Abstract
Current single-cell visualisation techniques project high dimensional data into ‘map’ views to identify high-level structures such as cell clusters and trajectories. New tools are needed to allow the transversal through the high dimensionality of single-cell data to explore the single-cell local neighbourhood. StarmapVis is a convenient web application displaying an interactive downstream analysis of single-cell expression or spatial transcriptomic data. The concise user interface is powered by modern web browsers to explore the variety of viewing angles unavailable to 2D media. Interactive scatter plots display clustering information, while the trajectory and cross-comparison among different coordinates are displayed in connectivity networks. Automated animation of camera view is a unique feature of our tool. StarmapVis also offers a useful animated transition between two-dimensional spatial omic data to three-dimensional single cell coordinates. The usability of StarmapVis is demonstrated by four data sets, showcasing its practical usability. StarmapVis is available at: https://holab-hku.github.io/starmapVis.
Keywords: Web application, Single-cell data visualisation, Spatial-single cell integration, Narrative visualisation
Graphical Abstract

1. Introduction
Advances in single-cell RNA-seq technology have enabled the profiling of the expression of hundreds of thousands of individual cells [1], [2], among which cell subpopulations can be identified. At the same time, spatial transcriptomics has developed to capture the distribution of cells sampled within larger tissue structures [3]. Domain-specific software and pipelines have standardised processing of such data, making generic analysis straightforward. However, tuning initial input parameters remains non-trivial, and thus conclusions based on pipeline-standard parameters add a source of heuristic bias. In the present study, the prime concern is that most processing pipelines present 2D analysis of high-dimensional data, relying on the "least bad" technique for projecting features into lower-dimensional space. Simultaneously, the data are plotted to form a ‘map’, excelling at showing proximity and providing the intuition of the route from point A to point B. However, these are not without controversy and often obfuscate intuition and interpretability for ease and simplicity. A recent publication suggests that the overall task error in immersive visualisation is reduced when using 3D compared to 2D visualisation because more information is available [4], even when not presented in a more complex manner. Under this paradigm, “data exploration” is aided early in the analysis workflow by allowing a viewer to assess the high-level spatial organisation of cells and compare results in different coordinate systems, molecular profiles of cells, etc.
Despite investigations suggesting interactive data exploration would benefit interpretability [4], [5], to our knowledge, this type of visualisation is not widely available using current tools. While there are many web-based data exploration tools developed for single-cell RNAseq data, most focus on 2D scatter plots of downstream results, i.e., selected gene expression and metadata. Several other visualisation tools utilising virtual reality (VR) for single-cell data have been developed in the past few years. CellexalVR [6], for example, places all projected data calculated by different dimension reduction tools such as tSNE, diffusion maps and UMAP in VR, allowing users to explore and compare different scRNA-seq analysing results. However, CellexalVR requires the use of dedicated commercial VR headsets, which limits widespread adoption in the field. Similar to StarmapVis, SinglecellVR [7] allows users to use a cheap ’cardboard’ VR headset through a web portal to present analysed data from common scRNA-seq platforms (i.e. Scanpy, Seurat). However, SinglecellVR and Squidpy + Napari [8], a spatial omics tool with no VR capability, may not be as scalable when it comes to visualising large data sets with more than a million cells. Theia [9] and vLume [10] do scale to millions of cells. However, they provide no animation support and are not web-based or compatible with the standard single-cell analysis packages such as Scanpy and Seurat. Furthermore, neither offers solutions for spatial transcriptomic data. Only very recently have computational methods [11] been developed to combine the techniques’ information to achieve single-cell resolutions in spatially related regions of the same tissue, etc. However, the matter of interpretability without necessarily tuning the input parameters of each individual technique persists.
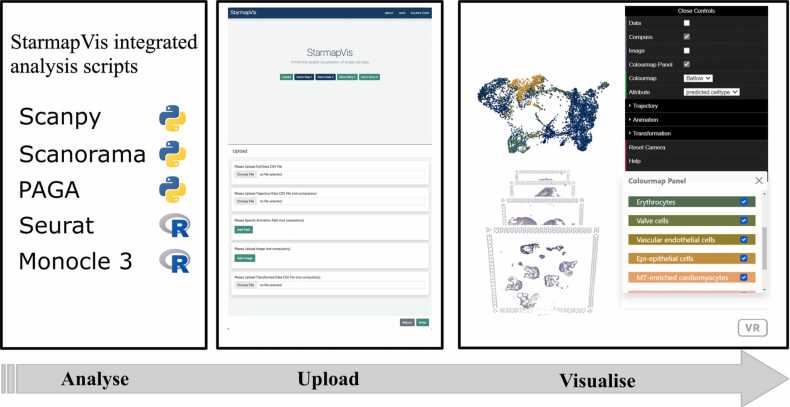
Inspired by the mature field of immersive web-based rendering, we find interactive visualisation combined with a temporally layered presentation of results can provide a novel solution for spatial, single-cell visualisation for exploration. In this manuscript, we present StarmapVis, a user-friendly visualisation tool designed specifically for integrated single-cell and spatial transcriptomic study for exploration and hypothesis generation from such transcriptomic data. Compared with other standard analysis tools packed into processing pipelines, StarmapVis organises the analysed results and presents them in a seamless user interface (UI), offering a spatially aware, single-cell trajectory visualisation. StarmapVis enables an interactive visualisation for the standard, fixed single-cell analysis pipeline analysis alluded to, including scatter plots with clustering information, connectivity network with trajectory, and cross-comparison among different coordinates. It introduces a scalable and new strategy to visualise single cells in the 3D space, allowing users to traverse through the single-cell environment just like experiencing a guided tour in a city. Users are able to ‘swim’ in cell space along the specific trajectories, assess the high-level clustering structure of cells and to further examine the interaction and connection between different coordinates, such as 3D projections and spatial coordinates. The included functions for formatting popular python and R analysis output, including Scanpy [12], Seurat [13], PAGA[14], Monocle3 [15] and Scanorama [16], make for seamless uploading and exploration to StarmapVis via the web application hosted at https://holab-hku.github.io/starmapVis. The concise user interface creates inline, publication-quality figures and is fully extendable via an open-source MIT licence. All source codes are stored in the repository: https://github.com/holab-hku/starmapVis (Fig. 1).
Fig. 1.
Overview of the StarmapVis pipeline.
1.1. Requirements
StarmapVis visualises single-cell analysis by formatting and directly interfacing with popular package output. The tool provides an intuitive interface for users to upload data, as well as four demo datasets. Five types of files let users upload different combinations of information via CSV files and PNG images (described in Table 1) for interactive downstream analysis. Users can choose to upload a single-cell CSV file or a combination of several files and information to generate multiple kinds of visualisation from single-cell RNA-seq data to a narrative of spatial transcriptomic data.
Table 1.
Overview of StarmapVis input file.
| File (s) | Details | Interpretation |
|---|---|---|
| Cell info (.csv) | x,y,z and labels | 3D coordinate points, cell labels (clusters, gene makers, pseudotime) |
| Trajectory (.csv) | name, x, y, z, children, and root | Names and 3D coordinates of edges and links |
| Animation path info (.csv) | edge list | Coordinates of animation path |
| Histology images (.png) | images | Histology data |
| Transformed coordinates (.csv) | alternative coordinates | Animate transformation from 2D to 3D, including 3D spatial transcriptomic data and 3D UMAP |
2. Application
2.1. Interactive 3D scatterplot
Single-cell analysis results are typically projected to the 2D or 3D space using different dimensionality reduction tools, i.e., UMAP, t-SNE, PCA etc., and coloured by user-selected labels. The labels can be based on any number of values, i.e., cluster labels, inference pseudotime, expression of selected genes, principal components, composite score, etc. Several VR tools exist to explore the 3D plots from different viewing angles, seamlessly zoom in and out, and change colours according to various labels and selected genes. While it is not our main ambition here, we include such functionality to satisfy user curiosity.
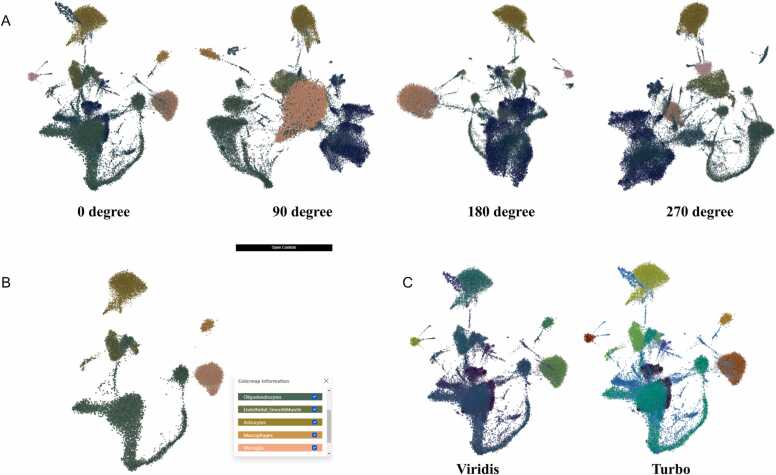
Using our first demo dataset of 65,539 mouse visual cortex single-cell RNA-seq sequences [17], we showcase the ability of StarmapVis to visualise the entire data set using a 3D UMAP projection, coloured by main cell type, subgroups and Leiden clusters. Nine main cell types are defined according to the known marker genes. Cells of two or more markers are removed because they most probably represent doublet artefacts and are grouped as ‘NaN’. As a fundamental requirement for single-cell visualisation, StarmapVis can achieve instance and smooth visualisation for even hundreds of thousands of cells. Users can select clusters of interest and choose the different colourmap for the best presentation. One can explore the 3D plots from different viewing angles, seamlessly zoom in and out, and change colours according to various labels and selected genes (Fig. 2). As a fundamental requirement for single-cell visualisation, StarmapVis can achieve instance and smooth visualisation for even hundreds of thousands of cells. Users can select clusters of interest and choose the different colourmap for the best presentation.
Fig. 2.
(A) 3D scatter plot visualised in the non-VR mode of single-cell RNA-seq data containing 65,539 mouse vertex cells with different viewing angles. (B) Customised selected clusters. (C) UMAP plot with different colour maps.
2.2. Trajectory visualisation
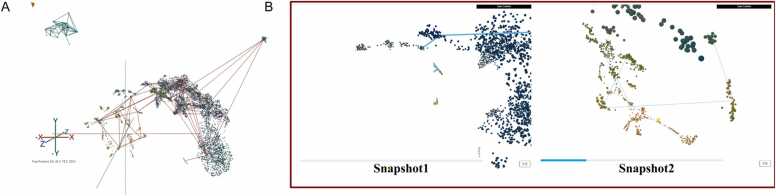
Trajectory analysis is important for studying the dynamic changes in gene expression. One of the main ambitions in trajectory analysis is to assign cells to branches. Determining the optimal assignment of those branches is nontrivial and can elucidate knowledge about the data topology. Traditionally, branches are presented as static 2D lines with arrows. Here we achieve dynamic visualisation of 3D trajectories by integrating StarmapVis with widely used trajectory inference tools, i.e., PAGA and Monocle3, to explore useful branches. In this way, StarmapVis provides a novel solution for branch visualisation by creating an interactive trajectory branch set to explore, as shown in Fig. 3. Dynamic visualisation in this way enables exploration of the information on relative spatial relations of cells along the trajectory path; the processing of any dataset proceeds by the following steps:
-
•
Get the 3D UMAP location and velocity of each cell.
-
•
Calculate PAGA and get the connectivity tree of each cluster.
-
•
Find the centroid of each cluster in 3D using the UMAP 3D positions.
-
•
Extract branches of interest.
Fig. 3.
(A) Overview of the connectivity tree of PAGA analysed results in the non-VR mode of Covid-19 single-cell RNA-seq data. (B) Dynamic visualisation of selected branch path (snapshot from animation).
Trajectory inference tools analyse high dimensional data and rely on methods of reduction to produce 2D inference trajectories, in the process reducing large amounts of data due to classical display restrictions. We were explicitly motivated to innovate in this space to overcome three limitations seen in the field: (1) visualisation of 3D projected data often has limited viewing angles, which makes it challenging to view peripheral/outlier populations, (2) difficulty comparing between projections from individual, static plots, and finally (3) the inability to interact with trajectories.
In our second demo dataset, fluid immune cells in patients with COVID-19 present enable trajectory analysis [18]. We inferred the trajectory path and calculated the progression of cells through geodesic distance along the graph as the pseudotime of each cell using PAGA. The connectivity network is presented as red lines which can be hidden, as shown in Fig. 3. Users can also specify branches they want to present, and the camera will follow the path from the root to the end. Dynamic visualisation enables the exploration of the relative position information of cells in corners along the trajectory path.
2.3. Integration and visualisation
Different analytic techniques have been developed to measure specific aspects of single-cell identity. Here we seek to marry two such analyses to leverage their mutual interpretation. The combination of single-cell and spatial transcriptomics enables the advanced study of the spatial-temporal status of cells. The integration analysis of multiple single-cell datasets is an essential part of identifying cell populations that are present across multiple datasets and enables information to be shared among different single-cell omics. For example, the integration of single-cell data and spatial transcriptomic data allows us to transfer cell type labels from scRNA-seq reference and to understand the spatial arrangement of different cell type and their interaction from the spatial transcriptomic data.
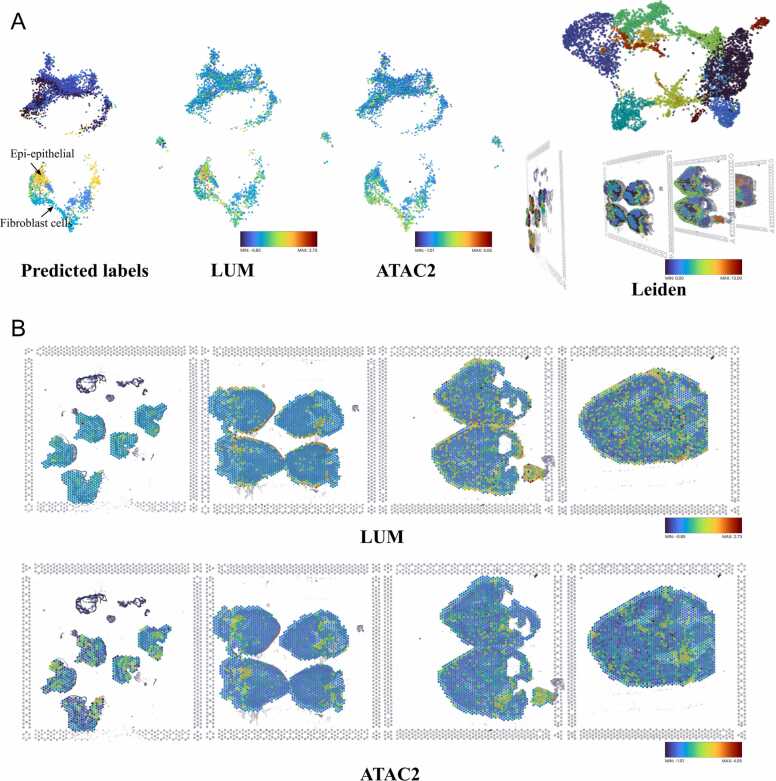
To demonstrate this capability, we investigate the spatial transcriptomic data of chicken hearts from the early to late four-chambered heart stages in our third demo dataset (Demo Story 2), [19]. Seurat helps to find integration anchors, which are pairs of cells in a matched biological state. By mapping spatial transcriptomic to scRNA-seq reference data, we can get 12 predicted cell types for chicken heart annotation. Using StarmapVis, we can select the cluster of interest in the colourmap panel. For example, as one of the important groups from the epicardial lineage, transferring “-epithelial” labels to spatial data, we can see that epithelial cells are spatially restricted to the outermost epicardial layer of the ventricular walls, as shown in Fig. 4. Lumican (LUM) is a marker gene known to be expressed in the outermost epicardial layer of embryonic hearts, which also corresponds to the epi-epithelial group (Fig. 4). Similarly, ATAC2 is highly related to “Fibroblast cell” predicted by scRNA-seq reference.
Fig. 4.
(A) UMAP of integration of the four transcriptomics datasets (left) and four spatial transcriptomics datasets with images of the chicken heart coloured by Leiden (right). (B) LUM and ATAC2 expression of the four transcriptomics datasets. Colour legends are provided for plots which are made up of continuous colour map.
2.4. Extracting meaning for storytelling
Spatial transcriptomics enables the study of the transcriptional activity of single cells in the context of the spatial organisation of these cells in the endogenous tissue environment. Spatial transcriptomic data analysis often involves multiple samples, which requires integration and cross-coordinates comparison.
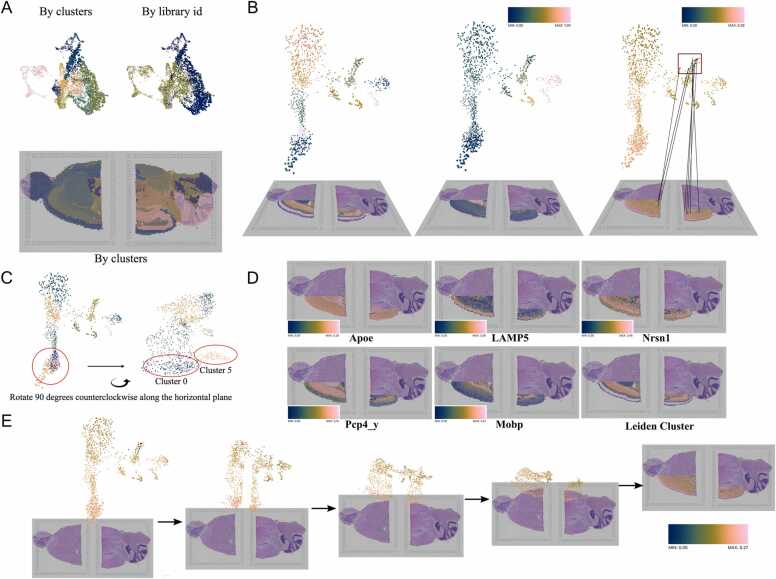
Finally, we repurpose the integration vignette dataset from Scanpy to visualise the analysed outcome and compare the integrated results. Two slices of mouse brain (anterior and posterior) data are used for our fourth demo (Demo Story 1), consisting of 22 Leiden clusters for the integrated data sets. Clusters 8, 11, and 19 of Fig. 5A belong to the posterior brain, and Cluster 10 and 17 belong to the anterior brain. Scanpy uses Scanorama [16] to integrate and remove batch effects between two spatial transcriptomics datasets of the mouse brain (Sagittal) provided by 10 × Visium. The lack of clear separation between the two groups is indicative that the integration works well. Furthermore, from the spatial location, we can see the Leiden clusters form stratification of the cortical layer in both tissues. Crucially, here StarmapVis highlights the necessity of multi-modal visualisation for interactive data exploration, wherein Fig. 5C clusters 0 and 5 present closely and even overlapping under standard parameters, and static analysis and yet are more correctly interpreted as being far apart when viewed at a different angle.
Fig. 5.
(A) Histological images with cells of the anterior mouse brain (bottom left) and posterior mouse brain (bottom right), and integration of the two transcriptomics datasets using Scanorama (top). (B) 3D transformation of cortex subset from 2D spatial location to 3D UMAP, coloured by Leiden clustering, Pseudotime and gene Apoe expression level from left to time. (C) Different angles of 3D UMAP. (D) Differentially expressed genes from Leiden group 5, 0, 2, 1, 3. (E) Transformation process from UMAP to spatial coordinates, coloured by mt-Nd5 expression level. Colour legends are provided for plots which are made up of continuous colour map.
StarmapVis can utilise meaningful label information and selected gene markers’ expressions with spatial coordinates of cells and different projections. We can further study the cortex spatial transcriptome as a static snapshot for information related to different temporal stages by integrating the subset of spatial data (only spots belong to the cortex approximately). We can select only cortex cells from spatial transcriptome and study the temporal status separately, visualising them together interactively using StarmapVis. It is clear to see the pseudotime trajectory from the cortex subset UMAP projection, which corresponds to several clusters from the early to late stage. We can also observe the stratification from the spatial coordinates. StarmapVis makes it easy to present the information sequentially and record a video clip describing your findings within a dataset. Fig. 5D shows the top-ranked gene expression from selected clusters, which corresponds to the developing stage. Sequential layers of cortical neurons can be correctly identified and presented, both in the anterior and posterior sagittal slide.
3. Implementation
The presentation of the StarmapVis is integrated into a web-based application to provide easy access and cross-platform usage. Users can use the application by either accessing the domain or fetching the source code locally and running it through any mainstream browser.
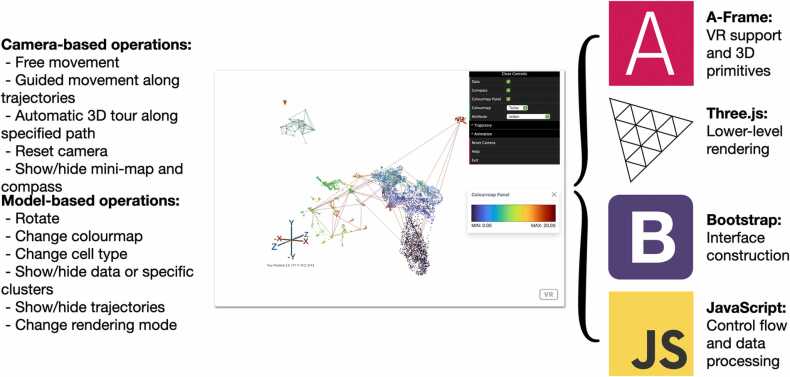
The web application is entirely written in JavaScript and HTML, and many popular and advanced frameworks and libraries, including A-Frame [20], Three.js [21] and Bootstrap [22], are involved in the architecture of the application to guarantee functionality and usability (Fig. 6). Responsive components, including buttons and control panel, and informative components, including mini-map, compass, colourmap and pop-up window of basic instructions in the web application, are intuitive and enhance the ease of user interaction and manipulation. The camera moves and pans smoothly by either following/tracing the users’ movement or following the customised paths. Multiple user-defined colourmaps, including Batlow [23], Turbo and Viridis, are available in the application control panel to display and highlight variability among the presented data. Compared with a traditional rainbow-like colourmap, a colourmap like Batlow can provide perceptual uniformity and order without distortion while being acceptable and accessible to a wider audience, such as colour-blind individuals [24].
Fig. 6.
Our application is constructed on several open-source libraries and frameworks to provide users with multiple operations to investigate or demonstrate the single cell and spatial omic data.
We designed and developed this web application in the direction of optimising the effect of visualisation and being compatible with devices with different hardware configurations. There are two options for rendering entities in the 3D space provided in the control panel so that users can choose to either render groups of entities concurrently to reach a higher frame rate or render each entity individually to enhance the fineness of the animation. Furthermore, data points are programmed to be visible or not by clusters so that users are able to hide certain clusters to reveal details in the overlapping space. While StarmapVis does not want to tie itself exclusively to high-end hardware, we include a VR mode within which users can explore single-cell data via camera and gyroscopes embedded within their mobile device-based VR headsets such as Google Cardboard and Daydream.
4. Discussion
A common analytical challenge for single-cell and spatial omics data is to explore the high dimensional embedding space of the samples. Single-cell data currently explores the global organisation of the cells by dimensionality-reduction of the expression space. Similarly, spatial omics explore the actual or inferred 2D or 3D organisation of cells to examine the cell status by comparing different coordinates. Here we present the lightweight, browser-based visualisation tool StarmapVis to visually explore both single-cell and spatial omics data spatially and temporally in an expandable open-source framework. The StarmapVis allows people to traverse biomedical data in a guided or semi-guided tour and interact with data in a 3D space.
The application was developed to handle single cell or spatial omic data at orders of magnitude of millions of cells, which is at least as many as other state-of-the-art 3D visualisation tools in the field. The upper limit of the number of cells that can be uploaded varies based on the configuration devices. We tested the software’s performance in visualising different input data scales by measuring the frame rate. The frame rate maintained stably at 60 fps when 2 million data points were rendered in the 3D space. However, the frame rate dropped to 40fps, which caused a small but perceptible difference in user experience when over 4 million data points were rendered. At the same time, the large data could be loaded and rendered in seconds by our application since we enabled the BufferGeometry and BufferAttribute in Three.js [21], which allows for more efficient passing of data to GPU. The testing was performed in a MacBook Air (M2, 2022) with 24 GB memory.
Traditional visualisation is less than ideal for high dimensionality analysis due to the limited and arbitrary viewing angles, which greatly affect the shape scope of any given projection. Different orientations may inform significantly different conclusions based on relative proximity in 3D space, e.g., the distance between clusters. Including 3D visualisation alongside 2D projections enables one to examine the relative position between cells, thereby enabling more rigorously drawn conclusions. A major innovation of StarmapVis is the ability to produce interactive animations to support single cell trajectory analysis and matching of single cell and spatial omic data. This type of animation is a unique feature of StarmapVis.
Another novel feature of StarmapVis is tracing the individual spot transformation from one coordinate system to another. For example, mt-Nd5 is one of seven mitochondrial genes encoding subunits of Complex I, which is differentially expressed in Leiden Cluster 5 (FDR corrected p-value = 2 × 10−17, picked randomly from the list of 34 from the dataset, 15 least significant with 20 genes being more DE, up to q-value 1 × 10−73), corresponding to the earliest stage. mt-Nd5 is highly expressed in most spots except for several spots (only 7th most highly expressed, as we see no correlation between DE q-value and expression), as shown in Fig. 5E. While these spots all belong to Cluster 5, there is a specific spot in the middle region of the cortex which is far from the majority of other spots in Cluster 5, indicating the possibility of a spatial-specific subgroup. This identification may be missed normally, but with StarmapVis, it is clearly seen (Table 2).
Table 2.
Comparison to 3D single-cell and spatial transcriptomics visualisation tools.
| StarmapVis | CellexalVR [6] | SinglecellVR [7] | Theia1 [9] | vLume [10] | Squidpy + napari [8] | |
|---|---|---|---|---|---|---|
| Animation | (1)Animated matching between 3D scatter plot and 2D spatial histology (2) Camera animation along trajectory | Arrow animation to represent RNA velocity | Arrow animation to represent RNA velocity | No | No | No |
| Data Type | scRNAseq ST SCP IMC |
scRNAseq | scRNAseq SCP |
scRNAseq STPT IMC |
SMLM | Mainly ST |
| Availability | Open source | Open source | Open source | Request access | Open source | Open Source |
| VR headset | any | specific hardware | any | any | any | NA |
|
Tech stack/ Web portal |
JS + browser/VR | C#, R | JS + browser/VR | Unity, C#, python | C# | python |
| Install requirement | NA (web portal) | Download | NA(web portal) | Download | Download | pip |
| Smartphone Support | Yes | No | Yes | No | No | No |
| Work with Scanpy and/or Seurat | Yes | Yes | Yes | No | No | Yes |
ST: Spatial transcriptomics; SCP: Single-cell proteomics; STPT: Serial two-photon tomography; IMC: Imaging Mass Cytometry; SMLM: Single-molecule localisation microscopy.
StarmapVis has the ability to display single-cell analysis results, single-cell trajectory branch information and spatial transcriptomic data in an immersive and interactive 3D space. We propose the idea of temporal visualisation of single-cell data with complete and versatile functions for user-custom presentation. Users can toggle and compare different coordinate systems such as UMAP, t-SNE, diffusion map and spatial coordinates. The labels can be based on any user-specified values, such as cluster labels, inferred pseudotime, expression of a handful of selected genes or some composite scores. Data with multiple projections (such as UMAP, t-SNE, spatial locations, etc.) can be visualised in a dynamic way and narratively guided by end-users.
StarmapVis can be launched on any device that has a web browser. When used on a mobile smartphone, StarmapVis can further utilise low-cost VR headsets, such as Google Cardboard and Daydream, to enable more immersive visualisation. We reason that a story-telling visual experience will likely spark public imagination and draws individuals to ask more questions and think about the analysis.
CRediT authorship contribution statement
JWKH and AY conceived the overall idea. JWKH supervised the study. AY, YY and JL developed the first version of the StarmapVis. The tool was significantly re-designed and re-implemented by SM and XF. Together with YX, CSMK, MCKL, DMDS and KKT, we extended the application of StarmapVis to spatial transcriptomic and single-cell image-based biophysical data analysis. DCM supported critical data interpretation and manuscript revision. JWKH, AY, SM, and XF wrote the paper. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Acknowledgements
This work was supported in part by AIR@InnoHK administered by the Innovation and Technology Commission of Hong Kong, the Research Grants Council of the Hong Kong Special Administrative Region of China (HKU 17208918, C7047-16G), and Innovation and Technology Support Programme (ITS/204/18). We thank Dr Chi Yan Ooi and Emily Nicola Estrada Stables for their critical feedback on the manuscript.
Consent for publication
All authors have approved the manuscript for submission.
Contributor Information
Andrian Yang, Email: ayang@ebi.ac.uk.
Joshua W.K. Ho, Email: jwkho@hku.hk.
References
- 1.Kiselev V.Y., Andrews T.S., Hemberg M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat Rev Genet. 2019;20:273–282. doi: 10.1038/s41576-018-0088-9. [DOI] [PubMed] [Google Scholar]
- 2.Chen G., Ning B., Shi T. Single-cell RNA-Seq technologies and related computational data analysis. Front Genet. 2019;10:317. doi: 10.3389/fgene.2019.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgess D.J. Spatial transcriptomics coming of age. Nat Rev Genet. 2019;20 doi: 10.1038/s41576-019-0129-z. [317–317] [DOI] [PubMed] [Google Scholar]
- 4.Wagner Filho JA, Rey MF, Freitas CMDS, Nedel L. Immersive visualization of abstract information: an evaluation on dimensionally-reduced data scatterplots. In: Proceedings of the 2018 IEEE conference on virtual reality and 3D user interfaces (VR); 2018, p. 483–90. 〈 10.1109/VR.2018.8447558〉. [DOI]
- 5.Ventura S., Brivio E., Riva G., Baños R.M. Immersive versus non-immersive experience: exploring the feasibility of memory assessment through 360° technology. Front Psychol. 2019:10. doi: 10.3389/fpsyg.2019.02509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Legetth O., Rodhe J., Lang S., Dhapola P., Wallergård M., Soneji S. CellexalVR: a virtual reality platform to visualize and analyze single-cell omics data. IScience. 2021 doi: 10.1016/j.isci.2021.103251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein D.F., Chen H., Vinyard M.E., Qin Q., Combs R.D., Zhang Q., et al. singlecellVR: interactive visualization of single-cell data in virtual reality. Front Genet. 2021;12:1992. doi: 10.3389/fgene.2021.764170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palla G., Spitzer H., Klein M., Fischer D., Schaar A.C., Kuemmerle L.B., et al. Squidpy: a scalable framework for spatial omics analysis. Nat Methods. 2022;19:171–178. doi: 10.1038/s41592-021-01358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bressan D., Mulvey C.M., Qosaj F., Becker R., Grimaldi F., Coffey S., et al. Exploration and analysis of molecularly annotated, 3D models of breast cancer at single-cell resolution using virtual reality. Cancer Biol. 2021 doi: 10.1101/2021.06.28.448342. [DOI] [Google Scholar]
- 10.Spark A., Kitching A., Esteban-Ferrer D., Handa A., Carr A.R., Needham L.-M., et al. vLUME: 3D virtual reality for single-molecule localization microscopy. Nat Methods. 2020;17:1097–1099. doi: 10.1038/s41592-020-0962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei R., He S., Bai S., Sei E., Hu M., Thompson A., et al. Spatial charting of single-cell transcriptomes in tissues. Nat Biotechnol. 2022 doi: 10.1038/s41587-022-01233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587. doi: 10.1016/j.cell.2021.04.048. [.e29] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf F.A., Hamey F.K., Plass M., Solana J., Dahlin J.S., Göttgens B., et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019;20:59. doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hie B., Bryson B., Berger B. Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat Biotechnol. 2019;37:685–691. doi: 10.1038/s41587-019-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hrvatin S., Hochbaum D.R., Nagy M.A., Cicconet M., Robertson K., Cheadle L., et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci. 2018;21:120–129. doi: 10.1038/s41593-017-0029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 19.Mantri M., Scuderi G.J., Abedini-Nassab R., Wang M.F.Z., McKellar D., Shi H., et al. Spatiotemporal single-cell RNA sequencing of developing chicken hearts identifies interplay between cellular differentiation and morphogenesis. Nat Commun. 2021;12:1771. doi: 10.1038/s41467-021-21892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neelakantam S., Pant T. Apress; Berkeley, CA: 2017. Introduction to A-Frame. Learning web-based virtual reality; pp. 17–38. [DOI] [Google Scholar]
- 21.Danchilla B. Apress; Berkeley, CA: 2012. Three.js framework. Beginning WebGL for HTML5; pp. 173–203. [DOI] [Google Scholar]
- 22.Krause J. Apress; Berkeley, CA: 2016. Introduction to bootstrap. Introducing bootstrap 4; pp. 23–32. [DOI] [Google Scholar]
- 23.Crameri, Fabio. Scientific colour maps 2021. 〈 10.5281/ZENODO.5501399〉. [DOI]
- 24.Crameri F., Shephard G.E., Heron P.J. The misuse of colour in science communication. Nat Commun. 2020;11:5444. doi: 10.1038/s41467-020-19160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]