Abstract
Abstract
An injured skin is rapidly restored in a manner of wound healing. We have previously shown that intact insulin signaling and glucose uptake are fundamental to proper wound closure. Consequently, under exacerbated inflammation, compromised insulin action and glucose uptake lead to impaired healing. However, in spite of the increased attention to cell metabolism during tissue regeneration, metabolic mediators that govern cellular and physiological processes throughout skin repair remained largely elusive. Through assessment of mRNA using real-time PCR and protein blot analysis, we report that healing of cutaneous wounds comprise a boosted expression of genes involved in glycolysis, oxidative phosphorylation, pentose phosphate shunt, and glutamine anaplerosis. We further focused on the functional role of pyruvate kinase M (PKM) isoenzymes that catalyze the final and rate-limiting step of glycolysis. Whereas the expression of the metabolic constitutively active Pkm1 isozyme remained almost unchanged, Pkm2 is augmented during the inflammatory phase of healing. The immunohistochemistry and RNA in situ hybridization analysis showed a confined Pkm2 expression to keratinocytes of the hyperproliferative epithelium and, to a lesser extent, infiltrating neutrophils and monocytes as well as later on in macrophages. Notably, the expression of Pkm2 in keratinocytes facing the wound bed side colocalized with VEGF expression. The in vitro knockdown of PKM2 in HaCaT keratinocytes using small interfering (si) RNA confirmed an acute role for PKM2 in facilitating the complete induction of VEGF mRNA and protein expression in keratinocytes; this function is mainly HIF-1α independent.
Key messages
• Wound healing involves activation of glycolysis, oxidative phosphorylation, pentos-phosphate shunt, and replenishment of tri-carboxylic acid (TCA) cycle through glutamine anaplerosis.
• The pyruvate kinase M2 (PKM2) isoform is upregulated during the inflammatory phase of cutaneous healing, mainly in keratinocytes of hyperproliferative epithelia.
• In vivo, the expression of VEGF in wound keratinocytes is colocalized with PKM2.
• PKM2 is required for full induction of VEGF in HaCaT keratinocytes in vitro.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00109-022-02280-6.
Keywords: Wound healing, Metabolic cues, Warburg phenomenon, PKM2, Keratinocytes
Introduction
Since the seminal work of Otto Warburg during the 1920s on ascites, the metabolic peculiarity of proliferating cells during pathological insults such as cancer and inflammation has been a matter of consistent scientific interest. Warburg’s observation that as opposed to resting cells, in rapidly growing tissues such as tumors, cells prefer to ferment glucose through the less efficient glycolysis pathway, even under aerobic conditions; an effect later coined the “Warburg phenomenon” [1]. Although this feature of fast-growing tumors to consume glucose and rapidly produce lactate was widely accepted, in the post-double helix era, recognizing that cancer is a genetic disease, researchers focused on genes rather than metabolites that promote malignancy. While Warburg’s conviction-aerobic fermentation may be caused by mitochondrial dysfunction turned out to be incorrect, Warburg’s view that metabolic processes drive malignancy [2] has revived [3, 4]. The notion that metabolic pathways could even determine the fate of tumorigenesis regained attention. Metabolites were proven to regulate oncogenic activity [5], as well as other pathophysiological processes like inflammation [6, 7] and immune response [8]. Similarly, it has emerged that metabolic conditions directly affect the outcome of cutaneous wound healing, an inflammatory-driven process that involves massive cell influx and outflow, proliferation, and remodeling. Accordingly, we have shown that intact insulin signaling and correct glucose uptake are essential to achieve wound closure [9, 10]. We proposed that sustained inflammation leads to shutting down insulin signaling in keratinocytes, also referred to as “insulin resistance” as manifested in obese mice [11]. Indeed, chronic energy surplus leads to adipose tissue expansion, and concomitant infiltration of inflammatory cells, in turn, induce a low-grade inflammation termed “metaflammation,” exacerbating the metabolic dysfunction [12]. In fact, we have shown that adipocytes exhibit a pro-inflammatory feature upon maturation in vitro, and contribute to healing impairment in obese mice [13]. Hence, given the central role of energy balance in the fate of cutaneous wound closure, we decided to journey into the metabolic landscape of normally healed wounds. Not surprisingly, skin injury induces the activation of the catabolic cascade of glycolysis as depicted by the expression of glyceraldehyde 3-phosphate dehydrogenase (Gapdh) and the glycolysis inducer, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (Pfkfb3), as well as mitochondrial oxidative phosphorylation (OXPHOS) as appeared from the expression of the mitochondria outer-membrane protein voltage-dependent anion channel 1 (Vdac1) and the mitochondrial-associated kinase hexokinase II (HkII). Induction of the anabolic pentose phosphate pathway (PPP) was also observed as manifested in gene expression of the first enzyme glucose-6-phosphate dehydrogenase (G6pdx), and the inducer of this shunt, the glycolytic phosphoglycerate mutase 1 (Pgam1). Glutamine anaplerosis of the mitochondrial tricarboxylic acid (TCA) cycle was also detected, as appears from the activation of the two pivotal genes that are involved: glutaminase (Gls) and glutamate dehydrogenase (Glud1). In this context, robust activation of glutamine synthase mRNA and protein, a hallmark for metabolically active tissue, was observed throughout the healing process. Of genes that were greatly induced during cutaneous repair and their product(s) primarily control metabolic processes, we were alerted to the temporal and spatial expression of the M2 isoform of pyruvate kinase (PK) PKM2. Pyruvate kinase is a homotetrameric enzyme that catalyzes the last and rate-limiting step of the glycolytic pathway, transferring the high-energy phosphate from phosphoenolpyruvate (PEP) to 2 ADP molecules, generating 2 ATP molecules and pyruvate. Two distinct genes, PKLR and PKM, code for PK in mammals. The PKLR gene codes for the L and R isoforms expressed in the liver and red blood cells, respectively, under the control of two different promoters [14]. Conversely, the PKM gene is expressed in the rest of the tissues and codes for the alternatively spliced exon 9 or exon 10 products, expressing variant M1 (PKM1) or variant M2 (PKM2), respectively [15]. PKM1 is a constitutively active tetrameric isoform, whereas PKM2 tetramerizes, and, hence, metabolic activation is allosterically regulated by metabolic cues such as the glycolytic intermediate fructose 1,6-bisphosphate (FBP) [16], amino acid serine [17], and the purine nucleotide synthesis intermediate SAICAR. Furthermore, PKM2 glycolytic activity is suppressed by growth signals directly through tyrosine phosphorylation of PKM2 [18], or indirectly by binding to a tyrosine-phosphorylated protein [19]. PKM1 is abundantly expressed in terminally differentiated and constantly high rate ATP-consuming tissues like muscles or neurons, whereas rapidly growing tissues like fetal tissues or tumors express mainly the regulated PKM2 isoform. This seeming disadvantageous expression of an enzyme having less pyruvate kinase activity can be explained by the existence of an alternative acceptor for the phosphate donated from PEP in the form of phosphoglycerate mutase (PGAM), thereby “rewiring” glycolysis and conveying glycolytic metabolite to replenish the metabolic demands of a growing tissue [20, 21]. A groundbreaking work of Christofk, Vander Heiden, Cantley, and their co-workers showed the significance of PKM2 expression for cancer metabolism and tumor growth [19]. Subsequently, the M2 isoform was considered as one of the key drivers of the Warburg phenomenon [22, 23]. Surfing into the ocean of PKM2 literature, a feature of multitasking, moonlighting protein has emerged. Though some of these features were revised [24], the metabolic requirements for PKM2 were confined to some tumors or stages of tumorigenesis [25, 26]. The parallelism of tumor biology and wound healing [27] motivated us to take a closer look at the role of Pkm2 isoforms in the murine model of skin excisional repair. Immunohistochemistry (IHC) staining revealed the expression of Pkm2 mainly confined to keratinocytes in the hyperproliferative epithelial tongue and, to a lesser extent, in infiltrating monocytes/macrophages. Additionally, we observed the co-localization of the pro-angiogenic mediator, vascular endothelial growth factor (Vegf) expression, to keratinocytes of the neo-epidermis, mainly adjacent to the wound bed, and this observation was reinforced by immunofluorescence (IF) and RNA in situ hybridization approaches. In accordance is the acute silencing of PKM2 expression by RNA interference in HaCaT-dampened VEGF expression upon growth factor stimulation in vitro. Remarkably, VEGF expression was not modified by altering PKM2 stoichiometry. The proangiogenic role of PKM2 in keratinocytes, which is independent of HIF-1α-expression, as opposed to previous observations in transformed cells, is described in this work.
Materials and methods
Materials
Aquatex, DNA oligonucleotide, duplex siRNA oligonucleotide for RNA interference (RNAi), 4′,6-diamidino-2-phenyl-indol-dihydrochlorid, 2-(4-amidinophenyl)-6-indolcarbamidin (DAPI), -dihydrochloridas (D9542-5MG), as well as reagents for molecular bacteriology were purchased from Sigma (Sigma Aldrich, Merck, Darmstadt, Germany). Deoxynucleotide triphosphates of PCR grade were obtained from Roche (Roche Diagnostics, Manheim, Germany). Calcium and magnesium free-Dulbecco’s phosphate buffered saline (PBS), Dulbecco’s modified Eagle medium (DMEM) (high glucose), DMEM/F12, OptiMEM, penicillin/streptomycin (5000 U/ml), sodium pyruvate (100 mM), and Trypsin EDTA (0.05%) were purchased from Gibco (Gibco, Thermo Fisher Scientific Darmstadt, Germany). Fetal calf serum (FCS), was from Biochrom (Biochrom KG, Berlin, Germany). Growth factors and cytokines were obtained from Peprotech (Peprotech, Hamburg, Germany). Lipofectamine 2000 was acquired from Invitrogen (Invitrogen, Thermo Fisher Scientific). Random hexameric primer was obtained from Qiagen (79236; QIAGEN, Hilden Germany), and 2 × qPCRBIO Probe mix Lo-ROX was obtained from Nippon Genetics (NIPPON Genetics Düren, Germany). PKM2 activator thieno-[3,2-b]pyrrole[3,2-d]pyridazinone NCGC00186528 (TEPP-46;ML265, PubChem CID 44246499) [28] was obtained from MedChemEexpress (Hoelzel-Biotech, Cologne, Germany).
Antibodies
Mouse monoclonal antibody (mAb) anti-ß-actin (ACTB) (AC-15) (A5441; Sigma), rabbit anti-Akt (9272; Cell Signaling Technology (CST), Frankfurt am Main, Germany), rabbit mAb anti phospho-Akt (Ser473) (D9E) XP® (4060; CST), rabbit mAb anti-eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) (53H11) (9644; CST), rabbit anti-phospho-4E-BP1 (Ser65) (9451; CST), rabbit anti-eukaryotic initiation factor 2 (eIF2) α (9722; CST), rabbit mAb anti-phosphor-eIF2α (Ser51) (119A11) (3597; CST), rabbit anti-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (9102; CST), rabbit mAb anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (20G11) (4376; CST), rat mAb anti-mouse F4/80 (MCA497; BioRad, Munich, Germany), mouse mAb anti-human GAPDH (GT239) (GTX627408; GeneTex, Biozol, Eching, Germany), mouse mAb anti-human glutamine synthase (6/GS) (610517; Becton Dickinson, Heidelberg, Germany), rabbit anti-mouse Hif-1α (NB100-479; Novusbio Biologicals, Wiesbaden, Germany), rabbit anti-human HIF-1α (10006421; Cayman Chemical Biomol, Hamburg, Germany), goat anti-human lamin B (sc-6216; Santa Cruz Biotechnology, Heidelberg, Germany), rat mAb anti-mouse Ly-6B.2 (7/4) (MCA771; BioRad), rat mAb anti mouse Ly-6G (1A8) (BE0075-1; Bio X Cell, Lebanon, NH, USA), rabbit mAb anti PKM1 (D30G6) XP® (7067; CST), rabbit mAb anti-PKM2 (D78A4) XP® (4053; CST), mouse mAb anti-PKM2 (1C11C7) (CoraLite (CL) 488–60268; Proteintech®, Chicago, USA), rabbit anti-phosphoPKM (Y105) (3827; CST), rabbit mAb anti-S6 ribosomal protein (5G10) (2217; CST), rabbit mAb anti-phospho-S6 ribosomal protein (Ser235/236) (2F9) (4856; CST), mouse mAb anti-Stat3 (124H6) (9139; CST), rabbit mAb anti-phospho-Stat3 (Tyr705) (D3A7) (9145; CST), goat anti-VEGF (sc-1836; Santa Cruz).
Alexa Fluor™ (AF) 594 conjugated donkey anti-goat IgG (H + L) (A-11058; Invitrogen, Thermo Fisher Scientific).
Animals
Female C57Bl/6 J (wild-type) mice were obtained from Charles River Laboratories (Charles River Laboratories, Sulzfeld, Germany). At the age of 12 weeks, the mice were caged individually, monitored for body weight, and wounded as described in the following subsection.
Wounding of mice
Wounding of mice was performed as described previously [29, 30]. Briefly, the mice were anesthetized. Subsequently, six full-thickness wounds (5 mm in diameter, 5 to 7 mm apart) were made on the backs of the mice by excising the skin and the underlying panniculus carnosus. The wounds were allowed to form a scab. An area of 7 to 8 mm in diameter, including the granulation tissue and complete epithelial margins, was excised at the indicated time points for analysis (Fig. S1A). When indicated, wounds were divided into the “wound edge” (“wound margin”), which harbors the hyper proliferative epithelia and the adjacent non-injured skin, and the “wound bed” (“inner wound”), which entails the developing granulation tissue, which consists mainly fibroblasts, polymorphonuclear granulocytes (PMN), monocytes-macrophages, and endothelial cells. The excised skins served as a control. Twelve wounds isolated from four animals were used for RNA analysis. For immunoblot analysis, eight wounds from four individual mice were used. All the animal experiments were performed according to the guidelines and approval of the local ethics animal review board (Regierungspräsidium Hessen, Darmstadt, Germany).
Cell culture
Cells were cultured in a humidified 5% CO2 environment at 37 ℃. Immortalized human keratinocyte HaCaT cell lines [31] were grown in complete DMEM (containing 10% (v/v) FCS and 1% (v/v) penicillin/streptomycin) to confluency before further passaging. Prior to stimulation, cells were brought to quiescence by serum withdrawal for 16–18 h. Human normal epidermal keratinocytes (NHEK) were isolated as previously described [32] and cultured in keratinocyte growth medium 2 (PromoCell, Heidelberg, Germany).
RNA isolation and real-time PCR analysis of gene expression
RNA isolation was carried out as described previously [30, 33]. Real-time PCR (RT-PCR) was performed using 2 × qPCRBIO Probe mix Lo-ROX (Nippon Genetics). The following pre-designed RT-PCR assays harboring the 6-carboxyfluorescein [Fam] fluorescent dye, and the 6-carboxytetramethylrhodamine quencher were purchased at Applied Biosystems (Thermo Fisher Scientific): Mm01257297_m1 (for mouse Gls), Mm00725701_s1 (for mouse Glul1), Mm00656735-g1 (for mouse G6PDx), Mm00443385_m1 (for mouse HKII), Mm00504650_m1 (for mouse Pfkfb3), Mm00437404_m1 (for mouse Vegf), Hs00173626_m1 (for human VEGF), and HS01000485g1 (for human SOCS3). RT-PCR was performed on ABI Prism 7500 Fast Sequence Detector (Applied Biosystems) as follows: 95 ℃ (2 min); 40 cycles: 95 ℃ (5 s) and 62 ℃ (30 s). Mouse Gapdh quencher (4352339E, VIC) has been use to assess the integrity of the samples. To assess the expression of mouse Pfkfb3, Pgam1, Vdac1, Glud1, and Gapdh as well as the two mouse and human Pkms isoforms by the intercalating fluorescent dye SYBR Green, 5 pmol of the respective forward and reverse primers (see Table 1) and the 2 × SYBR Select mix (4472908; Applied Biosystems) were used. RT-PCR was performed on ABI Prism 7500 as follows: 50 ℃ (2 min) and 95 ℃ (2 min); 40 cycles: 95 ℃ (15 s), 55 ℃ (30 s), and 72 ℃ (60 s). A dissociation stage, 95 ℃ (15 s), 60 ℃ (60 s) 95 ℃ (15 s), and 60 ℃ (15 s), has been added. The specificity of the PCR amplicons was confirmed by melting curve analysis for each amplification product. Analyses of RT-PCR runs were performed by the Sequence Detector software. Relative changes in the respective mRNA expression were normalized to human RPLPO (large ribosomal protein) (4310879E, VIC, human cDNA) and quantified by the 2−ΔCt or 2−ΔΔCt method.
Table 1.
Oligonucleotide sequence for real-time (RT)-PCR (SYBR Green), cloning, sequencing, and splicing assay, related to material and methods
| Oligonucleotides used for RT-PCR (SYBR Green) | |
|---|---|
| Mus musculus (mm)GAPDH | Fw: 5′ GTGTGAACGGATTTGGCCGT 3′ |
| Rev: 5′ GAGTGGAGTCATACTGGAACA 3′ | |
| mm PKM1 a | Fw: 5′ TTGTGCGAGCCTCCAGTC 3′ |
| Rev: 5′ ACTCCGTGAGAACTATCAAAGC 3′ | |
| mm PKM2 a | Fw: 5′ TTGCAGCTATTCGAGGAACTCCG 3′ |
| Rev: 5′ CACGATAATGGCCCCACTGC 3′ | |
| Homo sapiens (hs)PKM1 | Fw: 5′ TGAGGCAGCCATGTTCCAC 3′ |
| Rev: 5′ ACTCCGTCAGAACTATCAAAGC 3′ | |
| hsPKM2 | Fw: 5′ TTGCAATTATTTGAGGAACTCCG 3′ |
| Rev: 5′ GACGATTATGGCCCCACTGC 3′ | |
| mm Pgam1 | Fw: 5′ GCACTGCCCTTCTGGAATGA 3′ |
| Rev: 5’ CCTCTTCTGAGAGACCCTCCA 3′ | |
| mm VDAC1 | Fw: 5’ GGAAGACCAGCTTGCTCGT 3′ |
| Rev: 5’ CCAGCGATGTCAAAGTCCA 3′ | |
| mm Glud1 | Fw: 5’ ATCATCAAGCCCTGCAACCA 3′ |
| Rev: 5’ GCTGTAACGGATACCTCCCT 3′ | |
| siPKM2 sequence | |
| si156 b | Sense [5′ CCAUAAUCGUCCUCACCAA 3’]RNA[TT]DNA |
| Antisense [5′ UUGGUGAGGACGAUUAUGG 3′]RNA | |
Preparation of whole protein lysates
For whole protein lysate preparation, tissue biopsies and cultured cells were homogenized in a Triton X-100-lysis buffer (1% (v/v) Triton X-100, 20 mM Tris/HCl (pH 8.0), 137 mM NaCl, 10% (v/v) glycerol, 1 mM dithiothreitol, 5 mM EDTA, 10 mM sodium fluoride, 2 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 5 ng/ml aprotinin, 5 ng/ml leupeptin, and 50 nM okadaic acid). Extracts were cleared by centrifugation, and protein concentrations were determined using the BCA Protein Assay Kit (Pierce, Thermo Fisher Scientific).
Silencing of PKM2 expression by siRNA
HaCaT keratinocytes were cultured in 12-well plates (3E5 cells per well) and grown for 18 h to reach 40–60% confluency. Cells were subsequently transfected with si156 [34] (15 nM) using Lipofectamine 2000 and OptiMEM as described by the manufacturer (see Table 1).
Pharmacological activation of PKM2
HaCaT keratinocytes were cultured in 6-well plates (5.6E5 cells per well) and grown to confluency. Cells were brought to quiescence by a serum deprived medium as described above. One hour prior to stimulation, the quiescence medium was replaced with a medium containing 50 µM or 100 µM TEPP-46. A medium containing 0.2% DMSO (vehicle) has been used as a control.
Western blot analysis
Twenty-five to fifty micrograms of protein lysates were analyzed by SDS polyacrylamide gel electrophoresis (PAGE). After transfer to a 0.45-µm nitrocellulose membrane (GE Healthcare, Amersham, Thermo Fisher Scientific) and blocking in 10 mM Tris–HCl (pH 8.0), 150 mM NaCl, and 0.05% (v/v) Tween 20 (TBST) containing 2.5% (w/v) skim milk, specific proteins were identified using primary antibodies for 16 to 18 h at 4 ℃. A secondary antibody [donkey anti-goat (sc-2020; Santa Cruz), goat anti-rabbit (1706515; Bio-Rad), or goat anti-mouse (1706516; Bio-Rad)] conjugated to horseradish peroxidase, and a luminol enhancer detection system (32106, Pierce, Thermo Fisher Scientific) were used to visualize the proteins.
Blue native gel electrophoresis separation of PKM isoforms
Protein lysates (25 µg) in the Triton X-100 lysis buffer were adjusted to a blue native (BN) PAGE solution, and protein complexes were separated according to their mass on a linear 4 to 13% gradient gel for BN-PAGE as previously described [35].
Enzyme-linked immunosorbent assay (ELISA)
Cell culture supernatants were collected and cleared by brief centrifugation (300 × g, 5 min at 4 ℃). Fifty microliters of culture supernatants was used for analysis. Quantification of secreted VEGF was performed using human VEGFDuoSet ELISA kits (R&D Systems, Wiesbaden, Germany) according to the manufacturer’s instructions.
Heidenhain’s AZAN trichrome staining
Wound biopsies were isolated and fixed overnight at 4 ℃ in a 4.0% (v/v) formaldehyde solution, pH 7.0 (ROTI® Histofix 4%, Carl Roth, Karlsruhe, Germany) and embedded in paraffin. Three-micron wound sections were rehydrated and stained in 0.1% (w/v) azocarmine G (Sigma) in 1% (v/v) acetic acid for 7 min, washed briefly with 1% (v/v) acetic acid and followed by oxidation in 5% (w/v) phosphotungstic acid (Sigma) for 20 min and then briefly washed in water. The sections were then counterstained in 1.5% (w/v) aniline blue/orange G (Sigma) for 4 min, briefly washed in water, dehydrated, and mounted in Entellan (Merck, Darmstadt, Germany).
Immunohistochemistry (IHC)
The three-micron waxed wound sections were rehydrated, and targeted epitopes were unmasked by heat-induced epitope retrieval (HIER) at 100 °C for 20 min (see Table 2) and incubated at 4 °C with primary antibodies overnight. Primary antibodies were detected using either biotinylated rabbit anti-rat antibody (E0468; Dako, Hamburg, Germany), or the goat anti-rabbit sc-2018 or donkey anti-goat sc-2023 antibody of the avidin–biotin-peroxidase ImMunoCruz ABC staining system (Santa Cruz) (see Table 2). Antigens were stained by 3,3-diaminobenzidine-tetrahydrochloride (Sigma Aldrich) or by streptavidin–alkaline phosphatase (D0396; Dako) via Permanent AP Red, ZUC001-125 (Zytomed, Berlin, Germany) as chromogenic substrates. Finally, the sections were counterstained with hematoxylin (Mayer’s hematoxylin, Applichem. 254,766.1611 (Applichem, Darmstadt, Germany) and mounted in an aqueous mounting medium, Aquatex. For PKM2 immunostaining, rehydrated tissue sections were washed in IHC-Tris buffer saline (300 mM NaCl, 50 mM Tris–HCl pH 7.6) containing 0.1% (v/v) Tween 20 (TBST), and epitopes were unmasked by boiling in 10 mM citrate (pH 6.0) for 20 min. Tissue’s peroxidases were inactivated by 3% (v/v) H2O2, nonspecific epitopes were blocked using an animal-serum free blocking solution (CTS 15,019), and the sections were incubated with rabbit mAb anti-PKM2 diluted 1:300 in SignalStain® antibody diluent (CST) at 4 °C overnight. PKM2 epitopes were detected with SignalStain® HRP, Rabbit detection reagents (CST), and stained with SignalStain® DAB substrate (CST). The stained sections were counterstained as above or processed for double immunostaining. For Hif-1αIHC staining, tissue sections were blocked using a biotin/avidin block solution (X0590; Dako) before incubation with a primary antibody and treated by a Dakosignal amplification solution (K1500; Dako) prior to staining by DAB.
Table 2.
Antibodies used for immunohistochemistry
| Antigen | Epitope retrieval procedure | Primary antibody | Biotinylated secondary antibody/solution |
|---|---|---|---|
| F4/80 | HIER, 10 mM citrate (pH 6.0) | 1:200 | Rabbit anti-rat 1:200/DCSb |
| HIF-1α Cayman 10006421 | HIER, Dako 10 mM citrate (pH 6.0) | 1:10,000 | Goat anti-rabbit 1:1000 Vector BA-100/TBST |
| Ly-6B.2 | HIER, 10 mM citrate (pH 6.0) | 1:400 | Rabbit anti-rat 1:200/DCS |
| Ly-G | HIER, 10 mM citrate (pH 6.0) | 1:150 | Rabbit anti-rat 1:200/DCS |
| PKM2 | HIER, 10 mM citrate (pH 6.0) | 1:300 | CST 8114 |
| VEGF | HIER, Dako TRS a S1699 | 1:100 | Donkey anti-goat 1:200 ABC |
aTarget retrieval solution; DCS
bantibody diluent solution (AL120R; DCS, Hamburg, Germany)
Immunofluorescence (IF)
Three-micron tissue sections of day 5 wound biopsies were unmasked by HIER using a Dako-target retrieval solution (S1699; Dako), as described above, and incubated with CL 488 labeled mouse mAb anti PKM2 (1:100) and goat anti VEGF (1:100) at 4 °C overnight. Goat anti VEGF antibodies were detected using AF 594 conjugated donkey anti-goat (1:200). Nuclei were stained with DAPI (Sigma) at 1 µg/ml for 60 s, and sections were mounted in a Dako fluorescent mounting medium (S3023; Dako).
RNA Scope in situ hybridization
RNAScope in situ hybridization on paraffin sections of day 5 wound biopsies was performed using a BaseScope™ duplex detection reagent kit (323810, ACD, Bio-Techne, Minneapolis, MN, USA) and RNAScope H2O2 and protease reagents (322381, ACD). Tissue sections were unmasked by heat, succeeded by protease IV treatment, each for 15 min exactly according to the manufacturer’s instructions (document number 323800-USM, ACD). The BA-Mm- Vegfa-E2E3 (706421, ACD) and BA-Mm-Pkm-tv2-CE9E10 (714,131-C2, ACD) probes were used to codetect the mRNA expression of Vegf and Pkm2, respectively, in the wound sections.
Assessment of PKM2-expressing Ly6B.2 and F4/80 cells in wound sections
Sections of day 3 or day 5 wound biopsies co-stained against activated PMN and monocytes (Ly6B.2) or macrophages (F4/80) and PKM2 were ascertained in four distinct regions of wound edge and wound bed. The percentages of PKM2 expressing cells out of Ly6B.2 or F4/80 positive cells have been assessed.
Statistical analysis
Data are shown as mean ± SD or ± SEM as indicated. To compare two conditions, unpaired Student’s t-test was used for normally distributed homoscedastic data. To compare more than two conditions with unpaired samples, the following statistical tests were used: one-way ANOVA with Bonferroni’s (multiple comparison test) or Dunnett’s (comparison of each condition with control) post-hoc tests for normally distributed homoscedastic data and Kruskal–Wallis test with Dunn’s post-hoc test for data without normal distribution. Data were analyzed using GraphPad Prism software version 5 or 8 (GraphPad Software Inc., San Diego, CA). A p-value < 0.05 was considered statistically significant. Significances were indicated as the following: * P < 0.05, ** P < 0.01, and *** P < 0.001.
Results
Temporal and spatial activation of metabolic flows during cutaneous wound healing
The healing of a cutaneous wound is a controlled acute inflammatory process [9]. With minor differences according to the location of the wound (anterior, medial posterior; Fig. S1B and C), the injured skin is nearly perfectly healed within 13 days (Fig. S1A–C). As inflammation and immunity are instructed by metabolic cues [37–39] and inflammatory response is required for proper healing, attention has been drawn in recent years to the metabolic aspects of cutaneous wound repair [39]. Whereas most of our previous works, as well as the majority of other studies dealing with the involvement of various metabolites and/or their respective enzymatic mediators, a mechanistic look at the spatial and temporal roles of metabolic mediator(s) during skin repair is yet inadequately addressed and therefore attracted our attention. We first observed an overall increase in the mRNA expression of enzymes related to glycolysis: Gapdh and Pfkfb3, as well as oxidative phosphorylation: Vdac and the associated hexokinase HkII, which reflected mitochondrial expansion, during the early to the late inflammatory phases i.e., days 3 to 7 after injury (Fig. S2A and B, left panel). Interestingly, the expression of the M2 splice variant of the last glycolytic enzyme, pyruvate kinase, was continuously induced even on day 13 after injury when wound closure is attained, whereas the M1 variant remained unaltered during the whole healing process (Fig. 1A, left panel; Fig. 1B and C). Besides, an increase in glutaminase and glutamate dehydrogenase gene expression reflects an enhanced glutamine anaplerosis of the TCA pathway (Fig. S2C, left panel), which points to arise in anabolic demand of the injured skin. Shunting metabolites toward the pentose phosphate pathway (PPP) seems confined to day 5, as both central genes of this pathway, G6pdx and Pgam1, were found to be specifically expressed at this time point (Fig. S2D, left panel), indicating an increase in oxidative stress during this stage [40]. Markedly, glutamine synthase mRNA and protein were robustly induced immediately after injury and remained at a high level throughout the healing process (Fig. S2E). This fits well with the central role of glutamine metabolism as a carbon source, enabling proliferating cells to utilize glycolysis and TCA intermediates as a source of building blocks for cellular anabolic pathways [41]. Tissue sampling reveals a higher expression of metabolic genes at the wound bed subset than in the wound edge at least until day 5 post wounding, indicating that inflammatory cells are the main source for this pattern of metabolic gene expression. However, 5 days after wounding, an increased expression of these messengers was observed at the wound edge, parallel to the proliferation boost in keratinocytes at the migrating tongue around this time point [9] (Fig. S2A–D, middle and right panels).
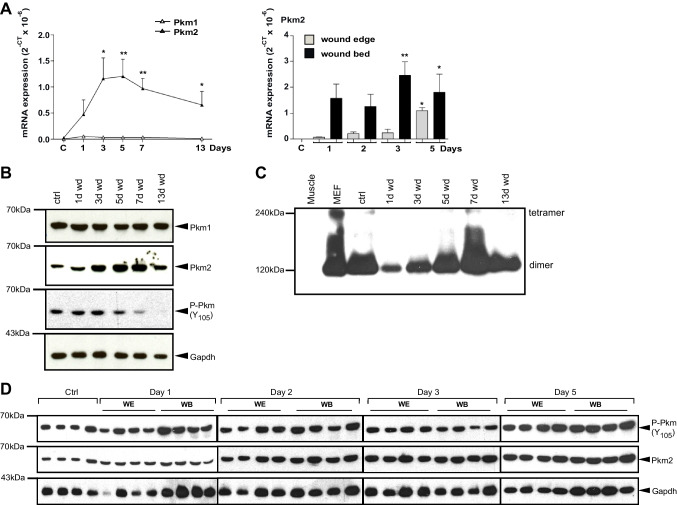
Fig. 1.
Expression of Pkm isoforms during cutaneous wound healing. A Quantification of Pkm1 and 2 mRNA expression in murine wounds as assessed by RT-PCR (left panel) and Pkm2 mRNA expression in wound edge (wound margin) and wound bed (inner wound granulation tissue) compartments (right panel) at the indicated time points after injury. Non-wounded skin served as a control (ctrl skin). Bars indicate means ± SD obtained from three wounds (n = 3) isolated from four individual animals (n = 4). **P < 0.01, *P < 0.05 (ANOVA) as compared to control skin (Dunnett’s post-hoc test). B Immunoblot analysis for Pkm isoforms and phosphorylated Pkm(Y105) protein expression in murine wounds (wd) at the indicated time points after injury. C The stoichiometry of PKM2 as determined by immunoblot analysis of BN-PAGE at the indicated time point. Protein lysates prepared from mouse muscle tissue (Muscle) or mouse embryonal fibroblasts (MEF) were used as a control for Pkm2 expression and stoichiometry. D Immunoblot analysis of Pkm2 and phosphorylated Pkm (Y105) protein expression in separated wound edge (WE) and wound bed (WB) compartments at the indicated time points after injury. Each individual time point constitutes of wounds (n = 8) from individual mice (n = 4). The expression of GAPDH was used as a loading control in B and D. C/ctrl: control skin
Increased expression of Pkm2 in mononuclear phagocytes and wound keratinocytes
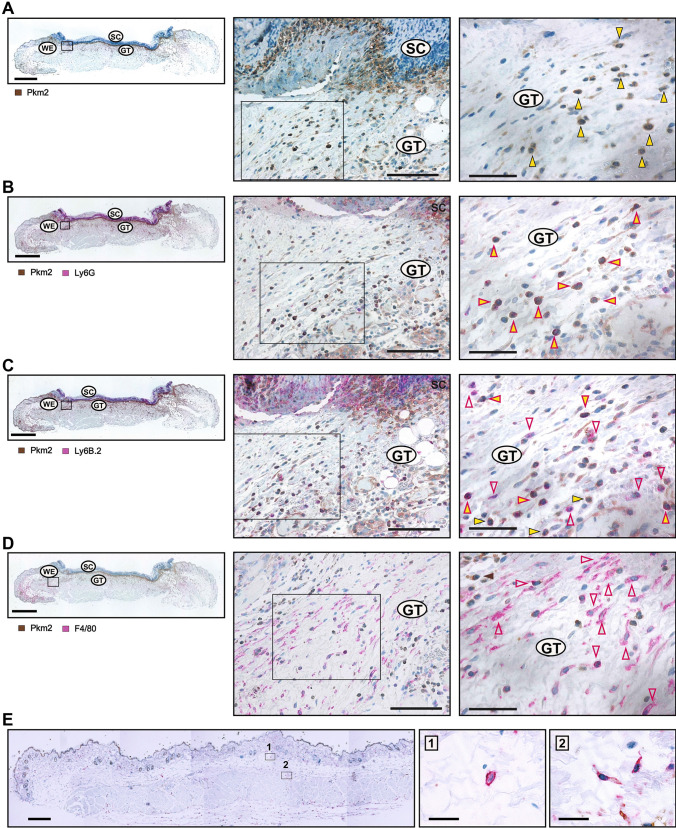
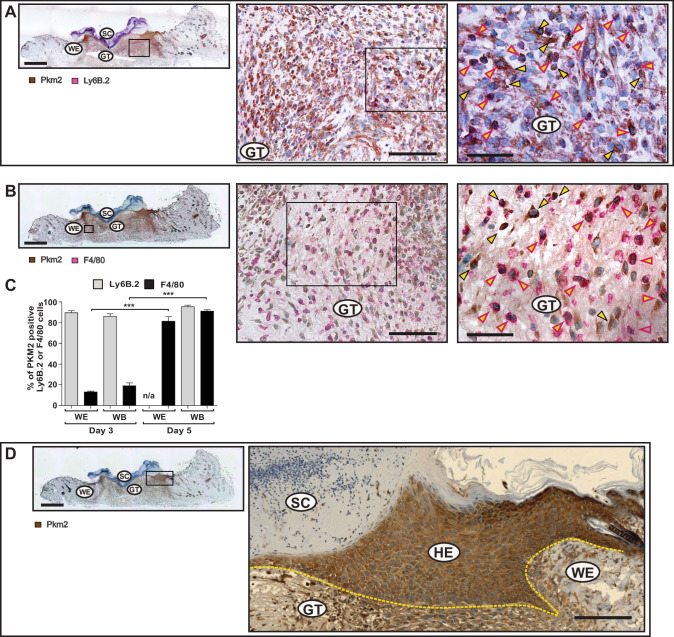
The divergent expression of Pkm2 as compared to the Pkm1 splice variant in the wound and its various proposed metabolic and non-metabolic activities [42, 43] prompted us to focus on the function of Pkm2 in cutaneous wound healing. A pro-angiogenic role as a neutrophile secreted factor was indeed suggested for Pkm2 [44]. Yet, our results show an increase and sustained expression of Pkm2 mRNA and protein in later inflammatory stages, when neutrophils decline, whereas proliferation, differentiation, and polarization of keratinocytes, fibroblasts, and macrophages, respectively, take over [45] (Fig. 1B and C). Indeed, in the wound, Pkm2 is mostly expressed as a dimer and hence is metabolically less active (Fig. 1C and Fig. S3A). Furthermore, on day 1 after the injury, stronger phosphorylation on tyrosine 105 (Y105) and hence less activity of Pkm2 have been confined to the wound bed (Fig. 1D). This compartment is mainly populated through migrated neutrophils and platelets at this early inflammatory stage [9]. In this context, it should be noted that Y105 phosphorylation may broaden to the M1 isoform as well (Fig. S3B). Overall, an increasing Pkm2 mRNA and protein expression is detected in the wound edge, which is comprised in part of the hyperproliferative epithelium (Fig. 1A, right panel, and D). Immunohistochemistry staining displays that on day 3 post wounding, leukocytes expressing PKM2 are mainly represented by inflammatory neutrophils and monocytes, which populate the complete wound (Fig. 2A–C). In this early inflammatory phase, wound macrophages scarcely express Pkm2 (Fig. 2D), similar to their counterparts in non-injured tissue, the skin resident macrophages (Fig. 2E). On day 5, inflammatory neutrophils/monocytes are merely at the wound bed (Fig. 3A and C). However, in this mid inflammatory phase, macrophages throughout the whole wound express Pkm2 (Fig. 3B and C). The significance of this transformation of expression pattern is not clear and may reflect a part of polarization process of wound macrophages. Importantly, this staining reinforced that proliferating keratinocytes of the migrating epithelial tongue are a dominant source of PKM2 (Fig. 3D).
Fig. 2.
Pkm2 Expression in neutrophils/activated monocytes in early inflammatory wound tissue. Serial sections from day 3 wounds isolated from C57BL/6 J mice using antibodies directed against Pkm2 (brown signals, field yellow arrows) alone (A) or in conjugation with antibodies directed against: Ly6G (neutrophils; red signals, red arrows) (B), Ly6B.2 (activated PMN/monocytes; red signals, red arrows) (C), or F4/80 (macrophage; red signals, red arrow) (D). A full wound section is shown at the left side (scale bars: 1000 µm) and the marked region is shown in the middle part (scale bars: 100 µm). A detailed image at higher magnification is shown at the right side (scale bars: 50 µm). WE, wound edge; GT, granulation tissue; SC, scab. A non-injured control-skin, co-stained for Pkm2 and F4/80 is shown in (E) (left, scale bar: 500 µm). Higher magnification of resident skin macrophages at [1] and [2] are shown at the right (scale bar: 20 µm)
Fig. 3.
Increasing spread of Pkm2 expression in macrophages and keratinocytes during healing of cutaneous wound. Serial sections from 5-day wounds isolated from C57BL/6 J mice using antibodies directed against Pkm2 (brown signals, field yellow arrows) in conjugation with antibodies directed against: Ly6B.2 (activated PMN/monocytes; red signals, red arrows) (A), or in conjugation with antibodies directed against F4/80 (macrophage; red signals, red arrow) (B). A full wound section is shown at the left side (scale bars: 1000 µm) and the marked region is shown in the middle part (scale bars: 100 µm). A detailed image at higher magnification is shown at the right side (scale bars: 50 µm). (C) Evaluation of Pkm2 expression in neutrophils and mononuclear phagocytes in wound sections obtained from three different mice (n = 3) on days 3 and 5 post injury. Ly6B.2 or F4/80 cells were assessed for expressing Pkm2 in 0.72 mm2. At least four images taken from wound edge (WE) and two from wound bed (WB) were evaluated for each section. ***P < 0.001; (one-way ANOVA) (Bonferroni’s post-hoc test). Ly6B.2 positive cells are rarely detectable in the wound edge on day 5 post injury and hence not assessed (n/a). Staining of Pkm2 alone is shown in (D). A full wound section is shown at the left side (scale bars: 1000 µm) and a detailed image of the marked region is shown at the right side (scale bars: 100 µm). GT, granulation tissue; SC, scab; HE, hyperproliferative epithelia; WE, wound edge; WB, wound bed. The HE is marked by a yellow dash line
Concomitant expression of Vegf and Pkm2 in wound keratinocytes
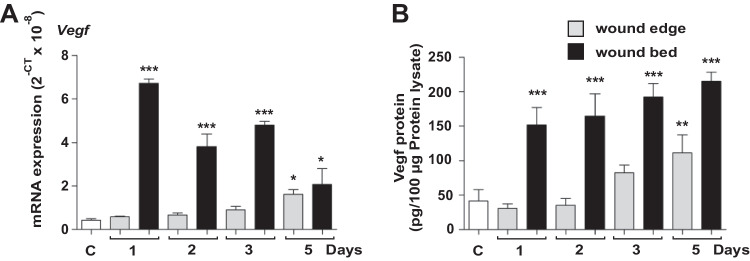
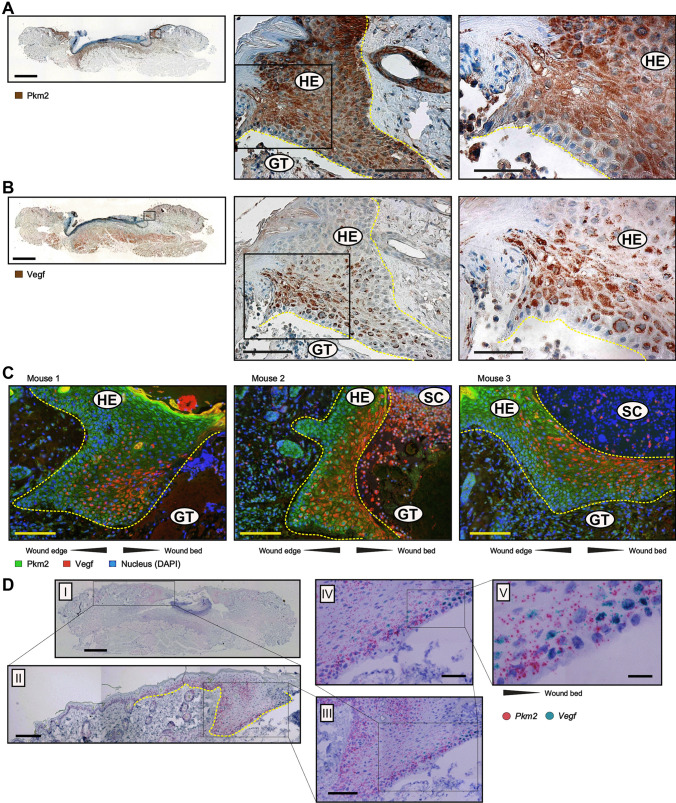
The expression of Vegf mRNA and protein was temporally and spatially congruent with Pkm2 expression in both wound edge and wound bed sites (Fig. 1A, right panel, Fig. 4A and B, and Fig. S4). During mid-inflammation (mainly on day 5), macrophages in the wound bed are a key source for Vegf in this wound compartment [45]. Of note, on day 5, Vegf expression is significantly increased in the wound edge (Fig. 4). We have previously described keratinocytes of the hyperproliferative epithelium as a major source of VEGF in wound tissue [32]. Immunostaining of serial sections from a wound on day 5 after injury shows Vegf expression co-localization with Pkm2-positive keratinocytes on the wound edge, albeit adjacent to the front of the migrating epithelial tongue (Fig. 5B–D). Similar results were obtained already on day 3 post wounding (not shown). Noteworthy, this expression pattern of Vegf in wound keratinocytes is reminiscent of the suppressor of cytokine signaling (Socs)3 expression in this compartment [46]. We further applied immunofluorescence (IF) staining to gain more insight on how Vegf production is interconnected to Pkm2 expression in keratinocytes of the proliferative epithelium. We used a CoraLite 488-conjugated PKM2-specific mouse monoclonal antibody in combination with an Alexa Fluor 594-conjugate donkey anti goat antibody to co-stain Pkm2 and Vegf, respectively. Co-IF staining of day 5 wound sections from three different mice confirmed the enhanced expression of Pkm2 in the proliferative epithelium and reveals the consistent expression of Vegf in this compartment, mainly in keratinocytes lying toward the wound bed (Fig. 5C). Of note, the patchy expression of the proangiogenic transcription factor Hif-1α is mostly detected in keratinocytes lying adjacent to the wound edge (Fig. S6A), as has been shown, also by Gilles Ponzio and co-workers [47]. Finally, mRNA in situ hybridization using BaseScope™ duplex technology further strengthened this observation and validated the continuously increased expression of Pkm2 along the rolled proliferative tongue together with Vegf mRNA expression in the keratinocytes sidewise to the wound bed (Fig. 5D).
Fig. 4.
Spatial expression of VEGF during early and mid-wound-inflammation. Quantification of Vegf mRNA (A) and Vegf165 protein (B) expression in murine wounds as assessed by RT-PCR (A) and ELISA (B), respectively, in separated wound edge (WE) and wound bed granulation tissue (WB) compartments at the indicated time points after injury. Non-wounded back skin served as a control (“C”, left-most columns in A and B). Bars indicate means ± SD obtained from wounds (n = 3) isolated from four individual animals (n = 4). ***P < 0.001, **P < 0.01, and *P < 0.05 (ANOVA) as compared to control skin (Dunnett’s post-hoc test). C: control skin
Fig. 5.
Co-expression of PKM2 and VEGF in keratinocytes of the hyperproliferative epithelial wound tissue. Serial sections from 5-day wounds isolated from C57BL/6 J mice using antibodies directed against Pkm2 A or antibodies directed against Vegf B (brown signals). A full wound section is shown at the left side (scale bars: 1000 µm) and the marked region is shown in the middle part (scale bars: 100 µm). A detailed image at higher magnification is shown at the right side (scale bars: 50 µm). C Immunofluorescence (IF) co-staining of Pkm2 (CL 488, green) and Vegf (AF594, red) in wound sections of day 5 taken from three different mice (scale bars: 100 µm). Nuclei were assessed by DAPI staining (blue). The hyperproliferative epithelium (HE) is marked by yellow dash line. Note that Vegf staining is confined toward wound bed in all three epithelia. GT, granulation tissue; SC, scab. D RNA in situ hybridization of Pkm2 (red dots) and Vegf (turquoise dots) mRNA on day 5 wound. Complete wound is shown in I (scale bar: 1000 µm). Enlargement of the left part of the wound is shown in II (scale bar: 200 µm). Expansion of the left hyperproliferative epithelium is shown in III (scale bar: 120 µm). The innermost part of the hyperproliferative epithelium is depicted in IV (scale bar: 60 µm) and a detailed image at higher magnification is shown in V (scale bare: 20 µm)
The requirement of PKM2 in keratinocytes for growth factor activation of VEGF expression
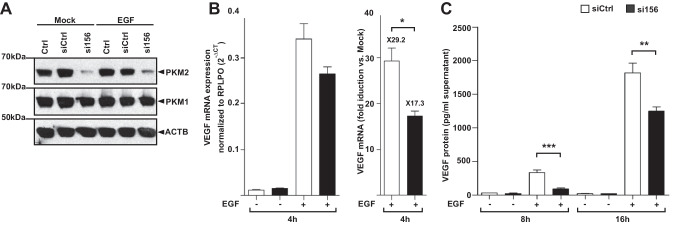
Elevated expression of PKM2 in the hyperproliferative epithelium may suggest a functional role for PKM2 driving VEGF expression in keratinocytes. Similar to normal human epidermal keratinocytes (NHEK), HaCaT cells express higher levels of PKM 1 and 2 isoforms, of which PKM2 is more strongly expressed in both strains, and this phenomenon is intensified in HaCaT as emerged from RNA and protein data (Fig. S5A and B). In cells expressing HIF-1α under hypoxic conditions or due to malfunctioning HIF-1α degradation signal, as in von-Hippel-Lindau (VHL) null renal carcinoma cells (RCC4), PKM2 has been shown to regulate the HIF-1α metabolic and angiogenic settings via enhancing HIF-1α binding to hypoxia response elements (HREs) within metabolic regulatory genes such as LDHA and PDK1 as well as VEGF [36]. However, the expression of Hif-1α is uneven in keratinocytes of the hyperproliferative epithelium tongue (Fig. S6A) [47], implying an additional mode to accelerate VEGF expression in this compartment, apart from HIF transcriptional program. Along this line, the expression of HIF-1α in HaCaT cells is restricted to hypoxia or hypoxia mimicry by CoCl2 [48] (Fig. S6B). To ascertain the contribution of PKM2 in keratinocyte-driven angiogenic program, we have performed a knockdown of PKM2 using small interfering (si) RNA designed to target PKM2 specifically [34] without affecting PKM1 levels (Fig. 6A), and this approach has been employed to study the role of PKM2 in VEGF expression by HaCaT keratinocytes. To this end, we stimulated HaCaT cells with epidermal growth factor (EGF), a key effector of epithelial cells during wound repair [49] and a potent activator of VEGF165 splice variant (hereafter VEGF) expression and secretion in cultured keratinocytes [50, 51]. Partial knockdown of PKM2 dampened EGF to induce VEGF mRNA (Fig. 6B), and this effect was more pronounced at the VEGF protein level (Fig. 6C). Increased PKM2 dimer and tetramer formation by PKM2 activator TEPP-46 has no detectable effect on VEGF expression (Fig. S7), indicating the importance of the total PKM protein rather than their overall stoichiometry for maximal induction of VEGF in HaCaT keratinocytes.
Fig. 6.
Silencing of PKM2 impedes EGF-induced VEGF expression in HaCaT keratinocytes. A HaCaT keratinocytes were transfected with siRNA targeting exon 10 (si156) or control siRNA (siCtrl), and harvested after 48 h; 25 µg of cell lysates were analyzed by immunoblot for the expression of PKM isoforms. Whereas PKM2 protein expression is markedly reduced in si156 transfected cells (upper blot), the expression of PKM1 remained unchanged (middle blot). The expression of ß-actin was used as a loading control. Expression levels of both PKM-isoforms are not affected by EGF (30 ng/ml) stimulation. B HaCaT keratinocytes transfected with siRNA against PKM2 (si156) or control siRNA (siCtrl) were treated with EGF (30 ng/ml) for 4 h, and VEGF mRNA was quantified as compared to RPLPO housekeeping gene by RT-PCR (left). EGF stimulation of VEGF mRNA expression in HaCaT keratinocytes is decreased upon silencing of PKM2 as compared to stimulation of control siRNA transfected cells (right). Bars indicate means ± SD obtained from three independent experiments (n = 3) performed in triplicate; *P < 0.05 (unpaired Student’s t-test). C The release of VEGF protein in culture supernatants of HaCaT cells transfected with control siRNA (siCtrl) or siRNA against PKM2 (si156) and stimulated by EGF (30 ng/ml) for the indicated time points was assessed by ELISA. VEGF protein is expressed as pg per ml supernatant. Bars indicate means ± SD obtained from supernatants of three independent cell culture experiments performed in triplicate; ***P < 0.001 and **P < 0.01 (unpaired Student’s t-test)
Silencing of PKM2 partially alters EGF signal transduction in keratinocytes
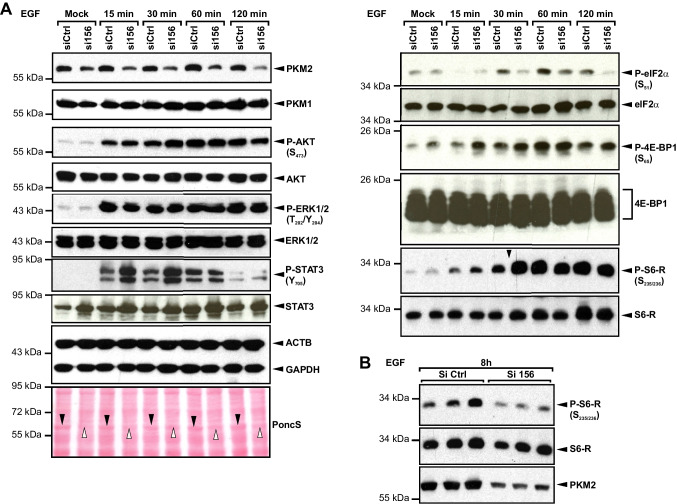
As pointed out, PKM2 has been shown to act through modulation of the transcriptional program to regulate glucose metabolism, Warburg effect, growth, and survival [52, 53]. In our setting, transient reduction of PKM2 expression in keratinocytes hampers full induction of transcription and, to a greater extent, translation of VEGF by EGF. Therefore we examined the effect of PKM2 attenuation on the activation of key signal transducers that were shown to regulate VEGF production in keratinocytes, following growth factor stimulation in vitro and during wound healing in vivo [54]. The silencing of PKM2 neither affected AKT nor ERK activation; however, it increased STAT3 phosphorylation and, to a minor degree, its expression (Fig. 7A, left panel). Unexpectedly, the eukaryotic initiation factor 2 (eIF2)α, which stimulates the translation of capped mRNA (e.g. VEGF), was found to be hypophosphorylated and hence activated. On the other hand, the repressor of cap-dependent, 4E-BP1 was slightly more phosphorylated and therefore inhibited upon knockdown of PKM2 (Fig. 7A, right panel). Impressively, the activation of the initiation of translation machinery-accompanied intense STAT3 phosphorylation was much stronger upon stimulation of PKM2 knockdown cells with bona fide wound healing cytokines IL-6 and IL-22 (Fig. S8). The higher activation of the translational machinery could represent a compensatory response in PKM2 knockdown cells. Recently, activation of AKT has been proposed to protect cancer cells from growth inhibition induced by PKM2 knockdown [55]. As indicated here, AKT activation remained unchanged upon PKM2 silencing but additional compensation mechanisms cannot be ruled out. Even more surprising was the earlier phosphorylation of S6 ribosomal protein (S6RP) upon EGF stimulation in cells with knockdown of PKM2, which peaked already at 30 min (Fig. 7A, right panel, penultimate blot). However, this activation state of S6RP declined more rapidly in PKM2 knockdown cells than in the control cells (Fig. 7A, right panel, penultimate blot, and Fig. 7B). Earlier deactivation of S6RP halts the ribosome in PKM2 knockdown cells and could be accounted for the pronounced reduction of VEGF protein level in these cells. Of note, visualization of the blotted protein lysates of control- and PKM2-siRNA transfected cells using Ponceau S staining revealed the abundance of PKM2 in keratinocytes. The weakening of a 60-kDa protein band, corresponding to PKM2, could be easily seen (Fig. 7A, left panel, last blot and Fig. S8 left and right panels, last blots).
Fig. 7.
Silencing of PKM2 dysregulates growth factor activation of STAT3 and translational regulators. A HaCaT keratinocytes were transfected with PKM2-specific siRNA (si156) or control siRNA (siCtrl), and stimulated after 48 h by 30 ng/ml EGF for the indicated time points, and 25 µg of cell lysates were analyzed by immunoblot to assess the phosphorylation state of PKB (AKT) (Ser473), ERK1/2 (Thr202/Tyr204), STAT3 (Tyr705), eIF2α (Ser51), 4E-BP1 (Ser65), and S6 ribosomal protein (S6RP) (Ser235/236), and the expression level of the corresponding total protein. The silencing of PKM2 expression was verified by immunostaining of PKM2 and PKM1 proteins (top and second blots, respectively, left panel). The expression of ß-actin and GAPDH was used as a loading control. Ponceau S staining (PoncS, left panel, last blot) shows a decrease of a 60-kDa band (black downward arrowheads), subsequent to transfection with si156 (white upward arrowheads). A representative blot from three independent experiments is shown. B The phosphorylation state of S6RP (Ser235/236) at 8 h of stimulation by 30 ng/ml EGF was assessed in 25-µg cell lysate. Staining of total S6RP has been used as a loading control. PKM2 expression is shown in the lower blot. Results from three (n = 3) independent experiments are presented
Silencing of PKM2 augments STAT3 signaling in keratinocytes
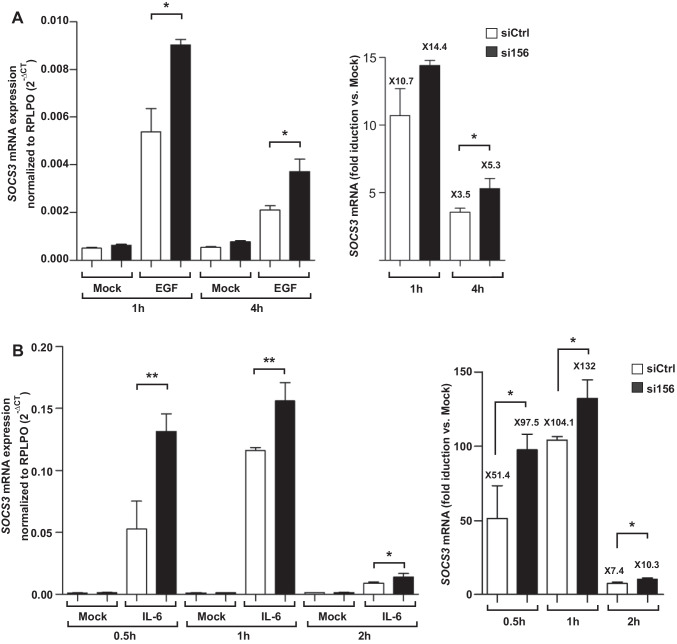
The enhanced phosphorylation of STAT3 by growth factor impetus or inflammatory stimuli is a part of an intact activated STAT3 signaling cascade. Nevertheless, the pronounced phosphorylation of STAT3 following stimulation of PKM2-silenced keratinocytes is not explicable. Therefore, to confirm that this amplified phosphorylation is also functionally manifested by increasing STAT3 transcriptional activity, we analyzed the expression of the suppression of cytokine signaling (SOCS)3, a surrogate marker for STAT3 activation. Remarkably, the silencing of PKM2 significantly augmented the activation of SOCS3 mRNA transcription in response to EGF (Fig. 8A). Stimulation of SOCS3 expression is only secondary to EGF signaling cascade, ensuing STAT3-activation. As a rapid immune modulator, SOCS3 is primarily induced by inflammatory cytokines (via STAT3) in the course of a negative feedback control mechanism. Indeed, aligned with the increased induction of STAT3 phosphorylation by pro-inflammatory cytokines in PKM2 knockdown cells (Fig. S8), silencing of PKM2 sharply enhances IL-6 stimulation of SOCS3 mRNA expression (Fig. 8B). This raised the induction of SOCS3, as opposed to the reduced stimulation of VEGF implying a specific effect of PKM2 to accomplish VEGF production in keratinocytes. Generally, inflammatory stimuli escalate wound angiogenesis [56] and IL-6 amplifies growth factor-stimulation of VEGF expression in keratinocytes (Fig. S9), ruling out an inhibitory effect of SOCS3 and probably other STAT3-regulated gene products on VEGF expression.
Fig. 8.
Enhanced activation of SOCS3 mRNA in PKM2 knockdown keratinocytes. A The expression of SOCS3 mRNA in EGF-stimulated HaCaTs was assessed by RT-PCR (left), and the fold induction of enhanced expression of SOCS3 in PKM2 knockdown cells as compared to stimulation of control siRNA transfected cells was evaluated by the 2−ΔΔCt method (right). Bars indicate means ± SD obtained from three independent experiments (n = 3) performed in triplicate; *P < 0.05 (unpaired Student’s t-test). B The expression of SOCS3 mRNA upon stimulation with IL-6 (20 ng/ml) was quantified by RT-PCR (left). IL-6 stimulation of SOCS3 mRNA expression is markedly increased upon targeting PKM2 expression as compared to stimulation of control siRNA transfected cells (right). Bars indicate means ± SD obtained from three independent experiments (n = 3) performed in triplicate; **P < 0.01 and *P < 0.05 (one-way ANOVA) (Bonferroni’s post-hoc test). The fold of stimulation in A and B is indicated on top of the bars (A and B)
Discussion
Wound healing is a multi-stage process orchestrated by complicated interactions between cellular and humoral inflammatory cues. It involves cell infiltration, proliferation, expulsion, and death as well as destruction of damaged structures and assembly of scaffolds for tissue repair [57]. In a way, wound healing reminisces neoplasia, but resolved wound healing is a well-coordinated process, whereas neoplasia is uncontrolled growth [27]. The altered metabolic landscape of cancer cells as compared to normal tissue has drawn the attention of researchers in the field of tumor biology, since the early studies of Otto Warburg almost a century ago [2, 58]. In analogy to cancer, altered metabolism was shown to affect the healing outcome, and along with this finding, injured tissue and neoplasia have been found to share common nodes [10, 54, 59]. Initially, this work tended to gain an overall idea on the metabolic nature of wound inflammation. Through following the expression profile of central enzymes involved in glycolysis, oxidative phosphorylation, the pentose phosphate shunt, and glutamine anaplerosis, a mixed picture has emerged, suggesting a spatially and temporarily regulated co-activation of metabolic pathways that support various polarization modes and not a particular one as we understand it today in other inflammatory conditions [60]. Notably, the expression of enzymes connected to the pentose phosphate shunt was confined to day 5 after injury, indicating a sharp rise in oxidative stress and the demand to counterbalance it at this stage. Of interest, we analyzed the expression mode of Pkm2, which, as opposed to its constantly expressed alternative splice variant and metabolic active Pkm1, was markedly induced up to day 3 after wounding. Whereas the relatively high phosphorylated state of Pkm2 at an early time point implies low activity, the injured tissue is governed by neutrophils. Its increased expression in macrophages and keratinocytes of the hyperproliferative epithelia, later on during mid-inflammatory phase as detected histologically, indicates an acute role of Pkm2 in inflammation and re-epithelialization of the damaged skin. Accordingly, the dimer stoichiometry of Pkm2 (Fig. 1) implies a reduced pyruvate kinase activity that is connected to intense cell proliferation [4]. The hyperproliferative epithelium was indicated as a source for Vegf to drive angiogenesis during acute healing [32]; hence, the co-localization of Vegf expression in a subset of keratinocytes in this compartment was not unexpected (Fig. 5). However, as opposed to the patchy expression pattern of the pro-angiogenic transcription factor Hif-1α, proximal to the wound edge (Fig. S6A),Vegf is homogenously expressed in keratinocytes proximal to the wound bed, as confirmed by IF and RNA in situ hybridization (Fig. 5C and D). This further supports the notion that the factors released from the wound bed (e.g., cytokines/chemokines) are required to stimulate Vegf expression in keratinocytes of the hyperproliferative epithelium. A strong association between PKM2 and VEGF expression has been proposed by Luo et al. in VHL null renal cell carcinoma a decade ago [36]. This important work provided an exo-metabolic mechanistic explanation that connects PKM2 to the enhancement of HIF-1α-dependent genomic reprogramming, including VEGF expression. However, as noted above, Vegf expression in wound keratinocytes does not necessarily correlate with the expression of Hif-1α (Fig. S6A) [48], and HaCaT keratinocytes do not express HIF-1α unless stimulated by hypoxic or inflammatory signals (Fig. S6B) [61]. In this context, we may recall the research by Vander Heiden and his co-workers who showed that in mouse tumor-derived cell lines, PKM2 was dispensable for hypoxia-induced gene expression [62]. In addition, Luo and colleagues targeted exon 6 to knockdown PKM gene product(s) and hence silenced both PKM1 and 2. Acute silencing of PKM2 in HaCaT by siRNA, specifically targeting exon 10, reveals a reduction in stimulation of VEGF mRNA and, to a greater extent, protein expression upon induction by growth factors. Importantly, the silencing of PKM2 per se leads to defined changes in signaling molecules, which we do not fully understand. Whereas STAT3 and eIF2α were induced, 4E-BP1 was inhibited, and the activation of S6RP was dampened, albeit at a later time point. While the reduction of S6RP activation parallel to PKM2 silencing matches the drop in VEGF-stimulated levels, induction of eIF2α concomitant to suppression of 4E-BP1 contradicts this result. Counting the inducing versus suppressing roles of eIF2α and 4E-BP1, respectively, in the translation of capped mRNAs, it was suggested that the above modification may reflect a cellular stress response, which leads at a later time point to attenuation of S6RP activation.
Still, another ambiguity is the enhancement of STAT3 phosphorylation following growth factor or inflammatory stimuli of PKM2 knockdown cells. The enhanced activation of STAT3 due to acute silencing of PKM2 manifests in higher transcriptional activation of SOCS3 mRNA by EGF or inflammatory stimuli. However, decreasing of VEGF production due to increased expression of SOCS3 can be ruled out as IL-6 rises the stimulation of VEGF expression by EGF, concomitantly to induction of SOCS3 mRNA expression (Figs. 8B and S9). STAT3 was among the first proteins that were appeared to be phosphorylated and activated by nuclear PKM2 [63]. Later on, this capability of PKM2 to phosphorylate effector proteins and thereby modulate their activity has been revised [24]. Nevertheless, activation of STAT3 by PKM2has been further suggested in many works. Recently, Alves-Filho and his co-workers showed that Pkm2 contribute to autoimmune inflammation in experimental autoimmune encephalomyelitis (EAE) via fine-tuning Stat3 activation [64]. Pharmacological activation of Pkm2 metabolic action via induction of tetramer formation by the small molecule activator TEPP-46 reduces Pkm2 nuclear translocation and Stat3 phosphorylation, thereby limiting CD4+T helper 17 (Th17) cell formation and ameliorating EAE. Similarly, Luke O’Neill and his co-workers provided evidence that tetramerization of Pkm2 by TEPP 46 limits the development of both Th17 and Th1 cells [65] and thereby restrains aggravating inflammation in EAE. However, their finding was independent of Stat3 or Stat1 activation. Similarly, metabolic activation of Pkm2 has been shown to restrain pro-inflammatory activation of murine macrophages by reducing the formation of Hif-1α-Pkm2 ternary complex at IL-1β promoter [66] or induce mitochondrial biogenesis [67]. Interestingly, activation of PKM2 tetramer formation TEPP-46 did not alter VEGF production in EGF-stimulated HaCaT cells (Fig. S7). EGF alone does not alter the stoichiometry of PKM2 (Fig. S7B) and barely induces phosphorylation of tyrosine 105 as the inflammatory cytokine IL-6 does (not shown). It is therefore tempting to assume that in keratinocytes the acute amount of PKM protein determines the full activation of VEGF expression. Curiously, on examining Ponceau S staining blotted HaCaT protein, we were surprised to note the weakening of a 60-kDa band, corresponding to the size of PKM2 of cell lysates, transiently knockdown PKM2 (lower blots in Fig. 7A, right panel, and Fig. S8A and B). Considering this 60-kDa species as the M2 splice variant of pyruvate kinase suggests an unusually high level of PKM2 protein expression in HaCaT keratinocytes. Along this line, in an accompanied study, we have shown that gene editing-mediated inactivation of the splicing of exon 10 leads to abandoned expression of PKM2 and yet does not result in alteration of VEGF induction, in part due to nearly sevenfold concomitant induction of PKM1 expression [68].
In summary, our study proposes a supportive role for PKM2 to facilitate the induction of angiogenesis by growth factors in keratinocytes, which is independent of HIF-1α. Furthermore, acute silencing of PKM2 in keratinocytes causes altered activation of transcriptional and translational factors stimulated by EGF and inflammatory cytokines. This culminated in the reduced stimulation of VEGF expression in keratinocytes.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. M. Wagenblast and Dr. A. Theissen for their help with the animal experiments.
Abbreviations
- PKM2
Pyruvate kinase 2
- OXPHOS
Oxidative phosphorylation
- HE
Hyperproliferative epithelium
- VEGF
Vascular endothelial growth factor
Author contribution
Study design and experiment plan: IG and SF; resources: JP and SF; experiments: IG, KS, SPN, RV, IW, and JM; animal experiments: IG and SF; data analysis and result interpretation: IG, SPN, KS, and RV; manuscript writing and editing: IG, KS, SPN, RV, and JP; manuscript reading and comments: all the authors.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the state of Hessen, to the Institute of General Pharmacology and Toxicology at the Johann Wolfgang-Goethe-University, Frankfurt.
Availability of data and materials
The data presented in this work are available upon request from the corresponding author.
Declarations
Ethics approval and consent to participate
The animal experiments were performed according to the guidelines and approval of the local ethics animal review board (Regierungspräsidium Darmstadt, D-64278 Darmstadt, Germany). The approval number to this project was V54-19c20/15-FU/1055.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/20/2023
Supplementary materials captions is missing and uploaded.
References
- 1.Warburg O, Posener K, Negelein E (1924) Über den Stoffwechsel der Carcinomzelle. Biochem Z.152 309–344
- 2.Warburg O (1956) On the origin of cancer cells. Science 123(80):309–314 [DOI] [PubMed]
- 3.Shlomi T, Benyamini T, Gottlieb E, Sharan R, Ruppin E. Genome-scale metabolic modeling elucidates the role of proliferative adaptation in causing the warburg effect. PLoS Comput Biol. 2011;7:1–8. doi: 10.1371/journal.pcbi.1002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vander Heiden M, Cantley L, Thompson C (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324(80):1029–1033 [DOI] [PMC free article] [PubMed]
- 5.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell. 2005;7:77–83. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 6.Tannahill GM, Curtis AM, Adamik J, Palsson-Mcdermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, O’Neill LAJ. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–243. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ip WE, Hoshi N, Shouval DS, Snapper S, Medzhitov R (2017) Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356(80):513–519 [DOI] [PMC free article] [PubMed]
- 8.Ganeshan K, Chawla A. Metabolic regulation of immune responses. Annu Rev Immunol. 2014;32:609–634. doi: 10.1146/annurev-immunol-032713-120236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singer AJ, Clark RAF. Cutaneous wound healing. N Engl J Med. 1999;341:738–746. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 10.Goren I, Müller E, Pfeilschifter J, Frank S. Severely impaired insulin signaling in chronic wounds of diabetic ob/ob mice. Am J Pathol. 2006;168:765–777. doi: 10.2353/ajpath.2006.050293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goren I, Müller E, Schiefelbein D, Christen U, Pfeilschifter J, Mühl H, Frank S. Systemic anti-TNFα treatment restores diabetes-impaired skin repair in ob/ob mice by inactivation of macrophages. J Invest Dermatol. 2007;127:2259–2267. doi: 10.1038/sj.jid.5700842. [DOI] [PubMed] [Google Scholar]
- 12.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annu Rev Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 13.Siebert A, Goren I, Pfeilschifter J, Frank S. Anti-inflammatory effects of rosiglitazone in obesity-impaired wound healing depend on adipocyte differentiation. PLoS ONE. 2016;11:1–25. doi: 10.1371/journal.pone.0168562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noguchi T, Yamada K, Inoue H, Matsuda T, Tanaka T. The L- and R-type isozymes of rat pyruvate kinase are produced from a single gene by use of different promoters. J Biol Chem. 1987;262:14366–14371. doi: 10.1016/S0021-9258(18)47947-1. [DOI] [PubMed] [Google Scholar]
- 15.Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807–13812. doi: 10.1016/S0021-9258(18)67091-7. [DOI] [PubMed] [Google Scholar]
- 16.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 17.Chaneton B, Hillmann P, Zheng L, Martin ACL, Maddocks ODK, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, Frezza C, O’Reilly M, Gottlieb E. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, Xie J, Gu TL, Polakiewicz RD, Roesel JL, Boggon TJ, Khuri FR, Gilliland DG, Cantley LC, Kaufman J, Chen J. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2(ra73):1–10. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 20.Locasale JW, Vander Heiden MG, Cantley LC. Rewiring of glycolysis in cancer cell metabolism. Cell Cycle. 2010;9(21):4253. doi: 10.4161/cc.9.21.13925. [DOI] [PubMed] [Google Scholar]
- 21.Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, Cantley LC (2010) Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 329(80):1492–1499 [DOI] [PMC free article] [PubMed]
- 22.Yang W, Lu Z. Nuclear PKM2 regulates the Warburg effect. Cell Cycle. 2013;12:3154–3158. doi: 10.4161/cc.26182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosios AM, Fiske BP, Gui DY, Vander Heiden MG. Lack of evidence for PKM2 protein kinase Activity. Mol Cell. 2015;59:850–857. doi: 10.1016/j.molcel.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hillis AL, Lau AN, Devoe CX, Dayton TL, Danai LV, Di Vizio D, Vander Heiden MG (2018) PKM2 is not required for pancreatic ductal adenocarcinoma. Cancer Metab 6 [DOI] [PMC free article] [PubMed]
- 26.Dayton TL, Gocheva V, Miller KM, Bhutkar A, Lewis CA, Bronson RT, Vander Heiden MG, Jacks T. Isoform-specific deletion of PKM2 constrains tumor initiation in a mouse model of soft tissue sarcoma. Cancer Metab. 2018;6:1–9. doi: 10.1186/s40170-018-0179-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 28.Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, Yang H, Mattaini KR, Metallo CM, Fiske BP, Courtney KD, Malstrom S, Khan TM, Kung C, Skoumbourdis AP, Veith H, Southall N, Walsh MJ, Brimacombe KR, Leister W, Lunt SY, Johnson ZR, Yen KE, Kunii K, Davidson SM, Christofk HR, Austin CP, Inglese J, Harris MH, Asara JM, Stephanopoulos G, Salituro FG, Jin S, Dang L, Auld DS, Park HW, Cantley LC, Thomas CJ, Vander Heiden MG. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stallmeyer B, Kämpfer H, Kolb N, Pfeilschifter J, Frank S. The function of nitric oxide in wound repair: inhibition of inducible nitric oxide-synthase severely impairs wound reepithelialization. J Invest Dermatol. 1999;113:1090–1098. doi: 10.1046/j.1523-1747.1999.00784.x. [DOI] [PubMed] [Google Scholar]
- 30.Frank S, Stallmeyer B, Kämpfer H, Kolb N, Pfeilschifter J (1999) Nitric oxide triggers enhanced induction of vascular endothelial growth factor expression in cultured keratinocytes (HaCaT) and during cutaneous wound repair. FASEB J 13:2002–2014 [PubMed]
- 31.Boukamp P. Normal keratinization in a spontaneously immortalized. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiefelbein D, Seitz O, Goren I, Dißmann JP, Schmidt H, Bachmann M, Sader R, Geisslinger G, Pfeilschifter J, Frank S. Keratinocyte-derived vascular endothelial growth factor biosynthesis represents a pleiotropic side effect of peroxisome proliferator-activated receptor-γ agonist troglitazone but not rosiglitazone and involves activation of p38 mitogen-activated protein k. Mol Pharmacol. 2008;74:952–963. doi: 10.1124/mol.108.049395. [DOI] [PubMed] [Google Scholar]
- 33.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 34.Goldberg MS, Sharp PA. Pyruvate kinase M2-specific siRNA induces apoptosis and tumor regression. J Exp Med. 2012;209:217–227. doi: 10.1084/jem.20111487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittig I, Braun HP, Schägger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 36.Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buck MD, Sowell RT, Kaech SM, Pearce EL. Metabolic instruction of immunity. Cell. 2017;169:570–586. doi: 10.1016/j.cell.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caputa G, Flachsmann LJ, Cameron AM. Macrophage metabolism: a wound-healing perspective. Immunol Cell Biol. 2019;97:268–278. doi: 10.1111/imcb.12237. [DOI] [PubMed] [Google Scholar]
- 39.Eming SA, Wynn TA, Martin P (2017) Inflammation and metabolism in tissue repair and regeneration. Science 356(80):1026–1030 [DOI] [PubMed]
- 40.Kuehne A, Emmert H, Soehle J, Winnefeld M, Fischer F, Wenck H, Gallinat S, Terstegen L, Lucius R, Hildebrand J, Zamboni N. Acute activation of oxidative pentose phosphate pathway as first-line response to oxidative stress in human skin cells. Mol Cell. 2015;59:359–371. doi: 10.1016/j.molcel.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 41.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB (2007) Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA 104:19345–19350 [DOI] [PMC free article] [PubMed]
- 42.Alquraishi M, Puckett DL, Alani DS, Humidat AS, Frankel VD, Donohoe DR, Whelan J, Bettaieb A (2019) Pyruvate kinase M2: a simple molecule with complex functions. Free Radic Biol Med 176–192 [DOI] [PMC free article] [PubMed]
- 43.Dayton TL, Jacks T, Vander Heiden MG. PKM 2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17:1721–1730. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Li L, Liu Y, Liu ZR. PKM2 released by neutrophils at wound site facilitates early wound healing by promoting angiogenesis. Wound Repair Regen. 2016;24:328–336. doi: 10.1111/wrr.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goren I, Allmann N, Yogev N, Schürmann C, Linke A, Holdener M, Waisman A, Pfeilschifter J, Frank S. A transgenic mouse model of inducible macrophage depletion. Am J Pathol. 2009;175:132–147. doi: 10.2353/ajpath.2009.081002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goren I, Linke A, Müller E, Pfeilschifter J, Frank S. The suppressor of cytokine signaling-3 is upregulated in impaired skin repair: implications for keratinocyte proliferation. J Invest Dermatol. 2006;126:477–485. doi: 10.1038/sj.jid.5700063. [DOI] [PubMed] [Google Scholar]
- 47.Fitsialos G, Bourget I, Augier S, Ginouvès A, Rezzonico R, Odorisio T, Cianfarani F, Virolle T, Pouysségur J, Meneguzzi G, Berra E, Ponzio G, Buscà R. HIF1 transcription factor regulates laminin-332 expression and keratinocyte migration. J Cell Sci. 2008;121:2992–3001. doi: 10.1242/jcs.029256. [DOI] [PubMed] [Google Scholar]
- 48.Cho YS, Bae JM, Chun YS, Chung JH, Jeon YK, Kim IS, Kim MS, Park JW. HIF-1α controls keratinocyte proliferation by up-regulating p21(WAF1/Cip1) Biochim Biophys Acta - Mol Cell Res. 2008;1783:323–333. doi: 10.1016/j.bbamcr.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 49.Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev. 2003;83:835–870. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- 50.Goren I, Lee SY, Maucher D, Nüsing R, Schlich T, Pfeilschifter J, Frank S (2017) Inhibition of cyclooxygenase-1 and -2 activity in keratinocytes inhibits PGE2 formation and impairs vascular endothelial growth factor release and neovascularisation in skin wounds. Int Wound J 14:53–63 [DOI] [PMC free article] [PubMed]
- 51.Frank S, Hubner G, Breier G, Longaker MT, Greenhalgh DG, Werner S. Regulation of vascular endothelial growth factor expression in cultured keratinocytes. Implications for normal and impaired wound healing. J Biol Chem. 1995;270:12607–12613. doi: 10.1074/jbc.270.21.12607. [DOI] [PubMed] [Google Scholar]
- 52.Luo W, Semenza GL. Pyruvate kinase M2 regulates glucose metabolism by functioning as a coactivator for hypoxia-inducible factor 1 in cancer cells. Oncotarget. 2011;2:551–556. doi: 10.18632/oncotarget.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harris I, McCracken S, Mak TW. PKM2: a gatekeeper between growth and survival. Cell Res. 2012;22:447–449. doi: 10.1038/cr.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goren I, Müller E, Schiefelbein D, Gutwein P, Seitz O, Pfeilschifter J, Frank S. Akt1 controls insulin-driven VEGF biosynthesis from keratinocytes: Implications for normal and diabetes-impaired skin repair in mice. J Invest Dermatol. 2009;129:752–764. doi: 10.1038/jid.2008.230. [DOI] [PubMed] [Google Scholar]
- 55.Qin X, Du Y, Chen X, Li W, Zhang J, Yang J. Activation of Akt protects cancer cells from growth inhibition induced by PKM2 knockdown. Cell Biosci. 2014;4:1–5. doi: 10.1186/2045-3701-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin P, Leibovich SJ. Inflammatory cells during wound repair: the good, the bad and the ugly. Trends Cell Biol. 2005;15:600–607. doi: 10.1016/j.tcb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Rodrigues M, Kosaric N, Bonham CA, Gurtner GC. Wound healing: a cellular perspective. Physiol Rev. 2019;99:665–706. doi: 10.1152/physrev.00067.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 59.Clark RAF, Pavlis M. Dysregulation of the mTOR pathway secondary to mutations or a hostile microenvironment contributes to cancer and poor wound healing. J Invest Dermatol. 2009;129:529–531. doi: 10.1038/jid.2008.441. [DOI] [PubMed] [Google Scholar]
- 60.Galván-Peña S, O’Neill LAJ. Metabolic reprogramming in macrophage polarization. Front Immunol. 2014;5:1–6. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Torales-Cardeña A, Martínez-Torres I, Rodríguez-Martínez S, Gómez-Chávez F, Cancino-Díaz JC, Vázquez-Sánchez EA, Cancino-Díaz ME. Cross talk between proliferative, angiogenic, and cellular mechanisms orchestred by HIF-1 in psoriasis. Mediators Inflamm. 2015;2015:1–11. doi: 10.1155/2015/607363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, Burga LN, Xie J, Jurczak MJ, Depinho RA, Clish CB, Jacks T, Kibbey RG, Wulf GM, Di Vizio D, Mills GB, Cantley LC, Vander Heiden MG. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao X, Wang H, Yang JJ, Liu X, Liu ZR (2012) Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell 45 [DOI] [PMC free article] [PubMed]
- 64.Damasceno LEA, Prado DS, Veras FP, Fonseca MM, Toller-Kawahisa JE, Rosa MH, Públio GA, Martins TV, Ramalho FS, Waisman A. PKM2 promotes Th17 cell differentiation and autoimmune inflammation by fine-tuning STAT3 activation. J Exp Med. 2020;217(e20190613):1–16. doi: 10.1084/jem.20190613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Angiari S, Runtsch MC, Sutton CE, Palsson-McDermott EM, Kelly B, Rana N, Kane H, Papadopoulou G, Pearce EL, Mills KHG, O’Neill LAJ. Pharmacological activation of pyruvate kinase M2 inhibits CD4+ T cell pathogenicity and suppresses autoimmunity. Cell Metab. 2020;31:391–405. doi: 10.1016/j.cmet.2019.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MA, Sheedy FJ, Gleeson LE, van den Bosch MW, Quinn SR, Domingo-Fernandez R, Johnston DG, Jiang JK (2015) Pyruvate kinase M2 regulates hif-1α activity and il-1β induction and is a critical determinant of the Warburg effect in LPS-activated macrophages. Cell Metab 21 [DOI] [PMC free article] [PubMed]
- 67.Yi Z, Wu Y, Zhang W, Wang T, Gong J, Cheng Y, Miao C. Activator-mediated pyruvate kinase M2 activation contributes to endotoxin tolerance by promoting mitochondrial biogenesis. Front Immunol. 2021;11:1–14. doi: 10.3389/fimmu.2020.595316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nold SP, Sych K, Imre G, Fuhrmann DC, Pfeilschifter J, Vutukuri R, Schnutgen F, Wittig I, Meisterknecht J, Frank S, Goren I (2022) Reciprocal abrogation of PKM isoforms: contradictory outcomes and differing impact of splicing signal on CRISPR/Cas9 mediates gene editing in keratinocytes. FEBS J 1–28 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this work are available upon request from the corresponding author.