Abstract
Background
Monkeypox DNA has been detected in skin lesions, saliva, oropharynx, urine, semen, and stool of patients infected during the 2022 clade IIb outbreak; however, the viral dynamics within these compartments remain unknown. We aimed to characterise the viral load kinetics over time in various parts of the body.
Methods
This was an observational, prospective, multicentre study of outpatients diagnosed with monkeypox in two hospitals and two sexual health clinics in Spain between June 28, 2022, and Sept 22, 2022. Men and women aged over 18 years were eligible if they reported having symptom onset within the previous 10 days of presentation, and were ineligible if disease was severe enough to be admitted to hospital. Samples were collected from five body locations (skin lesions, oropharynx, rectum, semen or vagina, and a dried blood spot) at six time points up to 57 days after the screening visit. Samples were analysed by quantitative PCR and a subset by cell culture. The primary endpoint was time from symptom onset to viral DNA clearance.
Findings
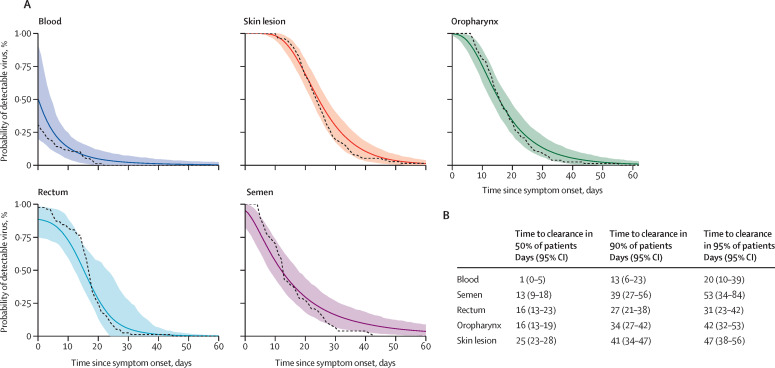
Overall, 1663 samples were collected from 77 study participants. 75 (97%) participants were men, the median age was 35·0 years (IQR 29·0–46·0), and 39 (51%) participants were living with HIV. The median time from symptom onset to viral clearance was 25 days (95% CI 23–28) in the skin lesions, 16 days (13–19) in the oropharynx, 16 days (13–23) in the rectum, 13 days in semen (9–18), and 1 day in blood (0–5). The time from symptom onset to viral clearance for 90% of cases was 41 days (95% CI 34–47) in skin lesions and 39 days (27–56) in semen. The median viral load in skin lesions was 7·3 log10 copies per mL (IQR 6·5–8·2) at baseline, compared with 4·6 log10 copies per mL (2·9–5·8) in oropharyngeal samples, 5·0 log10 copies per mL (2·9–7·5) in rectal samples, 3·5 log10 copies per mL (2·9–4·7) in semen samples, and 4·0 log10 copies per mL (4·0–4·0) in blood specimens. Replication-competent viruses were isolated in samples with high DNA levels (>6·5 log10 copies per mL).
Interpretation
In immunocompetent patients with mild monkeypox disease, PCR data alone would suggest a contact isolation period of 3 to 6 weeks but, based on detection of replication-competent virus, this time could be reduced. Based on findings from this cohort of patients, semen testing and prolonged use of condoms after recovery from monkeypox might not be necessary.
Funding
University Hospital Germans Trias i Pujol and the YoMeCorono.
Translation
For the Spanish translation of the abstract see Supplementary Materials section.
Introduction
Monkeypox, a zoonotic illness caused by the monkeypox virus, has affected rural communities in west and central Africa since 1970.1 In July 2022, WHO declared monkeypox as a public health emergency of international concern on account of an unprecedented global spread of the disease outside previously endemic countries in Africa. The multi-country outbreak has involved extensive human-to-human transmission in Europe and North America.2
The transmission of monkeypox virus between humans has historically been thought to occur primarily through respiratory droplets.3 However, during the 2022 clade IIb outbreak, direct contact with infectious material from skin lesions, lesions on mucous membranes, and body fluids occurring during sexual or close intimate contact is believed to constitute the primary mode of transmission.4, 5, 6, 7, 8 However, the frequency and duration with which viral DNA and viable virus is found in each body location and fluid and the relative contribution each makes to transmission remain unclear.
Viral DNA has been detected in skin lesions, saliva, oropharynx, urine, semen, and stool of patients infected during the 2022 outbreak.4, 9 A study from France observed that samples taken from 24 patients 14 days after diagnosis had a highly reduced proportion of positive PCR results as compared with baseline samples.10 However, the study collected specimens at only two time points, had no samples collected beyond day 14 of illness, and did not collect viral culture data. Another study on eight patients hospitalised with monkeypox in the UK reported prolonged viral shedding (22–39 days) in a range of samples but was limited by the small sample size.11 Finally, a study from the Democratic Republic of the Congo, of patients with clade I virus, showed higher viral loads in skin lesions than in other body sites, and also indicated the virus DNA might be detectable in blood and pharyngeal samples before the appearance of the rash.12
Research in context.
Evidence before this study
We searched PubMed for articles on monkeypox viral dynamics published from inception to Oct 10, 2022. We used the terms “Monkeypox” AND (“viral detection”, “viral dynamics”, “viral load”, “viral shedding”). Only eight papers reported quantitative measurements of viral DNA. Five studies had small sample sizes, inconsistent sampling, and short follow-up periods. One study from France, done during the 2022 clade IIb outbreak, included samples from 50 patients at enrolment and 24 patients after 14 days. The authors observed a reduction in the proportion of positive PCR results on day 14 after diagnosis. However, the study collected specimens at only two timepoints, had no samples collected after day 14 of illness, and did not report viral culture data. A UK study on eight hospitalised patients reported prolonged viral shedding in a range of samples, including blood, urine, lesions, and the respiratory tract, but was the study had a small sample size, included mostly patients with severe disease, and did not include viral culture data. A study from the Democratic Republic of the Congo, of clade I virus, examined an undetermined number of patients using PCR and found higher viral loads in skin lesions than in other body sites, and that viral DNA was detectable in blood and pharyngeal samples before the appearance of a rash. Several small studies have cultured monkeypox virus from lesion swabs, anal, urethral, or seminal samples, but all were small in size and had tested only a few timepoints. Overall, there is a substantial gap in knowledge on the dynamics of monkeypox viral clearance in the 2022 outbreak.
Added value of this study
In this prospective analysis, we collected 26 specimens from each of the 77 individuals with monkeypox over time, resulting in 1663 PCR results, providing a substantial amount of data for describing viral kinetics. We systematically collected specimens from five body locations at six different times and analysed them using PCR quantification of viral DNA and viral culture for a comprehensive assessment of viral dynamics. Our results indicate that swabs of skin lesions have higher viral loads and longer time to clearance than other locations, including oropharynx, rectum, semen, and blood. We found that viral DNA remains detectable in skin lesions for a median of 25 days, and most patients no longer have detectable viral DNA after 41 days. Most positive samples on viral culture were collected before day 15 of illness. Although qPCR can detect DNA for up to 6 weeks, the absence of culture viability could indicate a shorter infectious period than that indicated by PCR alone. Importantly, only 3 (1%) of 219 semen samples had a viral load higher than the threshold at which culture was assumed to be positive. These data suggest prolonged transmission in semen is unlikely.
Implications of all the available evidence
Based on PCR results, immunocompetent patients with mild monkeypox disease might require an isolation period of 3–6 weeks. However, if our findings regarding the shorter duration of replication-competent virus detected on viral cultures are supported, the contact isolation period could be reduced. Although sufficient data is not yet available to be completely conclusive, our data suggest testing semen after recovery or prolonged use of condoms might not be needed. Further studies in patients with severe disease or marked immunosuppression are required to understand viral dynamics in these patient groups.
In the absence of high-quality empirical evidence regarding the period of infectiousness, current infection prevention and control measures consist of respiratory isolation of patients with monkeypox virus infection until all lesion scabs have fallen off and the skin underneath has re-epithelialised, which could take up to 3 weeks. Also, in the absence of empirical data on persistence in semen over time, WHO has made a recommendation on condom use during any sexual activity for 12 weeks after recovery.13 There is an urgent need to better understand how the virus is transmitted and when it is cleared from each body compartment to inform isolation requirements and precautions after recovery. Therefore, we aimed to characterise the viral load kinetics over time in various parts of the body, combining both PCR and viral culture data.
Methods
Study design and participants
This was an observational, prospective, multicentre cohort study of patients diagnosed with monkeypox in Spain between June 28, 2022, and Sept 22, 2022. Participating centres included two tertiary hospitals and two sexual health clinics in Madrid and Barcelona.
Consecutive patients seen in the outpatient medical department were invited to participate in the study if they were women and men aged 18 years or older, who presented with signs of monkeypox infection, and who reported having symptom onset within the previous 10 days. Participants were ineligible if they had severe disease (defined as requiring admission to hospital). Participants without a confirmed diagnosis of monkeypox after initial molecular testing were subsequently withdrawn from the study. The study protocol was approved by the Ethics Committee of the Hospital Germans Trias i Pujol and written informed consent was obtained from all participants before enrolment.
Procedures
Patients who consented to participate were interviewed for demographic, epidemiological, and clinical characteristics at baseline (day 0). Data collected included information on the number and location of monkeypox lesions, the presence of systemic symptoms, lymphadenopathies, and proctitis. Physical examination and diagnostic testing (by quantitative PCR [qPCR]) for monkeypox virus were performed on day 0 by a sexually transmitted infection (STI) specialist. Patients underwent STI screening, including for HIV, Chlamydia trachomatis, Neisseria gonorrhoea, Treponema pallidum, and Herpes simplex virus. A symptom diary card was provided, and participants were asked to report their list of symptoms over time, along with symptom onset and resolution dates. On day 29, the research team interviewed participants by telephone to assess the clinical evolution of symptoms and lesions.
As part of the follow-up process, participants were provided with six packs of five sample self-collection devices, one for each day the participant was required to self-collect samples. Participants were asked to collect samples from their skin lesions (vesicle fluid or dry scraping of scabs or scars), oropharynx (swab), and blood (dried blood spot) on days 1, 8, 15, 22, 29, and 57 after the screening visit, and samples from their rectum (swab), semen (collection container), and vagina (swab) on days 1, 15, 29, and 57. The swabs (item number 310202) and viral transport medium (item number 304305KF) were purchased from Deltalab (Barcelona, Spain). The details regarding training and instructions given to patients for self-collection of samples are provided in appendix 2 (p 3), and the accuracy of self-sampling using these methods is reported elsewhere.14 The swabs were immediately placed in 3 mL of viral transport medium. For the dried blood spot, participants were asked to completely fill a circle with a diameter of 1·2 mm, which equates to an estimated blood volume of 50 μL.15
A courier service was used to transport self-collected samples from participants' homes to the microbiology laboratory at the University Hospital Germans Trias i Pujol (Badalona, Spain) under optimal conditions for sample stability. During transport, samples were kept at 4°C; time from sample collection to laboratory receipt was not allowed to exceed 24 h; once received, samples were processed immediately or stored at –80°C until processing. All samples were analysed for the detection of monkeypox virus DNA by qPCR. Detailed protocols on nucleic acid extraction and qPCR are in appendix 2 (p 4). Copy number per mL was determined using a linear dilution series of a quantified monkeypox virus DNA standard (AMPLIRUN Monkeypox virus DNA control, Vircell Spain, Santa Fe, Granada, Spain). Three replicates were included for each concentration. Viral loads from body fluids (ie, blood and semen) were expressed in DNA copies per mL, whereas the amount of virus in swab-collected samples (ie, skin lesions, oropharynx, and rectum) were expressed as copies per mL of viral transport medium.
MDF and MAM cultured the cells using a subgroup of specimens. Criteria for specimen selection and cell culture protocols are in appendix 2 (p 4). Upon observation of the cytopathic effect and at the end of the cell culture incubation period, culture supernatants were collected from each well, and RT-PCR was done, which was considered positive if it was at least three cycle thresholds (Ct) lower than the original sample. All cell culture-related procedures were done at a biosafety level 3 facility.
Study variables and outcomes
The primary outcome was the median time to viral clearance for each body region, defined as the interval between the onset of symptoms and the date when samples tested negative on PCR. For each sample, we converted the study day into the number of days from symptom onset. As a post-hoc analysis, we assessed the time to viral clearance for 90% and 95% of patients.
Prespecified secondary outcomes were viral load at each body location at each timepoint, presence of replication-competent viruses based on viral culture, and association between clinical features and the time to viral clearance. Other secondary outcomes included: the association between clinical features and demographic and epidemiological factors; behavioural factors associated with monkeypox acquisition, and barriers and facilitators to health services; monkeypox-specific humoral and cellular responses in a subset of individuals and associations with time to viral clearance or reinfection; and intrahost viral evolution in distinct locations during infection evolution. These four prespecified secondary outcomes will be reported elsewhere as they were related to ancillary substudies. As an exploratory analysis, we assessed the effect of age and HIV infection status on the predicted time to viral clearance in all compartments but did not incorporate this in our modelling approach.
Statistical analysis
The demographic and clinical characteristics of study participants were described using the median and IQR (defined by the 25th and 75th percentiles) or the number and percentage of available data. We used a linear mixed effect models to describe the log viral load of individuals with monkeypox and infer the time to viral clearance and describe the viral load over time (appendix 2 p 4). We assumed all bodily fluid compartments (skin lesions, rectum, oropharynx, semen, and blood) to be independent and fitted them separately. Values lower than the limit of detection of the assay were considered right censored. A limit of detection of 4·04 log10 copies per mL was used in the blood compartment and of 2·9 log10 copies per mL for all other samples. Model parameters were estimated using the stochastic approximation expectation maximisation algorithm implemented in Monolix 2019 R2.17 We used the Conditional Sampling use for Stepwise Approach on Correlation tests (COSSAC) to identify potential effects on both log viral load and time to viral clearance.18 The probability of detectable virus in each compartment was calculated by simulations, sampling 500 population parameters in their asymptotic estimation distribution accounting for parameter uncertainty. Then, for each set of population parameters, we sampled 300 individual parameters (leading to a total 150 000 individual parameters per compartment), and calculated the time to viral load clearance in each individual. We then derived the proportion of detectable virus in each dataset at each time, and obtained the mean and the 95% CI from the distribution observed in the simulated datasets. No imputation methods were used for missing values. For all hypothesis tests, the significance threshold was set at a two-sided alpha value of 0·05.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
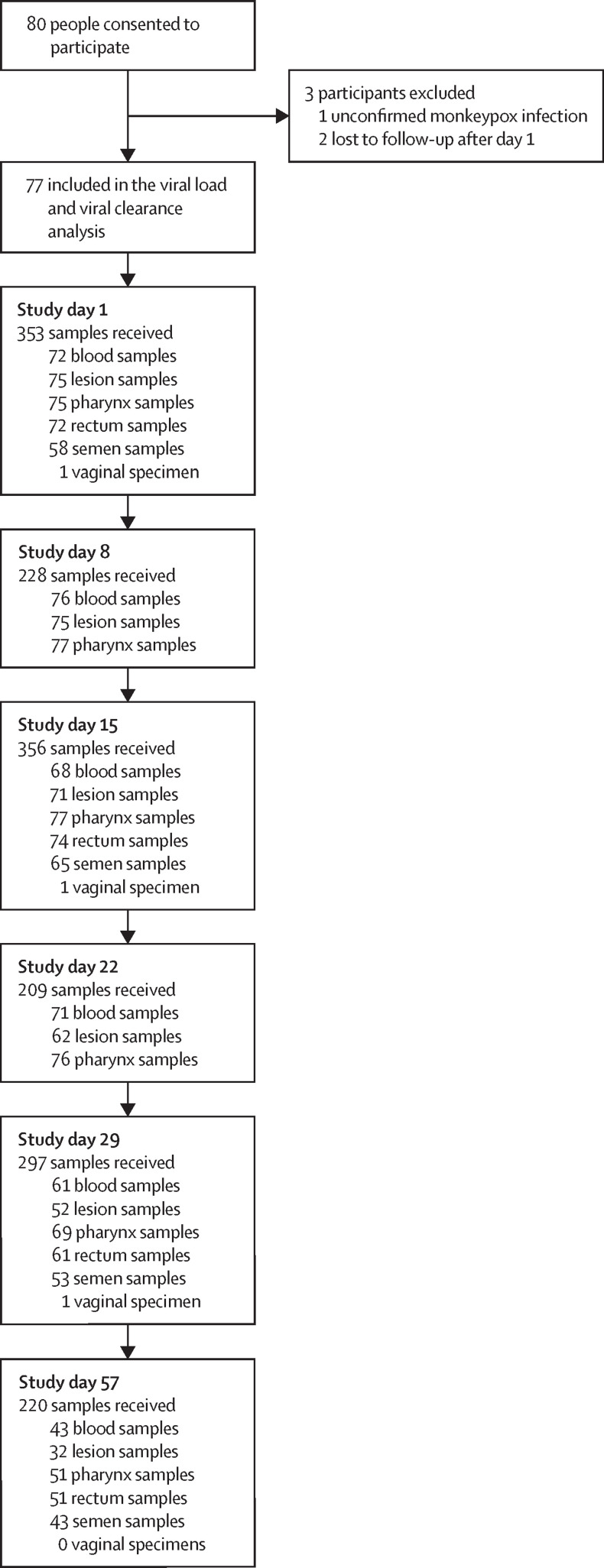
We analysed 1663 clinical samples collected at six time points from 77 people with confirmed monkeypox who were enrolled between June 28, 2022, and Sept 22, 2022 (figure 1 ). Two participants were recruited more than 10 days after symptom onset (day 11 and 14) and were included in the group of 77 analysed people. 75 (97%) of 77 participants were men, the median age was 35·0 years (IQR 29·0–46·0), and 39 (51%) of 77 participants were living with HIV (table ). Further demographic and clinical characteristics are in appendix 2 (p 6). Among participants living with HIV, 38 (97%) of 39 were taking antiretroviral medication, 34 (87%) had undetectable viral loads, 9 (23%) had CD4 counts lower than 500 cells per μL, two (5%) had CD4 counts lower than 300 cells per μL, and none had CD4 counts lower than 100 cells per μL. 73 (95%) of 77 patients presented with systemic symptoms (appendix 2 p 6), 69 (90%) of 77 patients had a rash on the anogenital or perioral region (or both regions), and 54 (70%) of 77 patients had a rash on a distant site from the point of inoculation. 46 (60%) participants presented with ulcerated skin lesions. The systemic illness lasted for a median of 5·0 days (IQR 3·0–8·0), whereas local site rashes lasted for a median of 21 days (13·0–26·0) and distant rashes for a median of 12·0 days (8·8–18·3). The median duration of lymphadenopathy was 11·0 days (7·8–19·0) and median duration of proctitis was 12·0 days (7·0–16·0; appendix 2 p 10). Only one patient required hospitalisation (due to a pulmonary venous thromboembolism), and no patients received systemic antiviral therapy.
Figure 1.
Trial flowchart.
Table.
Baseline and clinical characteristics
| N=77 | ||
|---|---|---|
| Gender | ||
| Men | 75 (97%) | |
| Women | 2 (3%) | |
| Age, years | 35·0 (29·0–46·0) | |
| Country of birth | ||
| South America or Latin America | 31 (40%) | |
| Spain | 36 (47%) | |
| People living with HIV* | 39 (51%) | |
| Concomitant Neisseria gonorrhoea Infection | 9 (12%) | |
| Concomitant Treponema pallidum infection | 4 (5%) | |
| Concomitant Chlamydia trachomatis infection | 3 (4%) | |
| Clinical characteristics | ||
| Recent smallpox vaccination | 2 (3%) | |
| Incubation period | 6·0 (4·0–8·0) | |
| Time from symptoms onset to first study PCR, days | 7·0 (5·0–8·0) | |
| Systemic illness | 73 (95%) | |
| Lymphadenopathies | 64 (83%) | |
| Proctitis | 24 (31%) | |
| Anogenital or perioral skin rash, or both | 69 (90%) | |
| Number of localised lesions† | ||
| 0 | 8 (10%) | |
| 1 | 17 (22%) | |
| 2–5 | 32 (42%) | |
| >5 | 19 (25%) | |
| Lesion location | ||
| Perioral | 13 (17%) | |
| Perianal | 21 (27%) | |
| Genital | 36 (47%) | |
| Oral mucosa | 5 (6%) | |
| Other location | 6 (8%) | |
| Skin rash at a distant site from the inoculation point | 54 (70%) | |
| Number of distant lesions† | ||
| 0 | 17 (22%) | |
| 1–4 | 20 (26%) | |
| 5–20 | 30 (39%) | |
| >20 | 6 (8%) | |
| Lesion location | ||
| Trunk, armpit, or neck | 46 (60%) | |
| Upper extremities | 47 (61%) | |
| Lower extremities | 36 (47%) | |
| Face | 31 (40%) | |
| Complications‡ | 29 (38%) | |
| Required hospitalisation | 1 (1%) | |
Data are n (%) or median (IQR).
Nine (23%) of 39 participants living with HIV had a CD4 cell count lower than 500 cells per μL with two (5%) having a CD4 cell count lower than 300 cells per μL. No participants had a CD4 cell count lower than 100 cells per μL.
Highest count of lesions during the follow-up period.
Complications were recorded in 64 participants on day 29.
Among the 77 participants, we collected a total of 367 swabs of skin lesions, 425 oropharyngeal samples, 258 rectal samples, 391 blood samples, 219 semen samples, and 3 vaginal specimens (collected from the same patient at three time points). The median time from symptoms onset to first study PCR was 7 days (IQR 5–8). Within the first 10 days following the onset of symptoms, 75 (100%) of 75 swab samples of skin lesion tested positive by PCR, 57 (76%) of 75 oropharyngeal samples, 51 (77%) of 66 rectal samples, 33 (67%) of 49 semen samples, and 17 (24%) of 72 blood samples tested positive by PCR. In samples collected more than 25 days after symptom onset, 36 (26%) of 138 skin lesion samples were positive by PCR, 21 (11%) of 187 oropharyngeal samples, five (4%) of 113 rectal samples, four (4%) of 98 semen samples, and five (3%) of 168 blood samples (appendix 2 p 11). One woman enrolled in the study tested positive in her vaginal sample on study day 1. Intermittent shedding (ie, negative PCR results in a specific body location that became positive at a later timepoint) was observed in all sample types (appendix 2 p 8).
The model of viral clearance replicated the observed data across all sample types (appendix 2 p 12). We found that skin lesions had the longest median time to viral clearance from symptom onset: 25 days (95% CI 23–28). The corresponding value for the other body locations were as follows: 16 days (13–19) for oropharyngeal samples, 16 days (13–23) for rectal samples, 13 days (9–18) for semen samples, and 1 day (0–5) for blood samples. According to the model, 90% of individuals would have undetectable viral DNA in skin lesions 41 days (95% CI 34–47) after symptom onset. The corresponding estimates for other samples was 34 days (27–42) in oropharyngeal samples, 27 days (21–38) in rectal samples, 39 days (27–56) in semen specimens, and 13 (6–23) days in the blood (figure 2 ). Time to viral clearance for 95% of patients was 47 days (38–56) for skin lesions, 42 days (32–53) for oropharyngeal samples, 31 days (23–42) for rectal samples, 53 days (34–84) for semen samples, and 20 days (10–39) for blood samples. Our exploratory analysis showed that age was not associated with time to viral clearance (appendix 2 p 13), and that the time to viral clearance in skin, throat, rectal, or blood samples did not differ but the time to viral clearance in semen appeared to be possibly faster in individuals living with HIV than in those without HIV (appendix 2 p 13). Individual viral load profiles for each sample type, correlations between compartments, and diagnostic plots for each compartment are shown in appendix 2 (pp 14–16).
Figure 2.
Time to viral clearance in all patients (A) and in 50%, 90%, and 95% of patients (B)
In A, the simulated means and 95% CIs of time to viral clearance are shown as a solid lines and shaded area, respectively. The dashed line represents the cumulative incidence calculated on the observed data. Model fits for each sample type are shown the appendix (p 12).
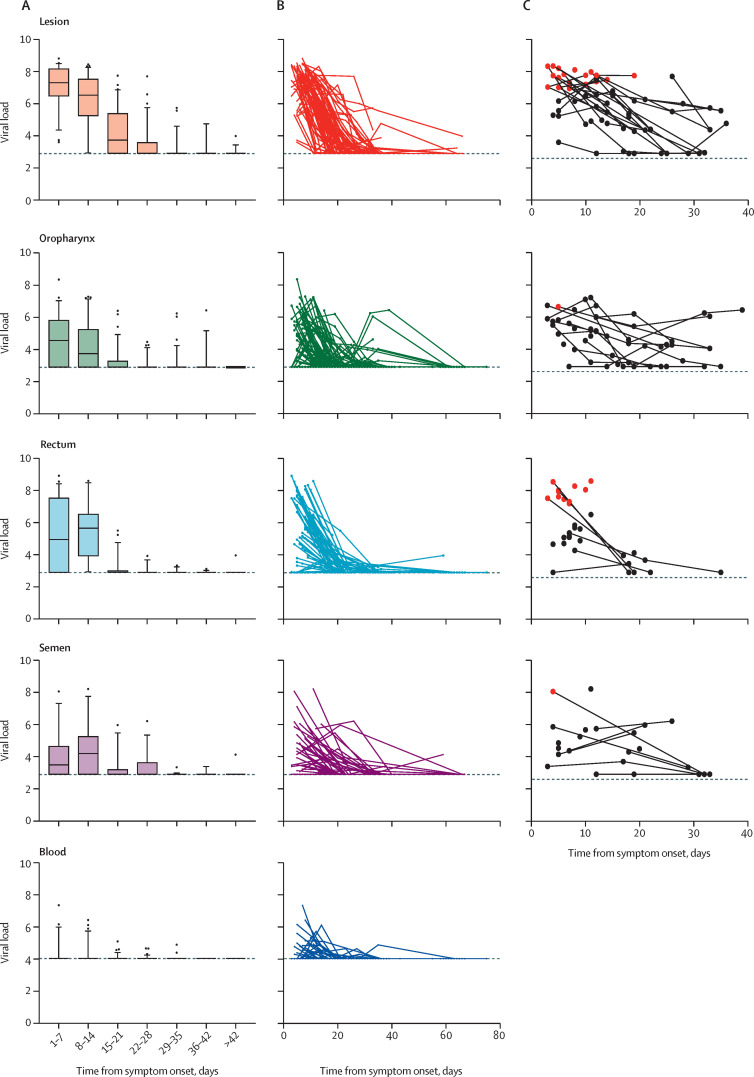
Median viral load at baseline was highest in swabs of skin lesions (7·3 log10 copies per mL [IQR 6·5–8·2]), followed by rectal (5·0 log10 copies per mL [2·9–7·5]) and oropharyngeal samples (4·6 log10 copies per mL [2·9–5·8]). Semen had median viral loads of 3·5 log10 copies per mL (2·9–4·7) and blood had 4·0 log10 copies per mL (4·0–4·0). The viral load decreased markedly in all sample types over time (figure 3A, B ). Four (5%) participants shed viral DNA more than 57 days from symptom onset: 3 (5%) from skin lesions (with viral loads of 3·2 log10 copies per mL, 3·2 log10 copies per mL, and 4·0 log10 viral copies per mL), and 1 (2%) from the rectum and semen (both samples with 4 log10 viral copies per mL).
Figure 3.
Median viral load (A), individual participant viral load (B), and viral culture (C) in lesions, oropharyngeal, rectal, and semen samples by time from symptom onset
Viral load measures are expressed as log10 copies per mL (mL of viral transport media for lesion, pharynx, and rectum, and mL of blood and semen). The limit of detection was 4·0 log10 copies per mL for the blood samples and 2·9 log10 copies per mL for the rest of the samples, both of which are indicated with dashed lines. For each patient, the sample collection day was adjusted for the date of onset of symptoms. In A, box plots represent median, IQR, and minimum and maxiumum viral load. In C, samples in red had replication-competent viruses detected by culture.
Viral culture was done on 174 samples in total (65 skin lesion samples, 52 oropharynx samples, 34 rectum samples, and 23 semen samples). The result was positive, as detected by cytopathic effect, in 33 (70%) of 47 samples with viral loads of 6·5 log10 copies per mL or higher (approximately a Ct value of 26). The 14 (30%) negative samples included eight lesional swabs, four oropharyngeal swabs, one rectal swab, and one semen sample (figure 3C). None of the 127 samples with viral loads below 6·5 log10 copies per mL were positive. Overall, three (1%) of 219 semen samples had a viral load of 6·5 log10 copies per mL or higher, and out of two tested only one was positive on viral culture.
The presence of monkeypox virus was confirmed by real-time qPCR on DNA purified from cell growth medium collected after 5 days in all the samples that showed a cytopathic effect. Only one sample collected after day 15 from symptom onset tested positive on viral culture, and this sample was from a skin lesion (figure 3C). Based on the viral load culture results (ie, threshold of 6·5 log10 copies per mL to determine viability) we did an exploratory analysis to calculate the time until the viral load was lower than 6·5 log10 copies per mL. For 90% of individuals to achieve a viral load below this threshold the corresponding values for each body location were 14 days (95% CI 11–17) for skin lesion samples, 5 days (0–10) for oropharyngeal samples, 10 days (8–14) for rectal samples, 2 days (0–11) for semen, and 0 days (0–0) for blood (appendix 2 p 9).
Discussion
In this prospective study, we have characterised the time to viral clearance of outpatients diagnosed with mild monkeypox disease, without substantial immunosuppression, in a real-world community setting using longitudinal data. Our findings indicate that viral DNA detected by qPCR remains present in swab samples of skin lesions for a median period of 25 days from symptom onset, and that most patients no longer have detectable viral DNA after 41 days. Furthermore, culture viability data suggests that the infectious period could be shorter than the one established by PCR data alone. However, both DNA positivity and culture as indicators of infectiousness have limitations, as PCR cannot distinguish viable and non-viable virus and culture could have inadequate sensitivity.19
We found that viral DNA was detectable by qPCR in skin lesions for a median time of 25 days from symptom onset and in other body locations or fluids for slightly shorter periods (between 1 and 16 days). According to our model, it would take until day 41 for 90% of individuals to have undetectable levels of viral DNA in swabs of skin lesions. Taken alongside data from previous studies,6, 9, 10, 11, 20, 21 we conclude that monkeypox DNA is nearly always found in skin lesions in the early illness, and it is detected less frequently (30–70%) and at lower viral loads in other samples. During the course of the disease, lesions on the skin had concentrations of viral DNA that were at least 2 orders of magnitude higher than all other samples, and remained above the threshold for a positive viral culture for more than 1 week from symptom onset. We did not find convincing evidence that HIV status or age affected viral kinetics.
A key question that remained unanswered during the first months of the 2022 global monkeypox outbreak was whether monkeypox virus could be transmitted through semen after recovery, as previously observed in other zoonotic viruses.22 Small studies of patients with monkeypox had found a reportedly high prevalence of PCR positivity in semen specimens but the data on the time to viral clearance or the presence of viable viruses in these samples was scarce.5, 9, 10, 23
We found that the median time to viral clearance from semen was 13 days from symptom onset, the time for 90% of individuals to have undetectable viral DNA in semen was 39 days, and viral loads in semen were generally low throughout the course of infection, with only 1% of samples having a viral load of 6·5 log10 copies per mL or higher (above the limit for successful viral culture). Moreover, only a single sample, collected early in the illness, was positive on culture. Based on these findings, the overall risk of transmission through seminal fluid is probably low and the time to clearance of replication-competent virus appears shorter than the duration of viral detection by qPCR. However, for many viruses, shedding in semen can be intermittent and extended follow-up will be of value to establish how long it takes for monkeypox DNA to be permanently undetectable in semen.
We found that samples from the oropharynx and rectum contained replication-competent viruses and could therefore be sources of infection. However, exposure to oropharyngeal secretions might be associated with a lower risk of infection than exposure to skin lesions, as viral DNA loads were lower and detection of replication-competent virus was less frequent from oropharyngeal samples than skin lesion samples in our study. The median time of viral shedding from blood was only 1 day from symptom onset and viral loads in blood were generally low. This finding is much shorter than as reported in a previous study that included individuals who required hospital admission, often with more severe disease, than typically described for most community cases in the 2022 outbreak.11 The short duration seen in our study probably reflects the relatively mild clinical course of our patient cohort, in which only one patient required hospitalisation. Of note, the viral dynamics could have differed in the very first days of infection, which could have implications for viable transmission routes, even though we recruited patients within 10 days of symptom onset.12
A potential limitation of our study is that the self-sampling strategy, although facilitating the study implementation and reducing loss to follow-up, might have resulted in samples of lower accuracy than those collected by health-care professionals. However, self-collected samples have been shown to be highly accurate for a variety of pathogens, and we have previously shown that lesion and oropharyngeal swabs collected by patients with monkeypox are comparable to those collected by health-care professionals.14 Second, we used dried blood samples, which might not be as accurate as samples collected by venepuncture. Third, we did not include saliva samples, which have been suggested to have a potential role in disease transmission.9, 24 Additionally, there is currently no international, standardised methodology for analysing monkeypox virus viability in cell culture, which could have hindered comparisons of our analysis with other studies evaluating viral viability, even though we successfully isolated replication-competent viral particles from all body locations. Finally, our study cohort consisted solely of outpatients, almost all of whom were young men. Viral dynamics might differ, in particular in severely unwell patients who might shed virus for longer, but our results should be applicable to the vast majority of patients who are managed outside of the hospital.
In summary, our study provides a comprehensive perspective of the dynamics of monkeypox viral clearance within the first 2 months following symptom appearance in a cohort of patients with relatively mild disease, managed as outpatients and without substantial immunocompromise. For this group of patients, our findings could help inform isolation and post-recovery precautions. Current recommendations, for all patients regardless of immune status or disease course, consist of respiratory isolation for up to 3 weeks and condom use during any sexual activity for 12 weeks after recovery. If isolation decisions are made on PCR results alone, our data would suggest a requirement for contact isolation lasting from 3 to 6 weeks in immunocompetent patients with mild monkeypox disease. However, integrating our data on the shorter duration of replication-competent virus detected on viral cultures, suggests the time to ending isolation could be reduced. Additionally, if further research supports our findings, semen testing after recovery and prolonged use of condoms might not be necessary for this group of patients. Further studies are needed to better inform decisions on infection prevention and control, particularly in individuals with marked immunosuppression and with asymptomatic infection.
Data sharing
De-identified participant data collected for the study, including individual participant data and a data dictionary defining each field in the set, will be made available from the corresponding author on reasonable request.
Declaration of interests
We declare no competing interests.
Acknowledgments
Acknowledgments
The study was funded by emergency response funds of the University Hospital Germans Trias i Pujol and the YoMeCorono crowdfunding campaign. We thank Gerard Carot-Sans for editing the final draft and Roser Escrig for medical writing assistance with the study documentation. We also thank Laia Bertran, Sergi Gavilan, and Miquel Angel Rodríguez for the operational and financial management of the project. OM, CS, MU, and AA, are supported by the European Research Council grant agreement number 850450 (European Union's Horizon 2020 Research and Innovation Program, ERC-2019-STG funding scheme). RP has grants or contracts with Gilead and ViiV, and received consulting fees from Gilead, MSD, ViiV, Lilly, Theratechnologies, and GSK.
Contributors
OM, MM, CS, MU and EJT-V conceived and designed the study. CS, MU, EJT-V, AMe, AA, AH-R, CC, VD, ARi, PC, XO, JMC, MV-M, MDF, MAM, MA-D, EG-C, APL, ARa, VB, acquired the data. CS, MU, EJT-V, DO, AMA, JG, MM and OM analysed and interpreted the data. CS, MU, DO, AMa, JG, MM and OM did the statistical analysis. CS, MU, EJT-V, MM, and OM drafted the manuscript. All authors reviewed and approved the manuscript; vouched for the accuracy and completeness of the data and for the adherence of the study to the protocol; and were responsible for the final decision to submit for publication. CS, MU, EJT-V, MM, and OM had full access and verified all the data in the study.
Contributor Information
The Movie Group:
José Ramón Santos, Lucía Bailón, Susana Benet, Jorge Arroyo Andres, Lorena Calderón Lozano, María Carrasco Díaz, Carla Budria Serrano, Enola Crespillo Galán, Ana Isabel Parra Manzano, Pamela Nef Rabadán, Laura Muntané, Cristina Sánchez-Lafuente Doncel, Yesinei Marina Marrero Pueo, Aroa Muñoz Quinto, Marlon Acosta, Patricia Alvarez, Maider Arando, Jorge N García, Arnau Monforte, Yolanda Maltas Hidalgo, Ramona Hervas Perez, and Laura Clotet Romero
Supplementary Materials
References
- 1.McCollum AM, Damon IK. Human monkeypox. Clin Infect Dis. 2014;58:260–267. doi: 10.1093/cid/cit703. [DOI] [PubMed] [Google Scholar]
- 2.WHO Monkeypox fact sheets. 2022. https://www.who.int/news-room/fact-sheets/detail/monkeypox
- 3.Centers for Disease Control and Prevention Potential exposure to person with confirmed human monkeypox infection—USA. Emergency preparedness and response. 2021. https://emergency.cdc.gov/han/2021/han00446.asp
- 4.Antinori A, Mazzotta V, Vita S, et al. Epidemiological, clinical, and virological characteristics of four cases of monkeypox support transmission through sexual contact, Italy, May 2022. Euro Surveill. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.22.2200421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thornhill JP, Barkati S, Walmsley S, et al. Monkeypox virus infection in humans across 16 countries—April–June 2022. N Engl J Med. 2022;387:679–691. doi: 10.1056/NEJMoa2207323. [DOI] [PubMed] [Google Scholar]
- 6.Tarín-Vicente EJ, Alemany A, Agud-Dios M, et al. Clinical presentation and virological assessment of confirmed human monkeypox virus cases in Spain: a prospective observational cohort study. Lancet. 2022;400:661–669. doi: 10.1016/S0140-6736(22)01436-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Català A, Clavo-Escribano P, Riera-Monroig J, et al. Monkeypox outbreak in Spain: clinical and epidemiological findings in a prospective cross-sectional study of 185 cases. Br J Dermatol. 2022;187:765–772. doi: 10.1111/bjd.21790. [DOI] [PubMed] [Google Scholar]
- 8.Patel A, Bilinska J, Tam JCH, Fontoura DDS, Mason CY, Daunt A, et al. Clinical features and novel presentations of human monkeypox in a central London centre during the 2022 outbreak: descriptive case series. BMJ. 2022;378 doi: 10.1136/bmj-2022-072410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peiró-Mestres A, Fuertes I, Camprubí-Ferrer D, et al. Frequent detection of monkeypox virus DNA in saliva, semen, and other clinical samples from 12 patients, Barcelona, Spain, May to June 2022. Euro Surveill. 2022;27 doi: 10.2807/1560-7917.ES.2022.27.28.2200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palich R, Burrel S, Monsel G, et al. Viral loads in clinical samples of men with monkeypox virus infection: a French case series. Lancet Infect Dis. 2022 doi: 10.1016/S1473-3099(22)00586-2. published online Sept 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adler H, Gould S, Hine P, et al. Clinical features and management of human monkeypox: a retrospective observational study in the UK. Lancet Infect Dis. 2022;22:1153–1162. doi: 10.1016/S1473-3099(22)00228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pittman PR, Martin JW, Kingebeni PM, et al. Clinical characterization of human monkeypox infections in the Democratic Republic of the Congo. medRxiv. 2022 doi: 10.1101/2022.05.26.22273379. published online May 29. (preprint). [DOI] [Google Scholar]
- 13.WHO Monkeypox questions and answers. 2022. https://www.who.int/news-room/questions-and-answers/item/monkeypox
- 14.Ubals M, Tarin-Vicente EJ, Oller X, et al. Evaluating the accuracy of self-collected swabs for the diagnosis of monkeypox. medRxiv. 2022 doi: 10.1101/2022.09.19.22280087. published online Sept 20. (preprint). [DOI] [PubMed] [Google Scholar]
- 15.Moat SJ, George RS, Carling RS. Use of dried blood spot specimens to monitor patients with inherited metabolic disorders. Int J Neonatal Screen. 2020;6:26. doi: 10.3390/ijns6020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lixoft Monolix. https://lixoft.com/products/monolix/
- 18.Ayral G, Si Abdallah JF, Magnard C, Chauvin J. A novel method based on unbiased correlations tests for covariate selection in nonlinear mixed effects models: the COSSAC approach. CPT Pharmacometrics Syst Pharmacol. 2021;10:318–329. doi: 10.1002/psp4.12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutson CL, Damon IK. Monkeypox virus infections in small animal models for evaluation of anti-poxvirus agents. Viruses. 2010;2:2763–2776. doi: 10.3390/v2122763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girometti N, Byrne R, Bracchi M, et al. Demographic and clinical characteristics of confirmed human monkeypox virus cases in individuals attending a sexual health centre in London, UK: an observational analysis. Lancet Infect Dis. 2022;22:1321–1328. doi: 10.1016/S1473-3099(22)00411-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nörz D, Brehm TT, Tang HT, et al. Clinical characteristics and comparison of longitudinal qPCR results from different specimen types in a cohort of ambulatory and hospitalized patients infected with monkeypox virus. J Clin Virol. 2022;155 doi: 10.1016/j.jcv.2022.105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention How it spreads: Monkeypox. 2022. https://www.cdc.gov/poxvirus/monkeypox/if-sick/transmission.html
- 23.Lapa D, Carletti F, Mazzotta V, et al. Monkeypox virus isolation from a semen sample collected in the early phase of infection in a patient with prolonged seminal viral shedding. Lancet Infect Dis. 2022;22:1267–1269. doi: 10.1016/S1473-3099(22)00513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernaez B, Muñoz-Gómez A, Sanchiz A, et al. Monitoring monkeypox virus in saliva and air samples in Spain: a cross-sectional study. Lancet Microbe. 2022 doi: 10.1016/S2666-5247(22)00291-9. published online Nov 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Uncited References
- 16.Olson VA, Laue T, Laker MT, et al. Real-time PCR system for detection of orthopoxviruses and simultaneous identification of smallpox virus. J Clin Microbiol. 2004;42:1940–1946. doi: 10.1128/JCM.42.5.1940-1946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
De-identified participant data collected for the study, including individual participant data and a data dictionary defining each field in the set, will be made available from the corresponding author on reasonable request.