Abstract
Background
Genetic variability in the cytochrome P450 CYP2C9 constitutes an important predictor for efficacy and safety of various commonly prescribed drugs, including coumarin anticoagulants, phenytoin and multiple non-steroidal anti-inflammatory drugs (NSAIDs). A global map of CYP2C9 variability and its inferred functional consequences has been lacking.
Results
Frequencies of eight functionally relevant CYP2C9 alleles (*2, *3, *5, *6, *8, *11, *13 and *14) were analyzed. In total, 108 original articles were identified that included genotype data from a total of 81,662 unrelated individuals across 70 countries and 40 unique ethnic groups. The results revealed that CYP2C9*2 was most abundant in Europe and the Middle East, whereas CYP2C9*3 was the main reason for reduced CYP2C9 activity across South Asia. Our data show extensive variation within superpopulations with up to tenfold differences between geographically adjacent populations in Malaysia, Thailand and Vietnam. Translation of genetic CYP2C9 variability into functional consequences indicates that up to 40% of patients in Southern Europe and the Middle East might benefit from warfarin and phenytoin dose reductions, while 3% of patients in Southern Europe and Israel are recommended to reduce starting doses of NSAIDs.
Conclusions
This study provides a comprehensive map of the genetic and functional variability of CYP2C9 with high ethnogeographic resolution. The presented data can serve as a useful resource for CYP2C9 allele and phenotype frequencies and might guide the optimization of genotyping strategies, particularly for indigenous and founder populations with distinct genetic profiles.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40246-023-00461-z.
Keywords: Pharmacogenomics, Precision medicine, Allele frequency, Metabolizer phenotype, Precision public health
Introduction
Inter-individual variability in drug response that leads to either adverse drug reactions (ADRs) or low drug efficacy is commonly observed in clinical practice and poses significant burden to patient health and health care system. Insufficient drug efficacy and drug-related toxicity occur in up to 50% of the clinical treatments and ADRs accounts for 6–9% of all hospital admission, of which up to 40% are life threatening [1–3]. Importantly, it is estimated that 20–30% of the variability in drug response can be explained by genetic polymorphisms that are primarily localized in genes involved in drug absorption, distribution, metabolism and excretion (ADME), as well as in drug target genes and immune-related genes [4].
The human cytochrome P450 (CYP) superfamily, comprises 57 functional genes and constitutes the largest family of enzymes involved in phase I drug metabolism [5]. Among them, CYP2C9 is the most abundantly expressed CYP2C isoform in the liver and accounts for around 20% of the hepatic CYP proteins quantified by mass spectrometry [6]. It metabolizes a variety of commonly prescribed drugs, including coumarin anticoagulants, NSAIDs and sulfonylureas, as well as endogenous substrates, such as arachidonic acid [7, 8]. Genetic polymorphisms in CYP2C9 have long been recognized as a determinant of inter-individual CYP2C9 variability. The most well-studied CYP2C9 alleles are CYP2C9*2 (NC_000010.11:g.94942290C > T, p.R144C, rs1799853) and *3 (NC_000010.11:g.94981296A > C, p.I359L, rs1057910). In vitro, CYP2C9*2 reduces enzyme activity by 50–70% whereas CYP2C9*3 almost completely abrogates enzyme function (reduction of 75–99%) [9, 10]. Both alleles have also been associated with decreased metabolism of many CYP2C9 substrates in vivo, including S-warfarin and phenytoin [11]. Besides *2 and *3, multiple other variant alleles can affect CYP2C9 activity, including the decreased function alleles CYP2C9*5 (NC_000010.11:g.94981301C > G, p.D360E, rs28371686), *8 (NC_000010.11:g.94942309G > A, p.R150H, rs7900194), *11 (NC_000010.11:g.94981224C > T, p.R335W, rs28371685), *14 (NC_000010.11:g.94942234G > A, p.R125H, rs72558189) and the loss-of-function (LOF) alleles *6 (NC_000010.11:g.94949283del, p.Lys273fs, rs9332131) and *13 (NC_000010.11:g.94941958 T > C, p.L90P, rs72558187) [12]. Due to the significant impact of CYP2C9 variations, the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) include CYP2C9 genotyping in the drug labels or summary of product characteristics of 19 drugs. Specifically, testing is required for the sphingosine-1-phosphate receptor modulator siponimod in multiple sclerosis and CYP2C9 genotype is also considered as actionable information for dosage of warfarin, phenytoin and several non-steroidal anti-inflammatory drugs (NSAIDs) [13].
Considerable variation in CYP2C9 allele frequencies across different populations and ethnicities has been observed. Previous studies reported that CYP2C9*2 was most abundant in European populations (minor allele frequency, MAF between 11.1 and 14.4%), whereas the highest frequency of CYP2C9*3 was found in Asians (MAF up to 13%) [14–17]. However, frequencies were commonly extrapolated from subpopulations within the same geographic group and the generalizability of these results remains questionable [14]. In addition, evaluation of CYP2C9 allele frequencies with higher resolution is required to account for the complex patterns of ethnogeographic variability that are not reflected when only aggregated populations are considered. To better understand the global distribution of CYP2C9 variability, we here systematically analyzed CYP2C9 allele frequency data from the literature, covering in total 81,662 unrelated individuals across 70 countries and 40 ethnogeographic groups. In addition, we translate these allele frequencies into functional metabolic consequences, thus providing the first comprehensive overview of genetic and inferred functional variability at a global scale.
Results
Geographic distribution of functionally important CYP2C9 alleles
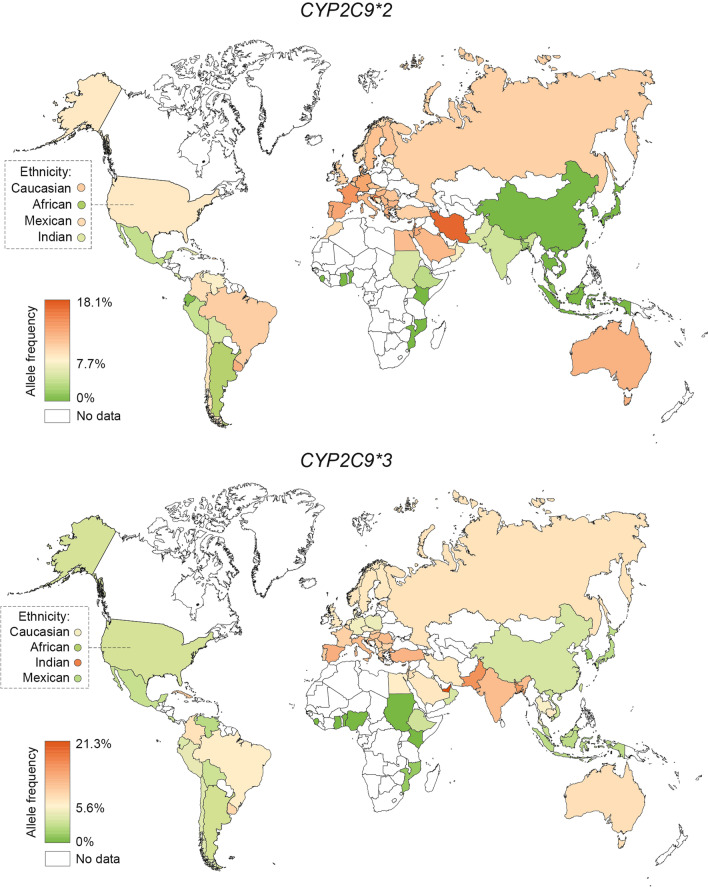
Analysis of the frequencies of functionally relevant CYP alleles across 70 countries showed that CYP2C9*2 was most abundant in Europe and across the Middle East (Fig. 1; Table 1). The prevalence was overall highest in Iran (minor allele frequency; MAF = 18.1%), followed by Croatia (MAF = 16.5%), Lebanon (MAF = 15.4%) and France (MAF = 15%). In contrast, CYP2C9*2 was absent in East Asian populations and low in South Asia with frequencies pivoting around 5%. In Africa, CYP2C9*2 was generally absent in Sub-Saharan Africa, but relatively high in North African populations (up to 12%). Notably, data about the genetic variability of CYP2C9 is still lacking for many African countries, suggesting that further exploration in these ethnogeographic groups remains to be important. In the Americas, high CYP2C9*2 frequencies were observed in Brazil (10.7%), but not in Ecuador (0.5%), Mexico (3.7%) and Peru (3.8%).
Fig. 1.
Global distribution of CYP2C9*2 and *3 alleles. Frequencies of 65 countries were color-coded with the highest frequency in red, the average frequency across all countries in yellow, and the lowest frequency in green. Countries with no frequency information available are colored white
Table 1.
National allele frequencies of CYP2C9*2 and *3
| Country | N (individuals) | *2 (in %) | *3 (in %) |
|---|---|---|---|
| Europe | |||
| South Europe | |||
| Bosnia and Herzegovina | 81 | 9.0 | - |
| Croatia | 200 | 16.5 | 9.5 |
| Greece | 283 | 12.9 | 8.1 |
| Italy | 792 | 12.4 | 9.4 |
| Portugal | 135 | 13.2 | 8.0 |
| Serbia | 716 | 12.3 | 7.9 |
| Spain | 596 | 13.8 | 10.1 |
| Turkey | 584 | 10.5 | 9.8 |
| North Europe | |||
| Denmark | 276 | 12.1 | 5.3 |
| Estonia | 2411 | 8.4 | 7.2 |
| Finland | 12,665 | 11.4 | 6.3 |
| Norway | 309 | 9.9 | 6.5 |
| Sweden | 13,495 | 11.7 | 6.5 |
| Central and East Europe | |||
| Bulgaria | 1503 | 12.5 | 7.5 |
| Czech | 341 | 11.6 | 5.9 |
| Germany | 118 | 14.0 | 5.0 |
| Hungary | 535 | 12.5 | 8.8 |
| North Macedonia | 308 | 12.4 | 7.1 |
| Poland | 85 | - | 4.7 |
| Romania | 332 | 11.3 | 9.3 |
| Russia | 290 | 10.5 | 6.7 |
| Slovak | 112 | 10.0 | 8.0 |
| Slovenia | 129 | 12.2 | 6.3 |
| West Europe | |||
| Belgium | 121 | 10.0 | 7.4 |
| France | 151 | 15.0 | 8.0 |
| Netherlands | 1252 | 13.0 | 6.0 |
| UK | 397 | 10.2 | 5.7 |
| Africa | |||
| Benin | 111 | 0.0 | 0.0 |
| Egypt | 247 | 12.0 | 6.0 |
| Ethiopia | 239 | 3.3 | 3.2 |
| Gambia | 179 | 0.4 | 0.0 |
| Ghana | 204 | 0.0 | 0.0 |
| Morocco | 290 | 9.5 | - |
| Mozambique | 103 | 0.0 | 1.0 |
| Nigeria | 293 | 0.0 | 0.0 |
| Sudan | 383 | 5.0 | 0.0 |
| Middle East | |||
| Iran | 693 | 18.1 | 6.5 |
| Israel | 438 | 14.3 | 8.2 |
| Jordan | 263 | 13.5 | 6.8 |
| Kuwait | 108 | 14.0 | 5.0 |
| Lebanon | 176 | 15.4 | 7.8 |
| Oman | 641 | 7.2 | 3.5 |
| Saudi Arabia | 323 | 12.4 | 6.3 |
| United Arab Emirates | 160 | 7.2 | 21.3 |
| Asia | |||
| Southeast Asia | |||
| Cambodia | 122 | - | 6.6 |
| Indonesia | 412 | 0.0 | 2.4 |
| Malay | 565 | 1.0 | 2.9 |
| Thai | 1205 | 0.1 | 5.3 |
| Vietnam | 381 | 0.0 | 3.0 |
| East Asia | |||
| Bhutan | 443 | 0.3 | 0.0 |
| China | 894 | 0.1 | 3.5 |
| Japan | 2105 | 0.0 | 2.4 |
| Korea | 574 | 0.0 | 1.1 |
| South Asia | |||
| Bangladesh | 144 | 1.7 | 11.6 |
| India | 2435 | 4.5 | 9.7 |
| Pakistan | 1762 | 5.0 | 11.9 |
| South America | |||
| Argentina | 101 | 2.6 | 3.0 |
| Bolivia | 778 | 4.8 | 3.0 |
| Brazil | 721 | 10.7 | 6.0 |
| Chile | 279 | 9.0 | 4.0 |
| Columbia | 375 | 9.3 | 6.8 |
| Ecuador | 297 | 0.5 | 3.7 |
| Peru | 340 | 3.8 | 4.4 |
| Uruguay | 103 | 13.0 | 7.6 |
| Venezuela | 680 | 7.4 | 2.3 |
| North America | |||
| Costa Rica | 443 | 4.7 | 2.8 |
| Cuba | 128 | 6.0 | 9.0 |
| Mexico | 1378 | 3.7 | 2.8 |
| USA | 1411 | 9.0 | 4.4 |
| Oceania | |||
| Australia | 5408 | 12.8 | 6.9 |
Global frequency distributions of CYP2C9*3 align with patterns of CYP2C9*2. European and Middle Eastern populations feature high CYP2C9*3 frequencies particularly in Spain (10.1%) and Turkey (9.8%), whereas the allele was absent or rare in Sub-Saharan Africa and East Asia. A recent study furthermore revealed very high frequencies of CYP2C9*3 in the United Arab Emirates (21.3%), which is in stark contrast to other Middle Eastern populations where CYP2C9*3 frequencies pivot around 6%. In contrast to CYP2C9*2 however, CYP2C9*3 was very common in South Asia with frequencies as high as 11.9% in Pakistan and 11.6% in Bangladesh. In South America, CYP2C9*3 frequencies are relatively higher in Uruguay (7.6%), Columbia (6.8%) and Brazil (6%) but lower than 5% in all other reported countries. Notably, frequency data for Australia was mostly derived from a pan-ethnic Australian population (n = 2,509), which results in an overall close alignment with data from European populations. In contrast, the indigenous Australian Tiwi population exhibited very high frequency of CYP2C9*3 (36%) whereas CYP2C9*2 was absent.
Besides the variants defining CYP2C9*2 and *3, more than 700 additional CYP2C9 variant alleles have been described, of which approximately 40% have been estimated to impact gene function [18]. While the vast majority of these variants are very rare and their ethnogeographic distribution has not been investigated, the frequencies of six additional functionally relevant CYP2C9 variant alleles (*5, *6, *8, *11, *13 and *14) have been investigated in multiple populations (Additional file 1: Table S1). Overall, the six alleles were observed in 16 populations, of which *8 and *11 were prevalent in African and South American populations with highest frequency found in Mozambican (14.6%) and Guarani populations (4.4%), respectively. *13 was identified in East Asia and African Americans with frequencies between 0.4 and 1.5%, whereas *5 and *6 were most abundant in the United Arab Emirates (7.8%) and Sudan (2%), respectively.
Frequencies of CYP2C9*2 and CYP2C9*3 across ethnic groups
In addition to geographic patterns, we analyzed CYP2C9*2 and CYP2C9*3 frequencies across 40 ethnic groups (Table 2). CYP2C9*2 was high in Sephardi Jews (MAF = 19.4%), a Jewish diaspora population originating from the Iberian Peninsula, as well as in Ashkenazim (MAF = 13.5%) who are of Middle Eastern origin with evidence of European admixture [19]. CYP2C9*2 prevalence was very high in Kosovars (MAF = 17.5%), whereas frequencies in neighboring Serbian (MAF = 12.3%) and North Macedonian (12.4%) populations were considerably lower. While CYP2C9*2 is mostly absent or rare in South and East Asia, specific subpopulations, such as Uyghurs (MAF = 7.8%) from Northwest China and the Kannadiga ethnic group (MAF = 6%) from southwest India, feature considerably high frequencies.
Table 2.
Allele frequencies of CYP2C9*2 and *3 across 35 ethnogeographic groups
| Ethnic group | N (individuals) | *2 (in %) | *3 (in %) |
|---|---|---|---|
| Europe | |||
| Faroese | 312 | 8.8 | 5.3 |
| Hungarian Roma | 465 | 11.8 | 15.5 |
| Kosovar | 234 | 17.5 | 10.9 |
| Slav | 479 | 10.0 | 12.0 |
| Africa | |||
| Luhya in Webuye, Kenya | 120 | 0.0 | 0.0 |
| Mende in Sierra Leone | 128 | 0.0 | 0.0 |
| Middle East | |||
| Ashkenazi Jewish | 5775 | 13.5 | 8.4 |
| Baloch | 110 | 11.8 | 7.3 |
| Greek Cypriot | 148 | 16.2 | 11.2 |
| Khorasan | 120 | 9.1 | 10.0 |
| Iranian Persian | 110 | 11.0 | 6.0 |
| Sephardi Jewish | 80 | 19.4 | 14.4 |
| Sistani | 140 | 16.1 | 7.8 |
| Turkmen | 110 | 8.0 | 4.0 |
| Asia | |||
| Southeast Asia | |||
| Jahai people | 155 | 0.0 | 36.2 |
| East Asia | |||
| Mongolian in China | 280 | 0.0 | 3.0 |
| Tibetan | 96 | 0.0 | 5.7 |
| Uygur | 96 | 7.8 | 0.0 |
| South Asia | |||
| Andhra Pradesh | 116 | 4.0 | 9.0 |
| Karnataka | 110 | 6.0 | 8.0 |
| Kerala | 120 | 2.0 | 8.0 |
| Tamilian | 270 | 2.8 | 6.9 |
| South America | |||
| Guarani | 90 | 1.1 | 0.0 |
| North America | |||
| African American | 982 | 2.1 | 1.7 |
| American from India | 109 | 4.9 | 13.1 |
| American from Mexico | 71 | 10.2 | 2.3 |
| European American | 249 | 11.2 | 5.1 |
| Huichol | 73 | 0.0 | 0.0 |
| Inuit from Canada | 151 | 0.0 | 0.0 |
| Mestizo from Chile | 253 | 6.0 | 4.0 |
| Mestizo from Ecuador | 297 | 0.5 | 3.7 |
| Mestizo from Mexico | 947 | 5.1 | 3.9 |
| Mestizo from Peru | 218 | 4.6 | 6.2 |
| Nahua | 212 | 0.7 | 0.4 |
| Native Indian from Canada | 153 | 3.0 | 6.0 |
| Puerto Rican | 314 | 12.1 | 4.3 |
| Teenek | 98 | 0.5 | 0.5 |
| Oceania | |||
| Maori | 60 | 1.7 | 0.8 |
| Polynesian | 1072 | 3.1 | 1.6 |
| Tiwi | 187 | 0.0 | 36 |
For CYP2C9*3, highest frequencies (MAF = 36.2%) were found in the Jahai people, an indigenous population living in Malaysia. This prevalence is higher than in any other population analyzed and is in stark contrast to national CYP2C9*3 frequencies in Malaysia (2.9%) and other Southeast Asian countries, such as Indonesia (2.4%), Vietnam (3%) and Thailand (5.3%), demonstrating the importance to consider ethnic backgrounds in addition to geographic factors for pharmacogenomic mapping studies.
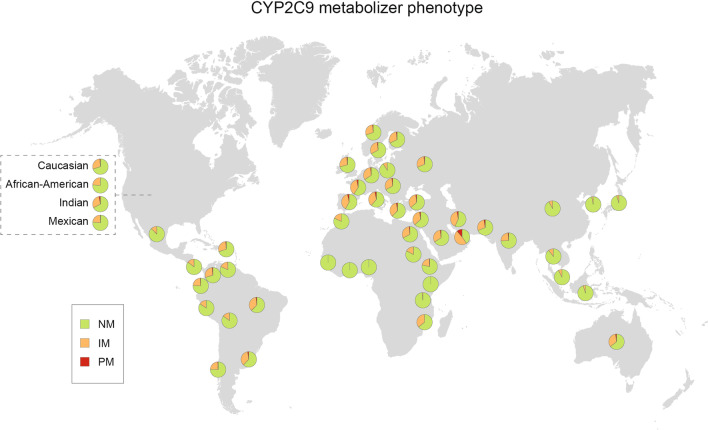
CYP2C9 phenotype distribution across different countries and regions
Next, we extrapolated the functional consequences of the observed genetic differences by inferring CYP2C9 metabolic phenotypes based on the frequencies of functionally important CYP2C9 alleles. To this end, we considered CYP2C9*2 and *3, as well as the less common variant alleles *5, *6, *8, *11, *13 and *14 that decrease or abolish enzyme function. Globally, the prevalence of CYP2C9 poor metabolizers (PMs) is relatively low, ranging from 3–4% in Southern Europe and the Eastern Mediterranean coast to < 1% in Asian and African populations, with the exception of Emiratis (11.1%) due to high frequencies of CYP2C9*3 and *5 (Table 3; Fig. 2). Similar patterns were found for CYP2C9 intermediate metabolizers (IMs), which were overall most common in the United Arab Emirates (48.7%), Croatia (41.2%) and Iran (40.3%) where almost every second individual was estimated to exhibit reduced CYP2C9 metabolism. In Africa where the prevalence of CYP2C9 IM is generally low, we found that around 35.5% were IMs in Mozambique due to the exceptionally high frequency of the decreased function allele CYP2C9*8 in this population (MAF = 14.6%). In contrast, reduced CYP2C9 metabolism was virtually absent in Ghana and Nigeria (Table 3).
Table 3.
Frequencies of inferred CYP2C9 metabolizer phenotype
| Country | NM (in %) | IM (in %) | PM (in %) |
|---|---|---|---|
| Europe | |||
| South Europe | |||
| Bulgaria | 64.0 | 33.6 | 2.4 |
| Croatia | 54.8 | 41.2 | 4.0 |
| Greek | 62.4 | 34.9 | 2.8 |
| Italy | 61.2 | 35.6 | 3.2 |
| North Macedonia | 64.9 | 32.9 | 2.3 |
| Portugal | 62.1 | 35.2 | 2.8 |
| Serbia | 63.7 | 33.7 | 2.6 |
| Spain | 58.0 | 38.2 | 3.8 |
| Turkey | 63.5 | 33.5 | 3.0 |
| North Europe | |||
| Denmark | 68.2 | 30.2 | 1.6 |
| Estonia | 71.1 | 27.1 | 1.7 |
| Finland | 67.8 | 30.4 | 1.8 |
| Norway | 69.9 | 28.4 | 1.7 |
| Sweden | 67.0 | 31.1 | 1.9 |
| Central and East Europe | |||
| Czech | 68.2 | 30.1 | 1.7 |
| Germany | 65.6 | 32.7 | 1.7 |
| Hungary | 61.9 | 35.1 | 3.0 |
| Romania | 63.0 | 34.0 | 3.0 |
| Russian | 68.6 | 29.6 | 1.9 |
| Slovak | 67.2 | 30.5 | 2.2 |
| Slovenia | 66.4 | 31.6 | 1.9 |
| West Europe | |||
| Belgium | 67.6 | 30.3 | 2.1 |
| France | 59.3 | 37.7 | 3.0 |
| Netherlands | 65.6 | 32.5 | 1.9 |
| UK | 70.8 | 27.7 | 1.5 |
| Africa | |||
| Benin | 91.2 | 8.8 | 0.0 |
| Egypt | 67.2 | 31.0 | 1.8 |
| Ethiopia | 78.3 | 21.0 | 0.6 |
| Gambia | 99.2 | 0.8 | 0.0 |
| Ghana | 100.0 | 0.0 | 0.0 |
| Morocco | 81.9 | 18.1 | 0.0 |
| Mozambique | 64.2 | 35.5 | 0.4 |
| Nigeria | 100.0 | 0.0 | 0.0 |
| Sudan | 82.8 | 16.9 | 0.3 |
| Middle East | |||
| Iran | 56.9 | 40.3 | 2.8 |
| Israel | 60.0 | 37.0 | 3.0 |
| Jordan | 63.5 | 34.2 | 2.3 |
| Kuwaiti | 65.6 | 32.7 | 1.7 |
| Lebanon | 59.0 | 38.0 | 3.0 |
| Oman | 79.9 | 19.5 | 0.6 |
| Saudi Arabia | 66.1 | 31.9 | 2.0 |
| United Arab Emirates | 40.2 | 48.7 | 11.1 |
| ASIA | |||
| Southeast Asia | |||
| Indonesia | 95.3 | 4.7 | 0.1 |
| Malay | 92.4 | 7.5 | 0.1 |
| Thai | 89.6 | 10.1 | 0.3 |
| Vietnam | 94.2 | 5.7 | 0.1 |
| East Asia | |||
| Bhutan | 99.3 | 0.7 | 0.0 |
| China | 92.9 | 7.8 | 0.2 |
| Japan | 95.2 | 4.8 | 0.1 |
| Korea | 97.8 | 3.0 | 0.0 |
| South Asia | |||
| Bangladesh | 75.2 | 23.1 | 1.7 |
| Indian | 74.3 | 24.0 | 1.7 |
| Pakistani | 69.0 | 28.4 | 2.6 |
| South America | |||
| Argentina | 89.1 | 10.6 | 0.3 |
| Bolivia | 85.0 | 14.6 | 0.4 |
| Brazil | 62.6 | 35.2 | 2.1 |
| Chile | 75.7 | 23.4 | 0.9 |
| Columbia | 70.4 | 27.9 | 1.7 |
| Ecuador | 91.8 | 8.0 | 0.2 |
| Peru | 84.3 | 15.2 | 0.5 |
| Uruguay | 63.0 | 34.4 | 2.6 |
| Venezuela | 81.6 | 18.0 | 0.4 |
| North America | |||
| Costa Rica | 85.5 | 14.1 | 0.3 |
| Cuba | 62.4 | 34.6 | 3.0 |
| Mexico | 87.6 | 12.2 | 0.3 |
| Oceania | |||
| Australia | 64.5 | 33.3 | 2.2 |
NM, normal metabolizers; IM, intermediate metabolizers; PM, poor metabolizers
Fig. 2.
CYP2C9 metabolizer phenotype across different countries and regions. Pie charts illustrate the percentage of normal metabolizer (NM, in green), intermediate metabolizer (IM, in orange) and poor metabolizer (NM, in red) for representative countries
Discussion
Genetic variations in CYP2C9 are major determinants of impaired CYP2C9 enzyme activity. In this study, we presented country-specific frequencies of functionally important CYP2C9 alleles at a global scale by integrating available genotype data from 108 studies comprising > 81,000 individuals in total. Our analysis showed that CYP2C9*2 is most prevalent in Middle Eastern populations (up to 18.1%), followed by South European populations (up to 16.5%), whereas CYP2C9*3 is most abundant in Emiratis (21.3%) and South Asian populations (up to 11.9%), followed by South European populations (up to 10.1%). These results are overall in accordance with previous studies that analyzed frequency data aggregated by continent or major ethnic groups [14, 15].
Our data indicate extensive variation within superpopulations, as evidenced by frequencies of CYP2C9*3 in the Jahai people that were up to tenfold higher than in geographically adjacent populations in Malaysia, Thailand and Vietnam. This population is considered to be among the earliest settlers entering the Malaysian peninsula from Africa over 50,000 years ago and endogamy results in a distinct genetic profile and high frequency founder mutations [20]. Similar results were observed for the Uygur population from Xinjiang, China, that harbors high frequencies of CYP2C9*2 (7.8%), an allele which is otherwise very rare in East Asia (< 0.1%). Previous studies showed that the Uygurs are a highly admixed population with admixture mapping suggesting a European ancestry contribution of 47% [21]. In South America, we observed large frequency differences of CYP2C9*2 (between 0.5% in Ecuador to 6% in Chile) but not *3 across different mestizo populations (Table 2). This is not surprising given the well-documented fluidity between Amerindian and European ancestral contributions [22] and similar results in different mestizo populations in Mexico [23].
Besides differences between ethnic groups, we also observed heterogeneity between allele frequencies of countries within the same macrogeographical region. For example, the frequency of CYP2C9*2 in Turkey (10.5%) were considerably lower than in its neighboring countries Bulgaria (12.5%), Greece (12.9%), Lebanon (15.4%) and Iran (18.1%). Similarly, CYP2C9*3 prevalence was graded across Europe with overall lower frequencies in North and Central Europe (4.7%-7.2%) compared to South and East Europe (7.9%-10.1%) in agreement with previous reports [24]. These results were corroborated by a large aggregate study in Scandinavia, which confirmed *3 frequencies of 6% in a large sample of 3,503 individuals from Norway, Sweden, Denmark and Finland [25]. Most pronounced differences were observed between the indigenous Tiwi population and groups of European ancestry in Australia with frequencies of CYP2C9*2 (12.8% and 0% in Europeans and Tiwi, respectively) and CYP2C9*3 (6.9% and 36%, respectively) differing by more than fivefold [26]. Genetic variability profiles of Tiwi were moreover drastically different from indigenous Polynesian and Maori populations. Substantial variability in prevalence of CYP2C9*2 (0.5–13%) and CYP2C9*3 (2.3–7.6%) was also observed across South America likely due to differing admixture between Amerindians, Europeans and Sub-Saharan Africans [27]. Therefore, genetic information of small geographically defined groups cannot provide accurate estimations of national allele frequencies particularly in countries with high population diversity [28]. Furthermore, our analyses conclude that there remains a pronounced underrepresentation of ethnogeographic groups, particularly in Central and Southeast Asia, Oceania and parts of Africa. Combined, these results demonstrate that pharmacogenomic studies require high ethnogeographic resolution to maximize its socioeconomic benefits, particularly for indigenous or founder populations with distinct genetic profiles.
Genetic profiles constitute important factors to infer metabolic phenotypes and CYP2C9 genotypes were sufficient to correctly identify the majority of individuals with impaired CYP2C9 activity [29, 30]. Estimation of metabolizer status at the population-scale, requires the aggregation of prevalence data from as many as possible functionally relevant alleles. While distribution patterns of CYP2C9*2 and *3 have been extensively profiled, considerably less information is available about frequencies of alleles considered to be population-specific, such as, *5, *6, *8, *11 for Africans and *13, *14 for Asians. Consequently, the calculated IM and PM frequencies could be underestimated for countries with missing data. Besides star alleles, also rare variants without functional information can contribute to altered CYP2C9 metabolism and have been associated with hypersensitivity to CYP2C9 substrates [31]. Overall, rare variants that have not been assigned star alleles have been estimated to account for 3.4% of the genetically encoded functional variability in CYP2C9 and could thus be an additional, but minor source of underestimation [18]. In addition, co-medication with inhibitors or inducers of CYP2C9 or impaired liver function can also affect CYP2C9 metabolic phenotypes, indicating that prescribers must integrate both genetic and non-genetic factors to guide treatment decisions.
CYP2C9 functionality is clinically relevant for a variety of commonly prescribed drugs. CYP2C9*2 and *3 have been associated with mean weekly warfarin dose [32] and carrier status of reduced function CYP2C9 alleles impacts the risk of warfarin-induced bleeding complications [33–35]. Consequently, guidelines recommend the use of pharmacogenetically guided dosing algorithms that consider CYP2C9*2 and *3, with adjustments for patients with African ancestry based on carrier status of CYP2C9*5, *6, *8 and *11 [36]. Reduced CYP2C9 metabolizer status is furthermore linked to phenytoin related neurotoxicity [37] and our data indicate that up to 40% of patients in Southern Europe and the Middle East might benefit from maintenance dose reductions of 25–50% [38]. Furthermore, reduced CYP2C9 activity is associated with higher drug exposure and increased risk of gastrointestinal bleeding, hypertension and myocardial infarction upon treatment with multiple NSAIDs, including oxicams, celecoxib, flurbiprofen and ibuprofen [39–42]. The current treatment recommendations are to reduce starting doses in poor CYP2C9 metabolizers by 50–75% and to titrate upwards with caution after steady-state concentrations are reached NSAIDs [43]. Based on global CYP2C9 allele distributions, these recommendations apply to more than 3% of patients in Croatia, Italy, Spain, France and Israel.
In order to implement CYP2C9 genotyping in a clinical setting, it is essential to consider the cost-effectiveness of preemptive CYP2C9 genotyping followed by treatment adjustment for decreased function allele carriers. For warfarin, previous cost-effectiveness analyses indicated that dosing guided by both CYP2C9 and VKORC1 genotypes was likely to be cost-effective compared to conventional dosing regimens [44, 45]. We are not aware of studies that evaluate the cost-effectiveness of preemptive CYP2C9 genotyping to guide therapy with phenytoin or NSAIDs. However, previous studies have shown that allele frequencies are one of the most important determinants of the cost-effectiveness of preemptive pharmacogenomic testing at the national level [46]. The data presented here might thus be useful for policy makers to evaluate whether CYP2C9 genotyping, most likely in a panel together with variants in other genes, might provide added value for national healthcare systems.
Conclusions
In summary, this study presents the worldwide distribution of CYP2C9 alleles and inferred metabolizer phenotypes with high ethnogeographic resolution. The results reveal global patterns as well as unexpected disparities of CYP2C9 genotype variability. Reduced CYP2C9 activity is most prevalent in South Europe and the Middle East, as well as in specific founder populations in Southeast Asia. The data presented here can serve as a valuable resource for population-specific CYP2C9 allele and phenotype frequencies that can provide important information for the guidance of personalized drug therapy and inform precision public healthcare at the global scale.
Methods
Data sources
We performed a systematic literature search in PubMed database covering publications before February 2023. All studies reporting frequencies of CYP2C9*2 (rs1799853) and CYP2C9*3 (rs1057910) in defined populations with cohort sizes ≥ 50 were included. We also included studies reporting frequencies of the functional alleles *5 (rs28371686), *6 (rs9332131), *8 (rs7900194), *11 (rs28371685), *13 (rs72558187) and *14 (rs72558189). In addition to published studies, we included population frequency data from the Genome Aggregation Database [47] and the 1000 Genomes Project [48]. As a result, we identified a total of 108 original articles reporting studied CYP2C9 allele frequencies from 81,662 unrelated individuals using a variety of methods for variant detection (Additional file 1: Table S1 and Additional file 2: Table S2). Frequency data for countries and ethnogeographic groups were aggregated using a weighted average approach using the cohort sizes as weighting factors.
Phenotype analyses
CYP2C9 allele function was defined based on the PharmVar consensus classifications [16]. CYP2C9 metabolizer phenotypes were defined according to the CPIC guideline [43]. Phenotype assignment of diplotypes is provided in Additional file 3: Table S3. Based on these definitions, frequencies of analyzed CYP2C9 alleles, i.e., decreased function allele *2, *5, *8, *11, *14 and LOF allele *3, *6 and *13, were used to calculate phenotype frequencies based on the Hardy–Weinberg equation. Frequencies of the CYP2C9 reference allele (*1) were calculated as f*1 = 1 − Σi fi, with fi being the frequency of each analyzed variant allele i.
Supplementary Information
Additional file 1. National frequencies of other six functionally relevant CYP2C9 variant alleles.
Additional file 2. Original articles reporting CYP2C9 allele frequencies.
Additional file 3. CYP2C9 metabolizer phenotypes defined by different diplotypes.
Acknowledgements
The work received support by the Swedish Research Council [Grant Agreement Number: 2019-01837] and by the Robert Bosch Foundation, Stuttgart, Germany.
Abbreviations
- ADME
Administration, distribution, metabolism and excretion
- ADR
Adverse drug reaction
- CPIC
The Clinical Pharmacogenetics Implementation Consortium
- CYP
Cytochrome P450
- EMA
The European Medicines Agency
- FDA
The US Food and Drug Administration
- IM
Intermediate metabolizer
- LOF
Loss-of-function
- MAF
Minor allele frequency
- NM
Normal metabolizer
- NSAID
Non-steroidal anti-inflammatory drug
- PM
Poor metabolizer
Author contributions
YZ and LN collected and analyzed the frequency data. YZ wrote the main manuscript text and prepared figures and tables. VML and EE reviewed the and revised manuscript. All authors read and approved the final manuscript.
Funding
Open access funding provided by Karolinska Institute.
Declarations
Competing interests
YZ and VML are co-founders and shareholders of PersoMedix AB. In addition, VML is CEO and shareholder of HepaPredict AB. EE is vice-chair of the Genomic Medicine Sweden Pharmacogenomics work package, supported by grants from The Swedish Innovation Agency. EE also received funding from the Stockholm Region (CIMED). The other authors do not disclose competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pirmohamed M, James S, Meakin S, Green C, Scott AK, Walley TJ, et al. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. BMJ. 2004;329(7456):15–19. doi: 10.1136/bmj.329.7456.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Impicciatore P, Choonara I, Clarkson A, Provasi D, Pandolfini C, Bonati M. Incidence of adverse drug reactions in paediatric in/out-patients: a systematic review and meta-analysis of prospective studies. Br J Clin Pharmacol. 2001;52(1):77–83. doi: 10.1046/j.0306-5251.2001.01407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Insani WN, Whittlesea C, Alwafi H, Man KKC, Chapman S, Wei L. Prevalence of adverse drug reactions in the primary care setting: a systematic review and meta-analysis. PLoS ONE. 2021;16(5):e0252161. doi: 10.1371/journal.pone.0252161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lauschke VM, Ingelman-Sundberg M. Prediction of drug response and adverse drug reactions: From twin studies to Next Generation Sequencing. Eur J Pharm Sci. 2019;130:65–77. doi: 10.1016/j.ejps.2019.01.024. [DOI] [PubMed] [Google Scholar]
- 5.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Zhang HF, Wang HH, Gao N, Wei JY, Tian X, Zhao Y, et al. Physiological content and intrinsic activities of 10 cytochrome P450 isoforms in human normal liver microsomes. J Pharmacol Exp Ther. 2016;358(1):83–93. doi: 10.1124/jpet.116.233635. [DOI] [PubMed] [Google Scholar]
- 7.Daly AK, Rettie AE, Fowler DM, Miners JO. Pharmacogenomics of CYP2C9: functional and clinical considerations. J Pers Med. 2017;8(1):1. doi: 10.3390/jpm8010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasar U, Bennet AM, Eliasson E, Lundgren S, Wiman B, De Faire U, et al. Allelic variants of cytochromes P450 2C modify the risk for acute myocardial infarction. Pharmacogenetics. 2003;13(12):715–720. doi: 10.1097/00008571-200312000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Rettie AE, Haining RL, Bajpai M, Levy RH. A common genetic basis for idiosyncratic toxicity of warfarin and phenytoin. Epilepsy Res. 1999;35(3):253–255. doi: 10.1016/S0920-1211(99)00017-0. [DOI] [PubMed] [Google Scholar]
- 10.Niinuma Y, Saito T, Takahashi M, Tsukada C, Ito M, Hirasawa N, et al. Functional characterization of 32 CYP2C9 allelic variants. Pharmacogenomics J. 2014;14(2):107–114. doi: 10.1038/tpj.2013.22. [DOI] [PubMed] [Google Scholar]
- 11.Kirchheiner J, Brockmöller J. Clinical consequences of cytochrome P450 2C9 polymorphisms. Clin Pharmacol Ther. 2005;77(1):1–16. doi: 10.1016/j.clpt.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Caudle KE, Rettie AE, Whirl-Carrillo M, Smith LH, Mintzer S, Lee MT, et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and HLA-B genotypes and phenytoin dosing. Clin Pharmacol Ther. 2014;96(5):542–548. doi: 10.1038/clpt.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Booven D, Marsh S, McLeod H, Carrillo MW, Sangkuhl K, Klein TE, et al. Cytochrome P450 2C9-CYP2C9. Pharmacogenet Genomics. 2010;20(4):277–281. doi: 10.1097/FPC.0b013e3283349e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sistonen J, Fuselli S, Palo JU, Chauhan N, Padh H, Sajantila A. Pharmacogenetic variation at CYP2C9, CYP2C19, and CYP2D6 at global and microgeographic scales. Pharmacogenet Genomics. 2009;19(2):170–179. doi: 10.1097/FPC.0b013e32831ebb30. [DOI] [PubMed] [Google Scholar]
- 15.Céspedes-Garro C, Fricke-Galindo I, Naranjo ME, Rodrigues-Soares F, Fariñas H, de Andrés F, et al. Worldwide interethnic variability and geographical distribution of CYP2C9 genotypes and phenotypes. Expert Opin Drug Metab Toxicol. 2015;11(12):1893–1905. doi: 10.1517/17425255.2015.1111871. [DOI] [PubMed] [Google Scholar]
- 16.Sangkuhl K, Claudio-Campos K, Cavallari LH, Agundez JAG, Whirl-Carrillo M, Duconge J, et al. PharmVar GeneFocus: CYP2C9. Clin Pharmacol Ther. 2021;110(3):662–676. doi: 10.1002/cpt.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nizamuddin S, Dubey S, Singh S, Sharma S, Machha P, Thangaraj K. CYP2C9 variations and their pharmacogenetic implications among diverse South Asian populations. Pharmgenomics Pers Med. 2021;14:135–147. doi: 10.2147/PGPM.S272015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Lauschke VM. The genetic landscape of major drug metabolizing cytochrome P450 genes-an updated analysis of population-scale sequencing data. Pharmacogenomics J. 2022. [DOI] [PMC free article] [PubMed]
- 19.Costa MD, Pereira JB, Pala M, Fernandes V, Olivieri A, Achilli A, et al. A substantial prehistoric European ancestry amongst Ashkenazi maternal lineages. Nat Commun. 2013;4:2543. doi: 10.1038/ncomms3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill C, Soares P, Mormina M, Macaulay V, Meehan W, Blackburn J, et al. Phylogeography and ethnogenesis of aboriginal Southeast Asians. Mol Biol Evol. 2006;23(12):2480–2491. doi: 10.1093/molbev/msl124. [DOI] [PubMed] [Google Scholar]
- 21.Xu S, Jin L. A genome-wide analysis of admixture in Uyghurs and a high-density admixture map for disease-gene discovery. Am J Hum Genet. 2008;83(3):322–336. doi: 10.1016/j.ajhg.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva-Zolezzi I, Hidalgo-Miranda A, Estrada-Gil J, Fernandez-Lopez JC, Uribe-Figueroa L, Contreras A, et al. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc Natl Acad Sci USA. 2009;106(21):8611–8616. doi: 10.1073/pnas.0903045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saldaña-Cruz AM, León-Moreno LC, Sánchez-Corona J, Santiago DA, Mendoza-Carrera F, Castro-Martínez XH, et al. CYP2C9 and CYP2C19 Allele and haplotype distributions in four mestizo populations from Western Mexico: An interethnic comparative study. Genet Test Mol Biomarkers. 2016;20(11):702–709. doi: 10.1089/gtmb.2016.0115. [DOI] [PubMed] [Google Scholar]
- 24.Mizzi C, Dalabira E, Kumuthini J, Dzimiri N, Balogh I, Başak N, et al. A European spectrum of pharmacogenomic biomarkers: implications for clinical pharmacogenomics. PLoS ONE. 2016;11(9):e0162866. doi: 10.1371/journal.pone.0162866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nordestgaard BG, Kontula K, Benn M, Dahlöf B, de Faire U, Edelman JM, et al. Effect of ACE insertion/deletion and 12 other polymorphisms on clinical outcomes and response to treatment in the LIFE study. Pharmacogenet Genomics. 2010;20(2):77–85. doi: 10.1097/FPC.0b013e328333f70b. [DOI] [PubMed] [Google Scholar]
- 26.Jaya Shankar A, Jadhao S, Hoy W, Foote SJ, Patel HR, Scaria V, et al. Pharmacogenomic analysis of a genetically distinct Indigenous population. Pharmacogenomics J. 2022;22(2):100–108. doi: 10.1038/s41397-021-00262-4. [DOI] [PubMed] [Google Scholar]
- 27.Suarez-Kurtz G, Parra EJ. Population diversity in pharmacogenetics: a latin American perspective. Adv Pharmacol. 2018;83:133–154. doi: 10.1016/bs.apha.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Suarez-Kurtz G. Pharmacogenetics in the Brazilian population. Front Pharmacol. 2010;1:118. doi: 10.3389/fphar.2010.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fekete F, Mangó K, Déri M, Incze E, Minus A, Monostory K. Impact of genetic and non-genetic factors on hepatic CYP2C9 expression and activity in Hungarian subjects. Sci Rep. 2021;11(1):17081. doi: 10.1038/s41598-021-96590-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varshney E, Saha N, Tandon M, Shrivastava V, Ali S. Genotype-phenotype correlation of cytochrome P450 2C9 polymorphism in Indian National Capital Region. Eur J Drug Metab Pharmacokinet. 2013;38(4):275–282. doi: 10.1007/s13318-013-0124-2. [DOI] [PubMed] [Google Scholar]
- 31.Ciccacci C, Falconi M, Paolillo N, Oteri F, Forte V, Novelli G, et al. Characterization of a novel CYP2C9 gene mutation and structural bioinformatic protein analysis in a warfarin hypersensitive patient. Pharmacogenet Genomics. 2011;21(6):344–346. doi: 10.1097/FPC.0b013e328344c340. [DOI] [PubMed] [Google Scholar]
- 32.Borgiani P, Ciccacci C, Forte V, Romano S, Federici G, Novelli G. Allelic variants in the CYP2C9 and VKORC1 loci and interindividual variability in the anticoagulant dose effect of warfarin in Italians. Pharmacogenomics. 2007;8(11):1545–1550. doi: 10.2217/14622416.8.11.1545. [DOI] [PubMed] [Google Scholar]
- 33.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353(9154):717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 34.Pirmohamed M, Burnside G, Eriksson N, Jorgensen AL, Toh CH, Nicholson T, et al. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med. 2013;369(24):2294–2303. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 35.Mega JL, Walker JR, Ruff CT, Vandell AG, Nordio F, Deenadayalu N, et al. Genetics and the clinical response to warfarin and edoxaban: findings from the randomised, double-blind ENGAGE AF-TIMI 48 trial. Lancet. 2015;385(9984):2280–2287. doi: 10.1016/S0140-6736(14)61994-2. [DOI] [PubMed] [Google Scholar]
- 36.Johnson JA, Caudle KE, Gong L, Whirl-Carrillo M, Stein CM, Scott SA, et al. Clinical pharmacogenetics implementation consortium (CPIC) Guideline for pharmacogenetics-Guided Warfarin dosing: 2017 update. Clin Pharmacol Ther. 2017;102(3):397–404. doi: 10.1002/cpt.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franco V, Perucca E. CYP2C9 polymorphisms and phenytoin metabolism: implications for adverse effects. Expert Opin Drug Metab Toxicol. 2015;11(8):1269–1279. doi: 10.1517/17425255.2015.1053463. [DOI] [PubMed] [Google Scholar]
- 38.Karnes JH, Rettie AE, Somogyi AA, Huddart R, Fohner AE, Formea CM, et al. Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2C9 and HLA-B genotypes and phenytoin dosing: 2020 update. Clin Pharmacol Ther. 2021;109(2):302–309. doi: 10.1002/cpt.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Figueiras A, Estany-Gestal A, Aguirre C, Ruiz B, Vidal X, Carvajal A, et al. CYP2C9 variants as a risk modifier of NSAID-related gastrointestinal bleeding: a case-control study. Pharmacogenet Genomics. 2016;26(2):66–73. doi: 10.1097/FPC.0000000000000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grosser T, Theken KN, FitzGerald GA. Cyclooxygenase inhibition: pain, inflammation, and the cardiovascular system. Clin Pharmacol Ther. 2017;102(4):611–622. doi: 10.1002/cpt.794. [DOI] [PubMed] [Google Scholar]
- 41.Bhala N, Emberson J, Merhi A, Abramson S, Arber N, Baron JA, et al. Vascular and upper gastrointestinal effects of non-steroidal anti-inflammatory drugs: meta-analyses of individual participant data from randomised trials. Lancet. 2013;382(9894):769–779. doi: 10.1016/S0140-6736(13)60900-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lundblad MS, Ohlsson S, Johansson P, Lafolie P, Eliasson E. Accumulation of celecoxib with a 7-fold higher drug exposure in individuals homozygous for CYP2C9*3. Clin Pharmacol Ther. 2006;79(3):287–288. doi: 10.1016/j.clpt.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 43.Theken KN, Lee CR, Gong L, Caudle KE, Formea CM, Gaedigk A, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2C9 and nonsteroidal anti-inflammatory drugs. Clin Pharmacol Ther. 2020;108(2):191–200. doi: 10.1002/cpt.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pink J, Pirmohamed M, Lane S, Hughes DA. Cost-effectiveness of pharmacogenetics-guided warfarin therapy vs. alternative anticoagulation in atrial fibrillation. Clin Pharmacol Ther. 2014;95(2):199–207. doi: 10.1038/clpt.2013.190. [DOI] [PubMed] [Google Scholar]
- 45.You JH. Pharmacogenetic-guided selection of warfarin versus novel oral anticoagulants for stroke prevention in patients with atrial fibrillation: a cost-effectiveness analysis. Pharmacogenet Genomics. 2014;24(1):6–14. doi: 10.1097/FPC.0000000000000014. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Y, Krebs K, Milani L, Lauschke VM. Global frequencies of clinically important HLA alleles and their implications for the cost-effectiveness of preemptive pharmacogenetic testing. Clin Pharmacol Ther. 2021;109(1):160–174. doi: 10.1002/cpt.1944. [DOI] [PubMed] [Google Scholar]
- 47.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. National frequencies of other six functionally relevant CYP2C9 variant alleles.
Additional file 2. Original articles reporting CYP2C9 allele frequencies.
Additional file 3. CYP2C9 metabolizer phenotypes defined by different diplotypes.