Abstract
The diversity of a microbial community covering the surface of a marine nematode was analyzed by performing a 16S ribosomal DNA (rDNA) restriction cutting and sequencing analysis. In two clone libraries constructed by using individual nematodes, 54 and 85 restriction patterns were identified, and only 13 of these patterns were common to both libraries. Sequence analysis indicated that the common patterns belonged to four groups related to sequences of cytophagas, sulfate-reducing bacteria, members of the gamma subclass of the class Proteobacteria, and caulobacters. At least two groups appeared to be permanent members of the community as they were also detected in a 16S rDNA library constructed 3 years previously by using 100 pooled nematode specimens. A surprising outcome was that very dominant filamentous bacteria were apparently not represented in the clone libraries, as quantitative probing showed that none of the common operational taxonomic unit groups displayed the expected overwhelming dominance. Nevertheless, our analysis revealed both an unexpectedly high level of bacterial diversity and heterogeneity in samples representing presumably very similar microenvironments.
Virtually all environments harbor greater bacterial diversity than that previously estimated by cultivation techniques. This is one of the central results of the adaptation of molecular approaches to studies of microbial community structure (7). Perhaps not surprisingly, soils and sediments have turned out to contain an unrivalled diversity of microorganisms regardless of the molecular techniques used (28). For example, reassociation kinetics analyses of total bacterial DNAs extracted from soils and sediments have suggested that more than 10,000 bacterial species are present (27). Similarly, analysis of 16S ribosomal DNA (rDNA) from soils by denaturing gradient gel electrophoresis (5) or sequencing of clones (1, 2) has revealed great and previously unknown diversity. However, recent investigations have indicated that finite microbial communities can be identified on a relatively large scale despite the seemingly overwhelming diversity in each individual sample. In one study, denaturing gradient gel electrophoresis fingerprinting performed with total DNA extracted from 1 g of grassland soil resulted in similar and reproducible patterns in samples taken a few meters to hundreds of meters apart (5). This suggests that most potential microenvironments are present in as little as 1 g of soil. But to what extent microenvironments harbor defined communities of bacteria cannot be determined by simply reducing sample sizes because of the need to reproducibly sample defined biogeochemical settings on a microscale.
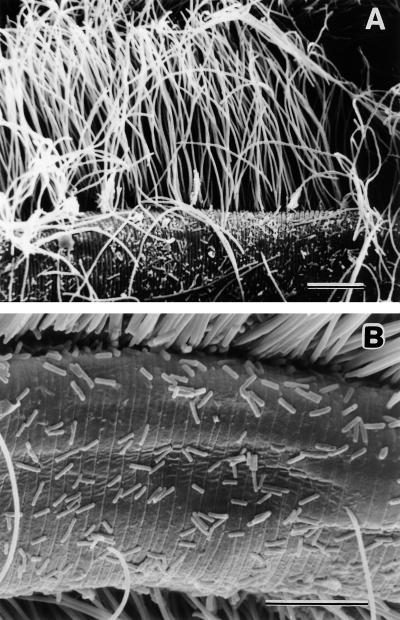
In an attempt to sample and compare the microbial diversity in a specific microenvironment from a marine sediment, we analyzed the bacterial community growing on the surface of the nematode Eubostrichus dianae (20). This microscopic animal lives in the pore space of the sediment and has been shown to actively seek out high sulfide concentrations around the chemocline (31). The entire body of this worm is covered by an epibiotic bacterial community, which is dominated by large, filamentous bacteria at a density and in an arrangement reminiscent of fur (20) (Fig. 1). These large bacteria (length, up to 50 μm) serve as the main, if not exclusive, source of nutrition for the nematodes as only bacteria with the same morphology were detected in electron microscopic gut sections (14, 15); however, the identity of these filamentous bacteria has remained unresolved. In the past, they have been compared to sulfur-oxidizing, chemoautotrophic bacteria growing in highly specific and exclusive associations on some marine nematode species (19, 20). In addition to these filaments, a number of morphologically distinct, smaller bacteria are attached to the nematode cuticle, as determined by scanning (20) (Fig. 1) or transmission (14) electron microscopy. It is thought that these epibiotic bacteria live under relatively defined and stable microhabitat conditions because of the stability of the cuticle as a substrate and the predictability of the physicochemical conditions actively sought by the animals (31). Here, we estimated the diversity of bacteria living on the nematode surface and determined the extent of overlap between the bacterial communities associated with individual specimens by using amplified rDNA restriction analysis (ARDRA) (29) and comparative 16S rDNA sequencing. The importance of individual sequence types in the epibiotic community was assessed by quantitative oligonucleotide probing (24) of total nucleic acids extracted from pooled nematode specimens.
FIG. 1.
Scanning electron micrographs of the bacterial community attached to the E. dianae cuticle. (A) Overview of the numerically dominant, large filaments and interspersed smaller cells. (B) higher magnification of the small cells. The scanning electron microscopy procedure used has been described previously (20). Bars = 10 μm.
16S rDNA clone library construction.
Sediment cores containing E. dianae specimens were collected from Tuckerstown Cove, a shallow sandflat in Bermuda (20). Cores were sectioned, and the meiofauna was extracted by using standard methods (8). Individual nematodes were collected, washed in freshly filtered (pore size, 0.2 μm) seawater, rinsed by brief immersion in Milli-Q-purified water (Millipore), and prepared for PCR amplification of the 16S rDNA either as individuals or as batches containing 100 specimens. No evidence of cell lysis was detected during the washing procedures, and even the nematodes, which are extremely sensitive to osmotic shock, remained viable. Individual worms were transferred to 0.5-ml centrifuge tubes containing 20 μl of Genereleaser (Bio Ventures, Inc.), a reagent which is used for performing PCR with whole cells. These samples were vortexed, lysed in a microwave oven used according to manufacturer’s instructions, and stored frozen until they were used for PCR. Batches of 100 worms were treated with lysozyme and proteinase K, and nucleic acids were purified by the standard phenol-chloroform extraction procedure. For both treatments, completeness of cell lysis was checked by microscopically examining subsamples. The PCR mixtures (final volume, 100 μl) contained 1× PCR buffer (Promega), each deoxynucleoside triphosphate at a concentration of 200 μM, 2.0 mM MgCl2, each of the domain Bacteria-specific primers 27F and 1492R (9) at a concentration of 100 nM, and 0.025 U Taq polymerase (Promega) μl−1. For amplification, we used an initial denaturation step consisting of 94°C for 3 min, followed by 35 cycles consisting of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C. The PCR products were purified on agarose gels and were cloned into pCRII vectors (Invitrogen). Three 16S rDNA libraries, one from 100 pooled specimens (ED 1) and two from single specimens (ED 2 and ED 3), were constructed.
ARDRA and phylogenetic analysis.
For ARDRA, all insert-containing colonies on a given culture plate were sampled. The inserts were amplified, the sizes of amplification products were determined on agarose gels, and double digestion was performed by using the tetrameric restriction enzymes HhaI and BstUI. These enzymes have previously been shown to cut 16S rDNA sequences into fragments that allow optimal resolution (13). Restriction patterns were analyzed on 3% Metaphor agarose gels (FMC Biochemicals) stained with ethidium bromide. Identical patterns were grouped into operational taxonomic units (OTUs) (12), and one representative clone of OTUs common to ED 2 and ED 3 was sequenced either in its entirety or partially at the 5′ end. A phylogenetic comparison was performed by using the SIMILARITY_RANK program provided by the Ribosomal Database Project (11) and distance and parsimony programs contained in the Phylip 3.4 package, as described previously (16).
Diversity of OTUs in the clone libraries.
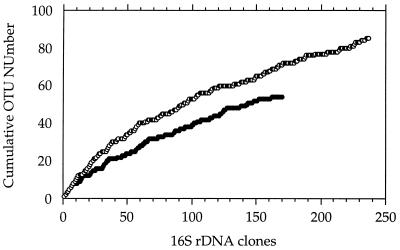
A total of 537 clones were picked, and 170 (ED 2) and 237 (ED 3) of these clones were used for further analysis because they contained inserts of the expected size (1.5 kb). Using restriction digests of PCR-amplified clone inserts, we identified 54 and 85 distinguishable restriction patterns (OTUs) in ED 2 and ED 3, respectively. The distribution of clones was heavily skewed. The three most abundant OTUs amounted to 50% and 34% of the total clones in ED2 and ED3, respectively. In both libraries, only roughly one-sixth of the OTUs were represented by more than one clone. To determine how well the sampling captured the total diversity of 16S rDNAs in the libraries, the cumulative number of OTUs was plotted as a function of appearance during the sampling of clones (Fig. 2) (12). Populations are thought to be well-sampled if continuing effort does not produce new OTUs (that is, the cumulative OTU curve reaches an asymptotic value). The shape of the curves indicated that ED 2 was more completely sampled than ED 3 was (Fig. 2). To estimate the asymptotic value or maximum diversity in each of the libraries, the two curves were linearized as double-reciprocal plots, which was analogous to a Lineweaver-Burk analysis (25). From the reciprocals of the y intercepts, a total diversity of 57 (ED 2) and 118 (ED 3) OTUs was estimated, which confirmed that the OTUs were indeed almost completely sampled in ED 2.
FIG. 2.
Estimated bacterial diversity in two clone libraries constructed from single specimens of E. dianae. OTUs are graphed in sequence of their detection as a function of the cumulative 16S rDNA clone numbers screened. Symbols: ●, clone library ED 2; ○, clone library ED 3.
OTUs common to both clone libraries.
Of all of the OTUs in ED 2 and ED 3, only 13 occurred in both libraries. However, we found almost all of the numerically dominant types among these 13 common OTUs; these types included OTUs 5, 6, 8, 10, 25, 37, and 38 in ED 2 and OTUs 1, 2, 7, 8, 15, 20, and 31 in ED 3 (Table 1). The clones of all of the common OTUs accounted for 68 and 50% of the total clones in ED 2 and ED 3, respectively. To further characterize the bacterial types common to both epibiotic communities, clones were sequenced and subjected to a phylogenetic analysis. One clone of the most abundant OTUs of each group was sequenced completely, while clones of the remaining common OTUs were sequenced only partially (300 nucleotides at the 5′ end).
TABLE 1.
OTUs common to both clone library ED 2 and clone library ED 3, ranked by the numbers of clones that each OTU was represented by
| OTU groupa | Clone library ED 2
|
Clone library ED 3
|
|||
|---|---|---|---|---|---|
| OTU no. | Total clones | OTU no. | Total clones | ||
| I | A | 5 | 36 | 20 | 46 |
| B | 8 | 21 | 2 | 8 | |
| C | 23 | 1 | 31 | 7 | |
| D | 1 | 1 | 33 | 2 | |
| II | A | 6 | 30 | 8 | 19 |
| B | 10 | 5 | 36 | 2 | |
| C | 38 | 5 | 25 | 1 | |
| D | 37 | 4 | 44 | 1 | |
| III | A | 25 | 6 | 1 | 15 |
| B | 9 | 3 | 15 | 4 | |
| C | 32 | 1 | 47 | 2 | |
| D | 51 | 1 | 4 | 1 | |
| IV | A | 27 | 1 | 7 | 1 |
OTU groups were defined on the basis of levels of sequence identity to one another and on the basis of similarity rank analysis data obtained with bacteria in the Ribosomal Database Project (11).
OTUs were defined on the basis of 16S rDNA inserts with identical restriction patterns and were numbered in order of detection in ED 2 and ED 3.
Phylogeny of common OTUs.
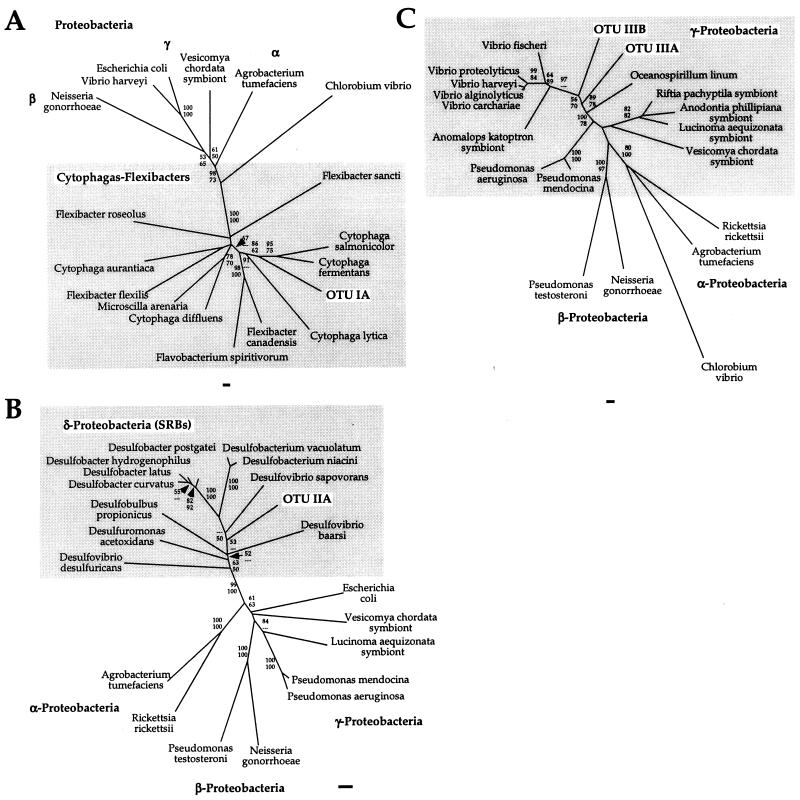
Four OTU groups were identified based on phylogenetic relationships (Table 1). Groups I through III each consisted of four OTUs, while group IV consisted of a single OTU. The four OTUs of group I exhibited at least 99.3% sequence identity and were associated with the genus Cytophaga (Fig. 3A). The sequences of group II were also >99% identical and were closely related to sulfate-reducing bacteria (SRBs), such as Desulfobacter postgatei and Desulfobacter latus (Fig. 3B). The four OTUs of group III were only distantly related to one another (levels of sequence identity, 79.9 to 89.8%), but all of them clustered with members of a group of bacteria in the γ subclass of the class Proteobacteria (γ-Proteobacteria) containing many Alteromonas, Aeromonas, and Vibrio species (Fig. 3C). Group IV consisted of a single OTU which was similar to Caulobacter spp. and other members of the α-Proteobacteria. The two major sequence groups (OTU groups I and II) were also dominant in a 16S rDNA library constructed from 100 pooled nematodes (ED 1) collected 3 years before the ED 2 and ED 3 nematodes were collected. This library was screened by partially sequencing 26 clones; 5 and 6 of these clones were identical to OTU groups I and II, respectively. Other clones were also similar to members of the genus Vibrio but were not identical to any sequences in OTU group III.
FIG. 3.
Evolutionary distance trees based on the 16S rRNA sequences of the most abundant OTUs of groups I through III and representative symbiotic and free-living bacteria. (A) OTU IA in the Flexibacter-Cytophaga-Bacteroides division. The final data set included 896 nucleotide positions. (B) OTU IIA in the SRB group. The final data set included 696 nucleotide positions. (C) OTUs IIIA and IIIB in the γ-Proteobacteria. The final data set included 810 nucleotide positions. The bootstrap values obtained in distance (upper values) and parsimony (lower values) analyses refer to species distal to the associated node. Bars = 1% nucleotide difference per sequence position.
Discussion.
It was surprising that the morphologically uniform, filamentous bacteria that densely covered the entire surfaces of the nematodes (14, 20) (Fig. 1) were apparently not detected by our PCR-based analysis. This conclusion was first indicated by a lack of clear dominance of a single OTU group in the clone libraries. Since PCR bias can skew amplification from multiple templates (6, 17, 26), quantitative probing of rRNAs extracted from approximately 200 nematode specimens was carried out. Oligonucleotide probes specific for the most abundant OTUs of groups I through III were designed (21) (data not shown), and the hybridization signals obtained were compared to the signals obtained with universal bacterial probe Eub338 when in vitro transcribed rRNAs were used as standards (18). OTUs IA and IIA comprised only 8.6 and 6.9%, respectively, of the total bacterial rRNA, while OTU IIIA could not be quantified due to low template abundance. At this time, it cannot be ascertained with certainty why the abundant filamentous bacteria were apparently not amplified despite the fact that they were lysed, as determined microscopically; however, mismatches with the domain-specific primers and extreme PCR bias are among the possible reasons. Regardless of the cause, this study confirmed potential shortcomings of quantitative interpretation of PCR-based analysis that have previously been demonstrated only by simplified experimental setups rather than in real communities (6, 17, 26).
Despite the potential biases, the results of the analysis of the two libraries from individual worms suggest that a relatively small space can contain a surprisingly high level of diversity of bacteria. The surface area of an average specimen of E. dianae is only 0.1 mm2, yet one worm appears to be populated by dozens of independent populations, as indicated by the numbers of OTUs detected (Fig. 1). When the combined approach of ARDRA and rarefaction (Fig. 2) analysis was used, OTUs appeared to provide good representation of the diversity of 16S rDNA sequences in the libraries. Sequences of OTUs of both group I and group II were >99% identical but were differentiated by the restriction enzymes used. Since a 16S rDNA sequence identity value of 97% is generally used to approximate species boundaries (23), the OTUs appeared to provide strain level differentiation of the community. However, the diversity may have been exaggerated by chimeric 16S rDNA molecules formed during the amplification process (30) that were considered independent OTUs. Thus, the analysis provided only a rough approximation of the numbers of independent populations, but the results suggested that a larger-than-expected community is associated with individual specimens of E. dianae.
The phylogenetic comparison showed that all of the sequences in ED 1, ED 2, and ED 3 are related to bacteria commonly found in marine environments and, in addition, provided further support for the view that the 16S rDNA of the morphologically dominant filaments was not amplified. None of the sequences was unambiguously associated with bacteria capable of chemolithoautotrophic sulfur oxidation, the hypothesized mode of metabolism of the filamentous bacteria (14, 19). OTU group II was related to SRBs (Fig. 3B), which form a phylogenetically coherent cluster with anaerobic metal and sulfate reducers (10). OTU groups I and III were associated with cytophagas and γ-Proteobacteria (Fig. 3A and C), both of which are composed of bacteria that are metabolically very diverse. Members of the genus Cytophaga often possess the ability to degrade polymers, and at least some of these organisms have been found to reduce N2O in the presence of sulfide, which may indicate that they have unrecognized lithotrophic capabilities (22). Cytophagas are frequently associated with surfaces, a finding which is supported by the detection of Cytophaga-like sequences in communities attached to particles (4). Sequences in group III were only distantly related to known genera and fell in a metabolically diverse group within the γ-Proteobacteria, which contains heterotrophic bacteria, as well as sulfide-oxidizing and chemolithoautotrophic ecto- and endosymbiotic bacteria (3). Thus, with the exception of the SRB-associated sequences, only limited speculation about the metabolic diversity of the populations on the nematode surface is possible without additional data.
At this time, little is known about the diversity, stability, and specificity of communities that inhabit microenvironments in sediments. The analysis presented here indicates that PCR-based techniques can be powerful tools for estimating the diversity of microbial populations but that only limited quantitative conclusions are possible. This was exemplified by the quantitative probing results, which strongly suggest that even dominant populations may be entirely missed by PCR amplification. Nevertheless, the almost complete screening of two libraries resulted in interesting conclusions about the bacterial community on the nematode surface. There is apparently great diversity on a small scale. However, only a few populations are constitutive, perhaps dominant, in this surface community, while the majority of populations are only transiently present. One possible explanation for this is that bacterial populations in sediment are extremely inhomogeneously distributed. Although E. dianae seeks out defined physicochemical conditions, it migrates only in a limited area in the pore space of the sediment. During the migrations, members of locally abundant populations may become passively or actively associated with the nematode. Alternatively, there may be an element of chance in the initial colonization of the nematode surface, and communities may remain relatively unchanged once they are established. Regardless of its cause, the large degree of incongruity in the populations associated with apparently very similar microenvironments is a surprising outcome of our analysis of the complex community inhabiting the surface of E. dianae.
Nucleotide sequence accession numbers.
The sequences of OTUs I, II, IIIA, and IIIB have been deposited in the GenBank database under accession no. AF154059, AF154058, AF154057, and AF154060, respectively.
Acknowledgments
This work was supported by grants from the National Science Foundation and the Office of Naval Research to C.M.C. and by a travel grant from the Department of Organismic and Evolutionary Biology of Harvard University to M.F.P.
Wolfgang Sterrer of the Museum of Natural History, Bermuda, provided facilities, lodging, food, and drink; his hospitality is gratefully acknowledged. We also thank Monika Bright and Werner Urbancik of the University of Vienna, Austria, for providing the photographs in Fig. 1.
REFERENCES
- 1.Borneman J, Austin S, Triplett E W. Molecular microbial diversity on an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavanaugh C M. Microbial symbiosis: patterns of diversity in the marine environment. Am Zool. 1994;34:79–89. [Google Scholar]
- 4.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacteria assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 5.Felske A, Akkermans A D L. Spatial homogeneity of abundant bacterial 16S rRNA molecules in grassland soils. Microb Ecol. 1998;36:31–36. doi: 10.1007/s002489900090. [DOI] [PubMed] [Google Scholar]
- 6.Hansen M C, Tolker-Nielsen T, Givskov M, Molin S. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol. 1998;26:141–149. [Google Scholar]
- 7.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 8.Higgins R P, Thiel H, editors. Introduction to the study of meiofauna. Washington, D.C: Smithsonian Institution Press; 1988. [Google Scholar]
- 9.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 10.Lonergan D J, Jenter H J, Coates J D, Phillips E J P, Schmidt T M, Lovley D R. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The Ribosomal Database Project (RDP) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moyer C L, Dobbs F C, Karl D M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1994;60:871–879. doi: 10.1128/aem.60.3.871-879.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moyer C L, Tiedje J M, Dobbs F C, Karl D M. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl Environ Microbiol. 1996;62:2501–2507. doi: 10.1128/aem.62.7.2501-2507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ott J A, Novak R. Living at an interface: meiofauna at the oxygen/sulide boundary of marine sediments. In: Ryland J S, Tyler P A, editors. Reproduction, genetics and distribution of marine organisms. Fredensbourg, Denmark: Olsen & Olsen; 1989. pp. 415–421. [Google Scholar]
- 15.Ott J A, Novak R, Schiemer F, Hentschel U, Nebelsick M, Polz M. Tackling the sulfide gradient: a novel strategy involving marine nematodes and chemoautotrophic ectosymbionts. PSZN I Mar Ecol. 1991;12:261–279. [Google Scholar]
- 16.Polz F M, Odintsova E V, Cavanaugh C M. Phylogenetic relationships of the filamentous sulfur-bacterium Thiothrix ramosa based on 16S rRNA sequence analysis. Int J Syst Bacteriol. 1996;46:94–97. doi: 10.1099/00207713-46-1-94. [DOI] [PubMed] [Google Scholar]
- 17.Polz M F, Cavanaugh C M. Bias in product-to-template ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polz M F, Cavanaugh C M. A simple method for the quantification of uncultured microorganisms in the environment based on in vitro transcription of 16S rRNA. Appl Environ Microbiol. 1997;63:1028–1033. doi: 10.1128/aem.63.3.1028-1033.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polz M F, Distel D L, Zarda B, Amann R, Felbeck H, Ott J A, Cavanaugh C M. A highly specific association between ectosymbiotic, sulfur-oxidizing bacteria and a marine nematode and its phylogenetic relationship to endosymbionts and free-living bacteria. Appl Environ Microbiol. 1994;60:4461–4467. doi: 10.1128/aem.60.12.4461-4467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polz M F, Felbeck H, Novak R, Nebelsick M, Ott J A. Chemoautotrophic, sulfur-oxidizing symbiotic bacteria on marine nematodes: morphological and biochemical characterization. Microb Ecol. 1992;24:313–329. doi: 10.1007/BF00167789. [DOI] [PubMed] [Google Scholar]
- 21.Polz M F, Harbison C, Cavanaugh C M. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Symbiosis and microbial community structure, abstr. N-176; p. 352. [Google Scholar]
- 22.Reichenbach H. The order Cytophagales. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer; 1992. pp. 3631–3675. [Google Scholar]
- 23.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation kinetics and sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 24.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stryer L. Biochemistry. W. H. New York, N.Y: Freeman and Company; 1988. [Google Scholar]
- 26.Suzuki M, Giovannoni S J. Bias caused by template annealing in the amplification mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torsvik V, Goksøyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torsvik V, Sørheim R, Goksøyr J. Total bacterial diversity in soil and sediment communities—a review. J Ind Microbiol. 1996;17:170–178. [Google Scholar]
- 29.Vaneechoutte M, Rossau R, De Vos P, Gillis M, Janssens D, Paepe N, De Rouck A, Fiers T, Claeys G, Kersters K. Rapid identification of bacteria of the Comamonadaceae with amplified ribosomal DNA-restriction analysis (ARDRA) FEMS Microbiol Lett. 1992;93:227–234. doi: 10.1111/j.1574-6968.1992.tb05102.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang G C-Y, Wang Y. Frequency and formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl Environ Microbiol. 1997;63:4645–4650. doi: 10.1128/aem.63.12.4645-4650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westphalen D. Stromatolid microbial nodules from Bermuda—a special micro habitat for meiofauna. Mar Biol. 1993;117:145–157. [Google Scholar]