Abstract
Emerging evidence demonstrates that some metabolic enzymes that phosphorylate soluble metabolites can also phosphorylate a variety of protein substrates as protein kinases to regulate cell cycle, apoptosis and many other fundamental cellular processes. However, whether a metabolic enzyme dephosphorylates protein as a protein phosphatase remains unknown. Here we reveal the gluconeogenic enzyme fructose 1,6-biphosphatase 1 (FBP1) that catalyzes the hydrolysis of fructose 1,6-bisphosphate (F-1,6-BP) to fructose 6-phosphate (F-6-P) as a protein phosphatase by performing a high-throughput screening of metabolic phosphatases with molecular docking followed by molecular dynamics (MD) simulations. Moreover, we identify IκBα as the substrate of FBP1-mediated dephosphorylation by performing phosphoproteomic analysis. Mechanistically, FBP1 directly interacts with and dephosphorylates the serine (S) 32/36 of IκBα upon TNFα stimulation, thereby inhibiting NF-κB activation. MD simulations indicate that the catalytic mechanism of FBP1-mediated IκBα dephosphorylation is similar to F-1,6-BP dephosphorylation, except for higher energetic barriers for IκBα dephosphorylation. Functionally, FBP1-dependent NF-κB inactivation suppresses colorectal tumorigenesis by sensitizing tumor cells to inflammatory stresses and preventing the mobilization of myeloid-derived suppressor cells. Our finding reveals a previously unrecognized role of FBP1 as a protein phosphatase and establishes the critical role of FBP1-mediated IκBα dephosphorylation in colorectal tumorigenesis.
Subject terms: Cancer metabolism, Phosphorylation, Cell signalling
Introduction
Phosphorylation plays critical roles in the regulation of many cellular processes including cell cycle, growth, apoptosis and signal transduction pathways. The reversible phosphorylation of proteins is accomplished by opposing activities of kinases and phosphatases. Metabolic enzymes, such as pyruvate kinase isoform M2, phosphoglycerate kinase 1, hexokinase, and nucleoside diphosphate kinase 1 and 2, that phosphorylate soluble metabolites can also function as protein kinases and phosphorylate a variety of protein substrates to regulate the Warburg effect, gene expression, cell cycle progression and proliferation, apoptosis, autophagy, exosome secretion, T cell activation, iron transport, ion channel opening, and many other fundamental cellular functions.1 However, whether a metabolic enzyme functions as a protein phosphatase and if so, in which cellular process it is involved has not been explored.
Fructose-1,6-bisphosphatase 1 (FBP1) catalyzes the hydrolysis of fructose 1,6-bisphosphate (F-1,6-BP) to fructose 6-phosphate (F-6-P) in the presence of divalent cations, acting as a rate-limiting enzyme in gluconeogenesis. FBP1 has been increasingly implicated as a tumor suppressor by antagonizing glycolysis through its metabolic activity, inhibiting the transcriptional activity of hypoxia-inducible factors in the nucleus, or limiting the activation of hepatic stellate cells,2–4 and is silenced in basal-like breast cancer, clear cell renal cell carcinoma and hepatocellular carcinoma. However, in contrast to its metabolic function of F-1,6-BP dephosphorylation, whether FBP1 functions as a protein phosphatase to dephosphorylate protein remains unknown.
It has been well recognized that tissue inflammation is associated with the development of cancer.5 Patients with inflammatory bowel disease are at higher risk of colorectal cancer (CRC).6 Inflammation involves interaction between various immune cells, inflammatory cells, chemokines, and cytokines, which may lead to signaling toward tumor cell proliferation, survival, migration, and invasion.7 Tumor necrosis factor α (TNFα) and interleukin 1β (IL1β) are two major proinflammatory cytokines produced by immune cells during inflammatory bowel disease and are responsible for a diverse range of signaling events within cells.7,8
Nuclear factor-κB (NF-κB) has a crucial and complex function in orchestrating multiple aspects of inflammation, and controls the expression of important regulatory genes that drive cell activation and proliferation during immunity, inflammation, and oncogenesis.9 In the canonical pathway, NF-κB/Rel proteins are bound and inhibited by inhibitory IκB proteins. Proinflammatory cytokines, lipopolysaccharide, growth factors, and antigen receptors activate an IKK complex (IKKβ, IKKα, and NEMO), which phosphorylates serine (S) 32 and 36 of IκB proteins. Phosphorylation of IκB leads to its ubiquitination and proteasomal degradation, freeing NF-κB/Rel complexes. Active NF-κB/Rel complexes translocate to the nucleus where they act, alone or in combination with other transcription factors, such as activator protein 1 and signal transducer and activator of transcription, to induce target gene expression.10
In this study, we performed a high-throughput screening of metabolic phosphatases that also function as protein phosphatases by adopting molecular docking followed by molecular dynamics (MD) simulations and identified FBP1 as a protein phosphatase. Moreover, we investigated the biological role of such activity of FBP1 and revealed that FBP1 dephosphorylates IκBα and plays a suppressive role in colorectal tumorigenesis by compromising tumor cell survival in inflammatory microenvironment and preventing myeloid-derived suppressor cells (MDSCs) mobilization.
Results
FBP1 dephosphorylates IκBα as a protein phosphatase
In order to investigate whether metabolic phosphatases can catalyze the dephosphorylation of proteins, molecular modeling was adopted to study the binding affinity of single-phosphorylated residue models, phosphorylated serine (pSer), phosphorylated threonine (pThr), and phosphorylated tyrosine (pTyr), with 57 human metabolic phosphatases (Fig. 1a). The docking results and rank were shown in Supplementary information, Table S1. The top 10 metabolic phosphatases with high average Vina scores for each dipeptide model were selected for further study (Supplementary information, Table S2). After visual inspection considering the docking poses, three candidates, including FBP1, inositol polyphosphate-5-phosphatase E (INPP5E), and enolase-phosphatase 1 (ENOPH1), were further employed for MD simulation to verify the docking results. The initial complexes of three dipeptide models with FBP1, INPP5E, and ENOPH1 were obtained from the results of Autodock Vina. The total binding free energies of the pSer dipeptide model with FBP1, INPP5E, and ENOPH1 were –80.21 kcal/mol, –28.38 kcal/mol, and –55.34 kcal/mol, respectively; pThr dipeptide model with FBP1, INPP5E, and ENOPH1 were –75.89 kcal/mol, –21.83 kcal/mol, and –54.95 kcal/mol, respectively; pTyr dipeptide model with FBP1, INPP5E, and ENOPH1 were –64.42 kcal/mol, –23.29 kcal/mol, and –40.93 kcal/mol, respectively (Fig. 1b; Supplementary information, Fig. S1a). Therefore, the binding free energies of phosphorylated residues with FBP1 were strongest among the three proteins. Collectively, molecular modeling and MD simulation results indicate that the phosphorylated residues are most favorable to bind FBP1, suggesting that among these metabolic phosphatases, FBP1 is most likely to function as a protein phosphatase.
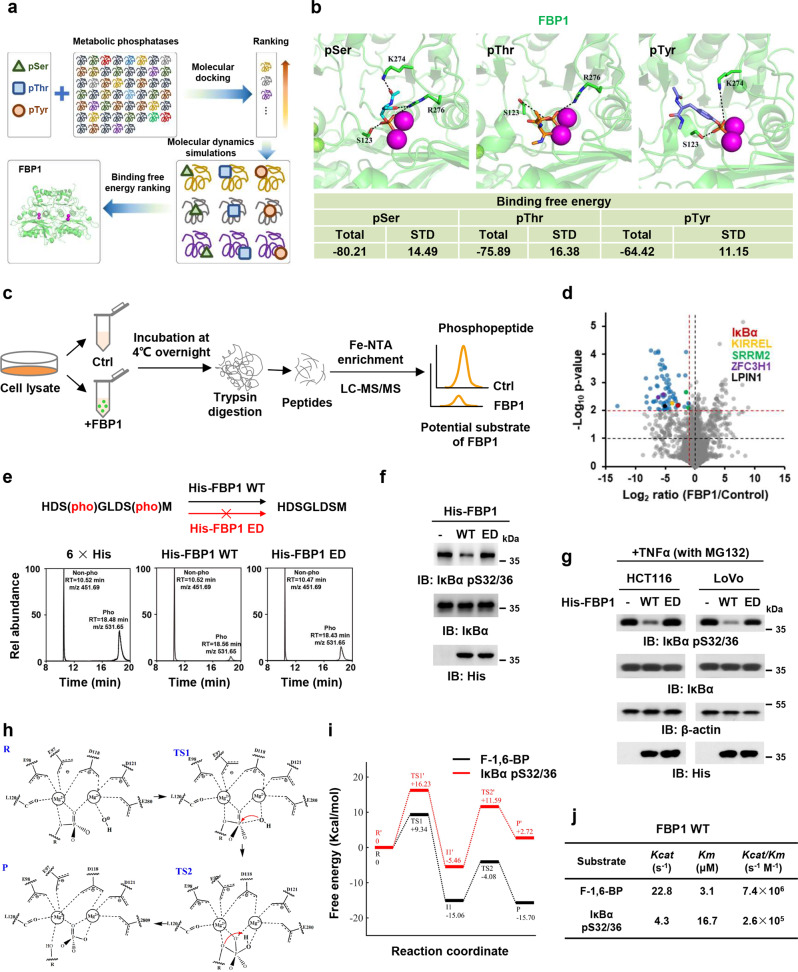
Fig. 1. FBP1 dephosphorylates IκBα as a protein phosphatase.
a The workflow of identification of metabolic phosphatases as potential protein phosphatases. b The binding modes and binding free energies of phosphorylated serine (pSer), phosphorylated threonine (pThr), and phosphorylated tyrosine (pTyr) models with FBP1 were shown (all energies are in units of kcal/mol). c Experimental workflow. For quantitative phosphoproteomic analysis, HCT116 cell lysates with or without FBP1 incubation (12 h) were subjected to trypsin digestion, Fe-NTA enrichment and LC-MS/MS analyses. d Mass spectrometry analyses of phosphopeptides in HCT116 cell lysates incubated with or without purified recombinant His-FBP1. The data are presented as a volcano plot for two biologically independent experiments (two-tailed Student’s t-test). Proteins with P < 0.01 and FC (fold change) >2.0 were regarded as the candidate proteins that showed a significant decrease of phosphorylation levels. e In vitro dephosphorylation of IκBα pS32/36 peptide with purified recombinant His-FBP1 WT or ED mutant were analyzed with mass spectrometry. f In vitro dephosphorylation of IκBα with purified recombinant His-FBP1 WT or ED were analyzed by immunoblotting assay. The phosphorylated IκBα was obtained by incubating purified recombinant IκBα with recombinant active IKKβ. g HCT116 or LoVo cells were pretreated with 50 μM MG132 for 1 h and then treated with 10 ng/mL TNFα for 3 h. Cell lysates were incubated with or without purified recombinant His-FBP1 WT or ED. Immunoblotting assay was performed. h Proposed proton transfer pathway and two-metal-ion mechanism of the P–O bond cleavage reaction catalyzed by FBP1. i Free energy profile. The reactant state (R), transition state 1 (TS1), transition state 2 (TS2), and product formation state (P) are highlighted. j The catalytic efficiency of FBP1 with different substrates was measured.
To validate the hypothesis that FBP1 dephosphorylates proteins, we performed the in vitro phosphatase assay by mixing bacteria-purified recombinant FBP1 and the lysates of HCT116 CRC cells, followed by mass spectrometry analysis of phosphorylated proteins (Fig. 1c), showing that the levels of 78 phosphorylated residues in 72 proteins were dramatically reduced by FBP1 incubation (Supplementary information, Table S3). Notably, among more than 2000 identified phosphorylated proteins, only a fraction of the proteins (72 proteins) were dephosphorylated after the incubation with FBP1, suggesting the substrate specificity for FBP1 (Supplementary information, Table S4). Moreover, according to the sequences of those dephosphorylated peptides, we further identified a conserved motif [S/T]P in FBP1 substrate (Supplementary information, Fig. S1b). Among the 72 proteins, 5 proteins contained two and more dephosphorylated residues, including Kin of IRRE-like protein 1 (KIRREL), Phosphatidate phosphatase LPIN1 (LPIN1), NF-kappa-B inhibitor alpha (IκBα), Serine/arginine repetitive matrix protein 2 (SRRM2) and Zinc finger C3H1 domain-containing protein (ZFC3H1) (Fig. 1d). The dephosphorylation of IκBα serine (S) 32/36 was further validated by incubating a synthesized IκBα peptide containing phosphorylated S32/36 (pS32/36) with purified recombinant His-FBP1 wild type (WT) or enzymatic dead (ED) mutant, in which glycine (G) 260 was mutated into arginine (R).2 The mass spectrometry analysis of IκBα peptide showed that FBP1 WT dephosphorylated IκBα peptide, while FBP1 ED lost the majority of its activity to dephosphorylate IκBα peptide (Fig. 1e; Supplementary information, Fig. S1c). Moreover, we purified recombinant His-IκBα and carried out the in vitro kinase assay by mixing it with recombinant active IKKβ to obtain IκBα pS32/36, followed by the incubation of IκBα pS32/36 with or without His-FBP1 WT or ED. The in vitro phosphatase assay confirmed that FBP1 dephosphorylated IκBα pS32/36 (Fig. 1f).
The NF-κB family of inducible transcription factors is activated in response to a variety of stimuli. TNFα is one of the most potent physiological inducers of the nuclear transcription factor NF-κB.11 Thus, we stimulated MG132-pretreated HCT116 and LoVo cells with TNFα. The phosphatase assay was carried out by mixing the lysates of these cells with purified recombinant His-FBP1 proteins, which showed that FBP1 WT, but not FBP1 ED, dephosphorylated TNFα-induced IκBα pS32/36 (Fig. 1g). Taken together, these results support that FBP1 indeed functions as a protein phosphatase to dephosphorylate IκBα, the key inhibitory regulator of NF-κB signaling.
The catalytic mechanism of FBP1-mediated IκBα dephosphorylation
To understand the mechanism of FBP1-catalyzed IκBα dephosphorylation, we carried out MD simulations on the models of FBP1 with IκBα pS32/36 peptide (L25–M37) or F-1,6-BP to construct the Quantum Mechanics (QM) conformation for QM calculation, and QM calculation provided the reaction barriers and processes. Seven initial molecular models of FBP1 and IκBα peptide complexes were obtained from the results of ZDOCK,12 and the seven conformations were divided into two classes, in which the S32 (named Conformation A) or S36 (named Conformation B) was located at the Mg2+ catalytic reaction center. The binding affinity of IκBα pS32/36 with FBP1 was estimated by MM-GBSA calculations. The total binding free energy of IκBα pS32/36 peptide with FBP1 was –119.88 kcal/mol, when S32 was located at the Mg2+ catalytic reaction center. However, when S36 was located at reaction center, total binding free energy of the IκBα peptide with FBP1 was –66.73 kcal/mol, suggesting that the most favorable binding mode of IκBα peptide with FBP1 is pS32 at the catalytic reaction center (Supplementary information, Fig. S1d).
Based on the conformation constructed from the MD simulations, QM calculation showed that hydrolysis reaction mechanism of FBP1 followed the common two-metal-ion catalytic mechanism proposed previously.13 The hydroxide ion coordinated with Mg2+, and its ionization was accomplished with the help of the Mg2+. In both models of F-1,6-BP with FBP1 and IκBα with FBP1, the nucleophile hydroxide ion coordinated to one Mg2+, and the oxygen of the P–O bond coordinated to the other Mg2+. The hydroxide ion first attacked on the phosphorus of the scissile phosphate (TS1). Then the pentacovalent intermediate was formed, followed by the proton transfer from hydroxide ion to the oxygen of the scissile phosphate (TS2), which is accompanied by the bond breakage of P–O (Fig. 1h). It should be noted that the barriers in the pathway of F-1,6-BP (9.34 kcal/mol, 10.98 kcal/mol) dephosphorylated by FBP1 were lower than those for IκBα pS32/36 (16.23 kcal/mol, 17.05 kcal/mol), suggesting that the F-1,6-BP were more easily hydrolyzed by FBP1 than IκBα pS32/36 (Fig. 1i).
Moreover, we performed the in vitro dephosphorylation assay by incubating FBP1 with F-1,6-BP or IκBα pS32/36 peptide and analyzed the catalytic rate constant (Kcat), Michaelis constant (Km), and the catalytic efficiency (Kcat/Km) of these reactions. Consistent with the results of QM calculation, FBP1-catalyzed reaction had higher catalytic rate, substrate affinity and efficiency with F-1,6-BP than with IκBα pS32/36 (Fig. 1j). Collectively, these results indicate that the hydrolysis reaction mechanisms of IκBα pS32/36 and F-1,6-BP catalyzed by FBP1 are similar, while the energy barriers in the pathway of F-1,6-BP with FBP1 are lower than those for IκBα pS32/36 with FBP1.
FBP1 interacts with IκBα and inhibits the NF-κB pathway in tumor cells after TNFα treatment
In order to identify the protein substrate of FBP1 in cells, we carried out mass spectrometry analysis of FBP1-associated proteins in HCT116 cells after TNFα treatment and noticed that IκBα is among the proteins that strongly interacted with FBP1 after TNFα treatment (Fig. 2a; Supplementary information, Fig. S2a). The interaction of these two proteins was further validated by performing co-immunoprecipitation (Co-IP) experiment in HCT116 and LoVo cells expressing Flag-tagged FBP1 (Flag-FBP1) and HA-tagged IκBα (HA-IκBα). The result showed that FBP1 interacted with IκBα in tumor cells upon TNFα stimulation (Fig. 2b). To determine whether endogenous FBP1 interacts with endogenous IκBα, we performed the Co-IP experiments in human colonic epithelial cells (HCoEpiC) that has much higher levels of FBP1 proteins than HCT116 cells and found that these two endogenous proteins could also interact with each other after TNFα stimulation (Fig. 2c; Supplementary information, Fig. S2b). These results raise the possibility that FBP1 might inhibit TNFα-induced NF-κB pathway by directly dephosphorylating IκBα.
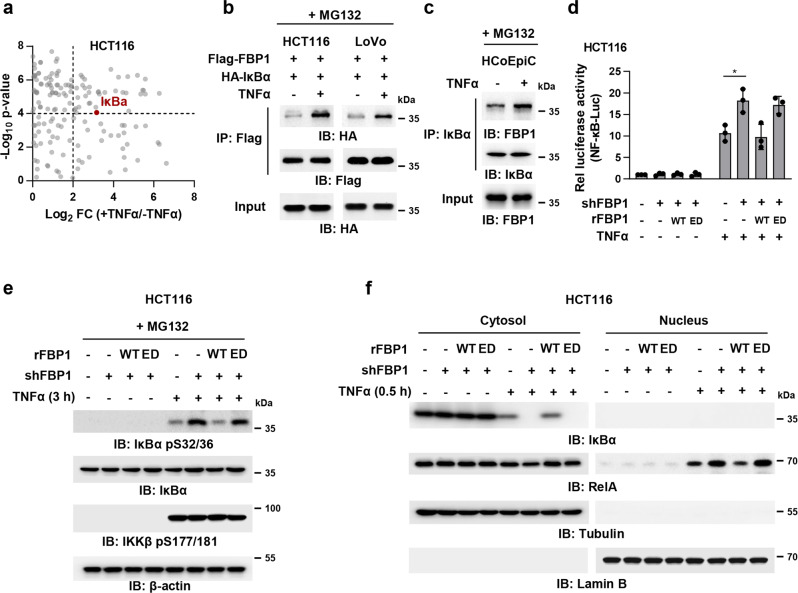
Fig. 2. FBP1 interacts with IκBα and inhibits TNFα-induced NF-κB signaling.
a HCT116 cells stably expressing Flag-FBP1 were pretreated with 50 μM MG132 for 1 h and treated with or without TNFα (10 ng/mL) for 3 h. Cells were harvested and Flag-FBP1 proteins were immunoprecipitated using anti-Flag agarose beads. FBP1-associated proteins were analyzed by mass spectrometry analyses. The data are presented as a volcano plot for two biologically independent experiments (two-tailed Student’s t-test). Proteins with P < 0.0001 and log2-transformed FC in expression of > 2.0 were regarded as the candidate proteins that showed strong interactions with FBP1. b HCT116 or LoVo cells co-transfected with Flag-FBP1 and HA-IκBα were pretreated with 50 μM MG132 for 1 h and treated with or without TNFα (10 ng/mL) for 3 h. Flag-FBP1 was immunoprecipitated. c HCoEpiC cells were pretreated with 50 μM MG132 for 1 h and treated with or without TNFα (10 ng/mL) for 3 h. IκBα was immunoprecipitated. d FBP1-depleted HCT116 cells rescued with rFBP1 WT or ED were transfected with NF-κB reporter and then treated with or without TNFα (10 ng/mL) for 3 h. The relative luciferase activities were normalized to those of the cells expressing shNT and to Renilla controls. Data represent the mean ± s.d. of three biologically independent experiments (two-tailed Student’s t-test). e FBP1-depleted HCT116 cells were rescued with rFBP1 WT or ED. Cells were pretreated with 50 μM MG132 for 1 h and treated with or without TNFα (10 ng/mL) for 3 h. Immunoblotting assay was performed. f FBP1-depleted HCT116 cells were rescued with rFBP1 WT or ED. Cells were treated with or without TNFα (10 ng/mL) for 0.5 h. Cells were harvested and cell fraction assay was performed.
To test the hypothesis, we firstly examined the levels of FBP1 proteins in a series of CRC cell lines, including Caco-2, HCT116, LoVo, RKO, SW480, SW620, HCT-8, and HT29. HCT116, LoVo and HCT-8 cells had relatively higher FBP1 expression than the others (Supplementary information, Fig. S2c). We then depleted FBP1 in HCT116 or LoVo cells with short hairpin RNA (shRNA) and rescued these cells with shRNA-resistant (r) FBP1 WT or ED (Supplementary information, Fig. S2d), followed by the transfection with a luciferase reporter construct containing NF-κB responsive elements (NF-κB-Luc). The measurement of luciferase activities indicated that FBP1 depletion enhanced TNFα-induced NF-κB activation. Rescued rFBP1 WT expression abrogated such enhancement of NF-κB activation in FBP1-depleted cells, excluding the off-targeting possibility of FBP1 shRNA, while rescued rFBP1 ED expression only slightly attenuated such enhancement (Fig. 2d), underscoring the importance of FBP1 enzymatic activity in FBP1-mediated regulation of NF-κB activation. Consistently, FBP1 depletion enhanced IκBα pS32/36 and its subsequent degradation in HCT116 or LoVo cells upon TNFα stimulation without affecting IKKβ pS177/181. Rescued expression of rFBP1 WT, but not that of rFBP1 ED, reduced the level of IκBα pS32/36 and restored IκBα protein level in FBP1-depleted cells (Fig. 2e, f; Supplementary information, Fig. S2e–g).
Collectively, these results indicate that FBP1 interacts with IκBα and subsequently inhibits TNFα-induced NF-κB activation.
FBP1 N213K mutation disrupts FBP1/IκBα interaction and enhances NF-κB activation
To understand the structural basis underlying FBP1-mediated IκBα dephosphorylation, we performed the binding free energy analysis based on the conformation of IκBα pS32/36 with FBP1 constructed from the MD simulations. The binding mode of IκBα pS32/36 with FBP1 is similar to that of F-1,6-BP with FBP1 (Fig. 3a). The dominant residues in FBP1 responsible for its binding with IκBα pS32/36 were R244, lysine (K) 275, S124, R314, K270, G123, R277, asparagine (N) 213 and K208 (Fig. 3b). Among these residues, R244, K275, N213, K270, and K208 directly interacted with pS36, which was not located in the Mg2+ catalytic reaction center.
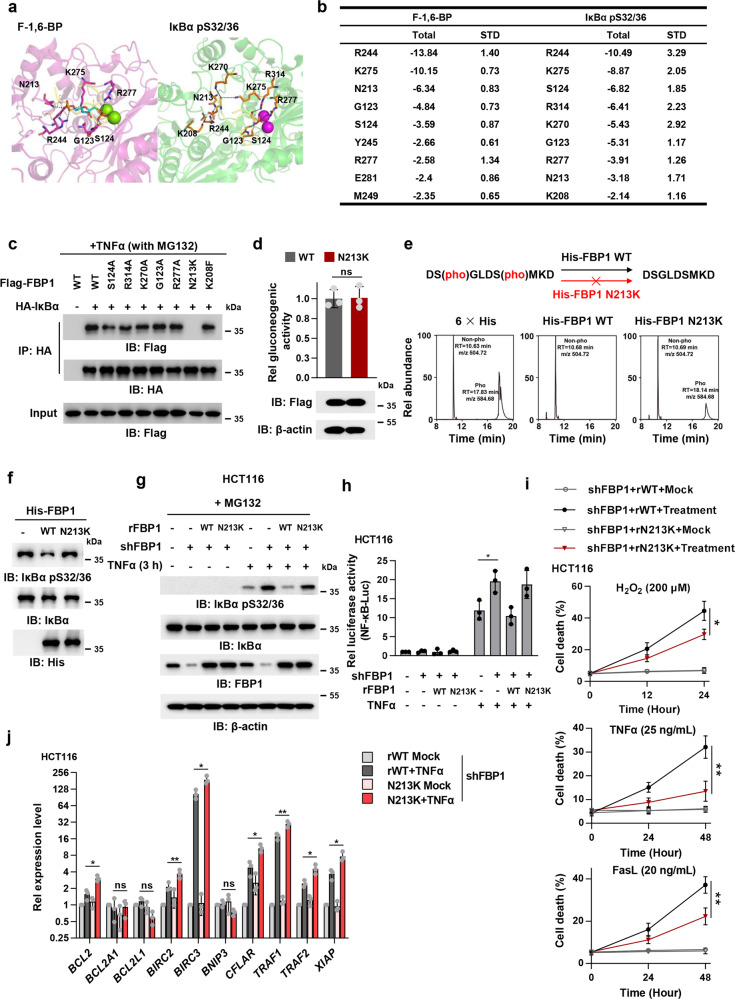
Fig. 3. FBP1 N213K mutation disrupts FBP1/IκBα interaction and activates NF-κB to restrain inflammation-induced cell death.
a, b The binding modes of F-1,6-BP or IκBα pS32/36 with FBP1. The FBP1 proteins binding with F-1,6-BP or IκBα are shown in cartoon and are colored in magenta and green, respectively. pS32, pS36, and dominate residues are shown in sticks, and Mg2+ are shown in spheres. FBP1-IκBα is superimposed to FBP1–F-1,6-BP complex (a). The dominant amino acid residues of FBP1 in binding with F-1,6-BP and IκBα pS32/36 are listed (b), and all energies are in units of kcal/mol. c HCT116 cells were co-transfected with Flag-FBP1 (WT or mutations) and HA-IκBα. Cells were pretreated with 50 μM MG132 for 1 h and treated with TNFα (10 ng/mL) for 3 h. Cells were harvested and HA-IκBα was immunoprecipitated. d HCT116 cells were overexpressed with Flag-FBP1 WT or N213K. Gluconeogenic activity in these cells was measured. ns, not significant. e In vitro dephosphorylation of phospho-IκBα peptide by purified recombinant His-FBP1 WT or N213K were analyzed with mass spectrometry. f In vitro dephosphorylation of IκBα with purified recombinant His-FBP1 WT or N213K were analyzed by immunoblotting assay. The phosphorylated IκBα was obtained by incubating purified recombinant IκBα with recombinant active IKKβ. g FBP1-depleted HCT116 cells were rescued with rFBP1 WT or N213K. Cells were pretreated with 50 μM MG132 for 1 h and treated with or without TNFα (10 ng/mL) for 3 h. h FBP1-depleted HCT116 cells rescued with rFBP1 WT or N213K were transfected with NF-κB reporter and then treated with or without TNFα (10 ng/mL) for 3 h. The relative luciferase activities were normalized to those of the cells expressing shNT and to Renilla controls. i FBP1-depleted HCT116 cells were rescued with rFBP1 WT or N213K. Cells were pretreated with 5 ng/mL TNFα before treated with 200 μM H2O2, 25 ng/mL TNFα or 20 ng/mL FasL. Cell death was determined by trypan blue staining. j FBP1-depleted HCT116 cells were rescued with rFBP1 WT or N213K. Cells were treated with or without 10 ng/mL TNFα for 12 h. Cells were harvested and subjected to real-time qPCR. Data represent the mean ± s.d. of the indicated values from three biologically independent experiments (d, h–j, two-tailed Student’s t-test).
In order to specifically disrupt the interaction between FBP1 and IκBα without influencing that between FBP1 and F-1,6-BP, we pinpointed the exact residue specifically required for the interaction between FBP1 and IκBα. As R244 and K275 were the top two residues required for the association of FBP1 with both IκBα and F-1,6-BP, we generated seven FBP1 mutants, including FBP1 S124A, R314A, K270A, G123A, R277A, N213K and K208F, in which seven dominant residues other than R244 and K275 were mutated into alanine (A), K, or phenylalanine (F). Co-IP experiments were performed with anti-HA antibody in HCT116 cells transfected with HA-IκBα and Flag-FBP1 WT or mutants, which showed that FBP1 N213K, but not the other FBP1 mutants, almost completely lost its binding with IκBα (Fig. 3c). Of note, FBP1 N213K had similar F-1,6-BP binding affinity and gluconeogenic activity to FBP1 WT (Fig. 3d; Supplementary information, Fig. S3a).
We next examined whether the N213K mutation affected FBP1-dependent dephosphorylation of IκBα. To test the hypothesis, we performed a phosphatase assay by incubating a synthesized IκBα pS32/36 peptide with recombinant His-FBP1 WT or N213K. The mass spectrometry analysis of IκBα peptide showed that the N213K mutation dramatically abrogated FBP1-mediated dephosphorylation of IκBα (Fig. 3e; Supplementary information, Fig. S3b). Besides, we purified recombinant His-IκBα and carried out the in vitro kinase assay by mixing it with recombinant active IKKβ to obtain IκBα pS32/36, followed by the incubation of IκBα pS32/36 with or without His-FBP1 WT or N213K. The in vitro phosphatase assay confirmed that the N213K mutation abolished FBP1-mediated IκBα dephosphorylation (Fig. 3f). Similarly, FBP1 depletion increased the phosphorylation of IκBα S32/36 in HCT116 or LoVo cells upon TNFα stimulation. Rescued expression of rFBP1 WT, but not that of rFBP1 N213K, abrogated the increased phosphorylation of IκBα S32/36 in FBP1-depleted cells (Fig. 3g; Supplementary information, Fig. S3c, d). The luciferase assays using NF-κB-Luc also confirmed that compared to that of rFBP1 WT, rescued expression of rFBP1 N213K enhanced TNFα-induced NF-κB activation (Fig. 3h). Collectively, these results indicate that FBP1 N213K mutation disrupts the interaction between FBP1 and IκBα and therefore abrogates FBP1-mediated IκBα dephosphorylation in tumor cells.
FBP1-catalyzed IκBα dephosphorylation sensitizes tumor cells to inflammation-associated cell death by downregulating the expression of prosurvival genes
It is well known that patients with inflammatory bowel disease are at higher risk of CRC. Molecular pathobiology of CRC implicates pro-inflammatory conditions promote the tumor malignant transformation, progression, invasion, and metastasis.14 Inflammation is driven by the accumulation of various immune and inflammatory cells and soluble inflammatory mediators, such as cytokines, chemokines, growth factors, lipid molecules, reactive oxygen, and nitrogen species. By means of direct contact or cytokine and chemokine production and acting in autocrine and paracrine manners, the diverse immune cells communicate with each other to control and shape tumor growth.
Cytokines produced by tumor-infiltrating immune cells activate key transcription factors, such as NF-κB or STAT3, in premalignant cells to control numerous pro-tumorigenic processes, including survival, proliferation, growth, angiogenesis, and invasion.15 Thus, we investigated how FBP1-mediated IκBα dephosphorylation influenced tumor cell survival in inflammatory microenvironment. We examined the viability of FBP1-depleted HCT116 or LoVo cells rescued with rFBP1 WT or N213K pretreated with TNFα (5 ng/mL), followed by the treatment of reactive oxygen species (ROS, 200 μM H2O2) and TNFα (25 ng/mL), often produced by inflammatory cells, and death ligand FasL (20 ng/mL), frequently adopted by cytotoxic lymphocytes to kill cancer cells.16 Compared to rescued expression of rFBP1 WT, that of rFBP1 N213K dramatically attenuated H2O2, TNFα or FasL-induced tumor cell death (Fig. 3i; Supplementary information, Fig. S3e). Of note, the proliferation of tumor cells was not influenced by rescued expression of rFBP1 N213K (Supplementary information, Fig. S3f).
Extensive evidence supports a role for NF-κB in regulation of anti-apoptotic gene expression and promotion of cell survival.17 To uncover the mechanism by which FBP1-mediated IκBα dephosphorylation promotes tumor cell death in inflammatory microenvironment, we examined the expression of well-established NF-κB-regulated prosurvival genes18 in FBP1-depleted HCT116 cells rescued with rFBP1 WT or N213K after TNFα treatment. Quantitative PCR (qPCR) analysis showed that among the 10 tested genes, the expression levels of BCL2, BIRC2, BIRC3, CFLAR, TRAF1, TRAF2, and XIAP, were upregulated in tumor cells rescued with rFBP1 N213K than in tumor cells rescued with rFBP1 WT after TNFα treatment (Fig. 3j). These results strongly suggest that FBP1 suppressed the expression of NF-κB-regulated prosurvival genes upon TNFα stimulation by dephosphorylating IκBα, thereby promoting tumor cell death under oxidative and inflammatory stresses.
FBP1-catalyzed IκBα dephosphorylation suppresses colorectal tumorigenesis by promoting inflammation-associated cell death and preventing MDSCs mobilization
Among the chemically induced CRC models, the combination of azoxymethane (AOM) with the exposure to the inflammatory agent dextran sodium sulfate (DSS) in rodents has been proven to dramatically shorten the latency time for induction of CRC and to rapidly recapitulate acute inflammation-promoted aberrant crypt foci-adenoma-carcinoma sequence that occurs in human CRC. To investigate the role of FBP1-mediated IκBα dephosphorylation in ulcerative colitis-associated CRC development, we generated knock-in (KI) mice harboring a point mutation of N213K in the mouse Fbp1 gene (Supplementary information, Fig. S4a, b). The observation of KI mice expressing Fbp1 N213K/N213K (Fbp1N213K/N213K mice) showed that Fbp1 N213K mutation did not influence the viability of embryo and the sex ratio of the mice (Data not shown). Fbp1+/+ or Fbp1N213K/N213K mice were treated with AOM and DSS to induce CRC. Ten weeks after AOM application, the mice were sacrificed. Examination of the colon morphology and hematoxylin and eosin (H&E) staining of dissected colon tissues showed that Fbp1N213K/N213K mice developed much more and bigger tumors than Fbp1+/+ mice (Fig. 4a–e). Moreover, Fbp1N213K/N213K mice exhibited much shorter survival duration than Fbp1+/+ mice during CRC progression (Fig. 4f).
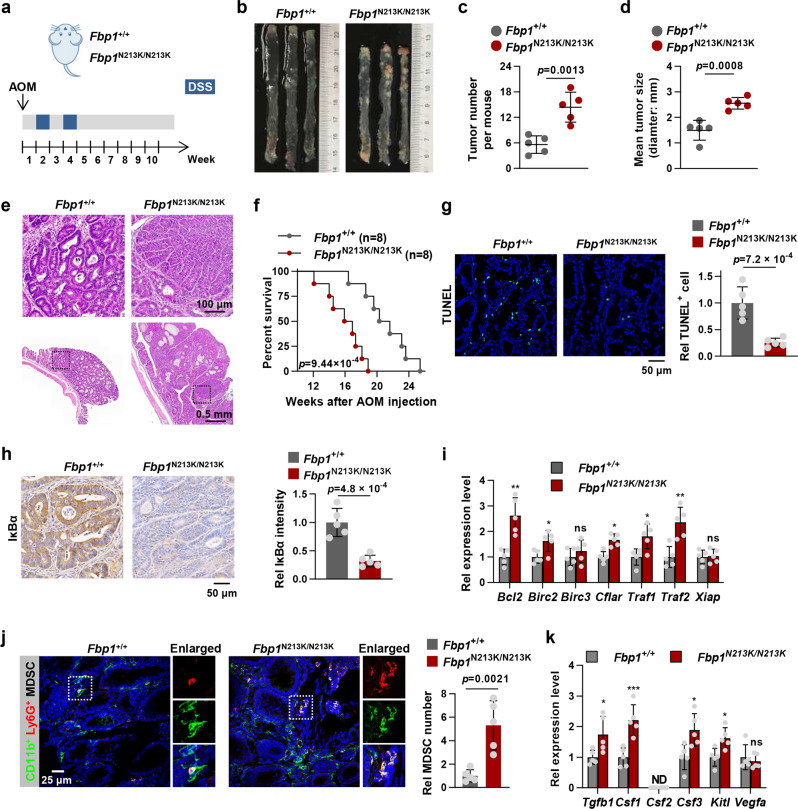
Fig. 4. FBP1-catalyzed IκBα dephosphorylation suppresses colorectal tumorigenesis by promoting inflammation-associated cell death and preventing MDSCs mobilization.
a A scheme of AOM/DSS-induced colon cancer model. b Representative images of colon tumors in Fbp1+/+ and Fbp1N213K/N213K mice at week 10 after injection of AOM. c Number of colon tumors in Fbp1+/+ and Fbp1N213K/N213K mice (n = 5). d Tumor size in Fbp1+/+ and Fbp1N213K/N213K mice (n = 5). e H&E staining of the intestinal sections with tumor from d. f Kaplan–Meier survival of Fbp1+/+ and Fbp1N213K/N213K mice (n = 8) that were induced with colon cancer (two-tailed log-rank test). g Representative images of TUNEL staining in tumor sections from Fbp1+/+ and Fbp1N213K/N213K mice (n = 5) were shown (left panel). The statistical analysis was shown on the right panel. h Representative images of IκBα staining in tumor sections from Fbp1+/+ and Fbp1N213K/N213K mice (n = 5) were shown (left panel). The statistical analysis was shown on the right panel. i The mRNA levels of indicated genes from Fbp1+/+ and Fbp1N213K/N213K intestinal tumor tissues (n = 5) were measured by qPCR analyses. Each symbol represents an individual mouse. j IF staining with anti-CD11b and anti-Ly6G antibodies was performed in AOM/DSS-induced tumor sections from Fbp1+/+ and Fbp1N213K/N213K mice. Representative images of CD11b and Ly6G staining were shown (left panel). The statistical analysis was shown on the right panel. k The mRNA levels of indicated genes from Fbp1+/+ and Fbp1N213K/N213K intestinal tumor tissues (n = 5) were measured by qPCR analyses. Each symbol represents an individual mouse. Data represent the mean ± s.d. of the indicated values obtained from five mice (c, d, g–k, two-tailed Student’s t-test).
We performed terminal deoxynucleotidyl transferase-mediated dUTP nick end labelling (TUNEL) assay in dissected CRC tumors, indicating that tumors from Fbp1N213K/N213K mice contained less apoptotic cells than the tumors from Fbp1+/+ mice (Fig. 4g). Immunohistochemistry (IHC) staining of IκBα (IκBα degradation representing NF-κB activation) in dissected CRC tumors indicated that tumors from Fbp1N213K/N213K mice had much stronger NF-κB activation than those from Fbp1+/+ mice (Fig. 4h). Consistently, qPCR analysis of canonical NF-κB-regulated prosurvival genes in dissected CRC tumors showed that most of genes, including Bcl2, Birc2, Cflar, Traf1 and Traf2 had higher expression in the tumors from Fbp1N213K/N213K mice than in the tumors from Fbp1+/+ mice (Fig. 4i). Collectively, these results suggest that FBP1-mediated IκBα dephosphorylation impairs ulcerative colitis-associated colorectal tumorigenesis by suppressing NF-κB-activated prosurvival genes and subsequent tumor cell survival.
In addition, as both canonical and noncanonical NF-κB pathways play important role in CRC development,19 we tested whether FBP1 also regulates the noncanonical NF-κB pathways in CRC tumors. In contrast to the activation of p50, RelA and c-REL by the canonical pathway, the non-canonical NF-κB pathway selectively activates p100-sequestered NF-κB members, predominantly p52 and RelB.20 Thus, we performed the IHC analysis of RelB in dissected CRC tumors and found that tumors from Fbp1N213K/N213K mice had the comparable levels of nuclear RelB to those from Fbp1+/+ mice (Supplementary information, Fig. S4c), suggesting that FBP1 does not regulate the noncanonical NF-κB pathway in vivo.
NF-κB transcription factor regulates expression of numerous components of the immune system, including innate immune response and the specific immune response.21 MDSCs are a diverse population of immature myeloid cells with immunosuppressive properties that accumulate under pathological conditions including specific types of cancer and infections.22 High levels of circulating MDSCs in cancer patients have been shown to correlate with clinical stage, metastatic burden, and resistance to both chemotherapy and immunotherapy.23 MDSCs mobilization is mediated by high levels of tumor- or niche-derived growth factors, such as granulocyte-macrophage colony-stimulating factor (GM-CSF) and granulocyte-colony stimulating factor (G-CSF), which promote the expansion of immature myeloid cells and inhibit their terminal differentiation, and the presence of pro-inflammatory cytokines, such as interferon γ (IFNγ) and IL1β, which promote the pathological activation of MDSCs.24 Thus, we examined the expression of a series of MDSCs mobilization-associated cytokines, including TGFB1, CSF1 (encoding M-CSF), CSF2 (encoding GM-CSF), CSF3 (encoding G-CSF), KITL and VEGFA,25 in FBP1-depleted HCT116 cells rescued with rFBP1 WT or N213K. qPCR analysis showed that the expression of CSF1 and CSF2 was upregulated in tumor cells rescued with rFBP1 WT after TNFα treatment and further enhanced in the cells rescued with rFBP1 N213K (Supplementary information, Fig. S4d).
Moreover, we examined MDSCs infiltration in the tumors dissected from Fbp1+/+ or Fbp1N213K/N213K mice by performing immunofluorescence (IF) staining of CD11b and Ly6G, the well-established markers for MDSCs, which showed that much more MDSCs were observed in Fbp1N213K/N213K tumors than in Fbp1+/+ tumors (Fig. 4j). In contrast, the infiltration of CD8+ tumor-infiltrating cytotoxic T lymphocytes (CTLs) was dramatically decreased in Fbp1N213K/N213K tumors (Supplementary information, Fig. S4e). Similarly, the abundance of NKp46-positive natural killer (NK) cells was also slightly decreased in Fbp1N213K/N213K tumors (Supplementary information, Fig. S4f). Consistently, the expression levels of Tgfb1, Csf1, Csf3 and Kitl were upregulated in the tumors from Fbp1N213K/N213K mice than those from Fbp1+/+ mice (Fig. 4k). These results suggest that disrupting Fbp1-mediated IκBα dephosphorylation recruits MDSCs in tumor microenvironment and promotes tumor immune evasion.
FBP1 expression inversely correlates with NF-κB activation, cell survival, and MDSCs infiltration in human CRC tumors
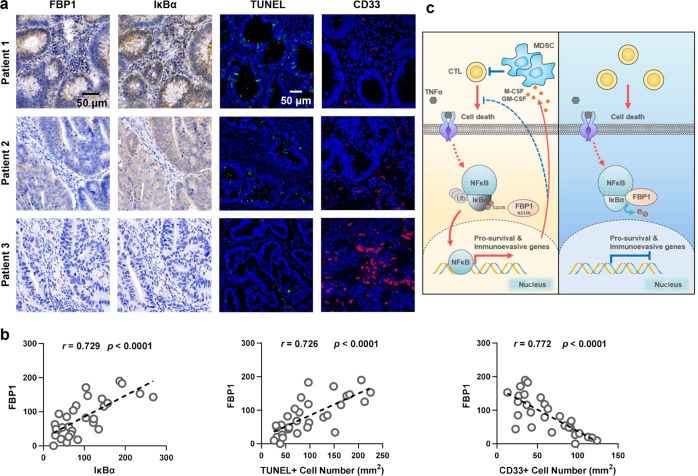
To determine the relationship between FBP1 protein levels, NF-κB activation, cell apoptosis and MDSCs infiltration in human colorectal adenocarcinoma, we performed IHC analysis of FBP1 and IκBα, IF staining of CD33 (a myeloid marker for MDSCs) and TUNEL assay in primary CRC tumors, which showed the strong correlation between FBP1 expression and NF-κB activation (indicated by IκBα degradation), FBP1 expression and the percentage of apoptotic cells, FBP1 expression and MDSCs abundance (Fig. 5a, b). The specificities of anti-FBP1 and anti-IκBα antibodies for IHC staining were validated by purified protein blocking assays (Supplementary information, Fig. S5a, b). These results indicate the clinical association among FBP1-regulated NF-κB activation, cell survival and MDSCs abundance in human CRC patients.
Fig. 5. FBP1 expression inversely correlates with NF-κB activation, cell survival, and MDSC infiltration in human CRC tumors.
a, b IHC staining with anti-FBP1 and anti-IκBα antibodies, IF staining with anti-CD33 antibody and TUNEL staining of 30 specimens from patients with CRC were performed. Representative images of IHC, IF and TUNEL staining of three specimens are shown (a). Semi-quantitative scoring (using a scale from 0 to 300) was carried out between FBP1 and IκBα. IHC staining scores and TUNEL+ cells or CD33+ cells of the CRC specimens were compared (b, Pearson’s correlation test). c Schematic model of the suppressive role of FBP1-mediated IκBα dephosphorylation in tumorigenesis. Upon TNFα stimulation, FBP1 interacts with and dephosphorylates IκBα, thereby inhibiting NF-κB signaling. Inactivated NF-κB suppresses tumorigenesis by sensitizing tumor cells to inflammatory stresses produced by CTLs and preventing the mobilization of MDSCs (Right panel). N213K mutation disrupts the interaction between FBP1 and IκBα, thereby restoring NF-κB activation and subsequent expression of prosurvival genes and MDSC-mobilizing genes, which promotes cell survival, MDSCs mobilization and tumorigenesis (Left panel).
Discussion
Emerging evidence demonstrates that many metabolic enzymes function as protein kinases to phosphorylate a variety of protein substrates, thereby regulating a set of fundamental cellular processes.1 However, in addition to the function as a protein kinase, whether a metabolic enzyme dephosphorylates proteins as a protein phosphatase remains unknown. To address the question, we performed a high-throughput virtual screening of potential protein phosphatase among 57 metabolic phosphatases by adopting molecular docking and MD simulations and identified FBP1 as a potential protein phosphatase. Further mechanistic investigation demonstrates that upon TNFα stimulation, FBP1 directly interacts and dephosphorylates IκBα to inhibit NF-κB signaling. Inactivated NF-κB in tumor cells accelerates inflammation-associated cell death and enhances antitumor immunity by dampening MDSCs mobilization, ultimately impeding colorectal tumorigenesis (Fig. 5c). These findings not only reveal the protein phosphatase activity of FBP1 and identify IκBα as its protein substrate, but also establish the suppressive role of FBP1-mediated IκBα dephosphorylation in CRC development.
FBP1 is the second rate-limiting enzyme in gluconeogenesis that catalyzes the hydrolysis of F-1,6-BP to F-6-P. FBP1 has been increasingly implicated as a tumor suppressor by antagonizing glycolysis through its cytosolic catalytic activity4,26 or directly interacting with hypoxia inducible factors (HIFs) in clear cell renal carcinoma, and inhibiting their transcriptional activity independent of its enzymatic properties.2 Additionally, a more recent study demonstrates that hepatocyte-specific loss of FBP1 promotes the activation and senescence of hepatic stellate cells and subsequent senescence-associated secretory phenotype development and tumor progression by releasing high mobility group protein B1.3 However, whether FBP1 can be a protein phosphatase remains undiscovered. In this study, we revealed FBP1 as a protein phosphatase for the first time and that IκBα pS32/36 is dephosphorylated directly by FBP1. Moreover, consistent with the findings of previous studies, FBP1-dependent IκBα dephosphorylation also plays a suppressive role in colorectal tumorigenesis. However, in contrast to the inhibition of glycolysis and HIF activity, FBP1 dephosphorylates IκBα and inhibits NF-κB signaling, thereby suppressing tumor cell survival in inflammatory microenvironment and preventing MDSCs mobilization. Taken together, our finding not only reveals the new molecular function of FBP1 as a protein phosphatase, but also uncovers a new mechanism of the tumor-suppressive activity of FBP1. In addition, we compared the hydrolysis reaction mechanism, substrate affinity, catalytic rate and efficiency of FBP1 to F-1,6-BP and IκBα pS32/36 and found that the hydrolysis reaction mechanisms of IκBα pS32/36 and F-1,6-BP catalyzed by FBP1 are similar, while the barriers in the pathway of F-1,6-BP with FBP1 are lower than those for IκBα pS32/36 with FBP1.
Notably, as a rate-limiting enzyme in gluconeogenesis, FBP1 plays an important role in cell metabolism. Just like most metabolic enzymes, the activity of FBP1 is influenced by metabolic signals of the cells. For example, FBP1 activity can be allosterically or competitively regulated by AMP and fructose-2,6-bisphosphate, respectively.27 Thus, a metabolic enzyme that moonlights as a protein phosphatase, such as FBP1, might possess the special capability to integrate the metabolic signals to the network of signal transduction in the cells, which definitely requires further exploration.
The canonical NF-κB signaling plays critical roles in controlling anti-apoptotic response, inflammation and immune response. Cell stimulation leads to phosphorylation of IκBα and subsequently rapid ubiquitination and degradation of IκBα through the 26S proteasome. Degradation of IκBα liberates NF-κB and allows its translocation to the nucleus, where it controls the expression of the target genes. Phosphorylation of IκBα is attained by the activated IKK complex, while dephosphorylation of IκBα is rarely reported.28 Tsuchiya et al. reported that protein phosphatase 2A subunit ABC trimer dephosphorylates IκBα but not IKKβ and RelA in cells.29 Wei et al. found that protein tyrosine phosphatase L1 inhibits NF-κB through dephosphorylating and stabilizing IκBα.30 Pons et al. reported that serine phosphatase calcineurin dephosphorylates IκBα.31 Naki et al. found that IκBα is a substrate of Fas-associated phosphatase-1.32 However, no rigorous in vitro dephosphorylation assays were performed in the above studies to provide the direct evidence for IκBα dephosphorylation. In contrast, we provide both in vitro and in vivo evidence to support that FBP1 directly interacts with and dephosphorylates of IκBα pS32/36. And such FBP1-mediated IκBα dephosphorylation inhibits subsequent NF-κB signaling and impairs colorectal tumorigenesis.
As both canonical and noncanonical NF-κB pathways play important role in CRC development,19 we tested whether FBP1 also regulates the noncanonical NF-κB pathways in CRC tumors. We revisited the mass spectrometry data of FBP1-associated protein dephosphorylation (Supplementary information, Table S4) and found that FBP1 did not affect the phosphorylation of p100, the central protein whose phosphorylation controls the activation of non-canonical NF-κB pathway, suggesting that FBP1 might not influence the noncanonical NF-κB pathway. To confirm the hypothesis, we performed IHC analysis of RelB in dissected CRC tumors and found that tumors from Fbp1N213K/N213K mice had similar levels of nuclear RelB with those from Fbp1+/+ mice (Supplementary information, Fig. S4c), suggesting that FBP1 may not regulate the noncanonical NF-κB pathway in the mouse CRC model.
Inflammatory microenvironment is an essential component of all tumors and plays decisive roles at different stages of tumor development, including initiation, promotion, malignant conversion, invasion, and metastasis. Tumor promotion is the process of tumor growth from a single initiated cell into a fully developed primary tumor. Initial tumor growth depends on increased cell proliferation and reduced cell death, both of which are stimulated by inflammation-driven mechanisms. In our study, we established a tumor-suppressive role of FBP1 in initial tumor growth of CRC by sensitizing tumor cells to inflammatory stress-induced cell death. Mechanistically, FBP1 interacts with and dephosphorylates IκBα upon TNFα stimulation, which prevents IκBα degradation and attenuates subsequent NF-κB activation. Inactivated NF-κB in the initiated malignant cells decreases their survival in inflammatory microenvironment by downregulating cell survival genes. Meanwhile, the expression levels of Tgfb1, Csf1, Csf3, and Kitl also decline in NF-κB-inactivated cells, which promotes the infiltration of CTLs and antitumor immunity by preventing MDSCs mobilization. Both mechanisms are involved in the tumor-suppressive activity of FBP1.
Materials and methods
Materials
Antibodies
Primary antibodies were used against: Flag (F3165, Sigma-Aldrich), HA (3724, Cell Signaling Technology), His-tag (66005-1, Proteintech), FBP1 (HPA005857, Sigma-Aldrich), FBP1 (PA5-76734, Thermo Fisher Scientific), Tubulin (sc-5286, Santa Cruz Biotechnology), β-actin (60008-1-lg, Proteintech), IκBα (ab32518, abcam), RelA (6956 and 8242, Cell Signaling Technology), IκBα S32/36 phosphorylation (9246, Cell Signaling Technology), IKKβ S177/181 phosphorylation (2697, Cell Signaling Technology), Lamin B1 (13435, Cell Signaling Technology), CD8a (98941, Cell Signaling Technology), NKp46 (PA5-102860, Thermo Fisher Scientific), CD11b (ab133357, abcam), Ly-6G/Ly-6C (Gr-1) (MAB1037, R&D Systems), CD33 (PA5-120758, Thermo Fisher Scientific), RelB (sc-226, Santa Cruz Biotechnology). The following secondary antibodies were used: goat anti-mouse IgG H&L (HRP) (ab6789, abcam), goat anti-rabbit IgG H&L (HRP) (ab6721, abcam), goat anti-rabbit IgG (H + L), Alexa Fluor 568 (A-11011, Thermo Fisher Scientific), goat anti-rat IgG (H + L), Alexa Fluor 568 (A-11077, Thermo Fisher Scientific), donkey anti-rabbit IgG (H + L), Alexa Fluor 488 (A-21206, Thermo Fisher Scientific). All the antibodies were used following the manufacturer’s recommended protocols.
Reagents
Anti-Flag M2 affinity gel (A2220, Sigma-Aldrich), Protein A/G PLUS-Agarose (sc-2003, Santa Cruz Biotechnology). Recombinant human TNFα (210-TA-020) and recombinant human Fas Ligand (126-FL-010) were bought from R&D Systems. The DeadEnd Fluorometric TUNEL System (G3250) and the dual luciferase reporter assay system were bought from Promega. Puromycin and hygromycin were bought from EMD Biosciences. Two phosphorylated IκBα peptides (#1, HDS(pho)GLDS(pho)M; #2, DS(pho)GLDS(pho)MKD) used for in vitro dephosphorylation assay were purchased from GenScript.
DNA constructs and mutagenesis
PCR-amplified human Fbp1, RelA, and IκBα were cloned into pCDH-Flag, pcDNA3.0-HA, pFlag-CMV, pLVX, or pCold I-His. Fbp1 mutations were constructed using the QuikChange site-directed mutagenesis kit (Stratagene). The pGIPZ shNT was generated with the control oligonucleotide 5′-GCTTCTAACACCGGAGGTCTT-3′. pGIPZ human FBP1 shRNAs was generated with 5′-ATCTTCACATCCTTGTCCA-3′ oligonucleotide.
Cell culture and transfection
293T, HCT116, LoVo, HCoEpiC, HCT-8, HT29, RKO, SW480, SW620 and Caco-2 cells were obtained from the Type Culture Collection of the Chinese Academy of Sciences. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) or Roswell Park Memorial Institute 1640 medium, and the culture medium was supplemented with 10% fetal bovine serum (FBS, Gibco, Thermo Fisher Scientific) and 1% antibiotics (Gibco, Thermo Fisher Scientific). All cell lines were authenticated using the short tandem repeat method and were tested negative for mycoplasma. Cells were seeded in 35 mm, 60 mm or 100 mm plates and transfected with the indicated plasmids using PolyJet according to the manufacturer’s instructions.
Purification of recombinant proteins
His-FBP1 (WT or mutants) was expressed in bacteria and purified. Briefly, the vectors expressing His-FBP1 (WT or mutants) were used to transform BL21/DE3 bacteria. Then 0.5 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) was used to induce protein expression at 16 °C for 20 h. Cell pellets were collected and sonicated in PBS with the addition of proteasome inhibitors before centrifugation at 12,000× g for 30 min at 4 °C. Cleared lysates were then bound to Ni-NTA resin (GenScript) with rolling at 4 °C for 4 h. Beads were washed extensively before eluting for 1 h in His elution buffer (PBS, pH 7.4, plus 500 mM imidazole). Eluted proteins were then dialyzed against PBS.
Immunoprecipitation and immunoblotting analysis
Extraction of proteins from cultured cells with a modified buffer was followed by immunoblotting or immunoprecipitation with corresponding antibodies, as described previously.33
Mass spectrometry analysis
Flag-FBP1 was immunoprecipitated from HCT116 cells. The precipitated complexes were boiled at 95 °C for 10 min for denaturation. Flag-FBP1-associated proteins were separated from the complexes using SDS-PAGE. The samples were processed as described before.33 The peptides were then analyzed by liquid chromatography-tandem mass spectrometry on a Q Exactive mass spectrometer (Thermo Fisher Scientific). Proteins were identified by a database search of the fragment spectra against the National SwissProt protein database (EBI) using Mascot Server 2.4 (Matrix Science).
For phosphopeptide analyses, phosphopeptide matches were analyzed using MaxQuant v.1.5.2.8 implemented in Proteome Discoverer and manually curated.34 For phosphoproteomic analysis, the enriched phosphopeptides were processed and analyzed as described before.35 The sequence motif enrichment analysis was performed using the motifeR software.36
Subcellular fractionation assay
HCT116 or LoVo cells were harvested and washed three times with cold PBS. Cytoplasmic and nuclear fractions were isolated using the Nuclear Extract Kit (ab113474, abcam) according to the manufacturer’s instructions.
Luciferase reporter assay
HCT116 cells were co-transfected with pGL3-NF-κB reporter and control Renilla plasmids. One day after transfection, cells were treated with or without 10 ng/mL TNFα for 3 h and harvested for the measurement of luciferase activity. Luciferase activities were measured using a Dual Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s instructions.
Real-time qPCR analysis
Total RNA was extracted with Trizol reagent (Life Technologies). 1 μg total RNA was used for complementary DNA (cDNA) synthesis by a HiScript II Q RT SuperMix for qPCR Kit (Vazyme), according to the manufacturer’s instructions. Quantitative real-time PCR analysis was performed using a Roche LightCycler96. Data were normalized to expression of a control gene (GAPDH) for each experiment.
Isothermal titration calorimetry (ITC) assay
Thermodynamic parameters of the binding of FBP1 WT or N213K to F-1,6-BP were measured by the ITC assays using the ITC200 Micro-calorimeter (MicroCal) at 25 °C. In all experiments, the initial injection of 0.5 mL of F-1,6-BP solution was discarded to eliminate the effect of titrant diffusion across the syringe tip during the equilibration process, and each dataset consisted of 20 injections of 2 μL each of 1 mM F-1,6-BP into the sample cell containing 250 μL of 0.05 mM FBP1. The heat of dilution was negligible in all cases. The binding constant and other thermodynamic parameters were determined by fitting the integrated titration data using the single binding site model by the nonlinear least-squares method implemented in MicroCal Origin software (version 7.0).
Molecular docking
Totally 57 human phosphatase structures, in which 28 phosphatase (PDB code 1D5R for PTEN, 1EW2 for ALPP, 1FIT for FHIT, 1IMD for IMPA1, 1L8L for PSPH, 1YNS for ENOPH1, 2CAR for ITPA, 2CZI for IMPA2, 2I3B for NTPCR, 2OYC for PDXP, 2VH7 for ACYP1, 2W4M for NANP, 2X4D for LHPP, 2XSW for INPP5E, 3ARA for DUT, 3DCY for TIGAR, 3IFC for FBP2, 3TVL for THTPA, 4CMN for OCRL, 4LQY for ENPP4, 4ZG9 for ENPP2, 5OKM for INPPL1, 5RWA for INPP5D, 5TCD for ENPP7, 5ZWK for FBP1, 6C01 for ENPP3, 6C45 for PPA1, 6WEU for ENPP1) were taken from the Protein Date Bank and the others (ADPRM, ACYP2, ALPI, ALPL, DCTP1, DOLPP1, ENPP6, G6PC1, G6PC2, G6PC3, IMPAD1, INPP1, INPP4A, INPP4B, INPP5A, INPP5B, INPP5J, INPP5K, MINPP1, PGP, PHOSPHO1, PHOSPHO2, PLPP1, PLPP2, PLPP3, PPA2, PRUNE, SGPP1, SGPP2) were predicted by Alfafold2,37 were prepared as the receptor. A 30*30*30 Å box was set, the center of which was next to the Active site shown in Uniprot webserver (https://www.uniprot.org/). Then the dipeptide models of pSer, pThr, pTyr were used as the ligands. The three dipeptide models were assessed by the virtual screening with the 57 human phosphatase structures by Autodock Vina.38 The average Vina scores of the lowest binding free energies for each ligand were re-ranked to obtain the phosphatase for further MD simulation.
MD program
The conformation of pSer, pThr, pTyr dipeptide models with FBP1, INPP5E, and ENOPH1 was taken from molecular docking structures based on the Vina scores. The conformation of IκBα L25–M37 was predicted by I-TASSER method.39 The dimer conformation of FBP1 (PDB code 5ZWK) and the structure of IκBα were used to build the initial complex of the FBP1 and IκBα. The ZDOCK 3.0.240 was employed to construct the complex of FBP1 and IκBα. Seven different initial conformations of FBP1 and IκBα pS32/36 were chosen to perform the molecular simulation. The parameters were obtained from the parameters reported by Homeyer et al.41 for pSer, pThr, and pTyr.
The atomistic MD simulations of the initial model were carried out in the AMBER16 program42 using AMBER14SB force field.43 Each system was neutralized with a number of neutralized ions and then was immersed in a solvent box filled with TIP3P water molecules.44 The distance between the surface of each FBP1 with ligand model and the edge of water box was required, and it was set more than 10 Å. Energy minimization was performed by imposing a strong restraint on each system and was followed by minimizing the whole system for a few thousand steps. NVT simulations were carried out by heating the whole system slowly from 100 K to 300 K, and the Berendsen thermostat45 was applied to govern the temperature of the whole system. Subsequently, a NVT dynamics of 1 ns was performed and was followed by a 1 μs NPT production run. During the NPT production run, all bonds associated with hydrogen atoms were constrained by employing the SHAKE algorithm46 as the integration time step of 4 fs could be used. A cutoff value of 12 Å was set for non-bonded interactions. The Particle Mesh Ewald method47 was employed for treating electrostatic interactions. For each model, five independent MD simulations were implemented using different velocities that were randomly generated at the beginning of the simulations. The analysis of each MD trajectory was performed the cpptraj module in AMBER 16.48 To understand the binding mode between ligands with FBP1, the binding free energies were calculated via the MM-GBSA method. For each complex, 500 snapshots were extracted from the last 50 ns of the MD trajectory at an interval of 100 ps.
All geometries and energies were computed using the Semi-empirical method with AM1 as implemented in the Gaussian09 program package.49 FBP1 catalytic reaction center and the substrate were truncated from the whole model, which contain two Mg2+, E97, E89, D118, L120, D121, E280, the pS32 or F-1,6-BP, and one hydroxide ion coordinated with Mg2+. The hydroxide ionʼs ionization is accomplished with the help of the Mg2+. Hessians were calculated to confirm the nature of the stationary points, and were also used for evaluation of zero-point vibrational effects. The energies reported in this study include both salvation and zero-point effects. In the geometry optimizations, certain truncation atoms were kept frozen to the position of the complex. This approach was used to keep the various groups in place to resemble the simulation results as much as possible.
Measurement of FBP1 activity and enzyme kinetics
The phosphatase activity of FBP1 (WT and N213K) against F-1,6-BP was measured spectrophotometrically using the Malachite Green reagent.50 The reactions were conducted using a mixed buffer system that consisted of 50 mM each of sodium acetate, MES, PIPES, HEPES, and TAPS and performed as described before.51 Briefly, the reaction mixture (150 μL) contained 50 mM mixed buffer, 0.5 mM substrate, and 200 ng of purified FBP1 protein. Following a 15 min incubation at 30 °C, the reaction was terminated by the addition of 40 μL of Malachite green reagent and the amount of released inorganic phosphate (Pi) was calculated based on absorbance at 630 nm. To determine Km and Kcat values, the initial rates were fitted to the Michaelis-Menten equation using GraphPad Prism Software (version 8.04). The protein phosphatase activity of FBP1 (WT and N213K) against immunoprecipitated IκBα pS32/36 was determined following a previous protocol.52 Initial phosphate hydrolysis was assessed using the Biomol Green kit (BML-AK111, Enzo Life Sciences) to detect total phosphate release, following the manufacturer’s protocol. The 200 μL assay mixture, containing 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, various concentrations of immunoprecipitated IκBα pS32/36 and 500 ng of purified FBP1 protein, was incubated for 30 min at 30 °C. The absorbance was noted at 620 nm using a visible wave spectrophotometer. To determine Km and Kcat values, the initial rates were fitted to the Michaelis-Menten equation using GraphPad Prism Software (version 8.04).
Cell viability assay
HCT116 or LoVo cells with or without genetic modifications were plated in DMEM (10% FBS). Cell viability was determined by trypan blue staining.
Cell proliferation assay
One thousand cells per well were plated in 96-well plates and cultured for indicated time. Cell density was determined by a Sulforhodamine B (SRB) colorimetric assay.53 In brief, cells were fixed with trichloroacetic acid and stained with SRB solution, followed by rinsing of the plates four times with 1% acetic acid and solubilization of the protein-bound dye with 10 mM Tris base solution. The absorbance was measured at 560 nm.
Generation of Fbp1N213K mice
Fbp1N213K KI mutant mice were generated using the CRISPR-Cas9 system from WT C57BL/6 background. The small guide RNA (sgRNA), donor DNA and Cas9 protein were mixed and injected into two blastomeres of 2-cell embryos. Then edited embryos were transferred into oviducts of pseudopregnant ICR females. The sgRNA sequence targeting Fbp1 is 5′-CCTTGGCATAACCCTCATTA-3′; the donor sequence for homology-directed repair is 5′-ATTATGGTGGACAGGGACGTGAAGATGAAGAAGAAAGGTAACATCTACAGTCTTAAGGAGGGTTATGCCAAGGACTTTGACCCTGCCATCAATGAGTATCTCCAGAG-3′.
AOM/DSS-induced colon cancer model
The Fbp1+/+and Fbp1N213K mouse lines in a C57BL/6 background were used and were maintained in a specific-pathogen-free facility. Mice were group-housed (4–5 mice per cage) except for < 5% of mice who were single-housed later because of death of cagemates. All mice were maintained under a 12 h light/dark cycle with free access to water and standard mouse chow. Littermates of the same sex were randomly assigned to experimental groups. The use of mice was following ethical regulations and was approved by the institutional review board at the Shanghai Institute of Biochemistry and Cell Biology. Six-week-old mice were used for AOM/DSS treatment. Mice were intraperitoneally injected with 10 mg per kg AOM (Sigma-Aldrich), followed by treatment of 2.5% DSS (Sigma-Aldrich) in drinking water for 7 d and in regular water for another 7 d. This cycle was repeated twice and mice were sacrificed 2–6 weeks after the end of the last DSS cycle. Colons were collected, washed and slit open longitudinally for further analyses.
H&E staining and IHC
Animals were sacrificed and the colons were dissected and fixed in 4% paraformaldehyde (PFA). Paraffin-embedded colon tissues were systematically sectioned with 5 μm thickness. H&E staining or IHC analyses were performed in these colon sections. The tissue sections from paraffin-embedded human CRC were stained with antibodies as indicated. The quantification and analysis of IHC signal intensity were described before.33 First, we quantitatively scored the tissue sections according to the percentage of positive cells and staining intensity. Then, the intensity of staining was rated on a scale of 0–3: 0, negative; 1, weak; 2, moderate; 3, strong. We assigned the following proportion scores: X indicates the percentage of the tumor cells that were stained (0 ≤ [X1 + X2 + X3] ≤ 100) where X3 indicates strong staining, X2 moderate staining and X1 weak staining. The score (H-score) was obtained using the formula: 1 × X1 + 2 × X2 + 3 × X3 giving a range of 0–300. The use of human CRC specimens was approved by the institutional review board at Shanghai Changhai Hospital (No. CHEC2021-026) and Shanghai Zhongshan Hospital (No. B2021-692R) and complied with all relevant ethical regulations. Informed consents were obtained from all patients.
IF staining
Animals were sacrificed and the colons were dissected, fixed in 4% PFA and embedded in optimum cutting temperature compound (Thermo Fisher Scientific) after dehydration by 30% sucrose solution. Tissues were sectioned with 10 μm thickness. The tissue sections were washed in PBS, treated with 0.2% Triton X-100 and blocked in PBS with 5% BSA followed by incubation with indicated antibodies. After washing, samples were incubated with corresponding secondary antibodies and then DAPI. Ten representative fields per tissue section crossing the whole tissue region were randomly selected and the confocal images were obtained using an Olympus FV3000 confocal laser scanning microscope. An average of the number of positive staining cells per mm2 was used to represent each case.
Statistical analysis
All statistics were performed using GraphPad Prism software (v.8.0.4). Specific statistical tests for each experiment are described in the Figure legends. *P < 0.05, **P < 0.01 and ***P < 0.001 was considered to be significant.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (92253305, 92053203 and 32025013) to W.Y.; the National Key R&D Program of China (2022YFA0806201 and 2019YFA0802000) to W.Y.; CAS Project for Young Scientists in Basic Research (YSBR-014) to W.Y.; Program of Shanghai Academic/Technology Research Leader (20XD1424400) to W.Y.; the Innovative Research Team of High-level Local Universities in Shanghai (SHSMU-ZLCX20212302) to W.Y.; the Strategic Priority Research Program of Chinese Academy of Sciences (XDB 37000000) to G.L.; the National Natural Science Foundation of China (21933010) to G.L.; the National Natural Science Foundation of China (21907094) to H.C.; the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2022265) to Y.Zhang; Shanghai Science and Technology Development Funds (22QA1409900) to Y.Zhang. We gratefully acknowledge the support of the Sanofi Scholarship Program. We also thank all the core facilities of Shanghai Institute of Biochemistry and Cell Biology for technical support.
Author contributions
W.Y. conceived and designed the study. W.Z., X.W., Y.Zhang, H.Y., and H.Z. performed the experiments. G.L. designed the molecular dynamics strategy. G.L., H.C., and Y.L. performed the simulations and data analysis. T.L. and Q.L. provided reagents and pathological assistance. H.G. assisted in reviewing the paper. Y.Zhao provided constructive suggestions. W.Y. and G.L. wrote the manuscript with comments from all authors.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Wencheng Zhu, Huiying Chu, Yajuan Zhang, Tianhang Luo, Hua Yu.
Contributor Information
Quanlin Li, Email: li.quanlin@zs-hospital.sh.cn.
Xiongjun Wang, Email: wangxiongjun@gzhu.edu.cn.
Guohui Li, Email: ghli@dicp.ac.cn.
Weiwei Yang, Email: wyang@sibcb.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-022-00773-0.
References
- 1.Lu Z, Hunter T. Metabolic kinases moonlighting as protein kinases. Trends Biochem. Sci. 2018;43:301–310. doi: 10.1016/j.tibs.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li B, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li F, et al. FBP1 loss disrupts liver metabolism and promotes tumorigenesis through a hepatic stellate cell senescence secretome. Nat. Cell Biol. 2020;22:728–739. doi: 10.1038/s41556-020-0511-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong C, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 6.Feagins LA, Souza RF, Spechler SJ. Carcinogenesis in IBD: potential targets for the prevention of colorectal cancer. Nat. Rev. Gastroenterol. Hepatol. 2009;6:297–305. doi: 10.1038/nrgastro.2009.44. [DOI] [PubMed] [Google Scholar]
- 7.Janakiram NB, Rao CV. The role of inflammation in colon cancer. Adv. Exp. Med. Biol. 2014;816:25–52. doi: 10.1007/978-3-0348-0837-8_2. [DOI] [PubMed] [Google Scholar]
- 8.Baker KJ, Houston A, Brint E. IL-1 Family members in cancer; two sides to every story. Front. Immunol. 2019;10:1197. doi: 10.3389/fimmu.2019.01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taniguchi K, Karin M. NF-kappaB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol. 2018;18:309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 10.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 11.Hayden MS, Ghosh S. Regulation of NF-kappaB by TNF family cytokines. Semin. Immunol. 2014;26:253–266. doi: 10.1016/j.smim.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS ONE. 2011;6:e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long AG, Lundsmith ET, Hamilton KE. Inflammation and colorectal cancer. Curr. Colorectal. Cancer Rep. 2017;13:341–351. doi: 10.1007/s11888-017-0373-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chavez-Galan L, Arenas-Del Angel MC, Zenteno E, Chavez R, Lascurain R. Cell death mechanisms induced by cytotoxic lymphocytes. Cell. Mol. Immunol. 2009;6:15–25. doi: 10.1038/cmi.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rath PC, Aggarwal BB. TNF-induced signaling in apoptosis. J. Clin. Immunol. 1999;19:350–364. doi: 10.1023/A:1020546615229. [DOI] [PubMed] [Google Scholar]
- 18.Van Antwerp DJ, Martin SJ, Verma IM, Green DR. Inhibition of TNF-induced apoptosis by NF-kappa B. Trends Cell Biol. 1998;8:107–111. doi: 10.1016/S0962-8924(97)01215-4. [DOI] [PubMed] [Google Scholar]
- 19.Patel M, Horgan PG, McMillan DC, Edwards J. NF-kappaB pathways in the development and progression of colorectal cancer. Transl. Res. 2018;197:43–56. doi: 10.1016/j.trsl.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Sun SC. The non-canonical NF-kappaB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017;17:545–558. doi: 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 22.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J. Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tavazoie MF, et al. LXR/ApoE activation restricts innate immune suppression in cancer. Cell. 2018;172:825–840.e18. doi: 10.1016/j.cell.2017.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37:208–220. doi: 10.1016/j.it.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li K, et al. Myeloid-derived suppressor cells as immunosuppressive regulators and therapeutic targets in cancer. Signal. Transduct. Target. Ther. 2021;6:362. doi: 10.1038/s41392-021-00670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirata H, et al. Decreased expression of fructose-1,6-bisphosphatase associates with glucose metabolism and tumor progression in hepatocellular carcinoma. Cancer Res. 2016;76:3265–3276. doi: 10.1158/0008-5472.CAN-15-2601. [DOI] [PubMed] [Google Scholar]
- 27.Hunter RW, et al. Metformin reduces liver glucose production by inhibition of fructose-1-6-bisphosphatase. Nat. Med. 2018;24:1395–1406. doi: 10.1038/s41591-018-0159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun W, et al. PPM1A and PPM1B act as IKKbeta phosphatases to terminate TNFalpha-induced IKKbeta-NF-kappaB activation. Cell. Signal. 2009;21:95–102. doi: 10.1016/j.cellsig.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuchiya Y, et al. Distinct B subunits of PP2A regulate the NF-kappaB signalling pathway through dephosphorylation of IKKbeta, IkappaBalpha and RelA. FEBS Lett. 2017;591:4083–4094. doi: 10.1002/1873-3468.12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei XM, et al. Protein tyrosine phosphatase L1 represses endothelial-mesenchymal transition by inhibiting IL-1beta/NF-kappaB/Snail signaling. Acta Pharmacol. Sin. 2020;41:1102–1110. doi: 10.1038/s41401-020-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pons S, Torres-Aleman I. Insulin-like growth factor-I stimulates dephosphorylation of ikappa B through the serine phosphatase calcineurin (protein phosphatase 2B) J. Biol. Chem. 2000;275:38620–38625. doi: 10.1074/jbc.M004531200. [DOI] [PubMed] [Google Scholar]
- 32.Nakai Y, Irie S, Sato TA. Identification of IkappaBalpha as a substrate of Fas-associated phosphatase-1. Eur. J. Biochem. 2000;267:7170–7175. doi: 10.1046/j.1432-1327.2000.01818.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, et al. UDP-glucose accelerates SNAI1 mRNA decay and impairs lung cancer metastasis. Nature. 2019;571:127–131. doi: 10.1038/s41586-019-1340-y. [DOI] [PubMed] [Google Scholar]
- 34.Wu YB, et al. Concurrent quantification of proteome and phosphoproteome to reveal system-wide association of protein phosphorylation and gene expression. Mol. Cell. Proteom. 2009;8:2809–2826. doi: 10.1074/mcp.M900293-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Q, et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179:561–577.e22. doi: 10.1016/j.cell.2019.08.052. [DOI] [PubMed] [Google Scholar]
- 36.Wang S, et al. motifeR: An integrated web software for identification and visualization of protein posttranslational modification motifs. Proteomics. 2019;19:e1900245. doi: 10.1002/pmic.201900245. [DOI] [PubMed] [Google Scholar]
- 37.Jumper J, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Compu. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan Y, et al. Ultrafast tracking of a single live virion during the invagination of a cell membrane. Small. 2015;11:2782–2788. doi: 10.1002/smll.201403491. [DOI] [PubMed] [Google Scholar]
- 40.Stratakis CA, Carney JA. The triad of paragangliomas, gastric stromal tumours and pulmonary chondromas (Carney triad), and the dyad of paragangliomas and gastric stromal sarcomas (Carney–Stratakis syndrome): molecular genetics and clinical implications. J. Intern. Med. 2009;266:43–52. doi: 10.1111/j.1365-2796.2009.02110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Homeyer N, Horn AH, Lanig H, Sticht H. AMBER force-field parameters for phosphorylated amino acids in different protonation states: phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J. Mol. Model. 2006;12:281–289. doi: 10.1007/s00894-005-0028-4. [DOI] [PubMed] [Google Scholar]
- 42.Case, D. et al. Amber 2016. San Francisco: University of California, 2016.
- 43.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for stimulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 45.Berendsen HJC, Postma JPM, Gunsteren WFV, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 46.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of cartesian equatios of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 47.Darden T, York D, Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 48.Roe DR, Cheatham TE. III PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 49.Liu P-S, et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 2017;18:985–994. doi: 10.1038/ni.3796. [DOI] [PubMed] [Google Scholar]
- 50.Proudfoot M, et al. High throughput screening of purified proteins for enzymatic activity. Methods Mol. Biol. 2008;426:331–341. doi: 10.1007/978-1-60327-058-8_21. [DOI] [PubMed] [Google Scholar]
- 51.Kuznetsova E, et al. Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae. J. Biol. Chem. 2010;285:21049–21059. doi: 10.1074/jbc.M110.118315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Debnath S, et al. A trapped human PPM1A-phosphopeptide complex reveals structural features critical for regulation of PPM protein phosphatase activity. J. Biol. Chem. 2018;293:7993–8008. doi: 10.1074/jbc.RA117.001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.