Abstract
Background
Aconitum transsectum Diels. (Ranunculaceae) is an important medicinal plant that is widely used in traditional Chinese medicine, but its morphological traits make it difficult to recognize from other Aconitum species. No research has sequenced the chloroplast genome of A.transsectum, despite the fact that phylogenetic analysis based on chloroplast genome sequences provides essential evidence for plant classification.
Results
In this study, the chloroplast (cp) genome of A. transsectum was sequenced, assembled, and annotated. A. transsectum cp genome is a 155,872 bp tetrameric structure including a large single copy (LSC, 87,671 bp) and a small single copy (SSC, 18,891 bp) section, as well as a pair of inverted repeat sequences (IRa and IRb, 25,894 bp each). 131 genes are encoded by the complete cp genome, comprising 86 protein-coding genes, 37 tRNAs, and 8 rRNAs. The most favored codon in the A. transsectum cp genome is AUG, and 46 repeats and 241 SSRs were also identified. The A. transsectum cp genome is similar in size, gene composition, and IR expansion and contraction to the cp genomes of seven Ranunculaceae species. Phylogenetic analysis of cp genomes of 28 plants from the Ranunculaceae family shows that A. transsectum is most closely related to A. vilmorinianum, A. episcopale, and A. forrestii of Subgen. Aconitum.
Conclusions
Overall, this study provides complete cp genome resources for A. transsectum that will be beneficial for identifying potential.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-023-09180-0.
Keywords: Aconitum transsectum, Sequencing, Chloroplast genome, SSR, Phylogenetic analysis
Background
Aconitum transsectum Diels. is a species of Aconitum in the Ranunculaceae family, mainly distributed in northwestern Yunnan, the roots are highly toxic and have medicinal value, and Chinese folk used them to treat rheumatism and other diseases [1]. Genus Aconitum belongs to the Ranunculaceae family, which has over 350 species worldwide and is found primarily in temperate parts of the Northern Hemisphere, particularly Asia, Europe, and North America [2]. China has recorded more than 200 species, the most abundant plant resources of the genus in the country [3]. The genus Aconitum is widely distributed in Taiwan and the mainland provinces of China except for Hainan provinces, mostly in the alpine zone of northern Yunnan, western Sichuan, and eastern Tibet, followed by a number of species in the northeastern provinces [1]. The genus Aconitum is a traditional Chinese medicinal plant with over 2000 years of use, it is classified into three subgenera: Aconitum, Gymnaconitum, and Paraconitum, with about 36 species available for medical use in China [4]. Genus Aconitum is widely used in traditional Chinese medicine, but it is not easy to distinguish among the species in terms of morphological characteristics, and there are huge differences in chemical composition, which makes it easy to endanger the lives of the wrong species during the actual use of medicine [5].
With the decrease in the cost of chloroplast whole-genome sequencing and the maturity of data analysis technology in latest years, a growing number of scholars have conducted chloroplast whole-genome studies, and chloroplast whole-genome data analysis has gradually become an efficient tool for species identification and species evolution studies [6]. Chloroplast (cp), a unique organelle of green plants and algae, is the location of photosynthesis in plants [7], and has its own genetic system consisting of a closed loop of double-stranded DNA molecules. The cp not only have their own genetic material, but are also a relatively independent genetic system capable of semi-autonomous replication under conditions where the nucleus provides genetic information [8]. Higher plants' cp genomes often have a tetrameric structure with a large single copy (LSC), a small single copy (SSC), and two inverted repeats (IRa and IRb) [9]. The cp genomes are small and highly conserved in sequence and structure, making them well suited for phylogenetic studies of complex plant populations [10–12].
It is the first time that the entire cp genome of A. transsectum was sequenced and analyzed, and the cp genome differences between A. transsectum and other related species were evaluated in this study. Based on cp genomic data, a phylogenetic tree of 26 Aconitum species and 2 Delphinium species was constructed to investigate the affinities between A. transsectum and other species, as well as to provide a theoretical foundation for understanding A. transsectum's cp genomic characteristics and phylogenetic relationships.
Result
Chloroplast genome characterization
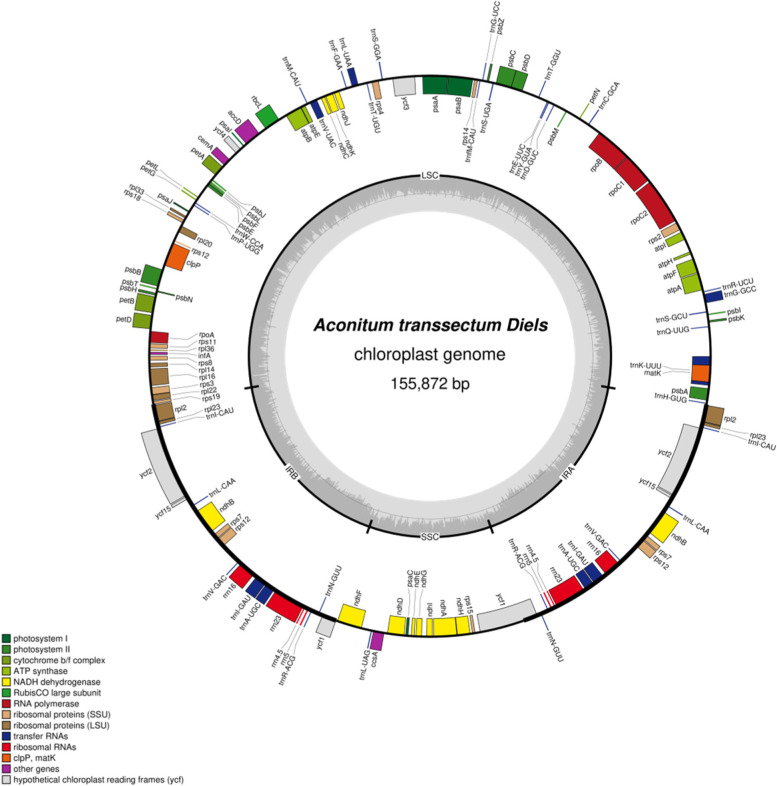
The cp genome of A. transsectum was 155,872 bp with a standard cyclic quadripartite structure, containing a couple of IR regions IRa and IRb (25,894 bp), an 18,891 bp SSC region and an 87,671 bp LSC region (Table 1 and Fig. 1). Overall GC content of the cp genome was 38.07%, while GC content was unevenly distributed across the cp genome, the IR region (42.97%) had a higher GC content than the LSC (36.19%) and SSC (32.54%) regions.
Table 1.
The Characteristics of A. transsectum cp genome
| Category | Item | Describe |
|---|---|---|
| Cp genome structure | Cp genome/bp | 155,872 |
| LSC/bp | 87,671 | |
| SSC/bp | 18,891 | |
| IRa/IRb/bp | 25,894 | |
| Gene composition | Cp gene | 131 |
| tRNA | 37 | |
| rRNA | 8 | |
| mRNA | 86 | |
| pseudo | 0 | |
| GC Content (%) | Cp gene | 38.07% |
| LSC | 36.19% | |
| SSC | 32.54% | |
| IRa/IRb | 42.97% |
Fig. 1.
Gene map of A. transsectum cp genome
The cp genome of A. transsectum has 131 predicted functional genes, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes, with no pseudogenes (Table 2). In the IR regions of the cp genomes, there were 19 duplicated genes, including 4 rRNA genes (rrn16, rrn23, rrn4.5, and rrn5), 8 protein-coding genes (ndhB, rpl2, rpl23, rps12, rps7, ycf15, ycf1 and ycf2), and 7 tRNA genes (trnL-CAA, trnI-CAU, trnI-GAU, trnN-GUU, trnA-UGC, trnR-ACG, trnV-GAC). In addition, among these 131 genes, 14 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, trnI-GAU, trnG-GCC, trnL-UAA, trnV-UAC trnA-UGC, trnK-UUU,) contained one intron and 3 genes (rps12, ycf3, clpP) contained two introns.
Table 2.
Genes in cp genome of A. transsectum
| Category | Gene group | Gene name |
|---|---|---|
| Photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of NADH dehydrogenase | ndhA*, ndhB*(2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Subunits of cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
| Large subunit of rubisco | rbcL | |
| Subunits photochlorophyllide reductase | - | |
| Self-replication | Proteins of large ribosomal subunit | rpl14, rpl16*, rpl2*(2), rpl20, rpl22, rpl23(2), rpl33, rpl36 |
| Proteins of small ribosomal subunit | rps11, rps12**(2), rps14, rps15, rps18, rps19, rps2, rps3, rps4, rps7(2), rps8 | |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | |
| Ribosomal RNAs | rrn16(2), rrn23(2), rrn4.5(2), rrn5(2) | |
| Transfer RNAs | trnA-UGC*(2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC*, trnG-UCC, trnH-GUG, trnI-CAU(2), trnI-GAU*(2), trnK-UUU*, trnL-CAA(2), trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC*, trnW-CCA, trnY-GUA, trnfM-CAU | |
| Other genes | Maturase | matK |
| Protease | clpP** | |
| Envelope membrane protein | cemA | |
| Acetyl-CoA carboxylase | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Translation initiation factor | infA | |
| other | - | |
| Genes of unknown function | Conserved hypothetical chloroplast ORF | ycf1(2), ycf15(2), ycf2(2), ycf3**, ycf4 |
Gene*: Gene with one intron; Gene**: Gene with two introns; Gene (2): Genes duplicated in the IR regions
Codon usage bias
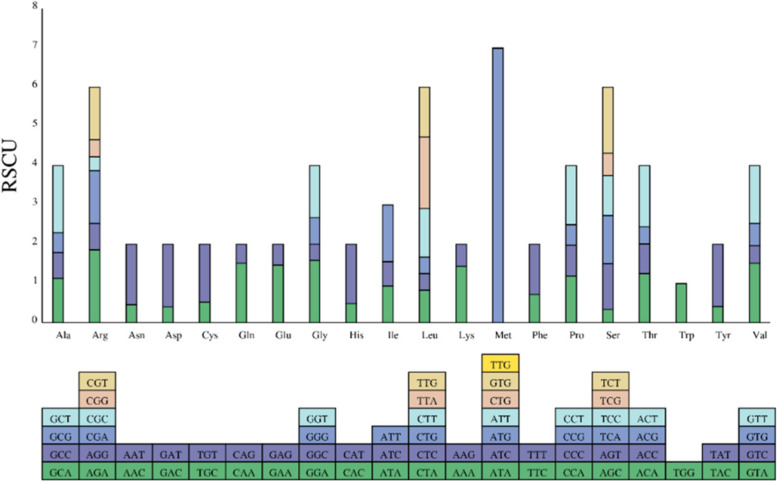
The codon usage bias (CUB) is a concept that describes to the differential frequency with which numerous synonymous codons encoding the same amino acid are seen [13]. CUB preferences are specific to different genes in different species or even within a particular species, a combination of mutation, selection and drift during the long-term evolution of genes and species [14]. We examined the codon usage frequency of protein-coding genes in the A. transsectum cp genome, finding that all proteins were encoded by 26,535 codons (Contains three termination codons) (Table 3 and Fig. 2). Leucine had the most codons (2752 codons, 10.37%), then isoleucine (2243 codons, 8.45%), and serine (2024 codons, 7.63%), while cysteine was the least common amino acid (308 codons, 1.16%). A total of 32 codons (73.4%) had relative synonymous codon usage (RSCU) greater than 1. The most favored codon was AUG, which encodes methionine (Met) and has an RSCU value of 6.9783, followed by AGA, which encodes arginine (Arg) and has an RSCU value of 1.8576.
Table 3.
Codon list of A. transsectum
| Codon | AminoAcid | Number | RSCU | Codon | AminoAcid | Number | RSCU |
|---|---|---|---|---|---|---|---|
| UAA | Ter(*) | 39 | 1.3605 | AUU | Met(M) | 0 | 0 |
| UAG | Ter(*) | 25 | 0.8721 | CUG | Met(M) | 0 | 0 |
| UGA | Ter(*) | 22 | 0.7674 | GUG | Met(M) | 2 | 0.0217 |
| GCA | Ala(A) | 395 | 1.1284 | UUG | Met(M) | 0 | 0 |
| GCC | Ala(A) | 230 | 0.6572 | AAC | Asn(N) | 298 | 0.4584 |
| GCG | Ala(A) | 177 | 0.5056 | AAU | Asn(N) | 1002 | 1.5416 |
| GCU | Ala(A) | 598 | 1.7084 | CCA | Pro(P) | 341 | 1.1912 |
| UGC | Cys(C) | 81 | 0.526 | CCC | Pro(P) | 223 | 0.7792 |
| UGU | Cys(C) | 227 | 1.474 | CCG | Pro(P) | 151 | 0.5276 |
| GAC | Asp(D) | 222 | 0.4018 | CCU | Pro(P) | 430 | 1.502 |
| GAU | Asp(D) | 883 | 1.5982 | CAA | Gln(Q) | 690 | 1.5132 |
| GAA | Glu(E) | 1006 | 1.4654 | CAG | Gln(Q) | 222 | 0.4868 |
| GAG | Glu(E) | 367 | 0.5346 | AGA | Arg(R) | 501 | 1.8576 |
| UUC | Phe(F) | 531 | 0.723 | AGG | Arg(R) | 182 | 0.675 |
| UUU | Phe(F) | 938 | 1.277 | CGA | Arg(R) | 362 | 1.3422 |
| GGA | Gly(G) | 725 | 1.5936 | CGC | Arg(R) | 95 | 0.3522 |
| GGC | Gly(G) | 187 | 0.4108 | CGG | Arg(R) | 118 | 0.4374 |
| GGG | Gly(G) | 306 | 0.6724 | CGU | Arg(R) | 360 | 1.335 |
| GGU | Gly(G) | 602 | 1.3232 | AGC | Ser(S) | 114 | 0.3378 |
| CAC | His(H) | 166 | 0.4896 | AGU | Ser(S) | 394 | 1.1682 |
| CAU | His(H) | 512 | 1.5104 | UCA | Ser(S) | 413 | 1.2246 |
| AUA | Ile(I) | 704 | 0.9417 | UCC | Ser(S) | 344 | 1.02 |
| AUC | Ile(I) | 459 | 0.6138 | UCG | Ser(S) | 193 | 0.5724 |
| AUU | Ile(I) | 1080 | 1.4445 | UCU | Ser(S) | 566 | 1.6776 |
| AAA | Lys(K) | 1005 | 1.444 | ACA | Thr(T) | 429 | 1.2508 |
| AAG | Lys(K) | 387 | 0.556 | ACC | Thr(T) | 260 | 0.758 |
| CUA | Leu(L) | 382 | 0.8328 | ACG | Thr(T) | 149 | 0.4344 |
| CUC | Leu(L) | 193 | 0.4206 | ACU | Thr(T) | 534 | 1.5568 |
| CUG | Leu(L) | 193 | 0.4206 | GUA | Val(V) | 548 | 1.516 |
| CUU | Leu(L) | 566 | 1.2342 | GUC | Val(V) | 162 | 0.448 |
| UUA | Leu(L) | 834 | 1.8186 | GUG | Val(V) | 205 | 0.5672 |
| UUG | Leu(L) | 584 | 1.2732 | GUU | Val(V) | 531 | 1.4688 |
| AUA | Met(M) | 0 | 0 | UGG | Trp(W) | 480 | 1 |
| AUC | Met(M) | 0 | 0 | UAC | Tyr(Y) | 201 | 0.4144 |
| AUG | Met(M) | 640 | 6.9783 | UAU | Tyr(Y) | 769 | 1.5856 |
Fig. 2.
The RSCU of amino acids in A. transsectum's cp genome. The color of the histogram is the same as the codon's color
Interspersed repeats and SSRs
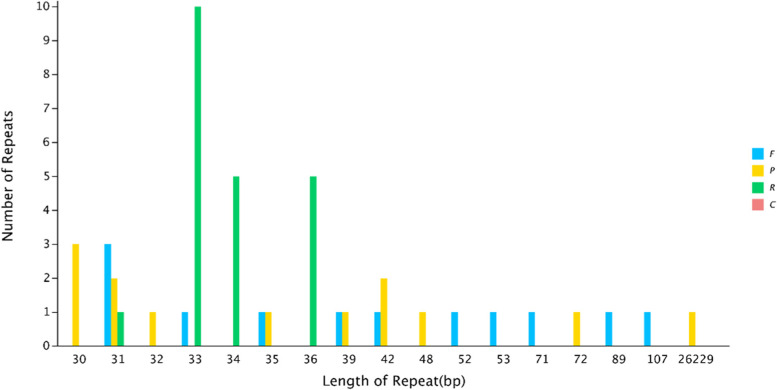
We discovered 46 interspersed repeats in the A. transsectum cp genome, including 21 reverse repeats, 13 palindromic repeats, and 12 forward repeats; complementary repeats were not discovered in the A. transsectum cp genome. The length of the repeats ranged from 30 to 26,229 bp, 35 repeats were between 30–39 bp, 10 repeats were between 42–107 bp, and only 1 repeat was 26,229 bp in length (Fig. 3). In the A. transsectum cp genome, we found 241 SSRs, 163 of which were found in the LSC region, 40 in the IRs region, and 38 in the SSC region (Fig. 4).
Fig. 3.
The A. transsectum cp genome's repeated sequences. The length of the repetition sequence is the abscissa, and the number of repeat sequences is the ordinate. Forward repetition is abbreviated as F, palindromic repetition is abbreviated as P, reverse repetition is abbreviated as R, and complementary repetition is abbreviated as C
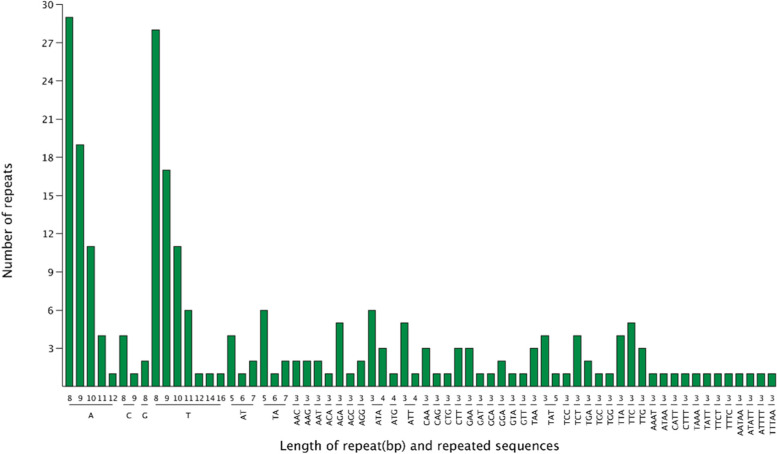
Fig. 4.
SSRs types in the cp genome of A. transsectum. The horizontal axis represents SSR repeats types and the vertical axis is the number of repeats
In addition, 137 SSRs were discovered in intergenic spaces, and 104 SSRs were discovered in genes like ycf4, ycf3, ycf2, ycf1, trnI-GAU, trnL-UAA, trnV-UAC, trnK-UUU, rrn23, rps3, rps19, rps18, rps14, rpoC2, rpoC1, rpoB rpoA, rpl22, rpl2, rpl16, rpl14, psbC, psbB, psaJ, psaB, psaA, petA, ndhH, ndhF, ndhD, ndhA, ndhB, matK, accD, ccsA, atpB, atpF, atpI, and other genes. These SSRs consist of 136 mononucleotides, 16 dinucleotides, 77 trinucleotides, 8 tetranucleotides and 4 pentanucleotides. The mononucleotide SSRs were dominated by polyadenine (PolyA) and polythymine (PolyT) repeats (94.86%), with fewer C and G mononucleotides (5.14%).
Comparative analysis of the cp genomes of A. transsectum and its related species
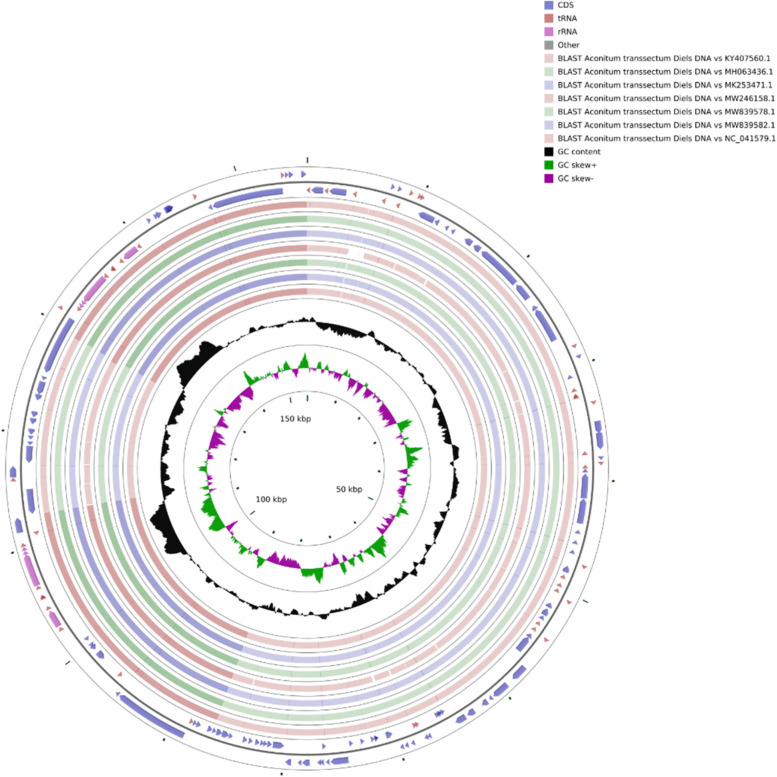
We acquired the cp genome sequences of 7 Ranunculacea species from NCBI to examine the divergence of the A. transsectum cp genome from its related specie, including 6 species of the genus Aconitum (A. flavum, A. pendulum, A. brachypodum, A. vilmorinianum, A. kusnezoffii, A. carmichaelii) and Delphinium yunnanense. The results of CGview analysis shown that the cp genomes of A. transsectum exhibited high similarity to the 6 species of the genus Aconite (Fig. 5). In addition, the cp genomes of A. transsectum also similarity to D. yunnanense, while it also shown some heterogeneity in LSC and SSC region, as shown by the partial deletion of D. yunnanense at 1–20 kbp, 60–90 kbp and 100–120 kbp.
Fig. 5.
Comparative analysis of cp genome structure. The two outermost circles in the figure represent the genome's gene length and orientation; the seven inner circles represent the similarity results compared to other reference genomes; the black circles represent the GC content; green represents GC-skew + and purple represents GC-skew-; and the black circles represent the GC content
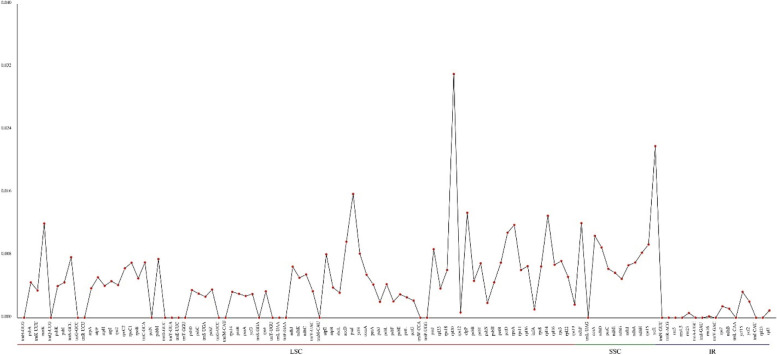
To inspection the degree of variation in DNA sequences, we examined the nucleotide diversity (PI) values of 112 loci in the cp genome, the PI values of cp genomic sequences ranged from 0–0.03106 with a value of 0.00438 on average (Fig. 6 and Supplementary Table S1). The mean PI value of SSC region was 0.00846, the mean PI value of LSC region was 0.00458, and the mean PI value of IR region was 0.00057, which indicated that the SSC region had the highest nucleotide diversity and the IR region had the lowest nucleotide diversity and was more conserved. In addition, 5 genes with high PI values were detected, including rpl20 (0.03106), ycf1 (0.02187), psaI (0.01577), clpP (0.01338) and rpl14 (0.01299), with rpl20, psaI, clpP and rpl14 in the LSC region and ycf1 in the SSC region. These results suggest that the rpl20, ycf1, psaI, clpP, and rpl14 loci were hypervariable loci (PI > 0.012) at the species level, which could also potentially be developed as barcodes for the identification of Aconitum.
Fig. 6.
Comparative analysis of Comparative analysis nucleotide diversity. The gene name is indicated by the horizontal coordinate, the PI value is indicated by the vertical coordinate
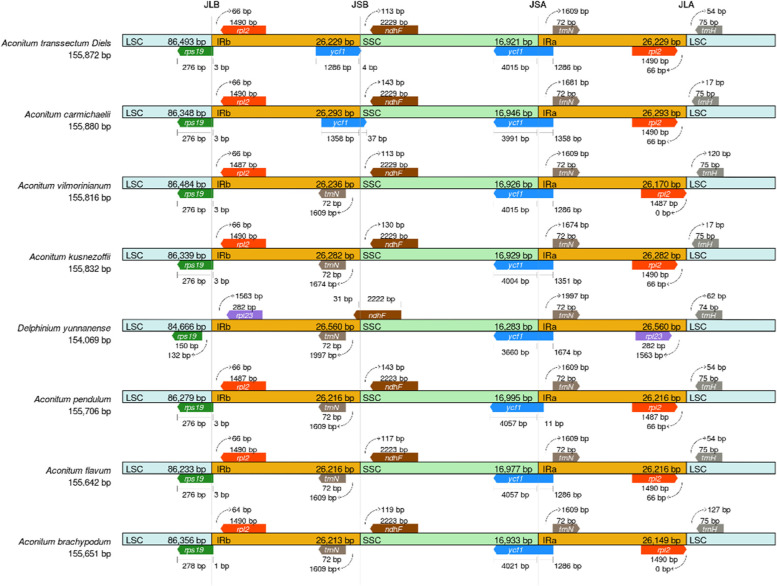
As illustrated in Fig. 7, we also investigated at the binding regions of IR/LSC and IR/SSC. The location of the rps19 gene was similar in all 7 Aconitum species, spanning the LSC and IRb binding regions, and 1–3 bp distant from the LSC and IRb binding regions, with the exception of D. yunnanense, where the rps19 gene was found within the LSC region. The TrnH genes of 7 Aconitum species are in the LSC region, 75 bp distant from the IRa/LSC boundary, and only the TrnH gene of D. yunnanense is 74 bases away from the IRa/LSC boundary. The ndhF genes of 7 Aconitum species are in the SSC region, 113–143 bp distant from the IRa/LSC boundary, and only the ndhF gene of D. yunnanense spanning the IRb and SSC binding regions. In addition, the trnN genes of 8 species are located within the IRa region. The above results demonstrated that the cp genome sequences of the 7 Aconitum are conserved.
Fig. 7.
Comparative analysis of IR Expansion and Contraction
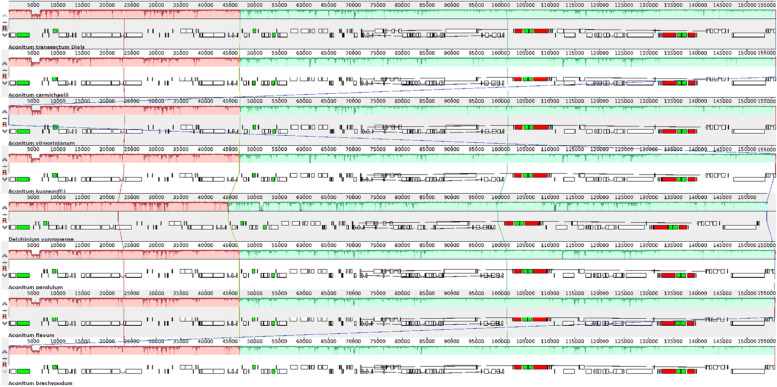
The results of the Mauve multiplex alignment analysis show that there are 3 locally collinear blocks (LCB) between the cp genomes of the eight species, indicating a high degree of similarity between the genomes of these 8 species (Fig. 8). Alignment results have shown no rearrangements or inversions between their genomes; however, mutations were observed in regions 5000 to 10,000, characterized by a high degree of gene sequence variation in aligned cp genomes.
Fig. 8.
MAUVE alignment of A. transsectum related species. As a reference, the cp genome of A. transsectum is presented at the top. The long squares show genomic similarity, while the lines connecting them represent a covariate association. Each genome's gene locations are represented by the short squares. CDS is represented by white, tRNA is represented by green, and rRNA is represented by red
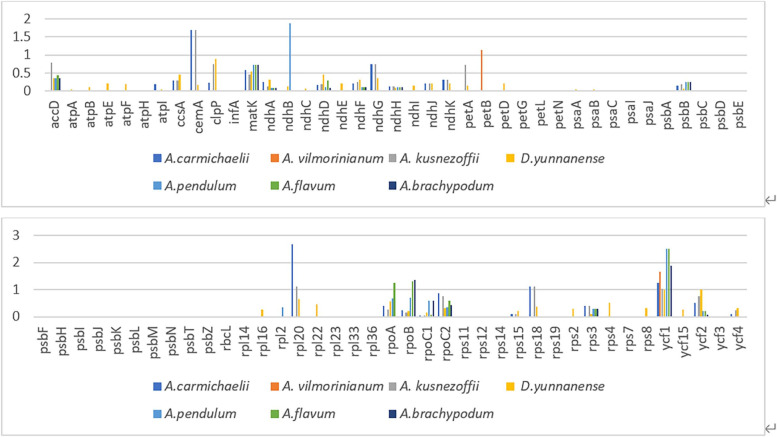
Using A. transsectum as a reference, synonymous and nonsynonymous changes in the cp genomes of A. transsectum were compared with those of 6 species of the genus Aconitum and one species of the genus Delphinium were investigated (Fig. 9). The Ka/Ks ratios of 78 protein-coding genes in these 7 cp genomes were found by comparison. The Ka/Ks ratios for the majority of the coding genes were below 1 or could not be determined since either the Ka or Ks values was zero, suggested that they were conserved. The ycf1 gene had Ka/Ks values greater than 1 in all seven species; the rpl20, cemA, and rps18 genes had Ka/Ks values greater than 1 in A. carmichaelii and A. kusnezoffii; and the rpoB gene had Ka/Ks values greater than 1 in A. flavum and A. brachypodum.
Fig. 9.
The Ka/Ks analysis was performed on 78 protein-coding genes from the A.transsectum cp genome and seven related species
Phylogenetic inference
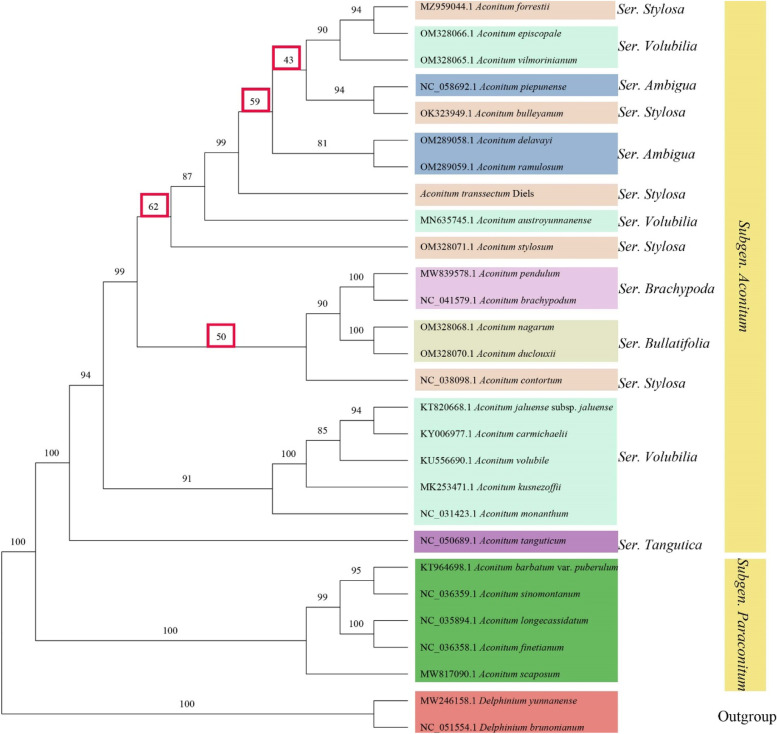
Plant phylogenetic studies frequently use cp genomes. The results of comparative analysis of the cp genomes of A. transsectum and its related species suggested that ycf1 has potential as a chloroplast DNA barcode for the genus Aconitum. Therefore, we utilized the maximum likelihood (ML) method to build a phylogenetic tree of ycf1 gene from 28 species (including 26 species of the genus Aconitum and 2 species of the genus Delphinium) to Whether ycf1 can be used for phylogenetic analysis within the genus Aconitum (Fig. 10). The phylogenetic tree comprised 26 nodes, and the support rate of most nodes was greater than 81 percent (4 nodes was less than 81 percent). The phylogenetic tree separates the subgenera Delphinium, Paraconitum and Aconitum with a very high support rate. However, the branching support rate is low and the classification is confusing when further delineating the subgenus Aconitum.
Fig. 10.
ML phylogenetic tree of 28 species of Ranunculaceae plants constructed with ycf1 gene sequences
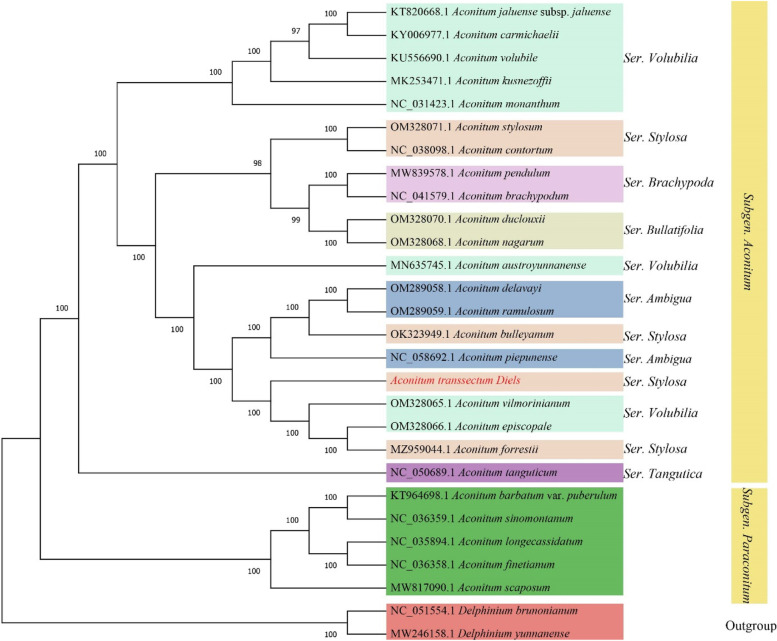
After that, we utilized the ML method to build a phylogenetic tree of cp sequences from 28 species (including 26 species of the genus Aconitum and 2 species of the genus Delphinium) to identify A. transsectum's phylogenetic position (Fig. 11). The phylogenetic tree comprised 25 nodes, and the support rate of all nodes was greater than 97 percent, with 22 nodes having a support rate of 100 percent, indicating that the clustering results were highly reliable. The 28 species might be grouped into three primary taxa on the evolutionary tree. The 21 species of Subgen. Aconitum was clustered into one major taxon, the 5 species of Subgen. paraconitum were grouped into another major taxon, and the 2 species of the outgroup genus Delphinium were grouped into one taxon. A. transsectum is located in the subgenus Aconitum, and is most closely related to A. vilmorinianum, A. episcopale and A. forrestii of Subgen. Aconitum. These results suggest that A. transsectum is highly homologous with Subgen. Aconitum.
Fig. 11.
ML phylogenetic tree of 28 species of Ranunculaceae plants constructed with cp genome sequences
Discussion
Using Illumina sequence data, we were successful in establishing the complete cp genome sequence of A. transsectum in this study. The cp genome of A. transsectum, like most land plant cp genomes, has a highly conserved structure and gene content. The size of the A. transsectum cp genome is 155872 bp, which is consistent with other members of Aconitum (150–157 kb) [15–19]. The A. transsectum cp genome is a typical tetrad structure, with four segments (LSC, SSC, and two IR) and highly conserved IR regions. A. transsectum is native to northwestern Yunnan and its cp genome size is similar to that of other Aconitum species [17], however, the SSC region is longer compared to other species.
Nucleotide diversity (PI) is an indicator that responds to the degree of variation in DNA sequences, and nucleotide diversity responds to the genetic diversity of the species [20]. In chloroplast genes of A. transsectum and related species, higher PI values for gene sequences in the LSC/SSC region were observed than in the IR region, which is coherent with other angiosperms [21, 22].
The expansion and contraction of the IR region of the cp genome is a common evolutionary phenomenon [23]. As the genome evolves, there is expansion and contraction of the IR region, when some genes enter the IR region or the LSC and SSC regions [23]. rps19 genes have a tendency to enter the IR region due to the expansion of the LSC/IR boundary in the genus Aconitum, while in D. yunnanense, the LSC-IR boundary is contracted and the rps19 genes are placed in the LSC region. The above findings indicated that the cp genome boundary genes of the genus Aconitum are different from D. yunnanense, and those of other genera in the Ranunculaceae family [17, 24].
Repeats and SSRs are widely present in plant cp genomes [25]. Repeats vary in type, number, and location from species to species, and they are used to identify mutational hotspots and phylogenetic links [26]. In this study, we found that A. transsectum has 46 repetitive sequences, the number of which is much higher than that of other species in the genus Aconitum. Furthermore, the majority of the repeats are found in genes, indicating that the A. transsectum cp genome preserves a lot of genetic information. SSR has been frequently utilized to determine phylogenetic relationships, genetic diversity research, and species identification due to their high variability and recessive inheritance [27]. The distribution characteristics of cp SSRs in Cyatheaceae have been shown to be useful for classification among genera [28]. The A. transsectum cp genome contained 241 SSRs, the majority of which were identified in the LSC region, which is consistent with observations of cp SSRs in other Aconitum species. Further analysis is needed in the future to see whether repeats and SSRs can be used for phylogenetic analysis of the genus Aconitum.
A synonymous mutation occurs when a base mutation results in an unmodified amino acid; otherwise, it is a nonsynonymous mutation, and nonsynonymous mutations are frequently affected by natural selection [29]. The counts of nonsynonymous substitutions at each nonsynonymous locus (Ka) and synonymous substitutions at each synonymous locus (Ks) are usually used to indicate the selection effect of a gene (Ks) [30]. When Ka/Ks is more than 1, a positive selection impact is present, and when Ka/Ks is less than 1, a purification selection effect is there [30]. The Ka/Ks of most genes (69 out of 78) were smaller than one in the comparison between A. transsectum and the other seven species, indicating that purification selection is essential in these species. In all species, however, the Ka/Ks of ycf1 genes were larger than 1, implying that ycf1 genes were positively selected to adapt to the living environment. The ycf1 gene, the largest chloroplast gene, encodes an ATP-binding cassette (ABC) protein in chloroplasts and generally evolves at a rapid mutation rate [31], as formalized in our study. ycf1 is the most potential chloroplast DNA barcode for land plants since it is very species-specific [32], the ycf1 phylogenetic analysis also shown that ycf1 has taxonomic potential at the subgenus level within the genus Aconitum.
Based on the cp genomes of 28 species, an ML phylogenetic tree was created. A. transsectum and other species of the subgenus Aconitum, such as A. vilmorinianum, A. episcopale and A. forrestii, constitute a monophyletic branch of the genus Aconitums. Phylogeographic results based on morphological features, nuclear DNA markers, and some cp genomes are congruent with our phylogenetic conclusions. Based on ITS sequences, a phylogenetic tree for 51 species of the genus Aconitum, including A. transsectum, A. vilmorinianum, A. episcopale, and A. forrestii, was previously created [33]. The phylogeny of this ITS sequence showed that A. transsectum, A. vilmorinianum, A. episcopale, and A. forrestii are in the same clade and belong to the same subgen. Aconitum. Another phylogenetic tree built on 27 cp genomes of Aconitums species demonstrates that A. vilmorinianum, A. episcopale, and A. forrestii are all members of the same clade, belonging to the subgen. Aconitum [34]. These results clearly reflect the phylogenetic relationships of A. transsectum within the genus Aconitum and provide reliable evidence for the phylogeny and molecular identification of this traditional medicinal plant.
Conclusions
In conclusion, the complete cp genome sequence of A. transsectum was sequenced and compared to that of other closely related species, providing a crucial reference for A. transsectum phylogeny. Although the cp genomes of A. transsectum and other Aconitum are essentially identical in terms of genome structure, gene content, and gene sequence, the IR region boundary section differs. Because it is exceedingly species-specific, ycf1 is the most promising chloroplast DNA barcode for land plants, and it will give informative markers for phylogenetic research of Aconitum. A close relationship has been discovered between A. transsectum and A. vilmorinianum, A. episcopale, and A. forrestii, according to phylogenetic research. The findings of this study not only contribute to the creation and utilization of A. transsectum, but also serve as a source of reference data for population genomics, phylogenetic analysis, and genetic engineering research.
Methods
Ethical statement
For the collection of samples for this study, no special licenses were needed. The relevant Chinese laws were followed as this research was conducted.
Preparation of materials
The plants were harvested from Machang village, Ludian Town, Yulong County, Lijiang City, and identified as A. transsectum by Prof. Su Tai of Yunnan Institute of Materia Medica. For sequencing, fresh young A. transsectum leaves were submitted to Genepioneer Biotechnologies in South China.
DNA extraction, genome sequencing, and annotation
The Cetyl Trimethyl Ammonium Bromide (CTAB) method was used to extract whole genomic DNA from 100 mg of fresh leaves. Paired-end (PE) sequencing with the Illumina NovaSeq 6000 platform, with 150 sequencing read lengths. To screen the raw data and obtain Clean Data of high quality, use the software fastp v 0.20.0. With k-mers of 55, 87, and 121, the cp genome of A. transsectum was assembled using SPAdes v3.10.1 [35]. Quality control was performed after assembly using the sequence of A. piepunense (accession number NC 058,692.1) [36]. To improve annotation accuracy, we used Prodigal v2.6.3 for cp-coding sequences (CDS), Hmmer v3.1b2 for ribosomal RNA (rRNA) prediction, and Aragorn v1.2.3 for transfer RNA prediction (tRNA). The assembled sequences were then checked using BLAST v2.6 to produce the second annotation results, which were based on sequences of related species that had been published in NCBI. To achieve the final annotation, the two annotation results were manually verified to remove any incorrect or redundant annotations and to establish the exon boundaries. Finally, using the OGDRAW software, the entire genome was mapped [37].
Codon usage and repeat sequence analysis
Due to codon simplicity, each amino acid has a minimum of 1 codon and a maximum of 6 codons. The genomic codon usage rate differs widely from species to species and organism to organism. Relative Synonymous Codon Usage refers to the inequality in the utilization of synonymous codons (RSCU). This preference is thought to be the outcome of a combination of natural selection, species mutation, and genetic drift. It is computed by dividing the actual codon usage frequency by the theoretical codon usage frequency. The unique CDSs (one copy of the CDS with numerous copies) were filtered using Perl scripts based on the CDSs of the 86 protein-coding genes, and the RSCU of each codon was computed using the software CodonW v1.4.2 [38].
Comparison of complete Cp genome
The cp genomes of 6 reported Aconitum species and 1 exogenous species were loaded from the NCBI website, which are, A. flavum (MW839582.1), A. pendulum (MW839578.1), A. brachypodum (NC_041579.1), A. vilmorinianum (MH063436.1), A. kusnezoffii (MK253471.1), A. carmichaelii (KY407560.1), and D. yunnanense (MW246158.1). CGView software was used to evaluate the cp genome structures of the eight plants [39]. Mauve v2.3.1 was used to analyze at the homology and covariance of cp sequences [40]. For broad comparison of homologous gene sequences from different plants, the MAFFT v7.310 (automatic mode) [41] was employed. Nucleotide diversity (PI) values for each gene were calculated using DNAsp v5.0 [42]. The IR, SSC, and LSC region boundary information was visualized using the SVG package in Perl. MAFFT v7.310 software was used to compare gene sequences, and Ka/Ks Calculator v2.0 software was utilized to calculate the Ka/Ks values of the genes.
Phylogenetic evaluation
Additional cp genome sequences and ycf1 gene sequences were obtained from the NCBI website for 25 Aconitum species and 2 Delphinium specie, which are, A. pendulum (MW839578.1), A. brachypodum (NC_041579.1), A. vilmorinianum (OM328065.1), A. kusnezoffii (MK253471.1), A. carmichaelii (KY006977.1), A. longecassidatum (NC_035894.1), A. piepunense (NC_058692.1), A. scaposum (MW817090.1), A. bulleyanum (OK323949.1), A. austroyunnanense (MN635745.1), A. tanguticum (NC_050689.1), A. episcopale (OM328066.1), A. delavayi (OM289058.1), A. contortum (NC_038098.1), A. sinomontanum (NC_036359.1), A. finetianum (NC_036358.1), A. volubile (KU556690.1), A. barbatum var. puberulum (KT964698.1), A. monanthum (NC_031423.1), A. jaluense subsp. jaluense (KT820668.1), A. stylosum (OM328071.1), A. duclouxii (OM328070.1), A. nagarum (OM328068.1), A. ramulosum (OM289059.1), A. forrestii (MZ959044.1), D. yunnanense (MW246158.1), and D. brunonianum (NC_051554.1). The sequence alignment was conducted by MAFFT [43] based on the cp genome sequences of 28 species, including A. transsectum; the alignment results were further optimized by trimAl software [44]. The maximum likelihood (ML) phylogenetic tree was constructed with IQ-TREE 1.6.12 [45] using D. yunnanense and D. brunonianum as outgroups, with Bootstrap value set to 1000, and the best tree building module was selected by the built-in Model Finder of IQ-TREE based on the optimized alignment result.
Supplementary Information
Acknowledgements
The authors sincerely thank Prof. Su Tai for his help in the identification of A. transsectum.
Abbreviations
- A.transsectum
Aconitum transsectum Diels
- cp
Chloroplast
- LSC
Large single copy
- SSC
Small single copy
- IR
Inverted repeat
- CUB
Codon usage bias
- RSCU
Relative synonymous codon usage
- Met
Methionine
- Arg
Arginine
- PolyA
Polyadenine
- PolyT
Polythymine
- LCB
Locally collinear blocks
- ML
Maximum likelihood
- PI
Nucleotide diversity
- Ka
Nonsynonymous locus
- Ks
Synonymous locus
- ABC
ATP-binding cassette
- CTAB
Cetyl Trimethyl Ammonium Bromide
- PE
Paired-end
- CDS
Cp-coding sequences
- rRNA
Ribosomal RNA
- tRNA
Transfer RNA
- ML
Maximum likelihood
Authors’ contributions
W.H. designed and directed this study. Y.Z. revised the paper. N.F. data analysis and·writing the original draft preparation. S.T. and N.F. Collect research materials. D.J. conduct experiments. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Major science and technology project-Biomedicine Major special project of Yunnan province, China (2018ZF011), Yunnan Province Science and Technology Department (202101BD070001-014) and the Key Laboratory of State Forestry and Grassland Administration on Highly-Efficient Utilization of Forestry Biomass Resources in Southwest China (2020-KF14).
Availability of data and materials
The datasets generated and analyzed in this study are available in the GenBank of NCBI, and the complete chloroplast genome sequence of Aconitum transsectum is deposited in GenBank of NCBI under accession number ON751949.1. The accession numbers for the remaining datasets used and analyzed in this study are listed in the Methods section.
Declarations
Ethics approval and consent to participate
For the collection of samples for this study, no special licenses were needed. The relevant Chinese laws were followed as this research was conducted.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Guan K, Xiao P, Pan K: Flora Reipublicae Popularis Sinicae: Ranunculaceae, vol. 27; 1979.
- 2.Been A. Aconitum: Genus of powerful and sensational plants. Pharm Hist. 1992;34(1):35–39. [PubMed] [Google Scholar]
- 3.Li H, Liu L, Zhu S, Liu Q. Case reports of aconite poisoning in mainland China from 2004 to 2015: a retrospective analysis. J Forensic Leg Med. 2016;42:68–73. doi: 10.1016/j.jflm.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 4.Li J. Flora of China. Harv Pap Bot. 2007;13(2):301–302. doi: 10.3100/1043-4534-13.2.301. [DOI] [Google Scholar]
- 5.He J, Wong K-L, Shaw P-C, Wang H, Li D-Z. Identification of the medicinal plants in Aconitum L. by DNA barcoding technique. Planta Med. 2010;76(14):1622–1628. doi: 10.1055/s-0029-1240967. [DOI] [PubMed] [Google Scholar]
- 6.Tonti‐Filippini J, Nevill PG, Dixon K, Small I: What can we do with 1000 plastid genomes? In., vol. 90: Wiley Online Library; 2017: 808–818. [DOI] [PubMed]
- 7.Kirchhoff H. Chloroplast ultrastructure in plants. New Phytol. 2019;223(2):565–574. doi: 10.1111/nph.15730. [DOI] [PubMed] [Google Scholar]
- 8.Bansal KC, Saha D. Chloroplast genomics and genetic engineering for crop improvement. Agricultural Research. 2012;1(1):53–66. doi: 10.1007/s40003-011-0010-6. [DOI] [Google Scholar]
- 9.Maier RM, Neckermann K, Igloi GL, Kössel H. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol. 1995;251(5):614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- 10.Daniell H, Lin C-S, Yu M, Chang W-J. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17(1):1–29. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci. 2007;104(49):19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui Y, Chen X, Nie L, Sun W, Hu H, Lin Y, Li H, Zheng X, Song J, Yao H. Comparison and phylogenetic analysis of chloroplast genomes of three medicinal and edible Amomum species. Int J Mol Sci. 2019;20(16):4040. doi: 10.3390/ijms20164040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian W, Yang JR, Pearson NM, Maclean C, Zhang J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8(3):e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parvathy ST, Udayasuriyan V, Bhadana V. Codon usage bias. Mol Biol Rep. 2022;49(1):539–565. doi: 10.1007/s11033-021-06749-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park I, Kim WJ, Yang S, Yeo SM, Li H, Moon BC. The complete chloroplast genome sequence of Aconitum coreanum and Aconitum carmichaelii and comparative analysis with other Aconitum species. PLoS ONE. 2017;12(9):e0184257. doi: 10.1371/journal.pone.0184257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park I, Yang S, Choi G, Kim WJ, Moon BC. The complete chloroplast genome sequences of aconitum pseudolaeve and aconitum longecassidatum, and development of molecular markers for distinguishing species in the aconitum subgenus lycoctonum. Molecules. 2017;22(11):2012. doi: 10.3390/molecules22112012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng J, Li X, Li H, Yang J, Wang H, He J. Comparative analysis of the complete chloroplast genomes of four aconitum medicinal species. Molecules. 2018;23(5):1015. doi: 10.3390/molecules23051015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong H, Liu W, Yao G, Gong W. A comparison of chloroplast genome sequences in Aconitum (Ranunculaceae): a traditional herbal medicinal genus. PeerJ. 2017;5:e4018. doi: 10.7717/peerj.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Li Q, Li Y, Qian J, Han J. Chloroplast genome of Aconitum barbatum var. puberulum (Ranunculaceae) derived from CCS reads using the PacBio RS platform. Front Plant Sci. 2015;6:42. doi: 10.3389/fpls.2015.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT, Coleman-Derr D, Conley EJ, Crossman CC, Deal KR, et al. Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genomics. 2010;11:702. doi: 10.1186/1471-2164-11-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Liu J, Su Y, Li M, Xie X, Su J. Complete chloroplast genome sequence of sonchus brachyotus helps to elucidate evolutionary relationships with related species of asteraceae. Biomed Res Int. 2021;2021:9410496. doi: 10.1155/2021/9410496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Song MF, Li Y, Sun HF, Tang DY, Xu AS, Yin CY, Zhang ZL, Zhang LX. Complete chloroplast genome analysis of two important medicinal alpinia species: alpinia galanga and alpinia kwangsiensis. Front Plant Sci. 2021;12:705892. doi: 10.3389/fpls.2021.705892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.H Hui Chao, Shi, Yuan, Liu, Shu-Yan, Mao, Li-Zhi, Gao: Thirteen camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships BMC Evol Biol 2014 ;14 1:151. [DOI] [PMC free article] [PubMed]
- 24.Li QJ, Su N, Zhang L, Tong RC, Zhang XH, Wang JR, Chang ZY, Zhao L, Potter D. Chloroplast genomes elucidate diversity, phylogeny, and taxonomy of Pulsatilla (Ranunculaceae) Sci Rep. 2020;10(1):19781. doi: 10.1038/s41598-020-76699-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin Z, Wang Y, Wang Q, Li A, Hou F, Zhang L. Evolution analysis of simple sequence repeats in plant genome. PLoS ONE. 2015;10(12):e0144108. doi: 10.1371/journal.pone.0144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powell W, Morgante M, McDevitt R, Vendramin GG, Rafalski JA. Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc Natl Acad Sci U S A. 1995;92(17):7759–7763. doi: 10.1073/pnas.92.17.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ping J, Feng P, Li J, Zhang R, Su Y, Wang T. Molecular evolution and SSRs analysis based on the chloroplast genome of Callitropsis funebris. Ecol Evol. 2021;11(9):4786–4802. doi: 10.1002/ece3.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu M, Feng P, Ping J, Li J, Su Y, Wang T. Phylogenetic significance of the characteristics of simple sequence repeats at the genus level based on the complete chloroplast genome sequences of Cyatheaceae. Ecol Evol. 2021;11(20):14327–14340. doi: 10.1002/ece3.8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmueller KE, Albrechtsen A, Li Y, Kim SY, Korneliussen T, Vinckenbosch N, Tian G, Huerta-Sanchez E, Feder AF, Grarup N. Natural selection affects multiple aspects of genetic variation at putatively neutral sites across the human genome. PLoS Genet. 2011;7(10):e1002326. doi: 10.1371/journal.pgen.1002326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nekrutenko A, Makova KD, Li W-H. The KA/KS ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res. 2002;12(1):198–202. doi: 10.1101/gr.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li ZS, Szczypka M, Lu YP, Thiele DJ, Rea PA. The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J Biol Chem. 1996;271(11):6509–6517. doi: 10.1074/jbc.271.11.6509. [DOI] [PubMed] [Google Scholar]
- 32.Dong W, Liu J, Yu J, Wang L, Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE. 2012;7(4):e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo Y. Zhang F-m, Yang Q-E: Phylogeny of Aconitum subgenus Aconitum (Ranunculaceae) inferred from ITS sequences. Plant Syst Evol. 2005;252(1):11–25. doi: 10.1007/s00606-004-0257-5. [DOI] [Google Scholar]
- 34.Ni X, Li J, Li Y, Zhang H, Duan B, Chen X, Xia C. The complete chloroplast genome of Aconitum piepunense (Ranunculaceae) and its phylogenetic analysis. Mitochondrial DNA B Resour. 2022;7(1):115–117. doi: 10.1080/23802359.2021.2011448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Takayama K, Youn J-S, Pak J-H, Kim S-C. Plastome characterization and phylogenomics of East Asian beeches with a special emphasis on Fagus multinervis on Ulleung Island, Korea. Genes. 2020;11(11):1338. doi: 10.3390/genes11111338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20(17):3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 38.Wong EH, Smith DK, Rabadan R, Peiris M, Poon LL. Codon usage bias and the evolution of influenza A viruses. codon usage biases of influenza virus. BMC Evol Biol. 2010;10(1):1–14. doi: 10.1186/1471-2148-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stothard P, Grant JR, Van Domselaar G. Visualizing and comparing circular genomes using the CGView family of tools. Brief Bioinform. 2019;20(4):1576–1582. doi: 10.1093/bib/bbx081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 43.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20(4):1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed in this study are available in the GenBank of NCBI, and the complete chloroplast genome sequence of Aconitum transsectum is deposited in GenBank of NCBI under accession number ON751949.1. The accession numbers for the remaining datasets used and analyzed in this study are listed in the Methods section.