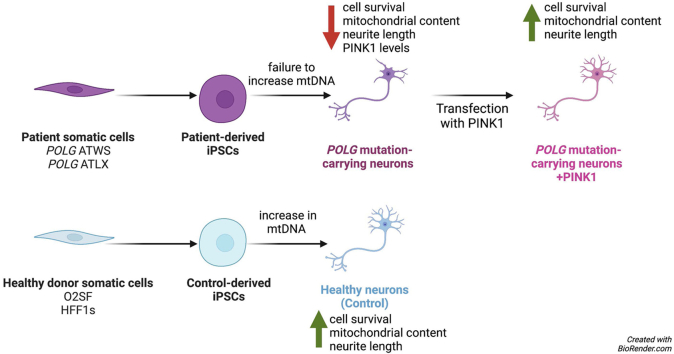
Abstract
Mutations in POLG, the gene encoding the catalytic subunit of DNA polymerase gamma, result in clinical syndromes characterized by mitochondrial DNA (mtDNA) depletion in affected tissues with variable organ involvement. The brain is one of the most affected organs, and symptoms include intractable seizures, developmental delay, dementia, and ataxia. Patient-derived induced pluripotent stem cells (iPSCs) provide opportunities to explore mechanisms in affected cell types and potential therapeutic strategies. Fibroblasts from two patients were reprogrammed to create new iPSC models of POLG-related mitochondrial diseases. Compared with iPSC-derived control neurons, mtDNA depletion was observed upon differentiation of the POLG-mutated lines to cortical neurons. POLG-mutated neurons exhibited neurite simplification with decreased mitochondrial content, abnormal mitochondrial structure and function, and increased cell death. Expression of the mitochondrial kinase PTEN-induced kinase 1 (PINK1) mRNA was decreased in patient neurons. Overexpression of PINK1 increased mitochondrial content and ATP:ADP ratios in neurites, decreasing cell death and rescuing neuritic complexity. These data indicate an intersection of polymerase gamma and PINK1 pathways that may offer a novel therapeutic option for patients affected by this spectrum of disorders.
Graphical abstract
Mitochondria are responsible for ATP generation through oxidative phosphorylation. Each mitochondrion may contain multiple copies of the 16,569 bp mitochondrial genome,1 encoding 13 proteins for the oxidative phosphorylation complex, 22 tRNAs, and 2 rRNAs.2 Polymerase gamma (Polγ), which is encoded by the nuclear POLG gene, replicates the mitochondrial genome and acts to correct mitochondrial DNA (mtDNA) errors.3 Decreased Polγ activity can lead to mtDNA deletions or mtDNA depletion. Over 160 mutations described in POLG (National Institute of Environmental Health Sciences, https://tools.niehs.nih.gov/polg, last accessed July 14, 2022), lead to a spectrum of predominantly pediatric-onset diseases.
The central nervous system is severely affected in patients with POLG mutations, with clinical features ranging from progressive external ophthalmoplegia and several syndromes associated with sensory ataxia to mitochondrial encephalopathy, lactic acidosis and stroke-like episodes (MELAS), and Alpers-Huttenlocher syndrome.4 A shared feature of many of these clinical syndromes is epilepsy, which commonly presents as status epilepticus5 and is believed to result from cortical glutamatergic hyperexcitability.6 However, the pathophysiological mechanisms that link POLG mutations to neurodegeneration are poorly understood, and there are no effective neuroprotective treatments.
Much effort has been put into modeling POLG mutations in experimental organisms. However, there is no strong mouse model for recessive POLG-linked disease. Homozygous mutations that affect exonuclease activity are embryonic lethal.7,8 The heterozygous Polg mutant mouse exhibits an accelerated aging phenotype with increased mtDNA mutations.9,10 A Polg mutation that affects proofreading activity without blocking replication elicits mtDNA deletions within substantia nigra neurons.11 However, Polg mouse lines fail to reproduce symptoms of recessive POLG-linked diseases, including the widely present neurologic phenotypes.9,11
Consequently, POLG-related central nervous system disorders have been difficult to study until the advent of induced pluripotent stem cell (iPSC) technology. iPSCs offer unique advantages for studying the impact of POLG mutations on affected human cell types that are otherwise inaccessible, such as neurons, and for modeling compound heterozygous mutations on the same genetic background as the patient from whom the tissue originated.12 Furthermore, it offers a replenishable resource of neural stem cells and neurons, thus allowing for the study of developmental aspects of these diseases. Finally, iPSC-derived neurons may be used for drug screening and future development of personalized medical therapies.13
For the current study, iPSC-derived cortical neurons (ic-neurons) from two patients bearing different compound heterozygous mutations in the POLG gene were characterized. The mutations, PgA467T/W748S (PgATWS) and PgA467T/L1173fsX (PgATLX),14,15 resulted in clinical diagnoses of myoclonic epilepsy myopathy sensory ataxia (MEMSA) and Alpers-Huttenlocher syndrome, respectively. Both patients exhibited epilepsy and severe neurologic changes such as developmental delay, cortical blindness, bipolar disease, and cognitive decline. To develop a model system to study neurologic pathogenesis in POLG-linked diseases, patient fibroblasts were reprogrammed into iPSCs and differentiated into cortical neurons. Compared with control subject–derived ic-neurons, POLG-mutated neurons showed cellular and molecular alterations in mtDNA content, neuronal morphology, and mitochondrial structure. Furthermore, mRNA levels of the mitochondrial kinase PTEN-induced kinase 1 (PINK1) were diminished, and restoration of PINK1 expression rescued several phenotypes in the patient-derived neurons, promoting dendritic arborization.
Materials and Methods
Generation of iPSC Lines
Frozen fibroblasts from two patients were obtained from existing residual clinical material using procedures reviewed by the University of Pittsburgh Institutional Review Board and designated as involving no human subjects according to federal regulations. At the time of skin biopsy, the first patient was a 16-year–old girl with compound heterozygous mutations p.A467T (c.1681 G→A) and p.W748S (c.2243 G→C) in the POLG gene. The second patient was a 14-year–old boy with Alpers-Huttenlocher syndrome, compound heterozygous for the p.A467T (c.1681 G→A) and p.L1173fsX (c.3800insGACT) mutations in the POLG gene. Both fibroblast samples were nucleofected with 3 μg each of four episomal plasmid vectors encoding OCT3/4 and p53 shRNA, SOX2 and KLF4, L-MYC and LIN28, and enhanced green fluorescent protein (GFP) using the Amaxa 4D-Nucleofector (Lonza, Basel, Switzerland). The generated iPSC lines PgATWS and PgATLX were cultured using mTeSR1 on human embryonic stem cell–qualified Matrigel-coated plates in a 4% oxygen incubator. Pluripotency markers were determined by using immunofluorescence staining of OCT4 (Cell Signaling Technology, Danvers, MA), SSEA4 (Thermo Fisher Scientific, Waltham, MA), and TRA-1-81 (Stemcell Technologies, Vancouver, BC, Canada). Chromosome analysis was performed through the WiCell Research Institute (Madison, WI).
The iPSC line 73-56010-02 (denoted O2SF) was generated from a healthy donor participating in the study entitled “Family-Based Genome-Wide Methylation Scan in Neurocognition and Schizophrenia.”16 The HFF1s line16 was generated from skin fibroblasts at the National Institute of Mental Health–funded Rutgers University Cell & DNA Repository. The iPSCs were established using a protocol approved by the University of Pittsburgh Institutional Review Board at the University of Pittsburgh Stem Cell Core and banked at the National Institute of Mental Health Center for Collaborative Studies of Mental Disorders (Rutgers University Cell & DNA Repository). The research subjects who provided these samples signed an informed consent that included permission for secondary analyses of samples.
Sequencing
Pellets from PgATWS and PgATLX iPSC cultures were collected, and DNA was extracted by using the Qiagen DNA extraction kit (Hilden, Germany). The concentration of DNA was determined, and PCR was performed by using the Phusion GC Master Mix (catalog no. F-532S; New England BioLabs, Ipswich, MA) and primers from Life Technologies (Carlsbad, CA) to amplify exons 7 to 8, 13, and 22. For amplification of exon 13, betaine was used in the PCR master mix. PCR products were resolved on a 0.7% agarose gel, and the amplified bands excised for DNA extraction using the Qiagen Gel extraction kit and sequenced in the Genomics Research Core Sanger Sequencing Facility at the University of Pittsburgh.
iPSC-Derived Neuron Differentiation
Frozen stocks of iPSCs were thawed and plated on Matrigel-coated plates with 10 μmol/L ROCK inhibitor (MilliporeSigma, Burlington, MA) in mTESR1 medium (Stemcell Technologies). Media were changed the next day without the ROCK inhibitor, and the colonies expanded until near confluent while ensuring that cells were still in the growth phase, as previously described.16 The cells were then cultured in NP selection media (Dulbecco’s modified Eagle’s medium/Nutrient Mixture F-12 with 0.5% N-2 Supplement, 1 mmol/L l-glutamine, 1% nonessential amino acids, 50 U/mL penicillin, and 50 mg/mL streptomycin) for 5 days before transfer to NP Expansion media (Dulbecco’s modified Eagle’s medium/Nutrient Mixture F-12 with 1% N-2 Supplement and 20 ng/mL rhFGFβ, 1 mmol/L l-glutamine, 1% nonessential amino acids, 50 U/mL penicillin, and 50 mg/mL streptomycin) in which they were cultured for an additional 7 days. Neurosphere-like structures (NLS) were identified and extracted by using a needle and cultured on ultra-low attachment plates in suspension. The next day, floating spherical cellular aggregates denoting neurosphere-like structures were identified and cultured for an additional 2 to 3 days. The neurosphere-like structures were then plated on a Matrigel-coated plate in the presence of NP expansion media. The following day, a needle was used to dissect and extract the neural rosettes, and a second round of purification was performed by replating them in ultra-low attachment plates. The rosettes again formed neurosphere-like structures, which were replated on Matrigel-coated plates to allow for expansion of neural progenitor cells (NPCs) and neural stem cells. The cells were allowed to expand in mTESR1 media, passaged and replated in Matrigel-coated plates. When the NPCs and neural stem cells were confluent, mTESR1 media were removed, and the cells were cultured in Neurobasal Media supplemented with 2% B-27 (Gibco, Waltham, MA), 10 ng/mL BDNF (PeproTech, Cranbury, NJ), 50 U/mL penicillin, and 50 mg/mL streptomycin. Half media changes were performed every other day for 6 to 8 weeks before the neurons were used for experiments.
Immunofluorescence
iPSC-derived NPCs and 8-week–old ic-neurons were fixed for 15 minutes with 4% paraformaldehyde followed by washes with phosphate-buffered saline (PBS). The cells were then permeabilized using 0.1% TritonX-100 in PBS for 15 minutes, followed by PBS washes. They were blocked using Superblock (Thermo Fisher Scientific) followed by overnight incubation with primary antibodies at 4°C (mouse anti-Nestin, 1:100; mouse anti-Map2; rabbit anti-Tuj1, 1:300; rabbit anti-GFP, 1:1000; Invitrogen, Waltham, MA). The cells were subjected to washes the following day, followed by incubation with species-specific Alexa Fluor antibodies at a 1:500 dilution for 1 hour at room temperature. Following this, washes were performed with PBS, and nuclei were stained with Hoechst (1:1000 for 2 minutes), and then washed and mounted on Superfrost slides (Thermo Fisher Scientific) using Gelvatol mounting medium. The staining was visualized with an Olympus 1X83 microscope equipped with a DP80 CCD camera and cellSens imaging software version 3.2 (Olympus America, Waltham, MA).
Western Blot Analysis
Cell pellets in duplicate were collected from control and patient-derived ic-neurons, and then lysed in cell lysis buffer. The Coomassie Plus Protein Assay (Thermo Fisher Scientific) was used to determine the protein concentration, and equal amounts of protein were loaded and resolved by SDS-PAGE. The proteins were then transferred to a polyvinylidene difluoride membrane and blocked with 5% milk solution. The membrane was probed with antibodies for POLG (Novus, Littleton, CO), PSD-95 (Postsynaptic Density 95; Invitrogen), synaptophysin (MilliporeSigma), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Abcam, Waltham, MA), and β-actin (Santa Cruz Biotechnology, Dallas, TX). The MitoBiogenesis Western Blot Cocktail (Abcam) was used to probe simultaneously for mitochondrial-encoded protein cytochrome c oxidase subunit I, succinate dehydrogenase A, and β-actin. Primary antibodies were incubated with gentle agitation at 4°C overnight. On the following day, anti-mouse or anti-rabbit secondary antibodies conjugated to horseradish peroxidase (GE Healthcare, Chicago, IL) were used, after which the membranes were developed using chemiluminescence following exposure to ECL solution. Chemiluminescence data were acquired using the LI-COR imaging system (LI-COR Biosciences, Lincoln, NE) and densitometry performed using the Image Studio analysis software version 3.1.4 (LI-COR Biosciences).
DNA and RNA Isolation, Quantitative PCR
DNA from control and mutant cell pellets were extracted by using the DNEasy kit (Qiagen, Germantown, MD). RNA was isolated using the RNeasy kit (Qiagen), followed by synthesis of cDNA from 1 μg of total RNA using the GeneAmp RNA PCR Core Kit (Invitrogen). Real-time PCR was performed using TaqMan probes to mitochondrial-encoded (ND1, ND4, and CYB) or nuclear-encoded (PINK1 and POLG) targets, normalized to housekeeping genes (ACTB, B2M, and GAPDH). mtDNA to nuclear DNA ratios were calculated using duplex real-time quantitative PCR. For ND1, the following primers (5′-GAGCGATGGTGAGAGCTAAGGT-3′, 5′-CCCTAAAACCCGCCACATCT-3′) and probe ([5HEX]-5′-CCATCACCC[ZEN]TCTACATCACCGCCC-3′-[3IABKFQ]) were used, and for B2M, the primers 5′-TCTCTCTCCATTCTTCAGTAAGTCAACT-3′, 5′-CCAGCAGAGAATGGAAAGTC-AA-3′ and probe [6FAM]-5′-ATGTGTCTG[ZEN]GG-TTTCATCCATCCGACA-3′-[3IABkFQ] were used, as previously described.17
Cell Viability Analysis
Eight-week–old ic-neurons from controls and PgATWS and PgATLX were cultured on a 24-well plate. On the day of the experiment, the neurobasal media were changed to neurobasal media containing 1:500 dilution of the 1.5 mmol/L stock of propidium iodide, and incubated at 37°C for 30 minutes, following which media were changed. After an equilibration period of 30 minutes, Hoechst was added to the media at a dilution of 1:2000 to stain the nuclei. After an incubation period of 10 minutes, media were changed again in preparation for imaging. The staining was visualized with an Olympus 1X71 microscope equipped with MicroSuite Basic Edition Five imaging software version 2.3 (Olympus America).
ATP Analysis
Eight-week–old ic-neurons from controls and PgATWS and PgATLX were cultured on 96-well plates. Intracellular ATP levels were measured by using a CellTiter-Glo Luminescent Cell Viability Assay kit (Promega, Madison, WI), according to the manufacturer’s protocol. Quantitative measurements of luminescence were determined by using the SpectraMax M2 plate reader (Molecular Devices, San Jose, CA). ATP content was normalized to the number of cells. To study the impact of increased PINK1 expression, ic-neurons were co-transfected with the PercevalHR reporter18 and either pcDNA3.1 vector control or PINK1-FLAG, and imaged 24 hours later on a NikonA1 confocal microscope (Nikon, Tokyo, Japan). PercevalHR was excited at 405 nm for the ADP-bound conformation and 488 nm for the MgATP-bound conformation. Clearly visualized somatic and neuritic regions of interest were analyzed by using ImageJ version 1.53c software (NIH, Bethesda, MD; http://imagej.nih.gov/ij) to calculate the ratio of integrated fluorescence intensity from the two excitation wavelengths (F488/F405) as a measure of intracellular ATP:ADP ratio.
Electron Microscopy
Control, PgATWS, and PgATLX ic-neurons were fixed with 2.5% glutaraldehyde in PBS for 1 hour, washed in PBS, and postfixed in aqueous 1% OsO4, 1%K3Fe(CN)6 for 1 additional hour. Following three washes with PBS, the cell pellets were dehydrated through graded ethanols and infiltrated with Poly/Bed 812 epoxy resin (Polysciences, Warrington, PA). After several 1-hour changes of 100% resin, the cells were embedded, cured, hardened, and ultrathin sections collected on 200 mesh copper grids; these were then stained and imaged by using a JEOL JEM-1400 Plus Transmission Electron Microscope (JEOL USA, Peabody, MA).
Transfection
Neurons were co-transfected with IRES-GFP or PINK1 IRES-GFP and Mito-dsRed plasmid constructs in Opti-MEM media using Lipofectamine 2000 (Invitrogen). The medium was changed 12 hours after transfection. The cells were cultured for an additional week and then fixed and analyzed for neurite length, mitochondrial content or rescue of neuronal phenotypes.
Neurite Tracing
To measure neurite length of transfected neurons, color images were opened using NIH ImageJ19 with the NeuronJ plugin (Erik Meijering, Biomedical Imaging Group Rotterdam, Rotterdam, the Netherlands). The dendrites of neurons whose entire arbors are visualized were traced and the total, summated dendrite length for each neuron was calculated following published protocols.20,21 Similarly, mitochondrial density per unit neurite length was measured and compared for each group of neurons.22,23
Statistical Analysis
Two-group data were analyzed by using the t-test. Multigroup data were analyzed by using one- or two-way analysis of variance followed by the post hoc Bonferroni multiple comparison test using GraphPad Prism version 9.3.1 (GraphPad Software, La Jolla, CA). P < 0.05 was considered significant. ANOVA and post-hoc P values, if applicable, are shown in the figure legends for comparisons of potential interest that resulted in nonsignificant trends.
Results
iPSC Reprogramming and Differentiation of iPSCs into Neurons
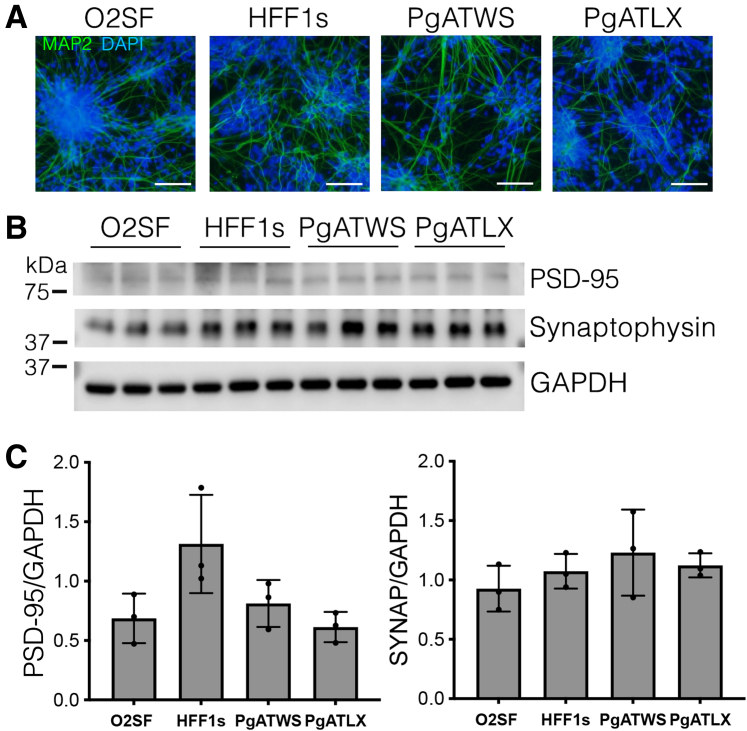
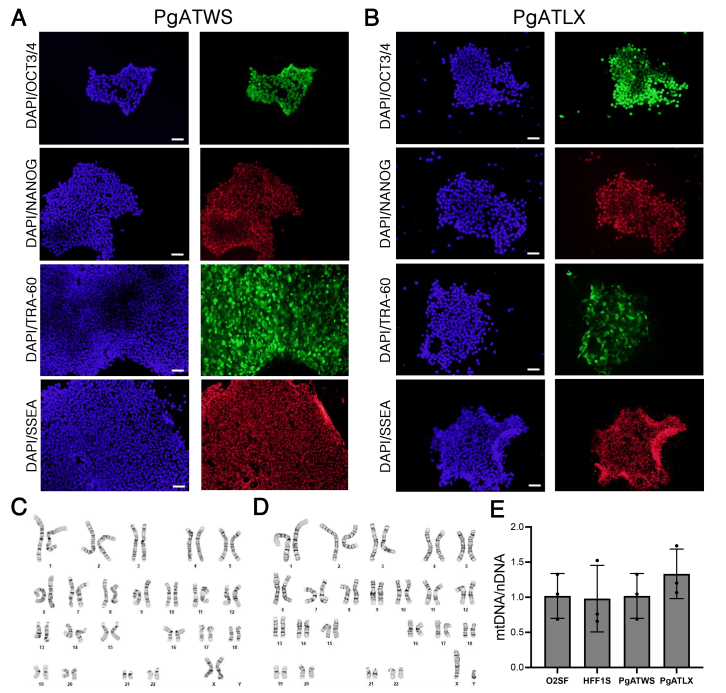
Patient-derived iPSC lines were generated from dermal fibroblasts and differentiated as previously described for control iPSC lines16 (see Materials and Methods). Immunofluorescence analysis of patient-derived iPSCs confirmed the expression of pluripotency-associated transcription factor OCT3/4 and Nanog, and the markers TRA-60 and SSEA-4 (Supplemental Figure S1, A and B). Karyotyping analysis showed that the patient-derived iPSC lines had normal karyotypes (Supplemental Figure S1, C and D). Sanger sequencing confirmed retention of the expected compound heterozygous mutations in each of the mutant lines (data not shown). An embryoid body protocol16 was used to differentiate iPSC (controls, O2SF and HFF1s; mutants, PgATWS and PgATLX) into NPCs, confirmed by Nestin staining of intermediate filaments as a marker for NPCs (Supplemental Figure S2). NPCs that were differentiated to cortical neurons stained positive for neuronal markers MAP2 (Figure 1A) and Tuj1 (Supplemental Figure S2). Neuronal differentiation and maturation of these neurons were further confirmed by determining the expression of the synaptic markers PSD-95 and synaptophysin (Figure 1, B and C).
Figure 1.
Induced pluripotent stem cell–derived cortical neurons express neuronal markers. A: Immunofluorescence images of MAP2 (green) and DAPI (blue) staining of neurons 8 weeks post-differentiation (see also Supplemental Figure S2). B: Western blot analysis of lysate from induced pluripotent stem cell–derived cortical neurons (ic-neurons) for synaptic markers PSD-95 (Postsynaptic Density 95) and synaptophysin, with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) loading control. C: Quantification of Western blots in panel B, with protein levels expressed as a ratio to GAPDH, normalized to the mean of both control lines. Data are expressed as means ± SD. n = 3 independent differentiation experiments (C). PSD-95, analysis of variance, P = 0.04, F∗(DFn, DFd) 4.43 (3, 8), post hoc HFF1s versus PgATLX, P = 0.065. Synaptophysin (SYNAP), analysis of variance, P = 0.46, F∗(DFn, DFd) 0.96(3, 8) (C). Scale bars = 100 μm (A).
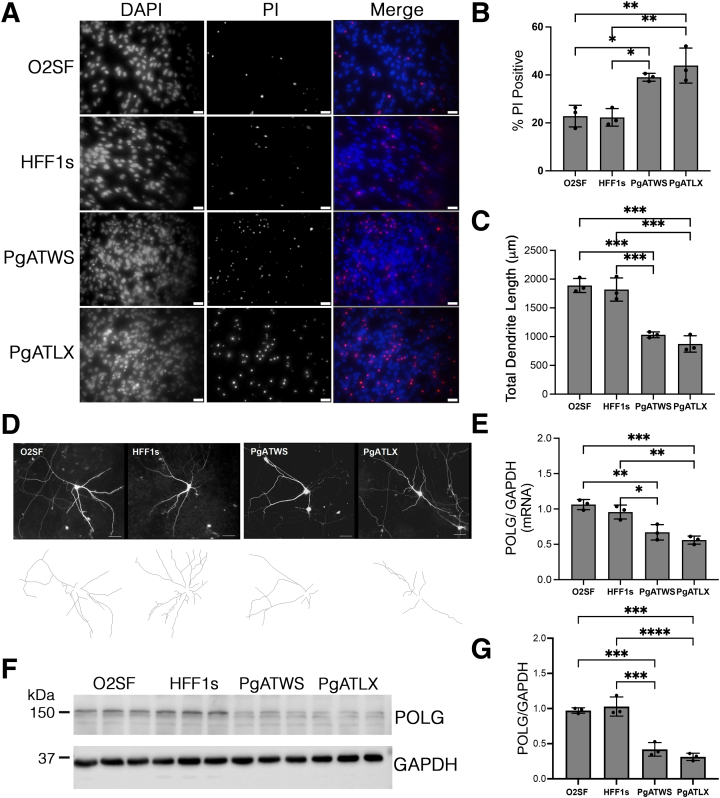
PgATWS and PgATLX ic-Neurons Exhibit Increased Basal Cell Death, Decreased Neurite Length, and Decreased POLG Expression
The ic-neurons carrying PgATWS and PgATLX mutations showed increased basal cell death as determined by increased numbers of propidium iodide–positive cells compared with control neurons (Figure 2, A and B). To examine whether there were morphologic differences among the iPSC-derived neuronal lines, ic-neurons were transfected with a plasmid expressing GFP, and neurite lengths analyzed using the NeuronJ plugin in ImageJ. There was a significant reduction in total dendrite lengths in the patient-derived neurons compared with control neurons (Figure 2, C and D).
Figure 2.
POLG mutant induced pluripotent stem cell–derived cortical neurons (ic-neurons) show increased cell death, decreased dendrite length, and decreased POLG expression. A: Representative immunofluorescence images of DAPI (blue in merged image) and propidium iodide (PI; red in merged image) staining of neurons 8 weeks post-differentiation. B: Quantification of percent PI-positive nuclei. C: Quantification of total dendrite length per neuron. D: Representative images of each line expressing green fluorescent protein to highlight individual neurons for neurite tracing. E: Determination of quantitative RT-PCR measurements of POLG normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). F: Western blot analysis of lysate from ic-neurons using antibodies for POLG and GAPDH. G: Quantification of Western blots in panel F, with protein levels expressed as a ratio to GAPDH and normalized to the mean of both control lines. Data are expressed as means ± SD. n = 3 independent differentiation experiments (B, C, E, and G). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. Scale bars = 25 μm (A and D).
POLG mRNA levels were significantly lower in mutant lines than in control lines (Figure 2E). Similarly, Polγ protein levels were also decreased in neurons expressing POLG mutations compared with control neurons as determined by Western blot analysis (Figure 2, F and G).
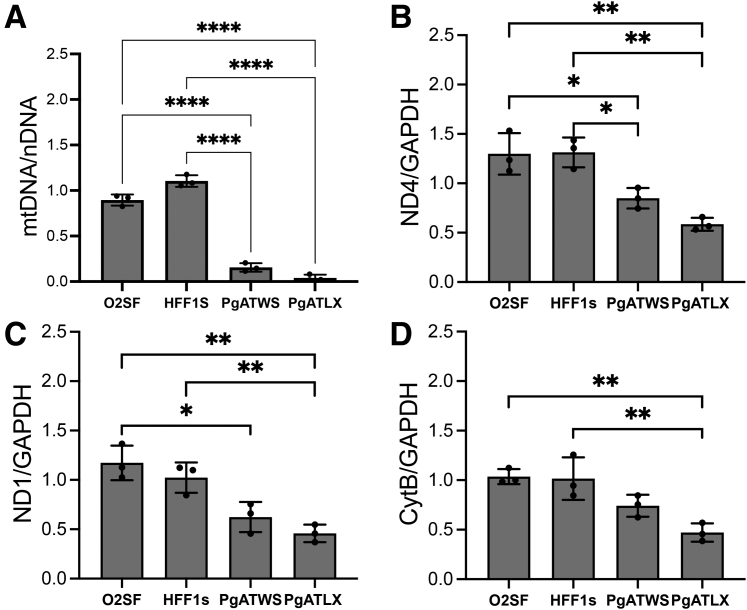
MtDNA Is Depleted in PgATWS and PgATLX Patient ic-Neurons
POLG mutations lead to mtDNA depletion in affected tissues.24 To determine whether PgATWS and PgATLX lines would recapitulate this phenotype, real-time quantitative PCR was used to study mtDNA content normalized to nuclear DNA. Interestingly, mtDNA content in mutant iPSC lines was not significantly different from control iPSC lines (Supplemental Figure S1E). However, neurons differentiated from both mutant lines showed significantly decreased mtDNA levels compared with control neurons (Figure 3A). In addition, there were decreases in mtDNA-encoded mRNA transcripts (ND4, ND1, and cytochrome B) in the patient-derived cortical neurons (Figure 3, B–D). Overall, the loss of mtDNA and mtDNA-encoded transcripts trended toward greater severity in the PgATLX line.
Figure 3.
POLG mutant-induced pluripotent stem cell–derived cortical neurons exhibit decreased mitochondrial DNA (mtDNA) content and decreased mRNA expression of mtDNA-encoded proteins. A: Quantitative PCR analysis of the mtDNA (ND1) to nuclear DNA (nDNA) (B2M) ratio in 6-week neurons, normalized to the mean of both control lines. B–D: Quantitative RT-PCR analysis of mtDNA-encoded mRNA for ND4, ND1, and CytB are expressed as a ratio to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data are expressed as means ± SD. n = 3 independent differentiation experiments (A–D). ND1/GAPDH, HFF1s versus PgATWS, P = 0.061 (C). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗∗P < 0.0001.
Mitochondrial Content Is Decreased in PgATWS and PgATLX Patient ic-Neurons
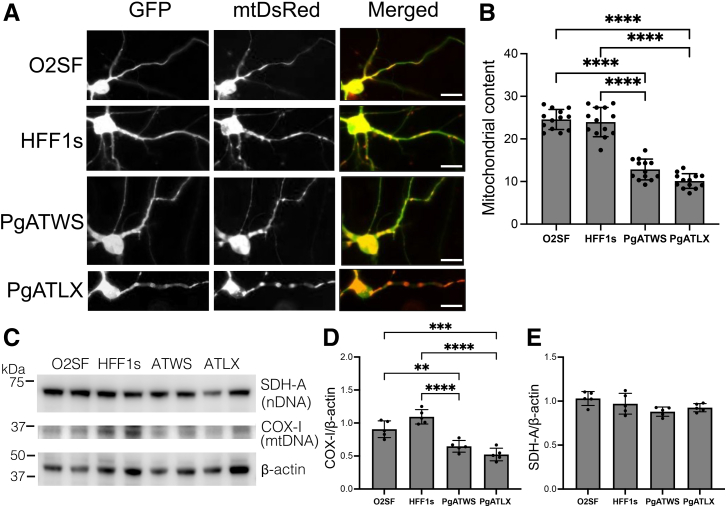
To determine whether loss of mtDNA content correlates with a decrease in mitochondrial content, mitochondrial content in control and POLG mutant ic-neurons was assessed by determining the percentage of neuritic length occupied by mitochondria using fluorescence microscopy (Figure 4, A and B) and levels of mitochondrial proteins using Western blot analysis (Figure 4, C–E). A significant decrease in mitochondrial content was observed in neurites of the mutant neurons compared with control neurons (Figure 4, A and B). Decreased mitochondrial content was also reflected by a decrease in the mtDNA-encoded protein cytochrome c oxidase subunit I, without significant changes in the levels of succinate dehydrogenase, which is encoded by nuclear DNA. These results indicate that decreased transcription and expression of mitochondrial-encoded proteins contribute to decreased mitochondrial content in POLG mutant neurons.
Figure 4.
POLG mutation decreases mitochondrial content in induced pluripotent stem cell–derived cortical neurons. A: Representative fluorescence microscopy images of 8-week–old neurons expressing green fluorescent protein (GFP) and mtDsRed. B: Quantification of mitochondrial content in neurites, expressed as a percentage of neurite length occupied by mitochondria. C: Western blot analysis of lysate from neurons using antibodies for nuclear-encoded protein succinate dehydrogenase A (SDH-A), mitochondrial-encoded protein cytochrome c oxidase 1 (COX-I), and β-actin. D: Quantification of COX-I, with protein levels expressed as a ratio to β-actin, normalized to the mean of both control lines. E: Quantification of SDH-A with protein levels expressed as a ratio to β-actin. Data are expressed as means ± SD. n = 13 neurons from three independent differentiation experiments (B); n = 5 differentiation experiments (D and E). PgATWS versus PgATLX, P = 0.057 (B). Analysis of variance, P = 0.0499, F (DFc, DFd) 3.242 (3, 16), post hoc O2SF versus PgATWS, P = 0.053 (E). ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. Scale bars = 10 μm (A).
Mitochondria in PgATWS and PgATLX Patient ic-Neurons Display Defects in Mitochondrial Ultrastructure and Membrane Polarization with Decreased ATP Content
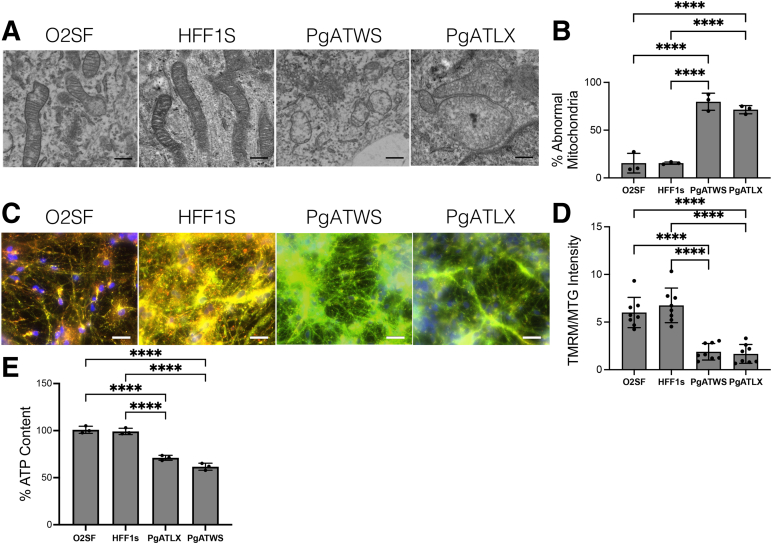
Transmission electron microscopy was used to study the structure of mitochondria in POLG patient-derived neurons. There were swollen mitochondria with multiple structural defects in POLG patient neurons (Figure 5A). Abnormal mitochondria represented a significantly higher percentage of mitochondrial profiles in POLG mutant neurons compared with controls (Figure 5B). To test whether mitochondrial function in POLG patient neurons was altered compared with controls, tetramethylrhodamine methyl ester was used to examine the polarization status of neuronal mitochondria in patient versus control neurons. There was a significant loss of mitochondrial membrane polarization in POLG patient neurons compared with control neurons (Figure 5, C and D). These structural and functional abnormalities were associated with significantly decreased levels of ATP in POLG patient neurons compared with control neurons (Figure 5E).
Figure 5.
POLG mutation alters mitochondrial structure, decreases mitochondrial membrane potential, and decreases intracellular ATP content. A: Representative transmission electron microscopy images of neurons, examining mitochondrial ultrastructure. B: Quantification of percentage of mitochondrial profiles assessed as abnormal (swollen, with disorganized cristae). C: Representative fluorescence microscopy images of neurons stained with tetramethylrhodamine methyl ester (TMRM) and MitoTracker Green (MTG). D: Quantification of mitochondrial membrane potential, expressed as a ratio of TMRM to MTG intensity. E: Quantification of induced pluripotent stem cell–derived cortical neuron ATP content normalized to cell number and expressed as percent control. Data are expressed as means ± SD. n = 3 independent differentiation experiments (B and E); n = 8 fields from three independent differentiation experiments (D). PgATWS versus PgATLX, P = 0.056 (E). ∗∗∗∗P < 0.0001. Scale bars: 500 nm (A); 50 μm (C).
PgATWS and PgATLX Patient ic-Neurons Show Decreased PINK1 mRNA Expression and PINK1 Elevation Improves Mitochondrial Content, Cell Survival, and Neurite Lengths
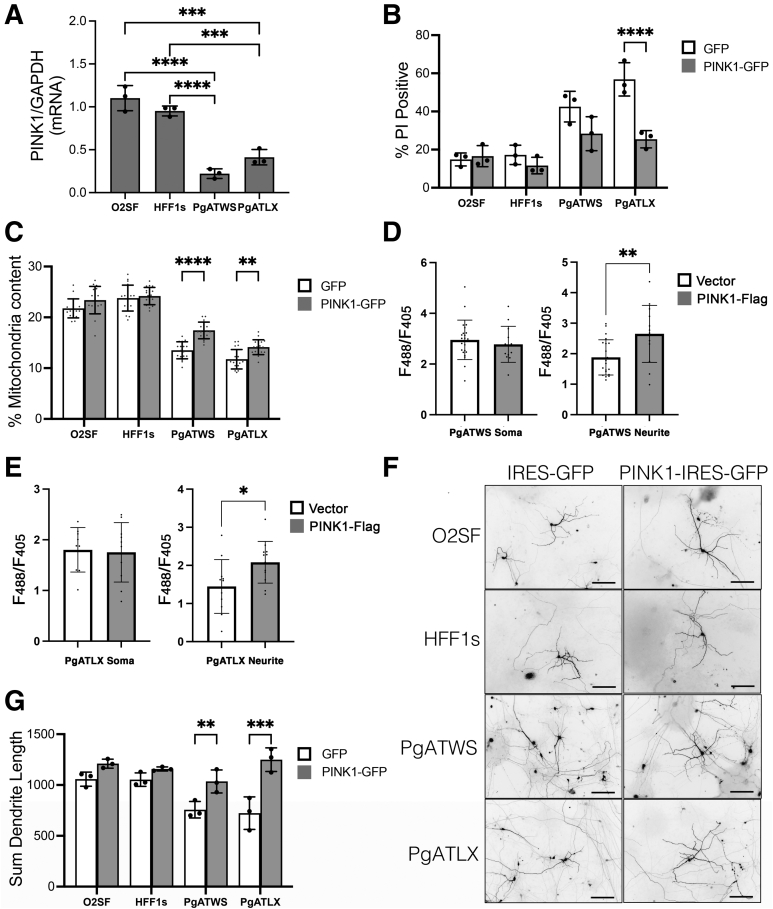
PINK1 regulates mitochondrial content in neurites25 and protects against cell death, as well as stress-induced neurite retraction.26 To test whether PINK1 is decreased in patient neurons with POLG mutations, PINK1 mRNA levels were studied using quantitative RT-PCR. PINK1 mRNA was significantly decreased in POLG mutant neurons compared with control neurons (Figure 6A).
Figure 6.
POLG mutant induced pluripotent stem cell–derived cortical neurons (ic-neurons) show decreased mRNA expression of PTEN-induced kinase 1 (PINK1), and PINK1 overexpression rescues cell death, mitochondrial content, and neuronal arborization. A: Determination of quantitative RT-PCR measurements of PINK1 normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). B: Quantification of percentage of DAPI-stained nuclei that are propidium iodide (PI) positive in ic-neuron cultures. C: Quantification of mitochondrial content in neurites, expressed as a percentage of neurite length occupied by mitochondria. D: Quantification of PercevalHR F488/F405 ratio as a measure of intracellular ATP:ADP in PgATWS ic-neurons. E: Quantification of PercevalHR F488/F405 ratio as a measure of intracellular ATP:ADP in PgATLX ic-neurons. F: Representative images of each line expressing GFP to highlight individual neurons for neurite tracing. G: Quantification of total dendrite length per neuron in microns. Data are expressed as means ± SD. n = 3 independent differentiation experiments (A, B, and G); n = 18 neurons per condition from three independent differentiation experiments (C); n = 13 to 22 neurons per condition from two independent differentiation experiments (D); n = 9 to 12 neurons per condition from two independent differentiation experiments (E). PgATWS, green fluorescent protein (GFP) versus PINK1-GFP, P = 0.06 (B). O2SF, GFP versus PINK1-GFP, P = 0.06 (C). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. Scale bars = 100 μm (F).
Interestingly, restoration of PINK1 expression by transfecting with a PINK1-IRES-GFP plasmid reduced cell death in both lines with significant protection of the more severe PgATLX line (Figure 6B). PINK1 overexpression elicited significant increases in mitochondrial content within the neurites of both patient lines (Figure 6C), accompanied by increased ATP:ADP ratios in the neuritic compartment of both lines (Figure 6, D and E). Increased expression of PINK1 also elicited full restoration of dendritic lengths (Figure 6, F and G).
Discussion
Polγ is the major polymerase responsible for the replication and maintenance of mtDNA.9 Mutations in POLG can result in several clinical syndromes associated with either mtDNA depletion in children or mtDNA deletions in adults.27,28 One of the key clinical features is intractable epilepsy,5,29 and >80% of pediatric patients with POLG mutations present with seizures.30 It is believed that seizures are a result of an imbalance between excitatory and inhibitory neurotransmission.31 However, there is still a gap in understanding the connection between mtDNA regulation and neuronal function. Until now, the cellular and molecular basis of neurologic syndromes that comprise POLG mutations in humans have been studied in postmortem brain tissues. Such studies are limited by the inability to perform functional studies and test interventions. Here, fibroblasts from two patients bearing different sets of compound heterozygous POLG mutations were reprogrammed into iPSCs, and differentiated to cortical neurons for cellular and molecular phenotyping.
Patients with POLG mutations have a varied onset of neurologic symptoms and deterioration, ultimately resulting in death. Some neuropathologic features observed in postmortem brain tissue from patients with POLG mutations are neuronal loss, neuronal simplification, cortical atrophy, and spongiosis.30,32, 33, 34 Consistent with these reports, there was an increase in neuronal cell death in iPSC-derived neurons bearing POLG mutations. Furthermore, mutant neurons exhibited significant decreases in neurite length, reminiscent of dendritic simplification described in postmortem brain tissues from patients.
As in the case of previously published postmortem reports, POLG mutant neurons exhibited swollen mitochondria with abnormal cristae and loss of inner membrane architecture as observed using electron microscopy.35 There are limited studies on mitochondrial function in POLG patient neurons, although prior studies have documented secondary accumulation of mtDNA deletions and decreased mitochondrial membrane potential in fibroblasts from POLG patients.34 This report may be the first to show defects in the maintenance of mitochondrial membrane potential in POLG mutant neurons compared with control neurons. Because iPSC-derived neurons from POLG patients accurately reflect several features seen in postmortem tissues from human patients, they represent a clinically relevant in vitro model system to understand the molecular and cellular basis of this devastating disease spectrum and to test new therapies.
Polγ has mtDNA polymerase and proofreading activities, and mtDNA encodes for 13 proteins that form part of the electron transport chain complex.36 Glycolysis is the predominant source of energy in stem cells, which is replaced by oxidative phosphorylation when stem cells are differentiated into neurons.37 Several studies have described the shift from glycolysis to mitochondrial oxidative phosphorylation with the onset of neurogenesis.38,39 Interestingly, mtDNA depletion relative to control lines was observed in neurons differentiated from the two patient-derived iPSC lines. The levels of mitochondrial-encoded mRNA and protein were also significantly decreased in mutant neurons compared with control neurons. Clinically, POLG-related disorders present at different stages postnatally, and mtDNA depletion is observed in affected organs. It is possible that symptoms arise in patients upon situations of increased bioenergetic demand, which is normally compensated by a significant mitochondrial reserve capacity in normal neurons.
In general, neurons bearing the PgATLX mutation trended toward a more severe cellular phenotype compared with neurons with the PgATWS mutation. This parallels the greater severity of clinical disease observed in this patient. The PgATLX line was derived from a 15-year–old boy with Alpers-Huttenlocher syndrome, who has been previously described clinically along with his similarly affected sister.14 He developed neurodegenerative changes at 18 months of age, with severe depletion of both gamma-DNA polymerase activity and mtDNA in the skeletal muscle by 9 years of age. He developed severe neurodegeneration with epilepsy and liver failure, dying at age 15 years. The PgATWS line was derived from a 16-year–old girl with aggression, irritability, depression, and memory deficits, exhibiting the clinical phenotype of myoclonic epilepsy myopathy sensory ataxia. The compound heterozygote state of A467T and W748S has been well-studied, typically involving a progressive neurologic disorder beginning in adolescence with epilepsy, headache, ataxia, neuropathy, myoclonus, ophthalmoplegia, and cognitive decline.33
PINK1 has been previously documented to ameliorate mtDNA copy number defects and mutations in other systems.40,41 In addition, loss of PINK1 elicits neuronal simplification, decreasing dendritic spine density in vitro and in vivo.42 Given the observation of decreased PINK1 expression in both POLG mutant lines, it is interesting that PINK1 overexpression was able to rescue neuronal complexity and ATP:ADP ratios in the neurites, implicating PINK1 as a potential target for exploring therapeutic benefits. Studies from multiple groups have shown that up-regulating PINK1 expression is protective in various neurodegenerative models.26,43 Parkin has also been shown to reduce heteroplasmic mutant mtDNA,44 as well as to regulate mitochondrial biogenesis.45 It would be interesting to explore these pathways in future studies.
In summary, these results show that POLG mutations lead to mtDNA depletion in differentiated cortical neurons. mtDNA depletion was correlated with significant loss of mitochondrial content in neurites, as well as abnormal mitochondrial ultrastructure, loss of membrane polarization, decreased ATP content, and increased cell death in POLG mutant neurons. PINK1, which has been shown to be neuroprotective against genetic or toxin-induced stress, showed partial rescue of cell viability and dendritic mitochondrial content in POLG mutant neurons, restoring dendritic complexity in both lines. Studying human neurons from patient-derived iPSC lines has the potential not only to advance understanding of epileptic and neurodegenerative symptoms in POLG-related mitochondrial disorders but also aid in identification of possible neuroprotective therapies.46
Author Contributions
M.V. performed experiments, generated figures, and analyzed data; L.F. and B.N.L. performed experiments, created figures, and wrote the manuscript; J.C. and G.F. performed experiments and developed and validated methodology; K.Z.-Q.W. performed experiments; B.A.K. developed and validated methodology and supervised data collection; L.D. developed methodology and provided resources; D.B.S. performed experiments; S.C.W. provided resources; V.L.N. contributed to methodology, resources, and supervision; A.S.-G. performed experiments, provided methodology, and acquired funding; A.G. conceptualized the project and provided resources; and C.T.C. conceptualized, administered, and supervised the project, acquired funding, analyzed data, prepared figures, and wrote the manuscript. All authors edited the manuscript.
Acknowledgment
We thank Noelle Toong for technical assistance in the early phases of characterizing neurite morphology in ic-neurons.
Footnotes
Supported in part by NIH grants R01 NS101628 (C.T.C.), R01 AG026389 (C.T.C.), and UH3 TR000503-05S1 (subaccount to C.T.C. and A.S.-G.). C.T.C. holds the A. Julio Martinez Endowed Chair in Neuropathology at the University of Pittsburgh School of Medicine.
M.V., L.F., and B.N.L. contributed equally to this work.
Disclosures: None declared.
Current address of M.V., Astellas Pharma Inc., Westborough, MA; of L.F., University of Texas Health Science Center at San Antonio, San Antonio, TX; of B.N.L., Cognition Therapeutics Inc., Pittsburgh, PA.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2022.11.002.
Supplemental Data
Supplemental Figure S1.
Characterization and validation of POLG patient-derived induced pluripotent stem cell (iPSC) lines. A and B: Immunocytochemical staining for the indicated pluripotency markers co-stained with DAPI (blue). C: Karyotype of patient iPSC line PgATWS. D: Karyotype of patient iPSC line PgATLX. E: Mitochondrial DNA (mtDNA)/nuclear DNA (nDNA) ratios in the patient iPSC lines (PgATWS and PgATLX) normalized to the mean of normal control iPSC lines (O2SF and HFF1s). Data are expressed as means ± SD. P = 0.6409, F (3, 8) = 0.5862, one-way analysis of variance (E). Scale bars: 100 μm (A and B).
Supplemental Figure S2.
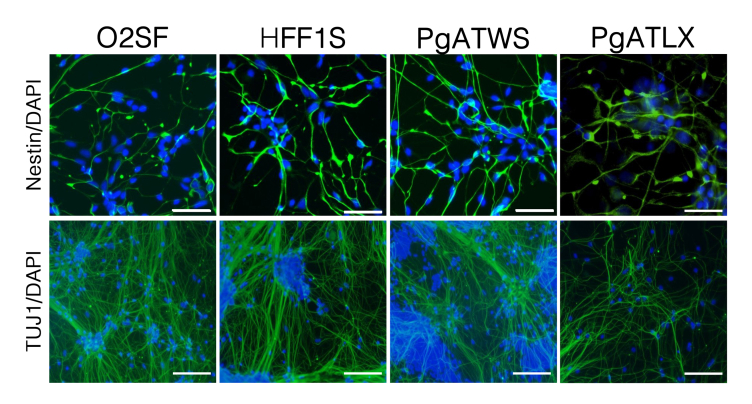
Validation of patient-induced pluripotent stem cell–derived neuronal differentiation (see also Figure 1). Upper panels: Immunostaining of control and patient neuronal progenitor cells for Nestin (green) and DAPI (blue). Lower panels: Immunostaining of 8-week differentiated control and patient cortical neurons for TUJ1 (green) and DAPI (blue). Scale bars: 50 μm (upper panels); 100 μm (lower panels).
References
- 1.Phillips N.R., Sprouse M.L., Roby R.K. Simultaneous quantification of mitochondrial DNA copy number and deletion ratio: a multiplex real-time PCR assay. Sci Rep. 2014;4:3887. doi: 10.1038/srep03887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearce S.F., Rebelo-Guiomar P., D’Souza A.R., Powell C.A., Van Haute L., Minczuk M. Regulation of mammalian mitochondrial gene expression: recent advances. Trends Biochem Sci. 2017;42:625–639. doi: 10.1016/j.tibs.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan S.S.L., Copeland W.C. DNA polymerase gamma and mitochondrial disease: understanding the consequence of POLG mutations. Biochim Biophys Acta. 2009;1787:312–319. doi: 10.1016/j.bbabio.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen B.H., Chinnery P.F., Copeland W.C. POLG-related disorders. In GeneReviews [Internet]. Copyright University of Washington, Seattle. 1993–2022. https://www.ncbi.nlm.nih.gov/books/NBK26471 Available at: (last revised March 1, 2018)
- 5.Anagnostou M.-E., Ng Y.S., Taylor R.W., McFarland R. Epilepsy due to mutations in the mitochondrial polymerase gamma (POLG) gene: a clinical and molecular genetic review. Epilepsia. 2016;57:1531–1545. doi: 10.1111/epi.13508. [DOI] [PubMed] [Google Scholar]
- 6.Zsurka G., Kunz W.S. Mitochondrial dysfunction and seizures: the neuronal energy crisis. Lancet Neurol. 2015;14:956–966. doi: 10.1016/S1474-4422(15)00148-9. [DOI] [PubMed] [Google Scholar]
- 7.Hance N., Ekstrand M.I., Trifunovic A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum Mol Genet. 2005;14:1775–1783. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- 8.Humble M.M., Young M.J., Foley J.F., Pandiri A.R., Travlos G.S., Copeland W.C. Polg2 is essential for mammalian embryogenesis and is required for mtDNA maintenance. Hum Mol Genet. 2013;22:1017–1025. doi: 10.1093/hmg/dds506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly Y.M., Gidlöf S., Oldfors A., Wibom R., Törnell J., Jacobs H.T., Larsson N.-G. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 10.Kujoth G.C., Hiona A., Pugh T.D., Someya S., Panzer K., Wohlgemuth S.E., Hofer T., Seo A.Y., Sullivan R., Jobling W.A., Morrow J.D., Van Remmen H., Sedivy J.M., Yamasoba T., Tanokura M., Weindruch R., Leeuwenburgh C., Prolla T.A. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 11.Perier C., Bender A., García-Arumí E., Melià M.J., Bové J., Laub C., Klopstock T., Elstner M., Mounsey R.B., Teismann P., Prolla T., Andreu A.L., Vila M. Accumulation of mitochondrial DNA deletions within dopaminergic neurons triggers neuroprotective mechanisms. Brain. 2013;136:2369–2378. doi: 10.1093/brain/awt196. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y., Inoue H., Wu J.C., Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen K.V., Sharief F.S., Chan S.S.L., Copeland W.C., Naviaux R.K. Molecular diagnosis of Alpers syndrome. J Hepatol. 2006;45:108–116. doi: 10.1016/j.jhep.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 15.Saneto R.P., Naviaux R.K. Polymerase gamma disease through the ages. Dev Disabil Res Rev. 2010;16:163–174. doi: 10.1002/ddrr.105. [DOI] [PubMed] [Google Scholar]
- 16.D’Aiuto L., Zhi Y., Kumar Das D., Wilcox M.R., Johnson J.W., McClain L., MacDonald M.L., Di Maio R., Schurdak M.E., Piazza P., Viggiano L., Sweet R., Kinchington P.R., Bhattacharjee A.G., Yolken R., Nimgaonkar V.L. Large-scale generation of human iPSC-derived neural stem cells/early neural progenitor cells and their neuronal differentiation. Organogenesis. 2014;10:365–377. doi: 10.1080/15476278.2015.1011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giordano L., Gregory A.D., Pérez Verdaguer M., Ware S.A., Harvey H., DeVallance E., Brzoska T., Sundd P., Zhang Y., Sciurba F.C., Shapiro S.D., Kaufman B.A. Extracellular release of mitochondrial DNA: triggered by cigarette smoke and detected in COPD. Cells. 2022;11:369. doi: 10.3390/cells11030369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tantama M., Martínez-François J.R., Mongeon R., Yellen G. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat Commun. 2013;4:2550. doi: 10.1038/ncomms3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cherra S.J., 3rd, Steer E., Gusdon A.M., Kiselyov K., Chu C.T. Mutant LRRK2 elicits calcium imbalance and depletion of dendritic mitochondria in neurons. Am J Pathol. 2013;182:474–484. doi: 10.1016/j.ajpath.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dagda R.K., Pien I., Wang R., Zhu J., Wang K.Z., Callio J., Banerjee T.D., Dagda R.Y., Chu C.T. Beyond the mitochondrion: cytosolic PINK1 remodels dendrites through protein kinase A. J Neurochem. 2014;128:864–877. doi: 10.1111/jnc.12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plowey E.D., Cherra S.J., 3rd, Liu Y.-J., Chu C.T. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu C.T., Plowey E.D., Dagda R.K., Hickey R.W., Cherra S.J., 3rd, Clark R.S.B. Autophagy in neurite injury and neurodegeneration: in vitro and in vivo models. Methods Enzymol. 2009;453:217–249. doi: 10.1016/S0076-6879(08)04011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart J.D., Schoeler S., Sitarz K.S., Horvath R., Hallmann K., Pyle A., Yu-Wai-Man P., Taylor R.W., Samuels D.C., Kunz W.S., Chinnery P.F. POLG mutations cause decreased mitochondrial DNA repopulation rates following induced depletion in human fibroblasts. Biochim Biophys Acta. 2011;1812:321–325. doi: 10.1016/j.bbadis.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Das Banerjee T., Dagda R.Y., Dagda M., Chu C.T., Rice M., Vazquez-Mayorga E., Dagda R.K. PINK1 regulates mitochondrial trafficking in dendrites of cortical neurons through mitochondrial PKA. J Neurochem. 2017;142:545–559. doi: 10.1111/jnc.14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y., Lear T.B., Verma M., Wang K.Z., Otero P.A., McKelvey A.C., Dunn S.R., Steer E., Bateman N.W., Wu C., Jiang Y., Weathington N.M., Rojas M., Chu C.T., Chen B.B., Mallampalli R.K. Chemical inhibition of FBXO7 reduces inflammation and confers neuroprotection by stabilizing the mitochondrial kinase PINK1. JCI Insight. 2020;5:e131834. doi: 10.1172/jci.insight.131834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hudson G., Chinnery P.F. Mitochondrial DNA polymerase-gamma and human disease. Hum Mol Genet. 2006;15(Spec No 2):R244–R252. doi: 10.1093/hmg/ddl233. [DOI] [PubMed] [Google Scholar]
- 28.Rahman S., Copeland W.C. POLG-related disorders and their neurological manifestations. Nat Rev Neurol. 2019;15:40–52. doi: 10.1038/s41582-018-0101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uusimaa J., Gowda V., McShane A., Smith C., Evans J., Shrier A., Narasimhan M., O’Rourke A., Rajabally Y., Hedderly T., Cowan F., Fratter C., Poulton J. Prospective study of POLG mutations presenting in children with intractable epilepsy: prevalence and clinical features. Epilepsia. 2013;54:1002–1011. doi: 10.1111/epi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hikmat O., Eichele T., Tzoulis C., Bindoff L.A. Understanding the epilepsy in POLG related disease. Int J Mol Sci. 2017;18:1845. doi: 10.3390/ijms18091845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueda K., Serajee F., Huq A.M. Clinical benefit of NMDA receptor antagonists in a patient with ATP1A2 gene mutation. Pediatrics. 2018;141(Suppl 5):S390–S394. doi: 10.1542/peds.2017-0852. [DOI] [PubMed] [Google Scholar]
- 32.Ferrari G., Lamantea E., Donati A., Filosto M., Briem E., Carrara F., Parini R., Simonati A., Santer R., Zeviani M. Infantile hepatocerebral syndromes associated with mutations in the mitochondrial DNA polymerase-gammaA. Brain. 2005;128(Pt 4):723–731. doi: 10.1093/brain/awh410. [DOI] [PubMed] [Google Scholar]
- 33.Tzoulis C., Tran G.T., Coxhead J., Bertelsen B., Lilleng P.K., Balafkan N., Payne B., Miletic H., Chinnery P.F., Bindoff L.A. Molecular pathogenesis of polymerase [gamma]-related neurodegeneration. Ann Neurol. 2014;76:66–81. doi: 10.1002/ana.24185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajakulendran S., Pitceathly R.D.S., Taanman J.-W., Costello H., Sweeney M.G., Woodward C.E., Jaunmuktane Z., Holton J.L., Jacques T.S., Harding B.N., Fratter C., Hanna M.G., Rahman S. A clinical, neuropathological and genetic study of homozygous A467T POLG-related mitochondrial disease. PLoS One. 2016;11:e0145500. doi: 10.1371/journal.pone.0145500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nolte K.W., Trepels-Kottek S., Honnef D., Weis J., Bien C.G., van Baalen A., Ritter K., Czermin B., Rudnik-Schonebörn S., Wagner N., Häusler M. Early muscle and brain ultrastructural changes in polymerase gamma 1-related encephalomyopathy. Neuropathology. 2013;33:59–67. doi: 10.1111/j.1440-1789.2012.01317.x. [DOI] [PubMed] [Google Scholar]
- 36.Gustafsson C.M., Falkenberg M., Larsson N.-G. Maintenance and expression of mammalian mitochondrial DNA. Annu Rev Biochem. 2016;85:133–160. doi: 10.1146/annurev-biochem-060815-014402. [DOI] [PubMed] [Google Scholar]
- 37.Mlody B., Prigione A. A glycolytic solution for pluripotent stem cells. Cell Stem Cell. 2016;19:419–420. doi: 10.1016/j.stem.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Almeida A.S., Vieira H.L.A. Role of cell metabolism and mitochondrial function during adult neurogenesis. Neurochem Res. 2017;42:1787–1794. doi: 10.1007/s11064-016-2150-3. [DOI] [PubMed] [Google Scholar]
- 39.Lopes C., Rego A.C. Revisiting mitochondrial function and metabolism in pluripotent stem cells: where do we stand in neurological diseases? Mol Neurobiol. 2017;54:1858–1873. doi: 10.1007/s12035-016-9714-8. [DOI] [PubMed] [Google Scholar]
- 40.Kandul N.P., Zhang T., Hay B.A., Guo M. Selective removal of deletion-bearing mitochondrial DNA in heteroplasmic Drosophila. Nat Commun. 2016;7:13100. doi: 10.1038/ncomms13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foote K., Reinhold J., Yu E.P.K., Figg N.L., Finigan A., Murphy M.P., Bennett M.R. Restoring mitochondrial DNA copy number preserves mitochondrial function and delays vascular aging in mice. Aging Cell. 2018;17:e12773. doi: 10.1111/acel.12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otero P.A., Fricklas G., Nigam A., Lizama B.N., Wills Z.P., Johnson J.W., Chu C.T. Endogenous PTEN-induced kinase 1 regulates dendritic architecture and spinogenesis. J Neurosci. 2022;42:7848–7860. doi: 10.1523/JNEUROSCI.0785-22.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K.Z.Q., Steer E., Otero P.A., Bateman N.W., Cheng M.H., Scott A.L., Wu C., Bahar I., Shih Y.-T., Hsueh Y.-P., Chu C.T. PINK1 interacts with VCP/p97 and activates PKA to promote NSFL1C/p47 phosphorylation and dendritic arborization in neurons. eNeuro. 2018;5 doi: 10.1523/ENEURO.0466-18.2018. ENEURO.0466–18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valenci I., Yonai L., Bar-Yaacov D., Mishmar D., Ben-Zvi A. Parkin modulates heteroplasmy of truncated mtDNA in Caenorhabditis elegans. Mitochondrion. 2015;20:64–70. doi: 10.1016/j.mito.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Shin J.-H., Ko H.S., Kang H., Lee Y., Lee Y.-I., Pletinkova O., Troconso J.C., Dawson V.L., Dawson T.M. PARIS (ZNF746) repression of PGC-1[alpha] contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamazaki T., El Rouby N., Fredette N.C., Santostefano K.E., Terada N. Concise review: induced pluripotent stem cell research in the era of precision medicine. Stem Cells. 2017;35:545–550. doi: 10.1002/stem.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]