Abstract
The consumption of healthy food, in order to strengthen the immune system, is now a major focus of people worldwide and is essential to tackle the emerging pandemic concerns. Moreover, research in this area paves the way for diversification of human diets by incorporating underutilized crops which are highly nutritious and climate-resilient in nature. However, although the consumption of healthy foods increases nutritional uptake, the bioavailability of nutrients and their absorption from foods also play an essential role in curbing malnutrition in developing countries. This has led to a focus on anti-nutrients that interfere with the digestion and absorption of nutrients and proteins from foods. Anti-nutritional factors in crops, such as phytic acid, gossypol, goitrogens, glucosinolates, lectins, oxalic acid, saponins, raffinose, tannins, enzyme inhibitors, alkaloids, β-N-oxalyl amino alanine (BOAA), and hydrogen cyanide (HCN), are synthesized in crop metabolic pathways and are interconnected with other essential growth regulation factors. Hence, breeding with the aim of completely eliminating anti-nutrition factors tends to compromise desirable features such as yield and seed size. However, advanced techniques, such as integrated multi-omics, RNAi, gene editing, and genomics-assisted breeding, aim to breed crops in which negative traits are minimized and to provide new strategies to handle these traits in crop improvement programs. There is also a need to emphasize individual crop-based approaches in upcoming research programs to achieve smart foods with minimum constraints in future. This review focuses on progress in molecular breeding and prospects for additional approaches to improve nutrient bioavailability in major crops.
Keywords: anti-nutritional factors, regulatory pathways, plant breeding, food processing, gene editing
Introduction
Consumption of foods for a sustainable diet has the potential to reduce hidden hunger in many countries. One of the major factors influencing nutrient absorption is the presence of anti-nutrients in foods (Thakur et al. 2019). These have largely been overlooked by research projects that aim to minimize nutritional deficiencies and toxicities in diets in the growing population (Gilani et al., 2012). Anti-nutritional factors in foods hinder digestion and reduce the bioavailability of the major nutrients. In some severe cases, they are a major contributor to serious disorders and, when intake is excessive, can even cause death (Frick et al., 2017). Hence, this has to be rectified in major food crops so that the mineral uptake from plant-based foods is unaltered. The major anti-nutritional factors in foods include phytic acid, raffinose, saponins, tannins, enzyme inhibitors, lectins, gossypol, glucosinolates, goitrogens, oxalic acid, erucic acid, alkaloids, β-N-oxalyl amino alanine (BOAA), and hydrogen cyanide (HCN) (Thakur et al., 2019; Samtiya et al., 2020). These factors play a major role in human health, as they hinder nutrient absorption and uptake via chelation and enzyme inhibition. Legumes are of particular concern, as they contain a comparatively higher proportion of anti-nutritional traits than other crops (Parca et al., 2018). This presumes that consumer favour less consumption of these crops despite their potential nutritive traits (Jaiswal, 2020).
Several traditional processing techniques, such as soaking, roasting, sprouting, fermentation, boiling, and extrusion, can reduce anti-nutritional components in grains. However, these techniques are adopted at a small scale in household cooking and in value-added products from agro-industries (Das et al., 2022). Industrial organizations utilize these processing methods to enhance the bioavailability of food grains in processed foods. Eliminating anti-nutrients in foods remains a major objective, and one that could be achieved by using advanced techniques, such as RNAi and gene editing, to develop high-nutrition crops. The reduction of anti-nutritional traits has been a progressively intense area of research since the 1950s, but there are several barriers to improving varieties by reducing anti-nutritional factors. The accumulation of anti-nutrients in crops is still to be completely explored for all the major traits (Tong et al., 2021). Some anti-nutrients have been explored more than others, and the genes responsible for their biosynthesis offer a major way of altering the concentrations of anti-nutrients in foods. Phytic acid, raffinose, glucosinolates, enzyme inhibitors, and erucic acid are the anti-nutrients that have been the predominant focus of breeding and transgenic approaches. Saponins, oxalic acid, alkaloids, HCN, goitrogens, and BOAA need to be further studied in the future (Thakur et al. 2019).
Another major factor in reducing these antinutrients in crops is their stable expression across locations. Anti-nutrients such as phytic acid, glucosinolates, and alkaloids are highly influenced by soil, fertilizer applications, and other edaphic factors (Zhuo et al., 2013; Frick et al., 2017; Pramitha et al., 2021). Therefore, alternate strategies involving advanced multi-omics accompanied by rapid estimation techniques and gene editing protocols play an essential role in optimizing the nutrient availability of major crops and developing non-toxic foods for human consumption. However, it is also important to monitor the effects of reduction of anti-nutrients in crops, as anti-nutrients such as saponins, raffinose, enzyme inhibitors, gossypol, glucosinolates, and phytic acid have a major role in plant growth metabolism (Sahu et al., 2020). Previous reports have shown that these compounds constitute a regulation on crop metabolism and growth (Rodríguez-Sifuentes et al., 2020; Pramitha et al., 2021; Elango et al., 2022). Thus, a focus on the reduction of negative pleiotropic effects on characteristics such as seed quality, seed yield, and stable expression, and on the influence of edaphic factors on nutrient accumulation, processing, and storage, are necessary to develop a high-value food crop with mineral availability in the near future (Coulibaly et al., 2011). Among all the major crops, soybean is the one that has been most explored for reducing anti-nutrients, followed by brassicas and cotton, which have been investigated to improve their overall acceptance for human and animal feed (Rathore et al., 2020; Le et al., 2020). Hence, this review highlights progress in research into breeding for anti-nutritional traits in major food crops and also predicts its future direction.
Major anti-nutritional traits in food crops and their effects on consumption
There are several anti-nutritional factors in cereal- and legume-based foods, and some of the major key anti-nutritional traits are elaborated here. The major factors that interrupt food digestion and absorption are phytic acids, gossypols, lectins, raffinose, enzyme inhibitors, goitrogens, saponins, tannins, oxalic acid, erucic acid, alkaloids, BOAA, and HCN. This section describes the effects of consumption of these anti-nutrients in foods and specifies levels of consumption in regular diets (Table 1).
Table 1.
The major role of anti-nutrients in consumption and plant growth regulation.
| S. no. | Anti-nutrient | Effects on consumption | Role in plant growth | Pathway | Reference |
|---|---|---|---|---|---|
| 1. | Phytic acid | i. Nutritional inhibitor in monogastric animals ii. Decreases the risk of colon cancer and inflammatory bowel disease iii. Lowers blood glucose level |
i. Phosphorus storage and chelation of micronutrients for growth and development | Myoinositol pathway |
Gupta et al., 2015; Dilworth et al., 2005; Graf and Eaton, 1990 |
| 2. | Raffinose | i. Raffinose not digested by humans and monogastric animals ii. Leads to flatulence in humans and animals iii. Prevents non-alcoholic fatty liver disease in humans iv. Reduces inflammation, diabetes, allergies, and obesity |
i. Acts as a cryoprotectant ii. Acts as a storage metabolite and is absorbed in seeds and roots iii. Acts as a source of energy for seed germination |
Inositol phosphate pathway |
Kannan et al., 2018; Elango et al., 2022 |
| 3. | Gossypol | i. Acute poisoning on ingestion ii. Causes iron deficiency known as erythropoiesis iii. Increases cytosolic Ca2+ activity iv. Decreases antioxidant levels in tissues |
i. Resistance to cotton bollworm | Sesquiterpenoid aldehyde pathway |
Soto-Blanco, 2008; Gadelha et al., 2011; Randel et al., 1996; Zhang et al., 2007; Mena et al., 2004; Zbidah et al., 2012; Bottger et al., 1964; Kovaci, 2003 |
| 4. | Saponins | i. Cause diarrhea and vomiting by damaging red blood cells ii. Affects the nutrient absorption by gut membranes iii. Negative impact on chick development and feed efficiency |
i. Act as phytoalexin during fruit and tuber development ii. Resistance against diseases in vegetables |
Cytosolic mevalonic acid pathway |
Akande et al., 2010; Ribera and Zuñiga, 2012; Cárdenas et al., 2015 |
| 5. | Goitrogen | i. Deficiency of thyroid hormone ii. Reduces growth and reproductive performance iii. Apoptotic and anti-proliferative effects in thyroid cancer cells |
– | Glycosyl transferase pathway |
Akande et al., 2010; Chatterjee et al., 2018; Boncompagni et al., 2018 |
| 6. | Glucosinolates | i. Cause rancidity ii. Prevent cardiovascular and neurodegenerative diseases |
– | Aliphatic glucosinolate pathway | Kamal et al., 2022 |
| 7. | Oxalic acid | i. Causes headache, coma, and kidney stones ii. Calcium oxalate has a severe impact on human nutrition and health iii. Leads to death due to oxalate poisoning |
i. Precursors of oxalic acid play a major role in climate resilience ii. Growth regulation of crops during pollination |
– | Egbuna, 2018; Awulachew, 2022 |
| 8. | Erucic acid | i. fat accumulation in heart muscles ii. cardiovascular diseases and myocardial lesions in the heart |
– | – | Wani et al., 2022 |
| 9. | Lectin | i. Agglutinates red blood cells ii. Anti-tumor agent iii. Antimicrobial, antifungal, antibacterial, antiviral iv. Alters the integrity of intestinal mucosa |
i. Regulation of cell signaling and plant response to biotic, abiotic, and symbiotic stimuli | – | López-Moreno et al., 2022 |
| 10. | Enzyme inhibitors | i. Trypsin inhibitors trigger pancreatic hyperplasia ii. Prevention of type 2 diabetes and obesity iii. Protease inhibitors reduce the activity of proteolytic enzymes during ingestion iv. Alpha-amylase inhibitors affect post-meal plasma glucose levels |
Confer biotic stress tolerance and act as biopesticides | – |
Bhutkar and Bhise, 2012; Battelino et al., 2019; do Amaral et al., 2022; Ribeiro et al. 2015 |
| 11. | Tannins | i. Inhibit digestive enzymes and cause intestinal damage ii. Have been associated with reduced feed intake, growth rate, feed efficiency, and protein digestibility iii. Enhance the food product’s oxidative stability iv. Improve the quality of the meat and milk. Act as a natural preservative |
i. Antiparasitic properties of plant tannins ii. Act against pathogenic bacteria, have antibacterial actions, and are antioxidants iii. Prevent neurodegenerative diseases and have anti-tumor, anti-inflammatory, and antibacterial properties |
Shikimate pathway |
Akande et al., 2010; Gemede and Ratta, 2014; Gilani et al., 2012 Tong et al., 2021; Mora et al., 2022 |
| 12. | HCN | i. In animals stops cellular respiration process due to asphyxia ii. Severe shortness of breath and frequent urination in animals |
– | – | Al-Beiruty et al., 2020 |
| 13. | BOAA | i. Causes neurolathyrism, a neurologic condition that is irreversible in both humans and animals | Act as an Antioxidant | Begins with the formation of BIA from O-acetyl- L-serine (OAS) | Das et al., 2021 |
Phytic acid
Phytic acid (C6H18O24P6) is a naturally occurring antioxidant that chelates positively charged minerals such as phosphorus, iron, and zinc (Raboy et al., 2000). It is found primarily in the grains, nuts, and seeds of cereals, legumes, and vegetables. Phytic acid is found in rice aleurone, and it is also abundant in the endosperm and embryo of maize (Raboy et al., 2000). Phosphorus is primarily stored in the form of phytic acid in seeds after pollination. During germination, it is degraded by the enzyme phytase to support plant growth and development (Pramitha et al., 2021). Monogastric animals lack the enzyme phytase in their digestive tract, and as a result phytic acid acts as a nutritional inhibitor by chelating the available micronutrients in foods (Gupta et al., 2015). The non-dissolvable form of phytic acid, i.e., the mineral-bound complex, and remains a problem, as its excretion in animal feces results in eutrophication and soil pollution (Raboy et al., 2001). Hence, reducing phytic acid in grains is a beneficial solution to enhance mineral availability following consumption (Pramitha et al., 2021). Despite these anti-nutritional features, dietary phytic acid has been found to reduce the risk of colon cancer and other inflammatory bowel diseases by acting as a beneficial antioxidant in foods. Its inclusion in foods thereby prevents lipid peroxidation, oxidative spoilage, discoloration, putrefaction, and syneresis. Hence, the reduction of phytic acid in foods should be optimized for normal growth and regulation of metabolism. The safest range for overall phytic acid consumption is reported to be around 250–800 mg (Graf and Eaton, 1990).
Gossypol
Gossypol (C30H30O8) is a group of polyphenols that can cause acute poisoning on ingestion (Stipanovic et al., 1975). Studies of gossypol report that cumulative toxic effects can occur after just 1–3 months of consumption (Soto-Blanco, 2008; Gadelha et al., 2011). It is safest to limit gossypol consumption to 20 mg of gossypol per kg of feed. Poisoning by gossypol has been reported in broiler chicks, pigs, dogs, sheep, and goats. However, gossypol toxicity is more severe in monogastric animals such as pigs, birds, fish, and rodents than in ruminants (Kenar, 2006; Alexander et al., 2008). The effect of gossypols is more severe in younger ruminants than in adults. The major impact of ingestion is anemia, which is frequently observed in cottonseed-fed animals. During ingestion, gossypol binds with iron in hemoglobin to form a gossypol–iron complex, which inhibits iron absorption, resulting in a deficiency known as erythropoiesis, i.e., erythrocyte fragility (apoptosis-like erythrocyte death) (Randel et al., 1996; Mena et al., 2004; Zhang et al., 2007). Further, this increases cytosolic Ca2+ activity, which causes cell membrane scrambling and contraction (Zbidah et al., 2012). In addition, clinical signs of gossypol poisoning are linked to decreased antioxidant levels in tissues (Kovaci, 2003). Hence, gossypol reduces energy generation from oxidative metabolism at high concentrations by interfering with enzymatic activity in the mitochondrial electron transport chain and oxidative phosphorylation. In addition, gossypol has an impact on both male and female gametogenesis and promotes embryo lesions linked to male infertility (Gadelha et al., 2011). Therefore, gossypol could be explored for its potential use as a male contraceptive in future pharmaceutical research (Soto-Blanco, 2008; Chang et al., 2011).
Lectins
Lectins (complex carbohydrate-binding proteins) are a type of glycoprotein with non-catalytic carbohydrate-binding sites that are classified into animal, algal, bacterial, fungal, and plant lectins (Mishra et al., 2019). Lectins are also known as hemagglutinins. These “anti-nutrients” have received a lot of attention because of their role in obesity, chronic inflammation, and autoimmune diseases. They are predominantly observed in raw legumes such as kidney beans, lentils, peas, soybeans, and peanuts, and in whole grains such as wheat. In leguminous plants, lectin content is higher in seeds than in bark, leaves, roots, or stem. Plant lectins are generally found in nuts, cereals, and leguminous seeds (El-Araby et al., 2020). Consumption of lectins in their active state, for example the consumption of even small amounts of raw or undercooked kidney beans, can cause severe adverse reactions in humans. Kidney beans contain phytohemagglutinin, a lectin that causes red blood cells to aggregate, leading to cause nausea, vomiting, stomach upset and diarrhea (Peumans and Van Damme, 1995). Bloating and flatulence are milder side effects. Active lectins have been found in animal cell studies to interfere with mineral absorption, affecting the concentrations of calcium, iron, phosphorus, and zinc in the digestive tract (Vasconcelos and Oliveira, 2004). Thus, 200–400 hemagglutinin units (hau) is considered a safe level for consumption of lectins from leguminous foods (Van Damme et al., 2008; Kobayashi et al., 2014). Despite their negative side effects, lectins have been shown to be useful for cancer treatment due to their antiangiogenic, antimetastatic, and antiproliferative activity (Bhutia et al., 2016; Panda et al., 2018; Sinha et al., 2019).
Raffinose
Pulses are rich in carbohydrates, proteins, dietary fiber, vitamins, minerals, and other bioactive substances in the human diet. However, their consumption and acceptance are constrained globally, particularly in industrialized countries, due to the high proportion of raffinose family oligosaccharides (RFOs). These are found in beans, cabbage, Brussels sprouts, broccoli, asparagus, and whole grains (Elango et al., 2022). RFOs (C18H32O16) is prevalent in the seeds of legume families such as chickpea (Cicer arietinum), lentil (Lens culinaris), and soybean (Glycine max). They are also found in the leaves and tubers of vegetables and in other specialized storage organs such as roots. Raffinoses are found in the tubers of Chinese artichoke (Stachys sieboldii) and in the leaves of a common bugle (Ajuga reptans). Defatted soy flour has an average range of raffinose from 1.15%-3.23% espectively. In lentil, RFOs level ranges from 4.5 to 5.5 mol 100 g–1 of flour, and in faba bean it ranges from 0.12% to 0.29% (Johnson et al., 2021).
Humans and monogastric animals cannot digest RFOs; instead they are fermented by the microflora of the large intestine. This fermentation produces carbon dioxide, hydrogen, and methane, causing flatulence and stomach discomfort (Kannan et al., 2018). However, RFOs also confer beneficial effects, such as antiallergic, anti-obesity, and anti-diabetic effects, the prevention of non-alcoholic fatty liver disease, and cryoprotection. They positively affect the gut microbiota and the health of the large intestine. Hence, RFOs could be used as therapeutic agents to reduce inflammation, diabetics, and allergies. As RFOs are considered the main cause of flatulence in humans and animals, there is a need to strike the right balance of RFOs content in crops if they are to be promoted as functional foods (Elango et al., 2022).
Enzyme inhibitors
Protease inhibitors are naturally occurring plant inhibitors that have become a focus of research due to their effective method of limiting enzyme activity through protein–protein interactions. They inhibit enzyme activity via the catalytic mode by blocking the enzymes’ active sites. Cereals contain substantially less of these digestive inhibitors than legumes (Nikmaram et al., 2017). Protease inhibitors substantially reduce the activity of proteolytic enzymes during ingestion (Troll and Wiesner, 1983). There are various enzyme inhibitors, among which trypsin inhibitors and alpha-amylase inhibitors are the major enzyme inhibitors in foods. Alpha-amylase primarily influences carbohydrates, namely polysaccharides, which are broken down to form oligosaccharides. Therefore, enzyme inhibitors that inhibit alpha-amylase activity will boost carbohydrate levels by slowing the digestion of carbohydrates, having an impact on the typical post-meal levels of plasma glucose (Bhutkar and Bhise, 2012). Speaking of the Trypsin inhibitors also enhance the production of hormones such as steatogenic hormone and cholecystokinin (CCK) and this would reduce food intake and body weight (Cristina Oliveira de Lima et al., 2019). In humans, consumption of trypsin inhibitors can reduce growth rate, slow protein digestion, and reduce amino acid availability, triggering pancreatic hyperplasia (Adeyemo and Onilude, 2013). Several studies have found that the inhibition of some enzymes, namely alpha-amylase, alpha-glucosidase, and lipase, is beneficial, increasing the digestibility of legume-based foods. Although it has health advantages associated with the prevention of type 2 diabetes and obesity, malfunctions relating to digestion have to be overlooked in the future (Li and Tsao, 2019).
Goitrogens
Goitrogens (C5H7NOS) got their name from “goiter,” which means “abnormal growth”. Goiter is the enlargement of the thyroid gland due to a deficiency of thyroid hormone. Soybean and cassava are cruciate vegetables of the genus Brassica and are rich in goitrogens. However, high goitrogen concentrations have also been reported in other cruciferous vegetables (Truong et al., 2010). Goitrogens interfere with iodine utilization and with thyroid hormone production. Deficiency of thyroid hormone thus results in reduced growth and reproductive performance of an individual. The effect of goitrogens can be reduced by iodine supplementation than by heat treatment (Akande et al., 2010). Foods containing goitrogens also contain different bioactive compounds that protect against thyroid cancer (Fiore et al., 2020). Crucifers contain sulforaphane, an isothiocyanate that has been observed to possess an apoptotic and antiproliferative effect in thyroid cancer cells (Chatterjee et al., 2018). Goitrogens have also been used in the treatment of COVID-19 to activate Nrf2-Keap1 and counteract the COVID-19-induced cytokine storm (Bousquet et al., 2021; Singh et al., 2021). Hence, safe consumption of these compounds needs to be ensured to avoid their negative side effects.
Saponins
Saponins (C58H94O27) are non-volatile, surface-active secondary metabolites found in soybeans, sugar beets, peanuts, spinach, asparagus, broccoli, potatoes, apples, eggplants, alfalfa, and ginseng root. Saponins are glycosidic triterpenoids that are widely distributed in the seed coat of crops (Faizal and Geelen, 2013). They are structurally diverse and chemically are known as triterpenes and steroid glycosides (Khodakov et al., 1996). The structural complexity of saponins is responsible for their varied physical, chemical, and biological properties, including sweetness, bitterness, and foaming and emulsifying properties. Hence, saponins have pharmacological, medicinal, hemolytic, antimicrobial, insecticidal, and molluscicidal activities (Sparg et al., 2004). Consumption of saponins often cause diarrhea and vomiting and also leads to the breakdown of red blood cells. It has also been demonstrated that saponins can attach to intestinal cells and influence nutrient absorption in gut membranes. Furthermore, it has been noted in the poultry sector that saponins have a negative impact on chicks’ development, feed efficiency, and ability to absorb dietary lipids, cholesterol, bile acids, and vitamins A and E (Akande et al., 2010).
Tannins
Tannins (C76H52O46) are plant polyphenolic compounds that bind to and precipitate proteins and other organic compounds such as amino acids and alkaloids. They combine with vitamin B12 to produce complexes during digestion. Hydrolyzable tannins and proanthocyanidins (PAs) are the two types of tannins (condensed tannins). Hydrolyzable tannins are more resistant to enzymatic and non-enzymatic hydrolysis than PAs, which are usually more water soluble (Chukwuebuka and Chinenye, 2015). Condensed tannins are abundant in leguminous forages and seeds. Thus, tannins combine with dietary proteins to form a digestible complex that binds to and thus inhibits endogenous proteins, including digestive enzymes (Moses et al., 2022). In addition, they have anti-nutritional effects that can lead to intestinal damage and interfere with iron absorption, and they can be carcinogenic (Akande et al., 2010). As tannic acid it is also used in the manufacture of rubber, inks, and dye fixatives. For consumption, reduction of tannins in foods leads to a healthier digestive tract.
Oxalic acid
Oxalic acid (C2H2O4) is the dicarboxylic acid that appears as a potassium and calcium salt in the cell sap of Oxalis and Rumex species of plants. After passing through the digestive system, insoluble compounds of oxalic acid (calcium oxalate) cannot be excreted via the urinary tract. This can result in kidney stones, and thus calcium oxalate can have a severe impact on human nutrition and health. Cruciferous vegetables such as kale, radishes, cauliflower, and broccoli, as well as chard, spinach, parsley, beets, black pepper, chocolate, nuts, berries, and beans, are rich in oxalates (Awulachew, 2022). Calcium supplements are suggested to be consumed with foods high in oxalic acid to expel calcium oxalate from the gut and reduce the levels of oxalates in blood. Although rare, consumption of oxalates can cause kidney disease or even death due to oxalate poisoning (Chukwuebuka and Chinenye, 2015).
Erucic acid
When triglycerides containing erucic acid in the lipids are digested, erucic acid is released into the bloodstream and distributed to tissues for release of energy through oxidation from mitochondrial cells in muscles. However, erucic acid oxidation in cardiac muscles are low. Thus, this results in the accumulation of fat in heart muscles, which causes cardiovascular diseases and myocardial lesions in the heart (Wani et al.2022).
Alkaloids
Alkaloids, especially quinolizidine, found in commercial legumes such as lupins (C10H19NO), are highly toxic when consumed. These secondary metabolites are specific to the genera Lupinus, Baptisia, Thermopsis, Genista, Cytisus, Echinosophora, and Sophora of the Leguminosae family. Consumption of these alkaloids at a high concentration leads to acute anticholinergic toxicity, the symptoms of which include blurry vision, headache, weakness, and nausea (Frick et al., 2017). It has also been also observed that the dose range of 11–25 mg/kg is lethal to children. However, so far, no fatalities in adults have been recorded (Daverio et al. 2014). Although Lupinus is a genus that has been domesticated only recently, four species containing toxic quinolizidine alkaloids (QAs) are cultivated. This is a major concern, and the threshold level of consumption considered safe is 0.02% alkaloid. Studies on QAs have been initiated and more should be carried out in the upcoming years. To date, only a few studies of alkaloids such as nicotine, vinblastine, vincristine, berberine, and morphine in economically important crops have been conducted (Frick et al., 2017).
Other anti-nutrients with health effects
Hydrogen cyanide (HCN) is a toxic chemical whose consumption has adverse effects in animals and humans. This is a major issue in fodder sorghum and sorghum during the earlier vegetative growth. Techniques to enable rapid detection of low HCN levels are being developed, and the latest advancements enable breeding of low-HCN types of sorghum (Fox et al., 2012; Al-Beiruty et al., 2020).
BOAA is a neurotoxin in seeds and leaves. BOAA is a by-product of nitrogen metabolism in plants and is a major problem in Lathyrus sativus, consumption of which causes a non-reversible neurologic disorder known as lathyrism. Although wide variations in the germplasm have been reported, further studies on the nature and actions of genes involved in BOAA biosynthesis are needed. Few molecular breeding techniques along with omic approach, intron based markers and gene editing are being standardized for reducing BOAA content in Lathyrus, as this is a major rice fallow crop in South Asian countries (Tripathy et al., 2015; Das et al., 2021). Varieties such as Pusa-24, Pusa-305, LSD-1, LSD-2, and LSD-3 are lower BOAA cultivars containing less than 0.2% BOAA (Gupta et al., 2021).
Regulatory role of anti-nutritional factors in crops and their biosynthesis
Anti-nutritional traits are compounds that interfere with the bioavailability of nutrients. They also serve as an integral part of growth and metabolism in plants. Hence, understanding their metabolism exhibits their role in regulation and facilitates genetic manipulation. The identification of anti-nutritional traits in crops, and of their wide range of pleiotropic effects, would provide a further basis for alternate strategies to overcome their constraining effects for developing high-nutritional crops (Table 1).
Phytic acid
Phytic acid is one of the most ubiquitous anti-nutritional factors, being present in the aleurone layer of cereals, maize embryo, and the cotyledon of legumes. It is synthesized by the myoinositol pathway, which is a part of starch and glucose metabolism in cells. The pathway is of two types: a lipid-independent pathway is found in seeds and a lipid-dependent pathway occurs in leaves. The lipid-independent pathway comprises the sequential phosphorylation of the six-carbon cyclic alcohol myoinositol (Ins) and soluble inositol phosphates (InsPs). However, the lipid-dependent pathway uses phosphatidylinositol (PtdIns) and PtdIns phosphates as precursors to synthesize phytic acid in leaves (Awad et al., 2012). These myoinositol phosphates play a major role in signal transduction and sugar metabolism for plant growth regulation and seed set. The major enzymes that are manipulated in breeding for lowering phytates are MIPS (myoinositol phosphate synthase), IPK (inositol phosphate kinase), and Mutli-drug Resistant Protein (transmembrane proteins). Genetic manipulation of MIPS was found to decrease phytic acid, resulting in a molar increase in free phosphate. Alteration in the IPK gene reduced phytic acid, accompanied by a limited increase in free phosphate and an increase in the content of lower InsPs. However, alteration of MRP genes lowered phytic acid, resulting in a molar increase in free phosphate in specific seed tissues. Thus, proper strategies have to be adopted to reduce the phytates in crops based on their distribution (Pramitha et al., 2021).
Raffinose
Raffinose (RF) is a trisaccharide composed of galactose, glucose, and fructose. RFOs is synthesized and stored in monocotyledonous seeds and protects the embryo from maturation. In addition, it acts as a storage metabolite and is observed in the seeds as well as the roots of beans, cabbage, Brussels sprouts, broccoli, and asparagus. Raffinose oligosaccharides (RFOs) act is an oligosaccharide that acts as a stachyose source of energy for seed germination, and its reduction in foods should be carried out in a proper way to substantiate seedling vigor. Furthermore, RFOs acts as a key desiccation protectant in seeds, playing a major role in sugar transport in phloem sap and sugar storage in tubers for active metabolism (Blochl et al. 2008). Hence, RFOs is sustained in plants to regulate storage and transport of sugar in crops and is also produced from a branching pathway of myoinositols that produces phytate. The precursor of raffinose is sucrose, and the key enzymes involved in its synthesis are galactosyl (Gol). FeGolS genes have been found to be involved in the synthesis of fagopyritols with the help of UDP-Galacytinol synthase (GolS) and d-chiro-inositol, which are also involved in the production of galactinol that produces raffinose along with sucrose (Tian et al., 2019). RFOs synthesis gene from Falcata medicago namely MfGolS1 enhances freezing and chilling tolerance in transgenic tobacco plants. Hence, RFOs could also be manipulated to enhance cold tolerance in plants (Zhuo et al., 2013).
Lectins
Lectins are unique among carbohydrates in having the ability to bind sugars. Some of the known lectins in crops include ricin, abrin, and favin. Plant lectins have a major role in host–pathogen interactions, as they have a major role in signaling. In addition, they are known to play a major role in establishing a symbiotic relationship with nitrogen fixers (Kobayashi and Kawagishi, 2014). Lectins are widely present in plants and they vary in their structure across families. They are widely used as antimicrobial, antifungal, and antiviral agents (Mishra et al., 2019). Lectins are predominantly synthesized in plants to selectively bind and detect glycans during a pathogenic infestation (Van Damme et al., 2008). Based on their synthesis in plasma membranes they are classified into G-type, C-type, and L-type lectin receptor kinases (LecRKs). In Arabidopsis, the chitin receptor kinases are the major chitin receptors and contain three Lys motifs. Few LecRKs are synthesized during ABA signaling and stomatal immunity (Singh et al., 2012). Tobacco plants express L-type LecRKs, which have a major role in plant immunity, whereas Medicago exhibits L-type LecRKs, which are involved in symbiosis (Navarro-Gochicoa et al., 2003; Gilardoni et al., 2011). The functional characterization of FIBexDB in flax seeds revealed the predominant role of lectins in cell wall biosynthesis, cytoskeleton functioning, and protein biosynthesis (Petrova and Mokshina, 2022).
Gossypol
Gossypol is yet another terpenoid observed in cotton seed, stem, flower, and root (Stipanovic et al. 1975). This is a part of the sesquiterpenoid aldehyde pathway, which is highly toxic to humans and offers resistance to various cotton pests, including bollworm (Bottger et al., 1964). During seed germination, the cotyledon acts as a primary site of gossypol accumulation; later gossypol is synthesized in the roots (Meng et al., 1999). δ-Cadinene acts a major precursor to produce different structured enzymes such as methylated hemigossypol, gossypol, hemigossypolone, or heliocides (Cai et al., 2010). Together with (+)-δ-cadinene synthase, P450 is involved in 7-hydroxy-(+)-δ-cadinene for the formation, of enzymes that convert farnesyl diphosphate (FPP) to hemigossypol (Wagner et al., 2015). Thus, gossypol is essential if cotton plants are to withstand bollworm attacks, as it confers host plant resistance.
Saponins
Triterpenoid saponins are synthesized from an isoprenoid pathway by cyclization of 2,3-oxidosqualene in the mevalonate pathway from acetyl-CoA. This further produces oleanane and its glycosylated forms (SGAs) in the Solanaceae and Liliaceae families. Saponins also act as a phytoalexin during fruit and tuber development in crops (Ribera and Zuñiga, 2012). Phytoalexins are synthesized from the cytosolic mevalonic acid pathway, which produces steroidal glycoalkaloids (SGAs) and cholesterol, which goes through several steps of hydroxylation, oxidation, transamination, and glycosylation (Haralampidis et al., 2002). The isoprenoid mevalonate pathway thus produces cholesterol from acetyl-CoA. Recent studies have revealed that acetate, mevalonate, lanosterol, cycloartenol and deuterium were categorized as cholesterol which are found to be the precursors for SGA in tomatoes (Itkin et al., 2013). Hence, these compounds could be manipulated in crops to confer resistance against diseases in vegetables.
Goitrogen and Glucosinolates
Another secondary metabolite, known as goitrogen, induces thyroid in tissue and is primarily found in rapeseed, cabbage, and canola seeds. Goitrin (l-5-vinyl-2-thiooxazolidone) is a water-soluble component in plants. Progoitrin is a precursor of goitrin and is produced by the enzyme thioglucosidase from cysteine and methionine (Chandra, 2010). Sufficient genetic variability in the pearl millet germplasm for goitrogens renders the identification and manipulation of genes related to C-C-glycosylfalvones (C-GFs) there by reducing goitrogens accumulation in grains. Glucosinolates are another group of unique secondary metabolites, and are found in the seeds of edible broccoli and plants of the genus Brassica. Methionine is also a precursor in the synthesis of glucosinolates, which include allylglucosinolate (sinigrin), glucotropaeolin, gluconasturtin, glucoraphanin, and sulforaphane. These are mainly converted to reactive isothiocyanates in mustard oils, which impart the mustard-like or garlic-like odors associated with horseradish and mustard (Banihani, 2017). Glucosinolates are synthesized from methionine, tryptophan, and seven additional protein amino acids. The pathways of goitrogen and glucosinolates are interconnected, as they are derived from similar precursors through a branched pathway. The breakdown of glycosylates often leads to a bitter taste owing to rancidity (Ishida et al., 2014). Hence, the degradation of amino acids in plants influences the storage quality of the produce.(Boncompagni et al., 2018).
Tannins
Tannins play a key role in the antioxidant activities of plants and are known to protect crops from pest infestations. They are classified into hydrolyzed tannins and condensed tannins (Khanbabaee and Van Ree, 2001). They are found in fruits such as bananas, blackberries, apples, and grapes. These foods are known to protect humans from cardiovascular diseases, cancer, and osteoporosis. Tannins are also utilized in industry as a natural preservative agent and are reported to possess antibacterial, antiviral, antiparasitic, anti-inflammatory, and anti-diarrheal activity (Tong et al., 2021). The synthesis of tannins takes place in plastids, and they are synthesized from l-phenylalanine via the shikimate pathway. The initial step involves the condensation of aldols and is catalyzed by 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase (DAHP), with phosphoenol pyruvate and erythrose-4-phosphate as substrates. The synthesis of tannins in plants is often triggered by mechanical wounding or insect attacks (Mora et al., 2022).
Oxalic acid
Oxalic acid is a secondary metabolite found in the leaves, fruits, and seeds of Rumex crispus, amaranthus, Chenopodium album, and sugar beet. It is poisonous and can cause headaches, coma, and even death. The oxalic acid metabolic pathway begins with glycine and ends with glyoxylate (Atanassova and Gutzow, 2013). Oxalate is synthesized from three precursors, namely glyoxylate, ascorbate, and oxaloacetate. Their accumulation takes place in the mature leaf lamina and leaf petiole (Cai et al., 2018). The maturing spike transcriptome of finger millet contains major genes of the oxalic acid precursors biosynthesis pathway (SGAT, GGAT, ICL, GLO, MHAR, APO, and OXO) (Akbar et al., 2018). Furthermore, it has been observed that these precursors play a major role in climate resilience and growth regulation of crops during pollination (Kobayashi et al., 2014).
Erucic acid
Erucic acid is a monounsaturated omega-9 fatty acid that is present in the seeds of plants of the genus Brassica. It is produced from the anabolic pathway initiating the synthesis of polyunsaturated C18 fatty acids via desaturation of VLCFAs (very long-chain fatty acids) involving elongation reactions (Venegas-Calerón et al., 2015). Acetyl fatty acid (acetyl-CoA) is synthesized in plastids, and erucic acid is formed from oleic acid by enzymes found in the endoplasmic reticulum. Thus, it is synthesized in the plastid and later exported to the cytosol. The seed lipids with FAD2 sense overexpression in embryos at mid-maturity exhibit an altered erucic acid content; thus, the FAD2 gene could be used to alter the erucic acid content of brassicas (Jadhav et al., 2005). Subsequently, Wu et al. (2008) identified that a particular gene, namely the fatty acid elongase 1 gene (FAE1), plays a major role in erucic acid synthesis in rapeseed. The sequencing of this gene from a zero erucic acid mutant revealed a four-basepair deletion between T1366 and G1369 that results in a frameshift mutation. This deletion leads to a premature stop of the translation at the 466th amino acid residue. This deletion is predominantly found in the C genome of Brassica napus. (Ghanevati and Jaworski, 2001).
Alkaloids
The quinolizidine alkaloids (QAs) comprise a ring structure and are classified into lupanine, angustifoline, lupinine, sparteine, multiflorine, aphylline, anagyrine, and cytisine. With the exception of anagyrine and cytisine, they are predominantly found in lupins. QAs have bitter taste when consumed and but confer resistance to pests and diseases. The biosynthesis of these alkaloids begins with the decarboxylation of l-lysine to produce cadaverine. This is then followed by oxidative deamination, regulated by copper amine oxidase (CuAO), to yield 5-aminopentanal, and this is further cyclized to Schiff’s base (Frick et al., 2017). The series of reactions after these processes include Schiff’s base formations, aldol-type reactions, hydrolysis, oxidative deamination, and coupling, thereby producing QAs. Until now, only two genes for the biosynthesis of alkaloids have been identified, one of which is La-L/ODC, which is a homolog of ODC, which is involved in the biosynthesis of a precursor of nicotine biosynthesis. In addition, other genes, namely MIA in Catharanthus roseus (vinblastine and vincristine) and BIA in Coptis japonica (berberine) and Papaver somniferum (morphine) serves as model pathways for identifying candidate genes for genetic manipulation in alkaloids (Bunsupa et al., 2012). Accumulation of alkaloids has also been observed in the aerial tissues and chloroplast in lupins (Frick et al., 2017). Recently, omics techniques have been used to develop low-alkaloid mutants that lead to a reduced alkaloid content in lupins. Gene editing approaches addressing source-to-sink transport in the metabolism of alkaloids are yet to be explored to manipulate alkaloid toxicity (Mancinotti et al., 2022).
Importance of traditional and processing techniques in overcoming the anti-nutrients in foods
Several traditional processing methods are being followed to enhance the bioavailability of micronutrients in plant-based diets. Today, a variety of methods are employed to counteract the effects of these food anti-nutrients, including milling, soaking, germination, autoclaving, and microwave treatment, as well as fermentation (Samtiya et al., 2020). This section focuses on the processing methods adopted to reduce anti-nutritional traits in crops (Table 2). Effective processing techniques adopted for reducing individual anti-nutritional traits are also described. Value-added products made using these techniques have recently become available on the market.
Table 2.
Effect of different processing techniques to minimize the anti-nutrients in foods.
| S. No | Anti-nutrient traits | Traditional methods | Effective method | Reference |
|---|---|---|---|---|
| 1. | Phytic acid | Milling Soaking Germination Fermentation Blanching |
Soaking Germination Fermentation |
Gupta et al., 2015; Udensi et al., 2008; Greiner and Konietzny, 2006; Coulibaly et al., 2011; Oghbaei and Prakash, 2016; Simwaka et al., 2017 |
| 2. | Lectins | Milling Boiling Soaking Fermentation |
Soaking Boiling Heating Fermentation |
Gupta et al., 2015; Maphosa and Jideani, 2017 |
| 3. | Tannins | Milling Soaking Autoclave Germination Fermentation Blanching Boiling |
Boiling Soaking |
Gupta et al., 2015; Ertaş and Türker, 2014; Patterson et al., 2017; Ogbonna et al., 2012; Simwaka et al., 2017 |
| 4. | Saponins | Boiling Washing Fermentation Roasting |
Fermentation |
Maphosa and Jideani, 2017; Samtiya et al., 2020 |
| 5. | Oxalic acid | Milling Blanching Boiling Soaking |
Boiling Soaking |
Suma and Urooj, 2014; Patel et al.2018 |
| 6. | Enzyme inhibitors | Soaking Autoclave Roasting Fermentation Boiling |
Fermentation Boiling |
Kumari, 2018; Patterson et al., 2017; Vagadia et al., 2017; Ogodo et al., 2019 |
| 7. | Polyphenols | Germination Soaking Fermentation |
Germination |
Singh et al., 2017; Simwaka et al., 2017 |
| 8. | Gossypols | Extrusion Fermentation |
Extrusion | Buser and Abbas, 2001 |
| 9. | Raffinose | De-hulling Germination Alcoholic extraction Microbial treatment |
Cooking | Kannan et al., 2018 |
| 10. | Goitrogens | Steaming Cooking Fermenting Milling Soaking Washing |
Soaking | Bajaj et al., 2016 |
| 11. | BOAA | Soaking Boiling Fermentation Cooking Autoclaving |
Soaking and cooking |
Srivastava et al., 2015; Hailu et al., 2015 |
| 12. | Alkaloids | Soaking Washing Germination Fermentation Aqueous thermal treatment Alkaline treatment |
Soaking, cooking fermentation, and alkaline treatment | Boschin and Resta, 2013 |
Milling
This is the most common technique for separating the bran layer from grains. Since anti-nutritional factors are mostly present in bran, this process removes anti-nutrients and reduces their distribution in grains. This procedure effectively eliminates anti-nutrients in bran, such as phytic acid, lectins, tannins, and enzyme inhibitors (Gupta et al., 2015). A study in pearl millet found that milling altered the chemical makeup and distribution of oxalic acid (Suma and Urooj, 2014). Hence, milling is effective in removing anti-nutrients from aleurone and bran.
Soaking
Soaking is yet another popular method for removing anti-nutrients from food. Soaking reduces the cooking time and enhances the release of endogenous phytases found in plant foods (Vashishth et al., 2017). Soaking provides essential moist conditions in nuts, grains, and other edible seeds that are required for germination and thereby also reduces trypsin inhibitors and phytic acid to improve digestibility by enhancing the nutritional value of grains (Kumari, 2018). Soaking, boiling and autoclaving was found to be effective to reduce tannins while soaking the seeds for 24 hours drastically reduced the hydrogen cyanide. Further soaking was found to be more helpful in reducing the stachyose and raffinose content with an average reduction of 51.20% and 21.20% respectively (Udensi et al., 2008). Soaking legumes in water overnight has been found to reduce phytate, protease inhibitors, lectins, and tannins. A 12-hour soaking was found to decrease the amount of phytate in peas by up to 9%, while soaking pigeon peas for 6–18 hours reduced the concentration of lectins, tannins, and protease inhibitors by 38–50%, 13–25%, and 30%, respectively (Ertaş and Türker, 2014). It has also been suggested that wheat and barley can be ingested after soaking for a length of time, preferably 12–24 hours (Onwuka, 2006). It has also been reported that soaking grains and beans can successfully enrich the amount of protein and minerals in grains (Coulibaly et al., 2011).
Boiling
Anti-nutrients such as lectins, tannins, and protease inhibitors can be ameliorated by high heat during boiling. One study found that boiling pigeon peas for 80 minutes reduced protease inhibitors by 70%, lectin by 79%, and tannin by 69% (Onwuka, 2006). It has also been reported that boiling of cooked green leafy vegetables further reduces calcium oxalate by (19–87%) and that boiling is be more efficient than baking and steaming (Amalraj and Pius, 2015). A study by Maphosa and Jideani (2017) found that boiling beans significantly improved their nutritional quality by reducing their lectin and saponin concentrations.
Autoclaving and Roasting
The majority of foods show health benefits when consumed after autoclaving. The cooking time required depends on the type of anti-nutrient and the cooking method. Generally, the longer the cooking time, the greater the reduction in anti-nutrients. According to earlier research, heating foods significantly increases their nutritional value by removing their content of anti-nutrients, especially tannins and trypsin inhibitors (Patterson et al., 2017). Trypsin inhibitor activity in soybean meal was dramatically reduced by roasting (Vagadia et al., 2017). Another study found that heating, soaking, and autoclaving of beans considerably reduced the amount of enzyme inhibitors and tannins in grains (Torres et al., 2016).
Sprouting
This is an effective process for lowering the anti-nutrient content in plant-based foods (Nkhata et al., 2018). During sprouting, anti-nutrients such as phytate and protease inhibitors are degraded. Lectins and protease inhibitors have also been found to be slightly reduced. Various kinds of grains and legumes have been enriched by sprouting, which reduced phytate by 37–81%. The enzyme phytase, which is often activated during seed germination, breaks down the phytate–mineral bound complex in grains. Hence, this approach is most usually employed to reduce the anti-nutritional content of cereals (Oghbaei and Prakash, 2016; Vashishth et al., 2017). Azeke et al. (2011) found that the phytate level of cereal grains was considerably lowered after 10 days of sprouting. Recent research also found that activation of beta-glucosidases during germination altered the isoflavone profile of soybeans, and this is significant for boosting nutritional value, as isoflavones have similar chelating effects (Yoshiara et al., 2018; Ida and de Camargo, 2022). In addition, it has been found that, in millets, the greatest reductions in polyphenol concentrations (up to 75%) are obtained by sprouting, exceeding those achieved by soaking, microwave treatment, and fermentation (Singh et al., 2017).
Fermentation
The metabolic process of fermentation is found to enhance the absorption of nutrients in grains. This also involves the oxidation of carbohydrates to produce energy. Grain nutritional value has been proven to be enhanced by fermentation that involves adding more critical amino acids, including lysine, methionine, and tryptophan (Mohapatra et al., 2019). The crucial process of fermentation dramatically reduces the amount of anti-nutrients such as phytic acid, tannins, and polyphenols in cereals (Simwaka et al., 2017). Tannin levels were found to be reduced by lactic acid fermentation, resulting in increased iron absorption (Ray et al., 2014). In a recent study, using typical fermentation techniques, maize flour was fermented with a mixture of lactic acid bacteria (LAB) for interval periods of 12 hours to examine the impact of fermentation on anti-nutritional components. It was found that anti-nutrients such as tannin, polyphenol, phytate, and trypsin inhibitor were significantly decreased by fermentation and that the reduction in anti-nutrients increased with increasing fermentation time. The results showed that, compared with spontaneous fermentation, anti-nutritional components were lowered more by LAB mixture fermentation (Ogodo et al., 2019).
Combination of methods
Combining several strategies can significantly reduce anti-nutrients. In some cases, anti-nutrients can be totally eliminated from foods. For example, soaking, sprouting, and lactic acid fermentation reduced phytate in quinoa by 98%. Similarly, sprouting and lactic acid fermentation of corn and sorghum entirely eliminated phytate. Furthermore, soaking and boiling pigeon peas reduced lectins, tannins, and protease inhibitors by 98–100% (Onwuka, 2006). Hence, combining multiple distinct elimination procedures is the most effective way to eliminate anti-nutrients in plant meals.
Extrusion
In the food industry, extrusion is a widely utilized processing method and has numerous benefits. A single screw or a series of screws are used to push food ingredients through a tiny aperture. It has been found that anti-nutrients such as phytic acid, tannins, phenols, alpha-amylase, and trypsin inhibitors are dramatically reduced by extrusion. Extrusion has also been found to reduce the proportion of phytic acid phosphorus to total phosphorus. Extrusion of legumes that have been previously soaked in water for 16 hours has been recommended to improve their nutritional value, and this has increased their utilization by humans and animals (Abd El-Hady and Habiba, 2003). Tannins in sesame oilseed meal were also reduced using a single-screw frying extruder (Mukhopadhyay and Bandyopadhyay, 2003). Based on the official standard procedures of the American Oil Chemists’ Society, test findings showed that around 71%–78% reduction in free gossypol levels was also effectively attained by extrusion (Buser and Abbas, 2001).
Breeding strategies to alter the anti-nutritional components for enhanced bioavailability of nutrients in foods
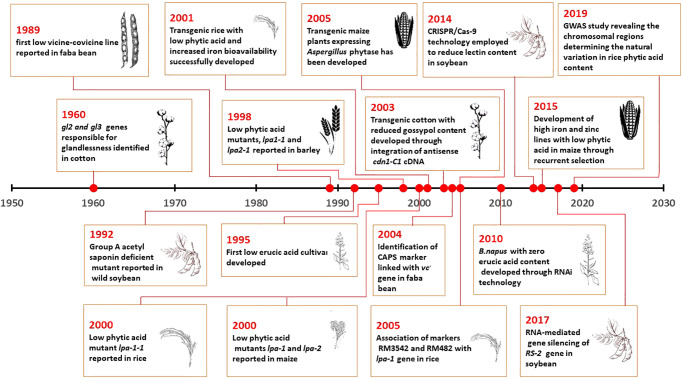
The reduction of anti-nutrients in crops is a crucial breeding strategy that plays a major role in enhancing the quality of the produce. Several breeding techniques, starting with selection, mutation, backcrossing, hybridization, and population improvement, have been implied with the natural and induced genetic resources. The breeding for reducing antinutrients in crops was intiated in the early 1960s with glandless cotton (Figure 1). More recently, gene silencing and editing techniques have been used to produce low anti-nutrient lines of major crops (Figure 1). Conventional breeding for anti-nutrient reduction began with the identification of reduced anti-nutrient accumulation in germplasm accessions. Genotypes with reduced gossypol content were selected in 1960, and McMichael (1960) reported that glandlessness in cotton is conferred by two genes, namely gl2 and gl3. As gossypol plays a major role in host plant resistance, these findings later led to the discovery of an ideal genotype with glandless seed-gossypol cum glanded plant (Dilday, 1986; Vroh Bi et al., 1999). This led to the identification reduced gossypols in seeds without manifesting their concentrations in the vegetative parts.
Figure 1.
Timeline of Anti-nutritional Breeding in major crops.
Subsequently, selection for reduced enzyme inhibitors from pulse germplasm was also observed to be an efficient way to identify potential donors with reduced inhibitors. Zero Kunitz inhibitor lines, namely PI 157-440 and PI 196-168, were identified in soybean (Orf and Hymowitz, 1979). These inhibitors were found to be controlled by a recessive gene, tj, which was later introgressed into an elite cultivar by Bernard and Hymowitz in 1986. Similarly, low-vicine and low-covicine lines were selected from the germplasm of 919 accessions in faba bean. The low vicine–covicine trait in pulses was found to be produced by a recessive gene, which was designated “vc”, and this was conferred for reducing the enzyme inhibitors (Duc et al., 1989; Duc et al. 2004; Gutiérrez et al., 2006; Webb et al. 2016; O’Sullivan et al. 2018).
Despite these adopted selection techniques, recurrent selection in maize with two synthetic populations, namely BS11 and BS3, was also performed. Three cycles of selection were successful in developing high-iron and high-zinc lines with low phytic acid in maize (Beavers et al., 2015). Similarly, selection for low saponin in quinoa after three cycles of pedigree breeding was found to reduce saponin accumulation in the population, but, due to the dominance of this trait, alternate strategies were required to reduce saponin content in polyploid and heterozygous crops (Ward, 2000). During the course of selection in the same period, there were investigations for induced mutations in the cultivar MACS 450 of soybean by gamma rays. These treatments were able to produce three mutants in M5 with lower lectin and normal germination rate. Further, this was also suggested to be used as a potential donor in improvising the soybean meal quality (George et al., 2008).
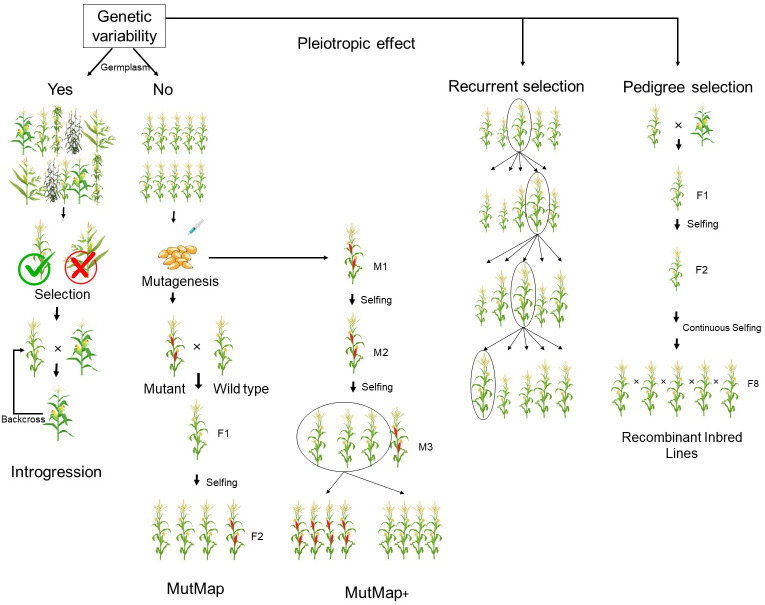
Backcrossing and mutation breeding are the strategies predominantly used to reduce anti-nutritional traits in crops (Figure 2) (Wilcox et al., 2000; Yuan et al., 2009). This is because several anti-nutritional traits play a major regulatory function in plants (Sureshkumar et al., 2014). Hence, their drastic reduction has been observed to have negative pleiotropic effects, affecting yield (Raboy et al., 2015). For this reason, phytic acid has been successfully reduced in potential donors identified from spontaneous and induced mutants in major crops (Pramitha et al., 2021). Raboy et al. (2000) identified three lpa mutants in maize. Among them, the lpa1 mutant was found to exhibit low phytic acid with meagre accumulation of myoinositol phosphates due to a mutation in the initial biosynthesis of phytic acid involving myoinositol. lpa2 had reduced phytic acid with accumulation of myoinositol phosphate intermediates, and lpa3 had reduced phytic acid with accumulation of myoinositol (Shi et al. 2003; Shi et al., 2005). Recent studies have shown that introgression of lpa 2 in the parents of a ruling hybrid, DMH 121, from the Indian Institute of Maize Research, by marker-assisted backcross was efficient in developing a better version of the released hybrid. The near isogenic lines (NILs) of the parents of DMH 121, namely BML 6 and BML 45, were observed to produce less phytate than the original lines. The newer versions of these parents were exactly the same as the earlier version, except for phytic acid content, and they could be further hybridized to produce low-phytic acid hybrids in maize (Yathish et al., 2022). This coincided with marker-assisted backcross of the null allele for Kunitz trypsin inhibitor (KTI) in DS9712 and DS9814 with a donor called P1542044 in soybean. In this case, in order to minimize linkage drag, three selections, foreground, background, and recombinant, were performed. This resulted in the development of six KTI-free lines in soybean with a maximum recovery percentage (Maranna et al., 2016).
Figure 2.
Breeding methods focused for reducing anti-nutritional factors.
Regarding RFOs, their amount of consumption and the ratio of balanced protein and oil profile in foods are yet to be determined (Elango et al., 2022). Selection for a lower RFOs version of high-RFOs foods such legumes found a negative correlation with protein and yield. In soybean, a significant negative correlation was observed between protein and RFOs, whereas RFOs was reported to have a positive correlation with oil content (Bueno et al., 2018). Studies on RFOs have identified a role for MIPS (myoinositol phosphate synthase) and galactinol synthase activity, which was exploited to manipulate RFOs levels in major crops (Elango et al., 2022). Further, genetic mapping for raffinose in recombinant inbred lines (RILs) of soybean produced from a cross of MD96-5722 and Spencer detected 14 major quantitative trait loci (QTLs) for raffinose which could be utilized to produce higher concentrations of sucrose and lower concentrations of raffinose and stachyose in the future (Akond et al., 2015).
Glucosinolates have also been similarly altered by breeding approaches. Glucosinolates were found to have a quantitative inheritance which was highly influenced by environmental factors. A high-density linkage map of the major genes involved in the synthesis of glucosinolates in Brassica oleracea has been created with sequences of BoGSL-ALK. In addition, comparative genomics studies of the glucosinolate biosynthesis pathway in Arabidopsis revealed significant QTLs and candidate genes to alter its profile in crops (Gao et al., 2007; Issa, 2010). This led to the development of high-glucoraphanin broccoli by marker-assisted selection involving an interspecific cross between B. oleraceae × B. villosa. In addition, marker-assisted selection for altered glucosinolate profiles was achieved between B. rapa × B. oleraceae (Hirani, 2011). Further projects have focused on developing super broccoli with higher isothiocyanate content by incorporating genes from wild species with the aim of developing pharmaceuticals (Ishida et al., 2014). Similar studies have investigated isothiocyanates and glucosinolates in Raphanus sativus, and QTL analyses using high genetic density mapping led to the development of candidate genes for glucosinolate synthesis in roots (Wang et al. 2013). These studies have improved the prospects of altering the profiles of glucosinolates and isothiocyanates (Zou et al., 2013).
Subsequently, breeding to achieve zero erucic acid, due to its serious health issues, was also effective in producing low erucic acid lines in brassicas (Sivaraman et al., 2004). The major gene that plays a role in erucic acid synthesis was observed to be FAE1. Sequencing of the FAE1 gene in high- and low-erucic acid cultivars revealed 28 base deletions containing 24 bases of AT-rich regions in a 1,300-bp section upstream of the promoter of the FAE1 start codon (Yan et al., 2015). Later, mutations in FAE1 were induced to identify low erucic acid lines, and introgression of these erucic acid mutant genes in elite cultivars was carried out by Karim et al. (2016). In that study, a genes named BnFAE1.1 and BnFAE1.2 in the A and C genome of rapeseed was introgressed to a turnip cultivar. The incorporation of the mutant gene bnfae1.1(e1) was monitored by a CAPS (cleaved amplified polymorphic sequence) primer. Early deteriorations in the seed set of backcross progenies were later observed to be improved in the advanced progenies. This suggested that the frequency of recombination events among progenies substantiated the negative effects on morphological traits in later generations (Pramitha et al., 2021).
The overall schemes adopted for marker-assisted breeding and QTLs detected for reducing the anti-nutrients are presented in Table 3. It can be seen that, in the earlier reports, anti-nutritional factors were manipulated either by introgression or by mutation breeding. The backcrosses also involved selfing in their intermittent process, as most of the reductions were controlled by recessive genes (Table 3). Genetic manipulation of anti-nutrients needs to be carefully monitored, as anti-nutrients play a major role in plant defense and abiotic stress tolerance (Guttieri et al. 2004). Hence, alternate strategies to minimize their negative pleiotropic effects with rapid selection among populations have to be further developed in the future using omics approaches. Upcoming projects involving transgenics and gene editing opens a new gateway to tissue-specific expression, an area that is gaining popularity (Wang et al., 2022).
Table 3.
Summary of the major QTL’s observed for the anti-nutrients in crops.
| S. no. | Crop | Anti-nutrient | QTL/marker | Location | Reference |
|---|---|---|---|---|---|
| 1 | Rice | Phytic acid | Chromosome 2L | Larson et al., 2000 | |
| Chromosome 2 | Andaya and Tai, 2005 | ||||
| Chromosome 5 | Stangoulis et al., 2007 | ||||
| Chromosome 12 | |||||
| qLPA8.1 | Chromosome 8 | Gyani et al., 2020 | |||
| 2 | Barley | Phytic acid | Chromosome 2H | Larson et al., 1998 | |
| 3 | Barley | Raffinose | QcRaf.2H | Chromosome 2H | Gudys et al., 2018 |
| 4 | Corn | Phytic acid | Chromosome 4 | Liu et al., 2013 | |
| Chromosome 6 | |||||
| Chromosome 1 | |||||
| Chromosome 7 | |||||
| Chromosome 2 | |||||
| 5 | Mung bean | Phytic acid | SDPAP4.1 | LG- 4A | Sompong et al., 2012 |
| SDPAP11.1 | LG-11A | ||||
| 6 | Pea | Phytic acid | LG-5 | Shanmugam et al., 2015 | |
| 7 | Rapeseed | Erucic acid | qEA.A8.1 | LG-A8 | Cao et al., 2010 |
| qEA.A8.2 | LG-A8 | ||||
| 8 | Indian mustard (Brassica juncea L.) | Erucic acid | ea-1 | LG-17 | Gupta et al., 2004 |
| ea-2 | LG-3 | ||||
| Eru-A8-1-EJ | LG-A08 | Rout et al., 2018 | |||
| Eru-A8-2-EJ | LG-A08 | ||||
| Eru-A8-3-EJ | LG-A08 | ||||
| Eru-A8-1-EPJ | LG-A08 | ||||
| Eru-A8-2-EPJ | LG-A08 | ||||
| Eru-A8-3-EPJ | LG-A08 | ||||
| Eru-A8-1-VH | LG-A08 | ||||
| Eru-A8-2-VH | LG-A08 | ||||
| Eru-A8-1-VH | LG-A08 | ||||
| Eru-B7-1-VH | LG-B07 | ||||
| Eru-B7-2-VH | LG-B07 | ||||
| Eru-B7-1-DE | LG-B07 | ||||
| Eru-B7-2-DE | LG-B07 | ||||
| Eru-B7-3-DE | LG-B07 | ||||
| Eru-A8-1-TD | LG-A08 | ||||
| Eru-A8–2-TD | LG-A08 | ||||
| Eru-A8-3-TD | LG-A08 | ||||
| 9 | Yellow mustard (Sinapis alba L.) | Erucic acid | Chromosome 3 | Javidfar and Cheng, 2013 | |
| 10 | Soybean | Gossypol | qGos1-c13-1 | Chromosome 13 | Yu et al., 2012 |
| qGos1-c19-1 | Chromosome 19 | ||||
| qGos2-c19-1 | Chromosome 19 | ||||
| 11 | Soybean | Raffinose | Chromosome 6 | Salari et al., 2021 | |
| Chromosome 6 | Skoneczka et al., 2009 | ||||
| qRAF001 | Chromosome 1 | Akond et al., 2015 | |||
| qRAF002 | Chromosome 3 | ||||
| qRAF003 | Chromosome 6 | ||||
| qRAF004 | Chromosome 9 | ||||
| qRAF005 | Chromosome 14 | ||||
| qRAF006 | Chromosome 14 | ||||
| qRAF007 | Chromosome 16 | ||||
| 12 | Soybean | Stachyose | qSTA001 | Chromosome 1 | Akond et al., 2015 |
| qSTA002 | Chromosome 6 | ||||
| qSTA003 | Chromosome 12 | ||||
| qSTA004 | Chromosome 14 | ||||
| 13 | Soybean | Group A saponin | Chromosome 15 | Sundaramoorthy et al., 2018 | |
| Group A saponin (hypocotyl) | Chromosome 5 (A1) | Teraishi et al., 2017 | |||
| Chromosome 8 (A2) | |||||
| Group A saponin (cotyledon) | Chromosome 6 (C2) | ||||
| 14 | Faba bean | Vicine–convicine | Chromosome 1 | Khazaei et al., 2015 | |
| 15 | Sorghum | HCN (Dhurrin) | Dhu1 | Chromosome 1 | Hayes et al., 2016 |
| Hydrocyanic acid | qPA7-1 | Chromosome 4 | Wu et al., 2022 | ||
| 16 | Sorghum | Tannin | Qsqr.t-2 | Chromosome 4 | Wu et al., 2012 |
| Qsqr.t-4 | Chromosome 4 | ||||
| 17 | Field mustard (Brassica rapa) | Glucosinolates | LG-A3 | Hirani, 2011 | |
| 18 | Rapeseed (Brassica napus L.) | Glucosinolates | GSL-1 | LG-20 | Toroser et al., 1995 |
| GSL-2 | LG-1 | ||||
| 19 | Indian mustard (Brassica juncea L.) | 3-Butenyl-glucosinolates | GSL-A2a | LG-2a (A) | Mahmood et al., 2003 |
| GSL-A2b | LG-2b(A) | ||||
| 2-Propenyl-glucosinolates | GSL-A2a | LG-2a (A) | |||
| GSL-F | unlinked segment | ||||
| GSL-B3 | LG-3 (B) | ||||
| 20 | Barbarea vulgaris | NAS, 2-phenylethylglucosinolate (gluconasturtiin) | qNAS-4-1 | LG-4 | Liu et al., 2019 |
| qNAS-4–2 | LG-4 | ||||
| BAR, (2S)-2-hydroxy-2-Phenylethylglucosinolate (glucobarbarin) | qBAR-3-1 | LG-3 | |||
| qBAR-4-1 | LG-4 | ||||
| qBAR-5-1 | LG-5 | ||||
| EBAR, (2R)2-hydroxy-2-phenylethylglucosinolate (epiglucobarbarin) | qEBAR-3–1 | LG-3 | |||
| qEBAR-4-1 | LG-4 | ||||
| qEBAR-5-1 | LG-5 | ||||
| IM, 3-indolylmethylglucosinolate (glucobrassicin) | qIM-4-1 | LG-4 | |||
| qIM-6-1 | LG-6 | ||||
| 4mIM, 4-methoxy-3-indolylmethylglucosinolate (4-methoxyglucobrassicin) | q4mIM-4-1 | LG-4 | |||
| q4mIM-5-1 | LG-5 | ||||
| 21 | Narrow leaf lupin (Lupinus angustifolius) | Quinolizidine alkaloids | iuc_RAP2-7-pauper loci | LG-07 | Kroc et al., 2019 |
| 22 | White lupin (Lupinus albus L.) | Quinolizidine alkaloids | 11 loci | LG-11 | Phan et al., 2007 |
| Pauper loci | LG-18 | Rychel and Książkiewicz, 2019 | |||
| 23 | Yellow lupin (Lupinus luteus L.) | Quinolizidine alkaloids | YL-06 loci | LG-06 | Iqbal et al., 2020 |
Advanced genetic approaches for developing sustainable food crops in future
Manipulating anti-nutritional traits to enhance the bioavailability of nutrients is a major concern in crops, as these traits have to be mitigated in such a way as to avoid negative influences on yield. The reduction of these traits in crops has been successfully carried out for major anti-nutrients and the various methods of altering their content are described in Table 4. Gene silencing using RNAi technology is an efficient way of optimizing the expression of these factors in crops and has been applied to the genes involved in the biosynthesis of these components in plants. Gossypol is one plant phytochemical that plays a major role in host plant resistance and is not needed in human nutrition. Therefore, ultra-low-gossypol cotton has been developed by silencing of δ-cadinene synthase gene. The knockdown of this gene reduced the accumulation of gossypol in seeds, foliage, and floral organs of transgenic cotton. The initial version of transgenic cotton showed on-par performance, in terms of yield and fiber quality, with stable expression. The transgenic cotton was also observed to exhibit a higher oil content than the control (Palle et al., 2013). Recently, selective RNAi knockout of the δ-cadinene gossypol gene in seeds of the cultivar TAM66274 effectively reduced the oil content by about 97%, and the cultivar also passed food safety tests conducted by the Food and Agricultural Organization of the USA (FAO) (Rathore et al., 2020). This method has also been found to be effective in controlling gossypol levels in seeds without affecting gossypol concentration in the vegetative parts, and the technique has been patented by Texas A & M university. Thus, transgenic cotton would contain either a δ-cadinene synthase gene or a δ-cadinene-8 hydroxylase gene, or both, linked to a seed-specific promoter gene for inducing RNA gene silencing when expressed in cottonseed of the plant (Rathore et al., 2009).
Table 4.
Major RNAi and gene editing techniques adopted in major crops.
| S. no. | Anti-nutrient | Crop | Gene/enzyme | Pathway | Technique | Reference |
|---|---|---|---|---|---|---|
| 1. | Phytic acid | Rice | IPK1 | Phosphorylation of Ins(3)P and phospholipase C mediated | Chromosome mapping | Ye et al., 2013 |
| lpa-1 | Lipid independent | Gene editing | Watanabe et al., 2018 | |||
| OsITP5/6K/1 | Lipid dependent phytic acid biosynthesis Lipid independent phytic acid biosynthesis pathway |
RNAi-mediated down-regulation | Karmakar et al., 2020 | |||
| OsIPK1 | Inositol phosphate pathway | RNAi-mediated seed-specific silencing | Ali et al., 2013 | |||
| ITPK, OsITP5 | Myoinositol pathway | RNAi-mediated down-regulation | Karmakar et al., 2020 | |||
| RINO1 | Myoinositol pathway and direct proanthocyanidin pathway | Antisense cDNA approach | Kuwano et al., 2009 | |||
| Wheat | TaIPK1 | Auxin signaling pathway | RNAi technology | Ibrahim et al., 2022 | ||
| TaABCC13 | Myoinositol pathway | RNAi technology | Bhati et al., 2016 | |||
| Soybean | GmMIPS-1 | Lipid dependent/salvage pathway | Gene silencing | Nunes et al., 2006; Kumar et al., 2019 | ||
| GmIPK1 | Both lipid independent and lipid dependent | CRISPR/Cas-9 genome editing | Song et al., 2022 | |||
| lpa1 | Myoinositol hexa-kis phosphate | EMS approach | Gillman et al., 2011 | |||
| Glyma.20G085100 | – | QTL mapping | Marsh et al., 2022; Jha et al., 2022 | |||
| Barley | lpa-1-1 | Signaling pathway | QTL mapping | Bregitzer and Raboy, 2006 | ||
| Corn | ZmMRP4 in lpa-2 | Supply pathway | C-T transition by mutation | Tamilkumar et al., 2014 | ||
| ZmIPK1 | Inositol phosphate pathway | ZFN approach | Shukla et al., 2009 | |||
| lpa2–1 | Myoinositol InsP6 pathway | EMS Mu insertion approach | Raboy et al., 2000 | |||
| 2.lpa1–1 | Myoinositol phosphate pathway | Gene silencing | Shi et al., 2007 | |||
| Rapeseed | BnITPK | Both lipid dependent and lipid independent | CRISPR/Cas-9 gene editing | Sashidhar et al., 2020 | ||
| Arabidopsis | AtIpk1-1 | – | T-DNA insertion method | Stevenson-Paulik et al., 2005 | ||
| AtITPK1, AtITPK4 | Inositol InsP6 pathway | Reverse genetic approach | Kim and Tai, 2011 | |||
| atips1, atips2 | – | T-DNA insertion method | Kim and Tai, 2011 | |||
| Lablab bean | dlMIPS | Myoinositol phosphate synthase | RT-PCR system | Jagal Kishore et al., 2020 | ||
| 2. | Erucic acid | Rapeseed | FAE1 | – | Gene editing | James et al., 1995; Yan et al., 2015 |
| FAD2 and FAE1 | Gene silencing | Peng et al., 2010 | ||||
| BnFAE1.1 | Ketoacyl-CoA synthase | RNAi silencing | Tian et al., 2011; Kaur, 2018 | |||
| BnFAE1 | RNAi silencing | Shi et al., 2015 | ||||
| BnFAE1 and BnFAD2 | Long-chain fatty acid biosynthesis | CRISPR/Cas9-mediated gene editing | Shi et al., 2022 | |||
| BnFAD2 and BnFAE1 | Fatty acid biosynthesis | RNAi silencing | Shi et al., 2017 | |||
| Indian mustard | BjFAE1 | Ketoacyl-CoA synthase | Agrobacterium-mediated transgenic method | Kanrar et al., 2006 | ||
| Ethiopian mustard | FAD2 and FAE | Fatty acid biosynthesis | Hairpin-RNA mediated silencing | Mietkiewska et al., 2008 | ||
| 3. | Gossypol | Cotton | Cad1-A | d-Cadinene synthase | RNAi technology | Davis et al., 1996; Luo et al., 2001 |
| gl2 and gl3 | d-Cadinene synthase | Southern analysis | Martin et al., 2003; Benedict et al., 2004; Sunilkumar et al., 2006 | |||
| gl1, gl2 , and gl3 | d-Cadinene synthase | RNAi silencing | Palle et al., 2013; Rathore et al., 2020 | |||
| GhMYB25 | – | Antisense gene silencing | Abdurakhmonov et al., 2016 | |||
| GhCLA1 | – | Temperature-sensitivity CRISPR/LbCpf1-mediated genome editing | Li et al., 2021 | |||
| GhCLA1 | – | CRISPR/Cas-9 technology | Wang et al., 2018 | |||
| CYP82D109 | Gossypol biosynthesis pathway | RNAi technology | Wagner et al., 2015 | |||
| 4. | Lectin | Soybean | P34 allergen | – | CRISPR/Cas-9 technology | Watanabe et al., 2018 |
| Peanut | Gly1 protein | NAD-dependent | 2-D gel electrophoresis | Kottapalli et al., 2008 | ||
| 5. | Saponin | Soybean | GmBAS1, GmBAS2 | β-Amyrin synthase | RNAi-mediated gene silencing | Takagi et al., 2011 |
| DeF26G1 | Flavonoid biosynthesis | Transcriptome profiling | Kuma A, et al., 2016 | |||
| Barrel medic | CYP93E2 | – | Agrobacterium-mediated transformation | Confalonieri et al., 2021 | ||
| Korean ginseng | CYP716A53v2 | PPT synthase | CRISPR/Cas9-mediated gene knockout | Choi et al., 2022 | ||
| CYP716A53v2 | PPT synthase | RNAi technology | Park et al., 2016 | |||
| 6. | Tannin | Quaking aspen | MYB134 | CT biosynthesis | RNAi suppression | Gourlay et al., 2020 |
| Peanut |
aflS/aflJ, aflR, aflC/pksA/pksL1, pes1, afelp |
– | RNAi silencing | Arias et al., 2015 | ||
| 7. | Oxalic acid | Soybean | b-ODAP | – | Transgenic production | Kumar V, et al., 2016 |
| Wheat | BoGSL-ELONG, BoGSL-PRO, and BoGSL-ALK | Glucosinolate biosynthesis pathway | Comparative genomic analysis (QTL mapping) | Ishida et al., 2014 | ||
| Tomato | FvOXDC | Oxalic acid biosynthesis pathway | Metabolic remodeling | Chakraborty et al., 2013 | ||
| Tobacco | Germin gf-2.8 | Co-A-dependent pathway, jasmonate pathways, and phenylpropanoid pathways | Transgenic approach | Kumar et al., 2019 | ||
| 8. | Vicine and convicine | Faba bean | vc, vcr | – | QTL mapping | Khazaei et al., 2019 |
| 9. | Enzyme inhibitors | Finger millet | Opaque2 | – | Random amplified polymorphic DNA (RAPD) and simple sequence repeat (SSR) profiling | Vinoth and Ravindhran, 2017 |
| Durum wheat | 0.28 ATI | ATI pathway | CRISPR/Cas-9 multiplex editing | Camerlengo et al., 2020 | ||
| Bread wheat | CM3, CM16 and 0.28 ATI | ATI pathway | RNAi silencing | Kalunke et al., 2020 | ||
| 10. | Glucosinolate | Wild cabbage | BjMYB28 | Aliphatic glucosinolate biosynthesis | RNAi targeted suppression | Augustine et al., 2013 |
| Chinese kale | BoaMYB28 | Aliphatic glucosinolate biosynthesis | RNAi approach | Yin et al., 2017 | ||
| Indian Mustard | BjuMYB28 | Aliphatic glucosinolate biosynthesis | Intron-spliced hairpin RNAi targeting | Augustine and Bisht, 2019 | ||
| BjuXLG | Aliphatic glucosinolate biosynthesis | RNAi based suppression | Tiwari et al., 2021 | |||
| Rapeseed | MAM | Aliphatic glucosinolate biosynthesis pathway | RNAi silencing | Liu et al., 2011 | ||
| BrGI | GSL biosynthetic pathway | RNAi knockdown | Kim et al., 2021 | |||
| Arabidopsis | HAG1/MYB28 | Aliphatic glucosinolate biosynthesis pathway | RNAi knockdown | Gigolashvili et al., 2007 | ||
| OBP2 | IAA biosynthetic pathway | RNAi mediated | Skirycz et al., 2006 | |||
| Garden cress | LcIND | Glucosinolate biosynthesis pathway | RNAi mediated | Karmakar et al., 2020 | ||
| 11. | Alkaloids | Potato and Tomato | Steroidal glycoalkaloids | Cytosolic mevalonic acid pathway | Silencing glycoalkaloid metabolism 4 | Itkin et al., 2013 |
| Steroidal glycoalkaloids (SSR2) | Cytosolic mevalonate pathway | Gene silencing | Cárdenas et al., 2015 | |||
| Tobacco and Catharanthus roeus | Steroidal glycoalkaloids (SSR2) | Mevalonate pathway | Gene editing | Cárdenas et al., 2016 | ||
| 12. | HCN | Cassava | MeCYP79D1 | Cyanogenic glycoside biosynthetic pathway | CRISPR/Cas9-mediated genome editing | Juma et al., 2022 |
| CYP79D1 and CYO79D2 | Cyanogenic glycoside biosynthetic pathway | CRISPR/Cas9-mediated knockout | Gomez et al., 2021 | |||
| Sorghum | CYP79A1 | Cyanogenic glycoside pathway | Antisense approach | Pandey et al., 2019 | ||
| 13. | BOAA | Grass pea | β-ODAP | β-ODAP biosynthesis pathway | CRISPR/Cas9-mediated gene editing | Das et al., 2021 |
Metabolite engineering for manipulating the concentration of raffinose in soybean was carried out by Valentine et al. (2017). For reducing the concentration of raffinose, the raffinose synthase 2 gene (RS2) was down-regulated by an RNAi construct. The silencing of this gene was further confirmed by qPCR and the total metabolizable energy for soybean meal in poultry was increased from 2,411 kcal/kg to 2,703 kcal/kg in the transgenic soybean. In contrast to this approach, the suppression of the cucumber stachyose synthase gene (CsSTS) by RNAi-mediated silencing had a significant impact on phloem loading, carbohydrate metabolism, and low-temperature stress tolerance (Lü et al., 2017). Recently, an advanced technique involving gene editing with two guide RNAs to knock out GmGoLS1A and GmGoLS1B (galactinol synthase genes) resulted in a reduction of raffinose from 64.70 mg/g to 41.95 mg/g (a 35% decrease) in soybean. The developed lines from these knockouts established a higher verbascose, protein, and fat content with no effect on plant growth, suggesting that they are potential targets for altering raffinose in soyabean genotypes (Le et al., 2020).
Adding to these findings, RNAi-mediated silencing of three amylase and trypsin inhibitor genes, namely CM3, CM16, and O.28 (α-amylase/trypsin inhibitors genes), revealed a higher trypsin inhibition which was acceptable to non-celiac wheat-allergic patients. Although there were some alterations in inhibitors, there were no changes in in the high-molecular-weight glutenin subunits or in yield (Kalunke et al., 2020). However, trypsin inhibitors such as TcTI from cocoa provide significant defense against Helicoverpa (do Amaral et al., 2022), and trypsin inhibitors that hinder digestion were also recently reported to be effective biopesticides (Rodríguez-Sifuentes et al., 2020). Advanced gene editing techniques targeting two seed-specific KTI genes, namely KTI1 and KTI3, resulting in small deletions and insertions in soybean open reading frames, offer an alternate strategy, by focusing on reducing trypsin inhibitors only in seeds for consumption, will be helpful in the future (Wang et al., 2022).
In addition to the above anti-nutrients, saponin has also been modified by RNAi-mediated silencing of two β-amyrin synthase genes (GmBAS1 and GmBAS2), and has a seed-specific promoter involved in the production of β-conglycinin, a seed storage protein in soybean (Takagi et al., 2011). Subsequently, metabolite remodeling of oxalate-by-oxalate decarboxylase (OXDC) effected a 90% reduction in oxalate, accompanied by with higher calcium, iron, and citrate, in transgenic tomatoes. Proteomic analysis of the OXDC leading to concerns that manipulation of this gene would also have undesirable effects unless tissue specific expression can be achieved (Chakraborty et al., 2013). Similarly, the use of RNAi to alter MYB134 to reduce tannins in poplar plants resulted in enhancing the susceptibility of the plant toward oxidative stress, emphasizing the importance of tissue-specific expression when reducing anti-nutrients in crops (Gourlay et al., 2020). Several approaches, including targeted silencing of IPK1 genes for lowering phytic acid in rice seeds (Ali et al., 2013), BjuMYB28 to reduce glucosinolates in brassicas (Augustine et al., 2013), OXDC in grass pea and soybean (Kumar V, et al., 2016), ITPK genes for reducing phytate in rice and wheat seeds with increased iron and zinc (Lucca et al. 2001) (Aggarwal et al. 2018; Pandey et al. 2021; Karmakar et al., 2020), have been successful in reducing anti-nutrients with minimum effects on morphological performance. Following the success of RNAi in IPK to reduce phytic acid, the CRISPR-Cas9 method has recently been used on a similar gene in soyabean, named GmIPK, to alter the phytate concentrations in soybean. This experiment was intended to standardize stable transformation of transgenic soybean lines with edited GmIPK2. This further emphasized the focus on implying more bioinformatic tools and study on transient expression which are necessary in future to further to improvise the soybean meal quality by CRISPR (Jose et al., 2022).
Considering the earlier observations for reducing the anti-nutrients, it can be observed that RNAi and gene editing are the two major techniques that are used in tissue-specific reduction in major crops (Figure 3) (Perera et al. 2018; Elkonin et al. 2021). Although the initial investigations have been conducted with reduced anti-nutrients, a standard protocol for strategic reduction of anti-nutrients is crucial in crops such as pulses needs to be reinforced in future. Legume-based foods are often reported to hinder the digestion process and, thus, standardization of protocols for seed-specific expression establishes a pathway to a sustainable diet in near future (Drakakaki et al. 2005). In addition, integrative omics will play an important role in the development of low-anti-nutrient versions of other major food crops and for detecting low anti-nutrient donors (Parca et al., 2018; Pandey et al. 2021).
Figure 3.
Future prospects of improving the quality of food crops.
Conclusion
Reducing anti-nutritional traits in crops is essential factor to achieve higher mineral bioavailability in foods. Although anti-nutrients pose a serious threat to human health, owing to their toxicity, some of them, such as phytic acid, raffinose, tannins and gossypol, are beneficial to growth and metabolism in plants. These anti-nutrients have both favorable and undesirable properties. On the one hand, they favor plant growth through regulatory activities such as biotic and abiotic stress tolerance. On the other hand, they hinder mineral absorption. This restrains any approach that focuses on a threshold reduction in anti-nutritional traits in major food crops. Despite this, a few anti-nutritional factors, such as Kunitz inhibitors, glucosinolates, tannins, alkaloids, and saponins, are being employed in the biopesticides and pharmaceutical industries. Therefore, a constitutive focus on manipulating this content for specific purposes needs to be ensured in future. This would facilitate safe consumption and processing of foods for the upcoming generation for specific anti-nutrients individually to avoid food allergies in future. Several techniques have been employed to alter the accumulation of anti-nutrients in grains, but the use of advanced omics techniques in genomics-assisted breeding, in the case of the majority of anti-nutrients, remain unused. Hence, omics offer a new gateway to understanding the regulatory pathways of crucial anti-nutritional traits in plants and their genetic manipulation. Recently, the use of mutation breeding, introgression, RNAi technology, and gene editing by CRISPR/Cas9 have enable us to achieve seed-specific expression in crops. Thereby, anti-nutrients that confer regulation of vegetative growth and their activity will remain unaffected. To conclude, we could observe that the expression of these anti-nutritional factors varies from crop to crop and, based on their intake, a specific strategy has to be adopted in major crops to provide high-value nutritional foods in future.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abd El-Hady E. A., Habiba R. A. (2003). Effect of soaking and extrusion conditions on antinutrients and protein digestibility of legume seeds. LWT-Food Sci. Technol. 36 (3), 285–293. doi: 10.1016/S0023-6438(02)00217-7 [DOI] [Google Scholar]
- Abdurakhmonov I. Y., Ayubov M. S., Ubaydullaeva K. A., Buriev Z. T., Shermatov S. E., Ruziboev H. S., et al. (2016). RNA Interference for functional genomics and improvement of cotton (Gossypium sp.). Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyemo S. M., Onilude A. A. (2013). Enzymatic reduction of anti-nutritional factors in fermenting soybeans by lactobacillus plantarum isolates from fermenting cereals. Nigerian Food J. 31 (2), 84–90. doi: 10.1016/S0189-7241(15)30080-1 [DOI] [Google Scholar]
- Aggarwal S., Kumar A., Bhati K. K., Kaur G., Shukla V., Tiwari S., et al. (2018). RNAi-mediated downregulation of inositol pentakisphosphate kinase (IPK1) in wheat grains decreases phytic acid levels and increases fe and zn accumulation. Front. Plant Sci. 9, 259. doi: 10.3389/fpls.2018.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akande K. E., Doma U. D., Agu H. O., Adamu H. M. (2010). Major antinutrients found in plant protein sources: their effect on nutrition. Pakistan J. Nutr. 9 (8), 827–832. doi: 10.3923/pjn.2010.827.832 [DOI] [Google Scholar]
- Akbar N., Gupta S., Tiwari A., Singh K. P., Kumar A. (2018). Characterization of metabolic network of oxalic acid biosynthesis through RNA seq data analysis of developing spikes of finger millet (Eleusine coracana): Deciphering the role of key genes involved in oxalate formation in relation to grain calcium accumulation. Gene 649, 40–49. doi: 10.1016/j.gene.2018.01.071 [DOI] [PubMed] [Google Scholar]
- Akond M., Liu S., Kantartzi S. K., Meksem K., Bellaloui N., Lightfoot D. A., Kassem M. A. (2015). Quantitative trait loci underlying seed sugars content in “MD96-5722” by “Spencer” recombinant inbred line population of soybean. Food Nutr. Sci. 6 (11), 964. doi: 10.4236/fns.2015.611100 [DOI] [Google Scholar]
- Al-Beiruty R. A., Cheyed S. H., Hashim M. H. (2020). Hazards of toxic hydrocyanic acid (HCN) in sorghum and ways to control it: A review. Plant Archives 20 (1), 2726–2731. [Google Scholar]
- Alexander J., Benford D., Cockburn A. (2008). Gossypol as undesirable substance in animal feed. EFSA J. 908, 1–55. doi: 10.2903/j.efsa.2009.908 [DOI] [Google Scholar]
- Ali N., Paul S., Gayen D., Sarkar S. N., Datta K., Datta S. K. (2013). Development of low phytate rice by RNAi mediated seed-specific silencing of inositol 1, 3, 4, 5, 6-pentakisphosphate 2-kinase gene (IPK1). PloS One 8 (7), e68161. doi: 10.1371/journal.pone.0068161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amalraj A., Pius A. (2015). Bioavailability of calcium and its absorption inhibitors in raw and cooked green leafy vegetables commonly consumed in India–an in vitro study. Food Chem. 170, 430–436. doi: 10.1016/j.foodchem.2014.08.031 [DOI] [PubMed] [Google Scholar]
- Andaya C. B., Tai T. H. (2005). Fine mapping of the rice low phytic acid (Lpa1) locus. Theor. Appl. Genet. 111 (3), 489–495. doi: 10.1007/s00122-005-2038-0 [DOI] [PubMed] [Google Scholar]
- Arias R. S., Dang P. M., Sobolev V. S. (2015). RNAi-mediated control of aflatoxins in peanut: method to analyze mycotoxin production and transgene expression in the peanut/Aspergillus pathosystem. J. Visualized Experiments 106), e53398. doi: 10.3791/53398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassova S. S., Gutzow I. S. (2013). Hippuric acid as a significant regulator of supersaturation in calcium oxalate lithiasis: The physiological evidence. BioMed. Res. Int. doi: 10.1155/2013/374950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine R., Bisht N. C. (2019). Targeted silencing of genes in polyploids: lessons learned from brassica juncea-glucosinolate system. Plant Cell Rep. 38 (1), 51–57. doi: 10.1007/s00299-018-2348-8 [DOI] [PubMed] [Google Scholar]
- Augustine R., Mukhopadhyay A., Bisht N. C. (2013). Targeted silencing of BjMYB28 transcription factor gene directs development of low glucosinolate lines in oilseed b rassica juncea. Plant Biotechnol. J. 11 (7), 855–866. doi: 10.1111/pbi.12078 [DOI] [PubMed] [Google Scholar]
- Awad E., Austin B., Lyndon A. (2012). Effect of dietary supplements on digestive enzymes and growth performance of rainbow trout (Oncorhynchus mykiss, walbaum). J. Am. Sci. 8 (12), 858–864. [Google Scholar]
- Awulachew M. T. (2022). A review of anti-nutritional factors in plant based foods. AdvNutr Food Sci. 7 (3), 223–236. doi: 10.33140/anfs.07.03.04 [DOI] [Google Scholar]
- Azeke M. A., Egielewa S. J., Eigbogbo M. U., Ihimire I. G. (2011). Effect of germination on the phytase activity, phytate and total phosphorus contents of rice (Oryza sativa), maize (Zea mays), millet (Panicum miliaceum), sorghum (Sorghum bicolor) and wheat (Triticum aestivum). J. Food Sci. Technol. 48 (6), 724–729. doi: 10.1007/s13197-010-0186-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj J. K., Salwan P., Salwan S. (2016). Various possible toxicants involved in thyroid dysfunction: a review. J. Clin. Diagn. Res. 10 (1), FE01. doi: 10.7860/JCDR/2016/15195.7092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banihani S. A. (2017). Radish (Raphanus sativus) and diabetes. Nutrients 9 (9), 1014. doi: 10.3390/nu9091014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battelino T., Danne T., Bergenstal R. M., Amiel S. A., Beck R., Biester T., et al. (2019). Clinical targets for continuous glucose monitoring data interpretation: recommendations from the international consensus on time in range. Diabetes Care 42 (8), 1593–1603. doi: 10.2337/dci19-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavers A. W., Goggi A. S., Reddy M. B., Lauter A. M., Scott M. P. (2015). Recurrent selection to alter grain phytic acid concentration and iron bioavailability. Crop Sci. 55 (5), 2244–2251. doi: 10.2135/cropsci2014.12.0807 [DOI] [Google Scholar]
- Benedict C. R., Martin G. S., Liu J., Puckhaber L., Magill C. W. (2004). Terpenoid aldehyde formation and lysigenous gland storage sites in cotton: variant with mature glands but suppressed levels of terpenoid aldehydes. Phytochemistry 65 (10), 1351–1359. doi: 10.1016/j.phytochem.2004.03.032 [DOI] [PubMed] [Google Scholar]
- Bernard R. L., Hymowitz T. (1986) Registration of L81-4590, L81-4871, and L83-4387 soybean germplasm lines lacking the kunitz trypsin inhibitor. Crop Sci. (USA). 26, 650–651. doi: 10.2135/cropsci1986.0011183X002600030058x [DOI] [Google Scholar]
- Bhati K. K., Alok A., Kumar A., Kaur J., Tiwari S., Pandey A. K. (2016). Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J. Exp. Bot. 67 (14), 4379–4389. doi: 10.1093/jxb/erw224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutia S. K., Behera B., Nandini Das D., Mukhopadhyay S., Sinha N., Panda P. K., et al. (2016). Abrus agglutinin is a potent anti-proliferative and anti-angiogenic agent in human breast cancer. Int. J. Cancer 139 (2), 457–466. doi: 10.1002/ijc.30055 [DOI] [PubMed] [Google Scholar]
- Bhutkar M. A., Bhise S. B. (2012). In vitro assay of alpha amylase inhibitory activity of some indigenous plants. Int. J. Chem. Sci. 10 (1), 457–462. doi: 10.31031/MAPP.2018.01.000518 [DOI] [Google Scholar]
- Blochl A., Peterbauer T., Hofmann J., Richter A. (2008). Enzymatic breakdown of raffinose oligosaccharides in pea seeds. Planta 228 (1), 99–110. doi: 10.1007/s00425-008-0722-4 [DOI] [PubMed] [Google Scholar]
- Boncompagni E., Orozco-Arroyo G., Cominelli E., Gangashetty P. I., Grando S., Kwaku Zu T. T., et al. (2018). Antinutritional factors in pearl millet grains: Phytate and goitrogens content variability and molecular characterization of genes involved in their pathways. PloS One 13 (6), e0198394. doi: 10.1371/journal.pone.0198394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschin G., Resta D. (2013). Alkaloids derived from lysine: quinolizidine (a focus on lupin alkaloids). Natural Products, 381–403. doi: 10.1007/978-3-642-22144-6_11 [DOI] [Google Scholar]
- Bottger G. T., Sheehan E. T., Lukefahr M. J. (1964). Relation of gossypol content of cotton plants to insect resistance. J. Economic Entomol 57 (2), 283–285. doi: 10.1093/jee/57.2.283 [DOI] [Google Scholar]
- Bousquet J., Anto J. M., Czarlewski W., Haahtela T., Fonseca S. C., Iaccarino G., et al. (2021). Cabbage and fermented vegetables: from death rate heterogeneity in countries to candidates for mitigation strategies of severe COVID-19. Allergy 76 (3), 735–750. doi: 10.1111/all.14549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregitzer P., Raboy V. (2006). Effects of four independent low-phytate mutations in barley (Hordeum vulgare l.) on seed phosphorus characteristics and malting quality. Cereal Chem. 83, 460–464. doi: 10.1094/CC-83-0460 [DOI] [Google Scholar]
- Bueno R. D., Borges L. L., God P. I. G., Piovesan N. D., Teixeira A. I., Cruz C. D., et al. (2018). Quantification of anti-nutritional factors and their correlations with protein and oil in soybeans. Anais da Academia Bras. Ciências 90, 205–217. doi: 10.1590/0001-3765201820140465 [DOI] [PubMed] [Google Scholar]
- Bunsupa S., Yamazaki M., Saito K. (2012). Quinolizidine alkaloid biosynthesis: recent advances and future prospects. Frontiers in plant science 3, 239. doi: 10.3389/fpls.2012.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser M. D., Abbas H. K. (2001). Mechanically processing cottonseed to reduce gossypol and aflatoxin levels. J. Toxicology: Toxin Rev. 20 (3-4), 179–208. doi: 10.1081/TXR-100108556 [DOI] [Google Scholar]
- Cai X., Ge C., Xu C., Wang X., Wang S., Wang Q. (2018). Expression analysis of oxalate metabolic pathway genes reveals oxalate regulation patterns in spinach. Molecules 23 (6)1286. doi: 10.3390/Molecules23061286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Xie Y., Liu J. (2010). Glandless seed and glanded plant research in cotton. a review. Agron. Sustain. Dev. 30 (1), 181–190. doi: 10.1051/agro/2008024 [DOI] [Google Scholar]
- Camerlengo F., Frittelli A., Sparks C., Doherty A., Martignago D., Larré C., et al. (2020). CRISPR-Cas9 multiplex editing of the α-amylase/trypsin inhibitor genes to reduce allergen proteins in durum wheat. Front. Sustain. Food Syst. 4. doi: 10.3389/fsufs.2020.00104 [DOI] [Google Scholar]
- Cao Z., Tian F., Wang N., Jiang C., Lin B., Xia W., et al. (2010). Analysis of QTLs for erucic acid and oil content in seeds on A8 chromosome and the linkage drag between the alleles for the two traits in brassica napus. J. Genet. Genomics 37 (4), 231–240. doi: 10.1016/S1673-8527(09)60041-2 [DOI] [PubMed] [Google Scholar]
- Cárdenas P. D., Sonawane P. D., Heinig U., Bocobza S. E., Burdman S., Aharoni A. (2015). The bitter side of the nightshades: Genomics drives discovery in solanaceae steroidal alkaloid metabolism. Phytochemistry 113, 24–32. doi: 10.1016/j.phytochem.2014.12.010 [DOI] [PubMed] [Google Scholar]
- Cárdenas P. D., Sonawane P. D., Pollier J., Vanden Bossche R., Dewangan V., Weithorn E., et al. (2016). GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat. Commun. 7 (1), 1–16. doi: 10.1038/ncomms10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty N., Ghosh R., Ghosh S., Narula K., Tayal R., Datta A., et al. (2013). Reduction of oxalate levels in tomato fruit and consequent metabolic remodeling following overexpression of a fungal oxalate decarboxylase. Plant Physiol. 162 (1), 364–378. doi: 10.1104/pp.112.209197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A. K. (2010). “Goitrogen in food: cyanogenic and flavonoids containing plant foods in the development of goiter,” in Bioactive foods in promoting health (Cambridge, Massachusetts, United States: Academic Press; ), 691–716. doi: 10.1016/B978-0-12-374628-3.00042-6 [DOI] [Google Scholar]
- Chang Q., Liu Z., Ma W. Z., Hei C. C., Shen X. S., Qian X. J., et al. (2011). Drug synergistic antifertility effect of combined administration of low-dose gossypol with steroid hormones in rats. Chin. Med. J. 124 (11), 1678–1682. doi: 10.3760/cma.j.issn.0366-6999.2011.11.014 [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Rhee Y., Chung P. S., Ge R. F., Ahn J. C. (2018). Sulforaphene enhances the efficacy of photodynamic therapy in anaplastic thyroid cancer through Ras/RAF/MEK/ERK pathway suppression. J. Photochem. Photobiol. B 179, 46–53. doi: 10.1016/j.jphotobiol.2017.12.013 [DOI] [PubMed] [Google Scholar]
- Choi H. S., Koo H. B., Jeon S. W., Han J. Y., Kim J. S., Jun K. M., et al. (2022). Modification of ginsenoside saponin composition via the CRISPR/Cas9-mediated knockout of protopanaxadiol 6-hydroxylase gene in panax ginseng. J. ginseng Res. 46 (4), 505–514. doi: 10.1016/j.jgr.2021.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukwuebuka E., Chinenye I. J. (2015). Biological functions and anti-nutritional effects of phytochemicals in living system. J. Pharm. Biol. Sci. 10 (2), 10–19. doi: 10.9790/3008-10231019 [DOI] [Google Scholar]
- Confalonieri M., Carelli M., Gianoglio S., Moglia A., Biazzi E., Tava A. (2021). CRISPR/Cas9-mediated targeted mutagenesis of CYP93E2 modulates the triterpene saponin biosynthesis in medicago truncatula. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.690231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulibaly A., Kouakou B., Chen J. (2011). Phytic acid in cereal grains: structure, healthy or harmful ways to reduce phytic acid in cereal grains and their effects on nutritional quality. Am. J. Plant Nutr. fertilization Technol. 1 (1), 1–22. doi: 10.3923/ajpnft.2011.1.22 [DOI] [Google Scholar]
- Cristina Oliveira de Lima V., Piuvezam G., Leal Lima Maciel B., Heloneida de Araújo Morais A. (2019). Trypsin inhibitors: promising candidate satietogenic proteins as complementary treatment for obesity and metabolic disorders? J. Enzyme inhibition medicinal Chem. 34 (1), 405–419. doi: 10.1080/14756366.2018.1542387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Parihar A. K., Barpete S., Kumar S., Gupta S. (2021). Current perspectives on reducing the β-ODAP content and improving potential agronomic traits in grass pea (Lathyrus sativus l.). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.703275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G., Sharma A., Sarkar P. K. (2022). Conventional and emerging processing techniques for the post-harvest reduction of antinutrients in edible legumes. Appl. Food Res. 12, 100112. doi: 10.1016/j.afres.2022.100112 [DOI] [Google Scholar]
- Daverio M., Cavicchiolo M. E., Grotto P., Lonati D., Cananzi M., Da Dalt L. (2014). Bitter lupine beans ingestion in a child: a disregarded cause of acute anticholinergic toxicity. Eur. J. Pediatr. 173 (12), 1549–1551. doi: 10.1007/s00431-013-2088-2 [DOI] [PubMed] [Google Scholar]
- Davis E. M., Tsuji J., Davis G. D., Pierce M. L., Essenberg M. (1996). Purification of (+)-δ-cadinene synthase, a sesquiterpene cyclase from bacteria-inoculated cotton foliar tissue. Phytochemistry 41 (4), 1047–1055. doi: 10.1016/0031-9422(95)00771-7 [DOI] [PubMed] [Google Scholar]
- Dilday R. H. (1986). Development of a cotton plant with glandless seeds, and glanded foliage and fruiting forms 1. Crop Sci. 26 (3), 639–641. doi: 10.2135/cropsci1986.0011183X002600030046x [DOI] [Google Scholar]
- Dilworth L. L., Omoruyi F. O., Simon O. R., Morrison E. Y., Asemota H. N. (2005). The effect of phytic acid on the levels of blood glucose and some enzymes of carbohydrate and lipid metabolism. West Indian Med. J. 54 (2), 102–106. doi: 10.1590/s0043-31442005000200003 [DOI] [PubMed] [Google Scholar]
- do Amaral M., Freitas A. C. O., Santos A. S., Dos Santos E. C., Ferreira M. M., da Silva Gesteira A., et al. (2022). TcTI, a kunitz-type trypsin inhibitor from cocoa associated with defense against pathogens. Sci. Rep. 12 (1), 1–16. doi: 10.1038/s41598-021-04700-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G., Marcel S., Glahn R. P., Lund E. K., Pariagh S., Fischer R., et al. (2005). Endosperm-specific co-expression of recombinant soybean ferritin and aspergillus phytase in maize results in significant increases in the levels of bioavailable iron. Plant Mol. Biol. 59 (6), 869–880. doi: 10.1007/s11103-005-1537-3 [DOI] [PubMed] [Google Scholar]
- Duc G., Sixdenier G., Lila M., Furstoss V. (1989). Search of genetic variability for vicine and convicine content in Vicia faba L.: a first report of a gene which codes for nearly zero-vicine and zero-convicine contents. In 1. International Workshop on 'Antinutritional Factors (ANF) in Legume Seeds', Wageningen (Netherlands), 23-25 Nov 1988. Pudoc.
- Duc G., Marget P., Page D., Domoney C. (2004). Facile breeding markers to lower contents of vicine and convicine in faba bean seeds and trypsin inhibitors in pea seeds. Publication-European Assoc. Anim. Production 110, 281–286. [Google Scholar]
- Egbuna C. (2018). “Phytochemicals as antinutrients,” in Phytochemistry, (New Jersey and Canada: Apple Academic Press; ), 557–564. [Google Scholar]
- Elango D., Rajendran K., van der Laan L., Sebastiar S., Raigne J., Thaiparambil N. A., et al. (2022). Raffinose family oligosaccharides: friend or foe for human and plant health? Front. Plant Sci. 13. doi: 10.3389/fpls.2022.829118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Araby D. A., Amer S. A., Khalil A. A. (2020). Effect of different feeding regimes on the growth performance, antioxidant activity, and health of Nile tilapia, oreochromis niloticus. Aquaculture 528, 735572. doi: 10.1016/j.aquaculture.2020.735572 [DOI] [Google Scholar]
- El-Araby M. M., El-Shatoury E. H., Soliman M. M., Shaaban H. F. (2020). Characterization and antimicrobial activity of lectins purified from three Egyptian leguminous seeds. AMB Express 10 (1), 1–14. doi: 10.1186/s13568-020-01024-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkonin L. A., Panin V. M., Kenzhegulov O. A., Sarsenova S. K. (2021). “RNAi-mutants of sorghum bicolor (L.) moench with improved digestibility of seed storage proteins,” in Grain and seed proteins functionality (London, UK: IntechOpen; ). doi: 10.5772/IntechOpen.96204 [DOI] [Google Scholar]
- Ertaş N., Türker S. (2014). Bulgur processes increase nutrition value: possible role in in-vitro protein digestability, phytic acid, trypsin inhibitor activity and mineral bioavailability. J. Food Sci. Technol. 51 (7), 1401–1405. doi: 10.1007/s13197-012-0638-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faizal A., Geelen D. (2013). Saponins and their role in biological processes in plants. Phytochem. Rev. 12 (4), 877–893. doi: 10.1007/s11101-013-9322-4 [DOI] [Google Scholar]
- Fiore M., Cristaldi A., Okatyeva V., Bianco S. L., Conti G. O., Zuccarello P., et al. (2020). Dietary habits and thyroid cancer risk: A hospital-based case–control study in Sicily (South Italy). Food Chem. Toxicol. 146, 111778. doi: 10.1016/j.fct.2020.111778 [DOI] [PubMed] [Google Scholar]
- Fox G. P., O’Donnell N. H., Stewart P. N., Gleadow R. M. (2012). Estimating hydrogen cyanide in forage sorghum (Sorghum bicolor) by near-infrared spectroscopy. J. Agric. Food Chem. 60 (24), 6183–6187. doi: 10.1021/jf205030b [DOI] [PubMed] [Google Scholar]
- Frick K. M., Kamphuis L. G., Siddique K. H., Singh K. B., Foley R. C. (2017). Quinolizidine alkaloid biosynthesis in lupins and prospects for grain quality improvement. Frontiers in plant science 8, 87. doi: 10.3389/fpls.2017.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadelha I. C. N., Rangel A. D. N., Silva A. R., Soto-Blanco B. (2011). Effects of gossypol on animal reproduction. Acta Veterinaria Brasilica 5 (2), 129–135. [Google Scholar]
- Gao M., Li G., Yang B., Qiu D., Farnham M., Quiros C. (2007). High-density Brassica oleracea linkage map: identification of useful new linkages. Theoretical and applied genetics 115 (2), 277–287. doi: 10.1007/s00122-007-0568-3 [DOI] [PubMed] [Google Scholar]
- Gemede H. F., Ratta N. (2014). Antinutritional factors in plant foods: Potential health benefits and adverse effects. Int. J. Nutr. Food Sci. 3 (4), 284–289. doi: 10.11648/j.ijnfs.20140304.18 [DOI] [Google Scholar]
- George M. A., Bhide S. V., Thengane R. J., Hosseini G. H., Manjaya J. G. (2008). Identification of low lectin mutants in soybean. Plant Breed. 127 (2), 150–153. doi: 10.1111/j.1439-0523.2007.01449.x [DOI] [Google Scholar]
- Ghanevati M., Jaworski J. G. (2001). Active-site residues of a plant membrane-bound fatty acid elongase β-ketoacyl-CoA synthase, FAE1 KCS. Biochim. Biophys. Acta 1530, 77–85. doi: 10.1016/S1388-1981(00)00168-2 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T., Yatusevich R., Berger B., Müller C., Flügge U. I. (2007). The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in arabidopsis thaliana. Plant J. 51 (2), 247–261. doi: 10.1111/j.1365-313X.2007.03133.x [DOI] [PubMed] [Google Scholar]
- Gilani G. S., Xiao C. W., Cockell K. A. (2012). Impact of antinutritional factors in food proteins on the digestibility of protein and the bioavailability of amino acids and on protein quality. Br. J. Nutr. 108 (S2), S315–S332. doi: 10.1017/S0007114512002371 [DOI] [PubMed] [Google Scholar]
- Gilardoni P. A., Hettenhausen C., Baldwin I. T., Bonaventure G. (2011). Nicotiana attenuata LECTIN RECEPTOR KINASE1 suppresses the insect-mediated inhibition of induced defense responses during manduca sexta herbivory. Plant Cell 23 (9), 3512–3532. doi: 10.1105/tpc.111.088229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillman J. D., Tetlow A., Lee J. D., Shannon J. G., Bilyeu K. (2011). Loss-of-function mutations affecting a specific glycine max R2R3 MYB transcription factor result in brown hilum and brown seed coats. BMC Plant Biol. 11 (1), 1–12. doi: 10.1186/1471-2229-11-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez M. A., Berkoff K. C., Gill B. K., Iavarone A. T., Lieberman S. E., Ma J. M., et al. (2021). CRISPR-Cas9-mediated knockout of CYP79D1 and CYP79D2 in cassava attenuates toxic cyanogen production. bioRxiv. doi: 10.1101/2021.10.08.462827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay G., Ma D., Schmidt A., Constabel C. P. (2020). MYB134-RNAi poplar plants show reduced tannin synthesis in leaves but not roots, and increased susceptibility to oxidative stress. J. Exp. Bot. 71 (20), 6601–6611. doi: 10.1093/jxb/eraa371 [DOI] [PubMed] [Google Scholar]
- Graf E., Eaton J. W. (1990). Antioxidant functions of phytic acid. Free Radical Biol. Med. 8 (1), 61–69. doi: 10.1016/0891-5849(90)90146-A [DOI] [PubMed] [Google Scholar]
- Greiner R., Konietzny U. (2006). Phytase for food application Vol. 44 (Germany: Food Technology and Biotechnology; ). [Google Scholar]
- Gudys K., Guzy-Wrobelska J., Janiak A., Dziurka M. A., Ostrowska A., Hura K., et al. (2018). Prioritization of candidate genes in QTL regions for physiological and biochemical traits underlying drought response in barley (Hordeum vulgare l.). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta D. S., Barpete S., Kumar J., Kumar S. (2021). Breeding for better grain quality in Lathyrus. In Breeding for enhanced nutrition and bio-active compounds in food legumes (pp. 131–156). Springer, Cham. doi: 10.1007/978-3-030-59215-8_6 [DOI] [Google Scholar]
- Gupta R. K., Gangoliya S. S., Singh N. K. (2015). Reduction of phytic acid and enhancement of bioavailable micronutrients in food grains. J. Food Sci. Technol. 52 (2), 676–684. doi: 10.1007/s13197-013-0978-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Mukhopadhyay A., Arumugam N., Sodhi Y. S., Pental D., Pradhan A. K. (2004). Molecular tagging of erucic acid trait in oilseed mustard (Brassica juncea) by QTL mapping and single nucleotide polymorphisms in FAE1 gene. Theor. Appl. Genet. 108 (4), 743–749. doi: 10.1007/s00122-003-1481-z [DOI] [PubMed] [Google Scholar]
- Gutiérrez N., Avila C. M., Duc G., Marget P., Suso M. J., Moreno M. T., et al. (2006). CAPs markers to assist selection for low vicine and convicine contents in faba bean (Vicia faba l.). Theor. Appl. Genet. 114 (1), 59–66. doi: 10.1007/s00122-006-0410-3 [DOI] [PubMed] [Google Scholar]
- Guttieri M., Bowen D., Dorsch J. A., Raboy V., Souza E. (2004). Identification and characterization of a low phytic acid wheat. Crop Sci. 44 (2), 418–424. doi: 10.2135/cropsci2004.4180 [DOI] [Google Scholar]
- Gyani P. C., Bollinedi H., Gopala Krishnan S., Vinod K. K., Sachdeva A., Bhowmick P. K., et al. (2020). Genetic analysis and molecular mapping of the quantitative trait loci governing low phytic acid content in a novel LPA rice mutant, PLM11. Plants 9 (12), 1728. doi: 10.3390/plants9121728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailu D., Abera S., Teka T. A., Box P. O., Jimma E. (2015). Effects of processing on nutritional composition and anti-nutritional factors of grass pea (Lathyrus sativus l): A. review. Food Sci. Qual. Manage. 36, 61–70. [Google Scholar]
- Haralampidis K., Trojanowska M., Osbourn A. E. (2002). Biosynthesis of triterpenoid saponins in plants. History Trends Bioprocessing Biotransformation 75, 31–49. doi: 10.1007/3-540-44604-4_2 [DOI] [PubMed] [Google Scholar]
- Hayes C. M., Weers B. D., Thakran M., Burow G., Xin Z., Emendack Y., et al. (2016). Discovery of a dhurrin QTL in sorghum: Co-localization of dhurrin biosynthesis and a novel stay-green QTL. Crop Sci. 56 (1), 104–112. doi: 10.2135/cropsci2015.06.0379 [DOI] [Google Scholar]
- Hirani A. H. (2011). QTL mapping, gene identification and genetic manipulation of glucosinolates in brassica rapa l (University of Manitoba (Canada; ). [Google Scholar]
- Ibrahim S., Saleem B., Rehman N., Zafar S. A., Naeem M. K., Khan M. R. (2022). CRISPR/Cas9 mediated disruption of inositol pentakisphosphate 2-kinase 1 (TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains. J. advanced Res. 37, 33–41. doi: 10.1016/j.jare.2021.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ida E. I., de Camargo A. C. (2022). “Soybean isoflavone profile: A new quality index in food application and health,” in Phytochemicals in soybeans (Boca Raton, Florida, United States: CRC Press; ), 45–76. [Google Scholar]
- Iqbal M. M., Erskine W., Berger J. D., Nelson M. N. (2020). Phenotypic characterisation and linkage mapping of domestication syndrome traits in yellow lupin (Lupinus luteus l.). Theor. Appl. Genet. 133 (10), 2975–2987. doi: 10.1007/s00122-020-03650-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida M., Hara M., Fukino N., Kakizaki T., Morimitsu Y. (2014). Glucosinolate metabolism, functionality and breeding for the improvement of brassicaceae vegetables. Breed. Sci. 64 (1), 48–59. doi: 10.1270/jsbbs.64.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa R. A. (2010). Identification of glucosinolate profile in brassica oleracea for quantitative trait locus mapping (England: Doctoral dissertation, University of Warwick; ). [Google Scholar]
- Itkin M., Heinig U., Tzfadia O., Bhide A. J., Shinde B., Cardenas P. D., et al. (2013). Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341 (6142), 175–179. doi: 10.1126/science.1240230 [DOI] [PubMed] [Google Scholar]
- Jadhav A., Katavic V., Marillia E. F., Giblin E. M., Barton D. L., Kumar A., et al. (2005). Increased levels of erucic acid in brassica carinata by co-suppression and antisense repression of the endogenous FAD2 gene. Metab. Eng. 7 (3), 215–220. doi: 10.1016/j.ymben.2005.02.003 [DOI] [PubMed] [Google Scholar]
- Jagal Kishore S., Mathew D., Shylaja M. R., Francies R. M., Sujatha R. (2020). Cloning and characterization of myo-inositol phosphate synthase gene (dlMIPS) and analysis of the putative structure of the enzyme responsible for the accumulation of anti-nutrient phytate in dolichos bean (Dolichos lablab l.). Plant Physiol. Rep. 25 (2), 370–375. doi: 10.1007/s40502-020-00507-7 [DOI] [Google Scholar]
- Jaiswal A. K. (2020). Nutritional composition and antioxidant properties of fruits and vegetables (Cambridge, Massachusetts, United States: Academic Press; ). [Google Scholar]
- James D. W., Lim E., Keller J., Plooy I., Ralston E., Dooner H. K. (1995). Directed tagging of the arabidopsis fatty acid elongation 1 (FAE1) gene with the maize transposon activator. Plant Cell 7, 309–319. doi: 10.1105/tpc.7.3.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javidfar F., Cheng B. (2013). Construction of a genetic linkage map and QTL analysis of erucic acid content and glucosinolate components in yellow mustard (Sinapis alba l.). BMC Plant Biol. 13 (1), 1–9. doi: 10.1186/1471-2229-13-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha U. C., Nayyar H., Parida S. K., Deshmukh R., von Wettberg E. J., Siddique K. H. (2022). Ensuring global food security by improving protein content in major grain legumes using breeding and ‘Omics’ tools. Int. J. Mol. Sci. 23 (14) 7710. doi: 10.3390/ijms23147710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N., Boatwright J. L., Bridges W., Thavarajah P., Kumar S., Shipe E., et al. (2021). Genome-wide association mapping of lentil (Lens culinaris medikus) prebiotic carbohydrates toward improved human health and crop stress tolerance. Sci. Rep. 11 (1), 1–12. doi: 10.1038/s41598-021-93475-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose J., Sachdev A., Jolly M., Krishnan V., Mehrotra U., Sahu S., et al. (2022). Efficient designing, validation, and transformation of GmIPK2 specific CRISPR/Cas9 construct for low-phytate soybean. Acta Sci. Agric. 6 (3), 24–32. doi: 10.31080/ASAG.2022.06.1105 [DOI] [Google Scholar]
- Juma B. S., Mukami A., Mweu C., Ngugi M. P., Mbinda W. (2022). Targeted mutagenesis of the CYP79D1 gene via CRISPR/Cas9-mediated genome editing results in lower levels of cyanide in cassava. Front. Plant Sci. 4236. doi: 10.3389/fpls.2022.1009860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalunke R. M., Tundo S., Sestili F., Camerlengo F., Lafiandra D., Lupi R., et al. (2020). Reduction of allergenic potential in bread wheat RNAi transgenic lines silenced for CM3, CM16 and 0.28 ATI genes. Int. J. Mol. Sci. 21 (16), 5817. doi: 10.3390/ijms21165817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal R. M., Abdull Razis A. F., Mohd Sukri N. S., Perimal E. K., Ahmad H., Patrick R., et al. (2022). Beneficial health effects of glucosinolates-derived isothiocyanates on cardiovascular and neurodegenerative diseases. Molecules 27 (3), 624. doi: 10.3390/Molecules27030624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan U., Sharma R., Gangola M. P., Chibbar R. N. (2018). Improving grain quality in pulses: strategies to reduce raffinose family oligosaccharides in seeds. Ekin J. Crop Breed. Genet. 4 (1), 70–88. [Google Scholar]
- Kanrar S., Venkateswari J., Dureja P., Kirti P. B., Chopra V. L. (2006). Modification of erucic acid content in Indian mustard (Brassica juncea) by up-regulation and down-regulation of the brassica juncea FATTY ACID ELONGATION1 (BjFAE1) gene. Plant Cell Rep. 25 (2), 148–155. doi: 10.1007/s00299-005-0068-3 [DOI] [PubMed] [Google Scholar]
- Karim M., Tonu N. N., Hossain M. S., Funaki T., Meah M. B., Hossain D. M., et al. (2016). Marker-assisted selection of low erucic acid quantity in short duration brassica rapa. Euphytica 208 (3), 535–544. doi: 10.1007/s10681-015-1596-8 [DOI] [Google Scholar]
- Karmakar A., Bhattacharya S., Sengupta S., Ali N., Sarkar S. N., Datta K., et al. (2020). RNAi-mediated silencing of ITPK gene reduces phytic acid content, alters transcripts of phytic acid biosynthetic genes, and modulates mineral distribution in rice seeds. Rice Sci. 27 (4), 315–328. doi: 10.1016/j.rsci.2020.05.007 [DOI] [Google Scholar]
- Kaur H. (2018). Influence of bna. FAD2 alleles on the erucic acid and polyunsaturates content in brassica napus oil (United Kingdom: Doctoral dissertation, University of York; ). [Google Scholar]
- Kenar J. A. (2006). Reaction chemistry of gossypol and its derivatives. J. Am. Oil Chemists’ Society. 83 (4), 269–302. doi: 10.1007/s11746-006-1203-1 [DOI] [Google Scholar]
- Khanbabaee K., Van Ree T. (2001). Tannins: classification and definition. Natural product Rep. 18 (6), 641–649. doi: 10.1039/b101061l [DOI] [PubMed] [Google Scholar]
- Khazaei H., O’Sullivan D. M., Jones H., Pitts N., Sillanpää M. J., Pärssinen P., et al. (2015). Flanking SNP markers for vicine–convicine concentration in faba bean (Vicia faba l.). Mol. Breed. 35 (1), 1–6. doi: 10.1007/s11032-015-0214-8 [DOI] [Google Scholar]
- Khazaei H., Purves R. W., Hughes J., Link W., O'Sullivan D. M., Schulman A. H., et al. (2019). Eliminating vicine and convicine, the main anti-nutritional factors restricting faba bean usage. Trends Food Sci. Technol. 91, 549–556. doi: 10.1016/j.tifs.2019.07.051 [DOI] [Google Scholar]
- Khodakov G. V., Akimov Y. A., Shashkov A. S., Kintia P. K., Grishkovets V. I. (1996). Triterpene and steroid saponins isolated from two melilotus species. Saponins Used Food Agric. 405, 211–222. doi: 10.1007/978-1-4613-0413-5_18 [DOI] [PubMed] [Google Scholar]
- Kim N. S., Kim S. J., Jo J. S., Lee J. G., Lee S. I., Kim D. H., et al. (2021). The BrGI circadian clock gene is involved in the regulation of glucosinolates in Chinese cabbage. Genes 12 (11), 1664. doi: 10.3390/genes12111664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. I., Tai T. H. (2011). Identification of genes necessary for wild-type levels of seed phytic acid in arabidopsis thaliana using a reverse genetics approach. Mol. Genet. Genomics 286 (2), 119–133. doi: 10.1007/s00438-011-0631-2 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Hattori T., Honda Y., Kirimura K. (2014). Oxalic acid production by citric acid-producing aspergillus niger overexpressing the oxaloacetate hydrolase gene oahA. J. Ind. Microbiol. Biotechnol. 41 (5), 749–756. doi: 10.1007/s10295-014-1419-2 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kawagishi H. (2014). Fungal lectins: a growing family. Lectins. 1200, 15–38. doi: 10.1007/978-1-4939-1292-6_2 [DOI] [PubMed] [Google Scholar]
- Kottapalli K. R., Payton P., Rakwal R., Agrawal G. K., Shibato J., Burow M., et al. (2008). Proteomics analysis of mature seed of four peanut cultivars using two-dimensional gel electrophoresis reveals distinct differential expression of storage, anti-nutritional, and allergenic proteins. Plant Sci. 175 (3), 321–329. doi: 10.1016/j.plantsci.2008.05.005 [DOI] [Google Scholar]
- Kovaci P. (2003). Mechanism of drug and toxic actions of gossypol: focus on reactive oxygen species and electron transfer. Curr. Medicinal Chem. 10 (24), 2711–2718. doi: 10.2174/0929867033456369 [DOI] [PubMed] [Google Scholar]
- Kroc M., Koczyk G., Kamel K. A., Czepiel K., Fedorowicz-Strońska O., Krajewski P., et al. (2019). Transcriptome-derived investigation of biosynthesis of quinolizidine alkaloids in narrow-leafed lupin (Lupinus angustifolius l.) highlights candidate genes linked to iucundus locus. Sci. Rep. 9 (1), 1–13. doi: 10.1038/s41598-018-37701-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Anju T., Kumar S., Chhapekar S. S., Sreedharan S., Singh S., et al. (2019). Integrating omics and gene editing tools for rapid improvement of traditional food plants for diversified and sustainable food security. Int. J. Mol. Sci. 22 (15), 8093. doi: 10.3390/ijms22158093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Chattopadhyay A., Ghosh S., Irfan M., Chakraborty N., Chakraborty S., et al. (2016). Improving nutritional quality and fungal tolerance in soya bean and grass pea by expressing an oxalate decarboxylase. Plant Biotechnol. J. 14 (6), 1394–1405. doi: 10.1111/pbi.12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S. (2018). The effect of soaking almonds and hazelnuts on phytate and mineral concentrations (Dunedin North, New Zealand: Doctoral dissertation, University of Otago; ). [Google Scholar]
- Kumar A., Metwal M., Kaur S., Gupta A. K., Puranik S., Singh S., et al. (2016). Nutraceutical value of finger millet [Eleusine coracana (L.) gaertn.], and their improvement using omics approaches. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano M., Mimura T., Takaiwa F., Yoshida K. T. (2009). Generation of stable ‘low phytic acid’transgenic rice through antisense repression of the 1d-myo-inositol 3-phosphate synthase gene (RINO1) using the 18-kDa oleosin promoter. Plant Biotechnol. J. 7 (1), 96–105. doi: 10.1111/j.1467-7652.2008.00375.x [DOI] [PubMed] [Google Scholar]
- Larson S. R., Rutger J. N., Young K. A., Raboy V. (2000). Isolation and genetic mapping of a non-lethal rice (Oryza sativa l.) low phytic acid 1 mutation. Crop Sci. 40 (5), 1397–1405. doi: 10.2135/cropsci2000.4051397x [DOI] [Google Scholar]
- Larson S. R., Young K. A., Cook A., Blake T. K., Raboy V. (1998). Linkage mapping of two mutations that reduce phytic acid content of barley grain. Theor. Appl. Genet. 97 (1), 141–146. doi: 10.1007/s001220050878 [DOI] [Google Scholar]
- Le H., Nguyen N. H., Ta D. T., Le T. N. T., Bui T. P., Le N. T., et al. (2020). CRISPR/Cas9-mediated knockout of galactinol synthase-encoding genes reduces raffinose family oligosaccharide levels in soybean seeds. Front. Plant Sci. 2033. doi: 10.3389/fpls.2020.612942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Liang S., Alariqi M., Wang F., Wang G., Wang Q., et al. (2021). The application of temperature sensitivity CRISPR/LbCpf1 (LbCas12a) mediated genome editing in allotetraploid cotton (G. hirsutum) and creation of nontransgenic, gossypol-free cotton. Plant Biotechnol. J. 19 (2), 221. doi: 10.1111/pbi.13470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Tsao R. (2019). UF-LC-DAD-MSn for discovering enzyme inhibitors for nutraceuticals and functional foods. J. Food Bioactives 7, 27–35. doi: 10.31665/JFB.2019.7195 [DOI] [Google Scholar]
- Liu Z., Hammerlindl J., Keller W., McVetty P. B., Daayf F., Quiros C. F., et al. (2011). MAM gene silencing leads to the induction of C3 and reduction of C4 and C5 side-chain aliphatic glucosinolates in brassica napus. Mol. Breed. 27 (4), 467–478. doi: 10.1007/s11032-010-9444-y [DOI] [Google Scholar]
- Liu J. C., Huang Y. Q., Zhou J. F., Bian F. R., Chen F. J., Mi G. H. (2013). Identification of quantitative trait loci for phytic acid concentration in maize grain under two nitrogen conditions. J. Integr. Agric. 12 (5), 765–772. doi: 10.1016/S2095-3119(13)60298-1 [DOI] [Google Scholar]
- Liu T. J., Zhang Y. J., Agerbirk N., Wang H. P., Wei X. C., Song J. P., et al. (2019). A high-density genetic map and QTL mapping of leaf traits and glucosinolates in barbarea vulgaris. BMC Genomics 20 (1), 1–14. doi: 10.1186/s12864-019-5769-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Moreno M., Garcés-Rimón M., Miguel M. (2022). Antinutrients: Lectins, goitrogens, phytates and oxalates, friends or foe? J. Funct. Foods 89, 104938. doi: 10.1016/j.jff.2022.104938 [DOI] [Google Scholar]
- Lucca P., Hurrell R., Potrykus I. (2001). Genetic engineering approaches to improve the bioavailability and the level of iron in rice grains. Theor. Appl. Genet. 102 (2), 392–397. doi: 10.1007/s001220051659 [DOI] [Google Scholar]
- Luo P., Wang Y. H., Wang G. D., Essenberg M., Chen X. Y. (2001). Molecular cloning and functional identification of (+)-δ-cadinene-8-hydroxylase, a cytochrome P450 mono-oxygenase (CYP706B1) of cotton sesquiterpene biosynthesis. Plant J. 28 (1), 95–104. doi: 10.1046/j.1365-313X.2001.01133.x [DOI] [PubMed] [Google Scholar]
- Lü J., Sui X., Ma S., Li X., Liu H., Zhang Z. (2017). Suppression of cucumber stachyose synthase gene (CsSTS) inhibits phloem loading and reduces low temperature stress tolerance. Plant Mol. Biol. 95 (1), 1–15. doi: 10.1007/s11103-017-0621-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood T., Ekuere U., Yeh F., Good A. G., Stringam G. R. (2003). Molecular mapping of seed aliphatic glucosinolates in brassica juncea. Genome 46 (5), 753–760. doi: 10.1139/g03-051 [DOI] [PubMed] [Google Scholar]
- Mancinotti D., Frick K. M., Geu-Flores F. (2022). Biosynthesis of quinolizidine alkaloids in lupins: mechanistic considerations and prospects for pathway elucidation. Natural Product Rep. 39, 1423–1437. doi: 10.1039/D1NP00069A [DOI] [PubMed] [Google Scholar]
- Maphosa Y., Jideani V. A. (2017). “The role of legumes in human nutrition,” in Functional food improve health through adequate food. Ed. Chavarri M. (London, UK: IntechOpen; ), 103–109. [Google Scholar]
- Maranna S., Verma K., Talukdar A., Lal S. K., Kumar A., Mukherjee K. (2016). Introgression of null allele of kunitz trypsin inhibitor through marker-assisted backcross breeding in soybean (Glycine max l. merr.). BMC Genet. 17 (1), 1–9. doi: 10.1186/s12863-016-0413-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J. I., Hu H., Petereit J., Bayer P. E., Valliyodan B., Batley J., et al. (2022). Haplotype mapping uncovers unexplored variation in wild and domesticated soybean at the major protein locus cqProt-003. Theor. Appl. Genet. 135 (4), 1443–1455. doi: 10.1007/s00122-022-04045-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. S., Liu J., Benedict C. R., Stipanovic R. D., Magill C. W. (2003). Reduced levels of cadinane sesquiterpenoids in cotton plants expressing antisense (+)-δ-cadinene synthase. Phytochemistry 62 (1), 31–38. doi: 10.1016/S0031-9422(02)00432-6 [DOI] [PubMed] [Google Scholar]
- McMichael S. C. (1960). Combined effects of glandless genes gl2 and gl3 on pigment glands in the cotton plant. Agron. J. 52 (7), 385–386. doi: 10.2134/agronj1960.00021962005200070005x [DOI] [Google Scholar]
- Mena H., Santos J. E. P., Huber J. T., Tarazon M., Calhoun M. C. (2004). The effects of varying gossypol intake from whole cottonseed and cottonseed meal on lactation and blood parameters in lactating dairy cows. J. Dairy Sci. 87 (8), 2506–2518. doi: 10.3168/jds.S0022-0302(04)73375-5 [DOI] [PubMed] [Google Scholar]
- Meng Y. L., Jia J. W., Liu C. J., Liang W. Q., Heinstein P., Chen X. Y. (1999). Coordinated accumulation of (+)-δ-Cadinene synthase mRNAs and gossypol in developing seeds of gossypium hirsutum and a new member of the cad 1 family from g. arboreum. J. Natural products 62 (2), 248–252. doi: 10.1021/np980314o [DOI] [PubMed] [Google Scholar]
- Mietkiewska E., Hoffman T. L., Brost J. M., Giblin E. M., Barton D. L., Francis T., et al. (2008). Hairpin-RNA mediated silencing of endogenous FAD2 gene combined with heterologous expression of crambe abyssinica FAE gene causes an increase in the level of erucic acid in transgenic brassica carinata seeds. Mol. Breed. 22 (4), 619–627. doi: 10.1007/s11032-008-9204-4 [DOI] [Google Scholar]
- Mishra A., Behura A., Mawatwal S., Kumar A., Naik L., Mohanty S. S., et al. (2019). Structure-function and application of plant lectins in disease biology and immunity. Food Chem. Toxicol. 134, 110827. doi: 10.1016/j.fct.2019.110827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra D., Patel A. S., Kar A., Deshpande S. S., Tripathi M. K. (2019). Effect of different processing conditions on proximate composition, anti-oxidants, anti-nutrients and amino acid profile of grain sorghum. Food Chem. 271, 129–135. doi: 10.1016/j.foodchem.2018.07.196 [DOI] [PubMed] [Google Scholar]
- Mora J., Pott D. M., Osorio S., Vallarino J. G. (2022). Regulation of plant tannin synthesis in crop species. Front. Genet. 13. doi: 10.3389/fgene.2022.870976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses C., Manyeula F., Radikara M. V., Mareko M. H.D., Madibela O. R. (2022). Carcass characteristics and meat quality of ross 308 broiler chickens fed malted red and white sorghum-based diets. Poultry 1 (3), 169–179. doi: 10.3390/poultry1030015 [DOI] [Google Scholar]
- Mukhopadhyay N., Bandyopadhyay S. (2003). Extrusion cooking technology employed to reduce the anti-nutritional factor tannin in sesame (Sesamum indicum) meal. J. Food Eng. 56 (2-3), 201–202. doi: 10.1016/S0260-8774(02)00250-9 [DOI] [Google Scholar]
- Navarro-Gochicoa M. T., Camut S., Timmers A. C., Niebel A., Hervé C., Boutet E., et al. (2003). Characterization of four lectin-like receptor kinases expressed in roots of medicago truncatula. structure, location, regulation of expression, and potential role in the symbiosis with sinorhizobium meliloti. Plant Physiol. 133 (4), 1893–1910. doi: 10.1104/pp.103.027680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikmaram N., Leong S. Y., Koubaa M., Zhu Z., Barba F. J., Greiner R., et al. (2017). Effect of extrusion on the anti-nutritional factors of food products: An overview. Food control 79, 62–73. doi: 10.1016/j.foodcont.2017.03.027 [DOI] [Google Scholar]
- Nkhata S. G., Ayua E., Kamau E. H., Shingiro J. B. (2018). Fermentation and germination improve nutritional value of cereals and legumes through activation of endogenous enzymes. Food Sci. Nutr. 6 (8), 2446–2458. doi: 10.1002/fsn3.846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes A., Vianna G. R., Cuneo F., Amaya-Farfan J., de Capdeville G., Rech E. L., et al. (2006). RNAi-mediated silencing of the myo-inositol-1-phosphate synthase gene (GmMIPS1) in transgenic soybean inhibited seed development and reduced phytate content. Planta 224 (1), 125–132. doi: 10.1007/s00425-005-0201-0 [DOI] [PubMed] [Google Scholar]
- O'Sullivan D. M., Angra D., Tagkouli V., Khamassi K., El-Rodeny W., Zeid M. (2018). “Gene identification in faba bean–to synteny and beyond,” in Plant and Animal Genome XXVI Conference, , January 13-17, 2018, PAG. [Google Scholar]
- Ogbonna A. C., Abuajah C. I., Ide E. O., Udofia U. S. (2012). Effect of malting conditions on the nutritional and anti-nutritional factors of sorghum grist. Ann. Univ. Dunarea Jos Galati. Fascicle VI-Food Technol. 36 (2), 64–72. [Google Scholar]
- Oghbaei M., Prakash J. (2016). Effect of primary processing of cereals and legumes on its nutritional quality: A comprehensive review. Cogent Food Agric. 2 (1), 1136015. doi: 10.1080/23311932.2015.1136015 [DOI] [Google Scholar]
- Ogodo A. C., Agwaranze D. I., Aliba N. V., Kalu A. C., Nwaneri C. B. (2019). Fermentation by lactic acid bacteria consortium and its effect on anti-nutritional factors in maize flour. J. Biol. Sci. 19 (1), 17–23. doi: 10.3923/jbs.2019.17.23 [DOI] [Google Scholar]
- Onwuka G. I. (2006). Soaking, boiling and antinutritional factors in pigeon peas (Cajanus cajan) and cowpeas (Vigna unguiculata). J. Food Process. preservation 30 (5), 616–630. doi: 10.1111/j.1745-4549.2006.00092.x [DOI] [Google Scholar]
- Orf J. H., Hymowitz T. (1979). Inheritance of the absence of the kunitz trypsin inhibitor in seed protein of soybeans 1. Crop Sci. 19 (1), 107–109. doi: 10.2135/cropsci1979.0011183X001900010026x [DOI] [Google Scholar]
- Palle S. R., Campbell L. M., Pandeya D., Puckhaber L., Tollack L. K., Marcel S., et al. (2013). RNA I-mediated ultra-low gossypol cottonseed trait: performance of transgenic lines under field conditions. Plant Biotechnol. J. 11 (3), 296–304. doi: 10.1111/pbi.12013 [DOI] [PubMed] [Google Scholar]
- Panda P. K., Naik P. P., Praharaj P. P., Meher B. R., Gupta P. K., Verma R. S., et al. (2018). Abrus agglutinin stimulates BMP-2-dependent differentiation through autophagic degradation of β-catenin in colon cancer stem cells. Mol. carcinogenesis 57 (5), 664–677. doi: 10.1002/mc.22791 [DOI] [PubMed] [Google Scholar]
- Pandey A. K., Aggarwal S., Meena V., Kumar A. (2021). Phytic acid reduction in cereal grains by genome engineering: Potential targets to achieve low phytate wheat. Genome Eng. Crop Improvement. 9, 146–153. doi: 10.1002/9781119672425.ch9 [DOI] [Google Scholar]
- Pandey A. K., Madhu P., Bhat B. V. (2019). Down-regulation of CYP79A1 gene through antisense approach reduced the cyanogenic glycoside dhurrin in [Sorghum bicolor (L.) moench] to improve fodder quality. Front. Nutr. 6. doi: 10.3389/fnut.2019.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parca F., Koca Y. O., Aydın U. N. A. Y. (2018). Nutritional and antinutritional factors of some pulses seed and their effects on human health. Int. J. Secondary Metabolite 5 (4), 331–342. doi: 10.21448/ijsm.488651 [DOI] [Google Scholar]
- Park S. B., Chun J. H., Ban Y. W., Han J. Y., Choi Y. E. (2016). Alteration of panax ginseng saponin composition by overexpression and RNA interference of the protopanaxadiol 6-hydroxylase gene (CYP716A53v2). J. ginseng Res. 40 (1), 47–54. doi: 10.1016/j.jgr.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K., Neeharika B., Suneetha J., Kumari B. A., Neeraja B., Prabhakar V., et al. (2018). Effect of blanching on anti-nutritional factors of bathua leaves. Pharma Innovation J. 7 (4), 214–216. [Google Scholar]
- Patterson C. A., Curran J., Der T. (2017). Effect of processing on antinutrient compounds in pulses. Cereal Chem. 94 (1), 2–10. doi: 10.1094/CCHEM-05-16-0144-FI [DOI] [Google Scholar]
- Peng Q., Hu Y., Wei R., Zhang Y., Guan C., Ruan Y., et al. (2010). Simultaneous silencing of FAD2 and FAE1 genes affects both oleic acid and erucic acid contents in brassica napus seeds. Plant Cell Rep. 29 (4), 317–325. doi: 10.1007/s00299-010-0823-y [DOI] [PubMed] [Google Scholar]
- Perera I., Seneweera S., Hirotsu N. (2018). Manipulating the phytic acid content of rice grain toward improving micronutrient bioavailability. Rice 11 (1), 1–13. doi: 10.1186/s12284-018-0200-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova N., Mokshina N. (2022). Using FIBexDB for in-depth analysis of flax lectin gene expression in response to fusarium oxysporum infection. Plants 11 (2), 163. doi: 10.3390/plants11020163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peumans W. J., Van Damme E. J. (1995). Lectins as plant defense proteins. Plant physiology. Oct;109(2):347. phillippy BQ. transport of calcium across caco-2 cells in the presence of inositol hexakisphosphate. Nutr. Res. 26, 146–149. doi: 10.1016/j.nutres.2006.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan H. T., Ellwood S. R., Adhikari K., Nelson M. N., Oliver R. P. (2007). The first genetic and comparative map of white lupin (Lupinus albus l.): identification of QTLs for anthracnose resistance and flowering time, and a locus for alkaloid content. DNA Res. 14 (2), 59–70. doi: 10.1093/dnares/dsm009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramitha J. L., Rana S., Aggarwal P. R., Ravikesavan R., Joel A. J., Muthamilarasan M. (2021). Diverse role of phytic acid in plants and approaches to develop low-phytate grains to enhance bioavailability of micronutrients. Adv. Genet. 107, 89–120. doi: 10.1016/bs.adgen.2020.11.003 [DOI] [PubMed] [Google Scholar]
- Raboy V., Gerbasi P. F., Young K. A., Stoneberg S. D., Pickett S. G., Bauman A. T., et al. (2000). Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiol. 124 (1), 355–368. doi: 10.1104/pp.124.1.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboy V., Peterson K., Jackson C., Marshall J. M., Hu G., Saneoka H., et al. (2015). A substantial fraction of barley (Hordeum vulgare l.) low phytic acid mutations have little or no effect on yield across diverse production environments. Plants 4 (2), 225–239. doi: 10.3390/plants4020225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboy V., Young K. A., Dorsch J. A., Cook A. (2001). Genetics and breeding of seed phosphorus and phytic acid. J. Plant Physiol. 158 (4), 489–497. doi: 10.1078/0176-1617-00361 [DOI] [Google Scholar]
- Randel R. D., Willard S. T., Wyse S. J., French L. N. (1996). Effects of diets containing free gossypol on follicular development, embryo recovery and corpus luteum function in brangus heifers treated with bFSH. Theriogenology 45 (5), 911–922. doi: 10.1016/0093-691X(96)00021-0 [DOI] [PubMed] [Google Scholar]
- Rathore K. S., Pandeya D., Campbell L. M., Wedegaertner T. C., Puckhaber L., Stipanovic R. D., et al. (2020). Ultra-low gossypol cottonseed: selective gene silencing opens up a vast resource of plant-based protein to improve human nutrition. Crit. Rev. Plant Sci. 39 (1), 1–29. doi: 10.1080/07352689.2020.1724433 [DOI] [Google Scholar]
- Rathore K. S., Sunilkumar G., Connell J. P., Reddy A. S. (2009). U.S. Patent No. 7,626,081. Washington, DC: U.S. Patent and Trademark Office.
- Ray R. C., Montet D., Zakhia-Rozis N. (2014). Lactic acid fermentation of vegetables and fruits. Microorganisms fermentation traditional foods, 108–140. doi: 10.1201/b17307-7 [DOI] [Google Scholar]
- Ribeiro J. A. D. N. C., Serquiz A. C., Silva P. F. D. S., Barbosa P. B. B. M., Sampaio T. B. M., Araújo Junior R. F. D., et al. (2015). Trypsin inhibitor from Tamarindus indica l. seeds reduces weight gain and food consumption and increases plasmatic cholecystokinin levels. Clinics 70, 136–143. doi: 10.6061/clinics/2015(02)11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribera A. E., Zuñiga G. (2012). Induced plant secondary metabolites for phytopatogenic fungi control: a review. J. Soil Sci. Plant Nutr. 12 (4), 893–911. doi: 10.4067/S0718-95162012005000040 [DOI] [Google Scholar]
- Rodríguez-Sifuentes L., Marszalek J. E., Chuck-Hernández C., Serna-Saldívar S. O. (2020). Legumes protease inhibitors as biopesticides and their defense mechanisms against biotic factors. Int. J. Mol. Sci. 21 (9), 3322. doi: 10.3390/ijms21093322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout K., Yadav B. G., Yadava S. K., Mukhopadhyay A., Gupta V., Pental D., et al. (2018). QTL landscape for oil content in brassica juncea: analysis in multiple bi-parental populations in high and “0” erucic background. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychel S., Książkiewicz M. (2019). Development of gene-based molecular markers tagging low alkaloid pauper locus in white lupin (Lupinus albus l.). J. Appl. Genet. 60 (3), 269–281. doi: 10.1007/s13353-019-00508-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu P., Tripathy B., Rout S. (2020). Significance of anti-nutritional compounds in vegetables. Agric. Rural Dev. Spat. Issues Chall. Approaches 98, 98–109. [Google Scholar]
- Salari M. W., Ongom P. O., Thapa R., Nguyen H. T., Vuong T. D., Rainey K. M. (2021). Mapping QTL controlling soybean seed sucrose and oligosaccharides in a single family of soybean nested association mapping (SoyNAM) population. Plant Breed. 140 (1), 110–122. doi: 10.1111/pbr.12883 [DOI] [Google Scholar]
- Samtiya M., Aluko R. E., Dhewa T. (2020). Plant food anti-nutritional factors and their reduction strategies: an overview. Food Production Process. Nutr. 2 (1), 1–14. doi: 10.1186/s43014-020-0020-5 [DOI] [Google Scholar]
- Sashidhar N., Harloff H. J., Potgieter L., Jung C. (2020). Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol. J. 18 (11), 2241–2250. doi: 10.1111/pbi.13380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam A. S. K., Liu X., Stonehouse R., Tar'An B., Bett K. E., Sharpe A. G., et al. (2015). Mapping seed phytic acid concentration and iron bioavailability in a pea recombinant inbred line population. Crop Sci. 55 (2), 828–836. doi: 10.2135/cropsci2014.08.0544 [DOI] [Google Scholar]
- Shi J., Lang C., Wang F., Wu X., Liu R., Zheng T., et al. (2017). Depressed expression of FAE1 and FAD2 genes modifies fatty acid profiles and storage compounds accumulation in brassica napus seeds. Plant Sci. 263, 177–182. doi: 10.1016/j.plantsci.2017.07.014 [DOI] [PubMed] [Google Scholar]
- Shi J., Lang C., Wu X., Liu R., Zheng T., Zhang D., et al. (2015). RNAi knockdown of fatty acid elongase1 alters fatty acid composition in brassica napus. Biochem. Biophys. Res. Commun. 466 (3), 518–522. doi: 10.1016/j.bbrc.2015.09.062 [DOI] [PubMed] [Google Scholar]
- Shi J., Ni X., Huang J., Fu Y., Wang T., Yu H., et al. (2022). CRISPR/Cas9-mediated gene editing of BnFAD2 and BnFAE1 modifies fatty acid profiles in brassica napus. Genes 13 (10), 1681. doi: 10.3390/genes13101681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wang H., Hazebroek J., Ertl D. S., Harp T. (2005). The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J. 42 (5), 708–719. doi: 10.1111/j.1365-313X.2005.02412.x [DOI] [PubMed] [Google Scholar]
- Shi J., Wang H., Schellin K., Li B., Faller M., Stoop J. M., et al. (2007). Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 25 (8), 930–937. doi: 10.1038/nbt1322 [DOI] [PubMed] [Google Scholar]
- Shi J., Wang H., Wu Y., Hazebroek J., Meeley R. B., Ertl D. S. (2003). The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 131 (2), 507–515. doi: 10.1104/pp.014258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla V. K., Doyon Y., Miller J. C., DeKelver R. C., Moehle E. A., Worden S. E., et al. (2009). Precise genome modification in the crop species zea mays using zinc-finger nucleases. Nature 459 (7245), 437–441. doi: 10.1038/nature07992 [DOI] [PubMed] [Google Scholar]
- Simwaka J. E., Chamba M. V. M., Huiming Z., Masamba K. G., Luo Y. (2017). Effect of fermentation on physicochemical and antinutritional factors of complementary foods from millet, sorghum, pumpkin and amaranth seed flours. Int. Food Res. J. 24 (5), 1869–1879. [Google Scholar]
- Singh L. H., Chandra A. K., Yumnam S. D., Sarkar D., Manglem R. K., Dhabali T., et al. (2021). Thiocyanate in excess develops goiter followed by auto immune thyroid diseases even after effective salt iodization in a rural community of north east India. Ecotoxicology Environ. Saf. 208, 111711. doi: 10.1016/j.ecoenv.2020.111711 [DOI] [PubMed] [Google Scholar]
- Singh A., Gupta S., Kaur R., Gupta H. R. (2017). Process optimization for anti-nutrient minimization of millets. Asian J. Dairy Food Res. 36 (4), 322–326. doi: 10.18805/ajdfr.DR-1215 [DOI] [Google Scholar]
- Singh P., Kuo Y. C., Mishra S., Tsai C. H., Chien C. C., Chen C. W., et al. (2012). The lectin receptor kinase-VI. 2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. The Plant Cell 24 (3), 1256–1270. doi: 10.1105/tpc.112.095778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N., Meher B. R., Naik P. P., Panda P. K., Mukhapadhyay S., Maiti T. K., et al. (2019). p73 induction by abrus agglutinin facilitates snail ubiquitination to inhibit epithelial to mesenchymal transition in oral cancer. Phytomedicine 55, 179–190. doi: 10.1016/j.phymed.2018.08.003 [DOI] [PubMed] [Google Scholar]
- Sivaraman I., Arumugam N., Sodhi Y. S., Gupta V., Mukhopadhyay A., Pradhan A. K., et al. (2004). Development of high oleic and low linoleic acid transgenics in a zero erucic acid brassica juncea L.(Indian mustard) line by antisense suppression of the fad2 gene. Mol. Breed. 13 (4), 365–375. doi: 10.1023/B:MOLB.0000034092.47934.d6 [DOI] [Google Scholar]
- Skirycz A., Reichelt M., Burow M., Birkemeyer C., Rolcik J., Kopka J., et al. (2006). DOF transcription factor AtDof1. 1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in arabidopsis. Plant J. 47 (1), 10–24. doi: 10.1111/j.1365-313X.2006.02767.x [DOI] [PubMed] [Google Scholar]
- Skoneczka J. A., Maroof M. A. S., Shang C., Buss G. R. (2009). Identification of candidate gene mutation associated with low stachyose phenotype in soybean line PI200508. Crop Sci. 49 (1), 247–255. doi: 10.2135/cropsci2008.07.0403 [DOI] [Google Scholar]
- Sompong U., Somta P., Raboy V., Srinives P. (2012). Mapping of quantitative trait loci for phytic acid and phosphorus contents in seed and seedling of mungbean (Vigna radiata (L.) wilczek). Breed. Sci. 62 (1), 87–92. doi: 10.1270/jsbbs.62.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. H., Shin G., Kim H. J., Lee S. B., Moon J. Y., Jeong J. C., et al. (2022). Mutation of GmIPK1 gene using CRISPR/Cas9 reduced phytic acid content in soybean seeds. Int. J. Mol. Sci. 23 (18), 10583. doi: 10.3390/ijms231810583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Blanco B. (2008). “Gossipol e fatores antinutricionais da soja,” in Toxicologia aplicada à MedicIna VeterInária. Eds. Spinosa H. S., Górniak S. L., Neto J. P. (Barueri, Brazil: Manole; ), 531–545. [Google Scholar]
- Sparg S., Light M. E., Van Staden J. (2004). Biological activities and distribution of plant saponins. J. ethnopharmacol 94 (2-3), 219–243. doi: 10.1016/j.jep.2004.05.016 [DOI] [PubMed] [Google Scholar]
- Srivastava R. P., Singh J., Singh N. P., Singh D. (2015). Neurotoxin and other anti-nutrients of khesari (Lathyrus sativus) genotypes and their reduction by water soaking and dehusking. Indian J. Agric. Biochem. 28 (2), 172–177. doi: 10.5958/0974-4479.2015.00012.x [DOI] [Google Scholar]
- Stangoulis J. C., Huynh B. L., Welch R. M., Choi E. Y., Graham R. D. (2007). Quantitative trait loci for phytate in rice grain and their relationship with grain micronutrient content. Euphytica 154 (3), 289–294. doi: 10.1007/s10681-006-9211-7 [DOI] [Google Scholar]
- Stevenson-Paulik J., Bastidas R. J., Chiou S. T., Frye R. A., York J. D. (2005). Generation of phytate-free seeds in arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. 102 (35), 12612–12617. doi: 10.1073/pnas.0504172102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipanovic R. D., Bell A. A., Mace M. E., Howell C. R. (1975). Antimicrobial terpenoids of gossypium: 6-methoxygossypol and 6, 6′-dimethoxygossypol. Phytochemistry 14 (4), 1077–1081. doi: 10.1016/0031-9422(75)85190-9 [DOI] [Google Scholar]
- Suma P., Urooj A. (2014). Nutrients, anti-nutrients and bio accessible mineral content (in vitro) of pearl millet as influenced by milling. J. Food Sci. Technol. 51 (4), 756–761. doi: 10.1007/s13197-011-0541-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy J., Park G. T., Mukaiyama K., Tsukamoto C., Chang J. H., Lee J. D., et al. (2018). Molecular elucidation of a new allelic variation at the sg-5 gene associated with the absence of group a saponins in wild soybean. PloS One 13 (1), e0192150. doi: 10.1371/journal.pone.0192150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunilkumar G., Campbell L. M., Puckhaber L., Stipanovic R. D., Rathore K. S. (2006). Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc. Natl. Acad. Sci. 103 (48), 18054–18059. doi: 10.1073/pnas.0605389103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureshkumar S., Tamilkumar P., Senthil N., Nagarajan P., Thangavelu A. U., Raveendran M., et al. (2014). Marker assisted selection of low phytic acid trait in maize (Zea mays l.). Hereditas 151 (1), 20–27. doi: 10.1111/j.1601-5223.2013.00030.x [DOI] [PubMed] [Google Scholar]
- Takagi K., Nishizawa K., Hirose A., Kita A., Ishimoto M. (2011). Manipulation of saponin biosynthesis by RNA interference-mediated silencing of β-amyrin synthase gene expression in soybean. Plant Cell Rep. 30 (10), 1835–1846. doi: 10.1007/s00299-011-1091-1 [DOI] [PubMed] [Google Scholar]
- Tamilkumar P., Senthil N., Sureshkumar S., Thangavelu A. U., Nagarajan P., Vellaikumar S., et al. (2014). Introgression of low phytic acid locus (‘lpa2-2’) into an elite maize (‘Zea mays’ l.) inbred through marker assisted backcross breeding. Aust. J. Crop Sci. 8, 1224–1231. [Google Scholar]
- Teraishi M., Tojo Y., Yamada N., Okumoto Y. (2017). Identification of environmentally stable QTLs controlling saponin content in glycine max. Breed. Sci. 67 (2), 123–128. doi: 10.1270/jsbbs.16086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A., Sharma V., Thakur A. (2019). An overview of anti-nutritional factors in food. Int. J. Chem. Stud. 7 (1), 2472–2479. [Google Scholar]
- Tian B., Wei F., Shu H., Zhang Q., Zang X., Lian Y. (2011). Decreasing erucic acid level by RNAi-mediated silencing of fatty acid elongase 1 (BnFAE1. 1) in rapeseeds (Brassica napus l.) . Afr. J. Biotechnol. 10 (61), 13194–13201. doi: 10.5897/AJB11.1465 [DOI] [Google Scholar]
- Tian C., Yang J., Zeng Y., Zhang T., Zhou Y., Men Y., et al. (2019). Biosynthesis of raffinose and stachyose from sucrose via an in vitro multienzyme system. Appl. Environ. Microbiol. 85 (2), e02306–e02318. doi: 10.1128/AEM.02306-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari R., Kaur J., Bisht N. C. (2021). Extra-large G-proteins influence plant response to sclerotinia sclerotiorum by regulating glucosinolate metabolism in brassica juncea. Mol. Plant Pathol. 22 (10), 1180–1194. doi: 10.1111/mpp.13096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z., He W., Fan X., Guo A. (2021). Biological function of plant tannin and its application in animal health. Front. Veterinary Sci. 8. doi: 10.3389/fvets.2021.803657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toroser D., Thormann C. E., Osborn T. C., Mithen R. (1995). RFLP mapping of quantitative trait loci controlling seed aliphatic-glucosinolate content in oilseed rape (Brassica napus l). Theor. Appl. Genet. 91 (5), 802–808. doi: 10.1007/BF00220963 [DOI] [PubMed] [Google Scholar]
- Torres J., Rutherfurd S. M., Muñoz L. S., Peters M., Montoya C. A. (2016). The impact of heating and soaking on the in vitro enzymatic hydrolysis of protein varies in different species of tropical legumes. Food Chem. 194, 377–382. doi: 10.1016/j.foodchem.2015.08.022 [DOI] [PubMed] [Google Scholar]
- Tripathy S. K., Ranjan R., Dash S., Bharti R., Lenka D., Sethy Y. D., et al. (2015). Genetic analysis of BOAA content in grasspea (Lathyrus sativus l.). Legume Res. 38 (4), 465–468. doi: 10.5958/0976-0571.2015.00028.4 [DOI] [Google Scholar]
- Troll W., Wiesner R. (1983). Protease inhibitors: possible anticarcinogens in edible seeds. Prostate 4 (4), 345–349. doi: 10.1002/pros.2990040404 [DOI] [PubMed] [Google Scholar]
- Truong T., Baron-Dubourdieu D., Rougier Y., Guénel P. (2010). Role of dietary iodine and cruciferous vegetables in thyroid cancer: a countrywide case–control study in new Caledonia. Cancer causes control 21 (8), 1183–1192. doi: 10.1007/s10552-010-9545-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udensi E. A., Arisa N. U., Maduka M. (2008). Effects of processing methods on the levels of some antinutritional factors in mucuna flagellipes. Nigerian Food J. 26 (2). doi: 10.4314/nifoj.v26i2.47437 [DOI] [Google Scholar]
- Vagadia B. H., Vanga S. K., Raghavan V. (2017). Inactivation methods of soybean trypsin inhibitor–a review. Trends Food Sci. Technol. 64, 115–125. doi: 10.1016/j.tifs.2017.02.003 [DOI] [Google Scholar]
- Valentine M. F., De Tar J. R., Mookkan M., Firman J. D., Zhang Z. J. (2017). Silencing of soybean raffinose synthase gene reduced raffinose family oligosaccharides and increased true metabolizable energy of poultry feed. Front. Plant Sci. 8, 692. doi: 10.3389/fpls.2017.00692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme E. J., Lannoo N., Peumans W. J. (2008). “Plant lectins,” in Advances in botanical research, vol. 48. (Cambridge, Massachusetts, United States: Academic Press; ), 107–209. doi: 10.1016/B978-044451967-2/00067-2 [DOI] [Google Scholar]
- Vasconcelos I. M., Oliveira J. T. (2004). Antinutritional properties of plant lectins. Toxicon 44 (4), 385–403. doi: 10.1016/j.toxicon.2004.05.005 [DOI] [PubMed] [Google Scholar]
- Vashishth A., Ram S., Beniwal V. (2017). Cereal phytases and their importance in improvement of micronutrients bioavailability. Biotech 7 (1), 1–7. doi: 10.1007/s13205-017-0698-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venegas-Calerón M., Troncoso-Ponce M. A., Martínez-Force E. (2015). “Sunflower oil and lipids biosynthesis,” in Sunflower (Elsevier: AOCS Press; ), 259–295. [Google Scholar]
- Vinoth A., Ravindhran R. (2017). Biofortification in millets: a sustainable approach for nutritional security. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroh Bi I., Maquet A., Baudoin J. P., Du Jardin P., Jacquemin J. M., Mergeai G. (1999). Breeding for” low-gossypol seed and high-gossypol plants” in upland cotton. analysis of tri-species hybrids and backcross progenies using AFLPs and mapped RFLPs. Theor. Appl. Genet. 99 (7), 1233–1244. doi: 10.1007/s001220051329 [DOI] [Google Scholar]
- Wagner T. A., Liu J., Puckhaber L. S., Bell A. A., Williams H., Stipanovic R. D. (2015). RNAi construct of a cytochrome P450 gene CYP82D109 blocks an early step in the biosynthesis of hemigossypolone and gossypol in transgenic cotton plants. Phytochemistry 115, 59–69. doi: 10.1016/j.phytochem.2015.02.016 [DOI] [PubMed] [Google Scholar]
- Wang Z., Shea Z., Rosso L., Shang C., Li J., Bewick P., et al. (2022). “CRISPR/Cas9-targeted mutagenesis of KTI1 and KTI3 to reduce trypsin inhibitors in soybean seeds,” in Plant and Animal Genome XXIX Conference, , January 8-12, 2022, PAG. [Google Scholar]
- Wang P., Zhang J., Sun L., Ma Y., Xu J., Liang S., et al. (2018). High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 16 (1), 137–150. doi: 10.1111/pbi.12755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Pan Y., Liu Z., Zhu X., Zhai L., Xu L., et al. (2013). De novo transcriptome sequencing of radish (Raphanus sativusl.) and analysis of major genes involved in glucosinolate metabolism. BMC genomics 14 (1), 1–13. doi: 10.1186/1471-2164-14-836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani I. A., ulAshraf Z., Muzzaffar S. (2022). “Erucic acid,” in Handbook of plant and animal toxins in food (Boca Raton, Florida, United States: CRC Press; ), 169–176. [Google Scholar]
- Ward S. M. (2000). Response to selection for reduced grain saponin content in quinoa (Chenopodium quinoa willd.). Field Crops Res. 68 (2), 157–163. doi: 10.1016/S0378-4290(00)00117-9 [DOI] [Google Scholar]
- Watanabe D., Lošák T., Vollmann J. (2018). From proteomics to ionomics: Soybean genetic improvement for better food safety. Genetika 50 (1), 333–350. doi: 10.2298/GENSR1801333W [DOI] [Google Scholar]
- Webb A., Cottage A., Wood T., Khamassi K., Hobbs D., Gostkiewicz K., et al. (2016). A SNP-based consensus genetic map for synteny-based trait targeting in faba bean (Vicia faba l.). Plant Biotechnol. J. 14 (1), 177–185. doi: 10.1111/pbi.12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox J. R., Premachandra G. S., Young K. A., Raboy V. (2000). Isolation of high seed inorganic p, low-phytate soybean mutants. Crop Sci. 40 (6), 1601–1605. doi: 10.2135/cropsci2000.4061601x [DOI] [Google Scholar]
- Wu G., Yu X., Yu Z., Lu Q., Yang D., Shi Y., et al. (2022). Fine mapping of a major QTL qPA7-1 for low hydrocyanic acid content in sorghum–sudangrass hybrid. Genome. 65 (12), 605–619. doi: 10.1139/gen-2021-0114 [DOI] [PubMed] [Google Scholar]
- Wu Y., Li X., Xiang W., Zhu C., Lin Z., Wu Y., et al. (2012). Presence of tannins in sorghum grains is conditioned by different natural alleles of Tannin1. Proceedings of the National Academy of Sciences. 109 (26), 10281–10286. doi: 10.1073/pnas.1201700109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Wu Y., Xiao L., Li X., Lu C. (2008). Zero erucic acid trait of rapeseed (Brassica napus l.) results from a deletion of four base pairs in the fatty acid elongase 1 gene. Theor. Appl. Genet. 116 (4), 491–499. doi: 10.1007/s00122-007-0685-z [DOI] [PubMed] [Google Scholar]
- Yan G., Li D., Cai M., Gao G., Chen B., Xu K., et al. (2015). Characterization of FAE1 in the zero erucic acid germplasm of. Breed. Sci. 65 (3), 257–264. doi: 10.1270/jsbbs.65.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yathish K. R., Karjagi C. G., Gangoliya S. S., Kumar A., Preeti P., Yadav H. K., et al. (2022). Introgression of the low phytic acid locus (lpa2) into elite maize (Zea mays l.) inbreds through marker-assisted backcross breeding (MABB) (Research Square; ) [Google Scholar]
- Ye H., Li C., Bellgard M., Lance R., Wu D. (2013). “Genes controlling low phytic acid in plants: identifying targets for barley breeding,” in Advance in barley sciences: Proceedings of 11th international 185, barley genetics symposium (Dordrecht: Springer; ). doi: 10.1007/978-94-007-4682-4_16 [DOI] [Google Scholar]
- Yin L., Chen H., Cao B., Lei J., Chen G. (2017). Molecular characterization of MYB28 involved in aliphatic glucosinolate biosynthesis in Chinese kale (Brassica oleracea var. alboglabra bailey). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiara L. Y., Mandarino J. M. G., Carrão-Panizzi M. C., Madeira T. B., da Silva J. B., de Camargo A. C., et al. (2018). Germination changes the isoflavone profile and increases the antioxidant potential of soybean. J. Food Bioactives 3, 144–150. doi: 10.31665/JFB.2018.3157 [DOI] [Google Scholar]
- Yuan F. J., Zhu D. H., Deng B., Fu X. J., Dong D. K., Zhu S. L., et al. (2009). Effects of two low phytic acid mutations on seed quality and nutritional traits in soybean (Glycine max l. merr). J. Agric. Food Chem. 57 (9), 3632–3638. doi: 10.1021/jf803862a [DOI] [PubMed] [Google Scholar]
- Yu J., Yu S., Fan S., Song M., Zhai H., Li X., et al. (2012). Mapping quantitative trait loci for cottonseed oil, protein and gossypol content in a gossypium hirsutum× gossypium barbadense backcross inbred line population. Euphytica 187 (2), 191–201. doi: 10.1007/s10681-012-0630-3 [DOI] [Google Scholar]
- Zbidah M., Lupescu A., Shaik N., Lang F. (2012). Gossypol-induced suicidal erythrocyte death. Toxicology 302 (2-3), 101–105. doi: 10.1016/j.tox.2012.09.010 [DOI] [PubMed] [Google Scholar]
- Zhang W. J., Xu Z. R., Pan X. L., Yan X. H., Wang Y. B. (2007). Advances in gossypol toxicity and processing effects of whole cottonseed in dairy cows feeding. Livestock Science. 111 (1-2), 1–9. doi: 10.1016/j.livsci.2007.03.006 [DOI] [Google Scholar]
- Zhuo C., Wang T., Lu S., Zhao Y., Li X., Guo Z. (2013). A cold responsive galactinol synthase gene from medicago falcata (MfGolS1) is induced by myo-inositol and confers multiple tolerances to abiotic stresses. Physiologia plantarum 149 (1), 67–78. doi: 10.1111/ppl.12019 [DOI] [PubMed] [Google Scholar]
- Zou Z., Ishida M., Li F., Kakizaki T., Suzuki S., Kitashiba H., et al. (2013). QTL analysis using SNP markers developed by next-generation sequencing for identification of candidate genes controlling 4-methylthio-3-butenyl glucosinolate contents in roots of radish, raphanus sativus l. PloS One 8 (1), e53541. doi: 10.1371/journal.pone.0053541 [DOI] [PMC free article] [PubMed] [Google Scholar]