Abstract
Long-term (>2.5 years) surveillance of SARS-CoV-2 RNA concentrations in wastewater was conducted within an enclosed university compound. This study aims to demonstrate how coupling wastewater-based epidemiology (WBE) with meta-data can identify which factors contribute toward the dissemination of SARS-CoV-2 within a local community. Throughout the pandemic, the temporal dynamics of SARS-CoV-2 RNA concentrations were tracked by quantitative polymerase chain reaction and analyzed in the context of the number of positive swab cases, the extent of human movement, and intervention measures. Our findings suggest that during the early phase of the pandemic, when strict lockdown was imposed, the viral titer load in the wastewater remained below detection limits, with <4 positive swab cases reported over a 14-day period in the compound. After the lockdown was lifted and global travel gradually resumed, SARS-CoV-2 RNA was first detected in the wastewater on 12 August 2020 and increased in frequency thereafter, despite high vaccination rates and mandatory face-covering requirements in the community. Accompanied by a combination of the Omicron surge and significant global travel by community members, SARS-CoV-2 RNA was detected in most of the weekly wastewater samples collected in late December 2021 and January 2022. With the cease of mandatory face covering, SARS-CoV-2 was detected in at least two of the four weekly wastewater samples collected from May through August 2022. Retrospective Nanopore sequencing revealed the presence of the Omicron variant in the wastewater with a multitude of amino acid mutations, from which we could infer the likely geographical origins through bioinformatic analysis. This study demonstrated that long-term tracking of the temporal dynamics and sequencing of variants in wastewater would aid in identifying which factors contribute the most to SARS-CoV-2 dissemination within the local community, facilitating an appropriate public health response to control future outbreaks as we now live with endemic SARS-CoV-2.
Keywords: SARS-CoV-2, Wastewater-based epidemiology, Nucleocapsid genes, Oxford Nanopore Technologies sequencing, University, Enclosed community
Graphical abstract
1. Introduction
SARS-CoV-2, a positive-sense enveloped single-stranded RNA virus, has S-proteins that bind to ACE-2 receptors, which are highly expressed in mucosal and cilial respiratory cells, type 2 alveolar pneumocytes, heart and vascular endothelium, and intestinal epithelial cells (Penninger et al., 2021). About 39 to 65 % of infected hosts, including asymptomatic carriers, shed the virus through their feces (Lavania et al., 2022; Pedersen et al., 2022; Zhang et al., 2021). By monitoring for RNA gene fragments associated with SARS-CoV-2 in wastewater and tracking the abundance of genes over time, various studies demonstrated the use of wastewater-based epidemiology (WBE) to infer the prevalence of infected cases in urban-dwelling populations served by sewer networks (Ahmed et al., 2020; Haramoto et al., 2020; Hopkins et al., 2022; Kumar et al., 2020; Medema et al., 2020; Randazzo et al., 2020; Sherchan et al., 2020). While the WBE approach is useful to track the extent of the outbreak and to determine how effective certain intervention measures were within a somewhat nonmobile population when shelter-in-place/lockdown was implemented in the early phase of the pandemic, it became increasingly more challenging to achieve the same aims when the population was in a constant state of movement, particularly as more countries are entering the endemic phase of COVID-19.
Subsequently, WBE was taken up as a tool by academic researchers who worked with campus stakeholders to guide universities to safe reopening (Wallace and Pappas, 2022) and as an early warning to quell emergent outbreaks (Korfmacher et al., 2021). The rationale behind doing so is that each university can be viewed as a closed boundary, where meta-data of key intervention measures and timelines of important events can be consolidated and matched against SARS-CoV-2 RNA abundance in wastewater collected within the university and potentially aid in public health decision making. A Scopus search using the keywords “university,” “SARS-CoV-2,” and “wastewater-based epidemiology” yielded at least 16 papers published in 2021 and 2022 that described RT-qPCR to track the temporal dynamics of SARS-CoV-2 RNA in wastewater (Table S1) (Anderson-Coughlin et al., 2022; Betancourt et al., 2021; Corchis-Scott et al., 2021; de Llanos et al., 2022; Fahrenfeld et al., 2022; Gibas et al., 2021; Jain et al., 2022; Kotay et al., 2022; Lee et al., 2022; Lu et al., 2022; Mangwana et al., 2022; Rainey et al., 2023; Reeves et al., 2021; Scott et al., 2021; Wang et al., 2022; Wright et al., 2022). Although most of these studies showed a good correlation between the genome copy numbers of SARS-CoV-2 RNA in wastewater against the number of SARS-CoV-2 infected cases (Table S1), the vast majority of these universities focused only on surveying individual dormitories and thus do not provide a monitoring resolution that represents the whole university community, which includes faculty members and other employees who do not reside in the surveyed buildings. Most of these studies also demonstrate the use of WBE data to inform decision makers on when to perform clinical swab testing, isolation, and contact tracing (Table S1). Carrying out such measures during the first 2 years of the COVID-19 pandemic was done with relative ease. However, as more countries adopt the strategy of living endemically with SARS-CoV-2, performing clinical swab testing, isolation, and contact tracing has become more challenging and is likely to face more resistance among community members. Instead, having a better understanding of which factors contribute the most to viral dissemination would perhaps aid local decision makers in developing relatively effective but minimally disruptive measures to curb future outbreaks.
In this study, we used an enclosed university compound (King Abdullah University of Science and Technology, KAUST) as a unique model. KAUST is a 36 km2 enclosed compound barricaded at all sides, with only two main entry/exit gates. The majority of the KAUST community, comprised of faculty, students, staff, and their family members, reside within the barricaded area that includes the core campus, residential area, recreational services, maintenance buildings, medical center, and pre-university schools (Fig. S1). Approximately 7500 residents live in the compound, and ~ 5000 registered service providers enter KAUST through the main gates daily but do not reside within the enclosed area. All wastewater generated by the community is treated by a decentralized wastewater treatment plant (WWTP) located within the compound. We surveyed wastewater for SARS-CoV-2 RNA over 2.5 years since the pandemic started (from 23 March 2020 until September 2022) and matched that data against clinical swab testing outcomes and key milestones/interventions that happened within the enclosed compound.
For example, the duration of this WBE study spans the period in which (i) strict intervention measures (e.g., shelter-in-place/lockdown, mandatory face coverings, online lessons, suspension of all flights, etc.) were put in place; (ii) rollout of vaccines given to all eligible community members; (iii) gradual easing of global travel; and (iv) removal of face-covering requirements. Considering the unique feature of this enclosed community in which meta-data of the key intervention measures and human movement were well-recorded against the number of clinical cases and that the WWTP only receives wastewater generated by the residents and service providers within the compound, examining the WBE over a long-term would allow us to identify which factors contribute most strongly to outbreaks. Such local-scale insights would aid decision makers in implementing appropriate intervention measures specifically tailored to the community.
2. Materials and methods
2.1. Important COVID-19 milestones of relevance to KAUST
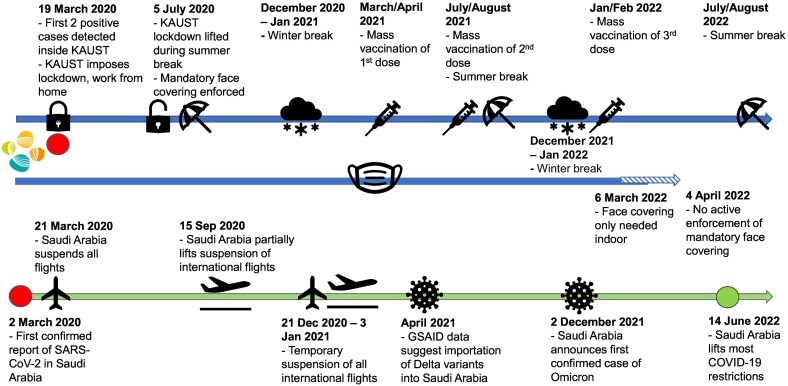
SARS-CoV-2 was first reported in Saudi Arabia on 2 March 2020, and the first two COVID-19 cases inside the enclosed KAUST compound were detected on 21 March 2020. The KAUST COVID task force responded by putting in place strict intervention measures to minimize the extent of SARS-CoV-2 dissemination within the enclosed compound (Fig. 1 ). These measures included first closing the campus's main gates and not allowing external visitors to enter KAUST. All service providers except those providing essential services were not permitted into the KAUST compound from 23 March until 5 July 2020. All KAUST community members were asked to work or study from home and were asked to observe a daily curfew from 3 pm to 6 am the next day. The curfew lasted from 23 March until 28 May 2020. The curfew was subsequently lifted, but KAUST community members were advised to continue to work from home. From 5 July 2020 onwards, all service providers were allowed to enter the KAUST compound, and a limited number of external visitors was allowed to enter, subject to approval. KAUST community members were also asked to gradually resume in-person working but to work in small shifts, practice social distancing, and wear a face covering at all times.
Fig. 1.
Important milestones that occurred during the 2.5 years surveillance. Blue line denotes the milestones related to KAUST. Green line denotes the milestones that happened in parallel within the kingdom of Saudi Arabia.
All lessons were conducted virtually from 2020 until the Fall of 2021. New incoming students (ca. 150 persons) who arrived within the compound in early August 2020 were mainly comprised of those who were already present in Saudi Arabia (KAUST Graduate Affairs, personal communication). International students could register for virtual classes and studied remotely in their home countries. In 2021 and 2022, new incoming students (>450 persons), comprising ~40 % locals and 60 % international students (comprised of students from South America, Europe, Asia and US) were onboarded in July–August before the Fall semester (KAUST Graduate Affairs, personal communication).
Mass vaccination programs were rolled out for the entire KAUST community in March/April 2021 when vaccines became available in Saudi Arabia, and then again in July/August 2021 and Jan/February 2022 for the booster doses, per Saudi government requirements (Ferrari, 2021; Khoja, 2021; Nihal, 2022).
Global travel was impossible for most of 2020 as Saudi Arabia suspended all flights (Naar, 2020a). Suspension of international flights was partially lifted on 15 Sep 2020 (Reuters, 2020) but resuspended again on 21 December 2020 for 3 weeks due to the detection of the SARS-Cov-2 Alpha variant (VUI 202012/01) (ArabNews, 2020; Naar, 2020b). After mid-Jan 2021, all international and domestic flights resumed in Saudi Arabia, but only travelers who had completed three vaccinations and could produce a negative COVID swab test result were eligible to travel (Nihal, 2022).
Saudi Arabia observes Foundation Day (22 Feb), National Day (22 Sep), Eid Al Fitr (end of the fasting month, Ramadan), and Eid Al Adha (end of the Hajj pilgrimage) as public holidays. Eid Al Fitr and Eid Al Adha follow the Islamic Hijri calendar. Eid Al Fitr break was observed by KAUST on 24–28 May 2020, 12–20 May 2021, and 1–5 May 2022, while Eid Al Adha was on 30 July until 6 Aug 2020, 18–22 July 2021, and 10–14 July 2022. KAUST community members generally use the Eid Al Fitr and Eid Al Adha breaks to travel domestically or overseas, except when global travel was not feasible due to flight suspensions.
Within Saudi Arabia, restrictions to curb the dissemination of SARS-CoV-2 were gradually loosened by first removing the requirement for face coverings in outdoor environments for fully vaccinated individuals in October 2021 (National, 2021). However, in December 2021–January 2022, the Kingdom was hit by an Omicron wave (Krimly, 2021). The Saudi government reimposed the requirement for face covering in indoor and outdoor environments, and KAUST followed suit in requiring community members to wear face coverings at all times (National, 2021). After the first wave of Omicron subsided, face covering was no longer required in the outdoor environment in March 2022 (Ferrari, 2022). KAUST first removed the requirement for face covering in outdoor environments on 6 March 2022 and then removed all COVID-19 intervention measures after 4 April 2022 (Fig. S2).
Important COVID-19 milestones relevant to KAUST are available in the dashboard managed by KAUST Health, Safety and Environment (HSE), which is accessible only through the KAUST intranet (Fig. S2, https://hse.kaust.edu.sa/keeping-kaust-safe/kaust-current-summary). The KAUST HSE also keeps a record of the number of rolling 14-day positive cases identified by swab testing and RT-qPCR from 21 March 2020 till 21 Sep 2022. The record stops after 21 Sep 2022 as the KAUST Medical Center switched to performing only rapid antigen test kits for symptomatic cases and could no longer provide KAUST HSE with a matching record.
2.2. Study site and sampling of influent from KAUST wastewater treatment plant (WWTP)
KAUST WWTP is situated within the barricaded compound and receives all municipal wastewater generated daily by the ~7500 residents and 5000 service providers. Grab samples were collected from the equalization tank twice, once in the morning (9 AM) and once in the afternoon (4 PM), every Sunday throughout the study. Samples were collected on Sunday, the start of the working week in Saudi Arabia. The samples were mixed in equal volumes to form the final sample for filtration. The WWTP plant operators recorded the total incoming volume of influent each day during the study period (Fig. S3), and the volume of wastewater influent is assumed to correlate to the population size inside the community at a given time since this WWTP exclusively serves the KAUST compound.
2.3. Processing of wastewater samples
Samples (300 mL) were individually concentrated for viral particles using the electronegative membrane method (Merck Millipore HAWP09000, Cork, Ireland) with slight modifications (Haramoto et al., 2004). Briefly, 2 mL of 2.5 M MgCl2 was added to every 100 mL of the sample, agitated for 3 min, and left to stand for 3 min. The samples were then filtered through the electronegative membrane. The electronegative membrane was then filtered and washed with 200 mL of 0.5 mM H2SO4 to adjust the isoelectric point of the viruses. The viral particles retained on the electronegative membrane were eluted with 10 mL of 1 mM NaOH into a sterile collection tube containing 100 μL of 100× Tris-EDTA buffer and 50 μL of 100 mM H2SO4. RNA was extracted from the eluate (140 μL) using QIAmp Viral RNA kits according to the manufacturer's protocol (ThermoFisher Scientific, Carlsbad, CA). The remaining eluate (about 9.8 mL) was stored at −80 °C. Extracted RNA was converted to complementary DNA (cDNA) using Superscript III First-Strand Synthesis System (ThermoFisher Scientific, Carlsbad, CA). Briefly, 6 μL viral RNA was mixed with 50 ng/μL random hexamers and 1 μL annealing buffer, then incubated in a thermal cycler at 65 °C for annealing before 5 min cooling on ice. The annealed product was mixed with 10 μL of 2× first-strand reaction mix and 2 μL superscript III/RNAase out enzyme mix before incubation at 25 °C for 10 min, 50 °C for 50 min and 85 °C for 5 min to terminate the reaction. The cDNA was kept at −20 °C before qPCR. The recovery efficiency was evaluated using bacteriophage MS2 as a surrogate (Supplementary Information S1 and Table S2).
2.4. Quantitative PCR for SARS-CoV-2
Genes associated with nucleocapsid (N) proteins N1 and N2 of SARS-CoV-2 were targeted for quantitative PCR (CDC, 2021). G-blocks that include the annealing regions of N1 and N2 primers-probes were synthesized based on the published sequences of SARS-CoV-2 (Forster et al., 2020) (Table S3). Six-point standard curves were generated for each primer-probe pair to determine their respective amplification efficiencies. Based on the standard curves, the average amplification efficiencies were 92.3 % and 89.8 %, and the R2 value ranged from 0.998 to 1 and 0.993 to 1 for N1 and N2, respectively. The limit of qPCR detection was estimated based on the number of copies per reaction with a detection rate of 95 % as recommended by the MIQE guidelines (Bustin et al., 2009) via probit regression analysis (CLSI, 2012). The 95 % LOD was estimated to be 5.9 and 6.9 copies/reaction for N1 and N2 annealed region targets. The positive control for qPCR was either cDNA derived from RNA of SARS-CoV-2-positive clinical swab specimens in the early phase of the study or with a 2019-nCoV_N positive control (Integrated DNA Technologies, Leuven, Belgium) when it became commercially available. A non-template control was prepared with aliquots of the same master mix used for qPCR assays to denote if reagents were contaminated. A negative control is prepared by replacing the cDNA template with sterile water added in volumes similar to that of the cDNA volume in positive controls. Positive controls have an average Cq value of 27.7 for N1 and 29.5 for N2. NTCs have non-detectable Cq values for both primer pairs targeting N1 and N2. qPCR was conducted by mixing 2.5 μL of sample cDNA with 12.5 μL TaqMan fast advance master mix (Applied Biosystems, Vilnius, Lithuania), 6.5 μL nuclease-free water, 400 nm of each forward and reverse primer, and 300 nm probe for all N1 and N2 for a total volume of 25 μL. All qPCR reactions were performed with a QuantStudio™ three real-time PCR system and 96-well plates (0.1 mL volume, Thermo Fisher Scientific, Carlsbad, California) and analyzed with QuantStudio design and analysis software v1.5.1. Thermal cycling conditions include 50 cycles of 95 °C for denaturation (3 s) and 55 °C for annealing and amplification (30 s). All samples, standards, and controls were performed in technical duplicates.
Concentrations of N1 and N2 genes were first averaged to obtain the overall SARS-CoV-2 nucleocapsid gene concentration (copies/L) of that sample. The SARS-CoV-2 gene concentrations were then divided by the pepper mild mottle virus (PMMoV) concentration (denoted in Section 2.5) and multiplied by a reference median PMMoV concentration value derived from the dataset (Wu et al., 2020) to obtain the normalized SARS-CoV-2 gene concentrations (copies/L). The ratio of normalized SARS-CoV-2 gene concentration (copies/L) to positive swab numbers registered by the KAUST Health and Safety Department over a 14-day period (WC ratio) (Xiao et al., 2022) was used as a metric for detecting differences between wastewater and clinical trends. Specifically, we defined a WC ratio near one to reflect that the testing capacity is sufficient to assess the extent of new infections within the compound. When the WC ratio increases compared to its preceding period and is >1, we defined this as an indication that the existing swab testing capacity has not kept up with the rising new cases within the compound but was detected by WBE.
2.5. Determination of PMMoV abundance
Quantification of PMMoV concentration (in terms of copies/L) was performed by RT-qPCR assay. PMMoV is the most abundant RNA virus in human feces. It was previously proposed as a potential viral indicator for human fecal contamination due to its ubiquitous global distribution without substantial seasonal fluctuations (Kitajima et al., 2018). Quantitative PCR for PMMoV was performed using forward primer (5′- GAGTGGTTTGACCTTAACGTTTGA-3′), reverse primer (5′-TTGTCGGTTGCAATGCAAGT-3′), and Taqman probe (5′-FAM-CCTACCGAAGCAAATG-ZEN/IBFQ-3′) (Integrated DNA Technologies, Leuven, Belgium). Gblock for PMMoV is listed in Table S3. A thermal cycler profile of 95 °C for 15 min, followed by 40 cycles of 95 °C for 15 s and 55 °C for 30 s was used (Haramoto et al., 2013). For PMMoV qPCR, a LOD of 10 copies/well and an amplification efficiency of 95.7 % was achieved.
2.6. Data analysis
Outlier identification in PMMoV abundance was performed using Grubbs' test with a significance level of 0.05 to detect qPCR inhibitors in the system. Correlation of the SARS-CoV-2 N gene (copies/L) against the number of 14-day rolling positive cases was performed using Pearson correlation in Microsoft Excel. One-way ANOVA was performed with OriginPro 2020, using the SARS-CoV-2 concentration (copies/L) as the input data. The different phases categorized accordingly in 3.2, 3.3, 3.4 were used as the factor for one-way ANOVA.
2.7. Oxford Nanopore Technologies sequencing and data analysis
Wastewater samples with SARS-CoV-2 RNA genes detected by RT-qPCR were retrospectively sequenced for SARS-CoV-2 on a GridION operated by KAUST Bioscience Core labs in October 2022. Fresh RNA was extracted from the eluate stored at −80 °C (Section 2.2), converted to cDNA, and amplified for the SARS-CoV-2 genes following the COVID-19 ARTIC-V3 protocol (https://github.com/artic-network/artic-ncov2019/blob/master/primer_schemes/nCoV-2019/V3/nCoV-2019.tsv) with 35 cycles prior to Nanopore sequencing. Sequenced datasets were subjected to base calling by guppy (v5.0.13) with the “High-accuracy basecalling” model (min_score 60 and min_qscore 7). Exported reads were trimmed by nanofilt (v2.8.0) (headcrop, 10 bp; read length, <100 bp) (De Coster et al., 2018) for quality control. Reads that passed quality control were mapped to the reference genome “hCoV-19/Wuhan/WIV04/2019|EPI_ISL_402124|2019-12-30|China” by minimap2 (v2.20) (Li, 2018) with default settings. Mapped reads with MAPQ >20 were retained by samtools (v1.9) (Danecek et al., 2021), and information for each nucleotide position (such as base counts, frequency of nucleotides) was summarized by bam-readcount (v0.8) (Khanna et al., 2021). According to a previous study that used Nanopore sequencing for SARS-CoV-2 genome analysis (Bull et al., 2020), single-nucleotide variants were called based on settings with a depth of coverage >60 hits and substituted nucleotide percentage > 25 % and further determined for nonsynonymous substitution (i.e., amino acid mutations). Specific variants of concern (VoC) from sequencing datasets were curated to determine the detection of VoC in wastewater samples. Raw Nanopore sequencing datasets were deposited into the sequence reads archive database of the European Nucleotide Archive under study accession number PRJEB56506.
2.8. Phylodynamic analysis and importation date estimation of SARS-CoV-2 genomes in Saudi Arabia
All Delta/Omicron genomes discovered in Saudi Arabia were collated from GISAID (last retrieved on 30 September 2022) (Shu and McCauley, 2017). To generate the phylogeny of these genomes, international Delta/Omicron variant genomes (with a complete sample collection date) were also downloaded from GISAID by Sep 30, 2022. A subset of 500 international genomes was selected for phylogeny analysis according to their genetic distance to genomes from Saudi Arabia: briefly, every international genome was aligned to genomes from Saudi Arabia by mafft (v7.508) (Katoh et al., 2002) and then Tamura Nei 93 distance between two genomes were computed by tn93 (v1.0.9) (https://github.com/veg/tn93). The most closely aligned 500 international genomes were selected as a subset for further phylogeny analysis. Based on the international genome subset, a maximum likelihood phylogeny based on the Hasegawa-Kishino-Yano substitution model for genomes from Saudi Arabia was computed by IQtree (v2.2.0) (Trifinopoulos et al., 2016). Additionally, short branches and polytomies of the phylogeny were then removed by ape (v5.6) (Paradis et al., 2004), using functions “multi2di” with setting random = 15 and “di2multi” with setting tol = 1e − 08) on R (v4.1.2). The sample collection date of genomes in phylogeny was labelled to estimate the time of common ancestry by treedater (v0.5.0) (Volz and Frost, 2017) with a strict molecular clock. Furthermore, the state of internal nodes of phylogeny was reconstructed by maximum parsimony with phangorn (v2.10.0) (Schliep, 2011) on R (v4.1.2). According to a previous study (Mourier et al., 2022), the importation events were determined based on the closest branch to the genome from Saudi Arabia. The importation time was determined by the midpoint of this branch, and the countries on the closest branch were inferred as the potential. The probability density of importation events for Delta and Omicron variants in Saudi Arabia were presented through the Gaussian kernel density estimator with the Silverman rule of thumb, implemented in the “geom_density” function of the ggplot2 (v3.3.6) (Wickham, 2016) with ggridges (v0.5.3) on R (v4.1.2).
3. Results and discussion
3.1. Viral titer load in wastewater when strict intervention measures were in place
There was no positive detection of N1 and N2 genes in the wastewater throughout the period when strict intervention measures were in place (23 March until 5 July 2020). This is despite an average 14-day positive swab test rate of 3.6 ± 2.9 cases reported in the compound (Fig. 2A) and the major Eid Al Fitr holiday (24–28 May 2020) during this period. All samples were positive for PMMoV at a concentration ranging from 5.5 to 7.9 log10 copies/L (Fig. S4A), which falls within the normal range of PMMoV abundance in wastewater (Kitajima et al., 2018) and with no significant outlier events (Fig. S4B).
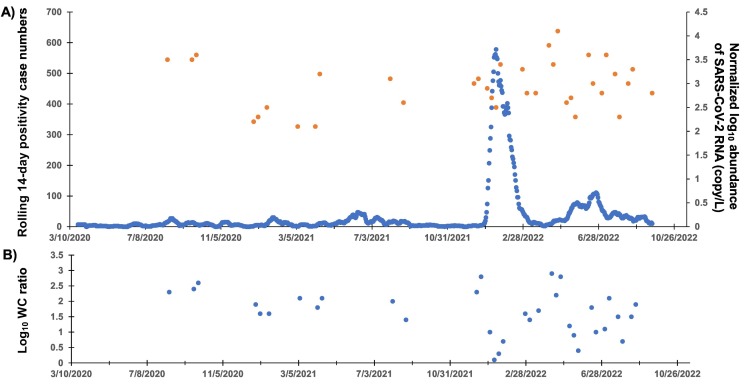
Fig. 2.
The temporal trend of COVID-19-related data mapped over the 2.5 years of surveillance. Top graph depicts the number of positive swab cases mapped over a 14-day rolling period in the primary axis, and the normalized log10 abundance of SARS-CoV-2 RNA in wastewater detected at certain time points, shown on the secondary axis. The bottom graph depicts the log10 WC ratio. The x-axis denotes dates in the format of MM/DD/YYYY.
The lack of SARS-CoV-2 in the wastewater during this period may be because the number of clinical cases within the community was too low for WBE to detect. Various parameters (e.g., sampling location, wastewater sampling and processing procedures, detection method) can affect detection sensitivity. The reported detection sensitivity ranged from one positive case (Feng et al., 2021; Medema et al., 2020; Wilder et al., 2021) to 26 positive cases in a 10,000-person community (Baldovin et al., 2021). A computational analysis of several WBE studies further listed the detection sensitivity at around 88 positive cases per 10,000 persons assuming fully homogenized wastewater and the absence of additional storm water or flow inputs to a sewer that would dilute the wastewater matrix (Hart and Halden, 2020). However, these studies report their estimated detection sensitivity based on information obtained from a non-enclosed community, and not all persons in the community were being swab-tested. In contrast, an earlier study showed that by monitoring wastewater in a confined hospital environment, in which the positive number of cases was known, at least 253 SARS-CoV-2-positive cases in a 10,000-person community were required to yield a positive occurrence by WBE (Hong et al., 2021). Another independent study adopted a similar approach in which the authors collected wastewater from a manhole connected to a nursing home, and making use of positive cases provided by the nursing home allowed the authors to place their WBE detection sensitivity at ~167 cases per 10,000 persons (Spurbeck et al., 2021). Lastly, the wide range in the number of positive cases denoted by different WBE studies can also be due to varying intra-person shedding rates of SARS-CoV-2 (Corchis-Scott et al., 2021; Schmitz et al., 2021).
3.2. Viral titer load in wastewater during a period with new incoming students, limited international travel, and mass vaccination campaigns
By 5 July 2020, the lockdown imposed on the KAUST community was lifted, and new students (~150), mainly comprised of those who were already physically residing inside Saudi Arabia, arrived at KAUST for the new semester. This new influx of residents was also shown by tracking the wastewater volume that was generated, captured, and entered into the KAUST WWTP during this period (Fig. S3). As the volume of wastewater increased during this period and the compound achieved 100 % capture of its sewage, this increase in volume indicated either an increase in resident population or activities after the major Eid Al Adha holiday (30 Jul till 6 Aug 2020).
Coincidentally, the first positive detection of both SARS-CoV-2 genes in wastewater occurred on 12 August 2020 and subsequently on 20 and 27 Sep 2020. The log10 normalized abundance of SARS-CoV-2 genes on these dates ranged from 3.5 to 3.6 copies/L (Fig. 2A). The log10 WC ratios were 2.3, 2.4, and 2.6 (Fig. 2B). Based on the high WC ratio measured during this period, we conclude that an outbreak was ongoing and that the existing testing capacity did not keep pace with the number of infected cases circulating within the community.
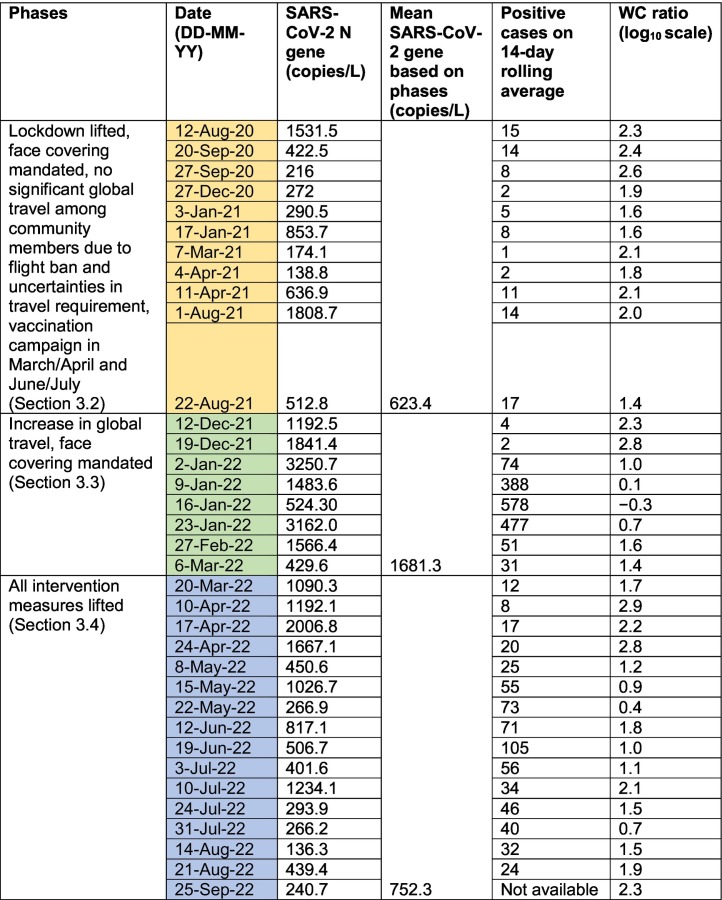
By 15 Sep 2020, the ban on international flights was partially lifted, and there was a gradual decrease in wastewater volumes entering the KAUST WWTP from then until late December 2020 (Fig. S3), suggesting a decrease in the resident population within the compound during that period and with positive detection of SARS-CoV-2 RNA in the wastewater on 27 December 2020 (log10 normalized abundance and WC ratio of 2.2 copies/L and 1.9, respectively) (Fig. 2A). Subsequently, as the wastewater volumes increased from early Jan until early Nov 2021, there were seven instances of positive detection of SARS-CoV-2 RNA on 3 and 17 Jan, 7 Mar, 4 and 11 April, and 1 and 22 August 2021 (Table 1 ). SARS-CoV-2 genes were detected during this period at a log10 normalized abundance that ranged from 2.1 to 3.2 copies/L (Fig. 2A). The log10 WC ratio on 3 and 17 Jan 2021 remained stable at 1.6. However, the log10 WC ratio then increased to an average of 2.0 between March and mid-April 2021 before decreasing back to 1.4 in August 2021 (Fig. 2B).
Table 1.
Summary of the dates on which SARS-CoV-2 RNA was detected in wastewater. The dates were categorized into three phases that saw an increasing relaxation of intervention measures and increasing global travel among community members.
The seven instances of positive detection did not occur immediately after the two major holidays, namely Eid Al Fitr on 12–20 May 2021 and Eid Al Adha on 18–22 Jul 2021. Instead, the comparatively high WC ratio during March and April 2021 coincided with the first round of mass vaccination (Fig. 1). The KAUST community achieved a vaccination rate (with at least one dose) of 77.6 % by 9 May 2021 and 93.1 % by 16 June 2021. With the second mass vaccination campaign, ~94 % of the KAUST community aged 12 and older and 100 % of all service providers were fully vaccinated (i.e., with both doses administered) as of 15 August 2021. Despite the high vaccination rates among members residing in the compound, the increase in the WC ratio suggests the occurrence of breakthrough infections among the community during this period when the strict lockdown was lifted.
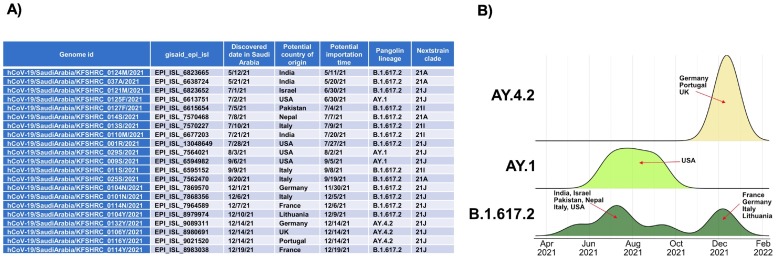
We wanted to assess if global travel was a contributing factor to the breakthrough infections since it accounted for the rapid dissemination of SARS-CoV-2 during the early period of the COVID-19 pandemic (Chinazzi et al., 2020). We made use of Nanopore sequencing data that was performed retrospectively. It was known that the period coincides with a change in the circulating SARS-CoV-2 strain to the Delta variant from late 2020 until November 2021 in Saudi Arabia (ArabNews, 2021a). By sequencing and aligning the sequences obtained within KAUST wastewaters to the SARS-CoV-2 Delta variants in Saudi Arabia, we can determine whether importation events occurred. We collated all available Delta variant genomes from Saudi Arabia deposited on GISAID to access the phylodynamic information (Fig. 3A). However, our Nanopore sequencing datasets did not find any traces of Delta variants in wastewater (Table S4), which was instead predominated by the original SARS-CoV-2 strain (hCoV19/Wuhan/WIV04/2019|EPI_ISL_402124|2019-12-30|China). Further importation event estimation did indicate the transition of Delta variants to Saudi Arabia but not within the KAUST community through global travel. The estimated importation duration started from April 2021 until February 2022, with different geographical locations contributing to the Delta variants within the population of Saudi Arabia (Fig. 3B).
Fig. 3.
Distribution of importation dates for Delta variants found in Saudi Arabia based on pangolin lineages. (A) Importation date for Delta variants into Saudi Arabia were estimated based on genomes deposited into GISAID. (B) Possible original countries of these importation events were estimated and labelled accordingly. The peak height of the probability curve represents magnitude of probability.
3.3. Viral titer load in wastewater during the Omicron surge
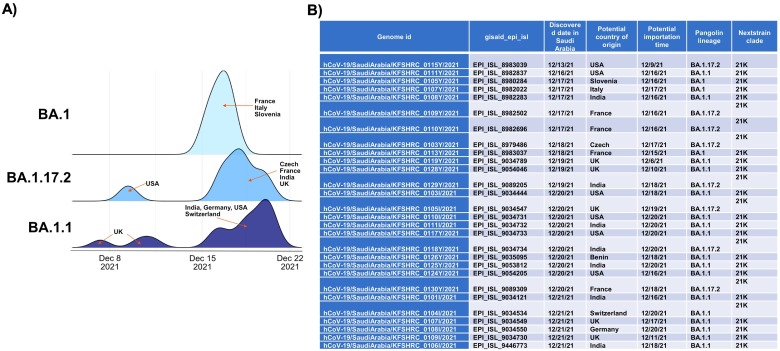
By late November 2021 until early Jan 2022, wastewater volume collected by the KAUST WWTP decreased (Fig. S3), indicating that a sizable portion of the community had left the compound during the winter semester break. SARS-CoV-2 genes were detected on 12 and 19 Dec 2021 at a log10 normalized abundance of 3 and 3.1 copies/L, respectively (Fig. 2A). The log10 WC ratio was 2.3 and 2.8, respectively (Fig. 2B), indicating a possible surge of infection within the community during this period. Subsequently, as more community members returned in January 2022, as indicated by the increase in wastewater volume from then until June 2022 (Fig. S3), SARS-CoV-2 genes were detected in most of the weekly samples collected in late December 2021 and in the month of January 2022. Specifically, in the month of January 2022, the number of 14-day positive swab tests performed during this period spiked to an average of 379 cases (Fig. 2A), which was at least 45 times higher than the past number of positive cases. This period of frequent detection of SARS-CoV-2 genes in the wastewater coincided with the Omicron surge that happened globally. Nanopore sequencing detected that the Omicron variant was present in two wastewater samples collected and sequenced during this surge (Table S4). The Omicron variant was first observed in our wastewater samples on 19 December 2021, which was shortly after the official Saudi announcement of the first case of Omicron (1 December 2021) (ArabNews, 2021b). It was also the period when we noted a high importation probability of Omicron from different geographical points throughout the month of December 2021 (Fig. 4 ). These results reflected a different situation compared to that described in Section 3.2, as the enclosed compound was affected by Omicron importation during the global surge that may have arisen due to the sizable number of community members traveling during this period.
Fig. 4.
Distribution of importation dates for Omicron variants found in Saudi Arabia based on pangolin lineages. (A) Possible original countries of these importation events were estimated and labelled accordingly. The peak height of the probability curve represents magnitude of probability. (B) Importation date for Omicron variants into Saudi Arabia were estimated based on genomes deposited into GISAID.
3.4. Viral titer load in wastewater after intervention measures were gradually removed
After 6 March 2022, KAUST relaxed several of its COVID-19 intervention measures by no longer requiring face coverings in outdoor environments and removing the need for physical distancing (Fig. 1). This decision was based on the decrease in the number of positive swab cases detected within the community (from a peak of 578 cases in January 2022 to a peak of 74 positive cases in the last week of February 2022) (Fig. 2A). However, positive detection of SARS-CoV-2 in wastewater was made in the samples collected on 6 and 20 March 2022, at a log10 normalized abundance of 2.8 copies/L (Fig. 2A) and at a log10 WC ratio of 1.4 and 1.7, respectively (Fig. 2B).
After 4 April 2022, all COVID-19 intervention measures were removed. That is, face coverings were no longer required outdoors or indoors except in the pre-university schools and medical facilities where the majority of occupants are unvaccinated young individuals or potentially immunocompromised. Subsequently, SARS-CoV-2 RNA was consistently detected in weekly wastewater samples in April and thereafter in at least two of the four weekly wastewater samples collected from May until August 2022 (Table 1). The log10 normalized abundance of SARS-CoV-2 in April 2022 ranged from 3.4 to 4.1 copies/L (Fig. 2A), with a log10 WC ratio from 2.2 to 2.9, which was higher than when intervention measures were actively enforced (Fig. 2B).
The detection of SARS-CoV-2 RNA in wastewater also coincided with the Eid holidays (Eid Al Fitr on 1–5 May 2022 and Eid Al Adha on 10–14 July 2022) when KAUST community members travel domestically and/or internationally. Nanopore sequencing further detected Omicron variants in 10 out of the 15 samples sequenced after intervention measures were no longer enforced (Table S5), a higher frequency of detection than before. Compared to other VoC, there are eight amino acid mutations found specifically in the Omicron variant (Table S5). These include the spike protein T547K (C23202A) and Q954H (A24424T), which improved the stability of the spike protein, subsequently increasing the persistence of the Omicron variant in the environment and creating a higher risk of environmental transmission (Cui et al., 2022; Pastorio et al., 2022). Amino acid mutations were also noted in the nucleocapsid N protein, for example, P13L (C28311T), which was reported to help the virus escape cellular immunity by T-cells (Jung et al., 2022). Both T547K and P13L were included in the diagnostic assay to distinguish Omicron from other VoCs (Bekliz et al., 2022; Ippoliti et al., 2022; Takemae et al., 2022). Other mutations of Omicrons were also noted; however, their functional relevance remains to be determined (Jung et al., 2022).
In summary, there was an increase in the frequency of positive detection of SARS-CoV-2 RNA in the wastewater once all intervention measures were lifted and community members resumed global travel. The more frequent detection of SARS-CoV-2 RNA in wastewater suggests a possible increased transmission of SARS-CoV-2 during this period (Table 1). In addition, there was a significant difference in the SARS-CoV-2 N gene copy number among the three periods discussed in 3.2, 3.3, 3.4 (One-way ANOVA, p = 0.006), with the mean viral load in this period specified in Section 3.4 significantly higher than the period specified in Section 3.2. However, the mean viral load was the highest in the period specified in Section 3.3 (Table 1), which coincided with the exceedingly high number of clinically diagnosed cases during this period (Fig. 2A).
3.5. Comparison of findings against that of earlier literature
Earlier reports of WBE in universities are collated in Table S1. A majority of the earlier studies were conducted in the US, with the remaining studies performed in other geographical locations like Canada, Spain, and South Africa. In addition, the majority of these studies sampled sewage generated from student residential buildings, and hence WBE findings reflect only a subset of the student population and not that of the entire university community. This was the first study to perform WBE in a university located in Saudi Arabia (KAUST), and because of the unique enclosed structure of this university with its own wastewater treatment plant, WBE findings were able to capture the extent of SARS-CoV-2 dissemination within the entire community.
The N gene is targeted in most studies, and it was observed that the maximum detected SARS-CoV-2 concentration enumerated in this study was lower than the maximum SARS-CoV-2 concentration detected in earlier studies by a magnitude of 2 to 3-log10 (Table S1). A further comparison of the correlation coefficient between the gene abundance in wastewater and that of positive clinical cases also revealed that the correlation obtained in this study was 0.28, lower than that reported elsewhere (Table S1). Normalization of the SARS-CoV-2 concentration against PMMoV did not improve the correlation coefficient, consistent with previous observations (Fahrenfeld et al., 2022; Lu et al., 2022). The lower concentration and correlation may be due to the dilution of sewage with other sources of municipal waste prior to sampling at the wastewater treatment plant. This suggests challenges in sampling at wastewater treatment plants versus a targeted sampling approach at manholes. While the advantage of sampling at a wastewater treatment plant is that it captures the viral load shed by a larger population, the lower concentration of SARS-CoV-2 in the wastewater may also mean that it is more prone to false-negative detection that does not accurately reflect the extent of an outbreak. This reiterates the need to improve the detection sensitivity of WBE approaches, presumably by sampling sludge instead of influent wastewater and by improving viral concentration methods.
Studies have shown that the concentration of SARS-CoV-2 in sludge exhibited significantly higher average positive proportion and viral RNA levels than in wastewater samples (Mantilla-Calderon et al., 2022). However, an issue with sampling sludge to relate to epidemiology is that most wastewater treatment plants tend to operate with a sludge retention time ranging from 3 to 18 days to achieve good nitrogen removal (Metcalf and Eddy, 2003). Therefore, positive detection of SARS-CoV-2 in the sludge may not accurately pinpoint the beginning of an outbreak. Alternatively, improvements can be made in the methods to concentrate viruses from wastewater. For example, the use of a targeted viral RNA concentration method via affinity capture magnetic hydrogel particles aids in viral recovery from samples with low viral loads, in turn revealing early cryptic SARS-CoV-2 variants in wastewater (Karthikeyan et al., 2022).
3.6. Facilitating science-based implementations of intervention measures with minimal disruptions to daily life
Although earlier WBE studies performed at residential buildings demonstrated clearly how lockdown, clinical swab testing, quarantine, and contact tracing could effectively mitigate the extent of the outbreak, such practices may not continue to gain acceptance in the endemic phase of COVID-19 as they were during the early phase of COVID-19. With changing attitudes toward COVID-19, authorities would be challenged to continue implementing hardline intervention strategies. Instead, finding a minimally disruptive measure that can still effectively mitigate the extent of infection is crucial.
The long sampling duration (2.5 years) conducted in this study encompassed the early phase of the pandemic when strict intervention measures were put in place, and then a gradual relaxation of intervention measures as vaccines were rolled out. Hence, we were able to assess the combinatory effect of different factors and determine which factor(s) possibly outweigh the others. For example, while earlier studies showed that public holidays (e.g., Labor Day and Halloween) resulted in a subsequent surge of viral load in wastewater and a spike in positive clinical diagnoses (Betancourt et al., 2021; de Llanos et al., 2022; Reeves et al., 2021), our studies showed that major public holidays did not result in a surge of viral load in wastewater when strict curfews were put in place. Neither did major public holidays result in a surge of viral load when face covering was adopted, and global travel remained limited among community members. This is despite the cease of curfew and a global surge of the Delta variant, which is more transmissible than the original SARS-CoV-2 strain (Li et al., 2022). Subsequently, when face covering remains in place and global travel was taken up extensively by the community, the Omicron variant produced a surge of positive cases within the KAUST university compound. These observations suggest that global travel contributes the most to dissemination within the compound when a highly transmissible variant is circulating.
As Saudi Arabia adopts the strategy of living with COVID-19, imposing lockdown, contact tracing, and quarantine can impose a tremendous burden on the mental and societal health of the community. Instead, WBE can be used to monitor the temporal dynamics of SARS-CoV-2 within the wastewater or sludge samples generated by a defined community. WBE within the KAUST community is ongoing and continually monitored for instances where the SARS-CoV-2 concentration in waste streams is increasing. If such instances occur, public health authorities will be informed, and advisories to community members to wear face coverings can be issued. In addition, Nanopore sequencing of the extracted RNA from waste streams can be used to determine the dominant SARS-CoV-2 strain. Using bioinformatic-driven importation estimates to analyze the sequencing data, we can inform public health authorities of the likely geographical source of the circulating strains in the community. A suggestion can then be made to issue advisories regarding best practices to community members who are planning to travel to those geographical locations that are contributing to the circulating strains. Such best practices can include, for example, keeping vaccination profiles up-to-date, wearing face coverings during travel, and practicing voluntary self-observation and/or quarantine upon return from travel.
4. Conclusions
A long-term WBE performed on an enclosed community revealed that strict lockdown during the early stages of the COVID-19 pandemic effectively controlled the widespread dissemination of SARS-CoV-2 in the compound. This is shown by the lack of positive detection of SARS-CoV-2 RNA in wastewater and the low number of 14-day positive swab cases. After the lockdown was lifted and international travel resumed, wearing a face covering in the indoor environment was necessary to control the dissemination of SARS-CoV-2 within the compound. However, significant global travel by community members and surges of SARS-CoV-2 variants, particularly the Omicron variant, are contributory factors that can lead to an uptick of positive infections within the compound. These insights denoted by WBE performed in the enclosed university can be particularly useful to pinpoint minimally disruptive intervention measures that can be adopted to mitigate a COVID-19 outbreak as it enters the endemic phase in most countries. By tracking the temporal dynamics and noting the presence of VoCs in wastewater, appropriate responses can be made (e.g., issuing advisories on when to wear a face covering and what are the best practices to adopt before and after travel to places that are importing VoCs to the local community). These concerted efforts can aid in keeping SARS-CoV-2 infection at a manageable level while imposing minimal disruption to daily life.
CRediT authorship contribution statement
TW, CW, YM, DMC: Experimentation, Formal analysis, Data curation. TW, CW: Writing and illustration – review & editing, ET: Conceptualization, Methodology, Writing - review & editing, Project administration. PYH: Supervision, Conceptualization, Methodology, Writing - original draft, review & editing, Project administration.
Funding
This work was partially funded by KAUST baseline grant BAS/1/1033-01-01 awarded to PYH and by the KAUST Smart Health Initiative for the KAUST Rapid Response Research Team.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgement
The authors would like to thank KAUST Facilities and Maintenance Utilities team for providing access to wastewater samples and for providing the wastewater volume data. The authors would also like to thank Professor Arnab Pain and his team for providing the RNA sample that serves as positive control, as well as members of the KAUST HSE (Mr. Rodion V. Gorchakov) and Community Life for collating data from swab testing and serological testing. Assistance from Dr. Andri Rachmadi in the early stages of the KAUST WBE and Ms. Elaf Alahdal in the latter stages is greatly appreciated.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2023.162466.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson-Coughlin B.L., Shearer A.E.H., Omar A.N., Litt P.K., Bernberg E., Murphy M., Anderson A., Sauble L., Ames B., Damminger O., Ladman B.S., Dowling T.F., Wommack K.E., Kniel K.E. Coordination of SARS-CoV-2 wastewater and clinical testing of university students demonstrates the importance of sampling duration and collection time. Sci. Total Environ. 2022;830 doi: 10.1016/j.scitotenv.2022.154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ArabNews Saudi Arabia suspends all international passenger flights for a week. 2020. https://www.arabnews.com/node/1780261/saudi-arabia (Last accessed 22 Jan 2023)
- ArabNews Delta variant reported among Saudi Arabia's new virus case. 2021. https://www.arabnews.com/node/1911526/saudi-arabia (Last accessed 30 Jan 2023)
- ArabNews Saudi Arabia, UAE confirm first omicron COVID-19 cases. 2021. https://www.arabnews.com/node/1978791/saudi-arabia (Last accessed 30 Jan 2023)
- Baldovin T., Amoruso I., Fonzo M., Buja A., Baldo V., Cocchio S., Bertoncello C. SARS-CoV-2 RNA detection and persistence in wastewater samples: an experimental network for COVID-19 environmental surveillance in Padua, Veneto Region (NE Italy) Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekliz M., Adea K., Puhach O., Perez-Rodriguez F., Marques Melancia S., Baggio S., Corvaglia A.R., Jacquerioz F., Alvarez C., Essaidi-Laziosi M., Escadafal C., Kaiser L., Eckerle I. Analytical sensitivity of eight different SARS-CoV-2 antigen-detecting rapid tests for omicron-BA.1 variant. Microbiol. Spectr. 2022;10(4) doi: 10.1128/spectrum.00853-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull R.A., Adikari T.N., Ferguson J.M., Hammond J.M., Stevanovski I., Beukers A.G., Naing Z., Yeang M., Verich A., Gamaarachchi H., Kim K.W., Luciani F., Stelzer-Braid S., Eden J.S., Rawlinson W.D., van Hal S.J., Deveson I.W. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat. Commun. 2020;11(1):6272. doi: 10.1038/s41467-020-20075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- CDC CDC 2019-Novel Coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. 2021. https://www.fda.gov/media/134922/download (Last accessed 4 August 2021) [DOI] [PMC free article] [PubMed]
- Chinazzi M., Davis J.T., Ajelli M., Gioannini C., Litvinova M., Merler S., Mu K., Rossi L., Sun K., Viboud C., Xiong X., Pastore y Piontti A., Yu H., Halloran M.E., Longini I.M., Vespignani A. The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak. Science. 2020;368(6489):395. doi: 10.1126/science.aba9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI . 2012. Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures; Approved Guideline—Second Edition, CLSI Document EP17-A2. [Google Scholar]
- Corchis-Scott R., Geng Q., Seth R., Ray R., Beg M., Biswas N., Charron L., Drouillard K.D., D'Souza R., Heath D.D., Houser C., Lawal F., McGinlay J., Menard S.L., Porter L.A., Rawlings D., Scholl M.L., Siu K.W.M., Tong Y., Weisener C.G., Wilhelm S.W., McKay R.M.L. Averting an outbreak of SARS-CoV-2 in a university residence hall through wastewater surveillance. Microbiol. Spectr. 2021;9(2) doi: 10.1128/Spectrum.00792-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Liu P., Wang N., Wang L., Fan K., Zhu Q., Wang K., Chen R., Feng R., Jia Z., Yang M., Xu G., Zhu B., Fu W., Chu T., Feng L., Wang Y., Pei X., Yang P., Xie X.S., Cao L., Cao Y., Wang X. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 omicron. Cell. 2022;185(5):860–871 e813. doi: 10.1016/j.cell.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10(2):1–4. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coster W., D'Hert S., Schultz D.T., Cruts M., Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 2018;34(15):2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Llanos R., Cejudo-Marín R., Barneo M., Pérez-Cataluña A., Barberá-Riera M., Rebagliato M., Bellido-Blasco J., Sánchez G., Hernández F., Bijlsma L. Monitoring the evolution of SARS-CoV-2 on a Spanish university campus through wastewater analysis: a pilot project for the reopening strategy. Sci. Total Environ. 2022;845 doi: 10.1016/j.scitotenv.2022.157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenfeld N.L., Morales Medina W.R., D'Elia S., Modica M., Ruiz A., McLane M. Comparison of residential dormitory COVID-19 monitoring via weekly saliva testing and sewage monitoring. Sci. Total Environ. 2022;814 doi: 10.1016/j.scitotenv.2021.151947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Roguet A., McClary-Gutierrez J.S., Newton R.J., Kloczko N., Meiman J.G., McLellan S.L. Evaluation of sampling, analysis, and normalization methods for SARS-CoV-2 concentrations in wastewater to assess COVID-19 burdens in Wisconsin communities. ACS ES&T Water. 2021;1(8):1955–1965. [Google Scholar]
- Ferrari M. Saudi Arabia's new COVID-19 vaccination rules come into effect. 2021. https://english.alarabiya.net/coronavirus/2021/10/10/Saudi-Arabia-s-new-COVID-19-vaccination-rules-come-into-effect Last accessed 22 Jan 2023.
- Ferrari M. Saudi Arabia drops COVID-19 measures including indoor masks, vaccine requirement. 2022. https://english.alarabiya.net/News/gulf/2022/06/13/Saudi-Arabia-drops-more-COVID-19-measures-including-indoor-mask-mandate Last accessed 22 Jan 2023.
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. 2020;117(17):9241. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C., Lambirth K., Mittal N., Juel M.A.I., Barua V.B., Roppolo Brazell L., Hinton K., Lontai J., Stark N., Young I., Quach C., Russ M., Kauer J., Nicolosi B., Chen D., Akella S., Tang W., Schlueter J., Munir M. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021;782 doi: 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Katayama H., Ohgaki S. Detection of noroviruses in tap water in Japan by means of a new method for concentrating enteric viruses in large volumes of freshwater. Appl. Environ. Microbiol. 2004;70(4):2154–2160. doi: 10.1128/AEM.70.4.2154-2160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Kishida N., Konno Y., Katayama H., Asami M., Akiba M. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Environ. Microbiol. 2013;79(23):7413–7418. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P.-Y., Rachmadi A.T., Mantilla-Calderon D., Alkahtani M., Bashawri Y.M., Al Qarni H., O'Reilly K.M., Zhou J. Estimating the minimum number of SARS-CoV-2 infected cases needed to detect viral RNA in wastewater: to what extent of the outbreak can surveillance of wastewater tell us? Environ. Res. 2021;195 doi: 10.1016/j.envres.2021.110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins L., Persse D., Caton K., Ensor K., Schneider R., McCall C., Stadler L.B. Citywide wastewater SARS-CoV-2 levels strongly correlated with multiple disease surveillance indicators and outcomes over three COVID-19 waves. Sci. Total Environ. 2022;158967 doi: 10.1016/j.scitotenv.2022.158967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippoliti C., De Maio F., Santarelli G., Marchetti S., Vella A., Santangelo R., Sanguinetti M., Posteraro B. Rapid detection of the omicron (B.1.1.529) SARS-CoV-2 variant using a COVID-19 diagnostic PCR assay. Microbiol. Spectr. 2022;10(4) doi: 10.1128/spectrum.00990-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N., Hamilton D., Mital S., Ilias A., Brinkmann M., McPhedran K. Long-term passive wastewater surveillance of SARS-CoV-2 for seven university dormitories in comparison to municipal surveillance. Sci. Total Environ. 2022;852 doi: 10.1016/j.scitotenv.2022.158421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Kmiec D., Koepke L., Zech F., Jacob T., Sparrer K.M.J., Kirchhoff F. Omicron: what makes the latest SARS-CoV-2 variant of concern so concerning? J. Virol. 2022;96(6) doi: 10.1128/jvi.02077-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan S., Levy J.I., De Hoff P., Humphrey G., Birmingham A., Jepsen K., Farmer S., Tubb H.M., Valles T., Tribelhorn C.E., Tsai R., Aigner S., Sathe S., Moshiri N., Henson B., Mark A.M., Hakim A., Baer N.A., Barber T., Belda-Ferre P., Chacón M., Cheung W., Cresini E.S., Eisner E.R., Lastrella A.L., Lawrence E.S., Marotz C.A., Ngo T.T., Ostrander T., Plascencia A., Salido R.A., Seaver P., Smoot E.W., McDonald D., Neuhard R.M., Scioscia A.L., Satterlund A.M., Simmons E.H., Abelman D.B., Brenner D., Bruner J.C., Buckley A., Ellison M., Gattas J., Gonias S.L., Hale M., Hawkins F., Ikeda L., Jhaveri H., Johnson T., Kellen V., Kremer B., Matthews G., McLawhon R.W., Ouillet P., Park D., Pradenas A., Reed S., Riggs L., Sanders A., Sollenberger B., Song A., White B., Winbush T., Aceves C.M., Anderson C., Gangavarapu K., Hufbauer E., Kurzban E., Lee J., Matteson N.L., Parker E., Perkins S.A., Ramesh K.S., Robles-Sikisaka R., Schwab M.A., Spencer E., Wohl S., Nicholson L., McHardy I.H., Dimmock D.P., Hobbs C.A., Bakhtar O., Harding A., Mendoza A., Bolze A., Becker D., Cirulli E.T., Isaksson M., Schiabor Barrett K.M., Washington N.L., Malone J.D., Schafer A.M., Gurfield N., Stous S., Fielding-Miller R., Garfein R.S., Gaines T., Anderson C., Martin N.K., Schooley R., Austin B., MacCannell D.R., Kingsmore S.F., Lee W., Shah S., McDonald E., Yu A.T., Zeller M., Fisch K.M., Longhurst C., Maysent P., Pride D., Khosla P.K., Laurent L.C., Yeo G.W., Andersen K.G., Knight R. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature. 2022;609(7925):101–108. doi: 10.1038/s41586-022-05049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna A., Larson D.E., Srivatsan S.N., Mosior M., Abbott T.E., Kiwala S., Ley T.J., Duncavage E.J., Walter M.J., Walker J.R., Griffith O.L., Griffith M., Miller C.A. Bam-readcount -- Rapid Generation of Basepair-resolution Sequence Metrics. 7 (69) 2021. p. 3722. ArXiv. [Google Scholar]
- Khoja S. Mandatory travel and vaccination policies in Saudi Arabia. 2021. https://www.clydeco.com/en/insights/2021/10/mandatory-travel-and-vaccination-policies-in-saudi Last accessed 22 Jan 2023.
- Kitajima M., Sassi H.P., Torrey J.R. Pepper mild mottle virus as a water quality indicator. npj CleanWater. 2018;1(1):19. [Google Scholar]
- Korfmacher K.S., Harris-Lovett S., Nelson K.L. Campus collaborations as a model for transforming SARS-CoV-2 wastewater surveillance research into public health action. Environ.Sci.Technol. 2021;55(19):12770–12772. doi: 10.1021/acs.est.1c03351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotay S.M., Tanabe K.O., Colosi L.M., Poulter M.D., Barry K.E., Holstege C.P., Mathers A.J., Porter M.D. Building-level wastewater surveillance for SARS-CoV-2 in occupied university dormitories as an outbreak forecasting tool: one year case study. ACS ES&T Water. 2022;2(11):2094–2104. doi: 10.1021/acsestwater.2c00057. [DOI] [PubMed] [Google Scholar]
- Krimly R. Saudi Arabia detects first case of COVID-19 omicron variant in Kingdom. 2021. https://english.alarabiya.net/coronavirus/2021/12/01/Saudi-Arabia-detects-first-case-of-COVID-19-Omicron-variant-in-Kingdom Last accessed 22 Jan 2023.
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavania M., Joshi M.S., Ranshing S.S., Potdar V.A., Shinde M., Chavan N., Jadhav S.M., Sarkale P., Mohandas S., Sawant P.M., Tikute S., Padbidri V., Patwardhan S., Kate R. Prolonged shedding of SARS-CoV-2 in feces of COVID-19 positive patients: trends in genomic variation in first and second wave. Front. Med. (Lausanne) 2022;9 doi: 10.3389/fmed.2022.835168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L., Valmond L., Thomas J., Kim A., Austin P., Foster M., Matthews J., Kim P., Newman J. Wastewater surveillance in smaller college communities may aid future public health initiatives. PLOS ONE. 2022;17(9) doi: 10.1371/journal.pone.0270385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Deng A., Li K., Hu Y., Li Z., Shi Y., Xiong Q., Liu Z., Guo Q., Zou L., Zhang H., Zhang M., Ouyang F., Su J., Su W., Xu J., Lin H., Sun J., Peng J., Jiang H., Zhou P., Hu T., Luo M., Zhang Y., Zheng H., Xiao J., Liu T., Tan M., Che R., Zeng H., Zheng Z., Huang Y., Yu J., Yi L., Wu J., Chen J., Zhong H., Deng X., Kang M., Pybus O.G., Hall M., Lythgoe K.A., Li Y., Yuan J., He J., Lu J. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. Nat. Commun. 2022;13(1):460. doi: 10.1038/s41467-022-28089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu E., Ai Y., Davis A., Straathof J., Halloran K., Hull N., Winston R., Weir M.H., Soller J., Bohrerova Z., Oglesbee M., Lee J. Wastewater surveillance of SARS-CoV-2 in dormitories as a part of comprehensive university campus COVID-19 monitoring. Environ. Res. 2022;212 doi: 10.1016/j.envres.2022.113580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangwana N., Archer E., Muller C.J.F., Preiser W., Wolfaardt G., Kasprzyk-Hordern B., Carstens A., Brocker L., Webster C., McCarthy D., Street R., Mathee A., Louw J., Mdhluli M., Johnson R. Sewage surveillance of SARS-CoV-2 at student campus residences in the Western Cape, South Africa. Sci. Total Environ. 2022;851 doi: 10.1016/j.scitotenv.2022.158028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantilla-Calderon D., Huang K., Li A., Chibwe K., Yu X., Ye Y., Liu L., Ling F. Emerging investigator series: meta-analyses on SARS-CoV-2 viral RNA levels in wastewater and their correlations to epidemiological indicators. Environ.Sci.: Water Res.Technol. 2022;8(7):1391–1407. [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ.Sci.Technol.Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Metcalf &. Eddy Inc. In: Wastewater Engineering: Treatment and Reuse. Fourth edition. Tchobanoglous George, Burton Franklin L., Stensel H. David., editors. McGraw-Hill; Boston: 2003. [2003] ©2003. [Google Scholar]
- Mourier T., Shuaib M., Hala S., Mfarrej S., Alofi F., Naeem R., Alsomali A., Jorgensen D., Subudhi A.K., Ben Rached F., Guan Q., Salunke R.P., Ooi A., Esau L., Douvropoulou O., Nugmanova R., Perumal S., Zhang H., Rajan I., Al-Omari A., Salih S., Shamsan A., Al Mutair A., Taha J., Alahmadi A., Khotani N., Alhamss A., Mahmoud A., Alquthami K., Dageeg A., Khogeer A., Hashem A.M., Moraga P., Volz E., Almontashiri N., Pain A. SARS-CoV-2 genomes from Saudi Arabia implicate nucleocapsid mutations in host response and increased viral load. Nat. Commun. 2022;13(1):601. doi: 10.1038/s41467-022-28287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar I. Saudi Arabia extends all flight suspensions and working from home. 2020. https://english.alarabiya.net/News/gulf/2020/03/29/Coronavirus-Saudi-Arabia-extends-all-flight-suspensions-workplace-attendance Last accessed 22 Jan 2023.
- Naar I. Saudi Arabia extends suspension of international flights by another week. 2020. https://english.alarabiya.net/coronavirus/2020/12/28/Coronavirus-Saudi-Arabia-extends-suspension-of-international-flights-by-another-week Last accessed 22 Jan 2023.
- National Saudi Arabia's new Covid-19 rules and what they mean. 2021. https://www.thenationalnews.com/gulf-news/saudi-arabia/2021/12/30/saudi-arabias-new-covid-19-rules-and-what-they-mean/ (Last accessed 22 Jan 2023)
- Nihal M. Saudi Arabia makes Covid-19 vaccine booster mandatory for travel abroad. 2022. https://www.thenationalnews.com/gulf-news/2022/02/03/saudi-arabia-makes-vaccine-booster-mandatory-for-travel-abroad/ Last accessed 22 Jan 2023.
- Paradis E., Claude J., Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pastorio C., Zech F., Noettger S., Jung C., Jacob T., Sanderson T., Sparrer K.M.J., Kirchhoff F. Determinants of spike infectivity, processing, and neutralization in SARS-CoV-2 omicron subvariants BA.1 and BA.2. Cell Host Microbe. 2022;30(9):1255–1268 e1255. doi: 10.1016/j.chom.2022.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen R.M., Tornby D.S., Bang L.L., Madsen L.W., Skov M.N., Sydenham T.V., Steinke K., Jensen T.G., Johansen I.S., Andersen T.E. Rectally shed SARS-CoV-2 in COVID-19 inpatients is consistently lower than respiratory shedding and lacks infectivity. Clin. Microbiol. Infect. 2022;28(2):304 e301–304 e303. doi: 10.1016/j.cmi.2021.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninger J.M., Grant M.B., Sung J.J.Y. The role of angiotensin converting enzyme 2 in modulating gut microbiota, intestinal inflammation, and coronavirus infection. Gastroenterology. 2021;160(1):39–46. doi: 10.1053/j.gastro.2020.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey A.L., Buschang K., O’Connor A., Love D., Wormington A.M., Messcher R.L., Loeb J.C., Robinson S.E., Ponder H., Waldo S., Williams R., Shapiro J., McAlister E.B., Lauzardo M., Lednicky J.A., Maurelli A.T., Sabo-Attwood T., Bisesi J.H., Jr. Retrospective analysis of wastewater-based epidemiology of SARS-CoV-2 in residences on a large college campus: relationships between wastewater outcomes and COVID-19 cases across two semesters with different COVID-19 mitigation policies. ACS ES&T Water. 2023;3(1):16–29. doi: 10.1021/acsestwater.2c00275. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves K., Liebig J., Feula A., Saldi T., Lasda E., Johnson W., Lilienfeld J., Maggi J., Pulley K., Wilkerson P.J., Real B., Zak G., Davis J., Fink M., Gonzales P., Hager C., Ozeroff C., Tat K., Alkire M., Butler C., Coe E., Darby J., Freeman N., Heuer H., Jones J.R., Karr M., Key S., Maxwell K., Nelson L., Saldana E., Shea R., Salveson L., Tomlinson K., Vargas-Barriga J., Vigil B., Brisson G., Parker R., Leinwand L.A., Bjorkman K., Mansfeldt C. High-resolution within-sewer SARS-CoV-2 surveillance facilitates informed intervention. Water Res. 2021;204 doi: 10.1016/j.watres.2021.117613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuters Saudi Arabia to lift some international flight restrictions on September 15. 2020. https://www.reuters.com/article/uk-health-coronavirus-saudi-travel-idUSKBN2640PG (Last accessed 22 Jan 2023)
- Schliep K.P. Phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27(4):592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz B.W., Innes G.K., Prasek S.M., Betancourt W.Q., Stark E.R., Foster A.R., Abraham A.G., Gerba C.P., Pepper I.L. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L.C., Aubee A., Babahaji L., Vigil K., Tims S., Aw T.G. Targeted wastewater surveillance of SARS-CoV-2 on a university campus for COVID-19 outbreak detection and mitigation. Environ. Res. 2021;200 doi: 10.1016/j.envres.2021.111374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;140621 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22(13):30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurbeck R.R., Minard-Smith A., Catlin L. Feasibility of neighborhood and building scale wastewater-based genomic epidemiology for pathogen surveillance. Sci. Total Environ. 2021;789 doi: 10.1016/j.scitotenv.2021.147829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemae N., Doan Y.H., Momose F., Saito T., Kageyama T. Development of new SNP genotyping assays to discriminate the omicron variant of SARS-CoV-2. Jpn. J. Infect. Dis. 2022;75(4):411–414. doi: 10.7883/yoken.JJID.2022.007. [DOI] [PubMed] [Google Scholar]
- Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44(W1):W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Frost S. Scalable relaxed clock phylogenetic dating. Virus Evol. 2017;3(2) [Google Scholar]
- Wallace M., Pappas G. Safe university: a guide for open academic institutions through the pandemic. Clin. Microbiol. Infect. 2022;28(5):634–636. doi: 10.1016/j.cmi.2022.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu P., Zhang H., Ibaraki M., VanTassell J., Geith K., Cavallo M., Kann R., Saber L., Kraft C.S., Lane M., Shartar S., Moe C. Early warning of a COVID-19 surge on a university campus based on wastewater surveillance for SARS-CoV-2 at residence halls. Sci. Total Environ. 2022;821 doi: 10.1016/j.scitotenv.2022.153291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; 2016. pp. 189–201. [Google Scholar]
- Wilder M.L., Middleton F., Larsen D.A., Du Q., Fenty A., Zeng T., Insaf T., Kilaru P., Collins M., Kmush B., Green H.C. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res.X. 2021;11 doi: 10.1016/j.wroa.2021.100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J., Driver E.M., Bowes D.A., Johnston B., Halden R.U. Comparison of high-frequency in-pipe SARS-CoV-2 wastewater-based surveillance to concurrent COVID-19 random clinical testing on a public U.S. university campus. Sci. Total Environ. 2022;820 doi: 10.1016/j.scitotenv.2021.152877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5(4) doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., Wu F., Bushman M., Zhang J., Imakaev M., Chai P.R., Duvallet C., Endo N., Erickson T.B., Armas F., Arnold B., Chen H., Chandra F., Ghaeli N., Gu X., Hanage W.P., Lee W.L., Matus M., McElroy K.A., Moniz K., Rhode S.F., Thompson J., Alm E.J. Metrics to relate COVID-19 wastewater data to clinical testing dynamics. Water Res. 2022;212 doi: 10.1016/j.watres.2022.118070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cen M., Hu M., Du L., Hu W., Kim J.J., Dai N. Prevalence and persistent shedding of fecal SARS-CoV-2 RNA in patients with COVID-19 infection: a systematic review and meta-analysis. Clin. Transl. Gastroenterol. 2021;12(4) doi: 10.14309/ctg.0000000000000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.