Abstract
tRNAs are key partners in ribosome-dependent protein synthesis. This process is highly dependent on the fidelity of tRNA aminoacylation by aminoacyl-tRNA synthetases and relies primarily on sets of identities within tRNA molecules composed of determinants and antideterminants preventing mischarging by non-cognate synthetases. Such identity sets were discovered in the tRNAs of a few model organisms, and their properties were generalized as universal identity rules. Since then, the panel of identity elements governing the accuracy of tRNA aminoacylation has expanded considerably, but the increasing number of reported functional idiosyncrasies has led to some confusion. In parallel, the description of other processes involving tRNAs, often well beyond aminoacylation, has progressed considerably, greatly expanding their interactome and uncovering multiple novel identities on the same tRNA molecule. This review highlights key findings on the mechanistics and evolution of tRNA and tRNA-like identities. In addition, new methods and their results for searching sets of multiple identities on a single tRNA are discussed. Taken together, this knowledge shows that a comprehensive understanding of the functional role of individual and collective nucleotide identity sets in tRNA molecules is needed for medical, biotechnological and other applications.
INTRODUCTION
tRNAs are the adapters that decode mRNAs into proteins in the ribosome-dependent translation apparatus, but this classical vision is expanding since tRNAs act in many biological processes (1–3). However, translation remains the cornerstone relying mainly on the fidelity of tRNA aminoacylation by aminoacyl-tRNA synthetases (aaRSs). How synthetases achieve fidelity/specificity was perhaps the first protein–RNA recognition problem to be seriously investigated. It led to the concept of tRNA identity, which refers to the amino acid ligated at the 3′ end (4,5). Currently, this issue is supported by a robust theoretical and experimental background. Due to their interaction with multiple components of the translation machinery, tRNAs have undergone significant constraints on their primary and secondary structures during evolution to retain structural similarity and the ability to interact with the ribosome-dependent translation machinery (Figure 1) (6). Early studies in the 1970s pinpointed the importance of the acceptor end and anticodon of tRNAs for recognition by aaRSs (7,8). It was also found that the specificity of aaRSs for amino acid activation and aminoacylation of tRNA is rather low (in other words, aaRSs catalyze amino acid misactivation and mischarging of tRNAs) and that accurate tRNA charging relies more on kinetic effects than on discrimination among cognate and non-cognate tRNAs through binding affinity (9). In addition, it was found that the binding of tRNAs to aaRSs, followed by the correct aminoacylation of tRNA, relies on a limited number of nucleotides called ‘identity elements’ supporting the ‘RNA operational code’ theory, associated with the idea of a second genetic code (10). The G3·U70 base pair in Escherichia coli tRNAAla was the first identity determinant experimentally validated (11,12). This pioneering result was followed by the characterization of determinant sets that specify the identity of all standard tRNA specificities, mainly in the tRNAs of E. coli and Saccharomyces cerevisiae. These early results were obtained mainly by in vitro methods and less often by in vivo methods that also measure the functional importance and strength of determinants (4,13). Two decades later, numerous results have enriched knowledge in the field of tRNA identity, shedding new light on the importance of the fidelity of the aminoacylation reaction. A full analysis incorporating these features and novelties is presented here.
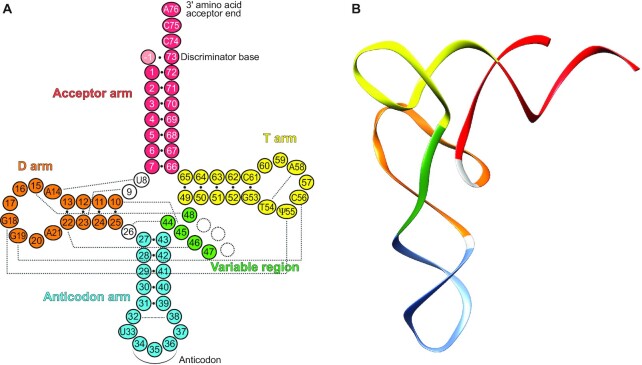
Figure 1.
Cloverleaf folding of tRNA and its three-dimensional L-shaped organization. The color code highlights the structural domains in tRNA. (A) The standard cloverleaf structure of cytosolic tRNAs and the conventional numbering system are used. Conserved nucleotides are explicitly indicated. The variable region (nucleotides 44–48) encompasses the long extra arm of tRNALeu, tRNASer and tRNATyr. The · symbol indicates Watson–Crick base pairings (including G·U pairs); dotted gray lines indicate other pairings important for tRNA L-shaped architecture. (B) Three-dimensional L-shaped structure of tRNAPhe (1ehz) showing the folding of the different arms.
IDENTITY ELEMENTS FOR tRNA AMINOACYLATION IN STANDARD CYTOSOLIC SYSTEMS
Positive identity elements in standard cytosolic tRNAs
General considerations
The panel of validated aminoacylation identity elements (nucleotides and structural features) also called ‘determinants’ has been substantially enriched over the last two decades. Typically, identification of determinants is achieved by comparing the aminoacylation capacities of mutant and native tRNAs determined by measuring their catalytic efficiencies which are related to the kcat/KM ratios, where kcat is the catalytic rate constant and KM is the Michaelis constant representing an approximation of the inverse of the tRNA’s affinity for an aaRS. Comparison of the aminoacylation capacities of mutant and native tRNAs defines the ‘loss’ parameter ‘L’ = (kcat/KM)native/(kcat/KM)mutant which reflects the loss of catalytic efficiency of the mutated tRNA. An ‘L’ <10 is generally considered a minor effect, while a larger ‘L’ >1000 is considered a major effect, with intermediate values being considered intermediate effects (4). Additionally, for E. coli tRNAs, strengths are also determined by tRNA suppressor-based genetic assays. However, despite its great potential, the in vitro SELEX approach that allows selection of aminoacylated tRNAs from a pool of randomized sequences is seldomly used (see below for Asp and Phe identities) (13,14). With the increasing number of studies in different organisms, the diversity of identity elements within each system has become apparent, challenging the idea of universal identity rules. The identity of mitochondrial tRNAs (mt-tRNAs) and atypical or standard tRNAs charged by aaRSs is covered in dedicated sections. Due to the abundance of literature covering the subject, some older references will only be cited sparingly; for additional references, see (4).
Below, the positive identity elements for tRNA aminoacylation, validated by functional assays, are shown in Table 1 and schematized in Figure 2. The identity determinants are discussed following the classification into classes and subclasses of aaRSs, namely subclass Ia (Arg-, Cys-, Ile-, Leu-, Met- and ValRS), subclass Ib (Glu- and GlnRS), subclass Ic (Trp- and TyrRS), subclass IIa (Ala-, Gly-, His-, Pro-, Ser- and ThrRS), subclass IIb (Asp-, Asn- and LysRS) and subclass IIc (PheRS). This classification is based on common structural characteristics, notably the oligomeric status of aaRSs, mainly monomeric for class I aaRSs and dimeric for class II aaRSs, which are considered among the most ancient in evolution (15). However, in some tRNA families, the oligomeric status of aaRSs is not conserved. Some class Ia Leu- and MetRSs and all class Ic Trp- and TyrRSs are dimers. Class IIa GlyRSs are dimers in Eukarya and Archaea and either dimers or heterotetramers in Bacteria. Tetrameric AlaRSs, GlyRSs and PheRSs are pseudodimers (15,16).
Table 1.
Positive identity determinants for aminoacylation experimentally characterized in tRNAs from Bacteria, Eukarya (cytosol) and Archaea
| tRNA families | tRNA domains | |||
|---|---|---|---|---|
| Acceptor branch | Core region | Anticodon branch | ||
| Arg a | (Bac) | A/G73, U73, G2–C71, C3–G70 | A20 | C35, U/G36 |
| (Euk) | —73b | U20c, C20a, U20a | A/C35, A/U/G36, A38 | |
| (Arc) | … | … | … | |
| Cys | (Bac) | U73, G2–C71, C3–G70 | TId | G34, C35, A36 |
| (Euk) | U73 | G15:G48, A13:A22 | G34, C35, A36 | |
| (Arc) | U73 | G15, A47 | G34, C35, A36, G37c | |
| Ile | (Bac) | A73, C4–G69 | U12–A23, G16, U20c, U21c | C29–G41, C/G34c, A35, U36, A37c, A38 |
| (Euk) | … | … | N34c, A35, U36 | |
| (Arc) | … | … | … | |
| Leu a | (Bac) | A73 | TId, A20a, largeVRe | … |
| (Euk) | A73, C3–G70, A4–U69, G5–C68 | largeVRe, U8:A14 | … | |
| (Arc) | A73 | largeVRe | … | |
| Met f | (Bac) | A73, (G2–C71) (C3–G70) U4–A69, A5–U68 | … | (C32) (U33) C34, A35, U36 (A37) |
| (Euk) | A73 | TId | C34, A35, U36 | |
| (Arc) | … | … | … | |
| Val | (Bac) | A73, G3–C70, U4–A69 | … | A35, C36 |
| (Euk) | A73 | … | A35 | |
| (Arc) | … | … | G34, A35, C36 | |
| Glu | (Bac) | –73b, G1–C72, U2–A71 | TId, Δ47 | U34c, U35, A37, C36 |
| (Euk) | … | … | … | |
| (Arc) | … | … | … | |
| Gln | (Bac) | G73, U1–A72, G2–C71, G3–C70 | G10 | U34c, U35, G36, A37, U38 |
| (Euk) | … | … | … | |
| (Arc) | … | … | … | |
| Trp | (Bac) | G73, A1–U72, G2–C71, G3–C70, G4–C69, G5–C68g | A9 | C34, C35, A36 |
| (Euk) | A73, G1–C72, U5·G68i/G5–C68 | … | C34, C35, A36 | |
| (Arc) | A73, G1–C72, G2–C71 | … | C34, C35, A36 | |
| Tyr | (Bac) | A73 | … | G34c, U35 |
| (Euk) | A73, C1–G72 | … | G34, U35c | |
| (Arc) | A73, C1–G72 | … | G34c, U35, A36 | |
| Ala | (Bac) | A73, G2–C71, G3·U70, G4–C69 | G20 | … |
| (Euk) | G3–U70 | … | … | |
| (Arc) | … | … | … | |
| Gly | (Bac) | U73, G1–C72, C2–G71, G3–C70 | … | C35, C36 |
| (Euk) | A/U73, G1–C72, C2–G71, G3–C70 | (G10:Y25):G45 | C35, C36 | |
| (Arc) | A73, C2–G71, G3–C70 | … | C35, C36 | |
| His | (Bac) | G-1, C73 | … | G34, U35, G36 |
| (Euk) | G-1, A73 | … | G34, U35 | |
| (Arc) | —-1b, C73 | C50–G64 | G29–C41 | |
| Pro | (Bac) | A73, G72 | G15:C48, U17a, G49 | G35, G36, G37 |
| (Euk) | C73 | … | G35, G36 | |
| (Arc) | A73, G1–C72, G2–C71, G3–C70 | … | G35, G36 | |
| Ser | (Bac) | G73h, C72, G2–C71, R4–Y69 | C11–G24; largeVRg | G30–C40 |
| (Euk) | G73 | largeVRe | … | |
| (Arc) | … | … | … | |
| Thr | (Bac) | G1–C72, C2–G71, G4–C69, G5–C68 | … | G34, G35, U36 |
| (Euk) | U73, G1–C72, U3–A70, G5–C68 | … | G35, U36 | |
| (Arc) | —73b/U73h, G1–C72, C2–G71, C3–U70 | … | G34, U35, C36 | |
| Asp | (Bac) | G73 | G10 | G34c, U35, C36, C38 |
| (Euk) | G73 | G10·U25 | G34, U35, C36 | |
| (Arc) | … | … | … | |
| Asn | (Bac) | G73 | … | G34, U35, U36, A37c |
| (Euk) | … | … | … | |
| (Arc) | … | … | … | |
| Lys | (Bac) | A73 | … | U34, U35, U36 |
| (Euk) | … | … | … | |
| (Arc) | … | … | … | |
| Phe | (Bac) | A73 | C10–G25, U20c, U45, U59 | A26:G44, G34, A35, A36 |
| (Euk) | A73 | G20 | G34, A35, A36, A37 | |
| (Arc) | A73 | C13–G22, G20 | G34, A35, A36 | |
The 20 tRNA families designated by their amino acid identity are displayed according to the class and subclass of their corresponding aaRSs. The position of the determinants in the sequence of the tRNA acceptor branch, the core region and the anticodon branch is shown, with standard numbering.
Bac, Bacteria; Euk, Eukarya; Arc, Archaea; N, nucleos/tide; R–Y or Y–R (with R for purine and Y for pyimidine), Watson–Crick pairs; G·U, non-Watson-Crick pair; modified residues are shown in standard abbreviations; TI, tertiary interaction (with atypical N:N pairing); Δ, missing residue or domain; VR, variable region; …, no data; n.d., not determined.
aInsertion of an additional N in the D loop of some tRNA isoacceptors; bposition not involved in identity; cmodified Ns; dspecific N as determinants in TIs; especific N as determinants in large VRs; fdeterminants of E. coli initiator tRNAMet are given in parentheses; gU5–G68 is determinant in Homo sapiens and G5–C68 in B. subtilis tRNATrp; hU73 is determinant in Haloferax volcanii tRNAThr but not in A. pernix tRNAThr.
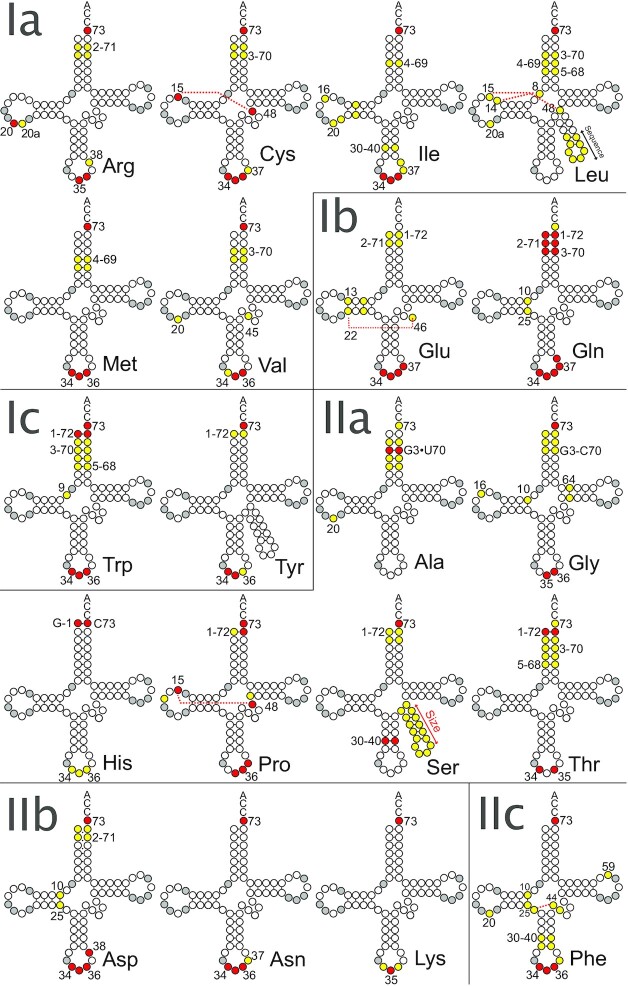
Figure 2.
Schematic representation of the distribution of identity elements across the 20 isoacceptor tRNA families. The tRNAs are presented by class and subclass of aaRSs. Invariant positions are represented in gray. The positions in red are the conserved or nearly conserved strong identity elements. Positions in yellow are weak determinants or determinants not conserved in the three domains of life. The red dashed lines indicate tertiary interactions involved in identity. For tRNALeu, the yellow double arrow indicates the low importance of the sequence elements in the extra arm; for tRNASer, the red double arrow indicates that the size rather than the sequence of the extra arm is the major identity element.
Due to the degeneracy of the genetic code, isoacceptor tRNAs can have different anticodons, for example up to six for tRNAArg, tRNALeu and tRNASer, but only one for tRNAMet and tRNATrp. This results in a variable distribution of identity elements in the tRNA structure, with or without selection of the anticodon as a main element. Overall, determinants in bacterial and eukaryal tRNAs are well documented, in contrast to poorly known determinants in archaeal tRNAs (Table 1). In the Appendix section at the end, we provide a list of definitions of common terms used throughout the review.
Conservation and diversity of identity sets
Identity sets recognized by subclass Ia aminoacyl-tRNA synthetases
Arginine identity
The arginine identity is mainly defined by A20 in the variable pocket of the D loop, C35 and U/C36 in the anticodon and discriminator base A/G73 (17–21). The arginine identity set was enriched by the addition of U20a and A38 elements in plant tRNAsArg (22). A20 is the main determinant of arginylation by E. coli ArgRS, as shown with a panel of E. coli and plant tRNAArg isoacceptors, despite variations in their frameworks (22). This is unique among the 20 tRNA families and is supported in the structures of the tRNAArg:ArgRS complexes (23).
In contrast, position 20 in the D loop of S. cerevisiae tRNAArg isoacceptors participates only marginally in identity, while C35 followed by G36 or U36 in the anticodon loop are the prominent determinants (24). Here, arginine identity is defined by two distinct combinations of determinants in the anticodon loop of the four yeast tRNAArg isoacceptors. Surprisingly, the arginine identity in yeast is related to that of aspartate and does not require the participation of the discriminator base at position 73. The cryptic aspartate identity was carefully studied in the minor tRNA4Arg(CCG) in which a specificity switch to aspartate was obtained with only the two mutations C38 and G73, thereby allowing discrimination between the three major tRNA1–3Arg and the minor tRNA4Arg isoacceptor (25). It is worth highlighting the importance of position 20 in controlling species specificity (A20 in E. coli and C20 in yeast) (26) and the association of A20 and C35 in hamster (27). The crystal structures of the yeast and Thermus thermophilus tRNAArg:ArgRS complexes (28,29) show how D20 and C20a in yeast and A20 in T. thermophilus control arginine identity. Finally, a comparative search of arginine identity elements performed in two related plant taxa, soybean and jack bean, concluded that the main arginine determinants are also A20 and C35 in plants, mammals and E. coli, but species variability occurs at other positions (22).
Cysteine identity
The discriminator U73 and the anticodon GCA are major identity determinants in E. coli and yeast tRNACys, although minor determinants occur in the yeast accepting stem and their strength is variable (30–32). The discriminator U73 alone is sufficient to confer cysteine identity to minihelices (33), suggesting an ancestral role in the evolution of cysteine identity. In addition, the atypical 15–48 trans-Watson–Crick Levitt pair (G15–G48 instead of R15–Y48) in the core region of E. coli tRNACys is a strong determinant for aminoacylation (34). This atypical pair affects both the structure and function of E. coli tRNACys (34,35) and is conserved in a few bacterial species, such as Haemophilus influenzae tRNACys (36). In Methanosarcina mazei, one of the few methanogenic Archaea encoding CysRS, serylation of tRNACys requires m1G37 that is also a serylation determinant for SepRS (37).
Isoleucine identity
The identity elements of the isoleucine system were among the first known and include strong determinants in the discriminator and anticodon positions (38–40). In E. coli, minor determinants in stem regions of tRNAIle (C4–G69, U12–G23 and C29–G41) (41) and discrete determinants in the D loop (G16, D20 and D21) are crucial for isoleucylation and editing (42,43). The determinants in the anticodon loop are of interest because of the closely related Ile (AUU, AUC and AUA) and Met (AUG) codons and the frequent modifications at wobble position 34. In E. coli, the minor tRNA2Ile with an anticodon mimicking the CAU anticodon of tRNAMet carries a lysine-substituted C34 named lysidine (L or k2C) which confers the ability to decode isoleucine AUA codons (38). This modified L34 residue in E. coli tRNA2Ile is an isoleucine identity determinant since its replacement by unmodified C impairs isoleucine-accepting activity and surprisingly confers methionine activity. In contrast, it is an antideterminant that prevents the decoding of the methionine AUG codon and its recognition by MetRS (38). This dual property is likely to be generalized, as lysidine or a lysidine mimic is found at position 34 in other Bacteria, such as in Lactobacilli tRNAsIle (44,45) and in Archaea where the modified base is called agmatidine (or C+) (46). In Eukarya, I34 in the anticodon of yeast tRNAIle is also a determinant (47). At position 37, t6A37 is a strong determinant for tRNA isoleucylation by bacterial-type but not eukaryal-type IleRSs (48) except human cyto-IleRS (and IleRSs in related mammalian taxa) (49). In Table 2, we have compiled the currently known post-transcriptional modifications that play a role in the expression of tRNA identity (50).
Table 2.
Modified tRNA nucleosides that act as identity elements for aminoacylation
| Short name | Full name | Nucleoside position | tRNA | Role in human diseasesd |
|---|---|---|---|---|
| I | Inosine | 34 | Ile | Yese |
| k2C or L | 2-Lysidine or 2-lysyl cytidine | 34 | Ile | – |
| mnm5s2U | 5-methylaminomethyl-2-thio uridine | 34 | Glu | Yes |
| C+ | Agmatidine (an L mimic) | 34 | Ile (Archaea) | – |
| Q | Queuosinea | 34 | Tyr | Yes |
| Ψ | Pseudouridineb | 35, 36 | Ile, Tyr | Yes |
| t6A | N 6-Threonylcarbamoyl adenosine | 37 | Ile | Yes |
| m1G | 1-Methyl guanosine | 37 | Asp, Cys, Pro | Yes |
| yW | Wybutosinec | 37 | Phe | Yes |
| m5C | 5-Methyl cytosine | 38 | Asp | Yes |
aHypermodified guanosine or 7-{[(4,5-cis-dihydroxy-2-cyclopenten-1-yl)amino]methyl}-7-deazaguanosine; bribose is linked to uracil position C5 instead of uracil position N1 in uridine; cheavily hypermodified guanosine; dfor an associated role in diseases due to aberrant mt- or cyto-tRNAs, see (422); eonly in cyto-tRNAIle (422)
Leucine identity
Cytosolic tRNAsLeu share with tRNASer and tRNATyr species a long variable arm typical of class II tRNAs. Given this unusual structural feature, the role of the variable arm in the identity was tested very early on. Nucleotide swap experiments resulted in identity changes from serine to leucine (51) and vice versa from leucine to serine (52). Leucine identity was further investigated in vitro or in vivo in five models covering the three domains of life, notably bacterial E. coli (53) and the primitive hyperthermophile Aquifex aeolicus (54), archaeal Haloferax volcanii (55), and eukaryal S. cerevisiae (56,57) and human (58,59) tRNAs.
In E. coli tRNALeu, the discriminator base A73 but not the anticodon, or the long variable arm, is crucial for leucine identity (60). The absence of an identity element in the anticodon was not surprising since tRNALeu belongs to the six codon families, and five isoacceptors of tRNALeu exist in E. coli with, however, a common A35 nucleotide. Using SELEX on a library of E. coli tRNA1Leu randomized in the D loop, T loop and variable arm revealed sequence elements crucial for leucylation in the hinge region. The trans-Levitt pair A15–U48, nucleotides G18G19 and A20a in the D loop were shown to be essential elements. Although the long variable arm is a characteristic feature of the tRNALeu structure, its sequence and folding were not correlated with the leucylation activity (61). A similar conclusion was obtained by additional studies performed in vitro and in vivo (62). Other studies showed that E. coli tRNALeu variants missing the anticodon arm and the long variable arm remain active. The aminoacylation of such truncated tRNAsLeu was abolished when the discriminator A73 was replaced with C73 or when the tertiary interactions between the D and T loops were disrupted, suggesting that these identity elements were still active in the minimal tRNAs (53). More focused mutagenesis of E. coli tRNALeu explicitly demonstrated the importance of tertiary interactions G18:U55, G19–C56 and U54:A58 in leucine identity (63). It can be noted that mutants of E. coli and A. aeolicus tRNALeu that remain active in charging also remain active in editing (53,54).
In S. cerevisiae tRNALeu(UAG), insertion of a U residue in the loop region of its long variable arm destroys the stable tetraloop U47aUCG47d, forming an additional U·G pair in the variable arm that confers serine acceptance to the variant (56). In another study performed on S. cerevisiae tRNALeu(UAG), seven nucleotides were shown to decrease steady-state levels of tRNA leucylation in vivo in a triple tRNALeu(UAG) knockout strain. Strong impact was observed with mutants G18, m1G37, Y55 and A73, and more moderate decreases with mutants of nucleotides G2, G30, C61 and C62. However, A35 or G36, which are essential for in vitro leucine identity, play only a marginal role in the leucylation of tRNALeu(UAG)in vivo, but are essential for decoding (57).
In the archaeon H. volcanii, leucine identity is defined by the discriminator A73 and the long variable arm of tRNALeu, especially the specific loop sequence A47CG47d and U47h at the base of the variable arm. Interestingly, the unmodified transcript maintains full cognate leucine acceptance (55).
Together, these results show a mosaic of different situations that can be summarized in a few points. (i) Minimal tRNA variants with the discriminator determinant A73 are leucylated in the presence of specific tertiary interactions between the D and T loops in E. coli and H. sapiens (53,64). (ii) Switches of leucine to serine identity can be achieved in different structural contexts after replacing the discriminator A73 with G73 (51,64). (iii) Structural variations are tolerated in the long variable arm of E. coli tRNALeu (53). (iv) Sequence-specific recognition of the long variable arm is unique to Archaea (55). From the perspective of aaRS recognition, aminoacylation of class II tRNAs predicts the importance of the discriminator base, tertiary nucleotides (at positions 15–48 and 59), the D loop (α and β subdomains), the number of bases and unpaired bases in the variable arm and position 37 in the anticodon loop (55), features that are currently conserved in known tRNA sequences (65).
Methionine identity
Early studies performed on variants of tRNAfMet enzymatically synthesized in vitro showed that recognition of tRNAfMet requires highly specific interactions of MetRS with functional groups on the nucleotide bases of the anticodon sequence (66). Mutant tRNA transcripts were later prepared that contained normal and interchanged anticodon sequences. It was confirmed that a tRNAVal variant with a CAU methionine anticodon is charged by E. coli MetRS with the same catalytic efficiency as native tRNAMet. This indicates that the anticodon contains sufficient information to distinguish methionine and valine tRNAs with high fidelity (67). The methionine recognition system is special since it governs the aminoacylation of elongator and initiator tRNAs which play different roles in protein synthesis and interact with different partners. Although the general principles of recognition of the elongator tRNA by MetRS and the translation machinery are similar in the three domains of life, those of the initiator tRNA show important phylogenetic differences. Formylation of the charged methionyl moiety exists in Bacteria, but not in Eukarya or in Archaea because of the lack of the tRNAfMet-formyltransferase recognition elements (the 1–72 mismatched pair and the R11:Y24 base pair disappeared) (68,69). However, the methionine anticodon CAU is important for in vivo methionylation of H. volcanii tRNAMet. Lastly, E. coli MetRS utilizes the anticodon nucleotides as determinants to mismethionylate E. coli tRNAArg(CCU) and tRNAThr(CGU), defining the concept of mischarging identity for MetRS (70).
Valine identity
Valine identity is closely related to the isoleucine and methionine identities, due to the common discriminator A73 and anticodon A35 determinants. This explains the efficient mischarging of tRNAIle and tRNAMet by ValRS (71). G20, in the variable pocket, and G45, in the central core of tRNA, are minor recognition elements. Mutations at either the G3–C70 or U4–A69 base pairs in the acceptor stem also affect the activity (72), and introduction of a G·U pair at the third or fourth position of the acceptor stem of E. coli tRNAVal significantly impairs its activity (73). Transplantation of A35, C36, A73, G20 and G45 in E. coli tRNAPhe containing a regular A-RNA acceptor helix and an tRNAVal anticodon stem is necessary to confer effective valine acceptance (74). The importance of the anticodon is also demonstrated in Archaea (68). Atomic group mutagenesis suggests that the unprotonated N1 position in A76 of E. coli tRNAVal, acting as a H-bond donor, is an essential valylation determinant (75).
Identity sets recognized by subclass Ib aminoacyl-tRNA synthetases
Glutamate identity
Glutamate identity is one of the few examples where a modified base is a major identity determinant, in particular the hypermodified thiol part of mnm5s2U34 at the wobble position of the anticodon (76–78). This role is reinforced by A37 next to the anticodon (76). In E. coli, the N73 discriminator position is not required, but the base pairs G1–C72 and U2–A71 in the acceptor stem are weak identity determinants. In summary, the strong identity determinants are U34, U35, C36 and A37 in the anticodon loop. In addition, the base pair U11–A24, the base-triple U13–G22:A46 and the absence of residue 47 serve as major identity determinants in tRNAGlu, presumably for the formation of structural features that are recognized by GluRS (79).
Glutamine identity
In early work, an E. coli tRNAfMet derivative (an amber suppressor tRNAfMet) containing an anticodon sequence altered with U34 showed a large increase in glutamine acceptance and a large decrease in methionine acceptance (80). The result was consistent with the previously reported aminoacylation with glutamine of the amber suppressor of tRNATrp (81). Therefore, it was concluded that the central position of the anticodon is involved in tRNA substrate recognition by GlnRS. Analysis of the amber tRNASer showed that the substitution of two base pairs in the acceptor helix changed the aminoacylation specificity from serine to glutamine. The importance of base pairs 1–72 and 3–70 for the identity of glutamine was thus established (82). Now, it is accepted that the elements of E. coli glutamine identity are located in the anticodon and acceptor stem regions, including the discriminating base (83). Comprehensive kinetics revealed that interactions with the acceptor stem act as strong determinants of tRNA specificity to correctly position the 3′-CCA end in the active site. The 10–25 base pair and central U35 are also important binding sites for GlnRS, with G36 contributing to both binding and recognition (84). Mutations of these determinants primarily affect kcat, resulting in up to a 105-fold loss of catalytic activity, far greater than in most other systems (83,85).
Many microorganisms such as the pathogenic bacterium Helicobacter pylori do not have a GlnRS but have two divergent glutamyl-tRNA synthetases: GluRS1 and GluRS2. While GluRS1 aminoacylates tRNAGlu as a canonical GluRS, GluRS2 has lost the ability to charge cognate tRNAsGlu and mischarges tRNAGln to form Glu-tRNAGln (86,87). GluRS2 rejects tRNAsGlu by predominantly looking at antideterminants located in the acceptor stem, at the first base pair position. The base pair U1–A72 is found in tRNAGln, while tRNA1Glu and tRNA2Glu both contain a G1–C72 base pair. The importance of this position is conserved throughout indirect aminoacylation (87). Amidotransferases that convert Glu-tRNAGln into Gln-tRNAGln also rely on the U1–A72 base pair for recognition of Glu-tRNAGln. In another study, identity elements were revealed in tRNAGln from Bacillus subtilis and E. coli for mischarging by B. subtilis GluRS, the main recognition element for GluRS being a modified U at the 34th position (88).
Identity sets recognized by subclass Ic aminoacyl-tRNA synthetases
Tryptophan identity
Data have been collected in five different taxa: Aeropyrum pernix, Arabidopsis thaliana, B. subtilis, E. coli and H. sapiens, covering the three domains of life. Early studies revealed that the discriminating base, the nucleotides of the anticodon and the first base pair of the acceptor stem are the major identity elements (89–93). More recent studies showed that in A. thaliana, the discriminator A73 and the C34C35A36 anticodon are also strong determinants. Mutation of the tRNATrp CCA anticodon to an amber CUA anticodon allows tRNATrp(CUA) to be mischarged in vitro and in vivo by plant LysRS (94). In H. sapiens, the major determinant is A73, in contrast to E. coli and B. subtilis where it is G73 (95). Three base pairs in the acceptor stem of B. subtilis tRNATrp (G2–C71, G3–C70 and G4–C69), selected from a random library, are required for efficient aminoacylation by cognate TrpRS (96). These pairs, found in the native sequence of B. subtilis tRNATrp, are strong identity determinants with a strength between that of the major G73 and the minor A1–U70 and G5–C68 elements (96). In A. pernix hyperthermophile aerobic archaeal tRNATrp, the main identity elements are the discriminator A73, the anticodon bases C34 and C35, and the base pair G1–C72. The G2–C71 pair only plays a minor role and acts by a KM effect (97). Together, these results suggest that species differences in tryptophan identity are modulated by minor determinants.
Tyrosine identity
tRNATyr is a type II tRNA with a long extra arm. While the two other type II tRNAs (tRNALeu and tRNASer) carry a long variable arm in all organisms and organelles except in animal mitochondria, the long variable arms of tRNAsTyr have been lost twice: early after the separation of Bacteria from Archaea and Eukarya, and later in parallel with comparable changes in tRNAsLeu and tRNAsSer in animal mitochondria (98). Studies carried out on the tyrosine identities of E. coli and S. cerevisiae have shown the critical importance of the A73 discriminator and the U35 anticodon for tyrosylation [reviewed in (4,99)]. These identity elements, also found in tRNAAla, are sufficient for the easy transformation of a tRNATyr into a tRNA accepting both alanine and tyrosine by simple substitution of the U3–A70 pair with the G3·U70 alanine identity pair (100). Mutated tRNA with ‘double identity’ was quantitatively aminoacylated with either of the two amino acids in vitro (100). As a class II tRNA, the identity of tRNATyr is close to that of tRNALeu and tRNASer, as demonstrated by identity nucleotide transplantation experiments of several tertiary elements between these tRNAs (55,60). Tyrosine identity is also close to phenylalanine identity since yeast tRNATyr with a phenylalanine GAA anticodon is mischarged by yeast PheRS, showing the major role of the anticodon in identity (101). Currently, determinants for tyrosylation are known for cytosolic tRNAs from five taxa (A. pernix, E. coli, Methanocaldococcus jannaschii, Pneumocystis carinii and S. cerevisiae) representing the three domains of life. In most of these systems, a small number of nucleotides in tRNAsTyr govern tyrosine identity in conjunction with adaptation of TyrRSs. Interestingly, the tyrosine system shows that identity elements are not always conserved in evolution. While the A73 discriminator is conserved as an identity element in all three domains of life, the first pair 1–72 is not conserved, although it has retained an identity function (102–106). In Archaea and Eukarya, the identity element is a C1–G72 pair, whereas in Bacteria it is a G1–C72 pair. This is accompanied by structural changes in TyrRSs to ensure recognition of the first base pair in the tRNA acceptor stem, notably in archaeal TyrRSs of A. pernix, Archaeoglobus fulgidus and Pyrococcus horikoshii (107), and in eukaryal TyrRS of S. cerevisiae (108).
Identity sets recognized by subclass IIa aminoacyl-tRNA synthetases
Alanine identity
The major identity determinant of tRNAAla was discovered in 1988 when it was shown that the G3·U70 wobble pair governs the specific charging of E. coli tRNAAla by alanine (11,12). This pioneering work was followed by numerous studies in several laboratories (109–122). The determinant role of G3·U70 is conserved in evolution, as shown by more recent studies. Conversion of G3·U70 to A3–U70 or G3–C70 eliminates alanylation by insect or human AlaRSs (123) and the introduction of a G3·U70 pair in a tRNATyr confers acceptance of alanine (100). In A. thaliana tRNAAla, conversion of the G3·U70 identity pair to G3–C70 blocks alanylation, while conversion of G3–C70 to G3·U70 in tRNAPhe allows this mutated plant tRNAPhe to be an efficient substrate of plant AlaRS (124). A genetic selection performed on an E. coli tRNAAla knockout strain revealed that tRNAAla mutants having a variety of sequence combinations in the acceptor stem region can support knockout cell growth. Several mutant tRNAs having substantial activity lacked the G·U wobble pair and instead contained mispairings C–C, C–U or G–A at the 3–70 position (125). In line with this result, atomic group mutagenesis was applied to discriminator A73 and base pair 2–76 that severely affected AlaRS recognition when mutated in the context of minihelices. The results revealed a subtle interplay between positive and negative effects on transition state stabilization of the alanylation reaction (126,127). It should be noted that although G3·U70 is the main identity element of tRNAAla, distinct modes of G·U pair recognition have been characterized by comparing bacterial AlaRS with eukaryotic/archaeal AlaRS (128–130). Finally, very rare exceptions occur in mitochondria, where the G3·U70 pair may be absent or translocated (131–133) (details below in the section dedicated to mitochondrial identities).
Glycine identity
Glycylation systems are very complex because neither the oligomeric structure of GlyRS (dimers in Eukarya and Archaea and dimers or heterotetramers in Bacteria) nor the discriminator bases (U73 in bacterial tRNAGly and A73 in eukaryal and archaeal tRNAsGly) are conserved in evolution. This diversity impacts glycine identity, since the consensus sequence of tRNAsGly only contains a G1–C72 pair in the acceptor arm, C35 and C36 in the anticodon and the (G10–Y25):G45 triple involved in tRNA folding (134). Early studies showed that mutation of the U73 discriminator base of E. coli tRNAGly reduced glycine acceptance, revealing that it acts as an identity element (135). A similar result was observed in vivo using the amber suppressor of tRNAGly (136). The reduction of the suppressor activity in vivo was even much more severe than that in the in vitro aminoacylation experiment (136). Mutation studies on tRNAGly of E. coli, T. thermophilus and the yeast S. cerevisiae showed that the identity set also contains the first and second base pairs, G1–C72 and C2–G71, in the acceptor stem, and the anticodon nucleotides C35 and C36. However, differences exist between yeast and the two bacteria in the acceptor stem. The first base pair, G1–C72, is important for glycylation in E. coli and T. thermophilus, whereas the second and the third base pairs are important in yeast (137). Thermus thermophilus GlyRS also recognizes the C50–G64 pairs together with the G10, U16, C35 and C36 single residues (138). The glycine identity set of archaeal A. pernix tRNAGly includes C35 and C36 from the anticodon and the C2–G71 and G3–C70 base pairs from the acceptor stem, but does not require discriminator base A73 (139). Currently, no examples of higher eukaryal tRNAGly identity elements are known.
Histidine identity
This identity is unique due to the presence of an additional residue G−1 in all three domains of life (although some organisms lack this residue; see below). The essential role of G–1 in identity and possibly of the G–1–C73 pair, together with that of the anticodon, was first demonstrated for E. coli (140) and S. cerevisiae tRNAHis (141). HisRS efficiently aminoacylates minihelix (13 bp) and microhelix (8 bp) RNAs resembling the tRNA acceptor stem which contain G–1–C73. Transplantation of this base pair is also sufficient to confer histidine acceptance to a tRNAAla minihelix (110). These identity conversions mediated by the G–1–C73 base pair were exploited to isolate secondary site revertants in E. coli HisRS which restore histidine identity to a tRNAHis suppressor carrying a G–1·U73 pair. The revertant substitutions were found in the anticodon-binding domain located in the C-terminal domain of HisRS, demonstrating that the anticodon of tRNAHis also plays an important role in tRNA selection in vivo (142). In the hyperthermophile archaeon A. pernix, G–1 but also A–1 and C–1 can be recognized by cognate HisRS together with a weak participation of the discriminator base C73, in contrast to the anticodon that is not recognized (143). Atomic group mutagenesis was carried out at the –1 to 73 position of chemically synthesized microhelixHis substrates. The results suggested that the G–1 base serves to position the 5′-monophosphate, which is critical for aminoacylation. Furthermore, the 6-keto oxygen of G–1 and the major groove amine of C73 contribute to HisRS recognition. This supports the existence of a canonical G–1–C73 pair (144).
In most Bacteria and Archaea, G–1 is encoded by the tRNA gene. In contrast, G–1 incorporation occurs post-transcriptionally in Eukarya and is catalyzed by tRNAHis-guanylyltransferase (Thg1), an enzyme that catalyzes the addition of nucleotides in the 3′–5′ direction, in contrast to all known DNA and RNA polymerases (145,146). Despite the lack of sequence similarity, Thg1 enzymes share structural homology with canonical 5′–3′ DNA polymerases and recognize the tRNAHis anticodon during the maturation process.
The crystal structure of T. thermophilus HisRS complexed with tRNAHis reveals that G–1 recognition is principally based on non-specific interactions with this base and is made possible by an enlarged binding pocket (for the extra base G–1) absent in other aaRSs, while the anticodon triplet makes additional specific contacts with the enzyme. The structural complementarity between the 5′ extremity of tRNA and the enzyme is probably a result of co-evolution of both tRNAHis and HisRS (147). Divergence of bacterial histidylation rules was observed in some groups of α-Proteobacteria. In this clade, neither the genetically encoded G–1 nor C73, which are essential for histidine identity in E. coli, is present. Instead, tRNAHis contains A73, which in yeast is a less essential but still important element of histidine identity. In parallel, the motif II loop in HisRS that recognizes the discriminator base 73 of tRNAHis covaries perfectly with the presence of C73 (148). This was experimentally validated in Caulobacter crescentus, a G–1-lacking α-Proteobacterium, whose in vitro identity is based on A73, U72 and the anticodon (149). Similarly, in some protists, tRNAHis lacks G–1 (150), and these organisms do not possess the tRNAHis-guanylyltransferase gene. In the case of the protists Trypanosoma brucei and Acanthamoeba castellanii, a non-canonical G−1-independent HisRS charges the atypical tRNAsHis (150).
Proline identity
Proline identity shows unexpected complexity, with deviations in the identity sets of the three E. coli tRNAPro isoacceptors and variations in the structures of ProRSs that are divided into two groups, prokaryal-like and eukaryal/archaeal-like (151,152). Beside strong determinants at the extremities of tRNA (G72, G35, A37 followed by G36 and A73) and the Levitt G15C48 pair, additional determinants were found in the major tRNAPro(CGG), notably G37, G72, G49 and U17a (153,154). In archaeal A. pernix tRNAPro, the G1–C72 identity pair on top of the acceptor stem completes the discriminator and anticodon determinants (155). Analysis of ProRS sequences in the three domains of life revealed that the sequences are divided into two evolutionarily distant groups (152). While A73 is strictly conserved in bacterial and archaeal tRNAsPro, a C73 pyrimidine is found in eukaryal tRNAsPro and the base pair 1–72 is inverted (G1–C72). Analysis of aminoacylation revealed that, while anticodon recognition has been maintained during evolution, significant changes in acceptor stem recognition have occurred. The C1–G72 pair is a strong determinant in E. coli tRNAPro, but the G1–C72 pair is without effect in human tRNAPro where identity relies predominantly on the anticodon branch. Atomic group mutagenesis was carried out to probe the role of sugar–phosphate backbone interactions in recognition of human tRNAPro. A network of interactions with the first base pair and the discriminator base was revealed in both E. coli and human tRNAPro. Therefore, unlike the bacterial system, backbone-specific interactions contribute much more to tRNA recognition by the human enzyme than base-specific interactions (156). Finally, in E. coli tRNAPro, m5 methylation of G37, which is known to suppress frameshift errors, also contributes to proline identity since its absence significantly affects prolylation efficiency (157).
Serine identity
Identity sets are known in various tRNASer isoacceptors of E. coli (51,82,158–160), S. cerevisiae (56), H. sapiens (98), Archaea (161) and Zea mays (162). Given that serine is assigned by six codons in the genetic code, the tRNASer anticodon can hardly play a major role in serine identity. Indeed, serine identity is of astonishing complexity, due to a phylogenetic divergence in size and orientation of the variable region and, more generally, to the variability of the tRNASer and SerRS sequences (163). For instance, the G1–C72 and G2–C71 pairs are absolutely conserved in E. coli and in most bacterial tRNASer isoacceptors, while the A27–U43 pair is conserved in yeast tRNASer and in some eukaryal (not human) but not in archaeal and bacterial tRNASer isoacceptors (164). Interestingly, the discriminator G73 in E. coli tRNASer acts as an identity determinant in vivo, but not in unmodified transcripts in vitro (165). The functional importance of sequence differences in the yeast and human tRNASer acceptor stems, which account for species-specific serylation, was confirmed in vivo in yeast (56). Overall, serine identity relies primarily on the variable arm and is independent of the anticodon (52).
Comparing the different domains of life, several differences are observable. The main determinants for tRNA serylation by human SerRS are the large variable arm and the G73 discriminator (58,59). In E. coli tRNASer, the moderate importance of the discriminator G73 is strengthened by determinants in the acceptor stem (165). Interestingly, E. coli SerRS selectively recognizes tRNASer on the basis of its characteristic tertiary structure rather than the nucleotides specific to tRNASer (165). Due to the co-existence of two dissimilar SerRSs in the archaea Methanosarcina barkeri (one bacterial-like, the other specific to methanogenic Archaea), two tRNA recognition modes with distinct overlapping identity sets co-exist in the three tRNASer isoacceptors (161). The discriminator base G73 followed by the weaker G30–C40 pair are strong identity elements in bacterial and archaeal SerRSs. Other determinants are required for serylation by methanogenic SerRS, including the G1–C72 pair and several unpaired nucleotides at the base of the extra stem in the variable region that control stem helicity and tertiary interactions (161). As a result, the serine identity elements are used differently in Archaea and Eukarya (166). In Z. mays plants, the discriminator base G73 is by far the strongest determinant of serylation by cytosolic SerRS, as is the case with human cytosolic SerRS (162). As the anticodon of tRNASer does not play a role in aminoacylation, variants of tRNASer with anticodon changes cause similarly high levels of mistranslation. tRNASer mutants with proline anticodons (UGG) remain serylated and therefore cause mistranslation in yeast (167).
Threonine identity
Threonine identity is unique, since the discriminator position 73 does not strongly participate in threonine identity, except in H. volcanii tRNAThr where U73 is an identity determinant (168) and in yeast where substitution of the discriminator base A73 by G73 or C73 impairs the threonine accepting activity (169). Threonine identity sets are known in five species, notably in tRNAThr from E. coli (170), T. thermophilus (171), H. volcanii (168), A. pernix (172) and S. cerevisiae (169). Identity elements consist of base pairs in the acceptor stem and nucleotides of threonine anticodons. Identity elements in the acceptor stem are not fully conserved between species and the identity sets alone are not sufficient to confer tRNAThr charging fidelity, as shown by the strong mischarging capacities of the tRNAThr isoacceptors (173). In Vertebra, the G4·U69-containing tRNAThr incorporates alanine, but mistranslation is prevented by a robust trans-editing activity of ThrRS towards alanyl-tRNAThr (173). Interestingly, in yeast, the G3·U70 wobble pair in tRNAAla acts as an antideterminant for ThrRS (169).
Identity sets recognized by subclass IIb aminoacyl-tRNA synthetases
Aspartate identity
Aspartate identity has been studied in S. cerevisiae, E. coli and T. thermophilus tRNAs (4,174). It is mainly based on five strong determinants (G73, G34, U35, C36 and C38) which are conserved during evolution. However, there are differences in the strength of the determinants in eukaryal and bacterial systems, with the anticodon determinants being strongest in Bacteria (175). The two pairs G1–C72 and G2–C71, strictly conserved in the bacterial tRNAAsp, are crucial for identity and act differentially. The G2–C72 pair is a minor determinant (175) while the C1–C72 pair helps to position the 3′ acceptor end in the catalytic site of E. coli AspRS (176). The charging specificity of yeast tRNAAsp is achieved by the modified base m1G37 which acts as an antideterminant against arginyl-tRNAAsp formation by yeast ArgRS (177). The iodine cleavage of yeast tRNAAsp transcripts substituted with phosphorothioates revealed the critical role of specific phosphates during AspRS recognition. The cognate AspRS protects the phosphate groups of four determinants (G34, U35, U25 and G73), and mutation of these nucleotides results in the loss of phosphate protection in the mutated regions while the overall protection pattern remains unchanged (178). In another study, active variants of yeast tRNAAsp lacking the D and T arms were constructed, leading to minimal active tRNAsAsp that mimic mt-tRNAs (179). In these minimal structures, the rules of identity are preserved, and aminoacylation activity remains strictly dependent on the discriminator G73 and the three anticodon nucleotides (179).
In yeast AspRS, the improved aminoacylation efficiency results from the acquisition of a lysine-rich N-terminal extension that interacts with the anticodon stem of tRNAAsp with, however, a loss of specificity and a risk of mischarging, especially of tRNAGlu(UUC) and tRNAAsn(GUU), which have identical G73 discriminator and anticodons close to the aspartate anticodon GUC (180,181). More generally, N-terminal extensions are conserved in eukaryal class IIb aaRSs where they can control cellular precision of tRNA charging. In yeast, the aspartate aminoacylation system appears to be connected to the arginine system, with the early observation that ArgRS aminoacylates tRNAAsp and more recently with the observation that a minor tRNAArg is a cryptic tRNAAsp (25,182). In yeast, the aspartate aminoacylation system appears to be connected to the arginine system as suggested by the early observation that ArgRS aminoacylates tRNAAsp (182). This hypothesis has been reinforced by the observation that a minor tRNAArg is a cryptic tRNAAsp (25). Finally, functional aspartylated tRNAAsp mutants were selected in vitro in a selection procedure applied to a yeast tRNAAsp library randomized at the anticodon triplet level. The active tRNAs mostly carried the original aspartate anticodon GUC, but one mutated tRNA had an alanine anticodon GGC (183). This mutated tRNA exhibits a 19-fold drop in catalytic efficiency resulting from a 4-fold reduction in affinity and a 5-fold drop in kcat (184). These substantial alterations in catalytic parameters certainly protect cells from effective in vivo aminoacylation of tRNAAla which, moreover, has an A73 discriminator not favorable to aspartylation.
Asparagine identity
A first study demonstrated that bacterial asparagine identity is transferable into a tRNALys by transplantation of the three base anticodon and the G73 discriminator of tRNAAsn (135). The aminoacylation levels in E. coli of tRNAAsn mutants confirmed that both the anticodon and the discriminator base are important for aminoacylation of tRNAAsn (185). Although early attempts to convert the E. coli tRNAAsn into an amber suppressor by modification of its anticodon sequence failed (186), several active glutamine-inserting suppressors were obtained using in vivo selection. The mutated suppressors all had substitutions in the first base pair 1–72 that reduced their stability and were all glutamine insertion suppressors (187). In the yeast S. cerevisiae, the identity conferred by the bases of the anticodon and discriminator is enhanced by the post-transcriptional modification t6A37 that prevents aspartylation (188). Despite similarity between Plasmodium falciparum and H. sapiens tRNAAsn sequences, cross-species aminoacylation is not observed with the corresponding enzymes. The human enzyme does not recognize the plasmodial transcript of tRNAAsn, and the P. falciparum enzyme charges the human transcript of tRNAAsn with an 8-fold reduction compared with its cognate tRNAAsn. Subtle differences in the two tRNAAsn sequences or the use of in vitro transcripts deprived of post-transcriptional modifications could prevent cross-recognition between species (189).
Lysine identity
The role of U35 in the lysine identity of tRNALys was initially suggested by the finding that several amber suppressor tRNAs with a U35 inserted lysine into the suppressed protein (19,190). Discriminator base A73 also played an important role in tRNALys identity since substitution to G73 reduced the suppression efficiency and the resulting tRNA became a partially glutamine-inserting suppressor (19). The unmodified E. coli tRNALys transcript showed a 140-fold lower lysine charging activity than the native tRNALys, suggesting the involvement of base modifications in recognition. Substitution of the discriminator base A73 by any of the other bases confirmed the decrease in lysine acceptor activity observed in vivo. Substitutions of anticodon nucleotides showed the involvement of all three bases in the lysine identity (20). Therefore, as with the aspartate and asparagine identities, the lysine identity relies on the anticodon triplet and the A73 discriminator, which differs from the G73 of the first two systems. In H. sapiens, LysRS aminoacylates an RNA minihelix that mimics the amino acid acceptor stem–loop domain of tRNALys, but without specificity of sequence. However, the continuity between the acceptor and anticodon domains is important for efficient lysylation (191,192). The UUU anticodon is sufficient for the acceptance of lysine of human tRNALys since its transplantation into tRNAAsp or the initiator tRNAMet confers lysine identity to these tRNAs. As in the aspartate system, the specificity of lysylation is facilitated by contacts with the N-terminal helical extension of human LysRS (192).
Identity sets recognized by subclass IIc aminoacyl-tRNA synthetase
Phenylalanine identity
Phenylalanine identity was investigated in tRNAPhe from five organisms (A. pernix, E. coli, T. thermophilus, S. cerevisiae and H. sapiens) covering the three domains of life. The common identity set of phenylalanine, revealed by different strategies, contains four strictly conserved elements which are the anticodon nucleotides G34, A35 and A36, and the discriminator base A73. Additional elements are found, such as G20 in S. cerevisiae and A. pernix or U20 and U59 in E. coli (193–195). In vitro selection from a random library was used in E. coli to isolate active tRNAPhe variants. Critical elements for phenylalanylation of tRNA were thus identified at the three positions of the anticodon G34, A35 and A36, and at the nucleotides of the variable pocket (U20 and U59) (196,197). The two lower base pairs (G30–C40 and A31–U39) in the anticodon stem of tRNAPhe are also recognition elements for human PheRS (198). The strength of determinants is significantly greater in human than in yeast tRNAPhe. Interestingly, tRNAPhe from bacteriophage T5 shows a non-identical mode of recognition by E. coli PheRS at low and high concentrations of Mg2+, suggesting that the local conformation of the tRNA is essential for recognition by bacterial PheRS (199). Finally, an amber suppressor derived from native plant tRNAPhe showed little suppressor activity in vivo in A. thaliana and was poorly phenylalanylated in vitro, suggesting that the anticodon is also a major identity determinant for tRNAPhe in plant cells (124).
Post-transcriptional modifications as identity signals for aminoacylation
In general, tRNA modifications do not participate in tRNA identity, as clearly shown for leucine identity in H. volcanii (55) and proline and tyrosine identities in S. cerevisiae (103,154). The main role of the modifications is in maintaining the structure of the tRNA and the decoding of the codons on the ribosome (200). However, in some cases, modifications are crucial and play an active role by acting as aminoacylation determinants (Table 2) or antideterminants (Table 3) (78). Decreases in the activity of in vitro transcribed tRNAs lacking modified bases have been observed for several identities in E. coli (cysteine, glutamate, isoleucine, lysine and phenylalanine) and S. cerevisiae (isoleucine and phenylalanine) (78). However, it is difficult to attribute the inhibitory effect to one or a combination of modifications. Finally, the low number of modified nucleotides essential for aminoacylation does not mean that only a few aaRSs use post-transcriptional modifications of tRNAs as identity signals, but rather reflects the fact that they have rarely been studied.
Table 3.
Antideterminants identified in tRNAs that prevent erroneous recognition of aaRS
| Antideterminant | tRNA (organism) | Against aaRS |
|---|---|---|
| A1–U72a | tRNATrp(E. coli) | MetRS |
| C2–G71 | tRNALeu (E. coli) | SerRS |
| G2·U71 | tRNALysb (B. burgdorferi)c | LysRS-1 |
| G3·U70 | tRNAAla (S. cerevisiae) | ThrRS |
| cyto-tRNAAla (D. melanogaster) | mt-AlaRS | |
| U30·G40 | tRNAIle (S. cerevisiae) | LysRS |
| k2C34 | tRNAIle (E. coli) | MetRS |
| C + C34 | tRNAIle (Archaea) | MetRS |
| A36 | tRNAArg (E. coli) | TrpRS |
| t6A37 | tRNAAsn (S. cerevisiae) | AspRS |
| G37 | tRNASer (S. cerevisiae) | LeuRS |
| m1G37 | tRNAAsp (S. cerevisiae) | ArgRS |
| A73 | tRNALeu (H. sapiens) | SerRS |
| G73 | tRNASer (E. coli) | LeuRS and TyrRS |
| tRNASer (H. volcanii) | LeuRS |
aA1–U72 is a context-dependent negative identity elemente; bexcept in Nanoarchaeota, found in all archaeal phyla; cprobably applies to Spirochetes (a bacterial phylum).
The base modifications that affect the identity of tRNA aminoacylation are mainly located in the anticodon loop (Table 2). At the anticodon wobble position, the thio group of s4U34 in E. coli tRNAGlu and the inosine I34 of S. cerevisiae tRNAIle are strong identity elements (47,77). In addition, Ψ36 is a weak aminoacylation determinant in tRNAIle where its main role is to prevent misreading of Ile codons (47). The yW37 residue has a dual identity role, weak for aminoacylation by PheRS and strong for decoding tRNAPhe on the ribosome (78). The weak tyrosylation activity of E. coli tRNATyr when Q34 is replaced by C34 also suggests its identity role (78). At position 37 adjacent to the anticodon, t6A37 in E. coli tRNAIle and mammalian cyto-tRNAIle as well as m1G37 in M. mazei tRNACys are determinants for aminoacylation (41,49,201). Interestingly, t6A37 that is crucial for isoleucylation in most prokaryotic IleRSs is not an identity element in Bacteria and its role in translation might vary greatly between organisms (48). t6A37 functions as a determinant of human cytoplasmic IleRS (49). In mouse tRNAAsp, m5C38 is essential for aminoacylation in vivo (202). Often, the modifications and hypermodifications involved in identity and located in anticodons have a dual function in both aminoacylation and codon reading (200,203). Some RNA modifications present in both tRNAs and tRNA-like structures (TLSs) of mRNAs suggest links between tRNA biology and mRNA regulation (204).
Distribution of identity elements on the cloverleaf structure of tRNAs
Identity elements are well known in bacterial and eukaryal tRNAs, in contrast to the poorly known determinants in archaeal tRNAs. Except for the E. coli and S. cerevisiae systems, which have 20 and 18 identity sets known to date, data from other organisms are still sparse. Several identity sets were established in T. thermophilus and H. sapiens tRNAs and in tRNAs from a dozen other organisms, notably four archaeal phyla. Identity elements were sometimes validated by chemical probing and crystallography. In general, a given identity is specified by a limited number of determinants (2–11 nt) (Table 1). All 20 tRNA families show determinants in both acceptor stems and anticodon loops, except the tRNAAla and tRNASer families. Discriminator and anticodon positions are by far the most represented (Figure 3). The highest diversity in determinants specifying aminoacylation occurs for tRNAs recognized by class IIa aaRSs. The eukaryal identities of glutamine, glutamate, asparagine and lysine are poorly documented (Table 1).
Figure 3.
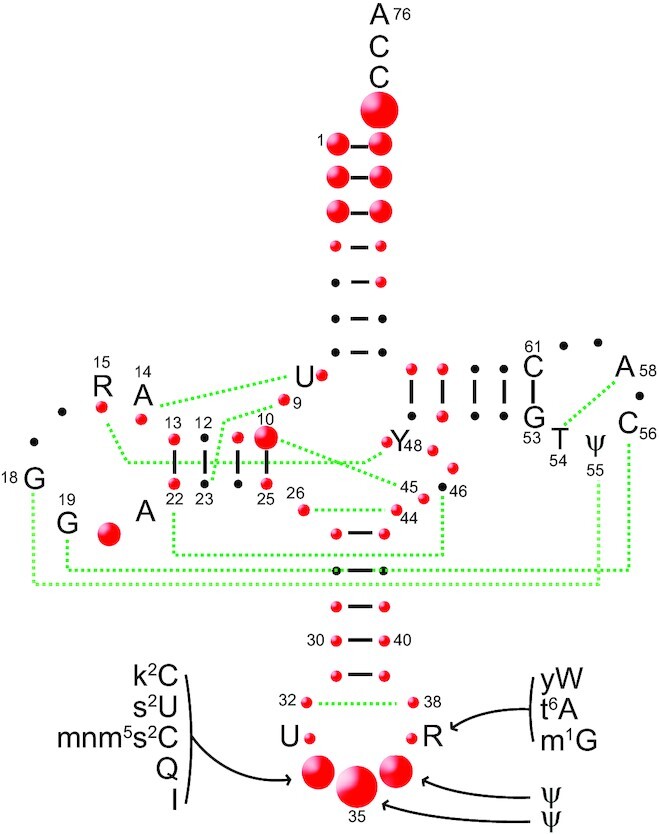
Positions occupied by identity elements in canonical tRNA cloverleaf folding, including identity-determining post-transcriptional modifications. Conserved and semi-conserved nucleotides, as well as tertiary interactions (broken green lines) are shown. The size of red bullets schematizes the extent of occupation in the canonical tRNA sequence (large for the four heavily occupied positions, medium for the eight significantly occupied positions and small for the 32 poorly occupied positions). The standard numbering of positions is used as in Figure 1. Variable region (nucleotides 44–48) with up to 16 nts for extra arms (47a to 47p). Modifications characterized as identity determinants for aminoacylation in anticodon loops are shown next to identity positions; they are displayed in standard abbreviations.
Only 11 identities utilize the complete anticodon triplet as a determinant (Cys, Ile, Met, Gln and Trp anticodons recognized by class I aaRSs, and His, Thr, Asp, Asn, Lys and Phe anticodons recognized by class II aaRSs). Identity sets can overlap (i.e. a given nucleotide at the same tRNA position can code for several identities in a given organism), such as A73 coding for 14 identities or C35 coding for 15 identities. The distribution of determinants varies when comparing aminoacylated tRNAs by class I or class II aaRSs and, more precisely, when comparing the same identity in Bacteria, Eukarya and Archaea. With the exception of the glutamine, glutamate, asparagine and lysine identities, the other 16 identities have conserved determinants in anticodons and the extremity of acceptor stems in model organisms from all three domains of life. Few determinants are found in the D loop due to sequence and structural constraints imposed by the formation of tertiary interactions with the T loop and the presence of determinants for the editing reaction catalyzed by class 1 aaRSs (63,205).
Many aminoacylation systems use determinants located in the central ‘core’ region of tRNA that forms tertiary atypical interactions, such as the non-Watson–Crick 15:48 interaction that connects the D loop to the variable arm and participates in the cysteine and proline identities in bacteria (34,154). However, data are still lacking for tRNAs specifying valine, tyrosine, threonine, asparagine and lysine identities. Likewise, the conserved U8:A14 tertiary pair in tRNALeu is crucial for recognition by the primitive bacterial A. aeolicus LeuRS (206). Altogether, this indicates the importance of the tRNA shape in aminoacylation identities.
Negative determinants or antideterminants
In addition to positive determinants, negative elements (antideterminants) contribute to identity by blocking false recognitions between aaRSs and non-cognate tRNAs, thus providing an additional layer of specificity control. Antideterminants in tRNA can be isolated nucleotides (59,207), modified residues (38,46,177,208) or base pairs (165,209–211). Table 3 shows a panel of antideterminants against identified aaRSs, located in the acceptor and anticodon arm of tRNA. The first modified nucleotides acting as antideterminants were found in the anticodon loop, such as k2C34 (i.e. lysidine or L34) in minor E. coli tRNA2Ile which blocks recognition by E. coli MetRS (38), and m1G37 in yeast tRNAAsp against yeast ArgRS (177,208).
In the acceptor stem, the A1–U72 base pair is a context-dependent negative identity element of E. coli tRNATrp (211). Likewise, the C2–G71 pair in E. coli tRNALeu is a negative identity element against E. coli SerRS (165) whereas in the archaeal H. volcanii tRNASer discriminator G73 acts as an antideterminant against LeuRS (55). In E. coli tRNASer, G73 is also an antideterminant for LeuRS and TyrRS (161,165). An amber suppressor corresponding to the S. cerevisiae tRNAIle carries a U30·G40 wobble pair in the anticodon stem that is a negative signal for the E. coli LysRS interaction under heterologous expression conditions (209).
In insects, including D. melanogaster, the base pairs 2–71 and 3–70 found in cytosolic tRNAAla behave as antideterminants for mitochondrial AlaRSs that cannot charge these tRNAsAla because of a shifted mode of recognition (131,132). Interestingly, the G3·U70 pair in yeast tRNAAla that blocks interaction with yeast ThrRS was predicted by an algorithm (212), confirming previous functional data (169). Many other putative antideterminants in yeast tRNAs have been predicted and await experimental validation (212).
Other examples are determinants in E. coli tRNAGln that are antideterminants in E. coli tRNAGlu and, conversely, determinants in tRNAGlu that are antideterminants in tRNAGln (213). In H. pylori, GluRS2 mischarges tRNAGln to form Glu-tRNAGln and rejects tRNAGlu by looking at the antideterminant base pair G1–C72 that is found in tRNAsGlu (87). In S. cerevisiae, tRNALeu becomes an efficient serine acceptor when unmodified (56). Several antideterminants can co-exist in a given tRNA, such as the base pair U28–A42 in the anticodon stem and the discriminator base A37 in unmodified yeast tRNATrp that act as negative elements for bovine TrpRS (214).
On the other hand, amino acids in aaRSs can play an equivalent antidetermining role against false aminoacylations of non-cognate tRNAs. For instance, several substitutions in E. coli MetRS induce recognition of nonsense suppressors, without affecting recognition of native tRNAMet (215). In Bacillus stearothermophilus TyrRS, Glu152 acts as an antideterminant for non-cognate tRNAs by electrostatic and steric repulsions (216). Some amino acids in aaRSs can also have dual positive and negative functions, as found in yeast AspRS (217,218) and E. coli GlnRS (219).
IDENTITIES IN ATYPICAL tRNA FOLDS AND ORGANELLAR tRNAS
Structural diversity in the tRNA world
Non-canonical cloverleaves and atypical folds in cytosolic tRNA and tRNA-like structures
A variety of non-canonical tRNA cloverleaves (220) and viral TLSs (221,222) aminoacylable by standard aaRSs have been described. Other tRNA mimics recognized by aaRSs are present in mRNAs (223). RNA fragments of the large tRF (tRNA-derived RNA fragment) family are additional tRNA mimics (224). They arise from individual transcription units or result from the processing of canonical tRNAs, and some interact with aaRSs (225).
Figure 4 shows four atypical tRNA folds that highlight deviations in helical regions and the presence of pseudoknots, but also shows conserved features, including anticodon loops. Selenocysteine-specific tRNAs, ubiquitously present in life, possess atypical secondary structures as in E. coli tRNASec (Figure 4A) (226). In M. barkeri, the D loop is small and the anticodon arm of tRNAPyl is unusually long (Figure 4B) (227). The presence of a pseudoknot at the 3′ terminus of many viral RNA genomes was discovered in the TLSVal of turnip yellow mosaic virus (TYMV) (Figure 4C) (222,228). Note the presence of standard anticodon loops in some viral TLSs with anticodon triplets that match the acceptor identity, such as the valine CAC anticodon in TYMV TLSVal (229). Other TLSs lack the anticodon, such as the intricate TLSTyr from brome mosaic virus (BMV) that is lacking a tyrosine anticodon in its short pseudo-anticodon loop of four nucleotides (230). tRNA mimicry is also responsible for the translational control of the ThrRS gene of E. coli. Its 5′-untranslated region contains a thrS operator which mimics a tRNA L-shape, with domain 2 corresponding to the anticodon loop and stem of tRNAThr, but lacking an equivalent to the acceptor arm replaced by a domain mimicking a second anticodon stem (Figure 4D) (231,232).
Figure 4.
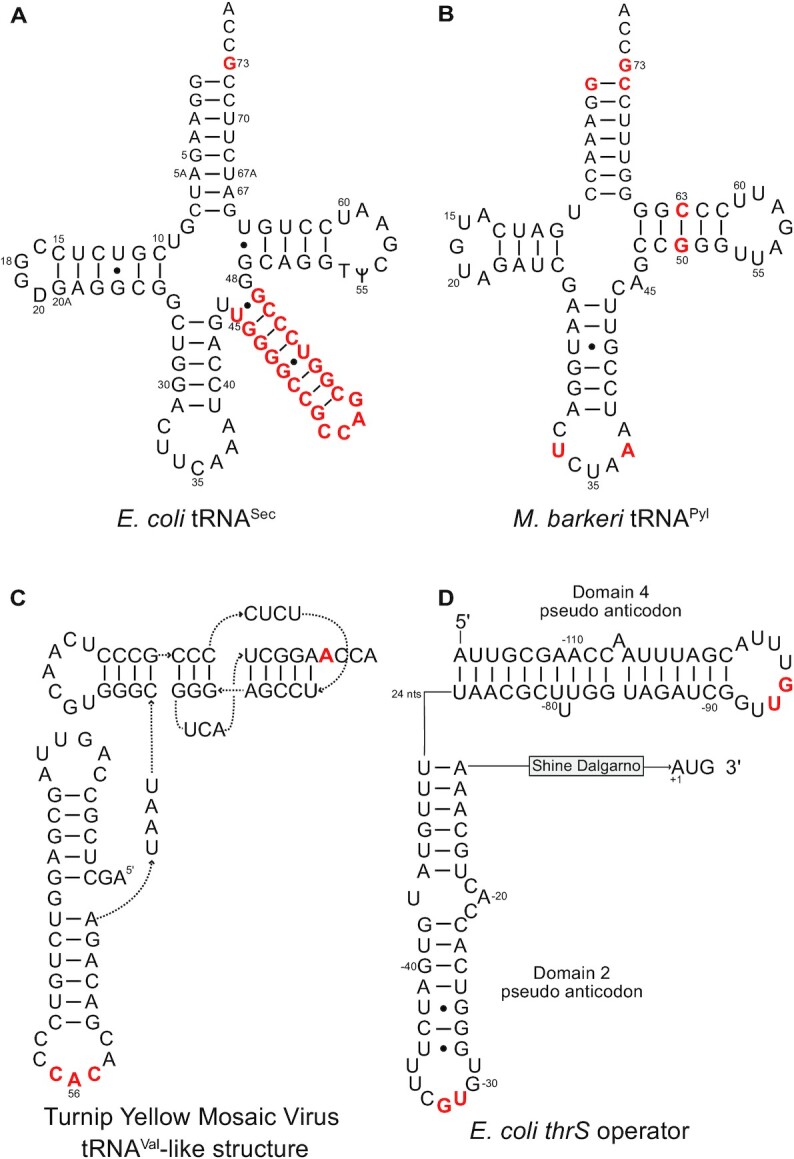
A panel of atypical RNA folds present in tRNAs aminoacylated by aaRSs or only recognized by aaRSs. (A) E. coli tRNASec. (B) M. barkeri tRNAPyl. (C) Turnip yellow mosaic virus (TYMV) tRNA-like structure (TLS) with valine-charging capacity. (D) E. coli thrS operator. Experimentally characterized identity elements are in red font. For easier comparison, the numbering of atypical tRNA folds is as in canonical tRNAs with, for example, positions 73 and 34–36 for discriminator bases and anticodons. For TYMV and mRNA TLSs, sequence numbering is from 3′ to 5′ ends of the molecules with starts at A1 (in the -CCAOH accepting end of the viral TLS and the A1UG triplet next to the Shine and Dalgarno sequence in the mRNA TLS).
Structural diversity in organellar tRNAs
In the world of tRNAs, organellar tRNAs show the greatest structural diversity as evidenced by mt-tRNAs (Figure 5). They include examples with altered cloverleaves such as human mt-tRNAAsp (Figure 5A) or missing either the D arm [e.g. Bos taurus mt-tRNASer(AGY)] (Figure 5B) or the T arm (e.g. Caenorhabditis elegans mt-tRNAAla) (Figure 5C) (233). Remarkably, armless structures deprived of both D and T arms are predicted in the nematode Enoplea (233). These bizarre armless structures, as first discovered in C. elegans (234), are common in nematodes, mites, arachnids and insects (235,236). The length of the sequences varies from 70 nt, for example for human mt-tRNAAsp, to 44 nt for Romanomermis culicivorax mt-tRNAArg (Figure 5D), the shortest experimentally characterized natural tRNA to date (233). This length is even predicted to be shorter for several spider mt-tRNAs (235,237) and for tRNAGln(UUG) from a common plant pest, the acariform mite Tetranychus urticae (238).
Figure 5.
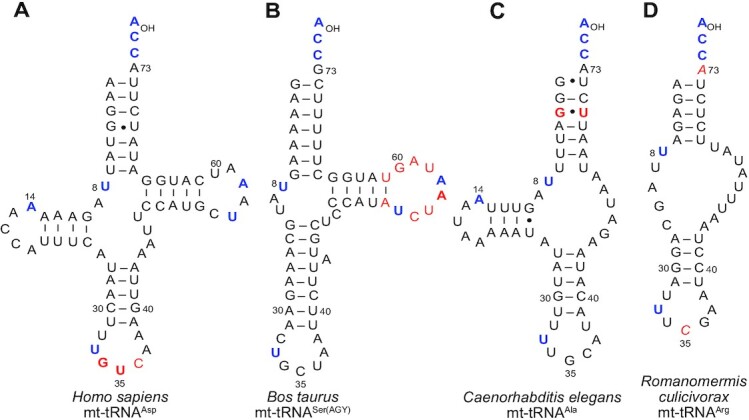
Typical 2D folds in the four mt-RNA structural families indicating partial conservation of universal features necessary for canonical 3D folding, as found in representative mammalian and/or nematode mt-RNAs. (A) Canonical-like cloverleaf folding, (B) folding missing D arm, (C) folding missing T arm, and (D) folding missing both D and T arms. The numbering is based on that of canonical tRNAs. Conserved and semi-conserved nucleotides are in blue bold scripts. Experimentally characterized identity determinants are in red font (in bold for major determinants). Predicted determinants in R. culicivorax tRNAArg, based on E. coli tRNAArg, are in red italic font.
Some of the conserved and semi-conserved nucleotides characterizing standard tRNAs are present in mt-tRNAs and therefore support their L-shaped conformation. Although they are almost absent in the miniature tRNAs, these tRNAs mimic an L shape as shown for R. culicivorax mt-tRNAArg by small angle X-ray scattering (233). The distribution and nature of modified nucleotides in organellar tRNA are essential for tRNA structure and activity (239). For instance, m1A9 has a structural role in human tRNALys by participating in the cloverleaf folding of the tRNA (240). m1A9 is also essential for binding of several T-armless Ascaris suum mt-tRNAs on the nematode elongation factor EF-Tu (241).
Surprising identities in atypical systems
Identity elements in atypical tRNAs and TLSs
Identity in tRNASec
These tRNAs allow the incorporation of selenocysteine into selenoproteins in response to a specific stop codon (226). Since there are no corresponding aaRSs, aminoacylation of tRNASec by selenocysteine occurs in a two-step indirect process involving in the first step serylation by a standard SerRS and in the second step conversion of seryl-tRNASec to selenocysteinyl-tRNASec by specialized factors, either selenophosphate synthetases (SelA/B) in Bacteria or other factors in a more complex process in Eukarya and Archaea (242). Consequently, tRNASec includes identity elements for recognition by SerRS, selenocysteine factors and other elements that prevent interactions with canonical partners of protein synthesis, notably canonical elongation factors. Many of these identity elements remain unknown.
In E. coli, serylation efficiency of tRNASec is only ∼1% that of the five canonical serine isoacceptors (226), probably due to its atypical solution conformation distinct from that of tRNASer (243). The discriminator base G73 in tRNASec is the strongest identity element for serylation by E. coli SerRS. In addition, the long extra arm and other elements of the atypical secondary structure of tRNASec contribute to a lesser extent to the serylation identity. These features are observed in the crystal structure of human SerRS in complex with tRNASec (244). On the other hand, three base pairs (C7–G66, G49·U65 and C50–G64) in the core of E. coli tRNASec are identity elements for SelB recognition and reject the standard elongation factor EF-Tu (245). Mutations in these pairs and elsewhere in E. coli tRNASec restore recognition by EF-Tu and improve serylation (246). In Eukarya and Archaea, the second pathway of synthesis of selenocysteinyl-tRNASec is dependent on the PSTK phosphorylation factor and the dedicated SepSecS conversion factor (242). However, most of the identity elements that control this pathway remain unknown.
Identity elements in viral TLS domains
TLSs are located at the 3′ end of viral genomic RNAs. They are found in several genera of plant viral RNAs and are recognized and aminoacylated (222) by either ValRSs (247), HisRSs (248) or TyrRSs (249). TLSs do not participate in protein synthesis but act in amplification of viral genomes. All TLSs contain the characteristic nucleotide N–1 of tRNAHis, so that the base pair N–1–N73 can form in all TLSs, including those aminoacylated by ValRS and TyrRS and therefore all are also substrates of HisRS.
The identity of TLSs follows the identity rules of tRNAs and includes determinants mimicking those from the canonical valine, tyrosine or histidine identity sets (222). The discriminator base N73 and the anticodon nucleotides are the conserved identity elements in all three plant viral TLS families, but they display functional idiosyncrasies. In the TYMV TLSVal (Figure 4C), the major valylation determinants are A35 and to a lesser extent A73 when the charging reaction is catalyzed by ValRS of yeast. To comprehensively uncover the full set of valine identity elements in this TLS and assess the role of the pseudoknot in aminoacylation, valylatable variants were selected from a pool of RNA molecules derived from the TYMV TLSVal. Selected sequences show strong conservation of C53, A56 and C57 in the pseudo-anticodon loop, but variability is allowed in the length of the L1 loop of the pseudoknot (250). In the presence of wheat germ ValRS, A35 and C36 of the CAC anticodon together with C38 are strong determinants, while A73 has no effect on valylation (251). Note that A73 and A35 are strong valylation identity elements in eukaryal tRNAsVal.
The tyrosine identity of bromoviral TLS includes the base A73 and anticodon nucleotides (U35 and A36), in addition to the pair C1–G72 in the acceptor helix. Interestingly, the tyrosine identity elements in the acceptor helix of TLSTyr are similar to those of eukaryal tRNATyr. The contacts seen in the cryo-electron microscopy (EM) structure of BMV-TLSTyr in complex with TyrRS of Phaseolus vulgaris (249) are essentially the same as in the crystal structure of the yeast tRNATyr:TyrRS complex (108) and involve the catalytic and anticodon-binding domains of TyrRS. However, the topology of the two complexes is different. As seen in the cryo-EM structures, the free BMV-TLSTyr RNA does not contain a classic L-shaped tRNA mimic. To bind the enzyme, the BMV-TLSTyr undergoes large conformational changes. The resulting complex resembles the overall configuration of the tRNATyr:TyrRS complex; however, there is substantially more space between the TLS and the surface of the enzyme (249). The determinants of histidine identity in tobamoviral TLS are the atypical pair N–1–N73 and two nucleotides that mimic the histidine anticodon nucleotides G34U35 (252).
The identity of TLSs can be engineered. For instance, replacement of the valine by the methionine anticodon in the TLSVal from TYMV results in an infectious virus with methionine acceptance (222). However, some TLSs are not aminoacylable, such as the one from tymoviral Erysimum latent virus (ELV) that lacks a mimic of the valine anticodon loop (253).
Identity of tRNA-like structures found in mRNAs
Such RNAs were discovered in the 5′-untranslated regions of mRNAs coding for aaRS (mRNAaaRS), first in mRNAThrRS (254) (Figure 4D) followed by S. cerevisiae mRNAAspRS (255). These two TLSs bind to their own aaRS through anticodon loop mimics, thereby regulating the translation of ThrRS and AspRS in processes under the control of tRNA identity rules. Functional and structural studies have discovered the mechanisms that lead to the regulation of the E. coli ThrRS gene (thrS) (232,256). Using tRNA identity rules, it was possible to switch the specificity of the translational control from ThrRS to MetRS by changing the threonine anticodon-like sequence to the methionine anticodon-like sequence (257). This happens because both ThrRS and MetRS recognize nucleotides in their anticodon loops, thereby allowing the switch of control of the mRNA. A more complex mechanism explains regulation of yeast AspRS by mRNAAspRS. Here, the N-terminal extension of each AspRS subunit anchors mRNAAspRS to AspRS via two distinct motifs, connected by a linker sequence, one mimicking the aspartate anticodon domain (255). Recently, several other yeast aaRSs (MetRS, GluRS, ValRS, GlnRS and HisRS) have been found to bind their own mRNAs at anticodon-like structures (223), suggesting that this type of regulation is widely used. Consistently, all these enzymes recognize their tRNAs through identity elements located in the anticodon.
Another TLS fold, called mascRNA, is a small cytoplasmic RNA derived from a long non-coding MALAT RNA that mediates multiple processes in mammalian cells. Once processed by tRNA-specific enzymes, mascRNA folds in a quasi-perfect cloverleaf, and interacts with GlnRS in the multi-aaRS complex and contains tRNAGlu identity determinants (258). However, mascRNA lacks a conserved anticodon loop, is inactive in aminoacylation and does not compete with tRNAGln for binding on GlnRS. MascRNA enhances global protein translation by increasing GlnRS stability and therefore provides a new paradigm for TLSs to regulate protein levels.
Identity rules in atypical aaRS systems
Five amino acid specificities appear in this group of aaRSs. These are monomeric class Ib LysRS (LysRS-1) and ND-GluRS, the dimeric α2 class IIb ND-AspRS and α2 class IIc PylRS, and the tetrameric class IIc SepRS. They are unevenly distributed in the phylogenies, particularly in archaeal and bacterial phyla (15).
Identity elements in tRNALys for LysRS-1
Atypical LysRS-1 is found mainly in Archaea and some Bacteria (in contrast to the standard class IIb LysRSs, which are present in all Eukarya, many Bacteria and some Archaea). To date, only the identity of the tRNALys lysylation of the Borrelia burgdorferi spirochete is known. While the anticodon bases U35 and U36 are determinants for both class I and class II LysRSs, the strength of U36 is more important for LysRS-1. In contrast, the discriminator base A73 plays a marginal role, but the nearby G2–U71 pair is essential for lysylation by LysRS-1. This pair is also an antideterminant that affects lysylation of E. coli class II LysRS (Table 3). Finally, the structural context of the acceptor stem is crucial, since a shift of the wild-type identity pair G2–U71 to another position in the acceptor stem has dramatic effects on lysine charging by B. burgdorferi LysRS (210).
Identity elements of tRNAs for ND-AspRS and ND-GluRS
AsnRS and GlnRS genes are missing in many bacterial and archaeal genomes. To overcome the absence of these enzymes, asparaginyl-tRNAAsn or glutaminyl-tRNAGln are produced via a two-step indirect process. First, a non-discriminating AspRS (ND-AspRS) aspartylates tRNAAsp and tRNAAsn,and a non-discriminating GluRS (ND-GluRS) glutamylates tRNAGlu and tRNAGln. Second, the mischarged aspartyl-tRNAAsn and glutamyl-tRNAGln are amidated in a dynamic supramolecular ternary complex (aaRS:tRNA:AdT) called the transamidosome (259), which comprises from five up to 14 macromolecular entities. Its mechanism is explained by the crystal structures of archaeal-type asparagine and glutamine transamidosomes from T. thermophilus (260) and Thermotoga maritima (261).
Quite similar identity sets exist in tRNAAsp/tRNAAsn and tRNAGlu/tRNAGln pairs (i.e. major determinants in anticodon triplets and discriminator base). In the case of the pair tRNAAsp/tRNAAsn, the anticodon determinant C36 in tRNAAsp is replaced by U36 in tRNAAsn. Consequently, the dual aminoacylation by ND-AspRSs is based primarily on the idiosyncratic features of their anticodon-binding domains. Similar considerations explain the dual glutamylation of tRNAGlu and tRNAGln by ND-GluRSs. In addition, the identity sets for aminoacylation of tRNAAsn or tRNAGln must co-exist with the identity elements for tRNA-dependent amidotransferases. The specificity of aspartyl-tRNAAsn amidation is conferred by the pair U1–A72 in tRNAAsn and prevented in aspartyl-tRNAAsp by its pair G1–C72 and U20a in the D loop that act as antideterminants (262). Although the same combination of nucleotides also determines specific tRNAGln-dependent formation of glutamine, the situation is more intricate in glutamine transamidosomes.
Helicobacter pylori and several other bacterial species possess two genes of GluRS. GluRS1 aminoacylates tRNAsGlu isoacceptors, while GluRS2 only misacylates tRNAGln to form glutamyl-tRNAGln (86,87). GluRS2 recognizes major identity elements clustered in the tRNAGln acceptor stem. An intermediate case is Acidithiobacillus ferrooxidans GluRS1 which charges the two tRNAsGlu isoacceptors and the tRNAGln species while GluRS2 preferentially charges tRNAGln. It appears that GluRS1 charges exclusively tRNAs with augmented D stem length and a deletion of nucleotide 47, which strongly suggests that these are the major identity elements for GlnRS1 (86). A similar situation could occur in mitochondria, as suggested by the discovery of mitochondrial ND-aaRSs in human fungal pathogens (263).
Identity of tRNAPyl for PylRS
The biology of the atypical PylRS/tRNAPyl system as present in Archaea and a few Bacteria is well documented (264). All tRNAsPyl possess an amber CUA stop codon, but behave towards bacterial elongation factors as typical elongator tRNAs (227). The major identity elements that guide charging are the discriminator base G73 and the G1–C72 pair in the acceptor stem (265,266). However, these determinants are not fully conserved in all tRNAsPyl and can be A73 in H. volcanii or U73 in tRNAPyl in some other bacterial species (267). In the extreme halophilic deep-rooted methanogen Candidatus Methanohalarchaeum thermophilum, two PylRS/tRNAPyl pairs are simultaneously present. The two pairs exhibit mutual orthogonality enabled by unique features. A G73 discriminator base specifies the identity of tRNA1Pyl whereas in tRNA2Pyl it is a A73 base. In addition, PylRS2 has evolved with a shorter motif 2 loop than PylRS1, which ensures the recognition specificity of A73 (268).
Pyrrolysine identity does not rely on anticodon determinants (Figure 4B) (266); therefore, the anticodon of tRNAPyl can be mutated to recognize codons other than UAG without affecting tRNA recognition by PylRS. Because of its high suppression efficiency, the PylRS/tRNAPyl system has been widely used for the construction of orthogonal aaRS/tRNA pairs with novel amino acid specificities to expand the genetic code. The crystal structure of the tRNAPyl:PylRS complex from Methanosarcina hafniense revealed that the anticodon branch of tRNAPyl does not contact PylRS, which is the basis for orthogonality engineering required for the synthesis of proteins encompassing abiotic amino acids (269).
Identity of tRNASer for SepRS
Synthesis of cysteinyl-tRNACys in the methanogenic Archaea missing CysRS is SepRS dependent. These organisms use a two-step route that is only partially elucidated (270,271). In M. jannaschii, phosphoserine is first charged on tRNACys by SepRS and subsequently transformed to cysteine by SepCysS (Sep-tRNACys-tRNA synthase). Interestingly, M. jannaschii SepRS differs from CysRS by recruiting the m1G37 modification as an aminoacylation determinant (272).
In M. mazei, one of the few Archaea encoding CysRS, serylation of tRNACys requires m1G37 that is a serylation determinant for both SepRS and CysRS. Aminoacylation kinetics reveal that M. mazei SepRS and CysRS prefer distinct tRNACys isoacceptors with specific determinants in their core region (201). Sequence analysis of tRNAsCys suggests that G37, A47 and A59 are additional minor identity elements specific to SepRS in addition to the strongest identity elements common to those of CysRS which are the anticodon G34C35A36 and the discriminator U73 and the weaker determinants G15 and A47 (273). Such features are in line with sequence variability of tRNACys in the three domains of life and are consistent with the idea that tRNACys identity is an ancient RNA record that depicts the emergence of the universal genetic code before the advent of modern aminoacylation systems (273).
Identity elements in organellar tRNAs are partially conserved
Understanding the identity rules explaining the specificity and efficiency of organelle tRNA aminoacylation is still a largely open field. In mt-tRNAs, identity elements have been experimentally validated in 13 of the 20 mt-tRNA families in a dozen taxa (Supplementary Table S1). To date, the identity of chloroplast and apicoplast tRNAs has been little studied. These tRNAs are structurally close to canonical tRNAs, but with some deviations from canonical sequences. For instance, atypical features are found in plant chloroplast tRNAs (e.g. tRNALeu, tRNASer and tRNATyr), revealing relatedness to cyanobacterial tRNAs, including tRNAMet and tRNAIle (274,275). In P. falciparum, the apicoplast tRNAs fold in canonical cloverleaves but have a nucleotide composition typical of mt-tRNAs (276). Identity elements are only known in P. falciparum tRNATyr, and the field of identities in chloroplast tRNAs remains unexplored.
As a rule, organellar tRNAs can be aminoacylated by bacterial, but not by eukaryal aaRSs, suggesting a relationship between organellar and bacterial identity elements. Identity determinants were first deduced by sequence comparison between bacterial and mt-tRNAs as, for example, for the 22 human mt-tRNAs (277) and other mt-tRNAs from many taxa (278). Due to the great structural diversity of mt-tRNAs, the search for identity elements has naturally been carried out on these idiosyncrasies, especially in Mammalia (278). The discovery of structural deviations of mt-tRNAs from canonical cloverleaf folding occurred first in human and bovine mt-tRNASer (279) and then in nematodes (234). It was followed by the discovery of the first pathogenic mutations in human mt-RNAs (280).
Seven mt-tRNA identities (arginine, leucine, tyrosine, alanine, serine, aspartate and phenylalanine) and the identity of apicoplast tRNATyr have been studied in detail. Partial data are available for tRNAs specific for Ile, Lys, Trp and His. All known identity elements are listed in Supplementary Table S2.
Mt-arginine identity
Mitochondrial arginine identity was investigated in tRNAs from the eumetazoan clade, notably in insects and sponges (281,282). For mt-tRNAArg(UCG) from the coleopteran insect Caryedes brasiliensis, the identity set for mt-ArgRS is restricted to the sole U34 and C35 anticodon determinants. However, in the dipteran D. melanogaster, transplantation of the arginine UCG anticodon into the structurally dissimilar mt-tRNAAsp(GUC) led to very low arginine acceptance. Complete transfer of arginine identity could only be achieved with transplantation of the entire anticodon arm of tRNAArg into tRNAAsp, suggesting that the arginine identity is sensitive to specific structural features and to m1A and other modified nucleotides present in mt-tRNAArg.
In eumetazoan organelles, the AGR (Arg) codons of the standard genetic code are reassigned to serine/glycine/termination. In C. brasiliensis and other evolutionarily related insects, the mitochondrial AGR codons are translated by tRNASer(UCU). Investigation of the arginine-accepting activity of tRNASer(UCU) from C. brasiliensis and other insects of related taxa revealed that the AGR reassignment does not result in arginine misaminoacylation and therefore the mitochondrial protein synthesis is not compromised by misincorporation of Arg residues. In other organisms, whose mitochondrial translation is dictated by the universal genetic code (e.g. Porifera/sponges), recognition of the two tRNAArg(UCG/UCU) resembles that of the yeast cytoplasmic system. The analysis of mt-tRNAArg variants representative of seven metazoan phyla shows that despite variations in secondary structures, the nucleotides and conformational identity elements (U34, C35 and anticodon environment) have been largely conserved in Eumetazoa that share the AGR codon reassignment but are complemented by specific structural features and/or by modified nucleotides present in insect mt-tRNAsArg (e.g. m1A or Ψ residues) (282).
Mt-leucine identity
As for E. coli tRNALeu, the identity of the unmodified human mt-tRNALeu(UUR) transcript is essentially specified by A73 and A14. Recognition by the mitochondrial LeuRS occurs despite the absence of a long variable arm and, surprisingly, with a partially unfolded anticodon branch, as revealed by probing the structure in solution. This floppy structure is the result of mismatched base pairs and the absence of modified bases in the transcript. Replacement of these mismatches in the anticodon arm by G–C base pairs restores the expected cloverleaf and improves the leucylation efficiency to a level similar to that of native mt-tRNALeu(UUR). These results suggest that mitochondrial LeuRS contacts mt-tRNALeu(UUR) in the acceptor and anticodon stems and in the D loop, what has never been observed in any leucine aminoacylation system, and shows a contribution of nucleotide modifications to structure and identity of the tRNA (283,284).
Mt-tyrosine identity
Human mt-TyrRS exhibits characteristics of both bacterial and archaeal TyrRS (285). Therefore, mt-TyrRS aminoacylates a tRNATyr with a G1–C72 base pair as efficiently as with an inverted C1–G72 pair. This is the first example of TyrRS lacking specificity with respect to N1–N72. This is due to the sequence of the mitochondrial enzyme, which has dual sequence features characteristic of bacterial and archaeal TyrRSs in the region recognizing the N1–N72 pair. Thus, human TyrRS disobeys general tyrosine identity rules, a behavior probably conserved in Vertebra as suggested by the phylogeny (285).
Mt-alanine identity
The first three base pairs of the acceptor stem (G1–C72, G2–C71 and G3·U70) and the discriminator A73 base are conserved in all known tRNAAla sequences from Prokarya, Archaea, eukaryal cytoplasm, chloroplast and plant mitochondria. However, strict conservation is not observed in all tRNAAla of animal and insect mitochondria. Thus, every animal mt-tRNAAla identified so far contains at least one sequence variation in these three base pairs (121,131), as found in C. elegans mt-tRNAAla where the G1–C72 pair is replaced by a G1·U72 pair (132). This results in C. elegans mt-tRNAAla carrying the identity pair G3·U70, the G2–G71 pair but not the G1–C72 pair replaced by G1·U72, being charged by C. elegans mt-AlaRS but not by E. coli AlaRS because of this G1·U72 pair that acts as an antideterminant (132). On the other hand, C. elegans mt-AlaRS charges both bacterial and mitochondrial microhelices, indicating that the G3·U70 pair remains active in a variety of structural contexts. Separate experiments confirmed that helix instability or irregularity in the acceptor stem is not important for the recognition of the G3·U70 pair by mt-AlaRS (121). In D. melanogaster, mt-tRNAAla has a G2·U71 but not a G3·U70 pair. In this case, the translocated pair G2·U71 and the pair G3–C70 are the two main identity elements for aminoacylation by D. melanogaster mt-AlaRS (131,132). A similar translocation of the G3·U70 identity pair to the 5–68 position is observed in human mt-tRNAAla (133). Furthermore, three weak identity pairs (A1–U72, A2–U71 and G4–C69) complete the identity set, suggesting that the interaction between human mt-AlaRS and its cognate mt-tRNAAla is different from that in other mt-alanine systems (133). A recent study suggests that because of high rates of mt-DNA sequence evolution in bilaterian animals, mt-tRNA genes have accumulated mutations at significantly higher rates than their cytoplasmic counterparts, resulting in foreshortened and fragile mt-tRNA structures. To compensate for this reduced structural complexity in mt-tRNAs and the loss of identity elements, bilaterians have developed sequence-independent induced-fit adaption mechanisms between cognate mitochondrial aaRSs and tRNAs. This would explain the loss of the G3·U70 determinants in the acceptor stem replaced by the U–A and G–C pairs (286).
Mt-serine identity
The T loop region is the main determinant for serylation and recognition by mt-SerRS in the bizarre animal mt-tRNASer(AGY) (279). Data on serine identity of the two serine mt-isoacceptors were collected on B. taurus and human mt-tRNASer isoacceptors. While mt-tRNASer(UGA) has a standard cloverleaf structure, mt-tRNASer(GCU) is missing the D arm. The striking feature of these two tRNAs is the absence of the long extra arm, which is the major identity element in cytosolic tRNASer. Instead, mammalian mt-SerRSs recognize the distinct shapes of the two mt-tRNAs by kinetic discrimination (287,288). In isoacceptor mt-tRNASer(UGA), the T loop and the interaction between T and D loops are required, while in the truncated mt-tRNASer(GCN), the T loop alone is sufficient with A57 and A58 as major identity elements (289). This implies a new tRNA binding mode to SerRS different from that in Bacteria and Eukarya and accounts for a dual-mode recognition employed to discriminate the two mt-tRNASer by alternative interaction sites (290). Shape recognition of the D-armless human mt-tRNASer(GCU) was recently visualized in cryo-EM structure with human mt-SerRS (291).
Mt-aspartate identity
In H. sapiens mt-tRNAAsp, the set of major elements of aspartate identity is restricted to only the three anticodon nucleotides, with U35 and C36 being the strongest and G34 being a moderate element (292,293). These elements are completed by structural determinants, notably the A9:A12–U23 triple interaction, since its disruption in the tRNAAsp core leads to a huge loss of aspartylation efficiency (294). In addition, changes in the conformation of the anticodon loop can modulate the aspartylation activity. Mutation of A38 (a minor determinant) to G38 leads to a decrease in activity, whereas C38 or U38 mutations increase it (292). However, the replacement of G73 by A73 in human mt-tRNAAsp is surprising considering that G73 is a major identity determinant of the aspartate system in all domains of life, including organelles, except in mammalian mitochondria. This is the consequence of a structural idiosyncrasy of mitochondrial AspRS in the peptide binding the acceptor end of human mt-AspRS (293). In marsupials, the mitochondrial aspartate identity is closely related to the glycine identity (see below).
Mt-aspartate/glycine identity in marsupials
The marsupial opossum Monodelphis domestica contains in its mitochondrial genome a typical tRNAAsp gene carrying the glycine anticodon GCC. After transcription, the incorrect anticodon is edited so that ∼50% of the transcripts carry the GUC sequence while the rest carry the GCC sequence GCC (295,296). The edited form functions as tRNAAsp whereas the unedited form serves as a tRNAGly (297). This is a unique case of a single tRNA gene that produces two tRNAs with different decoding specificities.
Mt-phenylalanine identity
Human mt-PheRS exhibits a minimalist monomeric structure. The enzyme exhibits a broad specificity for bacterial, eukaryal, chloroplastic and mitochondrial-derived tRNAPhe transcripts. The crystal structure of the human mitochondrial tRNAPhe:PheRS complex shows that mt-PheRS possesses a relatively simple recognition mechanism of tRNA (298). Unlike E. coli PheRS which recognizes an identity set scattered throughout the L-shaped tRNA (see above) (197), human mt-PheRS recognizes a restricted set consisting primarily of nucleotide C74 (from the CCA), the pair G1–C72 and the discriminator base A73. Recognition of the anticodon nucleotide G34 requires a significant rearrangement of the anticodon-binding domain (298). This recognition mode is probably preserved in Mammalia. Furthermore, m1A9 is an identity element of phenylalanine in A. suum mt-tRNAPhe (241).
The yeast mitochondrial PheRS, which is also a small monomeric enzyme, recognizes nucleotides from the anticodon and the acceptor end, including base A73 and the adjacent G1–C72 base pair or at least the C72 base. The small size of the monomeric yeast mitochondrial PheRS does not allow contacts with nucleotide 20 at the top corner of the L-shape as there are with the cytosolic enzyme of yeast, A. pernix and E. coli (299).
Diverse and poorly understood identities of mitochondrial tRNAs
In H. sapiens mt-tRNAIle, five modified nucleotides (m1G9, m22G26, Y27, Y28 and t6A37) collectively contribute to isoleucine identity since the unmodified transcript is less active than native mt-tRNAIle for isoleucylation by human mt-IleRS. Furthermore, A7 in the acceptor stem and A59 in the T loop are determinants of isoleucine identity, as suggested by two mutations of these residues that cause human pathologies (300).
Human mt-tRNALys deprived of modifications (m1A9, m2G10, Ψ27, Ψ28, tm5s2U34 and t6A37) folds in an inactive extended bulged hairpin that recovers activity and canonical folding upon insertion of m1A at position 9. This reveals the major contribution of m1A9 in the identity of lysine and the L-shaped folding of this mt-tRNA and indirectly to its identity (240). The T-armless mt-tRNAMet of A. suum also requires m1A9 for efficient methionine acceptance (241).
The aminoacylation of Oryza sativa mt-tRNATrp by TrpRS of human and B. subtilis reveals large changes in catalytic efficiency resulting from the presence of identity elements specific to the two taxa. However, the rice mt-tRNATrp identity elements are found in typical positions and include G73, G1–U72 and U5–A68 in the acceptor stem (301).
Caenorhabditis elegans is one of the few organisms that exhibits mt-tRNAHis that lacks nucleotide –1. Despite the lack of this canonical His identity element, mt-tRNAHis can be efficiently aminoacylated in vivo. However, C. elegans HisRS still prefers tRNAHis with G–1 but tolerates tRNAHis without G–1. Additionally, the results show that the anticodon has taken a leading role in the identity of tRNAHis instead of the almost universal –1 determinant (302).
Tyrosine identity in apicoplasts
In P. falciparum, apicoplast tRNAs fold into canonical cloverleaves but with a nucleotide content typical of mt-tRNAs (276). The identity nucleotide set of apicoplast tRNATyr is limited to only three weak identity elements for tyrosylation by cognate P. falciparum apicoplast TyrRS, namely G34 and U35 in the anticodon and the long variable region. The identity element commonly found in the acceptor stem of tRNATyr, notably the pair G1–C72 in Bacteria, is replaced by A1–U72 in apicoplast tRNATyr. This probably results from the high AT content of the Plasmodium apicoplast genome and the resulting mutational pressure on the tRNATyr gene. How efficiently and specifically tRNATyr is aminoacylated in the apicoplast could be explained by the presence of additional antideterminants in tRNATyr or additional idiosyncratic peptides in TyrRS (303).
MECHANISTIC AND EVOLUTIONARY ASPECTS OF THE tRNA AMINOACYLATION REACTION
Minimal structural requirements of tRNAs and modes of recognition by aaRSs
Positive identity elements for aminoacylation of cytosolic and/or organellar tRNAs have been searched and validated by functional assays in ∼35 taxa covering the three domains of life (Supplementary Table S1), but these taxa are unevenly distributed throughout the phylogeny. Only a few validated identity elements for tRNA aminoacylation are conserved in evolution. Most are concentrated in the distal parts of tRNAs: the amino acid acceptor branch and the anticodon (Table 4). All are strong identity elements. The number of phylogenetically conserved identity elements increases if the analysis is limited to bacterial and eukaryal elements (Table 1), showing the particular status of Archaea in evolution.
Table 4.
Experimentally validated positive identity elements that are almost strictly conserved in all three domains of life including organellar tRNAs and viral TLSsa
| tRNA families | tRNA domains | ||
|---|---|---|---|
| Acceptor branch | Core region | Anticodon branch | |
| Cys | U73 | … | G34, C35, A36 |
| Ile | … | … | A35 |
| Leu | A 73 | largeVR | … |
| Met | A73 | … | … |
| Val | A73b | … | A35b |
| Trp | … | … | C34, C35, A36 |
| Tyr | A73b | … | G34, U35 |
| Ala | G 3· U 70 | … | … |
| Gly | G3–C70 | … | C 35, C36 |
| Pro | … | … | G35, G36 |
| Thr | G1–C72 | … | G34, U36 |
| Phe | A 73 | … | G 34, A35, A36 |
Of the 20 tRNA families designated by their amino acid identity, eight do not contain conserved determinants (Arg, Gln, Glu, His, Ser, Asp, Asn, Lys). The position of the determinants in the sequence of the tRNA acceptor branch, the core region and the anticodon branch is shown with standard numbering. aRare exceptions can be found at certain positions such as G3·U70 in organellar tRNAsAla, or G34 in eukaryal tRNAsThr; bdeterminants in TLSs. ‘…’ no data available. VR, variable region.
If it is difficult to find common elements of identity, it is equally difficult to find a universal tRNA scaffold as the diversity of aminoacylable structures is great. For instance, in viral tRNA-like structures, the amino acid acceptor arm contains a pseudoknot and only the terminal -N73CCAOH sequence and the N1 residue are conserved. It has been known for a long time that a canonical acceptor arm is not mandatory for activity, since a fragment of yeast tRNAPhe with an excised 5′ quarter is efficiently aminoacylated by yeast PheRS provided the m7 group of G46 is removed (304). Likewise, a poly(U) (∼30 U residues) with an attached 3′-NCCAOH is aminoacylated by a mammalian LysRS (305). These mimics exhibit conformational flexibility and contain key anticodon identity determinants. In this context, it is noteworthy that arachnids contain remarkably short and unusual mt-tRNAs (235,306). Sequencing of mitochondrial genomes from several genera of spiders revealed that most tRNAs cannot be folded into a classical four-armed cloverleaf secondary structure. Most tRNAs lack the D or T arms, and at least four of them lack both D and T arms. In addition, the acceptor stems have multiple mismatches. In fact, the 39 bp tRNASer1 gene from Parachtes romandiolae is 3 bp shorter than the tRNAArg from the nematode Romanomermis culicivorax, setting a new record for the shortest tRNA gene ever described (235).
This highly diverse and aminoacylable set of RNAs leads to an L-shaped tripartite structural model (Figure 6) with a central region connecting the amino acid acceptor and anticodon branches. For translation, a 7 nt anticodon loop and a terminal 3′-NCCAOH are mandatory, but other tRNA regions may show high variability. The minimalist and necessary structural and functional requirements for tRNA aminoacylation can be summarized as follows.
Figure 6.
Generalized L-shaped structure of RNAs recognized and/or aminoacylated by aaRSs with structural requirements necessary for function. The drawing schematizes how the amino acid acceptor (7 bp) and anticodon (5 bp) branches are connected in canonical tRNAs by two linkers L1 (from U8 to N26 with the D arm) and L2 (N44 to N48 with the T arm and either the small or large variable region). In atypical and mt-tRNAs, this architecture shows important peculiarities, with distal stem regions of 5–8 and 4–9 bp for acceptor and anticodon branches, respectively, pseudoknotted acceptor stems and great diversity in L1 and L2 sequences. The conserved pairings are shown. The orientation of the two arms is variable and the distance between the CCAOH end and the anticodon triplet ranges between 45 and 75 Å.
For aminoacylation and more generally for ribosome-dependent protein synthesis, aaRSs recognize identity sets mainly located at the two distal ends of the tRNA.
AaRSs are platforms that anchor the RNAs in adapted active conformations.
Generally, mitochondrial aaRSs recognize minimalist identity sets in mt-tRNAs and sometimes altered sets, such as in metazoan animals, that can be restricted to elements located solely in the acceptor branch or the anticodon loop.
The architectural frameworks in mt-tRNAs and atypical tRNAs play crucial functional roles. Evolution adapted identity rules to the intrinsic conformational fragility of aminoacylable RNAs.
A simplistic view of the mechanism of identity expression would be that direct electrostatic interactions made by aaRS with discriminating functional groups on tRNA determine the specificity of aminoacylation following the general lock-and-key model of interaction. Although true to some extent, an additional mechanism of indirect reading completes the expression of identity. Revealed by structural and functional studies and called the ‘indirect readout’ mechanism, this mechanism senses sequence-dependent conformations of tRNA upon recognition of aaRS by contacts with the sugar–phosphate backbone and non-specific parts of the bases (307). In other words, the indirect readout suggests that the distribution of nucleotides is not random outside the set of identity elements. This explains previous studies showing that nucleotide changes outside of recognition sets are not neutral for aminoacylation, as shown, for example, for yeast tRNAPhe (308) or yeast tRNATyr (309).
Interactions with aaRS during tRNA aminoacylation
Direct contacts with identity elements of tRNA
Hydrogen bonding and stacking appear to be the straightforward way to recognize tRNA identity determinants during aminoacylation. Figure 7 displays examples of such contacts between strong determinants and aaRSs specifying arginine, aspartate and phenylalanine identities. Thus, A20, the strongest arginine determinant in E. coli tRNAArg, interacts with five amino acids of E. coli ArgRS through a network of direct H-bonds completed by a stacking interaction (Figure 7A). Interestingly, these direct contacts represent the initial recognition step before the intervention of strong anticodon determinants (23). In the yeast aspartate system, the major identity determinants G34, U35 and C36 interact with AspRS via six amino acids by H bonding and Phe127 by stacking (Figure 7B). Moreover, the adenine moiety of the discriminator base G73 establishes direct H-bonds with four amino acids of AspRS (Figure 7C) (310). The strongest identity determinant G34 of the T. thermophilus tRNAPhe makes direct contact with the β-subunit of the anticodon-binding domain B8 of PheRS, notably by H-bonds with three amino acids (Asp729, Ser742 and Arg780) and stacking with Tyr731 (Figure 7D) (311). The minor determinant A73 in T. thermophilus tRNAPhe does not contact PheRS, but forms intra-tRNA H-bonds with adjacent nucleotides, as seen in the ternary complex with a stable phenylalanyl-adenylate (312). Generally, most of the contact amino acids, especially those stacked on bases, are conserved in aaRSs. Interestingly, in the T. thermophilus tRNAHis:HisRS complex, only the two regions around the major G–1 and minor anticodon identity determinants are anchored on HisRS by a network of H-bonds (147).
Figure 7.
Examples of interactions between major identity determinants in tRNA with cognate aaRS as seen in crystal structures of tRNA:aaRSs complexes. (A) A20 in the E. coli tRNAArg:ArgRS complex (5b63). (B) U35 and C36 in the S. cerevisiae tRNAAsp:AspRS complex (1asy). (C) A73 in the S. cerevisiae tRNAAsp:AspRS complex (1asy). (D) G34 in the T. thermophilus tRNAPhe:PheRS complex (1eiy).
Sometimes mutations in aaRSs that disrupt interactions with tRNA identity elements are so critical that they induce lethality in vivo. A mutation within yeast AspRS (Glu188Lys) is lethal due to the disruption of the interaction of the tRNAAsp identity base G34 with the AspRS anticodon-binding β-barrel and thus inactivating the enzyme by relaxing the recognition specificity (217). Similar effects occur with other bacterial aaRSs, for example GlnRS, MetRS and TyrRS; see (4). In B. stearothermophilus TyrRS, Glu152 acts by electrostatic repulsion of non-cognate tRNAs (313). On the other hand, amino acids in aaRSs, although they do not make H-bonding contacts with tRNAs, may be critical for the specificity of aminoacylation, as is the case of Leu136 in E. coli GlnRS (314).
Direct contacts with the backbone of tRNA
It is generally expected that contacts with the standard sugar–phosphate backbone of tRNAs do not contribute to the specificity of recognition (although interactions of the same type can lead to the ‘indirect readout’ phenomenon). This is illustrated by the example of yeast AspRS where half of the tRNA/aaRS interactions do not participate in aspartate identity (310). These contacts are in the acceptor stem, the D stem and the anticodon branch of tRNAAsp, and include interactions with phosphate oxygens of the tRNA backbone. Mutation of the protein residues involved in these interactions only led to moderate changes in the affinity for tRNAAsp without relaxation of tRNA recognition when assayed with bulk tRNA. This confirms that they do not contribute to specific recognition but stabilize the tRNAAsp:AspRS complex (218). Similar contacts have been described in other systems, for example in the histidine T. thermophilus (147) and the E. coli arginine (23) systems. This type of interaction is widely used in tRNA:aaRSs complexes and represents more contacts than with the identity elements of tRNAs.
Structural adaptations required for recognition of identity elements
Many functional and crystallographic investigations have revealed conformational adaptations in tRNA:aaRS complexes. For example, the G10·U25 pair is an identity element in the yeast AspRS system despite the absence of interaction with the enzyme, indicating that the pair modulates the conformation and positioning of other identity elements in tRNA (310). On the other hand, the interaction of the tRNAAsp anticodon loop with the yeast AspRS results in a radical change in the L-shape, with the bases spreading in the β-barrel domain of the enzyme (315). This contrasts with T. thermophilus PheRS where the conformation of the anticodon loop is relatively similar to that of free tRNA. The only significant difference is that G34 is unwound outside the anticodon loop and stacks directly on Tyr731 in the B8 domain of PheRS (311).
As mentioned above, ‘indirect readout’ is an alternative way to specifically recognize tRNA (307). It takes advantage of global and local conformations, sequence elements and contacts with 2′-hydroxyl groups, as has been observed in several tRNA/aaRS systems (34,316). Indirect reading reinforced the importance of water-mediated H-bonds that complement direct macromolecular contacts. This is well illustrated in the complexes of E. coli AspRS with its cognate tRNAAsp (317) or with the heterologous yeast tRNAAsp (176), where H-bonds occur almost exclusively in the acceptor stem and often establish contacts with the phosphate oxygens and riboses of the ribophosphate backbone.
Kinetics in tRNA aminoacylation and allosteric effects
Strength of identity determinants
Steady-state kinetics accounting for the strength of identity determinants is well documented (9,318). The strength of the determinants is measured by the parameters kcat and KM, which reflect catalysis and binding, respectively. In general, the kcat parameter is most affected in mutational studies. This is illustrated by E. coli glutamine (83,85) and yeast aspartate identities (184). For glutamine identity, decreases of kcat can reach ∼104-fold upon mutation of anticodon identity determinants, while KM does not increase by more than 10-fold. The trend is similar for aspartate identity, although kcat is consistently less affected with decreases of at most ∼200-fold. A KM dependence characterizes the identity pair G10·U25 in yeast tRNAAsp and is explained by the absence of contact of this determinant with AspRS (184,310). Similar features characterize the U3–A69 pair in E. coli tRNAVal (73) and the G2–C71 pair in A. pernix tRNATrp (97). The case of E. coli tRNAGlu is remarkable, as the 100-fold reduction in catalytic efficiency of the transcript lacking mnm5s2 modification on the U34 determinant results solely from a KM effect, i.e. a loss of affinity (76). KM effects are regularly observed in transplanted tRNAs, leading, for example, to a moderate loss of catalytic efficiency when the yeast aspartate identity set is transplanted into the yeast tRNAPhe (184). The effect is much more pronounced when the aspartate set is transplanted in the E. coli tRNAGln, and vice versa when the E. coli glutamine set is inserted into the yeast tRNAAsp (319). This shows that the minor/major groove recognition modes of the tRNAs specific to each class of aaRS are not locked by the identity switches.
More generally, the conditions under which the aminoacylation parameters are measured in vitro can significantly modulate the catalytic efficiencies. One example is the tuning of the efficiency of phenylalanylation by Mg2+ concentration, which accounts for the non-identical sets of determinants in the E. coli tRNAPhe isoacceptor (199). Similarly, in vivo associations of aaRSs affect the efficiency of tRNA charging, as shown by the archaeal associations between LysRS, LeuRS and ProRS (320,321).
Allosteric communications in tRNA:aaRS complexes
Structural plasticity is an essential phenomenon occurring throughout the interaction of aaRSs with their substrates (15,322,323). Contacts of aaRSs with amino acid and ATP substrates occur mainly in the catalytic site, while with tRNAs they also occur with distant modules, located up to 75 Å from the catalytic site. These events involve long-distance information transfer across the structural body of aaRSs and/or tRNAs. It is now well established, through decades of experimental and theoretical studies, that aaRSs have adapted their catalytic mechanisms to achieve specificity with moderate affinity for their substrates (tight binding would make product release more difficult and decrease turnover) (324). The objective for aaRSs is to provide the highest affinity for adenylate intermediates that must remain bound to the enzyme as premature release would lead to their hydrolysis in solution and prevent the transfer of amino acids to the tRNA acceptor end. Given this need, allostery works through conformational changes or stabilization of flexible regions during substrate binding. Structural studies have revealed a wide range of substrate-binding modes, from a rigid lock-and-key model to various types of induced fit modes.
Examples of induced fit have been described in yeast AspRS. The ‘flipping loop’ of the AspRS active site, which is disordered in the absence of amino acid, becomes visible upon interaction with aspartic acid in the presence of tRNA (325,326). Differences in the conformation of the two AspRS subunits are also observed after binding of the tRNA nucleotides G73 and C74 to the catalytic domain (310). Upon binding, additional interactions are established between AspRS and the tRNAAsp anticodon that trigger the rotation of the anticodon and hinge domains, allowing the correct positioning of the 3′ end of the tRNA in the active site. Allosteric changes in the tRNAAsp:AspRS complex induce strong anticooperative effects on tRNAAsp mutants between the anticodon identity elements G34, U35 and C36 and the discriminator G73, all of which interact with AspRS at a distance of ∼75 Å (327). The triple mutant of the anticodon nucleotides probably loses interactions with the anticodon-binding domain of AspRS since it behaves kinetically as a minihelixAsp (328). Similar effects have been observed in other systems, notably in the proline system of T. thermophilus and in the mammalian arginine system. In the ProRS system, changes in the conformation of the Pro- and ATP-binding loops occur during binding, allowing functional binding of tRNAPro (329). In ArgRS, one of the three aaRSs that requires the presence of tRNA to catalyze the formation of arginyl-adenylate, the A76 terminal nucleotide, the D loop and the anticodon arm of tRNAArg together enable arginine activation in the active site (27). For some aaRSs, the induced adjustment mechanism resulting from substrate binding is probably precise enough not to require additional editing activity.
Currently, long-range allosteric communication pathways are documented in most aaRS families (324). For example, in the E. coli tRNACys:CysRS complex, a communication pathway between the anticodon-binding region and the catalytic site of CysRS has been identified in which specific amino acids provide functional coupling between the two sites and promote active tRNA conformation (330). In the case of the E. coli tRNAGln:GlnRS complex, pre-steady-state kinetics on E. coli GlnRS mutants revealed allosteric signaling pathways in the enzyme body that regulate glutamine binding and glutaminyl-tRNA formation (331).
Identity elements in the proofreading activities and ‘quality control’ of the aminoacylation reaction
The inaccuracy of amino acid and tRNA selection by aaRSs explains why the aminoacylation of tRNAs is not entirely specific. Aminoacylation errors increase the diversity of the proteome and appear to be a driving force of evolution (332). Errors are tolerated up to system-dependent thresholds. Too many errors could threaten cellular life and under certain circumstances lead to dysfunctions, which is why aaRSs have evolved correction mechanisms, also known as proofreading or editing mechanisms. Until recently, proofreading was thought to be absent from mitochondria, until the discovery of editing by human mt-AlaRS (333).
The concept of kinetic proofreading was proposed in the 1970s by Hopfield (334) and Ninio (335). It follows the observation of high levels of ATP consumption in the presence of non-cognate amino acids, as first demonstrated for valine mischarging on tRNAIle by E. coli IleRS (334). A two-step editing process was proposed by Fersht comprising pre-transfer editing (first sieve) that clears amino acids misactivated in the catalytic site, and post-transfer editing (second sieve) that edits mischarged tRNAs with the help of dedicated editing domains (336). More recently, trans-editing (third sieve) catalyzed by freestanding editing domains has been proposed (337). Currently, the editing mechanism is supported by a body of experimental data and is globally understood (338,339).
A balance between pre-transfer and post-transfer editing activities
The question of whether the editing mechanisms of IleRS, LeuRS and ValRS use a tRNA-independent or a tRNA-dependent pre-transfer pathway has been clarified. In E. coli IleRS, tRNA-dependent pre-transfer editing accounts for one-third of the total proofreading activity and uses a conserved tyrosine residue in the catalytic site for both editing and aminoacylation. This dual process is kinetically controlled and, in E. coli LeuRS, depends almost entirely on post-transfer tRNA editing in which the 3′-OH group of A76 in tRNALeu plays a crucial role (340).
Post-transfer editing deacylates mischarged tRNAs in editing domains distinct from the synthetic aminoacylation sites. This requires conformational changes in both aaRSs and tRNAs. The mechanism involves the use of partially distinct tRNA interactions in the editing and synthetic sites. In the isoleucine system, this results in the segregation of editing determinants in the L-shaped tRNA corner (G16, D20 and D21) while the anticodon of tRNAIle contains the aminoacylation identity elements (205). Similarly, the anticodon arm of tRNALeu is essential for LeuRS editing, but dispensable for aminoacylation (63).
In a comprehensive study, the functionality of editing domains was examined by domain exchanges between species. Deletion of the human LeuRS editing domain leads to a complete loss of synthetic activities (activation and transfer), and only the yeast editing domain can partially rescue the different functions of LeuRS. This demonstrates the structural interdependence of the synthesis and editing sites, which ensures the coordination of the two opposing activities (341–344).
Other aaRSs show different partitions of the pre- and post-transfer editing activities [for a review, see (339)], but little is known about the identity elements involved in these processes.
Editing in trans by freestanding editing proteins
AlaRSs, ProRSs and ThrRSs contain editing domains that are not strictly conserved through evolution and often involve trans-editing proteins [AlaXp, ProXp, YbaKp, ThrRS-ed and DTD (d-aminoacyl-tRNA-deacylase (345)]. These autonomous proteins recapitulate the editing function of the corresponding aaRSs. For example, ThrRS-ed proteins are truncated ThrRSs lacking the catalytic site, as found in some Archaea (346). ProXp proteins have relaxed specificities and recognize multiple mischarged tRNAs, even with non-proteinogenic amino acids (347). Sometimes a given aaRS mediates the trans-editing of its own tRNA mischarged by another aaRS (173). For example, in mammals, tRNAThr containing base pair G4·U69 is efficiently alanylated by AlaRS and is edited by ThrRS, thereby preventing the mistranslation of threonine to alanine. Interestingly, E. coli ThrRS has cross-editing capability, although alanyl-tRNAThr is not produced by AlaRS in bacteria (173).
Chiral proofreading catalyzed by DTD proteins deserves some attention (348). DTDs remove d-amino acids mischarged on tRNAs and achiral glycine mischarged on tRNAAla. This chiral trans-editing process appears to be ubiquitous. One type of DTD is probably the progenitor of archaeal ThrRS that contains a module homologous to the DTD fold (349). The alanine identity base pair G3·U70 is a universal determinant for DTD proteins, which explains the functional relationship of DTD with AlaRS. Bacterial DTDs efficiently remove non-cognate glycyl-tRNAAla but much less so the cognate glycyl-tRNAGly due to its discriminator base U73 which acts as an antideterminant. More generally, the discriminator base N73 in tRNA modulates the activity of DTD proteins (348) and may be specific to phyla and organelles (350). In higher Eukarya (i.e. Animalia), a paralog of DTD with relaxed specificity, named ADT, proofreads alanylated tRNAThr isoacceptors. ADT acts as a glycine deacylase and hydrolyzes mischarged glycyl-tRNAAla in Bacteria and Eukarya (351). Surprisingly, the high level of d-alanyl-tRNAAla synthesized by T. thermophilus AlaRS is not edited by a DTD protein but progressively deacylated by post-transfer editing in the synthetic site of AlaRS. This demonstrates the active role of AlaRS in controlling chirality (352). In summary, chiral proofreading DTD enzymes are a major cellular checkpoint, but other molecules prevent the infiltration of d-amino acids into the translation machinery. They include aaRSs, EF-Tu and ribosomes (348).
Finally, we can highlight the resistance to these proofreading activities of mischarged tRNAs by the non-discriminating aaRSs D-AspRS and ND-GluRS. This resistance is partly due to the channeling of the aminocylation and amidation processes that prevents the premature release of poorly charged products into the cell (260,261).
Relaxing the ‘quality control’ mechanism of tRNA aminoacylation can benefit the cell by reinterpreting the genetic code
It is generally accepted that the aaRSs provide the first checkpoint of ‘quality control’ of the translation by ensuring the accurate aminoacylation of tRNAs assisted by the proofreading activity. The accuracy of tRNA aminoacylation is generally considered to be better than 10−4, a value comparable with the typical accuracy of ribosome decoding, also of the order of 10−4.
These two reactions are the two key steps in maintaining the translational fidelity of the genetic message. In total, high-speed translation of a cell results in a translation error rate of approximately one error for every 103–105 amino acids (353–355). This concept of highly accurate protein synthesis is commonly accepted, as an error in tRNA aminoacylation would lead to ambiguity in the genetic code and produce statistically modified proteins. This concept has been challenged by the discovery in fungi of the Candida clade of ambiguous decoding of leucine codons that increases phenotypic diversity (356). New studies have shown that corrupted identities can produce misinterpretation of the genetic code, which can be an advantage under stressful conditions. However, excessive corruptions of identity rules produce toxic effects, especially in higher eukaryotes, leading to metabolic dysfunctions and diseases in humans (357–359). Therefore, controlling the quality of the aminoacylation reaction may lead to antagonistic effects in different cases or circumstances. For example, it has been shown that E. coli cells not only tolerate the presence of misacylated tRNAs but may even require them to grow under selective pressure. Up to 10% of mismade proteins are tolerated by E. coli, suggesting that the editing function of aaRS is not essential for survival in some circumstances. This triggers a heat shock response that stimulates non-optimized polypeptides to reach a native conformation or to be degraded (360).
On the other hand, a bacterial strain containing a defective PheRS in tyrosyl-tRNAPhe editing exhibits growth limitation when exposed to an excess of non-cognate amino acids and other stresses (361). Mischarging by human AlaRS of non-cognate tRNAs carrying a G4·U69 pair is beneficial to cells (362) whereas, conversely, errors mediated by E. coli AlaRS are not well tolerated and induce a global stress response that leads to a significant perturbation of the proteome, with potential catastrophic effects on fitness and viability (363). In agreement, a mouse with a ‘ticky’ mutation, which results in loss of cerebellar Purkinje cells and ataxia, carries a missense mutation in the AlaRS editing domain that results in low levels of misfolded tRNAs and accumulation of misfolded proteins in the neurons (357). Accordingly, different cell types tolerate different levels of mistranslation which is advantageous under certain physiological conditions due to a diversification of proteomes [for reviews, see (364,365)]. Loss of editing domains and ability to edit mischarged amino acids by LeuRS, ThrRS and PheRS in Mycoplasma results in tRNA mischarging, and the resulting mistranslation helps parasites evade host immune responses by increasing antigen diversity (366).
Finally, we cannot fail to mention the ultimate role of EF-Tu in the control of aminoacylated amino acids. The bacterial elongation factor EF-Tu binds to all tRNAs acylated with their correct amino acid (cognate) with almost uniform affinity. This occurs because the sequence of each tRNA has evolved to compensate for the variable thermodynamic contribution of the esterified amino acid. More precisely, each tRNA uses different combinations of three base pairs in the acceptor branch to adjust the affinity of aminoacyl-tRNAs for the EF-Tu elongation factor for optimized interaction (i.e. binding or rejection) with the ribosome (367). These base pairs are conserved in Bacteria (including tRNASec for recognition by SelB and rejection of EF-Tu), supporting the idea that EF-Tu has a safeguarding role in the quality control process. In conclusion, the control of translation errors strongly contributes to the quality control of the expression of the genetic code (368).
Comprehensive overview of tRNA identity expression processes
The two-step mechanism of tRNA aminoacylation is currently well understood. However, this mechanism is error prone in vitro and in vivo, revealing relatedness between identity sets, idiosyncrasies in catalytic processes and functions beyond aminoacylation. Strong identity elements in tRNA make direct and water-mediated indirect contacts with aaRSs, in processes requiring conformational changes in anticodon loops and acceptor termini. Major identity elements are generally conserved. However, idiosyncrasies occur in systems where aaRSs have a complex evolutionary history (AlaRSs, ArgRSs, ProRSs, ND-aaRSs and archaeal aaRSs). For example, AlaRSs use different ways to recognize the identity determinant G3·U70 (128), and the yeast ArgRS relies on different in vivo mechanisms, based on subtle relationships of nucleotides at identity positions 20, 34, 35 and 36 to charge the four tRNAArg isoacceptors (24). In addition, the use of sequence features outside canonical identity sets to adjust specificity is probably widespread but only occasionally documented (308,309).
The similarity of identity sets of certain specificities implies the possibility of identity changes. Such changes can be engineered in vitro by manipulation of identity elements, e.g. (112), and occur in vivo due to anticodon shifts (369), but are not obligatory as the identity of tryptophan is not altered in yeast amber suppressors (93).
Some unusual features of organelle tRNA/aaRS systems, such as minimalist or altered identity sets and sometimes miniaturized mt-tRNAs, add a new level of complexity. For example, although the structures of human mt-AspRS and E. coli AspRS are similar, the G73 discriminator is a major identity determinant in E. coli tRNAAsp but not in human mt-tRNAAsp, as a consequence of the enlarged catalytic groove, electropositive surface and reduced thermal stability of human mt-AspRS (293). Furthermore, in the human mitochondrial alanine system, the G3·U70-independent charging of tRNAAla requires minor elements in the acceptor stem (133) complemented by shape and folding features (308,309). Similar recognition mechanisms in which anticodons would play a crucial role are postulated for the recognition of the miniature tRNAs by their mt-aaRSs.
In conclusion, based on current knowledge, it can be deduced that the mechanism of tRNA aminoacylation by aaRSs is based on (i) plasticity and dynamic functioning of tRNA/aaRS systems with allosteric adaptations; (ii) specificity tuning by elements outside the canonical identity sets; (iii) error-prone mechanisms leading to frequent misactivation of the amino acid and mischarging of the tRNA; and (iv) proofreading strategies to overcome excessive functional errors. Most of these features are the result of an evolutionary process, which implies the current existence of idiosyncrasies in taxa deeply rooted in the tree of life.
EXPANDING THE WORLD OF tRNA IDENTITY
Towards cracking a challenging conundrum in life sciences
Sequence analysis by statistical methods provided the first functional signatures in tRNAs (370,371) that were rationalized with the proposal of universal identity rules for tRNA aminoacylation (4). Beyond this historical and fundamental function of tRNAs as essential components of translation, many recent studies suggest that the roles of tRNA in many biological processes go beyond this paradigm (3,372). How to deconvolute the different signatures specific to these functions and encrypted in tRNAs, tRFs (also known as tRNA-related RNA fragments or tRNA fragments) and TLSs is a new challenge (222,373). Finding reliable answers is difficult and requires new algorithms, given the increasing amount of tRNA sequences compiled in databases [∼170 000 for tRNAs, ∼106 for tRNA genes or pseudo-genes, ∼13 000 for tRFs, ∼3000 for mammalian mt-tRNA genes and ∼37 000 for plant photosynthetic tRNA genes (65,164,374–377)] as well as in TLSs and other tRNA-derived molecules (255).
Finally, the very notion of tRNA identity so often associated with the terms aminoacylation, positive and negative determinants and second genetic code (378,379) deserves to be revisited and refined, to include features and functions beyond aminoacylation. In what follows, extensive searches for ‘identity elements’ refer not only to signals that specify tRNA aminoacylation by aaRSs, but also to those that specify other functions of tRNAs.
Early bio-informatic predictions
The first studies aimed to find identity elements mainly for tRNA aminoacylation. The first computer-assisted comparison of tRNA sequences was conducted on 67 sequences of E. coli and Salmonella typhimurium tRNAs and showed the importance of anticodon positions 35 and 36 for decoding and putatively for identity (371). Interestingly, a second region in the tRNA acceptor branch was distinguished, including nucleotide 73, and the first four base pairs of the acceptor stem (371). Much later, the first candidate identity elements in archaeal tRNAs were derived from large-scale sequence analysis performed on ∼1100 tRNA genes from 22 archaeal species, mainly Euryarchaea, a few Crenarchaea and one Nanoarchaea (380). These candidate identity elements are found in anticodons, notably at three positions of Asn, Asp, Cys, Gln, Glu, Ile, Mete, Phe, Trp and Tyr anticodons, or two positions (Gly, Thr and Val anticodons), or one position in Lys and Pro anticodons. Many predicted determinants are outside the anticodons, notably the G3·U70 pair in tRNAsAla, the discriminator N73, A20 in tRNAsArg, various base pairs in stems of cloverleaves of tRNA and the variable region in tRNAsLeu and tRNAsSer. Interestingly, some identity candidates located in stem regions have no known homologs in Eukarya and Bacteria, such as C13–G22 and C50–C64 in tRNAsPhe and tRNAsHis, respectively. Similarly, some candidates validated by aminoacylation assays (see above) are specific to Archaea, such as C27–G43 in tRNAsIle, G22–U44 and A59 in tRNAsLeu, U11–A24 in tRNAsVal, G29–C41 and C50–G64 in tRNAsHis, and the first three G–C pairs on top of the acceptor stem in tRNAsPro. In contrast, the validated pair C1–G72 in A. pernix tRNAThr was not predicted to be a threonine determinant in the three domains of life (for comparisons, see Table 1). Later predictions conducted on larger sequence sets analyzed by standard routines revealed a co-evolution of tRNAHis identity rules (148) and found divergences in identity between tRNAs from Proteobacteria and Cyanobacteria (381). Other automated methods visualized determinants and antideterminants (212) and suggested a hierarchical organization of these elements according to ‘Class Informative Features’ (382).
Current bioinformatic-assisted searches of tRNA identity elements
Several bioinformatics studies have aimed to identify tRNA sequence elements characteristic of aaRS classes (383) or to predict identity elements for aminoacylation (384,385). Surprisingly, some of the predicted identity elements disagreed with known aminoacylation data, suggesting that they were not associated with recognition by aaRSs (385) but could act as antideterminants to prevent false recognition (386,387). Alternatively, they could encode identities unrelated to tRNA aminoacylation and contribute to the recognition of other tRNA-binding proteins, such as base-modifying enzymes (see below).
Ab initio searches of identity elements
Global search in tRNA sequences from the three domains of life
This was done from a set of ∼104 tRNA sequences analyzed by probalistic Bayesian methods (164,385). The predicted identity positions were classified by amino acid specificities in each of the three domains of life. This gives, for each tRNA family, unique ‘importance’ profiles, as illustrated for two patterns specifying arginine and alanine identity in Bacteria (Figure 8). The profiles conform to previously identified identity elements. Indeed, the most important positions 20 and 35 are occupied by the major A20 and C35 identity elements in bacterial arginine systems (Figure 8A). Likewise, the alanine profile confirms the identity elements at positions 3:70 and 20. The profile also highlights positions 35 and 36 of the anticodon, which have never been identified as alanine identity determinants (Figure 8B).
Figure 8.
Examples of position importance in bacterial tRNAs for (A) arginine and (B) alanine identities. Importance of positions in tRNA sequences is quantitatively evaluated by relative entropy (RE) values. Adapted from Branciamore, S. et al. (2018) Intrinsic properties of tRNA molecules as deciphered via Bayesian network and distribution divergence analysis. Life (Basel), 8, E5. (385). Profiles in (A) and (B) are adapted from panels c of Supplementary Figures S1 and S2 of reference (385). Arrows have been added to highlight the positions with the highest RE values associated with aminoacylation identity. According to the CC BY 4.0 license (https://creativecommons.org/licenses/by/4.0/).
Global data covering the conserved ‘identity positions’ in the three domains of life are summarized in Table 5. Several features are easily identifiable. (i) The number of conserved positions (nucleotides, base pairs and base-triples) in a given tRNA family is surprisingly low, with only two positions in the tRNAThr family and at most eight positions in the tRNAAsp family. (ii) Conserved identity positions in each of the 20 tRNA families always include anticodon positions. These positions are always found to be of medium importance and often correspond to experimentally proven strong aminoacylation determinants, meaning that additionally they could code for tRNA properties outside aminoacylation. (iii) About 59% of conserved identity positions relate to determinants for tRNA aminoacylation that are already validated in some of the presently 36 investigated model taxa (Supplementary Table S1). Only 13% map with positions outside the anticodon and mostly in tRNA families recognized by class II aaRSs. (iv) Only ∼13% of conserved positions show medium ‘relative entropies’. Altogether, and as expected, this highlights the importance of anticodons and the anticodon-encrypted genetic code in all extant tRNAs. It also shows the princeps status of tRNA aminoacylation determinants and suggests a primordial role for aaRSs in this process. (v) Significantly different distributions of conserved positions in the tRNA families agree with the functional and structural differences in class I and class II tRNA/aaRS systems. (vi) Many conserved positions sustain either specific tRNA conformation or presently uncharacterized new identities, in particular in tRNA families recognized by class Ib or class IIb aaRSs.
Table 5.
Importance of conserved ‘identity positions’ in tRNAs from the three domains of life, as measured by relative Eentropy according to (385)
| tRNA families | Nucleotides with important identity positions |
|---|---|
| Arg | 4–69; 20; 36 |
| Cys | 3–70; 13–22; 34; 35; 36 |
| Ile | 29–41; 34; 35; 36 |
| Leu | 12–23; 35; 36 |
| Met | 31–39; 34; 35; 36 |
| Val | 35 ; 36; 73 |
| Glu | 11–24; 12–23; 13, 34; 35; 36 |
| Gln | 12–23; 13–22; 34; 35; 38; 44; 73 |
| Trp | 31–39; 34; 35; 36 |
| Tyr | 12–23; 34; 35; 36 |
| Ala | 2–71; 3–70; 4–69; 20a; 35; 36 |
| Gly | 2–71; 3–70; 31–39; 35; 36 |
| His | 2–71; 32; 34; 35; 36; 38 |
| Pro | 2–71; 35; 36; 37 |
| Ser | 13–22; 35; 36; 46; 47; 73 |
| Thr | 35 ; 36 |
| Asp | 11–24; 20a; 25; 31–39; 34; 35; 36; 73 |
| Asn | 2–71; 31–39; 34; 35; 36; 51–63; 73 |
| Lys | 12–23; 34; 35; 36 |
| Phe | 12–23; 20a; 34; 35; 36; 73 |
tRNA families refer to the ensemble of isoacceptors and isodecoders specific to a given proteinogenic amino acid. Data are displayed according to the ranking of aaRSs in two classes, each subdivided into three subclasses. Relative entropy (RE) values are shown in regular black (RE 0.22–0.4) and bold black (RE ∼0.4–0.7). Note that RE values >0.7 are found in a few tRNA families from specific domains of life (see Supplementary Tables S3–S5). Positions of anticodons are underlined. Positions in italics are occupied in some taxa by validated determinants for tRNA aminoacylation.
More specific characteristics emerge when analyzing the predicted ‘identity positions’ in tRNAs from individual domains of life. In this case, the number of conserved positions in most tRNA families increases significantly, with up to 21, 25 and 22 positions in bacterial, eukaryal and archaeal tRNAs, respectively. This trend mainly concerns the tRNAMet, tRNAGlu, tRNAAsp and tRNAPhe families (Supplementary Tables S3–S5). However, in a few families, this number remains low, with only 3–7 important positions, notably three in the bacterial tRNAVal families and four in the eukaryal tRNAThr families. Finally, the variable distributions of predictions in the three domains of life associated with tRNA conformations or identity determinants support the particular evolutionary status of Archaea and of several tRNA families, notably the valine and threonine families that show well-differentiated importance profiles in the three domains of life.
A simplified global search
In this approach, the complexity of the problem was simplified by restricting the search to sequences of tRNAs whose lengths match the canonical cloverleaf (discarding the extra loop in the variable region and the 3′-CCAOH terminus). Overall ∼13 000 gene sequences retrieved from a curated database were analyzed by measuring information variations (374,384). The result of the study was the prediction of clusters of tRNA positions defining each specificity. A cluster contains a number of nucleotides whose presence is coordinated. In other words, each nucleotide present in a site is derived by the presence of a nucleotide found in another site of the cluster. In each cluster, the search differentiates the anticodon, Watson–Crick and non-Watson–Crick positions. The number of clusters for tRNAs recognized by class I aaRSs ranges from one (Arg, Met and Trp), two (Glu, Leu and Val), three (Gln), seven (Cys and Ile) and up to 13 (Tyr). For class II specificities, clusters range from two (Ala and Phe), three (Gly, Lys and Thr), four (Asp and His), five (Asn and Ser) to six (Pro) predicted identity positions. In each cluster, the number of positions varies from two to 13/14 in the clusters defining Cys, Ile or Tyr identities. Interestingly, in four large clusters (Arg, Gln, Ile and Leu), positions 8–14 and 54–58 (tertiary pairs) occur with anticodon position 35, an association that never occurs in clusters for class II aaRSs. In both classes, positions from the T loop are often associated with the anticodon. Except for anticodons that are always predicted, only a few base pairs correspond to conserved positions in the three domains of life, such as C3–G70 in 55% of tRNACys, or base pair 11–24 that is always Y11–R24 for Ile and often U11–A24 for Asp, in agreement with other results (385). Altogether, anticodon positions and positions associated with tertiary interactions are highlighted, but other predictions, such as the variable size of clusters, remain unexplained.
Other global comparative searches
Further tRNA searches in the three domains of life were performed using the Leipzig database (164) and the archaeal split tRNA databases (375), and on sequences specific to some phylogenetic groups (383). First, and as expected, positions in anticodon loops and at the top of the acceptor stems are predicted in Bacteria and Eukarya, with the highest probability in Bacteria (388). More interesting, positions 30–40 and 31–39 in the anticodon branch are predicted in several tRNAs as determinants, e.g. the pair U31–A39 in E. coli tRNATrp and S. cerevisiae tRNAMet and the pairs U30·G40 and G30·U40 in eukaryal tRNAsIle and S. cerevisiae tRNAAsp, respectively (383,388). With the exception of pair 30–40 in S. cerevisiae tRNAAsp slightly shifted from the predicted 31–39 pair, these findings are consistent with previous studies using a Bayesian search (385).
Secondly, identity clusters were searched in sequences of comparable length, omitting the variable region (389). Many of these clusters are related to position 35 of the anticodons. Furthermore, in Archaea, positions at the 5′ end of the acceptor stem are present in all clusters (except Leu). In Bacteria, position 8 is present in clusters for Ala, Arg, Gln, Gly, Thr, Trp and Val, and in Eukarya for Gln, Leu, Met, Phe, Ser, Trp, Tyr and Val, but never in Archaea. More generally, archaeal tRNAs possess the broadest spectrum of predicted identity elements, followed by eukaryal tRNAs. Some of the predicted positions are occupied by validated aminoacylation identity elements (e.g. the G3·U70 in tRNAAla is part of a cluster in the three domains of life). In addition, the G10·U25 base pair in S. cerevisiae tRNAAsp is either part of an eukaryal cluster or is associated with post-transcriptional modifications. Position 15, present in archaeal clusters, is probably associated with the identity of archaeosine tRNA guanine transglycosylase, the enzyme that inserts archaeosine derivatives into almost all archaeal tRNAs (390). However, the status of many positions (e.g. in the T branch) remains unknown and is probably associated with other tRNA functions.
Focused searches in individual kingdoms or specific tRNAs or derivatives
An extensive search for positions coding identity information in Archaea was performed on ∼4000 archaeal tRNA gene sequences from 86 species (instead of 22 previously) (386). The aim was to discover a minimal operational RNA code (moRNA) generalizing the operational RNA code for amino acids (10). The analysis was conducted on 43 potential informative positions outside the anticodon (since the anticodons alone perfectly predict the identity of the tRNAs). The six most informative positions are, respectively, G72, C73, U47, U73, A20 and G20 in the genes of tRNATyr, tRNAHis, tRNALeu, tRNAAla, tRNAArg and tRNAPhe (according to decreasing importance). These six positions are weakly predicted in other archaeal tRNAs, such as tRNAIle or tRNAVal. Interestingly, many of these statistically predicted positions were already known as archaeal identity elements for aminoacylation, for example in tRNALeu (55), tRNATyr (104), tRNAHis (143) and tRNAPhe (195). On the other hand, distinct phyla have different moRNA codes, suggesting that moRNA codes have evolved during speciation.
Other searches for identity determinants in mt-tRNAs have been performed in the ∼5000 mt-tRNA genes compiled in the Leipzig and Mamit databases (164,376). For example, the A14 and A73 determinants of human mt-tRNALeu(UUR) were found to be conserved in mammals (284). The A73 determinants and the GUC anticodon of human mt-tRNAAsp are conserved in primates but only partially in other mammalian families (293) (e.g. the GUC anticodon replaced by GCC in marsupials and A73 absent in insects but conserved in marsupials). The case of human mt-tRNAAla is notable for the shift of the determinant G3·U70 to position 5–68. The shift is found in primates, but only partially in other Mammalia and Eukarya that show a weak preference for G4·C69 wobble pairs. This results from sequence constraints relaxed in the mt-tRNAsAla of eumetazoans (286). In the human mt-tRNAPhe, the G1–C73 and A73 determinants are conserved in mammals but only partially in lower phyla (298,299).
Further focused searches on several hundred non-metazoan organisms largely confirmed the importance of A20 in the tRNAArg identity, but revealed notable exceptions in Stramenopiles and Diatoms, where A20 is replaced by C20 or U20 in most cytosolic tRNAs, whereas it is conserved in mitochondria and plastids (391).
The recent search for sequence covariations in viral genomic RNAs, complemented by biochemical studies, has expanded the world of viral TLSs (247,248). As for TLSsVal, 108 examples of in vitro valylatable TLSs were found in plant-infecting viruses and in some insect-infecting tetraviruses. Except for their 7 nt anticodon loops (with fully conserved A35CAC38) and their well-conserved anticodon stems, these molecules show great structural heterogeneity. In a few cases, the insertion of large stem–loops into the TLS core does not prevent valylation, and in one case an anticodon distant from the valine anticodon prevents valylation. In contrast to the canonical tRNAsVal where the discriminator base A73 is conserved and acts as a universal identity determinant, the discriminator of TLSsVal is not conserved, being A73 or C73 (247). However, the strong valine identity element A35 in the anticodon of tRNAVal is present in all TLSsVal and is complemented by C36 as characterized in TYMV TLSVal aminoacylated by wheat germ ValRS (392). Similarly, many new examples of histidinylatable TLSsHis have been added to the previously described TLSsHis (222). Like other viral TLSs, their sequences are organized in a consensus secondary structure in a pseudoknotted secondary structure (Figure 4C) that contains the identity elements previously characterized in tobamoviral TLSHis, including the atypical pair N−1–N73 and G34, U35 of the His anticodon (248).
Search of structural features in tRNA and tRNA:aaRS complexes related to tRNA identity
Evolutionary conservation of interacting regions
The binding surfaces between tRNAs and their respective aaRSs were studied in 16 crystal structures of tRNA:aaRS complexes, mainly from Bacteria and Eukarya (393). This interaction information was analyzed considering a sequence conservation analysis carried out on tRNA sequences of ∼400 organisms evenly distributed in the three domains of life. This allowed the identification of interaction regions in the tRNA:aaRS complexes in the three domains of life and the evolutionarily conserved ribonucleotides in the tRNA molecules (393). The resulting common features and heterogeneity between the tRNA and tRNA:aaRS complexes can be summarized as follows. (i) Three regions (anticodon loop, -CCAOH terminal region, followed by D stem) and two additional loop regions (D and T loops) involved in the formation of the tRNA L-shape are largely conserved. (ii) In tRNA:aaRS complexes, the regions of tRNA that interact most with aaRSs are the 3′-CCAOH and the anticodon loop, except in the bacterial Leu and Ser complexes. (iii) The 5′ half of tRNAs contains more interacting nucleotide than the 3′ half. (iv) In Bacteria, tRNA:aaRS complexes classified according to similarities in their tRNA interaction patterns have led to six classes. They occur in a mosaic pattern, in which the tRNA:aaRS complexes corresponding to each class are intermingled, suggesting that variations in the interaction characteristics between tRNAs and aaRSs are not always dependent on the aaRS class (393).
Structural signatures in tRNAs related with aminoacylation identity
Despite the large number of tRNA genes found in eumetazoans, specific tRNA sequence motifs are highly conserved and often span one or more sets of isoacceptors and isodecoders (394). This is the case for non-Watson–Crick base pairs in helical stems, notably G·U pairs and non-isosteric U·G pairs. The G10·U25 pair in the D stem is found in five families of tRNA isoacceptors, including tRNAAsp. The G10·U25 pair has been shown to participate indirectly in the aspartylation identity of S. cerevisiae tRNAAsp (184,327). Similarly, the U30·G40 pair is found in the anticodon stem of mammalian and insect tRNAsIle and other isoacceptors (394). The U30·G40 pair may be conserved because it interacts with the ribosome during translocation. For example, the suppression efficiency of the yeast amber tRNAIle in E. coli is modulated by the presence of the U30·G40 pair and is accompanied by an identity change revealing a different mode of recognition of the anticodon nucleotides (209). Similarly, although mutagenesis of the G30–U40 pair in the anticodon stem of S. cerevisiae tRNAAsp only marginally affects aspartylation (184), it results in a deviation of the anticodon arm and a conformational change in the anticodon loop required for recognition of identity elements by AspRS (315,326).
Primordial identities in ancestral systems
Early studies suggested that the two classes of primordial aaRSs or proto-aaRSs might originally be encoded by complementary strands of the same nucleic acid (395). Functional class I and II amino acid-activating peptides encoded by opposite strands of the same gene have been characterized more recently (396). This is a first step towards the characterization of proto-aaRSs with aminoacylation activity. This raises questions about the mechanism of recognition of ancestral tRNAs by proto-aaRSs. Regression methods have identified possible sequence motifs in modern aaRSs that are able to perform this discrimination. As a result, groove recognition rules have emerged based on the differential thermodynamic stability of tRNA helical -NCCAOH extensions (397).
Coding triplets in tRNA acceptor stems
The relationship between tRNA aminoacylation and the genetic code embedded in the tRNA acceptor arm has recently been re-investigated. A large-scale analysis of bacterial tRNA sequences revealed that for six amino acids (Ala, Asp, Gly, His, Pro and Ser), mainly those considered to be the oldest, the tRNA acceptor arm contains the corresponding coding triplets well beyond the statistical expectation (398). As relics and early identity elements, these coding triplets are suggested to have a primordial origin, being involved in the aminoacylation of pre-biotic tRNAs (399) and the establishment of the canonical codon set in agreement with the RNA operational code theory (10).
The unexplored identities of tRNA-derived fragments
A completely unexplored aspect in the emerging field of tRF (tRNA-derived RNA fragment) biology is the search for identity signatures for the recognition of the macromolecular partners interacting with these RNAs (224). These tRFs present in the three domains of life regulate many cellular processes, e.g. stress and immune responses, crosstalk with ribosomes, reverse transcriptases, tRNA modification enzymes and aaRSs. Recent work on tRF regulation of ribosome-associated aaRSs in S. cerevisiae provides information for a better understanding of identity signatures in tRNAs (225). Five tRFs [3′-tRFHis(GUG), 3′-tRFSer(AGA), 3′-tRFLeu(UAA), 3′-tRF1Thr(UGU) and 5′-tRFHis(GUG)] interact with their cognate yeast aaRSs and impact tRNA aminoacylation. Interestingly, 3′-tRFSer(AGA) includes part of the long variable arm important for serine identity, and both 3′-tRFLeu(UAA) and 3′-tRF1Thr(UGU) include, respectively, A73 and G71C72 known as identity determinants of aminoacylation in eukaryal tRNAs (Table 1). Such observations should stimulate further studies of tRFs, in particular to deconvolute the encrypted identity signatures in tRNAs.
tRNA identity for other tRNA-binding proteins
Fewer studies have been devoted to the study of identity elements governing tRNA biosynthesis or other tRNA functions. However, their number is increasing, starting with studies that focus on the identity elements of tRNA-modifying enzymes. For example, the G10–U25 base pair in the D arm was found as a major identity determinant for formation of m22G26 or m22G10 by archaeal Pyrococcus abyssi m2G transferases (400). In S. cerevisiae, the essential elements required for G10 methylation by the Trm11–Trm12 complex are the terminal A76 and the G10–C25 base pair in tRNAs with a variable region of regular size (e.g. tRNAs specific for Arg, Asn, Ile, Leu, Lys, Met, Phe, Thr, Trp, Tyr and Val). In addition, U38 in tRNAAla and the U32–A38 base pair in tRNACys are negative identity elements against this methylation (401). For t6A37 biosynthesis (e.g. in tRNAIle or tRNAThr), C32 and the D stem are essential determinants for the modification machinery (known as KEOPS complexes) in yeast, humans and nematodes (40). TrmL is the prokaryotic methyltransferase that catalyzes the 2′-O-methylation of base 34 of the isoacceptors tRNALeu(CAA) and tRNALeu(UAA). The anticodon loop of these tRNAs is critical for TrmL recognition. Nucleotide A35 is a key determinant, as is the A36A37A38 motif, which additionally requires the presence of the prior isopentenylation (i6) of A37. Only pyrimidine nucleotides at position 34 are substrates of TrmL (402). In another example, the human tRNA methylase DNMT2/TRDMT1 was shown to catalyze formation of m5C38 of several tRNAs thanks to a conserved identity sequence C32UNNCAC38 found in the anticodon loop (403).
Formylation of the tRNAMet initiator is another form of post-transcriptional modification. The formyl group is present in Prokarya and in organelles (chloroplasts and mitochondria) but not in Eukarya. Formylation is not dependent on the side chain of the esterified amino acid, and several misaminoacylated tRNAs have been shown to be formylatable, suggesting that the nucleotide sequence of the tRNA rather than the esterified amino acid carries the formylation identity. Indeed, the identity elements for methionyl-tRNA transformylase are clustered in the acceptor stem with a major role for the unpaired pair C1–A72 assisted by the minor determinants A73, G2–C71, C3–G70 and G4–C69 (404–406). Acetylation of the amino acid charged on the tRNA is another post-transcriptional modification. Acetylation occurs on the glycine charged on tRNAGly by TacT toxin of Salmonella typhimurium. The identity of the acetylation activity is specified by U73 and G71, a combination of nucleotides found only in tRNAGly isoacceptors (407).
In addition to their role in protein biosynthesis, aminoacyl-tRNAs are found to participate in various biochemical processes, such as cell wall formation, protein labeling for degradation, aminoacylation of cell membrane phospholipids or synthesis of peptides with antibiotic properties (408). For the synthesis of the peptidoglycan of the bacterial cell wall, there is a plethora of enzymes depending on the bacterium and the wall to be synthesized. Various amino acids are ligated to the peptidoglycan, but glycine, alanine and serine are the most frequently incorporated from the corresponding aminoacylated tRNAs. These tRNAs can be dedicated to synthesis of peptidoglycans. For example, there are three non-proteinogenic tRNAsGly in Staphylococcus aureus, in which sequence elements have been replaced to escape EF-Tu. The lost identity elements for EF-Tu include base pairs 49–65, 51–63, 49–65 and 51–63 in the T loop. The new determinants for these enzymes remain to be discovered (409,410).
To resist cationic antimicrobial peptides, many Bacteria have developed resistance mechanisms through MprF proteins that aminoacylate anionic phospholipids with l-lysine or l-alanine. The presence of positive charges on the membrane surface reduces the affinity for cationic antimicrobial peptides. How MprF and similar enzymes divert aminoacyl-tRNAs to membrane lipid modification remains an open question, as these tRNAs seem to have the same affinity for EF-Tu and MprF (411). The specificity of MprF was proposed to arise from direct recognition of the aminoacyl moiety of the aminoacyl-tRNA (411).
The aminoacyl-tRNAs also serve as amino acid donors in the synthetic pathways of significantly different compounds with antibiotic properties (412), including valanimycin, pacidamycin and cyclodipeptides (413). The identity elements for the enzymes responsible for these transformations are still unknown. Aminoacyl-tRNAs also serve as amino acid donors in protein degradation pathways through aminoacyl-tRNA-protein transferases, which recognize a secondary destabilizing residue at the N-terminus of proteins and attach a primary destabilizing residue. In Eukarya, this added residue is the arginine bound by the arginyl(R)-transferase. In Prokarya, leucine and phenylalanine are the primary destabilizing N-terminal residues for leucyl/phenylalanyl(L/F)-transferases. Aminoacyl-tRNA-protein transferases specifically recognize the aminoacyl moiety of aminoacyl-tRNAs and the constant unpaired CCA76 nucleotides of their acceptor ends (414,415).
The tRNAs also serve as substrates for various processing enzymes. RNase P and Z mature the 5′ and 3′ ends of primary transcripts regardless of their sequences. Similarly, tRNA nucleotidyltransferase adds the CCA end to all tRNAs whatever their identity and sequence. The tRFs are produced by cleavage in the anticodon loop by angiogenin or colicin nucleases. The removal of introns from Archaea and Eukarya tRNAs is orchestrated by several enzymatic activities. The process depends little on conserved sequence-specific recognition and is primarily based on proximity and base pair interactions between the intron and the tRNA body to form the proper structure for cleavage (416).
As exposed above, most Bacteria and Archaea do not possess AsnRS and/or GlnRS, and some methanogenic archaea do not possess CysRS. Furthermore, no aaRS for the rare amino acid selenocysteine has been found in any domain of life. Instead, these organisms use indirect pathways to synthesize these amino acids (Asn, Cys, Gln and Sec) directly on their corresponding tRNAs. After an initial step in which non-discriminating aaRSs form misacylated aminoacylated tRNAs, they are converted to the corresponding aminoacyl-tRNAs by various RNA-dependent modification enzymes (GatCAB, GatDE and SepCysS) that specifically recognize the corresponding identity determinants. How these misacylated aminoacyl-tRNAs are recognized by the modifying enzymes and diverted from the translation machinery, where they could be toxic, has been studied and is described in the preceding sections.
Together, the different enzymes that interact with tRNAs imply the presence on a given tRNA of distinct or overlapping sets of identities. By analogy with recognition by aaRSs, indirect reading of structural elements on tRNAs could also be used. The search for these new identity elements could benefit from the large-scale bioinformatics analyses currently being carried out in the three domains of life (385,386,389).
HUMAN DISEASES RESULTING FROM DISORDERS IN tRNA IDENTITY
Mutations in mt-tRNAs have been known for years to cause human diseases (417). More recently, cytoplasmic tRNA variants have also been identified (418,419). The toxicity of these disease-causing mutations often has pleiotropic effects that affect all stages from biogenesis, structure and function of tRNAs (420).
The number of diseases linked to dysfunction in human tRNAs is increasing. Diseases due to mutations in mt-tRNAs cover diverse pathologies, such as cardiopathies and neuropathies, with a wealth of clinical manifestations (277). A largely ignored cause of diseases relies on perturbed expression of genes encoding many tRNA modification enzymes, leading to aberrant tissue-specific profiles of tRNA modifications (421). Diseases, named ‘modopathies’, are associated with aberrant tRNA modifications (422). Today >350 mutations in human mt-tRNAs are compiled in the MITOMAP (http://www.mitomap.org) databases; among them about half are pathogenic (423). Their discovery, prediction, evolution and penetrance (presence in other organisms) result from a plethora of theoretical, experimental and clinical studies (424). Pathogenic mutations occur in the 22 human mt-tRNAs, but with uneven distributions. The two most common (A3243G and A8344G), which account for a majority of diseases, are expressed in mt-tRNALeu(UUR) and mt-tRNALys (Figure 9).
Figure 9.
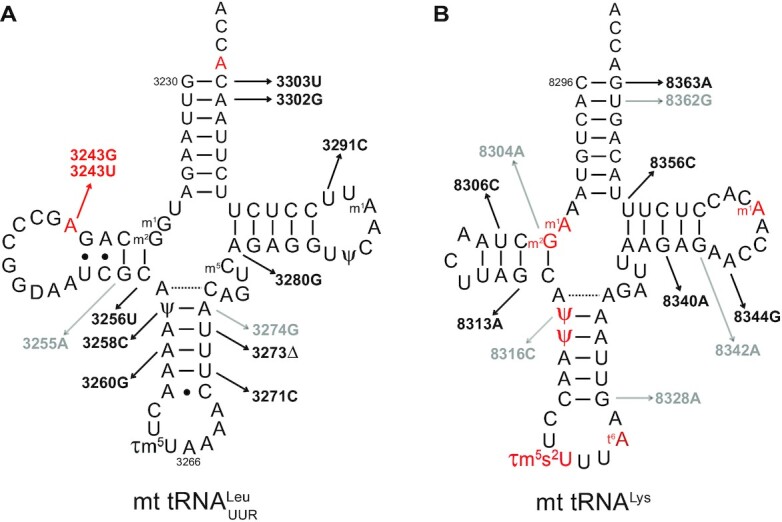
Pathogenic human mt-tRNAs, with most known mutations highlighted. (A) mt-tRNALeu(UUR) and (B) mt-tRNALys cloverleaves with base modifications indicated. The location of mutations is indicated by arrows (black arrows for mutations with confirmed pathogenetic status, gray arrows for mutations of ‘likely pathogenic’ status). Red arrows highlight confirmed pathogenic mutations affecting identity elements. Data and status are from MITOMAP.
In general, pathogenic mutations are mild, minimizing the risk of lethality, and rarely affect major identity positions in the tRNA. Most of the time, they are located in the vicinity of the positions occupied by the major identity determinants and often lead to mismatches in the helical regions that destabilize the conformations recognized by the aaRSs. Six pathogenic mutations leading to mismatches occur in the four stems of mt-tRNAAla (425), but only two (C6U and C69U) lead to G·U mismatches in the acceptor stem and are related to alanine identity (426). Thus, pathogeny is due to mutation of minor alanine identity elements.
Diseases associated with aminoacylation identity affect five other tRNA specificities (Ile, Leu, Asp, Ser and Lys). In mt-tRNAIle, the two isoleucine determinants A7 and A59 in the acceptor stem and T loop, found in two cardiopathy-causing variants with G7 and G59, are not located at the positions of known identity determinants and act by conformational effects (300). In well-studied human mt-tRNALeu(UUR), three disease-causing mutations are associated with MELAS syndrome (Mitochondrial Encephalomyopathy, Lactic Acidosis and Stroke-like episodes). These mutations either disrupt the conserved U8:A14 reverse Hoogsteen base pair or are associated with a perturbed base triple (G13–C22):G46 and a fragile anticodon stem constituted of four A–U pairs and a mismatched A41·C39 pair closing the 7 nt anticodon loop (Figure 9A). Mutations affect both the tRNA leucylation efficiency and the tRNA structure. Interestingly, this fragile anticodon stem participates in leucine identity. Collectively, this indicates that the pathogenicity of mt-tRNALeu(UUR) is sensitive to perturbed structural identity determinants and anticodon stem structure (284). In human mt-tRNAAsp, the A9G mutation in the D stem, associated with myopathy, disrupts the triple interaction A9:A12–U23 and reduces aspartylation efficiency. Therefore, both A9 and A9:A12–U23 may participate in aspartate identity (294). In mt-tRNASer(UCN), the G31A mutation associated with polycystic ovary syndrome and insulin resistance disrupts a G·U pair at the top of the anticodon stem and alters aminoacylation (427). In mt-tRNALys (Figure 9B), the absence of m1 methylation at A9 in the core of the tRNA impairs cloverleaf folding and alters aminoacylation (240). Finally, nine mitochondrial ‘modopathies’ affect nucleotides undergoing modification (428). Aberrant modification patterns (i.e. at i6A, m1A, Ψ, m1G, m22G, f5C, m5C and hypermodified U positions in anticodon loops) alter various steps of tRNA biology, including aminoacylation (240,300) and codon reading (203). Similarly, hypomodifications in pathogenic cytoplasmic tRNAs are common, particularly in the tRNA core and in the anticodon loops (i.e. at the positions mchm5U34, ms2t6A37 and yW37) (422). This probably perturbs tRNA conformations, impacting tRNA aminoacylation identity and/or codon reading on the ribosome.
Emerging evidence points to an increasing role for tRNA mutations in genetic disorders, cancer and neurological diseases. In this context, ∼600 pathogenic mutations have been identified in human tRNA genes, with consequences for tRNA function, mRNA translation and proteome composition. For example, a mutation at C65 in the extended acceptor stem of human tRNASec causes a complex phenotype of neurological disorders due to reduced expression of the tRNA and decreased 2′-O-methylribosylation at mcm5U34 in the anticodon of mutant tRNASec (418). As the C65 mutation occurs in one of the three base pairs that constitute identity elements for SelB, the elongation factor for selenoprotein translation in E. coli, it is tempting to propose that a disruption in recognition of the mammalian SelB homolog explains pathology (418).
CONCLUSION
A critical look at the methodologies used in identity studies
The determination of identity elements in tRNAs remains currently a technological issue that is not yet fully solved. Early approaches to constructing modified tRNAs by fragment ligation were complex and time consuming, e.g. (66). The use of suppressor tRNAs has proven to be an improvement due to its greater ease of implementation and the cellular context in which competition between aaRSs is ubiquitous, e.g. (51). However, the approach remains limited for some identities that use nucleotides in their anticodons that differ from those present in suppressor tRNAs. The synthesis of in vitro transcribed tRNA variants has been by far the most widely used (193). However, it is limited by the absence of modified bases, which can sometimes play a decisive role in identity. Modified bases may be involved in stabilizing the tertiary structure of tRNAs and thus play a role in indirect reading by ensuring the positioning of identity elements. In addition, standard nucleotide substitutions result in multiple atomic group changes relative to the wild-type base, leading to steric effects or repulsive effects. Interactions with the aaRS may disappear or be redistributed and generate amplifications or attenuations of the primary effects. Another drawback of the method is that aminoacylation assays are often performed in vitro with purified enzymes outside the context of competition, under experimental conditions that may be far from the cellular environment. Transplantation of the identified identity elements into another tRNA core is additional evidence that validates the identification, although it is also subject to some caveats due to the use of a different tRNA backbone.
Alternatively, mutagenesis of the side chains of aaRS residues that interact with identity elements can be used to remove interactions with identity elements (typically by Ala mutagenesis). Studies on well-known model systems have shown that loss of interactions involved in identity generally induces relaxed tRNA specificity with decreases in kcat in the presence of competing non-cognate tRNAs but not in the presence of the pure cognate tRNA. This loss of discrimination against non-cognate tRNAs revealed by this approach allows for a more certain identification of interactions involved in identity (215,308).
Another method not yet employed would be to create abasic tRNAs at defined positions that would allow precise measurement of the effect of the loss of a given interaction [inspired by the molecular surgery already used in the case of tRNAAla (126,316,429)]. Although molecular tools are still lacking to create such mutants, abasic sites are already known, for example in 28S rRNA where the base A4323 is removed by ricin (430). Abasic sites are also found in tRNAs where certain bases are labile, such as wybutosine (Y or yW) at position 37 in tRNAPhe (431), as well as the modified base m7G at position 46 in other tRNAs (432). Interestingly, removal of yW37 significantly impairs phenylalanine acceptance (433). It can be expected that in the near future new tools will be available. The recent characterization of methylpurine DNA glycosylase, which generates abasic RNAs on RNA–DNA hybrids (434), may become programmable in the future, opening up exciting prospects in the search for tRNA identity elements.
Future directions for tRNA identity research
Over the past two decades, the field of tRNAs has entered a new era in which the prototypical concept of tRNA identity has been challenged and progressively refined in response to new discoveries. The increasing importance of auxiliary protein factors alongside aaRSs in tRNA biology is significant.
This has produced a paradigm shift, with semantic changes illustrated by the concepts of ‘quality control’ underlying tRNA aminoacylation (361) and of ‘code biology’ (435) extending the ‘operational RNA code’ (10). More generally, tRNA is a pivotal marker in evolution, illustrated by breakthroughs in eukaryotic tRNA biology (2,3). On the other hand, large-scale searches of tRNA sequences by bioinformatics methods revealed patterns of importance of tRNA positions related to identities and suggested that ‘almost nothing in most tRNA positions is even close to random’ (385). Therefore, the long-term prospect will be to decipher globally in given tRNAs the functional role of nucleotides at these positions.
What are the most immediate prospects? Only a few aminoacylation identities are known in organelles, and the field is completely open for apicoplast and chloroplast tRNAs. Furthermore, it is crucial to better understand identities outside of aminoacylation, especially for tRNA maturation and splicing. To do this, known and predicted determinants must be deconvoluted in terms of aminoacylation and associated functions. In addition, much remains to be discovered in tRNA biology, including its dynamic regulation under specific environmental and ecological conditions. In particular, how the cellular tRNA pool varies in response to various metabolic contexts is poorly understood and needs to be quantified. Epistasic effects that cover combined effects of mild mutations in tRNAs and/or aaRSs are other challenges (387,436). To achieve these ambitious goals, appropriate biochemical, genetic, computational and deep sequencing tools are needed. Finally, it is becoming clear that the identity of tRNAs recapitulates the evolutionary history of protein synthesis and life, and this puzzle must be clarified to understand the origins of tRNAs.
DATA AVAILABILITY
All data are available from the authors upon request.
Supplementary Material
Appendix
Definitions
Catalytic efficiency: defined by the kcat/KM ratio of aminoacylation reactions, with kcat the catalytic rate constant and KM the Michaelis constant representing an approximation of the inverse of tRNA affinity for an aaRS, so that ‘L’ = (kcat/KM)native/(kcat/KM)mutant (with ‘L’ representing the loss of catalytic efficiency).
Classes of tRNAs: based on the presence of a small variable region of 4/5 nt (class I) or a large variable region with an extra loop (class II).
Classes and subclasses of aaRSs: defined based on the structure and peptide motifs present in their catalytic sites.
Identity determinant: nucleoside or chemical groups on a nucleoside, as well as structural elements (mainly in tRNAs) essential for biological activity (in proteins, this concerns amino acids).
Identity set: a set of key determinants for specific activities.
Identity rules: account for the operational functioning of identity determinants in taxonomic groups (originally they applied exclusively to the aminoacylation reaction of tRNAs and were considered ‘universal’ if they were conserved in the three domains of life).
Isoacceptor tRNAs: tRNAs that accept the same amino acid and may have different anticodons due to genetic code degeneracy.
Isodecoder tRNAs: tRNAs with the same anticodon sequence that decode the same codon. They belong to the same class of isoacceptors but have sequence differences elsewhere in the tRNA body.
Strength of aminoacylation identity determinants: measured in vitro by losses of catalytic efficiency of aminoacylation reactions of tRNA upon mutation at identity positions. Alternatively, strength can be measured in vivo by E. coli suppressor genetics that utilizes a reporter dihydrofolate reductase gene with the amber mutational position 10.
Tertiary interactions in tRNA: interactions that stabilize the structure of tRNA. They are mainly located in the core of the tRNA molecule and include, for example, interactions between the D stem and variable loop, T loop and anticodon loop, and U8 with A14.
tRNA domains: D stem/loops from N10 to N25 with loops from A14 to A21 divided into α and β subdomains (sizes from 2 to 4 nts) located down- and upstream of the constant G18 and G19); anticodon stem/loops from N27 to N43 with 7 nt loops (N32–N38); variable regions (VR) from N44 to Y48 with smallVRs (4 or 5 nt) or largeVRs (also named extra arm) (up to 16 nt from positions 47a to 47p); T stem/loops from N47 to N65 with 7 nt loops (T54–N60); connecting Ns: U8N9 and N26.
Contributor Information
Richard Giegé, Architecture et Réactivité de l’ARN, UPR9002 Centre National de la Recherche Scientifique, Université de Strasbourg, Institut de Biologie Moléculaire et Cellulaire, 2 allée Konrad Roentgen, 67084 Strasbourg, France.
Gilbert Eriani, Architecture et Réactivité de l’ARN, UPR9002 Centre National de la Recherche Scientifique, Université de Strasbourg, Institut de Biologie Moléculaire et Cellulaire, 2 allée Konrad Roentgen, 67084 Strasbourg, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank all our colleagues, both local, national, and worldwide, with whom we have had many discussions, exchanges of ideas, and collaborations in the field of tRNA over the years. Their names and contributions are listed in the references.
FUNDING
This work was supported by the French ‘Centre National de la Recherche Scientifique’; ‘University of Strasbourg’, and ‘Agence Nationale de la Recherche’ [ANR-17-CE11-0024].
Conflict of interest statement. None declared.
REFERENCES
- 1. Berg M.D., Brandl C.J.. Transfer RNAs: diversity in form and function. RNA Biol. 2021; 18:316–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rak R., Dahan O., Pilpel Y.. Repertoires of tRNAs: the couplers of genomics and proteomics. Annu. Rev. Cell Dev. Biol. 2018; 34:239–264. [DOI] [PubMed] [Google Scholar]
- 3. Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018; 19:45–58. [DOI] [PubMed] [Google Scholar]
- 4. Giegé R., Sissler M., Florentz C.. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998; 26:5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beuning P.J., Musier-Forsyth K.. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers. 1999; 52:1–28. [DOI] [PubMed] [Google Scholar]
- 6. Giegé R., Jühling F., Pütz J., Stadler P., Sauter C., Florentz C.. Structure of transfer RNAs: similarity and variability. Wiley Interdiscip. Rev. RNA. 2012; 3:37–61. [DOI] [PubMed] [Google Scholar]
- 7. Chambers R.W. On the recognition of tRNA by its aminoacyl-tRNA ligase. Prog. Nucleic Acid Res. Mol. Biol. 1971; 11:489–525. [DOI] [PubMed] [Google Scholar]
- 8. Mirzabekov A.D., Lastity D., Levina E.S., Bayev A.A.. Localization of two recognition sites in yeast valine tRNA I. Nat. New Biol. 1971; 229:21–22. [DOI] [PubMed] [Google Scholar]
- 9. Giegé R., Puglisi J.D., Florentz C.. tRNA structure and aminoacylation efficiency. Prog. Nucleic Acid Res. Mol. Biol. 1993; 45:129–206. [DOI] [PubMed] [Google Scholar]
- 10. Schimmel P., Giegé R., Moras D., Yokoyama S.. An operational RNA code for amino acids and possible relationship to genetic code. Proc. Natl Acad. Sci. USA. 1993; 90:8763–8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hou Y.M., Schimmel P.. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988; 333:140–145. [DOI] [PubMed] [Google Scholar]
- 12. McClain W.H., Foss K.. Changing the identity of a tRNA by introducing a G–U wobble pair near the 3′ acceptor end. Science. 1988; 240:793–796. [DOI] [PubMed] [Google Scholar]
- 13. Saks M.E., Sampson J.R., Abelson J.N.. The transfer RNA identity problem: a search for rules. Science. 1994; 263:191–197. [DOI] [PubMed] [Google Scholar]
- 14. Tuerk C., Gold L.. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990; 249:505–510. [DOI] [PubMed] [Google Scholar]
- 15. Rubio Gomez M.A., Ibba M.. Aminoacyl-tRNA synthetases. RNA. 2020; 26:910–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ibba M., Söll D.. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000; 69:617–650. [DOI] [PubMed] [Google Scholar]
- 17. McClain W.H., Foss K.. Changing the acceptor identity of a transfer RNA by altering nucleotides in a ‘variable pocket. Science. 1988; 241:1804–1807. [DOI] [PubMed] [Google Scholar]
- 18. Schulman L.H., Pelka H.. The anticodon contains a major element of the identity of arginine transfer RNAs. Science. 1989; 246:1595–1597. [DOI] [PubMed] [Google Scholar]
- 19. McClain W.H., Foss K., Jenkins R.A., Schneider J.. Nucleotides that determine Escherichia coli tRNAArg and tRNALys acceptor identities revealed by analyses of mutant opal and amber suppressor tRNAs. Proc Natl Acad. Sci. USA. 1990; 87:9260–9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tamura K., Himeno H., Asahara H., Hasegawa T., Shimizu M.. In vitro study of E. coli tRNAArg and tRNALys identity elements. Nucleic Acids Res. 1992; 20:2335–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sissler M., Giegé R., Florentz C.. Arginine aminoacylation identity is context-dependent and ensured by alternate recognition sets in the anticodon loop of accepting tRNA transcripts. EMBO J. 1996; 15:5069–5076. [PMC free article] [PubMed] [Google Scholar]
- 22. Aldinger C.A., Leisinger A.-K., Igloi G.L.. The influence of identity elements on the aminoacylation of tRNAArg by plant and Escherichia coli arginyl-tRNA synthetases. FEBS J. 2012; 279:3622–3638. [DOI] [PubMed] [Google Scholar]
- 23. Stephen P., Ye S., Zhou M., Song J., Zhang R., Wang E.-D., Giegé R., Lin S.-X.. Structure of Escherichia coli arginyl-tRNA synthetase in complex with tRNAArg: pivotal role of the D-loop. J. Mol. Biol. 2018; 430:1590–1606. [DOI] [PubMed] [Google Scholar]
- 24. McShane A., Hok E., Tomberlin J., Eriani G., Geslain R.. The enzymatic paradox of yeast arginyl-tRNA synthetase: exclusive arginine transfer controlled by a flexible mechanism of tRNA recognition. PLoS One. 2016; 11:e0148460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fender A., Geslain R., Eriani G., Giegé R., Sissler M., Florentz C.. A yeast arginine specific tRNA is a remnant aspartate acceptor. Nucleic Acids Res. 2004; 32:5076–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu W., Huang Y., Eriani G., Gangloff J., Wang E.-D., Wang Y.-L.. A single base substitution in the variable pocket of yeast tRNAArg eliminates species-specific aminoacylation. Biochim. Biophys. Acta. 1999; 1473:356–362. [DOI] [PubMed] [Google Scholar]
- 27. Guigou L., Mirande M.. Determinants in tRNA for activation of arginyl-tRNA synthetase: evidence that tRNA flexibility is required for the induced-fit mechanism. Biochemistry. 2005; 44:16540–16548. [DOI] [PubMed] [Google Scholar]
- 28. Delagoutte B., Moras D., Cavarelli J.. tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J. 2000; 19:5599–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shimada A., Nureki O., Goto M., Takahashi S., Yokoyama S.. Structural and mutational studies of the recognition of the arginine tRNA-specific major identity element, A20, by arginyl-tRNA synthetase. Proc. Natl Acad. Sci. USA. 2001; 98:13537–13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pallanck L., Li S., Schulman L.H.. The anticodon and discriminator base are major determinants of cysteine tRNA identity in vivo. J. Biol. Chem. 1992; 267:7221–7223. [PubMed] [Google Scholar]
- 31. Komatsoulis G.A., Abelson J.. Recognition of tRNACys by Escherichia coli cysteinyl-tRNA synthetase. Biochemistry. 1993; 32:7435–7444. [DOI] [PubMed] [Google Scholar]
- 32. Hou Y.M., Sterner T., Bhalla R.. Evidence for a conserved relationship between an acceptor stem and a tRNA for aminoacylation. RNA. 1995; 1:707–713. [PMC free article] [PubMed] [Google Scholar]
- 33. Hamann C.S., Hou Y.M.. Enzymatic aminoacylation of tRNA acceptor stem helices with cysteine is dependent on a single nucleotide. Biochemistry. 1995; 34:6527–6532. [DOI] [PubMed] [Google Scholar]
- 34. Hou Y.M., Westhof E., Giegé R.. An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc. Natl Acad. Sci. USA. 1993; 90:6776–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hauenstein S., Zhang C.-M., Hou Y.-M., Perona J.J.. Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2004; 11:1134–1141. [DOI] [PubMed] [Google Scholar]
- 36. Hou Y.M., Motegi H., Lipman R.S., Hamann C.S., Shiba K.. Conservation of a tRNA core for aminoacylation. Nucleic Acids Res. 1999; 27:4743–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hauenstein S.I., Perona J.J.. Redundant synthesis of cysteinyl-tRNACys in Methanosarcina mazei. J. Biol. Chem. 2008; 283:22007–22017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muramatsu T., Nishikawa K., Nemoto F., Kuchino Y., Nishimura S., Miyazawa T., Yokoyama S.. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988; 336:179–181. [DOI] [PubMed] [Google Scholar]
- 39. Normanly J., Kleina L.G., Masson J.M., Abelson J., Miller J.H.. Construction of Escherichia coli amber suppressor tRNA genes. III. Determination of tRNA specificity. J. Mol. Biol. 1990; 213:719–726. [DOI] [PubMed] [Google Scholar]
- 40. Pallanck L., Schulman L.H.. Anticodon-dependent aminoacylation of a noncognate tRNA with isoleucine, valine, and phenylalanine in vivo. Proc. Natl Acad. Sci. USA. 1991; 88:3872–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nureki O., Niimi T., Muramatsu T., Kanno H., Kohno T., Florentz C., Giegé R., Yokoyama S.. Molecular recognition of the identity-determinant set of isoleucine transfer RNA from Escherichia coli. J. Mol. Biol. 1994; 236:710–724. [DOI] [PubMed] [Google Scholar]
- 42. Farrow M.A., Nordin B.E., Schimmel P.. Nucleotide determinants for tRNA-dependent amino acid discrimination by a class I tRNA synthetase. Biochemistry. 1999; 38:16898–16903. [DOI] [PubMed] [Google Scholar]
- 43. Hendrickson T.L. Recognizing the D-loop of transfer RNAs. Proc. Natl Acad. Sci. USA. 2001; 98:13473–13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tomikawa C., Auxilien S., Guérineau V., Yoshioka Y., Miyoshi K., Hori H., Fourmy D., Takai K., Yoshizawa S.. Characterization of redundant tRNAsIle with CAU and UAU anticodons in Lactobacillus plantarum. J. Biochem. 2018; 163:233–241. [DOI] [PubMed] [Google Scholar]
- 45. Uesugi G., Fukuba Y., Yamamoto T., Inaba N., Furukawa H., Yoshizawa S., Tomikawa C., Takai K.. Recognition of tRNAIle with a UAU anticodon by isoleucyl-tRNA synthetase in lactic acid bacteria. FEBS J. 2022; 289:4888–4900. [DOI] [PubMed] [Google Scholar]
- 46. Ikeuchi Y., Kimura S., Numata T., Nakamura D., Yokogawa T., Ogata T., Wada T., Suzuki T., Suzuki T.. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat. Chem. Biol. 2010; 6:277–282. [DOI] [PubMed] [Google Scholar]
- 47. Senger B., Auxilien S., Englisch U., Cramer F., Fasiolo F.. The modified wobble base inosine in yeast tRNAIle is a positive determinant for aminoacylation by isoleucyl-tRNA synthetase. Biochemistry. 1997; 36:8269–8275. [DOI] [PubMed] [Google Scholar]
- 48. Thiaville P.C., El Yacoubi B., Köhrer C., Thiaville J.J., Deutsch C., Iwata-Reuyl D., Bacusmo J.M., Armengaud J., Bessho Y., Wetzel C.et al.. Essentiality of threonylcarbamoyladenosine (t6A), a universal tRNA modification, in bacteria. Mol. Microbiol. 2015; 98:1199–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang J.-T., Zhou J.-B., Mao X.-L., Zhou L., Chen M., Zhang W., Wang E.-D., Zhou X.-L.. Commonality and diversity in tRNA substrate recognition in t6A biogenesis by eukaryotic KEOPSs. Nucleic Acids Res. 2022; 50:2223–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boccaletto P., Stefaniak F., Ray A., Cappannini A., Mukherjee S., Purta E., Kurkowska M., Shirvanizadeh N., Destefanis E., Groza P.et al.. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022; 50:D231–D235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Normanly J., Ogden R.C., Horvath S.J., Abelson J.. Changing the identity of a transfer RNA. Nature. 1986; 321:213–219. [DOI] [PubMed] [Google Scholar]
- 52. Normanly J., Ollick T., Abelson J.. Eight base changes are sufficient to convert a leucine-inserting tRNA into a serine-inserting tRNA. Proc. Natl Acad. Sci. USA. 1992; 89:5680–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Larkin D.C., Williams A.M., Martinis S.A., Fox G.E.. Identification of essential domains for Escherichia coli tRNALeu aminoacylation and amino acid editing using minimalist RNA molecules. Nucleic Acids Res. 2002; 30:2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yao P., Zhu B., Jaeger S., Eriani G., Wang E.-D.. Recognition of tRNALeu by Aquifex aeolicus leucyl-tRNA synthetase during the aminoacylation and editing steps. Nucleic Acids Res. 2008; 36:2728–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Soma A., Uchiyama K., Sakamoto T., Maeda M., Himeno H.. Unique recognition style of tRNALeu by Haloferax volcanii leucyl-tRNA synthetase. J. Mol. Biol. 1999; 293:1029–1038. [DOI] [PubMed] [Google Scholar]
- 56. Himeno H., Yoshida S., Soma A., Nishikawa K.. Only one nucleotide insertion to the long variable arm confers an efficient serine acceptor activity upon Saccharomyces cerevisiae tRNALeuin vitro. J. Mol. Biol. 1997; 268:704–711. [DOI] [PubMed] [Google Scholar]
- 57. Huang Q., Yao P., Eriani G., Wang E.-D.. In vivo identification of essential nucleotides in tRNALeu to its functions by using a constructed yeast tRNALeu knockout strain. Nucleic Acids Res. 2012; 40:10463–10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Breitschopf K., Gross H.J.. The exchange of the discriminator base A73 for G is alone sufficient to convert human tRNALeu into a serine-acceptor in vitro. EMBO J. 1994; 13:3166–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Breitschopf K., Achsel T., Busch K., Gross H.J.. Identity elements of human tRNALeu: structural requirements for converting human tRNASer into a leucine acceptor in vitro. Nucleic. Acids. Res. 1995; 23:3633–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Asahara H., Himeno H., Tamura K., Hasegawa T., Watanabe K., Shimizu M.. Recognition nucleotides of Escherichia coli tRNALeu and its elements facilitating discrimination from tRNASer and tRNATyr. J. Mol. Biol. 1993; 231:219–229. [DOI] [PubMed] [Google Scholar]
- 61. Asahara H., Nameki N., Hasegawa T.. In vitro selection of RNAs aminoacylated by Escherichia coli leucyl-tRNA synthetase. J. Mol. Biol. 1998; 283:605–618. [DOI] [PubMed] [Google Scholar]
- 62. Tocchini-Valentini G., Saks M.E., Abelson J.. tRNA leucine identity and recognition sets. J. Mol. Biol. 2000; 298:779–793. [DOI] [PubMed] [Google Scholar]
- 63. Du X., Wang E.-D.. Tertiary structure base pairs between D- and TpsiC-loops of Escherichia coli tRNALeu play important roles in both aminoacylation and editing. Nucleic Acids Res. 2003; 31:2865–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Metzger A.U., Heckl M., Willbold D., Breitschopf K., RajBhandary U.L., Rösch P., Gross H.J.. Structural studies on tRNA acceptor stem microhelices: exchange of the discriminator base A73 for G in human tRNALeu switches the acceptor specificity from leucine to serine possibly by decreasing the stability of the terminal G1–C72 base pair. Nucleic Acids Res. 1997; 25:4551–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lin B.Y., Chan P.P., Lowe T.M.. tRNAviz: explore and visualize tRNA sequence features. Nucleic Acids Res. 2019; 47:W542–W547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schulman L.H., Pelka H.. Anticodon loop size and sequence requirements for recognition of formylmethionine tRNA by methionyl-tRNA synthetase. Proc. Natl Acad. Sci. USA. 1983; 80:6755–6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schulman L.H., Pelka H.. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988; 242:765–768. [DOI] [PubMed] [Google Scholar]
- 68. Ramesh V., RajBhandary U.L.. Importance of the anticodon sequence in the aminoacylation of tRNAs by methionyl-tRNA synthetase and by valyl-tRNA synthetase in an Archaebacterium. J. Biol. Chem. 2001; 276:3660–3665. [DOI] [PubMed] [Google Scholar]
- 69. Schmitt E., Coureux P.-D., Kazan R., Bourgeois G., Lazennec-Schurdevin C., Mechulam Y.. Recent advances in archaeal translation initiation. Front. Microbiol. 2020; 11:584152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jones T.E., Alexander R.W., Pan T.. Misacylation of specific nonmethionyl tRNAs by a bacterial methionyl-tRNA synthetase. Proc. Natl Acad. Sci. USA. 2011; 108:6933–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Giegé R., Kern D., Ebel J.P., Grosjean H., de Henau S., Chantrenne H.. Incorrect aminoacylations involving tRNAs or valyl-tRNA synthetase from Bacillus stearothermophilus. Eur. J. Biochem. 1974; 45:351–362. [DOI] [PubMed] [Google Scholar]
- 72. Tamura K., Himeno H., Asahara H., Hasegawa T., Shimizu M.. Identity determinants of E. coli tRNAVal. Biochem. Biophys. Res. Commun. 1991; 177:619–623. [DOI] [PubMed] [Google Scholar]
- 73. Liu M., Chu W.C., Liu J.C., Horowitz J.. Role of acceptor stem conformation in tRNAVal recognition by its cognate synthetase. Nucleic Acids Res. 1997; 25:4883–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Horowitz J., Chu W.C., Derrick W.B., Liu J.C., Liu M., Yue D.. Synthetase recognition determinants of E. coli valine transfer RNA. Biochemistry. 1999; 38:7737–7746. [DOI] [PubMed] [Google Scholar]
- 75. Tardif K.D., Horowitz J.. Functional group recognition at the aminoacylation and editing sites of E. coli valyl-tRNA synthetase. RNA. 2004; 10:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sylvers L.A., Rogers K.C., Shimizu M., Ohtsuka E., Söll D.. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry. 1993; 32:3836–3841. [DOI] [PubMed] [Google Scholar]
- 77. Madore E., Florentz C., Giegé R., Sekine S., Yokoyama S., Lapointe J.. Effect of modified nucleotides on Escherichia coli tRNAGlu structure and on its aminoacylation by glutamyl-tRNA synthetase. Predominant and distinct roles of the mnm5 and s2 modifications of U34. Eur. J. Biochem. 1999; 266:1128–1135. [DOI] [PubMed] [Google Scholar]
- 78. Giegé R., Lapointe J.. Transfer RNA aminoacylation and modified nucleosides. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. 2009; Georgetown, TX: 475–492. [Google Scholar]
- 79. Sekine S., Nureki O., Sakamoto K., Niimi T., Tateno M., Go M., Kohno T., Brisson A., Lapointe J., Yokoyama S.. Major identity determinants in the ‘augmented D helix’ of tRNAGlu from Escherichia coli. J. Mol. Biol. 1996; 256:685–700. [DOI] [PubMed] [Google Scholar]
- 80. Schulman L.H., Pelka H.. In vitro conversion of a methionine to a glutamine-acceptor tRNA. Biochemistry. 1985; 24:7309–7314. [DOI] [PubMed] [Google Scholar]
- 81. Yaniv M., Folk W.R., Berg P., Soll L.. A single mutational modification of a tryptophan-specific transfer RNA permits aminoacylation by glutamine and translation of the codon UAG. J. Mol. Biol. 1974; 86:245–260. [DOI] [PubMed] [Google Scholar]
- 82. Rogers M.J., Söll D.. Discrimination between glutaminyl-tRNA synthetase and seryl-tRNA synthetase involves nucleotides in the acceptor helix of tRNA. Proc. Natl Acad. Sci. USA. 1988; 85:6627–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jahn M., Rogers M.J., Söll D.. Anticodon and acceptor stem nucleotides in tRNAGln are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 1991; 352:258–260. [DOI] [PubMed] [Google Scholar]
- 84. Ibba M., Hong K.W., Sherman J.M., Sever S., Söll D.. Interactions between tRNA identity nucleotides and their recognition sites in glutaminyl-tRNA synthetase determine the cognate amino acid affinity of the enzyme. Proc. Natl Acad. Sci. USA. 1996; 93:6953–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hayase Y., Jahn M., Rogers M.J., Sylvers L.A., Koizumi M., Inoue H., Ohtsuka E., Söll D.. Recognition of bases in Escherichia coli tRNAGln by glutaminyl-tRNA synthetase: a complete identity set. EMBO J. 1992; 11:4159–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Salazar J.C., Ahel I., Orellana O., Tumbula-Hansen D., Krieger R., Daniels L., Söll D.. Coevolution of an aminoacyl-tRNA synthetase with its tRNA substrates. Proc. Natl Acad. Sci. USA. 2003; 100:13863–13868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chang K.-M., Hendrickson T.L.. Recognition of tRNAGln by Helicobacter pylori GluRS2—a tRNAGln-specific glutamyl-tRNA synthetase. Nucleic Acids Res. 2009; 37:6942–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kim S.I., Söll D.. Major identity element of glutamine tRNAs from Bacillus subtilis and Escherichia coli in the reaction with B. subtilis glutamyl-tRNA synthetase. Mol. Cells. 1998; 8:459–465. [PubMed] [Google Scholar]
- 89. Himeno H., Hasegawa T., Asahara H., Tamura K., Shimizu M.. Identity determinants of E. coli tryptophan tRNA. Nucleic Acids Res. 1991; 19:6379–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rogers M.J., Adachi T., Inokuchi H., Söll D.. Switching tRNAGln identity from glutamine to tryptophan. Proc. Natl Acad. Sci. USA. 1992; 89:3463–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pak M., Pallanck L., Schulman L.H.. Conversion of a methionine initiator tRNA into a tryptophan-inserting elongator tRNA in vivo. Biochemistry. 1992; 31:3303–3309. [DOI] [PubMed] [Google Scholar]
- 92. Xue H., Shen W., Giegé R., Wong J.T.. Identity elements of tRNATrp. Identification and evolutionary conservation. J. Biol. Chem. 1993; 268:9316–9322. [PubMed] [Google Scholar]
- 93. Yesland K.D., Johnson J.D.. Anticodon bases C34 and C35 are major, positive, identity elements in Saccharomyces cerevisiae tRNATrp. Nucleic Acids Res. 1993; 21:5079–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ulmasov B., Topin A., Chen Z., He S.H., Folk W.R.. Identity elements and aminoacylation of plant tRNATrp. Nucleic Acids Res. 1998; 26:5139–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Xu F., Chen X., Xin L., Chen L., Jin Y., Wang D.. Species-specific differences in the operational RNA code for aminoacylation of tRNATrp. Nucleic Acids Res. 2001; 29:4125–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xu M.-G., Chen J.-F., Martin F., Zhao M.-W., Eriani G., Wang E.-D.. Leucyl-tRNA synthetase consisting of two subunits from hyperthermophilic bacteria Aquifex aeolicus. J. Biol. Chem. 2002; 277:41590–41596. [DOI] [PubMed] [Google Scholar]
- 97. Tsuchiya W., Hasegawa T.. Molecular recognition of tryptophan tRNA by tryptophanyl-tRNA synthetase from Aeropyrum pernix K1. J. Biochem. 2009; 145:635–641. [DOI] [PubMed] [Google Scholar]
- 98. Achsel T., Gross H.J.. Identity determinants of human tRNASer: sequence elements necessary for serylation and maturation of a tRNA with a long extra arm. EMBO J. 1993; 12:3333–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bedouelle H. Recognition of tRNATyr by tyrosyl-tRNA synthetase. Biochimie. 1990; 72:589–598. [DOI] [PubMed] [Google Scholar]
- 100. Hou Y.M., Schimmel P.. Modeling with in vitro kinetic parameters for the elaboration of transfer RNA identity in vivo. Biochemistry. 1989; 28:4942–4947. [DOI] [PubMed] [Google Scholar]
- 101. Bare L., Uhlenbeck O.C.. Aminoacylation of anticodon loop substituted yeast tyrosine transfer RNA. Biochemistry. 1985; 24:2354–2360. [DOI] [PubMed] [Google Scholar]
- 102. Quinn C.L., Tao N., Schimmel P.. Species-specific microhelix aminoacylation by a eukaryotic pathogen tRNA synthetase dependent on a single base pair. Biochemistry. 1995; 34:12489–12495. [DOI] [PubMed] [Google Scholar]
- 103. Fechter P., Rudinger-Thirion J., Théobald-Dietrich A., Giegé R.. Identity of tRNA for yeast tyrosyl-tRNA synthetase: tyrosylation is more sensitive to identity nucleotides than to structural features. Biochemistry. 2000; 39:1725–1733. [DOI] [PubMed] [Google Scholar]
- 104. Fechter P., Rudinger-Thirion J., Tukalo M., Giegé R.. Major tyrosine identity determinants in Methanococcus jannaschii and Saccharomyces cerevisiae tRNATyr are conserved but expressed differently. Eur. J. Biochem. 2001; 268:761–767. [DOI] [PubMed] [Google Scholar]
- 105. Iwaki J., Asahara H., Nagaoka Y., Yokozawa J., Umehara T., Kawarabayasi Y., Koyama Y., Sako Y., Kuno A., Hasegawa T.. Differences in tyrosine tRNA identity between Escherichia coli and archaeon, Aeropyrum pernix K1. Nucleic Acids Res. Suppl. 2002; 2:225–226. [DOI] [PubMed] [Google Scholar]
- 106. Bonnefond L., Giegé R., Rudinger-Thirion J.. Evolution of the tRNATyr/TyrRS aminoacylation systems. Biochimie. 2005; 87:873–883. [DOI] [PubMed] [Google Scholar]
- 107. Kuratani M., Sakai H., Takahashi M., Yanagisawa T., Kobayashi T., Murayama K., Chen L., Liu Z.-J., Wang B.-C., Kuroishi C.et al.. Crystal structures of tyrosyl-tRNA synthetases from Archaea. J. Mol. Biol. 2006; 355:395–408. [DOI] [PubMed] [Google Scholar]
- 108. Tsunoda M., Kusakabe Y., Tanaka N., Ohno S., Nakamura M., Senda T., Moriguchi T., Asai N., Sekine M., Yokogawa T.et al.. Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms. Nucleic Acids Res. 2007; 35:4289–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. McClain W.H., Chen Y.M., Foss K., Schneider J.. Association of transfer RNA acceptor identity with a helical irregularity. Science. 1988; 242:1681–1684. [DOI] [PubMed] [Google Scholar]
- 110. Francklyn C., Schimmel P.. Aminoacylation of RNA minihelices with alanine. Nature. 1989; 337:478–481. [DOI] [PubMed] [Google Scholar]
- 111. Shi J.P., Francklyn C., Hill K., Schimmel P.. A nucleotide that enhances the charging of RNA minihelix sequence variants with alanine. Biochemistry. 1990; 29:3621–3626. [DOI] [PubMed] [Google Scholar]
- 112. Frugier M., Florentz C., Schimmel P., Giegé R.. Triple aminoacylation specificity of a chimerized transfer RNA. Biochemistry. 1993; 32:14053–14061. [DOI] [PubMed] [Google Scholar]
- 113. Hou Y.M., Sterner T., Jansen M.. Permutation of a pair of tertiary nucleotides in a transfer RNA. Biochemistry. 1995; 34:2978–2984. [DOI] [PubMed] [Google Scholar]
- 114. Liu H., Yap L.-P., Musier-Forsyth K.. Single atomic group in RNA helix needed for positive and negative tRNA synthetase discrimination. J. Am. Chem. Soc. 1996; 118:2523–2524. [Google Scholar]
- 115. McClain W.H., Gabriel K., Schneider J.. Specific function of a G·U wobble pair from an adjacent helical site in tRNAAla during recognition by alanyl-tRNA synthetase. RNA. 1996; 2:105–109. [PMC free article] [PubMed] [Google Scholar]
- 116. Frugier M., Schimmel P.. Subtle atomic group discrimination in the RNA minor groove. Proc. Natl Acad. Sci. USA. 1997; 94:11291–11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ramos A., Varani G.. Structure of the acceptor stem of Escherichia coli tRNAAla: role of the G3·U70 base pair in synthetase recognition. Nucleic Acids Res. 1997; 25:2083–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Beuning P.J., Yang F., Schimmel P., Musier-Forsyth K.. Specific atomic groups and RNA helix geometry in acceptor stem recognition by a tRNA synthetase. Proc. Natl Acad. Sci. USA. 1997; 94:10150–10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Hipps D., Schimmel P.. Cell growth inhibition by sequence-specific RNA minihelices. EMBO J. 1995; 14:4050–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. McClain W.H., Gabriel K., Bhattacharya S., Jou Y.Y., Schneider J.. Functional compensation by particular nucleotide substitutions of a critical G·U wobble base-pair during aminoacylation of transfer RNA. J. Mol. Biol. 1999; 286:1025–1032. [DOI] [PubMed] [Google Scholar]
- 121. Chihade J.W., Hayashibara K., Shiba K., Schimmel P.. Strong selective pressure to use G:U to mark an RNA acceptor stem for alanine. Biochemistry. 1998; 37:9193–9202. [DOI] [PubMed] [Google Scholar]
- 122. Shiba K., Ripmaster T., Suzuki N., Nichols R., Plotz P., Noda T., Schimmel P.. Human alanyl-tRNA synthetase: conservation in evolution of catalytic core and microhelix recognition. Biochemistry. 1995; 34:10340–10349. [DOI] [PubMed] [Google Scholar]
- 123. Hou Y.M., Schimmel P.. Evidence that a major determinant for the identity of a transfer RNA is conserved in evolution. Biochemistry. 1989; 28:6800–6804. [DOI] [PubMed] [Google Scholar]
- 124. Carneiro V.T., Dietrich A., Maréchal-Drouard L., Cosset A., Pelletier G., Small I.. Characterization of some major identity elements in plant alanine and phenylalanine transfer RNAs. Plant Mol. Biol. 1994; 26:1843–1853. [DOI] [PubMed] [Google Scholar]
- 125. Choi H., Otten S., McClain W.H.. Isolation of novel tRNAAla mutants by library selection in a tRNAAla knockout strain. Biochimie. 2002; 84:705–711. [DOI] [PubMed] [Google Scholar]
- 126. Fischer A.E., Beuning P.J., Musier-Forsyth K.. Identification of discriminator base atomic groups that modulate the alanine aminoacylation reaction. J. Biol. Chem. 1999; 274:37093–37096. [DOI] [PubMed] [Google Scholar]
- 127. Beuning P.J., Nagan M.C., Cramer C.J., Musier-Forsyth K., Gelpí J.-L., Bashford D.. Efficient aminoacylation of the tRNAAla acceptor stem: dependence on the 2:71 base pair. RNA. 2002; 8:659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Chong Y.E., Guo M., Yang X.-L., Kuhle B., Naganuma M., Sekine S.-I., Yokoyama S., Schimmel P.. Distinct ways of G:U recognition by conserved tRNA binding motifs. Proc. Natl Acad. Sci. USA. 2018; 115:7527–7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kumar A., Åqvist J., Satpati P.. Principles of tRNAAla selection by alanyl-tRNA synthetase based on the critical G3·U70 base pair. ACS Omega. 2019; 4:15539–15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Arutaki M., Kurihara R., Matsuoka T., Inami A., Tokunaga K., Ohno T., Takahashi H., Takano H., Ando T., Mutsuro-Aoki H.et al.. G:U-independent RNA minihelix aminoacylation by Nanoarchaeum equitans alanyl-tRNA synthetase: an insight into the evolution of aminoacyl-tRNA synthetases. J. Mol. Evol. 2020; 88:501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Lovato M.A., Chihade J.W., Schimmel P.. Translocation within the acceptor helix of a major tRNA identity determinant. EMBO J. 2001; 20:4846–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Lovato M.A., Swairjo M.A., Schimmel P.. Positional recognition of a tRNA determinant dependent on a peptide insertion. Mol. Cell. 2004; 13:843–851. [DOI] [PubMed] [Google Scholar]
- 133. Zeng Q.-Y., Peng G.-X., Li G., Zhou J.-B., Zheng W.-Q., Xue M.-Q., Wang E.-D., Zhou X.-L.. The G3–U70-independent tRNA recognition by human mitochondrial alanyl-tRNA synthetase. Nucleic Acids Res. 2019; 47:3072–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Mazauric M.H., Keith G., Logan D., Kreutzer R., Giegé R., Kern D.. Glycyl-tRNA synthetase from Thermus thermophilus–wide structural divergence with other prokaryotic glycyl-tRNA synthetases and functional inter-relation with prokaryotic and eukaryotic glycylation systems. Eur. J. Biochem. 1998; 251:744–757. [DOI] [PubMed] [Google Scholar]
- 135. Shimizu M., Asahara H., Tamura K., Hasegawa T., Himeno H.. The role of anticodon bases and the discriminator nucleotide in the recognition of some E. coli tRNAs by their aminoacyl-tRNA synthetases. J. Mol. Evol. 1992; 35:436–443. [DOI] [PubMed] [Google Scholar]
- 136. McClain W.H., Foss K., Jenkins R.A., Schneider J.. Rapid determination of nucleotides that define tRNAGly acceptor identity. Proc. Natl Acad. Sci. USA. 1991; 88:6147–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Nameki N., Tamura K., Asahara H., Hasegawa T.. Recognition of tRNAGly by three widely diverged glycyl-tRNA synthetases. J. Mol. Biol. 1997; 268:640–647. [DOI] [PubMed] [Google Scholar]
- 138. Mazauric M.H., Roy H., Kern D.. tRNA glycylation system from Thermus thermophilus. tRNAGly identity and functional interrelation with the glycylation systems from other phylae. Biochemistry. 1999; 38:13094–13105. [DOI] [PubMed] [Google Scholar]
- 139. Okamoto K., Kuno A., Hasegawa T.. Recognition sites of glycine tRNA for glycyl-tRNA synthetase from hyperthermophilic archaeon, Aeropyrum pernix K1. Nucleic Acids Symp. Ser. 2005; 49:299–300. [DOI] [PubMed] [Google Scholar]
- 140. Himeno H., Hasegawa T., Ueda T., Watanabe K., Miura K., Shimizu M.. Role of the extra G-C pair at the end of the acceptor stem of tRNAHis in aminoacylation. Nucleic Acids Res. 1989; 17:7855–7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Nameki N., Asahara H., Shimizu M., Okada N., Himeno H.. Identity elements of Saccharomyces cerevisiae tRNAHis. Nucleic Acids Res. 1995; 23:389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Yan W., Augustine J., Francklyn C.. A tRNA identity switch mediated by the binding interaction between a tRNA anticodon and the accessory domain of a class II aminoacyl-tRNA synthetase. Biochemistry. 1996; 35:6559–6568. [DOI] [PubMed] [Google Scholar]
- 143. Nagatoyo Y., Iwaki J., Suzuki S., Kuno A., Hasegawa T.. Molecular recognition of histidine tRNA by histidyl-tRNA synthetase from hyperthermophilic archaeon, Aeropyrum pernix K1. Nucleic Acids Symp. Ser. 2005; 49:307–308. [DOI] [PubMed] [Google Scholar]
- 144. Rosen A.E., Musier-Forsyth K.. Recognition of G-1:C73 atomic groups by Escherichia coli histidyl-tRNA synthetase. J. Am. Chem. Soc. 2004; 126:64–65. [DOI] [PubMed] [Google Scholar]
- 145. Gu W., Jackman J.E., Lohan A.J., Gray M.W., Phizicky E.M.. tRNAHis maturation: an essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev. 2003; 17:2889–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Heinemann I.U., Nakamura A., O’Donoghue P., Eiler D., Söll D.. tRNAHis-guanylyltransferase establishes tRNAHis identity. Nucleic. Acids. Res. 2012; 40:333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Tian Q., Wang C., Liu Y., Xie W.. Structural basis for recognition of G-1-containing tRNA by histidyl-tRNA synthetase. Nucleic Acids Res. 2015; 43:2980–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Ardell D.H., Andersson S.G.E.. TFAM detects co-evolution of tRNA identity rules with lateral transfer of histidyl-tRNA synthetase. Nucleic Acids Res. 2006; 34:893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Yuan J., Gogakos T., Babina A.M., Söll D., Randau L.. Change of tRNA identity leads to a divergent orthogonal histidyl-tRNA synthetase/tRNAHis pair. Nucleic Acids Res. 2011; 39:2286–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Rao B.S., Jackman J.E.. Life without post-transcriptional addition of G-1: two alternatives for tRNAHis identity in Eukarya. RNA. 2015; 21:243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Yaremchuk A., Cusack S., Tukalo M.. Crystal structure of a eukaryote/archaeon-like protyl-tRNA synthetase and its complex with tRNAPro(CGG). EMBO J. 2000; 19:4745–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Stehlin C., Burke B., Yang F., Liu H., Shiba K., Musier-Forsyth K.. Species-specific differences in the operational RNA code for aminoacylation of tRNAPro. Biochemistry. 1998; 37:8605–8613. [DOI] [PubMed] [Google Scholar]
- 153. Hasegawa T., Yokogawa T.. Escherichia coli proline tRNA: structure and recognition sites for prolyl-tRNA synthetase. Nucleic Acids Symp. Ser. 2000; 44:7–8. [DOI] [PubMed] [Google Scholar]
- 154. Liu H., Peterson R., Kessler J., Musier-Forsyth K.. Molecular recognition of tRNAPro by Escherichia coli proline tRNA synthetase in vitro. Nucleic Acids Res. 1995; 23:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Yokozawa J., Okamoto K., Kawarabayasi Y., Kuno A., Hasegawa T.. Molecular recognition of proline tRNA by prolyl-tRNA synthetase from hyperthermophilic archaeon, Aeropyrum pernix K1. Nucleic Acids Res. Suppl. 2003; 3:247–248. [DOI] [PubMed] [Google Scholar]
- 156. An S., Barany G., Musier-Forsyth K.. Evolution of acceptor stem tRNA recognition by class II prolyl-tRNA synthetase. Nucleic Acids Res. 2008; 36:2514–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Clifton B.E., Fariz M.A., Uechi G.-I., Laurino P.. Evolutionary repair reveals an unexpected role of the tRNA modification m1G37 in aminoacylation. Nucleic Acids Res. 2021; 49:12467–12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Himeno H., Hasegawa T., Ueda T., Watanabe K., Shimizu M.. Conversion of aminoacylation specificity from tRNATyr to tRNASerin vitro. Nucleic. Acids. Res. 1990; 18:6815–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Sampson J.R., Saks M.E.. Contributions of discrete tRNASer domains to aminoacylation by E. coli seryl-tRNA synthetase: a kinetic analysis using model RNA substrates. Nucleic Acids Res. 1993; 21:4467–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Saks M.E., Sampson J.R.. Variant minihelix RNAs reveal sequence-specific recognition of the helical tRNASer acceptor stem by E. coli seryl-tRNA synthetase. EMBO J. 1996; 15:2843–2849. [PMC free article] [PubMed] [Google Scholar]
- 161. Korencic D., Polycarpo C., Weygand-Durasevic I., Söll D.. Differential modes of transfer RNASer recognition in Methanosarcina barkeri. J. Biol. Chem. 2004; 279:48780–48786. [DOI] [PubMed] [Google Scholar]
- 162. Rokov-Plavec J., Lesjak S., Gruic-Sovulj I., Mocibob M., Dulic M., Weygand-Durasevic I.. Substrate recognition and fidelity of maize seryl-tRNA synthetases. Arch. Biochem. Biophys. 2013; 529:122–130. [DOI] [PubMed] [Google Scholar]
- 163. Lenhard B., Orellana O., Ibba M., Weygand-Durasević I.. tRNA recognition and evolution of determinants in seryl-tRNA synthesis. Nucleic Acids Res. 1999; 27:721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J.. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009; 37:D159–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Asahara H., Himeno H., Tamura K., Nameki N., Hasegawa T., Shimizu M.. Escherichia coli seryl-tRNA synthetase recognizes tRNASer by its characteristic tertiary structure. J. Mol. Biol. 1994; 236:738–748. [DOI] [PubMed] [Google Scholar]
- 166. Gruic-Sovulj I., Jaric J., Dulic M., Cindric M., Weygand-Durasevic I.. Shuffling of discrete tRNASer regions reveals differently utilized identity elements in yeast and methanogenic archaea. J. Mol. Biol. 2006; 361:128–139. [DOI] [PubMed] [Google Scholar]
- 167. Zimmerman S.M., Kon Y., Hauke A.C., Ruiz B.Y., Fields S., Phizicky E.M.. Conditional accumulation of toxic tRNAs to cause amino acid misincorporation. Nucleic Acids Res. 2018; 46:7831–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Ishikura H., Nagaoka Y., Yokozawa J., Umehara T., Kuno A., Hasegawa T.. Threonyl-tRNA synthetase of archaea: importance of the discriminator base in the aminoacylation of threonine tRNA. Nucleic Acids Symp. Ser. 2000; 44:83–84. [DOI] [PubMed] [Google Scholar]
- 169. Nameki N. Identity elements of tRNAThr towards Saccharomyces cerevisiae threonyl-tRNA synthetase. Nucleic Acids Res. 1995; 23:2831–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Hasegawa T., Miyano M., Himeno H., Sano Y., Kimura K., Shimizu M.. Identity determinants of E. coli threonine tRNA. Biochem. Biophys Res. Commun. 1992; 184:478–484. [DOI] [PubMed] [Google Scholar]
- 171. Nameki N., Asahara H., Hasegawa T.. Identity elements of Thermus thermophilus tRNAThr. FEBS Lett. 1996; 396:201–207. [DOI] [PubMed] [Google Scholar]
- 172. Nagaoka Y., Yokozawa J., Umehara T., Iwaki J., Okamoto K., Kawarabayasi Y., Koyama Y., Sako Y., Wakagi T., Kuno A.et al.. Molecular recognition of threonine tRNA by threonyl-tRNA synthetase from an extreme thermophilic archaeon, Aeropyrum pernix K1. Nucleic Acids Res. Suppl. 2002; 2:81–82. [DOI] [PubMed] [Google Scholar]
- 173. Chen M., Kuhle B., Diedrich J., Liu Z., Moresco J.J., Yates Iii J.R., Pan T., Yang X.-L.. Cross-editing by a tRNA synthetase allows vertebrates to abundantly express mischargeable tRNA without causing mistranslation. Nucleic Acids Res. 2020; 48:6445–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Giegé R., Florentz C., Kern D., Gangloff J., Eriani G., Moras D.. Aspartate identity of transfer RNAs. Biochimie. 1996; 78:605–623. [DOI] [PubMed] [Google Scholar]
- 175. Nameki N., Tamura K., Himeno H., Asahara H., Hasegawa T., Shimizu M.. Escherichia coli tRNAAsp recognition mechanism differing from that of the yeast system. Biochem. Biophys. Res. Commun. 1992; 189:856–862. [DOI] [PubMed] [Google Scholar]
- 176. Moulinier L., Eiler S., Eriani G., Gangloff J., Thierry J.C., Gabriel K., McClain W.H., Moras D.. The structure of an AspRS-tRNAAsp complex reveals a tRNA-dependent control mechanism. EMBO J. 2001; 20:5290–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Pütz J., Florentz C., Benseler F., Giegé R.. A single methyl group prevents the mischarging of a tRNA. Nat. Struct. Mol. Biol. 1994; 1:580–582. [DOI] [PubMed] [Google Scholar]
- 178. Rudinger J., Puglisi J.D., Pütz J., Schatz D., Eckstein F., Florentz C., Giegé R.. Determinant nucleotides of yeast tRNAAsp interact directly with aspartyl-tRNA synthetase. Proc. Natl Acad. Sci. USA. 1992; 89:5882–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Wolfson A.D., Khvorova A.M., Sauter C., Florentz C., Giegé R.. Mimics of yeast tRNAAsp and their recognition by aspartyl-tRNA synthetase. Biochemistry. 1999; 38:11926–11932. [DOI] [PubMed] [Google Scholar]
- 180. Frugier M., Moulinier L., Giegé R.. A domain in the N-terminal extension of class IIb eukaryotic aminoacyl-tRNA synthetases is important for tRNA binding. EMBO J. 2000; 19:2371–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Ryckelynck M., Giegé R., Frugier M.. Yeast tRNAAsp charging accuracy is threatened by the N-terminal extension of aspartyl-tRNA synthetase. J. Biol. Chem. 2003; 278:9683–9690. [DOI] [PubMed] [Google Scholar]
- 182. Gangloff J., Ebel J.P., Dirheimer G.. Isolation of a complex between yeast arginyl-tRNA synthetase and yeast tRNAAsp, and mischarging of tRNAAsp with arginine. Int. Res. Commun. Syst. 1973; 12:8. [Google Scholar]
- 183. Pütz J., Wientges J., Sissler M., Giegé R., Florentz C., Schwienhorst A.. Rapid selection of aminoacyl-tRNAs based on biotinylation of alpha-NH2 group of charged amino acids. Nucleic Acids Res. 1997; 25:1862–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Pütz J., Puglisi J.D., Florentz C., Giegé R.. Identity elements for specific aminoacylation of yeast tRNAAsp by cognate aspartyl-tRNA synthetase. Science. 1991; 252:1696–1699. [DOI] [PubMed] [Google Scholar]
- 185. Li S., Pelka H., Schulman L.H.. The anticodon and discriminator base are important for aminoacylation of Escherichia coli tRNAAsn. J. Biol. Chem. 1993; 268:18335–18339. [PubMed] [Google Scholar]
- 186. Normanly J., Abelson J.. tRNA identity. Annu. Rev. Biochem. 1989; 58:1029–1049. [DOI] [PubMed] [Google Scholar]
- 187. Martin F., Eriani G., Reinbolt J., Dirheimer G., Gangloff J.. Genetic selection for active E. coli amber tRNAAsn exclusively led to glutamine inserting suppressors. Nucleic Acids Res. 1995; 23:779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Kern D., Roy H., Becker H.D.. Ibba M., Francklyn C., Cusack S.. Asparaginyl-tRNA synthetases. The Aminoacyl-tRNA Synthetases. 2005; 18:Georgetown, TX: Landes Bioscience; 193–209. [Google Scholar]
- 189. Filisetti D., Théobald-Dietrich A., Mahmoudi N., Rudinger-Thirion J., Candolfi E., Frugier M.. Aminoacylation of Plasmodium falciparum tRNAAsn and insights in the synthesis of asparagine repeats. J. Biol. Chem. 2013; 288:36361–36371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Martin F., Reinbolt J., Dirheimer G., Gangloff J., Eriani G.. Selection of tRNAAsp amber suppressor mutants having alanine, arginine, glutamine, and lysine identity. RNA. 1996; 2:919–927. [PMC free article] [PubMed] [Google Scholar]
- 191. Stello T., Hong M., Musier-Forsyth K.. Efficient aminoacylation of tRNALys,3 by human lysyl-tRNA synthetase is dependent on covalent continuity between the acceptor stem and the anticodon domain. Nucleic Acids Res. 1999; 27:4823–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Francin M., Mirande M.. Identity elements for specific aminoacylation of a tRNA by mammalian lysyl-tRNA synthetase bearing a nonspecific tRNA-interacting factor. Biochemistry. 2006; 45:10153–10160. [DOI] [PubMed] [Google Scholar]
- 193. Sampson J.R., DiRenzo A.B., Behlen L.S., Uhlenbeck O.C.. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science. 1989; 243:1363–1366. [DOI] [PubMed] [Google Scholar]
- 194. Sampson J.R., DiRenzo A.B., Behlen L.S., Uhlenbeck O.C.. Role of the tertiary nucleotides in the interaction of yeast phenylalanine tRNA with its cognate synthetase. Biochemistry. 1990; 29:2523–2532. [DOI] [PubMed] [Google Scholar]
- 195. Tsuchiya W., Kimura M., Hasegawa T.. Determination of phenylalanine tRNA recognition sites by phenylalanyl-tRNA synthetase from hyperthermophilic archaeon, Aeropyrum pernix K1. Nucleic Acids Symp. Ser. 2007; 51:367–368. [DOI] [PubMed] [Google Scholar]
- 196. Peterson E.T., Uhlenbeck O.C.. Determination of recognition nucleotides for Escherichia coli phenylalanyl-tRNA synthetase. Biochemistry. 1992; 31:10380–10389. [DOI] [PubMed] [Google Scholar]
- 197. Peterson E.T., Blank J., Sprinzl M., Uhlenbeck O.C.. Selection for active E. coli tRNAPhe variants from a randomized library using two proteins. EMBO J. 1993; 12:2959–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Nazarenko I.A., Peterson E.T., Zakharova O.D., Lavrik O.I., Uhlenbeck O.C.. Recognition nucleotides for human phenylalanyl-tRNA synthetase. Nucleic Acids Res. 1992; 20:475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Kholod N.S., Pan’kova N.V., Mayorov S.G., Krutilina A.I., Shlyapnikov M.G., Kisselev L.L., Ksenzenko V.N.. Transfer RNAPhe isoacceptors possess non-identical set of identity elements at high and low Mg2+ concentration. FEBS Lett. 1997; 411:123–127. [DOI] [PubMed] [Google Scholar]
- 200. Agris P.F., Narendran A., Sarachan K., Väre V.Y.P., Eruysal E.. The importance of being modified: the role of RNA modifications in translational fidelity. Enzymes. 2017; 41:1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Hauenstein S.I., Hou Y.-M., Perona J.J.. The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity. J. Biol. Chem. 2008; 283:21997–22006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Shanmugam R., Fierer J., Kaiser S., Helm M., Jurkowski T.P., Jeltsch A.. Cytosine methylation of tRNAAsp by DNMT2 has a role in translation of proteins containing poly-Asp sequences. Cell Discov. 2015; 1:15010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Grosjean H., Westhof E.. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016; 44:8020–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Levi O., Arava Y.S.. RNA modifications as a common denominator between tRNA and mRNA. Curr. Genet. 2021; 67:545–551. [DOI] [PubMed] [Google Scholar]
- 205. Hale S.P., Auld D.S., Schmidt E., Schimmel P.. Discrete determinants in transfer RNA for editing and aminoacylation. Science. 1997; 276:1250–1252. [DOI] [PubMed] [Google Scholar]
- 206. Yao P., Zhu B., Jaeger S., Eriani G., Wang E.-D.. Recognition of tRNALeu by Aquifex aeolicus leucyl-tRNA synthetase during the aminoacylation and editing steps. Nucleic Acids Res. 2008; 36:2728–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Soma A., Kumagai R., Nishikawa K., Himeno H.. The anticodon loop is a major identity determinant of Saccharomyces cerevisiae tRNALeu. J. Mol. Biol. 1996; 263:707–714. [DOI] [PubMed] [Google Scholar]
- 208. Perret V., Garcia A., Grosjean H., Ebel J.P., Florentz C., Giegé R.. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature. 1990; 344:787–789. [DOI] [PubMed] [Google Scholar]
- 209. Büttcher V., Senger B., Schumacher S., Reinbolt J., Fasiolo F.. Modulation of the suppression efficiency and amino acid identity of an artificial yeast amber isoleucine transfer RNA in Escherichia coli by a G–U pair in the anticodon stem. Biochem. Biophys. Res. Commun. 1994; 200:370–377. [DOI] [PubMed] [Google Scholar]
- 210. Ambrogelly A., Frugier M., Ibba M., Söll D., Giegé R.. Transfer RNA recognition by class I lysyl-tRNA synthetase from the Lyme disease pathogen Borrelia burgdorferi. FEBS Lett. 2005; 579:2629–2634. [DOI] [PubMed] [Google Scholar]
- 211. Pak M., Willis I.M., Schulman L.H.. Analysis of acceptor stem base pairing on tRNATrp aminoacylation and function in vivo. J. Biol. Chem. 1994; 269:2277–2282. [PubMed] [Google Scholar]
- 212. Freyhult E., Moulton V., Ardell D.H.. Visualizing bacterial tRNA identity determinants and antideterminants using function logos and inverse function logos. Nucleic Acids Res. 2006; 34:905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Hadd A., Perona J.J.. Coevolution of specificity determinants in eukaryotic glutamyl- and glutaminyl-tRNA synthetases. J. Mol. Biol. 2014; 426:3619–3633. [DOI] [PubMed] [Google Scholar]
- 214. Carnicelli D., Brigotti M., Rizzi S., Keith G., Montanaro L., Sperti S.. Nucleotides U28–A42 and A37 in unmodified yeast tRNATrp as negative identity elements for bovine tryptophanyl-tRNA synthetase. FEBS Lett. 2001; 492:238–241. [DOI] [PubMed] [Google Scholar]
- 215. Schmitt E., Meinnel T., Panvert M., Mechulam Y., Blanquet S.. Two acidic residues of Escherichia coli methionyl-tRNA synthetase act as negative discriminants towards the binding of non-cognate tRNA anticodons. J. Mol. Biol. 1993; 233:615–628. [DOI] [PubMed] [Google Scholar]
- 216. Bedouelle H., Guez-Ivanier V., Nageotte R.. Discrimination between transfer-RNAs by tyrosyl-tRNA synthetase. Biochimie. 1993; 75:1099–1108. [DOI] [PubMed] [Google Scholar]
- 217. Ador L., Camasses A., Erbs P., Cavarelli J., Moras D., Gangloff J., Eriani G.. Active site mapping of yeast aspartyl-tRNA synthetase by in vivo selection of enzyme mutations lethal for cell growth. J. Mol. Biol. 1999; 288:231–242. [DOI] [PubMed] [Google Scholar]
- 218. Eriani G., Gangloff J.. Yeast aspartyl-tRNA synthetase residues interacting with tRNAAsp identity bases connectively contribute to tRNAAsp binding in the ground and transition-state complex and discriminate against non-cognate tRNAs. J. Mol. Biol. 1999; 291:761–773. [DOI] [PubMed] [Google Scholar]
- 219. Bullock T.L., Uter N., Nissan T.A., Perona J.J.. Amino acid discrimination by a class I aminoacyl-tRNA synthetase specified by negative determinants. J. Mol. Biol. 2003; 328:395–408. [DOI] [PubMed] [Google Scholar]
- 220. Krahn N., Fischer J.T., Söll D.. Naturally occurring tRNAs with non-canonical structures. Front. Microbiol. 2020; 11:596914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Florentz C., Giegé R.. tRNA-like structures in viral RNAs. tRNA: Structure, Biosynthesis, and Function. 1995; Washington, DC: American Society of Microbiology Press; 141–163. [Google Scholar]
- 222. Dreher T.W. Viral tRNAs and tRNA-like structures. Wiley Interdiscip. Rev. RNA. 2010; 1:402–414. [DOI] [PubMed] [Google Scholar]
- 223. Levi O., Arava Y.. mRNA association by aminoacyl tRNA synthetase occurs at a putative anticodon mimic and autoregulates translation in response to tRNA levels. PLoS Biol. 2019; 17:e3000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Magee R., Rigoutsos I.. On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res. 2020; 48:9433–9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Mleczko A.M., Celichowski P., Bąkowska-Żywicka K.. Transfer RNA-derived fragments target and regulate ribosome-associated aminoacyl-transfer RNA synthetases. Biochim. Biophys. Acta. 2018; 1862:647–656. [DOI] [PubMed] [Google Scholar]
- 226. Commans S., Böck A.. Selenocysteine inserting tRNAs: an overview. FEMS Microbiol. Rev. 1999; 23:335–351. [DOI] [PubMed] [Google Scholar]
- 227. Théobald-Dietrich A., Frugier M., Giegé R., Rudinger-Thirion J.. Atypical archaeal tRNA pyrrolysine transcript behaves towards EF-Tu as a typical elongator tRNA. Nucleic Acids Res. 2004; 32:1091–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Rietveld K., Van Poelgeest R., Pleij C.W., Van Boom J.H., Bosch L.. The tRNA-like structure at the 3′ terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res. 1982; 10:1929–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Colussi T.M., Costantino D.A., Hammond J.A., Ruehle G.M., Nix J.C., Kieft J.S.. The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA. Nature. 2014; 511:366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Rietveld K., Linschooten K., Pleij C.W., Bosch L.. The three-dimensional folding of the tRNA-like structure of tobacco mosaic virus RNA. A new building principle applied twice. EMBO J. 1984; 3:2613–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Moine H., Romby P., Springer M., Grunberg-Manago M., Ebel J.P., Ehresmann C., Ehresmann B.. Messenger RNA structure and gene regulation at the translational level in Escherichia coli: the case of threonine:tRNAThr ligase. Proc. Natl Acad. Sci. USA. 1988; 85:7892–7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Romby P., Caillet J., Ebel C., Sacerdot C., Graffe M., Eyermann F., Brunel C., Moine H., Ehresmann C., Ehresmann B.et al.. The expression of E. coli threonyl-tRNA synthetase is regulated at the translational level by symmetrical operator–repressor interactions. EMBO J. 1996; 15:5976–5987. [PMC free article] [PubMed] [Google Scholar]
- 233. Jühling T., Duchardt-Ferner E., Bonin S., Wöhnert J., Pütz J., Florentz C., Betat H., Sauter C., Mörl M.. Small but large enough: structural properties of armless mitochondrial tRNAs from the nematode Romanomermis culicivorax. Nucleic Acids Res. 2018; 46:9170–9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Wolstenholme D.R., Macfarlane J.L., Okimoto R., Clary D.O., Wahleithner J.A.. Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc. Natl Acad. Sci. USA. 1987; 84:1324–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Pons J., Bover P., Bidegaray-Batista L., Arnedo M.A.. Arm-less mitochondrial tRNAs conserved for over 30 millions of years in spiders. BMC Genomics. 2019; 20:665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Lu C., Huang X., Deng J.. The challenge of Coccidae (Hemiptera:coccoidea) mitochondrial genomes: the case of Saissetia coffeae with novel truncated tRNAs and gene rearrangements. Int. J. Biol. Macromol. 2020; 158:854–864. [DOI] [PubMed] [Google Scholar]
- 237. Li M., Chen W.-T., Zhang Q.-L., Liu M., Xing C.-W., Cao Y., Luo F.-Z., Yuan M.-L.. Mitochondrial phylogenomics provides insights into the phylogeny and evolution of spiders (Arthropoda:araneae). Zool. Res. 2022; 43:566–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Warren J.M., Sloan D.B.. Hopeful monsters: unintended sequencing of famously malformed mite mitochondrial tRNAs reveals widespread expression and processing of sense-antisense pairs. NAR Genom. Bioinform. 2021; 3:lqaa111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Suzuki T., Yashiro Y., Kikuchi I., Ishigami Y., Saito H., Matsuzawa I., Okada S., Mito M., Iwasaki S., Ma D.et al.. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 2020; 11:4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Helm M., Giegé R., Florentz C.. A Watson–Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry. 1999; 38:13338–13346. [DOI] [PubMed] [Google Scholar]
- 241. Sakurai M., Ohtsuki T., Watanabe K.. Modification at position 9 with 1-methyladenosine is crucial for structure and function of nematode mitochondrial tRNAs lacking the entire T-arm. Nucleic Acids Res. 2005; 33:1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Yuan J., Palioura S., Salazar J.C., Su D., O’Donoghue P., Hohn M.J., Cardoso A.M., Whitman W.B., Söll D.. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc. Natl Acad. Sci. USA. 2006; 103:18923–18927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Baron C., Westhof E., Böck A., Giegé R.. Solution structure of selenocysteine-inserting tRNASec from Escherichia coli. Comparison with canonical tRNASer. J. Mol. Biol. 1993; 231:274–292. [DOI] [PubMed] [Google Scholar]
- 244. Wang C., Guo Y., Tian Q., Jia Q., Gao Y., Zhang Q., Zhou C., Xie W.. SerRS–tRNASec complex structures reveal mechanism of the first step in selenocysteine biosynthesis. Nucleic Acids Res. 2015; 43:10534–10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Rudinger J., Hillenbrandt R., Sprinzl M., Giegé R.. Antideterminants present in minihelix(Sec) hinder its recognition by prokaryotic elongation factor Tu. EMBO J. 1996; 15:650–657. [PMC free article] [PubMed] [Google Scholar]
- 246. Fu X., Crnković A., Sevostyanova A., Söll D.. Designing seryl-tRNA synthetase for improved serylation of selenocysteine tRNAs. FEBS Lett. 2018; 592:3759–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Sherlock M.E., Hartwick E.W., MacFadden A., Kieft J.S.. Structural diversity and phylogenetic distribution of valyl tRNA-like structures in viruses. RNA. 2021; 27:27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Langeberg C.J., Sherlock M.E., MacFadden A., Kieft J.S.. An expanded class of histidine-accepting viral tRNA-like structures. RNA. 2021; 27:653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Bonilla S.L., Sherlock M.E., MacFadden A., Kieft J.S.. A viral RNA hijacks host machinery using dynamic conformational changes of a tRNA-like structure. Science. 2021; 374:955–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Wientges J., Pütz J., Giegé R., Florentz C., Schwienhorst A.. Selection of viral RNA-derived tRNA-like structures with improved valylation activities. Biochemistry. 2000; 39:6207–6218. [DOI] [PubMed] [Google Scholar]
- 251. Dreher T.W., Tsai C.H., Florentz C., Giegé R.. Specific valylation of turnip yellow mosaic virus RNA by wheat germ valyl-tRNA synthetase determined by three anticodon loop nucleotides. Biochemistry. 1992; 31:9183–9189. [DOI] [PubMed] [Google Scholar]
- 252. Rudinger J., Florentz C., Giegé R.. Histidylation by yeast HisRS of tRNA or tRNA-like structure relies on residues –1 and 73 but is dependent on the RNA context. Nucleic Acids Res. 1994; 22:5031–5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Dreher T.W., Goodwin J.B.. Transfer RNA mimicry among tymoviral genomic RNAs ranges from highly efficient to vestigial. Nucleic Acids Res. 1998; 26:4356–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Springer M., Graffe M., Dondon J., Grunberg-Manago M., Romby P., Ehresmann B., Ehresmann C., Ebel J.P.. Translational control in E. coli: the case of threonyl-tRNA synthetase. Biosci. Rep. 1988; 8:619–632. [DOI] [PubMed] [Google Scholar]
- 255. Ryckelynck M., Masquida B., Giegé R., Frugier M.. An intricate RNA structure with two tRNA-derived motifs directs complex formation between yeast aspartyl-tRNA synthetase and its mRNA. J. Mol. Biol. 2005; 354:614–629. [DOI] [PubMed] [Google Scholar]
- 256. Sankaranarayanan R., Dock-Bregeon A.C., Romby P., Caillet J., Springer M., Rees B., Ehresmann C., Ehresmann B., Moras D.. The structure of threonyl-tRNA synthetase-tRNAThr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999; 97:371–381. [DOI] [PubMed] [Google Scholar]
- 257. Graffe M., Dondon J., Caillet J., Romby P., Ehresmann C., Ehresmann B., Springer M.. The specificity of translational control switched with transfer RNA identity rules. Science. 1992; 255:994–996. [DOI] [PubMed] [Google Scholar]
- 258. Lu X., Huang J., Wu S., Zheng Q., Liu P., Feng H., Su X., Fu H., Xi Q., Wang G.. The tRNA-like small noncoding RNA mascRNA promotes global protein translation. EMBO Rep. 2020; 21:e49684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Bailly M., Blaise M., Lorber B., Becker H.D., Kern D.. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol. Cell. 2007; 28:228–239. [DOI] [PubMed] [Google Scholar]
- 260. Blaise M., Bailly M., Frechin M., Behrens M.A., Fischer F., Oliveira C.L.P., Becker H.D., Pedersen J.S., Thirup S., Kern D.. Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation. EMBO J. 2010; 29:3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Ito T., Yokoyama S.. Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions. Nature. 2010; 467:612–616. [DOI] [PubMed] [Google Scholar]
- 262. Bailly M., Giannouli S., Blaise M., Stathopoulos C., Kern D., Becker H.D.. A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine. Nucleic Acids Res. 2006; 34:6083–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Datt M., Sharma A.. Novel and unique domains in aminoacyl-tRNA synthetases from human fungal pathogens Aspergillus niger, Candida albicans and Cryptococcus neoformans. BMC Genomics. 2014; 15:1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Tharp J.M., Ehnbom A., Liu W.R.. tRNAPyl: structure, function, and applications. RNA Biol. 2018; 15:441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Herring S., Ambrogelly A., Polycarpo C.R., Söll D.. Recognition of pyrrolysine tRNA by the Desulfitobacterium hafniense pyrrolysyl-tRNA synthetase. Nucleic Acids Res. 2007; 35:1270–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Ambrogelly A., Gundllapalli S., Herring S., Polycarpo C., Frauer C., Söll D.. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc. Natl Acad. Sci. USA. 2007; 104:3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Guo L.-T., Amikura K., Jiang H.-K., Mukai T., Fu X., Wang Y.-S., O’Donoghue P., Söll D., Tharp J.M.. Ancestral archaea expanded the genetic code with pyrrolysine. J. Biol. Chem. 2022; 298:102521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Zhang H., Gong X., Zhao Q., Mukai T., Vargas-Rodriguez O., Zhang H., Zhang Y., Wassel P., Amikura K., Maupin-Furlow J.et al.. The tRNA discriminator base defines the mutual orthogonality of two distinct pyrrolysyl-tRNA synthetase/tRNAPyl pairs in the same organism. Nucleic Acids Res. 2022; 50:4601–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Nozawa K., O’Donoghue P., Gundllapalli S., Araiso Y., Ishitani R., Umehara T., Söll D., Nureki O.. Pyrrolysyl-tRNA synthetase-tRNAPyl structure reveals the molecular basis of orthogonality. Nature. 2009; 457:1163–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Sauerwald A., Zhu W., Major T.A., Roy H., Palioura S., Jahn D., Whitman W.B., Yates J.R., Ibba M., Söll D.. RNA-dependent cysteine biosynthesis in archaea. Science. 2005; 307:1969–1972. [DOI] [PubMed] [Google Scholar]
- 271. O’Donoghue P., Sethi A., Woese C.R., Luthey-Schulten Z.A.. The evolutionary history of Cys-tRNACys formation. Proc. Natl Acad. Sci. USA. 2005; 102:19003–19008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Zhang C.-M., Liu C., Slater S., Hou Y.-M.. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNACys. Nat. Struct. Mol. Biol. 2008; 15:507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Hohn M.J., Park H.-S., O’Donoghue P., Schnitzbauer M., Söll D.. Emergence of the universal genetic code imprinted in an RNA record. Proc. Natl Acad. Sci. USA. 2006; 103:18095–18100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Mohanta T.K., Khan A.L., Hashem A., Allah E.F.A., Yadav D., Al-Harrasi A.. Genomic and evolutionary aspects of chloroplast tRNA in monocot plants. BMC Plant Biol. 2019; 19:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Zhang T.-T., Yang Y., Song X.-Y., Gao X.-Y., Zhang X.-L., Zhao J.-J., Zhou K.-H., Zhao C.-B., Li W., Yang D.-G.et al.. Novel structural variation and evolutionary characteristics of chloroplast tRNA in Gossypium plants. Genes (Basel). 2021; 12:822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Pütz J., Giegé R., Florentz C.. Diversity and similarity in the tRNA world: overall view and case study on malaria-related tRNAs. FEBS Lett. 2010; 584:350–358. [DOI] [PubMed] [Google Scholar]
- 277. Florentz C., Sohm B., Tryoen-Tóth P., Pütz J., Sissler M.. Human mitochondrial tRNAs in health and disease. Cell Mol. Life Sci. 2003; 60:1356–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Helm M., Brulé H., Friede D., Giegé R., Pütz D., Florentz C.. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000; 6:1356–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. de Bruijn M.H., Schreier P.H., Eperon I.C., Barrell B.G., Chen E.Y., Armstrong P.W., Wong J.F., Roe B.A.. A mammalian mitochondrial serine transfer RNA lacking the ‘dihydrouridine’ loop and stem. Nucleic Acids Res. 1980; 8:5213–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Goto Y., Nonaka I., Horai S.. A mutation in the tRNALeuUUR gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990; 348:651–653. [DOI] [PubMed] [Google Scholar]
- 281. Leisinger A.-K., Janzen D.H., Hallwachs W., Igloi G.L.. Amino acid discrimination by the nuclear encoded mitochondrial arginyl-tRNA synthetase of the larva of a bruchid beetle (Caryedes brasiliensis) from northwestern Costa Rica. Insect Biochem. Mol. Biol. 2013; 43:1172–1180. [PubMed] [Google Scholar]
- 282. Igloi G.L., Leisinger A.-K.. Identity elements for the aminoacylation of metazoan mitochondrial tRNAArg have been widely conserved throughout evolution and ensure the fidelity of the AGR codon reassignment. RNA Biol. 2014; 11:1313–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Sohm B., Frugier M., Brulé H., Olszak K., Przykorska A., Florentz C.. Towards understanding human mitochondrial leucine aminoacylation identity. J. Mol. Biol. 2003; 328:995–1010. [DOI] [PubMed] [Google Scholar]
- 284. Sohm B., Sissler M., Park H., King M.P., Florentz C.. Recognition of human mitochondrial tRNALeuUUR by its cognate leucyl-tRNA synthetase. J. Mol. Biol. 2004; 339:17–29. [DOI] [PubMed] [Google Scholar]
- 285. Bonnefond L., Frugier M., Giegé R., Rudinger-Thirion J.. Human mitochondrial TyrRS disobeys the tyrosine identity rules. RNA. 2005; 11:558–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Kuhle B., Chihade J., Schimmel P.. Relaxed sequence constraints favor mutational freedom in idiosyncratic metazoan mitochondrial tRNAs. Nature Commun. 2020; 11:969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Kumazawa Y., Yokogawa T., Miura K., Watanabe K.. Bovine mitochondrial tRNAPhe, tRNASerAGY and tRNASerUCN: preparation using a new detection method and their properties in aminoacylation. Nucleic Acids Symp. Ser. 1988; 19:97–100. [PubMed] [Google Scholar]
- 288. Yokogawa T., Shimada N., Takeuchi N., Benkowski L., Suzuki T., Omori A., Ueda T., Nishikawa K., Spremulli L.L., Watanabe K.. Characterization and tRNA recognition of mammalian mitochondrial seryl-tRNA synthetase. J. Biol. Chem. 2000; 275:19913–19920. [DOI] [PubMed] [Google Scholar]
- 289. Shimada N., Suzuki T., Watanabe K.. Dual mode recognition of two isoacceptor tRNAs by mammalian mitochondrial seryl-tRNA synthetase. J. Biol. Chem. 2001; 276:46770–46778. [DOI] [PubMed] [Google Scholar]
- 290. Chimnaronk S., Gravers Jeppesen M., Suzuki T., Nyborg J., Watanabe K.. Dual-mode recognition of noncanonical tRNAsSer by seryl-tRNA synthetase in mammalian mitochondria. EMBO J. 2005; 24:3369–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Kuhle B., Hirschi M., Doerfel L.K., Lander G.C., Schimmel P.. Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA. Nat. Commun. 2022; 13:5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Fender A., Sauter C., Messmer M., Pütz J., Giegé R., Florentz C., Sissler M.. Loss of a primordial identity element for a mammalian mitochondrial aminoacylation system. J. Biol. Chem. 2006; 281:15980–15986. [DOI] [PubMed] [Google Scholar]
- 293. Neuenfeldt A., Lorber B., Ennifar E., Gaudry A., Sauter C., Sissler M., Florentz C.. Thermodynamic properties distinguish human mitochondrial aspartyl-tRNA synthetase from bacterial homolog with same 3D architecture. Nucleic Acids Res. 2013; 41:2698–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Messmer M., Gaudry A., Sissler M., Florentz C.. Pathology-related mutation A7526G (A9G) helps in the understanding of the 3D structural core of human mitochondrial tRNAAsp. RNA. 2009; 15:1462–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Janke A., Pääbo S.. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res. 1993; 21:1523–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Mörl M., Dörner M., Pääbo S.. C to U editing and modifications during the maturation of the mitochondrial tRNAAsp in marsupials. Nucleic Acids Res. 1995; 23:3380–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Börner G.V., Mörl M., Janke A., Pääbo S.. RNA editing changes the identity of a mitochondrial tRNA in marsupials. EMBO J. 1996; 15:5949–5957. [PMC free article] [PubMed] [Google Scholar]
- 298. Klipcan L., Moor N., Finarov I., Kessler N., Sukhanova M., Safro M.G.. Crystal structure of human mitochondrial PheRS complexed with tRNAPhe in the active ‘open’ state. J. Mol. Biol. 2012; 415:527–537. [DOI] [PubMed] [Google Scholar]
- 299. Aphasizhev R., Senger B., Rengers J.U., Sprinzl M., Walter P., Nussbaum G., Fasiolo F.. Conservation in evolution for a small monomeric phenylalanyl-tRNA synthetase of the tRNAPhe recognition nucleotides and initial aminoacylation site. Biochemistry. 1996; 35:117–123. [DOI] [PubMed] [Google Scholar]
- 300. Degoul F., Brulé H., Cepanec C., Helm M., Marsac C., Leroux J., Giegé R., Florentz C.. Isoleucylation properties of native human mitochondrial tRNAIle and tRNAIle transcripts. Implications for cardiomyopathy-related point mutations (4269, 4317) in the tRNAIle gene. Hum. Mol. Genet. 1998; 7:347–354. [DOI] [PubMed] [Google Scholar]
- 301. Jin X., Tao Z., Jia J., He X., Jin Y.. Species-specific aminoacylation of Oryza sativa mitochondrial tRNATrp. Chin. Sci. Bull. 2006; 51:824–829. [Google Scholar]
- 302. Lee Y.-H., Lo Y.-T., Chang C.-P., Yeh C.-S., Chang T.-H., Chen Y.-W., Tseng Y.-K., Wang C.-C.. Naturally occurring dual recognition of tRNAHis substrates with and without a universal identity element. RNA Biol. 2019; 16:1275–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Cela M., Paulus C., Santos M.A.S., Moura G.R., Frugier M., Rudinger-Thirion J.. Plasmodium apicoplast tyrosyl-tRNA synthetase recognizes an unusual, simplified identity set in cognate tRNATyr. PLoS One. 2018; 13:e0209805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Renaud M., Ehrlich R., Bonnet J., Remy P.. Lack of correlation between affinity of the tRNA for the aminoacyl-tRNA synthetase and aminoacylation capacity as studied with modified tRNAPhe. Eur. J. Biochem. 1979; 100:157–164. [DOI] [PubMed] [Google Scholar]
- 305. Khvorova A.M., Motorin YuA null, Wolfson A.D., Gladilin K.L.. Anticodon-dependent aminoacylation of RNA minisubstrate by lysyl-tRNA synthetase. FEBS Lett. 1992; 314:256–258. [DOI] [PubMed] [Google Scholar]
- 306. Masta S.E., Boore J.L.. The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol. Biol. Evol. 2004; 21:893–902. [DOI] [PubMed] [Google Scholar]
- 307. Perona J.J., Hou Y.-M.. Indirect readout of tRNA for aminoacylation. Biochemistry. 2007; 46:10419–10432. [DOI] [PubMed] [Google Scholar]
- 308. Frugier M., Helm M., Felden B., Giegé R., Florentz C.. Sequences outside recognition sets are not neutral for tRNA aminoacylation. Evidence for nonpermissive combinations of nucleotides in the acceptor stem of yeast tRNAPhe. J. Biol. Chem. 1998; 273:11605–11610. [DOI] [PubMed] [Google Scholar]
- 309. Fukunaga J., Ohno S., Nishikawa K., Yokogawa T.. A base pair at the bottom of the anticodon stem is reciprocally preferred for discrimination of cognate tRNAs by Escherichia coli lysyl- and glutaminyl-tRNA synthetases. Nucleic Acids Res. 2006; 34:3181–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Cavarelli J., Rees B., Ruff M., Thierry J.C., Moras D.. Yeast tRNAAsp recognition by its cognate class II aminoacyl-tRNA synthetase. Nature. 1993; 362:181–184. [DOI] [PubMed] [Google Scholar]
- 311. Goldgur Y., Mosyak L., Reshetnikova L., Ankilova V., Lavrik O., Khodyreva S., Safro M.. The crystal structure of phenylalanyl-tRNA synthetase from Thermus thermophilus complexed with cognate tRNAPhe. Structure. 1997; 5:59–68. [DOI] [PubMed] [Google Scholar]
- 312. Moor N., Kotik-Kogan O., Tworowski D., Sukhanova M., Safro M.. The crystal structure of the ternary complex of phenylalanyl-tRNA synthetase with tRNAPhe and a phenylalanyl-adenylate analogue reveals a conformational switch of the CCA end. Biochemistry. 2006; 45:10572–10583. [DOI] [PubMed] [Google Scholar]
- 313. Bedouelle H., Nageotte R.. Macromolecular recognition through electrostatic repulsion. EMBO J. 1995; 14:2945–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Sherman J.M., Söll D.. Aminoacyl-tRNA synthetases optimize both cognate tRNA recognition and discrimination against noncognate tRNAs. Biochemistry. 1996; 35:601–607. [DOI] [PubMed] [Google Scholar]
- 315. Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J.C., Moras D.. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp. Science. 1991; 252:1682–1689. [DOI] [PubMed] [Google Scholar]
- 316. Musier-Forsyth K., Schimmel P.. Functional contacts of a transfer RNA synthetase with 2’-hydroxyl groups in the RNA minor groove. Nature. 1992; 357:513–515. [DOI] [PubMed] [Google Scholar]
- 317. Eiler S., Dock-Bregeon A., Moulinier L., Thierry J.C., Moras D.. Synthesis of aspartyl-tRNAAsp in Escherichia coli—a snapshot of the second step. EMBO J. 1999; 18:6532–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Guth E.C., Francklyn C.S.. Kinetic discrimination of tRNA identity by the conserved motif 2 loop of a class II aminoacyl-tRNA synthetase. Mol. Cell. 2007; 25:531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Frugier M., Söll D., Giegé R., Florentz C.. Identity switches between tRNAs aminoacylated by class I glutaminyl- and class II aspartyl-tRNA synthetases. Biochemistry. 1994; 33:9912–9921. [DOI] [PubMed] [Google Scholar]
- 320. Praetorius-Ibba M., Rogers T.E., Samson R., Kelman Z., Ibba M.. Association between Archaeal prolyl- and leucyl-tRNA synthetases enhances tRNAPro aminoacylation. J. Biol. Chem. 2005; 280:26099–26104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Praetorius-Ibba M., Hausmann C.D., Paras M., Rogers T.E., Ibba M.. Functional association between three archaeal aminoacyl-tRNA synthetases. J. Biol. Chem. 2007; 282:3680–3687. [DOI] [PubMed] [Google Scholar]
- 322. Crnković A., Vargas-Rodriguez O., Söll D.. Plasticity and constraints of tRNA aminoacylation define directed evolution of aminoacyl-tRNA synthetases. Int. J. Mol. Sci. 2019; 20:E2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Chan C.W., Badong D., Rajan R., Mondragón A.. Crystal structures of an unmodified bacterial tRNA reveal intrinsic structural flexibility and plasticity as general properties of unbound tRNAs. RNA. 2020; 26:278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Li R., Macnamara L.M., Leuchter J.D., Alexander R.W., Cho S.S.. MD simulations of tRNA and aminoacyl-tRNA synthetases: dynamics, folding, binding, and allostery. Int. J. Mol. Sci. 2015; 16:15872–15902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Rees B., Webster G., Delarue M., Boeglin M., Moras D.. Aspartyl tRNA-synthetase from Escherichia coli: flexibility and adaptability to the substrates. J. Mol. Biol. 2000; 299:1157–1164. [DOI] [PubMed] [Google Scholar]
- 326. Sauter C., Lorber B., Cavarelli J., Moras D., Giegé R.. The free yeast aspartyl-tRNA synthetase differs from the tRNAAsp-complexed enzyme by structural changes in the catalytic site, hinge region, and anticodon-binding domain. J. Mol. Biol. 2000; 299:1313–1324. [DOI] [PubMed] [Google Scholar]
- 327. Pütz J., Puglisi J.D., Florentz C., Giegé R.. Additive, cooperative and anti-cooperative effects between identity nucleotides of a tRNA. EMBO J. 1993; 12:2949–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Giegé R. Toward a more complete view of tRNA biology. Nat. Struct. Mol. Biol. 2008; 15:1007–1014. [DOI] [PubMed] [Google Scholar]
- 329. Yaremchuk A., Tukalo M., Grøtli M., Cusack S.. A succession of substrate induced conformational changes ensures the amino acid specificity of Thermus thermophilus prolyl-tRNA synthetase: comparison with histidyl-tRNA synthetase. J. Mol. Biol. 2001; 309:989–1002. [DOI] [PubMed] [Google Scholar]
- 330. Ghosh A., Sakaguchi R., Liu C., Vishveshwara S., Hou Y.-M.. Allosteric communication in cysteinyl tRNA synthetase: a network of direct and indirect readout. J. Biol. Chem. 2011; 286:37721–37731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Rodríguez-Hernández A., Perona J.J.. Heat maps for intramolecular communication in an RNP enzyme encoding glutamine. Structure. 2011; 19:386–396. [DOI] [PubMed] [Google Scholar]
- 332. Boniecki M.T., Martinis S.A.. Coordination of tRNA synthetase active sites for chemical fidelity. J. Biol. Chem. 2012; 287:11285–11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Hilander T., Zhou X.-L., Konovalova S., Zhang F.-P., Euro L., Chilov D., Poutanen M., Chihade J., Wang E.-D., Tyynismaa H.. Editing activity for eliminating mischarged tRNAs is essential in mammalian mitochondria. Nucleic Acids Res. 2018; 46:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Hopfield J.J. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl Acad. Sci. USA. 1974; 71:4135–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975; 57:587–595. [DOI] [PubMed] [Google Scholar]
- 336. Fersht A.R. Enzyme, Structure and Mechanism. Freeman. 1985; New York. [Google Scholar]
- 337. Ahel I., Korencic D., Ibba M., Söll D.. Trans-editing of mischarged tRNAs. Proc. Natl Acad. Sci. USA. 2003; 100:15422–15427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Jakubowski H. Quality control in tRNA charging. Wiley Interdiscip. Rev.RNA. 2012; 3:295–310. [DOI] [PubMed] [Google Scholar]
- 339. Yadavalli S.S., Ibba M.. Quality control in aminoacyl-tRNA synthesis its role in translational fidelity. Adv. Protein Chem. Struct. Biol. 2012; 86:1–43. [DOI] [PubMed] [Google Scholar]
- 340. Dulic M., Cvetesic N., Zivkovic I., Palencia A., Cusack S., Bertosa B., Gruic-Sovulj I.. Kinetic origin of substrate specificity in post-transfer editing by leucyl-tRNA synthetase. J. Mol. Biol. 2018; 430:1–16. [DOI] [PubMed] [Google Scholar]
- 341. Yao P., Zhou X.-L., He R., Xue M.-Q., Zheng Y.-G., Wang Y.-F., Wang E.-D.. Unique residues crucial for optimal editing in yeast cytoplasmic leucyl-tRNA synthetase are revealed by using a novel knockout yeast strain. J. Biol. Chem. 2008; 283:22591–22600. [DOI] [PubMed] [Google Scholar]
- 342. Tan M., Wang M., Zhou X.-L., Yan W., Eriani G., Wang E.-D.. The Yin and Yang of tRNA: proper binding of acceptor end determines the catalytic balance of editing and aminoacylation. Nucleic Acids Res. 2013; 41:5513–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343. Huang Q., Zhou X.-L., Hu Q.-H., Lei H.-Y., Fang Z.-P., Yao P., Wang E.-D.. A bridge between the aminoacylation and editing domains of leucyl-tRNA synthetase is crucial for its synthetic activity. RNA. 2014; 20:1440–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Yan W., Ye Q., Tan M., Chen X., Eriani G., Wang E.-D.. Modulation of aminoacylation and editing properties of leucyl-tRNA synthetase by a conserved structural module. J. Biol. Chem. 2015; 290:12256–12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Liu Z., Vargas-Rodriguez O., Goto Y., Novoa E.M., Ribas de Pouplana L., Suga H., Musier-Forsyth K.. Homologous trans-editing factors with broad tRNA specificity prevent mistranslation caused by serine/threonine misactivation. Proc. Natl Acad. Sci. USA. 2015; 112:6027–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346. Korencic D., Ahel I., Schelert J., Sacher M., Ruan B., Stathopoulos C., Blum P., Ibba M., Söll D.. A freestanding proofreading domain is required for protein synthesis quality control in Archaea. Proc. Natl Acad. Sci. USA. 2004; 101:10260–10265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347. Bacusmo J.M., Kuzmishin A.B., Cantara W.A., Goto Y., Suga H., Musier-Forsyth K.. Quality control by trans-editing factor prevents global mistranslation of non-protein amino acid α-aminobutyrate. RNA Biol. 2018; 15:576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348. Kumar P., Bhatnagar A., Sankaranarayanan R.. Chiral proofreading during protein biosynthesis and its evolutionary implications. FEBS Lett. 2022; 596:1615–1627. [DOI] [PubMed] [Google Scholar]
- 349. Rigden D.J. Archaea recruited D-Tyr-tRNATyr deacylase for editing in Thr-tRNA synthetase. RNA. 2004; 10:1845–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350. Gogoi J., Bhatnagar A., Ann K.J., Pottabathini S., Singh R., Mazeed M., Kuncha S.K., Kruparani S.P., Sankaranarayanan R.. Switching a conflicted bacterial DTD-tRNA code is essential for the emergence of mitochondria. Sci. Adv. 2022; 8:eabj7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Kuncha S.K., Kruparani S.P., Sankaranarayanan R.. Chiral checkpoints during protein biosynthesis. J. Biol. Chem. 2019; 294:16535–16548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352. Rybak M.Y., Rayevsky A.V., Gudzera O.I., Tukalo M.A.. Stereospecificity control in aminoacyl-tRNA-synthetases: new evidence of d-amino acids activation and editing. Nucleic Acids Res. 2019; 47:9777–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353. Loftfield R.B., Vanderjagt D.. The frequency of errors in protein biosynthesis. Biochem. J. 1972; 128:1353–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354. Ibba M., Söll D.. Quality control mechanisms during translation. Science. 1999; 286:1893–1897. [DOI] [PubMed] [Google Scholar]
- 355. Rodnina M.V. Quality control of mRNA decoding on the bacterial ribosome. Adv. Protein Chem. Struct. Biol. 2012; 86:95–128. [DOI] [PubMed] [Google Scholar]
- 356. Bezerra A.R., Simões J., Lee W., Rung J., Weil T., Gut I.G., Gut M., Bayés M., Rizzetto L., Cavalieri D.et al.. Reversion of a fungal genetic code alteration links proteome instability with genomic and phenotypic diversification. Proc. Natl Acad. Sci. USA. 2013; 110:11079–11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357. Lee J.W., Beebe K., Nangle L.A., Jang J., Longo-Guess C.M., Cook S.A., Davisson M.T., Sundberg J.P., Schimmel P., Ackerman S.L.. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006; 443:50–55. [DOI] [PubMed] [Google Scholar]
- 358. Liu Y., Satz J.S., Vo M.-N., Nangle L.A., Schimmel P., Ackerman S.L.. Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc. Natl Acad. Sci. USA. 2014; 111:17570–17575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359. Wang Y., Zhou X.-L., Ruan Z.-R., Liu R.-J., Eriani G., Wang E.-D.. A human disease-causing point mutation in mitochondrial threonyl-tRNA synthetase induces both structural and functional defects. J. Biol. Chem. 2016; 291:6507–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360. Ruan B., Palioura S., Sabina J., Marvin-Guy L., Kochhar S., Larossa R.A., Söll D.. Quality control despite mistranslation caused by an ambiguous genetic code. Proc. Natl Acad. Sci. USA. 2008; 105:16502–16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361. Bullwinkle T.J., Ibba M.. Translation quality control is critical for bacterial responses to amino acid stress. Proc. Natl Acad. Sci. USA. 2016; 113:2252–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362. Sun L., Gomes A.C., He W., Zhou H., Wang X., Pan D.W., Schimmel P., Pan T., Yang X.-L.. Evolutionary gain of alanine mischarging to noncognate tRNAs with a G4:U69 base pair. J. Am. Chem. Soc. 2016; 138:12948–12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363. Kelly P., Backes N., Mohler K., Buser C., Kavoor A., Rinehart J., Phillips G., Ibba M.. Alanyl-tRNA synthetase quality control prevents global dysregulation of the Escherichia coli proteome. Mbio. 2019; 10:e02921–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364. Pan T. Adaptive translation as a mechanism of stress response and adaptation. Annu. Rev. Genet. 2013; 47:121–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Schwartz M.H., Pan T.. Function and origin of mistranslation in distinct cellular contexts. Crit. Rev. Biochem. Mol. Biol. 2017; 52:205–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366. Li L., Boniecki M.T., Jaffe J.D., Imai B.S., Yau P.M., Luthey-Schulten Z.A., Martinis S.A.. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc. Natl Acad. Sci. USA. 2011; 108:9378–9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367. Schrader J.M., Chapman S.J., Uhlenbeck O.C.. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc. Natl Acad. Sci. USA. 2011; 108:5215–5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368. Moghal A., Mohler K., Ibba M.. Mistranslation of the genetic code. FEBS Lett. 2014; 588:4305–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369. Rogers H.H., Griffiths-Jones S.. tRNA anticodon shifts in eukaryotic genomes. RNA. 2014; 20:269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Atilgan T., Nicholas H.B., McClain W.H.. A statistical method for correlating tRNA sequence with amino acid specificity. Nucleic Acids Res. 1986; 14:375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371. McClain W.H., Nicholas H.B.. Differences between transfer RNA molecules. J. Mol. Biol. 1987; 194:635–642. [DOI] [PubMed] [Google Scholar]
- 372. Kanai A. Welcome to the new tRNA world!. Front. Genet. 2014; 5:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Kumar P., Kuscu C., Dutta A.. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem. Sci. 2016; 41:679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374. Abe T., Inokuchi H., Yamada Y., Muto A., Iwasaki Y., Ikemura T.. tRNADB-CE: tRNA gene database well-timed in the era of big sequence data. Front. Genet. 2014; 5:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375. Chan P.P., Lowe T.M.. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016; 44:D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Pütz J., Dupuis B., Sissler M., Florentz C.. Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. RNA. 2007; 13:1184–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377. Cognat V., Pawlak G., Pflieger D., Drouard L.. PlantRNA 2.0: an updated database dedicated to tRNAs of photosynthetic eukaryotes. Plant J. 2022; 112:1112–1119. [DOI] [PubMed] [Google Scholar]
- 378. de Duve C. Transfer RNAs: the second genetic code. Nature. 1988; 333:117–118. [DOI] [PubMed] [Google Scholar]
- 379. Schimmel P. Development of tRNA synthetases and connection to genetic code and disease. Protein Sci. 2008; 17:1643–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380. Mallick B., Chakrabarti J., Sahoo S., Ghosh Z., Das S.. Identity elements of archaeal tRNA. DNA Res. 2005; 12:235–246. [DOI] [PubMed] [Google Scholar]
- 381. Freyhult E., Cui Y., Nilsson O., Ardell D.H.. New computational methods reveal tRNA identity element divergence between Proteobacteria and Cyanobacteria. Biochimie. 2007; 89:1276–1288. [DOI] [PubMed] [Google Scholar]
- 382. Ardell D.H. Computational analysis of tRNA identity. FEBS Lett. 2010; 584:325–333. [DOI] [PubMed] [Google Scholar]
- 383. Jakó E., Ittzés P., Szenes A., Kun A., Szathmáry E., Pál G.. In silico detection of tRNA sequence features characteristic to aminoacyl-tRNA synthetase class membership. Nucleic Acids Res. 2007; 35:5593–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384. Zamudio G.S., José M.V.. Identity elements of tRNA as derived from information analysis. Orig. Life Evol. Biosph. 2018; 48:73–81. [DOI] [PubMed] [Google Scholar]
- 385. Branciamore S., Gogoshin G., Di Giulio M., Rodin A.S.. Intrinsic properties of tRNA molecules as deciphered via Bayesian network and distribution divergence analysis. Life (Basel). 2018; 8:E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386. Galili T., Gingold H., Shaul S., Benjamini Y.. Identifying the ligated amino acid of archaeal tRNAs based on positions outside the anticodon. RNA. 2016; 22:1477–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387. Collins-Hed A.I., Ardell D.H.. Match fitness landscapes for macromolecular interaction networks: selection for translational accuracy and rate can displace tRNA-binding interfaces of non-cognate aminoacyl-tRNA synthetases. Theor. Popul. Biol. 2019; 129:68–80. [DOI] [PubMed] [Google Scholar]
- 388. Szenes A., Pál G.. Mapping hidden potential identity elements by computing the average discriminating power of individual tRNA positions. DNA Res. 2012; 19:245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389. Zamudio G.S., Palacios-Pérez M., José M.V.. Information theory unveils the evolution of tRNA identity elements in the three domains of life. Theory Biosci. 2020; 139:77–85. [DOI] [PubMed] [Google Scholar]
- 390. Turner B., Burkhart B.W., Weidenbach K., Ross R., Limbach P.A., Schmitz R.A., de Crécy-Lagard V., Stedman K.M., Santangelo T.J., Iwata-Reuyl D.. Archaeosine modification of archaeal tRNA: role in structural stabilization. J. Bacteriol. 2020; 202:e00748-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391. Igloi G.L. Evolutionary adjustment of tRNA identity rules in Bacillariophyta for recognition by an aminoacyl-tRNA synthetase adds a facet to the origin of diatoms. J. Mol. Evol. 2022; 90:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392. Giegé R., Florentz C., Dreher T.W.. The TYMV tRNA-like structure. Biochimie. 1993; 75:569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393. Tamaki S., Tomita M., Suzuki H., Kanai A.. Systematic analysis of the binding surfaces between tRNAs and their respective aminoacyl tRNA synthetase based on structural and evolutionary data. Front. Genet. 2017; 8:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394. Westhof E., Thornlow B., Chan P.P., Lowe T.M.. Eukaryotic tRNA sequences present conserved and amino acid-specific structural signatures. Nucleic Acids Res. 2022; 50:4100–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395. Rodin S.N., Ohno S.. Two types of aminoacyl-tRNA synthetases could be originally encoded by complementary strands of the same nucleic acid. Orig. Life Evol. Biosph. 1995; 25:565–589. [DOI] [PubMed] [Google Scholar]
- 396. Martinez-Rodriguez L., Erdogan O., Jimenez-Rodriguez M., Gonzalez-Rivera K., Williams T., Li L., Weinreb V., Collier M., Chandrasekaran S.N., Ambroggio X.et al.. Functional class I and II amino acid-activating enzymes can be coded by opposite strands of the same gene. J. Biol. Chem. 2015; 290:19710–19725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397. Carter C.W., Wills P.R.. Class I and II aminoacyl-tRNA synthetase tRNA groove discrimination created the first synthetase–tRNA cognate pairs and was therefore essential to the origin of genetic coding. IUBMB Life. 2019; 71:1088–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398. Agmon I., Fayerverker I., Mor T.. Coding triplets in the tRNA acceptor–TΨC arm and their role in present and past tRNA recognition. FEBS Lett. 2021; 595:913–924. [DOI] [PubMed] [Google Scholar]
- 399. Möller W., Janssen G.M.. Statistical evidence for remnants of the primordial code in the acceptor stem of prokaryotic transfer RNA. J. Mol. Evol. 1992; 34:471–477. [DOI] [PubMed] [Google Scholar]
- 400. Urbonavicius J., Armengaud J., Grosjean H.. Identity elements required for enzymatic formation of N2,N2-dimethylguanosine from N2-monomethylated derivative and its possible role in avoiding alternative conformations in archaeal tRNA. J. Mol. Biol. 2006; 357:387–399. [DOI] [PubMed] [Google Scholar]
- 401. Nishida Y., Ohmori S., Kakizono R., Kawai K., Namba M., Okada K., Yamagami R., Hirata A., Hori H.. Required elements in tRNA for methylation by the eukaryotic tRNA (Guanine-N2-) methyltransferase (Trm11–Trm112 complex). Int. J. Mol. Sci. 2022; 23:4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402. Zhou M., Long T., Fang Z.-P., Zhou X.-L., Liu R.-J., Wang E.-D.. Identification of determinants for tRNA substrate recognition by Escherichia coli C/U34 2’-O-methyltransferase. RNA Biol. 2015; 12:900–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403. Li H., Zhu D., Wu J., Ma Y., Cai C., Chen Y., Qin M., Dai H.. New substrates and determinants for tRNA recognition of RNA methyltransferase DNMT2/TRDMT1. RNA Biol. 2021; 18:2531–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404. Seong B.L., RajBhandary U.L.. Mutants of Escherichia coli formylmethionine tRNA: a single base change enables initiator tRNA to act as an elongator in vitro. Proc. Natl Acad. Sci. USA. 1987; 84:8859–8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405. Lee C.P., Seong B.L., RajBhandary U.L.. Structural and sequence elements important for recognition of Escherichiacoli formylmethionine tRNA by methionyl-tRNA transformylase are clustered in the acceptor stem. J. Biol. Chem. 1991; 266:18012–18017. [PubMed] [Google Scholar]
- 406. Guillon J.-M., Meinnel T., Mechulam Y., Lazennec C., Blanquet S., Fayat G.. Nucleotides of tRNA governing the specificity of Escherichia coli methionyl-tRNAfMet formyltransferase. J. Mol. Biol. 1992; 224:359–367. [DOI] [PubMed] [Google Scholar]
- 407. Yashiro Y., Zhang C., Sakaguchi Y., Suzuki T., Tomita K.. Molecular basis of glycyl-tRNAGly acetylation by TacT from Salmonella typhimurium. Cell Rep. 2021; 37:110130. [DOI] [PubMed] [Google Scholar]
- 408. Raina M., Ibba M.. tRNAs as regulators of biological processes. Front. Genet. 2014; 5:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409. Roy H., Becker H.D., Mazauric M.-H., Kern D.. Structural elements defining elongation factor Tu mediated suppression of codon ambiguity. Nucleic Acids Res. 2007; 35:3420–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410. Sanderson L.E., Uhlenbeck O.C.. The 51–63 base pair of tRNA confers specificity for binding by EF-Tu. RNA. 2007; 13:835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411. Roy H., Ibba M.. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc. Natl Acad. Sci. USA. 2008; 105:4667–4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412. Nolan E.M., Walsh C.T.. How Nature morphs peptide scaffolds into antibiotics. Chem. Biol. Chem. 2009; 10:34–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413. Shepherd J., Ibba M.. Direction of aminoacylated transfer RNAs into antibiotic synthesis and peptidoglycan-mediated antibiotic resistance. FEBS Lett. 2013; 587:2895–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414. Abramochkin G., Shrader T.E.. Aminoacyl-tRNA recognition by the leucyl/phenylalanyl-tRNA-protein transferase. J. Biol. Chem. 1996; 271:22901–22907. [DOI] [PubMed] [Google Scholar]
- 415. Kim B.H., Kim M.K., Oh S.J., Nguyen K.T., Kim J.H., Varshavsky A., Hwang C.-S., Song H.K.. Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates. Proc. Natl Acad. Sci. USA. 2022; 119:e2209597119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416. Lopes R.R.S., Kessler A.C., Polycarpo C., Alfonzo J.D.. Cutting, dicing, healing and sealing: the molecular surgery of tRNA. Wiley Interdiscip Rev. RNA. 2015; 6:337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417. Wallace D.C. Diseases of the mitochondrial DNA. Annu. Rev. Biochem. 1992; 61:1175–1212. [DOI] [PubMed] [Google Scholar]
- 418. Lant J.T., Berg M.D., Heinemann I.U., Brandl C.J., O’Donoghue P. Pathways to disease from natural variations in human cytoplasmic tRNAs. J. Biol. Chem. 2019; 294:5294–5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419. Jiang L., Jones J., Yang X.-L.. Human diseases linked to cytoplasmic aminoacyl-tRNA synthetases. Enzymes. 2020; 48:277–319. [DOI] [PubMed] [Google Scholar]
- 420. Suzuki T., Nagao A., Suzuki T.. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011; 45:299–329. [DOI] [PubMed] [Google Scholar]
- 421. de Crécy-Lagard V., Boccaletto P., Mangleburg C.G., Sharma P., Lowe T.M., Leidel S.A., Bujnicki J.M.. Matching tRNA modifications in humans to their known and predicted enzymes. Nucleic Acids Res. 2019; 47:2143–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422. Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021; 22:375–392. [DOI] [PubMed] [Google Scholar]
- 423. Lott M.T., Leipzig J.N., Derbeneva O., Xie H.M., Chalkia D., Sarmady M., Procaccio V., Wallace D.C.. mtDNA variation and analysis using Mitomap and Mitomaster. Curr. Protoc. Bioinformatics. 2013; 44:1.23–1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424. Richter U., McFarland R., Taylor R.W., Pickett S.J.. The molecular pathology of pathogenic mitochondrial tRNA variants. FEBS Lett. 2021; 595:1003–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425. Lehmann D., Schubert K., Joshi P.R., Hardy S.A., Tuppen H.A.L., Baty K., Blakely E.L., Bamberg C., Zierz S., Deschauer M.et al.. Pathogenic mitochondrial mt-tRNAAla variants are uniquely associated with isolated myopathy. Eur. J. Hum. Genet. 2015; 23:1735–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426. Swalwell H., Deschauer M., Hartl H., Strauss M., Turnbull D.M., Zierz S., Taylor R.W.. Pure myopathy associated with a novel mitochondrial tRNA gene mutation. Neurology. 2006; 66:447–449. [DOI] [PubMed] [Google Scholar]
- 427. Ding Y., Zhuo G., Zhang C., Leng J.. Point mutation in mitochondrial tRNA gene is associated with polycystic ovary syndrome and insulin resistance. Mol. Med. Rep. 2016; 13:3169–3172. [DOI] [PubMed] [Google Scholar]
- 428. Chujo T., Tomizawa K.. Human transfer RNA modopathies: diseases caused by aberrations in transfer RNA modifications. FEBS J. 2021; 288:7096–7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429. Musier-Forsyth K., Usman N., Scaringe S., Doudna J., Green R., Schimmel P.. Specificity for aminoacylation of an RNA helix: an unpaired, exocyclic amino group in the minor groove. Science. 1991; 253:784–786. [DOI] [PubMed] [Google Scholar]
- 430. Endo Y., Mitsui K., Motizuki M., Tsurugi K.. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J. Biol. Chem. 1987; 262:5908–5912. [PubMed] [Google Scholar]
- 431. Thiebe R., Zachau H.G.. A specific modification next to the anticodon of phenylalanine transfer ribonucleic acid. Eur. J. Biochem. 1968; 5:546–555. [DOI] [PubMed] [Google Scholar]
- 432. Wintermeyer W., Zachau H.G.. A specific chemical chain scission of tRNA at 7-methylguanosine. FEBS Lett. 1970; 11:160–164. [DOI] [PubMed] [Google Scholar]
- 433. Bruce A.G., Uhlenbeck O.C.. Enzymatic replacement of the anticodon of yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1982; 21:855–861. [DOI] [PubMed] [Google Scholar]
- 434. Liu Y., Rodriguez Y., Ross R.L., Zhao R., Watts J.A., Grunseich C., Bruzel A., Li D., Burdick J.T., Prasad R.et al.. RNA abasic sites in yeast and human cells. Proc. Natl Acad. Sci. USA. 2020; 117:20689–20695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435. Barbieri M. What is code biology?. Biosystems. 2018; 164:1–10. [DOI] [PubMed] [Google Scholar]
- 436. Meiklejohn C.D., Holmbeck M.A., Siddiq M.A., Abt D.N., Rand D.M., Montooth K.L.. An Incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLos Genet. 2013; 9:e1003238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the authors upon request.