Abstract
The immense structural diversity of products and intermediates of plant specialized metabolism (specialized metabolites) makes them rich sources of therapeutic medicine, nutrients, and other useful materials. With the rapid accumulation of reactome data that can be accessible on biological and chemical databases, along with recent advances in machine learning, this review sets out to outline how supervised machine learning can be used to design new compounds and pathways by exploiting the wealth of said data. We will first examine the various sources from which reactome data can be obtained, followed by explaining the different machine learning encoding methods for reactome data. We then discuss current supervised machine learning developments that can be employed in various aspects to help redesign plant specialized metabolism.
Keywords: Plant specialized metabolism, Supervised machine learning, Retrobiosynthesis, Predicting enzyme promiscuity, Predicting reaction-feasibility, Reactome data-mining, Encoding reactome data, Neural-network encoders
1. Introduction
A substantial part of terrestrial plants' remarkable adaptability is attributed to specialized metabolism (also known as secondary metabolism) [1]. For instance, metabolites produced by specialized metabolism (termed specialized/secondary metabolites) such as saiginols, lignin and wax provide desiccation defense, mechanical support, and protective sunscreen against damaging UV-B radiation [2], [3], [4]. Because specialized metabolism does not play a direct role in the development, growth and reproduction of plants, the pathways, intermediates and products of specialized metabolism can be highly lineage-specific and diverse [5], as exemplified by betalains in Caryophyllales [6], steroidal glycoalkaloids in Solanaceae [7], and glucosinolates in Brassicales [8]. As a result, specialized metabolites far outnumber metabolites produced by primary metabolism (primary metabolites) with as many as 21,000 alkaloids, 5000 flavonoids, and 22,000 terpenoids identified to date, but this number is probably understated given that many plant metabolomes remain uncharacterized [1]. Since specialized metabolites make up more than one-third of human medications (including paclitaxel, vincristine, morphine and artemisinin) [9] and can be used to make our food healthier [10], specialized metabolites have a significant impact on our lives [9].
Despite the medicinal and industrial promise of plant specialized metabolites, their total chemical synthesis can be cost prohibitive or even unattainable due to their structural complexity [11]. Consequently, the majority of specialized metabolites are still harvested from plant sources. Firmoss (Huperzia serra, source of Huperzine A, a potential Alzheimer's disease treatment), the pacific yew (Taxus brevifolia, source of anti-cancer drug taxol), and golden root (used in traditional medicine for various ailments) are examples of plant sources that can be difficult to grow, leading in the overharvesting of these species from the wild [[12], [13]]. Furthermore, many beneficial specialized metabolites may be found in plants in low quantities, preventing the cost-effective manufacture of these valuable compounds. As a result, significant efforts are being made to uncover the biosynthesis pathways of specialized metabolites which may be engineered into more efficient microbial or plant hosts and/or further manipulated via enzyme / metabolic engineering to increase their yield or generate novel, more useful compounds [14], [15], [16], [17], [18]. However, this approach is often an arduous endeavor due to the large number of reaction steps involved, coupled with the low efficiency of some enzymes that require extensive enzyme / metabolic engineering. For example, the total chemical synthesis of anti-cancer drug taxol is non-commercially viable as it consists of up to 40 reaction steps [19]. However, the 19-step biosynthetic pathway of taxol is highly complex and still not fully elucidated since the discovery of taxol more than four decades ago [20]. This has prevented the complete transfer of the taxol pathway into microbial hosts amenable to metabolic engineering even till this day [17], [20]. Consequently, taxol is still mainly produced via plant tissue culture [17], [20]. This highlights the need for methods to redesign shorter and more efficient routes of biosynthesis for natural products.
Enzyme promiscuity can be useful to redesign biosynthetic pathways. Briefly, promiscuous enzymes can accept different substrates (substrate promiscuity) [21], generate different products from the same substrate (product promiscuity) [22] and catalyze different reactions depending on the substrate (catalytic promiscuity) [23], [24]. This promiscuity can be leveraged to produce novel compounds [25], [26], [27] and thus, allows for the exploration of new biosynthetic routes for valuable specialized metabolites using retrobiosynthetic approaches [28]. Additionally, promiscuous enzymes can also be used to catalyze the derivatization of these metabolites into compounds with desirable (therapeutic) qualities. The exponential accumulation of reactome data in the public domain and recent advances in machine learning has made it opportune to explore the uses of supervised machine learning in redesigning pathways.
This review will explain supervised machine learning basics in brief, discuss available sources for reactome data, and how they can be encoded for machine learning. Later sections will explain the concept of retrobiosynthesis and how supervised machine learning can be used to aid retrobiosynthetic route planning in redesigning plant specialized metabolism. Due to the multidisciplinary nature of the topics discussed in this review (i.e., Machine learning, Cheminformatics, bioinformatics), first occurrences of certain technical terms beyond the introduction are bolded in-text and can be referred to in the glossary for their definitions together with relevant reference material for further reading.
2. Supervised machine learning basics
Despite encompassing many different algorithms under its umbrella, all supervised machine learning (SML) workflows essentially involve training a model with a training dataset containing input-output pairs. This “learning” is achieved as the model self-adjusts its parameters in an iterative fashion so that the accuracy of the predictions based on the input matches the output. The accuracy of the trained model is then tested on a never-before-seen testing dataset, by employing a train/test split of the data during model validation. Therefore, the algorithm’s predictive ability is exclusively data-driven and does not contain any defining rules of mechanistic understanding a priori. Consequently, a model’s performance is heavily influenced by the quality of the training set in a phenomenon known as “garbage in, garbage out” within the SML community. For example, the model’s performance can be heavily influenced by the size, sample bias, and data labeling/annotation of the training dataset.
A large training dataset can prevent overfitting [29], especially for models with many parameters, such as structural information of proteins and substrates. Besides a large dataset size, credible labeling of the training data is also paramount as mislabeled data will ultimately degrade the model’s performance [30], [31]. As such, best practices in SML often call for manual and expert curation to be involved in labeling training data (for further discussion on the topic in the context of metabolic studies please see [32]).
3. Publicly available data sources for machine learning
The machine learning use-cases relevant to the redesigning of plant specialized metabolism predominantly involves the utilization of data describing enzymatic reactions (reactome data). Reactome data should comprise information describing the substrate, enzymes, and products of enzymatic reactions. Aggregating enzymatic reaction data from KEGG [33], [34], [35], Reactome [36], [37], MetaCyc [38], [39], EcoCyc [40], [41] and M-CSA [42], Rhea [43] is debatably the most comprehensive database that hosts expert-curated and experimentally validated reactions, satisfying the size and label credibility requirements for a good training dataset. In the most recent release (release 122 of May 2022), Rhea hosts data for 14,583 unique reactions, with 12,601 unique reactants and supported by 16,520 unique citations of PubMed literature (https://www.rhea-db.org/statistics), while reactions link to at least 222,000 UniProtKB/Swiss-Prot and 32.2 million UniProtKB/TrEMBL accessions [43]. In addition, Rhea unifies various databases by providing (1) identifier mappings of reactions to aforementioned reaction databases, (2) UniProtKB accessions, Enzyme Commision (EC) numbers and Gene Ontology (GO) terms for enzymes, and (3) Chemical Entities of Biological Interest (ChEBI) ontology and InChIKeys for reactants/substrates. This allows one to select relevant features and compile a dataset tailored to the task of interest. For example, UniProtKB accessions can be used to acquire protein 3D structures of enzymes from AlphaFoldDB [45], [44], EC numbers can be used to acquire heuristically generated reaction rules from RetroRules [46], and InChIKeys can be used to obtain toxicity information of reactants from PubChem [47].
While mining chemical and biological “knowledge(data)bases” is an efficient and inexpensive way of gathering a large reactome dataset for machine learning, this approach also confers a set of unique challenges that have yet to be thoroughly investigated. Despite featuring information that are supported experimentally, these databases often lack experimental metadata by virtue of their design as well as intended utility. Although the spectrum of physiological conditions that facilitate enzymatic reactions is much narrower than chemical synthetic reactions, permitting-conditions for enzymatic reactions can differ greatly due to the heterogeneous environments between cellular compartments. As a case in point, proton concentrations in plastids (pH 7.2) are two orders of magnitude lower than in vacuoles (pH 5.2) of Arabidopsis thaliana [48]. Reaction databases like MetaCyc and Rhea do not include experimentally validated conditions for catalysis (e.g., substrate/enzyme concentrations, ionic environment, and pH). This hinders the identification and exclusion of enzymatic reactions that are specific to niche physiological conditions. Moreover, this may confound SML models as associations between features and ground truths to be learned by the model might be different for these reactions (e.g. protonation states of active site residues, and therefore catalytic activities of enzymes might be determined to be unfavorable in one pH and favorable in another [49]). The lack of these niche reaction subsets also precludes the use of a transfer-learning strategy to generate models specific to these “niches” by repurposing general models pre-trained on all enzymatic reactions.
Another issue with publicly available data mined from knowledge(data)bases is a bias towards positive relationships between substrate-enzyme pairs (e.g., an enzyme acts on a given substrate), while negative relationships (e.g., experimentally validated lack of catalysis between substrate and enzyme) are underrepresented. This is an issue for SML models, as the training data should represent positive and negative relationships equally, to avoid bias. If one were to use data from these sources exclusively, negative samples would have to be assigned unconfirmed data labels randomly, resulting in a dataset without true negative samples to train robust classification models [50].
Furthermore, experimental data sourced from the scientific community might also be biased towards well-studied pathways or chemical classes (composition bias), which can limit the predictive scope, as well as the performance of the resultant model [52], [51]. For example, the number of enzymatic reactions covered by the Rhea database (https://www.rhea-db.org/statistics) involving macromolecules and polymers is one and two orders of magnitude smaller than small molecule reactions, respectively. To give another example, 8 % of entries in UniProtKB/TrEMBL are represented by only 20 species, while the rest are made up of sequence data from more than 1.2 million species (https://www.ebi.ac.uk/uniprot/TrEMBLstatsl).
Due to the above-stated inadequacies, it is important for data drawn from public databases to be specifically curated in chemi- and bioinformatic SML workflows according to the problem at hand [53], which is contingent on metadata availability. This calls for the need of relevant databases, especially reaction databases, to include more metadata or the establishment of SML benchmarking datasets for enzymatic reaction data, similar to ones generated for chemical toxicity [54], bioactivity [55], and molecular docking [56].
4. Encoding enzymes as features for machine learning
To train machine-learning models to make predictions relating to different aspects of enzymatic pathway design, information on enzymes, substrates, products, and details about the reactions they catalyze (reaction data) have to be provided as input in a machine-readable format. Since the conception of SML, many studies have investigated and demonstrated the effect of features on the performance of SML [57]. To recapitulate the finding of these studies briefly, the ideal input for SML should consist of a small (low-dimensionality) and condensed set of features that encapsulate all information useful for learning. This is due to the well-established phenomenon of “the curse of dimensionality”, where any unnecessary increase of feature dimensions degrades a model’s generalization ability (overfitting) given the same dataset size. As such, deriving and selecting features from raw data (feature engineering) is a sophisticated process that often constitutes a considerable portion of SML workflows [58].
Many predictions in the SML use-cases for enzymatic pathway design (e.g. compound toxicity, substrate-enzyme binding, the feasibility of a particular catalytic reaction on a compound) are essentially extensions of an enzyme’s / compound’s intrinsic properties that are not unlike their molecular weights and solvent solubilities. Therefore, all useful information should theoretically be self-contained within the enzyme/compound’s molecular structure. As a case in point, the prediction of protein 3D structure using features exclusively derived from sequence data has been practically realized using supervised machine learning by DeepMind’s AlphaFold [44]. This was possible as all instructions needed for protein folding can be found in a protein’s primary sequence, as was established more than four decades ago [59].
Enzymes can be represented by protein-level global descriptors, sequence-based feature vectors, and learned protein embeddings (Fig. 1). Global descriptors capture the biophysical and sequence-derived properties of the proteins as a whole (e.g., amino acid composition, isoelectric point) but are poor in predicting protein function, structure, and interaction when they are used as input features [60], [61]. This is because these predictions are almost always mediated by specialized regions (protein domains) that might not be captured by global descriptors [61]. Conversely, sequence-based feature vectors can encapsulate region-specific information at amino-acid resolution and are more commonly used for the stated predictions than their global counterparts. The most direct form of generating these vectors is through one-hot encoding, where the protein sequence is vectorized into bits (binary values) within an L x 20 matrix where L is the length of the protein and each column indicates one of the 20 residues for each amino-acid position in the protein [62], [61], [63] (Fig. 1A). However, one-hot encoding is highly dimensional and assumes similarity between amino acids to be equidistant, leading to large data requirements in order to train a robust model (see Curse of dimensionality, Table 1) [61]. To mitigate this, several approaches have been developed to encode each positional amino acid as numerical values which represent their physicochemical properties [65], [64]. One such encoding scheme, VHSE8 (principal components score Vectors of Hydrophobic, Steric, and Electronic properties) encodes proteins into a L x 8 matrix with residues represented as scores across 8 principal components that were derived from more than 50 hydrophobic, steric, and electrochemical properties [66]. Despite reducing dimensions effectively, there are drawbacks when using VHSE8-like encoding schemes to construct sequence-based vectors. For one, the reliance on expert intuition in property selection fails to account for important physicochemical properties that are yet unknown. Furthermore, because full-length proteins do not readily yield vectors of fixed length, sequence-based vectors are often used to only encode overlapping peptide regions between input proteins via multiple-sequence alignment (MSA), thus limiting the analysis to proteins that share a certain degree of homology [63].
Fig. 1.
Protein representations for machine learning. A) Simplified schematic showing how different protein representations can be derived from raw sequence data. Formats for sequence-based vectors using one-hot encoding and physiochemical properties encoding are colored green and blue, respectively, while amino-acid level embeddings and protein/domain embeddings are colored red and purple, respectively. VHSE8 and ESM-1b were used to exemplify approaches in physiochemical properties encoding and learned embeddings, respectively. B) Format of distance/contact maps. Cells shaded with deep yellow within the adjacency matrix represent high proximity amino acids and correspond to similarly coloured amino-acid contact points observed in the protein structure illustration (left). C) Hierarchical categorization of different types of protein representations for machine learning. Coloured labels within the different types of representations correspond to their formats in panels A and B. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Glossary of terms.
| Term | Description | Further reading / reference material |
|---|---|---|
| Chem- / bioinformatics related terms | ||
| Chemical Entities of Biological Interest (ChEBI) ontology | Controlled vocabulary used to classify small molecules used to intervene in the processes of living organisms, based on e.g, their biological role, chemical properties. | Degtyarenko et al. [67] |
| Computer-aided synthesis planning (CASP) | Computational planning of steps to synthesize a target chemical compound from available starting materials. | Engkvist et al. [68], Ravitz [69], Warr [70] |
| Enzyme Commission (EC) numbers | Numerical classification system for enzymes based on the reaction they catalyze. The first three numbers (levels) describe the type of catalytic activity while the fourth level specifies the substrate. | McDonald and Tipton [71] |
| Gene Ontology (GO) | Controlled vocabulary that can be used to classify the function of gene products (e.g. proteins) based on biological process, molecular function and site of cellular localization. | Gene Ontology Consortium [72] |
| InChIKeys | Widely-used and unique identifiers for chemical compounds that are derived from hashing InChI (International IUPAC Identifiers) notations. | Goodman et al. [73] |
| Orphan enzymatic reactions | Reactions not known to be catalyzed by enzymes. | - |
| Reaction rules | A scheme that describes how reactants are converted to products. Useful for cheminformatic tasks such as retrosynthesis route-planning to transform reactants into products. | Plehiers et al. [74] |
| Retrosynthesis | A way of synthesis route planning that begins with the target chemical product and searches for the best possible synthetic route, arriving at reactants that are inexpensive and easily obtainable. | Klucznik et al. [75]; https://www.elsevier.com/solutions/reaxys/predictive-retrosynthesis) |
| Retrobiosynthesis | An approach of route planning for biochemical synthesis. Biosynthetic routes that arrive at abundant reactants (cellular metabolites) are prioritized in order to maximize biosynthetic yield. | de Souza et al. [28], Mohammadi Peyhani et al. [76], Probst et al. [29] |
| Structure-activity relationship (SAR) | Relationship that describes how structural properties of molecules relate to their (bio)activities. | Guha [77] |
| Machine-learning related terms | ||
| Data labels | Targeted output to train a supervised machine learning model. | - |
| Data labeling / annotation | Generation of data labels for sample data that are otherwise unlabelled. Labeling / annotation can be achieved via manual, semi-automatic or automatic means. | - |
| Dimensionality (of features) | The number of features. | - |
| Features | Refers to Input that has been preprocessed from sample data (often into numerical or binary values) to be fed directly into a machine learning model in order to generate an output value. Feature (singular) refers to a single numerical / binary value from the set of input. | - |
| Machine learning | Use of data and algorithms that learns iteratively to improve its accuracy of predictions or make decisions that can give the best outcome. | Greener et al. [78] |
| Model | An algorithm that can recognize patterns, make predictions or make decisions based on given input. | - |
| Model validation | Process of using the model to predict the output of samples outside of the training dataset to evaluate the predictive performance of a model. | - |
| Reinforcement learning | A machine learning method that improves iteratively to maximize reward. | [79] |
| Neural Network | A type of supervised machine learning algorithm that comprises an input layer, a hidden layer (can be more than one) and an output layer. Each layer consists of nodes that are connected to every node in adjacent layers via edges. Data is fed into the network via the input layer and processed as it propagates through the hidden layer(s) towards the output layer to give an output (often a prediction). Each edge is associated with weights (parameters) that transform the data from one node to another and can be adjusted during learning to improve accuracy of the prediction. Neural networks are also known as artificial neural networks (ANN). | Greener et al. [78]; https://playground.tensorflow.org) |
| One-hot encoding | A way of converting one column of categorical features into multiple binary columns. | Greener et al. [78]; https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.OneHotEncoder.html) |
| Overfitting | A phenomenon where a model learns irrelevant information from the training dataset resulting in the degradation of predictive performance on never-before-seen data. This can be caused by having a model that has too many parameters (too complex) or not having enough sample data to train the model. | Chicco [80], Greener et al. [78] |
| Parameters | Variables within the model that governs how input data is transformed into the output. Machine learning models self-adjust these parameters during training to minimize the error between output and labels. | - |
| Self-supervised (machine learning) | A subset of unsupervised machine learning algorithms that are able to take on tasks which are traditionally tackled by supervised machine learning, without using data labels. | Spathis et al. [81] |
| Sparsity (of features) | The number of features with zero values. | - |
| Supervised (machine learning) | A machine learning approach that uses a labeled dataset to improve the its prediction of outcomes. | - |
| The curse of dimensionality | The relationship between the predictive performance of machine learning models and dimensionality of input features. Performance first improves with increased dimensionality but starts to degrade past a certain point if the number of training samples remains the same. This is attributed to the exponential increase in training data needed to prevent overfitting as dimensions increase. | (https://deepai.org/machine-learning-glossary-and-terms/curse-of-dimensionality) |
| Training dataset and testing dataset | Subsets of sample data generated after a train/test split. Training data are used to train the model while testing dataset will be used to evaluate the model performance. | - |
| Transfer-learning | An approach of applying knowledge learned for one task to a different but related task to improve sample efficiency. This can effectively be achieved by training an already pre-trained model instead of building a model from scratch. | Cai et al. [82] |
| Unsupervised (machine learning) | A machine learning approach that uses algorithms to infer patterns from a dataset without the use of data labels. | - |
Consequently, recent models have transitioned from heuristically encoding proteins to using neural-network architectures designed for natural language processing (NLP) known as transformers that are pre-trained using unsupervised (more specifically, self-supervised) learning to predict representations of proteins (learned protein embeddings). The use of unsupervised learning allows embeddings to be learned from the vast amount of unlabeled protein sequence data (i.e., proteins without functional, structural, and/or interaction annotations) that have exploded in recent years due to the increasing affordability of sequencing technologies, as well as advances in gene prediction [83]. As such, protein embeddings excel at capturing all manners of biological information and can be generalized across a range of applications. This allows for pre-trained transformers to be deployed to embed proteins for SML [84], [85], [86], [87], [63]. ESM-1b [85], a transformer trained using 250 million protein sequences, can convert protein sequences into either amino-acid level embeddings of variable lengths (L x 1280 matrix similar to sequence-based feature vectors) or protein-/domain-level embeddings of fixed length (1280 features) by amino-acid level embeddings along the length-dimension. Embeddings generated by ESM-1b have been shown to predict regional and global protein properties [85] and outperform other embedding models in several metrics when utilized in fold prediction [86].
Besides sequence-derived representations, proteins can also be represented based on their 3D structure in distance maps in the form of an L x L adjacency matrix. Such matrices contain proximity values for each amino acid pair, or contact maps where binary values indicate a close contact between amino acid pairs (Fig. 1B). These maps can be used together with sequence-based vectors or amino-acid level embeddings to enrich input features for supervised learning predictions [88]. However, the usage of structural data to encode proteins for SML is still comparatively rare due to the highly dimensional nature of distance maps and that some level of structural information is already present in embeddings [85]. The chief reason for the rare use of protein structural data is undoubtedly due to its scarcity relative to sequence data. This predicament has recently (July 2022) been alleviated with the release of predicted structures for over 200 million proteins by AlphaFold [44]. As such, we anticipate seeing further development of structural representations and even learned embeddings predicted from both sequence and structural data. Moreover, the underlying neural network architecture employed by AlphaFold might allow for an interesting transfer-learning approach in tackling the dimensionality and sparsity problem of conventional protein structural representations. Instead of using readily-interpretable but highly-dimensional distance/contact maps, node values of hidden layers generated within AlphaFold during the forward propagation of protein sequence data can be vectorized into compact structural protein representations for machine learning although the effectiveness and practicality of this approach has yet to be evaluated given the relatively recent development of AlphaFold.
Despite the utility of currently available protein representations, they fail to account for post-translational modifications (PTM) and binding to non-protein entities (e.g., co-factors), both of which have been known to confer biological activity. However, the success of current representations used in SML models reinforces the theory that this information are sequence-embedded along with protein function. This is also supported by the fact that PTM and binding information can be predicted by protein sequence [89], [90]. Current protein representations are also limited by their very design to represent a single continuous polypeptide, thereby failing to represent multimeric proteins.
5. Encoding substrates as features
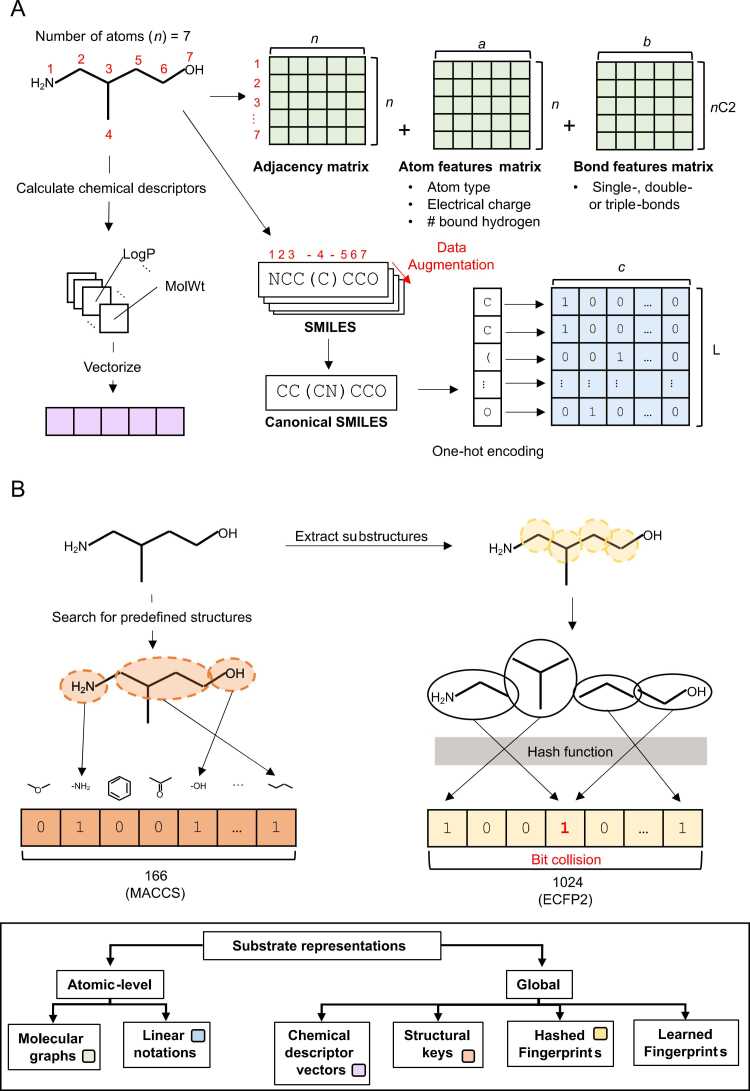
Enzyme substrates are small molecules that can be encoded for machine learning as molecular graphs, linear notations, chemical descriptor vectors, structural keys, hashed fingerprints, and learned fingerprints (Fig. 2) [91], [92]. For more in-depth information about molecular representations for machine learning, we refer readers to excellent resources by [91], [93], [94].
Fig. 2.
Substrate representations for machine learning. A) Simplified schematic showing how molecular graphs (green), linear notations (blue) and chemical descriptor vectors (purple) can be used to encode small molecule substrates for machine learning. Different linear notations (exemplified using SMILES notation) and molecular graph representations can be generated from the same molecule by changing atom numbering order (red), allowing for the use of data augmentation to enrich the dataset. B) Simplified schematic highlighting the difference between structural keys and hashed fingerprints. MACCS and ECFP representations were used to exemplify structural keys and hashed fingerprints respectively. An example of a bit collision (red) resulting in information loss in hashed fingerprints is also shown. C) Hierarchical categorization of different types of Substrate representations for machine learning. Coloured labels within the different types of representations correspond to their formats in panels A and B. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Despite being smaller than proteins, small molecules can be highly diverse in structure due to the different permutations of atoms and bonds that can make up their structure. Molecular graphs (Fig. 2A) describing bond connectivities between every atom within small molecules consists of an adjacency matrix (n x n, where n is the number of atoms present in the molecule), a one-hot matrix of atom features (n x a, where a is the number of atom features) and a one-hot matrix of bond features (nC2 x b, where b is the number of bond features) [93]. Features used to describe atoms include atom type, formal charge, and number of implicit hydrogens bound to the atom, while bonds can be described as single, double, or triple covalent bonds [93]. Small molecules can also be represented with linear notations such as Simplified Molecular Input Line Entry System (SMILES) [95] or SMILES Arbitrary Target Specification (SMARTS) (https://www.daylight.com/dayhtml/doc/theory/theory.smarts.html), where bonds and atoms are directly encoded by characters on a linear vector and formatted into a feature matrix (L x c, where c is the number of different characters in the notation system and L is the length of the linear notation) (Fig. 2A) [91].
Since information is captured at the atomic level, both molecular graphs and linear encodings have the benefit of being unambiguous and unique to one molecule while encapsulating structural stereochemistry. In addition, molecules can be represented in more than one way depending on atom numbering which allows for data augmentation in ML [97], [96]. However, these atomic-level representations are sparse and highly dimensional due to the use of one-hot encodings which precludes their usage in cases without a large labeled dataset (curse of dimensionality, Table 1). Furthermore, these representations vary in size depending on molecular complexity and are limited to represent molecules with only covalent bonds [98]. As such, it is more common for molecules to be represented indirectly based on their global (molecular) properties (e.g. molecular weight, number of hydrogen-bond acceptors, octanol–water partition coefficient etc.) as opposed to direct representations at the atomic-level. This is despite the ambiguous and non-unique nature of molecular-level representations where they cannot be decoded back into the molecule it represents, and not being unique to a specific molecule.
RD-kit (https://www.rdkit.org/docs/GettingStartedInPython.html #list-of-available-descriptors), a popular chemoinformatics package for python, can be used to calculate chemical descriptors that describe different chemical and structural properties of a molecule at the molecular level. These chemical descriptors can then be encoded as a molecular-level vector representation of a molecule (Fig. 2A) [99], [93], [100]. Other molecular-level representations like structural keys and hashed fingerprints are designed to describe the substructure of a molecule as a fixed-length bit-vector where each position denotes the presence/absence of a particular structural fragment [91], [93]. The key difference between these two representations lies in the selection of substructures to be encoded in each position of the bit vector. While structural keys like MACCS (Molecular ACCess System) [101] look for predefined substructures (up to 960), substructures to be extracted are determined algorithmically from the molecule itself and mapped to a position in the bit vector using a hash function in hashed fingerprint representations (Fig. 2B) [91], [93]. Because extracted substructures are not predefined, hashed fingerprint representations can capture properties that are useful for different types of predictions without any prior structure-activity relationship (SAR) knowledge which sees their widespread use in many different chemoinformatics prediction models [102], [103], [104], [105], [106]. However, the use of a hashing function in the generation of bit-vectors may result in the loss of substructural information in bit-collisions where the different substructures are mapped to the same bit (Fig. 2B). Although bit-vector length can be increased to minimize bit-collisions, doing so also increases the dimensionality and sparsity of the fingerprint significantly (long bit-vector of mostly zeros). Commonly used hash fingerprints include ECFP (Extended Connectivity Fingerprints) [107] and Morgan’s fingerprints [108] which extracts substructures based on different atomic neighborhoods of non-hydrogen atoms (up to a given diameter/radius), and Daylight fingerprints (https://www.daylight.com/dayhtml/doc/theory/theory.finger.html) which extracts substructures based on different paths that connect atoms (up to a given length).
Similar to protein representations, heuristically-generated molecular representations (i.e. chemical descriptors) might fail to account for certain properties that are important for different scenarios, limiting the universal applicability of these representations. As a result, transformers pre-trained using self-supervision have also been developed to generate learned fingerprints [92], [94]. However, these learned representations have not been shown to perform better than traditional representations in various drug-design tasks [109] and learned representations dedicated to represent SM substrates have yet to be developed, but we envision that future developments might change this. For more in-depth information about molecular representations for machine learning, we refer readers to excellent resources [91], [93], [94].
6. Designing new compounds and pathways with retrobiosynthesis
Retrosynthesis is a method of deconstructing a target molecule into its starting materials, that can be employed in the route-planning to chemically synthesize various organic compounds [75], [110], [111]. In retrosynthesis, molecules are recursively deconstructed into precursors, expanding the pathway from a target molecule until the entry into the pathway are starting materials that are relatively inexpensive and easily procurable. Similarly, retrobiosynthesis also involves working backwards from a target molecule but uses biochemical reactions instead of chemical reactions for route-planning. As such, retrobiosynthesis can be used to create/redesign metabolic pathways that occur in nature and have been used extensively to produce a variety of specialized metabolites. As opposed to economically- and logistically-efficient starting materials, retrobiosynthesis seeks to define the ideal entry into the pathway from the perspective of metabolic availability. The optimal starting materials should be abundant enough to prevent a large change in their molecular pool which can adversely affect the biological system [28]. In addition, their regeneration pathways should be high in flux such that the rate limitation can be confined to the design space and controlled by the designer, and rapid synthesis of the target molecule can be accommodated.
There are several robust programs developed for computer-aided synthesis planning (CASP) of retrosynthetic [75], [112], [113] and more recently, retrobiosynthetic route-planning [114], [28], [115], [116]. However, the exponential increase in possible pathway configurations with each enzymatic reaction step results in a large combinatorial search space that is not only computationally intensive to simulate but also impossible for experts to evaluate manually. Furthermore, the majority of retrobiosynthetic CASP approaches are based on reaction rules [107], [46], [118], which require expert input. Consequently, these approaches are typically poorly scalable and inflexible due to the reliance on a select group of enzymes with manually defined reaction rules that may not account for various aspects of promiscuity and thus, limits the discovery of new biosynthetic routes [119].
7. Predicting outcomes of enzymatic reactions for single-step retrobiosynthesis using SML
Retrobiosynthetic CASP involves mapping out biosynthetic pathways from a natural compound and needs to derive substrates of enzymatic reactions from a given product (termed single-step retrobiosynthesis) in a recursive manner. To facilitate this, reaction rules of enzymes that describe how reactants can be converted to products (and vice versa), were typically used to computationally expand biosynthetic pathways in route-planning. As such, the library of enzymes considered in retrobiosynthetic CASP is subject to the availability of their corresponding reaction rules and fails to encompass many specialized metabolism enzymes [107], [46], [118].
Traditionally, the establishment of enzymatic reaction rules requires extensive and expert knowledge of the enzyme, such as its promiscuity to act on different substrates [74]. Recent efforts have seen the automatic extraction of reaction rules from reactome data [46] where heuristics were used to identify the reaction center of enzymatic reactions. However, the reaction rules obtained from these approaches were still less than ideal as the exact reaction rules yielded are largely dependent on an arbitrarily selected promiscuity threshold (i.e. minimum diameter of the reaction center [denoted as d]) [46] which might not truly reflect on the true substrate/product specificities of enzymes. To expound upon this drawback, if reaction rules are extracted using the lowest possible promiscuity threshold (d = ∞), the retrobiosynthetic search space will be severely limited to enzymatic reactions that yielded the exact same product as the query compound, thereby failing to incorporate any kind of promiscuity in route planning. On the other hand, using the highest possible promiscuity threshold (d = 2) to extract reaction rules for use in retrobiosynthesis will result in an astronomical search space that is too large to evaluate computationally. Additionally, it is not intuitive to use a single blanket threshold to extract reaction rules across all enzymatic reactions as enzymes of different classes have been known to possess varying degrees of promiscuity [26], [24], [121].
Despite using chemical reactions for route-planning instead of enzymatic reactions, traditional retrosynthesis methods also rely on chemical reaction rules, for which several issues have been identified [119]. This has prompted the development of retrosynthetic approaches that use SML methods to circumvent the need to rely on reaction rules. These methods, termed template-free retrosynthesis [119], use sequence-based [122], [123], [124], [125] and graph-based [126], [127], [128] SML models trained on chemical reaction datasets that can be used to predict chemical reactants of a given compound, in order to reverse engineer chemical synthesis pathways for a target compound. Likewise, a template-free method for retrobiosynthesis, BioNavi-NP [129], has also been recently developed by training an end-to-end transformer neural network with 33,710 enzymatic reactions mined from MetaNetX [130]. BioNavi-NP uses this SML model in place of enzymatic reaction rules, to suggest a set of possible enzymatic reactions for a given product in retrobiosynthetic route-planning [129]. BioNavi-NP is able to predict the biosynthetic pathway of test compounds with an accuracy of 90.2 % which is 1.7 times higher than retrobiosynthetic methods using reactions-rules [129]. However, unlike approaches using reaction rules, BioNavi-NP is unable to assign enzymes to enzymatic reaction steps. Instead, enzymes for catalyzing orphan enzymatic reactions in the biosynthetic routes proposed by BioNavi-NP have to be suggested using third-party enzyme selection tools like Selenzyme [131] or E-zyme 2 [132] which assigns known enzymes that catalyzes similar reactions. As such, while BioNavi-NP claims to be fully data-driven as it does not rely on heuristically generated reaction rules [129], the assignment of enzymes to reactions is not data-driven. Even without considering the heuristic biases in how chemical similarity of reactions is calculated (for e.g., assuming all atoms are equidistant regardless of chemical properties), the basis of using chemical similarity of reactants and products to suggest enzymes might be intrinsically flawed based on the fact that different enzymes can possess varying degrees of substrate specificity as previously discussed. To address this, it is conceptually possible to train a separate SML model for the purpose of suggesting enzymes for orphan enzymatic reactions. Recently, SML has been used to predict metabolite-protein interactions [134], [133] and enzyme-substrate pairs [53], [135] using proteins and substrates as input. The Enzyme Substrate Prediction model (ESP; [135]) in particular, trained with 18,351 experimentally confirmed enzyme-substrate pairs obtained from the GO annotation database, is able to predict enzyme-substrate relationships with an accuracy of> 90 %. Briefly, ESP is based on a gradient-boosted decision tree model that accepts latent representations of enzymes generated by the ESM-1b transformer [85] and ECFP representations of substrates [107] as input. Therefore, it stands to reason that a similar model trained using enzyme-substrate and enzyme-product pairs can be developed in the future to suggest known enzymes or even predict sequences of novel enzymes to catalyze orphan enzymatic reactions in template-free retrobiosynthesis.
8. Reducing the combinatorial search space of retrobiosynthesis route-planning using SML
In retrobiosynthetic CASP, one-step retrobiosynthesis is applied on reactants recursively to generate sets of reactants, which results in the exponential increase in possible pathways too computationally intensive to explore. Since route-planning involves exploring the large combinatorial space of different reaction permutations to find routes of biosynthesis, it can be phrased as a combinatorial search problem [136]. Consequently, retrobiosynthetic (as well as retrosynthetic) route-planning uses heuristical tree-searching methods [119], [137] such as depth-first and best-first, as well as reinforcement-learning methods such as Monte Carlo tree-search [138], [112], [139] and others [129], [140], to evaluate only an optimal subset of possible routes. The use of such methods relies on calculating a score (usually based on the chemical similarity between reactant and product) to evaluate the feasibility of proposed enzymatic reactions and biosynthetic pathways [140], [138], [112], [139], [129]. However, while this method of quantifying reaction feasibility has shown to be effective in reducing the combinatorial search space for retrobiosynthesis route-planning [138], this measure might not account for other factors that might influence reaction feasibility, such as enzyme sequence availability [140]. Recently, DeepRFC [141] a deep-learning approach has been developed to evaluate the feasibility of enzymatic reactions generated by retrobiosynthesis. DeepRFC is able to evaluate the feasibility of enzymatic reactions from inputted SMILES strings of a substrate-product pair [141] and achieves an accuracy of 0.73 which is 1.2 fold higher than similarity-based methods[141]. Trained on only 4626 manually-curated positive reactions substrate-product pairs from KEGG [35], [33], [34] and 4626 artificially generated negative substrate-product pairs reactions, it is likely that the predictive performance of models similar to DeepRFC will increase with more data. Future models trained on an even larger and diverse reaction dataset might be able to generalize novel enzymatic reactions for the biosynthesis of increasingly novel natural products.
Besides reducing the retrobiosynthetic search space in the context of evaluating reaction feasibility, the search space can also be reduced by penalizing / rewarding certain qualities of intermediates. Since pathways are to be expressed in biological systems, exploration of biosynthetic pathways that involve intermediates that are toxic should be avoided. SML approaches to predict the toxicity of reactants in silico have been developed to aid hit-screening in small molecule drug-discovery [142], [143] and can potentially be used to eliminate biosynthetic pathways involving toxic intermediates as possible solutions for retrobiosynthetic route-planning. However, because drug-development is mainly concerned about a compound’s toxicity to humans, it might not be ideal to use these models directly for redesigning pathways for plant specialized metabolism. This calls for a need to develop models trained on predicting compound toxicity to heterologous hosts.
9. Concluding remarks and future perspectives
The wealth of publicly available reactome data as well as recent advances in SML has made it opportune to incorporate SML methods into retrobiosynthetic route-planning approaches. Importantly, neural-network transformers developed for NLP have for the first time been implemented in retrobiosynthetic route-planning to transform products into reactants, thereby eliminating the need to rely on expertly-crafted reaction rules. This method is especially important for the redesigning of plant specialized metabolism as it can take advantage of the rapidly-accumulating knowledge of experimentally validated enzymatic reactions to uncover better route-planning solutions. Besides predicting reactions outcomes to derive reactants from products, SML models trained on specific tasks such as (1) assigning known enzymes for orphan reactions, (2) evaluating the feasibility of reactions and (3) predicting the toxicity of pathway intermediates, can also be used in an ensemble manner to improve the predictive and computation performance of current template-free retrobiosynthesis methods. Cutting edge methods to represent substrates, enzymes and reactions are constantly evolving, while more sophisticated and accurate SML models are being developed at a dizzying pace. The research community will be able to build on the legacy of the present work to retrain models with higher accuracy and broader scope without the limitation of humanly curating reaction rules. This is contingent on the research community embracing the generation of open, well annotated data, which will provide the much-needed increase in quantity and quality training data for SML. This is especially important for plant sciences, as plant specialized metabolism evolved independently from the metabolism of non-plant species, resulting in poor overlap between specialized metabolism enzymes between plants and other organisms [144]. We envisage that the coming years will see a boom in SML-driven retrobiosynthetic approaches, allowing us to generate novel, useful compounds.
CRediT authorship contribution statement
Peng Ken Lim: Conceptualization, Visualization, Writing - original draft. Irene Julca: Conceptualization, Supervision. Marek Mutwil: Conceptualization, Funding acquisition, Supervision, Writing - review & editing.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
P.K.L. is supported by a Nanyang Technological University PhD stipend. I.J. is supported by Nanyang Biologics. M.M. is supported by Singapore Food Agency grant SFS_RND_SUFP_001_05 and Singaporean Ministry of Education grant MOE Tier 2 022580-00001.
References
- 1.Wink M. In: Annual plant reviews volume 40: biochemistry of plant secondary metabolism. John Wiley & Sons, Ltd,; 2010. Introduction: biochemistry, physiology and ecological functions of secondary metabolites; pp. 1–19. [DOI] [Google Scholar]
- 2.Kunst L., Samuels A.L. Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res. 2003;42(1):51–80. doi: 10.1016/s0163-7827(02)00045-0. [DOI] [PubMed] [Google Scholar]
- 3.Tohge T., Fernie A.R. Leveraging natural variance towards enhanced understanding of phytochemical sunscreens. Trends Plant Sci. 2017;22(4):308–315. doi: 10.1016/j.tplants.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. Lignin biosynthesis and structure. Plant Physiol. 2010;153(3):895–905. doi: 10.1104/pp.110.155119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milo R., Last R.L. Achieving diversity in the face of constraints: lessons from metabolism. Science. 2012;336(6089):1663–1667. doi: 10.1126/science.1217665. [DOI] [PubMed] [Google Scholar]
- 6.Brockington S.F., Yang Y., Gandia-Herrero F., Covshoff S., Hibberd J.M., Sage R.F., et al. Lineage-specific gene radiations underlie the evolution of novel betalain pigmentation in Caryophyllales. New Phytol. 2015;207(4):1170–1180. doi: 10.1111/nph.13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wink M. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry. 2003;64(1):3–19. doi: 10.1016/s0031-9422(03)00300-5. [DOI] [PubMed] [Google Scholar]
- 8.Halkier B.A., Gershenzon J. Biology and biochemistry of glucosinolates. Annu Rev Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- 9.Chakraborty P. Herbal genomics as tools for dissecting new metabolic pathways of unexplored medicinal plants and drug discovery. Biochim Open. 2018;6:9–16. doi: 10.1016/j.biopen.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hefferon K.L. Nutritionally enhanced food crops; progress and perspectives. Int J Mol Sci. 2015;16(2):3895–3914. doi: 10.3390/ijms16023895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chemler J.A., Koffas M.A.G. Metabolic engineering for plant natural product biosynthesis in microbes. Curr Opin Biotechnol. 2008;19(6):597–605. doi: 10.1016/j.copbio.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Busing R.T., Halpern C.B., Spies T.A. Ecology of Pacific Yew (Taxus brevifolia) in Western Oregon and Washington. Conserv. Biol. 1995;9:1199–1207. doi: 10.1046/j.1523-1739.1995.9051189.x-i1. [DOI] [PubMed] [Google Scholar]
- 13.Lan X., Chang K., Zeng L., Liu X., Qiu F., Zheng W., et al. Engineering salidroside biosynthetic pathway in hairy root cultures of Rhodiola crenulata based on metabolic characterization of tyrosine decarboxylase. PLoS One. 2013;8 doi: 10.1371/journal.pone.0075459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown S., Clastre M., Courdavault V., O’Connor S.E. De novo production of the plant-derived alkaloid strictosidine in yeast. Proc Natl Acad Sci USA. 2015;112(11):3205–3210. doi: 10.1073/pnas.1423555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cravens A., Payne J., Smolke C.D. Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat Commun. 2019;10(1):2142. doi: 10.1038/s41467-019-09848-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paddon C.J., Westfall P.J., Pitera D.J., Benjamin K., Fisher K., McPhee D., et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496(7446):528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 17.Sabzehzari M., Zeinali M., Naghavi M.R. Alternative sources and metabolic engineering of Taxol: advances and future perspectives. Biotechnol Adv. 2020;43 doi: 10.1016/j.biotechadv.2020.107569. [DOI] [PubMed] [Google Scholar]
- 18.Thodey K., Galanie S., Smolke C.D. A microbial biomanufacturing platform for natural and semisynthetic opioids. Nat Chem Biol. 2014;10(10):837–844. doi: 10.1038/nchembio.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holton R.A., Somoza C., Kim H.B., Liang F., Biediger R.J., Boatman P.D., et al. First total synthesis of taxol. 1. Functionalization of the B ring. J Am Chem Soc. 1994;116(4):1597–1598. doi: 10.1021/ja00083a066. [DOI] [Google Scholar]
- 20.Howat S., Park B., Oh I.S., Jin Y.-W., Lee E.-K., Loake G.J. Paclitaxel: biosynthesis, production and future prospects. New Biotechnol. 2014;31(3):242–245. doi: 10.1016/j.nbt.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka Y., Brugliera F. Flower colour and cytochromes P450. Philos Trans R Soc Lond Ser B Biol Sci. 2013;368(1612) doi: 10.1098/rstb.2012.0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakayama T., Yonekura-Sakakibara K., Sato T., Kikuchi S., Fukui Y., Fukuchi-Mizutani M., et al. Aureusidin synthase: a polyphenol oxidase homolog responsible for flower coloration. Science. 2000;290(5494):1163–1166. doi: 10.1126/science.290.5494.1163. [DOI] [PubMed] [Google Scholar]
- 23.Akashi T., Aoki T., Ayabe S.-I. Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis. Plant Physiol. 2005;137(3):882–891. doi: 10.1104/pp.104.056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waki T., Takahashi S., Nakayama T. Managing enzyme promiscuity in plant specialized metabolism: a lesson from flavonoid biosynthesis: Mission of a “body double” protein clarified. Bioessay New Rev Mol Cell Dev Biol. 2021;43(3) doi: 10.1002/bies.202000164. [DOI] [PubMed] [Google Scholar]
- 25.Atsumi S., Hanai T., Liao J.C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 26.Khersonsky O., Roodveldt C., Tawfik D.S. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr Opin Chem Biol. 2006;10(5):498–508. doi: 10.1016/j.cbpa.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez G.M., Tashiro Y., Atsumi S. Expanding ester biosynthesis in Escherichia coli. Nat Chem Biol. 2014;10(4):259–265. doi: 10.1038/nchembio.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Souza R.O.M.A., Miranda L.S.M., Bornscheuer U.T. A retrosynthesis approach for biocatalysis in organic synthesis. Chem Eur J. 2017;23(50):12040–12063. doi: 10.1002/chem.201702235. [DOI] [PubMed] [Google Scholar]
- 29.Probst D., Manica M., Nana Teukam Y.G., Castrogiovanni A., Paratore F., Laino T. Biocatalysed synthesis planning using data-driven learning. Nat Commun. 2022;13(1):1. doi: 10.1038/s41467-022-28536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pleiss G., Zhang T., Elenberg E., Weinberger K.Q. Identifying mislabeled data using the area under the margin ranking. Adv Neural Inf Process Syst. 2020;33:17044–17056. 〈https://proceedings.neurips.cc/paper/2020/hash/c6102b3727b2a7d8b1bb6981147081ef-Abstract.html〉 [Google Scholar]
- 31.Zhang C., Bengio S., Hardt M., Recht B., Vinyals O. Understanding deep learning (still) requires rethinking generalization. Commun ACM. 2021;64(3):107–115. doi: 10.1145/3446776. [DOI] [Google Scholar]
- 32.Lawson C.E., Martí J.M., Radivojevic T., Jonnalagadda S.V.R., Gentz R., Hillson N.J., et al. Machine learning for metabolic engineering: a review. Metab Eng. 2021;63:34–60. doi: 10.1016/j.ymben.2020.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillespie M., Jassal B., Stephan R., Milacic M., Rothfels K., Senff-Ribeiro A., et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50(D1):D687–D692. doi: 10.1093/nar/gkab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi-Tope G., Gillespie M., Vastrik I., D’Eustachio P., Schmidt E., de Bono B., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33(Database Issue):D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caspi R., Billington R., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018;46(D1):D633–D639. doi: 10.1093/nar/gkx935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krieger C.J., Zhang P., Mueller L.A., Wang A., Paley S., Arnaud M., et al. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2004;32(Database issue):D438–D442. doi: 10.1093/nar/gkh100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keseler I.M., Collado-Vides J., Gama-Castro S., Ingraham J., Paley S., Paulsen I.T., et al. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33(Database issue):D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keseler I.M., Gama-Castro S., Mackie A., Billington R., Bonavides-Martínez C., Caspi R., et al. The EcoCyc database in 2021. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.711077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribeiro A.J.M., Holliday G.L., Furnham N., Tyzack J.D., Ferris K., Thornton J.M. Mechanism and catalytic site Atlas (M-CSA): a database of enzyme reaction mechanisms and active sites. Nucleic Acids Res. 2018;46(D1):D618–D623. doi: 10.1093/nar/gkx1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bansal P., Morgat A., Axelsen K.B., Muthukrishnan V., Coudert E., Aimo L., et al. Rhea, the reaction knowledgebase in 2022. Nucleic Acids Res. 2022;50(D1):D693–D700. doi: 10.1093/nar/gkab1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50(D1):D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duigou T., du Lac M., Carbonell P., Faulon J.-L. RetroRules: a database of reaction rules for engineering biology. Nucleic Acids Res. 2019;47(D1):D1229–D1235. doi: 10.1093/nar/gky940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49(D1):D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen J., Zeng Y., Zhuang X., Sun L., Yao X., Pimpl P., et al. Organelle pH in the Arabidopsis endomembrane system. Mol Plant. 2013;6(5):1419–1437. doi: 10.1093/mp/sst079. [DOI] [PubMed] [Google Scholar]
- 49.Dissanayake T., Swails J.M., Harris M.E., Roitberg A.E., York D.M. Interpretation of pH-activity profiles for acid-base catalysis from molecular simulations. Biochemistry. 2015;54(6):1307–1313. doi: 10.1021/bi5012833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurczab R., Smusz S., Bojarski A.J. The influence of negative training set size on machine learning-based virtual screening. J Chemin- 2014;6:32. doi: 10.1186/1758-2946-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hussin S.K., Abdelmageid S.M., Alkhalil A., Omar Y.M., Marie M.I., Ramadan R.A. Handling imbalance classification virtual screening big data using machine learning algorithms. Complexity. 2021;2021 doi: 10.1155/2021/6675279. [DOI] [Google Scholar]
- 52.Sieg J., Flachsenberg F., Rarey M. In need of bias control: evaluating chemical data for machine learning in structure-based virtual screening. J Chem Inf Model. 2019;59(3):947–961. doi: 10.1021/acs.jcim.8b00712. [DOI] [PubMed] [Google Scholar]
- 53.Goldman S., Das R., Yang K.K., Coley C.W. Machine learning modeling of family wide enzyme-substrate specificity screens. PLoS Comput Biol. 2022;18(2) doi: 10.1371/journal.pcbi.1009853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang R., Xia M., Nguyen D.-T., Zhao T., Sakamuru S., Zhao J., et al. Tox21challenge to build predictive models of nuclear receptor and stress response pathways as mediated by exposure to environmental chemicals and drugs. Front Environ Sci. 2016:3. https://www.frontiersin.org/articles/10.3389/fenvs.2015.00085. [Google Scholar]
- 55.Keshavarzi Arshadi A., Salem M., Firouzbakht A., Yuan J.S. MolData, a molecular benchmark for disease and target based machine learning. J Chemin. 2022;14(1):10. doi: 10.1186/s13321-022-00590-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang N., Shoichet B.K., Irwin J.J. Benchmarking sets for molecular docking. J Med Chem. 2006;49(23):6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu L., Liu H. Efficient feature selection via analysis of relevance and redundancy. J Mach Learn Res. 2004;5:1205–1224. [Google Scholar]
- 58.Singh, A., Thakur, N., Sharma, A.2016. A review of supervised machine learning algorithms. In: Proceedings of the third international conference on computing for sustainable global development (INDIACom). p. 1310–15.
- 59.Anfinsen C.B. Principles that govern the folding of protein chains. Science. 1973;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 60.Ofer D., Linial M. ProFET: feature engineering captures high-level protein functions. Bioinformatics) 2015;31(21):3429–3436. doi: 10.1093/bioinformatics/btv345. [DOI] [PubMed] [Google Scholar]
- 61.Xu Y., Verma D., Sheridan R.P., Liaw A., Ma J., Marshall N.M., et al. Deep dive into machine learning models for protein engineering. J Chem Inf Model. 2020;60(6):2773–2790. doi: 10.1021/acs.jcim.0c00073. [DOI] [PubMed] [Google Scholar]
- 62.ElAbd H., Bromberg Y., Hoarfrost A., Lenz T., Franke A., Wendorff M. Amino acid encoding for deep learning applications. BMC Bioinform. 2020;21(1):235. doi: 10.1186/s12859-020-03546-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang K.K., Wu Z., Bedbrook C.N., Arnold F.H. Learned protein embeddings for machine learning. Bioinformatics. 2018;34(15):2642–2648. doi: 10.1093/bioinformatics/bty178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawashima S., Pokarowski P., Pokarowska M., Kolinski A., Katayama T., Kanehisa M. AAindex: amino acid index database, progress report 2008. Nucleic Acids Res. 2008;36:D202–D205. doi: 10.1093/nar/gkm998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandberg M., Eriksson L., Jonsson J., Sjöström M., Wold S. New chemical descriptors relevant for the design of biologically active peptides. A multivariate characterization of 87 amino acids. J Med Chem. 1998;41(14):2481–2491. doi: 10.1021/jm9700575. [DOI] [PubMed] [Google Scholar]
- 66.Mei H., Liao Z.H., Zhou Y., Li S.Z. A new set of amino acid descriptors and its application in peptide QSARs. Biopolymers. 2005;80(6):775–786. doi: 10.1002/bip.20296. [DOI] [PubMed] [Google Scholar]
- 67.Degtyarenko K., de Matos P., Ennis M., Hastings J., Zbinden M., McNaught A., et al. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;365(Database Issue):D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Engkvist O., Norrby P.-O., Selmi N., Lam Y., Peng Z., Sherer E.C., et al. Computational prediction of chemical reactions: current status and outlook. Drug Discov Today. 2018;23(6):1203–1218. doi: 10.1016/j.drudis.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 69.Ravitz O. Data-driven computer aided synthesis design. Drug Discov Today Technol. 2013;10(3):e443–e449. doi: 10.1016/j.ddtec.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 70.Warr W.A. A short review of chemical reaction database systems, computer-aided synthesis design, reaction prediction and synthetic feasibility. Mol Inform. 2014;33(6–7):469–476. doi: 10.1002/minf.201400052. [DOI] [PubMed] [Google Scholar]
- 71.McDonald, A.G., Tipton, K.F.n.d.. Enzyme nomenclature and classification: the state of the art. FEBS J, n/a(n/a). Available from: 10.1111/febs.16274. [DOI] [PubMed]
- 72.Gene Ontology, Blake J.A., Dolan M., Drabkin H., Hill D.P., Li N., et al. Gene ontology annotations and resources. Nucleic Acids Res. 2013;41(Database issue):D530–D535. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goodman J.M., Pletnev I., Thiessen P., Bolton E., Heller S.R. InChI version 1.06: now more than 99.99% reliable. J Chemin. 2021;13:40. doi: 10.1186/s13321-021-00517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Plehiers P.P., Marin G.B., Stevens C.V., Van Geem K.M. Automated reaction database and reaction network analysis: extraction of reaction templates using cheminformatics. J Chemin. 2018;10(1):11. doi: 10.1186/s13321-018-0269-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klucznik T., Mikulak-Klucznik B., McCormack M.P., Lima H., Szymkuć S., Bhowmick M., et al. Efficient syntheses of diverse, medicinally relevant targets planned by computer and executed in the laboratory. Chem. 2018;4(3):522–532. doi: 10.1016/j.chempr.2018.02.002. [DOI] [Google Scholar]
- 76.MohammadiPeyhani H., Hafner J., Sveshnikova A., Viterbo V., Hatzimanikatis V. Expanding biochemical knowledge and illuminating metabolic dark matter with ATLASx. Nat Commun. 2022;13(1):1. doi: 10.1038/s41467-022-29238-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guha R. On exploring structure activity relationships. Methods Mol Biol. 2013;993:81–94. doi: 10.1007/978-1-62703-342-8_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Greener J.G., Kandathil S.M., Moffat L., Jones D.T. A guide to machine learning for biologists. Nat Rev Mol Cell Biol. 2022;23(1):1. doi: 10.1038/s41580-021-00407-0. [DOI] [PubMed] [Google Scholar]
- 79.Sutton R.S., Barto A.G. Reinforcement Learning, second edition: An Introduction. MIT Press; 2018. https://books.google.com.sg/books?id=5s-MEAAAQBAJ [Google Scholar]
- 80.Chicco D. Ten quick tips for machine learning in computational biology. BioData Min. 2017;10(1):35. doi: 10.1186/s13040-017-0155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Spathis D., Perez-Pozuelo I., Marques-Fernandez L., Mascolo C. Breaking away from labels: the promise of self-supervised machine learning in intelligent health. Patterns. 2022;3(2) doi: 10.1016/j.patter.2021.100410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cai C., Wang S., Xu Y., Zhang W., Tang K., Ouyang Q., et al. Transfer learning for drug discovery. J Med Chem. 2020;63(16):8683–8694. doi: 10.1021/acs.jmedchem.9b02147. [DOI] [PubMed] [Google Scholar]
- 83.UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heinzinger M., Elnaggar A., Wang Y., Dallago C., Nechaev D., Matthes F., et al. Modeling aspects of the language of life through transfer-learning protein sequences. BMC Bioinforma. 2019;20(1):723. doi: 10.1186/s12859-019-3220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rives A., Meier J., Sercu T., Goyal S., Lin Z., Liu J., et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc Natl Acad Sci USA. 2021;118(15) doi: 10.1073/pnas.2016239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Villegas-Morcillo A., Gomez A.M., Sanchez V. An analysis of protein language model embeddings for fold prediction. Brief Bioinform. 2022;23(3):bbac142. doi: 10.1093/bib/bbac142. [DOI] [PubMed] [Google Scholar]
- 87.Weissenow K., Heinzinger M., Rost B. Protein language-model embeddings for fast, accurate, and alignment-free protein structure prediction. Structure. 2022;30(8):1169–1177.e4. doi: 10.1016/j.str.2022.05.001. [DOI] [PubMed] [Google Scholar]
- 88.Gligorijević V., Renfrew P.D., Kosciolek T., Leman J.K., Berenberg D., Vatanen T., et al. Structure-based protein function prediction using graph convolutional networks. Nat Commun. 2021;12(1):3168. doi: 10.1038/s41467-021-23303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramazi S., Zahiri J. Posttranslational modifications in proteins: resources, tools and prediction methods. Database J Biol Databases Curation. 2021;2021 doi: 10.1093/database/baab012. baab012. https://doi.org/10.1093/database/baab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang D., Liu D., Yuchi J., He F., Jiang Y., Cai S., et al. MusiteDeep: a deep-learning based webserver for protein post-translational modification site prediction and visualization. Nucleic Acids Res. 2020;48(W1):W140–W146. doi: 10.1093/nar/gkaa275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.David L., Thakkar A., Mercado R., Engkvist O. Molecular representations in AI-driven drug discovery: a review and practical guide. J Chemin. 2020;12(1):56. doi: 10.1186/s13321-020-00460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koge D., Ono N., Huang M., Altaf-Ul-Amin M., Kanaya S. Embedding of molecular structure using molecular hypergraph variational autoencoder with metric learning. Mol Inform. 2021;40(2) doi: 10.1002/minf.202000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grisoni F., Ballabio D., Todeschini R., Consonni V. Molecular descriptors for structure-activity applications: a hands-on approach. Methods Mol Biol. 2018;1800:3–53. doi: 10.1007/978-1-4939-7899-1_1. [DOI] [PubMed] [Google Scholar]
- 94.Wang H., Kaddour J., Liu S., Tang J., Kusner M., Lasenby J., et al. Evaluating self-supervised learning for molecular graph embeddings (arXiv:2206.08005) arXiv. 2022 doi: 10.48550/arXiv.2206.08005. [DOI] [Google Scholar]
- 95.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Comput Sci. 1988;28(1):31–36. doi: 10.1021/ci00057a005. [DOI] [Google Scholar]
- 96.Arús-Pous J., Johansson S.V., Prykhodko O., Bjerrum E.J., Tyrchan C., Reymond J.-L., et al. Randomized SMILES strings improve the quality of molecular generative models. J Chemin. 2019;11(1):71. doi: 10.1186/s13321-019-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bjerrum E.J. SMILES enumeration as data augmentation for neural network modeling of molecules. CoRR. 2017 〈http://arxiv.org/abs/1703.07076〉 abs/1703.07076. [Google Scholar]
- 98.Dietz A. Yet another representation of molecular structure. J Chem Inf Comput Sci. 1995;35(5):787–802. doi: 10.1021/ci00027a001. [DOI] [Google Scholar]
- 99.Comesana A.E., Huntington T.T., Scown C.D., Niemeyer K.E., Rapp V.H. A systematic method for selecting molecular descriptors as features when training models for predicting physiochemical properties. Fuel. 2022;321 doi: 10.1016/j.fuel.2022.123836. [DOI] [Google Scholar]
- 100.Orosz Á., Héberger K., Rácz A. Comparison of descriptor- and fingerprint sets in machine learning models for ADME-Tox targets. Front Chem. 2022;10 doi: 10.3389/fchem.2022.852893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Durant J.L., Leland B.A., Henry D.R., Nourse J.G. Reoptimization of MDL keys for use in drug discovery. J Chem Inf Comput Sci. 2002;42(6):1273–1280. doi: 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- 102.Capecchi A., Probst D., Reymond J.-L. One molecular fingerprint to rule them all: drugs, biomolecules, and the metabolome. J Chemin. 2020;12(1):43. doi: 10.1186/s13321-020-00445-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mayr A., Klambauer G., Unterthiner T., Hochreiter S. DeepTox: toxicity prediction using deep learning. Front Environ Sci. 2016;3 doi: 10.3389/fenvs.2015.00080. [DOI] [Google Scholar]
- 104.Merget B., Turk S., Eid S., Rippmann F., Fulle S. Profiling prediction of kinase inhibitors: toward the virtual assay. J Med Chem. 2017;60(1):474–485. doi: 10.1021/acs.jmedchem.6b01611. [DOI] [PubMed] [Google Scholar]
- 105.Riniker S., Fechner N., Landrum G.A. Heterogeneous classifier fusion for ligand-based virtual screening: or, how decision making by committee can be a good thing. J Chem Inf Model. 2013;53(11):2829–2836. doi: 10.1021/ci400466r. [DOI] [PubMed] [Google Scholar]
- 106.Sorgenfrei F.A., Fulle S., Merget B. Kinome-wide profiling prediction of small molecules. ChemMedChem. 2018;13(6):495–499. doi: 10.1002/cmdc.201700180. [DOI] [PubMed] [Google Scholar]
- 107.Rogers D., Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50(5):742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 108.Morgan H.L. The generation of a unique machine description for chemical structures-a technique developed at chemical abstracts service. J Chem Doc. 1965;5(2):107–113. doi: 10.1021/c160017a018. [DOI] [Google Scholar]
- 109.Sabando M.V., Ponzoni I., Milios E.E., Soto A.J. Using molecular embeddings in QSAR modeling: does it make a difference. Brief Bioinforma. 2022;23(1):bbab365. doi: 10.1093/bib/bbab365. [DOI] [PubMed] [Google Scholar]
- 110.McCowen S.V., Doering N.A., Sarpong R. Retrosynthetic strategies and their impact on synthesis of arcutane natural products. Chem Sci. 2020;11(29):7538–7552. doi: 10.1039/d0sc01441a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sun Y., Sahinidis N.V. Computer-aided retrosynthetic design: fundamentals, tools, and outlook. Curr Opin Chem Eng. 2022;35 doi: 10.1016/j.coche.2021.100721. [DOI] [Google Scholar]
- 112.Lin K., Xu Y., Pei J., Lai L. Automatic retrosynthetic route planning using template-free models. Chem Sci. 2020;11(12):3355–3364. doi: 10.1039/C9SC03666K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang Z., Zhang W., Liu B. Computational analysis of synthetic planning: past and future. Chin J Chem. 2021;39(11):3127–3143. doi: 10.1002/cjoc.202100273. [DOI] [Google Scholar]
- 114.Coley C.W., Green W.H., Jensen K.F. Machine learning in computer-aided synthesis planning. Acc Chem Res. 2018 doi: 10.1021/acs.accounts.8b00087. [DOI] [PubMed] [Google Scholar]
- 115.Finnigan W., Hepworth L.J., Flitsch S.L., Turner N.J. RetroBioCat as a computer-aided synthesis planning tool for biocatalytic reactions and cascades. Nat Catal. 2021;4(2):2. doi: 10.1038/s41929-020-00556-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kumar A., Wang L., Ng C.Y., Maranas C.D. Pathway design using de novo steps through uncharted biochemical spaces. Nat Commun. 2018;9(1):184. doi: 10.1038/s41467-017-02362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Baranwal M., Magner A., Elvati P., Saldinger J., Violi A., Hero A.O. A deep learning architecture for metabolic pathway prediction. Bioinformatics) 2020;36(8):2547–2553. doi: 10.1093/bioinformatics/btz954. [DOI] [PubMed] [Google Scholar]
- 118.Hadadi N., Hafner J., Shajkofci A., Zisaki A., Hatzimanikatis V. ATLAS of biochemistry: a repository of all possible biochemical reactions for synthetic biology and metabolic engineering studies. ACS Synth Biol. 2016;5(10):1155–1166. doi: 10.1021/acssynbio.6b00054. [DOI] [PubMed] [Google Scholar]
- 119.Dong J., Zhao M., Liu Y., Su Y., Zeng X. Deep learning in retrosynthesis planning: datasets, models and tools. Brief Bioinform. 2022;23(1):bbab391. doi: 10.1093/bib/bbab391. [DOI] [PubMed] [Google Scholar]
- 121.Weng J.-K., Noel J.P. The remarkable pliability and promiscuity of specialized metabolism. Cold Spring Harb Symp Quant Biol. 2012;77:309–320. doi: 10.1101/sqb.2012.77.014787. [DOI] [PubMed] [Google Scholar]
- 122.Baylon J.L., Cilfone N.A., Gulcher J.R., Chittenden T.W. Enhancing retrosynthetic reaction prediction with deep learning using multiscale reaction classification. J Chem Inf Model. 2019;59(2):673–688. doi: 10.1021/acs.jcim.8b00801. [DOI] [PubMed] [Google Scholar]
- 123.Karpov P., Godin G., Tetko I.V. In: Artificial neural networks and machine learning – ICANN 2019: workshop and special sessions. Tetko I.V., Kůrková V., Karpov P., Theis F., editors. Springer International Publishing; 2019. A transformer model for retrosynthesis; pp. 817–830. [DOI] [Google Scholar]
- 124.Liu B., Ramsundar B., Kawthekar P., Shi J., Gomes J., Luu Nguyen Q., et al. Retrosynthetic reaction prediction using neural sequence-to-sequence models. ACS Cent Sci. 2017;3(10):1103–1113. doi: 10.1021/acscentsci.7b00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zheng S., Rao J., Zhang Z., Xu J., Yang Y. Predicting retrosynthetic reactions using self-corrected transformer neural networks. J Chem Inf Model. 2020;60(1):47–55. doi: 10.1021/acs.jcim.9b00949. [DOI] [PubMed] [Google Scholar]
- 126.Shi, C., Xu, M., Guo, H., Zhang, M., Tang, J.2020. A graph to graphs framework for retrosynthesis prediction. In: Proceedings of the thirty seventh International Conference on Machine Learning. p. 8818–27.
- 127.Somnath V.R., Bunne C., Coley C.W., Krause A., Barzilay R. Learning graph models for retrosynthesis prediction (arXiv:2006.07038) arXiv. 2021 doi: 10.48550/arXiv.2006.07038. [DOI] [Google Scholar]
- 128.Yan C., Ding Q., Zhao P., Zheng S., YANG J., Yu Y., et al. In: Larochelle H., Ranzato M., Hadsell R., Balcan M.F., Lin H., editors. Vol. 33. Curran Associates, Inc; 2020. RetroXpert: decompose retrosynthesis prediction like a chemist; pp. 11248–11258. (Advances in neural information processing systems). https://proceedings.neurips.cc/paper/2020/file/819f46e52c25763a55cc642422644317-Paper.pdf. [Google Scholar]
- 129.Zheng S., Zeng T., Li C., Chen B., Coley C.W., Yang Y., et al. Deep learning driven biosynthetic pathways navigation for natural products with BioNavi-NP. Nat Commun. 2022;13(1):3342. doi: 10.1038/s41467-022-30970-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Moretti S., Tran V.D.T., Mehl F., Ibberson M., Pagni M. MetaNetX/MNXref: unified namespace for metabolites and biochemical reactions in the context of metabolic models. Nucleic Acids Res. 2021;49(D1):D570–D574. doi: 10.1093/nar/gkaa992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Carbonell P., Wong J., Swainston N., Takano E., Turner N.J., Scrutton N.S., et al. Selenzyme: enzyme selection tool for pathway design. Bioinformatics. 2018;34(12):2153–2154. doi: 10.1093/bioinformatics/bty065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moriya Y., Yamada T., Okuda S., Nakagawa Z., Kotera M., Tokimatsu T., et al. Identification of enzyme genes using chemical structure alignments of substrate-product pairs. J Chem Inf Model. 2016;56(3):510–516. doi: 10.1021/acs.jcim.5b00216. [DOI] [PubMed] [Google Scholar]
- 133.Jones D., Kim H., Zhang X., Zemla A., Stevenson G., Bennett W.F.D., et al. Improved protein-ligand binding affinity prediction with structure-based deep fusion inference. J Chem Inf Model. 2021;61(4):1583–1592. doi: 10.1021/acs.jcim.0c01306. [DOI] [PubMed] [Google Scholar]
- 134.Tsubaki M., Tomii K., Sese J. Compound-protein interaction prediction with end-to-end learning of neural networks for graphs and sequences. Bioinformatics. 2019;35(2):309–318. doi: 10.1093/bioinformatics/bty535. [DOI] [PubMed] [Google Scholar]
- 135.Kroll A., Ranjan S., Engqvist M.K.M., Lercher M.J. The substrate scopes of enzymes: a general prediction model based on machine and deep learning. bioRxiv. 2022 doi: 10.1101/2022.05.24.493213. 2022.05.24.493213. [DOI] [Google Scholar]
- 136.Kalé L.V., Jetley P. In: Encyclopedia of parallel computing. Padua D., editor. Springer; US: 2011. Combinatorial search; pp. 334–341. [DOI] [Google Scholar]
- 137.Menon A., Krdzavac N.B., Kraft M. From database to knowledge graph—using data in chemistry. Curr Opin Chem Eng. 2019;26:33–37. doi: 10.1016/j.coche.2019.08.004. [DOI] [Google Scholar]
- 138.Koch M., Duigou T., Faulon J.-L. Reinforcement learning for bioretrosynthesis. ACS Synth Biol. 2020;9(1):157–168. doi: 10.1021/acssynbio.9b00447. [DOI] [PubMed] [Google Scholar]
- 139.Segler M.H.S., Preuss M., Waller M.P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature. 2018;555(7698):604–610. doi: 10.1038/nature25978. [DOI] [PubMed] [Google Scholar]
- 140.Chen B., Li C., Dai H., Song L. Retro*: Learning Retrosynthetic Planning with Neural Guided A* Search (arXiv:2006.15820) arXiv. 2020 doi: 10.48550/arXiv.2006.15820. [DOI] [Google Scholar]
- 141.Kim Y., Ryu J.Y., Kim H.U., Jang W.D., Lee S.Y. A deep learning approach to evaluate the feasibility of enzymatic reactions generated by retrobiosynthesis. Biotechnol J. 2021;16(5) doi: 10.1002/biot.202000605. [DOI] [PubMed] [Google Scholar]
- 142.Wang M.W.H., Goodman J.M., Allen T.E.H. Machine learning in predictive toxicology: recent applications and future directions for classification models. Chem Res Toxicol. 2021;34(2):217–239. doi: 10.1021/acs.chemrestox.0c00316. [DOI] [PubMed] [Google Scholar]
- 143.Wu Y., Wang G. Machine learning based toxicity prediction: from chemical structural description to transcriptome analysis. Int J Mol Sci. 2018;19(8):2358. doi: 10.3390/ijms19082358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mutwil M. Computational approaches to unravel the pathways and evolution of specialized metabolism. Curr Opin Plant Biol. 2020;55:38–46. doi: 10.1016/j.pbi.2020.01.007. [DOI] [PubMed] [Google Scholar]