Abstract
Background
Commensal Escherichia coli residing in the guts of humans and animals are reservoirs of multidrug resistance (MDR) genes, including quinolone resistance genes, in humans and poultry. This study aimed to characterize quinolones resistance in E. coli recovered from poultry workers, chickens, and poultry farm/market environments in Abuja, Nigeria.
Methods
This was a cross-sectional study conducted between December 2018 and April 2019 comprising poultry workers, chickens and their poultry farm/market environments. This study characterized E. coli isolates from stool, faecal and environmental samples using antimicrobial susceptibility testing and whole-genome sequencing methods. Core-genome multilocus sequences-based phylogeny was used to determine the relatedness between quinolone-resistant E. coli isolates. Data were analyzed using descriptive statistics.
Results
Of 110 E. coli isolates, quinolone-resistant phenotypes were observed in 68.2% (n = 75) isolates. Whole-genome sequencing detected plasmid-mediated quinolone resistance (PMQR) genes in 63.6% (n = 70) isolates. The most prevalent PMQR gene detected in 56 of these 70 E. coli isolates was qnrS1, followed by qnrB19 in 14 isolates and aac(6’)-lb-cr in two isolates. Fifteen ciprofloxacin and 19 nalidixic acid-resistant isolates respectively showed double mutations in the quinolone-resistance determining regions (QRDRs) of gyrA, with single or double mutations in parC, and a single mutation in parE. The most prevalent amino-acid substitutions observed were S83L + D87N in gyrA (46.5%, n = 20), S80I in parC (51.2%, n = 22) and S458A in parE (14%, n = 6). About 2.9% (2/70) of PMQR isolates were extended-spectrum beta-lactamase (ESBL) producers while 2.9% (2/70) had plasmid-mediated colistin resistance (PMCR) genes.
Conclusions
PMQR genes were prevalent in E. coli isolates recovered from healthy humans, chickens and poultry farm/market environments. PMCR genes (mcr-1.1) occurred in PMQR-positive isolates recovered from manure and drinking water originating from poultry farm/market environments. It was found that the gene encoding ESBL coexisted with qnrS-positive isolates of human and avian origin. Horizontal transfer of PMQR genes among E. coli isolates in the human-poultry-environment interface has public health implications for the spread of antimicrobial resistance. Relevant government agencies should enforce regulations to restrict the use of critically important antimicrobials in poultry production.
Keywords: Escherichia coli, Plasmid-mediated quinolone resistance, Plasmid-mediated colistin resistance, Poultry workers, Chicken, Poultry environment, Nigeria
Background
Globally, commensal E. coli is known as an inhabitant of the intestinal microflora of warm-blooded humans and animals including poultry [1–3]. Although commensal E. coli is known not to be harmful to the host, studies have reported that the bacteria can acquire resistance and become a reservoir for multidrug resistance (MDR) genes including plasmid-mediated quinolone resistance (PMQR) genes [4, 5], hence serve as a useful indicator organism for measuring antimicrobial resistance (AMR) [4, 6, 7]. In most developing economies including Nigeria, the lack of enforcement on access, sale, and use of antimicrobials in humans and food-producing animals has resulted in the abuse of antimicrobials [8–10].
Fluoroquinolones are one of the available therapeutic options for E. coli infections in humans [11]. Quinolones and colistin have been classified as the highest-priority important antimicrobials for use in humans by the World Health Organization (WHO) because a strong correlation exists between use and an increase in drug resistance [11]. Several studies have reported resistance to ciprofloxacin and nalidixic acid in E. coli isolates recovered from food-producing animals particularly poultry as well as in humans and the environment [12–16]. However, the emergence of quinolone resistance has been attributed to the fact that fluoroquinolones are one of the most commonly prescribed antimicrobial classes in humans and food-producing animals in Nigeria [17–19]. Previous studies have reported that there is a paucity of information on PMQR and its importance in Africa, although resistance has been reported to evolve quickly in Nigeria [20]. E. coli isolates have enzymes important for bacterial replication which function as target sites for fluoroquinolones. These enzymes include DNA gyrase which acts as the primary site (gyrA and gyrB) and topoisomerase IV as the secondary site (parC and parE) [14]. Previous studies reveal that chromosomal mutations which modify the target enzymes are responsible for quinolone resistance in E. coli strains [21, 22]. Although quinolones and colistin have been classified as antimicrobials of last resort, co-occurrence of PMQR and plasmid-mediated colistin resistance (PMCR) have been reported in E. coli isolates recovered from poultry farm environments [23]. Evidence shows that PMQR genes may be found on conjugative plasmids with the ability for horizontal transfer between bacteria [24, 25]. Few studies have reported that plasmids carrying PMQR genes can harbor genes that can confer resistance to other antimicrobial agents including colistin [23, 24, 26].
Hypothesis: Poultry harboring PMQR E. coli isolates can become potential sources of horizontal transfer of resistance genes to poultry workers as well as to the poultry farm or market environments. We characterized quinolones resistance in commensal E. coli recovered from poultry workers, chickens, and selected poultry farms/market environments in Abuja, Nigeria using disk diffusion, broth microdilution, and whole-genome sequencing.
Methods
Research design
This cross-sectional study was conducted in Abuja, North Central Nigeria from December 2018 – April 2019. The study population comprised poultry farmers, poultry sellers, and chickens in farms/markets as well as the poultry farm/market environments. The sample size formula for cross-sectional studies was used to calculate the minimum sample size of 384 for the study with the following assumptions: expected proportion of 50%, with a precision of 5% and type 1 error of 5% [27]. A total of 429 samples were randomly selected from 52 commercial poultry farms and eight poultry markets in Abuja Municipal and Kuje Area Councils of the Federal Capital Territory, Abuja. The samples comprised 122 freshly collected human stool samples from poultry workers, 111 chicken faecal samples, and 196 environmental samples (poultry litter and drinking water), respectively. After collection, all samples were transported in ice to the National Reference Laboratory, Gaduwa, Abuja for processing within hours of collection.
Isolation and identification of bacterial strains
A total of 110 E. coli strains were isolated from stool samples of apparently healthy individuals working with chickens in farms or markets, faecal samples of chickens, poultry litter, and poultry drinking water from the farm or markets as described previously [28]. Briefly, about one gram of human stool, one gram of chicken faeces and 30 g of poultry litter, respectively, were placed in 9 mL of buffered peptone water (BPW) and incubated for 24 h at 37 °C. Thereafter, a 10µL loopful of BPW was plated on MacConkey agar plates and incubated for 24 h at 37 °C. Suspected pink to red E. coli colonies were transferred on Eosin Methylene Blue (EMB) agar plates for 24 h at 37 °C. Subsequently, isolates with a greenish metallic sheen were plated on Tryptic Soy agar for biochemical tests (indole, methyl red, Voges-Proskauer and Citrate utilization) and confirmed using commercially available kits, Microbact GNB 24E (Oxoid, UK) following the manufacturer’s instructions. For each 100 ml of a drinking water sample, the membrane filtration technique was employed using single sterile 0.45 µm pores filter disks. Thereafter the filter membranes were placed on EMB plates and incubated for 24 h at 37 °C. All E. coli isolates were subsequently tested as mentioned above.
Antimicrobial susceptibility testing
The Kirby Bauer disk diffusion method was used to determine the antimicrobial susceptibility patterns of the E. coli isolates against a panel of 14 antimicrobial agents namely ampicillin (10 μg), amoxicillin/clavulanic acid (20/10 μg), tetracycline (30 μg), gentamicin (10 μg), cefuroxime (30 μg), streptomycin (10 μg), chloramphenicol (30 μg), nalidixic acid (30 μg), sulfamethoxazole-trimethoprim (10 μg), nitrofurantoin (300 μg), ceftriaxone (30 μg), imipenem (10 μg), ceftazidime (30 μg) and cefotaxime (30 μg) as previously described [28]. Furthermore, the minimum inhibitory concentrations (MIC) against a panel of 16 antimicrobial agents for all E. coli isolates were determined by broth microdilution assay methods using the Gram-negative Sensititre™ (ESBL) plate according to the manufacturer’s instructions. These antimicrobials comprised ampicillin, cefazolin, ceftriaxone, cefepime, cefoxitin, cefotaxime, cefpodoxime, ceftazidime, cephalothin, ciprofloxacin, gentamicin, imipenem, meropenem, piperacillin/tazobactam, cefotaxime/clavulanic acid and ceftazidime/clavulanic acid. The recommendations of the Clinical and Laboratory Standards Institute (CLSI) M100 31st Edition were used to interpret the results [29]. E. coli ATCC 25922 and K. pneumoniae ATCC 700603 were used for internal quality control. Multidrug resistance (MDR) was defined as resistance to three or more drug classes.
Whole-genome sequencing (WGS) of E. coli isolates
The DNA of all the E. coli isolates was extracted using the Lucigen MasterPure™ Gram Positive DNA Purification Kit (ScienceVision, Selangor Darul Ehsan, Malaysia) following the Whole-Genome DNA Isolation for Gram-Negative Bacteria protocol according to the instructions of the manufacturer. This was followed by DNA quantification using the Qubit 4.0 Fluorometer assay kit (Thermo Fisher Scientific, Waltham, MA). Thereafter, libraries for WGS were prepared for each E. coli isolate using a Nextera XT DNA Library Preparation kit (Illumina Inc., San Diego, CA). For each isolate, high-quality Illumina MiSeq (Illumina Inc., San Diego, CA) deep shotgun 2 × 250 bp paired-end reads were assembled de novo into the draft genome sequence using SPAdes assembler v.3.13.1 [30].
Characterization of antimicrobial resistance determinants of E. coli isolates
The in silico detection and characterization of AMR determinants was performed using the ResFinder 4.1 tool (database version 2022–04-24), which is freely available on the Center for Genomics Epidemiology (CGE) website (https://cge.food.dtu.dk/services/ResFinder/) [31]. The ResFinder tool was used to determine the acquired AMR genes and predict the chromosomal point mutations. A 90–100% identity, 60% minimum length, and 90% threshold were used to match individual genes for each isolate to an annotated resistance gene [31]. In silico analysis of the plasmid replicon types for each isolate was conducted using the PlasmidFinder 2.1 tool (2021–11-29) available on the CGE website [32].
Determination of E. coli phylogroups, MLST, cgMLST and phylogeny
Multilocus sequence typing (MLST) was performed using MLST 2.0 (2022–11-14) with the E. coli PubMLST database (https://pubmed.ncbi.nlm.nih.gov/30345391/) [33]. The MLST analyses was based on the scheme previously described by Achtman [34] which considered allelic variations amongst seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) to assign sequence types (STs).
We determined the phylogenetic classification of the E. coli genomes using ClermonTyping method as previously described [35]. The CATIBioMed (IAME UMR 1137) hosts the ClermonTyper web interface accessible at http://clermontyping.iame-research.center/. The clonal relationship between isolates was estimated by their core genome MLST (cgMLST) profile which was determined using the cgMLSTFinder 1.2 (2021–08-29) with the Enterobase scheme [36, 37].
The cgMLST-based phylogenetic tree was annotated and visualized using the FigTree version 1.4.4 tool (http://tree.bio.ed.ac.uk/software/figtree/) and interactive Tree of Life tool – iTOL version 6 (http://itol.embl.de/itol.cgi).
Data analyses
Data were analyzed using Epi Info™ version 7.2.5.0 (https://www.cdc.gov/epiinfo/index.html). Descriptive statistics were used to summarize the data obtained. The raw reads for each E. coli isolate have been deposited in the National Center for Biotechnology Information (NCBI) database (Genome Trakr project) with the accession number PRJNA293225.
Results
Antimicrobial Susceptibility profile of quinolone-resistant E. coli
Overall, 110 E. coli isolates were recovered from 429 samples originating from 122 poultry workers, 111 chickens and 196 poultry farm/market environments. Of these, 75 (68.2%) isolates were resistant to nalidixic acid (NA) by disk diffusion method and 32 (29.1%) were resistant to ciprofloxacin (CIP) by broth microdilution method (Table 1).
Table 1.
Prevalence of nalidixic acid or ciprofloxacin-resistant Escherichia coli isolates from different sources characterized by disk diffusion and broth microdilution methods
| Origin | No of samples n = 429 | No of E. coli isolates n = 110 n (%)a | No of NA-resistant E. coli n = 75 n (%)b | No of CIP- resistant E. coli n = 32 n (%)b |
|---|---|---|---|---|
| Human | 122 | 47 (42.7) | 27 (36.0) | 7 (21.9) |
| Poultry | 111 | 36 (32.7) | 29 (38.7) | 15 (46.9) |
| Environment | 196 | 27 (24.5) | 19 (25.3) | 10 (31.3) |
aThe percentage value indicates the frequency of E. coli isolates from different sources. bThe percentage value indicates the frequency of NA (nalidixic acid)- or CIP (ciprofloxacin)-resistant isolates from the collected E. coli isolates using disk diffusion and broth microdilution methods. Note that all isolates that showed resistance against ciprofloxacin also showed resistance against nalidixic acid
Most of the isolates were resistant to tetracycline (98.7%), trimethoprim-sulfamethoxazole (92%), streptomycin (88%), ampicillin (82.7%) and gentamicin (68%). A majority of the isolates were however, susceptible to cefotaxime (94.6%), ceftriaxone (93.3%), cefuroxime (92%), amoxicillin/clavulanic acid (89.3%), ceftazidime (88%), and imipenem (85.3%). Moreover, 98.7% of the quinolone-resistant E. coli isolates recovered from the different sources were observed to be MDR i.e. resistant to three or more classes of antibiotics. The antimicrobial susceptibility profile of the 75 quinolone-resistant isolates against a panel of 14 antimicrobial agents is shown in (Table 2).
Table 2.
Antimicrobial resistance profiles of 75 quinolone-resistant Escherichia coli isolates from humans, chickens, and poultry farm/market environments using the disk diffusion method
| Drug Class | Drug | Resistance breakpoint (mm) R ≤ | Human n = 27 (%) | Chicken n = 29 (%) | Environment n = 19 (%) | Total n = 75 (%) |
|---|---|---|---|---|---|---|
| Tetracyclines | Tetracycline | ≤ 11 | 26 (96.3) | 29 (100.0) | 19 (100.0) | 74 (98.7) |
| Folate Pathway antagonists | Sulfamethoxazole/Trimethoprim | ≤ 10 | 25 (92.6) | 26 (89.7) | 18 (94.7) | 69 (92.0) |
| Penicillins | Ampicillin | ≤ 13 | 22 (81.5) | 26 (89.7) | 14 (73.7) | 62 (82.7) |
| Aminoglycosides | Streptomycin | ≤ 11 | 23 (85.2) | 26 (89.7) | 17 (89.5) | 66 (88.0) |
| Gentamicin | ≤ 12 | 15 (55.6) | 24 (82.8) | 12 (63.2) | 51 (68.0) | |
| Phenicols | Chloramphenicol | ≤ 12 | 10 (37.0) | 14 (48.3) | 6 (31.6) | 30 (40.0) |
| Nitrofurans | Nitrofurantoin | ≤ 14 | 5 (18.5) | 11 (37.9) | 6 (31.6) | 22 (29.3) |
| Carbapenems | Imipenem | ≤ 19 | 3 (11.1) | 6 (20.7) | 2 (10.5) | 11 (14.7) |
| β-lactam inhibitors | Amoxicillin/clavulanic acid | ≤ 13 | 2 (7.4) | 5 (17.2) | 1 (5.3) | 8 (10.7) |
| 3rd and 4th Generation Cephalosporins | Ceftazidime | ≤ 17 | 3 (11.1) | 1 (3.5) | 4 (21.1) | 8 (10.7) |
| Cefuroxime | ≤ 14 | 3 (11.1) | 3 (10.3) | 0 (0) | 6 (8.0) | |
| Cefotaxime | ≤ 22 | 3 (11.1) | 1 (3.5) | 0 (0) | 4 (5.3) | |
| Ceftriaxone | ≤ 19 | 3 (11.1) | 1 (3.5) | 1 (5.3) | 5 (6.7) | |
| Resistance to three or more classes of antibiotics | MDR | n/a | 26 (96.3) | 29 (100.0) | 19 (100.0) | 74 (98.7) |
The resistance breakpoint for the interpretation of the disk diffusion results was obtained from the 31st edition of CLSI M100 guidelines
Characterization of PMQR E. coli isolates
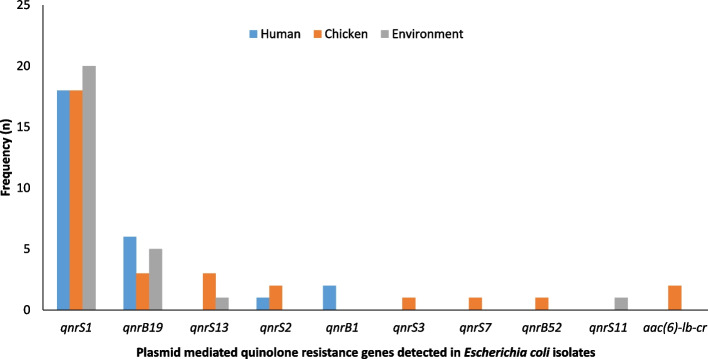
Of the 110 E. coli isolates, ten distinct quinolone-resistance genes were detected in 70 (63.6%) of the isolates. Out of the 70 PMQR E. coli isolates, the most prevalent PMQR gene was qnrS1, detected in 56 (80%) isolates recovered from all sources. (Fig. 1). In addition, only 43 out of 75 phenotypically quinolone-resistant E. coli isolates contained PMQR genes. E. coli carrying PMQR genes were most often detected in chickens (34.3%) followed by humans (32.9%) and poultry farm/market environments (32.9%).
Fig. 1.
Distribution of PMQR genes in 70 Escherichia coli isolates originating from humans, chickens, and the poultry farm/market environment
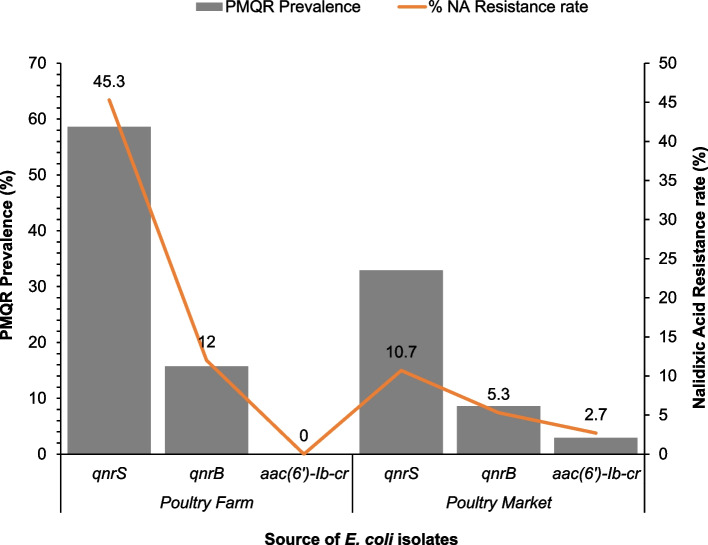
Thirteen PMQR E. coli isolates harbored two different PMQR gene combinations namely: qnrS1 + qnrB19 (n = 9); qnrS13 + qnrB19 (n = 1); qnrS1 + qnrB1 (n = 1); qnrS2 + aac(6’)-Ib-cr (n = 1) and qnrB52 + aac(6’)-Ib-cr (n = 1). The nine E. coli isolates harboring PMQR gene combination qnrS1 + qnrB19 were recovered from poultry environment (n = 4), humans (n = 3), and chickens (n = 2). PMCR (mcr-1.1) gene was detected in two E. coli isolates originating from the poultry environment. One PMCR isolate from a water sample harbored PMQR gene combination qnrS1 + qnrB19 while the second isolate from a poultry litter sample harbored only the qnrS1 gene. Of the 32 CIP-resistant isolates, PMQR genes were detected in 23 (71.9%) of the isolates with qnrS, qnrB and aac(6’)-Ib-cr genes identified in 21 (65.6%), four (12.5%) and one (3.1%) isolates, respectively. PMQR genes were detected in 48 (64%) of the 75 NA-resistant isolates with qnrS, qnrB and aac(6’)-Ib-cr genes identified in 42 (56%), 13 (17.3%) and two (2.7%) isolates, respectively (Fig. 2).
Fig. 2.
Distribution of 70 PMQR genes among 75 nalidixic acid-resistant Escherichia coli isolates originating from humans, chickens and poultry farms/ markets. This chart highlights the prevalence of PMQR among nalidixic acid-resistant E. coli isolates from all sample types originating from poultry farms/markets in Abuja, Nigeria, 2019. The prevalence of PMQR is plotted as bars on the primary axis while the nalidixic acid resistance rate in percentage is plotted as a line graph on the secondary axis. The various data points on the line graph are also displayed on the chart
Two (2.9%) genes encoding extended-spectrum beta-lactamase (ESBL); blaCTX-M-15 and blaCTX-M-65 co-existed with two qnrS-positive isolates recovered from a single chicken originating from a poultry farm and a poultry worker at the chicken market.
Point Mutations in the gyrA, parC, parE, and pmrB genes
Point mutations of quinolone resistance determining region (QRDR) in the DNA gyrase (gyrA) and DNA topoisomerase IV (parC and parE) of the E. coli isolates were determined by WGS. Antimicrobial susceptibility testing using the disk diffusion method showed that 75 (68.2%) isolates were resistant to NA and 43 (39.1%) of these showed point mutations. However, 55.8% (24/43) of the isolates that showed point mutations had CIP-resistant phenotypes. Overall, 43 isolates showed point mutations (Table 3), out of which 40 (93%) showed mutations in gyrA; 30 (69.7%) showed mutations in parC; and ten (23.3%) showed mutations in parE. One of the isolates (2.3%) showed mutations in pmrB which confers colistin resistance. Of the isolates that showed several point mutations, 19 (44.2%) were from chickens; 14 (32.6%) from humans, and ten (23.3%) from the poultry farm/market environments.
Table 3.
Characterization of 43 nalidixic acid-resistant Escherichia coli isolates from humans, chickens, and poultry farm/market environments
| Isolate number | Origin | Mutations to the QRDRs | Other mutations | ||
|---|---|---|---|---|---|
| gyrA | parC | parE | pmrB | ||
| ma 001 | Chicken (M) | S83L | S80I | - | - |
| ma 003 | Environment (M) | S83L | - | - | - |
| ma 008 | Chicken (M) | S83L | - | - | V161G |
| ma 010 | Chicken (M) | S83L | - | - | - |
| ma 013 | Chicken (M) | S83L + D87N | S80I | S458A | - |
| ma 021 | Environment (M) | S83L + D87N | S80I | S458A | - |
| ma 024 | Environment (M) | S83L | S80I | - | - |
| ma 038 | Human (M) | S83L | - | - | - |
| ma 055 | Chicken (M) | S83L | S80I | - | - |
| ma 069 | Human (M) | S83A | - | - | - |
| ma 078 | Human (M) | S83L | - | - | - |
| ma 105 | Human (F) | S83L | - | - | - |
| ma 120 | Environment (F) | S83L | S80I | - | - |
| ma 121 | Chicken (F) | S83L | - | - | - |
| ma 126 | Human (F) | S83L + D87N | S80I | L416F | - |
| ma 138 | Human (M) | S83L + D87N | S80I | - | - |
| ma 140 | Chicken (M) | S83L + D87N | S80I + A56T | - | - |
| ma 143 | Human (F) | S83L + D87N | S80I | - | - |
| ma 146 | Chicken (F) | S83L + D87N | S80I + E84G | - | - |
| ma 182 | Environment (F) | S83L + D87N | S80I | S458A | - |
| ma 197 | Chicken (F) | S83L + D87N | A56T + S80I + E84G | - | - |
| ma 218 | Environment (F) | S83L + D87N | S80I | - | - |
| ma 233 | Environment (F) | S83L | S80I | - | - |
| ma 234 | Chicken (F) | S83L + D87N | S80I | - | - |
| ma 236 | Human (F) | S83L + D87N | S80I | S458A | - |
| ma 238 | Chicken (F) | S83L | S80I | - | - |
| ma 246 | Chicken (M) | S83L + D87N | S80I | - | - |
| ma 249 | Environment (M) | S83L | - | - | - |
| ma 253 | Environment (F) | S83L + D87N | S80I | - | - |
| ma 261 | Human (M) | S83L + D87N | S80I | L416F | - |
| ma 263 | Chicken (F) | S83L + D87N | S80I | L416F | - |
| ma 281 | Chicken (F) | S83L | - | - | - |
| ma 282 | Environment (F) | S83L | - | - | - |
| ma 294 | Chicken (F) | S83L | - | - | - |
| ma 305 | Chicken (F) | S83L + D87N | S80I + A56T | - | - |
| ma 312 | Chicken (F) | S83L + D87N | S80I + A56T | - | - |
| ma 330 | Chicken (F) | S83L + D87N | S80I | S458A | - |
| ma 331 | Chicken (F) | S83L + D87Y | S80I | S458A | - |
| ma 384 | Human (F) | - | A56T | - | - |
| ma 390 | Human (F) | - | A56T | - | - |
| ma 392 | Chicken (F) | - | S57T | I355T | - |
| ma 415 | Human (F) | S83L + D87N | S80I | - | - |
| ma 420 | Human (F) | - | A56T | - | - |
| ma 421 | Human (F) | S83L | - | - | - |
*F – Poultry Farm; M – Poultry Market; QRDRs—quinolone-resistance determining region; Serine to Leucine at codon 83 (S83L); Aspartic acid to Asparagine at codon 87 (D87N); Serine to Alanine at codon 83 (S83A); Serine to Isoleucine at codon 80 (S80I); Alanine to Threonine at codon 56 (A56T); Serine to Threonine at codon 57 (S57T); Glutamic acid to Glycine at codon 84 (E84G); Serine to Alanine at codon 458 (S458A); Isoleucine to Threonine at codon 355 (I355T). “- “: represents the absence of mutations
There were no point mutations observed in gyrB as well as in the two isolates with PMCR genes. The single isolate that showed a mutation in pmrB (V161G) conferring colistin resistance was recovered from a chicken originating from a poultry farm.
Of 75 NA-resistant E. coli isolates, 43 (57.3%) showed point mutations in the topoisomerases while 48 (64%) had at least one PMQR gene. Twenty-three (30.7%) isolates showed both PMQR genes together with point mutations in the topoisomerases. Surprisingly, seven (9.3%) isolates (five from humans and two from chickens) showed neither point mutations nor PMQR genes (Table 4). The CIP MIC for these seven isolates was quite low (≤ 1).
Table 4.
Distribution of PMQR genes and QRDR mutations among 75 NAlidixic acid resistant Escherichia coli isolates
| PMQR genes | Mutations to the QRDRs | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample ID | qnrS | qnrB | aac(6')-Ib-cr | gyrA | parC | parE | pmrB | Location | Category |
| ma 001 | S83L | S80I | Market | Chicken | |||||
| ma 003 | S83L | Market | Environment | ||||||
| ma 008 | S83L | V161G | Market | Chicken | |||||
| ma 010 | qnrS | S83L | Market | Chicken | |||||
| ma 013 | qnrS | S83L + D87N | S80I | S458A | Market | Chicken | |||
| ma 021 | qnrS | S83L + D87N | S80I | S458A | Market | Environment | |||
| ma 024 | S83L | Market | Environment | ||||||
| ma 038 | S83L | Market | Human | ||||||
| ma 051 | qnrS | Market | Environment | ||||||
| ma 055 | qnrS | S83L | Market | Chicken | |||||
| ma 069 | S83A | Market | Human | ||||||
| ma 077 | Market | Human | |||||||
| ma 078 | qnrS | S83L | Market | Human | |||||
| ma 079 | qnrB | Market | Human | ||||||
| ma 088 | qnrB | Market | Human | ||||||
| ma 105 | S83L | Farm | Human | ||||||
| ma 120 | S83L | Farm | Environment | ||||||
| ma 121 | Farm | Chicken | |||||||
| ma 124 | qnrS | Farm | Human | ||||||
| ma 125 | qnrS | qnrB | Farm | Human | |||||
| ma 126 | S83L + D87N | S80I | L416F | Farm | Human | ||||
| ma 138 | S83L + D87N | S80I | Market | Human | |||||
| ma 140 | S83L + D87N | S80I + A56T | Market | Chicken | |||||
| ma 143 | S83L + D87N | S80I | Farm | Human | |||||
| ma 146 | qnrS | S83L + D87N | S80I + E84G | Farm | Chicken | ||||
| ma 158 | qnrS | Farm | Environment | ||||||
| ma159 | qnrS | Farm | Human | ||||||
| ma162 | qnrS | Farm | Chicken | ||||||
| ma 163 | qnrS | Farm | Environment | ||||||
| ma 167 | Farm | Chicken | |||||||
| ma 168 | qnrS | Farm | Chicken | ||||||
| ma 172 | qnrS | Farm | Human | ||||||
| ma 174 | qnrS | qnrB | Farm | Chicken | |||||
| ma 175 | qnrB | Farm | Environment | ||||||
| ma 179 | qnrS | qnrB | Farm | Chicken | |||||
| ma 182 | S83L + D87N | S80I | S458A | Farm | Environment | ||||
| ma 183 | Farm | Human | |||||||
| ma 197 | qnrS | S83L + D87N | A56T + S80I + E84G | Farm | Chicken | ||||
| ma 218 | qnrS | S83L + D87N | S80I | Farm | Environment | ||||
| ma 233 | qnrS | S83L | S80I | Farm | Environment | ||||
| ma 234 | qnrS | S83L + D87N | S80I | Farm | Chicken | ||||
| ma 236 | qnrS | S83L + D87N | S80I | S458A | Farm | Human | |||
| ma 237 | qnrS | Farm | Human | ||||||
| ma 238 | qnrS | S83L | S80I | Farm | Chicken | ||||
| ma 241 | qnrB | Farm | Human | ||||||
| ma 244 | qnrS | Farm | Environment | ||||||
| ma 245 | qnrS | aac(6')-Ib-cr | Market | Chicken | |||||
| ma 246 | qnrB | aac(6')-Ib-cr | S83L + D87N | S80I | Market | Chicken | |||
| ma 249 | qnrS | qnrB | S83L | Market | Environment | ||||
| ma 252 | qnrS | qnrB | Farm | Environment | |||||
| ma 253 | qnrS | S83L + D87N | S80I | Farm | Environment | ||||
| ma 257 | Market | Human | |||||||
| ma 261 | S83L + D87N | S80I | L416F | Market | Human | ||||
| ma 263 | qnrS | S83L + D87N | S80I | L416F | Farm | Chicken | |||
| ma 281 | qnrS | S83L | Farm | Chicken | |||||
| ma 282 | qnrS | S83L | Farm | Environment | |||||
| ma 286 | Farm | Human | |||||||
| ma 287 | qnrS | Farm | Chicken | ||||||
| ma 289 | qnrS | Farm | Environment | ||||||
| ma 293 | qnrS | Farm | Environment | ||||||
| ma 294 | S83L | Farm | Chicken | ||||||
| ma 305 | S83L + D87N | S80I + A56T | Farm | Chicken | |||||
| ma 312 | S83L + D87N | S80I + A56T | Farm | Chicken | |||||
| ma 314 | Farm | Human | |||||||
| ma 330 | S83L + D87N | S80I | S458A | Farm | Chicken | ||||
| ma 331 | qnrS | S83L + D87Y | S80I | S458A | Farm | Chicken | |||
| ma 367 | qnrS | Farm | Environment | ||||||
| ma 370 | qnrS | Farm | Chicken | ||||||
| ma 384 | qnrS | A56T | Farm | Human | |||||
| ma 390 | qnrS | A56T | Farm | Human | |||||
| ma 392 | S57T | I355T | Farm | Chicken | |||||
| ma 415 | qnrB | S83L + D87N | S80I | Farm | Human | ||||
| ma 420 | qnrS | qnrB | A56T | Farm | Human | ||||
| ma 421 | S83L | Farm | Human | ||||||
| ma 422 | qnrS | Farm | Chicken | ||||||
Substitutions in topoisomerase subunits: gyrA, parC, and parE
Of the 40 isolates showing mutations in gyrA, 45% (n = 18) presented single amino acid (AA) substitution from serine to leucine at codon 83 (S83L) while 50% (n = 20) possessed double substitution at S83L and aspartic acid to asparagine at codon 87 (D87N). Of the 30 isolates which showed mutations in parC, 73.3% (n = 22) presented single AA substitution of serine to isoleucine at codon 80 (S80I); 10% (n = 3) AA substitution alanine to threonine at codon 56 (A56T); and 3.2% (n = 1) AA substitution serine to threonine at codon 57 (S57T). Two (6.7%) isolates possessed double substitution at S80I and A56T; 3.2% (n = 1) double substitution at S80I and glutamic acid to glycine at codon 84 (E84G); and 3.2% (n = 1) possessed triple substitution at S80I, A56T, and E84G. Lastly, one (3.2%) isolate presented single AA substitution S57T as shown in (Table 5).
Table 5.
Distribution of substitutions in topoisomerase subunits detected in 43 PMQR Escherichia coli isolates from humans, chickens and poultry farm/market environments
| Target Enzymes | Mutation | Nucleotide change | Amino acid substitution | n = 43 (%) |
|---|---|---|---|---|
|
DNA gyrase gyrA |
S83L; D87N | TCG → TTG; GAC → AAC | S → L; D → N | 20 (46.5) |
| S83L | TCG → TTG | S → L | 18 (41.9) | |
| S83L; D87Y | TCG → TTG; GAC → TAC | S → L; D → Y | 1 (2.3) | |
| S83A | TCG → GCG | S → A | 1 (2.3) | |
| Topoisomerase IV parC | S80I | AGC → ATC | S → I | 22 (51.2) |
| S80I; A56T | AGC → ATC; GCC → ACC | S → I; A → T | 2 (4.7) | |
| S80I; E84G | AGC → ATC; GAA → GGA | S → I; E → G | 1 (2.3) | |
| S80I; E84G; A56T | AGC → ATC; GAA → GGA; GCC → ACC | S → I; E → G; A → T | 1 (2.3) | |
| A56T | GCC → ACC | A → T | 3 (7.0) | |
| S57T | AGC → ACC | S → T | 1 (2.3) | |
| parE | S458A | TCG → GCG | S → A | 6 (14.0) |
| L416F | CTT → TTT | L → F | 3 (7.0) | |
| I355T | ATC → ACC | I → T | 1 (2.3) | |
| pmrB | V161G | GTG → GGG | V → G | 1 (2.3) |
Serine to Leucine at codon 83 (S83L); Aspartic acid to Asparagine at codon 87 (D87N); Serine to Alanine at codon 83 (S83A); Serine to Isoleucine at codon 80 (S80I); Alanine to Threonine at codon 56 (A56T); Serine to Threonine at codon 57 (S57T); Glutamic acid to Glycine at codon 84 (E84G); Serine to Alanine at codon 458 (S458A); Isoleucine to Threonine at codon 355 (I355T); and pmrB- confers colistin resistance
Regarding parE, out of ten isolates that showed mutations, six (60%) showed a single AA substitution from serine to alanine at codon 458 (S458A); three (30%) showed a single AA substitution from leucine to phenylalanine at codon 416 (L416F) while one (10%) showed a single AA substitution from isoleucine to threonine at codon 355 (I355T).
Seven (16.3%) isolates with double AA substitutions in gyrA had point mutations in both parC and parE while one (2.3%) isolate with single AA substitution in gyrA had a point mutation in the pmrB gene, which confers colistin resistance (Table 5). Fifteen (15/32) CIP-resistant isolates showed double mutations in the QRDRs of gyrA, with single, double or triple mutations in parC and single mutation in parE.
Clonal relationship of PMQR E. coli isolates
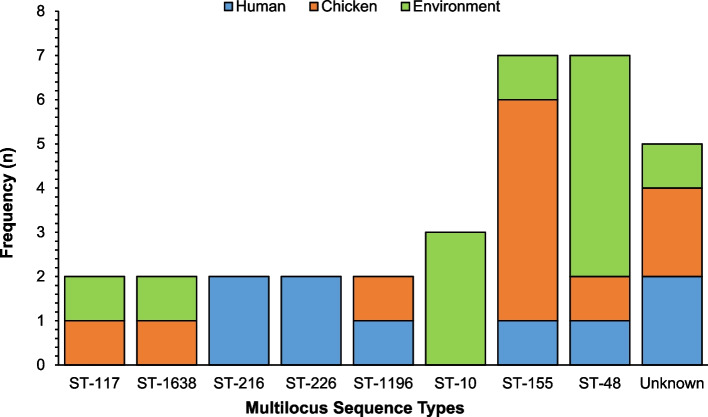
The 70 PMQR E. coli isolates belonged to 53 different sequence types (ST), out of which 9.4% (n = 5) were unknown. Eight major groups were identified by in silico analysis of PMQR-isolates (Fig. 3) namely: ST-48 (13.7%; n = 7), ST-155 (13.7%; n = 7), ST-10 (5.9%; n = 3), ST-1638 (3.9%; n = 2), ST-117 (3.9%; n = 2), ST-216 (3.9%; n = 2), ST-226 (3.9%; n = 2), and ST-1196 (3.9%; n = 2).
Fig. 3.
Multilocus Sequence Types for PMQR Escherichia coli isolates (n = 70) recovered from humans, chickens, and poultry environments. Each bar represents the various PMQR E. coli sequence types for isolates recovered from humans, chickens, and poultry farm/ market environments
In the ST-48 group, the most frequently represented isolates were recovered from poultry litter (5/7), followed by two isolates recovered each from a poultry farmer (1/7) and a chicken from a farm (1/7). Next was the ST-155 group from chickens at the poultry market (4/7); a chicken from a farm (1/7); a poultry farmer (1/7) and a poultry market environment (1/7). The ST-10 group carrying the qnrS1 or qnrS13 or the combination of qnrS1 + qnrB19 genes was from the poultry environment (3/3). The clustering of isolates belonging to the same phylogroup and sequence type was consistent.
The most common STs recovered from the poultry environment were ST-48 (5/7) and ST-155 (3/3). In the ST-48 group, 57.1% (n = 4) of the E. coli isolates harbored two different PMQR gene combinations namely: qnrS1 + qnrB19 (n = 3) and qnrS13 + qnrB19 (n = 1) while in ST-155 group, one isolate harbored PMQR gene combination: qnrS1 + qnrB19.
Two (28.6%) PMQR isolates in the ST-155 group showed point mutations in topoisomerase genes. One isolate from a chicken at the poultry market with single AA substitutions in gyrA (S83L) had a point mutation in parC (S80I) while the remaining isolate recovered from a poultry farmer had a single AA substitution in parC (A56T). One (14.3%) PMQR isolate in the ST-10 group from the poultry market environment harbored a PMCR gene (mcr-1.1).
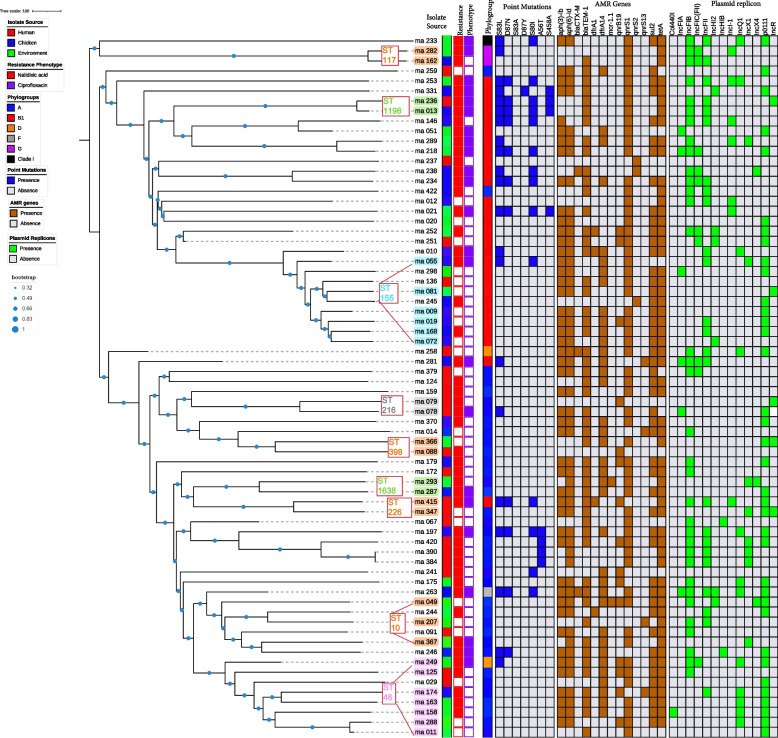
The plasmid replicon profiles of the PMQR E. coli isolates showed that the qnr and aac(6’)-Ib-cr genes were located on different plasmids with IncFIB, p0111, IncFII, IncQ1, IncHI2, IncI1-I, IncX1, and IncX4 being the most commonly shared replicons among humans, chickens and the poultry environment (Fig. 4).
Fig. 4.
Core genome MLST-based phylogeny of PMQR Escherichia coli isolates (n = 70) from humans, chickens, and poultry environments. cgMLST-based phylogenetic tree of PMQR E. coli isolates visualized in Interactive Tree Of Life tool (iTOL). The clustering of isolates was found to be following the core genome. The clustering of isolates belonging to the same phylogroup and sequence type was consistent. Shown for each isolate from left to right are the source, resistance phenotype, phylogroup, point mutations, AMR genes, and plasmid replicons
Determination of Phylogroups
Phylogenetic classification of the PMQR isolates from humans, chickens and the poultry environment showed that 36 (51.4%) of the isolates belonged to phylogroup A, 28 (40%) to phylogroup B1, two (2.9%) to phylogroup D, two (2.9%) to phylogroup G, and one each (1.4%) to phylogroups F and clade I, respectively (Fig. 4). Of the 36 PMQR E. coli isolates belonging to phylogroup A, 31 (86.1%) had the qnrS gene; 11 (30.6%) had the qnrB gene while one (2.8%) had the aac(6’)-Ib-cr gene. Of the 28 PMQR-positive isolates belonging to phylogroup B1, 27 (96.4%) had the qnrS gene; five (17.9%) had the qnrB gene while one (3.6%) had the aac(6’)-Ib-cr gene.
Antimicrobial resistance genes carried on plasmid replicon
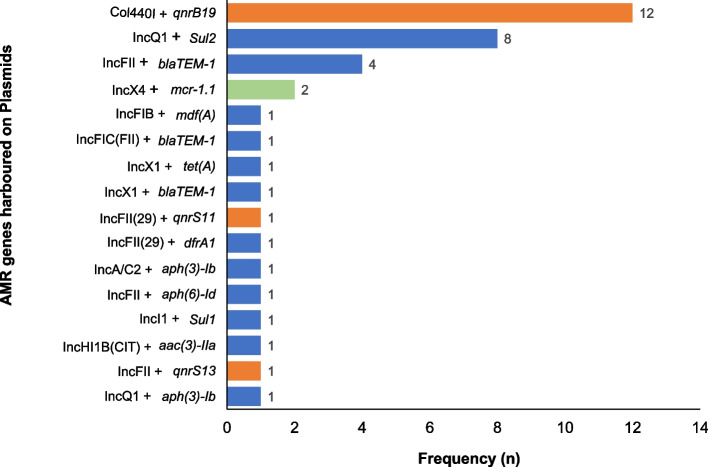
Thirty isolates harbored AMR genes on the plasmid replicon identified on the same assembly scaffold out of which 13 (43.3%) were recovered from the poultry farm/market environments, nine (30%) from chickens, and eight (26.7%) from humans. Five isolates harboring qnr genes also carried other AMR genes including the mcr-1.1 gene on plasmid replicon (Fig. 5). These include two isolates from poultry farm/market environment: ma 049 (IncX4 + mcr 1.1 and Col440l + qnrB19) and ma 288 (IncQ + sul2 and Col440l + qnrB19); two isolates from chickens: ma 174 (IncQ1 + sul2 and Col440l + qnrB19) and ma 179 (IncA/C2 + aph(3″)-lb and Col440l + qnrB19) and one human isolate: ma 415 (IncFII + aph(6)-ld and Col440l + qnrB19). We detected IncFIB (pLF82), a phage plasmid in one PMQR isolate originating from poultry market environment harbouring qnrS1 + qnrB19 gene combination as well as the mcr 1.1 gene.
Fig. 5.
Antimicrobial resistance genes carried on plasmid replicon detected in 70 PMQR Escherichia coli isolates from humans, chickens, and poultry farm/market environments. Each bar represents the resistance genes carried on the plasmid replicon. The orange bars represent PMQR genes (qnrB or qnrS) carried on specific plasmids. The green bar represents the PMCR gene (mcr-1.1) carried on specific plasmids. The blue bars represent other AMR genes carried on plasmids
Discussion
To the best of our knowledge, this study is the first in Nigeria to report the co-occurrence of PMQR genes with PMCR genes (mcr-1.1) carried on the backbones of Col440I and IncX4 plasmids in PMQR E. coli isolates from poultry market and farm environments. This finding may not be surprising as quinolones and colistin, which are considered critically important antimicrobial agents of last resort [11], are often used in poultry production in Nigeria [38]. Over-dependence on quinolones for therapeutic purposes in human and animal populations in Nigeria may be responsible for the selection pressure favoring the development of quinolone resistance in E. coli isolates recovered from different sources [18, 39].
It is also interesting to note that two genes encoding ESBL; blaCTX-M-15 and blaCTX-M-65 were found to co-exist with two qnrS-positive isolates from apparently healthy poultry workers and chickens respectively. This is consistent with findings of other studies reporting that beta-lactamase genes are usually detected along with qnr-positive isolates [14, 40, 41]. A possible explanation for the very few ESBL genes detected in qnr-positive isolates in the present study could be because third- and fourth-generation cephalosporins are rarely used in poultry production in Nigeria.
Previous studies have reported the horizontal spread of AMR through plasmid-mediated qnr genes [22, 25, 42]. The most prevalent PMQR gene from all sources in this study was qnrS1 which was found mainly in the ST-155 group while a combination of qnrB19 and qnrS1 was detected mainly in the ST-48 group. This is consistent with findings from similar studies conducted in Nigeria and other locations where the qnrS1 gene was reported to be the most prevalent [14, 17, 39, 43]. The variability of resistance genes detected in the isolates in the present study is an indication of the faecal carriage of resistance determinants in apparently healthy people, chickens, and the environment.
Fluoroquinolones, which are considered the highest priority important antimicrobials based on WHO classification are indicated for use in human medicine for infections that do not respond to other antimicrobials [11]. Our study shows that a majority of the isolates harboring PMQR genes had QRDR mutations and this is in agreement with reports of other studies [42, 44]. Our data points to the fact that PMQR genes in the human-poultry-environment interface may constitute a better means of assessing quinolone exposure as opposed to chromosomal mutations, an assumption that is consistent with the literature [45].
Quinolones are reported to inhibit cell replication, transcription, and DNA repair in bacteria by disabling essential enzymes DNA gyrase and topoisomerase IV, hence mutations in these enzymes result in quinolone-resistant strains being the main mechanism of quinolone resistance in bacteria [21, 41]. Studies have shown that the gyrA gene mutants carrying the S83L mutation play a major role in quinolone resistance observed in E. coli and are significantly higher than other quinolone resistance mechanisms [15, 42, 46]. It should be noted that the results of the present study showed that 90.7% of quinolone-resistant E. coli strains with the gyrA gene showed single or double mutations at codon 83 resulting in serine to leucine substitution and/or at codon 87 resulting in aspartic acid to asparagine substitution. Previous studies also reported that double mutations in gyrA at codons 83 and 87 have increased [19, 45]. The results presented show that parC mutations encoding the amino acid substitutions S80I and E84G were detected in some PMQR E. coli isolates. This is consistent with the findings of other studies [41, 45, 46]. Mutations in gyrA and parC genes are the main mechanisms of quinolone-resistant E. coli detected from poultry workers, chickens, and poultry farm/market environments in Nigeria in the present study. Our study, however, did not observe any mutations in gyrB. This is consistent with the report of other studies as mutations in gyrB less frequently occur in E. coli strains probably because the phenotypic expression of gyrB mutation is restricted to a narrow panel of bacteria strains [47, 48].
A similar study done in Korea reported that PMQR genes were detected in CIP-resistant E. coli isolates and all the CIP-resistant isolates were MDR [49]. This supports our study results revealing PMQR genes in 77.3% of the CIP-resistant isolates which were also 100% MDR. Our data revealed that the majority of CIP-resistant isolates from humans, chickens, and the poultry environment harbored point mutations in either the gyrA or parC or parE genes. This is consistent with the reports of other studies [41, 49]. The high level of multidrug resistance observed among CIP-resistant isolates in our study is worrisome and reflects the level of antimicrobial use in poultry, hence the need for close monitoring of antimicrobial usage in production.
In the present study, the aac(6’)-Ib-cr gene was very rare and detected in only two isolates recovered from chickens at the poultry market. The prevalence of this gene in NA-resistant E. coli isolates (2.7%) was much lower than observed in the CIP-resistant E. coli isolates (4.5%) and in agreement with a previous study which showed that the aac(6’)-Ib-cr determinant could act additively in generating CIP-resistance [50]. This observation suggests that the CIP-resistant isolates are able to withstand antimicrobial pressure for longer periods allowing for point mutations to occur.
Evidence shows that most quinolone resistant E. coli isolates belonged to phylogroup A and B1 [41] and this supports findings of the present study which observed that majority of the fluoroquinolone resistant E. coli isolates belonged to phylogroup A and B1. This observation suggests that apparently healthy people and chickens may be acting as reservoir of quinolone-resistant strains and this could have occurred as a result of inappropriate use of antimicrobials in humans as well as in poultry production.
The literature has shown that horizontal transfer of PMQR genes is usually accompanied by the presence of other AMR genes [44]. The current study demonstrates the co-occurrence of PMQR genes and other AMR genes most importantly the PMCR gene (mcr-1.1) which were detected in two isolates. However, only one isolate showed chromosomal mutations in the pmrB gene, which confers colistin resistance. This is in agreement with the findings of a similar study carried out in China where both the mcr-1 gene as well as mutations in the pmrB gene were detected in E. coli isolates recovered from food animals [51]. In a single isolate recovered from the poultry market environment, the qnrB19-carrying plasmid also co-harbored the mcr-1.1 gene which was identified on an IncX4 backbone. The qnrS1-positive isolate recovered from poultry manure originating from the farm environment, had two plasmids; IncX4 carrying the mcr-1.1 gene and IncX1 carrying the tet(A) gene. Previous studies [52–54] have identified mcr-1 gene on the backbone of IncX4 plasmids and this is in agreement with our study results. The reason is that the IncX4 plasmids are more prevalent carriers of the mcr-1 gene facilitating the spread of AMR genes in the poultry environments. Seven PMQR E. coli isolates carried more than one plasmid harboring AMR genes in the present study and this is in agreement with another study in Nigeria on quinolone-resistant isolates bearing multiple plasmids [39]. Our findings also showed that one PMQR isolate carried a phage plasmid with three resistant gene types. This is rather not surprising as AMR genes are often carried by phage-plasmids as reported by a recent study [55].
Studies have reported a clonal relationship between CIP-resistant E. coli isolates recovered from humans and chickens [56, 57]. However, the present study did not detect any clonal relationship between PMQR E. coli isolates from humans, chickens, and poultry environments. This is consistent with the findings of a similar study in the Czech Republic which reported that strains from different sources were not related [14]. The results of the present study showed that the qnrB19 genes were located on Col440I plasmids, while qnrS11 and qnrS13 were located on IncFII plasmids. This suggests that although the PMQR E. coli isolates were not clonally related, the high rate of plasmid carriage among the isolates may have been responsible for the emergence of fluoroquinolone resistance observed at the human-animal-environment interface [39, 58]. Previous studies in Nigeria and other locations have also reported that resistant bacteria strains often carry multiple plasmids similar to the findings in the present study [39, 58].
Our study have highlighted the potential for the plasmids to facilitate the horizontal spread of qnrS, qnrB, and mcr-1 genes at the human-animal-environment interface, hence should be considered a potential public health risk. The present study shows that higher levels of fluoroquinolone resistance were detected in E. coli isolates from chickens when compared to isolates from humans and the poultry environment and this is consistent with the literature [25]. This calls for close surveillance and monitoring of the use of fluoroquinolones as well as other critically important antimicrobials in poultry production.
Conclusions
PMQR E. coli isolates were prevalent amongst apparently healthy individuals, chickens, and the poultry farm/market environments in Abuja. ST-155 and ST-48 were the most prevalent STs detected in humans, chickens, and the poultry farm or market environments in this study. PMCR genes carried on IncX4 plasmids and genes encoding for ESBLs were detected in PMQR isolates. Horizontal transfer of PMQR genes among E. coli isolates at the human-poultry-environment interface has public health implications for the spread of AMR. The relevant government ministries and agencies should therefore enforce regulations necessary to restrict the use of critically important antimicrobials in poultry production in Nigeria.
Acknowledgements
The authors would like to acknowledge the intellectual contributions of Mrs. Mba Nwando, Dr. Abiodun Egwuenu, Miss Eme Ekeng, Mr. Akinpelu Afolabi, Mr. Micheal Popoola, and Mr. Chris Chukwu of the National Reference Laboratory (NRL), Nigeria Center for Disease Control (NCDC), Abuja. We appreciate the efforts of members of the Thakur Molecular Laboratory, North Carolina State University; Lyndy Harden, Dawn Hall, Nigatu Atlaw, Ayanna Glaize, Morgan Young, Luke Raymond, Bryson Staley, and Steven Branz towards the success of the research. Special appreciation goes to Dr. Muhammad Shakir Balogun (Nigeria FELTP) for his contributions and Dr. Olufemi Ajumobi (University of Nevada, Reno) for his review and intellectual contribution to the manuscript. We also acknowledge the contributions of Judit Szarvas and Pimlapas Leekitcharoenphon at the National Food Institute, Denmark Technical University towards the success of this research. This research was presented at the American Society for Tropical Medicine and Hygiene (ASTMH) Meeting in 2021.
Abbreviations
- AMR
Antimicrobial resistance
- CGE
Center for Genomics Epidemiology
- CIP
Ciprofloxacin
- CLSI
Clinical and Laboratory Standards Institute
- DNA
Deoxyribonucleic acid
- ESBL
Extended-Spectrum Beta-Lactamase
- FCT
Federal Capital Territory
- FDA
United States Food and Drug Administration
- MDR
Multi-drug Resistance
- MIC
Minimum Inhibitory Concentration
- MLST
Multi-locus Sequence Typing
- NCBI
National Center for Biotechnology Information
- NA
Nalidixic Acid
- PMCR
Plasmid-mediated colistin resistance
- PMQR
Plasmid-mediated quinolone resistance
- QRDRs
Quinolone-Resistance Determining Regions
- ST
Sequence Type
- WGS
Whole-genome Sequencing
- WHO
World Health Organization
Authors’ contributions
MA was the principal investigator, conceptualized the research idea, designed data collection tools, collected data, isolated the organism, performed antibiotic sensitivity testing on the isolates, analyzed and interpreted the data, and wrote the first draft of the manuscript. JK made substantial contributions to conception and design. JK and ST supervised the laboratory aspect of the research. EH performed broth microdilution assay methods, MA performed bioinformatic analyses and RSH interpreted the results. JK, RSH, EO, EH, and ST revised the article critically for important intellectual content. All authors read and approved the final manuscript.
Funding
The whole-genome sequencing was completed by the FDA Genome Trakr program-funded grant 1U18FD00678801.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request. All data generated or analyzed during this study are also included in this published article [and its supplementary information files]. The datasets generated and/or analysed during the current study are also available in the National Center for Biotechnology Information (NCBI) repository (Genome Trakr project) with the accession number to the dataset PRJNA293225.
Declarations
Ethics approval and consent to participate
Ethics approval for the study was obtained from the Federal Capital Territory (FCT) Health Research Ethics Committee (Approval Number: FHREC/2018/01/84/16–07-18) and the Ahmadu Bello University Committee on Animal Use and Care (Approval Number: ABUCAUC/2020/35). Permission was obtained from the management of the poultry farms and chicken markets before the commencement of the study. We assured participants of the confidentiality of all information obtained. All study participants who volunteered to participate in the study signed a written informed consent form detailing the purpose and benefits of the study before sample collection. All procedures were performed following the ethics committee’s guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Leimbach A, Hacker J, Dobrindt UE. coli as an all-rounder: The thin line between commensalism and pathogenicity. Curr Top Microbiol Immunol. 2013;358:3–32. doi: 10.1007/82_2012_303. [DOI] [PubMed] [Google Scholar]
- 2.Delmani F, Jaran AS, Tarazi Y Al, Masaadeh H, Zaki O. Characterization of Ampicillin Resistant Gene ( blaTEM-1 ) Isolated from E . coli in Northern Jordan. Asian J Biomed Pharm Sci. 2017;7:11–5. [Google Scholar]
- 3.Sarowska J, Futoma-Koloch B, Jama-Kmiecik A, Frej-Madrzak M, Ksiazczyk M, Bugla-Ploskonska G, et al. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathogens. 2019;11:10. doi: 10.1186/s13099-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odonkor ST, Addo KK. Prevalence of Multidrug-Resistant Escherichia coli Isolated from Drinking Water Sources. Int J Microbiol. 2018;2018:7204013. doi: 10.1155/2018/7204013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szmolka A, Nagy B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front Microbiol. 2013;4:258. doi: 10.3389/fmicb.2013.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katakweba AAS, Muhairwa AP, Lupindu AM, Damborg P, Rosenkrantz JT, Minga UM, et al. First Report on a Randomized Investigation of Antimicrobial Resistance in Fecal Indicator Bacteria from Livestock, Poultry, and Humans in Tanzania. Microb Drug Resist. 2018;24:260–268. doi: 10.1089/mdr.2016.0297. [DOI] [PubMed] [Google Scholar]
- 7.Arsène-Ploetze F, Chiboub O, Lièvremont D, Farasin J, Freel KC, Fouteau S, et al. Adaptation in toxic environments: comparative genomics of loci carrying antibiotic resistance genes derived from acid mine drainage waters. Environ Sci Pollut Res. 2018;25:1484–5. doi: 10.1007/s11356-017-0535-8. [DOI] [PubMed] [Google Scholar]
- 8.Ayukekbong JA, Ntemgwa M, Atabe AN. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob Resist Infect Control. 2017;6:47. doi: 10.1186/s13756-017-0208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adesokan HK, Akanbi IO, Akanbi IM, Obaweda RA. Pattern of antimicrobial usage in livestock animals in South-Western Nigeria: The need for alternative plans. Onderstepoort J Vet Res. 2015;82:1–6. doi: 10.4102/ojvr.v82i1.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alhaji NB, Isola TO. Antimicrobial usage by pastoralists in food animals in North-central Nigeria: The associated socio-cultural drivers for antimicrobials misuse and public health implications. One Heal. 2018;6:41–47. doi: 10.1016/j.onehlt.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization. WHO list of critically important antimicrobials for human medicine (WHO CIA list): World Health Organization; 2019. https://apps.who.int/iris/handle/10665/325036. License: CC BY-NC-SA 3.0 IGO.
- 12.Thorsteinsdottir TR, Haraldsson G, Fridriksdottir V, Kristinsson KG, Gunnarsson E. Prevalence and Genetic Relatedness of Antimicrobial-Resistant Escherichia coli Isolated From Animals, Foods and Humans in Iceland. Zoonoses Public Health. 2010;57:189–196. doi: 10.1111/j.1863-2378.2009.01256.x. [DOI] [PubMed] [Google Scholar]
- 13.Cho S, Nguyen HAT, McDonald JM, Woodley TA, Hiott LM, Barrett JB, et al. Genetic characterization of antimicrobial-resistant Escherichia coli isolated from a mixed-use watershed in northeast Georgia, USA. Int J Environ Res Public Health. 2019;16:1–14. doi: 10.3390/ijerph16193761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Röderova M, Halova D, Papousek I, Dolejska M, Masarikova M, Hanulik V, et al. Characteristics of Quinolone Resistance in Escherichia coli Isolates from Humans, Animals, and the Environment in the Czech Republic. Front Microbiol. 2017;7:2147. doi: 10.3389/fmicb.2016.02147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanni M, Meucci V, Tognetti R, Cagnardi P, Montesissa C, Piccirillo A, et al. Fluoroquinolone resistance and molecular characterization of gyrA and parC quinolone resistance-determining regions in Escherichia coli isolated from poultry. Poult Sci. 2014;93:856–863. doi: 10.3382/ps.2013-03627. [DOI] [PubMed] [Google Scholar]
- 16.Rabello RF, Bonelli RR, Penna BA, Albuquerque JP, Souza RM, Cerqueira AMF. Antimicrobial resistance in farm animals in Brazil: An update overview. Animals. 2020;10:552. doi: 10.3390/ani10040552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortini D, Fashae K, García-Ferná Ndez A, Villa L, Carattoli A. Plasmid-mediated quinolone resistance and b-lactamases in Escherichia coli from healthy animals from Nigeria. J Antimicrob Chemother. 2011;66:1269–72. doi: 10.1093/jac/dkr085. [DOI] [PubMed] [Google Scholar]
- 18.Ajayi AO, Oluduro AO, Olowe OA, Odeyemi AT, Famurewa O. Plasmid-mediated fluoroquinolone-resistance qnrA and qnrB genes among Escherichia coli from cattle in Ado-Ekiti. Nigeria West Indian Med J. 2012;61:784–788. [PubMed] [Google Scholar]
- 19.Ogbolu D, Alli A, Anorue M, Daini O, Oluwadun A. Distribution of plasmid-mediated quinolone resistance in Gram-negative bacteria from a tertiary hospital in Nigeria. Indian J Pathol Microbiol. 2016;59:322. doi: 10.4103/0377-4929.188108. [DOI] [PubMed] [Google Scholar]
- 20.Monárrez R, Braun M, Coburn-Flynn O, Botelho J, Odetoyin BW, Otero-Vera JI, et al. A large self-transmissible resistance plasmid from Nigeria contains genes that ameliorate a carrying cost. Sci Rep. 2019;9:1–13. doi: 10.1038/s41598-019-56064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooper DC, Jacoby GA. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci. 2015;1354:12–31. doi: 10.1111/nyas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyun J, Keun Cho J, Seuk KK. Prevalence and characterization of plasmid- mediated quinolone resistance genes in Salmonella isolated from poultry in Korea Prevalence and characterization of plasmid-mediated quinolone resistance genes in Salmonella isolated from poultry in Korea. Avian Pathol. 2013;42:221–229. doi: 10.1080/03079457.2013.779636. [DOI] [PubMed] [Google Scholar]
- 23.Dominguez JE, Redondo LM, Figueroa Espinosa RA, Cejas D, Gutkind GO, Chacana PA, et al. Simultaneous carriage of mcr-1 and other antimicrobial resistance determinants in Escherichia coli from poultry. Front Microbiol. 2018;9:1679. doi: 10.3389/fmicb.2018.01679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slettemeås JS, Sunde M, Ulstad CR, Norström M, Wester AL, Urdahl AM. Occurrence and characterization of quinolone resistant Escherichia coli from Norwegian turkey meat and complete sequence of an IncX1 plasmid encoding qnrS1. PLoS ONE. 2019;14:e0212936. doi: 10.1371/journal.pone.0212936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hricová K, Röderová M, Pudová V, Hanulík V, Halová D, Julínková P, et al. Quinolone-resistant Escherichia coli in poultry farming. Cent Eur J Public Health. 2017;25:163–167. doi: 10.21101/cejph.a4328. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Kuang X, Liu J, Sun RY, Li XP, Sun J, et al. Identification of the Plasmid-Mediated Colistin Resistance Gene mcr-1 in Escherichia coli Isolates From Migratory Birds in Guangdong, China. Front Microbiol. 2021;12:1–7. doi: 10.3389/fmicb.2021.755233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charan J, Biswas T. How to calculate sample size for different study designs in medical research? Indian J Psychol Med. 2013;35:121–126. doi: 10.4103/0253-7176.116232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aworh MK, Kwaga J, Okolocha E, Mba N, Thakur S. Prevalence and risk factors for multi-drug resistant Escherichia coli among poultry workers in the Federal Capital Territory, Abuja. Nigeria PLoS One. 2019;14:1–15. doi: 10.1371/journal.pone.0225379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 31st ed. CLSI supplement M100 (ISBN 978-1-68440-104-8 [Print]). USA: Clinical and Laboratory Standards Institute; 2021.
- 30.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carattoli A, Zankari E, Garciá-Fernández A, Larsen MV, Lund O, Villa L, et al. In Silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beghain J, Bridier-Nahmias A, Nagard H Le, Denamur E, Clermont O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genomics. 2018;4:e000192. doi: 10.1099/mgen.0.000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30:138–152. doi: 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clausen PTLC, Aarestrup FM, Lund O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018;19(1):307. doi: 10.1186/s12859-018-2336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oluwasile B, Agbaje M, Ojo O, Dipeolu M. Antibiotic usage pattern in selected poultry farms in Ogun state. Sokoto J Vet Sci. 2014;12:45. doi: 10.4314/sokjvs.v12i1.7. [DOI] [Google Scholar]
- 39.Sumrall ET, Gallo EB, Aboderin AO, Lamikanra A, Okeke IN. Dissemination of the Transmissible Quinolone-Resistance Gene qnrS1 by IncX Plasmids in Nigeria. PLoS ONE. 2014;9:e110279. doi: 10.1371/journal.pone.0110279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salah FD, Soubeiga ST, Ouattara AK, Sadji AY, Metuor-Dabire A, Obiri-Yeboah D, et al. Distribution of quinolone resistance gene (qnr) in ESBL-producing Escherichia coli and Klebsiella spp. in Lomé, Togo. Antimicrob Resist Infect Control. 2019;8:104. doi: 10.1186/s13756-019-0552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang F, Zhang S, Shang X, Wang L, Li H, Wang X. Characteristics of quinolone-resistant Escherichia coli isolated from bovine mastitis in China. J Dairy Sci. 2018;101:6244–6252. doi: 10.3168/jds.2017-14156. [DOI] [PubMed] [Google Scholar]
- 42.Shetty S, Deekshit V, Jazeela K, Vittal R, Rohit A, Chakraborty A, et al. Plasmid-mediated fluoroquinolone resistance associated with extra-intestinal Escherichia coli isolates from hospital samples. Indian J Med Res. 2019;149:192. doi: 10.4103/ijmr.IJMR_2092_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Literak I, Reitschmied T, Bujnakova D, Dolejska M, Cizek A, Bardon J, et al. Broilers as a source of quinolone-resistant and extraintestinal pathogenic Escherichia coli in the Czech Republic. Microb Drug Resist. 2013;19:57–63. doi: 10.1089/mdr.2012.0124. [DOI] [PubMed] [Google Scholar]
- 44.Kao CY, Wu HM, Lin WH, Tseng CC, Yan JJ, Wang MC, et al. Plasmid-mediated quinolone resistance determinants in quinolone-resistant Escherichia coli isolated from patients with bacteremia in a university hospital in Taiwan, 2001–2015. Sci Rep. 2016;6:1–8. doi: 10.1038/srep32281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnning A, Kristiansson E, Fick J, Weijdegård B, Larsson DG. Resistance mutations in gyrA and parC are common in Escherichia communities of both Fluoroquinolone-polluted and uncontaminated aquatic environments. Front Microbiol. 2015;6:1355. 10.3389/fmicb.2015.01355. [DOI] [PMC free article] [PubMed]
- 46.Sáenz Y, Zarazaga M, Briñas L, Ruiz-Larrea F, Torres C. Mutations in gyrA and parC genes in nalidixic acid-resistant Escherichia coli strains from food products, humans and animals. J Antimicrob Chemother. 2003;51:1001–1005. doi: 10.1093/jac/dkg168. [DOI] [PubMed] [Google Scholar]
- 47.Bhatnagar K, Wong A. The mutational landscape of quinolone resistance in Escherichia coli. PLoS ONE. 2019;14:1–18. doi: 10.1371/journal.pone.0224650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perault A, Turlan C, Eynard N, Vallé Q, Bousquet-Mélou A, Giraud E. Repeated Exposure of Escherichia coli to High Ciprofloxacin Concentrations Selects gyrB Mutants That Show Fluoroquinolone-Specific Hyperpersistence. Front Microbiol. 2022;13:908296. doi: 10.3389/fmicb.2022.908296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung YS, Hu YS, Shin S, Lim SK, Yang SJ, Park YH, et al. Mechanisms of quinolone resistance in Escherichia coli isolated from companion animals, pet-owners, and non-pet-owners. J Vet Sci. 2017;18:449–456. doi: 10.4142/jvs.2017.18.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang X, Li J, Zhang Y, Yan H, Wang Y, Shi L, et al. Brief Original Article Detection of plasmid-mediated quinolone resistance determinants and qnrS expression in Enterobacteriaceae clinical isolates. J Infect Dev Ctries. 2014;8:1625–1629. doi: 10.3855/jidc.4617. [DOI] [PubMed] [Google Scholar]
- 51.Huang X, Yu L, Chen X, Zhi C, Yao X, Liu Y, et al. High Prevalence of Colistin Resistance and mcr-1 Gene in Escherichia coli Isolated from Food Animals in China. Front Microbiol. 2017;8:562. doi: 10.3389/fmicb.2017.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flament-Simon SC, de Toro M, Mora A, García V, García-Meniño I, Díaz-Jiménez D, et al. Whole Genome Sequencing and Characteristics of mcr-1–Harboring Plasmids of Porcine Escherichia coli Isolates Belonging to the High-Risk Clone O25b:H4-ST131 Clade B. Front Microbiol. 2020;11:1–20. doi: 10.3389/fmicb.2020.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matamoros S, Van Hattem JM, Arcilla MS, Willemse N, Melles DC, Penders J, et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep. 2017;7:1–10. doi: 10.1038/s41598-017-15539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zając M, Sztromwasser P, Bortolaia V, Leekitcharoenphon P, Cavaco LM, Ziȩtek-Barszcz A, et al. Occurrence and Characterization of mcr-1-Positive Escherichia coli Isolated From Food-Producing Animals in Poland, 2011–2016. Front Microbiol. 2019;10:1753. doi: 10.3389/fmicb.2019.01753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfeifer E, Bonnin RA, Rocha EPC. Phage-Plasmids Spread Antibiotic Resistance Genes through Infection and Lysogenic Conversion. MBio. 2022;13:1–17. doi: 10.1128/mbio.01851-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson JR, Kuskowski MA, Menard M, Gajewski A, Xercavins M, Garau J. Similarity between Human and Chicken Escherichia coli Isolates in Relation to Ciprofloxacin Resistance Status. J Infect Dis. 2006;194:71–78. doi: 10.1086/504921. [DOI] [PubMed] [Google Scholar]
- 57.Agabou A, Lezzar N, Ouchenane Z, Khemissi S, Satta D, Sotto A, et al. Clonal relationship between human and avian ciprofloxacin-resistant Escherichia coli isolates in North-Eastern Algeria. Eur J Clin Microbiol Infect Dis. 2016;35:227–234. doi: 10.1007/s10096-015-2534-3. [DOI] [PubMed] [Google Scholar]
- 58.Salinas L, Cárdenas P, Johnson TJ, Vasco K, Graham J, Trueba G. Diverse Commensal Escherichia coli Clones and Plasmids Disseminate Antimicrobial Resistance Genes in Domestic Animals and Children in a Semirural Community in Ecuador. mSphere. 2019;4:e00316–19. doi: 10.1128/mSphere.00316-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request. All data generated or analyzed during this study are also included in this published article [and its supplementary information files]. The datasets generated and/or analysed during the current study are also available in the National Center for Biotechnology Information (NCBI) repository (Genome Trakr project) with the accession number to the dataset PRJNA293225.