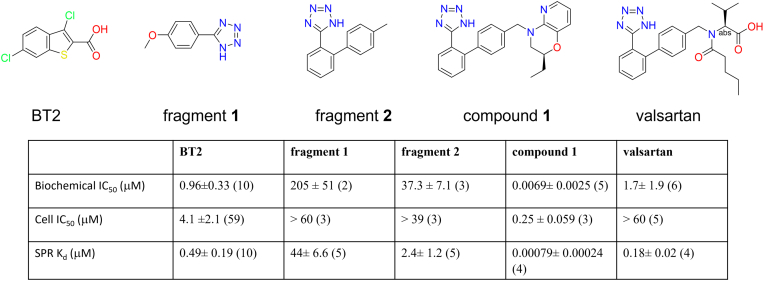
Abstract
The mammalian mitochondrial branched-chain ketoacid dehydrogenase (BCKD) complex is a multienzyme complex involved in the catabolism of branched-chain amino acids. BCKD is regulated by the BCKD kinase, or BCKDK, which binds to the E2 subunit of BCKD, phosphorylates its E1 subunit, and inhibits enzymatic activity. Inhibition of the BCKD complex results in increased levels of branched-chain amino acids and branched-chain ketoacids, and this buildup has been associated with heart failure, type 2 diabetes mellitus, and nonalcoholic fatty liver disease. To find BCKDK inhibitors for potential treatment of these diseases, we performed both NMR and virtual fragment screening and identified tetrazole-bearing fragments that bind BCKDK at multiple sites. Through structure-based virtual screening expanding from these fragments, the angiotensin receptor blocker class antihypertension drugs and angiotensin receptor blocker–like compounds were discovered to be potent BCKDK inhibitors, suggesting potential new avenues for heart failure treatment combining BCKDK inhibition and antihypertension.
Keywords: branched-chain ketoacid dehydrogenase, branched-chain ketoacid dehydrogenase kinase, inhibitors, angiotensin receptor blocker, structure
Abbreviations: ARB, angiotensin II type 1 receptor blocker; AT1R, angiotensin II type 1 receptor; BCAA, branched-chain amino acid; BCKA, branched-chain ketoacid; BCKD, branched-chain ketoacid dehydrogenase; BCKDK, BCKD kinase; BSA, bovine serum albumin; BT2, 3,6-dichlorobenzo[b]thiophene-2-carboxylic acid; DMSO, dimethyl sulfoxide; GST, glutathione-S-transferase; HF, heart failure; KD, α-ketoglutarate dehydrogenase; KIC, α-ketoisocaproate; MBP, maltose-binding protein; PD, pyruvate dehydrogenase; PDB, Protein Data Bank; PDK2, PD regulatory kinase 2; PDK3, PD regulatory kinase 3; SPR, surface plasmon resonance; STD, saturation transfer difference; TCEP, Tris(2-carboxyethyl)phosphine; TEV, tobacco etch virus
The mammalian branched-chain ketoacid (BCKA) dehydrogenase (BCKD) together with the pyruvate dehydrogenase (PD) and α-ketoglutarate dehydrogenase (KD) form the mitochondrial multienzyme complex family (1). These large multiple protein complexes consist of three basic subunits that are either homologous (E1 and E2) or shared (E3). These subunits have similar complex organization and carry out analogous enzymatic reactions in these complexes. Multiple copies of the dihydrolipoyl transacylase domain of E2 form the core of these complexes (2, 3, 4, 5), and the E1 and/or E3 subunit bind to E2 through a binding domain of E2(2). E1 is a substrate-specific dehydrogenase that decarboxylates α-ketoacids (branched-chain α-ketoacids, pyruvate, or α-ketoglutarate) and acylates the lipoyl moiety that is covalently bound to E2. The lipoyl domain of E2 then swings to the active site of E2, which catalyzes the transfer of the acyl group to coenzyme A. E3 is a dihydrolipoamide dehydrogenase that reoxidizes the reduced lipoyl residue of E2. In human, the E1 subunits of BCKD and PD consist of heterotetramers of α and β subunits, and the E1 of KD is a homodimer. The structures of the E1 α/β heterodimers of BCKD and PD and the E1 homodimer of KD are highly similar (4, 6, 7), so are those of the E2 core domains of BCKD, PD, and KD (8, 9, 10).
The activities of the BCKD and PD complexes are tightly regulated by specific protein kinases and/or protein phosphatases. These proteins bind to the E2 subunits through their lipoyl domains and phosphorylate (inhibit) or dephosphorylate (activate) specific residues in the E1 subunits (1, 11, 12). In the case of BCKD, the specific enzymes are BCKD kinase (BCKDK or BDK) and PP2Cm/PPM1K phosphatase (5, 11). PD is regulated by four isoforms of PD-specific kinases, PDK1–4. KD does not have a known kinase that regulates its activity. The 3D structures BCKDK and PDK1–4 are similar (13, 14, 15, 16, 17).
BCKD catalyzes the committed step of the catabolism of all three branched-chain amino acids (BCAAs; valine, isoleucine, and leucine), and its dysregulation is associated with multiple disorders (5, 18). Deficiency in BCKD activity can lead to maple syrup urine disease, also called branched-chain ketoaciduria (19). This disease is characterized by the accumulation of BCKA and BCAA in plasma and urine, which has severe clinical consequences, including often fatal acidosis, neurological derangement, and developmental delay (20). Recently, a gain-of-function BCKDK variant with a single mutation at its physiological ligand-binding site was reported to be associated with a mild form of maple syrup urine disease (21). Recent metabolomics studies have further reported that elevated blood BCAA concentration is associated with insulin resistance and nonalcoholic fatty liver disease (22, 23, 24, 25). Finally, deranged BCKA catabolism is also linked to congenital heart diseases and heart failure (HF) (26).
Because of its importance in the homeostasis of BCAA and BCKA, the activity of BCKDK is tightly regulated through a feed-forward inhibition mechanism by many physiological ligands, such as α-ketoisocaproate (KIC), phenylbutyrate, phenylpyruvate acid, thiamine pyrophosphate (12, 27, 28), and other nutrients in high fat food (27, 29, 30, 31, 32, 33). BCAAs are also essential amino acids for protein synthesis and brain neurotransmission (34, 35, 36). Deficiencies in BCKDK activity have been reported to lead to reduced BCAA in plasma and spinal fluids and were linked to epilepsy, autism, and intellectual disability in human (37, 38, 39) and defects in male reproduction in rats (40).
To find more potent BCKDK inhibitors, Tso et al. (32, 41) employed a high-throughput screen to identify the compound 3,6-dichlorobenzo[b]thiophene-2-carboxylic acid (BT2) as an allosteric BCKDK inhibitor that binds to the characterized physiological ligand-binding site. Administration of BT2 in mice inhibited BCKDK, promoted BCKA degradation, and preserved heart function (42). Inhibition of BCKDK using BT2 led to relief of key metabolic disease phenotypes in an obesity Zucker fatty rat model (43) as well as obesity-related insulin resistance in an ob/ob mouse model (44). The fact that BT2 is a fragment-sized compound inspired us to carry out both NMR and virtual fragment screening to find novel BCKDK leads. These efforts identified tetrazole-bearing fragments that are BCKDK inhibitors. X-ray crystal structures of BCKDK in complex with these fragments showed that they unexpectedly bind to multiple sites in BCKDK. Through structure-based fragment expansion, we identified potent BCKDK inhibitors that are angiotensin II type 1 receptor (AT1R) blocker (ARB)-like compounds and ARBs themselves, which are broadly used antihypertension drugs. The implications of ARBs on BCAA metabolism are discussed. Our discoveries provide potential leads to develop BCKDK inhibitors with or without antihypertension properties, and the use of these inhibitors may test the mechanism of modulating human disease by lowering BCKA and BCAA levels.
Results
Identification of fragment 1 as a BCKDK inhibitor
Because KIC and BT2 are fragment-sized BCKDK inhibitors that bind to a well-defined allosteric pocket in BCKDK (32, 41), we carried out an NMR-based fragment screen to identify additional fragments that can inhibit BCKDK. NMR fragment hits were first verified for their ability to bind to BCKDK in surface plasmon resonance (SPR) experiments. In these experiments, the SPR sensorgrams (the responses of reflection index at the SPR chip surface to the binding of the compounds in solution to immobilized BCKDK over time) were recorded and analyzed. These hits were then tested for inhibition activities in in vitro BCKDK and human skeletal myocyte cell–based functional inhibition assays. Fragment 1, 5-(4-methoxyphenyl)-1H-tetrazole, was identified to be a BCKDK inhibitor with an IC50 of 205 ± 51 μM (Figs. 1 and S1).
Figure 1.
Chemical structures and potencies of BCKDK inhibitors investigated in this study: BT2; fragment 1, identified in NMR-based fragment screen; fragment 2, identified by ligand-based virtual fragment screen; compound 1, identified by structure-based virtual expansion; valsartan, one of the most potent BCKDK inhibitors in the ARB family. The number of experiments performed are shown in parenthesis. Each experiment was a replicate of two inhibitor concentration series. The biochemical IC50 values were measurements at ATP concentration of 15 μM. The cell IC50 values were based on BCKDK-dependent phosphorylation of endogenous BCKD. SPR binding affinities were calculated from curve fitting to the responses of refractive index changes caused by inhibitors in solution binding to BCKDK immobilized on the SPR surface. Because of their fast off rates, the equilibrium Kd were calculated for BT2, fragment 1, and fragment 2. The more potent compound 1 and valsartan had measurable off rates, and their kinetic Kd were calculated. The absolute stereochemistry of compound 1 is undefined and arbitrarily assigned. ARB, angiotensin II type 1 receptor blocker; BCKDK, branched-chain ketoacid dehydrogenase kinase; BT2, 3,6-dichlorobenzo[b]thiophene-2-carboxylic acid; SPR, surface plasmon resonance.
To aid structure-based drug design, we determined the binding mode of fragment 1 in BCKDK using X-ray crystallography (Table 1). Fragment 1 bound BCKDK at three distinct sites, “KIC” or “BT2” site, site 2, and Pfz3 site, named after the homologous site in PD kinase 2 (PDK2) for inhibitor Pfz3 (Fig. 2). The BT2 site was formed by residues in helices α1 (residue 54–81), α3 (90–108), α4 (113–131), α5 (135–147), and α6 (151–181) in the regulatory domain of BCKDK. Site 2 was formed by residues in helices α0 (42–49, disordered in all previously published complex structures), α1 and α6 in the regulatory domain, as well as residues from the kinase domain of BCKDK. Pfz3 site was formed by residues in α2 (83–88), N terminus of α3 and α6, and the C terminus of α5.
Table 1.
Data collection and refinement statistics
| Crystala | Fragment 1 | Fragment 2 | Compound 1 | Valsartan |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P6422 | P6422 | P6422 | P6422 |
| Cells | ||||
| a, b, c (Å) | 111.96, 111.96, 137.18 | 111.40, 111.40 140.55 | 111.27 111.27 140.11 | 111.16 111.16 141.74 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å)b | 96.96–2.05 (2.19–2.05) | 96.50–1.85 (1.98–1.85) | 96.36–1.96 (2.14–1.96) | 96.27–1.92 (2.10–1.92) |
| Rmergec | 0.062 (2.01) | 0.053 (1.28) | 0.060 (1.489) | 0.081 (1.39) |
| I/σI | 26.9 (1.8) | 28.0 (1.5) | 24.8 (2.0) | 18.5 (1.8) |
| Completeness (%) | 85.2 (52.1) | 95.7 (74.0) | 88.6 (54.4) | 87.1 (42.0) |
| Redundancy | 19.1 (20.2) | 18.8 (11.1) | 19.5 (20.3) | 19.2 (16.9) |
| Refinement | ||||
| Resolution (Å) | 23.6–2.05 | 20.49–1.85 | 23.7–1.96 | 96.3–1.92 |
| No. of reflections | 24,779 (1224) | 36,650 (1892) | 27,571 (1441) | 29,906 |
| Rworkd | 0.239 (0.322) | 0.221 (0.213) | 0.226 (0.290) | 0.238 (0.263) |
| Rfreee | 0.256 (0.376) | 0.250 (0.287) | 0.267 (0.353) | 0.269 (0.373) |
| No. of atoms | ||||
| Protein | 2617 | 2599 | 2599 | 2593 |
| Ligand | 83 | 79 | 122 | 96 |
| Water | 48 | 88 | 175 | 160 |
| B-factors (Å2) | ||||
| Protein | 60.3 | 62 | 58.9 | 52.3 |
| Ligand | 71.5 | 64.0 | 72.4 | 64.5 |
| Water | 64.2 | 44.9 | 58.4 | 52.3 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.008 | 0.010 | 0.008 | 0.008 |
| Bond angles (°) | 0.86 | 0.97 | 0.88 | 0.88 |
| Ramachandran (favored, outlier [%])f | 96.9, 0.0 | 96.6, 0.0 | 97.5, 0.0 | 97.8, 0.3 |
Data sets were collected on one crystal for each complex. Values in parentheses are for highest resolution shell.
Statistics in the highest resolution bins are shown in parenthesis. Based on anisotropic scaling and merge.
Rmerge = ∑hkl∑i|Ii(hkl)−<I(hkl)>|/∑hkl∑iIi(hkl), where Ii (hkl) is the ith intensity measurement of reflection (hkl), and <I(hkl)> is the mean intensity from multiple observations of that reflection.
Rwork = ∑hkl||Fobs|−|Fcalc||/∑hkl∑|Fobs|, where Fobs and Fcalc are observed and calculated structure–factor amplitudes, respectively.
Rfree is calculated using 5% of reflections randomly excluded from refinement. For all crystals used in this study, the same set of reflections were chosen for exclusion.
Percentages of residues in the most favorable and disallowed regions of the Ramachandran plot.
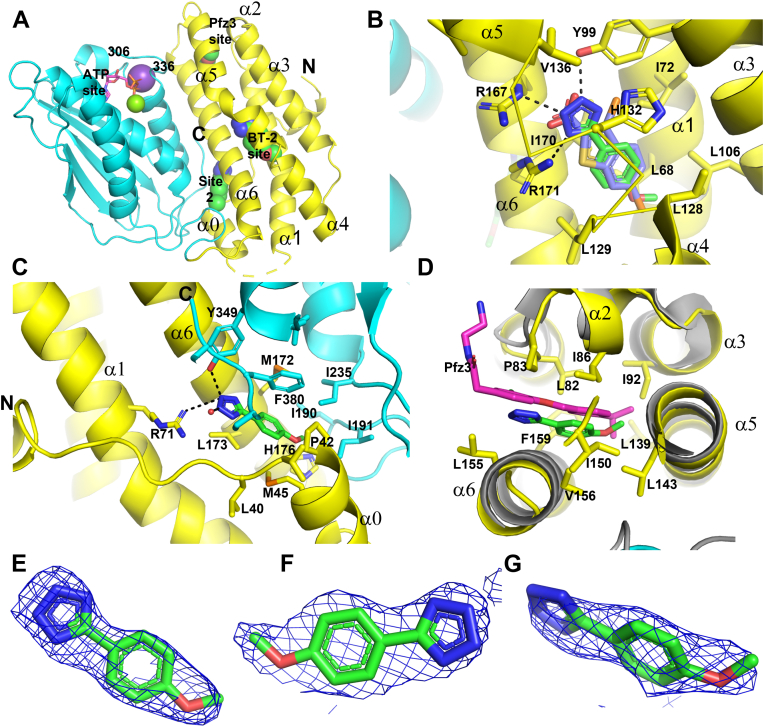
Figure 2.
Fragment 1 binds at multiple sites in BCKDK.A, locations of different binding sites for fragment 1 in BCKDK. The regulatory domain of BCKDK (1–184) in ribbon diagram is colored yellow, and the kinase domain (185-C terminus) is colored cyan. Fragment 1 molecules are shown as spheres, and bound ADP and Mg2+ and K+ are shown as sticks and spheres. The N and C termini of BCKDK, helices in the regulatory domain, and the flexible loop near the ATP site are labeled. B, at the BT2 site, fragment 1 (stick model with green colored C atoms) occupies the same position as BT2 (purple C atoms) and makes similar interactions with BCKDK (side chains of contacting residues are labeled and shown in sticks). Hydrogen bonds involving fragment 1 are shown in dashes. For clarity, main-chain ribbons for the link peptide between α4 and α5 (129–135) are removed, and only Cα traces are shown. C, site 2 for fragment 1 in BCKDK. Both the N- and C-terminal peptides are involved in the formation of site 2. D, Pfz3 site for fragment 1 in BCKDK. This site is named after a homologous site in PDK2 for PDK2 inhibitor Pfz3 (12). Superimposed is Pfz3 (magenta sticks) bound in PDK2 (gray ribbon) (Protein Data Bank ID: 2BU7). E–G, fragment 1 at the BT2, site 2, and Pfz3 sites, respectively, is shown in stick representation embedded in the initial 2Fo–Fc electron density map contoured at 1σ to illustrate the quality of model building. Figure 2, Figure 3, Figure 4 were generated using the graphic program PyMol. BCKDK, branched-chain ketoacid dehydrogenase kinase; BT2, 3,6-dichlorobenzo[b]thiophene-2-carboxylic acid.
The BT2 site is the binding site for BT2 (41) and many physiological BCKDK ligands, such as KIC and phenylpyruvate (32). At the BT2 site, fragment 1 interacted with BCKDK similarly to BT2 (Fig. 2B). The methoxyphenyl of fragment 1 overlapped with the benzothiophene of BT2 and made similar hydrophobic contacts with the compacted hydrophobic BT2-binding site. The tetrazole moiety, a carboxylic acid isostere, made polar interaction with R167, R171, and Y99, like the carboxylic acid of BT2. The acidic tetrazole was likely deprotonated, possessing a negative charge to make salt bridge interactions with the positively charged side chains of R167 and R171. In the BT2 complex structure, the side chain of H132 also made a hydrogen bond to the carboxylate. In the fragment 1 complex structure, the side chain of H132 rotated slightly and has characteristics of a π-stacking interaction with the tetrazole.
Unlike BT2, which only bound one site in BCKDK, fragment 1 was also found at site 2, which was formed by residues from both the regulatory domain and the kinase domain of BCKDK (Fig. 2C). In the BT2 complex, helix α0 and the C terminus of BCKDK, which interacted with fragment 1 at site 2, were disordered. Site 2 was separated from the BT2 site by helix α6. At this site, fragment 1 had interactions that to some degree resemble those observed at the BT2 site. The tetrazole made polar interactions with the side chains of R71 and Y349, from the regulatory and kinase domain, respectively. The methoxyphenyl of fragment 1 made hydrophobic interactions with hydrophobic side chains of residues from both domains of BCKDK. Although site 2 was not observed in previously reported BCKDK complex structures, a superimposition of BCKDK with the PD regulatory kinase 3 (PDK3)–L2 complex structure (Protein Data Bank [PDB] ID: 1Y8P) (17) (PDK3) suggested that occupying site 2 could also be inhibitory (Fig. S2). Based on this overlay, site 2 was next to the PDK-binding site for the lipoyl moiety of the E2 subunit of PD (45, 46). This lipoyl-binding site was also the site for potent PDK allosteric inhibitors, Nov3r and AZ12, to bind to PDKs (15) (Fig. S2).
Fragment 1 also bound BCKDK at a third site, a site that is homologous to that in PDK2 for the allosteric inhibitor Pfz3 (Fig. 2D) (15). At the Pfz3 site, the tetrazole moiety of fragment 1 was exposed to solvent, and the methoxyphenyl makes hydrophobic interactions with the protein. Pfz3 makes interactions that are exclusively hydrophobic with PDK2 at this homologous site. For PDK2, occupying this site leads to inhibition of PDK2 (15).
Identification of fragment 2 as BCKDK inhibitor
To expand our search for BCKDK inhibitors, we also carried out a ligand-based fragment virtual screen. Particularly, we searched our internal compound database for fragments bearing carboxylic acid and its isosteres with molecular weight <350 Da, then tested them in the biochemical in vitro assay, and verified hits for binding in SPR. One of the confirmed hits that went into crystallization for structural studies was fragment 2, 5-(4′-methyl-[1,1′-biphenyl]-2-yl)-1H-tetrazole (Fig. 1).
Fragment 2 bound BCKDK at two sites: a site that is adjacent to the BT2 site with small overlaps, and site 2 (Fig. 3). Surprisingly, fragment 2 does not bind at the BT2 site, although it shares structural similarity to fragment 1. Likely because of its shape, fragment 2 does not bind to BCKDK at Pfz3 site. Relative to fragment 1 binding, fragment 2 binding induces conformational changes in two peptide regions of BCKDK. Peptide 330 to 335 in the long flexible loop near the ATP site of BCKDK becomes ordered and extends into the active site cleft between the regulatory and kinase domains. This peptide interacts with fragment 2. The peptide from 375 to C terminus of BCKDK became disordered. In the fragment 1 complex, this segment of BCKDK is involved in fragment 1 binding to site 2 (Fig. 2).
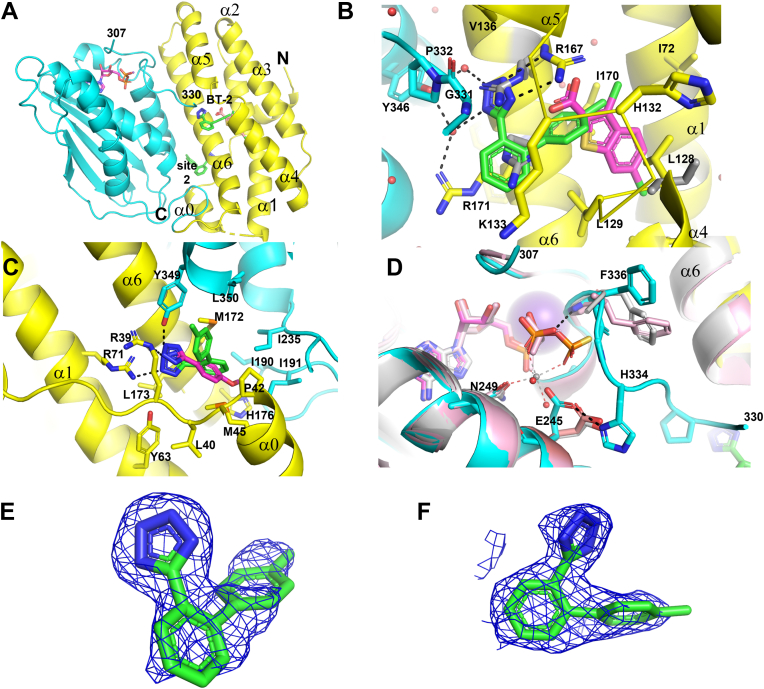
Figure 3.
Fragment 2 binds at two sites in BCKDK.A, locations of fragment 2 binding sites in BCKDK. To show an expanded BT2 site, BT2 is shown in magenta sticks. B, at the expanded BT2 site, fragment 2 binds to BCKDK adjacent to BT2. BT2 interacting residues that move significantly upon fragment 2 binding are shown in gray sticks. C, overlay of fragments 1 (magenta C atoms) and 2 in the BT2 site 2. D, comparisons of the ADP/ATP sites of the fragment 2 complex with those of the fragment 1 complex (pink C atoms) and the ATPγS-bound apo BCKDK (gray color, Protein Data Bank ID: 1GJV). ADP/ATP are shown in sticks. Bound Mg2+ is shown in sphere. E and F, fragment 2 at the expanded BT2 site and site 2, respectively, is shown embedded in the initial 2Fo–Fc maps contoured at 1.0 σ. BCKDK, branched-chain ketoacid dehydrogenase kinase; BT2, 3,6-dichlorobenzo[b]thiophene-2-carboxylic acid.
The most significant conformational changes at the BT2 site upon fragment 2 binding happened to the side chains of R167 and R171 (Fig. 3B). They adopted different rotamers, which created space for fragment 2 to bind between the BT2 site and the kinase domain. Their conformations were also different from those of the apo BCKDK structure (13). The side chain of R167 made a π-stacking interaction with the terminal phenyl of fragment 2 and formed salt bridge interactions with the tetrazole, which likely bore a negative charge. Unlike with BT2 or fragment 1, the side chain of R171 did not form polar interactions with fragment 2. Instead, its guanidinium group formed a π-stacking interaction with the central phenyl of fragment 2. L128 and H132 also adopted different rotamers upon fragment 2 binding. The tetrazole of fragment 2 also interacted with residues G331/P332 from the kinase domain, and with the side chain of Y346 via an extensive water-mediated hydrogen bond network. Interactions with G331/P332 apparently stabilized peptide 330 to 335. Bound fragment 2 only had small overlap with bound BT2 (and fragment 1) in space.
Fragment 2 bound to site 2 similarly to fragment 1, interacting with residues from both the regulatory and the kinase domains of BCKDK (Fig. 3C). However, the terminal phenyl in the biaryl of fragment 2 displaced F380 in the C-terminal peptide of BCKDK, a residue that interacted with fragment 1 (Fig. 2), resulting in the disorder of the C-terminal peptide of BCKDK. The tetrazole of fragment 2 had more polar interactions at site 2 than fragment 1, picking up an additional salt bridge interaction with the side chain of R71. As described for fragment 1, based on superposition with the PDK3–L2 structure, site 2 for fragment 2 was also next to the lipoyl-binding site in BCKDK for BCKD E2 subunit (Fig. S3).
Because binding of fragment 2 stabilizes the peptide 330 to 335, which is part of the so-called “ATP lid” that interacts with the phosphates of the bound nucleotides in BCKDK (13), we compared the ADP/ATP site structures of fragment 1 and fragment 2 complexes (both with ADP bound) with that of ATPγS-bound apo BCKDK (13) (Fig. 3D). Peptide 330 to 335 was disordered in fragment 1- (and BT2-) bound complexes as well as in the apo BCKDK structures. The more ordered “ATP lid” in the fragment 2 complex clashed with the γ-phosphate of ATPγS. The main-chain NH of F336 interacted with the β-phosphate of bound ADP but not ATP. These interactions favored the binding of ADP over ATP. Interestingly, the bound Mg2+ in ATPγS complex was displaced in fragment 2 complex by the side chain of residue E245 because of the conformational change of the “ATP lid,” in which the H334 side chain rigidified and formed a HB interaction with E245. The ordering of this “ATP lid” was proposed to be the mechanism for ADP trapping that leads to product inhibition of BCKDK (13).
Discovery of potent ARB and ARB-like BCKDK inhibitors
The observation that fragment 2 at the BT2 adjacent site left the hydrophobic pocket of the BT2 site largely unoccupied prompted us to carry out additional virtual screening to find higher molecular weight BCKDK inhibitors with improved potencies. During this ligand- and structure-based search, our internal compound database was queried for compounds that contained the tetrazole substructure attached to phenyl or any other five- or six-membered heterocycle to find substituents that could interact with the hydrophobic BT2 site. Based on this search, 158 virtual screening hits were tested in the biochemical assay, cell assay, and confirmed by SPR. One of the most potent hits, compound 1, demonstrated significantly improved potency compared with the original fragments (Fig. 1).
The chemical structure of compound 1 resembles those of the marketed antihypertension AT1R antagonists, also known as ARBs (30). As our virtual screening identified additional ARBs as potential BCKDK inhibitors, we tested the activities of all ARBs available to us (Table S1). Among them, valsartan and pratosartan are found to be the most active BCKDK inhibitors in the biochemical assay. During the time of our article preparation, Kitaura et al. (31) reported valsartan as a BCKDK inhibitor. In their study, they screened an existing drug library of ∼800 compounds for new potential compounds that inhibit BCKDK in assays using liver mitochondria protein extract and purified BCKD–BCKDK complex, respectively. Candesartan and irbesartan were also shown to inhibit BCKDK, but with potencies weaker than valsartan (31). In our hands, the BCKDK inhibition potencies of valsartan, candesartan, and irbesartan qualitatively agreed with their results and have the same rank order as reported. Despite their potencies in the BCKDK biochemical assay, none of the ARBs tested have an IC50 less than 60 μM in the cell assay (Table S1). This weak cell potency may imply that these ARBs may not display significant BCKDK pharmacology in human clinical practice, although additional work would be needed to inform if this is the case.
We determined the complex structures of both compound 1 and valsartan (Table 1). These two compounds, like fragment 2, bound BCKDK at two sites: an expanded BT2 site, and site 2 (Figs. 4 and S3). The overall structures of BCKDK in these two complexes are very similar to that of the fragment 2 complex, except for the linker peptide between helices α4 and α5. This linker peptide (H132–D134) contains the residue H132, the side chain of which directly interacted with BT2, fragments 1 and 2, and compound 1. In the valsartan complex, the linker peptide was displaced by the chiral branch of valsartan and became partially disordered. The “ATP lid” peptide 330 to 335 had the same conformation as in the fragment 2 complex.
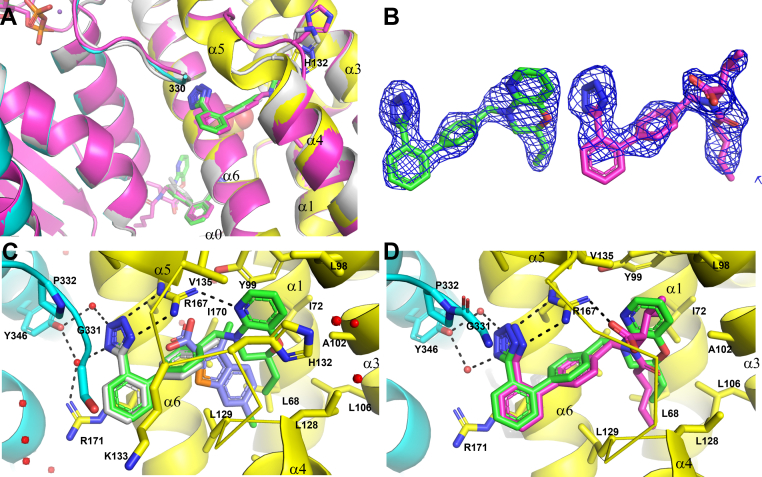
Figure 4.
Compound 1 and valsartan bind to BCKDK at the expanded BT2 site and site 2.A, a zoomed-in view of ligand-binding sites of compound 1, valsartan (main chain ribbon and ligand C atoms colored in magenta), and fragment 2 complexes (colored in light gray). The variable a4–α5 linker peptides containing H132 are highlighted. B, compound 1 and valsartan are shown embedded in their respective initial 2Fo–Fc maps contoured at 1.0 σ. Notice that the electron density is weak for the solvent-exposed edge of the first phenyl. C, compound 1 interactions with the expanded BT2 site. Superimposed are fragment 2 and BT2 (light blue C atoms). D, valsartan binds to the expanded BT2 site. BCKDK, branched-chain ketoacid dehydrogenase kinase; BT2, 3,6-dichlorobenzo[b]thiophene-2-carboxylic acid.
The 4′-methyl biphenyl tetrazole of compound 1 interacted with BCKDK similarly to fragment 2 at both sites, and it almost completely overlapped with fragment 2. The minimal positional shift between these two compounds indicated that the addition of the ethyl morpholino pyridine moiety in compound 1 caused minimal disturbance to its binding to BCKDK. The ethyl morpholino pyridine moiety extended into the BT2 pocket as predicted. Inside the BT2 pocket, the ethyl group made extensive hydrophobic contacts with the side chains of L68, L106, L128, and L129. The aromatic pyridyl ring was sandwiched between the side chains of H132 and Y99, making π-stacking interactions with both. The pyridyl N atom formed a hydrogen bond with the R167 side chain.
Compound 1 is a single diastereomer that was chirally separated from a racemic mixture of two diastereomers (1 and 1∗) with opposite stereochemistry. The absolute stereochemistry of compound 1 is currently undetermined. Compound 1∗ with the inverted chirality had slightly weaker in vitro biochemical activity (32 ± 0.02 nM; racemic: 11 ± 2.5 nM). Modeling showed that both diastereomers could fit into BCKDK, with only subtle differences in the pucker of the morpholine ring (data not shown). We could not deduce the absolute stereochemistry of compound 1 based on the electron density of the bound ligand. At site 2, the ethyl morpholino pyridine of compound 1 was exposed to solvent with minimum contacts with BCKDK and had very weak electron density (Fig. S3).
We also determined the crystal structure of BCKDK–valsartan complex at high resolution. As expected, valsartan bound to BCKDK at the expanded BT2 site (Fig. 4) as well as at site 2 (Fig. S2). The 4′-biphenyl tetrazole of valsartan interacted with BCKDK similarly to fragment 2. At the expanded BT2 site, the side chain of R167 made an additional salt bridge interaction with the carboxylate of valsartan. Relative to compound 1, valsartan moved further into the BT2 pocket, with the largest shift of 0.8 Å between the benzylic N atoms of two molecules. The branched isopropyl group of valsartan disrupted the H132 containing peptide, and the side chain of H132 did not interact with valsartan. The aromatic side chain of Y99 also rotated, unable to make the aromatic π-stacking interaction with valsartan. The disruption of the BT2 site and the loss of favorable interactions may well explain the weaker potency of valsartan, and other ARBs, in binding to BCKDK. Like compound 1, valsartan also binds to site 2 of BCKDK, with the moiety beyond the terminal phenyl group exposed to solvent (Fig. S3).
Mode of action of allosteric BCKDK inhibitors
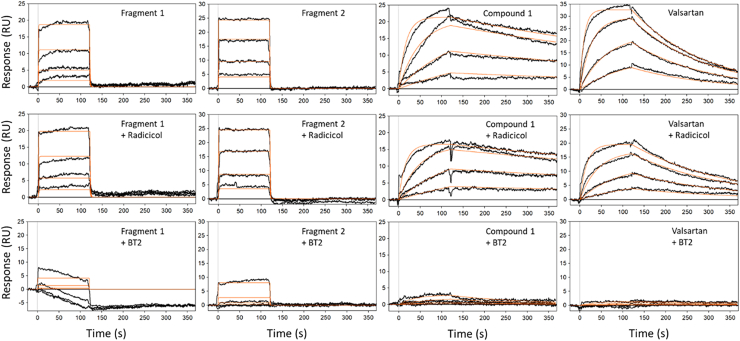
By virtual screening alone, we achieved potency improvement in BCKDK inhibition by more than 5000-fold from the initial fragment hits to compound 1 (Fig. 1). Importantly, we observed potent cellular activity of BCKDK inhibition of compound 1. The strategy we adopted was to optimize binding interactions to the expanded BT2 site. To confirm that this strategy worked as intended and to build confidence in designing novel compounds targeting this site, we further investigated the competitive binding of fragment 1, fragment 2, compound 1, and valsartan to BCKDK, using SPR in the presence of known single-site binders (Fig. 5). For these experiments, we chose 20 μM BT2 and 20 μM radicicol as the BT2 site and the ATP site blockers, respectively. Radicicol is a natural product that is known to be an ATP site binder (47, 48), and it was chosen over ATP because of improved stability. It also lacks the phosphates that may interfere with compound binding through conformation coupling between the “ATP lid” and the ligand-binding sites and may confound interpretations. Radicicol also bound BCKDK tighter than ATP (Kd = 1.47 μM versus 15 μM, respectively).
Figure 5.
Characterization of inhibitors binding to BCKDK by SPR.Top row, SPR sensorgrams of fragment 1, fragment 2, compound 1, and valsartan, respectively, binding to BCKDK superimposed with the equilibrium or kinetic curve fittings. Middle row, compounds binding in the presence of 20 μM ATP site competitor radicicol in the running buffer. Third row, competitive binding of ligands in the presence of 20 μM BT2. The compounds were tested in concentration series made with threefold dilution. The highest concentration tested was dependent on the potency of each compound: for fragment 1, it was 300 μM; for fragment 2, it was 20 μM; for valsartan, it was 2 μM; and for compound 1, it was 20 nM. The binding curves (black) were fit to a 1:1 Langmuir binding model (red) using Scrubber data analysis software. BCKDK, branched-chain ketoacid dehydrogenase kinase; BT2, 3,6-dichlorobenzo[b]thiophene-2-carboxylic acid; SPR, surface plasmon resonance.
As expected from their moderate binding affinities, fragment 1 and fragment 2 bound to BCKDK with diffusion limited fast-on and fast-off kinetics (Fig. 5), and only equilibrium binding constants could be derived from curve fittings to the SPR sensorgrams. On the other hand, consistent with their improved potencies, compound 1 and valsartan bound to BCKDK with measurable off rates, and kinetic fitting parameters could be calculated (Fig. 5). In addition, compound 1 had an off-rate visibly slower than that of valsartan, again agreeing with the functional potency differences observed. In the presence of 20 μM radicicol, the ATP site binder, all fragments, and compounds tested showed binding affinities similar to those in the absence of radicicol. When 20 μM BT2 was present in the SPR running buffer, the SPR-binding signals of all these ligands were significantly suppressed, and only fragment 2 and compound 1 at their highest concentrations showed some residual binding signals. Binding to other sites might happen at higher compound concentrations, but to distinguish these binding events from the residual binding at the BT2 site would be difficult. These results indicate that fragments 1 and 2 derive their binding affinity mostly from interactions with the allosteric BT2/BT2 adjacent site, and the large improvements in binding affinities of compound 1 and valsartan over these fragments are from additional interactions with the expanded BT2 sites.
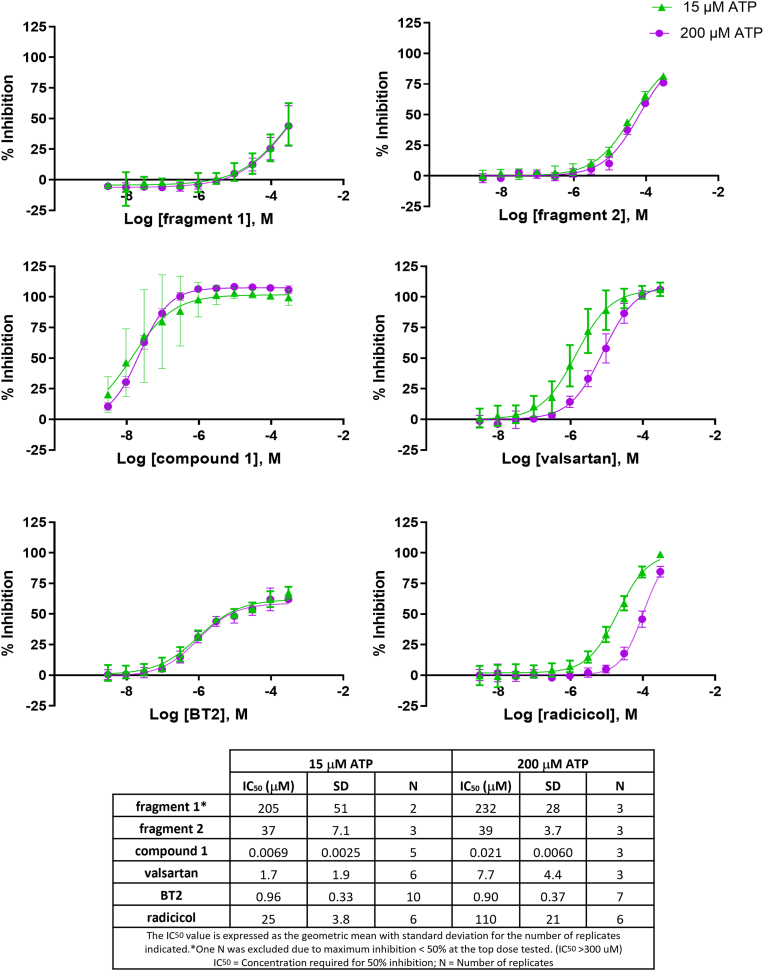
Since we observed conformational coupling between the ATP site and the 4′-biphenyl tetrazole site through rigidification of the “ATP lid” peptide (Fig. 3), we also investigated the influence of ATP concentrations on BCKDK inhibition by ligands. We chose 15 μM, close to the Km of ATP determined in our assay format (13.57 ± 0.32 μM) as the low ATP concentration, and 200 μM as the highest ATP concentration at which ATP fluorescence has minimum interference with detection. Fragment 1 is a weak inhibitor of BCKDK with both 15 and 200 μM ATP generating IC50 values of 205 ± 51 μM and 232 ± 28 μM, respectively (Fig. 6). The BCKDK inhibition curves of fragment 1 at 15 μM and 200 μM ATP overlap very well, indicating no significant differences between inhibition at these two ATP concentrations. The BCKDK inhibition IC50s of BT2 are essentially the same at 15 and 200 μM ATP (0.96 ± 0.33 μM versus 0.90 ± 0.37 μM, t test p = 0.73). The IC50 of fragment 2 at 15 μM ATP concentration is nearly identical to that at 200 μM (37.3 ± 7.1 μM versus 38.7 ± 3.7 μM, p = 0.78). Compound 1 inhibits BCKDK potently at both ATP concentrations. The IC50 of compound 1 in inhibiting BCKDK is significantly lower at 15 μM ATP than that at 200 μM ATP (0.0069 ± 0.0025 μM versus 0.021 ± 0.0060 μM, p = 0.003). The BCKDK inhibition IC50s of valsartan are also significantly different at 15 and 200 μM ATP (1.7 ± 1.9 μM versus 7.7 ± 4.4 μM, p = 0.02). As the positive control of a true ATP site binder, the BCKDK inhibition IC50 of radicicol at 15 μM ATP is significantly lower than that at 200 μM ATP (25.0 ± 3.8 μM versus 110 ± 21 μM, p < 0.001).
Figure 6.
BCKDK inhibition curves of compounds tested at 15 and 200 μM ATP concentrations.Curves are representative of all replicates averaged together. Error bars indicate ± standard deviation at each concentration. (Some may not be visible when they are small). Inhibition curves of BT2 (BT2 site binder) and radicicol (ATP site binder) are used as positive controls. BCKDK, branched-chain ketoacid dehydrogenase kinase; BT2, 3,6-dichlorobenzo[b]thiophene-2-carboxylic acid.
Interestingly, consistent with their apparent ATP competitive inhibition characteristics, compound 1 and valsartan achieve maximum BCKDK inhibition of 100% in the biochemical assay, like the true ATP competitive inhibitor radicicol (Fig. 6). On the other hand, inhibition of BCKDK by BT2 plateaus at ∼75%. This indicates a noncompetitive or uncompetitive mechanism of BT2. BCKDK inhibition of fragments 1 and 2 did not plateau at the highest concentration used (300 μM).
Combining SPR competitive binding results and BCKDK inhibition at different ATP concentrations, we can draw the conclusions that, although all compounds studied inhibit BCKDK through an allosteric mechanism by binding to the BT2/BT2 adjacent site/expanded BT2 site, compounds binding to the expanded BT2 site are ATP competitive. At the expanded BT2 site, the 4′-biphenyl tetrazole of compound 1 and valsartan rigidify the “ATP lid,” which disfavors ATP binding. Without the anchoring from BT2 site interactions, fragment 2 may bind to BCKDK more flexibly in solution and may not rigidify the ATP lid to the same extent as compound 1 and valsartan. This also explains why valsartan, which clearly does not bind to the ATP site, was reported to be an ATP-competitive BCKDK inhibitor (49). The ATP competitive nature of compound 1 and valsartan may also partly explain the large IC50 shift when tested in cells. ATP concentrations in cells (2–8 mM), especially in mitochondria, the site of BCKDK action, are much higher than the concentration used in our biochemical assay (50). There may also be differences in compound distribution that contribute to the disconnect between the in vitro and cellular potencies of compound 1 and valsartan.
Inhibition selectivity against PDK1–4
Because the structure of BCKDK is highly similar to those of PDK1–4 (13, 14, 15, 16, 17) and ligand sites homologous exist between BCKDK and PDK1–4 (Figs. 2 and S1–S3), we tested the inhibition selectivity of the compounds studied against the biochemical activities of PDK1–4. The activities of PDK1–4 were measured using fluorescence signals because of the phosphorylation of the artificial substrates of these kinases. The inhibitors are highly selective against PDK1–4 (Table S2). Only BT2 and valsartan showed weak activities, with an IC50 of 79 μM for BT2 against PDK1, and 78 μM for valsartan against PDK4, respectively. The IC50s of other BCKDK inhibitors for PDK1–4 were all greater than 100 μM, the highest inhibitor concentration tested.
Structure basis of crossactivity of ARBs and ARB-like compounds
ARBs are blockers of AT1R, which is the primary blood pressure regulator (51, 52, 53). AT1R is a G protein–coupled receptor on the cell surface, which is quite different from BCKDK, a soluble mitochondrial enzyme. It is interesting to explore the structural basis for the crossactivity between BCKDK inhibition and AT1R blocking. There are two AT1R complex crystal structures published (54, 55). We superimposed compound 1 in BCKDK complex with olmesartan acid bound in AT1R complex (PDB ID: 4ZUD) and observed both similarities and differences in their binding modes in the corresponding protein ligand-binding sites (Fig. S4). Olmesartan acid, the active species of olmesartan, is a potent AT1R blocker with Kd = 6.9 nM (54) but a weak BCKDK inhibitor with an in vitro IC50 of 43.7 μM (Table S1). In both BCKDK and AT1R ligand-binding sites, there is a key arginine, R167, the side chain of which makes essential polar interactions with the tetrazole on one side and a polar group on the other side of these two compounds. In BCKDK complexes, R167 makes the essential salt bridges with the carboxylate of BT2 or the tetrazoles of our BCKDK inhibitors. It also makes a hydrogen bond with the pyridyl N atom of compound 1 and the carboxylate of valsartan. It was reported that the R167A mutation in BCKDK completely abolishes ligand binding to the mutant BCKDK proteins (32). In the AT1R complex, R167 formed extensive networks of hydrogen bonds and salt bridges with the acidic tetrazole ring and the carboxyl group on the imidazole moiety of olmesartan acid. Mutations of R167 to alanine and glutamine in AT1R abolished both the angiotensin II and olmesartan acid binding to AT1R (54, 55).
Discussion
The tetrazole-bearing inhibitors studied here bound BCKDK at multiple sites. Although our study clearly indicated that for the inhibitors we investigated, the (expanded) BT2 site accounted for most of the BCKDK binding affinities, site 2 and Pfz3 sites could not be deemed unimportant. Further structure-based design focusing on site 2 and the Pfz3 site may lead to compounds with improved binding. For example, the biphenyl tetrazole 4′ substituents of compound 1 and valsartan contribute little to their binding at site 2, leaving space at the 3′ or 2′ position unexplored (Fig. S2). This empty space may provide additional affinity if occupied. Indeed, AZ3 and Nov3r are reported to be potent PDK2 and PDK1 inhibitors with nM potencies (56). Pfz3 was also reported to have 2 μM potency in inhibiting PDK2 (15, 57). Contrary to BCKDK, which has a larger BT2 site, the equivalent site in PDK is very small (15), and no known potent PDK inhibitors can bind there. Fragment screening against PDKs tended to identify other site binders (58, 59).
Based on our structure and competitive binding studies, valsartan is an allosteric inhibitor that does not directly compete with ATP for binding to BCKDK (Fig. 5). However, valsartan was reported to be an ATP competitive inhibitor of BCKDK, with a Ki of 1.36 μM (49), which is close to the IC50 we obtained at 15 μM ATP. Indeed, we also observed increases in the in vitro IC50s of compound 1 and valsartan at higher ATP concentrations (Fig. 5). The much more pronounced drop in cellular BCKDK inhibition by compound 1 and valsartan compared with BT2 may be partly related to high ATP concentration in cells, especially in the mitochondria (50). The structural basis for these discrepancies is the observed differences in the conformation of the “ATP lid” upon inhibitor binding (Fig. 3D). In fragment 2, compound 1 and valsartan complexes, the “ATP lid” is rigidified by interactions with the tetrazole of these ligands (Figs. 3B and 4, A and B). This rigidification of the “ATP lid” disfavors ATP binding and shows an apparent competition between these ligands and ATP. This rigidification of the “ATP lid” is much less apparent with fragment 1 and BT2 binding (Fig. 2B).
Besides the proposed inhibition mechanism through allosterically slowing the rate of the kinase reaction product ADP leaving the BCKDK ATP-binding site, KIC, BT2, and valsartan were also reported to be able to inhibit BCKDK through a mechanism of promoting the dissociation of BCKDK from the BCKD E2 subunit. This dissociation suppresses the E2-stimulated BCKDK activity (32, 41, 49). Dissociation of BCKDK from the E2 complex also reportedly leads to BCKDK degradation in different tissues (41). We could not reliably measure the binding of BCKDK with the E2 subunit with the reagents we had, using either SPR or isothermal titration calorimetry methods. Further work will be needed to determine if fragments 1 and 2, and compound 1, also promote the dissociation of BCKDK from the E2 subunit, and if this dissociation leads to the degradation of BCKDK in cells or in vivo.
Because AT1R is a primary blood pressure regulator, ARBs or AT1R blockers have been widely used in clinical settings as antihypertensive drugs. Most ARBs share the chemical scaffold of 4′-biphenyl tetrazole (or 4′-biphenyl carboxylic acid, Table S1), although even minor variations can lead to distinct therapeutic efficacies toward cardiovascular etiologies (53, 60, 61). Several ARBs, including candesartan, losartan, and valsartan, were approved for the treatment of HF (62). In clinical trials, candesartan, valsartan, and losartan significantly decrease the all-cause mortality rate and composite mortality and morbidity risk in HF patients (63, 64). One of the potential indications for BCKDK inhibitors is HF as well (42). From a HF treatment point of view and from our studies, it seems that ARB-like BCKDK inhibitors may have three possible different pharmacological profiles: selective BCKDK inhibitors without AT1R blocking activities, dual BCKDK inhibitors/AT1R blockers (such as valsartan), or AT1R blockers only (e.g., telmisartan). A complicating factor with dual profile inhibitors is the balance of the activity of BCKDK inhibition and AT1R blocking. For instance, valsartan has strong AT1R blocker activity, but its cellular BCKDK inhibition activity is weak. Consistent with our results, the in vitro IC50 of valsartan to inhibit the BCKD–BCKDK complex purified from rat liver was reported to be 37.3 μM at 500 μM ATP (49). On the other hand, valsartan administration was reported to increase hepatic BCKD activity and decrease plasma BCAA concentrations in rats, but no activation of BCKD by valsartan was observed in rat heart and kidney (49). The induction of rat hepatic BCKDK activity by valsartan could be due to active uptake of the compound in rat liver, which has a higher uptake rate than human liver (65). On the other hand, BT2 was able to activate BCKD in all the tissues inspected, including liver, kidney, muscle, and heart (32, 41). BCAA oxidation involves an extensive interplay between muscle and liver. In skeletal muscle, branched-chain aminotransferase activity is high. The reverse situation exists in the liver, with both cytosolic and mitochondrial aminotransferase activities undetectable in the rat liver. Further screening and testing may identify ARB-like compounds that block AT1R strongly and inhibit BCKDK in all tissues. Such dual inhibitors may activate BCKD in all tissues in situ. Administration of compounds with different BCKDK and AT1R inhibition profiles may provide clues on their therapeutic potential to treat HF. In doing so, the potential neuronal risks of BCAA deficiencies associated with BCKDK inhibition need monitoring, especially if these compounds can cross brain–blood barrier.
Experimental procedures
Chemicals
Fragment 1, fragment 2, the racemic precursor of compound 1, and valsartan are from corporate internal compound collections. Radicicol was purchased from Sigma. Dimethyl sulfoxide (DMSO) was purchased from EMD Millipore.
Protein reagents for biochemical and SPR assay
BCKDK protein was generated using a pET vector containing from N to C terminus: 6xHis, maltose-binding protein (MBP), a tobacco etch virus (TEV) protease site, a biotin acceptor peptide (GLNDIFEAQKIEWHE), and human BCKDK (residues 31–412 preprocessing or 1–382 postprocessing, NP_005872.2). In the paper, number is based on postprocessing numbering. Protein was coexpressed with GroEL–GroES in BL21(DE3) Escherichia coli in LB media and induced with 0.5 mM IPTG and 0.5 mg/ml l-arabinose for 16 h at 26 °C. Bacteria were lysed using a microfluidizer in 100 mM potassium phosphate (pH 7.5), 500 mM NaCl, 0.1 mM EDTA, 1% Tween-20, 0.25% Triton X-100, 10% glycerol, 1 mM DTT, and protease inhibitors. MBP-tagged protein was purified by affinity chromatography using amylose resin, and MBP was removed from BCKDK with overnight TEV protease incubation in the presence of 300 mM l-arginine followed by gel filtration chromatography in 50 mM Hepes (pH 7.5), 500 mM NaCl, 300 mM l-arginine, 2 mM MgCl2, 1 mM DTT, and 10% glycerol. For biotinylation of BCKDK for SPR assay, protein was incubated with BirA and D-biotin (BirA500; Avidity) during TEV protease treatment. This biotinylated protein was used as stock for NMR fragments screen.
For the biochemical assay, an artificial substrate consisting of a fusion protein of the E1α subunit and the lipoyl-binding domain of the E2 subunit of BCKD was used (41). The protein was expressed using a pET vector containing from N to C terminus: the lipoyl-binding domain of E2 (residues 62–160 preprocessing, NP_001909.4), a TEV protease site, residues 331 to 345 (preprocessing, NP_000700.1) from E1α, and 6xHis. The fusion substrate was expressed in BL21(DE3) E. coli in LB media and induced with 0.75 mM IPTG for 16 h at 30 °C. Bacteria were lysed using a microfluidizer in 50 mM sodium phosphate (pH 7.5), 350 mM NaCl, 10 mM imidazole, 10% glycerol, 1 mM DTT, and protease inhibitors. Fusion substrate was purified by nickel–nitrilotriacetic acid affinity chromatography followed by S75 gel filtration chromatography in 50 mM sodium phosphate (pH 7.5), 350 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, and 10% glycerol.
E. coli LplA used in lipolyation of the fusion protein substrate was expressed using a pET vector in BL21(DE3), and protein production was induced with 0.75 mM IPTG and grown for 16 h at 30 °C. Bacteria were lysed using a microfluidizer in 50 mM sodium phosphate buffer (pH 7.5), 350 mM NaCl, 1.5 mM MgCl2, and 1 mM DTT. LplA protein was precipitated from clarified lysate with 1 M ammonium sulfate and further purified by gel filtration chromatography of the same lysis buffer with 10% glycerol.
For lipoylation of the fusion substrate, purified protein was incubated with LplA at a 10:1 (substrate:LplA) ratio in 20 mM sodium phosphate (pH 7.4), 6 mM MgCl2, 4 mM ATP, 2 mM DTT, 3 mM dl-6,8-thioctic acid at 37 °C. The reaction was monitored using an Agilent 6530 Q-TOF coupled to an Agilent 1290 UPLC. The final lipoylated fusion substrate was purified by gel filtration chromatography in 50 mM Hepes (pH 7.5), 350 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, and 10% glycerol.
NMR fragment screening
To identify new chemical matter, 1D 1H NMR saturation transfer difference (NMR-STD) method was used to screen BCKDK against a proprietary library consisting of 3065 fragments. The screening samples contained 2 μM protein that was prepared from 80 μM BCKDK stock diluted 40× in 10 mM deuterated Hepes, pH 7.5, 150 μM NaCl, and 2 mM MgCl2 in D2O. A positive control, radicicol, was used at 250 μM to optimize the experimental parameters. Fragment library compounds were tested as mixtures of 10 or 12 fragments at a concentration of 250 μM. The NMR-STD data were collected at 298 K on a Bruker AVANCE spectrometer (Bruker) operating at a 1H Larmor frequency of 600 MHz, which was equipped with a 1.7 mm TCI micro-cryoprobe and a SampleJet automated sample handler (Bruker). Five hundred twelve scans with a data size of 8192 complex points were recorded per NMR-STD experiment, which needed about 30 min of acquisition time. On- and off-resonance saturation spectra were acquired in an interleaved fashion, and the difference spectra were generated using phase cycling. Gaussian bell-shaped pulses were used for saturation of the protein. The NMR data were processed and analyzed to identify hits using TopSpin 3.2 software (Bruker).
Biochemical assay
Compounds tested were solubilized in 100% DMSO to 30 mM. Compound stocks were then serially diluted by half log in 100% DMSO to create 11 concentrations ranging from 30 mM to 300 nM. Compound serial dilutions were spotted in duplicate into Corning 3824 assay plates as 0.1 μl spots. The final concentration of compounds in the assay was 300 μM to 3 nM with the final DMSO concentration at 1%. Initially, 5 μl of a 2× BCKDK mix in assay buffer (20 mM Tris–HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 0.02% [v/v] Tween-20, and 0.01% bovine serum albumin [BSA]) was added to the compound plates and preincubated for 15 min at room temperature. Final enzyme concentration (5 nM) was optimized to reflect the linear range of enzyme activity with less than 30% substrate conversion. This was followed by addition of 5 μl of 2× fusion substrate/ATP mix in assay buffer to initiate the reaction. Plates were covered with a lid and allowed to incubate at room temperature for 90 min. Final concentration of the fusion substrate was 50 nM, and final concentration of ATP was either 15 μM or 200 μM.
Following incubation, the enzyme reactions were stopped by adding 5 μl of 4× stop/primary antibody solution containing 100 mM (final) EDTA, 2 nM (final) anti-His Eu antibody (PerkinElmer), and 1:1000 anti-pE1 antibody (Cell Signaling Technology) in assay buffer. The plates were covered and incubated at room temperature for 30 min. Finally, 5 μl of 4× detection reagent consisting of 20 nM (final) anti-rabbit Ulight antibody (PerkinElmer) and 1× LANCE detection buffer (PerkinElmer) in deionized H2O was then added. The plates were centrifuged for 30 s at 1000 rpm. After a 60 min incubation, the plates were read on an EnVision MultiReader (Thermo Fisher Scientific) in time-resolved FRET mode (excitation 320 nm: emission: 665 nm).
The raw data from the EnVision MultiReader was expressed as a ratio and analyzed using proprietary software (ActivityBase, IDBS). The percent effect at each concentration of compound was calculated relative to the values for the uninhibited control wells (100% DMSO) and fully inhibited control wells (600 μM radicicol) on each assay plate. IC50 values were determined from the percent effect data using a four-parameter logistic dose–response model (ActivityBase).
PDK1–4 biochemical assays
The PDK1–4 enzymatic assays were run at AssayQuant (Marlboro). Compounds tested were solubilized in 100% DMSO to 30 mM. Compound stocks were then serially diluted by threefold in 100% DMSO to create 11 concentrations ranging from 5 mM to 85 nM. About 0.5 μl 50× inhibitor in 100% DMSO compound serial dilutions were added in duplicate into low-volume 384-well (catalog no.: 3824) plates. The final concentrations of compounds in the assay were 100 μM to 1.7 nM with the final DMSO concentration at 2%. About 19.5 μl reaction mix buffer containing substrates, ATP, and DTT were added to the wells, and the plates were preincubated at 30 °C for 30 min after sealing using optically clear adhesive film (TopSealA-Plus plate seal; PerkinElmer [catalog no.: 6050185]). The final ATP concentrations were at the Km of enzymes: PDK1, 6.0 μM; PDK2, 9.3 μM; PDK3, 4.8 μM; and PDK4, 97 μM. The substrates were 15 μM AQT0895 for PDK1, AQT0896 for PDK2 and PDK3, and AQT0872 for PDK4, respectively. After preincubation, 5.0 μl enzyme dilution buffer or 5× PDK kinases in enzyme dilution buffer were added to the reaction mixture to start the reaction. The enzyme dilution buffer contained 20 mM Hepes, pH 7.5, 0.01% Brij-35, 0.1 mM EGTA, 5% glycerol, 1.0 mg/ml BSA, and 1.0 mM DTT. Reaction was run at 30 °C for 240 min after sealing. The final enzyme concentrations were 10 nM PDK1 (amino acids 1–436 N-term glutathione-S-transferase [GST], SignalChem [catalog no.: and Lot no.: P57-10G/N221-2]), 40 nM PDK2 (amino acids 1–407 N-term GST; SignalChem [catalog and Lot nos.: P57-10G/N221-2]), 10 nM PDK3 [amino acids 1–406 N-term GST, SignalChem [catalog & Lot nos.: P57-10G/N221-2], and 10 nM PDK4 (amino acids 1–411 N-term GST, SignalChem [catalog & Lot nos.: P57-10G/N221-2], respectively). The final reaction buffer contained 54 mM Hepes, pH 7.5, ATP, substrates, enzymes, 1.2 mM DTT, 0.012% Brij-35, 0.52 mM EGTA, 1% glycerol, 0.2 mg/ml BSA, and 10 mM MgCl2.
Plates were read at a Synergy Neo2 Multiple Microplate Reader (BioTek) in kinetic mode (readings taken every 1 or 2 min), monitoring fluorescence intensity from the top using filters (excitation: 360 nm: emission: 485 nm, bandwidth of 40 and 20 nm, respectively), and a gain of 80. Data were processed using GraphPad Prism (GraphPad Software, Inc) following subtraction of the background fluorescence (fluorescence values obtained with all reaction components except enzyme) from total fluorescence at each time point for duplicate samples to determine corrected signal, which was plotted versus time. The slope of the initial linear region of resulting progress curve was used to determine the reaction rate (initial velocity) for each condition. IC50 values were determined from the percent effect data using a four-parameter logistic dose–response model (ActivityBase).
SPR assay
The binding affinities and kinetics of binding were measured using SPR-based binding assay detecting the reflection index changes at the interface when inhibitors in solution-bound BCKDK immobilized on the metal surface of SPR chips (66). These experiments were carried out on Biacore B3000 (Global Life Sciences Solutions) and Sierra Sensors (now Bruker) MASS-1 instruments. There was no significant difference in results obtained on both these instruments. Biotinylated bap-tagged BCKDK protein was captured on a streptavidin-coated sensor chip to achieve about 4000 to 4500 RUs of surface density. All the samples were prepared in buffer consisting of 10 mM Hepes, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 0.005% Tween-20, and 2% DMSO. The same buffer was used as the running buffer during the experiments. Compound samples were injected at a flow rate of 30 μl/min for 120 s of association time followed by at least 300 s of dissociation period. The compounds were tested in a concentration series consisting of at least six samples made with twofold dilution. The highest concentration was selected based on compound potency in the biochemical assay or binding affinity observed in a previous experiment. Multiple blank injections were run before and after each compound series to allow double reference subtraction during data processing and analysis. Radicicol was tested in every experiment as a positive control to assess activity of the captured protein on the surface. A DMSO curve was run during each experiment to properly correct for excluded volume. The data were processed and analyzed using Bruker Analyzer, Scrubber, and Biaeval software to calculate binding affinities and kinetics by fitting the data to 1:1 binding model.
Competition binding experiments were carried out to determine binding site specificity for compounds. The same concentration series for compounds was tested either in the presence of 20 μM radicicol (known ATP-site binder) or in the presence of 20 μM BT2 (known single-site allosteric binder) in the running buffer. Significant reduction in signal intensity was interpreted as compound competing for the same binding site as radicicol or BT2.
Cell assay
BCKDK activity and inhibition in human skeletal muscle cells was monitored by measuring BCKDK-dependent phosphorylation of endogenous BCKD using a customized AlphaLISA SureFire Ultra assay system. Prior to the assay, the total BCKD antibody (Bethyl Laboratories; catalog no.: A303-790A) was biotinylated using the ChromaLink One-Shot Antibody Biotinylation Kit (Trilink Technologies; catalog no.: B-9007-009K) and a phospho (S293) BCKDHA antibody (Bethyl Laboratories; catalog no.: A304-672A) was CaptSure tagged using the Lightning-Link CaptSure Conjugation Kit (TGR BioSciences; catalog no.: 6300007). Human skeletal myocytes (Gibco; catalog no.: A11440) were previously immortalized via the introduction of human telomerase reverse transcriptase into the cells. Immortalized human skeletal muscle cells were plated at 15,000 cells/well and grown in skeletal muscle growth media containing the media supplement and chick embryo extract (Promocell C-23060 and C-23160; MP92850145) in 384-well CulturPlate plates. After overnight incubation, media were removed, and compound treatments were performed in growth media diluted 10× in PBS for 60 min. Cells were washed with PBS and lysed in 1× lysis buffer (Cell Signaling; catalog no.: 9803) containing 2 nM biotinylated total BCKD antibody and 1× protease/phosphatase inhibitor cocktail (Cell Signaling; catalog no.: 5872). After an hour of incubation at room temperature, a 1× immunoassay buffer (PerkinElmer; catalog no.: AL000F) mix containing 1:400 CaptSure-tagged Phospho BCKD antibody and 40 μg/μl Anti CaptSure acceptor beads (PerkinElmer ALSU-ACAB) was added to the lysates and allowed to incubate for 60 min. The signal was then generated by the addition of streptavidin donor beads (PerkinElmer ALSU-ASDB) at 40 μg/μl in 1× immunoassay buffer and incubating at room temperature for an additional 30 min. The phospho BCKD signal was developed using EnVision multilabel reader (PerkinElmer) using Alphascreen settings (excitation: 680 nm; emission: 570 nm). The fluorescence emission was used to calculate the percent of effect relative to the HPE (30 μM BT2) and ZPE (DMSO). IC50 curves were generated using ActivityBase software.
Chiral separation of compound 1
About 100 mg of 4-((2'-(1H-tetrazol-5-yl)-[1,1′-biphenyl]-4-yl)methyl)-2-ethyl-3,4-dihydro-2H-pyrido[3,2-b][1,4]oxazine (racemic mixture) was purified using supercritical fluid chromatography (column: Chiral Technologies IG 30 × 250 mm, 5 μm; mobile phase A: liquid CO2; mobile phase B: methanol w/0.2% [v/v] ammonia [7 N in methanol]). Gradient 70% A isocratic. UV wavelength: 210 nm, flow rate: 80 ml/min, back pressure 120 bar, affording the separated diastereomers (R)-4-((2'-(1H-tetrazol-5-yl)-[1,1′-biphenyl]-4-yl)methyl)-2-ethyl-3,4-dihydro-2H-pyrido[3,2-b][1,4]oxazine and (S)-4-((2'-(1H-tetrazol-5-yl)-[1,1′-biphenyl]-4-yl)methyl)-2-ethyl-3,4-dihydro-2H-pyrido[3,2-b][1,4]oxazine. Absolute stereochemistry was not determined; the first-eluting diastereomer was designated as compound 1, and the second-eluting diastereomer was designated as compound 1∗. For each isomer, the residue from chromatography was taken up in a mixture of dichloromethane and heptanes and concentrated to afford white solids.
1—Yield: 35.4 mg, 89 μmol, 35.4%. 1H NMR (400 MHz, methanol-d4) δ 7.59 to 7.72 (m, 3H), 7.51 to 7.59 (m, 2H), 7.21 to 7.28 (m, 2H), 7.06 to 7.14 (m, 2H), 7.02 (dd, J = 1.37, 7.61 Hz, 1H), 6.58 (dd, J = 5.27, 7.61 Hz, 1H), 4.80 (ABq, J = 17.10 Hz, 2H [presumed—partially obscured by solvent peak]), 3.92 to 4.04 (m, 1H), 3.36 to 3.45 (m, 1H), 3.17 (dd, J = 8.00, 12.29 Hz, 1H), 1.54 to 1.76 (m, 2H), and 1.02 (t, J = 7.61 Hz, 3H). LCMS m/z 397.4 [M-H]+ retention time: 6.66 min (Analytical conditions; column: Chiral Technologies IG, 4.6 × 250 mm, 5 μm; mobile phase A: liquid CO2; mobile phase B: methanol w/0.2% [v/v] ammonia [7 N in methanol]; gradient: 5 to 60% B over 9 min; back pressure: 120 bar; UV wavelength: 210 nm; flow rate: 3.0 ml/min).
1∗–Yield: 32.4 mg, 81 μmol, 32.4%. 1H NMR (400 MHz, methanol-d4) δ 7.59 to 7.69 (m, 3H), 7.50 to 7.57 (m, 2H), 7.19 to 7.26 (m, 2H), 7.05 to 7.13 (m, 2H), 7.00 (dd, J = 1.37, 7.61 Hz, 1H), 6.57 (dd, J = 5.27, 7.61 Hz, 1H), 4.80 (ABq, J = 15.60 Hz, 2H [presumed-partially obscured by solvent peak]), 3.90 to 4.03 (m, 1H), 3.35 to 3.42 (m, 1H), 3.15 (dd, J = 8.00, 12.29 Hz, 1H), 1.53 to 1.74 (m, 2H), and 1.02 (t, J = 7.61 Hz, 3H). LCMS m/z 397.4 [M-H]+. Retention time: 7.00 min (analytical conditions identical to those used for 1).
Protein purification and crystallization
Based on previously published BCKDK purification strategies (13, 67), an MBP-TEV-ratBCKDK-His construct was coexpressed with GroEL/ES (pGro7) in E. coli BL21 (DE3) cells. Harvested cells were resuspended in 100 mM potassium phosphate, 1 mM Tris(2-carboxyethyl)phosphine (TCEP), 500 mM KCl, 0.1 mM EDTA, 10% glycerol, 1% Tween-20, 0.25% Triton X-100, pH 7.5 supplemented with Roche EDTA-free protease inhibitor tablets and benzonase. After lysis by Microfluidizer (Microfluidics), cellular debris was pelleted by centrifugation at 40,000g. The supernatant was applied to a 14 ml column of Amylose Resin High Flow (New England Biolabs), washed with 50 mM Hepes, 1 mM TCEP, 2 mM MgCl2, 250 mM KCl, 300 mM arginine, 10% glycerol, pH 7.5 and eluted with buffer containing 10 mM maltose. Because of phosphorylation of the protein at the TEV cleavage site, the pooled protein was dephosphorylated with lambda phosphatase (NEB) while simultaneously removing the MBP by TEV protease. Cleaved, dephosphorylated BCKDK was applied to a Superdex 200 column (Cytiva Life Sciences) equilibrated in 50 mM Hepes, 1 mM TCEP, 2 mM MgCl2, 1 M NaCl, 250 mM KCl, 300 mM arginine, 10% glycerol, pH 7.5. Fractions containing BCKDK were pooled and concentrated to at least 30 mg/ml.
Crystals of dephosphorylated rat BCKDK diffracted poorly. Rat BCKDK protein (20 mg/ml) was incubated overnight with 25 mM ATP and 65 mM MgCl2; autophosphorylation was verified by mass spectrometry. Crystals of rat BCKDK were grown by sitting drop vapor diffusion by mixing equal volumes of phosphorylated BCKDK and a well solution of 100 mM sodium citrate (pH 5.6), 800 mM lithium sulfate, and 750 mM ammonium sulfate. Crystals were soaked overnight in 100 mM sodium citrate (pH 5.6), 500 mM ammonium sulfate, 1400 mM lithium sulfate, 5% DMSO, and 1.5 mM fragment 1 and flash frozen in liquid nitrogen.
To generate crystals with fragment 2, rat BCKDK was purified as described previously, with an additional size exclusion chromatography step after autophosphorylation and stored in frozen aliquots. Rat BCKDK (20 mg/ml) was then cocrystallized with 25 mM ADP against a grid of 100 mM Bis–Tris (pH 5.6 to 6.5), 1 M lithium sulfate, and 400 to 840 mM ammonium sulfate. Crystals were soaked in 100 mM Bis–Tris (pH 5.6), 500 mM ammonium sulfate, 1300 mM lithium sulfate, 50 mM KCl, 50 mM MgCl2, 5% DMSO, and 0.3 mM fragment 2.
Subsequently, a new rat BCKDK-his construct was expressed as described previously. Harvested cells were resuspended and lysed in 100 mM potassium phosphate, 1 mM TCEP, 500 mM KCl, 10 mM imidazole, 10% glycerol, 1% Tween-20, 0.25% Triton X-100, pH 7.5. Clarified lysate was applied to a HisTrap FF (Cytiva) column, washed with 50 mM Hepes, 1 mM TCEP, 2 mM MgCl2, 10 mM imidazole, 250 mM KCl, 300 mM arginine, 10% glycerol, pH 7.5, and eluted with buffer containing 250 mM imidazole. BCKDK containing fractions were concentrated and purified on a Superdex 200 column as described previously. This BCKDK protein was crystallized as described previously. Crystals were soaked in 100 mM Bis–Tris (pH 5.7), 500 mM ammonium sulfate, 1250 mM lithium sulfate, 50 mM KCl, 50 mM MgCl2, 10% DMSO, and 3 mM of either compound 1 or valsartan.
Crystallographic data collection, structure determinations, and refinements
Crystal data sets were collected at beamline 17-ID at IMCA CAT, APS (Argonne National Laboratory). The wavelength used was 1 Å for all crystals. Crystals were kept at 100 K during data collection using cryo stream. Data were processed using the program autoPROC (68). Data sets were scaled and merged using anisotropic resolution cutoff, and ellipsoidal data completeness were reported (Table 1).
The initial BCKDK–fragment 1 complex structure was solved with molecular replacement method using the published BCKDK/BT2 crystal structure as model (PDB ID: 4E01), using the program Phaser (69). Subsequent complex structures were solved using rigid body refinements. Structure refinements were carried out using the program Buster (70) and manual model building using the program COOT (71). Final refinement statistics are listed in Table 1. For the preparation of structural figures in this article, the graphic program PyMol was used (72).
Data availability
The atomic coordinates and structure factors (PDB codes: 8EGD, 8EGF, 8EGQ, and 8EGU) have been deposited in the PDB, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Elizabeth Dushin and Noelle Ridgway for technical assistance in protein purification, David R. Derksen for establishing the initial biochemical assay, and Jane Withka for guidance of NMR fragment screening.
Author contributions
S. L. and K. J. F. conceptualization; S. L., B. L. K., J. D. K., P. V. S., A. R., R. F. S., A. R. R., J. W., and K. O. methodology; S. L., B. L. K., P. V. S., and A. R. formal analysis; J. D. K., P. V. S., A. R., R. F. S., A. R. R., X. W., L. M. B., M. R. R., S. K. B., and K. O. investigation; S. L. writing–original draft; B. L. K., R. F. S., R. J. R. F., and K. J. F. writing–review & editing; S. L., P. V. S., and A. R. visualization.
Reviewed by members of the JBC Editorial Board. Edited by Karen Fleming
Footnotes
Present address for Kiyoyuki Omoto: Neomorph, Inc, San Diego, CA 92121.
Contributor Information
Shenping Liu, Email: Shenping.liu@pfizer.com.
Kevin J. Filipski, Email: Kevin.Filipski@pfizer.com.
Supporting information
References
- 1.Reed L.J., Damuni Z., Merryfield M.L. In: Current Topics in Cellular Regulation. Shaltiel S., Chock P.B., editors. Academic Press; Cambridge, MA: 1985. Regulation of mammalian pyruvate and branched-chain α-keto acid dehydrogenase complexes by phosphorylation-dephosphorylation; pp. 41–49. [DOI] [PubMed] [Google Scholar]
- 2.Perham R.N. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry. 1991;30:8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- 3.Mattevi A., Obmolova G., Schulze E., Kalk K.H., Westphal A.H., de Kok A., et al. Atomic structure of the cubic core of the pyruvate dehydrogenase multienzyme complex. Science. 1992;255:1544–1550. doi: 10.1126/science.1549782. [DOI] [PubMed] [Google Scholar]
- 4.Aevarsson A., Chuang J.L., Max Wynn R., Turley S., Chuang D.T., Hol W.G.J. Crystal structure of human branched-chain α-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease. Structure. 2000;8:277–291. doi: 10.1016/s0969-2126(00)00105-2. [DOI] [PubMed] [Google Scholar]
- 5.Wynn R., Davie J., Meng M., Chuang D. Alpha-Keto Acid Dehydrogenase Complexes. Springer; New York, NY: 1996. Structure, function and assembly of mammalian branched-chain α-ketoacid dehydrogenase complex; pp. 101–117. [Google Scholar]
- 6.Ciszak E.M., Korotchkina L.G., Dominiak P.M., Sidhu S., Patel M.S. Structural basis for flip-flop action of thiamin pyrophosphate-dependent enzymes revealed by human pyruvate dehydrogenase. J. Biol. Chem. 2003;278:21240–21246. doi: 10.1074/jbc.M300339200. [DOI] [PubMed] [Google Scholar]
- 7.Bunik V.I., Degtyarev D. Structure–function relationships in the 2-oxo acid dehydrogenase family: substrate-specific signatures and functional predictions for the 2-oxoglutarate dehydrogenase-like proteins. Proteins. 2008;71:874–890. doi: 10.1002/prot.21766. [DOI] [PubMed] [Google Scholar]
- 8.Kato M., Wynn R.M., Chuang J.L., Brautigam C.A., Custorio M., Chuang D.T. A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain α-ketoacid dehydrogenase complex. EMBO J. 2006;25:5983–5994. doi: 10.1038/sj.emboj.7601444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang J., Baiesc F.L., Hiromasa Y., Yu X., Hui W.H., Dai X., et al. Atomic structure of the E2 inner core of human pyruvate dehydrogenase complex. Biochemistry. 2018;57:2325–2334. doi: 10.1021/acs.biochem.8b00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagy B., Polak M., Ozohanics O., Zambo Z., Szabo E., Hubert A., et al. Structure of the dihydrolipoamide succinyltransferase (E2) component of the human alpha-ketoglutarate dehydrogenase complex (hKGDHc) revealed by cryo-EM and cross-linking mass spectrometry: implications for the overall hKGDHc structure. Biochim. Biophys. Acta. 2021;1865:129889. doi: 10.1016/j.bbagen.2021.129889. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Y., Hawes J., Popov K.M., Jaskiewicz J., Shimomura Y., Crabb D.W., et al. Site-directed mutagenesis of phosphorylation sites of the branched chain alpha-ketoacid dehydrogenase complex. J. Biol. Chem. 1994;269:18583–18587. [PubMed] [Google Scholar]
- 12.Paxton R., Harris R.A. Regulation of branched-chain α-ketoacid dehydrogenase kinase. Arch. Biochem. Biophys. 1984;231:48–57. doi: 10.1016/0003-9861(84)90361-8. [DOI] [PubMed] [Google Scholar]
- 13.Machius M., Chuang J.L., Wynn R.M., Tomchick D.R., Chuang D.T. Structure of rat BCKD kinase: nucleotide-induced domain communication in a mitochondrial protein kinase. Proc. Natl. Acad. Sci. U. S. A. 2001;98:11218–11223. doi: 10.1073/pnas.201220098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato M., Li J., Chuang J.L., Chuang D.T. Distinct structural mechanisms for inhibition of pyruvate dehydrogenase kinase isoforms by AZD7545, dichloroacetate, and radicicol. Structure. 2007;15:992–1004. doi: 10.1016/j.str.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knoechel T.R., Tucker A.D., Robinson C.M., Phillips C., Taylor W., Bungay P.J., et al. Regulatory roles of the N-terminal domain based on crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands. Biochemistry. 2006;45:402–415. doi: 10.1021/bi051402s. [DOI] [PubMed] [Google Scholar]
- 16.Kukimoto-Niino M., Tokmakov A., Terada T., Ohbayashi N., Fujimoto T., Gomi S., et al. Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4. Acta Crystallogr D Biol Crystallogr. 2011;67:763–773. doi: 10.1107/S090744491102405X. [DOI] [PubMed] [Google Scholar]
- 17.Kato M., Chuang J.L., Tso S.-C., Wynn R.M., Chuang D.T. Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex. EMBO J. 2005;24:1763–1774. doi: 10.1038/sj.emboj.7600663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harper A.E., Miller R.H., Block K.P. Branched-chain amino acid metabolism. Annu. Rev. Nutr. 1984;4:409–454. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- 19.Dancis J., Hutzler J., Levitz M. Metabolism of the white blood cells in maple-syrup-urine disease. Biochim. Biophys. Acta. 1960;43:342–343. doi: 10.1016/0006-3002(60)90448-0. [DOI] [PubMed] [Google Scholar]
- 20.Menkes J.H., Hurst P.L., Craig J.M. A new syndrome: progressive familial infantile cerebral dysfunction associated with an unusual urinary substance. Pediatrics. 1954;14:462–467. [PubMed] [Google Scholar]
- 21.Maguolo A., Rodella G., Giorgetti A., Nicolodi M., Ribeiro R., Dianin A., et al. A gain-of-function mutation on BCKDK gene and its possible pathogenic role in branched-chain amino acid metabolism. Genes. 2022;13:233. doi: 10.3390/genes13020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newgard C.B., An J., Bain J.R., Muehlbauer M.J., Stevens R.D., Lien L.F., et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newgard C.B. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15:606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynch C.J., Adams S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 2014;10:723–736. doi: 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grzych G., Vonghia L., Bout M.A., Weyler J., Verrijken A., Dirinck E., et al. Plasma BCAA changes in patients with NAFLD are sex dependent. J. Clin. Endocrinol. Metab. 2020;105 doi: 10.1210/clinem/dgaa175. [DOI] [PubMed] [Google Scholar]
- 26.Sun H., Lu G., Ren S., Chen J., Wang Y. Catabolism of branched-chain amino acids in heart failure: insights from genetic models. Pediatr. Cardiol. 2011;32:305–310. doi: 10.1007/s00246-010-9856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paxton R., Harris R.A. Clofibric acid, phenylpyruvate, and dichloroacetate inhibition of branched-chain α-ketoacid dehydrogenase kinase in vitro and in perfused rat heart. Arch. Biochem. Biophys. 1984;231:58–66. doi: 10.1016/0003-9861(84)90362-x. [DOI] [PubMed] [Google Scholar]
- 28.Noguchi S., Kondo Y., Ito R., Katayama T., Kazama S., Kadota Y., et al. Ca(2+)-dependent inhibition of branched-chain α-ketoacid dehydrogenase kinase by thiamine pyrophosphate. Biochem. Biophys. Res. Commun. 2018;504:916–920. doi: 10.1016/j.bbrc.2018.09.038. [DOI] [PubMed] [Google Scholar]
- 29.Kadota Y., Toyoda T., Kitaura Y., Adams S.H., Shimomura Y. Regulation of hepatic branched-chain α-ketoacid dehydrogenase complex in rats fed a high-fat diet. Obes. Res. Clin. Pract. 2013;7:e439–e444. doi: 10.1016/j.orcp.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris R.A., Paxton R., DePaoli-Roach A. Inhibition of branched chain alpha-ketoacid dehydrogenase kinase activity by alpha-chloroisocaproate. J. Biol. Chem. 1982;257:13915–13918. [PubMed] [Google Scholar]
- 31.Kobayashi R., Murakami T., Obayashi M., Nakai N., Jaskiewicz J., Fujiwara Y., et al. Clofibric acid stimulates branched-chain amino acid catabolism by three mechanisms. Arch. Biochem. Biophys. 2002;407:231–240. doi: 10.1016/s0003-9861(02)00472-1. [DOI] [PubMed] [Google Scholar]
- 32.Tso S.C., Qi X., Gui W.J., Chuang J.L., Morlock L.K., Wallace A.L., et al. Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain α-ketoacid dehydrogenase kinase. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9728–9733. doi: 10.1073/pnas.1303220110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brunetti-Pierri N., Lanpher B., Erez A., Ananieva E.A., Islam M., Marini J.C., et al. Phenylbutyrate therapy for maple syrup urine disease. Hum. Mol. Genet. 2011;20:631–640. doi: 10.1093/hmg/ddq507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holeček M. Branched-chain amino acids in health and disease: metabolism, alterations in blood plasma, and as supplements. Nutr. Metab. (Lond) 2018;15:33. doi: 10.1186/s12986-018-0271-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monirujjaman M., Ferdouse A. Metabolic and physiological roles of branched-chain amino acids. Adv. Mol. Biol. 2014;2014:364976. [Google Scholar]
- 36.Conway M., Hutson S. The glutamate/GABA-Glutamine Cycle. Springer; New York, NY: 2016. BCAA metabolism and NH 3 homeostasis; pp. 99–132. [Google Scholar]
- 37.Boemer F., Josse C., Luis G., Di Valentin E., Thiry J., Cello C., et al. Novel loss of function variant in BCKDK causes a treatable developmental and epileptic encephalopathy. Int. J. Mol. Sci. 2022;23:2253. doi: 10.3390/ijms23042253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novarino G., El-Fishawy P., Kayserili H., Meguid N.A., Scott E.M., Schroth J., et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science. 2012;338:394–397. doi: 10.1126/science.1224631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.García-Cazorla A., Oyarzabal A., Fort J., Robles C., Castejón E., Ruiz-Sala P., et al. Two novel mutations in the BCKDK (branched-chain keto-acid dehydrogenase kinase) gene are responsible for a neurobehavioral deficit in two pediatric unrelated patients. Hum. Mutat. 2014;35:470–477. doi: 10.1002/humu.22513. [DOI] [PubMed] [Google Scholar]
- 40.Zigler J.S., Jr., Hodgkinson C.A., Wright M., Klise A., Sundin O., Broman K.W., et al. A spontaneous missense mutation in branched chain keto acid dehydrogenase kinase in the rat affects both the central and peripheral nervous systems. PLoS One. 2016;11 doi: 10.1371/journal.pone.0160447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tso S.C., Gui W.J., Wu C.Y., Chuang J.L., Qi X., Skvora K.J., et al. Benzothiophene carboxylate derivatives as novel allosteric inhibitors of branched-chain α-ketoacid dehydrogenase kinase. J. Biol. Chem. 2014;289:20583–20593. doi: 10.1074/jbc.M114.569251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun H., Olson K.C., Gao C., Prosdocimo D.A., Zhou M., Wang Z., et al. Catabolic defect of branched-chain amino acids promotes heart failure. Circulation. 2016;133:2038–2049. doi: 10.1161/CIRCULATIONAHA.115.020226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White P.J., McGarrah R.W., Grimsrud P.A., Tso S.-C., Yang W.-H., Haldeman J.M., et al. The BCKDH kinase and phosphatase integrate BCAA and lipid metabolism via regulation of ATP-citrate lyase. Cell Metab. 2018;27:1281–1293.e7. doi: 10.1016/j.cmet.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou M., Shao J., Wu C.-Y., Shu L., Dong W., Liu Y., et al. Targeting BCAA catabolism to treat obesity-associated insulin resistance. Diabetes. 2019;68:1730–1746. doi: 10.2337/db18-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green T., Grigorian A., Klyuyeva A., Tuganova A., Luo M., Popov K.M. Structural and functional insights into the molecular mechanisms responsible for the regulation of pyruvate dehydrogenase kinase 2. J. Biol. Chem. 2008;283:15789–15798. doi: 10.1074/jbc.M800311200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devedjiev Y., Steussy C.N., Vassylyev D.G. Crystal structure of an asymmetric complex of pyruvate dehydrogenase kinase 3 with lipoyl domain 2 and its biological implications. J. Mol. Biol. 2007;370:407–416. doi: 10.1016/j.jmb.2007.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roe S.M., Prodromou C., O'Brien R., Ladbury J.E., Piper P.W., Pearl L.H. Structural basis for inhibition of the hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J. Med. Chem. 1999;42:260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- 48.Besant P.G., Lasker M.V., Bui C.D., Turck C.W. Inhibition of branched-chain α-keto acid dehydrogenase kinase and Sln1 yeast histidine kinase by the antifungal antibiotic radicicol. Mol. Pharmacol. 2002;62:289–296. doi: 10.1124/mol.62.2.289. [DOI] [PubMed] [Google Scholar]
- 49.Kitaura Y., Shindo D., Ogawa T., Sato A., Shimomura Y. Antihypertensive drug valsartan as a novel BDK inhibitor. Pharmacol. Res. 2021;167:105518. doi: 10.1016/j.phrs.2021.105518. [DOI] [PubMed] [Google Scholar]
- 50.Greiner J.V., Glonek T. Intracellular ATP concentration and implication for cellular evolution. Biology (Basel) 2021;10:1166. doi: 10.3390/biology10111166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Criscione L., de Gasparo M., Bühlmayer P., Whitebread S., Ramjoué H.P.R., Wood J. Pharmacological profile of valsartan: a potent, orally active, nonpeptide antagonist of the angiotensin II AT1-receptor subtype. Br. J. Pharmacol. 1993;110:761–771. doi: 10.1111/j.1476-5381.1993.tb13877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siragy H. Angiotensin II receptor blockers: review of the binding characteristics. Am. J. Cardiol. 1999;84:3s–8s. doi: 10.1016/s0002-9149(99)00727-4. [DOI] [PubMed] [Google Scholar]
- 53.Balakumar P., Jagadeesh G. Structural determinants for binding, activation, and functional selectivity of the angiotensin AT1 receptor. J. Mol. Endocrinol. 2014;53:R71–R92. doi: 10.1530/JME-14-0125. [DOI] [PubMed] [Google Scholar]
- 54.Zhang H., Unal H., Desnoyer R., Han G.W., Patel N., Katritch V., et al. Structural basis for ligand Recognition and functional selectivity at angiotensin receptor. J. Biol. Chem. 2015;290:29127–29139. doi: 10.1074/jbc.M115.689000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang H., Unal H., Gati C., Han G.W., Liu W., Zatsepin N.A., et al. Structure of the angiotensin receptor revealed by serial femtosecond crystallography. Cell. 2015;161:833–844. doi: 10.1016/j.cell.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrell J.A., Orme J., Butlin R.J., Roche T.E., Mayers R.M., Kilgour E. AZD7545 is a selective inhibitor of pyruvate dehydrogenase kinase 2. Biochem. Soc. Trans. 2003;31:1168–1170. doi: 10.1042/bst0311168. [DOI] [PubMed] [Google Scholar]
- 57.Brough P.A., Baker L., Bedford S., Brown K., Chavda S., Chell V., et al. Application of off-rate screening in the identification of novel pan-isoform inhibitors of pyruvate dehydrogenase kinase. J. Med. Chem. 2017;60:2271–2286. doi: 10.1021/acs.jmedchem.6b01478. [DOI] [PubMed] [Google Scholar]
- 58.Akaki T., Bessho Y., Ito T., Fujioka S., Ubukata M., Mori G., et al. Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases. Bioorg. Med. Chem. 2021;44:116283. doi: 10.1016/j.bmc.2021.116283. [DOI] [PubMed] [Google Scholar]
- 59.Baker L.M., Aimon A., Murray J.B., Surgenor A.E., Matassova N., Roughley S.D., et al. Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures. Commun. Chem. 2020;3:122. doi: 10.1038/s42004-020-00367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaman M.A., Oparil S., Calhoun D.A. Drugs targeting the renin–angiotensin–aldosterone system. Nat. Rev. Drug Discov. 2002;1:621–636. doi: 10.1038/nrd873. [DOI] [PubMed] [Google Scholar]
- 61.Pitt B., Segal R., Martinez F.A., Meurers G., Cowley A.J., Thomas I., et al. Randomised trial of losartan versus captopril in patients over 65 with heart failure (Evaluation of Losartan in the Elderly Study, ELITE) Lancet. 1997;349:747–752. doi: 10.1016/s0140-6736(97)01187-2. [DOI] [PubMed] [Google Scholar]
- 62.Dézsi C.A. The different therapeutic choices with ARBs. Which one to give? When? Why? Am. J. Cardiovasc. Drugs. 2016;16:255–266. doi: 10.1007/s40256-016-0165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Piérard L. Clinical study of the month. Effects of valsartan in chronic heart failure: the VAL-HeFT study. Rev. Med. Liege. 2002;57:57–59. [PubMed] [Google Scholar]
- 64.McKelvie R.S., Yusuf S., Pericak D., Avezum A., Burns R.J., Probstfield J., et al. Comparison of candesartan, enalapril, and their combination in congestive heart failure: Randomized evaluation of strategies for left ventricular dysfunction (RESOLVD) pilot study. The RESOLVD pilot study Investigators. Circulation. 1999;100:1056–1064. doi: 10.1161/01.cir.100.10.1056. [DOI] [PubMed] [Google Scholar]
- 65.Poirier A., Cascais A.C., Funk C., Lavé T. Prediction of pharmacokinetic profile of valsartan in human based on in vitro uptake transport data. J. Pharmacokinet. Pharmacodyn. 2009;36:585–611. doi: 10.1007/s10928-009-9139-3. [DOI] [PubMed] [Google Scholar]
- 66.Szabo A., Stolz L., Granzow R. Surface plasmon resonance and its use in biomolecular interaction analysis (BIA) Curr. Opin. Struct. Biol. 1995;5:699–705. doi: 10.1016/0959-440x(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 67.Davie J.R., Wynn R.M., Meng M., Huang Y.S., Aalund G., Chuang D.T., et al. Expression and characterization of branched-chain alpha-ketoacid dehydrogenase kinase from the rat. Is it a histidine-protein kinase? J. Biol. Chem. 1995;270:19861–19867. doi: 10.1074/jbc.270.34.19861. [DOI] [PubMed] [Google Scholar]
- 68.Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S.M., Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 71.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schrodinger, L. Schrödinger, LLC; New York, NY: 2010. The PyMOL Molecular Graphics System. 2.3 Ed. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors (PDB codes: 8EGD, 8EGF, 8EGQ, and 8EGU) have been deposited in the PDB, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).