Abstract
Background:
A complex relationship between adipose tissue and malignancy, involving an inflammatory response, has been reported. We sought to assess the prevalence of white adipose tissue (WAT) inflammation in patients with endometrial cancer (EC), and the association with circulating inflammation markers. Furthermore, we aimed to characterize the pathways activated in and the cell type composition of adipose tissue in patients with EC.
Methods:
Adipose tissue and blood samples were prospectively collected from 101 patients with EC at initial surgery. WAT inflammation was determined based on adipocytes surrounded by macrophages forming crown-like structures. Circulating levels of metabolic syndrome–associated and inflammatory markers were quantified. RNA-sequencing was performed on adipose samples (n=55); differential gene expression, pathway, and cellular decomposition analyses were performed using state-of-the-art bioinformatics methods.
Results:
WAT inflammation was identified in 46 (45.5%) of 101 patients. Dyslipidemia, hypertension, and diabetes mellitus were significantly associated with WAT inflammation (p<0.05). WAT inflammation was associated with greater body mass index (BMI) (p<0.001) and higher circulating levels of leptin, high-sensitivity C-Reactive Protein (hsCRP), and interleukin-6 (IL-6), as well as lower levels of adiponectin and sex hormone–binding globulin (p<0.05). Transcriptomic analysis demonstrated increased levels of pro-inflammatory and pro-neoplastic–related gene expression in inflamed omental adipose tissue.
Conclusions:
WAT inflammation is associated with metabolic syndrome, obesity, and inflammatory markers, as well as increased expression of pro-inflammatory and pro-neoplastic genes.
Keywords: endometrial cancer, obesity, tissue inflammation, RNA-sequencing, white adipose
Precis
We analyzed adipose tissue and blood samples prospectively collected from 101 endometrial cancer patients at initial surgery and found approximately half had white adipose tissue inflammation, which was associated with metabolic syndrome, obesity, inflammatory markers, and increased expression of pro-inflammatory and pro-neoplastic genes. Our findings offer new insights into how adipose tissue inflammation correlates with clinical factors in endometrial cancer.
INTRODUCTION
Obesity is a recognized and growing epidemic, with rates as high as 60% in parts of the United States. Among the numerous detrimental effects on health, obesity has been shown to not only increase the risk of certain malignancies but has also been linked to worse cancer-specific outcomes in multiple tumor types, including breast, esophagus, ovary, and uterus (1-3). There is evidence to suggest that obesity is a trigger of chronic inflammation, which creates a pro-neoplastic environment via local effects in the tumor microenvironment as well as systemic effects in the host (4). Endometrial cancer (EC) has been strongly linked to obesity. Women with a body mass index (BMI) >40 kg/m2 have been shown to have a greater than 6-fold relative risk of death from uterine cancer compared to lean women (5). This association between obesity and EC has been largely attributed to increased circulating estrogen levels in obese women. However, in other hormonally driven cancers, such as breast cancer, local adipose tissue dysfunction contributes to disease progression in addition to bioavailable estrogens (6). Accordingly, we hypothesized that adipose tissue dysfunction mediates the relationship between obesity and EC. The identification of pathophysiological and/or molecular alterations that link metabolic syndrome/obesity to adverse EC outcomes could inform the development of novel therapeutic strategies that mitigate the growing burden of obesity-related EC mortality.
The link between inflammation and the promotion of neoplastic processes has been well described over the years (7-10). In the obesogenic state, adipose tissue hypertrophy can lead to adipocyte stress and death, ultimately triggering inflammation. Adipose tissue inflammation is histologically recognized when macrophages engulf dead or dying adipocytes, forming a crown-like structure (CLS) (11). The presence of CLS is indicative of inflammation in white adipose tissue (WAT) (2, 11). In breast and squamous cell carcinomas of the tongue, WAT inflammation represented by CLS not only correlates with multiple cardiometabolic disorders such as hyperlipidemia, hypertension, and diabetes, but also has a negative impact on progression-free survival (6, 12). In prostate cancer, the presence of CLS has been associated with higher grade disease (13). The mechanisms likely responsible for these associations of CLS with worse disease histology and course are thought to be at least in part mediated by the effects of WAT inflammation on insulin signaling, lipid metabolism, and circulating levels of steroid hormones (14, 15).
Given the well-recognized epidemiologic link between obesity and EC outcomes and the contribution of WAT inflammation to the obesity-cancer link in several other cancers, we investigated the effects of WAT inflammation in EC. The objectives of this study were to assess the prevalence of WAT inflammation in a prospectively accrued cohort of patients with EC, and to investigate whether WAT inflammation correlates with circulating blood markers in the setting of EC. Finally, we interrogated the omental microenvironment, a common site of EC metastasis, by examining relationships among obesity, WAT inflammation, and gene expression.
MATERIALS AND METHODS
Clinical data and biospecimen collection
This study was approved by the Institutional Review Board of Memorial Sloan Kettering Cancer Center (MSK), and written informed consents were obtained. Blood and non-malignant tissue specimens were prospectively collected from 101 patients with newly diagnosed EC, regardless of histology, consented to undergo surgical staging between 2015 and 2019. Non-neoplastic omental tissue samples collected during surgery were cut in half, of which one half was formalin-fixed and paraffin-embedded (FFPE) and the other half flash frozen on the day of surgery. Fasting blood samples were obtained preoperatively on the day of surgery, which were centrifuged and stored as plasma and serum within 3 h of collection at −80°C.
Clinicopathologic data, including age and BMI at the time of surgery, were obtained from the electronic medical records. EC histology was based on the final surgical pathology report. Stage was assigned based on the International Federation of Gynecology and Obstetrics (FIGO) classification system (16). Treatment information, such as adjuvant chemotherapy, radiation therapy, or hormonal therapy, was also collected. Menopausal status was categorized as either premenopausal or postmenopausal based on National Comprehensive Cancer Network criteria (17). Co-morbidities, including history of breast cancer, smoking status, alcohol use, hypertension, diabetes mellitus, and hyperlipidemia, were recorded.
Tissue assessment
Non-malignant omental WAT specimens were assessed for the presence of CLS. For the 101 cases, one 5-μm section was obtained from each of 5 FFPE blocks. In cases with fewer than 5 FFPE blocks, sections were obtained from all available blocks, and additional sections were obtained at 50-μm intervals until 5 sections were obtained from a given case. Sections were then subjected to hematoxylin & eosin (H&E) staining and immunohistochemical analysis of the macrophage marker CD68 (mouse monoclonal KP1 antibody; Dako; dilution, 1:4000), as previously described (6). CD68 immunohistochemical results were reviewed by the study pathologist (DDG) to detect the presence or absence of CLS, as previously described (6, 13). The total WAT area was calculated, after exclusion of epithelial and fibrotic tissues, using Image J Software (National Institutes of Health; Bethesda, MD) on the digital photographs of each slide. WAT inflammation was then quantified as number of CLSs per square centimeter of WAT (CLS/cm2), as previously described (13).
Blood measurements
Plasma levels of glucose (BioAssay Systems) and insulin (Mercodia), as well as leptin, adiponectin, sex hormone–binding globulin, high-sensitivity C-reactive protein (hcCRP), and interleukin-6 (IL6; R&D Systems) were measured by enzyme-linked immunosorbent assay (ELISA).
RNA extraction and RNA-sequencing (RNA-seq) analysis
Frozen sections of the omental tissue specimens were stained with H&E and reviewed by a pathologist (DDG) to verify they lacked neoplastic cells. These flash-frozen, non-neoplastic omental tissue specimens were placed in RNA lysis buffer, and total RNA was extracted using the RNeasy Lipid Tissue Mini kit (Qiagen; Hilden, Germany) according to the manufacturer’s instructions, and quantified using the Qubit Fluorometer (Invitrogen; Waltham, MA). We obtained sufficient RNA from 59 cases, which underwent polyA RNA-seq using validated protocols at MSK’s Integrated Genomics Operation (IGO), as previously described (18). RNA-seq data analysis was performed in R. Genes expressed at a count-per-million (CPM) below 0.5 in at least 2 samples were considered poorly expressed, which filtered out 5,832 genes from the analysis, resulting in a total of 17,497 genes. CPM were assessed using the cpm function from the edgeR R library. Normalization factor was estimated using calcNormFactors function to scale the raw library sizes. Counts were transformed to log2-counts per million (logCPM) and adjusted for mean-variance effects to be fit with a linear model in Limma using the voom function. After quality control, 4 samples were removed from the analysis due to lack of minimum standard of quality with a library size below the 5th percentile, totaling 55 samples that underwent analysis. Differential gene expression analysis was then performed between CLS-positive (n=27) and CLS-negative (n=28) samples fitting a linear model of each gene given a series of arrays using Limma. Genes found to be differentially expressed, with an absolute log 2-fold change >1.5 and a false discovery rate (FDR) <0.1, were subjected to gene ontology enrichment and pathway analyses; enrichment analysis was performed using GSVA R library, and the gene set variation analysis (gsva) function was used to estimate enrichment scores. We used ‘gsva’ method and a Poisson kernel for non-parametric estimation of the cumulative distribution function of expression levels across samples. A second differential gene expression analysis was performed between CLS-positive and CLS-negative samples, adjusting the model for BMI. Gene-sets were retrieved from MsigDB version 7.0 (19). The median value of the enrichment score was estimated for each pathway between the CLS-positive and CLS-negative cohorts. Pathways were considered enriched when the absolute difference between the average value of the two groups was greater than the 95th quantile. We performed a similar differential expression analyses focusing only on immune-related genes (IRGs). The list of IRGs was obtained from https://www.immport.org (July 2020).
Immune cell deconvolution
Immune cell deconvolution was performed using CIBERSORT absolute mode using the Immunedeconv R package (20, 21). Counts were transformed into transcripts per kilobase million (TPM). The CIBERSORT score was calculated as an estimate of the enrichment of a specific population of cells within a given sample.
Gene set variation analysis and antigen processing and presenting machinery score
To evaluate the antigen processing and presenting machinery as one determinant of tissue immunogenicity, a GSVA was performed to obtain an antigen processing and presenting machinery score (APS) measuring the overall expression enrichment of specific antigen processing and presenting machinery (APM) genes. APM gene expression was quantified using the GSVA method implemented in the R package GSVA (22). The GSVA evaluated the expression of a list of APM genes—PSMB5, PSMB6, PSMB7, PSMB8, PSMB9, PSMB10, TAP1, TAP2, ERAP1, ERAP2, CANX, CALR, PDIA3, TAPBP, B2M, HLA-A, HLA-B, and HLA-C— compared to all other genes. The enrichment score was then defined as the APS. This list of APM genes used to quantify the APS were retrieved from https://github.com/XSLiuLab/tumor-immunogenicity-score, as previously described by Wang et al. (23, 24).
Statistical Analysis
Wilcoxon rank sum tests, chi-square tests of independence, and the Fisher exact test were used to compare CLS-negative and CLS-positive patients according to their clinicopathologic features. We tested for differences in inflammation between levels of each categorical variable using Wilcoxon rank sum tests for dichotomous variables and Kruskal-Wallis tests when there were greater than 2 variable levels. We reported Spearman rank correlation coefficients to measure and characterize the strength and direction of any monotonic association between inflammation and continuous variables. Serology data on 98 of these patients was available, with two values for each measurement for a total of 196 measurements. We conducted clustered Wilcoxon rank sum tests using the Rosner-Glynn-Lee method (random permutation) to test for differences between CLS-negative and CLS-positive samples according to their blood measurement variables, while adjusting for multiple samples per patient (25). Statistical differences between groups across immune subpopulation in the CIBERSORT analysis was assessed using Wilcoxon test. A two-tailed P-value was reported and considered statistically significant if less than 0.05. Statistical computations were performed using R3.6.1 and SAS Software Version 9.4 (The SAS Institute, Cary, NC).
Data availability
RNA-seq data will be available in the Sequence Read Archive (SRA) upon acceptance of this manuscript.
RESULTS
Clinical components of metabolic syndrome and clinicopathologic features stratified by the presence of WAT inflammation
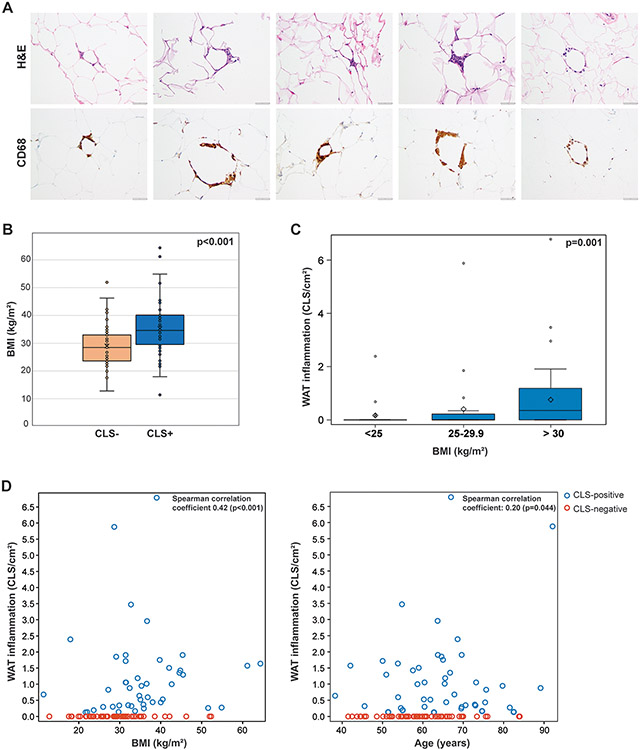
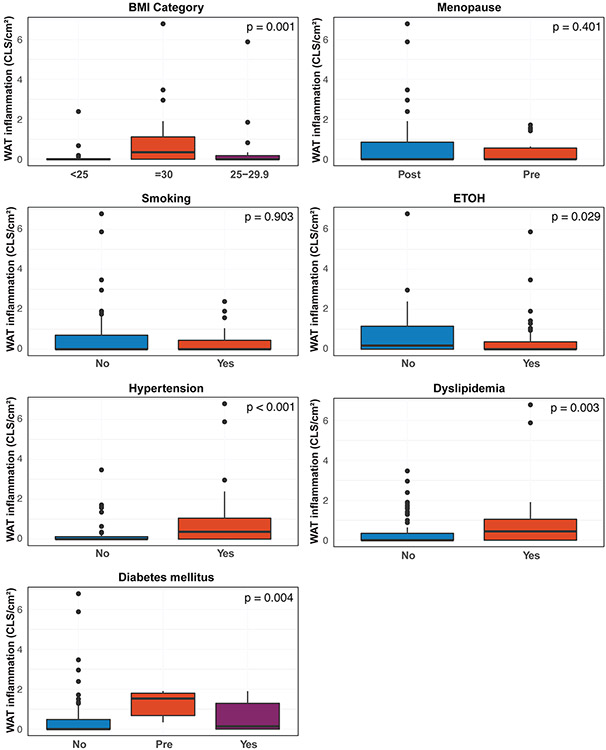
CLS was present in 46 (45.5%) and absent in 55 (54.5%) of the 101 omental biopsies from patients with newly diagnosed EC. The CLSs identified in these EC patients were similar to those described in breast cancer, characterized by the presence of dead/dying adipocytes surrounded by CD68+ macrophages (Figure 1A). Patients with CLS-positive omental tissue had elevated risk factors for metabolic syndrome, including a median age of 65.2 years (range, 38.2-92.1 years) at the time of initial surgery, compared to 58.6 years (range, 41.5-84.0 years) for patients with CLS-negative omental tissue (p=0.017). BMI was significantly greater among CLS-positive patients (median, 34.6 kg/m2; range, 11.4-64.3 kg/m2) compared to CLS-negative patients (28.5 kg/m2; range, 12.8-52.3 kg/m2; p<0.001; Figure 1B). Although a greater proportion of CLS-positive patients compared with CLS-negative patients were postmenopausal at EC diagnosis, the difference was not significant (81.8% vs. 72.7%, respectively; p=0.4). A comparison of clinical features of metabolic syndromes demonstrated that significantly more patients with CLS-positive tissue compared with CLS-negative tissue had hypertension (69.6% vs. 34.5%, respectively; p<0.001), dyslipidemia (39.1% vs. 12.7%, respectively; p=0.005), and/or pre-diabetes or diabetes mellitus (26.1% vs. 9.1%, respectively; p=0.009; Table 1).
Figure 1:
(A) Crown-like structures (CLSs) in endometrial cancer. Hematoxylin and eosin–stained sections of adipose tissue derived from patients with newly diagnosed endometrial cancer (left), and results of anti-CD68 immunohistochemical analysis of macrophages identified CLSs (right). (B) Box plot displaying the distribution of body mass index (BMI) based on the presence or absence of CLS/white adipose tissue (WAT) inflammation in patients with endometrial cancer. CLS-positive patients had a median BMI of 34.6 kg/m2 (range, 11.4-64.3), compared to 28.5 kg/m2 (range, 12.8-52.3) for CLS-negative patients (p<0.001). Wilcoxon rank sum test was performed. (C) Box plot displaying the quantity of WAT inflammation by BMI category among all patients. The median quantity of WAT inflammation was 0.00 CLS/cm2 (range, 0.00-2.39) for patients with a BMI <25 kg/m2, 0.00 CLS/cm2 (range, 0.00-5.88) for patients with a BMI of 25-29 kg/m2, and 0.35 CLS/cm2 (range, 0.00-6.79) for patients with a BMI ≥30 kg/m2. Kruskal-Wallis test was performed. (D) Scatterplots demonstrating measures of association between levels of WAT inflammation and BMI or age as continuous variables. Spearman rank correlation coefficients (nonparametric measures of association) were reported.
Table 1:
Clinicopathologic features of patients with endometrial cancer based on the presence or absence of inflammation
| Entire cohort (n=101) |
Cases with RNA-sequencing (n=55) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Characteristic | Overall n=101 |
CLS + n=46 |
CLS − n=55 |
p- value |
Overall n=55 |
CLS + n=27 |
CLS − n=28 |
p- value |
| Age, years (range) | 60.9 (38.2-92.1) | 65.2 (38.2- | 58.6 (41.5- | 0.017 | 58.6 (38.2- | 65.0 (38.2- | 58.6 (41.5- | 0.048 |
| BMI, kg/m2 | 31.1 (11.4-64.3) | 34.6 (11.4- | 28.5 (12.8- | <0.001 | 32.5 (11.4- | 34.8 (11.4- | 28.5 (12.8- | 0.013 |
| BMI category | 0.006 | 0.025 | ||||||
| Normal <25 kg/m2 | 22 (21.8) | 6 (13.0) | 16 (29.1) | 11 (20.0) | 2 (7.4) | 9 (32.1) | ||
| Overweight 25-29.9 kg/m2 | 24 (23.8) | 7 (15.2) | 17 (30.9) | 9 (16.4) | 4 (14.8) | 5 (17.9) | ||
| Obese ≥30 kg/m2 | 55 (54.5) | 33 (71.7) | 22 (40.0) | 35 (63.6) | 21 (77.8) | 14 (50.0) | ||
| Race | >0.9 | 0.631 | ||||||
| White | 88 (87.1) | 41 (89.1) | 47 (85.5) | 49 (89.1) | 25 (92.6) | 24 (85.7) | ||
| Black | 3 (3.0) | 1 (2.2) | 2 (3.6) | 1 (1.8) | 0 (0) | 1 (3.6) | ||
| Asian | 6 (5.9) | 2 (4.3) | 4 (7.3) | 3 (5.5) | 1 (3.7) | 2 (7.1) | ||
| Declined to answer | 4 (4.0) | 2 (4.3) | 2 (3.6) | 4 (3.6) | 1 (3.7) | 1 (3.6) | ||
| Histology | 0.12 | 0.123 | ||||||
| Endometrioid | 74 (73.3) | 35 (76.1) | 39 (70.9) | 44 (80.0) | 24 (88.9) | 20 (71.4) | ||
| Carcinosarcoma | 2 (2.0) | 0 (0.0) | 2 (3.6) | 2 (3.6) | 0 (0.0) | 2 (7.1) | ||
| Clear cell | 1 (1.0) | 1 (2.2) | 0 (0.0) | 1 (1.8) | 1 (3.7) | 0 (0) | ||
| Serous | 10 (9.9) | 7 (15.2) | 3 (5.5) | 2 (3.6) | 1 (3.7) | 1 (3.6) | ||
| Mixed endometrioid/clear | 5 (5.0) | 2 (4.3) | 3 (5.5) | 2 (3.6) | 1 (3.7) | 1 (3.6) | ||
| Mixed endometrioid/serous | 2 (2.0) | 0 (0.0) | 2 (3.6) | 2 (3.6) | 0 (0.0) | 2 (7.1) | ||
| No residual ca | 7 (6.9) | 1 (2.2) | 6 (10.9) | 2 (3.6) | 0 (0) | 2 (7.1) | ||
| Stage * | 0.9 | 0.217 | ||||||
| Stage I | ||||||||
| IA | 68 (70.8) | 34 (75.6) | 34 (66.7) | 39 (70.9) | 22 (81.5) | 17 (60.7) | ||
| IB | 15 (15.6) | 7 (15.6) | 8 (15.7) | 8 (14.5) | 4 (14.8) | 4 (14.3) | ||
| Stage II | 2 (2.1) | 1 (2.2) | 1 (2.0) | 0 (0) | 0 (0) | 0 (0) | ||
| Stage III | ||||||||
| IIIA | 6 (6.2) | 2 (4.4) | 4 (7.8) | 4 (7.3) | 1 (3.7) | 3 (10.7) | ||
| IIIB | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||
| IIIC | 4 (4.2) | 1 (2.2) | 3 (5.9) | 2 (3.6) | 0 (0) | 2 (7.1) | ||
| Stage IV | ||||||||
| IVA | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||
| IVB | 1 (1.0) | 0 (0.0) | 1 (2.0) | 0 (0) | 0 (0) | 0 (0) | ||
| Menopausal status ** | 0.4 | 0.746 | ||||||
| Pre | 23 (23.2) | 8 (18.2) | 15 (27.3) | 12 (21.8) | 5 (18.5) | 7 (25.0) | ||
| Post | 76 (76.8) | 36 (81.8) | 40 (72.7) | 43 (78.2) | 22 (81.5) | 21 (75.0) | ||
| Smoking | 28 (27.7) | 13 (28.3) | 15 (27.3) | >0.9 | 14 (25.5) | 8 (29.6) | 6 (21.4) | 0.547 |
| ETOH *** | 53 (53.0) | 18 (40.0) | 35 (63.6) | 0.031 | 29 (52.7) | 11 (40.7) | 18 (64.3) | 0.079 |
| Hypertension | 51 (50.5) | 32 (69.6) | 19 (34.5) | <0.001 | 27 (49.1) | 20 (74.1) | 7 (25.0) | <0.00 |
| Dyslipidemia | 25 (24.8) | 18 (39.1) | 7 (12.7) | 0.005 | 15 (27.3) | 13 (48.1) | 2 (7.1) | <0.00 |
| Diabetes mellitus | 0.009 | 0.011 | ||||||
| Pre | 6 (5.9) | 6 (13.0) | 0 (0.0) | 5 (9.1) | 5 (18.5) | 0 (0.0) | ||
| Yes | 11 (10.9) | 6 (13.0) | 5 (9.1) | 6 (10.9) | 4 (14.8) | 2 (7.1) | ||
| No | 84 (83.2) | 34 (73.9) | 50 (90.9) | 44 (80.0) | 18 (66.7) | 26 (92.9) | ||
CLS, crown-like structure; BMI, body mass index; ETOH, ethanol/ alcohol. Values are presented as numbers (%) or median (range).
1 missing in the CLS-positive group and 4 missing in the CLS-negative group.
2 missing in the CLS-positive group.
1 missing in the CLS-positive group.
There were no statistically significant differences in distribution of race, histological subtype, or FIGO stage between patients with and without CLS. Regarding comorbidities, smoking did not differ significantly between patients with CLS-positive and CLS-negative tissue (28.3% vs. 27.3%, respectively). Among patients who indicated regular alcohol intake, a significantly greater proportion were CLS-negative compared to CLS-positive (63.3% vs. 40.0%, respectively; p=0.031; Table 1). These findings persisted when analyzing the subset of 55 cases that underwent RNA-seq (Table 1).
The presence and severity of inflammatory foci in WAT based on BMI and other metabolic factors
Further investigation into the relationship between WAT inflammation and cardiometabolic disorders revealed that not only the presence of inflammation but also the degree of inflammation correlated with obesity (p<0.001), diabetes mellitus (p=0.004), hypertension (p<0.001), and dyslipidemia (p=0.003; Figure 1C, Figure 2). A further evaluation of BMI on a continuous scale showed there was an association between increasing levels of BMI with increasing levels of inflammation (Spearman coefficient=0.42, p<0.001). Age was also evaluated on a continuous scale, demonstrating that increasing age also correlated with increasing inflammation (Spearman coefficient=0.20, p=0.044; Figure 1D).
Figure 2: The severity of inflammatory foci in white adipose tissue (WAT) based on metabolic risk factors.
Box plots displaying the quantity of WAT inflammation based on the presence of diabetes mellitus, menopausal state, smoking status, alcohol use, presence of hypertension, and dyslipidemia across all samples. Wilcoxon rank sum test was used for dichotomous variables, and Kruskal-Wallis tests were used when there were greater than 2 variable levels.
Alterations in blood serologies of patients with WAT inflammation
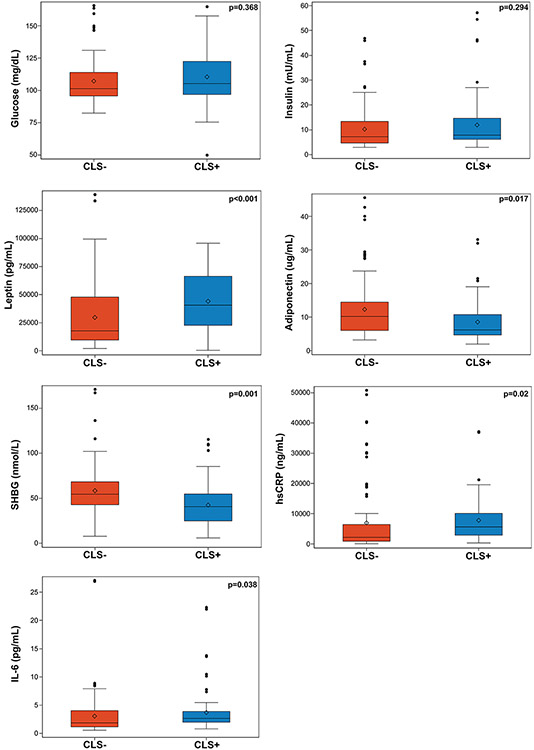
The presence versus absence of CLS in patients with EC was associated with elevated levels of circulating leptin (median [IQR]: 40,635 pg/mL [range, 22,809.5-66,333] vs. 17,776 pg/mL [range, 9,595.8-47,787], respectively; p<0.001) and lower levels of circulating adiponectin (median [IQR]: 6.2 ug/mL [range, 4.7-10.8] vs. 10.2 ug/mL [range, 6.1-14.4], respectively; p=0.017). Significantly lower levels of sex hormone–binding globulin, which is associated with higher risk of metabolic syndrome, insulin resistance, type II diabetes mellitus, and cardiovascular risk, was present in CLS-positive compared with CLS-negative patients (median [IQR]: 40.8 nmol/L [range, 25.3-54.7] vs. 54.5 nmol/L [range, 43.3-68.3], respectively; p=0.001) (26, 27). In addition, the presence versus absence of CLS was associated with increased levels of well-accepted inflammatory factors, including hsCRP (median [IQR]: 5,596.5 ng/mL [range, 2,909.2-10,036.8] vs. 2,201 ng/mL [range, 910.8-6,389.0], respectively; p=0.020) and IL-6 (median [IQR]: 2.6 pg/mL [range, 2.0-3.8] vs. 1.9 pg/mL [range, 1.1-4.0], respectively; p=0.038; Table S1; Figure 3).
Figure 3: Circulating blood biomarkers in relation to white adipose tissue (WAT) inflammation.
Box plots displaying levels of various circulating blood markers based on the presence or absence of crown-like structures (CLSs). WAT inflammation is determined by the presence of CLS. Statistical test performed was a clustered Wilcoxon rank sum test using Rosner-Glynn-Lee method (random permutation). SHBG, sex hormone–binding globulin.
WAT inflammation and corresponding changes in visceral tissue gene expression
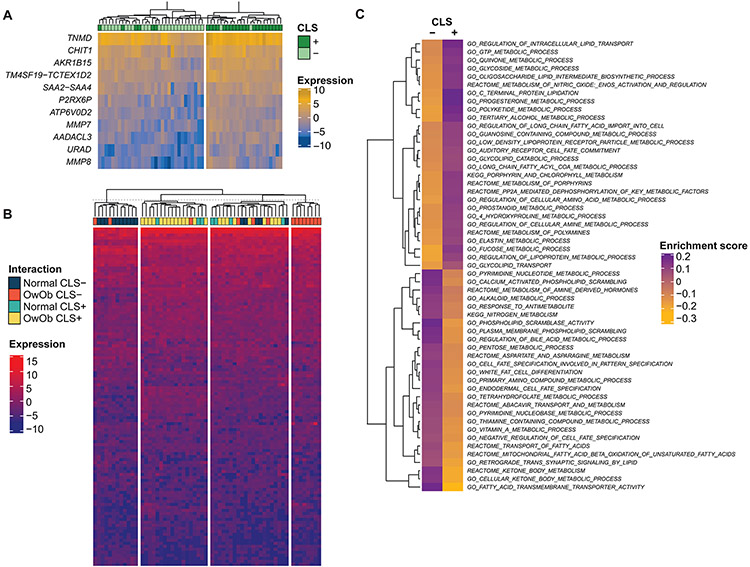
To understand the implications of WAT inflammation at the transcriptomic level, omental tissue from 55 patients underwent RNA-seq–based analysis. CLS was present in 27 (49%) and absent in 28 (51%) of these cases (Table 1). Differential gene expression analysis identified 11 genes—AADACLS, MMP8, AKR1B15, TNMD, CHIT1, P2RX6P, MMP7, SAA2-SAA4, TM4SF19-TCTEX1D2, URAD, and ATP6V0D2—that were significantly differentially expressed between CLS-positive and CLS-negative tissues (log2 fold change in gene expression greater than 1.5; FDR <0.1; Table S2; Figure 4A). A subset of these differentially expressed genes play a role in inflammation and tumorigenic response. For example, non-malignant omental tissues from CLS-positive patients displayed increased expression of MMP8 and MMP7 (LogFC 2.05, adj p-value 0.08, LogFC 1.77, adj p-value 0.07, respectively), which are involved in inflammatory responses and facilitate cancer invasion and angiogenesis through mechanisms such as degradation of extracellular matrix macromolecules (28-31). Consistent with the increase in the number of CLSs and macrophages in CLS-positive cases, we observed increased expression of CHIT1 and SAA2-SAA4, both of which are expressed in macrophages (LogFC 1.77, adj p-value 0.05; LogFC 1.60, adj p-value 0.06, respectively), and SAA2-SAA4 is also a marker of inflammation (32, 33).
Figure 4: Transcriptomic profiles and metabolic pathway gene sets associations with white adipose tissue inflammation as determined by the presence of crown-like structures (CLSs).
(A) Heatmap illustrating 11 genes differentially expressed as they relate to CLS status. Illustrated genes were selected based on an absolute log 2-fold change >1.5 and a false discovery rate (FDR) <0.1. (B) Heatmap illustrating an unsupervised clustering analysis of differentially expressed genes based on CLS status and body mass index (BMI) categorization (normal, <25 kg/m2; overweight/obese, >25 kg/m2). (C) Gene set enrichment analysis using MSigDB, fetching pathways related to metabolism. The median value of the enrichment score was estimated for each pathway between the CLS-positive and CLS-negative cohorts. The figure illustrates pathways that were considered enriched when the absolute difference between the average value of the two groups was greater than the 95th quantile.
Differential gene expression analysis based on CLS status was further stratified by BMI. Patients were categorized as normal weight if their BMI was <25 kg/m2 and overweight or obese if their BMI was >25 kg/m2. Unsupervised clustering of differentially expressed genes demonstrated no clear delineation based on BMI categorization (Figure 4B).
To further characterize the differentially expressed genes based on CLS status, a gene set enrichment analysis (GSEA) was performed. Using a Molecular Signatures Database classification scheme (see Methods), gene sets related to metabolism or lipid synthesis were found to be enriched in CLS-positive tissues compared to CLS-negative white adipose specimens in patients with EC (Figure 4C). Of the top 5% metabolic pathways with the greatest difference in enrichment scores based on CLS status, pathways involved in progesterone metabolism and regulation of lipoprotein metabolism were enriched among the CLS-positive cohort.
Changes in the immune microenvironment as it relates to WAT inflammation
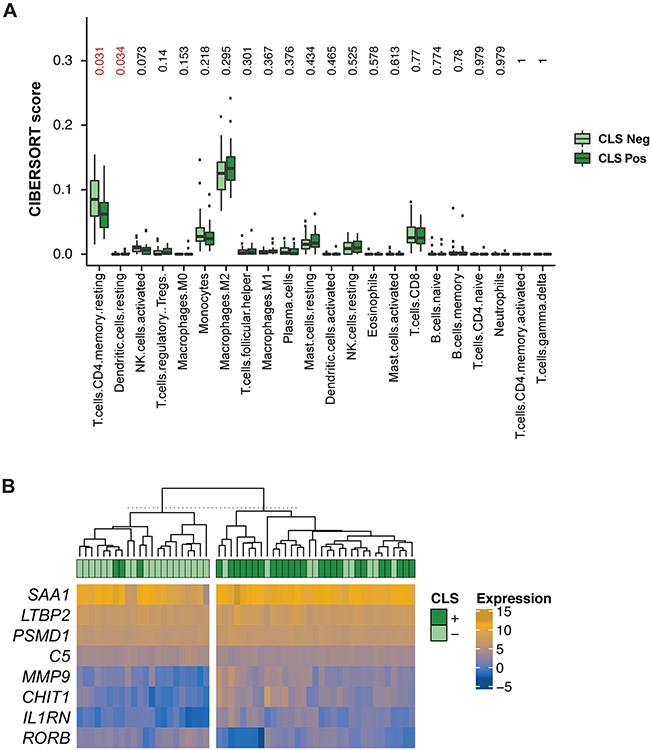
As an exploratory, hypothesis-generating analysis, we assessed the relation between WAT inflammation and immune response using CIBERSORT to characterize the cellular composition, including 22 types of infiltrating immune cells, of the tumor and normal tissue samples based on CLS status (20, 21). We identified that CD4 memory T cells were significantly more present in CLS-negative samples (CIBERSORT score: 0.063 CLS-positive vs. 0.085 CLS-negative; p=0.031; Figure 5A). In contrast, CLS-positive samples were enriched for dendritic cells compared to CLS-negative samples (CIBERSORT score: 0.006 CLS-positive vs. 0.002 CLS-negative; p=0.034; Figure 5A). It should be noted that upon multiple comparisons, these results no longer reached statistical significance (FDR 0.37).
Figure 5: Changes in the immune microenvironment as it relates to white adipose tissue inflammation as determined by the presence of crown-like structures (CLSs).
(A) Compositional differences of 22 immunocytes between CLS-negative (light green) and CLS-positive (dark green) tissue by box-plot. (B) Heatmap illustrating the expression levels of 8 immune-related genes (IRGs) found to be differentially expressed, with a false discovery rate (FDR) <0.1 between CLS-negative and CLS-positive tissue.
As a next step, given that immunotherapy is a rising promising treatment for patients with EC, a differential gene expression was performed to investigate the expression of IRGs to determine whether CLS status correlates with potential treatment response. Differential gene expression analysis identified 8 IRGs significantly differentially expressed between CLS-positive and CLS-negative samples with an FDR <0.1: LTBP2, CHIT1, SAA1, IL1RN, MMP9, RORB, C5, and PSMD1 (Figure 5B; Table S3). Among these, LTBP2, CHIT1, SAA1, IL1RN, MMP9, and PSMD1 had a significantly higher level of expression in CLS-positive compared to CLS-negative cases (FDR <0.1).
DISCUSSION
Approximately 50% of our patients with EC had visceral WAT inflammation, defined by the presence of CLS. In particular, visceral omental WAT inflammation was more common among patients with a higher BMI and cardiometabolic disorders. These findings are consistent with WAT inflammation studies in other cancers (1-3). Additionally, WAT inflammation in patients with EC was associated with risk factors, clinical features, and circulating factors characteristic of metabolic syndrome, as well as changes in gene expression indicative of an increased inflammatory and pro-tumorigenic environment.
We show that WAT inflammation is associated with potentially harmful systemic changes. Our results indicate that age and obesity, of which both are risk factors for metabolic syndrome, are associated with not only the presence but also increasing amounts of WAT inflammation. In addition, certain clinical features that are part of metabolic syndrome, including dyslipidemia, hypertension, and diabetes mellitus were among the variables that were associated with the presence of WAT inflammation. Our results also indicate that WAT inflammation is associated with levels of circulating factors, including leptin, adiponectin, and sex hormone–binding globulin, that all trended in a direction that portends greater risk of metabolic syndrome. These findings help to explain the growing body of evidence indicating that metabolic syndrome portends a worse outcome in several cancers, including colon, breast, gastric, and endometrial cancer (34-38). Of note, this association between components of metabolic syndrome, such as hypertension and impaired fasting glucose, have been associated with EC independent of obesity (39, 40). While obesity has been well established as a driver of a particular histologic type of EC (i.e., endometroid adenocarcinoma) secondary to higher levels of circulating estrogen, metabolic syndrome has been shown to increase the risk of EC across all histologic subtypes (39). Thus, our findings suggest that not only is there an association between obesity and increasing WAT inflammation, but that WAT inflammation is also associated with systemic metabolic alterations known to portend worse oncologic outcomes.
Our study also identified additional alterations in circulating factors that linked the WAT inflammation observed on the microscopic level to systemic inflammatory changes. This included elevated circulating levels of the inflammatory cytokines hsCRP and IL-6 among the patients with WAT inflammation. These findings are consistent with prior reports demonstrating an association between WAT inflammation and NFκB activation and increased levels of proinflammatory mediators (TNF-α, IL-1β, and cyclooxygenase-2 (COX-2)-derived prostaglandin E2) that are also known inducers of aromatase and thereby estrogen biosynthesis (14). This adipose tissue dysfunction causes a systemic inflammation that raises concern for worse oncologic outcomes given that proinflammatory mediators, including IL-6, have been shown to promote neoplasia and tumor progression (41).
Furthermore, WAT inflammation was associated with an altered transcriptomic landscape in the omentum. Among the genes most noticeably differentially expressed were the matrix metalloproteinases MMP8 and MMP7, for which expression has been shown to promote cancer cell growth and are thus associated with tumor invasion, recurrence, and poorer survival for various malignances (28-31). These data are consistent with study data of other cancer types, in which WAT inflammation is also associated with worse oncologic outcomes (6, 12, 13). In addition, the increased expression of CHIT1 and SAA2-SAA4 reflects the upregulation of genes implicated in the function of macrophages, which are the cornerstone of identifying WAT inflammation. Lastly, gene expression analysis demonstrated that the presence of WAT inflammation associated gene expression changes occurred in patients irrespective of whether they were normal weight or overweight/obese. This again indicates that although obesity has a role to play, it does not dictate the impact of WAT inflammation.
Given the rising use of immunotherapy for the treatment of advanced or recurrent EC, we chose to investigate whether the presence of WAT inflammation would be associated with potential markers of immunotherapy response on a transcriptomic level. The differential immune-related gene expression analysis identified 8 genes, including CHIT1. CHIT1 is a marker of macrophage activation, and has been reported to regulate many inflammatory processes through the direct stimulation of inflammatory mediators such as IL-8, MMP9, CCL2, CCL5, and CCL11, which increases the migratory capacity of many immunological cells, including T lymphocytes, macrophages, and eosinophils (42). The significantly greater gene expression of MMP9 present in the samples with WAT inflammation, compared to those without, aligns with the greater level of expression of CHIT1 also observed. In addition, levels of CHIT1 activity have been strongly correlated with levels of IL-1β and TNF-α (42). One may hypothesize that given immunotherapy works by activating a patient’s own immune system to attack cancer cells, patients with WAT inflammation with greater expression in visceral tissue at the time of diagnosis of genes such as CHIT1, involved in antigen presentation and immune activation, could signal susceptibility to immunotherapy. Further studies are warranted to define whether the observed increased expression of genes involved in antigen presentation in patients with WAT inflammation would corroborate a more immunogenic environment. By performing an exploratory deconvolution analysis, we found that CLS-positive samples were enriched for dendritic cells compared to CLS-negative samples. When activated, dendritic cells can lead to a cascade of pro-inflammatory cytokine release sometimes skewing T cell responses, which aligns with our other findings of a pro-inflammatory environment in CLS-positive patients. Although these results of the CIBERSORT analyses were not statistically significant upon correction for FDR, we believe the findings of this exploratory analysis warrant further study in larger datasets and potentially employing orthogonal methods such as multiplex immunoassays to assess the composition of infiltrating immune cells in these tissues.
This study has limitations. Given the high 5-year survival rates in patients with early-stage endometrial cancer, which comprised 89% of the study cohort, adequate median follow-up has not been reached at this point to assess patient outcome data. Follow-up data, however, are being collected to define the association of WAT inflammation with recurrence and disease-specific survival in patients with EC. In addition, due to insufficient RNA quality and/or quantity obtained from a portion of the WAT samples, gene expression analysis by RNA-seq could not be performed for all samples.
CONCLUSIONS
Taken together, through the prospective characterization of these 101 cases of EC, our work supports the concept that an inflammatory pro-tumorigenic state is present in the setting of obesity and cardiometabolic disorders. On the basis of these findings, further work to investigate the association of WAT inflammation and oncologic outcomes in EC is ongoing. Our findings help to explain the link between obesity and worse outcomes in EC and lay the groundwork for risk assessment, prevention, and therapeutic strategies for obesity-related cancers. Interventions that target WAT inflammation or its systemic and local effects warrant further investigation to improve EC outcomes.
Supplementary Material
Supplementary Table S1: Blood measurements based on presence or absence of white adipose inflammation in patients with endometrial cancer.
Supplementary Table S2: Differential gene expression of CLS-positive versus CLS-negative adipose tissue in patients with endometrial cancer.
Supplementary Table S3: Differential gene expression of immune-related genes (IRGs) between CLS-positive versus CLS-negative adipose tissue in patients with endometrial cancer.
ACKNOWLEDGEMENTS
Research reported in this publication was supported in part by a Cancer Center Support Grant of the NIH/NCI (P30 CA008748). NMI received research funding to the institution from the National Cancer Institute, Breast Cancer Research Foundation, American Cancer Society, and Conquer Cancer of ASCO. BW is funded in part by Breast Cancer Research Foundation and Cycle for Survival grants. GZ is funded in part by a Fondazione AIRC per la Ricerca sul Cancro Investigator Grant ID 21761, by a 5x1000 IRCCS grant ID C830B, by a University of Genoa CURIOSITY grant, and by anonymous patients’ liberal donations.
Footnotes
CONFLICT OF INTEREST STATEMENT
VM is a consultant and part of the advisory board for Clovis, GSK, Merck, Eisai, Karyopharm, Faeth, Deciphera, Astrazeneca, and received research funding to the institution from Merck, Eisai, Karyopharm, AstraZeneca, Clovis, Moreo, Takeda, Zymeworks, and Genentech, outside the submitted work. BW reports ad hoc membership of the advisory board of Repare Therapeutics, outside the scope of this study. NMI reports consulting for Novartis and SeaGen, and research funding to the institute from Novartis. GZ reports receiving travel grants from Roche, Novartis, and Pfizer, consultation fees from Pfizer, and reagents from ThermoFisher Scientific and Cytiva Life Sciences. NA reports grants from Stryker/Novadaq and GRAIL, outside the submitted work. The other authors declare no potential conflicts of interest.
REFERENCES
- 1.Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348(17):1625–38. [DOI] [PubMed] [Google Scholar]
- 2.Iyengar NM, Hudis CA, Dannenberg AJ. Obesity and inflammation: new insights into breast cancer development and progression. Am Soc Clin Oncol Educ Book. 2013:46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoon HH, Lewis MA, Shi Q, Khan M, Cassivi SD, Diasio RB, et al. Prognostic impact of body mass index stratified by smoking status in patients with esophageal adenocarcinoma. J Clin Oncol. 2011;29(34):4561–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6(1):24–37. [DOI] [PubMed] [Google Scholar]
- 5.Aune D, Navarro Rosenblatt DA, Chan DS, Vingeliene S, Abar L, Vieira AR, et al. Anthropometric factors and endometrial cancer risk: a systematic review and dose-response meta-analysis of prospective studies. Ann Oncol. 2015;26(8):1635–48. [DOI] [PubMed] [Google Scholar]
- 6.Iyengar NM, Zhou XK, Gucalp A, Morris PG, Howe LR, Giri DD, et al. Systemic Correlates of White Adipose Tissue Inflammation in Early-Stage Breast Cancer. Clin Cancer Res. 2016;22(9):2283–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–45. [DOI] [PubMed] [Google Scholar]
- 8.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–9. [DOI] [PubMed] [Google Scholar]
- 9.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–44. [DOI] [PubMed] [Google Scholar]
- 10.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isnaldi E, Richard F, De Schepper M, Vincent D, Leduc S, Maetens M, et al. Digital analysis of distant and cancer-associated mammary adipocytes. Breast. 2020;54:179–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyengar NM, Ghossein RA, Morris LG, Zhou XK, Kochhar A, Morris PG, et al. White adipose tissue inflammation and cancer-specific survival in patients with squamous cell carcinoma of the oral tongue. Cancer. 2016;122(24):3794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gucalp A, Iyengar NM, Zhou XK, Giri DD, Falcone DJ, Wang H, et al. Periprostatic adipose inflammation is associated with high-grade prostate cancer. Prostate Cancer P D. 2017;20(4):418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyengar NM, Hudis CA, Dannenberg AJ. Obesity and cancer: local and systemic mechanisms. Annu Rev Med. 2015;66:297–309. [DOI] [PubMed] [Google Scholar]
- 15.Iyengar NM, Gucalp A, Dannenberg AJ, Hudis CA. Obesity and Cancer Mechanisms: Tumor Microenvironment and Inflammation. J Clin Oncol. 2016;34(35):4270–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.AJCC Cancer Staging Manual. 8 ed: Springer International Publishing; 2017. 1032 p. [Google Scholar]
- 17.criteria NCCN. Breast Cancer January 15th, 2021. [Version 1.2021:[Available from: https://www.nccn.org/professionals/physician_gls/pdf/breast_blocks.pdf. [Google Scholar]
- 18.Pareja F, Lee JY, Brown DN, Piscuoglio S, Gularte-Merida R, Selenica P, et al. The Genomic Landscape of Mucinous Breast Cancer. J Natl Cancer Inst. 2019;111(7):737–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sturm G, Finotello F, Petitprez F, Zhang JD, Baumbach J, Fridman WH, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35(14):i436–i45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang SX, He ZK, Wang X, Li HM, Liu XS. Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leone P, Shin EC, Perosa F, Vacca A, Dammacco F, Racanelli V. MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. J Natl Cancer Inst. 2013;105(16):1172–87. [DOI] [PubMed] [Google Scholar]
- 25.Rosner B, Glynn RJ, Lee ML. Incorporation of clustering effects for the Wilcoxon rank sum test: a large-sample approach. Biometrics. 2003;59(4):1089–98. [DOI] [PubMed] [Google Scholar]
- 26.Wallace IR, McKinley MC, Bell PM, Hunter SJ. Sex hormone binding globulin and insulin resistance. Clin Endocrinol (Oxf). 2013;78(3):321–9. [DOI] [PubMed] [Google Scholar]
- 27.Goldstajn MS, Toljan K, Grgic F, Jurkovic I, Baldani DP. Sex Hormone Binding Globulin (SHBG) as a Marker of Clinical Disorders. Coll Antropol. 2016;40(3):211–8. [PubMed] [Google Scholar]
- 28.Fingleton B, Vargo-Gogola T, Crawford HC, Matrisian LM. Matrilysin [MMP-7] expression selects for cells with reduced sensitivity to apoptosis. Neoplasia. 2001;3(6):459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitsiades N, Yu WH, Poulaki V, Tsokos M, Stamenkovic I. Matrix metalloproteinase-7-mediated cleavage of Fas ligand protects tumor cells from chemotherapeutic drug cytotoxicity. Cancer Res. 2001;61(2):577–81. [PubMed] [Google Scholar]
- 30.Strand S, Vollmer P, van den Abeelen L, Gottfried D, Alla V, Heid H, et al. Cleavage of CD95 by matrix metalloproteinase-7 induces apoptosis resistance in tumour cells. Oncogene. 2004;23(20):3732–6. [DOI] [PubMed] [Google Scholar]
- 31.Dejonckheere E, Vandenbroucke RE, Libert C. Matrix metalloproteinase8 has a central role in inflammatory disorders and cancer progression. Cytokine Growth F R. 2011;22(2):73–81. [DOI] [PubMed] [Google Scholar]
- 32.Tans R, van Diepen JA, Bijlsma S, Verschuren L, Suppers A, Stienstra R, et al. Evaluation of chitotriosidase as a biomarker for adipose tissue inflammation in overweight individuals and type 2 diabetic patients. Int J Obesity. 2019;43(9):1712–23. [DOI] [PubMed] [Google Scholar]
- 33.Jumeau C, Awad F, Assrawi E, Cobret L, Duquesnoy P, Giurgea I, et al. Expression of SAA1, SAA2 and SAA4 genes in human primary monocytes and monocyte-derived macrophages. PLoS One. 2019;14(5):e0217005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, Mauldin PD, Ebeling M, Hulsey TC, Liu BR, Thomas MB, et al. Effect of metabolic syndrome and its components on recurrence and survival in colon cancer patients. Cancer. 2013;119(8):1512–20. [DOI] [PubMed] [Google Scholar]
- 35.Shen ZL, Ye YJ, Bin LA, Yin MJ, Yang XD, Jiang KW, et al. Metabolic syndrome is an important factor for the evolution of prognosis of colorectal cancer: survival, recurrence, and liver metastasis. Am J Surg. 2010;200(1):59–63. [DOI] [PubMed] [Google Scholar]
- 36.Berrino F, Villarini A, Traina A, Bonanni B, Panico S, Mano MP, et al. Metabolic syndrome and breast cancer prognosis. Breast Cancer Res Tr. 2014;147(1):159–65. [DOI] [PubMed] [Google Scholar]
- 37.Hu DA, Peng F, Lin XD, Chen G, Zhang HJ, Liang BY, et al. Preoperative Metabolic Syndrome Is Predictive of Significant Gastric Cancer Mortality after Gastrectomy: The Fujian Prospective Investigation of Cancer (FIESTA) Study. Ebiomedicine. 2017;15:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin JX, Dalwadi SM, Masand RP, Hall TR, Anderson ML, Ludwig MS. Association Between Metabolic Syndrome and Endometrial Cancer Survival in a SEER-Medicare Linked Database. Am J Clin Oncol-Canc. 2020;43(6):411–7. [DOI] [PubMed] [Google Scholar]
- 39.Trabert B, Wentzensen N, Felix AS, Yang HP, Sherman ME, Brinton LA. Metabolic syndrome and risk of endometrial cancer in the united states: a study in the SEER-medicare linked database. Cancer Epidemiol Biomarkers Prev. 2015;24(1):261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bjorge T, Stocks T, Lukanova A, Tretli S, Selmer R, Manjer J, et al. Metabolic syndrome and endometrial carcinoma. Am J Epidemiol. 2010;171(8):892–902. [DOI] [PubMed] [Google Scholar]
- 41.Hursting SD, Digiovanni J, Dannenberg AJ, Azrad M, Leroith D, Demark-Wahnefried W, et al. Obesity, energy balance, and cancer: new opportunities for prevention. Cancer Prev Res (Phila). 2012;5(11):1260–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elmonem MA, van den Heuvel LP, Levtchenko EN. Immunomodulatory Effects of Chitotriosidase Enzyme. Enzyme Res. 2016;2016:2682680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: Blood measurements based on presence or absence of white adipose inflammation in patients with endometrial cancer.
Supplementary Table S2: Differential gene expression of CLS-positive versus CLS-negative adipose tissue in patients with endometrial cancer.
Supplementary Table S3: Differential gene expression of immune-related genes (IRGs) between CLS-positive versus CLS-negative adipose tissue in patients with endometrial cancer.
Data Availability Statement
RNA-seq data will be available in the Sequence Read Archive (SRA) upon acceptance of this manuscript.