Abstract
Many replicative DNA polymerases couple DNA replication and unwinding activities to perform strand displacement DNA synthesis, a critical ability for DNA metabolism. Strand displacement is tightly regulated by partner proteins, such as single-stranded DNA (ssDNA) binding proteins (SSBs) by a poorly understood mechanism. Here, we use single-molecule optical tweezers and biochemical assays to elucidate the molecular mechanism of strand displacement DNA synthesis by the human mitochondrial DNA polymerase, Polγ, and its modulation by cognate and noncognate SSBs. We show that Polγ exhibits a robust DNA unwinding mechanism, which entails lowering the energy barrier for unwinding of the first base pair of the DNA fork junction, by ∼55%. However, the polymerase cannot prevent the reannealing of the parental strands efficiently, which limits by ∼30-fold its strand displacement activity. We demonstrate that SSBs stimulate the Polγ strand displacement activity through several mechanisms. SSB binding energy to ssDNA additionally increases the destabilization energy at the DNA junction, by ∼25%. Furthermore, SSB interactions with the displaced ssDNA reduce the DNA fork reannealing pressure on Polγ, in turn promoting the productive polymerization state by ∼3-fold. These stimulatory effects are enhanced by species-specific functional interactions and have significant implications in the replication of the human mitochondrial DNA.
INTRODUCTION
Human mitochondrial DNA polymerase, Polγ, is the main polymerase responsible for replication and repair of the mitochondrial DNA (mtDNA) (1,2). Polγ is a heterotrimeric holoenzyme that consists of a catalytic subunit (PolγA) and a dimeric accessory subunit (PolγB), Figure 1A (1,3). Pathogenic mutations in the genes encoding both the catalytic and accessory subunits have been identified in association with numerous human diseases (4). The catalytic subunit exhibits a finely tuned balance of polymerase (pol) and 3’-5’ exonuclease (exo) activities that ensures efficiency as well as fidelity of DNA synthesis (3,5–7). Polγ also exhibits DNA strand displacement activity, in which the holoenzyme displaces downstream DNA encountered during synthesis. This activity is relevant for replication through stable secondary structures (8–10), maintenance of the D-loop DNA structure at the origin of replication of the heavy strand (11), and removing RNA/DNA primers in coordination with primer processing factors (nucleases, helicases and/or mtSSBs) (12–14). As in the case of other replicative DNA polymerases (DNApols) (15–17), strand displacement DNA synthesis is usually limited to a few nucleotides (13,18–21). According to previous single-molecule manipulation studies on related DNApols, the regression or reannealing pressure of the DNA fork during strand displacement would inhibit polymerization-driven forward motion and promote the partition of the primer to the exo domain (22,23). Under these conditions Polγ is prone to enter an idling state, during which it undergoes intramolecular cycles of excising and adding a single nucleotide (6,20). In fact, mutations impeding the exo activity of Polγ stimulate the strand displacement activity of the holoenzyme (20,24). Excessive strand displacement by Polγ exo-deficient variants has been linked with the formation of double stranded breaks during mtDNA replication and development of progeroid phenotypes (18,21,25). These findings imply that the strand displacement activity of Polγ is intrinsically restricted. On the other hand, studies on other related DNA replication systems (26,27) would subject that the ability of Polγ to couple DNA synthesis and unwinding may provide the driving force necessary to open the DNA helix during leading strand DNA replication. Therefore, under these conditions, the holoenzyme partners within the replisome, the mitochondrial single-stranded DNA-binding protein (mtSSB) and the DNA helicase (Twinkle) (28–30), are expected to decrease the propensity of Polγ to idle at the fork junction (or favor its DNA unwinding activity).
Figure 1.
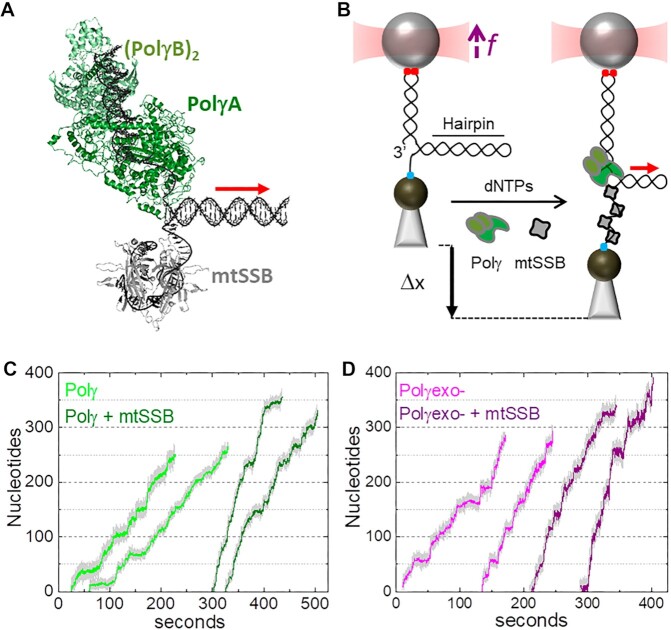
Experimental set up. (A) Schematic of Polγ (PDB: 3IKM) and mtSSB (PDB: 3ULL) at the DNA fork. Polγ holoenzyme is composed by the catalytic subunit, PolγA (dark green) and a dimer of the accessory subunit, PolγB, (light green). mtSSB (grey) binds the displaced ssDNA as a tetramer. (B) In the optical tweezers, a single DNA hairpin (559 bp) is tethered between two functionalized beads and held at constant tension (f). One strand of the hairpin is connected to the bead in the optical trap (red cone) through a ∼2.6-kb dsDNA handle via digoxigenin-antibody connections (red dots). The other strand is attached to a bead on a micropipette by biotin–streptavidin linkages (blue dot). The dsDNA handle includes a 3’ end for polymerase loading, and the 5’ end of the hairpin includes a poly-(dT)30 site for SSB binding. At constant tension, strand displacement DNA synthesis by Polγ (or Polγexo-) increases the end-to-end extension of the hairpin (Δx). Experiments were performed without and with several concentrations of cognate and noncognate SSBs (grey) in solution. In A) and B) red arrow indicates the 5’ to 3’ direction of DNA polymerase translocation along the hairpin. (C) Representative experimental traces of Polγ (2 nM) without (light green) and with (olive) mtSSB (50 nM) in solution (f= 6 pN). (D) Representative experimental traces of Polγexo- (2 nM) without (magenta) and with (purple) mtSSB (50 nM) in solution (f= 6 pN). Traces were displaced along the time axis for clarity of display.
Despite the putative relevance of the ability of Polγ to couple DNA synthesis and unwinding activities for mtDNA replication, little is known about the kinetics and mechanistic aspects of this reaction and it's modulation by proteins partners, such as the mtSSB. Human mtSSB is essential for mtDNA synthesis in vitro and in vivo (9,30,31). It binds ssDNA in a sequence independent manner (32,33) and forms the central nucleo-protein complex substrate upon which the mitochondrial polymerase must act (9,34). Although Polγ and mtSSB appear not to interact physically (35), mtSSB can stimulate the primer-extension activity of Polγ by imposing an optimal organization of the DNA template (8,36). In other DNA replication systems SSBs have been shown to stimulate the strand displacement activity of DNApols (15,16,37–39). However, the effect of mtSSB on the strand displacement activity of Polγ, a situation in which the two proteins bind to opposite strands of the DNA fork (Figure 1), remains largely unexplored.
Here, we present single-molecule manipulation optical tweezers assays, supported by ensemble biochemical experiments, to quantify the intrinsic strand displacement mechanism of Polγ and its modulation by mtSSB. We compare the real-time kinetics of the wild-type holoenzyme Polγ with that of an exo deficient variant, D198A/E200A (Polγexo-) (40), in order to determine the contribution of the exo reaction on the strand displacement activity (without interference of idling events). Overall, our results show that Polγ presents a robust DNA displacement mechanism that is limited by reannealing of the parental strands, which shifts its activity equilibrium towards the exo state. We demonstrate that mtSSBs use two key mechanisms to stimulate the strand displacement DNA synthesis by Polγ; i.e. mtSSB binding to the displaced ssDNA imposes additional destabilization energy on the DNA junction, which increases the pause-free rate, and reduces the DNA fork regression pressure on the holoenzyme. This, in turn decreases the residence time in the pause state per nucleotide. Interestingly, additional experiments with non-cognate Escherichia coli (EcoSSB) and phage T7 (gp2.5) SSBs revealed that species-specific interplay between the two mitochondrial partners may be critical under suboptimal mtSSB concentrations or stress conditions. Our results shed new light on the mechanism by which accessory proteins, such as SSBs, can enhance strand displacement DNA synthesis.
MATERIALS AND METHODS
Proteins and DNA constructs
Recombinant catalytic subunits (Polγ A) of wild-type and mutant (D198A/E200A) Polγexo-variants were prepared from Sf9 cells (41). The accessory subunit of the holoenzyme (Polγ B) was prepared from bacterial cells (41). The catalytic and the accessory subunits were combined in a 1:3 molar ratio to reconstitute the holoenzyme. Recombinant mtSSB was prepared from bacterial cells as described previously (36). Recombinant EcoSSB and Sequenase© were purchased from Thermofisher. T7DNAp was purchased from NEB. Recombinant gp2.5 was purchased from LSBio and Monserte Biotechnology. The hairpin construct was synthesized as described previously (23). The construct consists of a 2686 base pairs (bp) DNA ‘handle’ (pUC19 vector, Novagen) labeled with digoxigenin at one end, a 5’ (dT)35 end functionalized with biotin, and a 559 bp stem capped by a (dT)4 loop. The final hairpin construct contains a unique 3’ end loading site for the DNA polymerase. The hairpin stem sequence is described in (23) and contains a 75% AT bp. Considering the free energy formations of AT and GC bp on naked DNA as ΔGAT ∼ 1.5 kBT and ΔGGC ∼ 2.9 kBT (under ionic conditions similar to those used in this work) the average free energy of bp formation of the hairpin stem was ΔGbp ∼ 1.8 kBT (42). For primer extension experiments, we used a gapped DNA template, consisting of ∼900 nucleotides (nt) of single-stranded DNA (ssDNA) flanked by ∼3550 bp dsDNA handles labeled with biotin and digoxigenin, as described in (43) (Supplementary Figure S1).
Optical tweezers experiments
We used a miniaturized counter propagating dual-beam optical tweezers instrument (44) to manipulate individual DNA hairpins tethered between a streptavidin-coated bead (2.1 μm, Kisker Biotech) immobilized on top of a micropipette and an anti-digoxigenin-coated bead (3.0 μm diameter, Kisker Biotech) held in the optical trap, Figure 1B (45). Proteins were introduced inside the flow cell after dilution in the replication buffer containing 50 mM Tris pH 8.5, 30 mM KCl, 10 mM DTT, 4 mM MgCl2, 0.2 mg/ml BSA and the four dNTPs (50 μM). Unless otherwise indicated, DNApols were diluted to 2 nM in the reaction buffer containing the indicated amounts of SSBs. Polymerase exchange experiments were performed in a mixture of 2 nM Polγ or Polγexo- and 1 or 2 nM T7DNAp. For proper interpretation of the effect of mechanical tension on strand displacement activities, we performed independent measurements of the effect of tension on the primer extension replication kinetics of Polγ (8) and Polγexo- (Supplementary Figure S1) holoenzymes. Primer extension activities of Polγexo- diluted to 2 nM in the replication buffer were recorded on the gapped DNA construct, as described elsewhere (8,43). In all cases, data was monitored at 500 Hz at 22 ± 1°C using a feedback loop to maintain a constant force or constant mechanical tension on the DNA. Force ranged explored were 1–11 picoNewtons (pN) in strand displacement assays and 1–16 pN in primer extension assays. The trap stiffness calibrated for 3.0 μm beads was 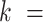 0.135 ± 0.0043 pN nm−1.
0.135 ± 0.0043 pN nm−1.
Bulk biochemical experiments
Polγ strand displacement processivity and fork residence time assays in bulk were carried out in the replication buffer on a forked DNA substrate resembling the organization of the hairpin used in optical tweezers (Supplementary Information (SI) and Supplementary Figure S2).
Data analysis
Processivity
The number of replicated nucleotides (processivity) in individual strand displacement assays was obtained by dividing the increase of the tether extension (Δx in Figure 1B) by the change in extension at a given tension accompanying during each catalytic step the generation of one new bp and one SSB-free or SSB-bound single-stranded nucleotide. The number of nucleotides incorporated in primer extension assays were obtained by dividing the change in tether extension by the change in extension due to the conversion of one single-stranded nucleotide into its double-stranded counterpart at a given tension (46,47). The extension of the dsDNA was approximated with the worm-like chain model for polymer elasticity with a persistent length of P = 53 nm and stretch modulus S = 1200 pN/ nm (48). The average extensions per nucleotide as a function of tension of free-ssDNA and ssDNA bound to mtSSB-, EcoSSB- or gp2.5-, under experimental conditions identical to those used in this work, were reported by us previously (8,49–51).
Average replication rates with and without pauses
The average replication rate at each tension (Vmean(f)) was determined by a line fit to the traces showing the number of replicated nucleotides versus time. The final average rate at each tension was obtained by averaging over all of the traces taken within similar tension values (±0.5 pN). Average replication rate without pauses at each tension (pause-free velocity, V(f)) was determined with an algorithm that computes the instantaneous velocities of the trajectory, averaging the position of the holoenzyme along the DNA over sliding time windows, as described previously (8). Tension dependent pause-free velocities were fitted to the strand displacement model described in SI and (52).
Average residence time at the pause state per nucleotide
The intrinsic flexibility of ssDNA together with the slow average strand displacement rates of the mitochondrial holoenzymes used in this work hindered the accurate identification of pause events (8). Nevertheless, identification of pause-free velocities (V(f)) allowed us to calculate the average residence time at the pause state per nucleotide at each tension, 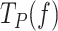 , as the difference between the average total residence time per nucleotide (
, as the difference between the average total residence time per nucleotide (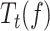 = 1/ Vmean(f)) and the residence time in the active state (
= 1/ Vmean(f)) and the residence time in the active state (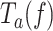 = 1/ V(f)). The tension dependencies of
= 1/ V(f)). The tension dependencies of 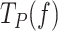 of each polymerase under study were fitted with Eq. 1.
of each polymerase under study were fitted with Eq. 1.
Moving probabilities
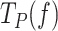 can be expressed in terms of moving probability (MP(f)), or the probability of finding the holoenzyme moving through the DNA hairpin as a function of tension. MP(f) was calculated as the ratio between the average replication rates with and without pauses at each tension, MP(f) = Vmean(f)/ V(f), which is equivalent to MP(f) = Ta(f)/ Tt(f) (see SI and (52)).
can be expressed in terms of moving probability (MP(f)), or the probability of finding the holoenzyme moving through the DNA hairpin as a function of tension. MP(f) was calculated as the ratio between the average replication rates with and without pauses at each tension, MP(f) = Vmean(f)/ V(f), which is equivalent to MP(f) = Ta(f)/ Tt(f) (see SI and (52)).
Maximum average processivities
In the absence of tension were estimated by multiplying the average residence time each holoenzyme spends at the fork (calculated from single turn over bulk experiments, 1/koff, Table 1) by their corresponding average velocities in the absence of tension, 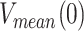 . The latter value was assessed from values of V(0) and
. The latter value was assessed from values of V(0) and 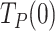 obtained from the fits to the data with the strand displacement and 2-state models, respectively,
obtained from the fits to the data with the strand displacement and 2-state models, respectively, 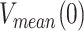 = 1/ (
= 1/ (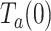 +
+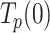 ).
).
Table 1.
Apparent detachment rates (koff) and maximum number of replicated nucleotides (Nt) measured in strand displacement bulk assays. ΔGint/M are the minimum values of the free-parameters yielded by least squared error fits of strand displacement model to pause-free velocity data. K(0) and d are the values of free-parameters of Eq. 1 obtained upon fitting the tension dependent average residence times in pause state data (Tp(f)) with the least mean squared error. Tp(f), average residence times at pause state at the indicated tensions (superscript). In all cases, uncertainties are shown as standard errors
| koff (s−1) (bulk) | Nt (bulk) | ΔGint(kBT)/M | K(0) | d(nm) | Tp(f) (s nt−1) | |
|---|---|---|---|---|---|---|
| Polγ | (6.26 ± 0.05) × 10−3 | 37 ± 15 | 0.90 ± 0.10 /1 | 24 ± 3 | 1.4 ± 0.4 | 3.10 ± 0.500pN |
| Polγ mtSSB | (8.21 ± 0.14) × 10−3 | 108 ± 42 | 1.40 ± 0.04 /1 | 10 ± 3 | 1.2 ± 0.3 | 1.23 ± 0.420pN |
| Polγ EcoSSB | (3.28 ± 0.04) × 10−3 | ≥150 | 1.38 ± 0.11 /1 | N.A. | N.A. | 0.4 ± 0.13pN |
| Polγexo- | (7.78 ± 0.29) × 10−3 | 69 ± 17 | 1.00 ± 0.10 /1 | 13 ± 2 | 1.2 ± 0.4 | 1.22 ± 0.280pN |
| Polγexo- mtSSB | (8.80 ± 0.50) × 10−3 | ≥150 | 1.23 ± 0.05 /1 | N.A. | N.A. | 0.23 ± 0.032pN |
| Polγexo-EcoSSB | (4.04 ± 0.05) × 10−3 | ≥150 | 1.39 ± 0.06 /1 | N.A. | N.A. | 0.20 ± 0.042pN |
RESULTS
Single-molecule strand displacement DNA synthesis assays
We used optical tweezers to follow the strand displacement DNA synthesis activity of Polγ holoenzyme (2 nM) and its exo deficient variant, Polγexo- (D198A/E200A, 2 nM), on individual DNA hairpins, in the absence or presence of varying concentrations of mtSSB, EcoSSB, or gp2.5, Figures 1A and B (Materials and Methods). Strand displacement activities were monitored at constant mechanical tension below 12 pN. In the absence of the holoenzyme, the hairpin remained stably closed below 12 pN, even in the presence of either SSB in solution. In the presence of the holoenzyme (and 50 μM dNTPs) the end-to-end distance between the beads increases gradually as the mitochondrial holoenzyme replicates through the hairpin stem converting each DNA bp unwound to 1 dsDNA bp and 1 ssDNA or ssDNA-SSB bound nucleotide (nt) (Figures 1B–D and Materials and Methods). We note that mechanical tension applied to the complementary strands of the hairpin modulates the strand displacement kinetics of replicative DNApols (22,23) and the binding properties of SSBs to ssDNA (51,53–55). Therefore, tension constitutes a useful variable to interrogate the interplay between these two proteins at the fork during active DNA synthesis.
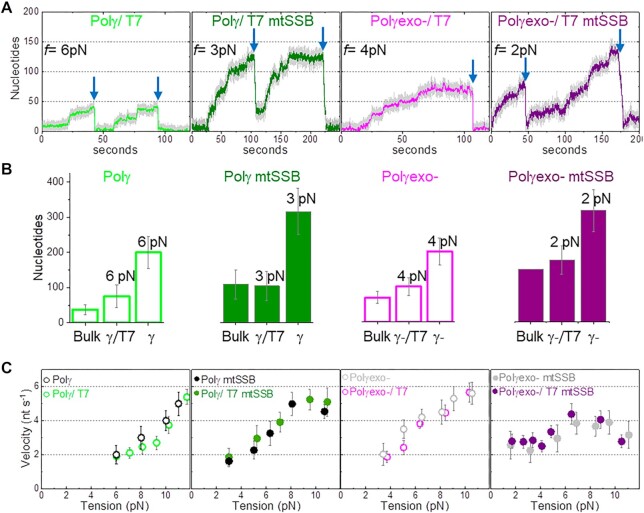
At the lowest tensions that allowed detection of activities, ∼3 and ∼6 pN for Polγexo- and Polγ, respectively, both holoenzymes replicated ∼200 nt (Figures 1C and D), which contrasted with the maximum processivity measured for each holoenzyme in bulk single turn over assays, 37 ± 15 nt for Polγ and 69 ± 17 nt for Polγexo- (Figures 2B and S2, Table 1). These results suggested that the replication traces measured in the optical tweezers may correspond to the consecutive action of several holoenzyme molecules that would exchange at the DNA fork. We tested this possibility using a ‘competitor polymerase method’ described previously (56,57). This method uses a mixture of DNA polymerases with similar affinities for the DNA but different kinetic rates. In this approach, sudden variations in the instantaneous replication rate can be readily attributed to the exchange of DNA polymerases at the DNA fork. For these experiments, we used the bacteriophage T7 DNA polymerase (T7DNAp) as a competitor and reporter. This polymerase shares high structural and sequence homology with PolγA and almost identical DNA binding affinity (∼3 nM) (58,59). However, at the DNA fork (at f < 8 pN) T7DNAp does not exhibit strand displacement synthesis on its own, and instead exerts fast, processive exonucleolysis, which was detected as a continuous decrease in the end-to-end distance of the DNA as the hairpin reanneals (22). These characteristic exo events, which were not observed for the mitochondrial holoenzymes in the absence of T7DNAp at any tension, were used as reporters to identify the exchange of Polγ or Polγexo- actively replicating the DNA with competing T7DNAp in solution in the absence or presence of mtSSB, Figure 2A. Supplementary Figure S3 shows detection of exchange events at f > 8 pN.
Figure 2.
Polymerase exchange is not rate limiting. (A) Representative strand displacement traces of Polγ and Polγexo- with competitor T7DNApol in solution in the absence and presence of mtSSB (50 nM). Exchange of Polγ and Polγexo- by T7DNAp was monitored as fast exo events (blue arrow) not observed in the absence of T7DNAp in solution (Figures 1C and D). Displayed traces were taken at the lowest tension (f) at which activity could be detected in each condition. (B) For all plots. First column (Bulk) shows the maximum number of replicated nucleotides obtained in single turn-over bulk experiments (Supplementary Figure S2). The second and third columns show the average number of replicated nucleotides measured in optical tweezers assays in the presence (γ/T7 and γ-/T7) and absence (γ, γ-) of T7DNApol in solution at the lowest detection tension in each case. Bulk Polγexo- mtSSB data does not include error bar because the enzyme generated full length product (150 nt) in each experiment. The average number of nucleotides replicated by Polγ and Polγexo- as a function of tension in the absence and presence of T7DNApol are shown in Figure S3. (C) Tension dependent average strand displacement rates (velocity, nt s−1) of Polγ and Polγexo- in the absence and presence of mtSSB without and with competitor T7DNApol in solution. The similarities between the average rates of Polγ (and Polγexo-) before exchange with T7DNAp with those measured in the absence of T7DNAp suggest that the polymerase exchange reaction is not rate limiting. Polymerase exchange events at f > 8 pN are shown in Figure S3. For all figures error bars show standard errors.
At a molar ratio of two mitochondrial:one phage holoenzymes, ∼80% of Polγ and Polγexo- replication traces (in the absence and presence of mtSSB) were interrupted by long exo events at the lowest detection tension for each holoenzyme or holoenzyme/mtSSB couple (Figure 2A). These results indicate that the mitochondrial holoenzyme at the DNA fork exchanged with T7DNAp in solution. Before the exchange with T7DNAp, the average processivities of Polγ and Polγexo- (in the absence and presence of mtSSB), were 2–3 times shorter than those in the absence of T7DNAp and approached those measured in the bulk single turnover assays (Figure 2B). These results suggest that in the absence of T7DNAp, replication traces, such as those shown in Figures 1C and 1D could correspond to the consecutive activity of 2–3 mitochondrial holoenzymes that may exchange at the DNA fork. We further tested that the effects of T7DNAp competition on the processivities of Polγ and Polγexo- were apparent up to ∼10 pN (Supplementary Figure S3). Remarkably, we observed that in the presence of T7DNAp the average strand displacement rates of Polγ and Polγexo- (with and without mtSSB) before exchange with T7DNAp were identical to those measured in the absence of T7DNAp in solution at all tensions, Figure 2C. These observations indicate that polymerase exchange at the fork is not rate limiting and does not contribute significantly to the kinetics of the frequent pause events characteristic of the replication traces (Figures 1C and 1D). Overall our results are in excellent agreement with previous bulk studies suggesting that Polγ can exchange with Polγ variants in solution during primer extension DNA synthesis conditions (60) and single-molecule reports from polymerase exchange kinetics in other DNA replication systems (24,56).
Polγ and Polγexo- present identical fork destabilization energies
Our bulk single turn-over assays showed that Polγexo- presents a strand displacement activity higher than that of Polγ (∼69 versus ∼37 nucleotides, respectively, Figure S2, Table 1). In the optical tweezers the strand displacement activity of Polγexo- was detected consistently at tension lower than that of Polγ (∼3 versus 6 pN, respectively) and at all tensions presented faster average velocities than the wild-type holoenzyme (Figure 2C). Overall these data are in agreement with preliminary bulk studies suggesting a higher efficiency of strand displacement synthesis by the mutant variant (13,18,20,21). To further investigate the differences in the strand displacement activities of the two holoenzymes, we measured the effect of mechanical tension (or mechanical destabilization of the DNA fork) on the moving and pause kinetics of each holoenzyme through the DNA fork.
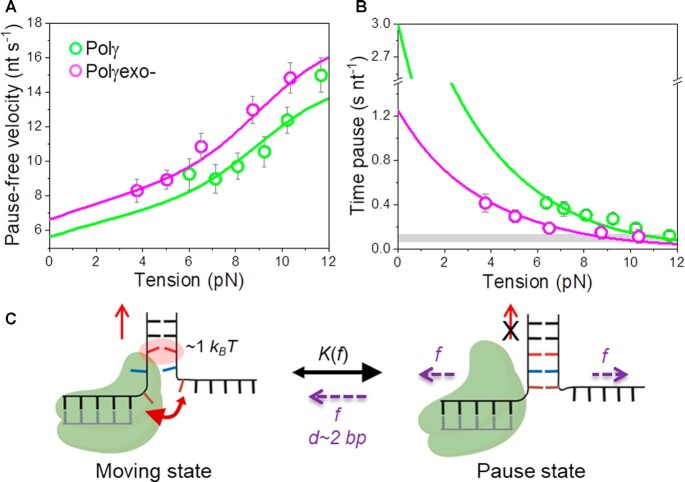
To examine the impact of mechanical destabilization of the fork on the moving kinetics (or moving state) we calculated the strand displacement rates without pauses, or pause-free velocity, at all tensions, Figure 3A (Materials and Methods). For both polymerases, pause-free velocity increased with tension continuously towards values found during primer extension conditions (Figure 3A and Supplementary Figure S1C), indicating that DNA unwinding is the rate limiting step of the reaction. Pause-free velocity was ∼12% faster for Polγexo- than for Polγ, in agreement with the difference between the maximum primer extension rates of each holoenzyme (Supplementary Figure S1C). Next, we fit the tension dependent pause-free velocity of each holoenzyme to a model (referred as strand displacement model) to calculate the interaction energy of the holoenzyme with the DNA fork. This model is an extension of the theoretical framework described by Betterton and Julicher to quantify the ‘unwinding activeness’ of nucleic acid helicases adapted to the case of replicative DNA polymerases (SI) (23,52,61,62). According to the strand displacement model, the pause-free strand displacement rate is governed, among other factors, by the average stability of the base pairs of the DNA fork ahead of the polymerase (ΔGbp), and the reduction of this free energy by the interaction energy of the polymerase with the fork, ΔGint (and the range on this interaction, M), and the mechanical tension applied to the DNA, ΔGf (SI). These considerations are in agreement with recent studies showing that the presence of replication proteins close to the DNA fork increases the breathing kinetics, or decreases the base pair stability, of the fork junction (63–65). According to (42), the average free energy of bp formation of our hairpin stem can be estimated as ΔGbp∼1.8 kBT (Materials and Methods), while ΔGf can be obtained from DNA elasticity (SI). Therefore, the interaction energy of the polymerase with the fork, ΔGint, and the range on this interaction, M are the only two free-parameters of the model. These parameters were fixed by least squares fits of the model to the pause-free velocity values of Polγ and Polγexo-, Figure 3A. The fits yielded ΔGint= 0.9 ± 0.1 kBT and M = 1 for Polγ, and ΔGint= 1.0 ± 0.1 kBT and M = 1 for Polγexo- (Table 1). We note that the model assumes that effective interaction energy of the holoenzyme with the dsDNA fork junction (ΔGint) decreases equally the binding energy of A–T and G–C base pairs. These results indicated that both holoenzymes decrease the activation energy of the nearest bp of the fork equally by ∼1 kBT. Therefore, other factors should account for the different ability of each holoenzyme to perform strand displacement DNA synthesis.
Figure 3.
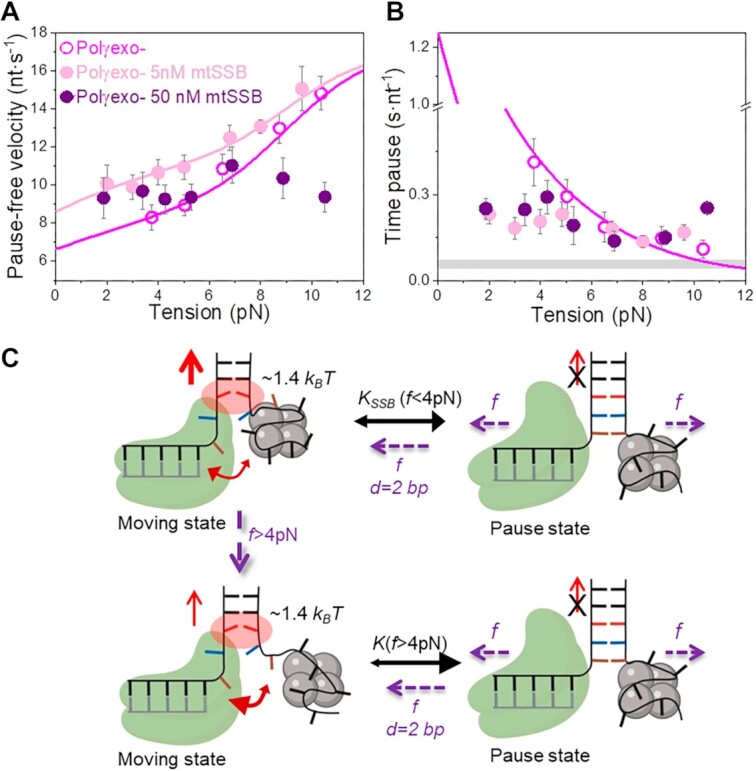
Effect of tension on Polγ and Polγexo- strand displacement kinetics. For all plots: Polγ green symbols (N = 80), Polγexo- magenta symbols (N = 71). Error bars show standard errors. (A) For both holoenzymes pause-free velocities (nt s−1) increased with tension continuously towards values measured during primer extension (Figure S1C). Green and magenta lines are the fits of the strand displacement model (SI) to Polγ and Polγexo- data, respectively. (B) Tension dependencies of the average residence times at the pause state per nucleotide (Tp(f), s nt−1). Green and magenta lines are the fits to Polγ and Polγexo- data, respectively, with a two state model (Eq. 1). Grey box show average Tp(f) values measured under primer extension conditions in the absence of mtSSB, Figure S1D and (16). (C) Diagram illustrating the two-state model in which the holoenzyme alternates between moving and pause or non-productive states during the strand displacement reaction. In the moving state, two template nucleotides (brown and blue) are bound to the pol site and the holoenzyme advances through the dsDNA destabilizing partially the first base pair of the junction (in red) with interaction energy of ΔGint ∼1 kBT per dNTP incorporation step. In the absence of tension, the regression pressure of the dsDNA fork outcompetes the holoenzyme for the template (two headed arrow), which shifts the equilibrium towards the pause or non-productive state strongly (K(0) >1, Table 1) and restricts the probability of finding Polγ and Polγexo- in the moving state to ∼4 and 12%, respectively (SI). Application of tension (f) to the hairpin decreases the rewinding kinetics and/or favors the unwinding of first ∼2 bp of the fork (d), which shifts the equilibrium towards the moving state.
Polγ spends longer times in a non-productive state than Polγexo-
Next, we studied the effect of the mechanical destabilization of the fork on the pause kinetics of each holoenzyme by quantifying the effect of tension on their average residence times in pause state per nucleotide, 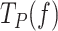 (Methods). Note that
(Methods). Note that  includes pause frequency and duration. The results showed that
includes pause frequency and duration. The results showed that  was higher for Polγ than for Polγexo- and for both holoenzymes decreased exponentially with tension towards values found during primer extension (Figure 3B). Previous studies on strand displacement DNA synthesis proposed that reannealing of the newly unwound bases (the fork regression pressure) would pause polymerase advancement by competing for template binding and promoting partition of the primer end from the pol to the exo sites (22,23,66). In the case of Polγexo-, the intramolecular transfer of the primer end (6) to the inactive exo site does not lead to excision and the primer returns intact to the pol site, favoring DNA synthesis. Conversely, in Polγ, the intramolecular transfer of the primer end to the exo site favors the excision of the newly incorporated nucleotide, effectively rendering the holoenzyme prone to idle at the fork in recurrent pol and exo events (20). We note that we did not detect exo events of Polγ at any tension, suggesting that this reaction, and/or associated idling, involves few nucleotides not resolved by our current resolution limit. Therefore, although different in nature, the events triggered by the fork regression pressure on each holoenzyme would be detected as pauses under our experimental conditions. In a simplified two-state scenario, in which each holoenzyme alternates between a moving and a pause states (Figure 3C), the effect of tension on the average residence time in pause state per nucleotide during strand displacement can be quantified as (SI):
was higher for Polγ than for Polγexo- and for both holoenzymes decreased exponentially with tension towards values found during primer extension (Figure 3B). Previous studies on strand displacement DNA synthesis proposed that reannealing of the newly unwound bases (the fork regression pressure) would pause polymerase advancement by competing for template binding and promoting partition of the primer end from the pol to the exo sites (22,23,66). In the case of Polγexo-, the intramolecular transfer of the primer end (6) to the inactive exo site does not lead to excision and the primer returns intact to the pol site, favoring DNA synthesis. Conversely, in Polγ, the intramolecular transfer of the primer end to the exo site favors the excision of the newly incorporated nucleotide, effectively rendering the holoenzyme prone to idle at the fork in recurrent pol and exo events (20). We note that we did not detect exo events of Polγ at any tension, suggesting that this reaction, and/or associated idling, involves few nucleotides not resolved by our current resolution limit. Therefore, although different in nature, the events triggered by the fork regression pressure on each holoenzyme would be detected as pauses under our experimental conditions. In a simplified two-state scenario, in which each holoenzyme alternates between a moving and a pause states (Figure 3C), the effect of tension on the average residence time in pause state per nucleotide during strand displacement can be quantified as (SI):
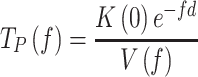 |
(1) |
where, 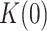 is the equilibrium constant of the transition between moving and pause state during strand displacement in the absence of tension. f is the mechanical tension that promotes the mechanical destabilization of the hairpin. d is the tension-induced conformational change along the pulling coordinate that shifts the equilibrium towards the moving state and
is the equilibrium constant of the transition between moving and pause state during strand displacement in the absence of tension. f is the mechanical tension that promotes the mechanical destabilization of the hairpin. d is the tension-induced conformational change along the pulling coordinate that shifts the equilibrium towards the moving state and 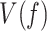 is the tension dependent pause-free velocity defined by the strand displacement model described above (SI). The two free variables,
is the tension dependent pause-free velocity defined by the strand displacement model described above (SI). The two free variables, 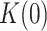 and d, were fixed upon least-squares fitting of Eq. (1) to
and d, were fixed upon least-squares fitting of Eq. (1) to 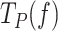 data, Figure 3B and Table 1.
data, Figure 3B and Table 1.
On one hand, extrapolation of the fits to 0 pN revealed that the average residence times at pause state per nucleotide in the absence of tension, 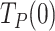 , of Polγ and Polγexo- were 3.10 ± 0.50 s nt−1 and 1.22 ± 0.28 s nt−1, respectively (Table 1). These
, of Polγ and Polγexo- were 3.10 ± 0.50 s nt−1 and 1.22 ± 0.28 s nt−1, respectively (Table 1). These 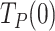 values were ∼30 and ∼10 times higher than those during primer extension (∼0.1 s nt−1 for both holoenzymes (8), Figure 3B and Supplementary Figure S1D), showing that stability and/or regression pressure of the DNA fork has a strong effect on increasing
values were ∼30 and ∼10 times higher than those during primer extension (∼0.1 s nt−1 for both holoenzymes (8), Figure 3B and Supplementary Figure S1D), showing that stability and/or regression pressure of the DNA fork has a strong effect on increasing 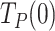 . In terms of moving probabilities, the
. In terms of moving probabilities, the 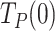 values indicate that the probabilities of finding Polγ and Polγexo- moving through the DNA hairpin are as low as ∼4 and ∼12%, respectively (Materials and Methods, SI). This data is in line with the higher efficiency of the strand displacement synthesis by the mutant variant (20).
values indicate that the probabilities of finding Polγ and Polγexo- moving through the DNA hairpin are as low as ∼4 and ∼12%, respectively (Materials and Methods, SI). This data is in line with the higher efficiency of the strand displacement synthesis by the mutant variant (20).
On the other hand, the values of the tension-induced conformational change that shifts the equilibrium towards the moving state (d) obtained from the fits were very similar for Polγ and Polγexo- 1.4 ± 0.4 and 1.2 ± 0.4 nm, respectively (Table 1). This conformational change is concomitant with release of ∼4 nt along the pulling coordinate or the unwinding of the first ∼2 bp of the DNA fork. This result is in agreement with a mechanism in which destabilization of the ∼2 first bp of the DNA fork by tension diminishes the fork regression pressure and creates two template nucleotides that could be accommodated at the polymerase template-binding pocket (67) shifting equilibrium towards the moving state, Figure 3C.
Differential effects of mtSSB on Polγ and Polγexo- strand displacement activities under mechanical tension
Next, we aimed to investigate the effect of mtSSB interaction with the displaced strand on the tension dependent strand displacement replication kinetics of Polγ and Polγexo- variants. We tested the effects of 5, 50 and 100 nM mtSSB, which under our current experimental conditions have been shown to cover respectively ∼85%, ∼98% or are expected to oversaturate individual ssDNA molecules stretched under mechanical tension (51).
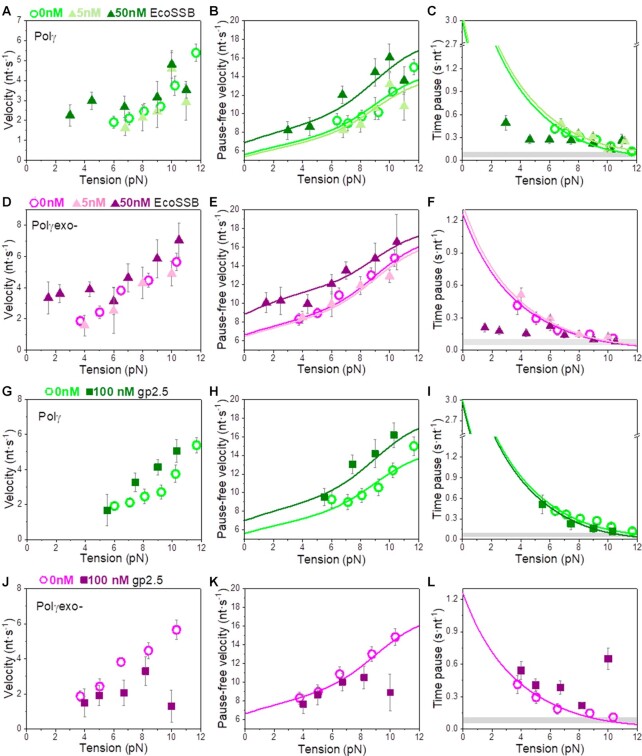
In the case of Polγ, 5 nM mtSSB favored detection of activities at slightly lower tensions (∼4–5 pN), decreased 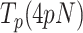 by 2–3-fold with respect to conditions in the absence of mtSSB, but did not alter pause-free rates, Figures 4A and B. Increasing mtSSB concentration to 50 nM promoted the strand displacement activity of Polγ more strongly; it dropped the minimum tension required to detect individual activities to lower values, ∼3 pN, stimulated the pause-free rates by a ∼25% at all tensions, and decreased
by 2–3-fold with respect to conditions in the absence of mtSSB, but did not alter pause-free rates, Figures 4A and B. Increasing mtSSB concentration to 50 nM promoted the strand displacement activity of Polγ more strongly; it dropped the minimum tension required to detect individual activities to lower values, ∼3 pN, stimulated the pause-free rates by a ∼25% at all tensions, and decreased 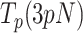 by ∼2–3 times with respect to conditions in the absence of mtSSB (Figures 4A and B).
by ∼2–3 times with respect to conditions in the absence of mtSSB (Figures 4A and B).
Figure 4.
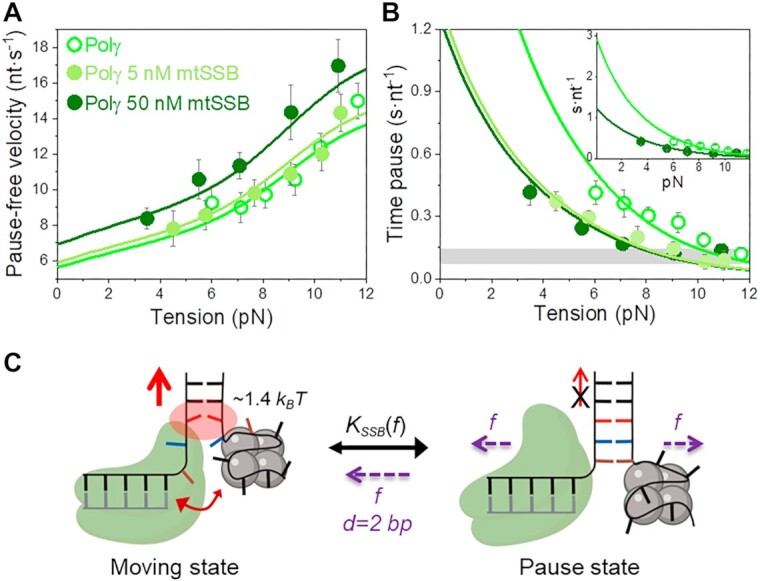
Effect of mtSSB on the tension dependent strand displacement kinetics of Polγ. (A) 50 nM (N = 40) but not 5 nM (N = 20) mtSSB stimulated the pause-free velocity of Polγ at all tensions. Green lines correspond to the fits of the strand displacement model to data in the absence and presence of mtSSB. (B) 5 and 50 nM concentrations of mtSSB decreased average residence time at pause state per nucleotide (Tp(f), s nt−1) of Polγ at all tensions. Grey box shows average Tp(f) values obtained under primer extension conditions in the absence of mtSSB (16). Green lines are the fits of two-state model (Eq. 1) to data in the absence and presence of mtSSB. mtSSB binding to the displaced strand decreases ∼2–3 times the average residence time of Polγ at a pause or non-productive state. For both figures error bars show standard errors. Inset shows the intersection of the fits with the Y-axis. (C) Diagram illustrating the two-state model in the presence of mtSSB. Polγ alternates between moving and pause or non-productive states. In the moving state, two template nucleotides (brown and blue) are bound at the pol active site and the holoenzyme-mtSSB complex destabilizes partially the first base pair of the DNA hairpin with interaction energy a ∼40% higher than in the absence of mtSSB (ΔGint ∼1.4 kBT per dNTP incorporated). In addition, mtSSB decreases the fork regression kinetics (represented by a two-headed arrow), which in turn, increases the probability of finding the holoenzyme at the moving state from ∼4 to ∼12% (SI). Even in the presence of mtSSB, the equilibrium is shifted towards the pause or inactive-state (KSSB(0) > 1, Table 1). Destabilization of ∼2 base pairs (d) of the DNA junction by application of mechanical tension (f) is required to shift the equilibrium towards the moving state.
In the case of Polγexo-, 5 nM and 50 nM mtSSB had both strong stimulatory effects on the strand displacement activity of the variant at tension below 5 pN; mtSSB dropped the minimum tension required to detect individual activities, from ∼4 to ∼2 pN, increased the average pause-free velocity by ∼25%, and decreased 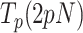 by ∼4 times, with respect to the values predicted by the strand displacement and the two-state models for conditions in the absence of mtSSB (Figures 5A and B). At the lowest tension (∼2 pN), these stimulatory effects resulted in a higher number of replicated nucleotides and faster average replication rates than in the absence of mtSSB (Figures 2B and C). Interestingly, as tension increased to 4–5 pN mtSSB lost the ability to decrease the residence time at pause state (
by ∼4 times, with respect to the values predicted by the strand displacement and the two-state models for conditions in the absence of mtSSB (Figures 5A and B). At the lowest tension (∼2 pN), these stimulatory effects resulted in a higher number of replicated nucleotides and faster average replication rates than in the absence of mtSSB (Figures 2B and C). Interestingly, as tension increased to 4–5 pN mtSSB lost the ability to decrease the residence time at pause state (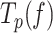 ) of the mutant holoenzyme (Figure 5B) and in turn, to stimulate the activity of the mutant variant (Figures 2C and S3). Moreover, at 50 nM mtSSB tension above ∼5–7 pN was detrimental for Polγexo- activity; under these conditions the average replication rates (with and without pauses) were lower and the residence times at pause state were significantly higher than those measured in the absence of mtSSB (Figures 5A and B).
) of the mutant holoenzyme (Figure 5B) and in turn, to stimulate the activity of the mutant variant (Figures 2C and S3). Moreover, at 50 nM mtSSB tension above ∼5–7 pN was detrimental for Polγexo- activity; under these conditions the average replication rates (with and without pauses) were lower and the residence times at pause state were significantly higher than those measured in the absence of mtSSB (Figures 5A and B).
Figure 5.
Effect of mtSSB on the tension dependent strand displacement kinetics of Polγexo-. Effects of 5 nM (N = 44) and 50 nM (N = 78) mtSSB on the tension dependent (A) pause-free velocity, and (B) average residence times at pause state per nucleotide (Tp(f), s nt−1) of Polγexo-. Both mtSSB concentrations stimulated the strand displacement activity of Polγexo- below 4–6pN. However, the stimulatory effects diminished above 4–6 pN. Even more, as tension increased above ∼8 pN, 50 nM mtSSB had detrimental effects on the pause-free rates (A) and residence times at pause state per nucleotide (B) of the mutant holoenzyme variant. In (A) magenta and pink lines correspond to the fits of the strand displacement model to the tension dependent pause-free rates in the absence and presence of 5 nM mtSSB, respectively. In (B), the magenta line is the fit of the two-state model (Eq.1) to the tension dependent average residence time at pause state per nucleotide in the absence of mtSSB. Grey box shows the average Tp(f) values measured under primer extension conditions in the absence of SSB, Figure S1. For (A) and (B) error bars show standard errors. (C) Diagram illustrating the effect of tension on the moving-pause state equilibrium of Polγexo- in the presence of mtSSB. At tension f< 4 pN, Polγexo- would alternate between moving and pause state with an equilibrium constant (KSSB(f< 4pN)) leading to a residence time in the pause state ∼3–4-times shorter than that in the absence of mtSSB. Application of tension above 4 pN promotes the release of ssDNA nucleotides from the mtSSB (brown and blue), which in turn could decrease its ability to counteract the fork regression kinetics. Under these conditions, the mutant holoenzyme would alternate between moving and pause states with an equilibrium constant similar to that in the absence of mtSSB (K(f > 4 pN)). In both situations, mtSSB binding energy and kinetics would help the holoenzyme to destabilize the first base pair of the DNA fork (ΔGint∼ 1.4 kBT per dNTP incorporated). Mechanical destabilization of the ∼2 first base pairs (d) of the DNA junction by tension (f) will further shift the equilibrium towards the moving state in the two situations.
Further increase of mtSSB concentration to 100 nM resulted in lack of stimulation and further inhibition of Polγ and Polγexo- strand displacement activities, Figure S4. These results are in line with previous bulk biochemical assays that showed deleterious effects of oversaturating mtSSB concentrations on the DNA synthesis activity of the human mitochondrial holoenzyme (36).
Overall, our results show that mtSSB stimulates the strand displacement activity of both holoenzymes by favoring detection of strand displacement activity at lower forces, increasing pause-free rates and decreasing the time at non-productive or pause state. These stimulatory effects resulted in a higher number of replicated nucleotides than in the absence of mtSSB, which at the lowest tensions (∼2 and ∼3 pN) agree well with those measured in bulk assays 108 ± 42 and ≥150 nt for Polγ and Polγexo respectively (Figure 2B, and Table 1). Interestingly, the real-time kinetics of each holoenzyme responded to the combined effect of mtSSB concentration and tension differently, Polγexo- being more prone to stimulation by lower mtSSB concentrations (10-fold) but also to inhibition by tension.
Species specific effects favor strand displacement DNA synthesis
To determine whether the stimulation of the tension dependent strand displacement activities of Polγ and Polγexo- by mtSSB depend on their coordination at the fork, or result from a passive binding of SSB to the displaced strand, we assessed the kinetic parameters of these two holoenzymes in the presence of various concentrations of homologous EcoSSB and heterologous phage T7 SSB, gp2.5. EcoSSB shares significant sequence and structural homology to mtSSB (68,69). Both proteins bind preformed ssDNA as tetramers with similar affinities (KD ∼2 nM) and footprints (number of nucleotides wrapped per tetramer) (32,33,46,51,70–72). In contrast, the multifunctional gp2.5 is organized as a dimer, shows smaller ssDNA binding footprint and lower affinity for ssDNA (KD ∼0.8 μM) than mtSSB and EcoSSB (73,74). EcoSSB and gp2.5 present intrinsically disordered acidic C-terminal tails that mediate interactions with other proteins including some of their respective replisomes (71,75). An analogous C-terminal tail is absent on mtSSB.
Bulk biochemical assays show that saturating concentrations of EcoSSB stimulated the strand displacement processivity of Polγ and Polγexo- in a way similar to that measured for its mtSSB homolog (Supplementary Figure S2, Table 1). However, notable differences were apparent at the single-molecule level between the effects of the two homologous SSB proteins on the real-time kinetics of each holoenzyme. At the lowest concentration (5 nM), EcoSSB did not have significant effects on the initiation force, tension dependent average rates (with and without pauses) and times at pause states of both holoenzymes (Figure 6A–F). These results contrast with the stimulatory effects of 5 nM mtSSB on the strand displacement kinetics of the two holoenzyme variants (Figures 4 and 5). The increase of EcoSSB concentration to 50 nM stimulated the activity of the two holoenzymes significantly at tension below 6–7 pN (Figure 6A–F). Under these conditions, the minimum tensions required to detect activities decreased (to ∼3 and ∼1 pN for Polγ and Polγexo-, respectively), and the average rates of the two holoenzymes increased as a consequence of stimulation of the pause-free velocities (∼25% increase) and the decrease of the time at pause state per nucleotide (2–4 times). However, above 6–7 pN EcoSSB (50 nM) lost the ability of decreasing Tp(f) of both holoenzymes (Figures 6C and F). This effect of tension on Tp(f) is similar to that measured for Polγexo- in the presence of mtSSB. However, it was not observed in the case of the wild-type holoenzyme in the presence of mtSSB. Interestingly, EcoSSB (50 nM) did not present inhibitory effects on Polγexo- activity at high tensions (Figure 6D–F). As in the case of high mtSSB concentrations, no stimulation or inhibition of the strand displacement activities of both holoenzymes was measured with 100 nM EcoSSB at high tensions (f > 8 pN), Supplementary Figure S4.
Figure 6.
Effects non-cognate SSB on Polγ and Polγexo- tension dependent strand displacement kinetics. (A–C) 5 nM EcoSSB had no apparent effects on the strand displacement kinetics of Polγ (N = 28). In contrast, 50 nM EcoSSB (N = 32) stimulated pause-free velocity at all tensions (B), and the average rates (A), and residence times at pause state, Tp(f), (C) below 5 pN. (D–F) 5 nM EcoSSB (N = 27) had no significant effects on the strand displacement kinetics of Polγexo-, whereas 50 nM EcoSSB (N = 42) stimulated the pause-free velocity at all tensions (E), and the average rates (D) and residence time at the pause state (F) at tension below 5 pN. (G–I) gp2.5 (100 nM, N = 15) stimulated the pause-free velocities of Polγ to a similar extend than 50 nM mtSSB and EcoSSB (H), but did not decrease the residence time at pause state of the wild-type holoenzyme at any tension (I). (J–L) gp2.5 (100 nM, N = 17) did not stimulate the strand displacement kinetics of Polγexo- and was inhibitory at tension above ∼8 pN. Lines correspond to the fits of the strand displacement model to the pause-free data (B, E, H, K) and Eq. 1 to the residence time at pause state data (C, F, I, L). Grey boxes show the average Tp(f) values obtained during primer extension conditions in the absence of SSBs (Figure S1 and (16)). For all plots error bars show standard errors.
In the case of non-cognate heterologous gp2.5, bulk biochemical assays showed that the phage SSB stimulated the processivity of Polγ but not that of Polγexo- (Supplementary Figure S2). At the single-molecule level, only the highest gp2.5 concentration used in our experiments (100 nM) had significant effects on the activity of the two holoenzymes, probably reflecting the lower affinity of this protein for ssDNA. Under these conditions, gp2.5 did not favor the detection of Polγ and Polγexo- activities at tensions significantly lower than those in the absence of SSB (Figure 6G–L). However, gp2.5 (100 nM) stimulated the pause-free velocities of Polγ to the extent similar as in the cases of 50 mM mtSSB and EcoSSB, but did not decrease the residence time at pause state of the wild-type variant at any tension, Figures 6H and I. In contrast, gp2.5 (100 nM) did not stimulate and even inhibited the strand displacement activity of Polγexo- (especially at high tensions), Figure 6J–L. These results showed again that the strand displacement activity of Polγexo- is more sensitive to the combined effect of tension and SSB than Polγ.
Although effects of SSBs on the two holoenzymes at hand differ in details, the results generally showed that at concentrations ∼10 to 20 times lower than those of noncognate SSBs, mtSSB has greater ability to stimulate the strand displacement kinetics of the two Polγ variants under mechanical tension. These results argue that efficient strand displacement DNA synthesis entails a specific interplay between the holoenzyme and the mtSSB. Notably, the lack of stimulation and/or inhibition of Polγexo- average strand displacement rate by the three SSBs under study at tensions above ∼5 pN, suggest that under mechanical stress conditions the mutant holoenzyme cannot correctly couple with these SSB proteins during strand displacement replication.
Finally, we sought to determine whether our observations of the behavior of mitochondrial holoenzymes in response to various SSB proteins and tension can be extrapolated to other polymerase-SSB systems. To this end, we measured the effects of cognate (gp2.5) and non-cognate (mtSSB and EcoSSB) SSBs on the kinetics of the strand displacement replication by the wild-type (T7DNAp) and the exo-deficient (Sequenase©) variants of phage T7 DNA polymerase under increasing mechanical tensions (SI and Supplementary Figure S5). Overall, the effects of cognate and non-cognate SSBs on the tension dependent kinetics of strand displacement replication by T7DNAp and Sequenase© were in line with those measured on the mitochondrial holoenzymes. Together, these results confirmed that i) cognate SSBs are more efficient than non-cognate SSBs in promoting pause-free velocities and decreasing time at pause state during stand displacement reaction, and ii) both cognate and non-cognate failed to decrease the residence times at the pause state characteristic of the exo deficient variants at high tensions.
Quantification of SSB effects on strand displacement kinetics
Under conditions favoring strand displacement replication, cognate and non-cognate SSBs promoted the pause-free velocities of the mitochondrial holoenzyme variants to a similar extent, by ∼20–25%. We fitted these data sets to the strand displacement model to quantify the energetic contribution of SSBs to DNA unwinding. Least square fits yielded similar values of the DNA fork interaction/destabilization energies of ΔGint∼ 1.4 kBT (M = 1) for all holoenzyme-SSB couples (Table 1): Polγ/50nM mtSSB (Figure 4A), Polγexo-/5nM mtSSB (Figure 5A), Polγ/50nM EcoSSB (Figure 6B), Polγexo-/50nM EcoSSB (Figure 6E), and Polγ/gp2.5 (Figure 6H). These results imply that, under conditions that allow stimulation of pause-free velocities, cognate and noncognate SSBs contribute non-specifically an extra ∼0.4 kBT to decrease the activation barrier of the first bp of the fork during the strand displacement activities.
Next, we quantified the contributions of SSBs to decrease the residence times at the pause state(s) ( ) by fitting to the two-state model (Eq. 1) the
) by fitting to the two-state model (Eq. 1) the  values of Polγ in the presence of 5 or 50 nM mtSSBs (Figure 4B). Note that these were the two only conditions where an SSB decreased the
values of Polγ in the presence of 5 or 50 nM mtSSBs (Figure 4B). Note that these were the two only conditions where an SSB decreased the  of the holoenzyme at all tensions. The fits yielded the equilibrium constant between pausing and polymerization (
of the holoenzyme at all tensions. The fits yielded the equilibrium constant between pausing and polymerization (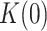 ) and the magnitude of the conformational change that shifts equilibrium towards moving state (d) in the presence of mtSSB (Table 1 and Supplementary Table S2). Extrapolation of the fits to 0 pN indicated that 5 and 50 nM mtSSB decreased the average residences time at pause state in the absence of tension by ∼2.5-fold. On the other hand, the values of d that resulted from the fits were identical for both mtSSB concentrations, ∼1.2 nm (Table 1). This conformational change indicates that, as in the absence mtSSB, unwinding of the first ∼2 bp of the DNA fork by tension is necessary to shift the equilibrium towards the moving state. Note that the fit of Polγ
) and the magnitude of the conformational change that shifts equilibrium towards moving state (d) in the presence of mtSSB (Table 1 and Supplementary Table S2). Extrapolation of the fits to 0 pN indicated that 5 and 50 nM mtSSB decreased the average residences time at pause state in the absence of tension by ∼2.5-fold. On the other hand, the values of d that resulted from the fits were identical for both mtSSB concentrations, ∼1.2 nm (Table 1). This conformational change indicates that, as in the absence mtSSB, unwinding of the first ∼2 bp of the DNA fork by tension is necessary to shift the equilibrium towards the moving state. Note that the fit of Polγ 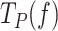 data in the presence of mtSSB (5 nM) to Eq. 1 explained well the effect of non-cognate EcoSSB (50 nM) on the
data in the presence of mtSSB (5 nM) to Eq. 1 explained well the effect of non-cognate EcoSSB (50 nM) on the 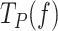 at tensions below ∼6 pN, Supplementary Figure S4C. Above this tension, EcoSSB had no longer an effect on the
at tensions below ∼6 pN, Supplementary Figure S4C. Above this tension, EcoSSB had no longer an effect on the 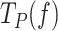 of Polγ. Similarly, cognate and non-cognate SSBs lost their ability to decrease the residence time at the pause state of Polγexo- at tension higher than ∼5 pN (Figures 5B, 6F and L). This effect limited the number of data points that changed with tension continuously and therefore, hindered consistent fits with the two-state model (Eq. 1). Nevertheless, from the data at the lowest tension (∼1–2 pN) we infer that cognate and non-cognate SSBs lowered the
of Polγ. Similarly, cognate and non-cognate SSBs lost their ability to decrease the residence time at the pause state of Polγexo- at tension higher than ∼5 pN (Figures 5B, 6F and L). This effect limited the number of data points that changed with tension continuously and therefore, hindered consistent fits with the two-state model (Eq. 1). Nevertheless, from the data at the lowest tension (∼1–2 pN) we infer that cognate and non-cognate SSBs lowered the 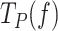 of Polγexo- ∼4-fold, with respect to the values predicted by the fit of the two-state model to data in the absence of SSB. As mentioned above, previous single molecule experiments suggested DNA fork regression pressure (or regression kinetics) as the main factor that pauses the polymerase advance through dsDNA and therefore, increases
of Polγexo- ∼4-fold, with respect to the values predicted by the fit of the two-state model to data in the absence of SSB. As mentioned above, previous single molecule experiments suggested DNA fork regression pressure (or regression kinetics) as the main factor that pauses the polymerase advance through dsDNA and therefore, increases 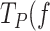 ). We next performed additional single-molecule experiments to check the consequence of SSBs binding to ssDNA on the reannealing rate or regression kinetics of the DNA hairpin. These experiments showed that all three SSBs (50 nM) decreased the reannealing rate of the complementary strands of the hairpin by ∼100- to 1000-fold, Figure S6, as previously shown for other SSBs proteins (76,77). These results explain, at least in part, the significant effect of SSBs on decreasing the average residence time that the holoenzyme spends in pause state.
). We next performed additional single-molecule experiments to check the consequence of SSBs binding to ssDNA on the reannealing rate or regression kinetics of the DNA hairpin. These experiments showed that all three SSBs (50 nM) decreased the reannealing rate of the complementary strands of the hairpin by ∼100- to 1000-fold, Figure S6, as previously shown for other SSBs proteins (76,77). These results explain, at least in part, the significant effect of SSBs on decreasing the average residence time that the holoenzyme spends in pause state.
The values of 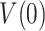 and
and 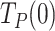 predicted by fits of the strand displacement and the two-state models (Eq. 1) to the single-molecule data (Figures 3-5, Table 1), together with the residence times at the DNA fork of each holoenzyme determined in bulk (1/koff, Table 1), can be used to estimate maximum average processivities in the absence of tension (Methods). These calculations yielded values of 49 ± 8 nt for Polγ, 91 ± 28 nt for Polγ with mtSSB (50 nM), and 92 ± 19 nt for Polγexo-. Remarkably, given the differences between bulk and single-molecule approaches, these values are within error with the maximum number of replicated nucleotides measured in bulk studies, 37 ± 15 for Polγ, 108 ± 42 for Polγ with mtSSB, and 69 ± 17 for Polγexo- (Figure 2B, Table 1), supporting the models used to explain the data.
predicted by fits of the strand displacement and the two-state models (Eq. 1) to the single-molecule data (Figures 3-5, Table 1), together with the residence times at the DNA fork of each holoenzyme determined in bulk (1/koff, Table 1), can be used to estimate maximum average processivities in the absence of tension (Methods). These calculations yielded values of 49 ± 8 nt for Polγ, 91 ± 28 nt for Polγ with mtSSB (50 nM), and 92 ± 19 nt for Polγexo-. Remarkably, given the differences between bulk and single-molecule approaches, these values are within error with the maximum number of replicated nucleotides measured in bulk studies, 37 ± 15 for Polγ, 108 ± 42 for Polγ with mtSSB, and 69 ± 17 for Polγexo- (Figure 2B, Table 1), supporting the models used to explain the data.
Finally, we note that cognate and non-cognate SSB presented stimulatory effects on T7DNApol and Sequenase© strand displacement kinetics of the same magnitude as those measured on the mitochondrial holoenzymes (Supplementary Figure S5 and Supplementary Table S1).
DISCUSSION
In this study, we demonstrated that Polγ and its exo deficient variant (D198A/E200A) present substantial strand displacement DNA synthesis and provided further insights into the mechanisms by which mtSSB assisted this reaction. Upon initiation of the strand displacement reaction, the two mitochondrial holoenzymes decreased the activation energy of the nearest bp of the fork equally, by ΔGint ∼1 kBT per dNTP incorporated, indicating that the intrinsic strand displacement mechanism of Polγ is independent from its exo activity. According to current models of the mechanism of coupling DNA synthesis with unwinding, the sharp bending of template (∼90o) induced characteristically by DNA polymerases within their polymerization domains (67,78) would impose mechanical stress at the DNA fork junction, which in turn lowers the energy barrier for DNA unwinding during each nucleotide incorporation cycle, Figure 3C (22,23,27,52). Under our experimental conditions, the ΔGint of the mitochondrial holoenzyme is ∼45% lower than the average stability of the next bp of the hairpin, ΔGbp(0pN) ∼1.8 kBT (SI). According to the strand displacement model, in the absence of tension the strand displacement rate depends on the difference between ΔGbp(0pN) and ΔGint (SI). Therefore, the fact that ΔGbp(0pN) is significantly higher than ΔGint explains the slower pause-free strand displacement rate of the holoenzyme (V(0pN)) as compare to the rate of primer extension over ssDNA template, i.e. ∼6 versus ∼24 nt s−1, respectively (Supplementary Figure S1).
The moderate effect of bp stability of the strand displacement rate (∼4-fold reduction) cannot explain the extended residence times of the holoenzyme variants in pause or non-productive states, which in the absence of tension are 30- (for Polγ) and 10- (for Polγexo-) times longer than during primer extension conditions. These long residence times at pause state constitute the main event limiting the elongation phase of the strand displacement replication. Previous single-molecule studies suggested that the principal factor that pause the advance of the polymerase through the DNA fork is the fork regression pressure, which outcompetes eventually the pol active site for binding to the template, Figure 3C (22,23,66). Previous structural studies showed that the active site of Polγ, and other related DNA polymerases, bind ∼2 template nucleotides stably (67,78,79). In agreement with these observations, fits of Eq. (1) to 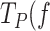 ) indicate that mechanical unwinding of ∼2 bp of the DNA fork rescues the holoenzyme from the pause state. These results suggest that application of tension decreases the fork regression pressure on the holoenzyme, and releases ∼2 template nucleotides which could be accommodated back at the template-binding pocket favoring in this way DNA synthesis (Figure 3C). The loss of the template by the holoenzyme against the fork regression pressure will induce the intramolecular kinetic partitioning of the primer between pol and exo sites (22,23,66). This process would have differential effects on the kinetics of Polγ and its exo-deficient variant. The active exo site of Polγ will lead to removal of one or few, newly incorporated nucleotides from the primer (20), while in the case of Polγexo-, the frayed primer end will return intact to the pol site (5,6). In practice, because individual exo events are below our current resolution limit, they would be included in the pause kinetics of wild-type Polγ, resulting in significantly higher
) indicate that mechanical unwinding of ∼2 bp of the DNA fork rescues the holoenzyme from the pause state. These results suggest that application of tension decreases the fork regression pressure on the holoenzyme, and releases ∼2 template nucleotides which could be accommodated back at the template-binding pocket favoring in this way DNA synthesis (Figure 3C). The loss of the template by the holoenzyme against the fork regression pressure will induce the intramolecular kinetic partitioning of the primer between pol and exo sites (22,23,66). This process would have differential effects on the kinetics of Polγ and its exo-deficient variant. The active exo site of Polγ will lead to removal of one or few, newly incorporated nucleotides from the primer (20), while in the case of Polγexo-, the frayed primer end will return intact to the pol site (5,6). In practice, because individual exo events are below our current resolution limit, they would be included in the pause kinetics of wild-type Polγ, resulting in significantly higher 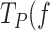 ) values compared to the Polγexo-. Indeed, this is consistent with our observation that Polγ spends 2–3 times longer residence times at the pause state per nucleotide than the exo deficient variant (Figure 3B) and is generally ∼3-fold less likely to be found in the moving state (Materials and Methods). This observation, in turn, also explains the higher processivity (2–3-fold) of Polγexo- on the dsDNA hairpin (note that the two enzymes have similar dissociation rates from DNA, Table 1).
) values compared to the Polγexo-. Indeed, this is consistent with our observation that Polγ spends 2–3 times longer residence times at the pause state per nucleotide than the exo deficient variant (Figure 3B) and is generally ∼3-fold less likely to be found in the moving state (Materials and Methods). This observation, in turn, also explains the higher processivity (2–3-fold) of Polγexo- on the dsDNA hairpin (note that the two enzymes have similar dissociation rates from DNA, Table 1).
Our work shows that mtSSB has a strong stimulatory effect on Polγ and Polγexo- strand displacement activities. We note that, in contrast to other SSBs, mtSSB lacks the C-terminal tail required for inter-protein cooperativity (71,80) and to date; there is no experimental evidence for cooperative binding of mtSSB to ssDNA (33). Therefore, we can rule out cooperative binding of mtSSB to the displaced ssDNA as a potential factor that promotes strand displacement activity of the mitochondrial holoenzyme. In addition, mtSSB did not affect the apparent dissociation rates of Polγ and Polγexo- from the DNA fork (Supplementary Figure S2 and Table 1) therefore; stimulation cannot be attributed to increased residence times on the DNA fork. According to our data, the stimulatory effects of mtSSB on the strand displacement activities of the mitochondrial holoenzymes can be attributed to two main mechanisms: increasing the pause-free rates and decreasing the residence time at pause state per nucleotide.
On the one hand, mtSSB stimulated the pause–free rate (∼25%) by decreasing the energy barrier for the unwinding of the first bp of the DNA fork, by additional ∼0.4 kBT (Figure 4C). This contribution is in excellent agreement with the average binding energies per nucleotide measured for mtSSB under similar experimental conditions (51,55). Overall, stimulation of pause-free velocities depended on SSB concentration but not on the external mechanical tension exerted to the system. On the other hand, at the lowest tension, mtSSB decreased the residence times in the non-productive or pause state (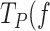 )) of Polγ and Polγexo- by ∼2–4 times, respectively. This stimulatory effect is stronger than that on the pause-free velocity, which makes it the main factor promoting the strand displacement activity of the holoenzyme. The cause of this stimulatory effect could be attributed, at least in part, to the reduction of the DNA fork reannealing rate (regression kinetics) imparted by SSB binding to ssDNA (Supplementary Figure S6). Remarkably, the stimulatory effect of SSB on
)) of Polγ and Polγexo- by ∼2–4 times, respectively. This stimulatory effect is stronger than that on the pause-free velocity, which makes it the main factor promoting the strand displacement activity of the holoenzyme. The cause of this stimulatory effect could be attributed, at least in part, to the reduction of the DNA fork reannealing rate (regression kinetics) imparted by SSB binding to ssDNA (Supplementary Figure S6). Remarkably, the stimulatory effect of SSB on 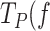 ) faded away as tension raised to ∼5 pN, specially for Polγexo- in the presence of cognate and non-cognate SSBs and Polγ with non-cognate SSBs. Previous single-molecule manipulation studies showed that mtSSB wraps 35 nt of the displaced strand when binding is couple to DNA replication (51) however, mechanical tension applied to the SSB-ssDNA complexes promotes the gradual unwrapping of ssDNA nucleotides from the SSBs, which affects the SSB binding footprint on ssDNA (51,55). Taken together, these results suggest that: i) the binding energy and kinetics but not the binding footprint or wrapping mode of mtSSB to/on ssDNA are relevant to stimulate pause-free velocities. ii) On the contrary, the binding footprint of the SSB (and/or its physical proximity to the fork junction), would be relevant to counter act the fork regression kinetics on the holoenzyme, or in other words, decrease the strong competition between the DNA fork and the holoenzyme for template binding (Figures 4C and 5C).
) faded away as tension raised to ∼5 pN, specially for Polγexo- in the presence of cognate and non-cognate SSBs and Polγ with non-cognate SSBs. Previous single-molecule manipulation studies showed that mtSSB wraps 35 nt of the displaced strand when binding is couple to DNA replication (51) however, mechanical tension applied to the SSB-ssDNA complexes promotes the gradual unwrapping of ssDNA nucleotides from the SSBs, which affects the SSB binding footprint on ssDNA (51,55). Taken together, these results suggest that: i) the binding energy and kinetics but not the binding footprint or wrapping mode of mtSSB to/on ssDNA are relevant to stimulate pause-free velocities. ii) On the contrary, the binding footprint of the SSB (and/or its physical proximity to the fork junction), would be relevant to counter act the fork regression kinetics on the holoenzyme, or in other words, decrease the strong competition between the DNA fork and the holoenzyme for template binding (Figures 4C and 5C).
Generally, mtSSB stimulated pause-free velocities and decreased time at pause state of Polγ and/or Polγexo- at concentrations 10-times lower than those of non-cognate SSBs. These results suggest that a specific interplay between the two mitochondrial partners is relevant for efficient DNA synthesis at the fork. Because the mitochondrial holoenzyme and SSB do not interact physically (35), coordination of their activities at the DNA fork could rely on functional interactions. For example, we found that local electrostatic repulsion between the two proteins facilitates primer extension over a ssDNA template (8). Future studies will help to identify the nature of and the specific residues responsible for the functional interplay between these two mitochondrial proteins. Remarkably, cognate and non-cognate SSBs stimulated the tension dependent strand displacement kinetics of T7DNApol and Sequenase© in ways similar to those found on Polγ (Supplementary Figure S5), suggesting that our findings on mitochondrial DNA replication system could be extrapolated to related DNA replication systems.
The potent strand displacement or DNA unwinding activity of the mitochondrial holoenzyme and its enhancement by mtSSB likely has significant physiological implications. (i) The bp destabilization energy of Polγ is similar or even higher to that reported for replicative helicases previously (0.05–1.6 kBT (62,81,82)) arguing that the holoenzyme may contribute substantially in promoting active DNA unwinding when assemble into the replisome. However, its poor ability to counter act the fork regression pressure (which restricts its moving probability through dsDNA to 4%) suggests that during leading (or H-strand) DNA synthesis a critical role of the mitochondrial helicase Twinkle may be to prevent fork regression pressure on the holoenzyme, ensuring robust DNA replication. A distribution of labor between these two enzymes has been proposed for other DNA replication systems (17,26,37,83). (ii) Because strand displacement activity is inversely related to polymerase slippage (15), the strand displacement ability of Polγ would generally favor its fidelity by decreasing the probability of deletions (and/or insertions) during replication of the lagging or L-strand of the mtDNA. (iii) Polγ-mtSSB coupling would be relevant for the processing of primers at the origins of replication of the mtDNA; i.e, generating long flaps upon reaching the origins of the two mtDNA strands, enabling their further processing by dedicated nucleases (84). In this context, the substantial strand displacement activity of Polγexo- in the presence of mtSSB would generate excessively long flaps precluding their processing by the associated nucleases and, in turn, maturation of mtDNA. 4) Reports to date indicate that ∼95% of all the mtDNA synthesis events initiated at the origin of replication (i.e. OH) are terminated prematurely at the termination associated sequence (TAS), likely due to the absence of mtDNA helicase Twinkle (24). In this scenario, Polγ would reside at the fork junction (the end of the D-loop, near TAS) accompanied only by the displaced strand-bound mtSSB, which resembles our study model. It is therefore possible that strand displacement activity is exerted at TAS. The likely occurrence of strand displacement mtDNA synthesis at TAS under physiological conditions is supported by the fact that in mice expressing the Polγexo- variant (which presents a more potent strand displacement activity) a dsDNA stretch terminated at TAS (i.e. 7S DNA) is significantly longer compared to that in the presence of wild-type Polγ (24). Interestingly, the mice exhibit progeroid phenotype with accumulation of mtDNA deletions (85), pointing out again to the deleterious effects of excessive strand displacement activity under physiological conditions.
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding authors (GLC, BI) upon reasonable request.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to members of B. Ibarra and G. Ciesielski labs for useful discussions.
Notes
Present address: Kateryna M. Lemishko, Department of Physics, King's College London, Strand Campus, London WC2R 2LS, UK.
Contributor Information
Ismael Plaza-G.A., Instituto Madrileño de Estudios Avanzados en Nanociencia, IMDEA Nanociencia, Faraday 9, 28049 Madrid, Spain.
Kateryna M Lemishko, Instituto Madrileño de Estudios Avanzados en Nanociencia, IMDEA Nanociencia, Faraday 9, 28049 Madrid, Spain.
Rodrigo Crespo, Departamento de Estructura de la Materia, Física Térmica y Electrónica, Universidad Complutense de Madrid, Pza. de Ciencias, 1, 28040 Madrid, Spain.
Thinh Q Truong, Department of Chemistry, Auburn University at Montgomery, Montgomery, AL 36117, USA.
Laurie S Kaguni, Department of Biochemistry and Molecular Biology and Center for Mitochondrial Science and Medicine, Michigan State University, East Lansing, MI 48823, USA.
Francisco J Cao-García, Departamento de Estructura de la Materia, Física Térmica y Electrónica, Universidad Complutense de Madrid, Pza. de Ciencias, 1, 28040 Madrid, Spain.
Grzegorz L Ciesielski, Department of Chemistry, Auburn University at Montgomery, Montgomery, AL 36117, USA; Department of Biochemistry and Molecular Biology and Center for Mitochondrial Science and Medicine, Michigan State University, East Lansing, MI 48823, USA.
Borja Ibarra, Instituto Madrileño de Estudios Avanzados en Nanociencia, IMDEA Nanociencia, Faraday 9, 28049 Madrid, Spain; Nanobiotecnología (IMDEA-Nanociencia), Unidad Asociada al Centro Nacional de Biotecnología (CSIC), 28049 Madrid, Spain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitiveness (SMEC) [RTI2018-095802-B-I00 to F.J.C.G., PGC2018-099341-B-I00 to B.I.]; National Institutes of Health [GM45925 to L.S.K., GM139104 to G.L.C.]; Comunidad de Madrid [NanoMagCOST P2018 INMT-4321]; I.P.G.-A. and K.M.L. were supported by fellowships PRE2019-088885 and FPU2014/06867, respectively; IMDEA Nanociencia acknowledges support from the Severo Ochoa Program for Centers of Excellence in R&D [CEX2020-001039-S]; University of Alabama at Birmingham, Vision Science Research Center (UAB VSRC), Molecular & Cellular Analysis Core for allowing us to use their GE Typhoon Trio + Variable Mode Imager; the UAB VSRC cores are supported by NIH grant [P30 EY003039]. Funding for open access charge: Spanish Ministry of Economy and Competitiveness (SMEC): PID2021-126755NB-I00.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kaguni L.S. DNA polymerase gamma, the mitochondrial replicase. Annu. Rev. Biochem. 2004; 73:293–320. [DOI] [PubMed] [Google Scholar]
- 2. Krasich R., Copeland W.C.. DNA polymerases in the mitochondria: a critical review of the evidence. Front. Biosci. (Landmark Ed.). 2017; 22:692–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee H.R., Johnson K.A.. Fidelity of the human mitochondrial DNA polymerase. J. Biol. Chem. 2006; 281:36236–36240. [DOI] [PubMed] [Google Scholar]
- 4. Stumpf J.D., Saneto R.P., Copeland W.C.. Clinical and molecular features of POLG-related mitochondrial disease. Cold Spring Harb. Perspect. Biol. 2013; 5:a011395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnson A.A., Johnson K.A.. Fidelity of nucleotide incorporation by human mitochondrial DNA polymerase. J. Biol. Chem. 2001; 276:38090–38096. [DOI] [PubMed] [Google Scholar]
- 6. Johnson A.A., Johnson K.A.. Exonuclease proofreading by human mitochondrial DNA polymerase. J. Biol. Chem. 2001; 276:38097–38107. [DOI] [PubMed] [Google Scholar]
- 7. Lee Y.S., Johnson K.A., Molineux I.J., Yin Y.W.. A single mutation in human mitochondrial DNA polymerase Pol gammaA affects both polymerization and proofreading activities of only the holoenzyme. J. Biol. Chem. 2010; 285:28105–28116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cerrón F., de Lorenzo S., Lemishko K.M., Ciesielski G.L., Kaguni L.S., Cao F.J., Ibarra B.. Replicative DNA polymerases promote active displacement of SSB proteins during lagging strand synthesis. Nucleic Acids Res. 2019; 47:5723–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miralles Fusté J., Shi Y., Wanrooij S., Zhu X., Jemt E., Persson Ö., Sabouri N., Gustafsson C.M., Falkenberg M.. In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet. 2014; 10:e1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sullivan E.D., Longley M.J., Copeland W.C.. Polymerase γ efficiently replicates through many natural template barriers but stalls at the HSP1 quadruplex. J. Biol. Chem. 2020; 295:17802–17815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takamatsu C., Umeda S., Ohsato T., Ohno T., Abe Y., Fukuoh A., Shinagawa H., Hamasaki N., Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002; 3:451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nicholls T.J., Zsurka G., Peeva V., Schöler S., Szczesny R.J., Cysewski D., Reyes A., Kornblum C., Sciacco M., Moggio M.et al.. Linear mtDNA fragments and unusual mtDNA rearrangements associated with pathological deficiency of MGME1 exonuclease. Hum. Mol. Genet. 2014; 23:6147–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uhler J.P., Thörn C., Nicholls T.J., Matic S., Milenkovic D., Gustafsson C.M., Falkenberg M.. MGME1 processes flaps into ligatable nicks in concert with DNA polymerase γ during mtDNA replication. Nucleic Acids Res. 2016; 44:5861–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng L., Zhou M., Guo Z., Lu H., Qian L., Dai H., Qiu J., Yakubovskaya E., Bogenhagen D.F., Demple B.et al.. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell. 2008; 32:325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Canceill D., Viguera E., Ehrlich S.D.. Replication slippage of different DNA polymerases is inversely related to their strand displacement efficiency. J. Biol. Chem. 1999; 274:27481–27490. [DOI] [PubMed] [Google Scholar]
- 16. Koc K.N., Stodola J.L., Burgers P.M., Galletto R.. Regulation of yeast DNA polymerase δ-mediated strand displacement synthesis by 5'-flaps. Nucleic Acids Res. 2015; 43:4179–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stano N.M., Jeong Y.J., Donmez I., Tummalapalli P., Levin M.K., Patel S.S.. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005; 435:370–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Farge G., Pham X.H., Holmlund T., Khorostov I., Falkenberg M.. The accessory subunit B of DNA polymerase gamma is required for mitochondrial replisome function. Nucleic Acids Res. 2007; 35:902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farr C.L., Wang Y., Kaguni L.S.. Functional interactions of mitochondrial DNA polymerase and single-stranded DNA-binding protein. Template-primer DNA binding and initiation and elongation of DNA strand synthesis. J. Biol. Chem. 1999; 274:14779–14785. [DOI] [PubMed] [Google Scholar]
- 20. He Q., Shumate C.K., White M.A., Molineux I.J., Yin Y.W.. Exonuclease of human DNA polymerase gamma disengages its strand displacement function. Mitochondrion. 2013; 13:592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Macao B., Uhler J.P., Siibak T., Zhu X., Shi Y., Sheng W., Olsson M., Stewart J.B., Gustafsson C.M., Falkenberg M.. The exonuclease activity of DNA polymerase γ is required for ligation during mitochondrial DNA replication. Nat. Commun. 2015; 6:7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Manosas M., Spiering M.M., Ding F., Bensimon D., Allemand J.F., Benkovic S.J., Croquette V.. Mechanism of strand displacement synthesis by DNA replicative polymerases. Nucleic Acids Res. 2012; 40:6174–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morin J.A., Cao F.J., Lázaro J.M., Arias-Gonzalez J.R., Valpuesta J.M., Carrascosa J.L., Salas M., Ibarra B.. Active DNA unwinding dynamics during processive DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:8115–8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jemt E., Persson Ö., Shi Y., Mehmedovic M., Uhler J.P., Dávila López M., Freyer C., Gustafsson C.M., Samuelsson T., Falkenberg M.. Regulation of DNA replication at the end of the mitochondrial D-loop involves the helicase TWINKLE and a conserved sequence element. Nucleic Acids Res. 2015; 43:9262–9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Falkenberg M., Gustafsson C.M.. Mammalian mitochondrial DNA replication and mechanisms of deletion formation. Crit. Rev. Biochem. Mol. Biol. 2020; 55:509–524. [DOI] [PubMed] [Google Scholar]
- 26. Manosas M., Spiering M.M., Ding F., Croquette V., Benkovic S.J.. Collaborative coupling between polymerase and helicase for leading-strand synthesis. Nucleic Acids Res. 2012; 40:6187–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pandey M., Patel S.S.. Helicase and polymerase move together close to the fork junction and copy DNA in one-nucleotide steps. Cell Rep. 2014; 6:1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ciesielski G.L., Oliveira M.T., Kaguni L.S.. Animal mitochondrial DNA replication. Enzymes. 2016; 39:255–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gustafsson C.M., Falkenberg M., Larsson N.G.. Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 2016; 85:133–160. [DOI] [PubMed] [Google Scholar]
- 30. Korhonen J.A., Pham X.H., Pellegrini M., Falkenberg M.. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004; 23:2423–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiang M., Xie X., Zhu X., Jiang S., Milenkovic D., Misic J., Shi Y., Tandukar N., Li X., Atanassov I.et al.. The mitochondrial single-stranded DNA binding protein is essential for initiation of mtDNA replication. Sci. Adv. 2021; 7:eabf8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaur P., Longley M.J., Pan H., Wang H., Copeland W.C.. Single-molecule DREEM imaging reveals DNA wrapping around human mitochondrial single-stranded DNA binding protein. Nucleic Acids Res. 2018; 46:11287–11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qian Y., Johnson K.A.. The human mitochondrial single-stranded DNA-binding protein displays distinct kinetics and thermodynamics of DNA binding and exchange. J. Biol. Chem. 2017; 292:13068–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shereda R.D., Kozlov A.G., Lohman T.M., Cox M.M., Keck J.L.. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008; 43:289–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ciesielski G.L., Kim S., de Bovi Pontes C., Kaguni L.S.. Physical and functional interaction of mitochondrial single-stranded DNA-binding protein and the catalytic subunit of DNA polymerase gamma. Front Genet. 2021; 12:721864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ciesielski G.L., Bermek O., Rosado-Ruiz F.A., Hovde S.L., Neitzke O.J., Griffith J.D., Kaguni L.S.. Mitochondrial single-stranded DNA-binding proteins stimulate the activity of DNA polymerase γ by organization of the template DNA. J. Biol. Chem. 2015; 290:28697–28707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nandakumar D., Pandey M., Patel S.S.. Cooperative base pair melting by helicase and polymerase positioned one nucleotide from each other. Elife. 2015; 4:e06562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stephens K.M., McMacken R.. Functional properties of replication fork assemblies established by the bacteriophage λ O and P replication proteins. J. Biol. Chem. 1997; 272:28800–28813. [DOI] [PubMed] [Google Scholar]
- 39. Yuan Q., McHenry C.S.. Strand displacement by DNA polymerase III occurs through a tau-psi-chi link to single-stranded DNA-binding protein coating the lagging strand template. J. Biol. Chem. 2009; 284:31672–31679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Longley M.J., Ropp P.A., Lim S.E., Copeland W.C.. Characterization of the native and recombinant catalytic subunit of human DNA polymerase gamma: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 1998; 37:10529–10539. [DOI] [PubMed] [Google Scholar]
- 41. Oliveira M.T. Mitochondrial DNA: Methods and Protocols. 2009; NJ Humana Totowa. [Google Scholar]
- 42. Markham N.R., Zuker M.. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005; 33:W577–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ibarra B., Chemla Y.R., Plyasunov S., Smith S.B., Lázaro J.M., Salas M., Bustamante C.. Proofreading dynamics of a processive DNA polymerase. EMBO J. 2009; 28:2794–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith S.B., Cui Y., Bustamante C.. Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 2003; 361:134–162. [DOI] [PubMed] [Google Scholar]
- 45. Bocanegra R., Ismael Plaza G.A., Pulido C.R., Ibarra B.. DNA replication machinery: insights from in vitro single-molecule approaches. Comput. Struct. Biotechnol. J. 2021; 19:2057–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cerrón F., Ibarra B.. Measurements of real-time replication kinetics of DNA polymerases on ssDNA templates coated with single-stranded DNA-binding proteins. Methods Mol. Biol. 2021; 2281:289–301. [DOI] [PubMed] [Google Scholar]
- 47. Morin J.A., Cao F.J., Lázaro J.M., Arias-Gonzalez J.R., Valpuesta J.M., Carrascosa J.L., Salas M., Ibarra B.. Mechano-chemical kinetics of DNA replication: identification of the translocation step of a replicative DNA polymerase. Nucleic Acids Res. 2015; 43:3643–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith S.B., Cui Y., Bustamante C.. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996; 271:795–799. [DOI] [PubMed] [Google Scholar]
- 49. Jarillo J., Morín J.A., Beltrán-Heredia E., Villaluenga J.P., Ibarra B., Cao F.J.. Mechanics, thermodynamics, and kinetics of ligand binding to biopolymers. PLoS One. 2017; 12:e0174830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Morin J.A., Cerrón F., Cao-García F.J., Ibarra B.. Optical tweezers to investigate the structure and energetics of single-stranded DNA-binding protein-DNA complexes. Methods Mol. Biol. 2021; 2281:273–288. [DOI] [PubMed] [Google Scholar]
- 51. Morin J.A., Cerrón F., Jarillo J., Beltran-Heredia E., Ciesielski G.L., Arias-Gonzalez J.R., Kaguni L.S., Cao F.J., Ibarra B.. DNA synthesis determines the binding mode of the human mitochondrial single-stranded DNA-binding protein. Nucleic Acids Res. 2017; 45:7237–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morin J.A., Cao F.J., Valpuesta J.M., Carrascosa J.L., Salas M., Ibarra B.. Manipulation of single polymerase-DNA complexes: a mechanical view of DNA unwinding during replication. Cell Cycle. 2012; 11:2967–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shokri L., Marintcheva B., Eldib M., Hanke A., Rouzina I., Williams M.C.. Kinetics and thermodynamics of salt-dependent T7 gene 2.5 protein binding to single- and double-stranded DNA. Nucleic Acids Res. 2008; 36:5668–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shokri L., Marintcheva B., Richardson C.C., Rouzina I., Williams M.C.. Single molecule force spectroscopy of salt-dependent bacteriophage T7 gene 2.5 protein binding to single-stranded DNA. J. Biol. Chem. 2006; 281:38689–38696. [DOI] [PubMed] [Google Scholar]
- 55. Suksombat S., Khafizov R., Kozlov A.G., Lohman T.M., Chemla Y.R.. Structural dynamics of E. coli single-stranded DNA binding protein reveal DNA wrapping and unwrapping pathways. Elife. 2015; 4:e08193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Johnson D.E., Takahashi M., Hamdan S.M., Lee S.J., Richardson C.C.. Exchange of DNA polymerases at the replication fork of bacteriophage T7. Proc. Natl. Acad. Sci. U. S. A. 2007; 104:5312–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Loparo J.J., Kulczyk A.W., Richardson C.C., van Oijen A.M.. Simultaneous single-molecule measurements of phage T7 replisome composition and function reveal the mechanism of polymerase exchange. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:3584–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ciesielski G.L., Hytönen V.P., Kaguni L.S.. Biolayer interferometry: a novel method to elucidate protein-protein and protein-DNA interactions in the mitochondrial DNA replisome. Methods Mol. Biol. 2016; 1351:223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huber H.E., Tabor S., Richardson C.C.. Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J. Biol. Chem. 1987; 262:16224–16232. [PubMed] [Google Scholar]
- 60. Bratic A., Kauppila T.E., Macao B., Grönke S., Siibak T., Stewart J.B., Baggio F., Dols J., Partridge L., Falkenberg M.et al.. Complementation between polymerase- and exonuclease-deficient mitochondrial DNA polymerase mutants in genomically engineered flies. Nat. Commun. 2015; 6:8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Betterton M.D., Jülicher F.. Opening of nucleic-acid double strands by helicases: active versus passive opening. Phys. Rev. E. 2005; 71:011904. [DOI] [PubMed] [Google Scholar]
- 62. Johnson D.S., Bai L., Smith B.Y., Patel S.S., Wang M.D.. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007; 129:1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Heussman D., Kittell J., Kringle L., Tamimi A., von Hippel P.H., Marcus A.H.. Measuring local conformations and conformational disorder of (Cy3)(2) dimer labeled DNA fork junctions using absorbance, circular dichroism and two-dimensional fluorescence spectroscopy. Farad. Discuss. 2019; 216:211–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jose D., Datta K., Johnson N.P., von Hippel P.H.. Spectroscopic studies of position-specific DNA “breathing” fluctuations at replication forks and primer-template junctions. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:4231–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Phelps C., Lee W., Jose D., von Hippel P.H., Marcus A.H.. Single-molecule FRET and linear dichroism studies of DNA breathing and helicase binding at replication fork junctions. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:17320–17325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Singh A., Pandey M., Nandakumar D., Raney K.D., Yin Y.W., Patel S.S.. Excessive excision of correct nucleotides during DNA synthesis explained by replication hurdles. EMBO J. 2020; 39:e103367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Szymanski M.R., Kuznetsov V.B., Shumate C., Meng Q., Lee Y.S., Patel G., Patel S., Yin Y.W.. Structural basis for processivity and antiviral drug toxicity in human mitochondrial DNA replicase. EMBO J. 2015; 34:1959–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Raghunathan S., Kozlov A.G., Lohman T.M., Waksman G.. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000; 7:648–652. [DOI] [PubMed] [Google Scholar]
- 69. Yang C., Curth U., Urbanke C., Kang C.. Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 Å resolution. Nat. Struct. Biol. 1997; 4:153–157. [DOI] [PubMed] [Google Scholar]
- 70. Curth U., Urbanke C., Greipel J., Gerberding H., Tiranti V., Zeviani M.. Single-stranded-DNA-binding proteins from human mitochondria and Escherichia coli have analogous physicochemical properties. Eur. J. Biochem. 1994; 221:435–443. [DOI] [PubMed] [Google Scholar]
- 71. Lohman T.M., Ferrari M.E.. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994; 63:527–570. [DOI] [PubMed] [Google Scholar]
- 72. Naufer M.N., Morse M., Möller G.B., McIsaac J., Rouzina I., Beuning P.J., Williams M.C.. Multiprotein E. coli SSB-ssDNA complex shows both stable binding and rapid dissociation due to interprotein interactions. Nucleic Acids Res. 2021; 49:1532–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hernandez A.J., Richardson C.C.. Gp2.5, the multifunctional bacteriophage T7 single-stranded DNA binding protein. Semin. Cell Dev. Biol. 2019; 86:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kim Y.T., Tabor S., Bortner C., Griffith J.D., Richardson C.C.. Purification and characterization of the bacteriophage T7 gene 2.5 protein. A single-stranded DNA-binding protein. J. Biol. Chem. 1992; 267:15022–15031. [PubMed] [Google Scholar]
- 75. Marintcheva B., Hamdan S.M., Lee S.J., Richardson C.C.. Essential residues in the C terminus of the bacteriophage T7 gene 2.5 single-stranded DNA-binding protein. J. Biol. Chem. 2006; 281:25831–25840. [DOI] [PubMed] [Google Scholar]
- 76. Hatch K., Danilowicz C., Coljee V., Prentiss M.. Direct measurements of the stabilization of single-stranded DNA under tension by single-stranded binding proteins. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2007; 76:021916. [DOI] [PubMed] [Google Scholar]
- 77. Hatch K., Danilowicz C., Coljee V., Prentiss M.. Measurement of the salt-dependent stabilization of partially open DNA by Escherichia coli SSB protein. Nucleic Acids Res. 2008; 36:294–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Doublié S., Tabor S., Long A.M., Richardson C.C., Ellenberger T.. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998; 391:251–258. [DOI] [PubMed] [Google Scholar]
- 79. Sohl C.D., Szymanski M.R., Mislak A.C., Shumate C.K., Amiralaei S., Schinazi R.F., Anderson K.S., Yin Y.W.. Probing the structural and molecular basis of nucleotide selectivity by human mitochondrial DNA polymerase γ. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:8596–8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lee W., Gillies J.P., Jose D., Israels B.A., von Hippel P.H., Marcus A.H.. Single-molecule FRET studies of the cooperative and non-cooperative binding kinetics of the bacteriophage T4 single-stranded DNA binding protein (gp32) to ssDNA lattices at replication fork junctions. Nucleic Acids Res. 2016; 44:10691–10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lionnet T., Spiering M.M., Benkovic S.J., Bensimon D., Croquette V.. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:19790–19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Manosas M., Xi X.G., Bensimon D., Croquette V.. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010; 38:5518–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Delagoutte E., von Hippel P.H.. Molecular mechanisms of the functional coupling of the helicase (gp41) and polymerase (gp43) of bacteriophage T4 within the DNA replication fork. Biochemistry. 2001; 40:4459–4477. [DOI] [PubMed] [Google Scholar]
- 84. Al-Behadili A., Uhler J.P., Berglund A.-K., Peter B., Doimo M., Reyes A., Wanrooij S., Zeviani M., Falkenberg M.. A two-nuclease pathway involving RNase H1 is required for primer removal at human mitochondrial OriL. Nucleic Acids Res. 2018; 46:9471–9483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Vermulst M., Wanagat J., Kujoth G.C., Bielas J.H., Rabinovitch P.S., Prolla T.A., Loeb L.A.. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat. Genet. 2008; 40:392–394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors (GLC, BI) upon reasonable request.