Abstract
Bacillus subtilis is currently used as an oral probiotic. We examined two commercial B. subtilis probiotic preparations, Enterogermina and Biosubtyl. Surprisingly, physiological and genetic characterization of the bacteria contained in each of these preparations has shown that neither contains B. subtilis.
Bacillus subtilis is currently being used for oral bacteriotherapy and bacterioprophylaxis of gastrointestinal disorders (mostly as a direct result of antibiotic treatment), many of which lead to diarrhea. Ingestion of significant quantities of B. subtilis is thought to restore the normal microbial flora following extensive antibiotic use or illness (for a review see reference 16). Probiotic preparations of B. subtilis are sold commercially in most European countries, although little is understood about how these bacteria exert their therapeutic benefit. B. subtilis is a gram-positive, nonpathogenic, spore-forming organism normally found in the soil, and the robustness of spores is thought to enable passage across the gastric barrier, where a proportion of spores germinate in the small intestine and populate, albeit briefly, the intestinal tract (16). In addition, the clinical effects of B. subtilis as an immunostimulatory agent in a variety of diseases (10, 18, 20, 24), as an in vitro and in vivo stimulant of secretory immunoglobulin A (2, 10), and as an in vitro mitogenic agent (6) have been documented. Other topical examples of such probiotic bacteria are the gram-positive lactobacilli and lactococci, which are sold commercially for both human and veterinary use.
Two commercial B. subtilis probiotic preparations are Enterogermina (Sanofi Winthrop, Milan, Italy), sold in Europe, and Biosubtyl (Biophar Co. Ltd., Nha Trang, Vietnam), sold in Southeast Asia. Although numerous reports have documented the clinical effects of oral administration of B. subtilis spores, the bacteria contained in these preparations have not been characterized. In this article we report the preliminary characterization of the bacteria contained in the commercial preparations Enterogermina and Biosubtyl.
Preliminary characterization of strains.
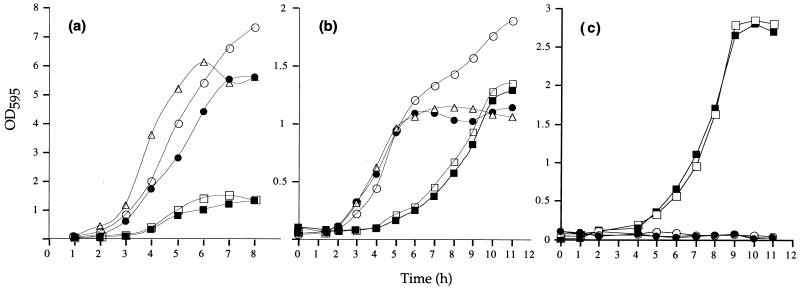
Bacteria were recovered from both the Enterogermina and Biosubtyl commercial preparations and were found to contain 2.9 × 108 spores/ml and 1 × 107 spores/g, respectively. Enterogermina is reportedly derived from the penicillin-resistant B. subtilis ATCC 9799 (5), and this strain was used here as a reference in addition to the genetically characterized prototrophic B. subtilis PY79 (27), which is a derivative of the Marburg type strain 168 (4). Initial observation of colonies grown on Luria-Bertani (LB) or Difco sporulation medium (DSM) solid agar showed the recovered Enterogermina or Biosubtyl bacteria to be homogeneous but revealed significant differences from PY79 and ATCC 9799. Biosubtyl produced intensely white, smooth, circular colonies which were markedly mucoid. Enterogermina produced rhizoid colonies, in contrast to PY79 and ATCC 9799, which produced smooth, circular colonies. Growth of Enterogermina in LB medium was found to be significantly reduced, both in growth rate and in maximum cell densities attained, compared to PY79, Biosubtyl, or ATCC 9799 (Fig. 1A).
FIG. 1.
Growth of probiotic strains. Bacterial strains were grown at 37°C in LB medium (a), DSM (b), or medium at pH 10.1 (c). B. subtilis PY79 (●), Biosubtyl (○), Enterogermina (two isolates [□ and ■]) and ATCC 9799 (▵) were used. For growth in alkaline medium, sodium carbonate was used to adjust the pH to 10.1 in liquid and solid media as described by Horikoshi and Teruhiko (11). OD595, optical density at 595 nm.
As shown in Table 1, we found that the probiotic strains differed from PY79 in production of amylase, maximum growth temperature, and growth at high pH. Biosubtyl was unable to produce amylase, which is an established marker for B. subtilis (23). As will be shown below, an important finding was that Enterogermina can grow in both solid and liquid medium at pH 10.1 (Table 1 and Fig. 1C); neither B. subtilis PY79 nor ATCC 9799 was able to grow under these conditions, although Biosubtyl grew weakly on solid agar but not in liquid media.
TABLE 1.
Differential characteristics of probiotic strains
| Characteristic | Strain
|
|||
|---|---|---|---|---|
| PY79 | Biosubtyl | Enterogermina | ATCC 9799 | |
| Hydrolysis of starcha | + | − | (+) | + |
| Maximum growth temperatureb | 54°C | 54°C | 48°C | 51°C |
| Growth at pH 10.1 | − | (+)c | + | − |
| Sporulation efficiencyd | 71% | 48% | 47% | 0.84% |
| Penicillin resistancee | S | S | R | R |
| Erythromycin resistancee | S | S | R | S |
| Lincomycin resistancee | S | S | R | S |
| Rifampin resistancee | S | S | R | S |
| Chloramphenicol resistancee | S | S | R | S |
| Neomycin resistancee | S | S | R | S |
| Tetracycline resistancee | S | S | S | S |
Hydrolysis of starch by amylase was measured as described previously (7). Parentheses indicate low levels of starch hydrolysis.
Maximum temperature at which growth could be sustained on LB agar plates.
Weak growth on solid medium only.
Sporulation was induced by nutrient exhaustion in DSM, and samples were examined at T24 (24 h after the onset of spore formation) for viable counts and the number of survivors of heat treatment (65°C, 45 min) (19).
Strains were streaked or plated on LB agar plates containing erythromycin (1 μg/ml), lincomycin (25 μg/ml), penicillin G (1.25 μg/ml), rifampin (100 μg/ml), chloramphenicol (100 μg/ml), neomycin (100 μg/ml), or tetracycline (100 μg/ml). R, resistant; S, sensitive.
Sporulation.
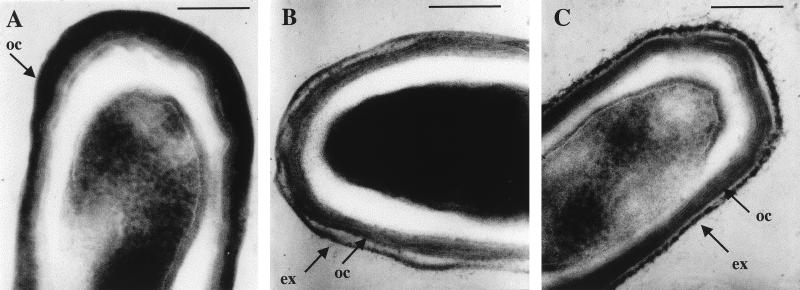
Strains were grown in the sporulation medium DSM, which induces spore formation by nutrient exhaustion (19). In this medium, Enterogermina grew at a rate indistinguishable from that of the other strains, although a lag of 2 to 3 h was always observed (Fig. 1B). However, the reference strain ATCC 9799 produced extremely low levels of spores (Table 1), and this finding was verified by observation of sporulating colonies grown on DSM agar. Spores of all strains were ellipsoidal and positioned mid-center. We used electron microscopy of uranyl acetate-stained thin sections (21) to examine mature spores of each strain (Fig. 2). Spores of B. subtilis PY79 possess a well-defined coat morphology comprising a lamellar inner layer and an electron-dense outer coat (1, 8). Our results show clearly that both Enterogermina and Biosubtyl spores exhibited a very different coat structure. Both Enterogermina and Biosubtyl spores possess an outer layer, which appeared loose and was unevenly associated with the electron-dense outer coat. This layer probably constitutes an exosporium, a complex and poorly understood spore structure (1).
FIG. 2.
Electron micrographs of mature spores. (A) B. subtilis PY79; (B) Biosubtyl; (C) Enterogermina. Bar, 0.2 μm. oc, outer coat; ex, exosporium.
Antibiotic resistances of Enterogermina.
Enterogermina is reported to contain a mixture of four antibiotic strains, each containing a unique spectrum of antibiotic resistance markers (5, 15). Since bacterial therapy is sometimes combined with the administration of antibiotics, these markers have been introduced by Sanofi Winthrop. Enterogermina reportedly originates from B. subtilis ATCC 9799, which is described as a producer of penicillinase. One strain was isolated by single- and multistep selection methods, which conferred chromosomal-borne resistance to erythromycin, lincomycin, cephalosporins, and cycloserine (5). Further derivatives, resistant to chloramphenicol (derivative O/C), novobiocin and rifampin (derivative N/R), tetracycline (derivative T), and streptomycin and neomycin (derivative SIN) were subsequently isolated from this strain (5, 15, 17). The commercial preparation of Enterogermina contains a mixture of equal amounts of all four derivatives (O/C, N/R, T, and SIN). We serially diluted the Enterogermina preparation directly onto selective plates (Oxoid Isosensitest agar), using the MICs defined by Ciffo (5), and were able to isolate individual colonies which carried unique antibiotic resistances to chloramphenicol, rifampin, and neomycin and presumably corresponded to derivatives O/C, N/R, and SIN (Table 1). All individual antibiotic-resistant isolates appeared to be phenotypically identical. However, we were unable to identify derivative T, which confers resistance to tetracycline, and it is possible that this chromosomal-borne mutation has been lost by reversion through repeated passaging. We also confirmed (Table 1) that all Enterogermina isolates are resistant to erythromycin, lincomycin, and penicillin G and that strain ATCC 9799 is resistant to penicillin G.
Deposition of strains.
The following strains, classified in this work, were deposited at the Bacillus Genetic Stock Centre, Department of Biochemistry, The Ohio State University: 14A1 (Biosubtyl), 15A1 (Enterogermina O/C), 15A2 (Enterogermina N/R), 15A3 (Enterogermina SIN) and 3A15 (ATCC 9799).
Analysis of 16S rRNA gene sequences.
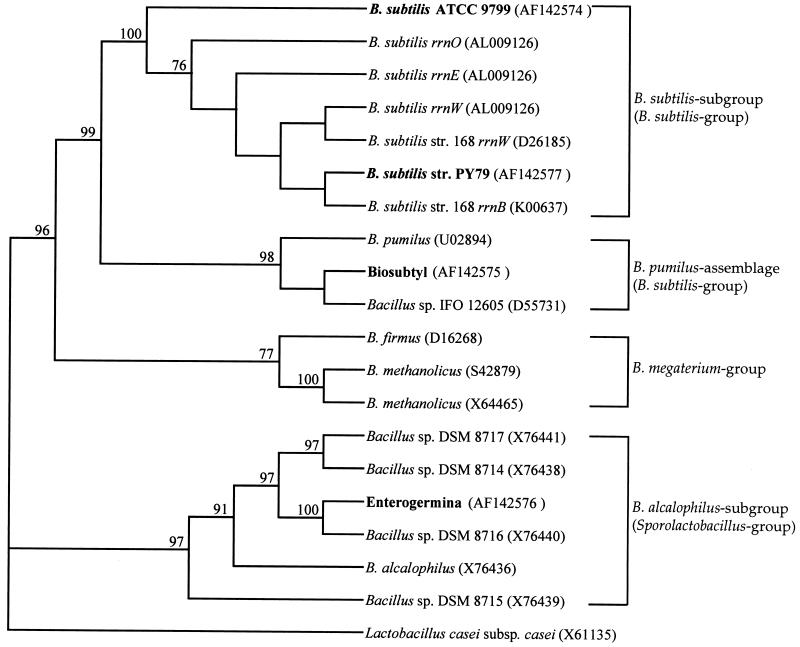
Our preliminary characterizations, based on colony morphology, growth, and sporulation, suggested that Biosubtyl and Enterogermina were significantly different from B. subtilis and might constitute alternative Bacillus species. To establish the relatedness of these strains at the genetic level, we sequenced the entire 16S rRNA genes from strains PY79 and ATCC 9799 and from Biosubtyl and Enterogermina. Sequence analysis of 16S rRNA has been increasingly relied upon to analyze species similarity (3, 14, 25, 26). We used two oligonucleotides to amplify the entire 16S rRNA as follows: P1 (5′-GCGGCGTGCCTAATACATGC anneals to nucleotides 40 to 59) and P2 (5′-CACCTTCCGATACGGCTACC anneals to nucleotides 1532 to 1513 of B. subtilis rrnE). The 1,400-base PCR product was sequenced in its entirety by using an automated sequencer. Phylogenetic analysis (Fig. 3) showed that ATCC 9799 (accession no. AF142574) was a member of the B. subtilis subgroup, although it was distinct from our laboratory type strain PY79. Biosubtyl (accession no. AF142575) was within the B. subtilis group but was more closely aligned with members of the Bacillus pumilus assemblage (13). Interestingly, this association was supported by the failure of Biosubtyl to produce amylase; a marker for discriminating B. subtilis from B. pumilus (23). We suggest that Biosubtyl is more likely to be a strain of B. pumilus. Enterogermina (accession no. AF142576) was unrelated to B. subtilis and its purported parent strain, ATCC 9799, and was aligned instead with members of the Sporolactobacillus group (13) (n. b., we have sequenced three independent isolates of Enterogermina). Enterogermina was most closely related to members of the subgroup Bacillus alcalophilus (13), which can tolerate alkaline environments (23). Our finding that Enterogermina can grow well in an alkaline medium suggests that this probiotic species may be a strain of B. alcalophilus. It was reported previously that Enterogermina cannot be transformed with chromosomal DNA prepared from another B. subtilis strain (15, 17). This result was attributed to the poor competence of Enterogermina, but we suggest that it more likely reflects an interspecies barrier. Our sequence analysis of ATCC 9799 also demonstrates that Enterogermina has rather obscure origins and that it clearly cannot have originated from strain ATCC 9799.
FIG. 3.
Phylogenetic relationship of Biosubtyl and Enterogermina. Phylogenetic relatedness of Biosubtyl and Enterogermina compared to representative Bacillus species. The branching pattern, rooted with Lactococcus casei as the outgroup, was generated by distance-matrix alignment (12) and neighbor joining (22) by using the PHYLIP suite of computer programs (9). Bootstrap values are given for each node having 70% or greater agreement. Group, subgroup, and assemblage associations are derived from sequence identity to the Ribosomal Database Project (13).
In conclusion, we found that two commercial preparations of probiotic bacteria purported to contain B. subtilis contain instead Bacillus species that are closely (Biosubtyl) and distantly (Enterogermina) related to B. subtilis. This finding is medically important and raises the question of whether any nonpathogenic, gram-positive microorganism can serve as a probiotic agent.
Acknowledgments
This work was supported by grants from the Medical Research Council (S.C.), The Wellcome Trust (S.C.), and the European Union (S.C. and E.R.).
REFERENCES
- 1.Aronson A I, Fitz-James P. Structure and morphogenesis of the bacterial spore coat. Bacteriol Rev. 1976;40:360–402. doi: 10.1128/br.40.2.360-402.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonomo R, Luzi G, Frielingsdorf A, Aiuti F. Ruolo delle IgA secretorie nelle funzioni dell’immunita locale dell’apparato digerente. Impiego di spore di B. subtilis in alcune forme morbose con deficit di IgA e ipogammaglobulinemia. Chemioter Antimicrob. 1980;3:237–240. [Google Scholar]
- 3.Brunk C F, Avaniss-Aghajani E, Brunk C A. A computer analysis of primer and probe hybridization with bacterial small-subunit rRNA sequences. Appl Environ Microbiol. 1996;62:872–879. doi: 10.1128/aem.62.3.872-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkholder P R, Giles N H. Induced biochemical mutants in Bacillus subtilis. Am J Bot. 1947;34:345–348. [PubMed] [Google Scholar]
- 5.Ciffo F. Determination of the spectrum of antibiotic resistance of the Bacillus subtilis strains of Enterogermina. Chemioterapia. 1984;3:45–52. [PubMed] [Google Scholar]
- 6.Ciprandi G, Scordamaglia A, Venuti D, Caria M, Canonica G W. In vitro effects of Bacillus subtilis on the immune response. Chemioterapia. 1986;5:404–407. [PubMed] [Google Scholar]
- 7.Cutting S M, Vander-Horn P B. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons Ltd.; 1990. pp. 27–74. [Google Scholar]
- 8.Driks A. Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 1999;63:1–20. doi: 10.1128/mmbr.63.1.1-20.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein J. PHYLIP (Phylogeny Inference Package). Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 10.Fiorini G, Cimminiello C, Chianese R. II B. subtilis come stimolatore selettivo delle IgA linfocitarie di membrana. Farmaci. 1985;9:331–334. [Google Scholar]
- 11.Horikoshi K, Teruhiko A. Alkalophilic microorganisms. New York, N.Y: Japan Scientific Societies Press; 1982. [Google Scholar]
- 12.Jukes T H, Cantor C R. Evolution of protein molecules. In: Murano N H, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 13.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchesi J R, Sato T, Weightman A J, Martin T A, Fry J C, Hiom S J, Wade W G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazza G. Genetic studies on the transfer of antibiotic resistance genes in Bacillus subtilis strains. Chemioterapia. 1983;2:64–72. [Google Scholar]
- 16.Mazza P. The use of Bacillus subtilis as an antidiarrhoeal microorganism. Boll Chim Farm. 1994;133:3–18. [PubMed] [Google Scholar]
- 17.Mazza P, Zani F, Martelli P. Studies on the antibiotic resistance of Bacillus subtilis strains used in oral bacteriotherapy. Boll Chim Farm. 1992;131:401–408. [PubMed] [Google Scholar]
- 18.Meroni P L, Palmieri R, Barcellini W, De Bartolo G, Zanussi C. Effect of long-term treatment with B. subtilis on the frequency of urinary tract infections in older patients. Chemioterapia. 1983;2:142–144. [Google Scholar]
- 19.Nicholson W L, Setlow P. Sporulation, germination and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1990. pp. 391–450. [Google Scholar]
- 20.Novelli A, Ulivelli A, Reali E F, Mannelli F, Trombi-Belcari L, Spezia R, Periti P. Bacillus subtilis spores as a natural pro-host oral agent. Preliminary data in children. Chemioterapia. 1984;3:152–155. [PubMed] [Google Scholar]
- 21.Page A M, Lagnado J R, Ford T W, Place G. Calcium alginate encapsulation of small specimens for transmission electron microscopy. J Microsc. 1994;175:166–170. [Google Scholar]
- 22.Saitou N, Nei M. The neighbor-joining method: a new method for reconstruction of phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Sneath P H A. Endospore-forming gram-positive rods and cocci. In: Holt J G, editor. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams & Wilkins; 1986. pp. 1104–1207. [Google Scholar]
- 24.Vacca A, Pantaleo G, Ronco M, Dammacco F. Chemoimmunotherapy for multiple myeloma using an intermittent combination drug schedule (melphalan + prednisone) and alternating course of B. subtilis spores. Chemioterapia. 1983;2:300–305. [Google Scholar]
- 25.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson K H, Blitchington R B, Greene R C. Amplification of bacterial 16S ribosomal DNA with the polymerase chain reaction. J Clin Microbiol. 1990;28:1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Youngman P, Perkins J, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]