Abstract
HIV-1 infection of macrophages is a multistep and multifactorial process that has been shown to be altered by exposure to methamphetamine (Meth). Identifying the resulting dynamic changes in the proteome of these cells, offers novel insights into the effect of Meth on HIV-1 infection. In this study, we measured the effect of Meth on a subset of the proteome of HIV-1-infected human monocyte-derived macrophages (hMDM) using Sequential Windowed Acquisition of All Theoretical Fragment Ion Mass Spectra (SWATH-MS) and multiple reaction monitoring (MRM). Cells were exposed to Meth either prior to, or after infection, and then maintained throughout the duration of the experiment. In discovery phase we used SWATH-MS full unbiased profiling followed by bioinformatic analyses which were further validated using Multiple Reaction Monitoring (MRM). This is the first study to use MRM approach in such biological setting. We found that Meth affects expression of multiple molecular pathways including Rab proteins, which are master regulators of intracellular trafficking. Besides highly dynamic changes, our data show high donor-to-donor variability of responses. Our results shed new light on the molecular mechanisms underlying cellular pathology of hMDM resulting from viral infection and a drug of abuse — Meth.
Keywords: HIV, macrophages, MRM, Rab proteins, SWATH-MS
Introduction
Mononuclear phagocytes (MPs) are immune cells that survey their milieus and react to contain the pathological environment, including infections or other sources of inflammation. The macrophage is one of the prime targets of HIV-1 infection, serving as a reservoir of productive viral replication and a vehicle to spread infection throughout the body, including the central nervous system [1, 2]. Thus, the impact of macrophages on the course of various diseases is central.
The complexity of HIV infection and treatment is exacerbated by drugs of abuse, where use of illicit drug and HIV infection pose fundamentally different yet interconnected health concerns [3]. The use of illicit drugs such as Meth is common in people living with HIV (PLWH) and can affect various stages of HIV infection [4–6]. Macrophages are impaired in their protective functions by exposure to toxic substances, such as Meth. It has been shown that both HIV-1 infection and Meth individually result in changes to protein expression in MP therefore, the presence of Meth may further impact the role of the innate immune system as a whole [7–9].
Quantitative proteomics of complex samples, such as whole cell lysates, poses a number of challenges. Several technology platforms have been developed, i.e., iTRAQ, TMT, SILAC, SWATH-MS and others and each of these platforms have strengths and weaknesses. In additions, proteomic samples contain peptides of very diverse properties, (e.g. post-translationally modified, highly hydrophobic and highly hydrophilic proteins that vary in size from small to very large). The dynamic range of cellular concentrations of proteins, thus peptides derived from these proteins, may reach as high as seven orders of magnitude [10]. We began this work by employing SWATH-MS for full unbiased quantification of proteomes of HIV-1 infected and/or Meth treated hMDM. This was followed by MRM-based targeted quantification of Ras-related proteins (Rab proteins) and actin that we intended to use as a reference. MRM is a well-established method for quantitative mass spectrometry (MS) adopted by the field of proteomics more than a decade ago [11, 12]. Ever since MRM gained popularity in quantitation of proteins, it has been used for targeted quantitative investigations in systems biology. Although its throughput is limited, MRM remains attractive due to the high sensitivity of assays and capability to perform measurements of the investigated targets in a broad dynamic range. In fact, it has been proposed to replace western blot analysis as the method of choice for validation of unbiased proteomic studies [13]. We anticipate that the application of MRM for the detection of selected protein targets, Rab proteins in our case, or the validation of candidates from unbiased proteomic analysis provides the highest possible sensitivity and the highest dynamic range among currently available MS-based approaches for complex biological samples.
Herein, we provide a thorough description of MRM assay design for the quantification of actin as a presumed control, and numerous Rab proteins, which have been proposed to act sequentially in autophagy [14], an essential process disrupted by HIV-1 infection [15]. The results presented here discuss some of the technical challenges of applying MRM as a quantitative method for discovery and validation of protein levels in highly complex biological samples. When MRM is combined with other MS techniques, such as data-independent acquisition approach (DIA) to monitor changes in protein abundances and fluctuations in the functional components of the cell, it provides critical insights into the underlying molecular mechanisms of biological processes.
Materials and Methods
Patients and samples
Monocytes from seven healthy human donors were obtained from HIV-1, HIV-2, and hepatitis seronegative donors and differentiated to hMDM. The University of Nebraska Medical Center Institutional Review Board has determined that leukapheresis from normal donors utilizing core facilities does not constitute human subject research as defined in 45CFR46.102 of U.S. Federal Policy for the Protection of Human Subjects. Therefore, this leukapheresis procedure is not subject to federal regulation of human subject research and has been classified as exempt.
Cell cultures and Infections
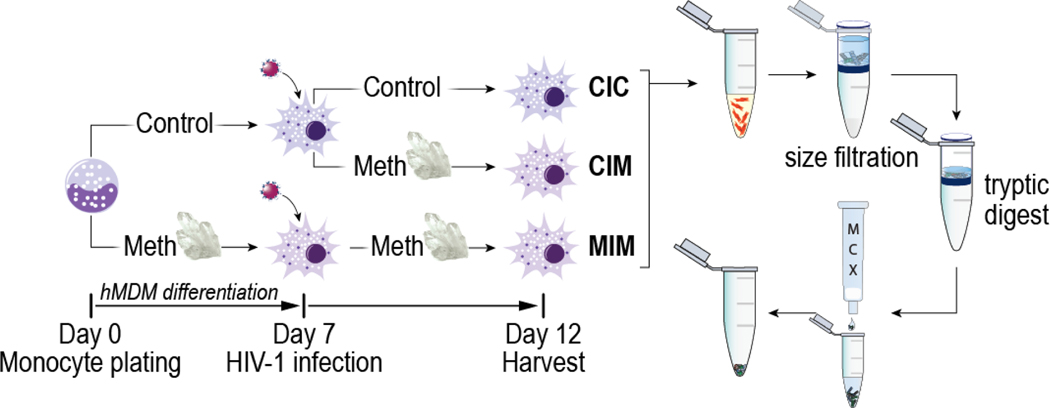
A graphical representation of the experimental design is presented in Figure 1. Monocytes were obtained via leukapheresis and purified by counter-current centrifugal elutriation as previously described [16]. The cells were placed into a serum-free media consisting of Macrophage-SFM supplemented with 10 mM HEPES, 50 mg/mL gentamicin, 2 μg/mL ciporflaxin (all from Invitrogen, Carlsbad, CA), 1% Nutridoma-SP (Sigma, St. Lois, MO), and 10 ng/mL of recombinant human macrophage colony stimulating factor (MCSF; PeproTech, Rocky Hill, NJ). Complete culture media was half-exchanged on day three and completely exchanged on day five. Cells were differentiated in this media for seven days +/− 100 μM Meth (Sigma, St Louis, MO, USA) before harvest.
Figure 1. An overview of experimental design.
CIC denotes condition in which infection only occurs; CIM denotes Meth treatment post-infection. MIM represents Meth treatment before and after infection. Since we compared infected macrophages in all three settings, addition of Meth prior to and after infection treatment (MIM) and addition of Meth after infection (CIM) represent longer and shorter exposure to Meth respectively. For protein expression measured by MRM, we present levels according to experimental settings of CIC, CIM, and MIM.
To study the effects of HIV and Meth on the human macrophages, human monocytes were differentiated in media conditions as described above. At day seven, media was removed, and cells were exposed to serum-free inoculum containing 1000 particles per cell of HIVADA for four hours. After four hours, all media containing the virus was removed, and monocytes were washed with serum-free media four times. HIV-1 inoculated differentiated macrophages were placed into a serum-free media consisting of Macrophage-SFM supplemented with 10 mM HEPES, 50 mg/mL gentamicin, 2 μg/mL ciprofloxacin (all from Invitrogen, Carlsbad, CA), 1% Nutridoma-SP (Sigma, St. Louis, MO) +/− 100 μM Meth. A full media exchange was completed on day three post-HIVADA infection with the supernatant reserved for a p24 assay (see Figure S1). On day five post-HIVADA infection, all cells and supernatants were harvested. In brief, media was removed, and adherent cells were washed with ice-cold Dulbecco’s phosphate-buffered saline (DPBS; Corning, Manassas, VA). DPBS with protease inhibitor was added to each well and cells were scraped and transferred to 15 mL tubes. Cells were pelleted and washed in DPBS supplemented with a protease inhibitor (Thermo Fisher Scientific). Pellets were transferred to Eppendorf tubes, pelleted again and all supernatant removed. Dry pellets were transferred to −80°C for storage.
Human primary T cells and Jurkat cells used along with hMDM to create complex biological matrix for the LC-MRM method validation were cultured as described in [17, 18].
Sample preparation for mass spectrometry
Cells were lysed using 0.3% (w/v) NP-40 (EMD Millipore, Billerica, MA) with 0.1M Tris-HCl, pH 7.6, with protease inhibitor (Sigma #P2714) to 1X final concentration. Proteins were treated with DTT at a final concentration of 25 mM and heated at 95°C for 5 min. Samples were cooled and digested using the Filter Aided Sample Preparation (FASP) protocol as previously described [19]. Following digestion, peptides were cleaned using Oasis mixed cation exchange according to the manufacturer’s protocol (Waters; Milford, MA). Cleaned peptides were quantified at 205 nm using a NanoDrop 2000 (Thermo Scientific, Wilmington, DE).
Aliquots of hMDM, human primary T cells and Jurkat cells whole cell lysates were pooled together to create a complex biological matrix used in LC-MRM method validation. They were spiked with various amounts of Trypsin-digested BSA MS standard (New England Biolabs), which resulted in 1, 10, 50, 100, 200, 500 and 1000 fmol/μL dilutions.
NanoLC-SWATH-MS quantification:
Label-free quantification of proteins present in hMDM whole cell extracts from the investigated CIC, CIM and MIM conditions was performed in a DIA SWATH mode on a nanoLC-MS/MS system comprising of Eksigent 415 nanoLC (Eksigent, Redwood, USA) coupled with TripleTOF 6600 (SCIEX, Framingham, MA, USA) hybrid Q-TOF mass spectrometer, which was equipped with ESI OptiFlow TurboV Ion Source and SteadySpray NanoProbe (SCIEX), and controlled by Analyst TF 1.7 software (SCIEX). The same nanoLC-MS/MS system was used for data-dependent (DDA) analyses of pooled CIC-CIM-MIM sample to create spectral ion library used during SWATH data extraction. The nanoLC-MS/MS system calibrations were performed between the samples runs using the SCIEX PepCalMix from SWATH Acquisition Performance Kit and the protocol described by the producer [20]. Two μg of the peptide mixtures was loaded onto the RP-1 (General RP, 10 × 0.075mm; Phenomenex, Torrance, CA, USA) Trap Column for 10 min at flow rate of 2 μL/min. Then they were transferred through SecurityLINK Sgl (25μm x 75 cm, Phenomenex) and separated on the bioZen Peptide Polar-C18 column (150 × 0.075 mm, 3 μm; Phenomenex), at the flow rate of 300 nL/min, with the use of the mobile phases A (0.1% formic acid in water) and B (0.1% formic acid in ACN (all solvents LC-MS grade from Honeywell Inc., Muskegon, MI, USA) and the following gradient program: from 3 to 8% B (0–2 min), then linearly increased to 30% B (2–90 min), 40% B (90–100 min) and 80% B (100–105 min), then kept at 80% B (105–110 min), and followed by column re-equilibration at 3% B (112–135 min). Portions of the separated peptide mixture were ionized by ESI at 3 kV ion spray voltage, 25 psi curtain gas, 12 psi ion source gas 1, 0 psi ion source gas 2, 150 °C temperature, 75 de-clustering potential and analyzed in a positive ion mode for 130 min.
For spectral library creation, we used a pooled sample consisting of aliquots of all samples from four of the investigated donors in all CIC, CIM and MIM conditions and spiked with iRT Kit peptides (Biognosys AG, Zurich, Switzerland). It was acquired in a DDA mode using a TOF-MS scan from 400–1250 Da in 250 ms accumulation time, after which up to 30 precursor ions, +2 to +5-charged, were then selected for fragmentation in the m/z range of 100–1500, using 50 ms accumulation time, rolling collision energy and high sensitivity mode of acquisition. The precursor ions were excluded from re-selection for 15 s, which resulted in cycle time of 1.8 s. The acquired MS/MS spectra for four technical replicates of pooled sample were searched together with the use of Paragon algorithm incorporated in ProteinPilot 5.0.1 software (SCIEX) against a Human and HIV reviewed database from UniProt [21] (downloaded on 09.04.2020). The database search parameters included: sample type – identification, cysteine modification – iodoacetamide, enzyme – trypsin, instrument – TripleTOF 6600, ID focus – biological modifications, search effort – thorough, and automatic FDR analysis. The summary of detected proteins/peptides can be found in Table S1 A and B provided in Supporting Information_2. These tables represent output of ProteinPilot searches of DDA and are considered as metadata.
DIA SWATH analyses were performed in five technical replicates for separately pooled: (1) CIC, (2) CIM and (3) MIM samples, which were composed of aliquots of hMDM whole cell extracts from four of the investigated donors for each of the conditions and spiked with iRT Kit peptides (Biognosys). Based on the precursors’ density observed in the investigated samples, the m/z range of TOF-MS MS1 scan in the SWATH analyses was adjusted to 400–1000 Da, divided into 100 overlapping variable precursor isolation windows, calculated using SCIEX variable windows calculator [22]. The MS1 scan was acquired with 250 ms accumulation time and followed by MS2 scans acquired in a looped product ion mode in 100–1500 m/z range, with collision energy calculated for +2- to +5-charged ions centered for each window, collision energy spread of 5 and accumulation time of 30 ms, which resulted in the 3.3 s cycle time. The acquired SWATH data were extracted with SWATH 2.0 MicroApp in PeakView 2.2 software (SCIEX) using the experimentally obtained spectral ion library and the following processing settings: up to 30 peptides per protein and up to 6 transitions per peptide, with peptide confidence threshold of 99% and 1% FDR rate, shared peptides were excluded, XIC extraction window set to 10 min and XIC width to 75 ppm. The retention times were recalibrated using 10 well performing peptides from iRT Kit. Representative MS/MS spectra and XICs for Rab proteins detected and then quantified based on single peptide of confidence threshold of 99% are shown in Supporting Information Figure S2 A–G. The processed peak areas were exported to MarkerView 1.2.1 software (SCIEX). The protein peak areas were then normalized using Total Area Sum approach and a t-Test was performed to compare CIC vs. CIM, CIC vs MIM and CIM vs MIM conditions. The fold changes were also calculated and Log transformed. The differences were considered as statistically significant when the p value was <0.05. The t-Test results and Log10(Fold changes) of proteins quantified in hMDM in CIC vs. CIM, CIM vs. MIM and CIM vs. MIM conditions are presented in Supporting Information_2 Tables S2 A–C, respectively. Normalized and averaged protein peak areas from the SWATH-MS analyses of CIC, CIM and MIM samples were used to compare protein expression patterns in bioinformatic analysis using Heatmapper [23] ([http://www.heatmapper.ca/expression/; accessed on 10.03.2021] data are presented in the Table S3, Supporting Information_2). The DDA and SWATH mass spectrometry proteomics data from the presented study have been deposited to the ProteomeXchange Consortium [24] via the PRIDE partner repository [25] with the dataset identifier PXD023291.
LC-MRM quantification:
Samples from seven human donors included in this study (D160, D164, D172, D222, D410, D421, D423) as well as BSA-spiked whole cell lysate digests for method validation were analyzed by reverse phase ultra-high-performance LC-MS/MS technique using a Nexera X2 UHPLC (Shimadzu, Kyoto, Japan) coupled with a 6500 QTRAP mass spectrometer (SCIEX) and controlled by Analyst software (SCIEX). Samples were kept at 8 °C and 2, 4, and 6-μL-volumes were injected via a SIL-30AC Autosampler (Shimadzu) into the Phenomenex reverse phase HPLC Omega (1.6 μm, PS C18, 100 × 2.1mm, 100 Å) column with SecurityGuard™ ULTRA Cartridge. The peptide mixtures were separated in the column with a solvent A (water containing 0.1% (v/v) formic acid) and a solvent B (ACN containing 0.1% (v/v) formic acid) (both LC-MS grade from Honeywell Inc.) using a linear gradient of 5% to 50% solvent B in 25 min, then increased to 95% in 3 min and followed by column re-equilibration with 5% solvent B for 10 min by the end of gradient at a flow rate of 0.2 mL/min. The eluting peptides were ionized by ESI in TurboV Ion Source (SCIEX) at a positive 5 kV spray voltage, 30 psi curtain gas, 50 psi for both ion source gas 1 and 2, 450 °C temperature, and then analyzed in multiple reaction monitoring (MRM) mode of acquisition. The MRM assays for quantification of the proteins selected in this study were developed by combining the in silico theoretical transition prediction and their empirical verification using the pooled donors’ hMDM sample. The pooled sample was comprised of aliquots of all the investigated donors’ hMDM samples of the CIC, CIM and MIM conditions. The use of combined samples allowed for a broad observation of peptides and transitions that would be high-responding in our system in the studied biological matrix. Following terminology adopted by proteomics literature, we used the term “high-responding” to describe those peptides that were suitable for MRM quantification and “non-responding” to describe those peptides that were not suitable for MRM quantification [26]. According to recommendations found in published literature, our MRM-based protein quantification used at least two peptides per protein and at least two transitions per peptide. Compound parameters for initial measurement of actin and BSA peptides for validation purposes were set to: de-clustering potential (DP) 80, collision energy (CE) 35, entrance potential (EP) 10 and cell exit potential (CXP): 13. We used Peptide Cutter tool from Expasy [27] for preselection of actin peptides for MRM development (Table S4, Supporting Information_2). In order to speed up the generation of working MRM assays to measure the investigated proteins in the samples from particular donors, save analyzed hMDM material and limit experimental verification efforts, we utilized Skyline software [28] for generation of their theoretical MRM transitions. Skyline settings for the peptides included peptides with 6 to 25 amino acids and Trypsin enzyme [KR|P] with a maximum of three missed cleavages. Peptides were narrowed down using library spectra from Human NIST and PRIDE libraries. The peptides were ranked by picked intensity. Under structural modifications, cysteine carbamidomethylation was selected. Canonical FASTA formatted sequences for proteins were obtained from UniProt [21]. Skyline settings for transitions used monoisotopic precursor and product masses. For predicted collision energy, ABI 5500 QTRAP was used with ABI de-clustering potential. For filtering the peptides, +2 and +3-charged precursor ions, +1 and +2-charged y and b product ions, from ion 1 to last ion were chosen. Special ions included N-terminal proline. The mass match tolerance for ions was set at 1 m/z. Product ions were chosen from library spectrum available using the most intense ions from filtered ion charges and types. A range of 300 m/z to 1500 m/z with a method match tolerance charge of 0.055 m/z was chosen for the instrument. The theoretical transitions were then experimentally verified. The final compound parameters for investigated protein peptide transitions included 10 V entrance potential (EP), 13 V collision cell exit potential (CXP), high collision gas (CAD), and 25ms dwell time. DP and CE were individually optimized for each of the precursor ions by Skyline software as described above. The transitions for each precursor ion are in Table S5 included in Supporting Information_2. The Skyline software was also used for extraction of the peak areas from the resulting LC-MS/MS raw data. The functions available in Excel package (Microsoft Inc.) were then used for calculation of peptides’ peak areas as a sum of peak areas of MRM transitions of their corresponding ions. Then, the peptide areas were summed to give peak areas for their corresponding proteins. The Log2 transformed peak areas of the investigated proteins in each of the studied conditions (CIC, CIM, and MIM) and concentrations (2, 4, or 6 μg injection) were compared using a two-sample unequal variance T-test. The differences were considered as statistically significant when the p value was <0.05. The MRM dataset from the presented study was submitted to PASSEL repository (PeptideAtlas SRM Experiment Library) [29] and is available under the accession number PASS01591.
RNA isolation and RNAseq
To isolate RNA, media was removed from each well, cells washed with ice-cold DPBS, and attached hMDMs were dissolved in Trizol (Invitrogen, Carlsbad, CA). MDM-Trizol extracts were transferred to tubes and stored at −80°C. Once all samples were collected, MDM extracts were thawed, chloroform was added to each tube, shaken and then allowed to rest at room temperature for 10 minutes before being separated by centrifugation. The aqueous phase was transferred to a new tube and RNA was precipitated by adding isopropyl alcohol, by incubating for 20 minutes at −20°C, and RNA recovered by centrifugation at 12,000 G at 4° C for 10 min. The resulting pellet was dried and dissolved in RNAse-free water. RNA was cleaned using the RNAeasy clean-up kit (Qiagen, Germantown, MD), quantified by 260 nm absorbance, checked for purity by absorbance at 230 and 280 nm, and intactness assessed on a Fragment Analyzer (Agilent Technologies, Ankeny, IA). A DNA library was constructed from the RNA using TrueSeq mRNA sample prep V2 (Illumina, San Diego, CA) and Next Generation Sequencing performed on the Illumina NextSeq550.
Bioinformatic and statistical analysis of gene expression
The fastq format reads were trimmed by fqtirm tool [30] to remove adapters, terminal unknown bases (Ns) and low quality 3’ regions (Phred score < 30). The trimmed fastq files were then processed utilizing STAR [31] as the aligner and RSEM [32] as the tool for annotation and quantification at both gene and isoform levels. The trimmed fastq files were mapped to human reference genome. Transcripts Per Kilobase Million (TPM) was calculated [33] and these values imported into the Partek Flow genomic analysis software package (Partek Inc., St. Louis, MO) for expression calculation and ANOVA analysis.
Results
Technical analysis of SWATH-MS data
Primary objective of this discovery study based on full unbiased quantitative profiling was to get insight into the extend of Meth exposure has effect on intracellular processes of already HIV-1 infected primary human macrophages. We used primary human monocytes differentiated to macrophages (hMDM) a critical component of the innate immune system. We choose to use SWATH-MS label free proteomic platform for full unbiased quantification. As presented in Figure 1, our experimental design sets HIV-1 infected hMDM (with no Meth treatment) as control (CIC) while experimental settings were when Meth was added after viral infection (CIM) or before and after infection (MIM). We used four biological replicates – four donors of monocytes – and five injections of each sample as technical replicates. The hMDM reference spectral library for SWATH-MS data extraction was generated experimentally with the use of DDA mass spectrometry (list of detected proteins/peptides is in Table S1 A and B in Supporting Information_2). We used non-fractionated trypsin digest of pooled CIC, CIM and MIM samples from four donors to generate a library that contained 1241 proteins identified with 99% confidence at the 1% critical false discovery rate (FDR) resulting from global FDR from fit analysis. This reference spectral library was applied for targeted data extraction from SWATH analyses of separately pooled for CIC, CIM and MIM conditions samples from the same four donors as for DDA analysis. The SWATH data processing with FDR analysis, enabled us to detect and quantify 1034 proteins based on one or more proteotypic peptides (Tables 2 A-C, Supporting Information_2).
Bioinformatic analyses of SWATH-MS data
SWATH-MS as full unbiased quantification of proteins in biological system provides a wide breath of changes occurring at the time of sample collection from manipulated biological system. To unravel biological/molecular consequences of these changes are possible when bioinformatic analyses are employed. To exploit information provided by our SWATH-MS analysis we performed several rounds of bioinformatic investigations.
Heatmapper
At first, we created a heat map for visual representation of changes in expression of proteins detected and quantified by SWATH-MS experiment in hMDM under different Meth exposed conditions. We applied on-line available Heatmapper Expression tool [23] with the following settings: Average Linkage clustering method and Euclidean distance measurement method, and, used normalized and averaged peak areas from SWATH analyses of the CIC, CIM and MIM samples as the input data. The resulting heat map is presented in Figure S3 (Supporting Information).
PANTHER
The PANTHER [34] [(https://pantherdb.org)] is based on ontology analysis providing families of genes/proteins that is used to classify and identify the function of gene products. Results of our PANTHER analyses are presented in Figures S4 and S5 (Supporting Information). Based on GO-Slim Molecular Function, the most enriched functions due to Meth treatment are catalytic activity (GO:0003824) and interaction (GO:0005488). GO Slim Biological Process shows that majority of differential proteins are associated with cellular processes (GO:0009987) and metabolic processes (GO:0008152). Results of PANTHER analyses do not further specify which particular pathways are affected. To answer such questions, we performed two other bioinformatic analyses using Reactome [35] (https://reactome.org/) and Cytoscape [36] [(https://cytoscape.org/)].
Reactome
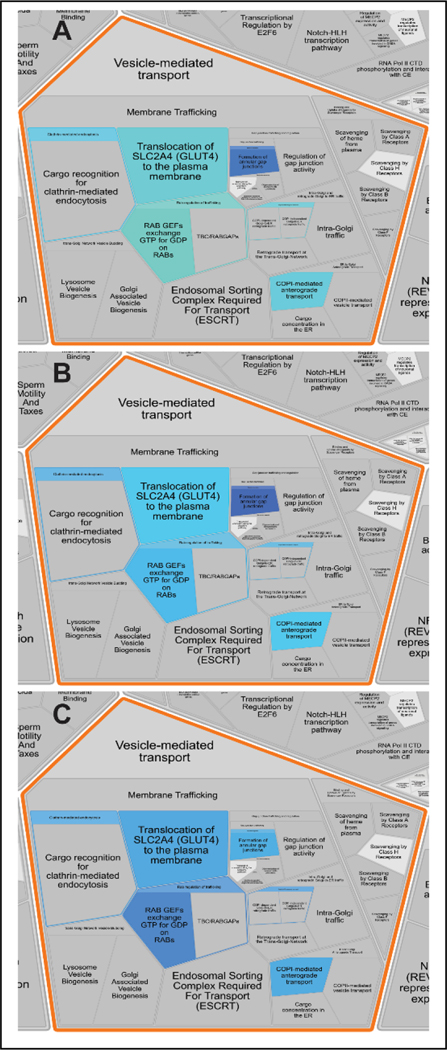
We investigated the relative expression of proteins detected and quantified in our SWATH-MS experiment in connection to the molecular pathways that they are involved in. The Log10(Fold Change) values for the proteins in CIC vs CIM, CIC vs MIM and CIM vs MIM conditions of hMDM (Table 2A, 2B and 2C in Supporting Information_2, respectively) were loaded and analyzed using “Analyze gene list” tool in Reactome Pathway Database version 76 [https://reactome.org/]. The analysis features “Project to humans” and “Include Interactors” were selected. There were 1894 pathways, where at least one identifier from our dataset was assigned, and, 199 pathways were statistically significantly (p<0.05) over-represented in our dataset (Table S6, Supporting Information_2). The detected proteins were mainly related to the pathways of: immune system (e.g. innate and adaptive immune system), metabolism (e.g. TCA cycle and respiratory electron transport; metabolism of lipids, amino acids, RNA and proteins), vesicle-mediated transport, DNA replication, but also those related to: cell cycle, HIV infection, signal transduction, transport of small molecules, localization, apoptosis or response to stimuli. The pathways overrepresented in our dataset in CIC vs CIM, CIC vs MIM and CIM vs MIM hMDM are depicted as Reactfoam (Voronoi) diagrams in Figures S6, S7 and S8 (Supporting Information), respectively. Figure 2 shows part of the Reactfoam diagrams focused on Vesicle-mediated transport.
Figure 2. Reactome analysis of SWATH-MS data – sub-pathways included in vesicle mediated transport pathway.
The presented magnification of Reactfoam (Voronoi diagrams) show overrepresentation of sub-pathways included in vesicle mediated transport molecular pathway in (A) CIC vs CIM, (B) CIC v MIM and (C) CIC vs MIM comparison. The depicted color range spans from yellow to blue, for the up-regulation to down-regulation, respectively. The more intense color shows degree of changes (the higher as the color is more intense). Full Reactfoam diagrams for each studied condition comparisons are in the Supporting Information (Figures S6, S7 and S8). The diagrams coverage shows consistent overrepresentation of the same molecular pathways for all three conditions, however the degree of the overrepresentation changes between the studied conditions. Vesicle-mediated transport which includes Rab proteins is one of most affected pathways by Meth exposure regardless exposure was for shorter (CIM) or longer (MIM) time.
Cytoscape
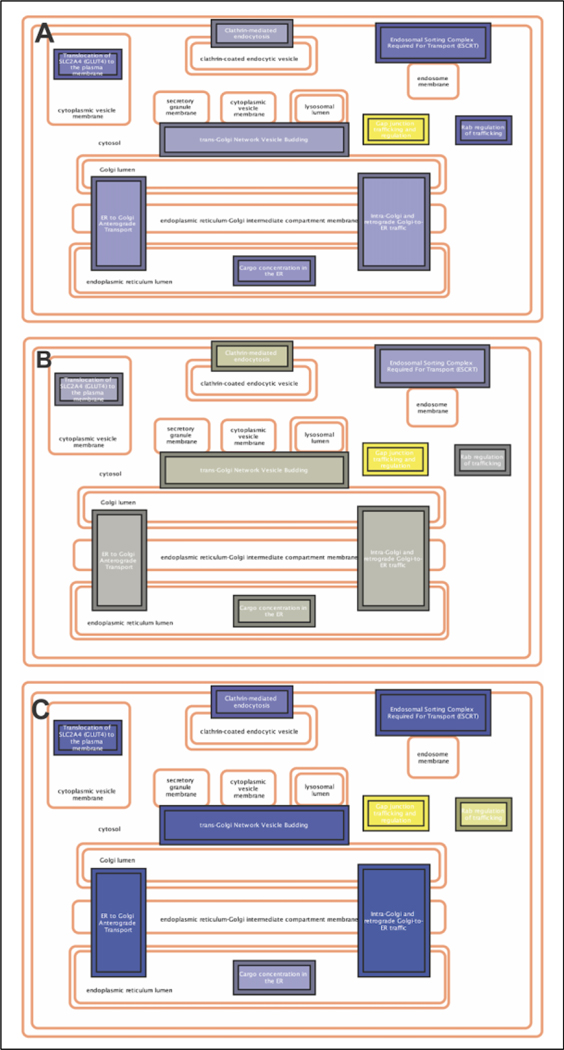
Finally, we employed Cytoscape 3.8.2 with ReactomeFIVIz App (accessed April 6, 2021) to perform pathway enrichment analysis. The protein names were transformed to corresponding gene names and used as an input data for pathway enrichment analysis. The selected significantly enriched pathways were then overlaid with the corresponding to these proteins Log10(Fold Change) values for CIC vs CIM, CIC vs MIM and CIM vs MIM conditions. Performed analysis showed enrichment of 349 pathways with statistical significance of p<0.05 (Table S7, Supporting Information_2). As expected, the innate immunity system represented by 1141 proteins pathway enrichment was highly significant (p=1.11E-16, FDR=5E-15), to which 211 proteins were classified from our dataset. The enrichment of pathway(s) associated with Innate Immunity System was expected because our biological system was hMDM, a central part of this system. This was followed with enrichment of Adaptive Immune System (p=1.8896E-9, FDR=4.9807E-4, 828 proteins in the pathway of which 96 proteins in our dataset). This is closely connected to innate immunity system. Another enriched pathways included: Membrane Trafficking (p=6.9482E-5, FDR=5.6688E-8, 611 proteins in the pathway of which 92 proteins in our dataset), Cytokine Signaling in Immune System (p=4.8723E-5, FDR=3.8978E-4, 704 proteins in the pathway of which 85 proteins in our dataset) and Respiratory Electron Transport (p=7.3748E-6, FDR=8.1123E-5, 101 proteins in the pathway of which 23 proteins in our dataset) (Table S7, Supporting Information_2). We expected that the latter pathway would be enriched based on our previous studies of cytokine production by THP-1 cells exposed to Meth [8]. Changes in intracellular trafficking was our special interest since this system is involved in HIV-1 infection as well as processing of nanoparticles used for long term anti-retroviral treatment [37]. The comparison of the protein’s expression in hMDM detected in our dataset for CIC vs CIM, CIC vs MIM and CIM vs MIM conditions shows their dynamic changes upon Meth-treatment and HIV infection (Figure 3A–C, respectively). Closer insight into this pathway shows that Ras-related Rab proteins constitute important part of this pathway, especially in the sub-pathway of RAB regulation of trafficking (Figures S9, S10 and S11 in Supporting Information, for CIC vs CIM, CIC vs MIM and CIM vs MIM, respectively). Figure 4 shows effect of Meth on expression of Rab proteins within Membrane Trafficking Pathway showing communication between ER and Golgi compartments which are regulated by Rab-1A especially in Intra-Golgi and retrograde Golgi-to-ER traffic and ER Golgi Anterograde Transport molecular pathways (Table S7, Supporting Information_2)
Figure 3. Molecular pathways within Membrane Trafficking Pathway.
Presented pathway was significantly enriched in our dataset, the colored sub-pathways depict the ones, that were found in our dataset as indicated by the results of Pathway Enrichment Analysis performed using Cytoscape 3.8.3 with Reactome FIVIz app in CIC vs CIM (A), CIC vs MIM (B) and CIM v MIM (C).The bluest color indicates the up-regulation, while the most yellow color shows down-regulation of the specific pathway. The genes of proteins detected in our SWATH-MS experiment, that are involved in this pathway included 92 proteins out of which 19 were Ras-related Rab proteins group contributing to the Membrane Trafficking Pathway: RAB31, RAB35, RAB5B, RAB5C, RAB6A, RAB5A, RAB1A, RAB1B, RAB21, RAB10, RAB14, RAB13, RAB18, RAB8A, RAB8B, RAB7B, RAB7A, RAB11B, RAB9A (Table S7, Supporting Information_2).
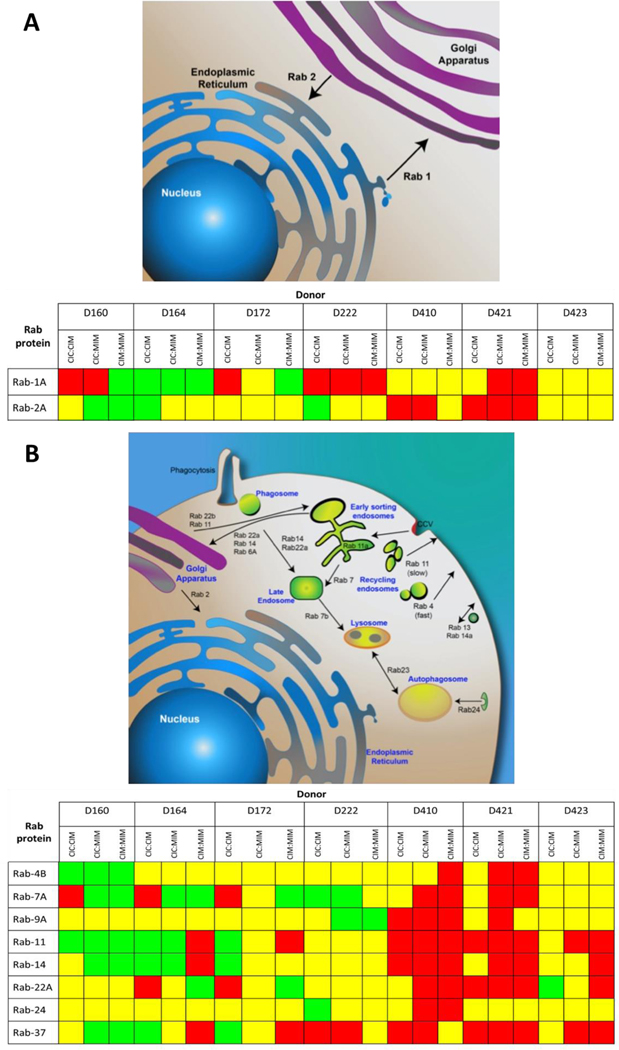
Figure 4. Statistically significant changes in Rab proteins in HIV-1-infected hMDM upon exposure to Meth.
The summary depicts only statistically significant changes in expression of Rab-1A, −2A, −4B, −7A, −9A, −11, −14, −22A, −24, and −37 between CIC, CIM and MIM conditions based on 2 μg-injection MRM measurements. Red squares represent downregulation, green upregulation, while yellow shows no significant differences. While the mechanism of the effect of Meth on the expression of Rab proteins is not known, it shows substantial variability of responses between donors of monocytes at the time of performing this experiment as depicted in Figure 1.
MRM Efficiency
To assess the linearity of MRM quantification we spiked-in a tryptic digest of bovine serum albumin (BSA) as an internal standard. Table S8 in Supporting Information shows BSA-derived peptides and their MS/MS transitions used to establish linearity curve of the concentration of this protein spiked in highly complex mixture of peptides obtained from whole cell lysates of hMDM, human primary T cells, and Jurkat cells. Figure S12, in the Supporting Information, presents linearity of BSA peptides spiked into a complex matrix. No interferences were found within a range of 1 to 1000 fmol/μL in the presence of the biological matrix as measured by R2; however, as presented in Table S8, out of ten BSA peptides, three of them did not provide a signal required for reliable quantification (r2 < 0.5). We designated these as non-responding peptides and eliminated them from the MRM method.
Actin expression in hMDM under Meth exposure
Actin is one of the most abundant intracellular proteins, and as such, is commonly used as a normalization control in analytical investigations such as western-blot analysis and/or MS. However, its assembly inside the cell is a dynamic process [38], and it is represented by several entries in the UniProt database. For example, Ravenscroft et al. (29) quantified differences in expression of common striated actin, skeletal muscle α-actin, and cardiac α-actin in human, sheep, and pig tissues, exploiting differences in sequences. Their MRM results showed substantial variation [39]. In this study we used the cytoskeletal 1 form (UniProt # P60709) (ACTB) of actin. We performed in silico tryptic digestion of actin and selected 10 peptides to perform MRM analysis. Results of actin cleavage by trypsin using Peptide Cutter tool from Expasy are presented in the Supporting Information Table S4, while peptides and transitions used for MRM are summarized in Table S9. Figure S13 showing a representative chromatogram of MRM analysis of 10 peptides ranging from low to high intensity is presented in Supporting Information.
Overall, MRM quantification showed that actin levels decreased in hMDM upon exposure to Meth irrespective of the time of its addition (before or after HIV infection). This effect was statistically significant in the cell lysates of all seven donors as presented in Figure S14, Supporting Information). Moreover, the effect was reproducible whether 2, 4, or 6 μg of peptides were loaded onto an RP-LC column. However, in some instances the 6 μg technical replicates (injections) resulted in increased variation. Therefore, in future analyses we used only 2 and 4 μg per injection. Important to note is that this observed effect is consistent across all seven donors of hMDM.
The effect of Meth on reducing intracellular pool of actin in hMDM is novel observation and may have several implications. Lower levels of actin may lead to alterations in biological functions of the cytoskeleton. At this time, we do not know the exact biological effect; however, some literature reports suggest that phagocytosis, which is one of the major functions of MP, might be altered in a negative manner by quantitative alterations in actin [40].
Effect of HIV and Meth on expression of Rab (Ras-related proteins) regulators – MRM investigation
A fraction of SWATH-MS extracted data shown in Table 1 represent results for actin and set of Ras-related (Rab) proteins. Actin and Rab proteins are followed up by targeted quantification using MRM platform. Heat map presenting changes in expression of proteins detected and quantified in nanoLC-SWATH-MS experiment between the CIC, CIM and MIM conditions is shown as Figure S3 in supplemental material. Results of PANTHER [34] analyses including Molecular Function and Biological process are shown as Figures S4 and S5, respectively.
Table 1.
Expression differences in actin cytoplasmic 1 and Ras-related proteins detected and quantified in hMDM in CIC, CIM and MIM conditions in SWATH experiment. The difference was considered as statistically significant when the p-value<0.05 and are marked in a bold text.
| Protein | CIC vs CIM | CIM vs MIM | CIC vs MIM | ||||
|---|---|---|---|---|---|---|---|
| UniProt accession number | Name | p-value | Log10(Fold Change) | p-value | Log10(Fold Change) | p-value | Log10(Fold Change) |
| sp|P60709|ACTB_HUMAN | Actin, cytoplasmic 1 | 3.09E-03 | −2.99E-01 | 4.37E-02 | 1.62E-01 | 5.49E-02 | −1.37E-01 |
| sp|P61026|RAB10_HUMAN | Ras-related protein Rab-10 | 3.49E-03 | 2.21E-01 | 2.43E-03 | −1.85E-01 | 4.93E-01 | 3.57E-02 |
| sp|Q15907|RB11B_HUMAN | Ras-related protein Rab-11B | 5.00E-01 | 4.54E-02 | 4.12E-01 | −6.13E-02 | 7.60E-01 | −1.58E-02 |
| sp|P51153|RAB13_HUMAN | Ras-related protein Rab-13 | 3.82E-03 | 3.76E-01 | 5.80E-01 | −1.01E-01 | 3.49E-02 | 2.75E-01 |
| sp|P61106|RAB14_HUMAN | Ras-related protein Rab-14 | 8.42E-01 | 1.29E-02 | 9.75E-01 | −1.51E-03 | 8.05E-01 | 1.14E-02 |
| sp|Q9NP72|RAB18_HUMAN | Ras-related protein Rab-18 | 1.93E-01 | −1.16E-01 | 6.24E-02 | −1.36E-01 | 3.94E-03 | −2.52E-01 |
| sp|P62820|RAB1A_HUMAN | Ras-related protein Rab-1A | 1.05E-01 | −6.01E-02 | 4.91E-02 | −6.43E-02 | 3.90E-04 | −1.24E-01 |
| sp|Q9H0U4|RAB1B_HUMAN | Ras-related protein Rab-1B | 2.47E-01 | 1.40E-01 | 2.90E-04 | −5.63E-01 | 6.00E-04 | −4.24E-01 |
| sp|Q9UL25|RAB21_HUMAN | Ras-related protein Rab-21 | 5.57E-01 | 1.55E-01 | 4.15E-01 | −1.77E-01 | 8.73E-01 | −2.18E-02 |
| sp|P61019|RAB2A_HUMAN | Ras-related protein Rab-2A | 3.15E-01 | −8.44E-02 | 5.01E-01 | −5.14E-02 | 7.60E-02 | −1.36E-01 |
| sp|Q13636|RAB31_HUMAN | Ras-related protein Rab-31 | 4.63E-01 | 8.83E-02 | 9.03E-01 | 1.66E-02 | 4.61E-01 | 1.05E-01 |
| sp|Q15286|RAB35_HUMAN | Ras-related protein Rab-35 | 9.83E-02 | 2.28E-01 | 7.29E-03 | −3.15E-01 | 2.34E-01 | −8.72E-02 |
| sp|P20339|RAB5A_HUMAN | Ras-related protein Rab-5A | 7.82E-03 | 9.22E-02 | 4.92E-01 | −1.80E-02 | 6.28E-03 | 7.41E-02 |
| sp|P61020|RAB5B_HUMAN | Ras-related protein Rab-5B | 1.99E-02 | 3.23E-01 | 5.56E-03 | −3.54E-01 | 6.41E-01 | −3.02E-02 |
| sp|P51148|RAB5C_HUMAN | Ras-related protein Rab-5C | 2.75E-02 | 7.18E-02 | 2.76E-02 | −7.30E-02 | 9.54E-01 | −1.18E-03 |
| sp|P20340|RAB6A_HUMAN | Ras-related protein Rab-6A | 4.56E-01 | −1.11E-01 | 2.91E-01 | −1.62E-01 | 9.76E-02 | −2.72E-01 |
| sp|Q53S08|RAB6D_HUMAN | Ras-related protein Rab-6D | 4.75E-02 | 2.12E-01 | 1.12E-03 | −3.29E-01 | 9.68E-02 | −1.17E-01 |
| sp|P51149|RAB7A_HUMAN | Ras-related protein Rab-7a | 5.56E-01 | −2.67E-02 | 1.30E-01 | 3.87E-02 | 7.77E-01 | 1.20E-02 |
| sp|Q96AH8|RAB7B_HUMAN | Ras-related protein Rab-7b | 3.91E-01 | 1.19E-01 | 1.79E-01 | 1.76E-01 | 8.24E-02 | 2.95E-01 |
| sp|P61006|RAB8A_HUMAN | Ras-related protein Rab-8A | 1.39E-02 | 2.62E-01 | 3.35E-03 | −3.08E-01 | 5.26E-01 | −4.55E-02 |
| sp|Q92930|RAB8B_HUMAN | Ras-related protein Rab-8B | 4.87E-02 | 3.24E-01 | 3.10E-04 | −5.10E-01 | 5.28E-02 | −1.86E-01 |
The Rab family of proteins is a member of the Ras superfamily of small G proteins [41]. Many Rab proteins are ubiquitously expressed; however, the expression of some is restricted to specific cell types. More importantly Rab proteins also show some degree of promiscuity by playing diverse functions when expressed in different cell types. These regulatory proteins have been studied as regulators in various models and conditions as well as during HIV-1 infection [42–46]; however, much less is known of their function in macrophages or whether they are altered by exposure to drugs of abuse, such as Meth.
Quantification of Rab proteins by MS can be challenging. For example, Rab-11A (UniProt # P62491) and Rab-11B (UniProt # Q15907) share a majority of their amino acid sequence with differences towards the C-terminus. Unfortunately, none of the peptides derived from that region provide high-responding peptides, which appear useful for MRM based quantification. Thus, if there are differences in their biological functions, it would be difficult to differentiate them. Thus, all our quantification measurements include both Rab-11A and Rab-11B proteins combined and should be interpreted as such. Since Rab proteins cooperate and play sequential roles in many functions, they were analyzed for patterns of expression changes. Our approach was to analyze the MRM data of each Rab protein individually to identify any patterns of change due to the exposure of Meth during HIV infection. Moreover, since CIM represents a short(er) term of exposure and MIM represents a long(er) time of exposure to Meth, we analyzed patterns of responses among seven donors.
Overall, the patterns of responses to Meth were reproducible whether 2 μg or 4 μg of peptides were used for MRM, and in the most instances, differences were statistically significant. Pattern A shows a drop in the levels of Rab-1A (Figure S15, Supporting Information, CIM) and rebounded levels in MIM cultures that had long(er) term exposure to Meth. We observed similar patterns of Rab-2A expression (Figure S16, Supporting Information). Both, Rab-1A and Rab-2A play similar roles in regulating vesicular protein transport from the endoplasmic reticulum to the Golgi compartment. Interestingly, when we compared the expression of Rab-1A (Figure S15, Supporting Information) and Rab-2A (Figure S16, Supporting Information) between same donors at the same time points of exposure to Meth, we posit that these two Rab proteins are working in sequence rather than in tandem. This further suggests that their roles in cellular function might be slightly different.
The majority of donors did not exhibit statistically different levels of Rab-4B upon Meth treatment (Figure S17, Supporting Information). Only one donor responded to Meth treatment with a significant increase in expression, while two other donors showed decreases after exposure to Meth. Four other donors did not show any significant changes. Similar to Rab-1A, Rab-7A exhibited variable responses to Meth exposure among the donors (Figure S18, Supporting Information). Two donors showed decreases only in MIM, two showed decreases in CIM, but rebounded levels in MIM, two showed increases in both CIM and MIM, and one was not responsive.
Levels of Rab-9A (Figure S19, Supporting Information) were similar in the hMDMs in four out of seven donors. In the remaining three cases, the observed changes were not consistently significant. Similarly, significant downregulation of Rab-4B was observed in hMDM from one out of seven donors (see Figure S17, Supporting Information). The response of Rab-22A to Meth treatment was similar to Rab-4B, as it was generally decreased upon Meth exposure, but the effect was variable across the donors (Figure S20, Supporting Information).
Quantification of Rab-24 in Meth-treated and HIV-1-infected hMDM indicated that Meth does not appear to substantially alter Rab-24 levels (Figure S21, Supporting Information); No effect of Meth was seen in three out of seven donors. In the other four donors, the differences behaved in an erratic manner as presented in Figure S21, Supporting Information. This might be explained by a low level of differences and/or higher standard deviation of MRM measurements despite the use of five replicates for each condition. hMDM exposed to Meth from some donors show decreases in levels of Rab-37 (Figure S22, Supporting Information) after long(er) exposure to Meth (CIC compared to MIM, see Figure 1), while in samples from other donors, no significant effect of Meth has been observed.
RNA gene expression levels of characterized proteins
Changes in protein levels can occur through a number of processes including alterations in transcription, translation, and degradation. To examine potential alterations at the transcriptional levels, RNA was purified from the cells of six of the donors at harvest and subjected to next-generation sequencing. Despite the differences observed by SWATH-MS and MRM, the expression of RAB1A, 2A, 4A, 7A, 9A, 11A, 11B, 14, 22A, and 24 mRNA was not observed to significantly vary between conditions (Figure S23, Supporting Information). Similarly, there was no difference in the expression of the ACTB gene, which encodes the protein analyzed here.
Discussion
Investigation of intracellular processes on global scale require reliable and robust methods of protein quantification and data analysis. Four factors are in play: high complexity of samples, dynamic changes in time, diversity of responses within population of primary cells and degree of changes which can be relatively small, yet they may play significant role in regulatory processes. This is in particular important when regulatory or signaling processes are being investigated. As much as analytical platforms are very helpful, in many instances changes which are small can be missed. In such instances, low throughput, nevertheless more precise, targeted proteomic profiling is necessary.
Full unbiased proteomic profiling is a holistic approach to unravel complexity of interaction of components in convoluted and highly dynamic biological system under investigation. Though, it is expected that such profiling approach will provide information or new connection between system’s elements that are not available at the time of performing experimentation. Thus, the specific outcome(s) of unbiased profiling is mostly unknown, however, we expect that a number of changes/alterations in metabolic pathways will be outlined. Once analytical data are collected biological meaning needs to be derived by means of bioinformatic analyses. In our current report we have utilized several bioinformatic tools (Heatmapper, Panther, Reactome and Cytoscape) to analyze SWATH-MS data. As expected, by inserting recorded changes in protein expression into bioinformatic tools directed us to changes of several pathways. In the second follow-up part of full unbiased SWATH-MS investigation we used MRM-based quantification focusing on changes in Ras related proteins expression as a validation step of SWATH-MS [47].
The Heatmapper analysis (Figure S3, Supporting Information) did not show distinct patterns grouping specific sets of proteins. Nevertheless, CIM comparison, also considered as short-term Meth exposure, has more suppressive effect on hMDM than longer exposure (MIM). This observation, qualitative rather than quantitative per se, is consistent with our previous data [8] indicating that macrophages respond quickly to insult of Meth exposure and then adjust to environment containing this toxin. This is also consistent with our hypothesis that, as previously measured by cytokine release [8], Meth quickly triggers multiple responses rather than initiate a single signaling pathway via putative receptor. Thus, we postulate that Meth exposure generates a cascade of metabolic intertwining changes at many levels. Our bioinformatic data presented here support such view. There are also some indications (our unpublished data) that some effects might be mediated by epigenetic mechanisms. Interestingly, we did not find evidence of changes in DNA methylation in hMDM upon exposure to Meth (unpublished data). This direction of investigations remains to be explored.
As expected, the Reactome analysis presented in Figures S6–S8, show major overrepresentation of pathways within Immune System, Metabolism, Metabolism of Proteins and, more importantly, Vesicle-mediated Transport categories. The latter group of metabolic pathways contains regulation of Rab proteins which play important role in immune system including their role during HIV-1 infection of macrophage which is one of two major targets of this retroviral infection [37, 48–51]. Ras-related Rab proteins are master regulators of large number of intracellular processes [42, 50] such as vesicle formation, vesicle and organelle motility, regulation to membrane transport, ER to Golgi transport including delivery of post-Golgi vesicles to the plasma membrane. The structural heterogeneity shown by Rab effectors suggests that Rab proteins are highly specialized factor which activities are exclusively designed for individual organelles and transport systems [52]. These proteins are involved in autophagy is another example of an intercellular process which is regulated by Rab proteins [53]. Intracellular trafficking is also exploited by HIV-1 for transport viral proteins to the cell surface. As such they are highly significant to all intracellular processes including viral infections [37, 51]. Our studies also indicate that modulation of Rab-regulated endolysosomal pathways by antiretroviral nanoparticles provides a strategic approach to contain HIV-1 infection [49].
Cytoscape is an open source software platform providing a basic set of features for data integration, analysis, and visualization and integrating these networks with annotations, gene expression profiles and other state data [54]. Cytoscape analysis showed enrichment of molecular pathways within Membrane Trafficking Pathway in particular in communication between ER and Golgi. This process, as shown in Figure 4A is regulated by Rab 1 and Rab 2 proteins again indicating that Meth at concentrations correlated with levels after taking this drug has a profound effect on intracellular trafficking without killing cells.
To our surprise, exposure of macrophages to Meth led to a consistent across all donors to reduction of intracellular pool of actin. This change in protein abundance occurred without alteration the level of RNA encoding actin and mechanism of this change is not known. This change in actin protein amount is novel observation and may have substantial consequences on how some intracellular processes work in macrophages. For example, Aslanyan et. al [40] investigated the effect of Meth on the role of actin as a key protein regulator of actin polymerization during the formation of the phagocytic pockets during phagocytosis of cryptococcal cells.
Thus, this study is the first attempt to quantify members of Rab proteins in HIV-1-infected hMDM exposed to Meth. As presented in Figure 2, Rab 1 and 2 are responsible for communication between endoplasmic reticulum (ER) and Golgi apparatus [55]. Other Rab proteins investigated in this study participate in endosomal, lysosomal and authophagosomal regulation [55], all playing role in HIV-1 infection. One drawback of MRM-based approach is that the similarities (identity) of amino acid sequences containing useable peptides for MRM quantification make it very challenging to distinguish similar proteins or different proteoforms. We experienced this challenge while quantifying some members of Rab family.
One of great challenges in global profiling using primary human material, i.e. ex vivo cells of the immune system is variability of responses. Macrophages are polarized creating classically activated M1 and alternatively activated M2 subsets and constitute extreme phenotypic heterogeneity in physiological and pathological conditions. This polarization is a continuous process when macrophages acquire specialized functional phenotypes [56] in response to external stimuli such as bacterial, viral infections [57]. This is also reflected in variety of responses of human body exposed toxins such as Meth [58] as well in other models, i.e. non-human primates [59]. Therefore, a population of macrophages obtained from individual donors have different functional phenotypes thus responding differently at any time point [60]. When cells are harvested at specific time point of response, the diversity seems to be quite obvious and is reflected by experimental variability, in particular in high throughput profiling experiments. As a consequence, patterns of responses as well statistical significance might be borderline when results obtained from investigation of cells from various donors are considered. Last, but not least, quantification of Rab proteins in cells from seven donors of monocytes show great variety of individual responses, which is summarized for 2 μg injections in Figure 4. hMDM from donors 410 and 421 show downregulation most of the Rab proteins, while hMDM from donors D160 and D 164 show majority of the Rab proteins to be up-regulated. Cells from other donors show a mixture of response close to equal up- down- or no change response. The fact that Meth affects expression of Rab proteins may indicate that there is unknown, at this time, molecular mechanism(s) underlying observed changes. Undoubtfully, cells from different donors react, as expected, differently to the same insult, which is clearly shown in Figure 4 including insult from HIV-1 infection [7]. This aspect of high-throughput studies using primary cells has been also discussed as part of our previous study [61].
We may expect that, since cells from seven different donors respond somewhat differently in vivo situation, responses might also be variable. This observation has been reported previously [62]. Obtained protein patterns may be thus related to different time points in the cellular responses of different donors at the time of sample collection. Various processes would be dynamically modified leading to heterogeneity cell states in an entire population of macrophages in an HIV-1-infected individual under the insult of Meth. Nevertheless, such variability will pose challenges to biological interpretation of any analytics, whether MS-based or by other methods.
While the responses of the biological system might be profound with multifold increases/decreases of change, or small in quantity, both may represent significant biological effects. Thus, precision and accuracy of quantification is of the upmost importance to avoid false positive results. Obtaining proper results also benefits from data normalization, which can be highly challenging with samples such as plasma, urine or culture supernatants using profiling methods that quantify unlabeled proteins on a large scale (i.e. “shotgun” methods). As much as these types of experiments provide novel insights, their validation remains a challenge. For several decades, western blot analyses have been used for validation [63]. In some instances, spiked-in heavy peptides were used as a shotgun reference to serve as a validation of accuracy and precision of SWATH-MS data [64]. As much as spiking in external standards into complex biological sample helps with normalization for processing and instrumentation variability, it does not normalize for variation due to biological diversity (e.g. donor-to-donor variability). Exposure of living cells to various environmental or experimental conditions may have quite unpredictable quantitative effects on the proteome. This is much more profound in a situation where not all cells are affected by viral infection at the same time; thus, the ratio of infected to uninfected (control) cells varies from culture to culture.
Conclusions
Several important and biologically relevant conclusions can be drawn from presented studiy. While SWATH-MS based full unbiased proteomic profiling shows quantitative profiling and changes in biological system under investigation, MRM platform provides more precise and accurate differences. This is important in particular when regulatory, thus short-lived factors, are investigated.
Meth treatment of hMDM leads to suppression of the levels of intracellular actin. It is novel observation and similar across all cell’s donors used in this study. Decrease of the levels of intracellular actin shows consistent pattern.
Bioinformatic analysis showed that Meth exposure also leads to alteration of many intracellular metabolic pathways. Based on bioinformatic analyses of SWATH-MS data we observe that Meth exposure, whether applied before and after infection (MIM) or only after infection (CIM) leads to consistent alterations of the same cellular processes.
Several molecular pathways such as vesicle-mediated transport is more affected than others. Considering that Rab proteins support many intracellular processes it is expected that many phenotypic functions of hMDM will be affected. To further advance research in this area, more precise than SWATH-MS method of protein quantification is necessary. Because RNA gene expression data do not always correlate with the changes of proteins within the cells and global profiling proteomic methods might not be precise enough, we employed mass spectrometry-based MRM. We demonstrate here that MRM can be successfully used for quantification of small changes in concentrations of proteins of interest present in complex mixtures, such as whole cell lysate.
Our study also shows the first attempt to systematically quantify Rab proteins in primary macrophages infected with HIV-1 and treated with Meth. These proteins are regulators of many intercellular processes such as communication between ER and Golgi and intracellular trafficking.
We hypothesize that addition of Meth to already HIV-1 infected hMDM further alters these cells and will likely have broader impact on the function of the innate immune system. Because no changes were found at the RNA level, thus emphasis the need for protein quantification to discern changes across conditions is even more important. Taken together, presented data are fundamental for future studies, which should include experimental designs that consider collecting samples at various time points. Presented results form a foundation for future, more focused and mechanism-oriented studies.
Supplementary Material
Significance of the study.
Macrophages, key components of the innate immune system, play an important role in HIV-1 infection. Along with CD4-positive T cells, they are important targets of the virus and participate in viral transfer to different organs such as brain. There is a significant overlap between those individuals who are HIV infected and those who use drugs of abuse. Many of these drugs, such as Meth, can have toxic effects on macrophages and other cell types and further impair function of the immune system. Following SWATH-MS unbiased profiling we employed MRM protocol, the most sensitive mass spectrometry approach, to quantify changes in expression of a set of preselected proteins in HIV-1 infected macrophages exposed to Meth. We made the novel finding that Meth has an effect on the levels of actin in macrophages. We also describe a unique study in which Ras-related Rab proteins (Rab-1A, −2A, −4B, −7A, −9A, −11, −14, −22A, −24 and −37) were quantified using MRM. While the decrease of actin levels among donors was consistent, expression of the Rabs varies. The analytical methodology developed in this study along with bioinformatic analyses give a novel insight into the cellular pathology of macrophages resulting from viral infection and drugs of abuse at the molecular level, which can be further applied to investigate molecular mechanisms in macrophages.
Acknowledgements
The authors would like to thank Dr. Katarzyna Lech for her help in preparing graphics for Figure 1, Mr. Andrew Schissel for technical assistance, Dr. James Eudy (sequencing by the UNMC Genomics Core Facility, which receives partial support from the Nebraska Research Network In Functional Genomics NE-INBRE P20GM103427-14 and the Fred & Pamela Buffett Cancer Center - P30CA036727), and Drs. Peng Xiao, Meng Niu and Babu Guda (bioinformatic pipeline from the UNMC Bioinformatics and Systems Biology Core). The mass spectrometry analyses were performed at the University of Nebraska Medical Center Mass Spectrometry and Proteomics Core Facility, which is administrated through the Office of the Vice Chancellor for Research and supported by state funds from the Nebraska Research Initiative (NRI).
Funding
Financial support was provided by National Institutes of Health grants: R01 DA043258 and P30 MH062261.
List of abbreviations:
- ACTB
actin, the cytoskeletal 1 form (UniProt # P60709)
- CAD
collision gas
- CE
collision energy
- CIC
a condition in which HIV-1 infection of macrophage only occurs without Meth treatment
- CIM
a condition in which macrophages were treated with Meth post-HIV-1 infection
- CXP
cell exit potential
- DIA
data-independent acquisition
- DDA
data-dependent acquisition
- DP
de-clustering potential
- DPBS
Dulbecco’s phosphate-buffered saline
- EP
entrance potential
- FASP
filter-aided sample preparation
- HIV
human immunodeficiency virus
- HIVADA
laboratory-adapted HIV-1 strain ADA
- hMDM
human monocyte-derived macrophages
- Meth
methamphetamine
- MIM
a condition in which the macrophages were Meth-treated before and after HIV-1 infection
- MP
mononuclear phagocyte
- Rab
Ras-related protein
- RP
reversed phase
- SWATH
Sequential Windowed Acquisition of All Theoretical Fragment Ion Mass Spectra
Footnotes
Declarations:
Ethics approval and consent to participate
Not applicable, no human subjects or animals used in this study.
Consent for publication
All co-authors read and agreed with data and data presentation to be published.
Associated data
The MRM datasets generated and analyzed during the current study are available in the PeptideAtlas PASSEL repository, which is a member of ProteomeXChange Consortium dedicated for MRM data, with accession number PASS01591 and can be accessed using the link: https://db.systemsbiology.net/sbeams/cgi/PeptideAtlas/PASS_View?identifier=PASS01591. The DDA and SWAH datasets were deposited to PRIDE repository, which is alo a member of ProteomeXChange Consortium, with accession number PXD023291. The RNAseq datasets have deposited in NCBI GEO, accession number GSE160323
Competing interests
The authors declare that they have no competing interest neither what may appear as conflict of interest.
References
- [1].Norris GT, Kipnis J, Immune cells and CNS physiology: Microglia and beyond. J Exp Med 2019, 216, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kierdorf K, Masuda T, Jordao MJC, Prinz M, Macrophages at CNS interfaces: ontogeny and function in health and disease. Nat Rev Neurosci 2019, 20, 547–562. [DOI] [PubMed] [Google Scholar]
- [3].Brecht ML, Stein J, Evans E, Murphy DA, Longshore D, Predictors of intention to change HIV sexual and injection risk behaviors among heterosexual methamphetamine-using offenders in drug treatment: a test of the AIDS Risk Reduction Model. J Behav Health Serv Res 2009, 36, 247–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nair MP, Saiyed ZM, Nair N, Gandhi NH, et al. , Methamphetamine enhances HIV-1 infectivity in monocyte derived dendritic cells. J Neuroimmune Pharmacol 2009, 4, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liang H, Wang X, Chen H, Song L, et al. , Methamphetamine enhances HIV infection of macrophages. Am J Pathol 2008, 172, 1617–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Talloczy Z, Martinez J, Joset D, Ray Y, et al. , Methamphetamine inhibits antigen processing, presentation, and phagocytosis. PLoS Pathog 2008, 4, e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ciborowski P, Kadiu I, Rozek W, Smith L, et al. , Investigating the human immunodeficiency virus type 1-infected monocyte-derived macrophage secretome. Virology 2007, 363, 198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Burns A, Ciborowski P, Acute exposure to methamphetamine alters TLR9-mediated cytokine expression in human macrophage. Immunobiology 2016, 221, 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Reynolds JL, Mahajan SD, Sykes DE, Schwartz SA, Nair MP, Proteomic analyses of methamphetamine (METH)-induced differential protein expression by immature dendritic cells (IDC). Biochim Biophys Acta 2007, 1774, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Beck M, Schmidt A, Malmstroem J, Claassen M, et al. , The quantitative proteome of a human cell line. Mol Syst Biol 2011, 7, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kuhn E, Wu J, Karl J, Liao H, et al. , Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics 2004, 4, 1175–1186. [DOI] [PubMed] [Google Scholar]
- [12].Anderson L, Hunter CL, Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics 2006, 5, 573–588. [DOI] [PubMed] [Google Scholar]
- [13].Aebersold R, Burlingame AL, Bradshaw RA, Western blots versus selected reaction monitoring assays: time to turn the tables? Mol Cell Proteomics 2013, 12, 2381–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Webster CP, Smith EF, Grierson AJ, De Vos KJ, C9orf72 plays a central role in Rab GTPase-dependent regulation of autophagy. Small GTPases 2018, 9, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Spector SA, Zhou D, Autophagy: an overlooked mechanism of HIV-1 pathogenesis and neuroAIDS? Autophagy 2008, 4, 704–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gendelman HE, Orenstein JM, Martin MA, Ferrua C, et al. , Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J Exp Med 1988, 167, 1428–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].DeBoer J, Madson CJ, Belshan M, Cyclophilin B enhances HIV-1 infection. Virology 2016, 489, 282–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kadiu I, Wang T, Schlautman JD, Dubrovsky L, et al. , HIV-1 transforms the monocyte plasma membrane proteome. Cell Immunol 2009, 258, 44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wisniewski JR, Zougman A, Nagaraj N, Mann M, Universal sample preparation method for proteome analysis. Nat Methods 2009, 6, 359–362. [DOI] [PubMed] [Google Scholar]
- [20].[https://sciex.com/Documents/protocols/SWATH_Acquisition_Performance_Kit_Protocol.pdf].
- [21]. https://www.uniprot.org/.
- [22].([https://sciex.com/sw-downloads-form?d=SWATH_Variable_Window_Calculator_v1.1.zip&asset=software&softwareProduct=SWATH%20Variable%20Window%20Calculator%20v1.1].
- [23].Babicki S, Arndt D, Marcu A, Liang Y, et al. , Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res 2016, 44, W147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. http://proteomecentral.proteomexchange.org.
- [25].Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, et al. , The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2019, 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Searle BC, Egertson JD, Bollinger JG, Stergachis AB, MacCoss MJ, Using Data Independent Acquisition (DIA) to Model High-responding Peptides for Targeted Proteomics Experiments. Mol Cell Proteomics 2015, 14, 2331–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. https://web.expasy.org/peptide_cutter/.
- [28].MacLean B, Tomazela DM, Shulman N, Chambers M, et al. , Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Farrah T, Deutsch EW, Kreisberg R, Sun Z, et al. , PASSEL: the PeptideAtlas SRMexperiment library. Proteomics 2012, 12, 1170–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. https://ccb.jhu.edu/software/fqtrim.
- [31].Dobin A, Davis CA, Schlesinger F, Drenkow J, et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Li B, Dewey CN, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN, RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 2010, 26, 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mi H, Ebert D, Muruganujan A, Mills C, et al. , PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res 2021, 49, D394–D403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sidiropoulos K, Viteri G, Sevilla C, Jupe S, et al. , Reactome enhanced pathway visualization. Bioinformatics 2017, 33, 3461–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Reimand J, Isserlin R, Voisin V, Kucera M, et al. , Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc 2019, 14, 482–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Guo D, Zhang G, Wysocki TA, Wysocki BJ, et al. , Endosomal trafficking of nanoformulated antiretroviral therapy facilitates drug particle carriage and HIV clearance. J Virol 2014, 88, 9504–9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lodish H, Berk A, Zipursky SL, Matsudaira P, et al. , The dynamics of actin assembly., New York: 2000. [Google Scholar]
- [39].Ravenscroft G, Colley SM, Walker KR, Clement S, et al. , Expression of cardiac alpha-actin spares extraocular muscles in skeletal muscle alpha-actin diseases--quantification of striated alpha-actins by MRM-mass spectrometry. Neuromuscul Disord 2008, 18, 953–958. [DOI] [PubMed] [Google Scholar]
- [40].Aslanyan L, Ekhar VV, DeLeon-Rodriguez CM, Martinez LR, Capsular specific IgM enhances complement-mediated phagocytosis and killing of Cryptococcus neoformans by methamphetamine-treated J774.16 macrophage-like cells. Int Immunopharmacol 2017, 49, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Stenmark H, Olkkonen VM, The Rab GTPase family. Genome Biol 2001, 2, REVIEWS3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Spearman P, Viral interactions with host cell Rab GTPases. Small GTPases 2018, 9, 192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lavin PT, Mc Gee MM, Cyclophilin function in Cancer; lessons from virus replication. Curr Mol Pharmacol 2015, 9, 148–164. [DOI] [PubMed] [Google Scholar]
- [44].Chu H, Wang JJ, Spearman P, Human immunodeficiency virus type-1 gag and host vesicular trafficking pathways. Curr Top Microbiol Immunol 2009, 339, 67–84. [DOI] [PubMed] [Google Scholar]
- [45].Kierczak M, Surmacz L, Wiejak J, Wyroba E, [Role of the adaptins, dynamin like GTP-ases and Rab proteins in metabolic disorders and various infections]. Postepy Hig Med Dosw 2003, 57, 727–737. [PubMed] [Google Scholar]
- [46].Agola JO, Jim PA, Ward HH, Basuray S, Wandinger-Ness A, Rab GTPases as regulators of endocytosis, targets of disease and therapeutic opportunities. Clin Genet 2011, 80, 305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gajbhiye A, Dabhi R, Taunk K, Vannuruswamy G, et al. , Urinary proteome alterations in HER2 enriched breast cancer revealed by multipronged quantitative proteomics. Proteomics 2016, 16, 2403–2418. [DOI] [PubMed] [Google Scholar]
- [48].Pritschet K, Donhauser N, Schuster P, Ries M, et al. , CD4- and dynamin-dependent endocytosis of HIV-1 into plasmacytoid dendritic cells. Virology 2012, 423, 152–164. [DOI] [PubMed] [Google Scholar]
- [49].Arainga M, Guo D, Wiederin J, Ciborowski P, et al. , Opposing regulation of endolysosomal pathways by long-acting nanoformulated antiretroviral therapy and HIV-1 in human macrophages. Retrovirology 2015, 12, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Prashar A, Schnettger L, Bernard EM, Gutierrez MG, Rab GTPases in Immunity and Inflammation. Front Cell Infect Microbiol 2017, 7, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kadiu I, Gendelman HE, Macrophage bridging conduit trafficking of HIV-1 through the endoplasmic reticulum and Golgi network. J Proteome Res 2011, 10, 3225–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zerial M, McBride H, Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2001, 2, 107–117. [DOI] [PubMed] [Google Scholar]
- [53].Ao X, Zou L, Wu Y, Regulation of autophagy by the Rab GTPase network. Cell Death Differ 2014, 21, 348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shannon P, Markiel A, Ozier O, Baliga NS, et al. , Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003, 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schwartz SL, Cao C, Pylypenko O, Rak A, Wandinger-Ness A, Rab GTPases at a glance. J Cell Sci 2007, 120, 3905–3910. [DOI] [PubMed] [Google Scholar]
- [56].Martinez FO, Gordon S, The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 2014, 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Glanzer JG, Enose Y, Wang T, Kadiu I, et al. , Genomic and proteomic microglial profiling: pathways for neuroprotective inflammatory responses following nerve fragment clearance and activation. J Neurochem 2007, 102, 627–645. [DOI] [PubMed] [Google Scholar]
- [58].Pottiez G, Jagadish T, Yu F, Letendre S, et al. , Plasma proteomic profiling in HIV-1 infected methamphetamine abusers. PLoS One 2012, 7, e31031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Niu M, Morsey B, Lamberty BG, Emanuel K, et al. , Methamphetamine Increases the Proportion of SIV-Infected Microglia/Macrophages, Alters Metabolic Pathways, and Elevates Cell Death Pathways: A Single-Cell Analysis. Viruses 2020, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Munoz MF, Arguelles S, Guzman-Chozas M, Guillen-Sanz R, et al. , Cell tracking, survival, and differentiation capacity of adipose-derived stem cells after engraftment in rat tissue. J Cell Physiol 2018, 233, 6317–6328. [DOI] [PubMed] [Google Scholar]
- [61].Grabowska K, Harwood E, Ciborowski P, HIV and Proteomics: What We Have Learned from High Throughput Studies. Proteomics Clin Appl 2021, 15, e2000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Reynolds JL, Law WC, Mahajan SD, Aalinkeel R, et al. , Nanoparticle based galectin-1 gene silencing, implications in methamphetamine regulation of HIV-1 infection in monocyte derived macrophages. J Neuroimmune Pharmacol 2012, 7, 673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhang X, Walsh T, Atherton JJ, Kostner K, et al. , Identification and Validation of a Salivary Protein Panel to Detect Heart Failure Early. Theranostics 2017, 7, 4350–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Luo Y, Mok TS, Lin X, Zhang W, et al. , SWATH-based proteomics identified carbonic anhydrase 2 as a potential diagnosis biomarker for nasopharyngeal carcinoma. Sci Rep 2017, 7, 41191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.