Abstract
Background
At present, the first-line treatment for advanced intrahepatic cholangiocarcinoma (ICC) is gemcitabine combined with cisplatin, but a considerable portion of ICC patients exhibit resistance to gemcitabine. Therefore, finding sensitisers for gemcitabine chemotherapy in ICC patients and predicting molecular markers for chemotherapy efficacy have become urgent needs.
Methods
In this study, PDX models were established to conduct gemcitabine susceptibility tests. The selected PDX tissues of the chemotherapy-sensitive group and drug-resistant group were subjected to transcriptome sequencing and protein chip technology to identify the key genes. Sixty-one ICC patients treated with gemcitabine chemotherapy were recruited for clinical follow-up validation.
Results
We found that thrombospondin-1 (TSP1) can predict gemcitabine chemosensitivity in ICC patients. The expression level of TSP1 could reflect the sensitivity of ICC patients to gemcitabine chemotherapy. Functional experiments further confirmed that TSP1 can increase the efficacy of gemcitabine chemotherapy for ICC. A mechanism study showed that TSP1 may affect the intake of oleic acid by binding to the CD36 receptor.
Conclusions
In summary, we found a key molecule—TSP1—that can predict and improve the sensitivity of ICC patients to gemcitabine chemotherapy, which is of great significance for the treatment of advanced cholangiocarcinoma.
Subject terms: Translational research, Predictive markers, Bile duct cancer
Background
The incidence of intrahepatic cholangiocarcinoma (ICC) has gradually increased in recent years, and this disease is characterised by insidious onset, rapid progression and a poor prognosis [1]. Surgical treatment is the best choice for patients with ICC, but only 30–40% of patients are suitable for radical surgery, and the recurrence rate is ~40–80% [2]. Systemic chemotherapy is still the preferred treatment for unresectable ICC and recurrent ICC, and it is also the basis of combined treatment. At present, the first-line treatment of patients with advanced ICCA is the combination of gemcitabine (GEM) and cisplatin, but the curative effect of this treatment is poor, and the total survival time of patients is only slightly improved [3]. The main reason for the poor efficacy of this drug is the problem of tumour resistance. A considerable number of ICC patients have a poor response to GEM chemotherapy in the clinic [4]. Some ICC patients may benefit from the targeted therapy and immunotherapy, but there is still no clear definition about the characteristics of potential beneficiaries. According to Response Evaluation Criteria In Solid Tumours, the evaluation of chemotherapy effect mainly depends on imaging examination. However, excessive imaging examinations increase physiological and economic burden on the patients, and cannot predict the curative effect in advance, which leads to delays in treatment plan adjustment. Therefore, it is urgent to find sensitisers and molecular markers for efficacy prediction to improve the chemotherapy effect of ICC.
Gemcitabine is a deoxycytidine nucleoside analogue that is transported into cells by nucleoside transporters, and then phosphorylated by deoxycytidine kinase to interfere with DNA synthesis, inhibit ribonucleotide reductase and block the cell cycle at the G1/S-phase transition, thereby playing a role in killing tumour cells [5]. The specific mechanisms of drug resistance in cancer patients include: nucleoside transporter- and nucleoside enzyme expression- mediated drug resistance [6]; the occurrence of epithelial-mesenchymal transition of tumour cells [7]; drug resistance caused by abnormal tumour energy metabolism, such as abnormal changes in glucose metabolism or lipid metabolism, which can affect the occurrence and development of cancer [8]; and noncoding RNA-mediated chemoresistance mechanisms [9]. Among patients with advanced cholangiocarcinoma, GEM resistance is an important factor affecting the survival of patients, and there is still a lack of effective intervention measures. Therefore, screening the markers that can be used to predict the GEM sensitivity of ICC patients and the potential targets of its drug resistance are crucial to improving the prognosis of advanced ICC patients.
Patient-derived tumour xenograft (PDX) models have been increasingly used in the research of various cancers. As preclinical models, they can truly simulate the situation in vivo, and are widely used in the research and development of tumour therapeutic drugs and the mechanism of drug resistance. PDX models capture the unique genomic and molecular characteristics of parental tumours, and enable the analysis of the specific clinical response of patients [10]. There have been a large number of studies involving different types of cancer in which the efficacy of drugs and chemotherapy resistance were studied by establishing PDX models [11–13].
TSP1 is a homotrimeric glycoprotein, and each TSP1 monomer is composed of n- and C-terminal globular domains and fine linkers. It is a multifunctional extracellular matrix protein that is generally secreted outside the cell to bind with related receptors to play a functional role [14, 15]. TSP1 not only plays an important role in normal cells but also shows important biological functions in different types of tumour cells. In hepatocellular carcinoma (HCC), TSP1 can act as an inhibitor of angiogenesis and inhibit the function of VEGF [16]. In addition, TSP1 mimics can inhibit angiogenesis in breast cancer by affecting VEGF [17]. TSP1 also inhibits invasion, migration and proliferation in multiple tumours [18–20]. However, the role of TSP1 in ICC is still unknown and worth exploring.
In this study, we established PDX models to carry out a GEM susceptibility tests. The selected PDX tissues of the chemotherapy-sensitive group and the drug-resistant group were subjected to transcriptome sequencing and protein chip technology and identifiedthrombospondin-1 (TSP1) as a gene that can predict the GEM chemosensitivity of ICC patients. At the clinical level, we verified whether the expression level of TSP1 can reflect the sensitivity of ICC patients to GEM chemotherapy. In vivo and in vitro functional experiments further confirmed that TSP1 can increase the sensitivity of cholangiocarcinoma to GEM chemotherapy. In the mechanistic study, we found that TSP1 may affect the intake of oleic acid (OA) by binding to the CD36 receptor on the surface of cholangiocarcinoma cells, reduce the levels of ROS and ATP in tumour cells, and further affect the chemotherapy effect of GEM. In short, we found a key molecule (TSP1) that can predict and improve the sensitivity of ICC patients to GEM chemotherapy, which is of great significance for the treatment of advanced cholangiocarcinoma.
Methods
Patients and tissue samples
All the samples were obtained from the Third Affiliated Hospital of Navy Medical University. The sample collection of all patients and healthy people was approved by the Ethics Committee of the Third Affiliated Hospital of Navy Medical University. Patient consent was obtained, and informed consent was signed before collection. More information about patients and samples is explained in the supplementary materials.
PDX models and drug susceptibility tests
The tumour tissues taken from different ICC patients during surgery were established by professional and technical personnel for the PDX model library, and the total rate of engraftment was ~30%. In this study, we randomly selected 20 PDX models derived from 20 patients with ICC in the library. The engraftment of the selected 20 models is 100% at third generation and GEM tests were performed. Model replicates from the same source patient were divided into GEM medication group (50 mg/kg) and a solvent group, with five nude mice (details in the “Animal study”) in each group. Drug treatment was started when the tumour size was ~150 cubic millimetres (the time was recorded from the first medication). Treatment was administered twice a week, and the tumour volume was measured twice a week. According to volume = l/2ab2 (where a represents the long diameter, and b represents the short diameter), the nude mice were sacrificed after 4 weeks, and the tumour tissue was properly preserved.
RNA transcriptome sequencing and protein chip technology
Three resistant PDX tissues and three sensitive PDX tissues were selected for high throughput RNA transcriptome sequencing. Sequencing and subsequent correlation analysis were carried out with the support of Shanghai Luming Biotechnology Co., Ltd. Two of the above resistant and sensitive tissues were selected for sequencing of the 2 vs. 2 protein chip in Shanghai Huaying Biomedical Technology Co., Ltd., and the target molecules were determined by comprehensive analysis of transcriptome sequencing and protein chip results.
Cell culture
Human ICC cells RBE and HCCC9810 were purchased from the Cell Bank of Shanghai Academy of Sciences. Penicillin and streptomycin (P/S) for cell culture were purchased from Beyotime Biotechnology Co., Ltd. RPMI-1640 cell culture medium and foetal bovine serum were purchased from Gibco, USA. The human hepatobiliary cancer cell lines RBE and HCCC9810 were cultured on RPMI 1640 medium (hereafter referred to as 1640 medium). Serum (FBS, 10%) was added to the medium, and 1% of streptomycin/ penicillin was added to prevent cell contamination. The conditions for cell growth in the incubator were 37 °C and 5% carbon dioxide. These cell lines were authenticated by short tandem repeat profiling and routinely tested as mycoplasma-free.
Animal study
All immunodeficient mice used in the experiment were purchased from Shanghai Experimental Animal Center of Chinese Academy of Sciences, which was approved by the Ethics Committee of the Third Affiliated Hospital of Navy Medical University and met the requirements of experimental ethics before the experiment. SPF grade 5-week-old male BALB/c nude mice were subjected to PDXs and subcutaneous tumour loading experiment. HCCC9810 ICC cells were cultured until cell growth showed a logarithmic growth phase. The cells were digested and resuspended to prepare a cell suspension with a density of 5 * 10 ^ 6/100 ul, and ~200 ul was injected into the subcutaneous tissue of nude mice. Then, 20 nude mice were randomly divided into four groups on average: (1) the MOCK group; (2) the TSP1 group; (3) the GEM group; and (4) the GEM + TSP1 group. Mice were administered 300 μl of vehicle (saline), recombinant human TSP1 protein (0.5 mg/kg), GEM (50 mg/kg) and recombinant human TSP1 protein (0.5 mg/kg) and GEM (50 mg/kg) via intraperitoneal injection twice a week for 4 weeks. The tumour volume was measured twice a week. The long and short diameters of the tumour were recorded, and the volume was calculated as follows: tumour volume = 0.5*a*b2. The investigators recording tumour growth were blinded to mouse allocation.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 and R version 4.2.0 software. We chose the appropriate statistical method according to the characteristics of the data. Statistical analysis methods included Student’s t test, the chi square test, the Mann‒Whitney test, the Wilcoxon signed rank test and the Pearson correlation test. The survival curve was calculated by the Kaplan‒Meier method. A P value < 0.05 was considered statistically significant. * P < 0.05, * * P < 0.01, * * * P < 0.001. The remaining methods are described in the Supplementary Materials.
Results
Preclinical models of PDXs preserved the patients’ pathological characteristics
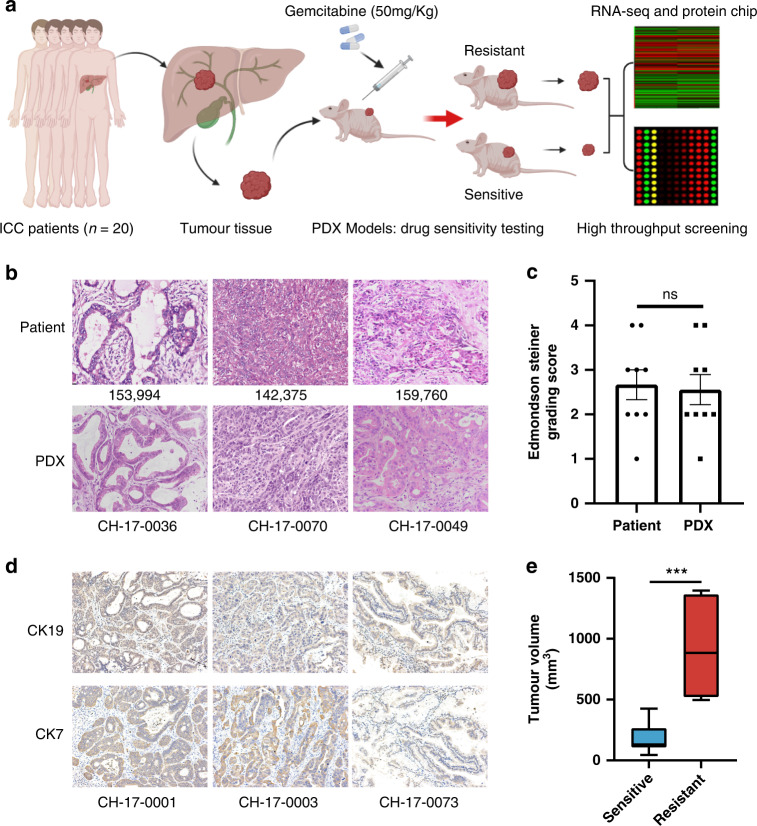
In this study, the PDX models were established using the cancer tissues of 20 patients with ICC from the PDX model library, and the key molecules mediating GEM resistance were screened by high-throughput technology (Fig. 1a). By pathological staining of tumour tissue, we found that the features of PDX tissues were highly consistent with those of patient tissues in terms of tumour morphology or tumour differentiation and atypia (Fig. 1b). According to the Edmondson score [21], there was no significant difference between the PDX models and patient tumour tissues (Fig. 1c). Immunohistochemical staining of CK19 and CK7 also confirmed that PDXs preserved ICC tumour characteristics (Fig. 1d). Therefore, the PDX models can accurately simulate the tumour state of clinical patients.
Fig. 1. Preclinical models of PDX preserved the patients’ pathological characteristics.
a The establishment of the PDX models and the screening process of research molecules in this study. b HE staining of clinical tumour tissue from patients and corresponding PDX model tumour tissue (200×). c Edmondson-Steiner grading of PDX (n = 9) and patients (n = 9) tumour tissues. ‘ns’ represents no significant difference. d Representative immunohistochemical images of CK19 and CK7 in PDX tumour tissues (200×). e Box plot of the tumour volumes at the end of treatment (day 30) obtained in PDX-sensitive tumour tissues (n = 13) and resistant tumour tissues (n = 7).
Thrombospondin-1 (TSP1) is significantly abundant in PDXs sensitive to gemcitabine
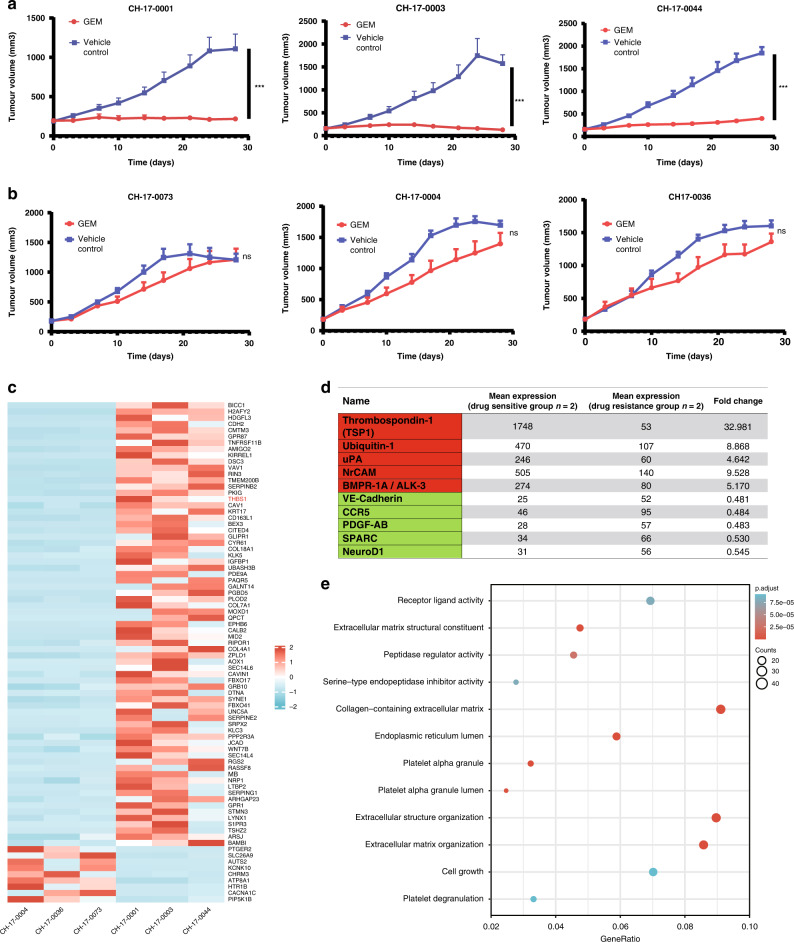
The PDX models established from 20 patients with ICC was used for the drug sensitivity tests (Fig. 1e), and 13 chemotherapy-sensitive models were selected, of which the representative models were CH-17-0001, CH-17-0003, and CH-17-0044 (Fig. 2a). Moreover, seven drug resistance models with poor effects on GEM were screened, and the representative models were CH-17-0073, CH-17-0004, and CH-17-0036 (Fig. 2b). The PDX models of the selected three pairs of sensitive and drug-resistant groups were subjected to transcriptome sequencing, and eighty genes with the highest expression difference (p < 1.02e−3, log2(Foldchange) > 2.5) were screened after the integration of the eligible gene analysis results. Seventy-one genes were highly expressed in the sensitive model, and the other nine genes were expressed at low levels in the drug-resistant model (Fig. 2c). Moreover, protein chip technology was used to screen the top ten secreted proteins (five upregulated proteins and five downregulated proteins) with significant differences in serum expression between the sensitive and resistant groups (Fig. 2d). We found that thrombospondin (TSP1) was highly expressed in the PDX models of the sensitive group and could be detected in serum as a secreted protein. TSP1 is a multifunctional extracellular matrix protein that is secreted into the extracellular matrix and binds to related receptors [15]. As an extracellular mediator of matrix mechanotransduction, TSP1 acts via integrin αvβ1 to establish focal adhesions [22]. Differential gene enrichment analysis showed that ‘receptor ligand activity’, ‘collagen−containing extracellular matrix’, ‘extracellular structure organization’ and ‘extracellular matrix organization’ were significantly enriched in the GO enrichment analysis (Fig. 2e). In the KEGG enrichment analysis, ‘focal adhesion’ was significantly enriched. (Fig. S1A). This further indicates that the functional pathways related to TSP1 may be significantly differentially enriched between the GEM-resistant and GEM-sensitive groups of GEM. The expression of TSP1 mRNA and protein in 20 PDX models was verified. The results showed that the mRNA expression of TSP1 in the 13 models of the sensitive group was significantly higher than that in 7 models of the resistant group (Fig. S1B), and the expression of TSP1 protein was significantly higher than that in the 7 models of resistant group (Fig. S1C). This further confirmed the consistency of the PDX models’ results and sequencing results.
Fig. 2. Thrombospondin-1 (TSP1) is significantly abundant in PDXs sensitive to gemcitabine.
a Drug sensitive group. b Drug resistance group. GEM represents gemcitabine (50 mg/kg), vehicle control represents solvent group, * represents p < 0.05, * * represents p < 0.01, * * * represents p < 0.001. c The top 80 differentially expressed genes in transcriptome sequencing. d The differentially expressed proteins in the protein chip assay. e GO enrichment analysis of differential genes.
High serum TSP1 levels correlate with better efficacy of gemcitabine-based chemotherapy for ICC
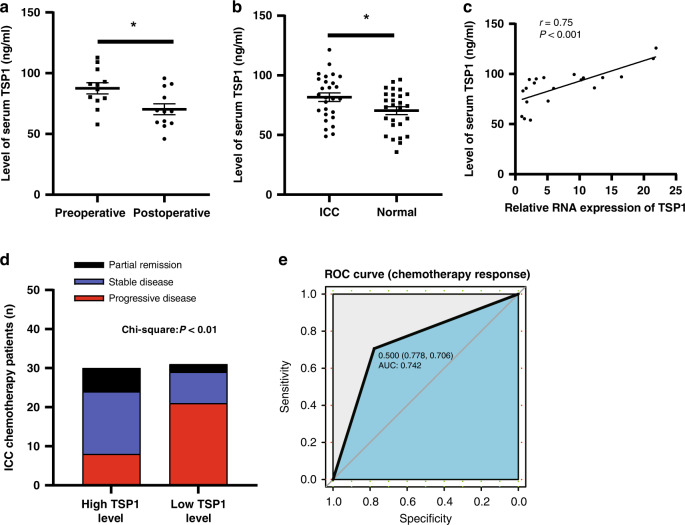
The expression of TSP1 in tumour tissues of different ICC patients was verified by immunohistochemistry (Fig. S2A). The serum samples of 12 ICC patients before and after the operation were randomly selected for the determination of TSP1. The overall level of TSP1 in the serum of patients after the operation was lower than that before the operation (Fig. 3a). Twenty-seven ICC patients and 27 healthy people were randomly selected to determine the expression level of TSP1 in the serum. The overall level of TSP1 expression in the serum of 27 ICC patients was higher than that in the serum of 27 normal people (Fig. 3b). We collected the tumour tissues and preoperative serum of 20 ICC patients, and detected the expression levels of TSP1 in the tissues and serum (Fig. S2B, S2C). We found that the two were highly positively correlated (Fig. 3c). This finding indicates that TSP1 in the serum of ICC patients may mainly come from tumour tissues, and the TSP1 level in serum before surgery can reflect the expression level of TSP1 in tumour tissues to some extent.
Fig. 3. High serum TSP1 levels correlate with better efficacy of gemcitabine-based chemotherapy for ICC.
a The difference in TSP1 expression between preoperative (n = 12) and postoperative ICC patients (n = 12). bThe expression level of serum TSP1 in ICC patients (n = 27) and healthy people (n = 27). c Correlation analysis of TSP1 expression levels in ICC patient tissues and serum (n = 20). * Represents P < 0.05. d Chemotherapy response of theTSP1 high expression group (n = 30) and low expression group (n = 31). PD represents disease progression, SD represents disease stability, PR represents disease remission. e The ROC curve between TSP1 and chemotherapy response.
To further explore whether the expression level of TSP1 in ICC patients can reflect their chemotherapy sensitivity, we detected the expression level of TSP1 in the serum of 61 patients with advanced ICC who met the inclusion criteria before chemotherapy, and selected the median expression level of TSP1 (82.851 ng/ml) as the cut-off value (Fig. S2D). According to this cut-off value, the patients were classified into the high expression group and the low expression group (n = 30 and n = 31, respectively). In patients with high TSP1 expression, the number of patients with a good chemotherapy effect of GEM was greater (16 cases of SD + 6 cases of PR), and the number of patients with a poor chemotherapy response was lower (8 cases of PD) than that in the low expression group; in this group, only 10 patients (8 SD + 2 PR) had a good response to chemotherapy, while a large proportion of patients exhibited a poor response to chemotherapy (21 PD) (Fig. 3d). The ROC curve showed that TSP1 had high predictive value for chemotherapy.response in patients with cholangiocarcinoma (AUC = 0.742) (Fig. 3e). We analysed the correlation between the basic pathological data of patients and the expression level of TSP1 and found that the expression level of TSP1 was related to the chemotherapy response (p < 0.001) but not related to other factors such as age, gender, hepatitis B surface antigen, chemotherapy selection and ECOG score (Tab. S2), which fully illustrated that the expression level of TSP1 could reflect the GEM chemotherapy sensitivity of ICC patients.
Serum TSP1 levels could predict the prognosis of patients with unresectable ICC
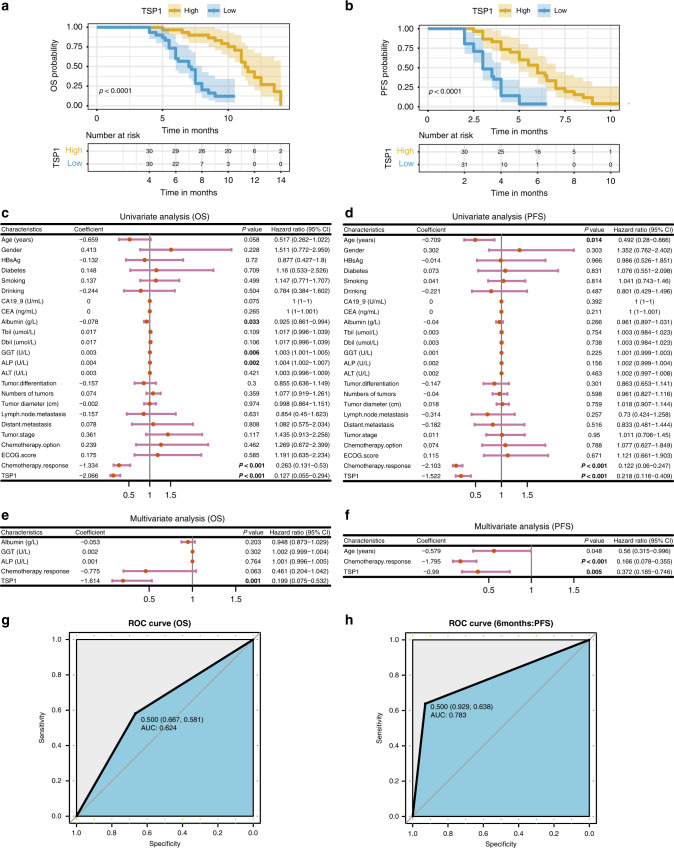
We followed up regarding the prognosis of 61 patients with advanced ICC. The overall survival (OS) time of the high TSP1 expression group was longer than that of the low TSP1 expression group (Fig. 4a), and the median survival times were 10.5 months and 7 months respectively. The progression-free survival (PFS) time in the high TSP1 expression group was also longer than that in the low TSP1 expression group (Fig. 4b). The median progression-free time was 6 months and 3 months, respectively. The difference was statistically significant. Cox univariate analysis showed that albumin, GGT, ALP, chemotherapy.response, and TSP1 were correlated with OS in patients with cholangiocarcinoma after chemotherapy (Fig. 4c); age, chemotherapy.response and TSP1 were correlated with PFS in patients with cholangiocarcinoma undergoing chemotherapy (Fig. 4d). Cox multivariate analysis showed that TSP1 was an independent risk factor for OS in patients with cholangiocarcinoma undergoing chemotherapy (Fig. 4e); age, chemotherapy.response and TSP1 were independent risk factors for PFS in patients with cholangiocarcinoma undergoing chemotherapy (Fig. 4f). This further indicates that TSP1 is a key factor affecting the prognosis of patients with cholangiocarcinoma after chemotherapy. During the follow-up period, it also had a good predictive value for the survival of patients (AUC = 0.624) (Fig. 4g); It is of great significance to predict whether the disease will progress in the first 6 months of chemotherapy (AUC = 0.783) (Fig. 4h).
Fig. 4. Serum TSP1 levels could predict the prognosis of patients with unresectable ICC.
a The expression level of TSP1 and the analysis of OS (overall survival). b The expression level of TSP1 and the analysis of PFS (progression-free survival). c Cox univariate analysis of OS in cholangiocarcinoma patients undergoing chemotherapy. d Cox univariate analysis of PFS in cholangiocarcinoma patients undergoing chemotherapy. e Cox multivariate analysis of OS in cholangiocarcinoma patients undergoing chemotherapy. f Cox multivariate analysis of OS in cholangiocarcinoma patients undergoing chemotherapy. g The ROC curve of OS prediction based on TSP1 expression. h The ROC curve showing the predictions for PFS withing 6 months based on TSP1 expression.
Thrombospondin-1 (TSP1) enhances the chemotherapy effect of gemcitabine in intrahepatic cholangiocarcinoma
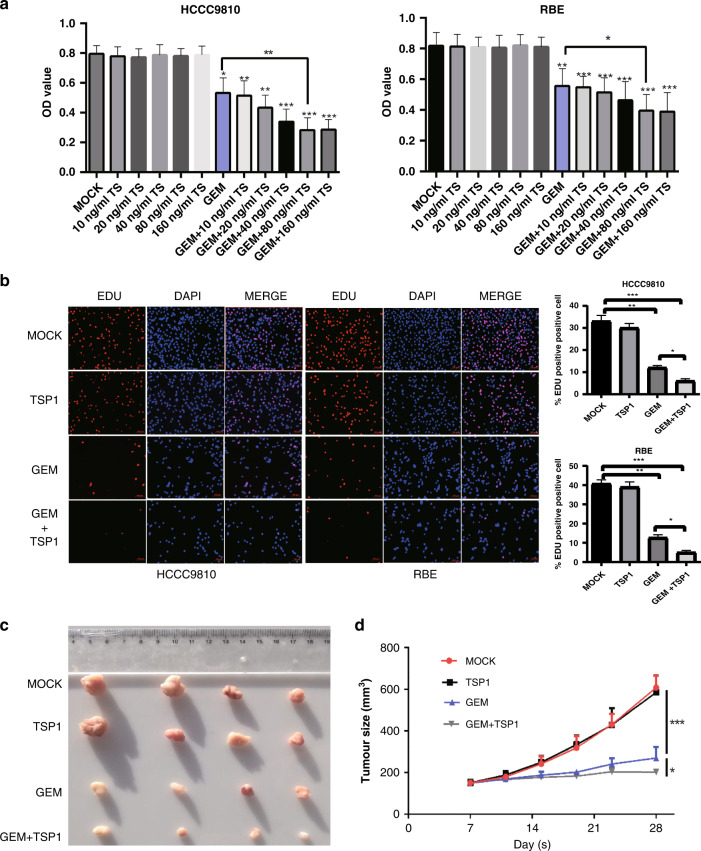
Next, we explored whether TSP1 had biological effects only as a molecular marker. We first screened out the half-maximal inhibitory concentration (IC50) of GEM in two bile duct cancer cell lines as the working concentration of GEM in subsequent experiments. The IC50 values of HCCC9810 and RBE were 2331 nM and 2044 nM, respectively (Fig. S3A). We set up different groups, including the MOCK group (blank control), GEM group, TSP1 group (different concentrations of TSP1 recombinant protein), and TSP1 + GEM group. After 72 h, the OD value of the cells was detected (OD value can represent cell proliferation activity). The results showed that in the HCCC9810 cell line, compared with the MOCK group, the addition of TSP1 recombinant protein did not significantly affect the proliferation of cholangiocarcinoma cells, and there was no significant difference in cell viability between groups with different concentrations of TSP1. In the GEM group, cell viability decreased significantly compared with that in the MOCK group, and the difference was statistically significant. With the increase in TSP1 concentration in a certain range, the cell viability of the TSP1 + GEM group also showed a downwards trend compared with that of the GEM group, and reached saturation when the concentration of TSP1 was ~80–160 ng/ml. Continuing to increase the concentration of TSP1 did not further decrease cell viability. The same phenomenon was also observed in RBE cholangiocarcinoma cells (Fig. 5a). Hence, we set the concentration of TSP1 recombinant protein to 100 ng/ml in the follow-up experiments. To further confirm the reliability of the results, we also conducted Edu experiments (Fig. 5b) and colony formation experiments (Fig. S3B) on HCCC9810 cells and RBE cells, which further confirmed the above conclusions.
Fig. 5. Thrombospondin-1 (TSP1) enhances the effect of gemcitabine chemotherapy on intrahepatic cholangiocarcinoma.
a The effect of CCK-8 on the proliferation of HCCC9810 and RBE cells under different treatments. b The proliferation ability of HCCC9810 and RBE cells under different treatments in the EdU experiment. c The detection of tumorigenicity of HCCC9810 cells by different treatment methods. d Changes in tumour growth curves corresponding to different treatments (n = 4 per group). Data are shown as mean ± s.d. of n = 3 independent experiments (a,b). MOCK is the blank control, TSP1 is the TSP1 recombinant protein, GEM is gemcitabine, GEM + TSP1 is gemcitabine + TSP1 recombinant protein * represents p < 0.05, * * represents p < 0.01, * * represents p < 0.001.
Then we subcutaneously xenografted HCCC9810 cells in nude mice (Fig. 5c, d). These results showed that there was no significant difference in tumour size or growth curve between the MOCK group and the TSP1 recombinant protein group. The GEM group (Gemcitabine alone) showed significantly inhibited tumour growth compared with the first two groups, with statistical significance. The tumours in the GEM + TSP1 group were further reduced compared with those in the GEM group, indicating that TSP1 recombinant protein could enhance the sensitivity of GEM in vivo.
TSP1 inhibits the uptake of oleic acid by intrahepatic cholangiocarcinoma cells, thereby increasing the therapeutic effect of gemcitabine
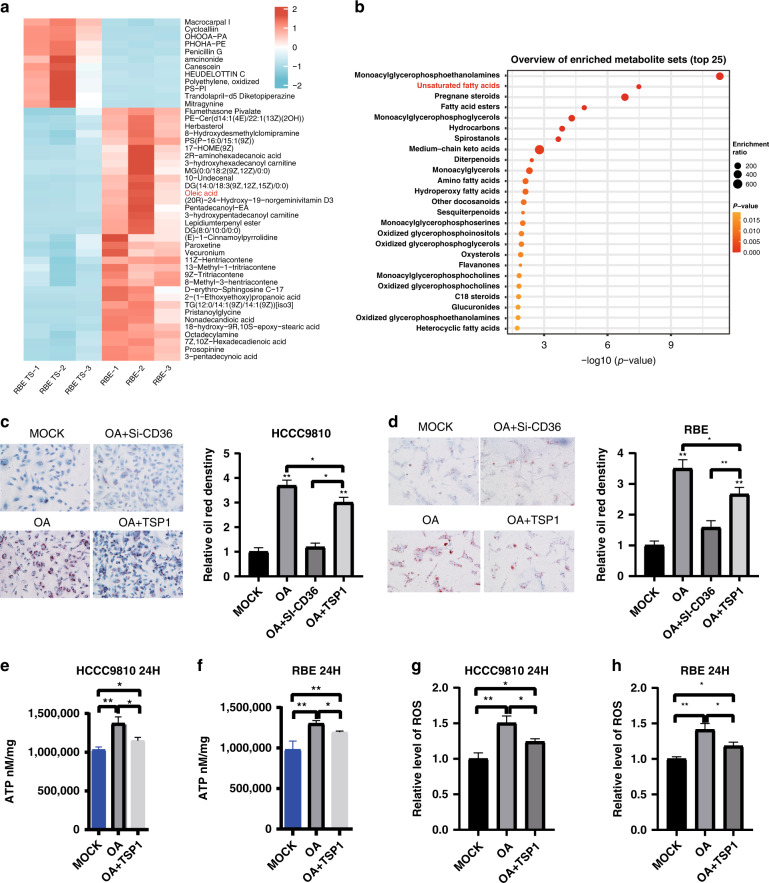
Studies have shown that changes in lipid metabolism can alter the effect of GEM on pancreatic cancer, resulting in chemotherapy resistance [23, 24]. Isenberg JS et al. found that TSP1 binds to CD36, a target cell surface receptor, and affects its uptake of certain fatty acids [25]. Therefore, we speculate that TSP1 may affect lipid metabolism by binding to CD36 on the cholangiocarcinoma cell surface, thereby affecting the chemotherapeutic effect of GEM. We detected the metabolites in RBE cell lines (the experimental group added TSP1 recombinant protein; the control group was saline). The metabolites with P values <0.05 were selected as the differentially expressed, and we screened 156 differentially expressed metabolites. The top 50 differentially (p < 0.05, log2(Foldchange) > 3.5) expressed metabolites are shown in the heatmap (Fig. 6a). Among these differential metabolites, OA, as an unsaturated fatty acid, has been reported to be associated with chemotherapy resistance in some tumours [26–29], with a significant research value. Functional enrichment of differential metabolites also showed significant enrichment of unsaturated fatty acids (Fig. 6b). Therefore, OA was taken as the research object. To further confirm the above hypothesis, we constructed a CD36 knockdown cholangiocarcinoma cell line (Fig. S3A, B). We found that in HCCC9810 cells, compared with that in the MOCK group, the oil red O staining level in the OA group was significantly enhanced after 24 h of OA addition. When CD36 was knocked out and OA was added (OA + si-CD36 group), the oil red O staining level of the cells was significantly decreased compared with that of the OA group, and there was no significant difference in the oil red O staining level between the OA group and the MOCK group. In OA + TSP1group (OA and TSP1recombinant protein), we found that compared to OA, TSP1decreased the oil red O staining density, but the staining was still stronger than that in the MOCK group and OA + Si-CD36 group, and the difference was statistically significant (Fig. 6c). Similar results were also verified in RBE cell lines (Fig. 6d). Based on the above experimental results, we speculated that OA mainly enters the cells through the membrane receptor CD36; after adding TSP1 recombinant protein, TSP1 can inhibited the uptake of OA by ICC cells. It is likely that TSP1 may affect the uptake of OA by binding to CD36 on the surface of cholangiocarcinoma cells, thereby affecting the chemotherapy effect of GEM. To further verify whether OA can alter the effect of GEM chemotherapy, a CCK-8 experiment was used to detect the cell viability of two ICC cell lines, RBE and HCCC9810. In RBE cells, the GEM group exhibited significantly decrease cell viability compared with the MOCK group; in the GEM + OA group (GEM and OA), although the cell viability of the GEM + OA group was lower than that of the MOCK group, the cell viability increased compared with that of the GEM group (Fig. S4C), indicating that OA weakened the chemotherapy effect of GEM. Similar results were obtained in HCCC9810 cells (Fig. S4F). Then, the above conclusions were verified in RBE and HCCC9810 cell lines by colony formation experiments (Fig. S4D, S4G) and EdU experiments (Fig. S4E, S4H). When fatty acid oxidation is enhanced, the contents of ROS and ATP, the related metabolites, will also increase, and these substances will promote the occurrence of drug resistance. Gemcitabine can cause DNA damage during cancer treatment, thereby inducing the formation of ROS. The increase in local ROS concentration can cause DNA damage and induce mutations, resulting in drug-resistant cancer cells. We also found that OA increased intracellular ROS and ATP levels, while TSP1 recombinant protein effectively reduced ATP to ROS levels (Fig. 6e–h; Fig. S3I–L). Therefore, we speculate that TSP1 promotes GEM chemosensitivity by inhibiting the entry of OA into the cell, leading to a decrease in ROS and ATP levels.
Fig. 6. TSP1 inhibits the uptake of oleic acid by intrahepatic cholangiocarcinoma cells, thereby increasing the therapeutic effect of gemcitabine.
a The LC-MS metabolomics differential metabolite map. b Enrichment analysis of differential metabolites. c The results of oil red O staining in different groups of HCCC9810 cells. d The results of oil red O staining in different groups of RBE cells. e ATP detection in HCCC9810 cells at 24 h. f ATP detection in RBE cells at 24 h. g Detection of ROS at 24 h in HCCC9810 cells. h Detection of ROS at 24 h in RBE cells. Data are shown as mean ± s.d. of n = 3 independent experiments (c–h). MOCK is the blank control, OA is oleic acid, OA + TSP1 is oleic acid + TSP1 recombinant protein, GEM is gemcitabine, OA + GEM is oleic acid + gemcitabine * represents p < 0.05, * * represents p < 0.01, * * represents p < 0.001.
Discussion
Although GEM is currently used as the first-line drug for chemotherapy in many cancer patients, the effect in most patients is poor and drug resistance is widespread [30, 31]. At present, many scholars are committed to studying the mechanism of GEM resistance in patients with ICC. Tiemin P et al. found that downregulation of miR-148a results in overexpression of GLUT1 in iCCA, leading to the progression of iCCA and GEM resistance [32]. Lu et al. found that LINC00665 plays an important role in the resistance of CCA cells to GEM, providing new biomarkers or therapeutic targets for CCA treatment [33]. Carotenuto P et al. found that MIR1249 is involved in GEM resistance in cholangiocarcinoma and is a potential therapeutic target [34]. Although these studies have explained the mechanism of resistance of cholangiocarcinoma to GEM chemotherapy from various aspects, it is difficult to translate these findings to the clinic in a short period of time. In addition, there is a lack of clinical markers that can effectively predict whether GEM chemotherapy is effective for a given cholangiocarcinoma case. This, to some extent, leads to blindness in the treatment of patients with advanced cholangiocarcinoma, which greatly shortens the survival time of patients. The protein encoded by the molecule TSP1 we studied is easy to detect in the serum, and we confirmed that the level of TSP1 in the serum of patients with ICCA can reflect their sensitivity to GEM treatment, indicating that TSP1 can be used as a serological indicator to predict the efficacy of chemotherapy in patients with ICC. Before chemotherapy for advanced cholangiocarcinoma patients, the level of TSP1 in serum can be detected, which can be more effective and accurate in formulating chemotherapy regimens than traditionally used markers.
Many studies have confirmed that TSP1 has great application prospects in tumour therapy. Uronis HE and other researchers have used the analogue of TSP1 (ABT-510) in combination with bevacizumab to conduct phase I clinical research on advanced solid tumours. The results show that compared with the single drug application of bevacizumab, the effect of combined treatment is significantly improved, and the inhibitory effect on tumours is significantly enhanced [35]. Campbell NE et al. found that TSP1 mimics increased the cellular uptake of cisplatin and paclitaxel in mice with epithelial ovarian cancer and improved the effectiveness of chemotherapy [36]. In this study, we confirmed that TSP1 plays a role in chemotherapy sensitisation in ICC by CCK8, EdU and colony formation experiments, indicating that TSP1 may be used as a chemotherapy sensitiser for patients with ICC in the future. Although studies have shown that high expression of TSP1 is associated with the invasion and progression of HCC, the expression of TSP1 was positively correlated with the expression of VEGF in HCC patients. It seems to be a proangiogenic factor that stimulates angiogenesis in HCC [37]. However, in this study we found that high expression of TSP1 in the serum of patients with cholangiocarcinoma indicated a better prognosis, and patients with high expression of TSP1 had a better response to GEM. In addition, we found that when TSP1 alone was applied to ICC cells, it had no significant effect on proliferation, indicating that TSP1 itself has no therapeutic effect on patients with ICC but can make cholangiocarcinoma cells sensitive to chemotherapy.
TSP1 needs to bind to its receptors to perform its corresponding biological functions, and one of its important receptors is CD36. CD36 is a scavenger receptor that mainly plays the role of uptake of long-chain fatty acids [38]. Studies have found that TSP1 inhibits fatty acid uptake by CD36 [25]. These findings are consistent with the conclusions of our study. In this study, we found that OA can be taken up by cholangiocarcinoma cells through CD36 mediation, and the addition of TSP1 reduces the uptake of OA by cholangiocarcinoma cells, indicating that TSP1 may reduce the intake of OA by acting on CD36 receptors in cholangiocarcinoma cells. Changes in lipid metabolism can affect chemosensitivity in many tumours, including GEM [39, 40]. When the oxidation of fatty acids is enhanced, the content of ROS and ATP will also increase, and these substances will promote the occurrence of drug resistance [28]. ATP is a ‘nutrient’ that cancer cells need to survive, and changes in its content will directly affect the growth of cancer cells [41]. Gemcitabine can cause DNA damage during cancer treatment, thereby inducing the formation of ROS. A high concentration of ROS can induce mutation of tumour cells, leading to the generation of drug-resistant cells [29]. In our study, we found that when OA was added, the inhibitory effect of GEM on tumour cells decreased, and intracellular ROS and ATP levels increased, while TSP1 recombinant protein effectively reduced ATP and ROS levels. Hence, we concluded that OA is a sort of competitor of TSP1 and that its presence reduces the effect of GEM. The addition of TSP1reduced intracellular ROS and ATP levels by inhibiting the uptake of OA in cholangiocarcinoma cells and restored the resistant conditions of tumour cells to a sensitive state. There are still some shortcomings in this study. Although the current results confirm that TSP1 has the potential to become a serological marker for predicting the effect of chemotherapy in cholangiocarcinoma, it does not mean that TSP1 has been clinically applied, and a rigorous and scientific verification process with real clinical translation is still needed. Moreover, the chemotherapy patients were enrolled from a single-centre, and the sample size is relatively small, so multicentre cohorts are needed for further validation. In the mechanism study, although the TSP1 - CD36 - OA connection is clear, this study mainly focuses on confirming that TSP1 can affect the entry of OA into ICC cells and that the entry of OA will affect the response to GEM. However, it remains unclear how TSP1 functions through CD36. We hypothesised that TSP1 competes with OA for binding to the CD36 receptor, which requires further experiments for confirmation. Furthermore, there is insufficient evidence that TSP1 mediates the entry of OA mainly through CD36. Therefore, the effect of abnormal OA and lipid metabolism on GEM chemotherapy sensitivity is still worthy of further study, which may be an important clue for disrupting the mechanism of GEM resistance.
In summary, based on the phenomenon of GEM resistance in patients with advanced cholangiocarcinoma, this study carried out a GEM drug sensitivity tests based on PDX models, used the tissue and serum samples of the drug-resistant group and sensitive group for transcriptome sequencing and protein chip detection, and finally identified that TSP1 is differentially expressed between the drug-resistant group and sensitive group. Then we confirmed in clinical studies that the expression level of TSP1 reflects the sensitivity of ICC patients to GEM chemotherapy. In vivo and in vitro experiments confirmed that TSP1 could enhance the chemotherapeutic effect of GEM on ICC. In the mechanistic study, we found that TSP1 may affect the intake of OA by binding to the CD36 receptor on the surface of cholangiocarcinoma cells, and then affect the chemotherapy effect of GEM. In conclusion, we found a key molecule (TSP1) that can predict and improve the sensitivity of ICC patients to GEM chemotherapy, which is of great significance for the treatment of advanced cholangiocarcinoma.
Supplementary information
Acknowledgements
Thank all participants for their support and help.
Author contributions
D-yD: Data curation, Writing-Original draft preparation; J-nZ and X-jG: Visualisation, Investigation; G-jH and Q-fT: Software; D-pS, WL and JY: Writing-Reviewing and Editing; YY, W-bD and LL: Software, Validation; S-XY and FY: Conceptualisation, Methodology; W-pZ: Supervision.
Funding
This study was supported by the National Natural Science Foundation of China (81972575,81903059, 81802983), the San Hang Program of the Second Military Medical University, and Shanghai Municipal Commission of Health and Family Planning Program (20174Y0085).
Data availability
All presented data are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was performed with the approval of the Ethics Committee of the Third Affiliated Hospital of Navy Medical University (ethical authorisation protocol numbers: EHBHKY201802012).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Dong-yang Ding, Xiao-jie Gan, Jia-ning Zhang, Guo-jun Hou.
Contributor Information
Lei Liu, Email: Dr_ray@foxmail.com.
Fu Yang, Email: yangfusq1997@smmu.edu.cn.
Wei-ping Zhou, Email: ehphwp@126.com.
Sheng-xian Yuan, Email: yuanshengx@126.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-02101-0.
References
- 1.Mavros MN, Economopoulos KP, Alexiou VG, Pawlik TM. Treatment and Prognosis for Patients With Intrahepatic Cholangiocarcinoma: Systematic Review and Meta-analysis. JAMA Surg. 2014;149:565–74. doi: 10.1001/jamasurg.2013.5137. [DOI] [PubMed] [Google Scholar]
- 2.Wirth TC, Vogel A. Surveillance in cholangiocellular carcinoma. Best Pract Res Clin Gastroenterol. 2016;30:987–99. doi: 10.1016/j.bpg.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Moeini A, Sia D, Bardeesy N, Mazzaferro V, Llovet JM. Molecular Pathogenesis and Targeted Therapies for Intrahepatic Cholangiocarcinoma. Clin Cancer Res. 2016;22:291–300. doi: 10.1158/1078-0432.CCR-14-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelley RK, Bridgewater J, Gores GJ, Zhu AX. Systemic therapies for intrahepatic cholangiocarcinoma. J Hepatol. 2020;72:353–63. doi: 10.1016/j.jhep.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Varamo C, Peraldo-Neia C, Ostano P, Basirico M, Raggi C, Bernabei P, et al. Establishment and Characterization of a New Intrahepatic Cholangiocarcinoma Cell Line Resistant to Gemcitabine. Cancers (Basel) 2019;11:519. doi: 10.3390/cancers11040519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rauchwerger DR, Firby PS, Hedley DW, Moore MJ. Equilibrative-sensitive nucleoside transporter and its role in gemcitabine sensitivity. Cancer Res. 2000;60:6075–9. [PubMed] [Google Scholar]
- 7.Adamska A, Elaskalani O, Emmanouilidi A, Kim M, Abdol RN, Metharom P, et al. Molecular and cellular mechanisms of chemoresistance in pancreatic cancer. Adv Biol Regul. 2018;68:77–87. doi: 10.1016/j.jbior.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Feng WW, Kurokawa M. Lipid metabolic reprogramming as an emerging mechanism of resistance to kinase inhibitors in breast cancer. Cancer Drug Resist. 2020;3:1–17. doi: 10.20517/cdr.2019.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun X, Zhou X, Zhang Y, Zhu X, Liu H. Systematic Review and Meta-Analysis of Diagnostic Accuracy of miRNAs in Patients with Pancreatic Cancer. Dis Markers. 2018;2018:6292396. doi: 10.1155/2018/6292396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosfjord E, Lucas J, Li G, Gerber HP. Advances in patient-derived tumor xenografts: from target identification to predicting clinical response rates in oncology. Biochem Pharmacol. 2014;91:135–43. doi: 10.1016/j.bcp.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Corso S, Isella C, Bellomo SE, Apicella M, Durando S, Migliore C, et al. A Comprehensive PDX Gastric Cancer Collection Captures Cancer Cell-Intrinsic Transcriptional MSI Traits. Cancer Res. 2019;79:5884–96. doi: 10.1158/0008-5472.CAN-19-1166. [DOI] [PubMed] [Google Scholar]
- 12.Beshiri ML, Tice CM, Tran C, Nguyen HM, Sowalsky AG, Agarwal S, et al. A PDX/Organoid Biobank of Advanced Prostate Cancers Captures Genomic and Phenotypic Heterogeneity for Disease Modeling and Therapeutic Screening. Clin Cancer Res. 2018;24:4332–45. doi: 10.1158/1078-0432.CCR-18-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada S, Vaeteewoottacharn K, Kariya R. Application of Highly Immunocompromised Mice for the Establishment of Patient-Derived Xenograft (PDX) Models. Cells. 2019;8:889. doi: 10.3390/cells8080889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galvin NJ, Dixit VM, O’Rourke KM, Santoro SA, Grant GA, Frazier WA. Mapping of epitopes for monoclonal antibodies against human platelet thrombospondin with electron microscopy and high sensitivity amino acid sequencing. J Cell Biol. 1985;101:1434–41. doi: 10.1083/jcb.101.4.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawler J, Derick LH, Connolly JE, Chen JH, Chao FC. The structure of human platelet thrombospondin. J Biol Chem. 1985;260:3762–72. doi: 10.1016/S0021-9258(19)83689-X. [DOI] [PubMed] [Google Scholar]
- 16.Bazzazi H, Zhang Y, Jafarnejad M, Isenberg JS, Annex BH, Popel AS. Computer Simulation of TSP1 Inhibition of VEGF-Akt-eNOS: An Angiogenesis Triple Threat. Front Physiol. 2018;9:644. doi: 10.3389/fphys.2018.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohrs JA, Sulistio CD, Finley SD. Predictive model of thrombospondin-1 and vascular endothelial growth factor in breast tumor tissue. NPJ Syst Biol Appl. 2016;2:16030. doi: 10.1038/npjsba.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuchida R, Osawa T, Wang F, Nishii R, Das B, Tsuchida S, et al. BMP4/Thrombospondin-1 loop paracrinically inhibits tumor angiogenesis and suppresses the growth of solid tumors. Oncogene. 2014;33:3803–11. doi: 10.1038/onc.2013.358. [DOI] [PubMed] [Google Scholar]
- 19.Nelius T, Filleur S, Yemelyanov A, Budunova I, Shroff E, Mirochnik Y, et al. Androgen receptor targets NFkappaB and TSP1 to suppress prostate tumor growth in vivo. Int J Cancer. 2007;121:999–1008. doi: 10.1002/ijc.22802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pal SK, Nguyen CT, Morita KI, Miki Y, Kayamori K, Yamaguchi A, et al. THBS1 is induced by TGFB1 in the cancer stroma and promotes invasion of oral squamous cell carcinoma. J Oral Pathol Med. 2016;45:730–9. doi: 10.1111/jop.12430. [DOI] [PubMed] [Google Scholar]
- 21.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::AID-CNCR2820070308>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 22.Yamashiro Y, Thang BQ, Ramirez K, Shin SJ, Kohata T, Ohata S, et al. Matrix mechanotransduction mediated by thrombospondin-1/integrin/YAP in the vascular remodeling. Proc Natl Acad Sci U S A. 2020;117:9896–905. doi: 10.1073/pnas.1919702117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller AL, Garcia PL, Yoon KJ. Developing effective combination therapy for pancreatic cancer: an overview. Pharmacol Res. 2020;155:104740. doi: 10.1016/j.phrs.2020.104740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin C, Yang G, Yang J, Ren B, Wang H, Chen G, et al. Metabolism of pancreatic cancer: paving the way to better anticancer strategies. Mol Cancer. 2020;19:50. doi: 10.1186/s12943-020-01169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isenberg JS, Jia Y, Fukuyama J, Switzer CH, Wink DA, Roberts DD. Thrombospondin-1 inhibits nitric oxide signaling via CD36 by inhibiting myristic acid uptake. J Biol Chem. 2007;282:15404–15. doi: 10.1074/jbc.M701638200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liotti A, Cosimato V, Mirra P, Cali G, Conza D, Secondo A, et al. Oleic acid promotes prostate cancer malignant phenotype via the G protein-coupled receptor FFA1/GPR40. J Cell Physiol. 2018;233:7367–78. doi: 10.1002/jcp.26572. [DOI] [PubMed] [Google Scholar]
- 27.Schlaepfer IR, Hitz CA, Gijon MA, Bergman BC, Eckel RH, Jacobsen BM. Progestin modulates the lipid profile and sensitivity of breast cancer cells to docetaxel. Mol Cell Endocrinol. 2012;363:111–21. doi: 10.1016/j.mce.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, et al. Chemotherapy-Resistant Human Acute Myeloid Leukemia Cells Are Not Enriched for Leukemic Stem Cells but Require Oxidative Metabolism. Cancer Discov. 2017;7:716–35. doi: 10.1158/2159-8290.CD-16-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Q, Wang JQ, Assaraf YG, Ren L, Gupta P, Wei L, et al. Modulating ROS to overcome multidrug resistance in cancer. Drug Resist Updat. 2018;41:1–25. doi: 10.1016/j.drup.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Ricciardiello F, Gang Y, Palorini R, Li Q, Giampa M, Zhao F, et al. Hexosamine pathway inhibition overcomes pancreatic cancer resistance to gemcitabine through unfolded protein response and EGFR-Akt pathway modulation. Oncogene. 2020;39:4103–17. doi: 10.1038/s41388-020-1260-1. [DOI] [PubMed] [Google Scholar]
- 31.Meng Q, Liang C, Hua J, Zhang B, Liu J, Zhang Y, et al. A miR-146a-5p/TRAF6/NF-kB p65 axis regulates pancreatic cancer chemoresistance: functional validation and clinical significance. Theranostics. 2020;10:3967–79. doi: 10.7150/thno.40566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tiemin P, Peng X, Qingfu L, Yan W, Junlin X, Zhefeng H, et al. Dysregulation of the miR-148a-GLUT1 axis promotes the progression and chemoresistance of human intrahepatic cholangiocarcinoma. Oncogenesis. 2020;9:19. doi: 10.1038/s41389-020-0207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu M, Qin X, Zhou Y, Li G, Liu Z, Geng X, et al. Long non-coding RNA LINC00665 promotes gemcitabine resistance of Cholangiocarcinoma cells via regulating EMT and stemness properties through miR-424-5p/BCL9L axis. Cell Death Dis. 2021;12:72. doi: 10.1038/s41419-020-03346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carotenuto P, Hedayat S, Fassan M, Cardinale V, Lampis A, Guzzardo V, et al. Modulation of Biliary Cancer Chemo-Resistance Through MicroRNA-Mediated Rewiring of the Expansion of CD133+ Cells. Hepatology. 2020;72:982–96. doi: 10.1002/hep.31094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uronis HE, Cushman SM, Bendell JC, Blobe GC, Morse MA, Nixon AB, et al. A phase I study of ABT-510 plus bevacizumab in advanced solid tumors. Cancer Med. 2013;2:316–24. doi: 10.1002/cam4.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell NE, Greenaway J, Henkin J, Moorehead RA, Petrik J. The thrombospondin-1 mimetic ABT-510 increases the uptake and effectiveness of cisplatin and paclitaxel in a mouse model of epithelial ovarian cancer. Neoplasia. 2010;12:275–83. doi: 10.1593/neo.91880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poon RT, Chung KK, Cheung ST, Lau CP, Tong SW, Leung KL, et al. Clinical significance of thrombospondin 1 expression in hepatocellular carcinoma. Clin Cancer Res. 2004;10:4150–7. doi: 10.1158/1078-0432.CCR-03-0435. [DOI] [PubMed] [Google Scholar]
- 38.Pepino MY, Kuda O, Samovski D, Abumrad NA. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu Rev Nutr. 2014;34:281–303. doi: 10.1146/annurev-nutr-071812-161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai CH, Wang Y, Chen P, Jiang Q, Lan T, Li MY, et al. Suppression of the FA pathway combined with CHK1 inhibitor hypersensitize lung cancer cells to gemcitabine. Sci Rep. 2017;7:15031. doi: 10.1038/s41598-017-15172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwamoto H, Abe M, Yang Y, Cui D, Seki T, Nakamura M, et al. Cancer Lipid Metabolism Confers Antiangiogenic Drug Resistance. Cell Metab. 2018;28:104–117.e5. doi: 10.1016/j.cmet.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y, Tozzi F, Chen J, Fan F, Xia L, Wang J, et al. Intracellular ATP levels are a pivotal determinant of chemoresistance in colon cancer cells. Cancer Res. 2012;72:304–14. doi: 10.1158/0008-5472.CAN-11-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All presented data are available from the corresponding author upon reasonable request.