Abstract
This was the first molecular study of the bacterial flora of the sheep scab mite (Psoroptes ovis). A sequence analysis of genes coding for 16S rRNA revealed that Serratia marcescens and bacteria closely related to Staphylococcus intermedius or Staphylococcus chromogens and Alloiococcus otitidis were present. These bacteria were associated with skin lesions, dermatitis, and otitis media caused by P. ovis.
Sheep scab is an allergic dermatitis that forms in response to the presence of the mite Psoroptes ovis on the skin of sheep (3, 13). P. ovis causes intense pruritis, weight loss, and fleece damage in infected animals, and P. ovis infestations can be fatal if they are not treated and thus have great economic impact and welfare importance to the world sheep-farming industry. Recent problems associated with traditional chemical means of mite control have led to an urgent need for new means of control (12). The digestive system of the mite is a possible target for novel control methods, including vaccines. It has been speculated that gut bacteria may be crucial to the mite as a food source (3, 17). These bacteria may also be the source of mite antigens (3, 4, 12). To enable further study of these possibilities, we identified the bacterial flora associated with P. ovis by analyzing the genes coding for 16S rRNA (rDNA).
Isolation of bacterial 16S rDNA.
A mixture of male and female P. ovis mites (Cornish strain, medium virulence) at all stages of development was obtained from an in vivo sheep culture at the Veterinary Laboratories Agency, Weybridge, United Kingdom. Approximately 80 mites were surface sterilized (17) and homogenized in 600 μl of sterile cell lysis buffer (50 mM Tris [pH 8.0], 10 mM EDTA, 2% sodium dodecyl sulfate), 60 μg of proteinase K was added, and the homogenate was incubated overnight at 55°C. Protein was precipitated with 5 M ammonium acetate, and the DNA was precipitated with isopropanol before it was resuspended in 25 μl of single-distilled sterile water. Bacterial 16S rDNA was amplified from the extracted genomic DNA by using the following universal bacterial 16S rDNA primers (5a, 15): forward primer 5′AGAGTTTGATCMTGGCTCAG3′ and reverse primer 5′AAGGAGGTGATCCANCCRCA3′. A PCR was performed with a 50-μl reaction mixture containing 1 μl (10 ng) of DNA extract as the template, each primer at a concentration of 0.5 μM, 1.5 mM MgCl2, and each deoxynucleoside triphosphate at a concentration of 50 μM, as well as 1 U of Gibco Taq polymerase and buffer used as recommended by the manufacturer. After the initial denaturation for 4.5 min at 95°C, there were 40 cycles consisting of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min and then a final extension step consisting of 5 min at 72°C; a Progene thermal cycler (Techne) was used. The amplified 1,500-bp product was purified from the gel slice by using a Sephaglass band prep kit (Pharmacia Biotech) and was ligated into PGEMT (Stratagene). PCR products obtained from 100 randomly selected clones by using M13 forward and reverse primers were digested with HAE3 and MSP1 (as recommended by the manufacturer), and this analysis resulted in 15 separate restriction fragment length polymorphism groups.
Nucleotide sequencing, alignment, and phylogeny.
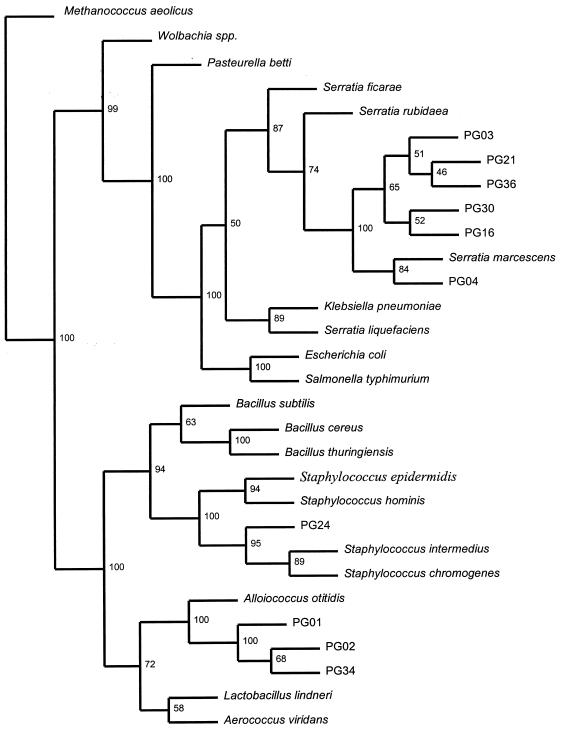
Qiagen-purified plasmids were sequenced by MWG Biotech (Ebersberg, Germany) and were edited to remove vector and universal 16S rDNA primer sequences. Sequences were preliminarily matched with previously published bacterial 16S rDNA sequences by using BLAST (2) and the similarity rank program of the Ribosomal Database Project (RDP) (16) and were checked for chimeras by using the RDP CHIMERA CHECK program. Five of the 15 nucleotide sequences were rejected as chimeras. The remaining 10 sequences were aligned with the most similar sequences and the sequences of other representative bacterial 16S rDNA regions by using Clustal W (20), and a phylogenetic analysis was performed by using PHYLIP (DNADIST, NEIGHBOUR, and SEQBOOT) (7). Based on the phylogenetic tree (Fig. 1), we identified three groups of bacteria in P. ovis; both gram-positive and gram-negative bacteria were present. Sequences PG16, PG04, PG30, PG03, PG36, and PG21 clustered with the gram-negative bacteria belonging to the genus Serratia (specifically, Serratia marcescens), which was consistent with the high levels of similarity to the previously published sequences of S. marcescens (BLAST level of similarity, 99%; RDP levels of similarity, 92 to 94%). One of the gram-positive bacterial sequences, PG24, clustered with the staphylococci close to Staphylococcus intermedius (RDP level of similarity, 93.3%) or Staphylococcus chromogens (BLAST level of similarity, 98%). Sequences PG01, PG34, and PG02 all clustered with Alloiococcus otitidis but exhibited low levels of similarity to previously published sequences for this species (BLAST levels of similarity, 86 to 87%; RDP levels of similarity, 81 to 82%). It is possible that the sequences of the alloiococci and staphylococci may represent as-yet-undescribed species of bacteria. The presence of S. marcescens is consistent with the results of a previous study (17) in which this bacterium was isolated by culturing P. ovis homogenates and feces. Transmission electron microscopic evidence suggested that the majority of bacteria associated with the mites were present in the intestinal tract (17). S. marcescens is a common colonizer of the midguts of a variety of arthropods (14) and is an opportunistic infectious agent of wounds (6, 10, 19). The presence of staphylococci is consistent with sheep scab as these bacteria are ubiquitous and have been associated with Psoroptes cuniculi and otitis media in rabbit ear infections (5). Staphylococci are also associated with the mange mites that cause cutaneous infections in cattle (8, 9) and with the Sarcoptes mites that cause acute and subacute dermatitis in pigs (11). Our identification of sequences that are closely related to A. otitidis sequences is unusual and especially interesting. A. otitidis, which was formerly named A. otitis (21), is normally associated with acute otitis media in human ears (1). The presence of this bacterium is of considerable interest as P. cuniculi also causes otitic conditions in rabbits (5). Also, when P. cuniculi is present in sheep ears, it is believed to be a cryptic form of P. ovis (3, 18). Our discovery of a bacterial sequence related to the A. otitidis sequence in a mite which does not live in vertebrate ears prompted us to speculate that mites may be the source of A. otitidis-like infections and possibly other organisms in otitic infections. This possibility should be studied further.
FIG. 1.
Phylogenetic relationships among bacterial 16S rDNA sequences isolated from P. ovis and 20 selected bacterial 16S rDNA sequences. Methanococcus aeolicus was used as an outgroup species; the other bacteria represent the gram-positive and gram-negative divisions. The numbers are maximum bootstrap values based on 100 replicates.
In summary, we describe the presence in P. ovis of sequences resembling 16S rDNA sequences of bacteria that are related to allergic conditions, such as dermatitis and otitis media (Staphylococcus spp. and A. otitidis), and also of bacteria which are known to colonize wounds in vertebrates and invertebrate guts (Serratia spp.). Our results confirmed the results of a previous bacterial study of P. ovis in which the authors proposed that more than one strain of S. marcescens was present in P. ovis and that there were also other, unidentified bacterial species (cocci) present (17). Whether the bacteria that have been described are initially associated with the mite and play a part in lesion formation in diseased sheep or whether they are primarily associated with the lesions and are only subsequently ingested by the mite is an intriguing but open question. By using the data gathered in this study it is now possible to address these hypotheses.
Nucleotide sequence accession numbers.
Nonchimeral sequences were deposited in the GenBank database under the following accession numbers: Pg01, AF124033; Pg02, AF124034; Pg03, AF124035; Pg04, AF124036; Pg16, AF124037; Pg21, AF124038; Pg24, AF124039; Pg30, AF124040; Pg34, AF124041; and Pg36, AF124042.
Acknowledgments
This work was supported by a grant from M.A.F.F.
REFERENCES
- 1.Aguirre M, Collins M D. Development of a polymerase chain reaction-probe test for identification of Alloiococcus otitis. J Clin Microbiol. 1992;30:2177–2180. doi: 10.1128/jcm.30.8.2177-2180.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Warren G, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Bates P G. Recent advances in the biology and control of sheep scab. Proc Sheep Vet Soc. 1991;15:23–27. [Google Scholar]
- 4.Beetham P K. Sheep scab mite (Acari: Psoroptidae): general morphology and immunocytochemistry using serum from infested animals. Ann Entomol Soc Am. 1997;90:202–207. [Google Scholar]
- 5.Bjotvedt G, Geib L W. Otitis media associated with Staphylococcus epidermis and Psoroptes cuniculi in a rabbit. Vet Med Small Anim Clin. 1981;76:1015–1016. [PubMed] [Google Scholar]
- 5a.Collingham, R. (Veterinary Laboratories Agency). Personal communication.
- 6.Edgar P, Mlcak R, Desai M, Linares H A, Phillips L G, Heggers J P. Containment of a multiresistant Serratia marcescens outbreak. Burns. 1997;1:15–18. doi: 10.1016/s0305-4179(97)81117-5. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein J. Estimating effective population-size from samples of sequences—a bootstrap monte-carlo integration method. Genet Res. 1993;60:209–220. doi: 10.1017/s0016672300030962. [DOI] [PubMed] [Google Scholar]
- 8.Hazarika R A, Mahanta P N, Dutta G N. Cutaneous infection associated with Staphylococcus hyicus in cattle. Res Vet Sci. 1991;50:374–375. doi: 10.1016/0034-5288(91)90146-f. [DOI] [PubMed] [Google Scholar]
- 9.Hazarika R A, Mahanta P N, Dutta G N. Association of Staphylococcus aureus in bovine dermatitis. Indian J Anim Sci. 1995;65:629–632. [Google Scholar]
- 10.Hume E B H, Willcox M D P, Sweeney D F, Holden B A. An examination of the clonal variants of Serratia marcescens that infect the eye during contact lens wear. J Med Microbiol. 1996;45:127–132. doi: 10.1099/00222615-45-2-127. [DOI] [PubMed] [Google Scholar]
- 11.Istvan R. Mass incidence of a skin disease in swine. Magy Allatorv Lapja. 1993;48:351–355. [Google Scholar]
- 12.Jayawardena K G I, Heller-Haupt A, Woodland R M, Varma M G R. Antigens of the sheep scab mite Psoroptes ovis. Folia Parasitol (Prague) 1998;45:239–244. [PubMed] [Google Scholar]
- 13.Kirkwood A C. History, biology and control of sheep scab. Parasitol Today. 1986;2:302–307. doi: 10.1016/0169-4758(86)90124-9. [DOI] [PubMed] [Google Scholar]
- 14.Klein M G, Kaya H K. Bacillus and Serratia species for scarab control. Mem Inst Oswaldo Cruz Rio de J. 1995;90:87–95. [Google Scholar]
- 15.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 16.Larsen N, Olsen G J, Maidak B L, McCaughey M J, Overbeek R, Macke T J, Marsh T L, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1993;21:3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathieson B R F, Lehane M J. Isolation of the gram-negative bacterium, Serratia marcescens, from the sheep scab mite, Psoroptes ovis. Vet Rec. 1996;138:210–211. doi: 10.1136/vr.138.9.210. [DOI] [PubMed] [Google Scholar]
- 18.Morgan K L. Parasitic otitis in sheep associated with Psoroptes infestation—a clinical and epidemiologic study. Vet Rec. 1992;130:530–532. doi: 10.1136/vr.130.24.530. [DOI] [PubMed] [Google Scholar]
- 19.Passoro D J, Waring L, Armstrong R, Bolding F, Bouvier B, Rosenberg J, Reingold A W, McQuitty M, Philpott S M, Jarvis W R, Werner S B, Tompkins L S, Vugia D J. Postoperative wound infections traced to an out of hospital source. J Infect Dis. 1997;175:992–995. doi: 10.1086/514008. [DOI] [PubMed] [Google Scholar]
- 20.Thompson J D, Higgins D G, Gibson T J. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vongraevenitz A. Revised nomenclature of Alloiococcus otitis. J Clin Microbiol. 1993;31:472. doi: 10.1128/jcm.31.2.472-472.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]