Abstract
Glioblastoma multiforme (GBM) is the most malignant and aggressive type of glioma. Non‐coding RNAs (ncRNAs) are RNAs that do not encode proteins but widely exist in eukaryotic cells. The common characteristics of these RNAs are that they can all be transcribed from the genome without being translated into proteins, thus performing biological functions, particularly microRNAs (miRNAs), long non‐coding RNAs (lncRNAs) and circular RNAs. Studies have found that ncRNAs are associated with the occurrence and development of GBM, and there is a complex regulatory network among ncRNAs, which can regulate cell proliferation, migration, apoptosis and differentiation, thus provide a basis for the development of highly specific diagnostic tools and therapeutic strategies in the future. The present review aimed to comprehensively describe the biogenesis, general features and functions of regulatory ncRNAs in GBM, and to interpret the potential biological functions of these ncRNAs in GBM as well as their impact on clinical diagnosis, treatment and prognosis and discusses the potential mechanisms of these RNA subtypes leading to cancer in order to contribute to the better design of personalized GBM therapies in the future.
Glioblastoma multiforme (GBM) is the most malignant and aggressive glioma. Non‐coding RNAs (ncRNAs) are a category of RNAs that do not encode proteins but widely exist in eukaryotic cells. In recent years, the functions of ncRNAs, especially microRNAs (miRNAs), lncRNAs and circRNAs and their biological mechanisms of action in GBM have been studied more and more deeply, these ncRNAs may function by modifying gene transcription at the epigenetic level in cis or trans form. In this review, we comprehensively describe the biogenesis, general features, functions and mechanisms of regulatory ncRNAs, namely miRNAs, lncRNAs and circRNAs, in GBM, and interpret the potential biological functions of these non‐coding RNAs in GBM and their impact on clinical diagnosis, treatment and prognosis.
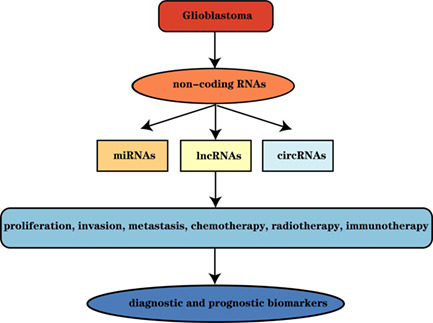
1. INTRODUCTION
Glioblastoma multiforme (GBM), also known as Grade IV glioma, is the most common tumour in the central nervous system (CNS) and one of the most lethal cancer types in humans, accounting for more than 30% of all CNS tumours, with a median survival time of only 12–15 months. 1 In addition to comprehensive treatment including surgery and chemoradiotherapy, new strategies have been carried out in clinical practice; however, the median survival time of patients with GBM has not been significantly improved. 2 This also highlights the urgent need to explore novel clinical diagnostic and treatment options for GBM, and investigate its pathogenic mechanism and functional targets to develop effective treatment and prevention measures for patients with GBM.
Non‐coding RNA (ncRNA) is a category of RNA with an extensive ability to regulate gene expression. It constitutes the majority of the transcriptome, while only ~3% of the genome contains protein‐coding RNA. 3 The majority of ncRNAs can be transcribed as various RNA products, but do not encode proteins and are mainly responsible for gene regulation at various different levels, including pre‐transcriptional or post‐translational level. 4 This study on ncRNAs in GBM mainly focuses on the general features and biological functions of microRNAs (miRNAs or miRs), long non‐coding RNAs (lncRNAs) and circular RNAs (circRNAs), in GBM, including cell proliferation, invasion, migration, apoptosis, angiogenesis, cell cycle, epithelial‐to‐mesenchymal transition (EMT) and changes in chemoradiotherapy sensitivity. In addition, their possible mechanism of action in the occurrence and development of GBM are discussed, which lays a foundation for the diagnosis, treatment and prognosis of patients with GBM in the future.
2. CLASSIFICATION AND CHARACTERISTICS OF ncRNAs
NcRNAs include various types and have several functions. NcRNAs can be divided into three types according to the length of nucleotide (nt): (i) <50 nt including miRNA, small interfering RNA and PIWI‐interacting RNA (piRNA); (ii) 50–500 nt, including ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), SLRNA and SRPRNA; and (iii) >500 nt, including long mRNA‐like ncRNAs and lncRNAs without a poly(A) tail. 5
According to their functions, ncRNAs can be divided into the following categories: (i) tRNAs with amino acid transport function; (ii) guide RNA with mRNA editing function; (iii) snRNAs with mRNA processing functions (cleavage and maturation); (iv) snoRNA with rRNA processing functions (cleavage and modification); (v) Telomerase RNA that has the function of DNA replication; (vi) signal recognition particles involved in protein transport and secretion; and (vii) regulation of piRNA in germ cells by combining with PIWI protein family members. 6 , 7 In addition, there are numerous RNAs whose functions have not been identified yet, and future research may reveal novel RNA functions. Thus, only the major ncRNAs are described in the present review.
3. MICRORNAs
3.1. Biogenesis of miRNAs
MiRNAs are endogenous ncRNAs of 18–22 nt in length that negatively regulate gene expression by interacting with the 3′ untranslated region (UTR) of mRNA targets. 8 The biogenesis of miRNA involves multiple step: the miRNA is preliminarily transcribed as a primary miRNA under the action of RNA polymerase II/III and then converted into a precursor miRNA (pre‐miRNAs) in the nucleus. 9 , 10 Under the action of Exportin5, GTP and Ran, pre‐miRNAs are transferred from the nucleus to the cytoplasm, and cleaved into mature double‐stranded miRNAs. 11 When the mature miRNAs' double helix is opened, one strand binds to the RNA‐induced silencing complex (RISC) and then binds to the target mRNA to negatively regulate gene expression, whereas the other strand is degraded 5 (Figure 1A).
FIGURE 1.
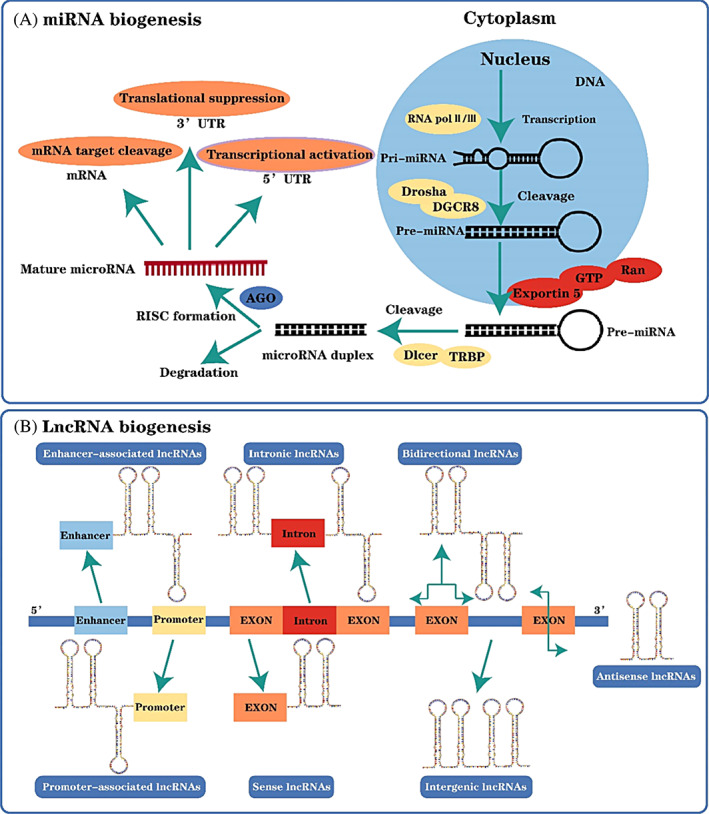
The biogenesis of microRNA (miRNA) and long non‐coding RNA (lncRNA). (A). The miRNA is preliminarily transcribed as a primary miRNA in the nucleus, then converted into a precursor miRNA under the action of Drosha and DGCR8, and transported to the cytoplasm under the action of Exportin5, GTP and Ran. After entering the cytoplasm, the precursor miRNA is transformed into double‐stranded miRNA under Dicer and TRBP processing. One miRNA is degraded, and the other miRNA becomes mature miRNA under the action of AGO and RISC, and then plays different biological roles. (B) LncRNA is transcribed by RNA polymerase II and has an mRNA‐like structure. After shearing, lncRNA has a polyA tail and promoter structure. During differentiation, lncRNA has dynamic expression and different splicing modes. (B) The different bio‐origins of lncRNAs, namely enhancer‐associated lncRNAs, intronic lncRNAs, bidirectional lncRNAs, promoter‐associated lncRNAs, sense lncRNAs and intergenic lncRNAs.
3.2. Functions of miRNAs
MiRNAs induce degradation at the mRNA level or translational repression by binding to mRNA transcripts in eukaryotes. MiRNAs may degrade mRNAs directly by labelling them and directing them to be degraded. 12 MiRNA‐induced silencing complex (miRISC) is considered to mediate the degradation of mRNAs under the action of enzymes and complexes, while argonaute in miRISCs targets mRNAs by binding to miRNAs. 13 In addition, miRNAs can reduce or inhibit gene expression by inhibiting the translation process. 14 Beilharz et al. found in mammalian cell experiments that miRNAs can enhance translation inhibition by mediating mRNA deadenylation and decay. Numerous studies have identified that miRNAs can negatively influence the expression of target genes through atypical mechanisms. For example, Matsui et al. 15 reported that miR‐589 could bind to the promoter RNA of cyclooxygenase‐2 (COX2) and cause COX2 transcriptional activation.
3.3. MiRNAs in GBM
MiRNAs have been shown to play diverse roles in GBM initiation, progression and treatment, including tumour diagnosis, treatment development and optimization, and improved patient prognosis, and are critical for regulating tumour growth, metastasis, drug resistance and metabolism 16 (Table 1).
TABLE 1.
Summarization of the mechanism and functions of miRNAs in tumorigenesis of
| MiRNAs | Role in GBM | Effect of altered expression | Target gene | References |
|---|---|---|---|---|
| miR‐1258 | Tumour suppressor | Inhibited proliferation, therapeutic resistance, migration and invasion | E2F1 | 25 |
| miR‐128 | Tumour suppressor | Decreased resistance to chemotherapy | — | 21 |
| miR‐342 | Tumour suppressor | Decreased resistance to chemotherapy | — | 21 |
| miR‐935 | Tumour suppressor | Inhibited proliferation | FZD6 | 23 |
| miR‐138 | Tumour suppressor | Inhibited proliferation, increased sensitivity to chemotherapy | Survivin | 137 |
| miR‐424 | Tumour suppressor | Inhibited proliferation, migration, promoted apoptosis and cell‐cycle arrest | RAF1, AKT1 | 49 |
| miR‐489‐3p | Tumour suppressor | Inhibited proliferation, migration, promoted apoptosis and cell‐cycle arrest | BDNF | 138 |
| miR‐128‐3p | Tumour suppressor | Increased sensitivity to chemotherapy | RUNX1 | 24 |
| miR‐138 | Tumour suppressor | Inhibited proliferation, migration, promoted cell‐cycle arrest | CD44 | 33 |
| miR‐370‐3p | Tumour suppressor | Increased sensitivity to temozolomide | FOXM1 | 28 |
| miR‐448 | Tumour suppressor | Inhibited cell viability, migration and invasion | ROCK1 | 139 |
| miR‐3928 | Tumour suppressor | Inhibited cell growth and invasion | MDM2, p53 | 32 |
| miR‐3189 | Tumour suppressor | Inhibited cell growth and promoted apoptosis | GLUT3 | 140 |
| miR‐181a‐5p | Tumour suppressor | Inhibited proliferation | ADAM8 | 50 |
| miR‐21‐5p | Tumour suppressor | Inhibited proliferation, migration and invasion | KANSL2 | 141 |
| miR‐4286 | Tumour suppressor | Inhibited invasion and mesenchymal transition | TGFB1, TGFBR2 | 142 |
| miR‐674‐5p | Tumour suppressor | Inhibited proliferation and migration | Cul4b | 143 |
| miR‐185‐5p | Tumour suppressor | Inhibited proliferation and promoted apoptosis | ANXA2 | 144 |
| miR‐21 | Oncogene | Increased resistance to chemotherapy | — | 145 |
| miR‐221/222 | Oncogene | Increased resistance to radiotherapy | — | 26 |
| miR‐133a | Oncogene | Increased proliferation, migration and invasion | TGFBR1 | 19 |
| miR‐542‐3p | Oncogene | Increased proliferation and glycolytic activity | HK2 | 27 |
| miR‐27a‐3p | Oncogene | Increased proliferation and resistance to temozolomide | BTG2 | 34 |
| miR‐601 | Oncogene | Increased proliferation | TINP1 | 35 |
Abbreviations: GBM, glioblastoma multiforme; miRNAs, microRNAs.
3.3.1. MiRNAs as diagnostic and prognostic biomarkers
MiRNAs are endogenous small ncRNAs that regulate various biological functions. 17 The majority of miRNAs have notably different expression levels between patients with GBM and normal controls. In addition, the abnormal expression of miRNAs can predict the progression of GBM, and detect the survival and prognosis of patients with GBM, which has attracted increasing attention in research. MiRNAs are expected to become biomarkers in future clinical practice. 5
As a hotspot for cancer biomarkers, circulating miRNAs are found in cell‐free body fluids, such as serum, tissue and cerebrospinal fluid. 18 , 19 ParvizHamidi et al. 20 reported that the expression of circulating miRNA‐21 and miRNA‐26a were higher in the serum and tumour tissue samples of patients with GBM than in the normal control group, and the levels of miRNA‐21 and miRNA‐26a were higher in patients with GBM before surgery than following surgery. Moreover, Wang et al. demonstrated that miR‐128 and miR‐342 expression in the plasma and tissue samples of patients with GBM was lower than that in normal controls, whereas their expression in patients tended to be normal after surgery and chemotherapy. 21 These findings suggest that miRNAs can be used as GBM‐specific diagnostic biomarkers and may be helpful in the clinical treatment of patients with GBM.
The average survival time of patients with glioma is 12–15 months, the 5‐year survival rate is <5%, and the survival and prognosis of patients with GBM are worse 22 ; thus, it is important to evaluate the patient's prognosis. High expression of miR‐1258, miR‐935 and miR‐128‐3p was associated with better overall survival (OS) in patients with GBM, 23 , 24 , 25 while low miR‐542‐3p and miR‐221/222 clusters expression was associated with better OS in patients with GBM. 26 , 27 Previous studies evaluated the effect of cell‐free circulating RNA on patients with GBM. Nadaradjane et al. 28 found that miR‐370‐3p could be treated in patients with GBM by enhancing temozolomide sensitivity, while it could not be used as a cell‐free circulating biomarker and was not associated with patient OS.
A number of researchers are currently developing prognostic models based on miRNA signatures. Cheng et al. 29 developed five miRNA signatures with prognostic value for patients with GBM. Among them, risky miRNAs included miR‐222, miR‐132 and miR‐129, whereas protective miRNAs included miR‐145 and miR‐20a. High‐risk patients expressed higher levels of risky miRNAs, as well as patients with a shorter OS, whereas low‐risk controls expressed higher levels of protective miRNAs, and had a longer OS. Moreover, a prognostic model based on miR‐125A‐5p, miR‐615‐5p, let‐7a‐5p and let‐7b‐5p expression effectively predicted the correlation with OS. 30 Santangelo et al. 31 identified alterations in the OS and progression‐free survival (PFS) of patients with GBM upon treatment with regorafenib by using miRNAs signatures, which are models that may provide potential treatment options for patients with GBM.
3.3.2. MiRNAs regulate cancer cell proliferation
The malignant proliferation of cancer cells is critical in the development of cancer. The proliferation of GBM cells is often accompanied by cell invasion, as well as oncogene activation and up‐regulation. Studies have shown that miRNAs may regulate GBM malignant proliferation by targeting multiple GBM‐related genes.
Transcription factors can enhance or inhibit gene expression by interacting with cis‐factors through their DNA‐binding domains. MiR‐1258 can inhibit GBM cell proliferation, invasion and migration and control cell cycle by targeting transcription factor E2F1. 25 Xu et al. 24 indicated that miR‐128‐3p could act on the transcription factor RUNX to enhance the sensitivity of patients with GBM to chemotherapeutic drugs and inhibit the proliferation of tumour cells. In addition, forkhead box M1 (FOXM1), a member of the Forkhead Box transcription factor family, controls cell cycle processes mediated by miR‐370‐3p, which enhanced the sensitivity of patients with GBM to temozolomide. 28 Moreover, p53, a tumour suppressor gene, forms a negative feedback loop with mouse double minute 2 homologue (MDM2) after targeted activation by miR‐3928, thus significantly inhibiting cell proliferation and invasion. 32 MiR‐138 can down‐regulate CD44 expression, thus reducing the heterogeneous adhesion between tumour cells and host matrix and inhibiting the proliferation, invasion and metastasis of GBM cells. 33
The carcinogenic role of miRNAs in GBM can also not be ignored, and its carcinogenic mechanism also needs to be further explored. MiR‐133a, miR‐27a‐3p and miR‐601 are significantly overexpressed in GBM, and they may facilitate the proliferation of GBM cells by downregulating TGFBR1, BTG anti‐proliferation factor 2 and TGF β‐inducible nuclear protein 1. 19 , 34 , 35 Previous studies have found that miR‐27a‐3p can also enhance the resistance of patients with GBM to temozolomide, which is closely associated with poor OS. 34 In addition, Kim et al. 27 found that miR‐542‐3p could induce the glycolytic activity of GBM cells by activating hexokinase 2, thus promoting cell proliferation and chemotherapy resistance, leading to reduced OS in patients.
3.3.3. MiRNAs influence cell invasion and metastasis
Thanks to the in‐depth study of the aetiology and pathogenesis of GBM, it has been found that cancer‐related miRNAs have a great impact on the metastasis and invasion of GBM cells, which also provides a new strategy for improving malignant biological activities and survival in patients with GBM. 36
EMT is markedly associated with cancer invasion and metastasis, and miRNAs affect the function of EMT in GBM, thus miRNAs may become potential diagnostic or therapeutic targets for GBM. 22 Previous studies found that miR‐451 inhibited the PI3K/AKT/Snail signalling pathway by activating calcium binding protein 39 in GBM, thereby inhibiting EMT and metastasis, 37 and miR‐200b‐3p promoted E‐cadherin expression by down‐regulating ERK5, resulting in reduced invasion ability of GBM cells. 38 Moreover, miR‐424, 39 miR‐940, 40 miR‐378 41 and miR‐139‐5p 42 inhibited the EMT of GBM by targeting the KIF23, ZEB2, IRG1 and Notch 1 genes, respectively, thus resulting in reduced invasion and metastasis of GBM cells. Furthermore, MTSS I‐BAR domain containing 1 (MTSS1) is important for inhibiting the proliferation and invasion of glioma cells, while TGF‐β1 induces EMT. MiRNAs can negatively influence MTSS1 expression, thereby facilitating the invasion and metastasis of glioma cells. 43
MMP2 and MMP9 are gelatinases of the MMP family, which degrade extracellular matrix and are involved in tumour invasion, metastasis and immune surveillance. 44 Overexpression of MMP2 and MMP9 often supported the proliferation and metastasis of GBM cells. For example, both miRNA‐146a and miRNA‐564 were able to weaken the viability, invasion and migration of GBM cells by downregulating MMP9 and EGFR expression. 45 , 46 In addition, miR‐373 can reduce the invasion and metastasis of GBM cells by negatively influencing HOXA cluster antisense RNA 2, and then inhibiting vascular endothelial cadherin expression and the activity of MMP9 and MMP2. 47
MiRNAs may also affect the metastasis and invasion of GBM cells by regulating protein expression in various signalling pathways. For instance, miR‐451 is a tumour suppressor that acts on inhibitor of NF‐κB kinase subunit β (IKKβ) to activate the NF‐κB signalling pathway and reduce MMP9, MMP2, proliferating cell nuclear antigen (PCNA), cyclin D1, IKKβ and phosphorylated p65 expression, so as to weaken the proliferation and invasion of glioma cells, 48 whereas Gheidari et al. 49 reported that miR‐489‐3p could activate the PI3K/AKT signalling pathway under the action of brain‐derived neurotrophic factor and inhibit the invasion of GBM cells. The metalloproteinase integrin ADAM metallopeptidase domain 8 can act on the STAT3 and MAPK signalling pathways to regulate the expression of MMP9, CAMP responsive element binding protein 1, MEK1 and ERK2, thus regulating the level of miR‐181a‐5p and promoting the invasiveness of GBM. 50
3.3.4. MiRNAs affect the sensitivity to radiotherapy and chemotherapeutic drugs
Apart from surgical treatment, radiotherapy and chemotherapy are also important means to treat GBM. However, GBM cells have been clinically shown in recent years to be resistant to radiotherapy and chemotherapy, resulting in poor prognosis of patients. 51 , 52 Previous studies have found that miRNAs play crucial roles in regulating the radiation and drug resistance of GBM cells.
It was found that both ionizing radiation‐induced miR‐494 and miR‐30 e can activate the AKT and ERK signalling pathways by acting on EGFR, thereby facilitating the invasion and migration of GBM cells. 53 , 54 This result provides a basis for the study of radiation resistance targets in tumour radiotherapy. Areeb et al. 55 found that EGFR expression was decreased in GBM cells resistant to radiation and temozolomide. MiRNA prediction software was used to find that miR‐221 could negatively regulate EGFR and mediate resistance to radiotherapy and chemotherapy, thus becoming a potential target for GBM treatment. It has also been confirmed that the exosome‐derived miR‐1238 of chemotherapy‐resistant GBM cells can activate EGFR signalling pathway by targeting caveolin 1 and induce chemotherapy resistance in GBM cells to resist chemotherapy. 56 In addition, miR‐21 inhibitors in combination with paclitaxel can increase GBM cell apoptosis by inhibiting the STAT3 signalling pathway and increase sensitivity to chemotherapy drugs. 57
3.3.5. Exosomal miRNAs in GBM
Exosomes are extracellular vesicles (EVs) derived from endosomes, which are involved in intercellular communication, molecular transfer and antigen presentation. 58 Tumour‐derived exosomes are involved in distant metastasis of tumour cells, remodelling of the tumour microenvironment and changes in drug resistance of tumour cells. 59 Qiu et al. 60 found that miR‐25‐3p exosomes can promote GBM cell proliferation, cyclin E expression and temozolomide resistance by targeting F‐box and WD repeat domain containing 7(FBXW7). 59 It has been found that exosome miR‐1246 isolated from cerebrospinal fluid, plasma and cells of patients with glioma promotesthe differentiation of bone marrow‐derived inhibitory cells (MDSCs) by activating the dual specificity protein phosphatase 3 (DUSP3)/ERK signalling pathway, while hypoxia can induce the transcription of exosomal miR‐1246 and promote the activation of MDSCs. In addition, numerous researchers have also explored serum exosomal miRNAs of patients with GBM and normal subjects through comprehensive gene expression databases or prospective studies. For example, Yang et al. 61 suggested that serum exosomal miR‐98‐5p, miR‐183‐5p, miR‐323‐3p and miR‐19b‐3p were potential biomarkers for GBM by using the GSE112462 and GSE122388 datasets, whereas Olioso et al. 62 found that patients with GBM with higher exosomal miRNA expression had relatively lower OS and PFS. In conclusion, exosome‐mediated miRNAs play a variety of roles in the biological functions of GBM cells and the prognosis of patients with GBM, which requires further investigation in the future.
4. LONG NON‐CODING RNAs
4.1. Biogenesis of lncRNAs
LncRNAs have been widely studied in the field of cancer. Their length exceeds 200 nt and they do not encode proteins but retain the function of protein‐coding genes. 63 LncRNAs are transcribed by RNA polymerase II, capped at the 5′ end and polyadenylated at the 3′ end, and most of them contain >2 exons, while >60% contain poly(A) tails. 64 LncRNAs can be divided into sense transcript, antisense transcript, bidirectional transcript, intergenic gene, promoter, intron or enhancer according to their genome origin and distribution 65 (Figure 1B). Although the mechanisms of lncRNAs involved in the regulation of GBM have been explored in depth recently, their biogenesis and pathogenesis remain to be further investigated due to their variety and complex mechanism.
4.2. Functions of lncRNAs
LncRNAs have a variety of regulatory functions, including chromatin remodelling, transcriptional and post‐transcriptional regulation, and their versatility may be the source of their functional diversity. 66 LncRNAs can regulate gene expression by participating in responses to various stimuli through different mechanisms. 67 It has been found that lncRNAs can form R‐loops on target gene promoters to regulate their transcription, 68 and can also bind to transcription factors and histone modification complexes to regulate the transcription process. 69 Previous studies have found that non‐translational lncRNAs can participate in the regulation of effector proteins expressed by genes. For example, lncRNAs MEG3 can negatively influence c‐Myc expression by promoting the translation of the PHLPP2 gene, thereby inhibiting the invasion of bladder cancer cells. 70 Furthermore, lncRNAs participate in chromatin modification and can aggregate in the nucleus to regulate chromatin structure, or interact with chromatin modification enzymes to catalyse covalent changes of histones or nucleic acids, thus regulating genetic information. 71 Recently, studies have found that lncRNAs can be used as miRNA sponges as competing endogenous RNAs (ceRNAs). Such LncRNAs can bind to specific binding sites of miRNAs and regulate miRNA expression and function. 72 For example, LINC01123 can sponge miR‐199a‐5p to upregulate c‐Myc expression in patients with non‐small cell lung cancer, thus leading to a poor prognosis. 73 In conclusion, lncRNAs have multiple functions, and can regulate chromatin modification complexes at the transcriptional or translational level, as well as the growth, proliferation, differentiation, epigenetic inheritance and genomic imprinting of tumour cells. 74 However, the biological functions and regulatory mechanisms of lncRNAs remain unclear to some extent, and further research is necessary.
4.3. LncRNAs in GBM
Increasing evidence indicates that lncRNAs can positively or negatively regulate the expression of GBM cells and the prognosis of patients with GBM. It has been found that lncRNAs play crucial roles in affecting GBM cell proliferation, apoptosis, invasion, metastasis and drug resistance to chemotherapy. 75 The pathogenic role of lncRNAs in GBM is increasingly known, and they may even be used as potential biomarkers in the diagnosis and treatment of GBM in the future.
4.3.1. LncRNAs influence cancer cell growth
The expression of lncRNA DLGAP1 antisense RNA 1 (AS1) is higher in GBM tissues and cells than in the corresponding normal controls. Wang et al. 76 found that silencing lncRNA DLGAP1‐AS1 inhibited GBM cell proliferation by targeting the miR‐515‐5p/Rho‐associated coiled‐coil containing protein kinase 1 (ROCK1)/NFE2 like BZIP transcription factor 1 (NFE2L1) axis by functional analysis. Similarly, Zhang et al. 77 reported that LncRNA MIR31HG could activate STAT1 and promote GBM cell proliferation and inhibit cell apoptosis via the Wnt/β‐catenin signalling pathway. Previous research has found that lncRNA can promote GBM tumorigenesis through the ubiquitin‐proteasome pathway. For instance, Liang et al. 78 demonstrated that lncRNA nuclear enriched abundant transcript 1 (NEAT1) deubiquitinated phosphoglycerate kinase 1 to synergistically promote GBM cell proliferation and glycolysis, whereas Lv et al. 79 reported that lncRNA plasmacytoma variant translocation 1 (PVT1) could recruit COP9 signalosome subunit 5 (COPS5) to deubiquitinate and stabilize tripartite motif containing 24 (TRIM24), thus promoting GBM cell proliferation (Table 2).
TABLE 2.
Summarization of the cellular functions of lncRNAs in tumorigenesis of GBM.
| LncRNAs | Role in GBM | Effect of altered expression | Molecular mechanism | References |
|---|---|---|---|---|
| DLGAP1‐AS1 | Oncogene | Promoted proliferation | Regulated miR‐515‐5p/ROCK1/NFE2L1 axis | 76 |
| MIR31HG | Oncogene | Promoted proliferation and inhibited apoptosis | Activated STAT1 and Wnt/β‐catenin signals | 77 |
| NEAT1 | Oncogene | Promoted proliferation and glycolysis | Deubiquitinate and stabilize PGK1 | 78 |
| PVT1 | Oncogene | Promoted proliferation | Deubiquitinate and stabilize TRIM24 | 79 |
| LINC00998 | Tumour suppressor | Inhibited proliferation | Regulated c‐Met/Akt/mTOR axis | 82 |
| SEMA3B | Tumour suppressor | Inhibited proliferation | Downregulated cyclin D1 | 83 |
| RBPMS‐AS1 | Tumour suppressor | Inhibited proliferation and enhanced radiosensitivity | Sponged miR‐301a‐3p/CAMTA1 axis | 84 |
| CHRM3‐AS2 | Oncogene | Promoted proliferation, invasion and migration | Regulated miRNA‐370‐5p/KLF4 axis | 85 |
| MIR210HG | Oncogene | Promoted proliferation, invasion | Interaction with OCT1 | 86 |
| LINC01057 | Oncogene | Promoted proliferation, invasion, migration and radio‐resistance | Activated NF‐κB signals | 87 |
| PRADX | Oncogene | Promoted proliferation, basal respiration, proton leak and ATP production | Activated STAT3 signals | 146 |
| NEAT1 | Oncogene | Promoted invasion and EMT | — | 89 |
| OXCT1‐AS1 | Oncogene | Promoted proliferation, invasion, migration; increased the G0/G1 phase cells and decreased the G2/M phase cells | Sponged miR‐195/CDC25A axis | 90 |
| HNF1A‐AS1 | Oncogene | Promoted proliferation, invasion and migration | Sponged miR‐22‐3p/ENO1 axis | 91 |
| HOXD‐AS2 | Oncogene | Promoted proliferation, invasion and migration | Sponged miR‐3681‐5p/MALT1 axis | 92 |
| HOTAIRM1 | Oncogene | Promoted proliferation, invasion and radiotherapy resistance | Sponged miR‐17‐5p/TGM1 axis | 93 |
| NONHSAT079852.2 | Oncogene | Promoted proliferation, invasion, migration; decreased the G1 phase cells and increased the G2 phase cells | Sponged miR‐10,401‐3p/HSPA1A axis | 147 |
| MUF | Oncogene | Promoted proliferation and radiotherapy resistance, decreased apoptosis rate | Sponged miR‐34a/Snail1 axis | 148 |
| TCONS‐00004099 | Oncogene | Promoted proliferation, invasion and migration | Regulated miRNA/PTPRF axis | 149 |
| LINC‐PINT | Tumour suppressor | Inhibited proliferation, invasion and migration | Regulated Wnt/β‐catenin signals | 94 |
| DGCR10 | Tumour suppressor | Inhibited invasion and migration | Regulated STAT5 and NF‐κB signals | 95 |
| HRA1B | Tumour suppressor | Inhibited invasion and migration | Regulated STAT5 and NF‐κB signals | 95 |
| OIP5‐AS1 | Oncogene | Promoted proliferation and temozolomide resistance | Sponged miR‐129‐5p/ IGF2BP2 axis | 96 |
| UCA1 | Oncogene | Promoted proliferation and temozolomide resistance, decreased apoptosis rate | Regulated miR‐182‐5p/MGMT axis | 97 |
| LINC00511 | Oncogene | Promoted temozolomide resistance | Sponged miR‐126‐5p and regulated Wnt/β‐catenin signals | 98 |
| DANCR | Oncogene | Promoted etoposide resistance | Interacted with FOXO1 and promoted FOXO2 ubiquitination | 100 |
| HOTAIR | Oncogene | Promoted proliferation, invasion and temozolomide resistance | Sponged miR‐526b‐3p/EVA1 axis | 101 |
Abbreviations: GBM, glioblastoma multiforme; lncRNAs, long non‐coding RNAs.
Numerous studies have shown that lncRNA as ceRNA regulates tumorigenesis and promotes the progression of GBM. Wang et al. 80 reported that lncRNA H19 was highly expressed in GBM compared with normal controls, and H19 promoted the proliferation and autophagy of GBM cells and inhibited apoptosis by sponging miR‐491‐5p. Lu et al. 81 found that lncRNA HAS2‐AS1 could sponge miR‐137 to promote GBM cell and tissue proliferation and reduce survival rate (Table 3). Conversely, multiple lncRNAs also play tumour‐suppressive roles in the development of GBM. LncRNA LINC00998 can bind to chromobox 3 (CBX3) to inhibit GBM cell proliferation through the c‐Met/AKT/mTOR axis and improve the survival rate of patients with GBM. 82 In U87‐MG and U251‐MG cells, lncRNA semaphoring 3B (SEMA3B) inhibited the proliferation of cyclin D1 by downregulating miR‐195. 83 In addition, lncRNA RBPMS‐AS1 enhanced calmodulin‐binding transcription activator (CAMTA) expression in GBM cells by sponging miR‐301a‐3p, thereby enhancing the radiosensitivity of GBM and inhibiting tumour proliferation and occurrence. 84
TABLE 3.
Summarization of the role of lncRNAs as ceRNA in GBM.
| LncRNAs | MiRNA | Expression of mRNA | Function | References |
|---|---|---|---|---|
| H19 | miR‐491‐5p | Upregulated ERN1 | Promoted proliferation and autophagy | 80 |
| HAS2‐AS1 | miR‐137 | Upregulated LSP1 | Promoted proliferation | 81 |
| RBPMS‐AS1 | miR‐301a‐3p | Upregulated CAMTA1 | Inhibited proliferation and enhanced radiosensitivity | 84 |
| OXCT1‐AS1 | miR‐195 | Upregulated CDC25A | Promoted proliferation, invasion, migration, induced cell cycle arrest | 90 |
| HNF1A‐AS1 | miR‐22‐3p | Upregulated ENO1 | Promoted proliferation, invasion and migration | 91 |
| HOXD‐AS2 | miR‐3681‐5p | Upregulated MALT1 | Promoted proliferation, invasion and migration | 92 |
| HOTAIRM1 | miR‐17‐5p | Upregulated TGM1 | Promoted proliferation, invasion and radiotherapy resistance | 93 |
| NONHSAT079852.2 | miR‐10,401‐3p | Upregulated HSPA1A | Promoted proliferation, invasion, migration; decreased apoptosis rate | 147 |
| MUF | miR‐34a | Upregulated Snail1 | Promoted invasion | 148 |
| OIP5‐AS1 | miR‐129‐5p | Upregulated IGF2BP2 | Promoted proliferation and temozolomide resistance | 96 |
| LINC00511 | miR‐126‐5p | Upregulated DVL3, WISP1 and WISP2 | Promoted temozolomide resistance | 98 |
| HOTAIR | miR‐526b‐3p | Upregulated EVA1 | Promoted proliferation, invasion and temozolomide resistance | 101 |
Abbreviations: ceRNA, competing endogenous RNAs; GBM, glioblastoma multiforme; lncRNAs, long non‐coding RNAs; miRNA, microRNA.
4.3.2. LncRNAs regulate migration and metastasis
Increasing evidence suggests that lncRNAs promote GBM cell invasion and metastasis in vitro and in vivo. It has been demonstrated that lncRNA CHRM3‐AS2 is upregulated in GBM, and enhances GBM cell viability while promoting GBM invasion and migration by targeting the miRNA‐370‐5P / KLF transcription factor 4 (KLF4) axis. 85 Ho et al. 86 found that hypoxia‐induced lncRNA‐MIR210HG promoted the proliferation and invasion of GBM cells by interacting with organic cation transporter 1 (OCT1). In addition, since a variety of lncRNAs facilitate the invasion and metastasis of GBM cells, numerous researchers have explored whether lncRNAs can maintain the mesenchymal phenotype of GBM cells. It has been found that lncRNA LINC01057 can promote the invasion and radio‐resistance of GBM cells, as well as inducing EMT by promoting the nuclear translocation of IKKα to activate the NF‐κB signalling pathway. 87 Yang et al. 88 constructed EMT‐related lncRNA prognostic signatures for patients with GBM, and confirmed that EMT and metastasis‐related pathways are risk indicators for patients with GBM. Moreover, by exploiting invasion‐related lncRNAs from single‐cell RNA sequencing data, it was found that patients with GBM exhibiting high NEAT1 expression had poor OS and DFS, and could promote the occurrence and progression of the malignant phenotype in patients with GBM. 89
Previous studies have used microarray analyses to construct a ceRNA network and to explore whether lncRNAs play roles of ceRNA in GBM as well as the specific molecular mechanism. It has been reported that lncRNA OXCT1‐AS1 competitively binds to miR‐195 and negatively regulates CDC25A to facilitate the proliferation, migration and invasion of GBM cells, while the number of cells in G0/G1 phase decreases and the number of cells in G2/M phase increases, which promotes the malignant progression of GBM. 90 Ma et al. 91 found that lncRNA HNF1A‐AS1 sponges miR‐22 and induces its degradation to promote the malignant behaviour of GBM cells. LncRNA HOXD‐AS2 maintains mucosa‐associated lymphoid tissue lymphoma translocation protein 1 (MALT1) expression by sponging miR‐3681‐5p, thereby inducing the proliferation, invasion and migration of GBM cells. 92 Ahmadov et al. 93 suggested that lncRNA HOTAIRM1 (HOXA transcript antisense RNA, myeloid‐specific 1) upregulated transglutaminase 2 (TGM2) by sponging miR‐17‐5p, thereby upregulating the viability and invasion ability of GBM cells and enhancing their radiotherapy resistance in vitro and in vivo.
In contrast, certain studies have demonstrated that lncRNAs also act as inhibitors of cell migration. LncRNA LINC‐PINT (long intergenic non‐protein coding RNA, p53‐induced transcript) is downregulated in GBM tissues and cells, and plays a tumour‐suppressive role by weakening the proliferation and viability of GBM cells. Zhu et al. 94 suggested that LINC‐PINT could inhibit the proliferation, invasion and EMT of GBM through the Wnt/β‐catenin signalling pathway. Huang et al. 95 constructed a prognostic model of GBM mesenchymal associated lncRNAs, and found that DiGeorge syndrome critical region gene 10 (DGCR10) and HRA1B could significantly prevent the invasion and migration of GBM cells according to The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/), and predicted that their tumour‐suppressive effect may be closely associated with the STAT5 and NF‐κB signalling pathways by Gene Set Enrichment Analysis (GSEA).
4.3.3. LncRNAs affect the sensitivity to chemotherapy
Previous studies have demonstrated that Opa interacting protein 5 (OIP5)‐AS1 is a highly expressed lncRNA in GBM. Inhibition of OIP5‐AS1 can promote the sensitivity of GBM cells to temozolomide and reduce the proliferation of tumour cells. The resistance mechanism of OIP5‐AS1 depends on its binding to miR‐129‐5p, thereby upregulating insulin‐like growth factor 2 mRNA binding protein 2 (IGF2BP2) expression. 96 In addition, upregulation of lncRNA UCA1 contributes to temozolomide resistance in GBM, while silencing urothelial cancer associated 1 (UCA1) attenuates temozolomide resistance in GBM cells by inhibiting O‐6‐methylguanine‐DNA methyltransferase (MGMT) protein levels. 97 It has also been found that LINC00511 expression is increased in temozolomide‐resistant GBM cells compared with that of parental cells and is associated with a low OS rate in patients with GBM. The resistance mechanism is that LINC00511 sponges miR‐126‐5p and activates the Wnt/β‐catenin signalling pathway. 98 RNA methylation modification (m6A) is the most usual modification in eukaryotic mRNAs. 99 IGF2BP2 family members can recognize and stabilize target RNAs. Han et al. 100 found that IGF2BP2 induced differentiation antagonizing non‐protein coding RNA (DANCR) to interact with forkhead box protein O1 (FOXO1) and promote FOXO2 ubiquitination by stabilizing lncRNA DANCR. Thus, the protein expression of FOXO3 was inhibited, and the resistance of GBM cells to etoposide was eventually promoted. In addition, Wang et al. 101 proposed that GBM serum EV‐derived lncRNA HOTAIR (HOX transcript antisense RNA) could induce the proliferation, invasion and temozolomide resistance of GBM cells, and its malignant characteristics were mainly induced by the upregulation of epithelial V‐like antigen 1 (EVA1) expression via sponging miR‐526b‐3p.
4.3.4. LncRNAs mediate immunotherapy
In recent years, the mechanism of the immune response in GBM has been identified, and immunotherapy strategies have a potential value in initiating and enhancing host anti‐tumour immunity. 102 However, tumour‐mediated immune suppression, including checkpoint suppression against PD‐1/PD‐L1, makes GBM difficult to eradicate. 103 Yi et al. 104 found that the novel RNA‐binding protein polymerase I and transcript release factor (PTRF) maintained the mRNA stability of lncRNA NEAT1 and inhibited UBX domain protein 1 (UBXN1) expression, consequently activating the NF‐κB signalling pathway, promoting the binding of NF‐κB to the PD‐L1 promoter region, and enhancing the transcription of PD‐L1, ultimately promoting the immune evasion of GBM cells. Previous studies focused on the transcriptome‐wide m6A methylation profile of lncRNAs in GBM. It was suggested that heat shock 70 kDa protein 7 (HSPA7) could be a possible prognostic risk factor for patients with GBM according to the analysis of GBM m6A sequencing data, and the results revealed that HSPA7 could upregulate yes‐associated protein 1 (YAP1) and lysyl oxidase (LOX) expression in GBM stem cells, thus promoting the recruitment of macrophages into the tumour microenvironment. However, silencing HSPA7 could inhibit the efficiency of PD‐1 therapy, making it possible to use HSPA7 as a new target for immunotherapy. 105 In addition, GBM data based on lncRNA high‐throughput sequencing obtained from TCGA database are currently being explored by various researchers. Li et al. 106 identified immune‐related lncRNAs by Pearson correlation and constructed an immune‐lncRNAs co‐expression network. A total of five immune‐related lncRNA signatures were obtained, and it was verified that the high‐risk group had lower OS and worse overall prognosis. Moreover, via GSEA, Gao et al. 107 analysed the transcriptome information of 144 GBM cases using TCGA database, and evaluated and identified six lncRNAs associated with immunophenotypes, including USP30‐AS1, LINC01684, PSMB8‐as1, AL133264.2, HCP5 and LINC01506. Reverse transcription‐quantitative PCR analyses confirmed that these six lncRNAs were highly expressed in GBM tissues compared with their expression in normal samples, and were crucial in tumour immune infiltration; thus, they may become diagnostic indicators of the GBM immunophenotype in the future. In conclusion, lncRNA‐dependent immune regulation and immune‐related lncRNAs in the immunophenotype and treatment strategies of GBM are promising research hotspot.
4.3.5. LncRNAs may serve as promising prognostic and diagnostic biomarkers
Due to the stable secondary structure of lncRNA, various studies have included it as a promising peripheral biomarker and prognostic indicator for GBM. 108 It has been found that the mRNA expression of lncRNA HOTAIR in the serum of patients with GBM is higher than that of low‐grade gliomas controls. Tan et al. 109 evaluated the serum HOTAIR level of patients with GBM and controls as a potential diagnostic marker, with an AUC (area under the curve) value of 0.913, and a sensitivity and specificity of 86.1% and 87.5%, respectively. Shen et al. 110 observed that HOTAIR drove the proliferation and invasion of GBM cells by analysing the serum of 106 patients with GBM, and HOTAIR expression was negatively correlated with the OS of patients, thus becoming an indicator of poor prognosis in GBM. In contrast, growth arrest specific 5 expression in GBM serum was lower than that in normal controls, and its downregulation was associated with decreased disease‐free survival and OS. In general, this type of minimally invasive liquid biopsy is expected to greatly improve the diagnosis and prognosis of patients with GBM. Wang et al. 111 proposed an exosome‐derived ceRNA network based on the Gene Expression Omnibus and TCGA databases by LASSO and multivariate Cox regression analysis, and recognized HOTAIR, SRY‐box transcription factor 21‐AS1 and six‐transmembrane epithelial antigen of prostate 3‐AS1 as possible prognostic exosomal lncRNAs. The nomogram was constructed based on the patients' age, isocitrate dehydrogenase status, MGMT promoter status, selection of chemoradiotherapy, and exosomal lncRNA, and the results showed that it had good recognition and prediction ability. In conclusion, increasing evidence has shown that lncRNA can be applied as possible diagnostic and therapeutic biomarkers for GBM, which plays important roles in detecting the effect of therapy and tumour recurrence.
5. CIRCULAR RNAs
5.1. Biogenesis of circRNAs
CircRNAs were initially found in RNA viruses by electron microscopy by Sanger et al. 112 In 1993, the structure of circRNAs was confirmed. 113 CircRNA is biosynthesized in the presence of precursor RNA transcribed by RNA polymerase II and RNA‐binding protein, 114 it can be divided into intronic circRNAs, exonic circRNAs and EciRNA. 115 The mechanism of circRNA maturation is not fully understood. Jeck et al. 116 have proposed three circRNA formation models, namely intron‐pairing‐driven circularization, RNA‐binding protein‐dependent circularization and lariat‐driven circularization. (Figure 2).
FIGURE 2.
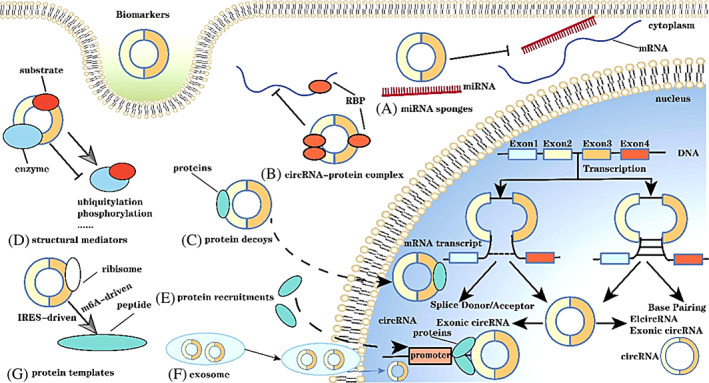
The biogenesis and biological functions of circular RNAs (circRNAs). (1) LARIAT‐driven circularization: the cleaved donor at the 3′ end of Exon 1 binds covalently to the cleaved acceptor at the 5′ end of Exon 4. Lariat is generated under exon hopping, and then the intron is removed to form circRNA. (2) Intron pairing‐driven circularization: successive introns form a circular structure, which is then cleaved to form exon circRNA. (A–G) systematically summarizes the general functions of circRNA.
5.2. Functions of circRNAs
The most common biological function of circRNAs is miRNA sponge (Figure 2), which means that the circRNA acts as a competitive inhibitor to competitively inhibit the binding between miRNA and target genes, so as to protect mRNA from miRNA degradation and maintain the stability of target genes. 117 For instance, circENTPD7 affects ROS1 expression by sponging miR‐101‐3p, thus facilitating the proliferation of GBM cells and enhancing cell viability. 117 Zhang et al. found that circFOXO3 acts as a ceRNA to upregulate the nuclear factor expression of nuclear factor of activated T cells 5 (NFAT5) by sponging miR‐138‐5p and miR‐432‐5p, so as to promote tumour invasion and metastasis. 118 CircRNA can bind to RNA‐binding proteins to influence the expression of target genes, and it can also inhibit the binding of miRNA and RNA‐binding proteins to indirectly regulate the function of RNA‐binding proteins. 119 In addition, circRNA can induce the transfer of target proteins to the nucleus to perform biological functions. It was found that circ‐AMOTL1 (angiomotin like 1) increased the expression and stability of c‐Myc in the nucleus by interacting with c‐Myc and circ‐AMOTL1 expression also enhanced the binding affinity of c‐Myc to multiple promoters. 120 Moreover, circRNA can promote the interaction between certain enzymes and substrates and influence the kinetics of reaction, such as ubiquitination and phosphorylation. 121 Since circRNA is mainly localized in the nucleus, it can recruit proteins to promoters or other specific sites at the transcriptional level, thereby enhancing the function of ribonucleoprotein and regulating gene expression. 122 Certain circRNAs can also be carried by exosomes to participate in functional interactions between cells. Finally, circRNA enters the inteRNAl ribosome entry site (IRES) and m6A sites via inteRNAl ribosomes in the 5′ UTR of mRNA to initiate the translation process, IRES initiates the circRNA translation process by recruiting ribosomes, whereas m6A initiates the translation process by binding to eukaryotic initiation factor 3 (EIF). 123 In conclusion, circRNA has a variety of functions, among which, the most often studied and reported is miRNA sponge, including in GBM. The findings of future studies will suggest new possibilities for the future diagnosis and treatment of GBM by circRNA.
5.3. CircRNAs in GBM
Increasing evidence considers circRNA as a new hotspot for the study of the association between ncRNAs and various cancer types. The mechanism of circRNAs in cancer, particularly in GBM, is still being explored. To date, the research on circRNAs in GBM has mainly focused on ‘miRNA sponges’, which refers to the participation of circRNA in the progression of GBM as a ceRNA, which can bind to downstream target genes of miRNA, thereby inhibiting the binding of miRNA and target mRNA, thus protecting mRNA from miRNA degradation. 117 Furthermore, numerous studies have suggested that circRNAs may have a potential role in the diagnosis and prognosis of GBM. The present review has summarized the oncogenic or anticancer effects of circRNAs as miRNA sponges in GBM and their mechanism of action (Figure 3).
FIGURE 3.
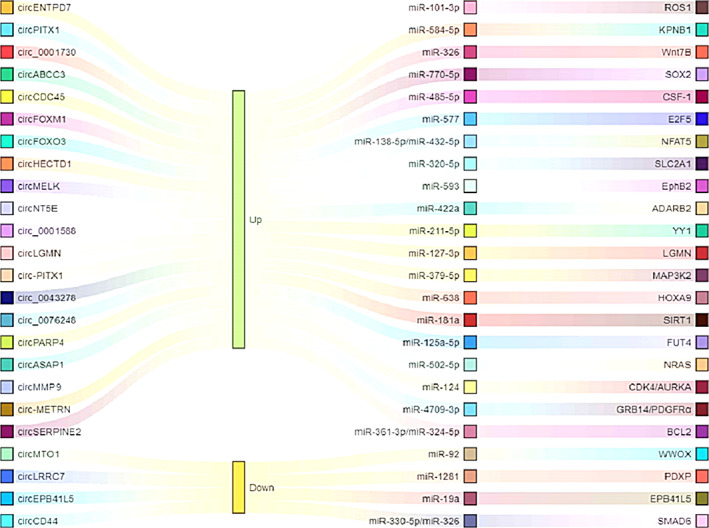
Some important circular RNAs (circRNAs) as microRNA (miRNA) sponges and corresponding target genes. circRNAs can be served as miRNAs sponge, thus targeting target genes, which positively or negatively participate in the interaction or crosstalk of various key pathways in glioblastoma, thus affecting the biological function of glioblastoma cells.
5.3.1. CircRNAs as tumour promoters
CircRNAs, as miRNA sponges, can promote numerous malignant behaviours in GBM tissues and cells, such as promoting tumour cell proliferation and sensitivity to chemotherapy drugs. It was found that circ‐MELK was upregulated in GBM, and its high expression promoted the proliferation and viability of GBM cells, while knockdown of circ‐MELK could significantly inhibit GBM growth in vitro and in vivo. A previous study indicated that circ‐MELK could promote the mesenchymal transition of GBM cells and maintain the roles of tumour stem cells by targeting EPH receptor B2 (EphB2) via sponging miR‐593. 124 CircNT5E (circ 5′‐nucleotidase ecto [NT5E]) derived from NT5E is formed by binding of adenosine deaminase RNA‐specific B2 (ADARB2) binding to the flanking site of the circRNA intron, and its high expression often causes the proliferation and distant metastasis of GBM cells in GBM. CircNT5E can restrain cell proliferation, invasion and migration by targeting PIK3CA and NT5E via sponging miR‐422a in GBM. 125 In addition, circ‐LGMN (legumain) may also promote the development of GBM by targeting the miR‐127‐3p /LGMN axis, and circ‐LGMN can upregulate LGMN expression by sponging miR‐127‐3p, thus facilitating the proliferation and invasion of tumour cells and tissues. 126 Wei et al. 127 also found that EIF4A3‐induced circASAP1 (ArfGAP with SH3 domain, ankyrin repeat and PH domain 1) was markedly upregulated in recurrent GBM tissues and temozolomide‐resistant cell lines, and circASAP1 upregulated NRAS expression by sponging miR‐502‐5p, increasing the proliferation of temozolomide‐resistant GBM cells and their resistance to temozolomide.
Previous studies indicated that circRNAs are important for GBM invasion, metastasis, and radio‐sensitization. It has been found that circFOXO3 expression is upregulated in GBM tissues. Functional experiments have found that circFOXO3 significantly enhances the invasion and migration of GBM cells, and biochemical analysis has shown that circFOXO3, as a ceRNA, can upregulate the expression of the nuclear factor of NFAT5 by sponging miR‐138‐59 and miR‐432‐5p, thus promoting the progression of GBM in vitro and in vivo. 128 Activated EGFR signalling drives the occurrence and progression of the majority of GBM tumours, and the secreted E‐cadherin protein variant encoded by E‐cadherin‐RNA plays an important role in activating EGFR signal transduction in GBM and promoting tumorigenicity of glioma stem cells. 129 Wang et al. 130 indicated that circMMP9 upregulated the protein expression of CDK4 and aurora kinase A (AURKA) by targeting miR‐124, and facilitated the proliferation, invasion and migration of GBM cells via the circMMP9/ miR‐124 axis. It has been previously found that low‐dose radiation‐induced exosomes (ldrEXOs) and ldrEXOs‐derived circ‐METRN (meteorin) affect the progression and radiotherapy sensitivity of GBM. Low‐dose radiation can increase the level of circ‐METRN by stimulating IdrEXOs secretion, thereby increasing the level of the DNA damage repair protein γ‐H2AX, and facilitating the proliferation, invasion, migration and radiation resistance of GBM cells by targeting growth factor receptor‐bound protein 14 (GRB14)/platelet‐derived growth factor receptor (PDGFRα) with miR‐4709‐3p. 131
5.3.2. CircRNAs as tumour suppressors
Although circRNAs are usually involved in gene regulation, tissue and cell carcinogenesis and other pathophysiological processes associated with high expression levels in GBM, certain circRNAs also play tumour suppressor roles in GBM. It was found that circ‐SHPRH (SNF2 histone linker PHD RING helicase) was downregulated in GBM but highly expressed in normal human brain, and SHPRH‐146AA increased the ability of SHPRH to ubiquitinate PCNA by protecting SHPRF from ubiquitin proteasome degradation, resulting in decreased tumour proliferation and reduced tumour‐genic ability. 132 CircAKT3 is an AKT transcriptional variant, and its expression level in GBM tissues is markedly lower than that in adjacent normal controls. AKT3‐174aa encoded by circAKT3 negatively regulates the PI3K/AKT signalling pathway by interacting with phosphorylated pyruvate dehydrogenase kinase 1 (PDK1) in GBM; thus, the proliferation, radiation resistance and tumorigenic ability of GBM cells could be inhibited, which would provide benefits for the long‐term prognosis of patients. 133 Lou et al. 134 found that the protein expression of circCDR1 (cerebellar degeneration‐related protein 1) was notably reduced in GBM, and may serve as a reliable predictor of OS in patients with GBM. Previous studies have found that circCDR1 as stabilizes p53 protein by de‐ubiquitination of p53, thus making p53 interact with the DNA‐binding domain, disrupting the formation of the p53/MDM2 complex, and finally protecting GBM cells from DNA damage, thus playing a tumour suppressor role. Yang et al. 135 found that the open reading frame of circ‐FBXW7 encodes a small functional protein FBXW7‐185aa driven by the ribosome entry site. The expression levels of circ‐FBXW7 and FBXW7‐185aa were low in GBM, and the up‐regulation of CIRC‐FBXW7 and FBXW7‐185aa could significantly inhibit cell proliferation and cell cycle accelerating, and improve the OS of GBM patients, thus becoming a potential functional protein with prognostic significance in GBM. In addition, it has been suggested that circCD44 can regulate SMAD6 expression by sponging miR‐326 and miR‐330‐5p, thereby downregulating circCD44 and causing reduced proliferation, invasion and migration ability of GBM cells. 136 In conclusion, the mechanism of the tumour‐suppressive effect of circRNA in GBM remains unclear; thus, it is necessary to further explore its specific function and mechanism in GBM in the future (Table 4).
TABLE 4.
Mechanism and functions of circRNAs in GBM tumorigenesis.
| CircRNAs | Role in GBM | Cancer phenotype | Sponge miRNAs | References |
|---|---|---|---|---|
| circENTPD7 | Oncogene | Promoted proliferation and viability | miR‐101‐3p | 118 |
| circPITX1 | Oncogene | Promoted proliferation, migration, invasion, angiogenesis and induced cell cycle arrest | miR‐584‐5p | 150 |
| circ_0001730 | Oncogene | Promoted proliferation, invasion and EMT | miR‐326 | 151 |
| circABCC3 | Oncogene | Promoted proliferation, migration, invasion, angiogenesis and apoptosis | miR‐770‐5p | 152 |
| circCDC45 | Oncogene | Promoted proliferation, migration and invasion | miR‐485‐5p | 153 |
| circFLNA | Oncogene | Promoted proliferation, and invasion | miR‐199‐3p | 154 |
| circFOXM1 | Oncogene | Promoted proliferation, migration and invasion | miR‐577 | 155 |
| circFOXO3 | Oncogene | Promoted migration and invasion | miR‐138‐5p/miR‐432‐5p | 128 |
| circHECTD1 | Oncogene | Promoted proliferation and migration | miR‐320‐5p | 156 |
| circMELK | Oncogene | Promoted proliferation, and invasion | miR‐593 | 124 |
| circNT5E | Oncogene | Promoted proliferation, migration and invasion | miR‐422a | 125 |
| circNUP98 | Oncogene | Promoted proliferation | miR‐519a‐3p | 157 |
| circSKA3 | Oncogene | Promoted proliferation | miR‐1 | 158 |
| circ_0001588 | Oncogene | Promoted proliferation, migration and invasion | miR‐211‐5p | 159 |
| circLGMN | Oncogene | Promoted proliferation and migration | miR‐127‐3p | 126 |
| circNF1 | Oncogene | Promoted proliferation | miR‐340 | 160 |
| circ‐PITX1 | Oncogene | Promoted proliferation and apoptosis | miR‐379–5p | 161 |
| circ_0043278 | Oncogene | Promoted migration and invasion | miR‐638 | 162 |
| circ_0076248 | Oncogene | Promoted proliferation and induced cell cycle arrest | miR‐181a | 163 |
| circPARP4 | Oncogene | Promoted proliferation, migration invasion and EMT | miR‐125a‐5p | 164 |
| circASAP1 | Oncogene | Promoted proliferation and TMZ resistance | miR‐502‐5p | 127 |
| circMMP9 | Oncogene | Promoted proliferation, migration and invasion | miR‐124 | 130 |
| circ‐METRN | Oncogene | Promoted proliferation, migration, invasion and apoptosis | miR‐4709‐3p | 131 |
| circSERPINE2 | Oncogene | Promoted proliferation and apoptosis | miR‐361‐3p/miR‐324‐5p | 165 |
| circMTO1 | Tumour suppressor | Inhibited proliferation, | miR‐92 | 166 |
| circ‐EPB41L5 | Tumour suppressor | Inhibited proliferation, migration and invasion | miR‐19a | 167 |
| circCD44 | Tumour suppressor | Inhibited proliferation, migration and invasion | miR‐330‐5p/miR‐326 | 136 |
Abbreviations: circRNA, circular RNA; GBM, glioblastoma multiforme; miRNA, microRNA.
6. CONCLUSION
In conclusion, ncRNAs play important roles in the occurrence and development of GBM. The present review summarized the roles of three typical regulatory ncRNAs, as well as their functions and mechanisms in GBM as shown in Tables 1, 2, 3, 4. To date, the function and mechanism of miRNAs and lncRNAs in GBM have been widely studied and consensus is gradually being reached among researchers. However, the mechanism of action of ncRNAs, including circRNAs, remains unclear. Future studies should provide further information on the association between ncRNAs and GBM, which may help to improve the clinical treatment and long‐term prognosis of GBM.
AUTHOR CONTRIBUTIONS
Lirui Durai wrote the article. Wulong Liang, Shaolong Zhou, Zimin Shi, Xiang Li, Weihua Hu and Zhou Yang revised it critically for important intellectual content and gave important advice. Xinjun Wang provided the overall idea of the article and revised the original article. All authors read and approved the article and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
FUNDING INFORMATION
This work was supported by the National Natural Science Foundation of China under Grant number (81972361 to Xinjun Wang) and Medical science and Technology project of Henan Province under Grant number (LHGJ20210487 to Wulong Liang). Wulong Liang revised the article critically for important intellectual content and gave important advice and Xinjun Wang provided the overall idea of the article and revised the original article.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Dai L, Liang W, Shi Z, et al. Systematic characterization and biological functions of non‐coding RNAs in glioblastoma. Cell Prolif. 2023;56(3):e13375. doi: 10.1111/cpr.13375
Funding information Medical science and Technology project of Henan Province, Grant/Award Number: LHGJ20210487; National Natural Science Foundation of China, Grant/Award Number: 81972361
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987‐996. doi: 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 2. Quesnel A, Karagiannis GS, Filippou PS. Extracellular proteolysis in glioblastoma progression and therapeutics. Biochim Biophys Acta Rev Cancer. 2020;1874:188428. doi: 10.1016/j.bbcan.2020.188428 [DOI] [PubMed] [Google Scholar]
- 3. Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170‐6175. doi: 10.1038/sj.onc.1209911 [DOI] [PubMed] [Google Scholar]
- 4. Ling H, Girnita L, Buda O, Calin GA. Non‐coding RNAs: the cancer genome dark matter that matters! Clin Chem Lab Med. 2017;55:705‐714. doi: 10.1515/cclm-2016-0740 [DOI] [PubMed] [Google Scholar]
- 5. Mei J, Hao L, Wang H, et al. Systematic characterization of non‐coding RNAs in triple‐negative breast cancer. Cell Prolif. 2020;53:e12801. doi: 10.1111/cpr.12801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Toden S, Zumwalt TJ, Goel A. Non‐coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188491. doi: 10.1016/j.bbcan.2020.188491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liang J, Zhang L, Cheng W. Non‐coding RNA‐mediated autophagy in cancer: a protumor or antitumor factor? Biochim Biophys Acta Rev Cancer. 2021;1876:188642. doi: 10.1016/j.bbcan.2021.188642 [DOI] [PubMed] [Google Scholar]
- 8. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202‐1207. doi: 10.1016/j.jaci.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051‐4060. doi: 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu B, Mei J, Ji W, et al. MicroRNAs involved in the EGFR pathway in glioblastoma. Biomed Pharmacother. 2020;134:111115. doi: 10.1016/j.biopha.2020.111115 [DOI] [PubMed] [Google Scholar]
- 11. Li J, Pu W, Sun HL, et al. Pin1 impairs microRNA biogenesis by mediating conformation change of XPO5 in hepatocellular carcinoma. Cell Death Differ. 2018;25:1612‐1624. doi: 10.1038/s41418-018-0065-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giraldez AJ et al. Zebrafish MiR‐430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75‐79. doi: 10.1126/science.1122689 [DOI] [PubMed] [Google Scholar]
- 13. Huntzinger E, Kuzuoğlu‐Öztürk D, Braun JE, Eulalio A, Wohlbold L, Izaurralde E. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res. 2013;41:978‐994. doi: 10.1093/nar/gks1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mathonnet G, Fabian MR, Svitkin YV, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap‐binding complex eIF4F. Science. 2007;317:1764‐1767. doi: 10.1126/science.1146067 [DOI] [PubMed] [Google Scholar]
- 15. Matsui M, Chu Y, Zhang H, et al. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 2013;41:10086‐10109. doi: 10.1093/nar/gkt777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen M, Medarova Z, Moore A. Role of microRNAs in glioblastoma. Oncotarget. 2021;12:1707‐1723. doi: 10.18632/oncotarget.28039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21:22‐36. doi: 10.1038/s41568-020-00306-0 [DOI] [PubMed] [Google Scholar]
- 18. Garcia CM, Toms SA. The role of circulating MicroRNA in glioblastoma liquid biopsy. World Neurosurg. 2020;138:425‐435. doi: 10.1016/j.wneu.2020.03.128 [DOI] [PubMed] [Google Scholar]
- 19. Wei L, Peng Y, Shao N, Zhou P. Downregulation of Tim‐1 inhibits the proliferation, migration and invasion of glioblastoma cells via the miR‐133a/TGFBR1 axis and the restriction of Wnt/β‐catenin pathway. Cancer Cell Int. 2021;21:347. doi: 10.1186/s12935-021-02036-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. ParvizHamidi M, Haddad G, Ostadrahimi S, et al. Circulating miR‐26a and miR‐21 as biomarkers for glioblastoma multiform. Biotechnol Appl Biochem. 2019;66:261‐265. doi: 10.1002/bab.1707 [DOI] [PubMed] [Google Scholar]
- 21. Birkó Z, Nagy B, Klekner Á, Virga J. Novel molecular markers in glioblastoma‐benefits of liquid biopsy. Int J Mol Sci. 2020;21:7522. doi: 10.3390/ijms21207522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Setlai BP, Hull R, Reis RM, et al. MicroRNA interrelated epithelial mesenchymal transition (EMT) in glioblastoma. Genes (Basel). 2022;13:244. doi: 10.3390/genes13020244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang D, Ma S, Zhang C, et al. MicroRNA‐935 directly targets FZD6 to inhibit the proliferation of human glioblastoma and correlate to glioma malignancy and prognosis. Front Oncol. 2021;11:566492. doi: 10.3389/fonc.2021.566492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu J, Song J, Xiao M, et al. RUNX1 (RUNX family transcription factor 1), a target of microRNA miR‐128‐3p, promotes temozolomide resistance in glioblastoma multiform by upregulating multidrug resistance‐associated protein 1 (MRP1). Bioengineered. 2021;12:11768‐11781. doi: 10.1080/21655979.2021.2009976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qin H, Gui Y, Ma R, et al. miR‐1258 attenuates tumorigenesis through targeting E2F1 to inhibit PCNA and MMP2 transcription in glioblastoma. Front Oncol. 2021;11:671144. doi: 10.3389/fonc.2021.671144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schnabel E, Knoll M, Schwager C, et al. Prognostic value of microRNA‐221/2 and 17‐92 families in primary glioblastoma patients treated with postoperative radiotherapy. Int J Mol Sci. 2021;22:29960. doi: 10.3390/ijms22062960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim J, Park MW, Park YJ, et al. miR‐542‐3p contributes to the HK2‐mediated high glycolytic phenotype in human glioma cells. Genes (Basel). 2021;12:633. doi: 10.3390/genes12050633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nadaradjane A, Briand J, Bougras‐Cartron G, et al. miR‐370‐3p is a therapeutic tool in anti‐glioblastoma therapy but is not an Intratumoral or cell‐free circulating biomarker. Mol Ther Nucleic Acids. 2018;13:642‐650. doi: 10.1016/j.omtn.2018.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheng W, Ren X, Cai J, et al. A five‐miRNA signature with prognostic and predictive value for MGMT promoter‐methylated glioblastoma patients. Oncotarget. 2015;6:29285‐29295. doi: 10.18632/oncotarget.4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Niyazi M, Pitea A, Mittelbronn M, et al. A 4‐miRNA signature predicts the therapeutic outcome of glioblastoma. Oncotarget. 2016;7:45764‐45775. doi: 10.18632/oncotarget.9945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Santangelo A, Rossato M, Lombardi G, et al. A molecular signature associated with prolonged survival in glioblastoma patients treated with regorafenib. Neuro Oncol. 2021;23:264‐276. doi: 10.1093/neuonc/noaa156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mulcahy EQX, Zhang Y, Colόn RR, et al. MicroRNA 3928 suppresses glioblastoma through downregulation of several oncogenes and upregulation of p53. Int J Mol Sci. 2022;23:3930. doi: 10.3390/ijms23073930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yeh M, Wang YY, Yoo JY, et al. MicroRNA‐138 suppresses glioblastoma proliferation through downregulation of CD44. Sci Rep. 2021;11:9219. doi: 10.1038/s41598-021-88615-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen L, Li Z, Hu S, Deng Q, Hao P, Guo S. Extracellular vesicles carry miR‐27a‐3p to promote drug resistance of glioblastoma to temozolomide by targeting BTG2. Cancer Chemother Pharmacol. 2022;89:217‐229. doi: 10.1007/s00280-021-04392-1 [DOI] [PubMed] [Google Scholar]
- 35. Chen L, Zhu S, Wang H, Pang X, Wang X. MiR‐601 promotes cell proliferation of human glioblastoma cells by suppressing TINP1 expression. Altern Ther Health Med. 2022;28:102‐108. [PubMed] [Google Scholar]
- 36. Duff D, Long A. Roles for RACK1 in cancer cell migration and invasion. Cell Signal. 2017;35:250‐255. doi: 10.1016/j.cellsig.2017.03.005 [DOI] [PubMed] [Google Scholar]
- 37. Nan Y, Guo L, Lu Y, et al. miR‐451 suppresses EMT and metastasis in glioma cells. Cell Cycle. 2021;20:1270‐1278. doi: 10.1080/15384101.2021.1933303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zou M, Zhu W, Wang L, et al. AEG‐1/MTDH‐activated autophagy enhances human malignant glioma susceptibility to TGF‐β1‐triggered epithelial‐mesenchymal transition. Oncotarget. 2016;7:13122‐13138. doi: 10.18632/oncotarget.7536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao C, Wang XB, Zhang YH, Zhou YM, Yin Q, Yao WC. MicroRNA‐424 inhibits cell migration, invasion and epithelial‐mesenchymal transition in human glioma by targeting KIF23 and functions as a novel prognostic predictor. Eur Rev Med Pharmacol Sci. 2018;22:6369‐6378. doi: 10.26355/eurrev_201810_16049 [DOI] [PubMed] [Google Scholar]
- 40. Xu R, Zhou F, Yu T, et al. MicroRNA‐940 inhibits epithelial‐mesenchymal transition of glioma cells via targeting ZEB2. Am J Transl Res. 2019;11:7351‐7363. [PMC free article] [PubMed] [Google Scholar]
- 41. Shi HZ, Wang D, Sun XN, Sheng L. MicroRNA‐378 acts as a prognosis marker and inhibits cell migration, invasion and epithelial‐mesenchymal transition in human glioma by targeting IRG1. Eur Rev Med Pharmacol Sci. 2018;22:3837‐3846. doi: 10.26355/eurrev_201806_15268 [DOI] [PubMed] [Google Scholar]
- 42. Li J, Li Q, Lin L, et al. Targeting the Notch1 oncogene by miR‐139‐5p inhibits glioma metastasis and epithelial‐mesenchymal transition (EMT). BMC Neurol. 2018;18:133. doi: 10.1186/s12883-018-1139-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Z, Zhang L, Liu Z, et al. miRNA‐182 regulated MTSS1 inhibits proliferation and invasion in glioma cells. J Cancer. 2020;11:5840‐5851. doi: 10.7150/jca.47588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang JF, Wang P, Yan YJ, et al. IL‐33 enhances glioma cell migration and invasion by upregulation of MMP2 and MMP9 via the ST2‐NF‐κB pathway. Oncol Rep. 2017;38:2033‐2042. doi: 10.3892/or.2017.5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lv S, Sun B, Dai C, et al. The downregulation of MicroRNA‐146a modulates TGF‐β signaling pathways activity in glioblastoma. Mol Neurobiol. 2015;52:1257‐1262. doi: 10.1007/s12035-014-8938-8 [DOI] [PubMed] [Google Scholar]
- 46. Jiang C, Shen F, du J, et al. MicroRNA‐564 is downregulated in glioblastoma and inhibited proliferation and invasion of glioblastoma cells by targeting TGF‐β1. Oncotarget. 2016;7:56200‐56208. doi: 10.18632/oncotarget.8987 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47. Gao Y et al. Long non‐coding RNA HOXA‐AS2 regulates malignant glioma behaviors and Vasculogenic mimicry formation via the MiR‐373/EGFR Axis. Cell Physiol Biochem. 2018;45:131‐147. doi: 10.1159/000486253 [DOI] [PubMed] [Google Scholar]
- 48. Nan Y, Guo L, Zhen Y, et al. miRNA‐451 regulates the NF‐κB signaling pathway by targeting IKKβ to inhibit glioma cell growth. Cell Cycle. 2021;20:1967‐1977. doi: 10.1080/15384101.2021.1969496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gheidari F, Arefian E, Adegani FJ, et al. miR‐424 induces apoptosis in glioblastoma cells and targets AKT1 and RAF1 oncogenes from the ERBB signaling pathway. Eur J Pharmacol. 2021;906:174273. doi: 10.1016/j.ejphar.2021.174273 [DOI] [PubMed] [Google Scholar]
- 50. Schäfer A, Evers L, Meier L, et al. The metalloprotease‐Disintegrin ADAM8 alters the tumor suppressor miR‐181a‐5p expression profile in glioblastoma thereby contributing to its aggressiveness. Front Oncol. 2022;12:826273. doi: 10.3389/fonc.2022.826273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cao H, Li X, Wang F, Zhang Y, Yang Q. Phytochemical‐mediated glioma targeted treatment: drug resistance and novel delivery systems. Curr Med Chem. 2020;27:599‐629. doi: 10.2174/0929867326666190809221332 [DOI] [PubMed] [Google Scholar]
- 52. Chistiakov DA, Chekhonin VP. Contribution of microRNAs to radio‐ and chemoresistance of brain tumors and their therapeutic potential. Eur J Pharmacol. 2012;684:8‐18. doi: 10.1016/j.ejphar.2012.03.031 [DOI] [PubMed] [Google Scholar]
- 53. Kwak SY, Kim BY, Ahn HJ, et al. Ionizing radiation‐inducible miR‐30 e promotes glioma cell invasion through EGFR stabilization by directly targeting CBL‐B. FEBS J. 2015;282:1512‐1525. doi: 10.1111/febs.13238 [DOI] [PubMed] [Google Scholar]
- 54. Kwak SY, Yang JS, Kim BY, Bae IH, Han YH. Ionizing radiation‐inducible miR‐494 promotes glioma cell invasion through EGFR stabilization by targeting p190B rhoGAP. Biochim Biophys Acta. 2014;1843:514‐516. doi: 10.1016/j.bbamcr.2013.11.021 [DOI] [PubMed] [Google Scholar]
- 55. Areeb Z, Stuart SF, West AJ, et al. Reduced EGFR and increased miR‐221 is associated with increased resistance to temozolomide and radiotherapy in glioblastoma. Sci Rep. 2020;10:17768. doi: 10.1038/s41598-020-74746-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yin J, Zeng A, Zhang Z, Shi Z, Yan W, You Y. Exosomal transfer of miR‐1238 contributes to temozolomide‐resistance in glioblastoma. EBioMedicine. 2019;42:238‐251. doi: 10.1016/j.ebiom.2019.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ren Y, Zhou X, Mei M, et al. MicroRNA‐21 inhibitor sensitizes human glioblastoma cells U251 (PTEN‐mutant) and LN229 (PTEN‐wild type) to taxol. BMC Cancer. 2010;10:27. doi: 10.1186/1471-2407-10-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang J, Li T, Wang B. Exosomal transfer of miR253p promotes the proliferation and temozolomide resistance of glioblastoma cells by targeting FBXW7. Int J Oncol. 2021;59:64. doi: 10.3892/ijo.2021.5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Qiu W, Guo X, Li B, et al. Exosomal miR‐1246 from glioma patient body fluids drives the differentiation and activation of myeloid‐derived suppressor cells. Mol Ther. 2021;29:3449‐3464. doi: 10.1016/j.ymthe.2021.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang Q, Wei B, Peng C, Wang L, Li C. Identification of serum exosomal miR‐98‐5p, miR‐183‐5p, miR‐323‐3p and miR‐19b‐3p as potential biomarkers for glioblastoma patients and investigation of their mechanisms. Curr Res Transl Med. 2022;70:103315. doi: 10.1016/j.retram.2021.103315 [DOI] [PubMed] [Google Scholar]
- 62. Olioso D, Caccese M, Santangelo A, et al. Serum Exosomal microRNA‐21, 222 and 124‐3p as noninvasive predictive biomarkers in newly diagnosed high‐grade gliomas: a prospective study. Cancers (Basel). 2021;13:3006. doi: 10.3390/cancers13123006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yousefi H, Maheronnaghsh M, Molaei F, et al. Long noncoding RNAs and exosomal lncRNAs: classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene. 2020;39:953‐974. doi: 10.1038/s41388-019-1040-y [DOI] [PubMed] [Google Scholar]
- 64. Naderi‐Meshkin H, Lai X, Amirkhah R, Vera J, Rasko JEJ, Schmitz U. Exosomal lncRNAs and cancer: connecting the missing links. Bioinformatics. 2019;35:352‐360. doi: 10.1093/bioinformatics/bty527 [DOI] [PubMed] [Google Scholar]
- 65. Castro‐Oropeza R, Melendez‐Zajgla J, Maldonado V, Vazquez‐Santillan K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol (Dordr). 2018;41:585‐603. doi: 10.1007/s13402-018-0406-4 [DOI] [PubMed] [Google Scholar]
- 66. Fico A, Fiorenzano A, Pascale E, Patriarca EJ, Minchiotti G. Long non‐coding RNA in stem cell pluripotency and lineage commitment: functions and evolutionary conservation. Cell Mol Life Sci. 2019;76:1459‐1471. doi: 10.1007/s00018-018-3000-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li J, Tian H, Yang J, Gong Z. Long noncoding RNAs regulate cell growth, proliferation, and apoptosis. DNA Cell Biol. 2016;35:459‐470. doi: 10.1089/dna.2015.3187 [DOI] [PubMed] [Google Scholar]
- 68. Arab K, Karaulanov E, Musheev M, et al. GADD45A binds R‐loops and recruits TET1 to CpG Island promoters. Nat Genet. 2019;51:217‐223. doi: 10.1038/s41588-018-0306-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv. 2017;3:eaao2110. doi: 10.1126/sciadv.aao2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huang C, Liao X, Jin H, et al. MEG3, as a competing endogenous RNA, binds with miR‐27a to promote PHLPP2 protein translation and impairs bladder cancer invasion. Mol Ther Nucleic Acids. 2019;16:51‐62. doi: 10.1016/j.omtn.2019.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Han P, Chang CP. Long non‐coding RNA and chromatin remodeling. RNA Biol. 2015;12:1094‐1098. doi: 10.1080/15476286.2015.1063770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhao Y, Wang H, Wu C, et al. Construction and investigation of lncRNA‐associated ceRNA regulatory network in papillary thyroid cancer. Oncol Rep. 2018;39:1197‐1206. doi: 10.3892/or.2018.6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hua Q, Jin M, Mi B, et al. LINC01123, a c‐Myc‐activated long non‐coding RNA, promotes proliferation and aerobic glycolysis of non‐small cell lung cancer through miR‐199a‐5p/c‐Myc axis. J Hematol Oncol. 2019;12:91. doi: 10.1186/s13045-019-0773-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li L, Chang HY. Physiological roles of long noncoding RNAs: insight from knockout mice. Trends Cell Biol. 2014;24:594‐602. doi: 10.1016/j.tcb.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Goyal B, Yadav SRM, Awasthee N, Gupta S, Kunnumakkara AB, Gupta SC. Diagnostic, prognostic, and therapeutic significance of long non‐coding RNA MALAT1 in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188502. doi: 10.1016/j.bbcan.2021.188502 [DOI] [PubMed] [Google Scholar]
- 76. Wang Z, Han Y, Li Q, Wang B, Ma J. LncRNA DLGAP1‐AS1 accelerates glioblastoma cell proliferation through targeting miR‐515‐5p/ROCK1/NFE2L1 axis and activating Wnt signaling pathway. Brain Behav. 2021;11:e2321. doi: 10.1002/brb3.2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang R, Wu D, Wang Y, Wu L, Gao G, Shan D. LncRNA MIR31HG is activated by STAT1 and facilitates glioblastoma cell growth via Wnt/beta‐catenin signaling pathway. Neurosci Res. 2021;S0168‐0102:00092‐4. doi: 10.1016/j.neures.2021.04.008 [DOI] [PubMed] [Google Scholar]
- 78. Liang J, Liu C, Xu D, Xie K, Li A. LncRNA NEAT1 facilitates glioma progression via stabilizing PGK1. J Transl Med. 2022;20:80. doi: 10.1186/s12967-022-03273-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lv T, Jin Y, Miao Y, et al. LncRNA PVT1 promotes tumorigenesis of glioblastoma by recruiting COPS5 to deubiquitinate and stabilize TRIM24. Mol Ther Nucleic Acids. 2022;27:109‐121. doi: 10.1016/j.omtn.2021.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80. Wang G, Lin X, Han H, et al. lncRNA H19 promotes glioblastoma multiforme development by activating autophagy by sponging miR‐491‐5p. Bioengineered. 2022;13:11440‐11455. doi: 10.1080/21655979.2022.2065947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lu Y, Guo G, Hong R, et al. LncRNA HAS2‐AS1 promotes glioblastoma proliferation by sponging miR‐137. Front Oncol. 2021;11:634893. doi: 10.3389/fonc.2021.634893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cai H, Yu Y, Ni X, et al. LncRNA LINC00998 inhibits the malignant glioma phenotype via the CBX3‐mediated c‐met/Akt/mTOR axis. Cell Death Dis. 2020;11:1032. doi: 10.1038/s41419-020-03247-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Liu K, Deng Y, Yang Y, Wang H, Zhou P. MicorRNA‐195 links long non‐coding RNA SEMA3B antisense RNA 1 (head to head) and cyclin D1 to regulate the proliferation of glioblastoma cells. Bioengineered. 2022;13:8798‐8805. doi: 10.1080/21655979.2022.2052646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li W, Cui Y, Ma W, Wang M, Cai Y, Jiang Y. LncRNA RBPMS‐AS1 promotes NRGN transcription to enhance the radiosensitivity of glioblastoma through the microRNA‐301a‐3p/CAMTA1 axis. Transl Oncol. 2022;15:101282. doi: 10.1016/j.tranon.2021.101282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang D, Chen Q, Liu J, Liao Y, Jiang Q. Silencing of lncRNA CHRM3‐AS2 expression exerts anti‐tumour effects against glioma via targeting microRNA‐370‐5p/KLF4. Front Oncol. 2022;12:856381. doi: 10.3389/fonc.2022.856381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ho KH, Shih CM, Liu AJ, Chen KC. Hypoxia‐inducible lncRNA MIR210HG interacting with OCT1 is involved in glioblastoma multiforme malignancy. Cancer Sci. 2022;113:540‐552. doi: 10.1111/cas.15240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tang G, Luo L, Zhang J, et al. lncRNA LINC01057 promotes mesenchymal differentiation by activating NF‐kappaB signaling in glioblastoma. Cancer Lett. 2021;498:152‐164. doi: 10.1016/j.canlet.2020.10.047 [DOI] [PubMed] [Google Scholar]
- 88. Yang X, Niu S, Liu JQ, et al. Identification of an epithelial‐mesenchymal transition‐related lncRNA prognostic signature for patients with glioblastoma. Sci Rep. 2021;11:23694. doi: 10.1038/s41598-021-03213-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pang B, Quan F, Ping Y, Hu J, Lan Y, Pang L. Dissecting the invasion‐associated Long non‐coding RNAs using single‐cell RNA‐Seq data of glioblastoma. Front Genet. 2020;11:633455. doi: 10.3389/fgene.2020.633455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhong C, Yu Q, Peng Y, et al. Novel LncRNA OXCT1‐AS1 indicates poor prognosis and contributes to tumorigenesis by regulating miR‐195/CDC25A axis in glioblastoma. J Exp Clin Cancer Res. 2021;40:123. doi: 10.1186/s13046-021-01928-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ma C, Wang H, Zong G, et al. EGR1 modulated LncRNA HNF1A‐AS1 drives glioblastoma progression via miR‐22‐3p/ENO1 axis. Cell Death Discov. 2021;7:350. doi: 10.1038/s41420-021-00734-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhong X, Cai Y. Long non‐coding RNA (lncRNA) HOXD‐AS2 promotes glioblastoma cell proliferation, migration and invasion by regulating the miR‐3681‐5p/MALT1 signaling pathway. Bioengineered. 2021;12:9113‐9127. doi: 10.1080/21655979.2021.1977104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ahmadov U, Picard D, Bartl J, et al. The long non‐coding RNA HOTAIRM1 promotes tumor aggressiveness and radiotherapy resistance in glioblastoma. Cell Death Dis. 2021;12:885. doi: 10.1038/s41419-021-04146-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhu H, Chen Z, Shen L, Tang T, Yang M, Zheng X. Long noncoding RNA LINC‐PINT suppresses cell proliferation, invasion, and EMT by blocking Wnt/beta‐catenin signaling in glioblastoma. Front Pharmacol. 2020;11:586653. doi: 10.3389/fphar.2020.586653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Huang K, Yue X, Zheng Y, et al. Development and validation of an mesenchymal‐related Long non‐coding RNA prognostic model in glioma. Front Oncol. 2021;11:726745. doi: 10.3389/fonc.2021.726745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wang X, Li X, Zhou Y, Huang X, Jiang X. Long non‐coding RNA OIP5‐AS1 inhibition upregulates microRNA‐129‐5p to repress resistance to temozolomide in glioblastoma cells via downregulating IGF2BP2. Cell Biol Toxicol. 2021;0742‐2091. doi: 10.1007/s10565-021-09614-z [DOI] [PubMed] [Google Scholar]
- 97. Cheng M, Wang Q, Chen L, et al. LncRNA UCA1/miR‐182‐5p/MGMT axis modulates glioma cell sensitivity to temozolomide through MGMT‐related DNA damage pathways. Hum Pathol. 2022;123:59‐73. doi: 10.1016/j.humpath.2022.02.016 [DOI] [PubMed] [Google Scholar]
- 98. Lu Y, Tian M, Liu J, Wang K. LINC00511 facilitates Temozolomide resistance of glioblastoma cells via sponging miR‐126‐5p and activating Wnt/beta‐catenin signaling. J Biochem Mol Toxicol. 2021;35:e22848. doi: 10.1002/jbt.22848 [DOI] [PubMed] [Google Scholar]
- 99. Huang H, Weng H, Sun W, et al. Recognition of RNA N(6)‐methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285‐295. doi: 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Han J, Yu X, Wang S, et al. IGF2BP2 induces U251 glioblastoma cell Chemoresistance by inhibiting FOXO1‐mediated PID1 expression through stabilizing lncRNA DANCR. Front Cell Dev Biol. 2021;9:659228. doi: 10.3389/fcell.2021.659228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wang X, Yu X, Xu H, et al. Serum‐derived extracellular vesicles facilitate temozolomide resistance in glioblastoma through a HOTAIR‐dependent mechanism. Cell Death Dis. 2022;13:344. doi: 10.1038/s41419-022-04699-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Turkowski K, Brandenburg S, Mueller A, et al. VEGF as a modulator of the innate immune response in glioblastoma. Glia. 2018;66:161‐174. doi: 10.1002/glia.23234 [DOI] [PubMed] [Google Scholar]
- 103. Jackson CM, Choi J, Lim M. Mechanisms of immunotherapy resistance: lessons from glioblastoma. Nat Immunol. 2019;20:1100‐1109. doi: 10.1038/s41590-019-0433-y [DOI] [PubMed] [Google Scholar]
- 104. Yi K, Cui X, Liu X, et al. PTRF/Cavin‐1 as a novel RNA‐binding protein expedites the NF‐kappaB/PD‐L1 Axis by stabilizing lncRNA NEAT1, contributing to tumorigenesis and immune evasion in glioblastoma. Front Immunol. 2021;12:802795. doi: 10.3389/fimmu.2021.802795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhao R, Li B, Zhang S, et al. The N(6)‐Methyladenosine‐modified pseudogene HSPA7 correlates with the tumor microenvironment and predicts the response to immune checkpoint therapy in glioblastoma. Front Immunol. 2021;12:653711. doi: 10.3389/fimmu.2021.653711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Li X, Sun L, Wang X, et al. A five immune‐related lncRNA signature as a prognostic target for glioblastoma. Front Mol Biosci. 2021;8:632837. doi: 10.3389/fmolb.2021.632837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Gao M, Wang X, Han D, et al. A six‐lncRNA signature for Immunophenotype prediction of glioblastoma Multiforme. Front Genet. 2020;11:604655. doi: 10.3389/fgene.2020.604655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Pastori C, Kapranov P, Penas C, et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci U S A. 2015;112:8326‐8331. doi: 10.1073/pnas.1424220112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Tan SK, Pastori C, Penas C, et al. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer. 2018;17:74. doi: 10.1186/s12943-018-0822-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Shen J, Hodges TR, Song R, et al. Serum HOTAIR and GAS5 levels as predictors of survival in patients with glioblastoma. Mol Carcinog. 2018;57:137‐141. doi: 10.1002/mc.22739 [DOI] [PubMed] [Google Scholar]
- 111. Wang Z, Ji X, Gao L, et al. Comprehensive In Silico analysis of a novel serum exosome‐derived competitive endogenous RNA network for constructing a prognostic model for glioblastoma. Front Oncol. 2021;11:553594. doi: 10.3389/fonc.2021.553594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single‐stranded covalently closed circular RNA molecules existing as highly base‐paired rod‐like structures. Proc Natl Acad Sci U S A. 1976;73:3852‐3856. doi: 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Farooqi AA, Attar R, Yulaevna IM, Berardi R. Interaction of long non‐coding RNAs and circular RNAs with microRNAs for the regulation of immunological responses in human cancers. Semin Cell Dev Biol. 2022;124:63‐71. doi: 10.1016/j.semcdb.2021.05.029 [DOI] [PubMed] [Google Scholar]
- 114. van Zonneveld AJ, Kölling M, Bijkerk R, Lorenzen JM. Circular RNAs in kidney disease and cancer. Nat Rev Nephrol. 2021;17:814‐826. doi: 10.1038/s41581-021-00465-9 [DOI] [PubMed] [Google Scholar]
- 115. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18:6. doi: 10.1186/s12943-018-0934-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141‐157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zhu F, Cheng C, Qin H, Wang H, Yu H. A novel circular RNA circENTPD7 contributes to glioblastoma progression by targeting ROS1. Cancer Cell Int. 2020;20:118. doi: 10.1186/s12935-020-01208-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Guo X, Piao H. Research Progress of circRNAs in glioblastoma. Front Cell Dev Biol. 2021;9:791892. doi: 10.3389/fcell.2021.791892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Yang Q, Du WW, Wu N, et al. A circular RNA promotes tumorigenesis by inducing c‐myc nuclear translocation. Cell Death Differ. 2017;24:1609‐1620. doi: 10.1038/cdd.2017.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Santer L, Bär C, Thum T. Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol Ther. 2019;27:1350‐1363. doi: 10.1016/j.ymthe.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ding L, Zhao Y, Dang S, et al. Circular RNA circ‐DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. 2019;18:45. doi: 10.1186/s12943-019-1006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Liu J, Li D, Luo H, Zhu X. Circular RNAs: the star molecules in cancer. Mol Aspects Med. 2019;70:141‐152. doi: 10.1016/j.mam.2019.10.006 [DOI] [PubMed] [Google Scholar]
- 124. Zhou F, Wang B, Wang H, et al. circMELK promotes glioblastoma multiforme cell tumorigenesis through the miR‐593/EphB2 axis. Mol Ther Nucleic Acids. 2021;25:25‐36. doi: 10.1016/j.omtn.2021.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Wang R, Zhang S, Chen X, et al. CircNT5E acts as a sponge of miR‐422a to promote glioblastoma tumorigenesis. Cancer Res. 2018;78:4812‐4825. doi: 10.1158/0008-5472.CAN-18-0532 [DOI] [PubMed] [Google Scholar]
- 126. Chen B, Wang M, Huang R, et al. Circular RNA circLGMN facilitates glioblastoma progression by targeting miR‐127‐3p/LGMN axis. Cancer Lett. 2021;522:225‐237. doi: 10.1016/j.canlet.2021.09.030 [DOI] [PubMed] [Google Scholar]
- 127. Wei Y, Lu C, Zhou P, et al. EIF4A3‐induced circular RNA ASAP1 promotes tumorigenesis and temozolomide resistance of glioblastoma via NRAS/MEK1/ERK1‐2 signaling. Neuro Oncol. 2021;23:611‐624. doi: 10.1093/neuonc/noaa214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Zhang S, Liao K, Miao Z, et al. CircFOXO3 promotes glioblastoma progression by acting as a competing endogenous RNA for NFAT5. Neuro Oncol. 2019;21:1284‐1296. doi: 10.1093/neuonc/noz128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Gao X, Xia X, Li F, et al. Circular RNA‐encoded oncogenic E‐cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR‐STAT3 signalling. Nat Cell Biol. 2021;23:278‐291. doi: 10.1038/s41556-021-00639-4 [DOI] [PubMed] [Google Scholar]
- 130. Wang R, Zhang S, Chen X, et al. EIF4A3‐induced circular RNA MMP9 (circMMP9) acts as a sponge of miR‐124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17:166. doi: 10.1186/s12943-018-0911-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wang X, Cao Q, Shi Y, et al. Identification of low‐dose radiation‐induced exosomal circ‐METRN and miR‐4709‐3p/GRB14/PDGFRalpha pathway as a key regulatory mechanism in glioblastoma progression and radioresistance: functional validation and clinical theranostic significance. Int J Biol Sci. 2021;17:1061‐1078. doi: 10.7150/ijbs.57168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805‐1814. doi: 10.1038/s41388-017-0019-9 [DOI] [PubMed] [Google Scholar]
- 133. Xia X, Li X, Li F, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide‐dependent Kinase‐1. Mol Cancer. 2019;18:131. doi: 10.1186/s12943-019-1056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lou J, Hao Y, Lin K, et al. Circular RNA CDR1as disrupts the p53/MDM2 complex to inhibit Gliomagenesis. Mol Cancer. 2020;19:138. doi: 10.1186/s12943-020-01253-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304‐315. doi: 10.1093/jnci/djx166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Feng J, Ren X, Fu H, et al. LRRC4 mediates the formation of circular RNA CD44 to inhibitGBM cell proliferation. Mol Ther Nucleic Acids. 2021;26:473‐487. doi: 10.1016/j.omtn.2021.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Yoo JY, Yeh M, Wang YY, et al. MicroRNA‐138 increases chemo‐sensitivity of glioblastoma through downregulation of Survivin. Biomedicine. 2021;9:780. doi: 10.3390/biomedicines9070780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Zheng B, Chen T. MiR‐489‐3p inhibits cell proliferation, migration, and invasion, and induces apoptosis, by targeting the BDNF‐mediated PI3K/AKT pathway in glioblastoma. Open Life Sci. 2020;15:274‐283. doi: 10.1515/biol-2020-0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Liu C, Zhu B, Zhong M, Bao J. miRNA‐448 regulates the development of glioblastoma (GBM) by regulating rho‐associated protein kinase 1. Comput Math Methods Med. 2022;2022:2502010. doi: 10.1155/2022/2502010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 140. Kwak S, Park SH, Kim SH, et al. miR‐3189‐targeted GLUT3 repression by HDAC2 knockdown inhibits glioblastoma tumorigenesis through regulating glucose metabolism and proliferation. J Exp Clin Cancer Res. 2022;41:87. doi: 10.1186/s13046-022-02305-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Deng Z, Jian Y, Cai H. Ropivacaine represses the proliferation, invasion, and migration of glioblastoma via modulating the microRNA‐21‐5p/KAT8 regulatory NSL complex subunit 2 axis. Bioengineered. 2022;13:5975‐5986. doi: 10.1080/21655979.2022.2037955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Ho KH et al. miR‐4286 is involved in connections between IGF‐1 and TGF‐β signaling for the mesenchymal transition and invasion by glioblastomas. Cell Mol Neurobiol. 2022;42:791‐806. doi: 10.1007/s10571-020-00977-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Li W, Liu J, Ji L, et al. MiR‐674‐5p suppresses the proliferation and migration of glioma cells by targeting Cul4b. Neurochem Res. 2022;47:679‐691. doi: 10.1007/s11064-021-03476-x [DOI] [PubMed] [Google Scholar]
- 144. Wu W, Yu T, Wu Y, Tian W, Zhang J, Wang Y. The miR155HG/miR‐185/ANXA2 loop contributes to glioblastoma growth and progression. J Exp Clin Cancer Res. 2019;38:133. doi: 10.1186/s13046-019-1132-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Wu L, Li G, Feng D, et al. MicroRNA‐21 expression is associated with overall survival in patients with glioma. Diagn Pathol. 2013;8:200. doi: 10.1186/1746-1596-8-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Xu C, Zhao J, Song J, et al. lncRNA PRADX is a mesenchymal glioblastoma biomarker for cellular metabolism targeted therapy. Front Oncol. 2022;12:888922. doi: 10.3389/fonc.2022.888922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Zhao N, Zhang J, Zhao L, et al. Long noncoding RNA NONHSAT079852.2 contributes to GBM recurrence by functioning as a ceRNA for has‐mir‐10401‐3p to facilitate HSPA1A upregulation. Front. Oncologia. 2021;11:636632. doi: 10.3389/fonc.2021.636632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Shree B, Tripathi S, Sharma V. Transforming growth factor‐Beta‐regulated LncRNA‐MUF promotes invasion by modulating the miR‐34a Snail1 Axis in glioblastoma Multiforme. Front Oncol. 2021;11:788755. doi: 10.3389/fonc.2021.788755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Wang Y, Shan A, Zhou Z, et al. LncRNA TCONS_00004099‐derived microRNA regulates oncogenesis through PTPRF in gliomas. Ann Transl Med. 2021;9:1023. doi: 10.21037/atm-21-2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Cao Y et al. CircPITX1 regulates proliferation, angiogenesis, migration, invasion, and cell cycle of human glioblastoma cells by targeting miR‐584‐5p/KPNB1 Axis. J Mol Neurosci. 2021;71:1683‐1695. doi: 10.1007/s12031-021-01820-y [DOI] [PubMed] [Google Scholar]
- 151. Lu Y, Deng X, Xiao G, Zheng X, Ma L, Huang W. circ_0001730 promotes proliferation and invasion via the miR‐326/Wnt7B axis in glioma cells. Epigenomics. 2019;11:1335‐1352. doi: 10.2217/epi-2019-0121 [DOI] [PubMed] [Google Scholar]
- 152. Zhang H, Xu W. CircABCC3 knockdown inhibits glioblastoma cell malignancy by regulating miR‐770‐5p/SOX2 axis through PI3K/AKT signaling pathway. Brain Res. 2021;1764:147465. doi: 10.1016/j.brainres.2021.147465 [DOI] [PubMed] [Google Scholar]
- 153. Liu R, Dai W, Wu A, Li Y. CircCDC45 promotes the malignant progression of glioblastoma by modulating the miR‐485‐5p/CSF‐1 axis. BMC Cancer. 2021;21:1090. doi: 10.1186/s12885-021-08803-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Sun Y, Ma G, Xiang H, et al. circFLNA promotes glioblastoma proliferation and invasion by negatively regulating miR1993p expression. Mol Med Rep. 2021;24:786. doi: 10.3892/mmr.2021.12426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Fan X, Liu M, Fei L, Huang Z, Yan Y. CircFOXM1 promotes the proliferation, migration, invasion, and glutaminolysis of glioblastoma by regulating the miR‐577/E2F5 axis. Bosn J Basic Med Sci. 2022;22:205‐216. doi: 10.17305/bjbms.2021.6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Li W, Wang S, Shan B, et al. CircHECTD1 regulates cell proliferation and migration by the miR‐320‐5p/SLC2A1 Axis in glioblastoma multiform. Front Oncol. 2021;11:666391. doi: 10.3389/fonc.2021.666391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Lu J, Lou G, Jiang L, Liu X, Jiang J, Wang X. CircNUP98 suppresses the maturation of miR‐519a‐3p in glioblastoma. Front Neurol. 2021;12:679745. doi: 10.3389/fneur.2021.679745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Zhou M, Li H, Chen K’, Ding W, Yang C, Wang X. CircSKA3 downregulates miR‐1 through methylation in glioblastoma to promote cancer cell proliferation. Cancer Manag Res. 2021;13:509‐514. doi: 10.2147/CMAR.S279097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Wang Q, Zhang D, Li Y, et al. Circular RNA circ_0001588 sponges miR‐211‐5p to facilitate the progression of glioblastoma via up‐regulating YY1 expression. J Gene Med. 2021;23:e3371. doi: 10.1002/jgm.3371 [DOI] [PubMed] [Google Scholar]
- 160. Liu L, Jia L, Shao J, Liu H, Wu Q, Wu X. Circular RNA circNF1 siRNA silencing inhibits glioblastoma cell proliferation by promoting the maturation of miR‐340. Front Neurol. 2021;12:658076. doi: 10.3389/fneur.2021.658076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Lv X, Wang M, Qiang J, Guo S. Circular RNA circ‐PITX1 promotes the progression of glioblastoma by acting as a competing endogenous RNA to regulate miR‐379‐5p/MAP3K2 axis. Eur J Pharmacol. 2019;863:172643. doi: 10.1016/j.ejphar.2019.172643 [DOI] [PubMed] [Google Scholar]
- 162. Wu Z, Zheng M, Zhang Y, et al. Hsa_circ_0043278 functions as competitive endogenous RNA to enhance glioblastoma multiforme progression by sponging miR‐638. Aging (Albany NY). 2020;12:21114‐21128. doi: 10.18632/aging.103603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Lei B, Huang Y, Zhou Z, et al. Circular RNA hsa_circ_0076248 promotes oncogenesis of glioma by sponging miR‐181a to modulate SIRT1 expression. J Cell Biochem. 2019;120:6698‐6708. doi: 10.1002/jcb.27966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Zhou J, Wang H, Hong F, et al. CircularRNA circPARP4 promotes glioblastoma progression through sponging miR‐125a‐5p and regulating FUT4. Am J Cancer Res. 2021;11:138‐156. [PMC free article] [PubMed] [Google Scholar]
- 165. Li D, Li L, Chen X, Yang W, Cao Y. Circular RNA SERPINE2 promotes development of glioblastoma by regulating the miR‐361‐3p/miR‐324‐5p/BCL2 signaling pathway. Mol Ther Oncolytics. 2021;22:483‐494. doi: 10.1016/j.omto.2021.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Zhang X, Zhong B, Zhang W, Wu J, Wang Y. Circular RNA CircMTO1 inhibits proliferation of glioblastoma cells via miR‐92/WWOX signaling pathway. Med Sci Monit. 2019;25:6454‐6461. doi: 10.12659/MSM.918676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Lv T, Miao Y, Xu T, et al. Circ‐EPB41L5 regulates the host gene EPB41L5 via sponging miR‐19a to repress glioblastoma tumorigenesis. Aging (Albany NY). 2020;12:318‐339. doi: 10.18632/aging.102617 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


