Abstract
Background
DNA methylation (DNAm) plays an important role in lipid metabolism, however, no epigenome-wide association study (EWAS) of lipid levels has been conducted among childhood cancer survivors. Here, we performed EWAS analysis with longitudinally collected blood lipid data from survivors in the St. Jude lifetime cohort study.
Methods
Among 2052 childhood cancer survivors of European ancestry (EA) and 370 survivors of African ancestry (AA), four types of blood lipids, including high-density lipoprotein (HDL), low-density lipoprotein (LDL), total cholesterol (TC), and triglycerides (TG), were measured during follow-up beyond 5-years from childhood cancer diagnosis. For the exposure EWAS (i.e., lipids measured before blood draw for DNAm), the DNAm level was an outcome variable and each of the blood lipid level was an exposure variable; vice versa for the outcome EWAS (i.e., lipids measured after blood draw for DNAm).
Results
Among EA survivors, we identified 43 lipid-associated CpGs in the HDL (n = 7), TC (n = 3), and TG (n = 33) exposure EWAS, and 106 lipid-associated CpGs in the HDL (n = 5), LDL (n = 3), TC (n = 4), and TG (n = 94) outcome EWAS. Among AA survivors, we identified 15 lipid-associated CpGs in TG exposure (n = 6), HDL (n = 1), LDL (n = 1), TG (n = 5) and TC (n = 2) outcome EWAS with epigenome-wide significance (P < 9 × 10−8). There were no overlapping lipids-associated CpGs between exposure and outcome EWAS among EA and AA survivors, suggesting that the DNAm changes of different CpGs could be the cause or consequence of blood lipid levels. In the meta-EWAS, 12 additional CpGs reached epigenome-wide significance. Notably, 32 out of 74 lipid-associated CpGs showed substantial heterogeneity (Phet < 0.1 or I2 > 70%) between EA and AA survivors, highlighting differences in DNAm markers of blood lipids between populations with diverse genetic ancestry. Ten lipid-associated CpGs were cis-expression quantitative trait methylation with their DNAm levels associated with the expression of corresponding genes, out of which seven were negatively associated.
Conclusions
We identified distinct signatures of DNAm for blood lipids as exposures or outcomes and between EA and AA survivors, revealing additional genes involved in lipid metabolism and potential novel targets for controlling blood lipids in childhood cancer survivors.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-023-01447-3.
Keywords: DNA methylation, Lipid levels, Childhood cancer survivors, EWAS, African ancestry, European ancestry
Background
Mounting evidence suggests that epigenetics, specifically DNA methylation (DNAm), plays an important role in lipid metabolism, and epigenome-wide association studies (EWAS) of blood lipid levels have identified robust 5′-cytosine-phosphate-guanine-3′ (CpG) sites and plausible underlying genes associated with lipid metabolism and related diseases [1]. However, an EWAS analysis of lipid levels has not been conducted among survivors of childhood cancer who experience early onset and a substantially higher burden of chronic health conditions (CHCs), compared to community controls without a history of childhood cancer [2, 3]. These health disparities are mostly attributable to genotoxic cancer treatment exposures at a young age with the most notable link being between cardiovascular diseases and exposures to anthracyclines and/or chest-directed radiation therapy (RT) [4]. Recognizing the high burden of CHCs among childhood cancer survivors [2, 3, 5], we have comprehensively analyzed DNAm variations among long-term survivors and conducted systematic investigations of potential casual pathways for treatment-associated CHCs [6]. Our previous findings provide compelling evidence of mediation effect of DNAm between abdominal-RT and dyslipidemia (triglycerides > 150 mg/dL or total cholesterol > 200 mg/dL) [6]. Dyslipidemia is highly prevalent within the broad spectrum of morbidities of childhood and adolescent cancer survivors [7], and a major risk factor for cardiac events, which are the leading cause of noncancer-related premature mortality and account for approximately 26% of deaths among survivors within 45 years of diagnosis [8].
In the general population, African American adults have higher prevalence of high low-density lipoprotein (LDL) and low high-density lipoprotein (HDL) levels but lower prevalence of high triglycerides (TG) than European American adults in both men and women [9, 10]. A study considering racial/ethnic differences among childhood cancer survivors in the St. Jude lifetime cohort study (SJLIFE) reported that childhood cancer survivors of African ancestry (AA) had higher risk of cardiovascular diseases overall including specific conditions such as stroke, heart attack, and heart failure than survivors of European ancestry (EA), potentially explained by the higher prevalence of obesity, diabetes, hypertension, and dyslipidemia among AA survivors [11]. Studies have reported notable population-specific DNAm differences in multiple physical functions (e.g., immunity and kidney development) [12, 13], suggesting that EWAS across populations is critical to the interpretation of health disparities [14]. However, there is a lack of diversity in currently available EWAS data, with most studies conducted in individuals of EA.
To further our understanding of the underlying biological mechanisms of different blood lipid levels among childhood cancer survivors and the differences between EA and AA populations as determined by their genetic ancestry, we employed a comprehensive and agnostic EWAS approach across these two populations. Taking advantage of longitudinal clinical assessments of SJLIFE survivors, we analyzed association of DNAm with blood lipids as exposures (i.e., blood lipids were measured before DNAm) and outcomes (i.e., blood lipids were measured after DNAm). Findings were compared between these two scenarios as well as between the two ancestral groups (i.e., EA and AA). The potential function of significant CpG sites were further demonstrated by their correlations with gene expression levels measured by RNA sequencing. We compared our findings among childhood cancer survivors with the known blood lipid-associated CpGs previously reported in non-cancer general populations. Clinically, the set of lipid-associated CpG sites (i.e., signatures) would facilitate the identification of survivors who have already experienced abnormal lipid levels or at higher risk of abnormal lipid levels in the future.
Results
Characteristics of the study population and EWAS analysis design
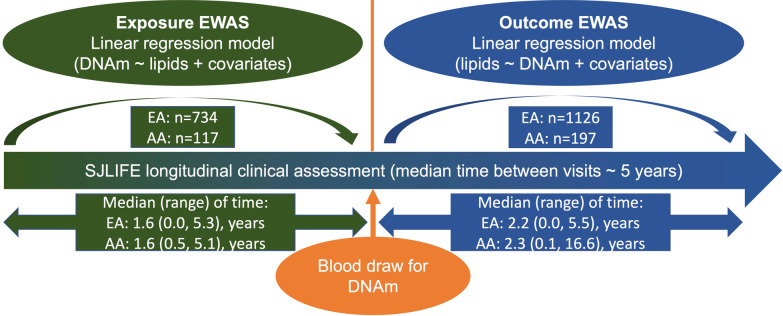
Adult survivors of childhood cancer from the SJLIFE study [15, 16] were included in this analysis (Table 1). Among 2052 EA survivors (median age at blood draw for DNAm = 32.3 years, interquartile range [IQR] = 26.5–40.1 years; 47.2% female), body mass index (BMI) was 9.9–67.7 kg/m2 (Table 1). Among 370 AA survivors (median age at blood draw for DNAm = 29.6 years, IQR = 23.8–37.0 years; 53.2% female), BMI was 12.6–58.9 kg/m2 (Table 1). The summary statistics of the weighted average levels of HDL, LDL, TG, and TC (including the number of survivors with multiple lipid measurements) before or after DNA sampling are shown in Table 1. Compared with survivors of EA, those of AA had lower mean of weighted average of TG as an exposure (81.2 vs. 125.3 mg/dL, P < 0.0001) and outcome (83.4 vs. 130.6, P < 0.0001), TC as an exposure (171.3 vs 179.2 mg/dL, P=0.02) and an outcome (163.8 vs. 182.3 mg/dL, P < 0.0001), and LDL as an outcome (95.0 vs. 106.1 mg/dL, P <0.0001). AA survivors also had higher mean of weighted average of HDL as an exposure (57.2 vs. 51.1 mg/dL, P < 0.0001). The correlations of weighted average levels of lipids before and after DNA sampling were shown in Additional file 1: Table S1. The percentage of survivors taking any lipid control medications before DNAm sampling was 8.04% in EA and 4.86% in AA. The median time and range between the DNAm and pre-lipid profiles are 1.6, 0.0–5.3, years for EA and 1.6, 0.5–5.1, years for AA, and the median time and range between DNAm and post-lipid profiles are 2.2, 0.0–5.5, years for EA, and 2.3, 0.1–16.6, years (Table 1).
Table 1.
Characteristics of the SJLIFE study population
| Characteristic | Survivors of European ancestry | Survivors of African ancestry | Pa | ||
|---|---|---|---|---|---|
| n | (%) | n | (%) | ||
| Total | 2052 | (100.0) | 370 | (100.0) | |
| Sex | 0.03 | ||||
| Male | 1084 | (52.8) | 173 | (46.8) | |
| Female | 968 | (47.2) | 197 | (53.2) | |
| Diagnosis | |||||
| Leukemia | 699 | (34.1) | 77 | (20.8) | < 0.0001 |
| Acute lymphoblastic leukemia | 644 | (31.4) | 67 | (18.1) | |
| Acute myeloid leukemia | 53 | (2.6) | 9 | (2.4) | |
| Other leukemia | 2 | (0.1) | 1 | (0.3) | |
| Lymphoma | 448 | (21.8) | 69 | (18.6) | 0.17 |
| Hodgkin lymphoma | 288 | (14.0) | 45 | (12.2) | |
| Non-Hodgkin lymphoma | 160 | (7.8) | 24 | (6.5) | |
| Sarcoma | 274 | (13.4) | 56 | (15.1) | 0.36 |
| Ewing sarcoma | 74 | (3.6) | 2 | (0.5) | |
| Osteosarcoma | 74 | (3.6) | 18 | (4.9) | |
| Rhabdomyosarcoma | 71 | (3.5) | 18 | (4.9) | |
| Soft tissue sarcoma | 55 | (2.7) | 18 | (4.9) | |
| CNS tumors | 231 | (11.3) | 45 | (12.2) | 0.61 |
| Astrocytoma or glioma | 93 | (4.5) | 18 | (4.9) | |
| Medulloblastoma or PNET | 56 | (2.7) | 10 | (2.7) | |
| Ependymoma | 26 | (1.3) | 5 | (1.4) | |
| Other CNS tumors | 56 | (2.7) | 12 | (3.2) | |
| Embryonal | 276 | (13.5) | 81 | (21.9) | < 0.0001 |
| Wilms tumor | 134 | (6.5) | 44 | (11.9) | |
| Neuroblastoma | 107 | (5.2) | 14 | (3.8) | |
| Germ cell tumor | 35 | (1.7) | 23 | (6.2) | |
| Other | 124 | (6.0) | 42 | (11.4) | 0.0002 |
| Retinoblastoma | 45 | (2.2) | 21 | (5.7) | |
| Hepatoblastoma | 13 | (0.6) | 2 | (0.5) | |
| Melanoma | 12 | (0.6) | 2 | (0.5) | |
| Carcinomas | 24 | (1.2) | 14 | (3.8) | |
| Others | 30 | (1.5) | 3 | (0.8) | |
| Chemotherapy | |||||
| Alkylating agent, classical | 1194 | (58.2) | 202 | (54.6) | 0.20 |
| Alkylating agent, heavy metal | 239 | (11.7) | 63 | (17.0) | 0.004 |
| Alkylating agent, nonclassical | 67 | (3.3) | 12 | (3.2) | 0.98 |
| Anthracyclines | 1190 | (58.0) | 180 | (48.6) | 0.0008 |
| Antimetabolites | 1024 | (49.9) | 133 | (35.9) | < 0.0001 |
| Asparaginase enzymes | 631 | (30.8) | 76 | (20.5) | < 0.0001 |
| Epipodophyllotoxins | 709 | (34.6) | 108 | (29.2) | 0.04 |
| Corticosteroids | 965 | (47.0) | 122 | (33.0) | < 0.0001 |
| Vinca alkaloids | 1482 | (72.2) | 236 | (63.8) | 0.0009 |
| Radiation therapy, region exposed | |||||
| Brain | 629 | (30.7) | 98 | (26.5) | 0.11 |
| Chest | 577 | (28.1) | 102 | (27.6) | 0.83 |
| Abdominal | 412 | (20.1) | 84 | (22.7) | 0.25 |
| Pelvic | 352 | (17.2) | 80 | (21.6) | 0.04 |
| Tobacco smoking status | 0.11 | ||||
| Never smoking | 956 | (46.6) | 183 | (49.5) | |
| Ever smoking | 349 | (17.0) | 51 | (13.8) | |
| Unknown | 747 | (36.4) | 136 | (36.8) | |
| Lipid control medication before DNA sampling | 0.03 | ||||
| Never used | 1887 | (92.0) | 352 | (95.1) | |
| Ever used | 165 | (8.0) | 18 | (4.9) | |
| Lipid control medication after DNA sampling | 0.06 | ||||
| Never used | 1958 | (95.4) | 361 | (97.6) | |
| Ever used | 94 | (4.6) | 9 | (2.4) | |
| Chronic health condition (Weighted average, mg/dL) | n | Mean ± SD | n | Mean ± SD | Pa |
|---|---|---|---|---|---|
| Triglycerides, exposure | 734 | 125.3 ± 92.9 | 117 | 81.2 ± 46.6 | < 0.0001 |
| Triglycerides, outcome | 1126 | 130.6 ± 98.0 | 197 | 83.4 ± 47.8 | < 0.0001 |
| Total cholesterol, exposure | 734 | 179.2 ± 34.9 | 117 | 171.3 ± 32.3 | 0.02 |
| Total cholesterol, outcome | 1124 | 182.3 ± 39.2 | 197 | 163.8 ± 48.0 | < 0.0001 |
| High-density lipoprotein, Exposure | 734 | 51.1 ± 14.7 | 117 | 57.2 ± 14.2 | < 0.0001 |
| High-density lipoprotein, Outcome | 1124 | 50.5 ± 16.3 | 197 | 52.3 ± 20.0 | 0.21 |
| Low-density lipoprotein, Exposure | 717 | 103.6 ± 29.6 | 116 | 98.3 ± 28.4 | 0.08 |
| Low-density lipoprotein, Outcome | 1092 | 106.1 ± 31.5 | 196 | 95.0 ± 35.3 | < 0.0001 |
| Body mass index, kg/m2 | 2051 | 28.5 ± 7.4 | 368 | 29.2 ± 8.0 | 0.13 |
| Median | Range | Median | Range | Pa | |
|---|---|---|---|---|---|
| Age at DNA sampling, years | 32.3 | 18.0, 66.4 | 29.6 | 18.4, 65.1 | < 0.0001 |
| Median age of multiple lipid measurements, yearsb | |||||
| Exposure | 30.0 | 14.6, 65.1 | 26.6 | 15.2, 56.6 | 0.03 |
| Outcome | 35.3 | 19.6, 67.8 | 34.0 | 20.0, 67.5 | 0.01 |
| Time duration between DNA sampling and median age of multiple lipid measurements, years | |||||
| Exposure | 1.6 | 0.0, 5.3 | 1.6 | 0.5, 5.1 | 0.41 |
| Outcome | 2.2 | 0.0, 5.5 | 2.3 | 0.1, 16.6 | 0.44 |
CNS central nervous system, IQR interquartile range, PNET primitive neuroectodermal tumor
aChi-square test for categorical variables, Student’s t-test for continuous variables. Distribution differences for both age and time duration were assessed by Wilcoxon rank sum test
bThe median age of multiple lipid measurements for each survivor was calculated, and the median across all survivors was subsequently derived
After quality control of DNAm data, a total of 689,414 CpGs were further advanced for EWAS. The associations between the DNAm level of each CpG and specific blood lipid level (HDL, LDL, TG, or TC) as an exposure or outcome were analyzed separately (Fig. 1). Quantile–quantile plots of each EWAS among survivors of EA and AA were shown in Additional file 1: Fig. S1 and Additional file 1: Fig. S2, respectively. EWAS of blood lipids among survivors of EA showed moderately low genomic inflation factors between 0.92 and 1.13 (Additional file 1: Fig. S1). EWAS of blood lipids among survivors of AA showed moderately low to high genomic inflation factors between 0.92 and 2.32 (Additional file 1: Fig. S2).
Fig. 1.
Schematic framework of study design. EWAS epigenome-wide association study, DNAm DNA methylation, SJLIFE St. Jude lifetime cohort study. Median (range) of time, the median/range of time between DNAm sampling age and median age of multiple lipid measurements
CpG sites associated with blood lipids among survivors of European ancestry
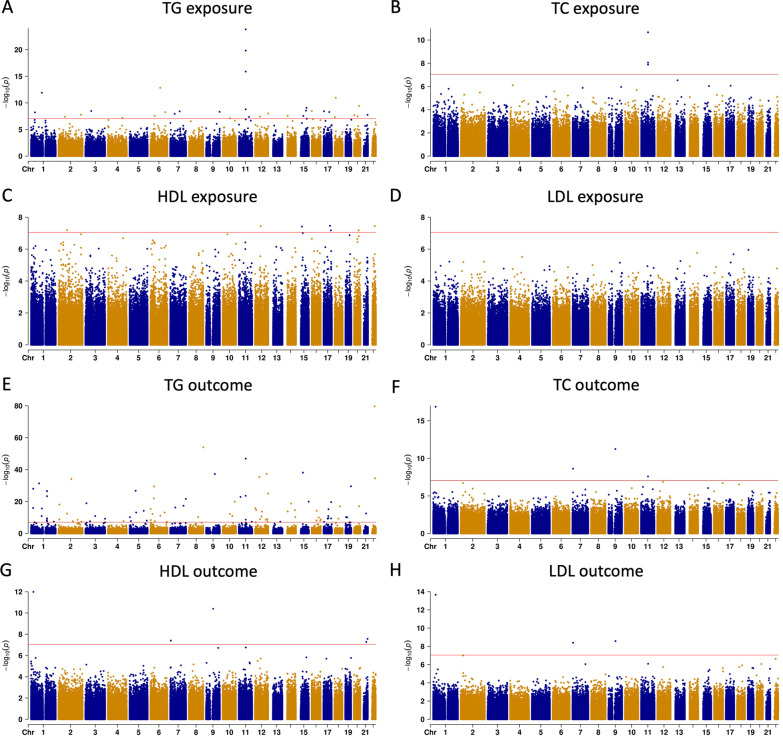
The landscapes of the overall association results among survivors of EA were shown in Fig. 2. Seven, three, and 33 epigenome-wide significant blood lipid-associated CpGs were identified for HDL, TC, and TG, respectively, in the exposure EWAS (P < 9 × 10−8, Fig. 2A–C). No significant CpG achieved epigenome-wide significance in the LDL exposure EWAS among survivors of EA (P < 9 × 10−8, Fig. 2D). Detailed estimates for the association between each CpG and specific blood lipid level as an exposure were provided in Additional file 1: Table S2. Notably, a cluster of three CpGs (cg00574958, cg05325763, and cg17058475), mapped to the 5′UTR of the CPT1A gene, were common in the TG and TC exposure EWAS (Table 2 and Additional file 1: Fig. S3). Five, three, four, and 94 CpGs were significantly associated with HDL, LDL, TC, and TG, respectively, in the outcome EWAS (P < 9 × 10−8, Fig. 2E–H and Additional file 1: Table S3). Three CpGs were common across HDL, LDL, and TC outcome EWAS, including ch.1.829344F mapped to the 5′UTR region of the SRPM1 gene, cg20935223 mapped to the 3′UTR region of the CYTH3 gene, and cg21750129 mapped to the 3′UTR region of the TRPM3 gene (Table 2 and Additional file 1: Fig. S3). No significant CpGs were common between blood lipid exposure and outcome EWAS (P < 9 × 10−8).
Fig. 2.
Manhattan plots of exposure and outcome EWAS of blood lipids among survivors of European ancestry in SJLIFE cohort. EWAS epigenome-wide association study, SJLIFE St. Jude lifetime cohort study, HDL high-density lipoprotein, LDL low-density lipoprotein, TG triglycerides, TC total cholesterol
Table 2.
Overlapping significant CpGs in exposure and outcome EWAS of blood lipid among survivors of European ancestry in SJLIFE cohort
| CpG | chr_hg38 | Start_hg38 | Nearby gene gengebnegene | EWAS | Lipid | Effect | SE | P |
|---|---|---|---|---|---|---|---|---|
| cg00574958 | 11 | 68840153 | CPT1A | Exposure | TC | − 2.25E−03 | 3.30E−04 | 2.15E−11 |
| TG | − 1.28E−03 | 1.20E−04 | 1.50E−24 | |||||
| cg05325763 | 11 | 68840250 | CPT1A | TC | − 1.84E−03 | 3.20E−04 | 1.31E−08 | |
| TG | − 1.13E−03 | 1.17E−04 | 1.48E−20 | |||||
| cg17058475 | 11 | 68840268 | CPT1A | TC | − 2.05E−03 | 3.52E−04 | 8.81E−09 | |
| TG | − 1.11E−03 | 1.31E−04 | 1.27E−16 | |||||
| ch.1.829344F | 1 | 24657005 | SRRM1 | Outcome | HDL | 6.39 | 0.89 | 1.03E−12 |
| LDL | 17 | 2.19 | 2.18E−14 | |||||
| TC | 25.51 | 2.94 | 1.36E−17 | |||||
| cg20935223 | 7 | 6168606 | CYTH3 | HDL | 8.30 | 1.50 | 3.96E−08 | |
| LDL | 22.27 | 3.75 | 3.99E−09 | |||||
| TC | 30.22 | 5.02 | 2.40E−09 | |||||
| cg21750129 | 9 | 71249778 | TRPM3 | HDL | 7.03 | 1.05 | 3.97E−11 | |
| LDL | 15.86 | 2.64 | 2.63E−09 | |||||
| TC | 24.53 | 3.52 | 5.73E−12 |
EWAS epigenome-wide association study, chr_hg38 chromosome in GRCh38/hg38, Start_hg38 CpG start position in GRCh38/hg38, SE standard error, HDL high-density lipoprotein, LDL low-density lipoprotein, TG triglycerides, TC total cholesterol
CpG sites associated with blood lipids among survivors of African ancestry
The overall landscape of CpG associations with blood lipid EWAS among survivors of AA were shown in the Additional file 1: Fig. S4. Six TG-associated CpGs were identified in the exposure EWAS (Additional file 1: Fig. S4A and Table S4), and five TG-associated CpGs, two TC-associated CpGs, one HDL-associated CpG, and one LDL-associated CpG were found in the outcome EWAS (Additional file 1: Fig. S4E–H and Table S4) (P < 9 × 10−8). No significant CpG was found in HDL, LDL, and TC exposure EWAS. Similarly, there was no significant CpGs common in exposure and outcome EWAS among survivors of AA (P < 9 × 10−8). In TG exposure EWAS, there were five significant CpGs mapping to nearby genes including cg26675329 and cg04747445 within 1500 bp upstream of the transcription start site of the IL18RAP gene and the BBX gene, respectively; cg05416955 and cg21376908 in the gene body of the CARD9 gene and the MSI2 gene, respectively; and cg16197879 in the 5’UTR region of the CLDN14 gene (P < 9 × 10−8, Additional file 1: Table S4). In TC outcome EWAS, cg16411101 in the 5’UTR of the SSCP3 gene was significant in both LDL and TC outcome EWAS, and the other significant CpG was cg23724016 in the 3’UTR of the CDHR5 gene (P < 9 × 10−8). In HDL outcome, one significant CpG cg14558275 is mapped to the PIK3CG gene. In TG outcome, cg05416345 and cg01111718 are in the gene body of the IFFO2 gene and the GRIA4 gene, respectively; cg04348872 and cg12686539 are in the first and second exon of the ADCY3 gene and the ZNF891 gene, respectively. We did not identify any overlapping significant CpG between survivors of EA and AA in SJLIFE cohort in any of the exposure or outcome EWAS of blood lipids (P < 9 × 10−8).
Trans-ethnic meta-analysis
In the meta-analysis of blood lipid EWAS among EA and AA survivors, we identified 74 significant lipid-CpG associations (70 unique CpGs, P < 9 × 10−8). Specifically, four, one, and 33 significant CpGs were associated with HDL, TC, and TG exposures, respectively; and two, one, two, and 31 were associated with HDL, LDL, TC, and TG outcomes, respectively (P < 9 × 10−8, Additional file 1: Table S5). Among these significant lipid-CpG associations, twelve did not reach epigenome-wide significance level in either EWAS among survivors of EA or AA alone, including three for HDL exposure, three for TG exposure, and six for TG outcome (Table 3). All 12 had homogeneous effects with the same direction of association in survivors of EA and AA (Phet > 0.1, Table 3). Among the remaining 62 lipid-CpG associations that were significant in EWAS among survivors of EA or AA alone (P < 9 × 10−8), twenty-two had homogeneous effects with the same direction between survivors of EA and AA (Phet > 0.1), twenty-four had opposite directions of association with significant heterogeneity between survivors of AA and EA (Phet < 0.1), and the remaining 16 significant lipid-CpG associations either had the same direction of association but with significant heterogeneity between survivors of EA and AA (Phet < 0.1) or had the opposite direction of association but with homogenous effect (Phet > 0.1) (Additional file 1: Table S5).
Table 3.
Additional significant CpGs identified in meta-analysis of blood lipids EWAS among survivors of European and African ancestry (P < 9 × 10−8)
| EWAS | Lipid | CpG | chr_hg38 | Start_hg38 | Nearby genes | Population | Effect | SE | P | Directions (EA/AA) | I2 | Phet |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exposure | HDL | cg27324117 | chr4 | 153793134 | NA | EA | − 0.0025 | 0.0005 | 2.11E−07 | |||
| AA | − 0.0016 | 0.0012 | 0.20 | |||||||||
| Meta | − 0.0024 | 0.0004 | 8.71E−08 | −/− | 0 | 0.4765 | ||||||
| cg02443467 | chr10 | 47191407 | NA | EA | − 0.0022 | 0.0004 | 1.17E−07 | |||||
| AA | − 0.0016 | 0.0010 | 0.09 | |||||||||
| Meta | − 0.0021 | 0.0004 | 2.30E−08 | −/− | 0 | 0.5916 | ||||||
| cg07373304 | chr16 | 998982 | RP11-161M6.3 | EA | − 0.0021 | 0.0004 | 2.26E−07 | |||||
| AA | − 0.0044 | 0.0017 | 0.01 | |||||||||
| Meta | − 0.0023 | 0.0004 | 1.41E−08 | −/− | 36 | 0.2115 | ||||||
| TG | cg01082498 | chr11 | 68840756 | CPT1A | EA | − 0.0006 | 0.0001 | 1.24E−07 | ||||
| AA | − 0.0005 | 0.0007 | 0.48 | |||||||||
| Meta | − 0.0006 | 0.0001 | 7.23E−08 | −/− | 0 | 0.8322 | ||||||
| cg06004232 | chr16 | 11293335 | RMI2 | EA | 0.0003 | 6.09E−05 | 1.20E−07 | |||||
| AA | 0.0007 | 0.0003 | 0.04 | |||||||||
| Meta | 0.0003 | 0.0001 | 1.79E−08 | +/+ | 5.7 | 0.3032 | ||||||
| cg11686214 | chr19 | 58573616 | CENPBD1P1;MZF1;MZF1-AS1 | EA | 0.0006 | 0.0001 | 1.45E−07 | |||||
| AA | 0.0006 | 0.0006 | 0.24 | |||||||||
| Meta | 0.0006 | 0.0001 | 5.20E−08 | +/+ | 0 | 0.929 | ||||||
| Outcome | TG | cg23550429 | chr2 | 30797935 | CAPN13 | EA | − 40.3640 | 20.2715 | 0.05 | |||
| AA | − 14.9963 | 2.8360 | 3.37E−07 | |||||||||
| Meta | − 15.4833 | 2.8087 | 3.54E−08 | −/− | 34.9 | 0.2152 | ||||||
| cg03260858 | chr3 | 49103337 | QARS | EA | 20.1158 | 13.6051 | 0.14 | |||||
| AA | 38.5596 | 6.9392 | 9.14E−08 | |||||||||
| Meta | 34.7521 | 6.1816 | 1.89E−08 | +/+ | 31.4 | 0.2272 | ||||||
| cg20601481 | chr11 | 67288314 | ANKRD13D | EA | − 27.9673 | 10.8875 | 0.01 | |||||
| AA | − 37.6708 | 7.6983 | 2.10E−06 | |||||||||
| Meta | − 34.4364 | 6.2857 | 4.29E−08 | −/− | 0 | 0.4668 | ||||||
| cg19590858 | chr14 | 34540645 | EAPP;RP11-671J11.5 | EA | − 31.9752 | 9.1836 | 5.17E−04 | |||||
| AA | − 27.2994 | 6.6324 | 5.73E−05 | |||||||||
| Meta | − 28.9022 | 5.3768 | 7.64E−08 | −/− | 0 | 0.6798 | ||||||
| cg20433103 | chr17 | 63941062 | SCN4A | EA | 43.8451 | 13.1675 | 8.98E−04 | |||||
| AA | 36.7039 | 8.0731 | 9.67E−06 | |||||||||
| Meta | 38.6549 | 6.8825 | 1.95E−08 | +/+ | 0 | 0.6438 | ||||||
| cg07439166 | chr19 | 35902502 | AD000864.6;HCST;NFKBID | EA | − 51.5564 | 15.2318 | 7.37E−04 | |||||
| AA | − 34.0310 | 7.4137 | 8.01E−06 | |||||||||
| Meta | − 37.3876 | 6.666 | 2.04E−08 | −/− | 6.6 | 0.3009 |
chr_hg38 chromosome in GRCh38/hg38, Start_hg38 CpG start position in GRCh38/hg38, HDL high-density lipoprotein, LDL low-density lipoprotein, TG triglycerides, TC total cholesterol, EA survivors of European ancestry, AA survivors of African ancestry
Association between DNAm levels of lipid–associated CpGs and gene expression
For each of the blood-lipid associated CpGs, we estimated the linear association between DNAm levels and gene expression levels (adjusting for DNA/RNA sampling age and sex). Among the nearby genes of the significant lipid-associated CpGs among EA, there were no count data (number of reads from RNA sequencing) for 52 genes. Of the remaining 56 CpG-gene pairs, there were ten CpG-gene pairs with significant (FDR < 0.05) associations between the DNAm level of lipid-associated CpGs and gene expression of their nearby genes, including HDAC7 (cg01620154), AXIN2 (cg23475474), ECE1 (cg01758046), TRERF1 (cg07507418), TNRC6B (cg00543524), MICU1 (cg08641767), NUDCD3 (cg01507280), NLN (cg01710244), AKAP1 (cg18807499), and LRP5 (cg24040155) among EA (Table 4). Most of the estimated effects of these CpGs were negative (i.e., increased methylation associated with decreased gene expression), except for ECE1 (cg01758046), NUDCD3 (cg01507280), and NLN (cg01710244). Among the 12 annotated genes of significant lipid-associated CpGs among AA survivors (Additional file 1: Table S4), nine gene had no count data (i.e., number of reads from RNA sequencing). Of the remaining three CpG-gene pairs (PIK3CG-cg14558275, BBX-cg04747445, and IFFO2-cg05416345), there was no significant association between DNAm levels of lipid-associated CpGs and the gene expression among AA.
Table 4.
Significant associations between significant CpGs identified in blood lipid (exposure/outcome) EWAS among survivors of European ancestry and the expression of their nearby genes (FDR < 0.05)
| CpG | Gene Ensembl ID | Nearby gene | Effect | SE | P | FDR |
|---|---|---|---|---|---|---|
| cg01620154 | ENSG00000061273 | HDAC7 | − 3.68 | 0.63 | 4.43E−08 | 1.24E−06 |
| cg23475474 | ENSG00000168646 | AXIN2 | − 4.30 | 0.72 | 2.54E−08 | 1.24E−06 |
| cg01758046 | ENSG00000117298 | ECE1 | 2.28 | 0.56 | 8.54E−05 | 1.34E−03 |
| cg07507418 | ENSG00000124496 | TRERF1 | − 3.49 | 0.87 | 9.55E−05 | 1.34E−03 |
| cg00543524 | ENSG00000100354 | TNRC6B | − 1.49 | 0.39 | 1.69E−04 | 1.89E−03 |
| cg08641767 | ENSG00000107745 | MICU1 | − 1.74 | 0.47 | 2.75E−04 | 2.57E−03 |
| cg01507280 | ENSG00000015676 | NUDCD3 | 3.13 | 0.92 | 8.48E−04 | 6.78E−03 |
| cg01710244 | ENSG00000123213 | NLN | 5.50 | 1.69 | 1.45E−03 | 1.01E−02 |
| cg18807499 | ENSG00000121057 | AKAP1 | − 3.88 | 1.45 | 8.27E−03 | 4.63E−02 |
| cg24040155 | ENSG00000162337 | LRP5 | − 6.12 | 2.28 | 8.09E−03 | 4.63E−02 |
SE standard error, FDR false discovery rate
Cross-reference with the EWAS Catalog
By comparing our findings with previously reported blood lipid-associated CpGs in the general population from the EWAS Catalog, only four overlapping CpGs were identified among EA survivors, including cg00574958 (associated with TG and TC exposure), cg09737197 (associated with TG exposure), and cg17058475 (associated with TG and TC exposure) in CPT1A, and cg03725309 (associated with TG exposure) in SARS (Additional file 1: Tables S6 and S7), and none among AA survivors. Among the remaining 136 novel lipid-associated CpGs in childhood cancer survivors of EA, 26 were mapped to 23 genes that have been previously reported as lipid-associated (Additional file 1: Table S8). Among 12 additional blood lipid-associated CpGs identified in the meta-EWAS, five CpGs were reported to be associated with other traits (e.g., sex, age, Schizophrenia, and ADHD (attention-deficit and hyperactivity disorder)) but only cg01082498 (in the 5’UTR region of the CPT1A gene) was associated with blood lipid level in the EWAS Catalog (Additional file 1: Table S9).
Discussion
Genetic and epigenetic (specifically, DNAm) studies have identified numerous genetic variants or CpG sites that are associated with blood lipids in the general population, hence at least 2572 genes have been implicated in lipid metabolism (Additional file 1: Fig. S5) [17, 18]. We conducted the first EWAS of blood lipids among childhood cancer survivors, including EA and AA survivors from the SJLIFE cohort. Among EA survivors, we identified 149 (140 unique CpGs) significant associations with blood lipid levels; 136 of these were novel findings. Among AA survivors, we found 14 novel significant blood lipid-associated CpGs. There was no overlapping CpGs between EA and AA survivors. A majority of these findings are unique to the survivor population, which may be attributable to childhood cancer diagnoses and/or treatments. For example, two TG exposure associated CpGs, cg24327132 and cg19120513, were associated with chest-RT and abdominal-RT [6].
In the meta-EWAS, twenty-four CpGs had opposite direction of association with significant heterogeneity between EA and AA survivors (Phet < 0.1), suggesting substantial disparity in lipid-associated CpGs between the two ancestral groups. Meta-EWAS yielded eight additional epigenome-wide significant CpGs with heterogeneity in effect size between EA and AA survivors (Phet < 0.1). However, future replication in EA or AA alone with independent data set is warranted to validate such findings.
TG outcome EWAS yielded the greatest number of significant CpGs among EA, with multiple novel lipid-associated CpGs mapped to the same nearby genes, including CDK5RAP3, FCGR2B, HSPA6, and HSPA7. CDK5RAP3, known to play important roles in liver development and hepatic function. Previous research showed that hepatocyte-specific Cdk5rap3 knockout mice suffered post-weaning lethality because of impaired lipid metabolism and serious hypoglycemia [19]. FCGR2B gene encodes FcγRIIb, with a novel role in CD11c+ cells in modulating serum cholesterol and triglyceride levels and maintaining liver cholesterol homeostasis [20]. HSPA6 and HSPA7 are family members of the HSP70 proteins, which are abundantly present in cancer and play crucial roles in cancer development, progression, and metastasis, clinically resulting in diverse outcomes for patient survival [21]. Moreover, cg20935223 was significantly associated with multiple lipid traits in the outcome EWAS and mapped to CYTH3 gene. CYTH3 gene encodes Cytohesin-3, which is essential for insulin receptor signaling and body fat regulation via lipid excretion [22]. Among novel genes (i.e., not genes with nearby lipid-associated CpGs in EWAS Catalog), four of them were reported as high-confidence genes that play a role in lipid levels, including LPIN2 (near cg07616376 associated with TG exposure among EA), SCARB1 (near cg08458758 associated with TG outcome among EA), MSI2 (near cg21376908 associated with TG exposure among AA), and SSBP3 (near cg16411101 associated with LDL and TC outcome among AA) [23].
We integrated gene expression levels from RNA sequencing to further characterize the associations between DNAm and blood lipid levels, which strengthened this study. For example, we demonstrated that ten blood lipid-associated CpGs were associated with levels of expression of the annotated genes, in which seven were inversely associated.
However, it is important to note that there are several limitations in this study. First, although we innovatively designed both exposure and outcome EWAS based on our longitudinal follow-up study, the cross-sectional nature of the data prevented us from disentangling the complex interplay between DNAm and blood lipid levels. Nevertheless, we demonstrated potential regulation of gene expression as plausible mechanisms for DNAm alterations by performing RNA-sequencing analysis. Second, the sample size of survivors of AA was limited, that led to the limited power and the exploratory nature of the AA EWAS (i.e., some findings might be identified by chance). However, differences between EA and AA populations as determined by their genetic ancestry were observed with no overlapping blood lipid levels associated CpG between survivors of EA and those of AA. To further validate the findings, a larger sample size of AA survivors is warranted in the future. Previous methodological work suggested that more than 1,000 subjects are required to achieve 80% power for detection of differential DNAm at nominal genome-wide significance with an odds ratio of 1.15 [24]. Third, we obtained DNAm data at only one time point. In the outcome EWAS, all the blood lipid levels were measured after blood draw for DNAm, so the DNAm may be predictive of blood lipid levels. However, to better assess and interpret the changes of blood lipids level in exposure EWAS, longitudinal DNAm measurement (ideally, after the first blood lipid level measurement) is required to correlate changes of DNAm between two time points with changes in blood lipid levels. Fourth, the follow-up of our cohort is limited and still-ongoing, so there was large proportion of missing data in the analytic setting of bi-directional association between DNAm and lipid levels which requires multiple clinical assessments of lipid levels. Lastly, we did not consider cell type-specific DNAm in the current work. Recent research identified that DNAm variation in diseases, such as type 1 diabetes, can be cell type-specific [25]. Therefore, in the future, we may deconvolute bulk DNAm measured in blood leukocytes into cell type–specific quantities and analyze the DNAm associations of each specific cell type.
Conclusions
Our findings demonstrated distinct DNAm signatures associated with blood lipid levels in EA and AA survivors, and that an additional set of genes may be implicated in lipid metabolism in the survivor population compared to the general population. Further longitudinal studies are warranted to replicate and validate DNAm biomarkers for blood lipid levels and other CHCs to facilitate the clinical translation for improved survivorship care.
Methods
Study population
SJLIFE is a retrospectively-constructed cohort with periodic evaluations of survivors beyond 5-years from childhood cancer diagnosis who were treated at St. Jude Children’s Research Hospital. The details of SJLIFE cohort study have been previously described [15, 16, 26]. Participants complete questionnaires assessing demographic and clinical factors, and receive comprehensive medical and laboratory assessments at each visit to determine health conditions. In this study, a total of 2,052 survivors of EA and 370 survivors of AA, with genome-wide DNAm profiling data, were included [27]. The ancestry for each survivor was determined using genotypes derived from whole-genome sequencing and population admixture analysis as previously described [28]. Primary childhood cancer diagnoses, exposure to chemotherapeutic agents and region-specific radiation dosimetry was obtained from medical records. All SJLIFE survivors completed at least one comprehensive clinical assessment that included a battery of laboratory tests including blood lipid measurement (HDL, LDL, TC, and TG) [26]. The blood lipid levels measured before blood sampling for DNAm were used for exposure EWAS and the blood lipid levels measured after were used in outcome EWAS (Fig. 1). Weighted average was calculated if there were multiple measurements, and time intervals between two consecutive measurements were used as weights. We excluded lipid measurements without fasting. Samples with only one lipid measurements (coinciding with the time point for the blood draw for DNAm) were excluded to ensure that our exposure and outcome EWAS examined the temporal association between DNAm and blood lipid levels. All participants provided written informed consent, with institutional review board approval at St. Jude Children’s Research Hospital.
DNAm profiling and data processing
Illumina Infinium® MethylationEPIC BeadChip array including 850K CpG sites was used to generate genome-wide DNAm profiling on DNA derived from peripheral blood mononuclear cells (PBMC) collected at each follow-up visit for SJLIFE survivors. Details about laboratory experimental processes, array scanning, and DNAm bioinformatics data analysis were previously described by Song et al. [6].
Genotyping based on whole-genome sequencing (WGS)
Genotyping was based on whole-genome sequencing data of blood derived DNA from 4402 SJLIFE survivors as previously described [29, 30]. Details about data processing, genotyping calling as well as additional genotype quality control criteria and procedures were previously described in Dong et al. [31].
Epigenome-wide association analysis
Bidirectional EWAS was conducted using a multivariable linear regression to test the association of DNAm levels at each CpG (M-value, continuous variable) with blood lipid levels (continuous variable). We performed principal components analysis of methylation levels of all CpG sites to quantify potential batch effects in the DNAm data. The top four principal components were determined by the change rate of eigenvalues [6] and were included as covariates in the regression model. We also performed principal components analysis of genotypes derived from WGS to quantify the population substructure in EA and AA survivors. The top four principal components were determined by the change rate of eigenvalues and were included as covariates in the regression model. In the exposure EWAS, a multivariable linear regression model was used with lipid level (weighted average was calculated if there were multiple measurements, and time intervals between two consecutive measurements were used as weights) prior to DNA sampling as an independent variable and DNAm as a dependent variable, adjusting for sex, age at DNA sampling, leukocyte subtype proportions, top four significant genetic principal components, top four methylation principal components, cancer treatments, median age of lipid measurement, BMI, cigarette smoking, and lipid lowering medicine use. All these covariates were potential confounding factors for DNAm level of each CpG, and hence were considered in the exposure EWAS. Cancer treatments included chemotherapy and radiation therapy within 5 years from primary childhood cancer diagnosis. The chemotherapy agents included classical alkylating agent, anthracyclines, corticosteroids, vinca alkaloids, asparaginase enzymes, antimetabolites, and epipodophyllotoxins. The region-specific RT included brain-RT, chest-RT, abdomen-RT, and pelvis-RT. For smoking status as a categorical variable, we included three levels (“never”, “ever”, and “unknown”) in the model. BMI was measured at the same time as DNAm sampling. CpGassoc R package was used for the exposure EWAS analyses [32]. In the outcome EWAS, a multivariable linear regression model was used for DNAm (age-, sex-, cell-type-, genotype principal components-, and methylation principal components- adjusted) as an independent variable and lipid level (weighted average was calculated if there were multiple measurements, and time intervals between two consecutive measurements were used as weights) after DNA sampling as a dependent variable. For EA survivors, a base model without DNAm level but including the complete set of covariate (i.e., sex, cancer treatments, median age of lipid measurement, BMI, smoking, lipid lowering medicine use, lipid level measured at DNA sampling, age at DNA sampling, and polygenic risk score for specific lipid level (in EA only) was fitted. Cancer treatment exposures that were not statistically significant (P > 0.05) in the base model were subsequently excluded. In the final model, DNAm level of each CpG was added for the EWAS analysis. For AA survivors, considering the smaller sample size and potential overfitting, a similar but slightly different variable selection approach was taken by additionally excluding BMI, smoking status, and lipid lowering medicine use if any of these was not statistically significant (P > 0.05) in the base model. Polygenic risk score for specific lipid level was constructed by following the same approach described previously [28] for EA survivors. Custom R code was used for the outcome EWAS analyses. A P value less than 9 × 10−8 was deemed as epigenome-wide significance level corresponding to 5% family-wise error [33].
RNA-sequence profiling and data processing
RNA was extracted from the same PBMC used for DNA methylation profiling. Details of library construction, sequencing, and data processing were described previously [27]. Briefly, paired-end 100 cycle sequencing was performed on a NovaSeq 6000 (Illumina). After quality control procedures, raw reads from the fastq files were aligned to the GRCh38.p13 version (v31) of the reference human genome from GENCODE through the automated internal pipeline [34]. The generated bam files were sorted and used to build an index using Samtools (version 1.9) [35] then used as inputs for counting reads using htseq-count [36] with GENCODE v31 gene annotation gtf file.
A total of 165 samples of RNA-seq data (135 EA survivors and 30 AA survivors) were available for further analysis. After removing transcripts with mean read counts across all 165 samples less than 10, a total of 12,882 genes were determined to be expressed in PBMC. Transcripts per million (TPM) [37] were calculated and transformed in the form of log2(TPM + 0.01). The function normalizeQuantiles in the limma package [38] in R (version 3.6.1) was used for quantile normalization [39] of the log-transformed values before further downstream analyses.
Expression quantitative trait methylation
We used the Infinium® MethylationEPIC BeadChip array annotations (v1.0 B5) provided by Illumina (https://webdata.illumina.com/downloads/productfiles/methylationEPIC/infinium-methylationepic-v-1-0-b5-manifest-file-csv.zip) to map CpGs to their annotated genes. For 135 RNA-seq samples from EA survivors (out of 165 in total), the normalized expression values of the nearest genes of each lipid-associated CpGs were extracted to fit a linear regression against the DNAm levels of each CpG.
Additional statistical and bioinformatic analyses
Quantile–Quantile plots were generated from P-values in each EWAS using the R lattice package. GenABEL R package [40] was used to estimate genomic inflation factor (i.e., lambda). We searched lipid-associated CpGs and nearby genes identified in our study and compared with those previously reported from the EWAS Catalog [17].
Supplementary Information
Additional file 1. Supplementary Information.
Acknowledgements
The authors thank all individuals who participated in this study.
Abbreviations
- AA
African ancestry
- CpG
5′-Cytosine-phosphate-guanine-3′
- CHCs
Chronic health conditions
- DNAm
DNA methylation
- EA
European ancestry
- HDL
High-density lipoprotein
- LDL
Low-density lipoprotein
- RT
Radiotherapy
- SJLIFE
St. Jude lifetime cohort study
- EA
European ancestry
- TG
Triglycerides
- TC
Total cholesterol
Author contributions
ZW designed the study. LLR and MMH and KKN assisted in or provided support for data collection and recruitment of study participants. JE, HM, EP, GN, EW, and JZ assisted in or supervised sample processing, performed DNA/RNA extraction and library preparation for whole-genome sequencing, the Infinium MethylationEPIC Array and RNA sequencing. QD, CC, NS, NQ, and ZW performed the methylation analysis and statistical analysis. QD and ZW wrote the first draft of the manuscript. All authors contributed to data interpretation and writing and approved the final manuscript for publication.
Funding
This work was supported by funding from the V Foundation [Grant # DT2020-014], the National Institutes of Health [Grant # CA021765, CA195547], and the American Lebanese Syrian Associated Charities (ALSAC). The funders of the study had no role in the design and conduct of the study and were not involved in collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or the decision to submit the manuscript for publication.
Availability of data and materials
The DNA methylation data is accessible through the St. Jude Cloud (https://stjude.cloud).
Declarations
Ethics approval and consent to participate
The SJLIFE study protocol was approved by the Institutional Review Board (IRB) at St. Jude Children’s Research Hospital.
Consent for publication
Not applicable.
Competing interests
We declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qian Dong and Cheng Chen contributed equally as co-first authors
References
- 1.Mittelstraß K, Waldenberger M. DNA methylation in human lipid metabolism and related diseases. Curr Opin Lipidol. 2018;29(2):116–124. doi: 10.1097/MOL.0000000000000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robison LL, Hudson MM. Survivors of childhood and adolescent cancer: life-long risks and responsibilities. Nat Rev Cancer. 2014;14(1):61–70. doi: 10.1038/nrc3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhakta N, Liu Q, Ness KK, Baassiri M, Eissa H, Yeo F, et al. The cumulative burden of surviving childhood cancer: an initial report from the St Jude lifetime cohort study (SJLIFE) Lancet. 2017;390(10112):2569–2582. doi: 10.1016/S0140-6736(17)31610-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armenian SH, Armstrong GT, Aune G, Chow EJ, Ehrhardt MJ, Ky B, et al. Cardiovascular disease in survivors of childhood cancer: insights into epidemiology, pathophysiology, and prevention. J Clin Oncol. 2018;36(21):2135–2144. doi: 10.1200/JCO.2017.76.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khanna A, Pequeno P, Gupta S, Thavendiranathan P, Lee DS, Abdel-Qadir H, et al. Increased risk of all cardiovascular disease subtypes among childhood cancer survivors: population-based matched cohort study. Circulation. 2019;140(12):1041–1043. doi: 10.1161/CIRCULATIONAHA.119.041403. [DOI] [PubMed] [Google Scholar]
- 6.Song N, Hsu CW, Pan H, Zheng Y, Hou L, Sim JA, et al. Persistent variations of blood DNA methylation associated with treatment exposures and risk for cardiometabolic outcomes in long-term survivors of childhood cancer in the St. Jude lifetime cohort. Genome Med. 2021;13(1):53. doi: 10.1186/s13073-021-00875-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hudson MM, Ness KK, Gurney JG, Mulrooney DA, Chemaitilly W, Krull KR, et al. Clinical ascertainment of health outcomes among adults treated for childhood cancer. JAMA. 2013;309(22):2371–2381. doi: 10.1001/jama.2013.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reulen RC, Winter DL, Frobisher C, Lancashire ER, Stiller CA, Jenney ME, et al. Long-term cause-specific mortality among survivors of childhood cancer. JAMA. 2010;304(2):172–179. doi: 10.1001/jama.2010.923. [DOI] [PubMed] [Google Scholar]
- 9.Frank AT, Zhao B, Jose PO, Azar KM, Fortmann SP, Palaniappan LP. Racial/ethnic differences in dyslipidemia patterns. Circulation. 2014;129(5):570–579. doi: 10.1161/CIRCULATIONAHA.113.005757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheet SF. Older Americans and cardiovascular diseases. Dallas: American Stroke Association; 2013. [Google Scholar]
- 11.Liu Q, Leisenring WM, Ness KK, Robison LL, Armstrong GT, Yasui Y, et al. Racial/ethnic differences in adverse outcomes among childhood cancer survivors: the childhood cancer survivor study. J Clin Oncol. 2016;34(14):1634–1643. doi: 10.1200/JCO.2015.66.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Husquin LT, Rotival M, Fagny M, Quach H, Zidane N, McEwen LM, et al. Exploring the genetic basis of human population differences in DNA methylation and their causal impact on immune gene regulation. Genome Biol. 2018;19(1):222. doi: 10.1186/s13059-018-1601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breeze CE, Batorsky A, Lee MK, Szeto MD, Xu X, McCartney DL, et al. Epigenome-wide association study of kidney function identifies trans-ethnic and ethnic-specific loci. Genome Med. 2021;13(1):74. doi: 10.1186/s13073-021-00877-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breeze CE, Wong JYY, Beck S, Berndt SI, Franceschini N. Diversity in EWAS: current state, challenges, and solutions. Genome Med. 2022;14(1):71. doi: 10.1186/s13073-022-01065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson MM, Ness KK, Nolan VG, Armstrong GT, Green DM, Morris EB, et al. Prospective medical assessment of adults surviving childhood cancer: study design, cohort characteristics, and feasibility of the St. Jude lifetime cohort study. Pediatr Blood Cancer. 2011;56(5):825–836. doi: 10.1002/pbc.22875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudson MM, Ehrhardt MJ, Bhakta N, Baassiri M, Eissa H, Chemaitilly W, et al. Approach for classification and severity grading of long-term and late-onset health events among childhood cancer survivors in the St. Jude lifetime cohort. Cancer Epidemiol Biomark Prev. 2017;26(5):666–674. doi: 10.1158/1055-9965.EPI-16-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battram T, Yousefi P, Crawford G, Prince C, Sheikhali Babaei M, Sharp G, et al. The EWAS catalog: a database of epigenome-wide association studies. Wellcome Open Res. 2022;7:41. doi: 10.12688/wellcomeopenres.17598.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–d1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang R, Wang H, Kang B, Chen B, Shi Y, Yang S, et al. CDK5RAP3, a UFL1 substrate adaptor, is crucial for liver development. Development. 2019;146(2):1–13. doi: 10.1242/dev.169235. [DOI] [PubMed] [Google Scholar]
- 20.Marvin J, Rhoads JP, Major AS. FcγRIIb on CD11c(+) cells modulates serum cholesterol and triglyceride levels and differentially affects atherosclerosis in male and female Ldlr(-/-) mice. Atherosclerosis. 2019;285:108–119. doi: 10.1016/j.atherosclerosis.2019.04.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albakova Z, Armeev GA, Kanevskiy LM, Kovalenko EI, Sapozhnikov AM. HSP70 multi-functionality in cancer. Cells. 2020;9(3):587. doi: 10.3390/cells9030587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jux B, Gosejacob D, Tolksdorf F, Mandel C, Rieck M, Namislo A, et al. Cytohesin-3 is required for full insulin receptor signaling and controls body weight via lipid excretion. Sci Rep. 2019;9(1):3442. doi: 10.1038/s41598-019-40231-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanoni S, Graham SE, Wang Y, Surakka I, Ramdas S, Zhu X, et al. Implicating genes, pleiotropy, and sexual dimorphism at blood lipid loci through multi-ancestry meta-analysis. Genome Biol. 2022;23(1):1–42. doi: 10.1186/s13059-022-02837-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai PC, Bell JT. Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. Int J Epidemiol. 2015;44(4):1429–1441. doi: 10.1093/ije/dyv041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul DS, Teschendorff AE, Dang MA, Lowe R, Hawa MI, Ecker S, et al. Increased DNA methylation variability in type 1 diabetes across three immune effector cell types. Nat Commun. 2016;7:13555. doi: 10.1038/ncomms13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howell CR, Bjornard KL, Ness KK, Alberts N, Armstrong GT, Bhakta N, et al. Cohort profile: the St. Jude lifetime cohort study (SJLIFE) for paediatric cancer survivors. Int J Epidemiol. 2021;50(1):39–49. doi: 10.1093/ije/dyaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song N, Sim JA, Dong Q, Zheng Y, Hou L, Li Z, et al. Blood DNA methylation signatures are associated with social determinants of health among survivors of childhood cancer. Epigenetics. 2022;17:1–15. doi: 10.1080/15592294.2022.2030883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Liu Q, Wilson CL, Easton J, Mulder H, Chang T-C, et al. Polygenic determinants for subsequent breast cancer risk in survivors of childhood cancer: the St Jude lifetime cohort study (SJLIFE) polygenic determinants for subsequent breast cancer risk. Clin Cancer Res. 2018;24(24):6230–6235. doi: 10.1158/1078-0432.CCR-18-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin N, Wang Z, Liu Q, Song N, Wilson CL, Ehrhardt MJ, et al. Pathogenic germline mutations in DNA repair genes in combination with cancer treatment exposures and risk of subsequent neoplasms among long-term survivors of childhood cancer. J Clin Oncol. 2020;38(24):2728. doi: 10.1200/JCO.19.02760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Wilson CL, Easton J, Thrasher A, Mulder H, Liu Q, et al. Genetic risk for subsequent neoplasms among long-term survivors of childhood cancer. J Clin Oncol. 2018;36(20):2078. doi: 10.1200/JCO.2018.77.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong Q, Song N, Qin N, Chen C, Li Z, Sun X, et al. Genome-wide association studies identify novel genetic loci for epigenetic age acceleration among survivors of childhood cancer. Genome Med. 2022;14(1):1–12. doi: 10.1186/s13073-022-01038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barfield RT, Kilaru V, Smith AK, Conneely KN. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics. 2012;28(9):1280–1281. doi: 10.1093/bioinformatics/bts124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansell G, Gorrie-Stone TJ, Bao Y, Kumari M, Schalkwyk LS, Mill J, et al. Guidance for DNA methylation studies: statistical insights from the Illumina EPIC array. BMC Genom. 2019;20(1):366. doi: 10.1186/s12864-019-5761-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature. 2014;506(7489):451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131(4):281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 38.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 40.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23(10):1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary Information.
Data Availability Statement
The DNA methylation data is accessible through the St. Jude Cloud (https://stjude.cloud).