Abstract
Background
As therapy for solid tumours, various tumour antigens have been selected as targets, but CAR-T cells targeting these antigens have shown limited efficacy, in contrast to the effectiveness of CAR-T cells targeting haematological malignancies. In a previous report, we identified a cancer-testis antigen, DNAJB8. DNAJB8 plays a major role in tumorigenicity in cancer stem-like cells/cancer-initiating cells (CSCs/CICs). Here, we report a DNAJB8-reactive CAR yielding anti-tumour effects against renal cell carcinoma (RCC) and osteosarcoma.
Methods
We constructed a second-generation chimeric antigen receptor (CAR) against HLA-A*24:02/DNAJB8-derived peptide (DNAJB_143) complex (B10 CAR). The reactivity of B10-CAR T cells against T2-A24 cells pulsed with the cognate peptide and an RCC and osteosarcoma cell lines were quantified. The effects of adoptive cell transfer (ACT) therapy were assessed using in vivo xenografted mice models.
Results
B10 CAR-T cells recognised DNAJB8_143-pulsed T2-A24 cells and HLA-A*24:02(+)/DNAJB8(+) renal cell carcinoma and osteosarcoma cell lines. Moreover, ACT using B10 CAR-T cells showed anti-tumour effects against RCC and osteosarcoma cells.
Conclusion
B10 CAR-T cells could show specific cytotoxicity against RCC and osteosarcoma cells in vitro and in vivo. B10 CAR-T cells targeting the CSC/CIC antigen DNAJB8 might be a candidate immunotherapy for carcinoma and sarcoma.
Subject terms: Tumour immunology, Immunotherapy, Sarcoma, Cancer stem cells
Background
Chimeric antigen receptor (CAR)-engineered T cells targeting CD19 were approved by the US Food and Drug Administration (FDA) for the treatment of certain B-cell malignancies in 2017 [1], and their remarkable effectiveness has been reported [2]. As a therapy for solid tumours, a variety of tumour antigens have been selected as targets, but CAR-T cells that target these antigens have shown limited efficacy, in contrast to the effectiveness of CAR-T cells targeting haematological malignancies [3, 4]. Unfortunately, most currently reported antigens are present at low levels in normal tissues, causing on-target/off-tumour toxicity [5]. In a previous report, we identified a cancer-testis antigen, DNAJB8 (heat shock protein Hsp40 [DNAJ] chaperone family, subfamily B, member 8 protein). DNAJB8 plays a major role in tumorigenicity in cancer stem-like cells/cancer-initiating cells (CSCs/CICs) [6]. To target the cancer stem cell antigen DNAJB8 using CAR, which localises in both nuclei and cytosol but not on the cytoplasmic membrane of CSCs/CICs, we previously identified DNAJB8-derived peptide (DNAJB8-143) recognised by a CTL clone in the context of HLA-A*24:02 molecules. The CTL clone could react with naturally presented DNAJB8-143 on CSCs/CICs [7]. Next, we developed scFv (single-chain variable fragment) reacting with HLA-A*24:02/DNAJB8-143 complex using scFv phage display library. The scFv clone (B10 scFv) could specifically recognise DNAJB8-143 presented by HLA-A*24:02 molecules on renal cell carcinoma (RCC) and osteosarcoma cells, similar to TCR. Therefore, we hypothesised that a new CAR using B10 scFv (B10 CAR) could target DNAJB8 expressed in CSCs/CICs and that B10 CAR-T cells might show anti-tumour effects efficiently on solid tumours. Here, we report a DNAJB8-reactive CAR (B10 CAR) T cell yielding anti-tumour effects against RCC and osteosarcoma.
Methods
Generation of the HLA-A*24:02/DNAJB8_143 complex specific chimeric antigen receptor (B10 CAR)
We generated a second-generation B10 CAR construct containing a CD28 costimulatory and CD3 zeta intracellular signalling domains. Kinetics analysis of protein interactions between the B10 scFv antibody and HLA-A24:02/DNAJB8_143 peptide complex was determined by surface plasmon resonance analysis (KD = 5.04 × 10−9 M) [8]. The B10 CAR was cloned into a retroviral vector with the truncated form of the low-affinity nerve growth factor receptor (ΔNGFR) marker (B10 CAR plasmid). In addition, a retroviral vector containing only ΔNGFR was synthesised (Mock plasmid).
CAR-T-cell production
Human T cells were isolated by density gradient centrifugation using Lymphoprep (Serumwerk, Bernburg, Germany, Cat. #04-03-9391) from the whole blood of HLA-A24-positive healthy donors in our laboratory. The bulk T cells were activated and cultured on day 0 in AIM-V medium (Gibco BRL, Grand Island, NY) with 10% human serum, the soluble anti-CD3 antibody clone OKT3 (BioLegend, San Diego, CA, Cat. #317326, RRID: AB_11150592) at 50 ng/mL, and 100 IU/mL recombinant human IL-2. Retroviral transduction of B10 CAR into T cells was performed on days 1, 2 and 3 using a RetroNectin (Takara Bio Inc., Shiga, Japan, Cat. #T100A/B)-coated plate on day 0 (B10 CAR-T). In parallel, the Mock gene was transduced into the residual T cells (Mock T). In experiments using untransduced T cells, residual T cells were cultured in the same manner without retroviral transduction. After transduction and expansion (about 10–15 days after the blood draw), B10 CAR-T and Mock T were stored in liquid nitrogen and used in a timely fashion in assays. Before any functional assay, they were cultured in AIM-V medium without any stimulants for at least 48 h.
Cell lines and culture
We used a human RCC cell line [CAKI-1 (RRID: CVCL_0234)], human osteosarcoma cell lines [KIKU (RRID: CVCL_D885), HOS (RRID: CVCL_0312), and Saos-2 (RRID: CVCL_0548)], retrovirus packaging cell lines [PLAT-A (RRID: CVCL_B489) and PG13 (RRID: CVCL_4273)], a HLA class I- and II-deficient CML cell line [K562 (RRID: CVCL_0004)], a human embryonic kidney cell line [293 T (RRID: CVCL_0063)] and a mutant TAP-deficient cell line T2 transduced with HLA-A*2402 (T2-A24). T2-A24 cells were obtained from K. Kuzushima [9, 10] (Aichi Cancer Research Institute, Nagoya, Japan). KIKU [11] cells were established in our laboratory. HOS cells transfected with HLA-A*2402 (HOS-A24 [8]) were also used. 293 T cells in which human B2M (beta 2 microglobulin) was knocked out using CRISPR/Cas9 KO Plasmid (Santa Cruz Biotechnology Inc., Dallas, TX, Cat. #sc-417704-KO-2) were transduced with HLA-A*2402 and used as 293T-A24 cells. The other cell lines were purchased from the Japanese Collection of Research Bio-resources Cell Bank (Tokyo, Japan) and the American Type Culture Collection (ATCC; Manassas, VA). CAKI-1, K562 and T2-A24 cells were maintained in RPMI 1640 (Sigma–Aldrich, St Louis, MO) supplemented with 10% foetal bovine serum (FBS). The other cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (Sigma–Aldrich) containing 10% FBS in a 5% CO2 incubator. G418 (0.8 mg/mL) was continuously added to the culture medium for T2-A24 cells. The proportion of the side population that contained the candidate CSCs was 0.75%, 0.01% and 0.05% in KIKU, HOS and Saos-2, respectively [12] and 0.2% in CAKI-1(unpublished data). All cell lines were authenticated by HLA typing.
Antibodies
The hybridomas for the anti-HLA class I monoclonal antibody (mAb) [W6/32 (RRID: CVCL_7872)] and the anti-HLA-DR mAb [L243 (RRID: CVCL_4533)] were purchased from ATCC. The hybridoma for anti-HLA-A24 mAb (C7709A2.6 [13]) was a gift from P. Coulie (Universite Catholique de Louvain, Brussels, Belgium). The produced mAbs were collected as described previously [14].
Synthetic peptides and pulsing onto T2-A24 cells
The DNAJB8_143 peptide (AFMEAFSSF) and the HIVenv584-594 peptide (RYLRDQQLL) were purchased from Sigma Genosys (Spring, TX). The affinity of the DNAJB8_143 peptide and the HIVenv584-594 peptide were similar [7, 15]. T2-A24 cells (1.5 × 106) were seeded in a flat-bottomed 12-well tissue culture plate in a volume of 3 mL AIM-V medium (without serum) containing the peptides prepared to the desired concentration per well and incubated for 1 h at room temperature. The T2-A24 cells were then washed, re-suspended in the same medium as the effector cells, and used for co-culture.
Flow cytometric analysis of CAR expression
To evaluate the frequency of CAR expression, the following antibodies and tetramers were used: Pacific Blue anti-hCD8 (Beckman Coulter, Brea, CA, Cat. #A82791), PE anti-hCD271 (NGFR) (BioLegend, Cat. #345106, RRID: AB_2152647) and FITC anti-hCD4 (BD Biosciences, San Jose, CA, Cat. #340133, RRID: AB_400007). The CAR-T cells were stained with fluorescent-labelled HLA class I tetramer: HLA-A*24:02 DNAJB8_143 tetramer APC (MBL, Nagoya, Japan, T-select TSCM-1TA). The CAR-T cells were stained with HLA-A*24:02 HIV env tetramer-RYLRDQQLL-FITC (MBL, T-select TS-M007-3) as a negative control (when the HIV env tetramer-FITC was used, FITC anti-hCD4 was not added). Dead cells were stained with the AmCyan Fixable Aqua Dead Cell Stain Kit (Thermo Fisher Scientific, Waltham, MA, Cat. #L34965) and discriminated. The stained cells were analysed with a FACSCanto II (BD Biosciences, RRID: SCR_018056).
Quantitative real-time PCR
Total RNA was isolated using an RNeasy Mini Kit (Qiagen, Hilden, Germany, Cat. #74104). Complementary DNA (cDNA) was synthesised from 2 μg of total RNA by reverse transcription using a RevertAid RT Kit (Thermo Fisher Scientific, Cat. #K1691). The SYBR Green qPCR assay was performed according to the manufacturer’s recommendations with the PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, Cat. #a25742). The primer pairs used were 5′-GCTACACCTT CCGTAACCCT GA-3′ and 5′-CACGGTCACT ATTGAATGGG CTG-3′ for DNAJB8 and 5′-ACCACAGTCC ATGCCATCAC-3′ and 5′-TCCACCACCC TGTTGCTGTA-3′ for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The thermal cycling conditions used were as follows: 50 °C for 2 min and 10 min denaturation at 95 °C, followed by 40 cycles at 95 °C for 15 s and 60° for 1 min, and a dissociation curve was subsequently plotted. All samples were assayed in triplicate, and the analysis was completed by determining the 2–∆∆Ct values of all amplicons generated in QuantStudio 3 (Applied Biosystems, Foster City, CA, RRID: SCR_018712).
Western blot analysis
The target cells were lysed in ice-cold RIPA buffer. The lysates were quantified using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Cat. #23225). The proteins were separated by 15% SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked overnight with 5% non-fat milk in PBS-Tween solution (0.05% Tween 20 in phosphate-buffered saline) at 4 °C. The blots were then washed with PBS-Tween solution, incubated with anti-DNAJB8 primary antibody (Abcam, Cambridge, UK, Cat. #ab168585) for 1 h at room temperature and then with peroxidase-labelled anti-mouse IgG secondary antibody (SeraCare Life Sciences, Milford, MA, Cat. #5220-0341, RRID: AB_2891080) for 1 h at room temperature, washed again with TBS-Tween, visualised with ECL western blotting detection reagents (Cytiva/Global Life Sciences Solutions, Marlborough, MA, Cat. #RPN2106) according to the manufacturer’s protocol, and analysed on an Odyssey Fc Imaging System (LI-COR Biosciences, Lincoln, NE). β-actin was used as a loading control and was detected with anti-β-actin mouse mAb (Sigma–Aldrich, Cat. #A-5441, RRID: AB_476744) used at 2000-times dilution.
Evaluation of HLA-A*24:02 expression on target cells
Cultured tumour cell lines or appropriate controls were washed twice in ice-cold 1× PBS containing 1% bovine serum albumin and then incubated for 90 min at 4 °C with 50 μL C7709A2.6 (anti-HLA-A24) or W6/32 (anti-HLA class I) mAb. Staining was performed with FITC goat anti-mouse IgG+IgM (SeraCare Life Sciences, Cat. #02-18-09) secondary antibody for 30 min at 4 °C. Fluorescence was analysed by a FACSCanto II. FACS data are presented as the difference in the mean fluorescence intensity (ΔMFI), which reflects the difference in MFI values between samples with and without the primary antibody. Peripheral blood mononuclear cells isolated from a HLA-A*24:02-positive donor were used as a positive control, and the K562 cell line was used as a negative control.
Analysis of cytokine production and T-cell activation
CD107a expression and IFN-γ production were evaluated as T-cell activation markers. CAR-T cells were co-cultured with peptide-loaded T2-A24 cells at 20 µM or the other target cell lines for 4 h at an E:T ratio of 1:1. Brefeldin A solution 1000× (BioLegend, Cat. #420601) was diluted in the culture medium at the start of the co-culture. Surface markers were stained with HLA-A*24:02 DNAJB8_143 tetramer APC (MBL, T-select TSCM-1TA), PE anti-hCD107a (BioLegend, Cat. #328608, RRID: AB_1186040) and Pacific Blue anti-hCD8 (Beckman Coulter, Cat. #A82791). Next, the cells were permeabilized using the Cytofix/Cytoperm Kit (BD Biosciences, Cat. #554722) and stained with FITC anti-hIFN-γ (BD Biosciences, Cat. #552887, RRID: AB_394516). The stained cells were analysed with a FACSCanto II. The B10 CAR-T cells were evaluated after gating by the double expression of B10 CAR and CD8. The untransduced T cells were analysed after CD8-positive gating. The frequencies of hCD107a (PE)-positive cells and hIFN-γ (FITC)-positive cells were measured.
Quantitative cytokine secretion measurement
CAR-T cells were co-cultured with peptide-loaded T2-A24 cells at 20 µM or the other target cell lines for 4 h at an E:T ratio of 1:1 without brefeldin A. The culture plate was centrifuged at 400 × g for 3 min at 4 °C. The concentration of human IFN-γ in the supernatant was measured with a V-PLEX Human IFN-γ Kit (Meso Scale Discovery [MSD] LLC, Rockville, MA, Cat. #K151QOD) and MESO QuickPlex SQ 120 (MSD, Cat. #AI0AA-0, RRID: SCR_020304) following the manufacturer’s instructions.
Lactate dehydrogenase release assay of cytotoxicity
B10 CAR-T cells were co-cultured with peptide-loaded T2-A24 cells at 20 µM or the other target cell lines (CAKI-1 and KIKU) at E:T ratios of 10:1, 3:1 and 1:1 for 6 h. Round-bottomed 96-well plates with 100 μL of the AIM-V medium containing 10% human serum were used. The target cell volumes were fixed at 1.0 × 104 cells. After 6 h of co-culture, the plate was centrifuged at 400 × g for 3 min at 4 °C. Cell lysis was quantified by measuring the release of lactate dehydrogenase (LDH) using the CyQUANT LDH Cytotoxicity Assay Kit (Thermo Fisher Scientific, Cat. #C20300) according to the manufacturer’s protocol. The concentration of LDH in the supernatant was measured as the absorbance at 490 nm (reference, 680 nm) with an Infinite M1000 Pro multiplate reader (Tecan, Zurich, Switzerland). The effector cells or the target cells incubated alone at each amount allowed for correction for spontaneous LDH release. The target cells without the effector cells lysed with the lysis buffer represented the maximal possible LDH release. The mean amount of LDH in the medium was subtracted from all measurements. The percentage of cytotoxicity was calculated according to the following formula:
Percent lysis = (experimental LDH release − spontaneous LDH release from effector cells − spontaneous LDH release from target cells)/(maximum − spontaneous LDH release from target cells)
DNAJB8_143 titration assay
T2-A24 cells were pulsed with a 10-fold serial dilution of DNAJB8_143 peptide (from 10−4 to 10−14 M). B10 CAR-T cells were co-cultured with each dilution (2.0 × 104 cells) at an E:T ratio of 1:1 for 4 h. The concentration of IFN-γ in the supernatant was measured with a V-PLEX Human IFN-γ Kit (MSD). The peptide concentrations were transformed into ordinary logarithms and used for statistical calculations. The initial values of the parameters were estimated by the Emax model, and then the parameter values were estimated by the nonlinear regression functionality in the statistical software package SPSS (IBM Corp., Armonk, NY, RRID: SCR_002865). The half-maximal effective concentration (EC50) was calculated from the fitted curve.
DNAJB8_143 alanine scanning
T2-A24 cells were loaded with DNAJB8_143 (AFMEAFSSF) or peptides in which each single amino acid residue was replaced by alanine at 20 µM (with alanine itself replaced by serine). After a 4 h co-culture with B10 CAR-T cells at 1:1, the concentration of IFN-γ was measured.
HLA-blocking assay
CAR-T cells (2.0 × 104) were co-cultured with CAKI-1, KIKU or K562 cells at an E:T ratio of 1:1. The target cell lines were treated with W6/32 (anti-HLA class I mAb), C7709A2.6 (anti-HLA-A24 mAb) or L243 (anti-HLA-DR mAb). After a 4 h co-culture, the concentration of IFN-γ in the supernatant was measured with a V-PLEX Human IFN-γ Kit (MSD) according to the manufacturer’s protocol.
DNAJB8 overexpression in 293 T cells
The Myc-tagged whole human DNAJB8 gene was cloned into pcDNA3.1 (RRID: Addgene_79663). The TransIT-293 reagent (Mirus Bio, Madison, WI, Cat. #MIR 2700) was used for transfection following the manufacturer’s protocol. DNAJB8 expression was confirmed using western blot analysis. The transfected cells were used in a co-culture assay 96 h after transfection.
Mouse models
All mouse procedures were carried out following the institutional protocol guidelines at Sapporo Medical University School of Medicine. NOD-SCID-IL2 rg (null) (NSG) female mice were purchased from Charles River Laboratory Japan at the age of 6–8 weeks. The tumour cells were harvested in the logarithmic growth phase and washed with PBS. The number of tumour cells depended on the cell culture. Tumour cells were injected subcutaneously into the back in 100 μL PBS on day 0. After 5–15 days, the mice were divided into three groups (five mice each) according to the average tumour size. Then, the effector cells were infused by orbital vein plexus infusion in 100 μL. Effector cell populations were normalised to contain 1 × 107 cells per infusion. In the RCC (CAKI-1) model, 15 NSG mice were injected with 6.5 × 106 CAKI-1 cells on day 0, 5 of which were given an intravenous infusion of B10 CAR-T cells on day 15, 5 of which were administered Mock-transduced T cells (transduced with a plasmid containing only the ΔNGFR marker), and 5 of which were not administered anything. In the osteosarcoma cell line (KIKU) model, in consideration of the tumour ossification, the experiment was conducted twice with different treatment timings: a late-stage treatment and an early treatment trial. The 20 mice in the KIKU late treatment test were injected with 1.0 × 106 KIKU cells on day 0, 5 of which were administered B10 CAR-T cells intravenously on day 12, 5 of which were administered Mock-transduced T cells, and 10 of which were untreated. The other 15 mice in the KIKU early treatment trial were inoculated with 2.0 × 106 KIKU cells on day 0, and an intravenous administration was given on day 5. In order to observe a clear difference, we doubled the number of KIKU cells in the early treatment trial. Tumour volume was assessed three times a week using a caliper and calculated using the following formula: tumour volume (mm3) = (longest diameter × shortest diameter2)/2. After the endpoint, the weight of the excised tumour was measured (calculated as zero for no tumour).
Statistical analysis
For statistical evaluations, analysis of variance followed by Turkey’s honestly significant difference post hoc test was used in SPSS. Where relevant, figures indicate statistical parameters, including the value of n, means ± SD, and statistical significance.
Results
Development of B10 CAR-T cells against HLA-A24:02/DNAJB8-derived peptide
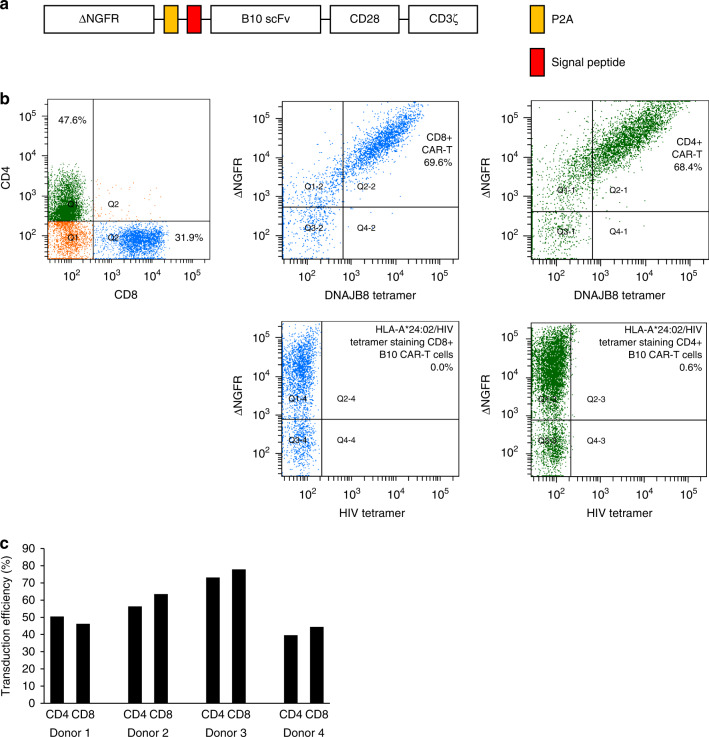
A single-chain variable fragment (scFv) named B10 that specifically recognises HLA-A*24:02/DNAJB8-derived peptide (DNAJB8_143) was isolated in our laboratory using the phage display method [8]. First, we generated a second-generation B10 CAR construct and transduced it into Jurkat/MA cells with an NFAT-luciferase reporter gene [16] and assessed CAR reactivity against various target cell lines (Fig. S1A and B). We confirmed that the reactivity of B10 CAR-Jurkat/MA cells against the cognate peptide-pulsed T2-A24 cells and an osteosarcoma cell line HOS-A24 was dependent on the expression of DNAJB8 in the context of HLA-A*24:02 molecules (Fig. S1C). Next, we generated a second-generation B10 CAR construct with ΔNFGR (Fig. 1a). Ten to fifteen days after retroviral transduction, flow cytometric evaluation was performed. The CAR-T cells were defined as being both ΔNGFR and HLA-A*24:02/DNAJB8_143 tetramer positive. The B10 CAR-T cells did not react with HLA-A*24:02/HIV env tetramer (Fig. 1b). The transduction efficiency of B10 CAR was ~60% among T cells from four independent donors and did not significantly differ between the CD4-positive cells and CD8-positive cells (Fig. 1c).
Fig. 1. Construction of B10 CAR-T cells recognising the HLA-A*24:02/DNAJB8_143 complex.
a Schematic representation of the B10 CAR construct. b Flow cytometric plots of B10 CAR-T cells. B10 CAR expression was defined as the population that was positive for both HLA-A*24:02/DNAJB8_143 tetramer (DNAJB8 tetramer) and ΔNGFR. CD8-positive cells and CD4-positive cells are coloured blue and green, respectively. The reactivity of HLA-A*24:02/HIV peptide control tetramer against B10 CAR is also shown. c Transduction efficacy of the four independent donors.
B10 CAR-T cells react against antigen-presenting cells exogenously pulsed with DNAJB8-derived peptide
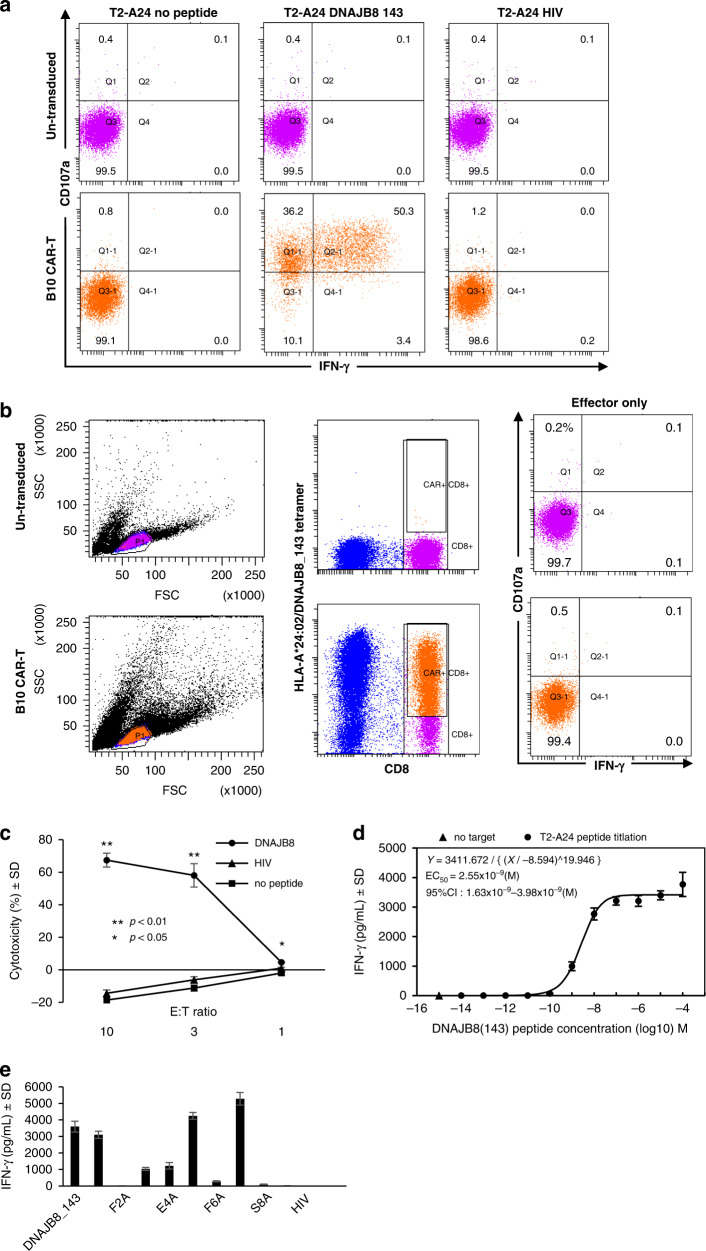
In the cytometric assessment of IFN-γ production and CD107a expression, B10 CAR-T cells specifically reacted to T2-A24 cells pulsed with DNAJB8_143 but not to those loaded with HIV peptide or unloaded, in contrast to untransduced T cells (Fig. 2a). B10 CAR-T cells alone did not show any reactivity (Fig. 2b). By measuring LDH resulting from cell membrane damage, B10 CAR-T cells were found to exert a concentration-dependent increase in LDH activity against DNAJB8_143-pulsed T2-A24 cells (Fig. 2c). The cytotoxicity was more than 65% at an E:T ratio of 10:1. To test the sensitivity of the ability of B10 CAR-T cells to recognise the HLA-A*24:02/DNAJB8_143 complex, we performed co-cultures with a series of T2-A24 cells loaded with various concentrations of DNAJB8_143 peptide. The amount of IFN-γ secreted by the B10 CAR-T cells decreased with a reducing amount of the peptide (Fig. 2d). Curve fitting was performed using the nonlinear regression function in SPSS, and the EC50 was extracted as 2.55 × 10−9 (M) [95% CI: 1.63 × 10−9–3.98 × 10−9(M)], suggesting high avidity of B10 CAR.
Fig. 2. Reactivity of B10 CAR-T cells against peptide-pulsed antigen-presenting cells.
a, b Flow cytometric CD107a expression and IFN-γ production analysis of CAR-T cells against T2-A24 cells pulsed with the indicated peptide (a). The gating strategy is shown (b). c LDH release cytotoxicity assay. The results are calculated as %cytotoxicity compared with lysis buffer. The B10 CAR-T cells were co-cultured with peptide-loaded T2-A24 cells at 20 µM. d DNAJB8_143 titration assay. B10 CAR-T cells were co-cultured with T2-A24 cells that were pulsed with a 10-fold serial dilution of DNAJB8_143 peptide (from 10−4 to 10−14 M). The concentrations of IFN-γ in the supernatant are shown. The half-maximal effective concentration (EC50) was calculated by fitting an Emax model. e DNAJB8 alanine scanning. The IFN-γ levels in the supernatant of the culture medium were measured when the B10 CAR-T cells were co-cultured with T2-A24 cells pulsed with DNAJB8_143 peptide or DNAJB8_143 peptides in which one of the amino acids was replaced by alanine, although alanine was itself replaced by serine.
Next, the specificity of B10 CAR-T cells was assessed by measuring IFN-γ produced against T2-A24 cells pulsed with single amino acid substitutions of DNAJB8_143 peptide (AFMEAFSSF). The results showed that position (P) 2, P3, P4, P6, P8 and P9 amino acids were critical for recognition (Fig. 2e). To further determine the specificity of DNAJB8_143, we used Protein BLAST [17]. No peptide sequence showed any single amino acid substitutions in Homo sapiens. This indicates sufficient specificity of B10 CAR.
B10 CAR-T cells specifically react against DNAJB8-expressing RCC and osteosarcoma cells in the context of HLA-A*24:02
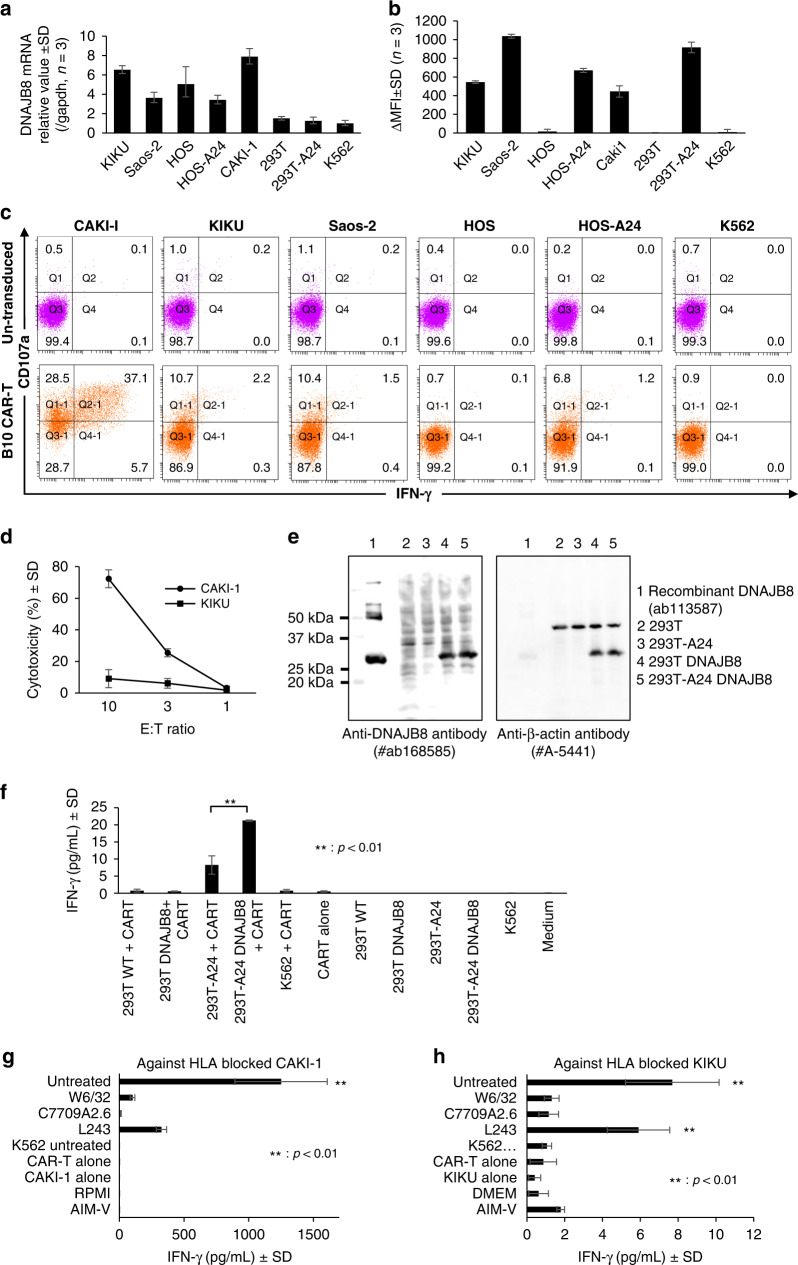
Similar experiments were conducted against cell lines with varying expression levels of DNAJB8 mRNA (Fig. 3a) and HLA-A*24:02 (Fig. 3b). In the cytometric analysis, B10 CAR-T cells expressed CD107a and produced IFN-γ against both the DNAJB8-positive and HLA-A*24:02-positive cell lines CAKI-1, KIKU, Saos-2 and HOS-A24 (Fig. 3c). Moreover, B10 CAR-T exhibited specific lysis against CAKI-1, KIKU, and Saos-2 cells using an LDH release assay and impedance-based assay (Fig. 3d, Fig. S2A–D).
Fig. 3. Reactivity of B10 CAR-T cells against various cell lines.
a Quantitative RT-PCR of DNAJB8 among the target cells. Data are shown as the mean 2–∆∆Ct ± SD normalised by comparison with GAPDH. b HLA-A*24:02 expression on the target cells defined by staining with HLA-A24-specific monoclonal antibody (C7709A2.6). Data are shown as the difference between samples and negative controls without primary monoclonal antibody in units of the mean fluorescence intensity (ΔMFI). c Flow cytometric CD107a expression and IFN-γ production of CAR-T cells against target cells. The experiments were performed using four healthy independent donors. Representative results are shown. d Lactate dehydrogenase release assay of cytotoxicity against CAKI-1 and KIKU cells. e Western blotting analysis of hDNAJB8 and β-actin. f–h IFN-γ levels in the supernatant after co-culture of B10 CAR-T cells with the indicated target cells. g, h IFN-γ production of B10 CAR-T cells after treatment with HLA-blocking antibodies. CAKI-1 (g) and KIKU (h) were used as the target cells.
Next, to confirm the antigen specificity and HLA-A24 restriction manner, we assessed the reactivity of B10 CAR-T cells against 293 T cells overexpressing DNAJB8 and against CAKI-1 and KIKU cells treated with HLA-blocking antibodies. The results showed that B10 CAR-T cells produced IFN-γ against 293 T cells with exogenous expression of HLA-A24 and DNAJB8 (293T-A24 DNAJB8) but not against the other cell lines lacking either HLA-A24 or DNAJB8 or both (Fig. 3e and f). Furthermore, the IFN-γ production of B10 CAR-T cells against CAKI-1 and KIKU cells was inhibited by anti-HLA-A24 and anti-HLA class I antibodies (Fig. 3g and h). These results suggested that B10 CAR-T cells specifically recognised endogenously presented DNAJB8_143 on the cell surface in the context of HLA-A24.
B10 CAR-T cells exhibit in vivo anti-tumour effects
Our in vitro experiments revealed strong anti-tumour activity of B10 CAR-T cells against the RCC cell line CAKI-1 and osteosarcoma cell line KIKU. Therefore, NSG mice xenografted with CAKI-1 or KIKU cells (CAKI-1 and KIKU models, respectively) were used for adoptive cell transfer therapy (ACT) experiments.
First, we performed ACT experiments on mice xenografted with or without CAKI-1 using T cells with a low population of B10 CAR (1.8% and 5.4% among CD8 + and CD4 + T cells, respectively) (Fig. S3A). Although the anti-tumour effects were not significantly different between B10 CAR and untransduced groups in mice with CAKI-1, a complete response was observed in two of the five B10 CAR group mice (Fig. S3B and C). The infused B10 CAR-T cells could be detected in the spleen of both mice with and without CAKI-1. The proportion of B10 CAR-T cells was higher in mice without CAKI-1 than in mice with CAKI-1 (Fig. S3D and E), suggesting that B10 CAR-T cells accumulated in the tumour microenvironment. There was no difference in the expression of PD-1 and LAG-3 among untransduced T cells, B10 CAR-positive cells, and B10 CAR-negative cells before ACT (Fig. S3F). After ACT, the increase in the expression of PD-1 and LAG-3 was similarly observed among all T cells in mice with and without CAKI-1. These findings suggested that ACT using T cells with a larger population of B10 CAR might show better in vivo effects.
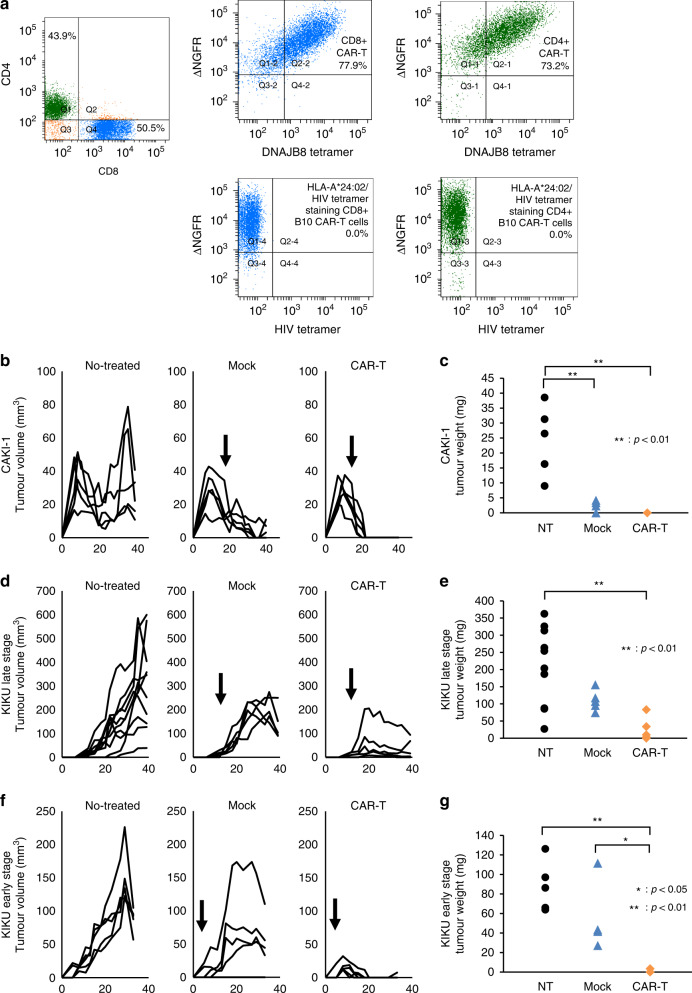
Next, we performed ACT experiments in the three experimental models. In the CAKI-1 model, the effector cells were administered on day 15 after the inoculation. The proportions of B10 CAR-positive T cells were 77.9% and 73.2% among CD8 + and CD4 + T cells, respectively (Fig. 4a). After day 23, tumours were no longer palpable on the skin in the B10 CAR-T group and no tumour tissue was observed at the time of sacrifice (Fig. 4b, Fig. S4A). There was a significant difference in the weight of the excised tumours between the B10 CAR-T group and the untreated group (p < 0.01, Fig. 4c). In the KIKU model, given the tumour ossification, the experiment was conducted twice with different treatment timings. When effector cells were transferred on day 12 (KIKU late-stage treatment model), the tumour growth began to be inhibited after day 20 in the B10 CAR-T group (Fig. 4d, Fig. S4B). The tumour weights were significantly lower in the CAR-T group than in the untreated group (p < 0.01) at the time of sacrifice (Fig. 4e). When effector cells were transferred on day 5 (KIKU early-stage treatment model), the tumour volumes began to decrease after day 11 and almost disappeared after day 20 in the B10 CAR-T group (Fig. 4f, Fig. S4C). The tumour weights were significantly lower in the B10 CAR-T group than in the Mock (p < 0.05) and untreated (p < 0.01) groups at the time of sacrifice (Fig. 4g). Finally, the residual tumours in the KIKU late-stage treatment model were histologically analysed. In the B10 CAR-T group, viable osteosarcoma cells had completely disappeared, and only hollowed-out osteoid components remained in all residual tumours. Therefore, the objective responses in the B10 CAR-T group could be considered a pathological complete response. In contrast, no similar findings were observed in the Mock and untreated groups (Fig. S4D–F).
Fig. 4. B10 CAR-T cells exert anti-tumour effects in a xenografted mouse model.
a Flow cytometric plots of B10 CAR-T cells administered to mice. This analysis was performed the day before administration. CD8-positive cells and CD4-positive cells are coloured blue and green, respectively. The reactivity of HLA-A*24:02/HIV peptide control tetramer against B10 CAR is also shown. b, d, f Tumour volume over time. Arrows indicate the date of administration of B10 CAR-T or mock T cells. c, e, g Weights of the removed tumours at the endpoint. b, c Adoptive transfer therapy of B10 CAR-T cells in mice xenografted with CAKI-1 cells (CAKI-1 model). d, e Model in which effector cells were administered on day 12 to mice implanted with KIKU cells (KIKU late-stage model). f, g Mice subcutaneously injected with KIKU received effector cells on day 5 (KIKU early-stage model). NT group of the KIKU late-stage model, n = 10; the other group, n = 5. NT no treatment. Mock: treated by T cells transduced with a plasmid containing only ΔNGFR.
Discussion
Several candidate target antigens for sarcomas have been reported. Against human epidermal growth factor receptor 2 (HER2), a phase I/II clinical study (NCT00924287) of HER2-targeted CAR-T cells for patients with recurrent or refractory sarcomas found that 4 of the 19 patients achieved stable disease lasting from 12 weeks to 14 months [18]. Disialoganglioside (GD2) is another target antigen overexpressed by almost all neuroblastomas and to various degrees by sarcomas [19]. The phase I clinical trial of GD2-specific CAR-T therapy in patients with advanced neuroblastoma conducted by Louis et al. (NCT00085930) showed that 3 of 19 patients achieved complete remission [20]. For osteosarcoma, neuroblastoma, and melanoma patients, a phase I trial of anti-GD2 CAR-T (NCT02107963) was completed in 2019 but has not reported any conclusive results. However, these target antigens are also expressed at low levels in normal tissues, which can be problematic, as in a case report by Morgan et al. [21].
We chose DNAJB8 as the target antigen for CAR-T cells in this study. Together with its family member DNAJB6, DNAJB8 acts to inhibit polyglutamine (polyQ) peptide aggregation [22]. However, unlike the ubiquitously expressed DNAJB6, the expression of DNAJB8 is localised to the testis [6, 23]. In a previous clinical trial, T-cell receptor (TCR) targeting NY-ESO-1 showed efficacy and a lack of transferred cell-derived toxicities [24]. This TCR-T is safe because the target is a cancer-testis antigen. Similarly, there is a high probability that anti-DNAJB8 CAR targeting a cancer-testis antigen is safe.
As a mechanism for enabling resistance to CAR-T, antigen loss has been a major problem [25]. Antigen expression in tumour cells falls below the threshold required for CAR-T-cell activity, causing relapse. DNAJB8, which we targeted in this report, is expressed in CSCs/CICs and has the ability to induce and maintain them [6]. DNAJB8-transduced RCC cells showed increasing tumour-initiating ability, while decreased expression of DNAJB8 has been reported to eliminate tumorigenicity [7, 26]. Therefore, DNAJB8 targeting means that cells with high proliferative capacity are attacked, and we believe that B10 CAR-T can effectively reject tumours.
The question arises as to whether the targeting of an antigen that is only expressed in CSCs/CICs can reject the entire tumour. CAR-T-cell concepts currently being developed emphasise the need to have a broader coverage of tumour cells [27]. In a previous study, we reported that CSCs were induced from non-CSCs by heat shock or oxidative stress. Even when only non-CSCs were isolated, CSCs were induced by cellular stress [28]. Therefore, we hypothesised that B10 CAR-T could reject the tumour by targeting only CSCs/CICs. The results of in vivo ACT experiments in the present study might support our hypothesis. However, the pros and cons of targeting only cancer stem cells rather than the entire tumour need further study. DNAJB8 is expressed not only in osteosarcoma and RCC cells, as shown here, but also in colorectal cancer [7]. Because it may be expressed in other histological types, B10 CAR-T could be applied to various tumours.
We did not analyse the difference in function between CD4 + and CD8 + B10 CAR-T cells because the transduction of CAR into activated T cells is generally performed using whole PBMCs derived from patients in the clinical setting. However, the importance of the T-cell subset in both CD4 + and CD8 + T cells in CD19 CAR-T therapy was previously reported [29]. In addition to the central memory subset of CD8 + CAR, the naïve subset of CD4 + CAR is also important to the superior efficiency of CAR therapy in vivo. Although it is possible that the cell viability of CAR-T cells decreases after the selection of each subset, combining the effective CAR-T-cell subsets among CD4 + and CD8 + CAR warrants further study.
In conclusion, we demonstrated that B10 CAR-T cells targeting the cancer stem cell antigen DNAJB8 showed strong anti-tumour activities against RCC and osteosarcoma cells in vitro and rejected xenografted tumours in vivo. These findings suggest that B10 CAR-T cells might be promising candidates for immunotherapy targeting solid tumours expressing DNAJB8.
Supplementary information
Acknowledgements
We thank Ms. Asuka Akamatsu for technical assistance.
Author contributions
T Tsukahara designed the studies, YW, T Tsukahara, KM and SH acquired data and prepared samples, ME provided materials, T Kubo., T Kanaseki, YH and MN interpreted data, YW and T Tsukahara wrote the manuscript, and AT, TY and T Torigoe supervised the study.
Funding
This work was supported by grants from the Japan Society for the Promotion of Science (KAKENHI 20H03807 to T Tsukahara and 17H01540 to T Torigoe), the Princess Takamatsu Cancer Research Fund (19-25125 to T Tsukahara), the Kobayashi Foundation for Cancer Research (15-9 to T Tsukahara), and AMED (21cm0106309h0006 to T Torigoe and 22ym0126801j0001 to T Tsukahara).
Data availability
All data generated or analysed during this study are included in this article and the supplementary files.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was performed by the guidelines established by the Declaration of Helsinki and was approved by the Ethics Committee of Sapporo Medical University. Healthy donors provided informed consent for the use of their blood samples in our research.
Consent for publication
Consent for publication was obtained.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-02100-1.
References
- 1.Mullard A. FDA approves first CAR T therapy. Nat Rev Drug Discov. 2017;16:669–669. doi: 10.1038/nrd.2017.196. [DOI] [PubMed] [Google Scholar]
- 2.Holstein SA, Lunning MA. CAR T-cell therapy in hematologic malignancies: a voyage in progress. Clin Pharm Ther. 2020;107:112–22. doi: 10.1002/cpt.1674. [DOI] [PubMed] [Google Scholar]
- 3.Umut Ö, Gottschlich A, Endres S, Kobold S. CAR T cell therapy in solid tumors: a short review. Memo. 2021;14:143–9. doi: 10.1007/s12254-021-00703-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kakarla S, Gottschalk S. CAR T cells for solid tumors: armed and ready to go? Cancer J Sudbury Mass. 2014;20:151–5. doi: 10.1097/PPO.0000000000000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner J, Wickman E, DeRenzo C, Gottschalk S. CAR T cell therapy for solid tumors: bright future or dark reality? Mol Ther. 2020;28:2320–39. doi: 10.1016/j.ymthe.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishizawa S, Hirohashi Y, Torigoe T, Takahashi A, Tamura Y, Mori T, et al. HSP DNAJB8 controls tumor-initiating ability in renal cancer stem–like cells. Cancer Res. 2012;72:2844–54. doi: 10.1158/0008-5472.CAN-11-3062. [DOI] [PubMed] [Google Scholar]
- 7.Morita R, Nishizawa S, Torigoe T, Takahashi A, Tamura Y, Tsukahara T, et al. Heat shock protein DNAJB8 is a novel target for immunotherapy of colon cancer-initiating cells. Cancer Sci. 2014;105:389–95. doi: 10.1111/cas.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tadano H, Tsukahara T, Mizushima E, Akamatsu A, Watanabe K, Nojima I, et al. Development of an artificial antibody specific for HLA/peptide complex derived from cancer stem-like cell/cancer-initiating cell antigen DNAJB8. Br J Cancer. 2020;123:1387–94. doi: 10.1038/s41416-020-1017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuzushima K, Hayashi N, Kimura H, Tsurumi T. Efficient identification of HLA-A*2402–restricted cytomegalovirus-specific CD8+ T-cell epitopes by a computer algorithm and an enzyme-linked immunospot assay. Blood. 2001;98:1872–81. doi: 10.1182/blood.V98.6.1872. [DOI] [PubMed] [Google Scholar]
- 10.Salter RD, Howell DN, Cresswell P. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics. 1985;21:235–46. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 11.Wada T, Uede T, Ishii S, Matsuyama K, Yamawaki S, Kikuchi K. Monoclonal antibodies that detect different antigenic determinants of the same human osteosarcoma-associated antigen. Cancer Res. 1988;48:2273–9. [PubMed] [Google Scholar]
- 12.Murase M, Kano M, Tsukahara T, Takahashi A, Torigoe T, Kawaguchi S, et al. Side population cells have the characteristics of cancer stem-like cells/cancer-initiating cells in bone sarcomas. Br J Cancer. 2009;101:1425–32. doi: 10.1038/sj.bjc.6605330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda H, Lethé B, Lehmann F, Van Baren N, Baurain JF, De Smet C, et al. Characterization of an antigen that Is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6:199–208. doi: 10.1016/S1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- 14.Miyamoto S, Kochin V, Kanaseki T, Hongo A, Tokita S, Kikuchi Y, et al. The antigen ASB4 on cancer stem cells serves as a target for CTL immunotherapy of colorectal cancer. Cancer Immunol Res. 2018;6:358–69. doi: 10.1158/2326-6066.CIR-17-0518. [DOI] [PubMed] [Google Scholar]
- 15.Hirohashi Y, Torigoe T, Maeda A, Nabeta Y, Kamiguchi K, Sato T, et al. An HLA-A24-restricted cytotoxic T lymphocyte epitope of a tumor-associated protein, survivin. Clin Cancer Res. 2002;8:1731–9. [PubMed] [Google Scholar]
- 16.Aarnoudse CA, Krüse M, Konopitzky R, Brouwenstijn N, Schrier PI. TCR reconstitution in Jurkat reporter cells facilitates the identification of novel tumor antigens by cDNA expression cloning. Int J Cancer. 2002;99:7–13. doi: 10.1002/ijc.10317. [DOI] [PubMed] [Google Scholar]
- 17.Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022;50:D20–6. [DOI] [PMC free article] [PubMed]
- 18.Ahmed N, Brawley VS, Hegde M, Robertson C, Ghazi A, Gerken C, et al. Human epidermal growth factor receptor 2 (HER2) –specific chimeric antigen receptor–modified T cells for the immunotherapy of HER2-positive sarcoma. J Clin Oncol. 2015;33:1688–96. doi: 10.1200/JCO.2014.58.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nazha B, Inal C, Owonikoko TK. Disialoganglioside GD2 expression in solid tumors and role as a target for cancer therapy. Front. Oncol. 2020;10:1000. [DOI] [PMC free article] [PubMed]
- 20.Louis CU, Savoldo B, Dotti G, Pule M, Yvon E, Myers GD, et al. Antitumor activity and long-term fate of chimeric antigen receptor–positive T cells in patients with neuroblastoma. Blood. 2011;118:6050–6. doi: 10.1182/blood-2011-05-354449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan RA, Yang JC, Kitano M, Dudley ME, Laurencot CM, Rosenberg SA. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther. 2010;18:843–51. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillis J, Schipper-Krom S, Juenemann K, Gruber A, Coolen S, van den Nieuwendijk R, et al. The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides. J Biol Chem. 2013;288:17225–37. doi: 10.1074/jbc.M112.421685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hageman J, Rujano MA, van Waarde MAWH, Kakkar V, Dirks RP, Govorukhina N, et al. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol Cell. 2010;37:355–69. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Robbins PF, Kassim SH, Tran TLN, Crystal JS, Morgan RA, Feldman SA, et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin Cancer Res. 2015;21:1019–27. doi: 10.1158/1078-0432.CCR-14-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majzner RG, Mackall CL. Tumor antigen escape from CAR T-cell therapy. Cancer Discov. 2018;8:1219–26. doi: 10.1158/2159-8290.CD-18-0442. [DOI] [PubMed] [Google Scholar]
- 26.Yamashita M, Hirohashi Y, Torigoe T, Kusumoto H, Murai A, Imagawa T, et al. Dnajb8, a member of the heat shock protein 40 family has a role in the tumor initiation and resistance to docetaxel but Is dispensable for stress response. PLoS ONE. 2016;11:e0146501. doi: 10.1371/journal.pone.0146501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei J, Han X, Bo J, Han W. Target selection for CAR-T therapy. J Hematol OncolJ Hematol Oncol. 2019;12:62. doi: 10.1186/s13045-019-0758-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kusumoto H, Hirohashi Y, Nishizawa S, Yamashita M, Yasuda K, Murai A, et al. Cellular stress induces cancer stem-like cells through expression of DNAJB8 by activation of heat shock factor 1. Cancer Sci. 2018;109:741–50. doi: 10.1111/cas.13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sommermeyer D, Hudecek M, Kosasih PL, Gogishvili T, Maloney DG, Turtle CJ, et al. Chimeric antigen receptor-modified T cells derived from defined CD8+ and CD4+ subsets confer superior antitumor reactivity in vivo. Leukemia. 2016;30:492–500. doi: 10.1038/leu.2015.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this article and the supplementary files.