Abstract
Objectives
V600E-BRAF kinase is an essential therapeutic target in melanoma and other types of tumors. Because of its resistance to known inhibitors and the adverse effects of some identified inhibitors, investigation of new potent inhibitors is necessary.
Methods
In the present work, in silico strategies such as molecular docking simulation, pharmacokinetic evaluation, and density functional theory (DFT) computations were used to identify potential V600E-BRAF inhibitors from a set of 72 anticancer compounds in the PubChem database.
Results
Five top-ranked molecules (12, 15, 30, 31, and 35) with excellent docking scores (MolDock score ≥90 kcal mol−1, Rerank score ≥60 kcal mol−1) were selected. Several potential binding interactions were discovered between the molecules and V600E-BRAF. The formation of H-bonds and hydrophobic interactions with essential residues of V600E-BRAF suggested the high stability of these complexes. The selected compounds had excellent pharmacological properties according to the drug likeness rules (bioavailability) and pharmacokinetic properties. Similarly, the energy for the frontier molecular orbitals, such as the HOMO, LUMO, energy gap, and other reactivity parameters, was computed with DFT. The frontier molecular orbital surfaces and electrostatic potentials were investigated to demonstrate the charge-density distributions potentially associated with anticancer activity.
Conclusion
The identified compounds were found to be potent hit compounds for V600E-BRAF inhibition with superior pharmacokinetic properties; therefore, they may be promising cancer drug candidates.
Keywords: ADMET, DFT, Drug likeness, Molecular docking, V600E-BRAF
المخلص
أهداف البحث
كيناز "ب.ر.أ. ف-في. 600.إي". هو هدف علاجي أساسي في سرطان الجلد وأنواع أخرى من الأورام. تستلزم مقاومته للمثبطات والآثار الجانبية المعروفة لبعض المثبطات المحددة الاستقصاء عن مثبطات جديدة وفعالة.
طرق البحث
في العمل الحالي، تم استخدام استراتيجيات في السيليكون مثل محاكاة الالتحام الجزيئي وتقييم الحرائك الدوائية وحسابات نظرية الكثافة الوظيفية لتحديد مثبطات "ب.ر.أ. ف-في. 600.إي" المحتملة من مجموعة من 72 مركبا مضادا للسرطان من قاعدة بيانات "بوبكيم".
النتائج
ما مجموعه خمسة جزيئات من الدرجة الأولى (12 ، 15 ، 30 ، 31 ، 35) مع درجات رائعة في الالتحام (درجة مولدوك:> 90 كيلو كالوري مول -1 ، ونقاط إعادة التصنيف:> 60 كيلو كالوري مول -1) تم اختيارها. تم اكتشاف العديد من تفاعلات الارتباط المحتملة بين الجزيئات المقترحة و "ب.ر.أ. ف-في. 600.إي". أثبت ظهور الروابط الهيدروجينية والتفاعلات الكارهة للماء مع المخلفات الأساسية لـ "ب.ر.أ. ف-في. 600.إي" الثبات العالي لهذه المجمعات. أظهرت المركبات المختارة خصائص دوائية فائقة وفقا لقواعد التشابه الدوائي (التوافر البيولوجي) وخصائص الحرائك الدوائية. وبالمثل، تم حساب الطاقة الخاصة بالمدارات الجزيئية الحدودية مثل المدار الجزيئي الأعلى المشغول، وأدنى المدار الجزيئي غير المشغول، وفجوة الطاقة، ومعلمات التفاعل الأخرى باستخدام نظرية الكثافة الوظيفية. تم فحص الأسطح المدارية الجزيئية الحدودية والإمكانات الكهروستاتيكية لإثبات توزيعات كثافة الشحنة التي قد تكون مرتبطة بالنشاط المضاد للسرطان.
الاستنتاجات
وبالتالي، تم التعرف على المركبات المختارة كضربات قوية لـ "ب.ر.أ. ف-في. 600.إي". بخصائص حركية دوائية فائقة، ويمكن اقتراحها كعقاقير مرشحة واعدة للسرطان.
الكلمات المفتاحية: ب.ر.أ. ف-في. 600.إي؛ الالتحام الجزيئي؛ شبيه-الدواء؛ نظرية الكثافة الوظيفية؛ امتصاص وتوزيع وتمثيل الدواء وإفرازه وسميته.
Introduction
MAPK signaling is a key regulator of fundamental cellular processes such as growth, proliferation, differentiation, migration, and apoptosis.1,2 Signal transmission is enabled by intracellular protein kinase phosphorylation cascades (e.g., MAPK, MKKK, and MKK).3 The abnormal regulation of MAPK cascades is associated with the onset of a variety of severe diseases, including Parkinson's disease, Alzheimer's disease, amyotrophic lateral sclerosis, and multiple types of cancer.4,5 Disruption of the Ras-Raf-MEKERK signaling pathway has been associated with human tumorigenesis.6 Raf kinases (A, B, and CRAF) play important roles in the ERK signaling branch of the MAPK cascade.7 RAF kinases bind RAS, the upstream activator, and mediate MAPK signaling transduction to MEK, thus activating downstream MEK1/2 and ERK1/2.8,9 BRAF has the highest basal activity among the three RAF kinases and is a key activator of MEK1/2. In comparison to A and CRAF,10,11 pathological mutations in RAF kinases typically affect the BRAF subtype.12 The most common mutation is V600E-BRAF, which occurs when the valine residue at position 600 is replaced with a glutamate residue.13 Because this mutation mimics the active phosphorylation state, the V600E-BRAF protein continues to activate downstream pathways without being regulated.14 This mutation is present in 8% of all cancers, such as colorectal cancer (10%), melanoma (60%), and thyroid cancer (30–70%).15,16 Thus, V600E-BRAF kinase is considered an important target for managing and treating cancer.17,18
Dabrafenib and vemurafenib are both selective inhibitors of V600E-BRAF; they induce automatic cell death in melanoma and have shown high effectiveness against melanoma cells. They have been endorsed by the US Food and Drug Administration for late-stage melanoma therapy.19,20 A single treatment with a V600E-BRAF inhibitor considerably improves patient lifestyle and survival rate. Unfortunately, despite the success of approved V600E-BRAF inhibitors, resistance to these selective inhibitors emerges after 5–8 months.21,22 The resistance to selective V600E-BRAF inhibitors makes the discovery and validation of novel candidates critical for the development of new treatments for V600E-BRAF cancers. Moreover, current understanding of tumor heterogeneity and the evolution of resistance suggests that the development of novel anticancer agents is critical. In drug discovery, the identification and confirmation of lead compounds and the evaluation of active binding sites of bioactive targets linked with specific lead compounds is performed through wet-laboratory investigations, which are costly and time-consuming.23 In silico strategies can effectively decrease the time needed to acquire valuable drugs and the accompanying financial costs, thus enabling new potent drugs to be identified with lower costs and selective targeting.24 Molecular docking is an excellent in silico approach for filtering large chemical libraries to detect prospective chemicals that may be used to determine the binding ability for a certain target. Over the past two decades, molecular docking has evolved as a model for structure-based virtual screening of several chemical databases.25 It is extensively used to select the most suitable alignment of a drug candidate in the active site of a protein, and to predict target affinity. Previously, comprehensive docking investigations were performed to study the biological activity of numerous chemical structures.25, 26, 27 In silico drug-like and pharmacokinetic analyses are additional (virtual) screening methods for the approval of compounds that might demonstrate physiological and drug-like ability. The procedures used to assess drug likeness and pharmacokinetic properties are based on a combination of the experimental findings reported in several drug databases.28 Density functional theory (DFT) is an extensive technique with lower computational costs than those of several other approaches. DFT computations currently produce the most reliable and accurate outcomes for various chemical systems, providing results well matched to experimental findings.29 In this investigation, a systematic computational investigation of a set of 72 anticancer compounds from the PubChem database was conducted with molecular docking simulation, DFT computations, and pharmacokinetic property prediction to explore their potential to inhibit the V600E-BRAF kinase. The objective of the work was to assess the anticancer potential of these chemical structures as likely drug candidates with desirable properties.
Materials and Methods
Retrieval of compounds and optimization
A series of 72 anticancer compounds was retrieved from the PubChem database (https://pubch em.ncbi.nlm.nih.gov). Structures of ligands were drawn in ChemDraw (Table S1), and minimization of energy was achieved with the MM2 forcefield in Spartan 14 to assist the docking program in detecting the bioactive conformer from the local minima. Optimization of the compounds was achieved with the DFT/B3LYP approach and 6–31G∗ basis set.
Docking preparation and simulation
The Protein Data Bank (http://www.rcsb.org/) was used to obtain the crystal structure of V600E-BRAF (PDB code: 3OG7)12,30,31 and the native ligand (vemurafenib). The target was prepared by extracting water molecules and detaching vemurafenib from the protein–ligand complex. The native ligand was re-docked to the target to validate the molecular docking with Molegro Virtual-Docker (MVD) 6.0.32 The active site of the target was determined with a cavity detection package in MVD 6.0. The binding cavity of X: 1.59, Y: −1.28, Z: −6.2, and r: 28 Å was set with 0.30 Å resolution.25,33 The prepared structures with vemurafenib were imported into MVD, and 1500 iterations were set for the docking algorithm. The docking simulation was run at least 50 times for each of the 10 poses, and the best poses were selected on the basis of predefined scoring functions.34 Inter-molecular interactions of the selected poses were visualized with Discovery-Studio.
Drug-like and pharmacokinetic biochemical evaluation
Drug-like behavior of the selected compounds was investigated with SwissADME (www.swissadme.ch/), an online server, and pharmacokinetic parameters were examined with pkCSM (https://biosig.unimelb.edu.au/pkcsm//).
DFT computation
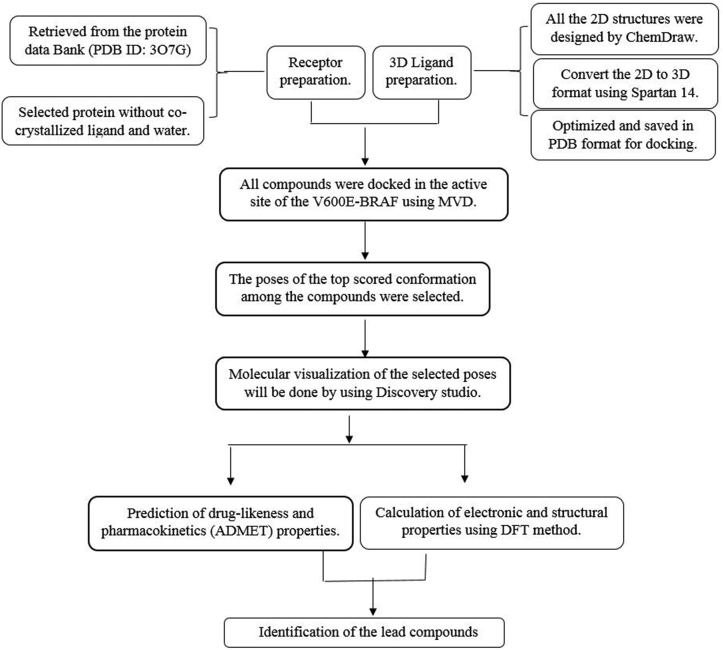
The structural and electronic properties of the four best compounds selected from the docking analysis were computed with DFT/B3LYP and the 6–31G∗ basis set in Spartan 14. The parameters calculated in this investigation are the energies of the frontier molecular orbitals (HOMO, LUMO, and energy gap) and other reactivity parameters: chemical hardness (η), softness (σ), electronegativity (χ) chemical potential (μ), and electrophilicity index (ω).35 The electrostatic potential (EP) surfaces for the molecules were achieved from the population-analysis computations and displayed with Spartan 14. Consequently, the importance of ligand/protein interactions in the active site of the target were clarified. The flowchart of the study design for identifying potent V600E-BRAF inhibitors is presented in Figure 1.
Figure 1.
Flowchart of the study design for identifying potent V600E-BRAF inhibitors.
Results
Molecular docking results of the compounds including vemurafenib are shown in Table S1. The superimposed alignments of the re-docked and original co-crystal ligands are presented in Fig. S1. The complete docking results for the best five ligands as well as vemurafenib, and the types of interactions involved in each of the selected complexes, are presented in Table 1, Table 2. Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7 depict the 3D and 2D interaction models of the selected docked ligands at the V600E-BRAF binding site. To further confirm that the selected compounds were potential drugs, we determined their drug likeness and pharmacokinetics. The predicted drug likeness and pharmacokinetic properties are shown in Table 3, Table 4. The optimized geometries and the frontier molecular orbitals (HOMO and LUMO illustrations) of the selected ligands, achieved through DFT computations, are shown in Figure 8, Figure 9. Table 5 shows the quantum descriptors, and Figure 10 depicts the EP surfaces of the studied ligands.
Table 1.
Results of docking for the four best-docked ligands in V600E-BRAF.
| SN | CID | Name | aMolDock score (kcal mol−1) | bRerank score | cE-inter (kcal mol−1) | dE–H-bond (kcal mol−1) |
|---|---|---|---|---|---|---|
| 12 | 72,742 | Dermocybin | −139.140 | −106.737 | −149.917 | −8.202 |
| 15 | 354,395 | 11-Hydroxymethyl-20(RS)-camptothecin | −116.449 | −17.752 | −133.922 | −4.628 |
| 30 | 5,477,796 | Anthra[1,9-cd]pyrazol-6(2H)-one der | −147.599 | −115.756 | −186.185 | −4.770 |
| 31 | 5,351,180 | Cytosine, monohydrochloride | −101.144 | −76.975 | −104.928 | −9.939 |
| 35 | 5,351,321 | Bisantrene hydrochloride | −134.615 | −105.511 | −152.904 | −3.563 |
| Vem. | – | Vemurafenib | −158.139 | −118.607 | −167.952 | −4.741 |
CID, compound identification number; SN, serial number; Vem., vemurafenib.
Table 2.
Interaction types and amino acids involved in each of the selected complexes.
| Complex | H-bond (HB) | Bond length (Å) for HB | C–HB | Alkyl | π–alkyl | π–π | π–cation | π–sulfur | Halo-bond |
|---|---|---|---|---|---|---|---|---|---|
| 12 | CYS532 CYS532 CYS532 GLN530 GLN530 |
2.38906 1.73263 1.92474 1.99728 1.6863 |
VAL471 ALA481 LEU514 CYS532 CYS532 |
TRP531 TRP531 PHE583 |
|||||
| 15 | ASP594 CYS532 GLU533 |
2.34313 2.09834 2.65953 |
ASP594 | VAL471 | TRP531 PHE583 VAL471 ALA481 LYS483 LEU514 ALA481 CYS532 |
TRP531 PHE583 |
CYS532(s) | ||
| 30 | LYS483 ALA481 |
2.48979 2.17008 |
VAL471 ALA481 VAL471 ALA481 CYS532 |
TRP531 PHE583 PHE583 PHE583 |
|||||
| 31 | LYS483 ASP594 ASP594 PHE595 GLY596 THR508 LEU514 THR529 |
2.44902 2.135 2.24205 2.36902 2.30303 2.9056 1.96262 2.6848 |
LYS483 | LEU505 LEU514 |
|||||
| 35 | GLY534 CYS532 |
1.64054 1.99523 |
LYS483 GLY534 ILE527 ALA481 THR529 ALA481 ILE527 |
VAL471 VAL471 ALA481 LEU514 CYS532 |
TRP531 PHE583 PHE583 |
||||
| Vem. | CYS532 ASP594 GLN530 |
1.788 1.980 1.761 |
GLY593 CYS532 THR529 |
LEU505 ILE527 |
VAL471 LYS483 ALA481 LEU514 CYS532 ALA481 CYS532 |
TRP531 PHE583 |
LYS483 | ALA481 |
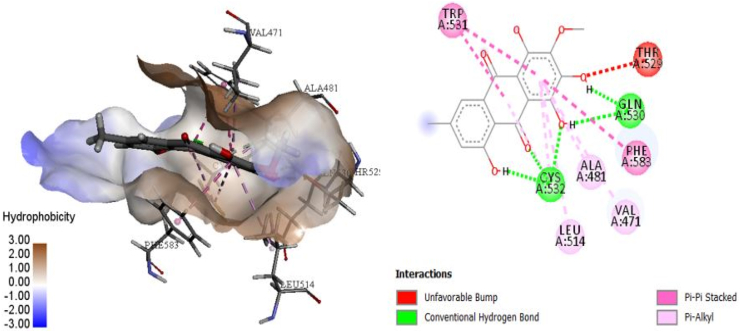
Figure 2.
3D and 2D models for the interaction of complex 12.
Figure 3.
3D and 2D models for the interaction of complex 15.
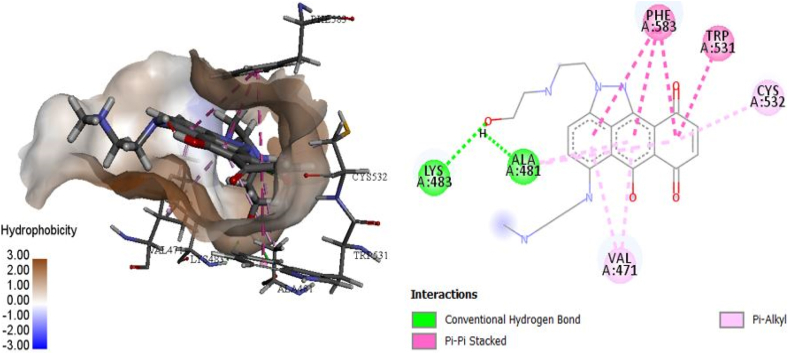
Figure 4.
3D and 2D models for the interaction of complex 30.
Figure 5.
3D and 2D models for the interaction of complex 3.
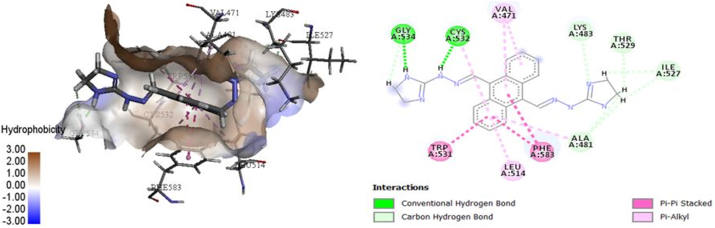
Figure 6.
The 3D and 2D models for the interaction of complex 35.
Figure 7.
The 3D and 2D models for the interaction of the complex with vemurafenib.
Table 3.
Predicted drug likeness parameters of the selected ligands.
| SN | Mol. wt. | HBA | HBD | log P | NRB | TPSA (Å2) | Bioavailability |
|---|---|---|---|---|---|---|---|
| 12 | 316.26 | 7 | 4 | 2.15 | 1 | 124.29 | 0.55 |
| 15 | 378.38 | 6 | 2 | 2.56 | 2 | 101.65 | 0.55 |
| 30 | 453.49 | 7 | 5 | 3.33 | 11 | 137.48 | 0.55 |
| 31 | 243.22 | 6 | 4 | 0.32 | 2 | 130.83 | 0.55 |
| 35 | 398.46 | 4 | 4 | 2.71 | 6 | 97.56 | 0.55 |
| Vem. | 489.92 | 6 | 2 | 4.97 | 7 | 100.30 | 0.55 |
HBA, hydrogen bond acceptors; HBD, hydrogen bond donors; NRB, number of rotatable bond; SN, serial number; Vem., vemurafenib.
Table 4.
Predicted pharmacokinetic properties of the selected ligands.
| Absorption |
Distribution |
Metabolism |
Excretion |
Toxicity |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Substrate |
Inhibitor |
||||||||||||
| CYP |
|||||||||||||
| SN | Intestinal absorption Numeric (% absorbed) | VDss (human) Numeric (log L kg−1) | BBB permeability Numeric (log BB) | CNS permeability Numeric (log PS) | 2D6 | 3A4 | 1A2 | 2C19 (Yes/no) | 2C9 | 2D6 | 3A4 | Total clearance Numeric (log mL min−1 kg−1) | AMES toxicity (Yes/no) |
| 12 | 87.842 | 0.201 | 0.152 | −2.236 | No | No | No | Yes | No | No | No | 0.051 | No |
| 15 | 88.004 | 0.241 | −0.584 | −3.086 | No | Yes | Yes | No | No | No | Yes | 0.607 | No |
| 30 | 55.695 | 1.900 | −1.689 | −3.447 | Yes | Yes | No | No | No | No | Yes | 1.543 | No |
| 31 | 43.155 | −0.025 | −0.97 | −4.023 | No | No | No | No | No | No | No | 0.562 | No |
| 35 | 80.488 | 1.483 | −0.198 | −2.482 | No | Yes | Yes | No | No | Yes | No | 0.496 | Yes |
| Vem. | 98.853 | −0.445 | −1.647 | −3.463 | No | Yes | No | Yes | Yes | No | Yes | 0.132 | No |
BBB, blood–brain barrier; CNS, central nervous system; CYP, cytochrome P; VDss, volume distribution in a stable state.
Figure 8.
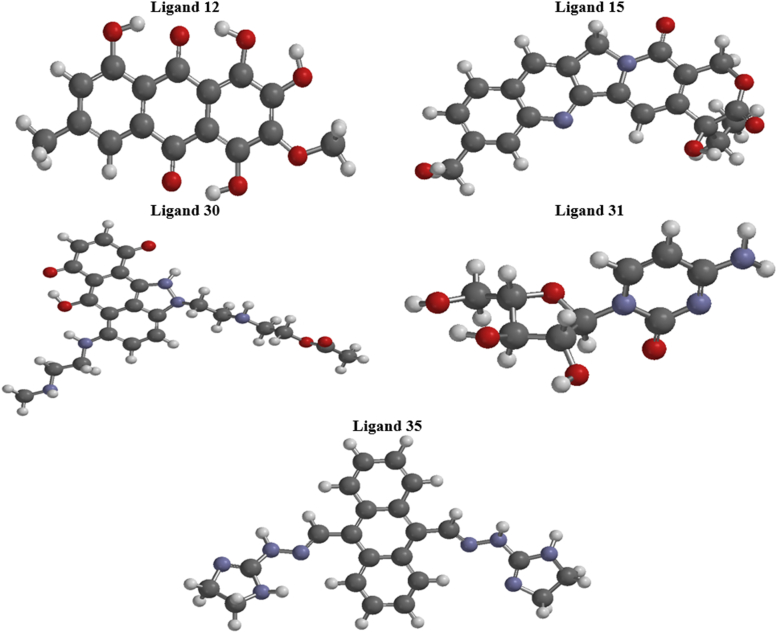
Optimized-geometric structures of the investigated ligands (12, 15, 30, 31, and 35).
Figure 9.
Frontier molecular orbital surfaces of the investigated ligands (12, 15, 30, 31, and 35).
Table 5.
Frontier molecular orbital energies and global reactivity descriptors of the studied ligands.
| S/N | E-HOMO (eV) | E-LUMO (eV) | ΔE | η | σ | χ | Μ | ω |
|---|---|---|---|---|---|---|---|---|
| 12 | −5.85 | −2.90 | 2.95 | 1.48 | 0.68 | 4.38 | −4.38 | 6.49 |
| 15 | −5.97 | −2.29 | 3.68 | 1.84 | 0.54 | 4.13 | −4.13 | 4.64 |
| 30 | −4.54 | −2.56 | 1.98 | 0.99 | 1.01 | 3.55 | −3.55 | 6.36 |
| 31 | −6.02 | −0.79 | 5.23 | 2.62 | 0.38 | 3.41 | −3.41 | 2.22 |
| 35 | −4.94 | −2.15 | 2.79 | 1.40 | 0.72 | 3.55 | −3.55 | 4.50 |
Figure 10.
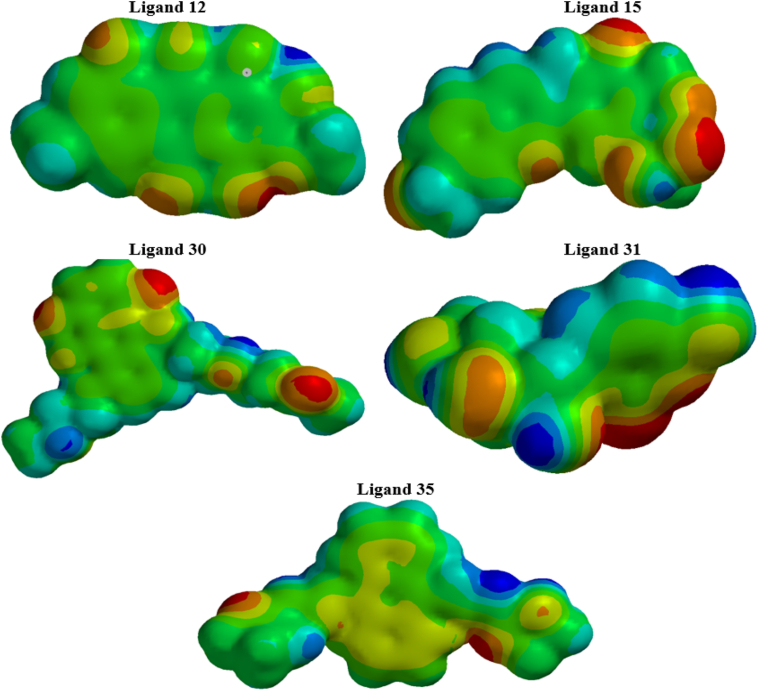
Electrostatic potential of the studied ligands (12, 15, 30, 31, and 35).
Discussion
Potential hit molecules were identified by docking all curated ligands from the PubChem database, including vemurafenib, into the binding pocket of the V600E-BRAF target. Before the docking was performed for the entire data set, the docking method was authenticated through a re-docking technique. Thus, vemurafenib (co-crystallized) was re-docked at the exact site where the co-crystal ligand was initially bound to the V600E-BRAF kinase. Re-docking of vemurafenib at the V600E-BRAF kinase receptor revealed an RMSD value of 1.413 Å, which met the validity standards of RMSD <2.0 Å.36 Hence, the docking procedure with MVD had acceptable precision in repositioning vemurafenib at the V600E-BRAF active sites. The superimposed alignment of the re-docked and actual co-crystal structures is shown in Fig. S1. The coordinates for the active site and the grid box size used in the established re-docking procedure were also used for docking of the studied compounds. The top inhibitors of V600E-BRAF were sorted according to docking score with respect to that of vemurafenib (reference), and the selected ligands were found to efficiently bind the target.
To provide superior performance with outstanding docking scores, we examined the molecular interactions of the five hit inhibitors from the data set. The identification of the main residues in the binding pocket of the V600E-BRAF interacting with the five selected complexes (12, 15, 30, 31, and 35) was achieved with Discovery-Studio Visualizer. The potency of the interaction between the ligands and protein was assessed with the Rerank score. The complete docking results for the best five ligands as well as vemurafenib are presented in Table 1. Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7 show the 3D and 2D interaction models of the selected docked poses in the binding site of V600E-BRAF. Table 2 presents the interaction types involved in each of the selected complexes. The selected ligands formed bonds and non-bonding interactions with the binding pocket of the V600E-BRAF target, as indicated by the inter-energy and H-bonding energy (Table 1). All ligands had a MolDock score <−90 kcalmol-1, thus indicating their potential to bind the protein efficiently.37 The complex structures of the five best poses, on the basis of the docking scores, are discussed in detail below and presented in Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7.
The complex structure of docked compound 12 with the target is shown in Figure 2. A MolDock score of −139.140 kcal mol−1 and Rerank score of −106.737 were determined. The E–H-bond was −8.202 kcal mol−1 (Table 1). The good docking scores suggested the potential for desirable interactions between this compound and the protein. The indicated binding mode in Figure 2 indicated that compound 12 establishes interactions through H-bonding with the backbone of the central amino acid residues CYS532 and GLN530; and shows favorable π–π interactions with the TRP531 and PHE583 residues, which are important for selectivity.38 Additionally, compound 12 forms π–alkyl interactions with VAL471, ALA481, LEU514, and CYS532 (2) at the binding cavity, in a similar pattern to that reported in the literature.25
Figure 3 depicts the binding mode of compound 15 on the V600E-BRAF receptor, with MolDock, and Rerank scores of −123.093 kcal mol−1 and −93.530, respectively, and E–H-bond of −5.170 kcal mol−1 (Table 1). Compound 15 binds the V600E-BRAF binding cavity through three H-bonds with ASP594, CYS532, and GLU533, one C–H-bond with ASP594, and two π–π stacked interactions with TRP531 and PHE583 residues. A π–sigma interaction with CYS532 is also formed. Furthermore, the good docking score of compound 15 might be associated with other weak interactions: alkyl with VAL471, and π–alkyl interactions with the TRP531, PHE583, VAL471, ALA481, LYS483, LEU514, ALA481, and CYS532 residues. The 3D protein surface in Figure 3 suggested that compound 15 has a high affinity toward the target.
The docked structure of compound 30 with the receptor (Figure 4) had good MolDock and Rerank scores of −147.599 kcal mol−1 and −115.756, respectively, and E–H-bond of −4.770 kcal mol−1 (Table 1). These findings demonstrated that a stable interaction between this molecule and the receptor was possible. LYS483 and ALA481 were identified to form H-bonds with V600E-BRAF. For compound 30, similarly to vemurafenib (Figure 7), the benzene ring moiety intercalates in the space, thus forming four π–π interactions with TRP531 and PHE583 (3). The complex's stability may be associated with an additional alkyl interaction and π–alkyl interactions with the VAL471, ALA481, VAL471, ALA481, and CYS532 residues.
Figure 5 depicts the complex structure of compound 31 docked with the receptor. The MolDock and Rerank scores of −101.144 kcal mol−1 and −76.975, respectively, and E–H-bond of −9.939 kcal mol−1 (Table 1) indicated the possibility of good interactions between this molecule and the receptor. The LYS483, ASP594 (2), PHE595, GLY596, THR508, LEU514, and THR529 residues form four H-bonds between the molecule and the receptor. In addition, one C–H-bond forms with LYS483. Additional π–alkyl interactions with LEU505 and LEU514 residues may contribute to the complex's stability.
Compound 35 docked in the binding site of V600E-BRAF (Figure 6) had a high docking score (Table 1), thus indicating its binding in the binding site of one of the protomers in the protein dimer via two H-bonds with GLY534 and CYS532 residues. One H-bond forms between the N-atom of the imidazole ring and GLY534, and the other forms between the N atom of the –NH linker and the CYS532 residue. Seven C–H bonds were also observed with the LYS483, GLY534, ILE527, ALA481 (2), THR529, and ILE527 residues. Furthermore, three π–π stacked interactions were observed between the active site of the target residues TRP531 and PHE583 and compound 35, owing to aromatic ring intercalation (Figure 6). Additional π–alkyl interactions with five residues (VAL471 (2), ALA481, LEU514, and CYS532) were observed.
H-bonding is a unique sign of robust interactions between proteins and ligands, and typically results in elevated binding affinity.39 In such interactions, the number of H-bonds usually increases the inhibitory potential toward the target. As indicated in Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, which show 3D and 2D models of the interaction modes in the target binding site, the appearance of conventional H-bonds between the selected ligands and the anticancer receptor resulted in suitable ligand binding. Vemurafenib, as a standard for comparison with the investigated ligands, was docked into the same protein, and all selected ligands were found to outperformed vemurafenib. Notably, the investigated ligands inhibited the melanoma target in a pattern similar to that of vemurafenib. Some selected ligands had higher E–H-bond scores than vemurafenib.
Drug likeness and pharmacokinetic investigations of the investigated ligands were performed through the SwissADME and pkCSM web servers40,41 (Table 3, Table 4). Lipinski's rule42 suggests that good absorption occurs only at a molecular weight <500 g mol−1, with fewer than five H bond donors, log P < 5, and fewer than 10 H bond acceptors. Table 3 shows that the molecular weights of the investigated compounds were in the range of 453.49 to 243.22 gmol−1; therefore, the selected compounds were within the permissible range of Lipinski's rule. Moreover, none of the studied compounds had more than 10 H-bond acceptors; the highest number was seven for ligands 12 and 30. In addition, the selected ligands had fewer than five H-bond donors. The value for log P was <5 for all selected ligands. Veber's rule43 recommends that the TPSA should be < 140 Å2 and that the TPSA of the selected ligands should not be >137.48 Å2. Veber has also proposed that the NRB in the ligand should not exceed 10. According to the results in Table 3, all ligands examined in this study had the highest NR of 6 with only ligand 30 that have 11, which agrees with the Veber's rule. The selected ligands were further filtered for an optimum permeability and bioavailability profile according to bioavailability score (ABS) standards. An ABS score of 0.55 implies compliance with Lipinski's rule.42
The intestinal absorption of the selected ligands had values above 50%, thus demonstrating their ease of absorption. VDss indicates a volume distribution in a stable state, thus demonstrating uniform distribution of the drug to all tissues. A VDss value >0.5 suggests that a drug candidate is sufficiently distributed in the plasma, whereas a value below−0.5 indicates that a drug has a low ability to cross the cell membrane. The VDss were in the range of 1.900 to−0.445, thus indicating that the investigated ligands had adequate distribution in the plasma. In addition, the blood–brain barrier (BBB) and central nervous system (CNS) penetrability are essential factors that must be considered for acquiring optimal pharmacological drugs. The BBB and CNS penetrability standard values were to >0.3 for log blood brain (BB), and <−3 to >−2 for log PS. In a given ligand, log BB <−1 indicates insufficient diffusion of the drug molecule to the brain, and log BB >0.3 indicates that the drug molecule can cross the BBB; a log PS value >−2 indicates that the drug molecule can enter the CNS, whereas a value <−3 suggests that the drug molecule cannot easily reach the CNS.44 The results in Table 4 revealed that the selected ligands demonstrated a high possibility of crossing these barriers.
Cytochrome (CYP450), a major metabolic enzyme in the human body, has five main isoforms: CYPA2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4. The results in Table 4 indicated promising inhibition ability toward the enzymes, with safe pharmacokinetic interactions. The dosing effect and bioavailability of a drug to reach the required concentrations is determined by the clearance. Low clearance suggests persistence of a drug molecule in the body. All selected ligands demonstrated good acceptability in the body. Toxicity is used to decide whether a drug candidate is toxic. As shown in Table 4, ligands 12, 15, 30, and 31 were non-toxic. Consequently, the selected ligands had favorable pharmacokinetic properties and might be used as V600E-BRAF inhibitors in the future.
The geometry-optimized structures of the selected ligands from DFT computations are shown in Figure 8. All geometry-optimized structures conformed to a global minimum. The frontier molecular orbitals (HOMO and LUMO) of the five best ligands indicated a crucial role in charge–transfer interactions between the ligand and the target's active site. In Figure 9, the blue and red colors indicate the positive and negative regions of the orbital.45 Additionally, the shapes of the frontier orbitals can be used as a guide for determining reactivity. In every ligand, the HOMO is delocalized onto the π-bonds. According to the pattern, the blue area denotes the highest value of HOMO, whereas the red area denotes the lowest value.46 The HOMO electron-density distribution of the investigated ligands indicated promising interactions of the ligands with V600E-BRAF. Similarly, the LUMO delocalized over several areas of the aryl ring of the ligands. Nonetheless, the conjugated bonds and hetero-atoms made to the binding interaction with the target. The HOMO and LUMO electronic surfaces indicated that the π-bonds and hetero-atoms interact with the target under favorable conditions. This typical behavior is suitable for donor-acceptor interactions which may be responsible for the excellent binding of the ligands to the V600E-BRAF.
Table 5 displays the energies of the HOMO and LUMO, including quantum chemical descriptors associated with the studied ligands. A good electron-donor molecule has a high HOMO energy, whereas a lower energy value indicates a weak electron acceptor.47 In addition, a low energy gap (LUMO–HOMO) substantially affects intermolecular charge transfer and molecule bioactivity. Consequently, the small energy gap observed in the hit ligands positively affected electron movement from the HOMO to the LUMO, thus resulting in a strong affinity of the inhibitor for V600E-BRAF. The Egap value increased as follows: 30 (1.98 eV) > 35 (2.79 eV) > 12 (2.95 eV) > 15 (3.68 V) > 31 (5.23 eV). Hence, the reactivity order increased, and the most reactive value was 30 (1.98 eV). The order of the reactivity increase matched the decreases in energy gap values.
The η (hardness) and σ (softness) are important reactivity variables for the performance of a ligand in a chemical system. Hard molecules have higher resistance to alteration of their electronic-dispersal during a chemical reaction, whereas soft molecules have lower resistance to alteration of the distribution of their electrons in a reaction.48,49 The results in Table 5 indicated high η and low σ values with respect to those of analogous reported molecules.50 The χ (electronegativity) of a molecule determines its electron-attraction capability.51 The χ was computed to be approximately 3.41–4.38 eV, describing the studied ligands as donor-electrons. The μ (chemical potential) had negative values for all studied ligands, which implies good stability, and the formation of a stable complex with the receptor. The ω (electrophilicity) of a molecule predicts the electrophilic nature and measures the tendency to accept an electron. Organic molecules’ ω values are classified as follows: ω < 0.8 eV indicates weak electrophiles; ω between 0.8 and 1.5 eV indicates moderate electrophiles; and ω > 1.5 eV indicates strong electrophiles.52 The computed ω for the studied ligands indicated that they were good electrophiles. Ligands with a high ω value have potential anticancer activity.53,54 Finally, comparison of the orbital energies (eV), global reactivity variables, and docking scores of the best four ligands from the data set indicated that the selected ligands may be considered potential V600E-BRAF inhibitors with desirable properties.
The molecular electrostatic-potential (MEP) surface designates the charge distribution, thus providing a good understanding of the physical and chemical properties of a molecule. The MEP predicts the electrophilic and nucleophilic active sites of a given molecule.45,54 When the point of charge is located in an area of surplus positive charge, the point charge ligand interaction becomes repulsive, and the EP therefore is positive. However, if the point of charge is situated in a region of a surplus negative charge, then an attractive interaction occurs, and the EP becomes negative.55,56 The MEP maps of the selected ligands are presented in Figure 10; red color shows the nucleophilic region, blue color indicates the electrophilic region, and intermediate colors indicate moderate values of MEP. Thus, the increase follows the order red < orange < yellow < green < blue.57 In most cases, the negative charges are situated on the O-atoms, and the positive areas of the molecules are the areas where the H-atoms bonded to O-atoms are situated. The positive and negative cores of the selected molecules participate in forming interactions with both bonded and non-bonded (particularly H-bonds) areas in the complexes during docking.58
Future research should include molecular dynamic simulations of the selected compounds to further investigate their ability to induce confrontational changes in the V600E-BRAF kinase. In addition, new molecules should be designed from the selected hit molecules, synthesized, and tested in vivo and in vitro, to establish their potency as V600E-BRAF inhibitors for the treatment of melanoma and other V600E-BRAF related cancers.
Conclusions
Molecular docking simulation, pharmacokinetic evaluation, and DFT computations were successfully performed to determine potential hit compounds against V600E-BRAF from a series of 72 anticancer compounds from the PubChem database. The five top-ranked compounds (12, 15, 30, 31, and 35) were identified to have excellent docking scores in the active site of the V600E-BRAF target. The docking results indicated that both hydrophobic interactions and H bonds play major roles in the binding interactions of the potential compounds with V600E-BRAF. The predicted physicochemical and pharmacokinetic parameters were in acceptable ranges for drug screening criteria. The quantum chemical parameters computed with the DFT approach indicated that the selected molecules have stable structures and are highly electrophilic, whereas the distribution of the MEP identified the potential sites for nucleophilic and electrophilic attack. The broad computational investigations indicated that the selected molecules may be considered potential hits against the V600E-BRAF target and may be promising as anticancer agents.
Source of funding
This work was fully sponsored by the Tertiary Education Trust Fund (TETFUND), grant number TETF/DR&D/UNI/ZARIA/IBR/2020/VOL./54.
Conflict of interest
The authors have no conflict of interest to declare.
Ethical approval
This research was fully sponsored by the Tertiary Education Trust Fund (NG) under IBR Project grant 2020 with project number TETF/DR&D/UNI/ZARIA/IBR/2020/VOL./54 dated 21st February, 2022.
Consent
Not relevant.
Authors’ contributions
ABU: Designed and performed the study, interpreted the results, and wrote the manuscript. AU: Contributed to designing the study, and supervised and edited the manuscript. All authors have critically reviewed and approved the final draft and are responsible for the content and similarity index of the manuscript.
Acknowledgments
We thank the TETFUND for the 2020 Institution-Based Research project grant (TETF/DR&D/UNI/ZARIA/IBR/2020/VOL.1/54) for this study.
Footnotes
Peer review under responsibility of Taibah University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtumed.2023.01.013.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Li Z.-L., Prakash P., Buck M. A “tug of war” maintains a dynamic protein–membrane complex: molecular dynamics simulations of C-Raf RBD-CRD bound to K-Ras4B at an anionic membrane. ACS Cent Sci. 2018;4(2):298–305. doi: 10.1021/acscentsci.7b00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z., Wang M., Wang H., Fang L., Gou S. Platinum-based modification of styrylbenzylsulfones as multifunctional antitumor agents: targeting the RAS/RAF pathway, enhancing antitumor activity, and overcoming multidrug resistance. J Med Chem. 2019;63(1):186–204. doi: 10.1021/acs.jmedchem.9b01223. [DOI] [PubMed] [Google Scholar]
- 3.Wang P.-F., Qiu H.-Y., Wang Z.-F., Zhang Y.-J., Wang Z.-C., Li D.-D., et al. Identification of novel B-RafV600E inhibitors employing FBDD strategy. Biochem Pharmacol. 2017;132:63–76. doi: 10.1016/j.bcp.2017.02.022. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z.F., Wang P.F., Ma J.T., Chai Y.Z., Hu H.M., Gao W.L., et al. Design of potent B-RafV600E inhibitors by multiple copy simulation search strategy. Chem Biol Drug Des. 2018;91(2):567–574. doi: 10.1111/cbdd.13121. [DOI] [PubMed] [Google Scholar]
- 5.Umar B.A., Uzairu A., Shallangwa G.A., Uba S. Rational drug design of potent V600E-BRAF kinase inhibitors through molecular docking simulation. J Eng Exact Sci. 2019;5(5):469–481. [Google Scholar]
- 6.McCubrey J.A., Steelman L.S., Abrams S.L., Lee J.T., Chang F., Bertrand F.E., et al. Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv Enzym Regul. 2006;46(1):249–279. doi: 10.1016/j.advenzreg.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Chappell W.H., Steelman L.S., Long J.M., Kempf R.C., Abrams S.L., Franklin R.A., et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget. 2011;2(3):135. doi: 10.18632/oncotarget.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H., Higgins B., Kolinsky K., Packman K., Go Z., Iyer R., et al. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res. 2010;70(13):5518–5527. doi: 10.1158/0008-5472.CAN-10-0646. [DOI] [PubMed] [Google Scholar]
- 9.Blasco R.B., Francoz S., Santamaría D., Cañamero M., Dubus P., Charron J., et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell. 2011;19(5):652–663. doi: 10.1016/j.ccr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin J., Xie P., Ventocilla C., Zhou G., Vultur A., Chen Q., et al. Identification of a novel family of BRAFV600E inhibitors. J Med Chem. 2012;55(11):5220–5230. doi: 10.1021/jm3004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang P.-F., Zhang Y.-J., Wang D., Hu H.-M., Wang Z.-C., Xu C., et al. Design, synthesis, and biological evaluation of new B-RafV600E kinase inhibitors. Bioorg Med Chem. 2018;26(9):2372–2380. doi: 10.1016/j.bmc.2018.03.038. [DOI] [PubMed] [Google Scholar]
- 12.Brose M.S., Volpe P., Feldman M., Kumar M., Rishi I., Gerrero R., et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62(23):6997–7000. [PubMed] [Google Scholar]
- 13.Long G.V., Stroyakovskiy D., Gogas H., Levchenko E., de Braud F., Larkin J., et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;371(20):1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 14.Ammar U.M., Abdel-Maksoud M.S., Oh C.-H. Recent advances of RAF (rapidly accelerated fibrosarcoma) inhibitors as anti-cancer agents. Eur J Med Chem. 2018;158:144–166. doi: 10.1016/j.ejmech.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Amin K.M., El-Badry O.M., Rahman D.E.A., Ammar U.M., Abdalla M.M. Design, synthesis, anticancer evaluation and molecular docking of new V600EBRAF inhibitors derived from pyridopyrazinone. Eur J Chem. 2016;7(1):19–29. [Google Scholar]
- 16.Prahallad A., Sun C., Huang S., Di Nicolantonio F., Salazar R., Zecchin D., et al. Unresponsiveness of colon cancer to BRAF (V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 17.Akhtar M.J., Siddiqui A.A., Khan A.A., Ali Z., Dewangan R.P., Pasha S., et al. Design, synthesis, docking and QSAR study of substituted benzimidazole linked oxadiazole as cytotoxic agents, EGFR and erbB2 receptor inhibitors. Eur J Med Chem. 2017;126:853–869. doi: 10.1016/j.ejmech.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Regad T. Targeting RTK signaling pathways in cancer. Cancers. 2015;7(3):1758–1784. doi: 10.3390/cancers7030860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilhelm S.M., Adnane L., Newell P., Villanueva A., Llovet J.M., Lynch M. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol Cancer Therapeut. 2008;7(10):3129–3140. doi: 10.1158/1535-7163.MCT-08-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryan M.C., Falsey J.R., Frohn M., Reichelt A., Yao G., Bartberger M.D., et al. N-substituted azaindoles as potent inhibitors of Cdc7 kinase. Bioorg Med Chem Lett. 2013;23(7):2056–2060. doi: 10.1016/j.bmcl.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Flaherty K.T., Puzanov I., Kim K.B., Ribas A., McArthur G.A., Sosman J.A., et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Infante J., Fecher L., Nallapareddy S., Gordon M., Flaherty K., Cox D., et al. Safety and efficacy results from the first-in-human study of the oral MEK 1/2 inhibitor GSK1120212. J Clin Oncol. 2010;28(15_suppl):2503. 2503. [Google Scholar]
- 23.Karthick T., Tandon P. Computational approaches to find the active binding sites of biological targets against busulfan. J Mol Model. 2016;22(6):1–9. doi: 10.1007/s00894-016-3015-z. [DOI] [PubMed] [Google Scholar]
- 24.Cumming J.G., Davis A.M., Muresan S., Haeberlein M., Chen H. Chemical predictive modelling to improve compound quality. Nat Rev Drug Discov. 2013;12(12):948–962. doi: 10.1038/nrd4128. [DOI] [PubMed] [Google Scholar]
- 25.Umar A.B., Uzairu A., Shallangwa G.A., Uba S. Molecular docking strategy to design novel V600E-BRAF kinase inhibitors with prediction of their drug-likeness and pharmacokinetics ADMET properties. Chem Africa. 2020:1–17. [Google Scholar]
- 26.Deghady A.M., Hussein R.K., Alhamzani A.G., Mera A. Density functional theory and molecular docking investigations of the chemical and antibacterial activities for 1-(4-Hydroxyphenyl)-3-phenylprop-2-en-1-one. Molecules. 2021;26(12):3631. doi: 10.3390/molecules26123631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussein R., Elkhair H. Molecular docking identification for the efficacy of some zinc complexes with chloroquine and hydroxychloroquine against main protease of COVID-19. J Mol Struct. 2021;1231 doi: 10.1016/j.molstruc.2021.129979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Opo F.A., Rahman M.M., Ahammad F., Ahmed I., Bhuiyan M.A., Asiri A.M. Structure based pharmacophore modeling, virtual screening, molecular docking and ADMET approaches for identification of natural anti-cancer agents targeting XIAP protein. Sci Rep. 2021;11(1):1–17. doi: 10.1038/s41598-021-83626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Umar B.A., Uzairu A. In-silico approach to understand the inhibition of corrosion by some potent triazole derivatives of pyrimidine for steel. SN Appl Sci. 2019;1(11):1413. [Google Scholar]
- 30.Bollag G., Hirth P., Tsai J., Zhang J., Ibrahim P.N., Cho H., et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467(7315):596. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi W.-K., El-Gamal M.I., Choi H.S., Baek D., Oh C.-H. New diarylureas and diarylamides containing 1, 3, 4-triarylpyrazole scaffold: synthesis, antiproliferative evaluation against melanoma cell lines, ERK kinase inhibition, and molecular docking studies. Eur J Med Chem. 2011;46(12):5754–5762. doi: 10.1016/j.ejmech.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 32.Molegro A. DK-8000 Aarhus C; Denmark: 2011. MVD 5.0 Molegro virtual docker. [Google Scholar]
- 33.Wang G.-m., Wang X., Zhu J.-m., Guo B.-b., Yang Z., Xu Z.-j., et al. Docking-based structural splicing and reassembly strategy to develop novel deazapurine derivatives as potent B-Raf V600E inhibitors. Acta Pharmacol Sin. 2017;38(7):1059–1068. doi: 10.1038/aps.2016.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomsen R., Christensen M.H. MolDock: a new technique for high-accuracy molecular docking. J Med Chem. 2006;49(11):3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 35.Tsuneda T., Song J.-W., Suzuki S., Hirao K. On Koopmans' theorem in density functional theory. J Chem Phys. 2010;133(17) doi: 10.1063/1.3491272. [DOI] [PubMed] [Google Scholar]
- 36.Yusuf D., Davis A.M., Kleywegt G.J., Schmitt S. An alternative method for the evaluation of docking performance: RSR vs RMSD. J Chem Inf Model. 2008;48(7):1411–1422. doi: 10.1021/ci800084x. [DOI] [PubMed] [Google Scholar]
- 37.Abdullahi M., Uzairu A., Shallangwa G.A., Arthur D.E., Umar B.A., Ibrahim M.T. Virtual molecular docking study of some novel carboxamide series as new anti-tubercular agents. Eur J Chem. 2020;11(1):30–36. [Google Scholar]
- 38.Hassan A.H., Lee K.-T., Lee Y.S. Flavone-based arylamides as potential anticancers: design, synthesis and in vitro cell-based/cell-free evaluations. Eur J Med Chem. 2020;187 doi: 10.1016/j.ejmech.2019.111965. [DOI] [PubMed] [Google Scholar]
- 39.Khan I.M., Islam M., Shakya S., Alam N., Imtiaz S., Islam M.R. Synthesis, spectroscopic characterization, antimicrobial activity, molecular docking and DFT studies of proton transfer (H-bonded) complex of 8-aminoquinoline (donor) with chloranilic acid (acceptor) J Biomol Struct Dyn. 2021:1–15. doi: 10.1080/07391102.2021.1969280. [DOI] [PubMed] [Google Scholar]
- 40.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7 doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pires D.E., Blundell T.L., Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58(9):4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23(1–3):3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 43.Veber D.F., Johnson S.R., Cheng H.-Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 44.Clark D.E. In Silico prediction of blood–brain barrier permeation. Drug Discov Today. 2003;8(20):927–933. doi: 10.1016/s1359-6446(03)02827-7. [DOI] [PubMed] [Google Scholar]
- 45.Khattab M., Al-Karmalawy A.A. Revisiting activity of some nocodazole analogues as a potential anticancer drugs using molecular docking and DFT calculations. Front Chem. 2021;9 doi: 10.3389/fchem.2021.628398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Celik S., Akyuz S., Ozel A.E. Structural and vibrational investigations and molecular docking studies of a vinca alkoloid, vinorelbine. J Biomol Struct Dyn. 2022:1–20. doi: 10.1080/07391102.2022.2145369. [DOI] [PubMed] [Google Scholar]
- 47.Manoj K., Elangovan N., Chandrasekar S. Synthesis, XRD, hirshfeld surface analysis, ESP, HOMO-LUMO, quantum chemical modeling and anticancer activity of di (p-methyl benzyl)(dibromo)(1, 10-phenanthroline) tin (IV) complex. Inorg Chem Commun. 2022;139 [Google Scholar]
- 48.Murulana L.C., Singh A.K., Shukla S.K., Kabanda M.M., Ebenso E.E. Experimental and quantum chemical studies of some bis (trifluoromethyl-sulfonyl) imide imidazolium-based ionic liquids as corrosion inhibitors for mild steel in hydrochloric acid solution. Ind Eng Chem Res. 2012;51(40):13282–13299. [Google Scholar]
- 49.Wazzan N.A. DFT calculations of thiosemicarbazide, arylisothiocynates, and 1-aryl-2, 5-dithiohydrazodicarbonamides as corrosion inhibitors of copper in an aqueous chloride solution. J Ind Eng Chem. 2015;26:291–308. [Google Scholar]
- 50.Fahim A.M., Farag A.M. Synthesis, antimicrobial evaluation, molecular docking and theoretical calculations of novel pyrazolo [1, 5-a] pyrimidine derivatives. J Mol Struct. 2020;1199 [Google Scholar]
- 51.Karton A., Spackman P.R. Evaluation of density functional theory for a large and diverse set of organic and inorganic equilibrium structures. J Comput Chem. 2021;42(22):1590–1601. doi: 10.1002/jcc.26698. [DOI] [PubMed] [Google Scholar]
- 52.Edim M.M., Enudi O.C., Asuquo B.B., Louis H., Bisong E.A., Agwupuye J.A., et al. Aromaticity indices, electronic structural properties, and fuzzy atomic space investigations of naphthalene and its aza-derivatives. Heliyon. 2021;7(2) doi: 10.1016/j.heliyon.2021.e06138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srivastava R. Theoretical studies on the molecular properties, toxicity, and biological efficacy of 21 new chemical entities. ACS Omega. 2021;6(38):24891–24901. doi: 10.1021/acsomega.1c03736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El-Bindary M.A., El-Desouky M.G., El-Bindary A.A. Metal–organic frameworks encapsulated with an anticancer compound as drug delivery system: synthesis, characterization, antioxidant, anticancer, antibacterial, and molecular docking investigation. Appl Organomet Chem. 2022;36(5) [Google Scholar]
- 55.Celik S., Vagifli F., Akyuz S., Ozkok F., Ozel A.E., Dosler S., et al. Synthesis, vibrational spectroscopic investigation, molecular docking, antibacterial and antimicrobial studies of a new anthraquinone derivative compound. Spectrosc Lett. 2022:1–19. [Google Scholar]
- 56.Singh P., Islam S., Ahmad H., Prabaharan A. Spectroscopic investigation (FT-IR, FT-Raman), HOMO-LUMO, NBO, and molecular docking analysis of N-ethyl-N-nitrosourea, a potential anticancer agent. J Mol Struct. 2018;1154:39–50. [Google Scholar]
- 57.Balachandran V., Karpagam V., Revathi B., Kavimani M., Ilango G. Conformational stability, spectroscopic and computational studies, HOMO–LUMO, NBO, ESP analysis, thermodynamic parameters of natural bioactive compound with anticancer potential of 2-(hydroxymethyl) anthraquinone. Spectrochim Acta Mol Biomol Spectrosc. 2015;150:631–640. doi: 10.1016/j.saa.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 58.Jordaan M.A., Ebenezer O., Damoyi N., Shapi M. Virtual screening, molecular docking studies and DFT calculations of FDA approved compounds similar to the non-nucleoside reverse transcriptase inhibitor (NNRTI) efavirenz. Heliyon. 2020;6(8) doi: 10.1016/j.heliyon.2020.e04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.