Abstract
The kinase ataxia telangiectasia mutated (ATM) plays a key role in the DNA damage response (DDR). It is thus essential to visualize spatiotemporal dynamics of ATM activity during DDR. Here, we designed a robust ATM activity reporter based on phosphorylation-inducible green fluorescent protein phase separation, dubbed ATM–SPARK (separation of phases-based activity reporter of kinase). Upon ATM activation, it undergoes phase separation via multivalent interactions, forming intensely bright droplets. The reporter visualizes spatiotemporal dynamics of endogenous ATM activity in living cells, and its signal is proportional to the amount of DNA damage. ATM-SPARK also enables high-throughput screening of biological and small-molecule regulators. We identified the protein phosphatase 4 that blocks ATM activity. We also identified BGT226 as a potent ATM inhibitor with a median inhibitory concentration of ~3.8 nanomolars. Furthermore, BGT226 sensitizes cancer cells to the radiomimetic drug neocarzinostatin, suggesting that BGT226 might be combined with radiotherapeutic treatment. ATM-SPARK achieves large dynamic range, bright fluorescence, and simple signal pattern.
A GFP phase separation-based ATM reporter visualizes spatiotemporal dynamics of endogenous ATM activity in living cells.
INTRODUCTION
The protein kinase ataxia telangiectasia mutated (ATM) is a key regulator of the DNA damage response (DDR) (1–3). It maintains genomic integrity in mammalian cells. The name of ATM comes from the fact that its mutations result in ataxia telangiectasia, a rare autosomal recessive disorder with phenotypes such as cerebellar degeneration (2). To understand key roles of ATM during DDR, it is essential to visualize the spatial and temporal dynamics of ATM activity in living cells. Genetically encoded reporters are ideal in enabling spatiotemporal imaging of cell signaling. However, previous reporters such as Förster resonance energy transfer (FRET)–based reporters suffer from poor signal due to small changes of the donor and acceptor fluorophores (4, 5). Furthermore, without tagging to chromosomal proteins such as histones, FRET-based reporters appear to have poor spatial resolution in imaging ATM activity in the subnuclear regions.
To address these limitations of FRET-based ATM reporters, we designed a robust ATM activity reporter by applying a different physical principle, phosphorylation-inducible green fluorescent protein (GFP) phase separation. The reporter is named ATM–SPARK (separation of phases-based Activity reporter of kinase), which contains GFP and an ATM-specific substrate peptide that has been used and characterized in the FRET-based ATM reporters. Upon phosphorylation by active ATM, the reporter undergoes phase separation, forming intensely bright green droplets. ATM-SPARK visualizes spatiotemporal dynamics of ATM activity in living cells, with no requirement of tagging with chromosomal proteins. Upon spatially localized DNA damage by laser irradiation, the punctate signal of ATM-SPARK is localized to the DNA damage site. Furthermore, ATM-SPARK droplets are colocalized with known DNA damage markers including p53-binding protein 1 (53BP1, in nonhomologous recombination–based DNA repair) and BReast CAncer gene 1 (BRCA1, in homologous recombination–based DNA repair). With ATM-SPARK, we were able to show that active ATM is associated with telomere after DNA damage induced by the radiomimetic drug neocarzinostatin (NCS).
Because of large dynamic range, bright fluorescence, and simple signal pattern, ATM-SPARK also enables high-throughput screening of its regulators in living cells, including biological regulators and small molecules. Loss or inactive mutations of ATM have been reported to confer radiosensitivity for tumors, in addition to cancer predisposition (6–9). ATM inhibition has thus been proposed and examined in a combination therapy with radiotherapeutic treatment (7, 10). Here, we demonstrate that ATM-SPARK not only allows us to identify biological regulators such as phosphatases but also enables us to identify small-molecule inhibitors that may be used as radiosensitizers in anticancer therapy.
RESULTS AND DISCUSSION
An ATM reporter is designed on the basis of phosphorylation-inducible GFP phase separation
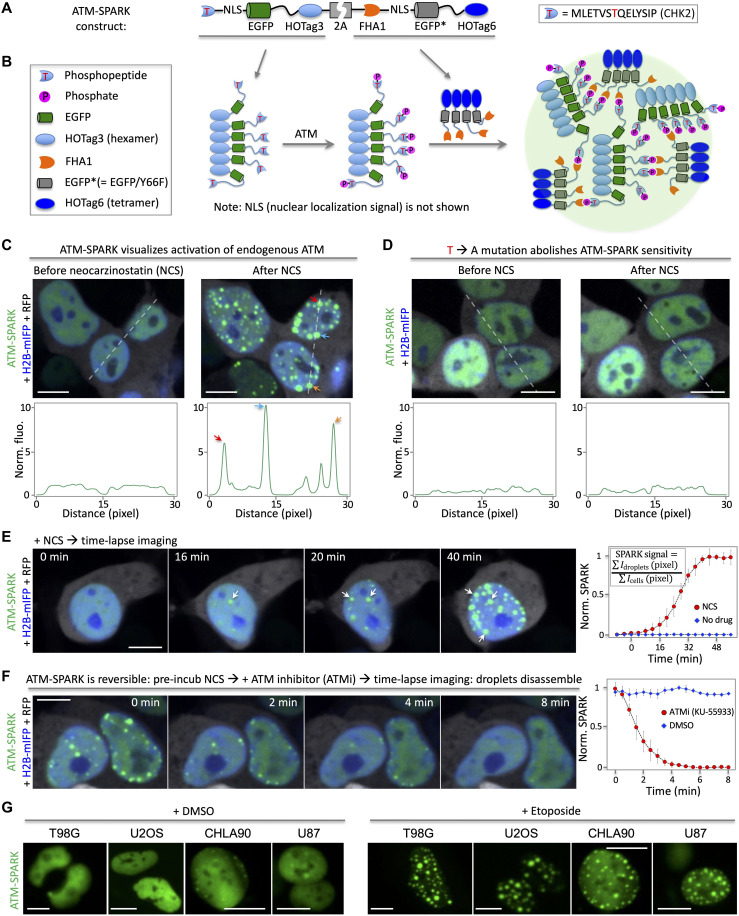
To develop an ATM reporter for imaging endogenous ATM activity in living cells, we engineered GFP so that upon phosphorylation by ATM, it undergoes phase separation forming intensely bright droplets. In particular, we designed a multivalent interaction system to drive ATM activity–dependent phase separation of GFP. First, we fused a Checkpoint kinase 2 (Chk2) kinase–derived substrate peptide to EGFP (enhanced GFP) (11, 12). We chose the T68-containing phosphopeptide derived from Chk2 because it has been used in previous FRET-based ATM activity reporters, and its specificity to ATM activity has been well characterized (5, 13). Next, we fused a phosphopeptide-binding domain Forkhead-associated protein 1 (FHA1) to a nonfluorescent GFP mutant (EGFP* containing Y66F mutation). We used the nonfluorescent EGFP* because it ensures that the reporter’s signal only comes from the EGFP fused to the phosphopeptide (see below about calculation of the SPARK signal). We did not remove EGFP because our previous work on the SPARK-based kinase sensors suggested that EGFP acts as a linker that improves phase separation and droplet formation of the reporter (14, 15). Third, to introduce multivalency, we fused a multivalent tag named homo-oligomeric tag (HOTag) to each of the above constructs (Fig. 1A). In particular, HOTag3, a hexameric tag, is fused to the phosphopeptide-EGFP. In addition, HOTag6, a tetrameric tag, is fused to the FHA1-EGFP*. Both HOTag3 and HOTag6 are de novo designed coiled coils (14, 15). To express the reporter in the nucleus, we also inserted a nuclear localization signal (NLS) to the reporter. Last, the two fusions were connected by a “self-cleaving” 2A sequence. Upon activation of ATM, the phosphopeptide is phosphorylated, which induces its interaction with FHA1 (Fig. 1B). This, together with HOTags, leads to multivalent interaction–driven phase separation of EGFP. Briefly, each hexameric phosphopeptide-NLS-EGFP-HOTag3 recruits six FHA1-NLS-EGFP*-HOTag6. Then, each tetrameric FHA1-NLS-EGFP*-HOTag6 recruits four phosphopeptide-NLS-EGFP-HOTag3, and so on. Eventually, the HOTag-introduced multivalency and the ATM activity–dependent phosphopeptide/FHA1 interactions lead to EGFP phase separation, forming highly fluorescent green droplets (Fig. 1B).
Fig. 1. Design, demonstration, and characterization of an ATM activity reporter based on phosphorylation-inducible GFP phase separation.
(A) Schematic showing constructs of the ATM reporter ATM-SPARK. (B) Schematic showing working principles. (C) Fluorescence images of ATM-SPARK–expressing HEK293 cells upon addition of a radiomimetic NCS. The histogram corresponds to the dashed line in the images. (D) Fluorescence images of the mutated reporter that cannot be phosphorylated by ATM. The phospho-threonine is mutated to alanine. (E) Time-lapse images of cells expressing ATM-SPARK upon addition of NCS. (F) Reversibility of ATM-SPARK. Cells expressing ATM-SPARK were preincubated with NCS, followed by addition of the ATM inhibitor KU-55933. (G) Application of ATM-SPARK in various cancer cells. The images were taken 1 hour after addition of the drugs. Data in (E) and (F) are means ± SE (n = 10 cells). RFP is shown in gray pseudo-color. Scale bars, 10 μm (C to G).
To demonstrate and characterize ATM-SPARK, we expressed the reporter in human embryonic kidney (HEK) 293 cells, which revealed punctate fluorescence in the nucleus upon DNA damage by NCS (Fig. 1C, fig. S1, and movie S1). We also coexpressed a monomeric infrared fluorescent protein (mIFP)–fused histone 2B (H2B) in HEK293 cells (16), which labeled the nucleus, and a red fluorescent protein (RFP; gray pseudo-color). NCS is well known to activate ATM by eliciting DNA damage. In contrast, a mutant reporter, in which the phospho–threonine (T) is mutated to alanine (A) so that it cannot be phosphorylated by ATM, showed homogeneously distributed green fluorescence (Fig. 1D). Our fluorescence imaging also showed that the ATM-SPARK signal is dependent on the doses of NCS from 5 to 400 ng/ml (fig. S2), although the reporter’s signal is saturated when NCS is above 135 ng/ml. These data suggest that ATM-SPARK reports ATM activity in the cells and that its signal is proportional to the amount of DNA damage.
ATM-SPARK achieves temporal resolution, reversibility, and specificity
To characterize the temporal resolution of ATM-SPARK, we conducted time-lapse imaging of HEK293 cells expressing the reporter after addition of NCS. Green fluorescent droplets appeared within 20 to 40 min after addition of NCS (Fig. 1E and movie S2) but not dimethyl sulfoxide (DMSO) (fig. S3 and movie S3), indicating that ATM-SPARK reports endogenous ATM activity. To quantitatively analyze the data, we defined SPARK signal as the percentage of the activated reporters over the total amount of the reporters, which is the ratio of the substrate phosphopeptide (fused to EGFP) in the droplets (condensed phase) over total (condensed plus dilute phase). In particular, we determined SPARK signal by calculating the ratio of “the sum of droplet’s pixel fluorescence intensity over the sum of the cells’ pixel intensity.” For the “normalized SPARK,” the SPARK values at different time points were normalized to the peak SPARK value (see details in Materials and Methods). Our data showed that the normalized SPARK signal plateaued at around 40 min. This is consistent with previous report using FRET-based ATM reporter, which shows ATM activation within 20 to 40 min upon addition of NCS, with FRET signal plateaued at around 40 min (5). Furthermore, single-cell analysis of ATM-SPARK signal showed that ATM was activated around 20 to 30 min after addition of NCS (fig. S4).
We also compared ATM-SPARK signal with endogenous ATM activity upon addition of NCS. We found that ATM was activated after addition of NCS, reaching maximal level at 20 to 30 min, based on phosphorylation of S1981 (fig. S5). This is consistent with the previous studies (17). Our reporter ATM-SPARK was also activated at 20 to 30 min after addition of NCS (Fig. 1E and figs. S4 and S6). Thus, ATM-SPARK has similar kinetics as the endogenous ATM activity following NCS-induced DNA damage.
Furthermore, we compared ATM-SPARK signal with the endogenous ATM substrate Chk2 by measuring Chk2 phosphorylation at T68 because ATM-SPARK is based on the T68-containing phosphopeptide derived from Chk2. We found that Chk2 was quickly phosphorylated at T68, reaching maximum at ~10 min after addition of NCS (fig. S7), which is consistent with the previous study (17). This is faster than ATM-SPARK that was activated at ~30 min after addition of NCS (fig. S4). Thus, our data show that activation of ATM-SPARK is slower than phosphorylation of T68 of Chk2, but it is similar to the endogenous ATM activation. Activation of the FRET-based ATM reporter has also been reported to be around 30 min, which is similar to ATM-SPARK. The FRET reporter also used the same T68-containing phosphopeptide from Chk2 as the ATM-SPARK. This suggests that the phosphorylation of T68 in both reporters is slower than that of the full-length Chk2. On the other hand, the activation of both reporters is similar to that of endogenous ATM activity.
In addition, we also found that pT68 of Chk2 was dephosphorylated at around 40 to 60 min after addition of NCS, which is consistent with previous studies on the kinetics of Chk2 phosphorylation and dephosphorylation after addition of NCS (17). ATM-SPARK signal also started to decrease at ~1.5 hours and disappeared at ~7 hours after addition of NCS (fig. S6). This suggests that the reporter inactivation kinetics follows that of Chk2, although it is slower than the dephosphorylation of pT68 of the full-length Chk2. Inactivation of FRET-based ATM activity reporter after addition of NCS has also been reported previously (4) Thus, ATM-SPARK has an activation and inactivation kinetics that is similar to the FRET-based reporter; both reporters show slower kinetics than that of the endogenous ATM substrate Chk2, likely because both reporters are based on the short phosphopeptide derived from Chk2, and lack of other motifs of Chk2 might result in the slower kinetics of phosphorylation and dephosphorylation.
Next, we demonstrated that the reporter is reversible. We preincubated the cells with NCS to activate ATM, forming ATM-SPARK droplets. Then, we added ATM inhibitor KU-55933, followed by time-lapse imaging. The green droplets rapidly disassembled over time (Fig. 1F, fig. S8, and movies S4 and S5). Quantitative analysis of SPARK signal indicated that the reporter was disassembled within 4 min, with half-time of ~1.5 min.
We also demonstrated that this ATM reporter is applicable to monitor ATM activity in various cancer cells (Fig. 1G). We engineered ATM-SPARK into a lentiviral construct. After packaging into the lentivirus, we infected several types of tumor cells, including glioma cells T98G and U87, osteosarcoma cell line U2OS, and cellosaurus cell line CHLA90. Expression of ATM-SPARK in these cell lines showed green fluorescence, which formed bright droplets upon addition of etoposide that induces DNA damage and activates ATM.
We also used the reporter to image whether different drugs, including camptothecin, etoposide, and NCS, could elicit same or different response of ATM activity in various types of cells including HEK293, HeLa, U2OS, and U87 cells (fig. S9). We found common and differential responses. First, most drugs activated ATM in all these cells, except that camptothecin failed to activate ATM in U87 cells. Second, in HEK293 and HeLa cells, all the drugs exhibited similar kinetics and amplitude in ATM activity. However, in U2OS, etoposide showed a slower kinetics in ATM activation than the other two drugs. On the other hand, etoposide showed larger amplitude of ATM activity than the others. In U87 cells, etoposide showed slower kinetics and smaller amplitude of ATM activation than NCS. Consistently, a previous study reported that etoposide is slower in ATM activation than NCS (4). Thus, our data indicate that different drugs induce different responses in different types of cells in ATM activation. These data indicate that ATM-SPARK can be used to monitor ATM activity in various cancer cells using the reporter-containing lentivirus, which will be useful for biological and therapeutic studies such as drug resistance mechanisms that may involve DNA damage and repair via ATM.
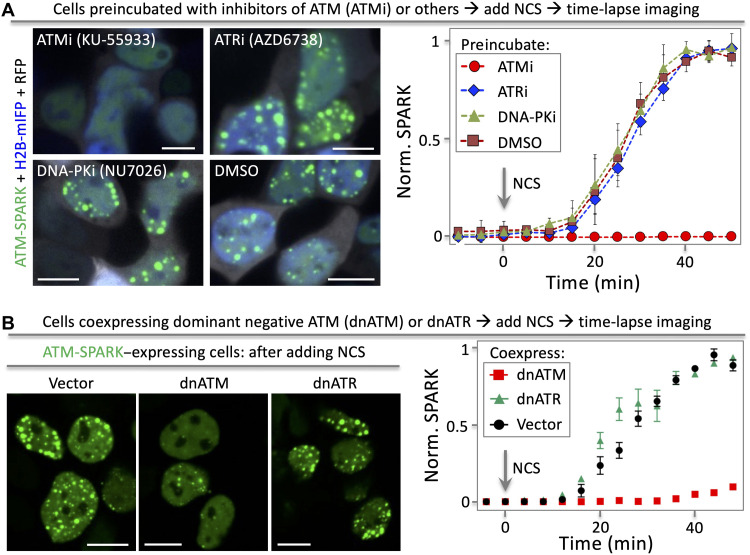
Last, we determined that ATM-SPARK specifically reports ATM activity, and it does not respond to the closely related Ataxia telangiectasia and Rad3-related (ATR) or DNA-dependent protein kinase (DNA-PK). Here, we first preincubated the cells with various inhibitors of ATM, ATR, or DNA-PK. Then, we applied NCS to induce DNA damage and ATM activation. Our time-lapse imaging data showed that the ATM inhibitor blocked ATM-SPARK droplet formation, whereas the ATR inhibitor or the DNA-PK inhibitor had no effect (Fig. 2A). Quantitative analysis of the time-lapse imaging data revealed that the ATM inhibitor blocked the reporter droplet formation, while the other inhibitors showed no difference from the DMSO control. Furthermore, we confirmed that the AKT inhibitor triciribine did not induce droplet formation of ATM-SPARK as expected (fig. S10). Other than the small-molecule inhibitors, we also used the kinase dead and dominant negative ATM (dnATM) or ATR (dnATR). Coexpression of dnATM blocked the droplet formation, whereas dnATR did not inhibit the ATM-SPARK droplet formation upon addition of NCS (Fig. 2B), indicating that ATM-SPARK reports ATM but not ATR activity. Together, these data demonstrate that ATM-SPARK specifically reports activity of endogenous ATM in living cells.
Fig. 2. ATM-SPARK specifically reports ATM activity but not the closely related ATR or DNA-PK.
(A) Fluorescence images of cells showing that ATM-SPARK response is blocked by inhibitors of ATM but not ATR or DNA-PK. HEK293 cells were cotransfected with ATM-SPARK and mIFP-H2B. The cells were preincubated with inhibitors of ATM, ATR, or DNA-PK. Then, the cells were treated with NCS, followed by time-lapse imaging. (B) Fluorescence images of cells showing that ATM-SPARK response is blocked by dnATM but not dnATR. Data are means ± SE (n = 30 cells). Scale bars, 10 μm.
ATM-SPARK achieves spatial resolution and visualizes ATM activity at telomere
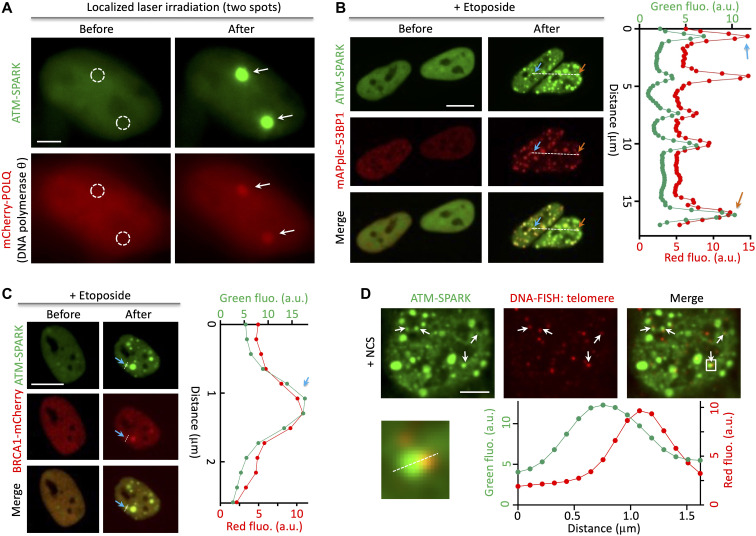
To demonstrate whether the ATM-SPARK has spatial resolution, we used laser to locally damage DNA. This led to two bright GFP droplets in the two irradiation sites (Fig. 3A). Furthermore, these two spots also accumulated DNA damage marker DNA polymerase θ (POLQ) that is fused to mCherry (mCherry-POLQ). In addition, we conducted laser irradiation in cells expressing the control of the sensor (T/A mutant) and found no formation of GFP droplets in this control (fig. S11).
Fig. 3. ATM-SPARK achieves spatial resolution and colocalizes with DNA damage markers and telomere DNA upon radiomimetic drug-induced DNA damage.
(A) Fluorescence images of cells upon laser irradiation on two spatially localized areas (circled by the white dashed lines). Cells were transfected with ATM-SPARK and mCherry-POLQ. (B) ATM-SPARK droplets were colocalized with mApple-53BP1 upon etoposide-induced DNA damage. a.u., arbitrary units. (C) ATM-SPARK droplets were colocalized with BRCA1-mCherry upon addition of etoposide. (D) ATM-SPARK droplets were associated with telomere DNA upon NCS-induced DNA damage. Scale bars, 5 μm (A and D) and 10 μm (B and C).
We next determined that upon DNA damage by etoposide, ATM-SPARK droplets colocalized with 53BP1, a DNA damage marker involved in nonhomologous recombination–based DNA repair (Fig. 3B). We conducted time-lapse imaging and found that many ATM-SPARK droplets colocalized with the mApple-fused 53BP1 foci and that when the mApple-53BP1 foci fused together, the ATM-SPARK droplets also fused together (fig. S12). A previous study reported that upon DNA damage, 53BP1 forms repair compartments that have liquid droplet property and that the 53BP1 droplets can fuse together (18). Thus, our results suggest that the fusion of ATM-SPARK droplets is likely due to the fusion of DNA repair foci.
In addition, we also examined whether the droplet size of ATM-SPARK is proportional to DNA damage. We coexpressed ATM-SPARK and mApple-53BP1, incubated the cells with NCS for 1 hour, then removed NCS, and conducted time-lapse imaging. Our data showed that when mApple-53BP1 foci decreased in size over time, the droplet size of ATM-SPARK also decreased (fig. S13). This suggests that the droplet size of ATM-SPARK is proportional to DNA damage. This is consistent with the laser irradiation experiment, where less intense laser irradiation induced smaller droplet size of ATM-SPARK, while more intense laser irradiation led to larger droplet size of ATM-SPARK (fig. S14). This is also consistent with our findings that the droplet size of ATM-SPARK appears to be correlated with ATM activity because the reporter’s droplet size reaches maximum at around 0.5 and 1.5 hours after addition of NCS (fig. S6), which is consistent with the activation kinetics of the endogenous ATM that also reaches maximum at ~30 min after addition of NCS (fig. S5). These results suggest that the droplet size is proportional to the ATM activity and the DNA damage.
We also found that some ATM-SPARK droplets colocalized with BRCA1 that is involved in homologous recombination–based DNA repair after addition of etoposide (Fig. 3C). Therefore, our data indicate that the ATM reporter achieves spatial resolution in imaging ATM activity and DNA damage.
Last, we characterized whether ATM-SPARK droplets are associated with DNA. It has been reported that NCS induces damages of telomere DNA (19). Therefore, we labeled the NCS-treated cells by DNA fluorescence in situ hybridization (FISH) of human telomere (20). Our data revealed that ATM-SPARK droplets were indeed associated with the telomere DNA (Fig. 3D). Thus, our work suggests that ATM is activated at the telomere DNA after NCS-induced DNA damage and that our reporter achieves spatial resolution in living cells.
Screening of phosphatases identifies protein phosphatase 4 that inhibits ATM activity
Because of its large dynamic range and bright fluorescence than other genetically encoded reporters such as FRET-based ones, we decided to apply ATM-SPARK to screen for phosphatases that block ATM activity. Here, we cloned 156 human phosphatases from the human ORFeome library (21) and coexpressed them with ATM-SPARK in HEK293 cells in 96-well plates (Fig. 4A). To ensure expression of each phosphatase, we linked the phosphatase to mCherry via a self-cleaving 2A (T2A) peptide (phosphatase-T2A-mCherry). Our data showed that each construct had positive mCherry fluorescence (fig. S15), suggesting that each plasmid was transfected and each phosphatase was transcribed. Twenty-four hours after transfection, we added etoposide to induce DNA damage and ATM activation. Fluorescence imaging was carried out at ~1 hour after addition of etoposide. Quantitative analysis of ATM-SPARK signal revealed several phosphatases that blocked ATM activity (Fig. 4B). These include previously known phosphatases that inhibit ATM, such as PPP2CA, PPP2CB, and Wip1 (1, 3, 22–24).
Fig. 4. Screening of ~150 phosphatases identifies PPP4C that inhibits ATM activity upon radiomimetic drug-induced DNA damage.
(A) Schematic of screening procedure. (B) Inhibitory activity of 156 phosphatases. Cells were transfected with ATM-SPARK and the phosphatases. Cells were then treated with etoposide, followed by imaging. ATM activity was calculated on the basis of the SPARK signal. Here, the SPARK value for each phosphatase was normalized to the plasmid vector without phosphatase. (C) Time-lapse imaging of cells expressing vector or PPP4C. Cells were treated with NCS. (D) Inactive PPP4C mutant (D82A) does not block ATM activity. Cells were transfected with ATM-SPARK and PPP4C or its inactive mutant, followed by treatment with NCS. Data are means ± SE (n = 5 imaging fields). Each imaging field contains ~20 cells. Scale bars, 10 μm (C and D). ***P < 0.001. N.S., not significant.
We also identified a previously unidentified phosphatase PPP4C, the catalytic unit of protein phosphatase 4 (PP4), which inhibited ATM upon etoposide induction. To further verify this, we added NCS to activate ATM. Time-lapse imaging showed that PP4 blocked ATM activity upon addition of NCS, whereas a control vector without PPP4C did not (Fig. 4C). We also introduced an inactive mutation D82A to PPP4C. This mutant lost its inhibitory activity against ATM (Fig. 4D). This indicates that the phosphatase activity is indeed required for the inhibitory activity of PPP4C against ATM. Last, we examined whether PP4 directly dephosphorylates and inactivates ATM. We conducted coimmunoprecipitation in HEK293 cells and found that PP4 (D82A) coimmunoprecipitated with ATM, suggesting that PP4 physically interacts with ATM (fig. S16). Our data thus indicates that PP4 directly acts on ATM. PP4 has been reported to play a role in DDR by dephosphorylating proteins including 53BP1 and Phosphorylated histone H2AX (γH2AX) (25–27). However, it was not clear whether PP4 also acts on ATM. Our data suggest that PP4 regulates DDR by directly acting on ATM. Together, our work demonstrates that ATM-SPARK enables high-throughput screening of biological regulators of ATM and that it can used for future studies in identifying more regulators including the rest of the human phosphatases that are not examined in this study.
High-throughput screening identifies BGT226 as a potent ATM inhibitor
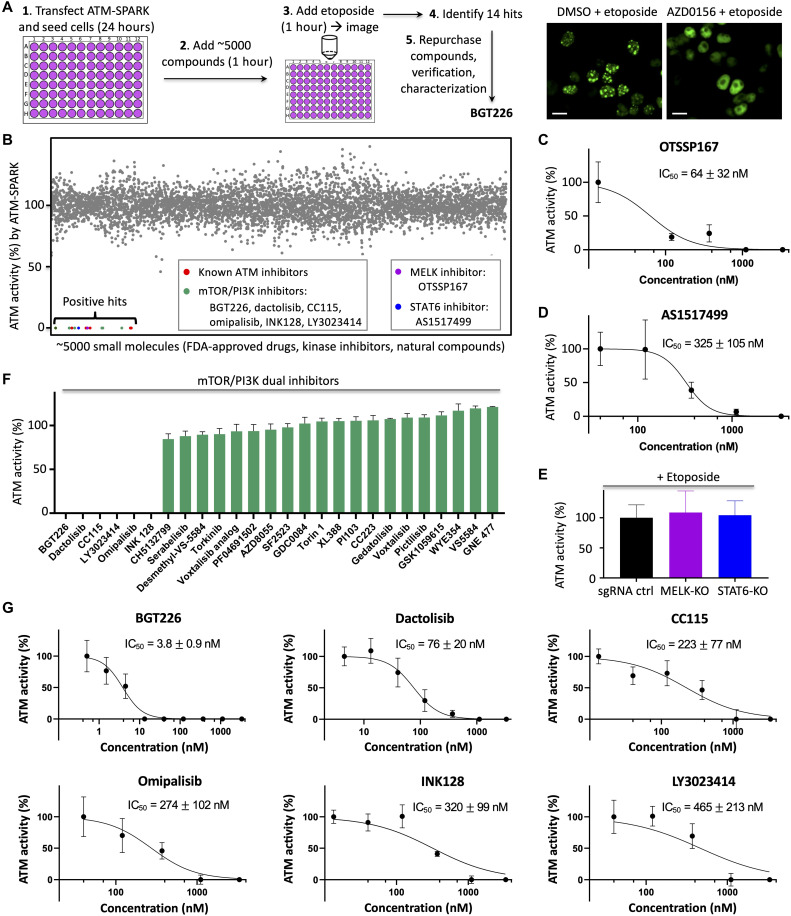
Following the successful screening of biologics-based regulators of ATM, we next decided to demonstrate whether ATM-SPARK can be used to screen for small molecule–based regulators of ATM. We selected ~5000 compounds from the Food and Drug Administration (FDA) drug library, the kinase inhibitor library, and a natural compound library. We chose the kinase inhibitor library because we reasoned that some kinase inhibitors might have activity against ATM in addition to their known target. We chose the FDA drug library because we hoped that some drugs might be repurposed against ATM. We chose the natural compound library because we hoped to identify previously unidentified small-molecule inhibitors against ATM.
We first expressed the reporter in HEK293 cells. Twenty-four hours after transfection, we added the compounds (final concentration at 1 μM) to the cells (Fig. 5A). One hour after incubation of the small-molecule compounds, we added etoposide for 1 hour, followed by fluorescence imaging. We demonstrated this platform using a positive control, an inhibitor of ATM, AZD0156 and a negative control, DMSO (Fig. 5A, right). Quantitative analysis of the SPARK signal of these 5000 compounds identified 14 hits based on their potent effect in inhibiting ATM activity. These hits were confirmed by a second round of imaging using newly purchased compounds (Fig. 5B). In addition to the known ATM inhibitors, our screening also identified several previously unknown inhibitors. These include six mammalian target of rapamycin (mTOR)/phosphatidylinositol 3-kinase (PI3K) dual inhibitors, a maternal embryonic leucine zipper kinase (MELK) inhibitor OTSSP167, and signal transducers and activators of transcription 6 (STAT6) inhibitor AS1517499. A common structural core was identified among the hits, which suggests that they likely bind to the adenosine 5′-triphosphate (ATP)–binding pocket of ATM (fig. S17).
Fig. 5. High-throughput screening identifies BGT226 as a potent ATM inhibitor.
(A) Schematic of the screening procedure. Right: Fluorescence images of negative (DMSO) and positive (AZD0156) control. (B) Percentage of ATM activity in small-molecule inhibitor–incubated cells after addition of etoposide. The ATM activity was determined by ATM-SPARK signal. The percentage of ATM activity was calculated by normalizing ATM activity with that of cells treated with DMSO. Gray dots indicate no or weak inhibitory activity. Colored dots indicate identified hits. (C and D) Dose-response curve of the inhibitors of MELK (C) or STAT6 (D) in inhibiting ATM after addition of etoposide. (E) Knockout (KO) of MELK and STAT6 has no effect on ATM activity after addition of etoposide. (F) Percentage of ATM activity in small-molecule inhibitor–incubated cells upon addition of etoposide. The ATM activity was determined by ATM-SPARK signal. The percentage of ATM activity was calculated by normalizing ATM activity with that of cells treated with DMSO. (G) Dose-response curve of the identified top hits in inhibiting ATM after addition of etoposide. Concentration (1 μM) of compound was used in (B) and (F). Data are means ± SE (n = 5). IC50 was represented as means ± SD (n = 5). Here, “n” represents the number of imaging fields with each field contains ~20 cells. Scale bars, 10 μm (A).
To characterize the identified mTOR/PI3K dual inhibitors, we also measured inhibitory activity of many other dual inhibitors of mTOR/PI3K. While the identified six dual inhibitors showed strong activity against ATM, no other inhibitors had activity against ATM (Fig. 5F). This strongly suggests that the identified inhibitors do not inhibit ATM via the mTOR/PI3K pathway. To identify the most potent inhibitors, we measured median inhibitory concentration (IC50) against ATM using ATM-SPARK in living cells. Our data indicated that BGT226 exhibited the most potent activity with IC50 = 3.8 ± 0.9 nM, whereas the other five inhibitors showed IC50 one to two orders of magnitude lower (Fig. 5G). A known ATM inhibitor AZD0156 showed similar or slightly weaker activity with IC50 = 8.9 ± 1.2 nM under our experimental conditions using etoposide and ATM-SPARK in living cells (fig. S18). This is further confirmed by measuring the endogenous ATM activity (phosphorylation at S1981) using Western blot analysis (fig. S19, A and B). Our data showed that BGT226 inhibited ATM with IC50 similar to that using ATM-SPARK, indicating that ATM-SPARK can be used in measuring IC50 of ATM inhibitors. Our analysis of the endogenous ATM activity also revealed similar IC50 of AZD0156 to that using ATM-SPARK. Thus, our measurement indicates that BGT226 is similar or slightly more potent than AZD0156 in inhibiting ATM in the cellular context (fig. S19). Last, we confirmed that Chk2 activity was also blocked upon inhibition of ATM by BGT226 or AZD0156 (fig. S19B).
We also characterized OTSSP167 and AS1517499, which showed IC50 of 64 and 325 nM, respectively (Fig. 5, C and D), which is one to two orders of magnitude lower than that of BGT226. OTSSP167 was developed as an inhibitor of MELK (28). We thus decided to further characterize whether OTSSP167’s activity against ATM is through the MELK pathway. We used a MELK-specific inhibitor MELK-8A (29), which showed no activity against ATM (fig. S20). This suggests that OTSSP167’s activity is not through MELK. To further verify this, we carried out clustered regularly interspaced short palindromic repeats (CRISPR)–based knockout of MELK in U2OS cells. In the MELK-knockout U2OS cells (fig. S21), etoposide evoked similar ATM activation (Fig. 5E), but OTSSP167 still showed potent activity against ATM, with similar IC50 as that in the control cells (fig. S22). These data strongly suggest that OTSSP167 inhibits ATM not by inhibiting MELK. Using similar procedure, we also showed that AS1517499 also blocked ATM not by the STAT6 pathway, because STAT6 knockout had no effect on etoposide-evoked ATM activation (Fig. 5E and figs. S21 and S22).
Recent studies showed that many kinase inhibitors including OTSSP167 are promiscuous with several tens of kinase targets (30), although their activity against ATM has not been reported. Furthermore, the common structural core analysis showed that the hits could bind to the ATP pocket of ATM. Therefore, we performed molecular docking of these two inhibitors to ATM and found that both molecules could dock to the ATP-binding pocket of ATM with binding affinity at ~170 and 870 nM for OTSSP167 and AS1517499, respectively (fig. S23). The relative binding affinity of the two inhibitors to ATM is consistent with their relative potency in inhibiting ATM. Thus, our work suggests that both molecules inhibit ATM likely via direct binding to the ATP pocket of ATM.
Our screening also identified one PI3K inhibitor, AS605240, which blocked ATM. Thus, we measured other PI3K inhibitors’ activity against ATM, which showed no effect (fig. S24A). This suggests that AS605240 does not inhibit ATM via the PI3K pathway. We determined IC50, which showed two orders of magnitude lower activity than BGT226 (fig. S24B). Last, we also determined other mTOR inhibitors’ activity against ATM, which showed no effect (fig. S24C). Thus, our screening identified BGT226 as a potent ATM inhibitor with similar or slightly higher activity than AZD0156.
BGT226 selectively blocks ATM but not ATR
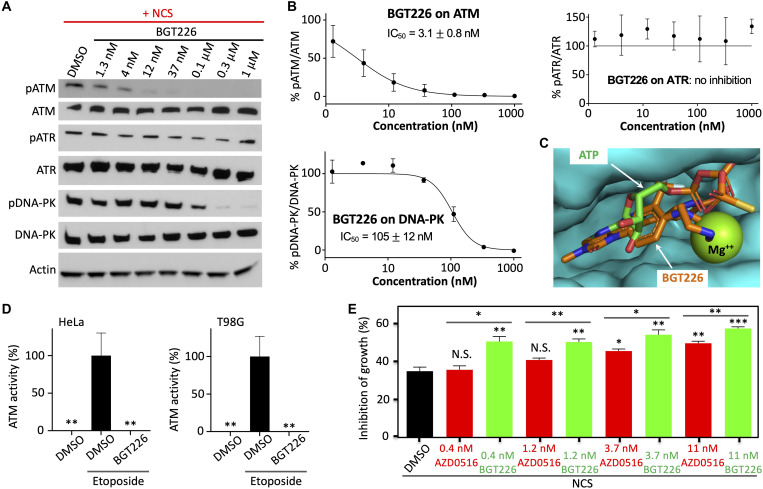
To further characterize the identified ATM inhibitor BGT226, we determined its specificity against ATM over the closely related ATR and DNA-PK. We induced ATM, ATR, and DNA-PK activity using NCS-induced DNA damage. We measured their activities using antibodies against phosphorylated ATM at residue 1981, phosphorylated ATR at residue 1989, phosphorylated DNA-PK at residue 2056, which have been widely used in determining activities of these kinases (4, 5). Our characterization revealed potent activity of BGT226 against ATM with IC50 = 3.1 ± 0.8 nM (Fig. 6, A and B). In contrast, BGT226 showed no inhibitory activity against ATR and 30-fold lower activity against DNA-PK. Structural modeling using SwissDock (31) showed that BGT226 binds to the kinase domain of ATM in the ATP-binding site (Fig. 6C), with large Gibbs free energy change and an estimated dissociation constant (Kd) of ~20 nM. We further demonstrated that BGT226 exhibited potent activity against ATM in various cancer cells including HeLa and T98G (Fig. 6D and fig. S25). Because ATM is a member of the PI3K-related kinase family, which also includes PI3K and mTOR, it might be expected that BGT226 has potent activity against ATM. BGT226 has great selectivity for ATM over the closely related ATR and DNA-PK.
Fig. 6. BGT226 selectively inhibits ATM and sensitizes NCS-induced inhibition of cancer cell proliferation.
(A) BGT226 blocks ATM activity more than ATR or DNA-PK. Western blot analysis was conducted using antibodies against pATM (1981), pDNA-PK (2056), or pATR (1989). (B) Dose-response curve of BGT226 inhibition on ATM, ATR, and DNA-PK. (C) Structural model of BGT226 bound to ATM, overlayed with ATP*Mg++. The model was built using SwissDock. (D) Percentage of ATM activity in HeLa or T98G cells preincubated with BGT226. ATM was activated using etoposide. (E) Percentage of proliferation inhibition in cells treated with NCS (180 ng/ml) and various concentrations of BGT226 or AZD0156. Data are means ± SE [n = 3 biological replicates for (B) and (E); n = 5 imaging fields with each field containing ~20 cells for (D)]. *P < 0.05, **P < 0.01, and ***P < 0.001.
BGT226 sensitizes cancer cells to radiomimetic drugs
Blocking DDR by inhibiting, e.g., ATM/ATR has been shown to sensitize cancer cells to radiotherapeutic treatments, because under radiotherapy, cancer cells accumulate large DNA damage and ATM/ATR plays an important role in DNA damage repair (32–34). Inhibition of ATM/ATR blocks DDR and reduces survival of the cancer cells. For example, the ATM inhibitor AZD0156 has been demonstrated to sensitize cancer cells for radiotherapy (35). Here, we determined whether the identified ATM inhibitor BGT226 is able to sensitize cancer cells to radiomimetic drug NCS and compared it side by side with AZD0156.
Combination of BGT226 with NCS substantially sensitized HeLa cells (Fig. 6E). BGT226 increased inhibition of the cancer cell growth by up to ~22%. In particular, NCS itself (180 ng/ml) blocked cancer cell proliferation by ~35%, which increased to ~57% when 11 nM BGT226 was combined. AZD0156 (11 nM) also increased growth inhibition by ~15%, consistent with previous reports (32, 34, 36). On the other hand, our data indicate that BGT226 sensitizes the cancer cells to NCS more strongly than AZD0156, which is also true when lower concentrations of the ATM inhibitors are combined with NCS. At 0.4 to 1.2 nM, BGT226 increased inhibition of cancer cell growth by ~15%. However, at these concentrations, AZD0156 did not increase inhibition of cancer cell growth. Therefore, our data demonstrate that BGT226 is able to sensitize cancer cells to the radiomimetic drug and that it has stronger effect than AZD0156, consistent with slightly more potent inhibitory activity of BGT226 than AZD0156 against ATM.
We next examined whether the observed effect of BGT226 is due to its inhibitory activity against ATM or mTOR/PI3K. We compared the effect of BGT226 with another PI3K/mTOR inhibitor VS-5584 that has similar or stronger effect in inhibiting PI3K and mTOR but no inhibition against ATM (37). We found that BGT226 but not VS-5584 sensitizes growth inhibition of cancer cells in response to NCS treatment (fig. S26), demonstrating that the observed effect of BGT226 is due to inhibition of ATM but not inhibition of PI3K/mTOR. Last, to further examine whether the effect of BGT is due to direct target engagement with ATM, we generated an ATM-deficient cell line (fig. S27, A and B) and found that the effect of BGT226 in growth inhibition when combined with NCS was lost in this ATM-deficient cell line (fig. S27C). This demonstrates that the effect of BGT226 is through direct engagement of ATM and confirms the target specificity of BGT226 via ATM in sensitizing cancer cells to NCS. Our results suggest that BGT226 may be combined with radiotherapeutic treatment.
In summary, we have designed a GFP phase separation–based activity reporter of ATM, which visualizes spatial and temporal dynamics of endogenous ATM activity in living cells. In the future, we can apply the engineering principles used here to design reporters of the other kinases in the DDR, including ATR, DNA-PK, Chk1, and Chk2. For example, we can use an ATR-specific phosphopeptide to sense kinase activity of ATR. Then, we can use the phosphopeptide binding domain (e.g., FHA1) and the multivalent tags (e.g., HOTag3 and HOTag6) to induce GFP phase separation upon ATR activation. Another direction in designing SPARK-based reporters is to develop multicolor sensors using red and near-infrared fluorescent proteins and orthogonal HOTag pairs, which will be useful for imaging multiple kinases in a signaling network such as in DDR. The GFP phase separation–based SPARK represents a new class of kinase activity reporters for robust imaging of kinase signaling in living cells.
MATERIALS AND METHODS
Plasmid construction and lentivirus production
All plasmid constructs were created by standard molecular biology techniques and confirmed by exhaustively sequencing the cloned fragments. To create ATM-SPARK, DNA fragments encoding LETVSTQELYSIP (for ATM-SPARK) with an NLS (PAAKRVKLD) were inserted to replace FK506 binding protein (FKBP) in the pcDNA3 FKBP-EGFP-HOTag3 construct (15). DNA sequence encoding FHA1 with NLS was inserted upstream of a nonfluorescent GFP mutant (EGFP* containing Y66F mutation), followed by HOTag6. A T2A sequence was further added to 3′-downstream of ATM-NLS-EGFP-HOTag3 via polymerase chain reaction (PCR), and the resulting ATM-NLS-EGFR-HOTag3-T2A fragment was then subcloned into 5′-upstream of FHA1-NLS-EGFP*-HOTag6 to generate the full ATM-SPARK–expressing vector. The threonine was changed to alanine (LETVSAQELYSIP) for the mutATM-SPARK via designed DNA oligonucleotide synthesis. For lentiviral vector, the DNA fragment encoding ATM-SPARK was cloned into pHR-SFFV-GFP1-10 to replace GFP1-10. Lentivirus production followed standard protocol of “Improve Lentiviral Production Using Lipofectamine 3000 Reagent” from Thermo Fisher Scientific.
Cell culture
The HEK293, Hela, U2OS, T98G, and U87 cells were passaged in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum (FBS), nonessential amino acids, penicillin (100 U/ml), and streptomycin (100 μg/ml). CHLA90 cells were following similar protocol, except that FBS is 20%. All culture supplies were obtained from the UCSF Cell Culture Facility.
Live-cell imaging
Cells were grown on Nunc Lab-Tek II chambered cover glass for imaging experiment.
To express ATM-SPARK, HEK293 cells were transiently transfected using calcium phosphate transfection reagent (15) with 300 ng of ATM-SPARK plasmid plus 50 ng of H2B-mIFP-T2A-mCherry plasmid, 100 ng of mCherry-POLQ plasmid, or 300 ng of dnATM (ATM kinase dead) or dnATR (ATR kinase dead). Hela cells were transiently transfected using Lipofectamine 3000 transfection reagent with 300 ng of ATM-SPARK and 100 ng of mApple-53BP1 or BRCA1-mCherry plasmids. HEK293 and Hela cell were imaged 1 day after transfection. U2OS, T98G, CHLA, and U87 cells were infected with medium containing ATM-SPARK lentivirus and imaged 3 days after infection.
All imaging was carried out on Nikon Eclipse Ti inverted microscope equipped with Yokogawa CSU-W1 confocal scanner unit (Andor), digital complementary metal-oxide semiconductor camera ORCA-Flash4.0 (Hamamatsu), and ASI MS-2000 XYZ automated stage (Applied Scientific Instrumentation). Imaging was performed in environmental control unit incubation chamber (In Vivo Scientific) maintained at 37°C and with 5% CO2. Fluorescence images were acquired using Nikon CFI Plan Apochromatic 20× dry [numerical aperture (NA), 0.75] objective or CFI apochromatic TIRF 60× oil objective (NA, 1.49) against GFP, mCherry, or mIFP per experiment settings. Imaging started before adding NCS (500 ng/ml) or etoposide (3 μM) and continued for around 1.3 hours. For experiments with inhibitors (KU-55933, AZD6738, and NU7026), cells were pretreated with inhibitors (20 μM) or DMSO before NCS treatment. To demonstrate sensor reversibility, cells were preincubated with NCS and then treated with KU-55933 or DMSO, and imaging started for several frames before adding KU-55933 or DMSO and continued for around 1 hour.
Laser-induced DNA damage
HEK293 cells grown on four-compartment glass-bottom dishes were transfected with ATM-SPARK and mCherry-POLQ plasmids. Twenty-four hours after transfection, the cells were irradiated on two spatially localized area by General Electric DeltaVision OMX-SR super-resolution microscope. The region of interest roughly 0.5 μm in diameter (60× 1.42 NA oil-immersion objective) was selected and irradiated with a 405-nm argon laser at 5% power for 0.25 s. Imaging were taken with 30-s interval for 30 min.
Telomere DNA FISH combined with immunofluorescence
Telomere DNA FISH were performed as described (20). Briefly, Hela cells grown on eight-well chambered slide (Nunc Lab-Tek II, 155409) were transfected with ATM-SPARK plasmid. Thirty minutes after challenged with NCS, cells were fixed in 4% (w/v) paraformaldehyde for 10 min at room temperature. After fixation, cells were permeabilized in 1× phosphate-buffered saline (PBS) with 0.5% Triton X-100 for 10 min at room temperature. Cells were incubated in 0.1 N of HCl for 5 min and washed twice in 2× saline-sodium citrate (SSC) with 0.1% Tween 20 (SSCT) for 2 min. After incubated in 2× SSCT with 50% formamide for 1 hour at 60°C, the well was loaded with 150 μl of ISH solution [2× SSCT, 50% (v/v) formamide, 10% dextran sulfate, ribonuclease A (400 ng/μl), and 100 nM probe]. After denaturation at 80°C for 3 min, cells were incubated overnight at 44°C.
After hybridization, cells were washed four times in 2× SSCT at 60°C and rinsed in 1× PBS for 1 min. Cells then incubated with 1 μM Atto 550–labeled imager in PBS and washed twice in 1× PBS. The cells were blocked with 2% bovine serum albumin and 10% goat serum in PBS. Then stained with anti-GFP antibody (1:200 dilution; Abcam, Ab290) 3 hours at room temperature. After washing three times with phosphate-buffered saline with 0.1% Tween 20 (PBST), the cells incubated with Alexa Fluor 488–conjugated secondary antibodies (1:200 dilution; Cell Signaling Technology, #4412) at room temperature for 1 hour. The telomere probe sequence is ACATCATCATGGGCCTTTTGGCCCATGATGATGTATGATGATG/3InvdT/, and the Imager sequence is ATGATGATGTATGATGATGT.
Phosphatase screening
To generate the phosphatase library, cDNAs of phosphatases were amplified from Human ORFeome cDNA clone library and cloned into pcDNA vector (pcDNA3-phosphatases). pcDNA3-phosphatases were aliquoted into 96-well plate at 200 ng/μl. For screening, HEK293 cells were seeded into 96-well plate and transfected with 100 ng of plasmid expressing ATM-SPARK and 100 ng of plasmid expressing phosphatases on the second day. Screening was carried out 1 day after transfection with snapshot imaging 1 hour after 3 μM etoposide treatment using 20× dry objective. Positive hits were identified, and further validation was carried out with time-lapse imaging under a 60× objective. D82A mutant PPP4C was generated through overlapping PCR using primers containing the mutation.
Screening small-molecule inhibitor
Inhibitor libraries were purchased from TargetMol and MedChemExpress (MCE) and then aliquoted into 96-well plate at 100 μM. For screening, HEK293 cells from a 10-cm plate were suspended in 19 ml of medium. Thirty micrograms of ATM-SPARK–expressing plasmid was mixed with 1.2 ml of 1× Hanks’ balanced salt buffer and 70 μl of 2.5 M of CaCl2 for 20 min in room temperature. The mixture was added into the cell suspension and then aliquoted in 96-well plate for 100 μl per well. On the second day, 1 μl of inhibitor (100 μM aliquots in the 96-well plate) was added into the 96-well plate with cells; 1 hour later, etoposide was further aliquoted into each well (final concentration of 3 μM). Imaging was carried out 1 hour after etoposide treatment under 20× objective.
Dose titration of identified hits followed similar procedures as in screening except for that inhibitor were added as 3× dilution series starting from 3 μM. Quantification was done by calculating ATM-SPARK value.
Generation of knockout cells
Guide RNAs for targeting human STAT6 (AGGGAATGGCGCACCGTTTG) and human MELK (CTATCTGACGGAAGACAACC) were designed using CHOPCHOP (http://chopchop.cbu.uib.no/) with nontargeted short-guide RNA (sgRNA) (ACGGAGGCTAAGCGTCGCAA) as control. DNA oligos were ordered, annealed, and cloned into LentiCRISPRv2 vector following standard protocol (38). Lentivirus were produced and used to infect U2Os cell. After puromycin selection, single clones were picked up, propagated, and then proceeded for Western blot analysis against STAT6 or MELK.
Generation of ATM knockdown cell lines
An inducible CRISPR interference (CRISPRi) system was used to repress ATM expression in MDA-MB231 cells. The MDA-MB231 cells were transduced to express catalytically inactive cas9 (dcas9) and Krüppel-associated box (KRAB) domain fusion protein that was further tagged to a reporter fluorescent protein mCherry. The expression of the dcas9-KRAB fusion protein was under the control of inducible Tet3 system (Clonetech; inducible in the presence of doxycycline). For the transduction of MDA-MB231 cells, lentiviral particles were generated in HEK293T cells. Packaging plasmids psPAX2 and envelope plasmid pMD2G were used. MDA-MB231 cells expressing dcas9-KRAB-mCherry fusion proteins were selected by cell sorting (Sony SH800) for mCherry-positive cells. The sorted cells were then further transduced with lentiviral particles with ATM-specific sgRNAs tagged to blue fluorescent protein (BFP) reporter gene. The sgRNA against ATM was selected from previously published and validated data from genome-scale libraries for CRISPRi-mediated loss-of-function genomic screens (39). The sgRNA was cloned into pU6-sgRNA Ef1α Puro-T2A-BFP vector (reporter protein BFP). BFP-positive cells were selected by single-cell sorting, and several cell lines were expanded from single-cell clones. TRE3G-KRAB-dcas9-P2A-mcherry and pU6-sgRNA Ef1α Puro-T2A-BFP vector plasmids were gifts from L. Gilbert and J. Weismann laboratories.
In validation of the clones, to induce ATM repression, the clones were cultured in the presence of doxycycline (0.5 μg/ml) for 72 hours. To validate repression of ATM, total RNA was isolated from the control cells and the clones using a QIAGEN RNeasy mini kit. The reverse transcription PCR (RT-PCR) assay was performed with the iScript cDNA Synthesis Kit (Bio-Rad). The expression levels of ATM (Hs00175892_m1) and control gene endogenous human β-actin (Hs1060665_g1) were determined by quantitative RT-PCR. Each gene was tested in the clone and the control cell line in triplicates in three independent experiments. The relative changes in gene expression were normalized to endogenous human β-actin gene expression levels respectively by the −2ΔΔCT method. On the basis of the results, the clone University of California Santa Cruz (UCSC) ATM A5 was selected as a clone that shows the repression of ATM expression.
Western blot
HEK293 cells were pretreated with BGT226 or AZD0156 for 1 hour with titration starting from 1 μM. Then, NCS (500 ng/ml) or etoposide (3 μM) was added for 1 hour. After that, cells were washed three times in cold PBS and lysed in 1× lysis buffer (with protease inhibitor and phosphatase inhibitor added). The lysate was further incubated at 4°C for 40 min and centrifuged at 15,000g at 4°C for 20 min. Supernatant was collected and mixed with NuPAGE LDS sample buffer (4×). Sample was further resolved in NuPAGE 4 to 12% bis-tris protein gels. After transferring, the membrane were blocked in PBST containing 5% nonfat milk and incubated with first antibodies (1:1000; except for anti–β-actin antibody at 1:5000) at 4°C overnight. On the following day, membrane was washed three times in PBST and then incubated in horseradish peroxidase–conjugated second antibodies (1:3000) for 1 hour at room temperature. After washing three times with PBST, signal was visualized using standard enhanced chemiluminesence substrate on film. Blots were quantified with ImageJ.
Cell growth assays
HeLa cells were seeded on 384-well plate. On the second day, NCS, BGT226, and AZD0156 were added at dose series at 3× dilution. NCS doses started from 180 ng/ml for five titrations, while doses for BGT226 and AZD0156 started from 100 nM for six titrations. Combinations between NCS and BGT226/AZD0156 were set for triplicates. Cell viability was tested 3 days after adding drugs with CellTiter-Glo 2.0 Cell Viability Assay following standard protocol. Cell growth inhibition was calculated by normalizing with DMSO-treated cell.
Docking
Docking of BGT226, OTSSP167, and AS1517499 to ATM was performed using SwissDock. The BGT226 mol2 file containing the atomic coordinates of BGT226 was downloaded from LigandBox database. The kinase domain of ATM from pdb file 7ni4 was used as the template for docking. Residues within the docking site were set to be flexible within 5 Å. Kd of BGT226 to the kinase domain of ATM was derived from the free energy change, which was obtained from SwissDock.
Image analysis
For quantitative analysis of the SPARK signal, images were processed in ImageJ. The sum of droplet’s pixel fluorescence intensity and the cells’ pixel intensity was scored using Analyze Particle function in ImageJ. In particular, to analyze the images, we used “Analyze Particle” function in ImageJ, which involve two “mask” process. In the first mask, droplet’s signal was masked using high threshold based on the distinct intensity from fluorescent signal in the cell, while in the second mark, the fluorescent signal of the whole cell was masked from background signal using low threshold (slightly above background). For each mask, Analyze Particle function was applied for calculating the sum of droplet’s pixel fluorescence intensity and the sum of the cells’ pixel intensity. Then, the SPARK signal was determined by calculating the ratio of “the sum of droplet’s pixel fluorescence intensity over the sum of the cells’ pixel intensity.” For the normalized SPARK value (e.g., in Fig. 1E), the SPARK values at different time points were normalized to the peak SPARK value.
Western blot analysis
For quantitative analysis of the Western blot signal, films were scanned as JPEG and imported to ImageJ. Bands were inverted and selected with region of interest (ROI) function. A region without band was selected as background. Mean gray value within each ROI was analyzed, and band signal was calculated after subtracting with background. Phosphorylation level was calculated by dividing the band value of phosphoband to total protein band. Normalized phosphorylation after inhibitor treatment was calculated by normalizing the value of phospho/total protein to that of DMSO-treated cells.
Data quantification and plotting
All data were quantified and plotted using GraphPad Prism 8. IC50 was calculated using nonlinear fit function. Comparison between groups was done using Student’s t test function.
Acknowledgments
The laser-based micro-irradiation was performed at the Center for Advanced Light Microscopy-Nikon Imaging Center at UCSF on an OMX-SR. We thank B. Beliveau for advice on the DNA FISH method SABER. We thank S. Menon, A. Tulpule, J. Mukherjee, B. Gini, T. Bivona, W. A. Weiss, and Z. Qadeer for discussion and sharing cells/reagents/antibodies. We thank H. J. Lee, Y.-W. Jun, and D. Ma for sharing cells. We thank X. Feng for analyzing the common structural motifs of the identified compounds.
Funding: This work was supported by the NIH/NIGMS R35 GM131766 and R01CA258327 (to X.S.), gift from Don and Ellie Knauss (to P.N.M.), NIH R35GM118119, the UCSF Program for Breakthrough Biomedical Research funded in part by the Sandler Foundation, the UCSF Research Resource Fund Award, and HHMI. .
Author contributions: X.S. initiated the project. X.L. performed experiments. X.L. and C.-I.C. conducted the laser irradiation. C.-I.C. conducted the DNA FISH. C.-I.C. and J.Y. constructed the phosphatase library. X.L., C-I.C., and J.Y. prepared the chemical libraries. S.C. and P.N.M. generated the ATM-deficient cell lines. X.S., X.L., and C.-I.C. analyzed data and wrote the manuscript. All the authors contributed to the final draft.
Competing interests: X.L. and X.S. have filed a patent on a new use of the identified compounds. The authors declare that they have no other competing interests.
Data and materials availability: The plasmid of the ATM-SPARK reporters will be deposited to Addgene as pcDNA-wtATMSPARK2 (195247), pcDNA-mutATMSPARK2 (195248), ssFv_lenti_wtATMSPARK2 (195249), ssFv_lenti_mutATMSPARK2 (195250), and pcDNA-BRCA1-mCherry (195275) (www.addgene.org/). Requests for plasmids can also be submitted to Xiaokun Shu (xiaokun.shu@ucsf.edu). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Key Resource Table
Figs. S1 to S27
Legends for movies S1 to S5
Other Supplementary Material for this : manuscript includes the following:
Movies S1 to S5
REFERENCES AND NOTES
- 1.J.-H. Lee, T. T. Paull, Cellular functions of the protein kinase ATM and their relevance to human disease. Nat. Rev. Mol. Cell Biol. 22, 796–814 (2021). [DOI] [PubMed] [Google Scholar]
- 2.P. Awasthi, M. Foiani, A. Kumar, ATM and ATR signaling at a glance. J. Cell Sci. 128, 4255–4262 (2015). [DOI] [PubMed] [Google Scholar]
- 3.T. T. Paull, Mechanisms of ATM activation. Annu. Rev. Biochem. 84, 711–738 (2015). [DOI] [PubMed] [Google Scholar]
- 4.H. Jaiswal, J. Benada, E. Müllers, K. Akopyan, K. Burdova, T. Koolmeister, T. Helleday, R. H. Medema, L. Macurek, A. Lindqvist, ATM/Wip1 activities at chromatin control Plk1 re-activation to determine G2 checkpoint duration. EMBO J. 36, 2161–2176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S. A. Johnson, Z. You, T. Hunter, Monitoring ATM kinase activity in living cells. DNA Repair (Amst). 6, 1277–1284 (2007). [DOI] [PubMed] [Google Scholar]
- 6.P. P. Tomasini, T. N. Guecheva, N. M. Leguisamo, S. Péricart, A.-C. Brunac, J. S. Hoffmann, J. Saffi, Analyzing the opportunities to target DNA double-strand breaks repair and replicative stress responses to improve therapeutic index of colorectal cancer. Cancer 13, 3130 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A. Schmitt, G. Knittel, D. Welcker, T. P. Yang, J. George, M. Nowak, U. Leeser, R. Büttner, S. Perner, M. Peifer, H. C. Reinhardt, ATM deficiency is associated with sensitivity to PARP1- and ATR inhibitors in lung adenocarcinoma. Cancer Res. 77, 3040–3056 (2017). [DOI] [PubMed] [Google Scholar]
- 8.A. M. Weber, A. J. Ryan, ATM and ATR as therapeutic targets in cancer. Pharmacol Ther. 149, 124–138 (2015). [DOI] [PubMed] [Google Scholar]
- 9.A. Fedier, M. Schlamminger, V. A. Schwarz, U. Haller, S. B. Howell, D. Fink, Loss of atm sensitises p53-deficient cells to topoisomerase poisons and antimetabolites. Ann. Oncol. 14, 938–945 (2003). [DOI] [PubMed] [Google Scholar]
- 10.S. T. Durant, L. Zheng, Y. Wang, K. Chen, L. Zhang, T. Zhang, Z. Yang, L. Riches, A. G. Trinidad, J. H. L. Fok, T. Hunt, K. G. Pike, J. Wilson, A. Smith, N. Colclough, V. P. Reddy, A. Sykes, A. Janefeldt, P. Johnström, K. Varnäs, A. Takano, S. Ling, J. Orme, J. Stott, C. Roberts, I. Barrett, G. Jones, M. Roudier, A. Pierce, J. Allen, J. Kahn, A. Sule, J. Karlin, A. Cronin, M. Chapman, K. Valerie, R. Illingworth, M. Pass, The brain-penetrant clinical ATM inhibitor AZD1390 radiosensitizes and improves survival of preclinical brain tumor models. Sci Adv. 4, eaat1719 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R. Y. Tsien, Constructing and exploiting the fluorescent protein paintbox (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 48, 5612–5626 (2009). [DOI] [PubMed] [Google Scholar]
- 12.R. Y. Tsien, The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 (1998). [DOI] [PubMed] [Google Scholar]
- 13.J. Y. Ahn, J. K. Schwarz, H. Piwnica-Worms, C. E. Canman, Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 60, 5934–5936 (2000). [PubMed] [Google Scholar]
- 14.X. Shu, Imaging dynamic cell signaling in vivo with new classes of fluorescent reporters. Curr. Opin. Chem. Biol. 54, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Q. Zhang, H. Huang, L. Zhang, R. Wu, C.-I. Chung, S.-Q. Zhang, J. Torra, A. Schepis, S. R. Coughlin, T. B. Kornberg, X. Shu, Visualizing dynamics of cell signaling in vivo with a phase separation-based kinase reporter. Mol. Cell. 69, 334–346.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D. Yu, M. A. Baird, J. R. Allen, E. S. Howe, M. P. Klassen, A. Reade, K. Makhijani, Y. Song, S. Liu, Z. Murthy, S.-Q. Zhang, O. D. Weiner, T. B. Kornberg, Y.-N. Jan, M. W. Davidson, X. Shu, A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat. Methods 12, 763–765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. Ouchi, T. Ouchi, Distinct DNA damage determines differential phosphorylation of Chk2. Cancer Biol. Ther. 15, 1700–1704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S. Kilic, A. A. Lezaja, M. Gatti, E. Bianco, J. Michelena, R. Imhof, M. Altmeyer, Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 38, e101379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.G. Hewitt, D. Jurk, F. D. M. Marques, C. Correia-Melo, T. Hardy, A. Gackowska, R. Anderson, M. Taschuk, J. Mann, J. F. Passos, Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat. Comm. 3, 708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.J. Y. Kishi, S. W. Lapan, B. J. Beliveau, E. R. West, A. Zhu, H. M. Sasaki, S. K. Saka, Y. Wang, C. L. Cepko, P. Yin, SABER amplifies FISH: Enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat. Methods 16, 533–544 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.X. Yang, J. S. Boehm, X. Yang, K. Salehi-Ashtiani, T. Hao, Y. Shen, R. Lubonja, S. R. Thomas, O. Alkan, T. Bhimdi, T. M. Green, C. M. Johannessen, S. J. Silver, C. Nguyen, R. R. Murray, H. Hieronymus, D. Balcha, C. Fan, C. Lin, L. Ghamsari, M. Vidal, W. C. Hahn, D. E. Hill, D. E. Root, A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.P. Schwertman, S. Bekker-Jensen, N. Mailand, Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 17, 379–394 (2016). [DOI] [PubMed] [Google Scholar]
- 23.A. Marechal, L. Zou, DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 5, a012716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Y. Shiloh, Y. Ziv, The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14, 197–210 (2013). [PubMed] [Google Scholar]
- 25.M. Isono, A. Niimi, T. Oike, Y. Hagiwara, H. Sato, R. Sekine, Y. Yoshida, S.-Y. Isobe, C. Obuse, R. Nishi, E. Petricci, S. Nakada, T. Nakano, A. Shibata, BRCA1 directs the repair pathway to homologous recombination by promoting 53BP1 dephosphorylation. Cell Rep. 18, 520–532 (2017). [DOI] [PubMed] [Google Scholar]
- 26.S. Nakada, G. I. Chen, A.-C. Gingras, D. Durocher, PP4 is a γH2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep. 9, 1019–1026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.P. T. W. Cohen, A. Philp, C. Vázquez-Martin, Protein phosphatase 4–From obscurity to vital functions. FEBS Lett. 579, 3278–3286 (2005). [DOI] [PubMed] [Google Scholar]
- 28.S. Chung, H. Suzuki, T. Miyamoto, N. Takamatsu, A. Tatsuguchi, K. Ueda, K. Kijima, Y. Nakamura, Y. Matsuo, Development of an orally-administrative MELK-targeting inhibitor that suppresses the growth of various types of human cancer. Oncotarget 3, 1629–1640 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.I. M. McDonald, G. D. Grant, M. P. East, T. S. K. Gilbert, E. M. Wilkerson, D. Goldfarb, J. Beri, L. E. Herring, C. Vaziri, J. G. Cook, M. J. Emanuele, L. M. Graves, Mass spectrometry-based selectivity profiling identifies a highly selective inhibitor of the kinase MELK that delays mitotic entry in cancer cells. J. Biol. Chem. 295, 2359–2374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.S. Klaeger, S. Heinzlmeir, M. Wilhelm, H. Polzer, B. Vick, P.-A. Koenig, M. Reinecke, B. Ruprecht, S. Petzoldt, C. Meng, J. Zecha, K. Reiter, H. Qiao, D. Helm, H. Koch, M. Schoof, G. Canevari, E. Casale, S. R. Depaolini, A. Feuchtinger, Z. Wu, T. Schmidt, L. Rueckert, W. Becker, J. Huenges, A.-K. Garz, B.-O. Gohlke, D. P. Zolg, G. Kayser, T. Vooder, R. Preissner, H. Hahne, N. Tõnisson, K. Kramer, K. Götze, F. Bassermann, J. Schleg, H.-C. Ehrlich, S. Aiche, A. Walch, P. A. Greif, S. Schneider, E. R. Felder, J. Ruland, G. Médard, I. Jeremias, K. Spiekermann, B. Kuster, The target landscape of clinical kinase drugs. Science 358, eaan4368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A. Grosdidier, V. Zoete, O. Michielin, SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39, W270–W277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J. M. Cleary, A. J. Aguirre, G. I. Shapiro, A. D. D’Andrea, Biomarker-guided development of DNA repair inhibitors. Mol. Cell 78, 1070–1085 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. J. O’Connor, Targeting the DNA damage response in cancer. Mol. Cell 60, 547–560 (2015). [DOI] [PubMed] [Google Scholar]
- 34.N. J. Curtin, DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer 12, 801–817 (2012). [DOI] [PubMed] [Google Scholar]
- 35.L. C. Riches, A. G. Trinidad, G. Hughes, G. N. Jones, A. M. Hughes, A. G. Thomason, P. Gavine, A. Cui, S. Ling, J. Stott, R. Clark, S. Peel, P. Gill, L. M. Goodwin, A. Smith, K. G. Pike, B. Barlaam, M. Pass, M. J. O’Connor, G. Smith, E. B. Cadogan, Pharmacology of the ATM inhibitor AZD0156: Potentiation of irradiation and olaparib responses preclinically. Mol. Cancer Ther. 19, 13–25 (2020). [DOI] [PubMed] [Google Scholar]
- 36.M. D. Rainey, M. E. Charlton, R. V. Stanton, M. B. Kastan, Transient inhibition of ATM kinase is sufficient to enhance cellular sensitivity to ionizing radiation. Cancer Res. 68, 7466–7474 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.C. Borsari, M. De Pascale, M. P. Wymann, Chemical and structural strategies to selectively target mTOR kinase. ChemMedChem 16, 2744–2759 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.N. E. Sanjana, O. Shalem, F. Zhang, Improved vectors and genome-wide libraries for CRISPR screening. Nat. Meth. 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.L. A. Gilbert, M. A. Horlbeck, B. Adamson, J. E. Villalta, Y. Chen, E. H. Whitehead, C. Guimaraes, B. Panning, H. L. Ploegh, M. C. Bassik, L. S. Qi, M. Kampmann, J. S. Weissman, Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Key Resource Table
Figs. S1 to S27
Legends for movies S1 to S5
Movies S1 to S5