Abstract
A poly(aspartic acid) degrading bacterium (strain KT-1 [JCM10459]) was isolated from river water and identified as a member of the genus Sphingomonas. The isolate degraded only poly(aspartic acid)s of low molecular masses (<5 kDa), while the cell extract hydrolyzed high-molecular-mass poly(aspartic acid)s of 5 to 150 kDa to yield aspartic acid monomer.
The poly(aspartic acid) (PAA), belonging to the family of synthetic polypeptides, is a biodegradable water-soluble polymer. PAA polymers attract attention as environmentally degradable water-soluble materials to be used as dispersants, as detergent builders, and in biomedical applications (13).
Recently, an attempt to produce PAA on large scale was made by the thermal polymerization of l-aspartic acid to yield polysuccinimide as prepolymer, followed by hydrolysis of polysuccinimide (4, 19). The thermal polymerization of l-aspartic acid with or without catalyst leads to the formation of a mixture of l- and d-succinimide units, and the resulting PAA after the hydrolysis of polysuccinimide is composed of 70% of β-amide and 30% of α-amide units (5, 11, 12, 20). Several research groups reported that the thermally synthesized PAA has a branched structure and several irregular end groups (8, 9, 20). The thermal polymerization of l-aspartic acid in the absence of catalyst gives low-molecular-mass PAA of <10 kDa. In contrast, high-molecular-mass PAA (10 to 90 kDa) can be prepared by the hydrolysis of the polysuccinimides synthesized from l-aspartic acid in the presence of phosphoric acid as a catalyst (7).
Several research groups have studied the biodegradability of PAA in activate sludge and reported that the biodegradability of PAA is affected by the structures of branching and irregular end groups in PAA (1, 3, 6, 16). The biodegradation of PAA chains must be caused by some bacterium in the natural environment. However, there has been no report on the isolation of PAA-degrading bacteria, which prompted us to isolate PAA-degrading bacteria from environments. In this study, we examined the biodegradability of thermally synthesized PAA polymers in natural river water, and we isolated a PAA-degrading bacterium.
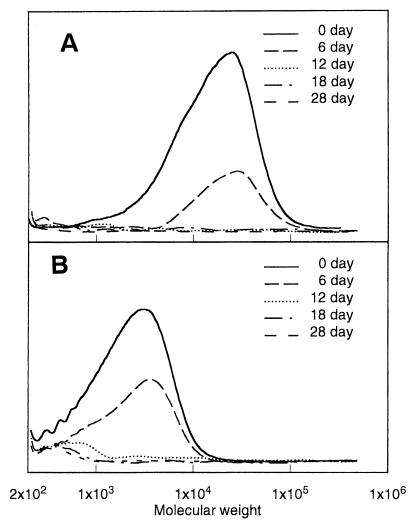
The biodegradabilities of PAA samples in river water (Arakawa River, Saitama, Japan) were measured by biochemical oxygen demand (BOD) assay and by gel permeation chromatography (GPC) analysis. Two types of PAA samples were used for the biodegradation test. PAA-T sample (number-average molecular weight [Mn], 2,100; weight-average molecular weight [Mw], 4,500; number of branched units/100 monomer units, 5.6) was obtained by hydrolyzing polysuccinimide prepared by the thermal polymerization of l-aspartic acid without catalyst at 180 to 240°C. PAA-P sample (Mn, 7,500; Mw, 20,000, number of branched units/100 monomer units, 3.1) was obtained by hydrolyzing polysuccinimide prepared by the thermal polymerization of l-aspartic acid with phosphoric acid as a catalyst at 160 to 200°C. The procedures of the BOD biodegradation test were previously reported (2, 21). The BOD biodegradabilities of PAA-P and PAA-T in river water at 25°C increased with time to reach ca. 78 and 70%, respectively, within 15 days. The time-dependent changes in molecular weights of PAA-P and PAA-T samples in river water at 25°C are shown in Fig. 1. PAA-P sample was completely hydrolyzed within 12 days, and no product was detected by GPC. In contrast, high-molecular-weight fractions of PAA-T sample that were greater than 1,000 were completely degraded within 12 days, while low-molecular-weight fractions of less than 1,000 were slowly degraded, and a portion remained after the test of 28 days. These results indicate that PAA-degrading microorganisms are present in river water.
FIG. 1.
Time-dependent changes in the molecular weights of PAA-P (A) and PAA-T (B) in fresh water. PAA at 0.175 mg/ml was incubated in fresh water at 25°C, and the molecular weight was analyzed by GPC.
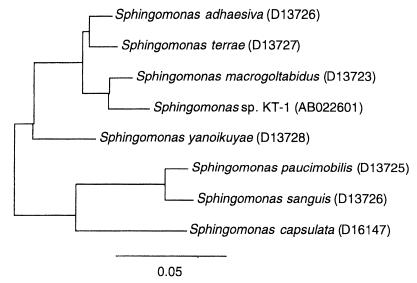
A PAA-degrading bacterium was isolated from Arakawa River water by an enrichment culture in the mineral medium containing 0.15% PAA-T sample as a substrate. The composition of mineral medium was as follows (per liter); 4.60 g of KH2PO4, 11.60 g of NaHPO4 · 12H2O, 1.00 g of NH4Cl, 0.50 g of MgSO4 · 7H2), 0.05 g of CaCl2 · 2H2O, and 0.01 g of FeCl3 · 6H2O. One bacterial strain capable of degrading PAA-T was isolated and designated strain KT-1. Strain KT-1 has been deposited in the Japan Collection of Microorganisms under accession number JCM10459. Strain KT-1 was a gram-negative, motile rod with more than one polar flagellum. Strain KT-1 grew aerobically, and oxidase and catalase activities were absent. No fermentative growth was observed under anaerobic conditions. Methanol did not support growth. The colony was circular and yellow. Cellular quinone type and (G+C) content of DNA were Q10 and 63%, respectively. On the basis of these phenotypic properties, strain KT-1 was identified as a member of the genus Sphingomonas belonging in the alpha subclass of Proteobacteria. To clarify the phylogenetic relationships between strain KT-1 and known strains belonging to the genus Sphingomonas, we determined the 16S ribosomal DNA sequence of strain KT-1 and compared with the sequences of six species belonging to the genus Sphingomonas. The sequence of 16S rDNA was determined by PCR (15) by using the MicroSeq 16S rRNA gene kit (PE Applied Biosystems). The 16S rDNA sequence of the PAA-degrading strain KT-1 has been deposited in the GenBank database (National Center for Biotechnology Information, National Library of Medicine) under accession number AB022601. The GenBank database was used to search for 16S rDNA sequences. The sequences were aligned, and phylogenetic tree was constructed with the Genetics Program (Software Development Co. Ltd.) by using the neighbor-joining method and the Jukes-Cantor distance correction method (14). Figure 2 illustrates the phylogenetic relationship of bacteria in the Sphingomonas group. The closest relative to strain KT-1 was Sphingomonas macrogoltabidus (97.3% similarity).
FIG. 2.
Phylogenetic tree based on the partial 16S rRNA gene sequence of Sphingomonas sp. KT-1 and the six species belonging to the genus of Sphingomonas. The bar insert represents 5% sequence divergence as determined by measuring the lengths of the horizontal lines connecting any species. Nucleotide sequence database accession numbers are shown in parentheses.
The effect of carbon source on the growth of strain KT-1 was investigated by measurement of turbidity at 660 nm of culture media (Table 1). PAA-degrading activities of strain KT-1 cell extract were evaluated from the time-dependent changes in the amount of NH2-terminal amino acid by measuring the fluorescence of products from the reaction with o-phthalaldehyde (10). As shown in Table 1, the growth of the bacteria and the PAA-degrading activity of cell extract were dependent of the kinds of carbon substrates. When l-aspartic acid, d-aspartic acid, or l-asparagine was used as sole carbon source, strain KT-1 grew well and a high PAA-degrading activity was detected. l-Aspartic acid and d-aspartic acid are the final end products of PAA degradation by cell extract of KT-1. These results suggest that there is no end product inhibition of the enzyme responsible for this activity. A weak growth was observed on PAA-P and PAA-T, but PAA-degrading activities were as high as the activities in the presence of aspartic acids. When strain KT-1 was incubated with l-glutamic acid, the strain grew well, but the PAA-degrading activity was relatively low.
TABLE 1.
Yields and PAA degradation activities of strain KT-1 grown on various carbon sources
| Carbon sourcea | OD660b | PAA degradation activityc (U/mg of protein) |
|---|---|---|
| Glucose | 0.05 | 0.27 ± 0.10 |
| Fructose | 0.04 | 0.29 ± 0.20 |
| Succinate | 0.05 | 0.17 ± 0.05 |
| Alanine | 0.06 | 0.19 ± 0.09 |
| Isoleucine | 0.04 | 0.58 ± 0.24 |
| Valine | 0.05 | 0.02 ± 0.01 |
| l-Aspartic acid | 0.50 | 6.32 ± 3.17 |
| d-Aspartic acid | 0.48 | 10.18 ± 4.66 |
| l-Asparagine | 0.53 | 1.79 ± 0.42 |
| l-Glutamic acid | 0.57 | 0.42 ± 0.12 |
| Oxalacetic acid | 0.03 | 0.03 ± 0.02 |
| PAA-P | 0.04 | 6.62 ± 1.25 |
| PAA-T | 0.04 | 7.35 ± 2.91 |
Strain KT-1 was grown in mineral medium containing a 0.15% (wt/vol) carbon source at 25°C for 3 days.
The optical density (OD) of the culture medium was measured at 660 nm.
PAA-degrading activity by cell extract was determined by measuring the amount of NH2-terminal amino acid.
Strain KT-1 was grown on PAA as the sole carbon source, and the degradation of PAA was analyzed by GPC. Low molecular weight components (Mr <5,000) of PAA-T sample were degraded by strain KT-1 during the test for 5 days at 25°C, while high-molecular-weight components of greater than 5,000 were hardly degraded for 5 days. When PAA-P was incubated with strain KT-1, only low-molecular-weight components of <5,000 were degraded, as was the case with PAA-T. Thus, strain KT-1 is capable of degrading only low-molecular-weight PAA polymer chains. In this study, strain KT-1 was isolated on the basis of the ability to grow on PAA-T. As shown in Fig. 1A, high-molecular-weight PAA chains were completely degraded in river water. Therefore, other bacteria capable of degrading high-molecular-weight PAA must be present in river water.
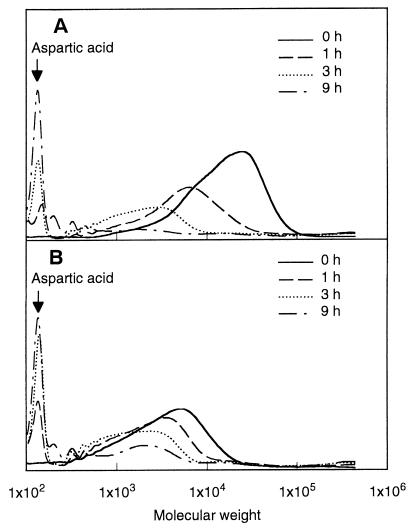
The hydrolysis of PAA samples by cell extract of strain KT-1 was analyzed by GPC. The changes in GPC profile of PAA-P and PAA-T samples by treatment with cell extract are shown in Fig. 3. It is of interest that the cell extract hydrolyzed the high-molecular-weight PAA-P sample to yield a low-molecular-weight product (Fig. 3A). The molecular weight of the PAA-P sample decreased gradually with time, and aspartic acid monomer was accumulated in solution as a final product. After 9 h, most of the PAA-P sample was converted into aspartic acid monomer by the cell extract. It is apparent that the cell extract can hydrolyze high-molecular-weight PAA chains of greater than 5,000, though strain KT-1 degraded only PAA polymers with low molecular weights (Mr <5,000). When PAA-T sample was incubated with cell extract, the molecular weight of the PAA-T sample decreased with time, as with PAA-P, and the amount of aspartic acid as final product increased with time (Fig. 3B).
FIG. 3.
Time-dependent changes in the molecular weights of PAA-P (A) and PAA-T (B) treated with cell extract of strain KT-1. Cells of strain KT-1 were disrupted by ultrasonic treatment, and the cell extract was incubated with 0.15% (wt/vol) PAA in carbonate buffer (pH 7.0) at 28°C. The solutions were then removed and analyzed by GPC.
The cell extract could hydrolyze PAA polymers with high molecular weights of up to 150,000, while the strain KT-1 degrades only low-molecular-weight PAA components (Mr <5,000). A study on the localization of PAA-degrading activity revealed that the activity was localized in the cytoplasmic membrane within the cell (data not shown). These results suggest that high-molecular-weight PAA polymers (Mr >5,000) are not transported within the cell. As shown in Fig. 2, strain KT-1 belongs to the group consisting of Sphingomonas adhaesivia, Sphingomonas terrae, and Sphingomonas macrogoltabidus, which utilize polyethylene glycol (PEG) (17, 18). S. macrogoltabidus, which was the most similar to strain KT-1, was reported to utilize only low-molecular-weight PEG (Mr <4,000), while an intracellular PEG dehydrogenase in the cytoplasm of S. macrogoltabidus was found to dehydrogenate high-molecular-weight PEGs (Mrs 6,000 and 20,000) (22). Thus, degradation of PAA by strain KT-1 showed some similar characteristics of the PEG degradation by S. macrogoltabidus.
The structure and properties of an intracellular PAA depolymerase in strain KT-1 are under investigation.
Acknowledgments
We gratefully acknowledge Bernhard Mohr of BASF for supplying PAA-T and PAA-P samples.
The work was supported by CREST (Core Research for Evolutional Science and Technology) of the Japan Science and Technology Corporation.
REFERENCES
- 1.Alford D D, Wheeler A P, Pettigrew C A. Biodegradation of thermally synthesized polyasparate. J Environ Polym Degrad. 1994;2:225–236. [Google Scholar]
- 2.Abe H, Doi Y. Enzymatic and environmental degradation of racemic poly(3-hydroxybutyric acid)s with different stereoregularities. Macromolecules. 1996;29:8683–8688. [Google Scholar]
- 3.Freedman M R, Paik Y H, Swift G, Wilczynski R, Wolk S K, Yocom K M. Biodegradability of polycarboxynates: structure-activity studies. Polymer Reprints. 1994;35:423–424. [Google Scholar]
- 4.Harada K. Polycondensation of thermal precursors of aspartic acid. J Org Chem. 1959;24:1662–1666. [Google Scholar]
- 5.Kovacs J, Kovacs H N, Konyves I, Csaszar J, Vajda T, Mix H. Chemical studies of polyaspartic acid. J Org Chem. 1961;26:1084–1091. [Google Scholar]
- 6.Nakato T, Toshitake M, Matsubara K, Tomida M, Kakuchi T. Relationship between structure and properties of poly(aspartic acid)s. Macromolecules. 1998;31:2107–2113. [Google Scholar]
- 7.Neri P, Antoni G, Benvenuti F, Cocola F, Gazzei G. Synthesis of α,β-poly[(2-hydroxyethyl)-dl-aspartamide], a new plasma expander. J Med Chem. 1973;16:893–897. doi: 10.1021/jm00266a006. [DOI] [PubMed] [Google Scholar]
- 8.Matsubara K, Nakato T, Tomida M. 1H and 13C NMR characterization of poly(succinimide) prepared by thermal polycondensation of l-aspartic acid. Macromolecules. 1997;30:2305–2312. [Google Scholar]
- 9.Matsubara K, Nakato T, Tomida M. Endogroup and irregular structure analysis in the thermally prepared sodium polyaspartate by 1H and 13C NMR spectroscopy. Macromolecules. 1998;31:1466–1472. [Google Scholar]
- 10.Peterson G L. Determination of total protein. Methods Enzymol. 1983;91:95–119. doi: 10.1016/s0076-6879(83)91014-5. [DOI] [PubMed] [Google Scholar]
- 11.Pivcova H, Saudek V, Dronbnik H. 13C NMR study of the structure of poly(aspartic acid) Polymer. 1982;23:1237–1241. [Google Scholar]
- 12.Pivcova H, Saudek V, Dronbnik J, Vlasak J. NMR study of poly(aspartic acid) Biopolymer. 1980;20:1605–1614. [Google Scholar]
- 13.Roweton S, Huang S J, Swift G. Poly(aspartic acid): synthesis, biodegradation, and current applications. J Environ Polym Degrad. 1997;5:175–181. [Google Scholar]
- 14.Saitou N, Nei M. The neighbor-jointing method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 15.Sakaki R K, Gelfand D H, Stoffe S, Scharf S J, Higuch R, Horn G T, Mullis K B, Erlich A. Primer-directed enzymatic application of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 16.Swift G, Freeman M B, Paik Y H, Simon E, Wolk S K, Yocom K M. Design and development of biodegradable polymeric poly(carboxylic acid) as co-builder for detergent. Macromol Symp. 1997;123:195–207. [Google Scholar]
- 17.Takeuchi M, Kawai F, Shimada Y, Yokota A. Taxonomic study of polyethylene glycol-utilizing bacteria: emended description of the genus Sphingomonas and new description of Sphingomonas macrogoltabidus sp. nov., Sphingomonas sanguis sp. nov. and Sphingomonas terrae sp. nov. Syst Appl Microbiol. 1993;16:227–238. [Google Scholar]
- 18.Takeuchi M, Sawada H, Oyaizu H, Yokota A. Phylogenetic evidence for Sphingomonas and Rhizomonas as nonphotosynthetic members of the alpha-4 subclass of Proteobacteria. Int J Syst Bacteriol. 1994;44:308–314. doi: 10.1099/00207713-44-2-308. [DOI] [PubMed] [Google Scholar]
- 19.Vegotsky A, Harada K, Fox S W. The characterization of polyaspartic acid and some related compounds. J Am Chem Soc. 1959;80:3361–3366. [Google Scholar]
- 20.Wolk S K, Swift G, Paik Y H, Yocom K M, Smith R L, Simon E S. One- and two-demensional nuclear magnetic resonance characterization of poly(aspartic acid) prepared by thermal polymerization of l-asparitic acid. Macromolecules. 1994;27:7613–7620. [Google Scholar]
- 21.Yakabe Y, Kitano M. Evaluation of biodegradability plastics in activated sludge. In: Doi Y, Fukuda K, editors. Biodegradable plastics and polymers. Amsterdam, The Netherlands: Elsevier; 1994. pp. 331–336. [Google Scholar]
- 22.Yamanaka H, Kawai F. Purification and characterization of constitutive polyethylene glycol (PEG) dehydrogenase of a PEG 4,000-utilizing Flavobacterium sp. No. 203. J Ferment Bioeng. 1989;67:324–330. [Google Scholar]