Abstract
The endometrial microbiota composition may be associated with implantation success. However, a ‘core’ composition has not yet been defined. This exploratory study analysed the endometrial microbiota by 16S rRNA sequencing (V1–V2 region) of 141 infertile women whose first IVF/ICSI cycle failed and compared the microbiota profiles of women with and without a live birth within 12 months of follow-up, and by infertility cause and type. Lactobacillus was the most abundant genus in the majority of samples. Women with a live birth compared to those without had significantly higher Lactobacillus crispatus relative abundance (RA) (p = 0.029), and a smaller proportion of them had ≤ 10% L. crispatus RA (42.1% and 70.4%, respectively; p = 0.015). A smaller proportion of women in the male factor infertility group had ≤ 10% L. crispatus RA compared to women in the unexplained and other infertility causes groups combined (p = 0.030). Women with primary infertility compared to secondary infertility had significantly higher L. crispatus RA (p = 0.004); lower proportions of them had ≤ 10% L. crispatus RA (p = 0.009) and > 10% Gardnerella vaginalis RA (p = 0.019). In conclusion, IVF/ICSI success may be associated with L. crispatus RA and secondary infertility with endometrial dysbiosis, more often than primary infertility. These hypotheses should be tested in rigorous well-powered longitudinal studies.
Subject terms: Microbiology, Infertility
Introduction
The uterus has long been considered a sterile environment. However, evidence for the presence of bacteria, albeit in low quantities, is mounting: next-generation sequencing (NGS) studies have detected bacteria in the uterine cavity belonging to a wide range of phyla, among which Actinobacteria, Firmicutes, Bacteroidetes, and Proteobacteria1–8. Embryo implantation occurs in the endometrium, and it has been hypothesized that the composition of the endometrial microbiota may affect implantation via modulation of local immune responses and tissues9,10. The endometrium is difficult to access and most studies to date have therefore used the vaginal microbiota as a proxy of the endometrial microbiota. In women undergoing assisted reproductive technologies (ART), a Lactobacillus-dominated vaginal microbiota profile has been associated with a higher pregnancy probability and the presence of vaginal bacterial vaginosis (BV)-associated anaerobic bacteria with a higher ART failure probability11–14. More recently, associations between endometrial microbiota that were not Lactobacillus-dominated and poor reproductive outcomes in patients undergoing ART have also been demonstrated8. While vaginal microbiota profiling has been proposed as a means to predict ART outcome15–17, further research is required to investigate its clinical utility. In addition, new evidence-based methods to improve reproductive outcomes in infertile women and couples are urgently needed because implantation failure accounts for more than 70% of all ART failures18.
In this exploratory study, we used 16S rRNA sequencing to characterise bacteria present in endometrial tissue samples of women who had one full failed in vitro fertilization (IVF)/intracytoplasmic sperm injection (ICSI) cycle and were about to undergo a second cycle. Our primary objective was to compare the microbiota profiles of women with and without a live birth within 12 months of follow-up. Our secondary objectives were to compare endometrial microbiota profiles of women with different causes and types of infertility.
Methods
Study population and design
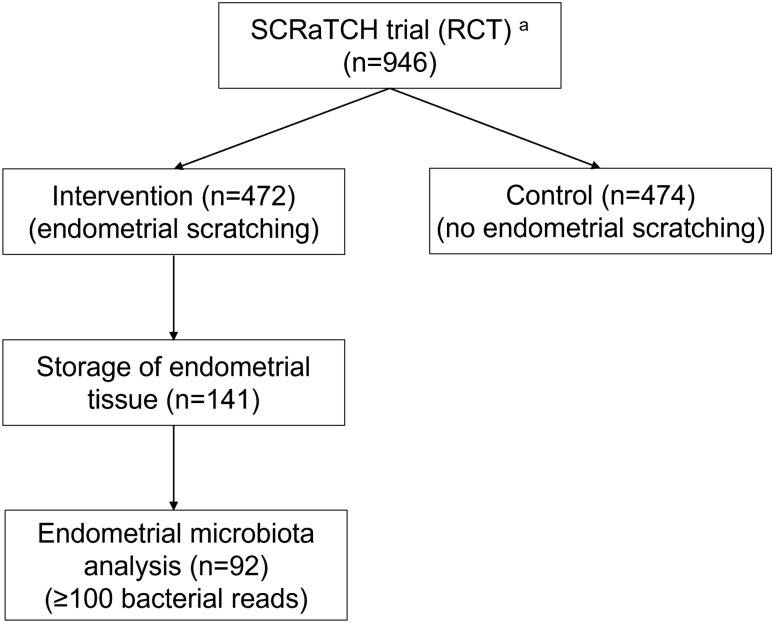
In the SCRaTCH trial, 472 infertile women were randomised to endometrial scratching and 474 to no intervention. No statistically significant differences in pregnancy outcomes were observed between the groups19. The full eligibility criteria of the trial have been described elsewhere19. Briefly, women were eligible if they were aged 18–44 years, had failed implantation after one full IVF/ICSI cycle, and were planning to undergo a second full IVF/ICSI cycle. Only 141 of the 472 women in the endometrial scratching group provided informed consent for endometrial tissue storage and future use in research (Fig. 1). We used the tissue samples from all 141 consenting women, which were obtained in the natural cycle prior to their second IVF/ICSI cycle. All women were followed-up for 12 months after randomisation. Women who reached an ongoing pregnancy (defined as a positive heartbeat on ultrasound at 10 weeks gestational age) within those 12 months were followed-up until delivery. The primary outcome was live birth, which was defined as the delivery of at least one live foetus after at least 24 weeks of gestation19.
Figure 1.
Flow chart of sample selection for the current endometrial microbiota study. RCT, randomised controlled trial. aEndometrial biopsies were obtained from women undergoing endometrial scratching in the SCRaTCH trial, a randomised controlled trial on endometrial scratching in women with a first failed IVF/ICSI cycle19.
Tissue sampling and storage
The endometrial biopsy was performed in the mid-luteal phase of the natural cycle prior to the second IVF/ICSI cycle, five to eight days after the luteinizing hormone surge was detected by urinary tests. Endometrial tissue was obtained with an endometrial biopsy catheter (e.g. a Pipelle or other similar catheter). Women were instructed to prevent pregnancy during that cycle by refraining from sexual intercourse or using condoms. They were asked whether they had adhered to this instruction prior to undergoing the endometrial biopsy, and if there was a chance that they could be pregnant, the biopsy was not performed.
Biopsies were performed in an outpatient setting in six hospitals in The Netherlands. All hospitals followed the same protocol for endometrial biopsy and subsequent tissue storage as described below. Physicians wore a sterile gown, sterile gloves and a hair cap to limit microbial contamination as much as possible. All instruments and materials were unpacked from their sterile casing right before the procedure and were put on a sterile field. After insertion of a Trelat speculum, the cervix was extensively washed with sterile water. The Pipelle catheter was then introduced through the cervix up to the uterine fundus, during which care was taken to avoid contact with the vaginal walls. The piston of the catheter was drawn back to the end of the catheter to create a vacuum, and the catheter was slowly retracted within 1–2 min while constantly rotating 360°. After the procedure, physicians put on new sterile gloves and divided the tissue over three tissue tubes (Brooks Life Sciences, USA) using a sterile scalpel and forceps. Within three minutes after taking the biopsy, the tubes were snap-frozen in liquid nitrogen and stored in a − 80 °C freezer as soon as possible.
DNA isolation, 16S rRNA sequencing and bioinformatics
Frozen endometrial tissue was thawed and homogenized, and total DNA was extracted by bead-beating and chemical lysis (“Supplementary Methods”). The V1-V2 region of the 16S rRNA gene was amplified and sequenced on a MiSeq platform (Illumina, USA). The QIIME2 microbial community analysis pipeline (version 2018.8)20 was used with DADA221 for amplicon sequence variant detection, and SILVA as the 16S rRNA reference gene database (SILVA 138)22. Reads classified as human DNA, mitochondria or ‘unassigned’ were removed, and data from all samples with < 100 bacterial reads were discarded. The pipeline included positive (mock community) and negative (blank) controls (“Supplementary Methods”).
Data analysis
In our primary analysis, we compared women with a live birth (from ongoing pregnancies occurring within 12 months after randomisation in the SCRaTCH trial) to women with no live birth. Within the no live birth group, we also compared women with no pregnancy to women with pregnancy loss. No pregnancy was defined as never having had beta-hCG detected in serum or urine. Pregnancy loss was defined as loss of an intrauterine pregnancy or loss of a pregnancy after beta-hCG had been detected in serum or urine but before ultrasound evaluation ≤ 24 weeks of gestation, excluding ectopic pregnancies. Our secondary analyses included comparisons of different infertility causes (categorised as male factor infertility, unexplained infertility, and other infertility causes) and infertility types (primary versus secondary infertility). Male factor infertility was defined as a semen analysis with total motile sperm count of < 10 million23. Unexplained infertility was defined as the inability to conceive after at least one year of unprotected intercourse while having ovulatory menstrual cycles (defined as a mean cycle length of 21–35 days), at least one patent fallopian tube (i.e. a negative Chlamydia trachomatis antibody titre and/or evidenced by hysterosalpingography or laparoscopy), and a partner with normal semen analysis by WHO criteria24. The group of other infertility causes included mostly women with a female cause, such as tubal factor, ovulatory disorder or endometriosis, but also included women with mixed causes (e.g. male and female causes). We hypothesized that the endometrial microbiota composition as a potential infertility cause was least likely in the male factor and other causes of infertility groups because the infertility cause was known in these cases, and most likely in the unexplained infertility group. Additionally, the group of other causes of infertility was small and heterogeneous. We therefore compared: (1) the male factor infertility group with the unexplained infertility group (excluding the other causes group); (2) the male factor infertility group with the unexplained infertility plus other causes groups combined; and (3) the unexplained infertility group with the male factor infertility plus other causes groups combined.
In addition to untargeted analysis of the entire taxonomic table, we also categorised the microbiota data into biologically meaningful ways for use in targeted analyses. First, we grouped the relative abundances (RAs) of related taxa together into the following bacterial groups: (1) Lactobacillus genus (consisting of the subgroups L. crispatus, L. iners, and other lactobacilli), (2) Gardnerella vaginalis, (3) other bacterial vaginosis (BV)-associated anaerobes, and (4) a residual group of ‘other bacteria’. Supplement 2 contains an overview of all taxa that were identified in the study and how we grouped them. Second, we categorised samples as containing an RA of ≤ 10%, 11–89%, or ≥ 90% total Lactobacillus or a Lactobacillus subgroup. The non-lactobacilli bacterial groups were categorized as ≤ 10% or > 10%.
Continuous data were compared using Wilcoxon rank sum tests because they were considered non-normally distributed. Categorical data were compared using Fisher’s exact tests. Alpha diversity was described by inverse Simpson and Chao1 indices and beta diversity by principal component analysis (PCA) based on centered log-ratio (clr) data transformation. ANCOM-BC with false discovery rate (FDR) adjustment for multiple testing was used for untargeted detection of differentially relatively abundant taxa25. Differences in RAs of prespecified bacterial groups in targeted analyses were determined by Wilcoxon rank sum tests. Multivariable modelling was not performed due to the small sample size. We used R 4.1.0 (phyloseq26 and microbiome27 packages) and IBM SPSS Statistics version 26.0 for all analyses.
Ethics approval
The SCRaTCH trial, a randomised controlled trial from which the endometrial tissue samples were obtained, was approved by the Institutional Review Board of the University Medical Center Utrecht (approval number 15-495, 30 November 2015) (12). For the current microbiota study, ethical approval was obtained from the Biobank Research Ethics Committee of the University Medical Centre Utrecht (approval number 19-520, 31 October 2019). All methods were performed in accordance with the relevant guidelines and regulations.
Results
Participant characteristics
The median age of the 141 women was 35.7 years and their median duration of infertility was 30 months (Table 1). During the 12-months follow-up period, 84/141 women (57.4%) conceived and 61/84 of them (72.6%) reached a live birth. Of the women who reached a live birth, 1/61 (1.6%) conceived spontanously and 60/61 (98.4%) with IVF/ICSI.
Table 1.
Characteristics of all participants and by number of bacterial reads (≥ 100 and < 100).
| All participants (n = 141) | ≥ 100 reads (n = 92) | < 100 readsa (n = 49) | P ≥ 100 vs. < 100 readsb | |
|---|---|---|---|---|
| Median female age in years (IQR) | 35.7 (31.9–39.4) | 34.9 (31.2–39.4) | 36.5 (33.9–39.8) | 0.055 |
| Median female BMI in kg/m2 (IQR) c | 23.8 (21.6–26.4) | 23.5 (21.8–25.6) | 24.1 (21.4–27.9) | 0.214 |
| Median duration of infertility in months (IQR) | 30.0 (22.5–43.0) | 29.5 (23.0–43.0) | 31.0 (20.0–43.5) | 0.952 |
| Female smokers, n (%) d | 18 (12.8) | 8 (8.7) | 10 (20.4) | 0.061 |
| Type of infertility of female, n (%) e | 0.157 | |||
| Primary | 73 (51.8) | 52 (56.5) | 21 (42.9) | |
| Secondary | 68 (48.2) | 40 (43.5) | 28 (57.1) | |
| Cause of infertility, n (%) | 0.929 | |||
| Male factor | 69 (48.9) | 46 (50.0) | 23 (46.9) | |
| Unexplained | 60 (42.6) | 38 (41.3) | 22 (44.9) | |
| Otherf | 12 (8.5) | 8 (12.7) | 4 (8.2) | |
| Median # previous embryo transfers per participant (IQR) | 2.0 (1.0–3.0) | 2.0 (1.0–3.0) | 2.0 (1.0–3.0) | 0.854 |
| Pregnancy outcome during follow-up, n (%) | 0.593 | |||
| Live birthg | 61 (43.3) | 38 (41.3) | 23 (46.9) | |
| No live birth | 80 (56.7) | 54 (58.7) | 26 (53.1) | |
| No pregnancy | 70 (87.5) | 46 (85.2) | 24 (92.3) | |
| Pregnancy loss | 10 (12.5) | 8 (14.8) | 2 (7.7) | |
| Median number of reads per sample (IQR) | 442 (45–6067) | 2167 (532–11,839) | 13 (2–45) |
BMI body mass index, IQR interquartile range.
aSeven samples had 0 reads. The clinical characteristics of this group did not differ from the other 42 women who had between 1 and 99 reads, with the exception of smoking.
bAll continuous variables were compared using the Wilcoxon rank sum test, whereas all categorical variables were compared using the Fisher’s exact test.
cData was missing for one participant in the ≥ 100 reads group.
dData was missing for one participant in the < 100 reads group.
ePrimary: female has never conceived before. Secondary: female has conceived before.
fOther causes of infertility are tubal factor (≥ 100 reads: n = 1, < 100 reads: n = 4), ovulatory disorder (≥ 100 reads: n = 3), endometriosis (≥ 100 reads: n = 1) and mixed causes (≥ 100 reads: n = 3).
gOne woman conceived spontaneously, and all other women conceived via IVF/ICSI. The sample of the woman who conceived spontaneously had < 100 bacterial reads and was therefore not included in the analyses.
Sequencing results
Most endometrial samples had low bacterial abundance: 7/141 samples (5.0%) had 0, 42/141 samples (29.8%) had 1–99, and 92/141 samples (65.2%) had ≥ 100 bacterial reads. Participant characteristics and pregnancy outcomes during follow-up were comparable between the women with < 100 and the women with ≥ 100 reads (Table 1). The 92 selected samples with ≥ 100 bacterial reads were selected for all subsequent analyses (Fig. 1). They generated 860,366 reads in total with a median of 2167 reads per sample (interquartile range (IQR) 532 – 11,839) (Table 1). The positive and negative controls did not give cause for concern (“Supplementary Results”, Fig. S1, Table S1). The most abundant genus present in all samples combined was Lactobacillus (mean RA 69.3%), followed by Gardnerella (mean RA 11.8%) (Fig. S2).
Endometrial microbiota compositions of women with and without a live birth
Among the 92 women with ≥ 100 bacterial reads, 38/92 (41.3%) had a live birth and 54/92 (58.7%) did not. Among the women with no live birth, 46/54 (85.2%) did not get pregnant at all and 8/54 (14.8%) suffered a pregnancy loss. Women with a live birth were significantly younger than women without (median age of 33.5 vs. 36.8 years, respectively; p = 0.016; Table 2). Among the latter group, women with a pregnancy loss had a significantly longer median duration of infertility than women who never conceived (40 vs. 28 months, respectively; p = 0.021). None of the other clinical characteristics differed significantly between the groups. Alpha and beta diversities did not differ between the groups (Table 3, Fig. S3a–d). Untargeted ANCOM-BC analysis did not identify any taxa with significantly different mean RAs between women with and without live birth (Table 3, Fig. S3e), but women who never conceived had significantly higher mean RAs of Aerococcus and Corynebacterium compared to women with a pregnancy loss (Table 3, Fig. S3f). These mean RAs were, however, only 3% or lower. In targeted Wilcoxon rank sum analyses (comparing prespecified bacterial groups), women with a live birth had a significantly higher L. crispatus RA (p = 0.029) and ‘other bacteria’ RA (p = 0.045) than women without a live birth (Table 3, Fig. 2a,b). In addition, a significantly smaller proportion of women with than without a live birth had ≤ 10% L. crispatus RA (42.1% and 70.4%, respectively; p = 0.015) (Table 3). For all other prespecified bacterial (sub)groups, the Wilcoxon analysis results and the proportions of women with > 10% RA did not differ between the birth outcome groups (Table 3, Table S2).
Table 2.
Participants characteristics by birth outcome.
| LB (n = 38) | NLBa (n = 54) | p LB vs. all NLBb | p NP vs. PLb | |||
|---|---|---|---|---|---|---|
| All NLB (n = 54) | NP (n = 46) | PL (n = 8) | ||||
| Median female age in years (IQR) | 33.5 (28.7–36.3) | 36.8 (31.8–39.8) | 36.8 (31.6–39.8) | 35.8 (32.5–40.6) | 0.016 | 0.990 |
| Median female BMI in kg/m2 (IQR)c | 23.0 (21.4–24.9) | 24.0 (21.8–26.6) | 23.8 (21.8–26.7) | 24.9 (21.3–27.0) | 0.136 | 0.951 |
| Median duration of infertility in mo (IQR) | 29.0 (22.5–43.5) | 30.0 (23.0–43.0) | 28.0 (21.8–39.8) | 40.0 (31.5–57.3) | 0.953 | 0.021 |
| Female smokers, n (%) | 3 (7.9) | 5 (9.3) | 4 (8.7) | 1 (12.5) | 1.000 | 0.567 |
| Type of infertility of the female, n (%)d | 0.835 | 0.063 | ||||
| Primary | 22 (57.9) | 30 (55.6) | 23 (50.0) | 7 (87.5) | ||
| Secondary | 16 (42.1) | 24 (44.4) | 23 (50.0) | 1 (12.5) | ||
| Cause of infertility, n (%) | 0.188 | 0.181 | ||||
| Unexplained | 15 (39.5) | 23 (42.6) | 17 (37.0) | 6 (75.0) | ||
| Male factor | 22 (57.9) | 24 (44.4) | 22 (47.8) | 2 (25.0) | ||
| Othere | 1 (2.6) | 7 (13.0) | 7 (15.2) | 0 | ||
| Median # previous embryo transfers per participant (IQR) | 2.0 (1.0–3.0) | 1.0 (1.0–3.0) | 1.5 (1.0–3.0) | 1.0 (1.0–2.0) | 0.352 | 0.322 |
Significant values are in [bold].
BMI body mass index, IQR interquartile range, LB live birth, mo months, NLB no live birth, NP no pregnancy, PL pregnancy loss.
Only samples with ≥ 100 16S rRNA sequencing reads (V1–V2 region) were included.
aThe NLB participants included participants with a pregnancy loss (PL) as well as participants who did not get pregnant at all (NP).
bContinuous variables were compared by Wilcoxon rank sum test and categorical variables by Fisher’s exact test.
cData was missing for one participant in the LB group.
dPrimary: female has never conceived before. Secondary: female has conceived before.
eOther causes of infertility are tubal factor (NLB n = 1), ovulatory disorder (LB n = 1, NLB n = 2), endometriosis (NLB n = 1) and mixed causes (NLB n = 3).
Table 3.
Endometrial microbiota characteristics by birth outcome.
| LB (n = 38) | NLBa (n = 54) | p LB vs. all NLBb | p NP vs. PLb | |||
|---|---|---|---|---|---|---|
| All NLB (n = 54) | NP (n = 46) | PL (n = 8) | ||||
| Alpha diversity | ||||||
| Mean inverse Simpson diversity (SD) | 2.7 (± 1.6) | 2.8 (± 2.4) | 2.6 (± 2.0) | 4.1 (± 3.9) | 0.654 | 0.154 |
| Mean Chao1 diversity (SD) | 15.9 (± 26.7) | 13.5 (± 15.9) | 12.7 (± 16.2) | 18.0 (± 14.0) | 0.769 | 0.073 |
| Untargeted ANCOM-BC resultsb,c | ||||||
| Mean RA Aerococcus genus (SD) | 0.2 (± 1.2) | 0.1 (± 0.5) | 0.2 (± 0.5) | 0 | NS | < 0.001 |
| Mean RA Corynebacterium genus (SD) | 3.3 (± 11.5) | 1.9 (± 6.0) | 2.0 (± 6.5) | 1.0 (± 1.7) | NS | < 0.001 |
| Targeted bacterial groups (specified a priori)b,c | ||||||
| Lactobacillus genus | 0.893 | 0.450 | ||||
| Median RA (IQR) | 94.2 (39.1–99.2) | 94.5 (18.9–99.6) | 95.8 (32.5–99.7) | 48.1 (7.6–98.4) | ||
| 95% CI | 80.1–97.7 | 69.9–98.8 | 69.9–99.3 | 1.5–100.0 | ||
| Mean RA (SD) | 73.5 (± 35.2) | 66.4 (± 41.5) | 69.0 (± 40.6) | 51.0 (± 46.5) | ||
| 95% CI | 61.2–77.4 | 55.0–77.7 | 57.0–81.1 | 12.2–89.9 | ||
| L. crispatus | 0.029 | 0.510 | ||||
| Median RA (IQR) | 37.4 (0–94.4) | 0 (0–55.7) | 0 (0–55.7) | 4.2 (0–37.8) | ||
| 95% CI | 0–91.5 | 0–0 | 0–0 | 0–99.5 | ||
| Mean RA (SD) | 45.8 (± 44.3) | 25.4 (± 41.4) | 25.0 (± 41.4) | 27.9 (± 44.1) | ||
| 95% CI | 31.2–60.4 | 14.1–36.7 | 12.7–37.3 | 0–64.8 | ||
| L. iners | 0.141 | 0.971 | ||||
| Median RA (IQR) | 0 (0–15.9) | 0.3 (0–71.7) | 0.2 (0–71.7) | 0.3 (0–22.3) | ||
| 95% CI | 0–5.5 | 0–21.5 | 0–63.1 | 0–92.8 | ||
| Mean RA (SD) | 16.5 (± 30.6) | 30.2 (± 40.5) | 31.6 (± 41.0) | 21.7 (± 39.0) | ||
| 95% CI | 6.4–26.5 | 19.1–41.2 | 19.5–43.8 | 0–54.3 | ||
| Other lactobacilli | 0.203 | 0.495 | ||||
| Median RA (IQR) | 0.7 (0–5.0) | 0 (0–4.4) | 0 (0–6.8) | 0.5 (0.2–1.4) | ||
| 95% CI | 0–2.6 | 0–0.6 | 0–0.6 | 0–6.6 | ||
| Mean RA (SD) | 11.2 (± 24.5) | 10.8 (± 27.1) | 12.4 (± 29.0) | 1.3 (± 2.2) | ||
| 95% CI | 3.1–19.3 | 3.4–18.1 | 3.8–21.0 | 0–3.2 | ||
| Gardnerella genus | 0.541 | 1.000 | ||||
| Median RA (IQR) | 0 (0–0) | 0 (0–1.6) | 0 (0–1.6) | 0 (0–9.3) | ||
| 95% CI | 0–0 | 0–0 | 0–0 | 0–98.3 | ||
| Mean RA (SD) | 8.6 (± 20.3) | 14.0 (± 28.7) | 13.4 (± 27.8) | 16.9 (± 35.4) | ||
| 95% CI | 2.0–15.3 | 6.1–21.8 | 5.2–21.7 | 0–46.5 | ||
| Other BV-anaerobes | 0.558 | 0.802 | ||||
| Median RA (IQR) | 0 (0–1.5) | 0 (0–2.6) | 0 (0–4.5) | 0.1 (0–0.6) | ||
| 95% CI | 0–2.4 | 0–1.4 | 0–0.6 | 0–58.6 | ||
| Mean RA (SD) | 4.4 (± 10.8) | 8.3 (± 18.8) | 8.4 (± 18.7) | 7.6 (± 20.6) | ||
| 95% CI | 0.9–8.0 | 3.2–13.4 | 2.9–14.0 | 0–24.8 | ||
| Other bacteriad | 0.045 | 0.233 | ||||
| Median RA (IQR) | 4.7 (0.3–13.2) | 0.5 (0–8.9) | 0.4 (0–5.9) | 1.2 (0.1–36.8) | ||
| 95% CI | 0.8–6.9 | 0.1–1.4 | 0–1.4 | 0–91.4 | ||
| Mean RA (SD) | 13.5 (± 23.7) | 11.4 (± 22.8) | 9.1 (± 18.6) | 24.5 (± 38.5) | ||
| 95% CI | 5.7–21.2 | 5.2–17.6 | 3.6–14.6 | 0–56.6 | ||
| L. crispatus RA subgroups, n (%) | 0.015 | 0.679 | ||||
| ≤ 10 | 16 (42.1) | 38 (70.4) | 33 (71.7) | 5 (62.5) | ||
| 11–89 | 9 (23.7) | 4 (7.4) | 3 (6.5) | 1 (12.5) | ||
| ≥ 90 | 13 (34.2) | 12 (22.2) | 10 (21.7) | 2 (25.0) | ||
Significant values are in [bold].
BV bacterial vaginosis, CI confidence interval, IQR interquartile range, LB live birth, mo months, NLB no live birth, NP no pregnancy, NS not significant, PL pregnancy loss, RA relative abundance, SD standard deviation.
Only samples with ≥ 100 16S rRNA sequencing reads (V1–V2 region) were included.
aThe NLB participants included participants with a pregnancy loss (PL) as well as participants who did not get pregnant at all (NP).
bANCOM-BC analyses were corrected for multiple testing and were used to identify individual taxa with significantly different RAs in comparison groups in an untargeted manner. In the targeted analyses (using prespecified bacterial groups and subgroups), we assumed that the data were not normally distributed. All p-values were calculated by Wilcoxon rank sum tests only, but we are showing both median and mean RAs for illustrative purposes.
cRelative abundances are presented as percentages of the number of reads of the taxon out of the total number of bacterial reads.
dThe group of “other bacteria” contains skin bacteria, unresolved bacteria and minority taxa that could not be assigned to any of the other categories.
Figure 2.
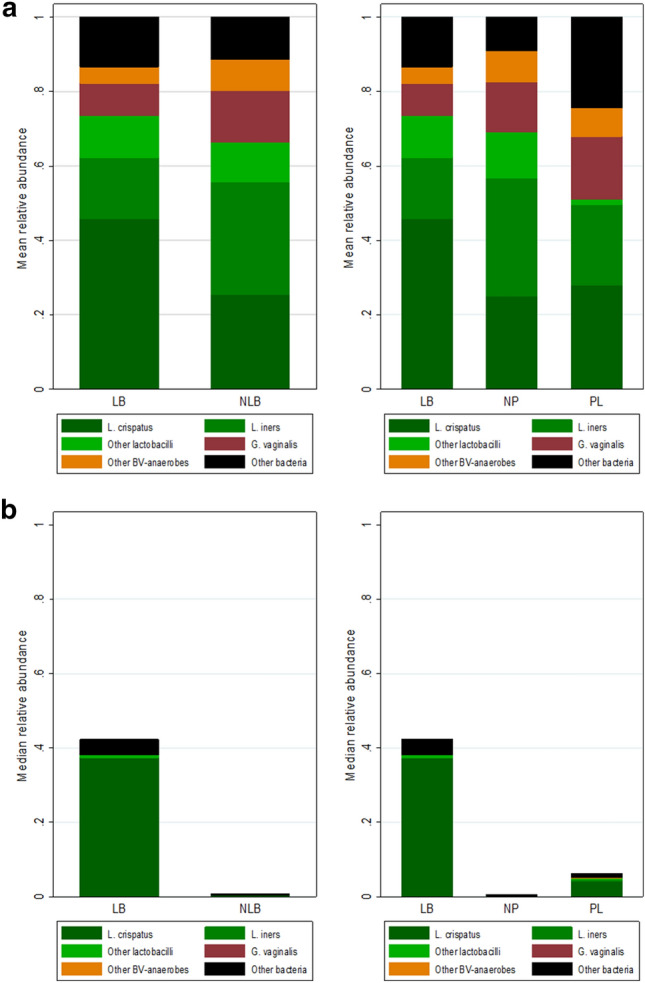
Relative abundance of prespecified bacterial groups by birth outcome. Stacked bar graphs of mean (a) and median (b) relative abundances of prespecified bacterial groups in the live birth (LB) versus no live birth (NLB) groups. The NLB group was further divided into the no pregnancy (NP) and pregnancy loss (PL) group. BV, bacterial vaginosis; G. vaginalis, Gardnerella vaginalis; L. crispatus, Lactobacillus crispatus; L. iners, Lactobacillus iners.
Endometrial microbiota compositions by infertility causes
Male factor infertility was most common (46/92; 50.0%), followed by unexplained infertility (38/92; 41.3%), and other causes (8/92; 8.7%). Women with unexplained and other causes of infertility were significantly older than women with male factor infertility (median ages of 37.7, 37.7, and 32.7 years, respectively), and also had a longer median duration of infertility (median infertility duration of 35.0, 37.0 and 27.5 months, respectively; Table S3). None of the other clinical characteristics differed significantly between the groups. Endometrial microbiota profiles did not differ in alpha and beta diversities (Table S4, Fig. S4a,b). Untargeted ANCOM-BC analysis and targeted Wilcoxon rank sum analyses did not identify any taxa or prespecified bacterial groups with significantly different RAs between the infertility causes groups (Table S4, Fig. S4c–e). However, the proportion of women with ≤ 10% L. crispatus was significantly lower in the male factor infertility group compared to in the other two groups (p = 0.030) (Table S4). For all other prespecified bacterial (sub)groups the proportions of women with > 10% RA, did not differ between the infertility causes groups (Table S4).
Endometrial microbiota compositions by primary vs. secondary infertility
Primary infertility was present in 52/92 (56.5%) women and secondary infertility in 40/92 (43.5%) women. Women with secondary infertility were significantly older than women with primary infertility (median age of 36.8 vs. 33.4 years, respectively; p = 0.001) (Table S5). The other clinical characteristics did not differ significantly between the two groups (Table S5). Alpha and beta diversities did not differ significantly either (Table S6, Fig. S5a,b). Untargeted ANCOM-BC analysis identified a significantly lower mean Gardnerella genus RA in women with primary compared to women with secondary infertility (6.1% vs. 19.2%, respectively; p = 0.030) (Table S6, Fig. S5c). In targeted Wilcoxon rank sum analyses, women with primary compared to secondary infertility had a significantly higher L. crispatus RA (p = 0.004), and a trend towards lower G. vaginalis RA (p = 0.051) (Table S6, Fig. S5d,e). The proportions of women with ≤ 10% L. crispatus and > 10% G. vaginalis also differed significantly between the groups (p = 0.009 and 0.019 respectively), with women with primary infertility being more likely to have higher levels of L. crispatus and lower levels of G. vaginalis than women with secondary infertility (Table S6).
Discussion
Lactobacillus was the most abundant genus in the majority of samples. While untargeted analysis did not identify any differentially relatively abundant taxa by birth outcomes, women with a live birth had significantly higher L. crispatus RA than women with no live birth in targeted analysis. Although untargeted and targeted analyses did not identify any differentially relatively abundant taxa between women with different infertility causes, we revealed that women with male factor infertility were significantly less likely to have endometrial microbiota containing ≤ 10% L. crispatus. We observed the largest differences in endometrial microbiota profiles between women with primary or secondary infertility, with the latter women showing several signs of endometrial microbiota dysbiosis (i.e. reduced L. crispatus and increased BV-anaerobes, particularly G. vaginalis).
Lactobacillus was the most relatively abundant genus in almost all endometrium samples in our study, which is consistent with several previous studies3,5–8,28–30. However, one previous study using endometrial tissue, obtained after hysterectomy, detected hardly any lactobacilli at all31. It is well-known that lactobacilli are by far the most common bacteria in the vagina and cervix, except in BV patients and in women with other less common types of vaginal dysbiosis2,32. Cervicovaginal bacteria can travel through the endocervical canal33. It would therefore seem unlikely for the endometrium to not contain lactobacilli. However, endometrial sample contamination may also explain their presence. When the endometrial sample is collected via the endocervical canal, the clinician might pick-up cervicovaginal bacteria during the sampling procedure. We were very much aware of this risk and tried to minimize such contamination (see “Methods”). We believe that the endometrium likely does contain lactobacilli in most women and offer an alternative explanation as to why Winters et al. may not have detected them31. While they did use endometrial tissue obtained after hysterectomy, as opposed to a sample through the endocervical canal (as we and others did), their study population consisted of 25 women whose endometrial tissue was abnormal due to fibroids or endometrial hyperplasia. Bacteria metabolize carbohydrates they obtain from tissues and mucus34, and altered endometrial tissue may therefore also alter the endometrial microbiota composition35.
Contamination may also be problematic for a second reason. The endometrium typically contains few micro-organisms. The introduction of small quantities of contaminants (for example, via reagents) might have a large impact on the sequencing results, making it difficult to identify true biological signals36. We therefore included controls in our DNA extraction and sequencing pipelines and did not detect any such contaminants.
We showed a beneficial association of endometrial L. crispatus with ART outcome, which is in agreement with findings by others who assessed cervicovaginal37–42 or endometrial microbiota compositions8 in relation to birth outcomes. While evidence for this beneficial association of L. crispatus is mounting, the clinical significance remains unclear. The ability to conceive, and the success of ART treatment, depends on many factors, of which the presence of specific bacteria and/or inflammation in the female genital tract is only one43,44. At the moment, it is still unclear whether optimizing the female genital tract microbiota might improve the ART success rate, and if yes, to what extent. Our own data shows this heterogeneity quite clearly: while women who conceived had a ‘healthier’ microbiota on average than women who did not conceive, not all L. crispatus-dominated women did conceive. Even if L. crispatus-domination is considered desirable, the next question is how to achieve it. A recent systematic review of lactobacilli-containing vaginal probiotics showed that the concept is promising, but currently available products require improvement, especially in terms of their ability to colonize the female genital tract45. To date, two Japanese studies have investigated endometrial microbiota manipulation as a means to improve ART success46,47. One study investigated combinations of oral and vaginal lactobacilli-containing probiotics and metronidazole46, while the other investigated oral lactoferrin supplementation as a prebiotic47. However, the results of these studies are difficult to interpret due to lack of no-intervention control groups. Once pre- or probiotics capable of increasing the colonization of lactobacilli in the female genital tract have been developed, randomised controlled clinical trials would have to be conducted to determine their effect on ART outcomes empirically.
Our data support our hypothesis that women with male factor infertility would have ‘healthier’ endometrial microbiota than women with unexplained or other causes of infertility. Other than the differences in L. crispatus RA, we did not find any evidence for severe female genital tract dysbiosis (e.g. high RA of BV-anaerobes or other bacteria) as a potential explanation for infertility in the total group of primary or secondary unexplained infertility cases. However, our data suggest that severe dysbiosis may be associated with secondary infertility. To our knowledge, this has not been described before. It should be noted that women with secondary infertility were also on average 3–4 years older than women with primary infertility. While age could act as a confounder, we would not normally expect an age-related decrease of lactobacilli in the female genital tract until menopause48,49. A recent study demonstrated that the vaginal and uterine microbiota composition remained stable with increasing age among women younger than 40 years50.
Our study is unique in that we could prospectively obtain endometrial biopsies as part of an intervention trial. Endometrial sampling is invasive and data on endometrial microbiota are therefore scarce. While our sample size was sufficient for detecting some important microbiota profile differences between our comparison groups, an even larger sample size would have been desirable to better accommodate the large variability in infertility causes and microbiota profiles and adjust for confounding. In addition, we did not have data on a few important potential confounders such as ethnicity, recent use of antibiotics or immunosuppressant drugs, and history of vaginitis, cervicitis, endometritis, pelvic inflammatory disease, and uterine surgery. As already noted, we tried to minimize cervicovaginal contamination during transcervical sampling, as well as contamination from other sources, by promoting strict sampling and hygiene procedures. We also included positive and negative (blank) controls in our sequencing pipeline to assess the presence of contaminant DNA. However, we cannot be sure that contamination was ruled out. Related to this, we had to exclude one-third of the samples that we sequenced, because they had fewer than 100 bacterial reads. Another limitation was that 16S rRNA gene sequencing only allows inferences to be made about bacteria, and not about other micro-organisms that are known to be common in the female reproductive tract (such as viruses and yeasts), which may also play a role in ART outcome51.
Conclusion
In summary, our data point towards a beneficial association of L. crispatus with IVF/ICSI success, and suggest that secondary infertility may be associated with endometrial dysbiosis more often than primary infertility. These hypotheses should be tested in rigorous well-powered longitudinal studies. The clinical implications of our study are unclear. Further research is needed to investigate mechanisms linking the endometrial microbiota to the process of implantation52,53, as well as clinical trials of potential interventions.
Supplementary Information
Acknowledgements
The authors are grateful to the women who donated tissue for research.
Abbreviations
- 16S rRNA
16S ribosomal ribonucleic acid
- ANCOM-BC
Analysis of compositions of microbiomes with bias correction
- ART
Assisted reproductive technology
- BV
Bacterial vaginosis
- Clr
Centered log ratio
- DNA
Deoxyribonucleic acid
- FDR
False discovery rate
- ICSI
Intracytoplasmic sperm injection
- IQR
Interquartile range
- IVF
In vitro fertilization
- LB
Live birth
- NGS
Next generation sequencing
- NLB
No live birth
- PCA
Principal component analysis
- RA
Relative abundance
- WHO
World Health Organization
Author contributions
Study design: B.B., S.M., G.S., F.B., F.P., J.W. Obtaining funding: F.B. Data collection: N.H., F.M., G.T., J.B., D.B., L.S. Tissue collection: N.H., F.M., G.T., J.B., D.B., L.S. DNA isolation and 16S rRNA sequencing: M.V. Data analysis: B.B., F.P., M.R., J.W. Manuscript writing: B.B., F.P., J.W. Preparation of tables and figures: B.B., F.P., J.W. Manuscript review and approval: All authors.
Funding
The randomised controlled trial was funded by the Netherlands Organisation for Health Research and Development, ‘ZonMw’ (ZonMW project number 843002601). The current microbiota project was funded by Merck and the Utrecht Exposome Hub of Utrecht Life Sciences (www.uu.nl/exposome), which was funded by the Executive Board of Utrecht University. None of the study donors had input in the study design, analysis, or data interpretation.
Data availability
Sequencing data has been made available on the European Nucleotide Archive under project code PRJEB53740 (https://www.ebi.ac.uk/ena/browser/view/PRJEB53740). R scripts are available on https://gitlab.com/PB_Stege/diet_microbiome_resistome/.
Competing interests
The SCRATCH study and the current endometrial microbiota substudy received funding from public sources and Merck (see Funding section below) but none of these donors had input in the study design and analysis. In addition, several authors report having received personal fees for consultations, conference attendance, and travel as follows: BB from Gedeon Richter Benelux and Guerbet; NvH from Organon; GT from Merck; GS from Guerbet and SM from Abbott Pharmaceuticals, Ferring, Abbott Pharmaceuticals, and IBSA. FB is a paid member of the advisory boards of Merck and Ferring, and also received a speakers’ fee from Besins Healthcare. All of these personal fees were unrelated to the study and substudy reported in this manuscript. The remaining authors declare no conflicts of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Fernanda L. Paganelli and Janneke H. H. M. van de Wijgert.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-30591-2.
References
- 1.Baker JM, Chase DM, Herbst-Kralovetz MM. Uterine microbiota: Residents, tourists, or invaders? Front. Immunol. 2018;9:208. doi: 10.3389/fimmu.2018.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C, et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat. Commun. 2017;8:875. doi: 10.1038/s41467-017-00901-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diaz-martínez MDC, et al. Impact of the vaginal and endometrial microbiome pattern on assisted reproduction outcomes. J. Clin. Med. 2021;10(18):4063. doi: 10.3390/jcm10184063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franasiak JM, et al. Endometrial microbiome at the time of embryo transfer: Next-generation sequencing of the 16S ribosomal subunit. J. Assist. Reprod. Genet. 2016;33:129–136. doi: 10.1007/s10815-015-0614-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ichiyama T, et al. Analysis of vaginal and endometrial microbiota communities in infertile women with a history of repeated implantation failure. Reprod. Med. Biol. 2021;20:334–344. doi: 10.1002/rmb2.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitaya K, Nagai Y, Arai W, Sakuraba Y, Ishikawa T. Characterization of microbiota in endometrial fluid and vaginal secretions in infertile women with repeated implantation failure. Mediators Inflamm. 2019;2019:4893437. doi: 10.1155/2019/4893437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lüll K, et al. Differences in microbial profile of endometrial fluid and tissue samples in women with in vitro fertilization failure are driven by Lactobacillus abundance. Acta Obstet. Gynecol. Scand. 2022 doi: 10.1111/aogs.14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno I, et al. Endometrial microbiota composition is associated with reproductive outcome in infertile patients. Microbiome. 2022;10:1–17. doi: 10.1186/s40168-021-01184-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benner M, Ferwerda G, Joosten I, van der Molen RG. How uterine microbiota might be responsible for a receptive, fertile endometrium. Hum. Reprod. Update. 2018;24:393–415. doi: 10.1093/humupd/dmy012. [DOI] [PubMed] [Google Scholar]
- 10.D’Ippolito S, et al. Endometrial microbes and microbiome: Recent insights on the inflammatory and immune “players” of the human endometrium. Am. J. Reprod. Immunol. 2018;80:1–8. doi: 10.1111/aji.13065. [DOI] [PubMed] [Google Scholar]
- 11.Bernabeu A, et al. Effect of the vaginal microbiome on the pregnancy rate in women receiving assisted reproductive treatment. J. Assist. Reprod. Genet. 2019;36:2111–2119. doi: 10.1007/s10815-019-01564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckert LO, Moore DE, Patton DL, Agnew KJ, Eschenbach DA. Relationship of vaginal bacteria and inflammation with conception and early pregnancy loss following in-vitro fertilization. Infect. Dis. Obstet. Gynecol. 2003;11:11–17. doi: 10.1155/S1064744903000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haahr T, et al. Abnormal vaginal microbiota may be associated with poor reproductive outcomes: A prospective study in IVF patients. Hum. Reprod. 2016;31:795–803. doi: 10.1093/humrep/dew026. [DOI] [PubMed] [Google Scholar]
- 14.Hyman RW, et al. The dynamics of the vaginal microbiome during infertility therapy with in vitro fertilization-embryo transfer. J. Assist. Reprod. Genet. 2012;29:105–115. doi: 10.1007/s10815-011-9694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koedooder R, et al. The vaginal microbiome as a predictor for outcome of in vitro fertilization with or without intracytoplasmic sperm injection: A prospective study. Hum. Reprod. 2019;34:1042–1054. doi: 10.1093/humrep/dez065. [DOI] [PubMed] [Google Scholar]
- 16.Eskew AM, et al. Association of vaginal bacterial communities and reproductive outcomes with prophylactic antibiotic exposure in a subfertile population undergoing in vitro fertilization: A prospective exploratory study. F S Sci. 2021;2:71–79. doi: 10.1016/j.xfss.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoenmakersa S, Laven NJ, Schoenmakersa S. The vaginal microbiome as a tool to predict IVF success. Curr. Opin. Obstet. Gynecol. 2020;32:169–178. doi: 10.1097/GCO.0000000000000626. [DOI] [PubMed] [Google Scholar]
- 18.Szamatowicz M. Assisted reproductive technology in reproductive medicine—Possibilities and limitations. Ginekol. Pol. 2016;87:820–823. doi: 10.5603/GP.2016.0095. [DOI] [PubMed] [Google Scholar]
- 19.van Hoogenhuijze NE, et al. Endometrial scratching in women with one failed IVF/ICSI cycle-outcomes of a randomised controlled trial (SCRaTCH) Hum. Reprod. 2020 doi: 10.1093/humrep/deaa268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.NVOG (The Dutch Society of Obstetrics and Gynaecology). Guidelines on Male Infertility. https://www.nvog.nl/wp-content/uploads/2017/12/Mannelijke-subfertiliteit-1.0-15-09-2010.pdf (2010).
- 24.Cooper TG, et al. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update. 2009;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 25.Lin H, Peddada SD. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMurdie PJ, Holmes S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lahti, L. & Shetty, S. Tools for microbiome analysis in R. http://microbiome.github.com/microbiome (2017).
- 28.Moreno I, et al. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am. J. Obstet. Gynecol. 2016;215:684–703. doi: 10.1016/j.ajog.2016.09.075. [DOI] [PubMed] [Google Scholar]
- 29.Bednarska-Czerwińska A, et al. Marking the profile of the microflora of the endometrium and uterine cervix in women as a potential factor determining the effectiveness of in vitro fertilization. J. Clin. Med. 2022;11:3348. doi: 10.3390/jcm11123348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao X, et al. Characterizing the endometrial microbiome by analyzing the ultra-low bacteria from embryo transfer catheter tips in IVF cycles: Next generation sequencing (NGS) analysis of the 16S ribosomal gene. Hum. Microbiome J. 2017;3:15–21. doi: 10.1016/j.humic.2017.01.004. [DOI] [Google Scholar]
- 31.Winters AD, et al. Does the endometrial cavity have a molecular microbial signature? Sci. Rep. 2019;9:1–17. doi: 10.1038/s41598-019-46173-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA. 2011;108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen LK, et al. The cervical mucus plug inhibits, but does not block, the passage of ascending bacteria from the vagina during pregnancy. Acta Obstet. Gynecol. Scand. 2014;93:102–108. doi: 10.1111/aogs.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armstrong, S. K. Bacterial metabolism in the host environment: pathogen growth and nutrient assimilation in the mammalian upper respiratory tract. Microbiol. Spectr.3(3). 10.1128/microbiolspec.MBP-0007-2014 (2015). [DOI] [PubMed]
- 35.Walther-António MRS, et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 2016;8:1–15. doi: 10.1186/s13073-016-0368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisenhofer R, et al. Contamination in low microbial biomass microbiome studies: Issues and recommendations. Trends Microbiol. 2019;27:105–117. doi: 10.1016/j.tim.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 37.van de Wijgert JHHM, et al. The vaginal microbiota: What have we learned after a decade of molecular characterization? PLoS One. 2014;9(8):e105998. doi: 10.1371/journal.pone.0105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haahr T, et al. Vaginal microbiota and in vitro fertilization outcomes: Development of a simple diagnostic tool to predict patients at risk of a poor reproductive outcome. J. Infect. Dis. 2019;219:1809–1817. doi: 10.1093/infdis/jiy744. [DOI] [PubMed] [Google Scholar]
- 39.Witkin SS, Linhares IM. Why do lactobacilli dominate the human vaginal microbiota? BJOG An Int. J. Obstet. Gynaecol. 2017;124:606–611. doi: 10.1111/1471-0528.14390. [DOI] [PubMed] [Google Scholar]
- 40.Kindinger LM, et al. The interaction between vaginal microbiota, cervical length, and vaginal progesterone treatment for preterm birth risk. Microbiome. 2017;5:6. doi: 10.1186/s40168-016-0223-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrova MI, Lievens E, Malik S, Imholz N, Lebeer S. Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front. Physiol. 2015;6:1–18. doi: 10.3389/fphys.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng N, et al. Lactobacillus iners is associated with vaginal dysbiosis in healthy pregnant women: A preliminary study. Biomed. Res. Int. 2019;2019:6079734. doi: 10.1155/2019/6079734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levi Setti PE, et al. Implantation failure in assisted reproduction technology and a critical approach to treatment. Ann. N. Y. Acad. Sci. 2004;1034:184–199. doi: 10.1196/annals.1335.021. [DOI] [PubMed] [Google Scholar]
- 44.Franasiak JM, et al. A review of the pathophysiology of recurrent implantation failure. Fertil. Steril. 2021;116:1436–1448. doi: 10.1016/j.fertnstert.2021.09.014. [DOI] [PubMed] [Google Scholar]
- 45.van de Wijgert JHHM, Verwijs M. Lactobacilli-containing vaginal probiotics to cure or prevent bacterial or fungal vaginal dysbiosis: A systematic review and recommendations for future trial designs. BJOG Int. J. Obstet. Gynaecol. 2020;127:287–299. doi: 10.1111/1471-0528.15870. [DOI] [PubMed] [Google Scholar]
- 46.Kadogami D, Nakaoka Y, Morimoto Y. Use of a vaginal probiotic suppository and antibiotics to influence the composition of the endometrial microbiota. Reprod. Biol. 2020;20:307–314. doi: 10.1016/j.repbio.2020.07.001. [DOI] [PubMed] [Google Scholar]
- 47.Kitaya K, Ishikawa T. Genital tract dysbiosis in infertile women with a history of repeated implantation failure and pilot study for reproductive outcomes following oral enteric coating lactoferrin supplementation. Arch. Gynecol. Obstet. 2022 doi: 10.1007/s00404-022-06755-2. [DOI] [PubMed] [Google Scholar]
- 48.Muhleisen AL, Herbst-Kralovetz MM. Menopause and the vaginal microbiome. Maturitas. 2016;91:42–50. doi: 10.1016/j.maturitas.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 49.Hickey RJ, Zhou X, Pierson JD, Ravel J, Forney LJ. Understanding vaginal microbiome complexity from an ecological perspective. Transl. Res. 2012;160:267–282. doi: 10.1016/j.trsl.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, et al. Translocation of vaginal microbiota is involved in impairment and protection of uterine health. Nat. Commun. 2021;12:1–15. doi: 10.1038/s41467-021-24516-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eskew A, et al. Association of the eukaryotic vaginal virome with prophylactic antibiotic exposure and reproductive outcomes in a subfertile population undergoing in vitro fertilisation: A prospective exploratory study. BJOG Int. J. Obstet. Gynaecol. 2020;127:208–216. doi: 10.1111/1471-0528.15951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molina NM, et al. Analysing endometrial microbiome: Methodological considerations and recommendations for good practice. Hum. Reprod. 2021;36:859–879. doi: 10.1093/humrep/deab009. [DOI] [PubMed] [Google Scholar]
- 53.Sola-Leyva A, et al. Mapping the entire functionally active endometrial microbiota. Hum. Reprod. 2021;36:1021–1031. doi: 10.1093/humrep/deaa372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data has been made available on the European Nucleotide Archive under project code PRJEB53740 (https://www.ebi.ac.uk/ena/browser/view/PRJEB53740). R scripts are available on https://gitlab.com/PB_Stege/diet_microbiome_resistome/.