecular mechanisms that are controlled by these two enzymes in physiological conditions. We also included another tubulin PTM, acetylation (L'Hernault & Rosenbaum, 1985), which takes place at the N‐terminally located lysine (K) 40 residue of α‐tubulin and has recently been shown to protect microtubules from mechanical ageing (Portran et al, 2017).
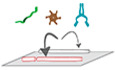
Figure 1. Purification of brain tubulin with altered posttranslational modifications.
- Schematic representation of the tubulin PTMs acetylation and polyglutamylation, both highly enriched on brain tubulin. The acetylation of α‐tubulin at lysine‐40 (K40) is catalysed by ATAT1 and is detected by the 6‐11B‐1 antibody (Appendix Table S2). Polyglutamylation takes place within the C‐terminal tubulin tails of both α‐ and β‐tubulin and is catalysed by TTLL1 and TTLL7, respectively. Glutamate chains of longer than three residues, predominantly found on α‐tubulin, are specifically detected by polyE antibody, while a β‐tubulin‐specific monoglutamylation at amino acid residue 435 (β2‐tubulin) is detected by β‐monoE antibody (Bodakuntla et al, 2021; Appendix Table S2).
- Overview of the PTM patterns of tubulin purified from knockout mouse models used in this study (Bodakuntla et al, 2021).
- Schematic overview of the tubulin purification pipeline from mouse brains using three cycles of temperature‐induced polymerisation‐depolymerisation, including a polymerisation step in high molarity buffer to remove microtubule‐associated proteins.
- SDS‐PAGE analysis of purified tubulin from brains of all mouse models used in the current study, stained with Coomassie brilliant blue. Alpha‐ and β‐tubulin are separated using a specific SDS‐PAGE protocol (Souphron et al, 2019).
- Immunoblot analysis of all purified tubulin variants. The molecular weight in (D) and (E) is indicated in kDa.
Tubulin was purified by cycles of polymerisation and depolymerisation from single brains of wild‐type, Atat1 −/−, Ttll1 −/−, Ttll7 −/− and Ttll1 −/− Ttll7 −/− mice (Fig 1B), with an alternation of low‐ and high‐molarity buffers (Fig 1C and Appendix Fig S1). Two micrograms of purified tubulin were run on SDS‐PAGE gels that allow for the separation of α‐ and β‐tubulin (Lacroix & Janke, 2011; Souphron et al, 2019) and stained with Coomassie brilliant blue. Apart from the two protein bands for α‐ and β‐tubulin, no other protein was detected in any of the purified tubulin samples (Figs 1D and EV1A). The presence of tubulin PTMs on the purified brain tubulin was characterised by a panel of PTM‐specific antibodies (Fig 1E) for three independently purified sets of tubulin (Fig EV1). Levels of α‐tubulin detected by 12G10 were similar in all samples, while K40 acetylation, detected with 6‐11B‐1 (Piperno & Fuller, 1985), was entirely absent from tubulin purified from Atat1 −/− brains. The polyE antibody, which recognises glutamate chains longer than three glutamate residues (Rogowski et al, 2010; Fig 1A) solely detects α‐tubulin, which confirmed our previous observations in brain extracts (Bodakuntla et al, 2021). This indicates that β‐tubulin is hardly modified with long glutamate chains, and shows that the polyE antibody allows for a selective detection of α‐tubulin polyglutamylation. Purified Ttll1 −/− tubulin entirely lacks the polyE signal, indicating strongly decreased α‐tubulin polyglutamylation while β‐tubulin polyglutamylation detected with β‐monoE is unchanged. β‐monoE detects a single glutamate modification at the GE*F motif (Fig 1A), which is unique in β‐tubulins, thus making this antibody a specific probe to detect β‐tubulin polyglutamylation (Bodakuntla et al, 2021). Tubulin from mice lacking TTLL7 (Ttll7 −/−) entirely lost β‐monoE reactivity, while α‐tubulin polyglutamylation (polyE) remained at wild‐type levels. Finally, tubulin purified from Ttll1 −/− Ttll7 −/− double‐knockout mice lacks both, α‐ and β‐tubulin polyglutamylation.
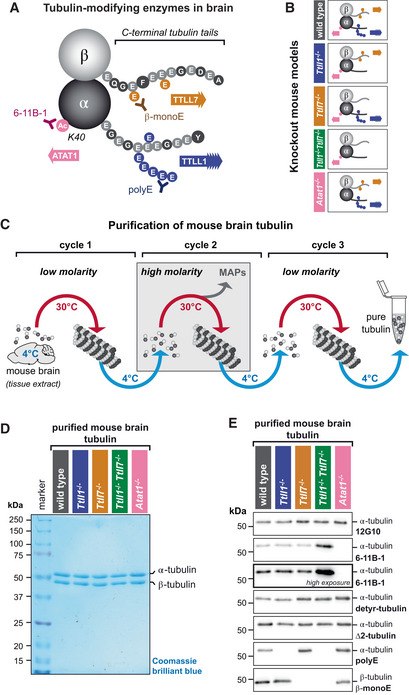
Figure EV1. Analyses of three independent sets of purified tubulin (complement to Fig 1D and E).
- Direct comparison of immunoblot analyses of all purified tubulin variants from three independent experiments. Experiment 1 is also shown in Fig 1E. The molecular weight in (A) and (B) is indicated in kDa. Source data are available.
Source data are available online for this figure.
To confirm that the lack of ATAT1 or TTLL enzymes does not affect another set of tubulin PTMs that take place on the C‐termini of α‐tubulin (Fig 1A), we tested the levels of both detyrosinated and ∆2‐tubulin (Paturle‐Lafanechere et al, 1994), and found that these are similar in all tubulin samples (Figs 1E and EV1B). Strikingly, we found an increase of acetylation on tubulin from Ttll1 −/− Ttll7 −/− double‐knockout mice, which might be related to a physiological compensation to the very low levels of polyglutamylation on tubulin from these mice. This intriguing observation will give rise to further investigation of the cross‐talk of tubulin PTMs. In our study, however, it has no impact on the interpretation of our data as will be discussed below.
Combining recently developed mouse models (Kalebic et al, 2013; Bodakuntla et al, 2021) with an adapted purification protocol (Souphron et al, 2019) allowed us to assemble a set of purified brain tubulins with well‐defined alterations in tubulin acetylation and polyglutamylation (Fig 1B). Importantly, wild‐type levels of these PTMs are physiologically relevant, and in the tubulin samples from knockout mice, only the PTMs catalysed by the inactivated enzymes are selectively affected. This then allowed us to directly test the impact of individual PTMs, or PTM patterns, on the function of three representative neuronal microtubule‐interacting proteins using in vitro reconstitution assays.
Tubulin polyglutamylation increases the affinity of Tau for microtubules
Tau is among the most intensely studied MAPs as its dysfunction is one of the central hallmarks of Alzheimer's disease and other neurodegenerative disorders, commonly known as tauopathies (Wang & Mandelkow, 2016; Goedert et al, 2017; Chang et al, 2021). Given that Tau is a prominent neuronal MAP, it is likely that it is sensitive to some of the tubulin PTMs that are present on neuronal microtubules. To directly measure the dynamics of Tau–microtubule interactions on microtubules carrying different tubulin PTM patterns, we purified the largest splice isoform (Tau(2N4R)) of human Tau (Goedert et al, 1991), C‐terminally tagged with a monomeric enhanced green fluorescent protein (Tau–meGFP). Tau can co‐exist on the microtubule surface in two distinct forms, as non‐cooperative, diffusing molecules, or as a cooperatively formed envelope shielding the microtubule surface from interactions with other proteins (Siahaan et al, 2019). To initially quantify the affinity of the diffusible Tau species for the microtubule surface, we polymerised microtubules from our panel of tubulin variants (Fig 1) in the presence of GMPCCP, which prevents the formation of Tau envelopes (Siahaan et al, 2019, 2022). We deposited these microtubules in a flow‐chamber under the microscope and visualised them with interference reflection microscopy (IRM; Mahamdeh et al, 2018). To distinguish two different microtubule species in the same flow‐chamber, we deposited the first type of microtubules (e.g. wild type), took a snapshot by IRM, and subsequently attached the second type of microtubules followed by another IRM snapshot. In the next step, 70 nM Tau–meGFP was added into the chamber, and its association with the microtubules was imaged via total internal reflection fluorescence (TIRF) microscopy at 488 nm for 5 min (Fig 2A and B, and Movie EV1), to allow Tau binding to reach a chemical equilibrium (Fig EV2B and C). Experiments were performed by pairs of two different microtubule variants per assay (Fig EV2A) and repeated for three independently purified batches of tubulins of each PTM variant. Intensities of meGFP along microtubules were measured to determine the binding affinity of Tau–meGFP to different microtubule PTM variants.
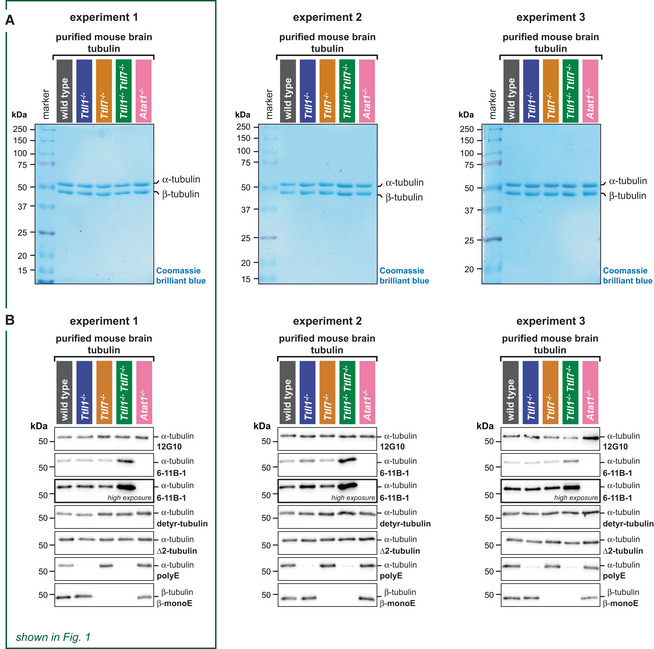
Figure 2. Polyglutamylation controls the affinity of Tau protein to microtubules.
- Schematic representation of the TIRF microscopy assay setup. Label‐free GMPCPP‐stabilised microtubules, polymerised from wild‐type and knockout‐mouse brain tubulin, are attached to a glass coverslip chamber sequentially and imaged in IRM mode. Their identity is retained by their positions on the coverslip (Steps 1 and 2). Next, 70 nM purified Tau‐meGFP is added to the chamber and its microtubule binding recorded in TIRF microscopy mode (Step 3). The fluorescence intensity of Tau‐meGFP on each microtubule in the chamber is measured in the subsequent affinity analysis (Step 4).
- Representative IRM and TIRF microscopy images showing microtubule positions and Tau‐meGFP intensities for different PTM variants (grey: wild type; blue: Ttll1 −/−; yellow: Ttll7 −/−; green: Ttll1 −/− Ttll7 −/−; pink: Atat1 −/−). In the last column, false‐coloured TIRF images highlight differences in Tau‐meGFP intensities on different types of microtubules (Movie EV1). Scale bars 5 μm.
- Quantification of the integrated intensity of Tau‐meGFP on GMPCPP microtubules in three independent sets of experiments performed with independently purified tubulin samples. Single assays (Fig EV2A) of one experiment were combined after normalisation to the wild‐type microtubule values (A.U., arbitrary units). Each data point represents the normalised fluorescence intensity value of one microtubule. Mean values and standard deviation are shown for each scatter plot. Student's t‐test; P‐values displayed.
- Summary quantification of the three independent experiments shown in (C). Each point is a normalised mean of the triplicates, and error bars represent standard deviation. Student's t‐test; P‐values displayed.
- Saturation curve of increasing concentration Tau‐mCherry binding to GMPCPP wild‐type and Ttll1 −/− Ttll7 −/− microtubules. Each data point is shown as mean ± standard deviation of the raw integrated intensity values (A.U. × 104) of three experiments with independently purified tubulin samples (Fig EV2D). Dissociation constants K d and R 2 coefficient from non‐linear fits of each microtubule type are indicated below the plot.
- Tau‐envelope formation on Taxol‐stabilised wild‐type and Ttll1 −/− Ttll7 −/− microtubules (grey and green arrowheads, respectively) 5 and 120 s after the addition of 40 nM Tau‐mCherry to the TIRF microscopy flow‐chamber (Movie EV2). Scale bars 5 μm.
- Analysis of percentage total microtubule length in the flow‐chamber covered by Tau envelopes. Pooled data points from three experiments (each data point represents tau coverage of microtubules within one field of view) with medians and interquartile ranges represented. Mann–Whitney test, P‐values displayed.
Figure EV2. Additional assays on Tau‐microtubule interactions (complement to Fig 2).
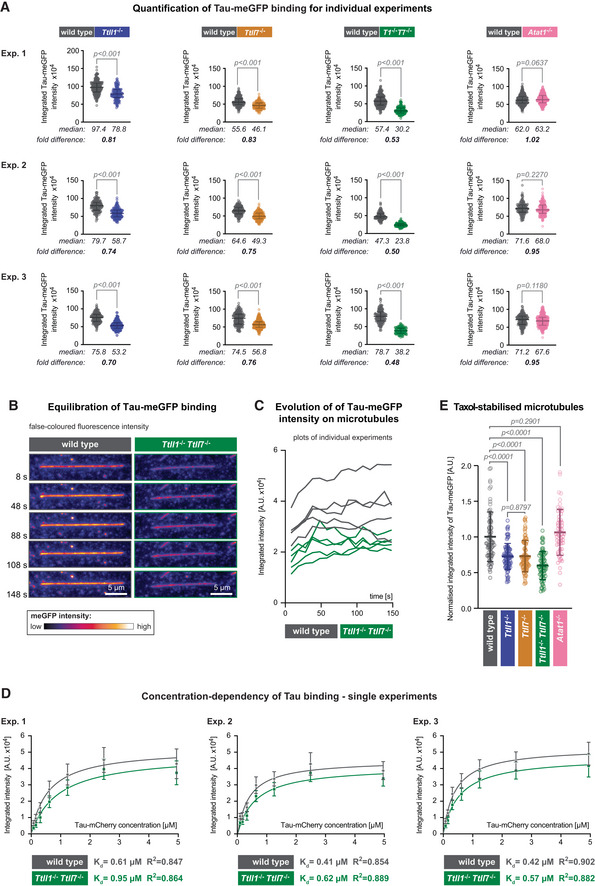
- Quantification of the integrated intensity of Tau‐meGFP on GMPCPP microtubules shown for the single assays that were combined in Fig 2C after normalisation to the median values measured on wild‐type microtubules. Three independent sets of experiments were performed with independently purified tubulin samples. Each data point represents the quantification of one microtubule. Median values and interquartile ranges are shown for each scatter plot. Mann–Whitney test, P‐values displayed.
- Representative still images from a time lapse of Tau‐meGFP binding to GMPCPP microtubules. The images are false coloured to better reveal the intensity evolution over time. Scale bars 5 μm.
- Raw integrated intensity values (A.U. × 104) of Tau‐meGFP binding to GMPCPP wild‐type and Ttll1 −/− Ttll7 −/− microtubules over a 150 s time interval (data points every 10 s). For each type 5 microtubules from the same assay were analysed.
- Saturation curves from individual experiments with microtubules from independently purified tubulin samples. Increasing concentration Tau‐mCherry binding to GMPCPP wild‐type and Ttll1 −/− Ttll7 −/− microtubules were quantified in one assay with each of the shown concentrations per experiment. Each data point is a mean (±standard deviation) of the raw integrated intensity values of single microtubules for the respective concentration (A.U. × 104). The derived dissociation constants K d and R 2 coefficient from the nonlinear fit for each microtubule type are indicated below the plots. The means of the three independent experiments shown here are combined in Fig 2E.
- Quantification of normalised integrated intensity of Tau‐meGFP fluorescent intensity on Taxol‐stabilised microtubules. Data of individual assays were normalised to mean values from wild‐type microtubules, and then combined as in Fig 2C. Each data point represents a single microtubule measurement from one experiment (n = 57–80 microtubules). Scatter plots with mean values (A.U.) and standard deviation. Student's t‐test, P‐values are shown.
Quantification of normalised microtubule‐associated meGFP intensities 5 min after the addition of Tau‐meGFP in three independent experiments revealed that Tau bound stronger to wild‐type microtubules, while partial or complete loss of polyglutamylation consistently led to a reduction of Tau‐meGFP signal along microtubules. Both microtubules lacking polyglutamylation on either α‐tubulin (Ttll1 −/−) or β‐tubulin (Ttll7 −/−) showed a reduction of about 25%, while absence of polyglutamylation on both tubulin subunits (Ttll1 −/− Ttll7 −/−) led to a decrease of about 50%. By contrast, the absence of acetylation on Atat1 −/− microtubules did not impact Tau–microtubule interactions (Fig 2C). These results were highly consistent between three sets of experiments (Fig 2D) performed with three independently purified sets of brain tubulin (Fig 1), thus demonstrating the reproducibility of our tubulin purifications.
Our results demonstrate that the levels and patterns of polyglutamylation present in wild‐type brain tubulin enhance the affinity of Tau for microtubules, while acetylation of K40 has no impact. The effect of polyglutamylation on the binding affinity of Tau was similar for each tubulin subunit, which was surprising given that α‐tubulin from the brain is polyglutamylated, that is, carrying more posttranslationally added glutamate residues than β‐tubulin, which carries only short glutamate chains (Fig 1A). This demonstrates that the presence of this PTM, but less so the number of incorporated glutamate residues controls the affinity of Tau to the microtubule surface. Consequently, the impact of polyglutamylation of α‐ and β‐tubulin in the affinity of Tau to microtubules is additive. This is consistent with recent structural data showing that Tau binds to microtubules along their protofilaments, thereby attaching its microtubule‐binding domains to both α‐ and β‐tubulin (Kellogg et al, 2018).
To determine whether microtubule polyglutamylation can control Tau‐microtubule interactions at a range of concentrations, we measured the binding of up to 5 μM Tau‐mCherry to wild‐type and Ttll1 −/− Ttll7 −/− microtubules. Fitting these intensities with a one‐site‐specific binding model (GraphPad Prism 9) yielded dissociation constants of 0.48 ± 0.1 μM (mean ± SD, N = 3 experiments with tubulin from independent purifications) for wild‐type, that is, glutamylated microtubules, and 0.71 ± 0.2 μM (mean ± SD, N = 3) for Ttll1 −/− Ttll7 −/−, that is, non‐glutamylated microtubules (Figs 2E and EV2D). Moreover, while the binding of Tau‐mCherry to microtubules saturated at about 2.5 μM for both PTM types, we persistently observed a higher fluorescent intensity of Tau on wild‐type compared with Ttll1 −/− Ttll7 −/− microtubules even at the maximum concentration of 5 μM, where a large part of Tau molecules remains unbound in the buffer surrounding the microtubules in the flow‐chamber. This indicates that an excess of Tau cannot fully compensate for the loss of microtubule polyglutamylation and that not only the affinity but also the degree of Tau occupancy on the microtubules is determined by the presence of this PTM. In general, this result implies that Tau–microtubule interactions in neurons can be regulated by polyglutamylation in a wide range of intracellular Tau concentrations.
Finally, to determine whether the effects of polyglutamylation that we found to affect Tau‐microtubule binding can also impact collective molecular processes, we determined how this PTM impacts the formation of cohesive Tau envelopes, a process in which Tau molecules condense into low‐dynamic patches that envelop microtubule surfaces (Siahaan et al, 2019; Tan et al, 2019). The formation of these cohesive envelopes is dependent on the nucleotide‐bound state of the tubulin, as they were shown to specifically enrich on the compacted GDP‐tubulin lattice. Consequently, these envelopes assemble on Taxol‐, but not GMPCPP‐stabilised microtubules (Siahaan et al, 2022).
We first compared the diffusive, non‐cohesive binding of Tau to Taxol‐stabilised microtubules made from all PTM variants previously analysed with GMPCPP‐stabilised microtubules (Fig 2). To minimise the formation of envelopes that conceal the diffuse binding mode, we used lower concentration of Tau‐meGFP (18 nM) in these experiments. Quantification of the microtubule‐associated meGFP signal revealed an about 25% decrease of Tau‐binding to Ttll1 −/− or Ttll7 −/− microtubules, as well as an almost 50% reduction of Tau‐binding to Ttll1 −/− Ttll7 −/− microtubules, while Atat1 −/− had no impact on Tau binding (Fig EV2E). This demonstrated that diffusive, non‐cohesive binding of Tau is similarly affected by tubulin polyglutamylation on Taxol‐ and GMPCPP‐stabilised microtubules (Fig 2B and C).
Following previous reports (Siahaan et al, 2019; Tan et al, 2019), we next increased the Tau concentration to 40 nM on Taxol‐stabilised microtubules, which readily led to the formation of envelopes on both wild‐type and Ttll1 −/− Ttll7 −/− microtubules (Fig 2F and G, and Movie EV2). As demonstrated before (Siahaan et al, 2019; Tan et al, 2019), Tau envelopes are dynamic and grow along the microtubules. To avoid a potential saturation of this process in our assays, we analysed only the active phase of Tau envelope formation, that is, 5 and 120 s after the addition of Tau‐mCherry. To determine whether polyglutamylation affects the efficiency of Tau‐envelope assembly, we quantified the relative microtubule length covered by Tau envelopes. At the two chosen time points, we observed a clear tendency for Tau envelopes to form preferentially on polyglutamylated (wild‐type) microtubules compared with non‐glutamylated (Ttll1 −/− Ttll7 −/−) microtubules (Fig 2F and G, and Movie EV2). This indicates that despite the important role of cohesive Tau–Tau interactions in the formation of Tau envelopes, the regulation of Tau–microtubule interactions by polyglutamylation also plays an important role in promoting the formation of those cohesive assemblies.
Polyglutamylation increases the microtubule severing rate of katanin
An enzymatic process known to be controlled by tubulin polyglutamylation is microtubule severing. In vitro evidence has so far been obtained for the microtubule‐severing enzyme spastin by two experimental approaches. Tubulin in cultured HeLa cells was glutamylated with TTLL4 or TTLL6. Both types of polyglutamylation increased the severing rate of spastin; however, TTLL6‐mediated generation of long glutamate chains had a much stronger effect than TTLL4, which generates only short chains (Lacroix et al, 2010). A second study used purified TTLL7 to generate a spectrum of tubulin polyglutamylation variants in vitro. They demonstrated that initial increase in the number of incorporated glutamates (≤ 8E) increased spastin‐mediated severing, whereas further incorporation of glutamate into tubulin had an inverse effect (Valenstein & Roll‐Mecak, 2016). Very recently, a similar approach has been used to assess the impact of polyglutamylation on katanin‐mediated microtubule severing (Szczesna et al, 2022). However, all experiments so far had used tubulin on which polyglutamylation had been added by overexpressed or recombinant enzymes; thus, the impact of physiological sites and levels of polyglutamylation on microtubule severing had remained unknown.
We, thus, used our panel of brain tubulin variants (Fig 1) in combination with the microtubule‐severing enzyme katanin (McNally & Vale, 1993) to determine whether physiological levels of tubulin polyglutamylation can affect microtubule severing. In our assays, we always compared two different PTM variants of Taxol‐stabilised microtubules side‐by‐side in the presence of 100 nM katanin. We purified a complex of the murine p60 subunit together with the truncated (amino acids 481–658) p80 subunit, which was C‐terminally tagged with two StrepII and one EGFP tag (Jiang et al, 2017; Rezabkova et al, 2017). After immobilising microtubules on the coverslip, the p60/p80 katanin complex was flushed into the chamber and the microtubules were recorded in IRM for 3 min (Fig 3A and B, and Movie EV3). To determine the severing rate, we analysed the change in the overall length of each microtubule throughout the experiment, which provided decay curves from which we determined the time required to lose 50% of the original overall microtubule length (Fig 3C). In each assay, we measured half‐life for multiple microtubules and determined their medians (Figs 3D and EV3). In our assays, the severing efficiencies of katanin could differ substantially between different sets of experiments (Fig EV3), most likely due to varying overall numbers of microtubules present in the reaction chambers. To allow statistical analyses between different sets of experiments, we cross‐normalised the measured half‐life values obtained from microtubules of the same PTM subtype, with wild‐type values set to 1 (Fig EV3).
Figure 3. Polyglutamylation controls the microtubule‐severing activity of katanin.
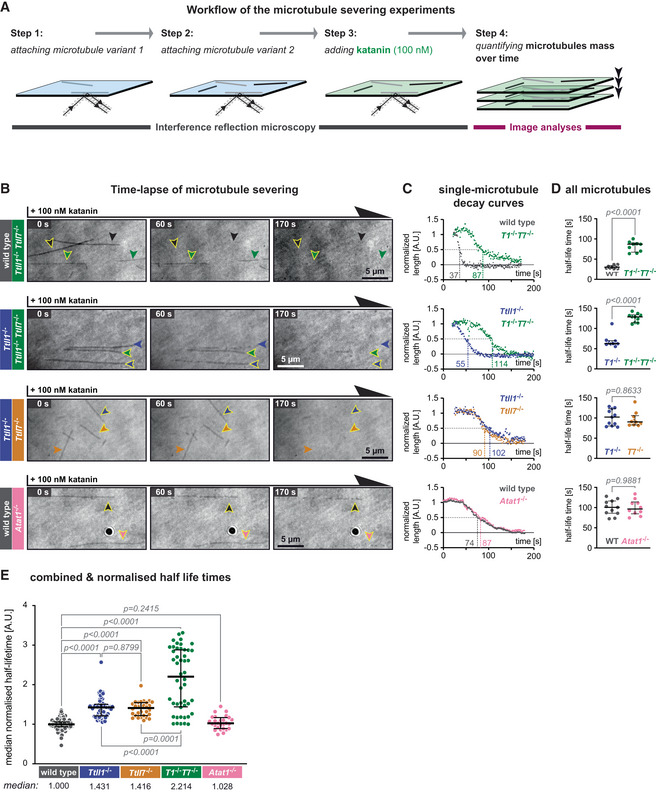
- Schematic representation of the katanin severing assay. Label‐free Taxol‐stabilised microtubules, polymerised from wild‐type and knockout‐mouse brain tubulin, are attached to a glass coverslip sequentially and imaged in IRM mode. Their identity is retained by their positions at the coverslip (Steps 1 and 2). Next, 100 nM purified p60/p80 katanin complex is added to the chamber (Step 3). The change in the microtubule length over time is recorded in IRM and quantified in subsequent analyses (Step 4).
- Representative IRM still images at different time points from time‐lapse following the addition of 100 nM p60/p80 katanin complex on microtubules (Movie EV3). Arrowheads indicate microtubules of a certain type (colour‐coded, as labels on the left). Scale bars 5 μm.
- Decay curves of normalised microtubule length plotted against time upon incubation with katanin (A.U., arbitrary units) for single microtubules highlighted by arrow heads with yellow borders in (B).
- Half‐life times (seconds) of all microtubules from the assays of which representative images are displayed in (B) and representative decay curves are shown in (C). Each data point represents the half‐life of a single microtubule. Medians with interquartile ranges are shown. Mann–Whitney test, P‐values displayed.
- Analysis of normalised microtubule half‐lifetimes from 14 independent experiments (Fig EV3). Each data point represents half‐life of a single microtubule. Medians with interquartile ranges shown, Mann–Whitney test and P‐values displayed.
Figure EV3. Individual katanin severing assays (complement to Fig 3).
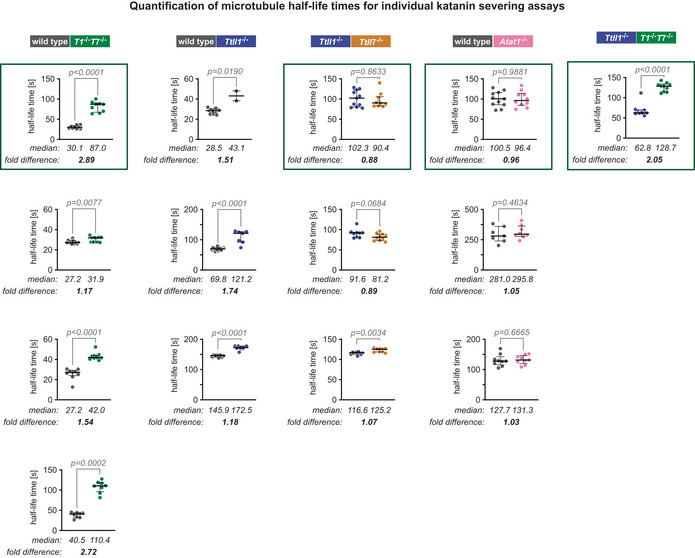
Representation of all individual quantifications of microtubule length half‐life times in the presence of 100 nM p60/p80 katanin separately for all 14 experiments. As shown in Fig 3A, two types of microtubules were always compared in the same chamber to mitigate the variability of katanin and microtubule concentrations in different measurement channels and, thus, ensure comparability between the samples. Each data point represents the half‐life time of one microtubule determined as shown for representative examples in Fig 3B, with bars representing medians and interquartile ranges. Mann–Whitney test, P‐values are shown. Selected plots (green frames) are shown in Fig 3D. To adjust all assays for a comparative analysis shown in Fig 3E, values were normalised to wild‐type microtubules. To normalise assays not including this type of microtubules, we used the weighted median fold difference between wild‐type and Ttll1 −/− microtubules from three independent experiments (second column) which provided the factor 1.45. This factor was used to normalise values in experiments omitting wild‐type, but including Ttll1 −/− microtubules.
Comparing the normalised half‐life values revealed that the strong reduction of polyglutamylation on α‐ and β‐tubulin (Ttll1 −/− Ttll7 −/−) decreased the severing activity of katanin on those microtubules more than twofold (Fig 3E). We further observed that the loss of either α‐ (Ttll1 −/−) or β‐tubulin polyglutamylation (Ttll7 −/−) alone led to an about 1.5‐fold decrease in severing activity. The lack of acetylation (Atat1 −/−), by contrast, did not affect the severing rate of katanin (Fig 3E), thus demonstrating that both α‐ and β‐tubulin polyglutamylation, but not acetylation, control katanin‐mediated severing.
A recent structural study has shown that katanin grips the C‐terminal tails of tubulin via electrostatic interactions. Using synthetic peptides, it further showed that the presence of greater numbers of glutamate residues in the amino acid sequence of the tails increases katanin activity, which suggested that polyglutamylation could activate katanin‐mediated severing (Zehr et al, 2020). Our work now demonstrates that posttranslational polyglutamylation of tubulin does stimulate katanin activity. Importantly, we show that physiological levels of polyglutamylation found on neuronal tubulin are sufficient to activate katanin. Our observation that polyglutamylation of both α‐ and β‐tubulin can activate katanin‐mediated severing, suggesting that both enzymes involved in tubulin polyglutamylation in the brain, TTLL1 and TTLL7, are able to control microtubule mass and architecture, two physiological roles of microtubule severing (Kuo & Howard, 2021).
β‐Tubulin polyglutamylation specifically decreases the run‐length of Kif5B
The motility of molecular motors may be controlled by subunit‐specific polyglutamylation of tubulin, as previously suggested by experiments with semi‐synthetic chimeric yeast tubulin (Sirajuddin et al, 2014). Moreover, we have recently shown that pathologically increased polyglutamylation in neurons negatively affects the mobility of several neuronal cargoes (Magiera et al, 2018; Bodakuntla et al, 2020b), while the inverse effect was observed in neurons from Ttll1 −/− mice that display decreased polyglutamylation levels (Bodakuntla et al, 2021). Although cargo transport is a complex process involving multiple molecular players, these observations strongly suggested that molecular motors can sense the polyglutamylation of the microtubule tracks. We, thus, aimed to determine whether polyglutamylation levels and patterns of brain tubulin directly impact the motility paraments of the kinesin‐1 motor Kif5B.
To measure the motility of Kif5B on our set of tubulin variants (Fig 1), we attached two microtubule variants side‐by‐side in the imaging chamber, after which we added 0.2 nM of human Kif5B (1–905), C‐terminally labelled with EGFP followed by a 6xHis‐tag (Kif5B‐EGFP). This truncated version is constitutively active, as it lacks the C‐terminal domain responsible for kinesin autoinhibition (Verhey et al, 1998; Coy et al, 1999), which allows for measuring single‐molecule motility. Single kinesin motors (Kif5B‐EGFP) moving along microtubules (Fig 4A and B, and Movie EV4) were analysed using the high‐precision tracking software FIESTA (Ruhnow et al, 2011) to obtain the instantaneous frame‐to‐frame velocities, average velocities, run lengths and interaction times (Appendix Fig S2).
Figure 4. Selective impact of polyglutamylation patterns on kinesin‐1 motility.
-
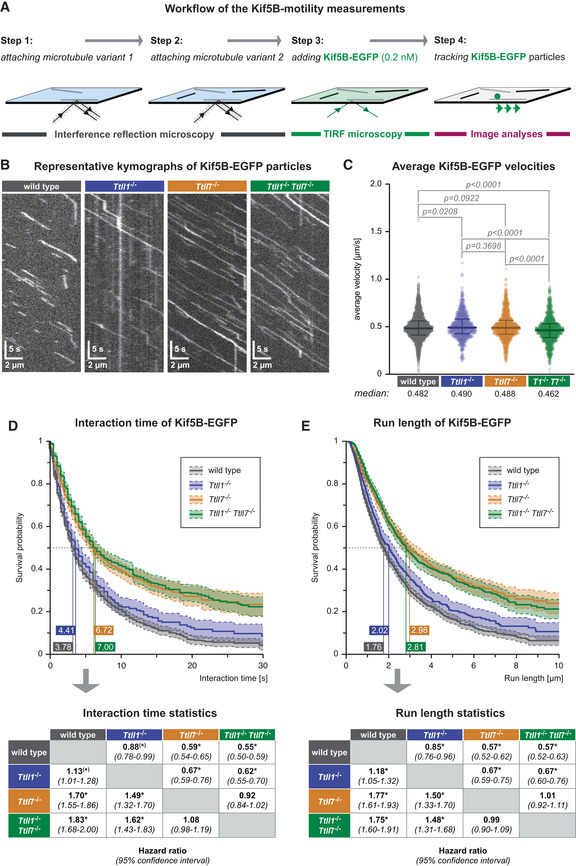
ASchematic representation of the single‐molecule kinesin‐1 motility assay. Taxol‐stabilised microtubules of two different types are subsequently attached in the flow‐chamber (Steps 1, 2), 0.2 nM Kif5b‐EGFP are added, the motility of single kinesin motors recorded in TIRF microscopy (Step 3) and subsequently analysed (Step 4).
-
BKymographs showing single Kif5B‐EGFP motor molecules moving on one single microtubule of known PTM state (Movie EV4). Horizontal scale bars 2 μm; vertical scale bars 5 s.
-
CScatter plots showing the average velocities (μm/s) exhibited by single Kif5b‐EGFP molecules on the different microtubule types (each data point represents one Kif5b‐EGFP molecule). Data pooled from 29 independent experiments (Appendix Fig S2). Medians with interquartile ranges shown, Mann–Whitney test and p‐values displayed.
-
D, ESurvival probability (Kaplan–Meier plots, taking varying microtubule lengths into account; see Materials and Methods) of the interaction time (D) and run length (E) of Kif5b‐EGFP molecules on different microtubule types. Data in (C–E) were pooled from individual experiments (Appendix Fig S2) without normalisation. The 50% probability values are shown in the plots. Statistical analysis displayed in the tables below: hazard ratios with 95% confidence intervals pairing all data from different microtubule types (*pairs differing with significance level P < 0.05).
While the average velocities (Fig 4C) and landing rates (Fig EV4A) of Kif5B‐EGFP molecules were almost identical on wild‐type, Ttll1 −/−, Ttll7 −/−, or Ttll1 −/− Ttll7 −/− microtubules of similar median lengths (Fig EV4B), the interaction time (Fig 4D), and the run length (Fig 4E) of the motors were significantly increased on microtubules without polyglutamylation (Ttll1 −/− Ttll7 −/−). The qualitative similarities of the run lengths and interaction times show that the motors spent most of their time on microtubules in the motile state, with only few pauses. Strikingly, both parameters appear to be specifically controlled by the polyglutamylation of β‐tubulin, as lack of TTLL7 (Ttll7 −/−), but not of TTLL1 (Ttll1 −/−) alone resulted in increased interaction time and run length to a similar extent as seen for entirely non‐polyglutamylated (Ttll1 −/− Ttll7 −/−) microtubules (Fig 4D and E).
Figure EV4. Additional parameters for kinesin‐1 motility assays (complement to Fig 4).
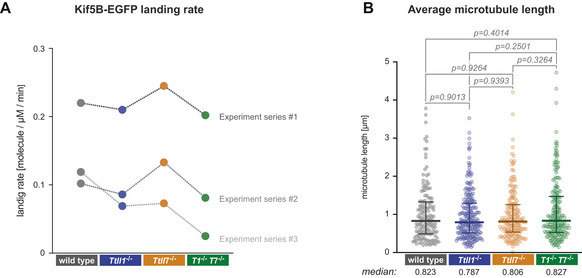
- Comparison of the Kif5B‐EGFP landing rates in three independent sets of experiments. Landing rates were calculated by summing up the total number of single‐molecule Kif5B‐EGFP landing events on microtubules relative to the total (summed up) length of microtubules in the field of view and the observation time. Note that landing rates differ between different sets of experiments, but are mostly similar for different PTM subtypes of microtubules within all experimental sets.
- Median microtubule length (each data point represents one microtubule) determined from all microtubules observed in three independent sets of experiments. Medians with interquartile ranges shown, Mann–Whitney test and P‐values displayed.
Our findings demonstrate that polyglutamylation of β‐tubulin, catalysed by TTLL7 under physiological conditions in neurons, can act as a specific regulator of Kif5B motility. Initial mass‐spectrometry analyses had shown that the two major β‐tubulin isotypes ‐ β2b and β3, are modified with 2–4 and 1–6 glutamates, respectively (Alexander et al, 1991; Rüdiger et al, 1992), which amounts to an average of three glutamates per β‐tubulin in the brain. That a modification with three glutamates on β‐tubulin can impact kinesin motility had previously been suggested from experiments with the genetically engineered chimera of yeast tubulin. Chemical coupling of three‐glutamates branch‐chains to chimeric β‐tubulin, and a 10‐glutamate branch chain to α‐tubulin decreased the processivity of Kif5B by almost 50% compared with non‐glutamylated control microtubules (Sirajuddin et al, 2014). While these experiments did not allow us to distinguish the impact of α‐and β‐tubulin polyglutamylation, they provided a first indication that polyglutamylation can impact the velocity of Kif5B at the single‐molecule level.
Mechanistically, the selective effect of β‐tubulin polyglutamylation on kinesin concurs with the fact that kinesin motor domains directly bind to the β‐tubulin subunits of the tubulin dimer (Hoenger et al, 2000; Uchimura et al, 2006), and in particular, the kinesin‐1 binding site lies across the interface between β‐ and α‐tubulin, where the C‐terminal tail of β‐tubulin is positioned (Gigant et al, 2013). Moreover, it had been demonstrated previously that loss of the negatively charged C‐terminal tubulin tails increases the binding of kinesin heads to tubulin (Skiniotis et al, 2004), which could explain why the further accumulation of negative charge on those tails by polyglutamylation decreases kinesin–microtubule interactions. The polyglutamylation of β‐tubulin by TTLL7 could, thus, be a specific regulatory mechanism of kinesin motility which acts independently of other functions controlled by TTLL1‐mediated polyglutamylation.
Discussion
Our work demonstrates that tubulin polyglutamylation modulates the function of three different types of microtubule‐interacting proteins: Tau as a neuron‐specific “structural” MAP; katanin, a microtubule‐severing enzyme, and a molecular motor from the kinesin‐1 family. All three proteins show a specific sensitivity to polyglutamylation patterns, which are generated at the C‐terminal tails of tubulin that are located at the microtubule surface and, thus, accessible to interacting proteins. By contrast, we and others (Walter et al, 2012; Kaul et al, 2014) found that Tau, katanin and kinesin‐1 are insensitive to tubulin acetylation, a tubulin PTM localised at the luminal microtubule face, which likely precludes direct contact with those proteins. This observation also excludes that the increase of tubulin acetylation in the absence of polyglutamylation, which we have observed, contributes to the data obtained on those microtubules.
The observation that different patterns of polyglutamylation had unique and specific impacts on the behaviour of the three proteins provided direct in vitro evidence for the tubulin code hypothesis, which predicts that specific PTM patterns can selectively control microtubule‐based functions. Importantly, we demonstrated that these effects can be observed at physiologically relevant levels of PTMs, given that the tubulin used in our assays was purified from the brain tissue of mice, thus representing average wild‐type levels of all tubulin PTMs, or selectively lacking a PTM or PTM subtype catalysed by the enzyme that is knocked out in the mouse models. Such experiments could only be performed now, as the required mouse models with specific polyglutamylation patterns in the brain have just recently become available (Bodakuntla et al, 2021).
A technical aspect that was essential to the success of our approach was the reproducibility of measurements performed with independently purified batches of tubulin. In the past, in vitro experiments with microtubules were classically performed with single, large batches of brain tubulin to assure reproducibility throughout entire studies. This, however, is impossible if the role of tubulin PTMs should be tested, as different, independent purifications of tubulin need to be compared. Here we demonstrated that multiple batches of tubulin purified from different mouse brains (Souphron et al, 2019; Bodakuntla et al, 2020a) give reproducible results in our in vitro reconstitution experiments, which enabled us to compare different PTM subtypes with biological replicates of tubulin of each type.
Our in vitro measurements revealed that multiple microtubule‐interacting proteins are regulated by polyglutamylation, thus raising the question about the functional relevance of this PTM in cells. One of the striking observations was that the kinesin‐1 motor Kif5B in single‐molecule settings was solely impacted by polyglutamylation generated on β‐tubulin by TTLL7, but not by TTLL1‐mediated α‐tubulin polyglutamylation. This was surprising as it had previously been shown that axonal transport in neurons, a process driven by kinesin‐1 motors was not affected by the loss of TTLL7‐, but rather by TTLL1‐mediated polyglutamylation (Bodakuntla et al, 2021). The most likely explanation for this discrepancy is that axonal cargoes are transported by multiple motor proteins. Therefore, the effect we observed with single kinesin‐1 motors – alteration of interaction time and run length – would be overridden when multiple motor molecules bound to a cargo collectively participate to its motility in cells. But why then did TTLL1‐mediated polyglutamylation, which in the single‐molecule experiments had no effect on Kif5B motility, systematically affect the transport of several cargoes in neurons (Bodakuntla et al, 2021)? Two possible answers could be provided based on our current findings that microtubule polyglutamylation impacts their interactions with Tau, the propensity of Tau to form envelopes, and the activation of katanin‐mediated severing. On one hand, the formation of more, or more stable Tau envelopes on glutamylated microtubules could hinder cargo transport by blocking the advancement of the molecular motors (Siahaan et al, 2019; Tan et al, 2019). Alternatively, the activation of katanin or other severing enzymes by microtubule polyglutamylation (Lacroix et al, 2010; Valenstein & Roll‐Mecak, 2016) could lead to a remodelling of the microtubule cytoskeleton in cells, to generate, for instance, more fragmented microtubule tracks. This could cause cargoes to change tracks more frequently, and lead to more frequent halts. Whether one of those, or other, yet unknown mechanisms controlled by tubulin polyglutamylation affect the transport of cargoes in living cells is an exciting question to be addressed next.
Our work illustrates that analysing how individual proteins sense changes in tubulin PTMs paves the way to a mechanistic understanding of complex intracellular processes, and must, thus, precede and accompany studies in cells to avoid their misinterpretation. For example, overexpression of the deacetylase HDAC6 and katanin in fibroblasts and neurons had initially suggested that microtubule acetylation increases the rate of katanin‐mediated severing (Sudo & Baas, 2010), which we could not confirm in our in vitro experiments. Similarly, pharmacological inhibition of HDAC6 led to increased microtubule acetylation and changes in kinesin‐1 behaviour in these cells. This suggested that tubulin acetylation controls kinesin‐1 transport in cells (Reed et al, 2006), which however was not confirmed in vitro (Walter et al, 2012; Kaul et al, 2014). Discrepancies between cell‐based and in vitro observations can have many reasons. Apart from multiple PTM‐sensitive mechanisms being affected simultaneously in cells, another possibility is that manipulation of PTMs in cells through acute overexpression, or chemical inhibition of tubulin‐modifying enzymes could have additional side‐effects that might affect the behaviour of microtubule interactors inside the cells.
Having shown that three key microtubule interactors are regulated by the same PTM—polyglutamylation—raises the question of the specificity by which this PTM regulates microtubule functions in cells. By showing that Kif5B was only sensitive to β‐tubulin polyglutamylation, while both Tau and katanin were sensitive to polyglutamylation of α‐ and β‐tubulin, we demonstrated that different polyglutamylation enzymes can independently control different microtubule interactors. Our current work, thus, provides the first snapshot of a complex regulatory network that might, by tightly regulating PTM levels of microtubules in a locally and temporally controlled manner choreograph the multiple cellular functions that depend on the microtubule cytoskeleton. Our data underpin that the impact of microtubule polyglutamylation on microtubule‐based processes is rather gradual, as none of them was abolished in the absence of this PTM, but only altered. This confirms and broadens the observation of previous studies, which showed that polyglutamylation acts as a graded controller of spastin, and very recently of katanin‐mediated microtubule severing (Valenstein & Roll‐Mecak, 2016; Szczesna et al, 2022). While such gradual regulatory mechanisms, in contrast to PTM switches that for instance control cell cycle progression (Nurse, 2002; Santos & Ferrell, 2008; Skotheim et al, 2008), might have small effects on cells we observe in culture dishes, they are likely to be essential for the maintenance of homeostasis and cellular functions throughout the life of an organism. That this could indeed be the case is illustrated by recent findings showing that deregulated tubulin PTMs can cause neurodegeneration (Magiera et al, 2018; Shashi et al, 2018), vision disorders (Kastner et al, 2015; Branham et al, 2016), male infertility (Vogel et al, 2010; Giordano et al, 2019; Gadadhar et al, 2021), ciliopathies (Lee et al, 2012) and cancer (Rocha et al, 2014).
Materials and Methods
Mouse breeding
Animal care and use for this study were performed in accordance with the recommendations of the European Community (2010/63/UE) for the care and use of laboratory animals. Experimental procedures were specifically approved by the ethics committee of the Institut Curie CEEA‐IC #118 (authorisation no. 04395.03 given by National Authority) in compliance with the international guidelines. Adult males and females (2–8 months) were used in this study.
Mouse lines
Ttll1 −/−: C57BL/6NTac‐Ttll1 tm1a(EUCOMM)Wtsi /IcsOrl mice were generated at PHENOMIN‐ICS (Institut Clinique de la Souris, Illkirch, France; www.ics‐mci.fr) with an ES‐cell clone obtained from the International Mouse Phenotyping Consortium (IMPC) (www.mousephenotype.org/data/alleles/MGI:2443047/tm1a(EUCOMM)Wtsi), and bred first to flp‐, then to cre‐recombinase‐expressing animals (Lallemand et al, 1998) to create the Ttll1 −/− line as described before (Bodakuntla et al, 2021).
Ttll7 −/−: The Ttll7 tm1Ics conditional mutant mouse line was established at PHENOMIN‐ICS (Institut Clinique de la Souris, Illkirch, France; www.ics‐mci.fr) as conditional (Lox‐P) line, and then bred to cre‐recombinase–expressing animals (Lallemand et al, 1998) to create the Ttll7 −/− line as described before (Bodakuntla et al, 2021).
Ttll1 −/− /Ttll7 −/−: The line was created by crossing Ttll1 −/+ to Ttll7 −/+ mice.
Atat1 −/−: These mice were described before (Kalebic et al, 2013).
Mouse genotyping
Genotyping of animals was performed as previously described (Magiera et al, 2018). The PCR primers, the amplification program and the expected PCR product size for mouse each line are presented in Appendix Table S1.
Tubulin purification from murine brains
Tubulin was purified from mouse brains via cycles of temperature‐dependent microtubule polymerisation and depolymerisation as described in detail in Souphron et al (2019) and Bodakuntla et al (2020a). Briefly, brains were extracted from the skulls of mice immediately after sacrificing the animals. For 1 g of brain tissue, 2 ml of ice‐cold lysis buffer consisting of BRB80 (80 mM K‐PIPES pH 6.8, 1 mM K‐EGTA, 1 mM MgCl2, 1 mM β‐mercaptoethanol, 1 mM PMSF, and 1× protease inhibitor cocktail composed of 20 μg/ml leupeptin, 20 μg/ml aprotinin and 20 μg/ml 4‐(2‐aminoethyl)‐benzenesulfonyl fluoride; Sigma‐Aldrich) were added, and brains were homogenised using an Ultra‐Turrax® blender. Lysates were cleared by centrifugation at 50,000 rpm in TLA‐55 fixed‐angle rotor (~112,000 × g; Beckman Coulter) for 30 min at 4°C, from now on referred to as “cold centrifugation”. This first supernatant (later referred to as “SN1”) was adjusted to 1 mM GTP and 1/3 volume pre‐warmed 100% glycerol, which after careful mixing was incubated for 20 min at 30°C for microtubule polymerisation. Polymerised microtubules were pelleted for 30 min at 112,000 × g, 30°C, referred to as “warm centrifugation”. The pellet was resuspended in BRB80 (1/10 of the initial SN1‐volume), and microtubules were disassembled by incubation on ice for 20 min, and occasionally pipetting up‐and‐down to accelerate their depolymerisation.
Solubilised tubulin was cleared via cold centrifugation and the supernatant SN3 was adjusted to final concentrations of 1 mM GTP and 0.5 M PIPES, and complemented with 1/3 volume pre‐heated 100% glycerol. After careful mixing and incubation for 20 min at 30°C the resulting microtubules were pelleted via warm centrifugation. Microtubules polymerised at high molarity do not contain associated proteins. The pellet was resuspended in 1 × BRB80 (1/40 of the initial SN1‐volume), depolymerised on ice for 20 min, and cleared via cold centrifugation. The resulting SN5 was adjusted to 1 mM GTP, supplemented with 1/3 volume of pre‐heated 100% glycerol, and incubated for 20 min at 30°C. Microtubules were pelleted by warm centrifugation and resuspended in ice‐cold BRB80 (1/40 of the initial SN1‐volume). After 15 min on ice, the soluble tubulin was cleared by a final cold centrifugation. The tubulin yield was estimated with a NanoDrop ND‐1000 spectrophotometer (Thermo Scientific; absorbance at 280 nm; MW = 110 kDa; ε = 115,000 M−1/cm−1). Samples were adjusted to 4 mg/ml, aliquoted, snap‐frozen in liquid N2 and stored at −80°C. Samples for SDS‐PAGE were collected and analysed for each purification cycle step.
Immunoblot analyses
Samples of the purified tubulin were mixed with Laemmli buffer (90 mM DTT, 2% SDS, 80 mM tris–HCl pH 6.8, 10% glycerol, Bromophenol blue) and denatured for 5 min at 95°C. Tubulin (60–80 ng per sample) was run on 10% SDS‐PAGE gels that allow the separation of α‐ and β‐tubulin (Magiera & Janke, 2013), and transferred onto a nitrocellulose membrane using Bio‐Rad Trans‐Blot® Turbo system according to manufacturer's protocol. Membranes were blocked in 5% fat‐free milk in PBS containing 0.1% Tween‐20 (PBST) for 1 h at room temperature. Primary antibodies were diluted in 5% milk in PBST and incubated with the blocked membranes overnight at 4°C. On the next day, the membrane was washed three times for 5 min with PBST and incubated with HRP‐conjugated secondary antibodies diluted in PBST for 1 h at room temperature. After three washes in PBST, membranes were incubated with Clarity™ Western ECL Substrate (Bio‐Rad), and the chemiluminescence signal was revealed using the Vilber Fusion FX imaging system. A list of all antibodies and dilutions is shown in Appendix Table S2.
Production of recombinant proteins
The longest human isoform of Tau with four microtubule‐binding repeats (hTau441, 2N4R), fused with C‐terminal meGFP or mCherry, 3C (PreScission) protease cleavage site and 6 × His‐tag, and human Kif5B‐EGFP (amino acids 1–905), containing a C‐terminal fluorescent‐tag followed by a 3C protease cleavage site and a 6 × His‐tag (Henrichs et al, 2020), were expressed and purified from Sf9 cells (Expression systems, Davis, CA, USA) using the opensource FlexiBAC baculovirus vector system for protein expression (Lemaitre et al, 2019). Cells were pelleted by centrifugation at 300 × g for 10 min at 4°C, snap‐frozen in liquid N2.
For purification, cells were resuspended in 1:1 ratio of lysis buffer (25 mM HEPES pH 7.4, 150 mM KCl, 1 mM DTT, 20 mM imidazole, 31.25 U Benzonase), and homogenised by repeated pipetting. The lysate was cleared for 1 h at 35,000 rpm (95,800 × g) in 45 Ti fixed‐angle rotor (Beckman Coulter) at 4°C. The supernatant was incubated with pre‐equilibrated HisPUR Ni‐NTA agarose resin (Thermo Fisher Scientific) for 2 h in a tightly closed gravity‐flow column (BioRad) at 4°C. Subsequently, the resin was washed once with low‐salt buffer (25 mM HEPES pH 7.4, 150 mM KCl, 1 mM DTT, 20 mM imidazole), once with high‐salt wash buffer (25 mM HEPES pH 7.4, 700 mM KCl, 1 mM DTT, 20 mM imidazole), and once again with low‐salt wash buffer. The protein was eluted from the resin after cleavage of the 6 × His‐tag with 3C HRV protease in low‐salt wash buffer at 4°C for 3.5 h. The eluted protein was further purified via size‐exclusion chromatography on Superdex® 200 10/300 GL column using ÄKTA Pure chromatography system (GE Healthcare) with gel filtration buffer (25 mM HEPES pH 7.4, 150 mM KCl, 1 mM DTT, 0.1 mM ATP, 1 mM EDTA). The collected peak fractions were analysed via SDS‐PAGE, pooled and the protein was concentrated with Amicon Ultra‐15 Centrifugal Filter tubes (Millipore). The final concentration was measured with NanoDrop ND‐1000 spectrophotometer (absorbance at 280 nm, MW and ε calculated according to the amino acid composition). Purified proteins were aliquoted, snap‐frozen in liquid N2, and stored at −80°C.
A complex of the katanin of subunits P60 and P80 (amino acids 1–481), C‐terminally tagged with two sequential StrepII tags and one EGFP tag, was expressed in HEK293 cells and purified as described previously (Jiang et al, 2017). Cells were harvested 36 h after transfection (500 × g for 5 min, 4°C), resuspended in 50 mM HEPES, 300 mM NaCl, 2 mM MgCl2, 1 mM EGTA, 0.5% Triton X‐100, pH 7.4 supplemented with protease inhibitors cocktail (cOmplete™, EDTA‐free, Roche, Basel, Switzerland), and lysed by sonication. Insoluble material was removed by centrifugation for 15 min at 9,000 × g, 4°C, and the supernatant was centrifuged again for 30 min at 30,000 × g, 4°C. The high‐speed supernatant was then incubated with 2 ml StrepTactin XT beads for 45 min at 4°C. Beads were subsequently washed with 50 ml of 50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, 1 mM EGTA, 0.05% Triton X‐100 and 10% glycerol. Finally, katanin was eluted with 50 mM HEPES, 150 mM NaCl, 1 mM EGTA, 2 mM MgCl2, 0.01% Triton X‐100 and 50 mM biotin. Purified protein was snap‐frozen and stored at −80°C.
Preparation of microscopy chambers
Microscopy chambers were prepared from two sandwiched silanised glass coverslips (18 and 24 mm, Marienfield High Precision) attached through parafilm strips (heated for 15–20 s at 60°C and gently pressed together) to form ~2.5 mm wide parallel flow channels. Prior to the chamber assembly, the coverslips were extensively cleaned in “piranha” solution (consisting of one part 30% hydrogen peroxide and 2.5 parts 95–97% sulfuric acid) and silanised with 200 μl dichlorodimethylsilane mixed in 350 ml trichloroethylene. To immobilise the microtubules at the glass surface, the flow channels of the microscopy chambers were incubated for 5 min with 30 μg/ml monoclonal anti‐β‐tubulin (TUBB) antibody (Sigma‐Aldrich T7816; Roach et al, 1998) at room temperature. The glass surface was subsequently blocked with 1% Pluronic‐F127 in PBS for at least 30 min before use.
Preparation of stabilised microtubules
For Tau binding experiments, microtubules were polymerised in the presence of the slowly hydrolysable GTP‐analogue GMPCPP (Guanosine‐5′‐[(α,β)‐methyleno]triphosphate; Jena Bioscience NU‐405). Tubulin from murine brains was mixed with 1.25 mM GMPCPP and 1.25 mM MgCl2 to final concentrations of 4 μM in BRB80, and incubated for 3–5 h at 37°C. The polymerised microtubules were pelleted by centrifugation at 18,000 × g for 30 min. The supernatant was discarded, and pellets were resuspended in 100 μl of warm (37°C) BRB80.
For Tau‐envelope formation, kinesin stepping assays and microtubule severing, Taxol‐stabilised microtubules were prepared in BRB80 following a previously described protocol (Nitzsche et al, 2010; Braun et al, 2011). We added 1.25 μl of the polymerisation mixture (25% DMSO, 20 mM MgCl2, 5 mM GTP in BRB80) to 5 μl of 4 mg/ml porcine tubulin. Microtubules were polymerised for 30 min at 37°C. Following centrifugation for 30 min at 18,000 × g and room temperature, the microtubule pellets were resuspended in 100 μl BRB80 containing 10 μM Taxol (Paclitaxel; Sigma‐Aldrich T7191).
GMPCPP‐ and Taxol‐stabilised microtubules were stored at room temperature and used for several days.
Immobilisation of microtubules in assay chambers
Stabilised microtubules were diluted in BRB80 and injected into a microscopy chamber. Dilutions were determined ad hoc for each experiment to obtain an optimal microtubule density. Importantly, all microscopy assays were performed in assay chambers containing two PTM types of microtubules concomitantly. These two different microtubule types were injected sequentially; each sample was incubated for 30–60 s, after which unbound microtubules were washed away with BRB80. The identity of the first set of microtubules was documented at several positions on the coverslip via IRM using 50:50 UVFS plate beamsplitter (ThorLabs BSW29R) and the position‐saving function of the NIS‐Elements imaging software (Nikon). The same procedure was repeated with the second type of microtubules, and images are taken for the previously saved positions (Fig 2A and B).
TIRF microscopy
Experiments were performed on inverted widefield microscope Nikon Eclipse Ti2 (Nikon, Tokyo, Japan) equipped with a motorised XY‐stage, a perfect focus system, Nikon Apo TIRF 60× Oil, NA 1.49 objective, and a Prime BSI Scientific CMOS (sCMOS) camera (Teledyne Photometrics, Tucson, AZ, USA), or with a Nikon Ti‐E equipped with a piezo Z‐stage, a motorised XY‐stage, a Perfect Focus System, an automatised H‐TIRF module, a 100× oil immersion TIRFM objective and an sCMOS Hamamatsu ORCA 4.0 V2 (Hamamatsu Photonics) camera, respectively. All experiments were carried out at stable room temperature and were repeated at least three times on different days, unless otherwise stated.
Tau‐binding: assay, image analyses and statistics
Seventy nanomolar Tau‐meGFP in Tau‐assay buffer (50 mM Hepes/KOH pH 7.3, 75 mM KCl, 10 mM DTT, 1 mM ATP, 20 μg/ml casein, 20 mM glucose, 220 μg/ml glucose oxidase, 20 μg/ml catalase) was injected into the imaging chamber containing two sets of immobilised microtubules, as video acquisition was started prior to the injection. Tau binding was recorded in TIRF mode at a rate of 0.5 fps for 5 min for one of the saved coverslip positions, after which single images were taken for all other positions. To determine the dissociation constant (K d) of Tau to wild‐type and Ttll1 −/− Ttll7 −/− microtubules, the following range of Tau‐mCherry concentrations was assayed as described above: 78, 156, 313, 625 nM; 1.25, 2.5 and 5 μM.
The binding affinity of Tau to microtubules was analysed with Fiji (ImageJ) software (Schindelin et al, 2012). To determine the fluorescence intensity of Tau‐meGFP or ‐mCherry bound to a given microtubule, a segmented line of 5 px (0.55 μm) width is drawn over parts or the entire microtubule. From this line, the area (A) and the raw integrated density (RID) of the fluorescent signal was determined. To determine the background, the line width was expanded to 12 px (1.32 μm) so that it covers parts of the surface surrounding the microtubule and the same measurements were repeated. To obtain the background value (B) the following calculation was applied:
To subtract the background signal (B), the following correction calculation was used:
where D is the corrected integrated fluorescence signal density of Tau on the microtubule.
In Fig 2C the value D for each wild‐type and mutant microtubule was normalised to the mean wild‐type D in the same channel of the microscopy chamber. Each experiment was performed with wild‐type and one differentially modified type of microtubules. For representation purposes, one normalised wild‐type data set was shown in the scatter plot for each experiment. At least 100 microtubules were measured for each condition in each biological replicate. Statistical analysis was performed using either unpaired t‐test or Mann–Whitney test in Prism 9 software (GraphPad), error bars represent mean with standard deviation (SD) and median with interquartile range, respectively.
To calculate the dissociation constants (Kd) of Tau, the experimental data were fitted with a built‐in model for one‐site specific binding using Prism 9 software (GraphPad) based on the following formula:
where D is the corrected integrated fluorescence signal density of Tau on the microtubule (calculated as described above), D max is the estimated maximum value of D and [Tau] is the concentration of Tau. Fig 2E shows the fitting of the mean D values from three independent experiments, as well as the calculated K d with SD. At least 40 microtubules were measured for each condition in each individual experiment.
Tau envelope assembly was recorded for 5 min and analysed at time points 5 and 120 s. The total length of the envelopes on all microtubules in the field of view was divided by the total length of those microtubules and expressed as a percentage in Fig 2G. The data are pooled from 17 assays, where tubulin from three independent purifications was used.
Katanin microtubule‐severing: assay, image analyses and statistics
One hundred nanomolar purified katanin‐eGFP in severing buffer (20 mM Hepes/KOH, pH 7.2, 75 mM KCl, 10 mM Paclitaxel, 0.5 mg/ml casein, 10 mM DTT, 3 mM ATP, 0.1% Tween‐20, 20 mM glucose, 110 μg/ml glucose oxidase, 20 μg/ml catalase) were injected into the imaging chamber. Fluorescent images were taken in TIRF mode at a rate of 0.5 fps for 5 min at all previously saved coverslip positions, image acquisition was started before injection.
Image analysis for estimating the microtubule disassembly was performed in Fiji (Schindelin et al, 2012). The integrated microtubule mass of each microtubule in each movie frame was determined by integrating the IRM intensity profile over time. This signal was background‐subtracted by subtracting the integrated IRM intensity profile of an equally sized region directly adjacent to the microtubule in each frame over time. The resulting signal was normalised to “1” at time 0, which is defined as the moment when katanin was added. Characteristic severing times were defined as the time it takes for the microtubule integrated intensity, reflecting the microtubule mass, to decrease to half of its initial value. For direct comparison in identical conditions, in each experiment, two types of microtubules were imaged simultaneously in one channel and one set of microtubules was taken as a reference. The mean value of the wild‐type severing rate was set to “1,” and values of other microtubule variants were normalised to the wild‐type value. If no wild‐type microtubules were included in an assay, we adjusted the values to the median values of one of the microtubule types obtained with the wild‐type microtubules. Statistical analysis was performed using Mann–Whitney t‐test in Prism 9 software (GraphPad), bars represent the median with an interquartile range.
Kinesin‐stepping: assay, image analyses and statistics
For Kif5B stepping assays, 0.2 nM Kif5B‐EGFP in kinesin motility buffer (BRB80, 10 μM Paclitaxel, 10 mM dithiothreitol, 20 mM D‐glucose, 0.5 mg/ml casein, 1 mM Mg‐ATP, 220 μg/ml glucose oxidase and 20 μg/ml catalase) were injected into the flow cell. Fluorescent images of single molecules were acquired at five frames per second for at least 1 min. Detailed acquisition information is indicated in the figure legends.
Motility parameters such as the interaction times, run length and velocities of Kif5B were determined by individually tracking the movement of single Kif5B‐EGFP molecules with the tracking software FIESTA (Ruhnow et al, 2011). The run length and interaction time were expressed as survival probability, determined by the Kaplan–Meier estimator in MATLAB (The MathWorks, Natick, MA, USA) and statistically compared as hazard ratios in JMP (Cary, NC, USA) as described previously (Henrichs et al, 2020). Kaplan–Meier analysis is a non‐parametric statistical test to estimate the survival function from lifetime data taking into account “censored events” that terminate due to causes unrelated to the kinesin walking mechanism (Ruhnow et al, 2017). Those events are processive runs prematurely terminated when Kif5B‐EGFP reaches microtubule ends, or move in‐ or out of the field of view. Including these events is important, as otherwise the analysis would be biased against long‐distance runs of kinesin molecules.
Using Kaplan–Meier analyses, thus, minimises the impact of possible variations in microtubule lengths (Ruhnow et al, 2017). Given that, in addition, we used microtubule sets of similar length distributions for our analyses (Fig EV4B) we can confidently exclude an influence of the microtubule lengths on the observed Kif5B‐EGFP run lengths and interaction times.
Author contributions
Mariya Genova: Data curation; formal analysis; funding acquisition; validation; investigation; visualization; methodology; writing – original draft; writing – review and editing. Lenka Grycova: Data curation; formal analysis; validation; investigation; visualization; methodology; writing – review and editing. Verena Puttrich: Resources; data curation; formal analysis; validation; investigation; visualization. Maria M Magiera: Conceptualization; resources; supervision; methodology; writing – original draft; project administration; writing – review and editing. Zdenek Lansky: Conceptualization; formal analysis; supervision; funding acquisition; validation; investigation; writing – original draft; project administration; writing – review and editing. Carsten Janke: Conceptualization; formal analysis; supervision; funding acquisition; validation; visualization; writing – original draft; project administration; writing – review and editing. Marcus Braun: Conceptualization; formal analysis; supervision; funding acquisition; validation; investigation; visualization; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Source Data for Expanded View
Review Process File
PDF+
Acknowledgements
This work was supported by the Agence Nationale de la Recherche ANR‐10‐IDEX‐0001‐02 and the LabEx Cell(n)Scale ANR‐11‐LBX‐0038. CJ is supported by the Institut Curie, the French National Research Agency (ANR) awards ANR‐17‐CE13‐0021 and ANR‐20‐CE13‐0011, and the Fondation pour la Recherche Medicale (FRM) grant DEQ20170336756. MMM is supported by the Fondation Vaincre Alzheimer FR‐16055p and the France Alzheimer grant 2023. MG is supported by Institut Curie 3‐i PhD Program (IC‐3i) and the European Molecular Biology Organisation short‐term fellowship 8843. We further acknowledge support by the Czech Science Foundation grants 19‐27477X to ZL and 22‐11753S to MB, institutional support from the CAS (RVO: 86652036), Centre of Molecular Structure supported by MEYS CR (LM2015043), the Imaging Methods Core Facility at BIOCEV supported by the MEYS CR (Large RI Project LM2018129 Czech‐BioImaging) and ERDF (project no. CZ.02.1.01/0.0/ 0.0/16_013/0001775). The Ttll7 −/− mouse line was established at the Mouse Clinical Institute (PHENOMIN‐Institut Clinique de la Souris, MCI/ICS) in the Genetic Engineering and Model Validation Department. We further thank C. Alberti, E. Belloir, K. Belloul, V. Dangles‐Marie, V. Henriot, C. Jouhanneau (Institut Curie) and V. Siahaan, V. Váňová and T. Šmídová (IBT CAS) for technical assistance. We are grateful to M.‐N. Soler and L. Besse from the imaging platform PICT‐IBiSA@Orsay (Institut Curie, Orsay) and J. Sabó (IBT CAS) for technical assistance in the use of the light microscopy systems.
The EMBO Journal (2023) 42: e112101
Contributor Information
Carsten Janke, Email: carsten.janke@curie.fr.
Marcus Braun, Email: marcus.braun@ibt.cas.cz.
Data availability
This study includes no data deposited in external repositories.
References
- Alexander JE, Hunt DF, Lee MK, Shabanowitz J, Michel H, Berlin SC, MacDonald TL, Sundberg RJ, Rebhun LI, Frankfurter A (1991) Characterization of posttranslational modifications in neuron‐specific class III beta‐tubulin by mass spectrometry. Proc Natl Acad Sci USA 88: 4685–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert S, Desbruyeres E, Gruszczynski C, Koulakoff A, Gros F, Denoulet P, Eddé B (1993) Reversible polyglutamylation of alpha‐ and beta‐tubulin and microtubule dynamics in mouse brain neurons. Mol Biol Cell 4: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodakuntla S, Jijumon AS, Janke C, Magiera MM (2020a) Purification of tubulin with controlled posttranslational modifications and isotypes from limited sources by polymerization‐Depolymerization cycles. J Vis Exp 10.3791/61826 [DOI] [PubMed] [Google Scholar]
- Bodakuntla S, Schnitzler A, Villablanca C, Gonzalez‐Billault C, Bieche I, Janke C, Magiera MM (2020b) Tubulin polyglutamylation is a general traffic‐control mechanism in hippocampal neurons. J Cell Sci 133: jcs241802 [DOI] [PubMed] [Google Scholar]
- Bodakuntla S, Yuan X, Genova M, Gadadhar S, Leboucher S, Birling M‐C, Klein D, Martini R, Janke C, Magiera MM (2021) Distinct roles of alpha‐ and beta‐tubulin polyglutamylation in controlling axonal transport and in neurodegeneration. EMBO J 40: e108498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branham K, Matsui H, Biswas P, Guru AA, Hicks M, Suk JJ, Li H, Jakubosky D, Long T, Telenti A et al (2016) Establishing the involvement of the novel gene AGBL5 in retinitis pigmentosa by whole genome sequencing. Physiol Genomics 48: 922–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M, Lansky Z, Fink G, Ruhnow F, Diez S, Janson ME (2011) Adaptive braking by Ase1 prevents overlapping microtubules from sliding completely apart. Nat Cell Biol 13: 1259–1264 [DOI] [PubMed] [Google Scholar]
- Castoldi M, Popov AV (2003) Purification of brain tubulin through two cycles of polymerization‐depolymerization in a high‐molarity buffer. Protein Expr Purif 32: 83–88 [DOI] [PubMed] [Google Scholar]
- Chang C‐W, Shao E, Mucke L (2021) Tau: enabler of diverse brain disorders and target of rapidly evolving therapeutic strategies. Science 371: eabb8255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy DL, Hancock WO, Wagenbach M, Howard J (1999) Kinesin's tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol 1: 288–292 [DOI] [PubMed] [Google Scholar]
- Eddé B, Rossier J, Le Caer JP, Desbruyeres E, Gros F, Denoulet P (1990) Posttranslational glutamylation of alpha‐tubulin. Science 247: 83–85 [DOI] [PubMed] [Google Scholar]
- Eshun‐Wilson L, Zhang R, Portran D, Nachury MV, Toso DB, Lohr T, Vendruscolo M, Bonomi M, Fraser JS, Nogales E (2019) Effects of alpha‐tubulin acetylation on microtubule structure and stability. Proc Natl Acad Sci USA 116: 10366–10371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadadhar S, Alvarez Viar G, Hansen JN, Gong A, Kostarev A, Ialy‐Radio C, Leboucher S, Whitfield M, Ziyyat A, Toure A et al (2021) Tubulin glycylation controls axonemal dynein activity, flagellar beat, and male fertility. Science 371: eabd4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigant B, Wang W, Dreier B, Jiang Q, Pecqueur L, Pluckthun A, Wang C, Knossow M (2013) Structure of a kinesin‐tubulin complex and implications for kinesin motility. Nat Struct Mol Biol 20: 1001–1007 [DOI] [PubMed] [Google Scholar]
- Gilmore‐Hall S, Kuo J, Ward JM, Zahra R, Morrison RS, Perkins G, La Spada AR (2019) CCP1 promotes mitochondrial fusion and motility to prevent Purkinje cell neuron loss in pcd mice. J Cell Biol 218: 206–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano T, Gadadhar S, Bodakuntla S, Straub J, Leboucher S, Martinez G, Chemlali W, Bosc C, Andrieux A, Bieche I et al (2019) Loss of the deglutamylase CCP5 perturbs multiple steps of spermatogenesis and leads to male infertility. J Cell Sci 132: jcs226951 [DOI] [PubMed] [Google Scholar]
- Goedert M, Crowther RA, Garner CC (1991) Molecular characterization of microtubule‐associated proteins tau and MAP2. Trends Neurosci 14: 193–199 [DOI] [PubMed] [Google Scholar]
- Goedert M, Eisenberg DS, Crowther RA (2017) Propagation of tau aggregates and neurodegeneration. Annu Rev Neurosci 40: 189–210 [DOI] [PubMed] [Google Scholar]
- Henrichs V, Grycova L, Barinka C, Nahacka Z, Neuzil J, Diez S, Rohlena J, Braun M, Lansky Z (2020) Mitochondria‐adaptor TRAK1 promotes kinesin‐1 driven transport in crowded environments. Nat Commun 11: 3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger A, Thormahlen M, Diaz‐Avalos R, Doerhoefer M, Goldie KN, Müller J, Mandelkow E (2000) A new look at the microtubule binding patterns of dimeric kinesins. J Mol Biol 297: 1087–1103 [DOI] [PubMed] [Google Scholar]
- Ikegami K, Mukai M, Tsuchida J‐i, Heier RL, Macgregor GR, Setou M (2006) TTLL7 is a mammalian beta‐tubulin polyglutamylase required for growth of MAP2‐positive neurites. J Biol Chem 281: 30707–30716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C (2014) The tubulin code: molecular components, readout mechanisms, and functions. J Cell Biol 206: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM (2020) The tubulin code and its role in controlling microtubule properties and functions. Nat Rev Mol Cell Biol 21: 307–326 [DOI] [PubMed] [Google Scholar]
- Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub J‐M, Temurak N, van Dijk J, Boucher D, van Dorsselaer A et al (2005) Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science 308: 1758–1762 [DOI] [PubMed] [Google Scholar]
- Jiang K, Rezabkova L, Hua S, Liu Q, Capitani G, Altelaar AFM, Heck AJR, Kammerer RA, Steinmetz MO, Akhmanova A (2017) Microtubule minus‐end regulation at spindle poles by an ASPM‐katanin complex. Nat Cell Biol 19: 480–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalebic N, Sorrentino S, Perlas E, Bolasco G, Martinez C, Heppenstall PA (2013) alphaTAT1 is the major alpha‐tubulin acetyltransferase in mice. Nat Commun 4: 1962 [DOI] [PubMed] [Google Scholar]
- Kastner S, Thiemann I‐J, Dekomien G, Petrasch‐Parwez E, Schreiber S, Akkad DA, Gerding WM, Hoffjan S, Gunes S, Gunes S et al (2015) Exome sequencing reveals AGBL5 as novel candidate gene and Additional variants for retinitis Pigmentosa in five Turkish families. Invest Ophthalmol Vis Sci 56: 8045–8053 [DOI] [PubMed] [Google Scholar]
- Kaul N, Soppina V, Verhey KJ (2014) Effects of alpha‐tubulin K40 acetylation and Detyrosination on Kinesin‐1 motility in a purified system. Biophys J 106: 2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EH, Hejab NMA, Poepsel S, Downing KH, DiMaio F, Nogales E (2018) Near‐atomic model of microtubule‐tau interactions. Science 360: 1242–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo Y‐W, Howard J (2021) Cutting, amplifying, and aligning microtubules with severing enzymes. Trends Cell Biol 31: 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, Janke C (2011) Generation of differentially polyglutamylated microtubules. Methods Mol Biol 777: 57–69 [DOI] [PubMed] [Google Scholar]
- Lacroix B, van Dijk J, Gold ND, Guizetti J, Aldrian‐Herrada G, Rogowski K, Gerlich DW, Janke C (2010) Tubulin polyglutamylation stimulates spastin‐mediated microtubule severing. J Cell Biol 189: 945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand Y, Luria V, Haffner‐Krausz R, Lonai P (1998) Maternally expressed PGK‐Cre transgene as a tool for early and uniform activation of the Cre site‐specific recombinase. Transgenic Res 7: 105–112 [DOI] [PubMed] [Google Scholar]
- Lee JE, Silhavy JL, Zaki MS, Schroth J, Bielas SL, Marsh SE, Olvera J, Brancati F, Iannicelli M, Ikegami K et al (2012) CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium. Nat Genet 44: 193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre RP, Bogdanova A, Borgonovo B, Woodruff JB, Drechsel DN (2019) FlexiBAC: a versatile, open‐source baculovirus vector system for protein expression, secretion, and proteolytic processing. BMC Biotechnol 19: 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard DV, Zinder OJ, Hotta T, Verhey KJ, Ohi R, Berger CL (2019) Polyglutamylation of tubulin's C‐terminal tail controls pausing and motility of kinesin‐3 family member KIF1A. J Biol Chem 294: 6353–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault SW, Rosenbaum JL (1985) Chlamydomonas alpha‐tubulin is posttranslationally modified by acetylation on the epsilon‐amino group of a lysine. Biochemistry 24: 473–478 [DOI] [PubMed] [Google Scholar]
- Magiera MM, Janke C (2013) Investigating tubulin posttranslational modifications with specific antibodies. In Methods cell biol, Correia JJ, Wilson L (eds), pp 247–267. Burlington: Academic Press; [DOI] [PubMed] [Google Scholar]
- Magiera MM, Bodakuntla S, Ziak J, Lacomme S, Marques Sousa P, Leboucher S, Hausrat TJ, Bosc C, Andrieux A, Kneussel M et al (2018) Excessive tubulin polyglutamylation causes neurodegeneration and perturbs neuronal transport. EMBO J 37: e100440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamdeh M, Simmert S, Luchniak A, Schaffer E, Howard J (2018) Label‐free high‐speed wide‐field imaging of single microtubules using interference reflection microscopy. J Microsc 272: 60–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally FJ, Roll‐Mecak A (2018) Microtubule‐severing enzymes: from cellular functions to molecular mechanism. J Cell Biol 217: 4057–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally FJ, Vale RD (1993) Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 75: 419–429 [DOI] [PubMed] [Google Scholar]
- Nitzsche B, Bormuth V, Brauer C, Howard J, Ionov L, Kerssemakers J, Korten T, Leduc C, Ruhnow F, Diez S (2010) Studying kinesin motors by optical 3D‐nanometry in gliding motility assays. Methods Cell Biol 95: 247–271 [DOI] [PubMed] [Google Scholar]
- Nurse P (2002) Cyclin dependent kinases and cell cycle control (nobel lecture). Chembiochem 3: 596–603 [DOI] [PubMed] [Google Scholar]
- Paturle‐Lafanechere L, Manier M, Trigault N, Pirollet F, Mazarguil H, Job D (1994) Accumulation of delta 2‐tubulin, a major tubulin variant that cannot be tyrosinated, in neuronal tissues and in stable microtubule assemblies. J Cell Sci 107: 1529–1543 [DOI] [PubMed] [Google Scholar]
- Piperno G, Fuller MT (1985) Monoclonal antibodies specific for an acetylated form of alpha‐tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol 101: 2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portran D, Schaedel L, Xu Z, Thery M, Nachury MV (2017) Tubulin acetylation protects long‐lived microtubules against mechanical ageing. Nat Cell Biol 19: 391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ (2006) Microtubule acetylation promotes Kinesin‐1 binding and transport. Curr Biol 16: 2166–2172 [DOI] [PubMed] [Google Scholar]
- Rezabkova L, Jiang K, Capitani G, Prota AE, Akhmanova A, Steinmetz MO, Kammerer RA (2017) Structural basis of katanin p60:p80 complex formation. Sci Rep 7: 14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach MC, Boucher VL, Walss C, Ravdin PM, Luduena RF (1998) Preparation of a monoclonal antibody specific for the class I isotype of beta‐tubulin: the beta isotypes of tubulin differ in their cellular distributions within human tissues. Cell Motil Cytoskeleton 39: 273–285 [DOI] [PubMed] [Google Scholar]
- Rocha C, Papon L, Cacheux W, Marques Sousa P, Lascano V, Tort O, Giordano T, Vacher S, Lemmers B, Mariani P et al (2014) Tubulin glycylases are required for primary cilia, control of cell proliferation and tumor development in colon. EMBO J 33: 2247–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowski K, van Dijk J, Magiera MM, Bosc C, Deloulme J‐C, Bosson A, Peris L, Gold ND, Lacroix B, Bosch Grau M et al (2010) A family of protein‐deglutamylating enzymes associated with neurodegeneration. Cell 143: 564–578 [DOI] [PubMed] [Google Scholar]
- Roll‐Mecak A (2020) The tubulin code in microtubule dynamics and information encoding. Dev Cell 54: 7–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdiger M, Plessman U, Kloppel KD, Wehland J, Weber K (1992) Class II tubulin, the major brain beta tubulin isotype is polyglutamylated on glutamic acid residue 435. FEBS Lett 308: 101–105 [DOI] [PubMed] [Google Scholar]
- Ruhnow F, Zwicker D, Diez S (2011) Tracking single particles and elongated filaments with nanometer precision. Biophys J 100: 2820–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhnow F, Klobeta L, Diez S (2017) Challenges in estimating the motility parameters of single Processive motor proteins. Biophys J 113: 2433–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SDM, Ferrell JE (2008) Systems biology: on the cell cycle and its switches. Nature 454: 288–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashi V, Magiera MM, Klein D, Zaki M, Schoch K, Rudnik‐Schoneborn S, Norman A, Lopes Abath Neto O, Dusl M, Yuan X et al (2018) Loss of tubulin deglutamylase CCP1 causes infantile‐onset neurodegeneration. EMBO J 37: e100540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siahaan V, Krattenmacher J, Hyman AA, Diez S, Hernandez‐Vega A, Lansky Z, Braun M (2019) Kinetically distinct phases of tau on microtubules regulate kinesin motors and severing enzymes. Nat Cell Biol 21: 1086–1092 [DOI] [PubMed] [Google Scholar]
- Siahaan V, Tan R, Humhalova T, Libusova L, Lacey SE, Tan T, Dacy M, Ori‐McKenney KM, McKenney RJ, Braun M et al (2022) Microtubule lattice spacing governs cohesive envelope formation of tau family proteins. Nat Chem Biol 18: 1224–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, Vale RD (2014) Regulation of microtubule motors by tubulin isotypes and post‐translational modifications. Nat Cell Biol 16: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiniotis G, Cochran JC, Muller J, Mandelkow E, Gilbert SP, Hoenger A (2004) Modulation of kinesin binding by the C‐termini of tubulin. EMBO J 23: 989–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotheim JM, Di Talia S, Siggia ED, Cross FR (2008) Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature 454: 291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souphron J, Bodakuntla S, Jijumon AS, Lakisic G, Gautreau AM, Janke C, Magiera MM (2019) Purification of tubulin with controlled post‐translational modifications by polymerization–depolymerization cycles. Nat Protoc 14: 1634–1660 [DOI] [PubMed] [Google Scholar]
- Sudo H, Baas PW (2010) Acetylation of microtubules influences their sensitivity to severing by katanin in neurons and fibroblasts. J Neurosci 30: 7215–7226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesna E, Zehr EA, Cummings SW, Szyk A, Mahalingan KK, Li Y, Roll‐Mecak A (2022) Combinatorial and antagonistic effects of tubulin glutamylation and glycylation on katanin microtubule severing. Dev Cell 57: 2497–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R, Lam AJ, Tan T, Han J, Nowakowski DW, Vershinin M, Simo S, Ori‐McKenney KM, McKenney RJ (2019) Microtubules gate tau condensation to spatially regulate microtubule functions. Nat Cell Biol 21: 1078–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura S, Oguchi Y, Katsuki M, Usui T, Osada H, Nikawa J‐i, Ishiwata SI, Muto E (2006) Identification of a strong binding site for kinesin on the microtubule using mutant analysis of tubulin. EMBO J 25: 5932–5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenstein ML, Roll‐Mecak A (2016) Graded control of microtubule severing by tubulin Glutamylation. Cell 164: 911–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB (1986) Reversible assembly purification of microtubules without assembly‐promoting agents and further purification of tubulin, microtubule‐associated proteins, and MAP fragments. Methods Enzymol 134: 89–104 [DOI] [PubMed] [Google Scholar]
- van Dijk J, Rogowski K, Miro J, Lacroix B, Eddé B, Janke C (2007) A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol Cell 26: 437–448 [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Gaertig J (2007) The tubulin code. Cell Cycle 6: 2152–2160 [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Lizotte DL, Abramson T, Barenboim L, Schnapp BJ, Rapoport TA (1998) Light chain‐dependent regulation of Kinesin's interaction with microtubules. J Cell Biol 143: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel P, Hansen G, Fontenot G, Read R (2010) Tubulin tyrosine ligase‐like 1 deficiency results in chronic rhinosinusitis and abnormal development of spermatid flagella in mice. Vet Pathol 47: 703–712 [DOI] [PubMed] [Google Scholar]
- Walter WJ, Beranek V, Fischermeier E, Diez S (2012) Tubulin acetylation alone does not affect kinesin‐1 velocity and run length in vitro. PLoS ONE 7: e42218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mandelkow E (2016) Tau in physiology and pathology. Nat Rev Neurosci 17: 22–35 [DOI] [PubMed] [Google Scholar]
- Zehr EA, Szyk A, Szczesna E, Roll‐Mecak A (2020) Katanin grips the beta‐tubulin tail through an electropositive double spiral to sever microtubules. Dev Cell 52: e116 [DOI] [PMC free article] [PubMed] [Google Scholar]


