Abstract
c-Fos is a useful marker gene of neuron activation for neuroscience and physiology research. The mechanism and function of neural networks have been elucidated using c-Fos reporter knock-in (KI) mice, but the small size of the mice makes it difficult to perform surgical procedures on specific brain regions. On the other hand, there is a large amount of accumulated data on behavioral studies using rats. Thus, the generation of c-Fos reporter rat is expected, but it is difficult to generate gene-modified rats. Furthermore, c-Fos gene abnormality is expected to be severe in rats, as shown in homozygous of c-Fos knockout (KO) mouse, but such analysis has rarely been performed and is not certain. This study generated c-Fos-deficient rats using CRISPR/Cas, with 1067 bp deletion including exon 1 of the c-Fos gene. Homozygous c-Fos KO rats had growth latency and the same tooth and bone abnormality as homozygous c-Fos KO mice but not heterozygous c-Fos KO rats. Therefore, the c-Fos gene in rats is expected to have the same function as that in mice, and the generation of c-Fos reporter KI rats is further anticipated.
Keywords: bone, c-Fos, gene modification, knockout rat
Introduction
Fos (c-Fos) [1, 2] is one of the Fos family proteins, including Fra1 [3], Fra2 [4], and FosB [5], and works as a part of the compartment of activator protein-1 (AP-1), a dimeric transcription complex regulating gene expression [6,7,8]. More than 30 years ago, c-Fos was identified as a homolog of v-fos, FBJ murine osteosarcoma virus proviral DNA, from a murine cell line [1]. In 1992, two c-Fos knockout (KO) mice were established, and these strains had severe phenotypes, including reduced body size, a lack of teeth, and abnormal bone development [9, 10]. c-Fos KO mice have been used as model mice for osteopetrosis because c-Fos deficiency disturbed the differentiation of osteoclasts responsible for bone resorption [11, 12]. In contrast, Morgan et al. demonstrated that c-Fos expression is rapidly and transiently induced by the provoking of a voltage-dependent calcium influx with PC12 cells, which can differentiate neuron-like cells [13]. In addition, they revealed that the c-Fos expression of neuron is a short period after stimulation in vivo [14]. Therefore, c-Fos is widely used as a marker of neural activation in neuroscience and physiology.
c-Fos reporter mice, which can indicate c-Fos expression as reporters, such as fluorescent protein, luciferase, and LacZ using the Cre/loxP system, are useful experimental animals for neural network research in neuroscience and physiology [15,16,17]. In particular, Guenthner et al. established a novel method, targeted recombination in active populations (TRAP), combined with c-Fos reporter knock-in (KI) mice [18]. The TRAP system allows marking neural cells activated by a certain stimulus in vivo. Due to these c-Fos reporter mice, it is unnecessary to sacrifice the experimental animal immediately after the experiment to detect c-Fos expression with immunostaining or in situ hybridization.
In neurophysiology research, rats have been used as a major experimental animal compared with mice because rats have strong advantages. First is the large size of their body, which has a sufficient quantity of tissue or blood for molecular analysis, and surgery on specific brain regions for neurophysiology experiments. Second is the accumulation of valuable knowledge about physiology studies, including behavioral tests, or pharmacological and nutritional studies. Third are the breeding advantages like those of mice, such as easy handling and breeding, short life cycle, and high fertility. On the basis of these advantages, the establishment of c-Fos reporter rats would be useful for physiological studies.
To generate KI rats with a reporter gene inserted under the promoter of the endogenous c-Fos gene, one allele of c-Fos is inevitably lost in the generation of these rats. However, there are few in vivo studies about c-Fos deficiency in rats. There is a strong concern that a c-Fos defect in rats would disturb the physiological experiment because c-Fos-deficient mice have severe phenotypes described above [9, 10]. In this study, we generated a c-Fos KO rat strain with the CRISPR/Cas system, and observed the phenotype of c-Fos KO rats, focusing on growth and bone development as shown in c-Fos KO mice.
Materials and Methods
Ethics
The experimental protocol was approved by the Institutional Animal Care and Use Committee and the Safety Committee for Recombinant DNA Experiment at Tottori University.
Animals
Slc:Wistar and Slc:SD rats were purchased from Japan SLC (Japan) and kept under the standard conditions (room temperature: 23°C, light cycle: 12 h light/12 h dark, light from 7:00 AM). c-Fos KO rats were fed powder diets (CE-2, powder; CLEA Japan, Inc., Tokyo, Japan) after weaning.
CRISPR/Cas system
The rat genome sequence assembly Rnor_6.0 was used. The target sequence of guide RNA (gRNA) was designed by CRISPRdirect (“https://crispr.dbcls.jp”) [19]. The 1,000 bp DNA sequence from 500 bp upstream of the 1st ATG was input in CRISPRdirect, and two target sequences were selected for gRNAs. The target sequences are shown in Fig. 1a. crRNAs containing the target sequences were synthesized (Alt-R® CRISPR-Cas9 crRNA; Integrated DNA Technologies, Inc., Coralvill, IA, USA). Ribonucleoprotein (RNP) was made of 12.5 ng/µl of each crRNA, 25 ng/µl Alt-R® CRISPR-Cas9 tracrRNA (1072532; Integrated DNA Technologies), and 20 ng/µl Alt-R® S.p. Cas9 Nuclease V3 (1081058; Integrated DNA Technologies) in sterilized water (W1503; Sigma-Aldrich Co., Burlington, MA, USA) and filtered with Ultrafree-MC (UFC30GV25; Merck KGaA, Darmstadt, Germany).
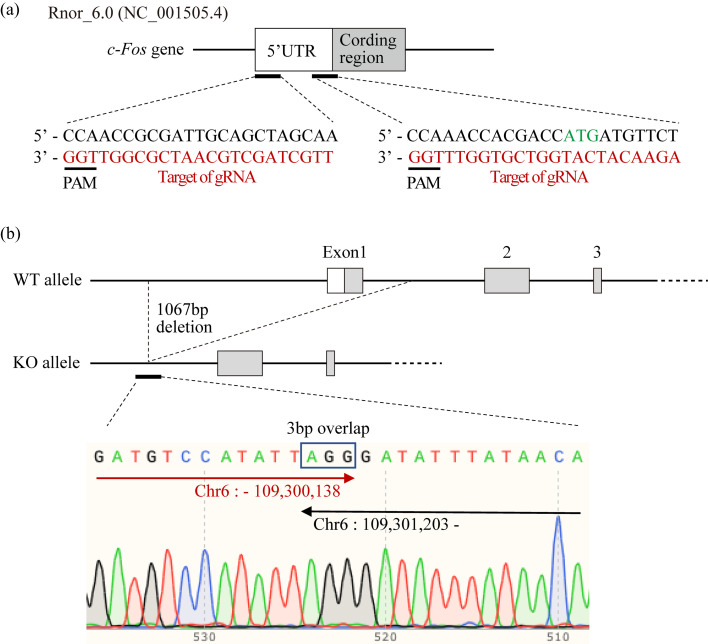
Fig. 1.
Generation of c-Fos KO rats. (a) CRISPR/Cas design for c-Fos KO rats. Two gRNA were used for the generation of c-Fos KO rats. The red characters show the target sequence of gRNA, and the green characters show the first ATG of the c-Fos gene. (b) DNA sequence analysis of c-Fos KO rats. The genome of c-Fos KO rats was amplified with the sense primer 5′-ggcgagctgttcccgtcaatccc and the antisense primer 5′-gtccagaatcgctactcacctgctctac. The PCR product was sequenced with the antiprimer.
Generation of c-Fos KO rats
Thirteen- to 20-week-old female Slc:SD rats were superovulated with pregnant mare serum gonadotropin (PMSG; 150 IU/kg body weight, intraperitoneally, Serotropin; ASKA Animal Health Co., Ltd., Tokyo, Japan) at 11:00 AM on the day of metestrus, and human chorionic gonadotropin (hCG; 75 IU/kg body weight, intraperitoneally, Puberogen; Novartis Animal Health K.K., Tokyo, Japan) injection was administered 48 h after PMSG administration. After hCG injection, the female rats were crossed with sexually matured male Slc:SD rats. The next day, mating was confirmed by the vaginal plug and sperm in their vagina, and zygotes were collected from the oviduct after sacrificing by cervical dislocation under 5% isoflurane anesthesia. RNP was microinjected into the cytoplasm and pronuclei of zygotes, and injected zygotes were cultured in vitro with KSOM-R [20]. Injected zygotes were transferred into the oviduct of pseudo-pregnant Slc:Wistar rats, which were mated with vasectomized Slc:Wistar rats. c-Fos KO rats were deposited at the National BioResource Project-Rat (NBRP-Rat) [21], and the NBRP-Rat no. is 0968.
Genotyping
The genome of rats was purified from the tail, and PCR genotyping was performed with KOD FX Neo (KFX-201; Toyobo Co., Ltd., Osaka, Japan) according to the manufacturer’s instructions. The PCR genotyping primers 5′-gactcgctaactagagcctgggagg and 5′-cctgaattccgcagctcagcctttc were used to detect the c-Fos Wild-type (WT) allele, 5′-ggcgagctgttcccgtcaatccc and 5′-gtccagaatcgctactcacctgctctac were used to detect the c-Fos KO allele. The PCR product was purified with Wizard® SV Gel and PCR Clean-Up System (A9281; Promega Co., Madison, WI, USA) and sequenced with Value Read DNA sequence (Eurofins Genomics Co., Ltd., Tokyo, Japan).
Histological analysis
Under anesthesia with 3.5% isoflurane inhalation, 4-week-old c-Fos KO rats were perfused with saline, followed by Mildform 10N (131-10317; FUJIFILM Wako Pure Chemical, Osaka, Japan). Tissue samples were decalcified in 10% EDTA (pH 7) for 1 week. Decalcified tissues were embedded in paraffin. Paraffin‐embedded tissues were sliced into 5-µm-thick sections and analyzed using hematoxylin and eosin (H&E) staining. Safranin O (Waldeck GmbH & Co., Havixbecker, KG)‐Fast Green (FUJIFILM Wako Pure Chemical) staining was performed to observe the morphology of cartilage tissues. The images were captured by Nanozoomer S60 (Hamamatsu Photonics, Hamamatsu, Japan). The quantification of the epiphysis and medullary cavity area with the images was performed using ImageJ 1.53k software (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
All error bars show the standard deviation. Normality and equal variances were calculated by the Shapiro-Wilk test and Levene tests using R [22]. The Wilcoxon rank-sum test was performed using the “exactRankTests” package.
Results
Generation of c-Fos KO rats
The generation of c-Fos KO rats is shown in Fig. 1. The CRISPR/Cas system was used to generate c-Fos KO rats, and two target sequences of gRNA were designed (Fig. 1a). The most effective condition of RNP was examined for the modification of the c-Fos gene, and 55 founder rats were generated. Among all c-Fos founder rats, founders KO7 and KO12 had no incisors, and founder KO22 lacked the lower incisors. Founder KO12 was bred with female Slc:SD rats, and an incisor-less rat again was found from mating between offspring. The DNA sequence of the toothless offspring revealed that they had a 1067 bp deletion, including exon 1 (Fig. 1b). Therefore, the c-Fos KO strain was established from the founder rat KO12. The gene expression of c-Fos in homozygous c-Fos KO rats was examined by quantitative PCR (qPCR), and it was significantly lower than that in the WT and heterozygous c-Fos KO rats (Supplementary Fig. 1).
Appearance feature of c-Fos KO rats
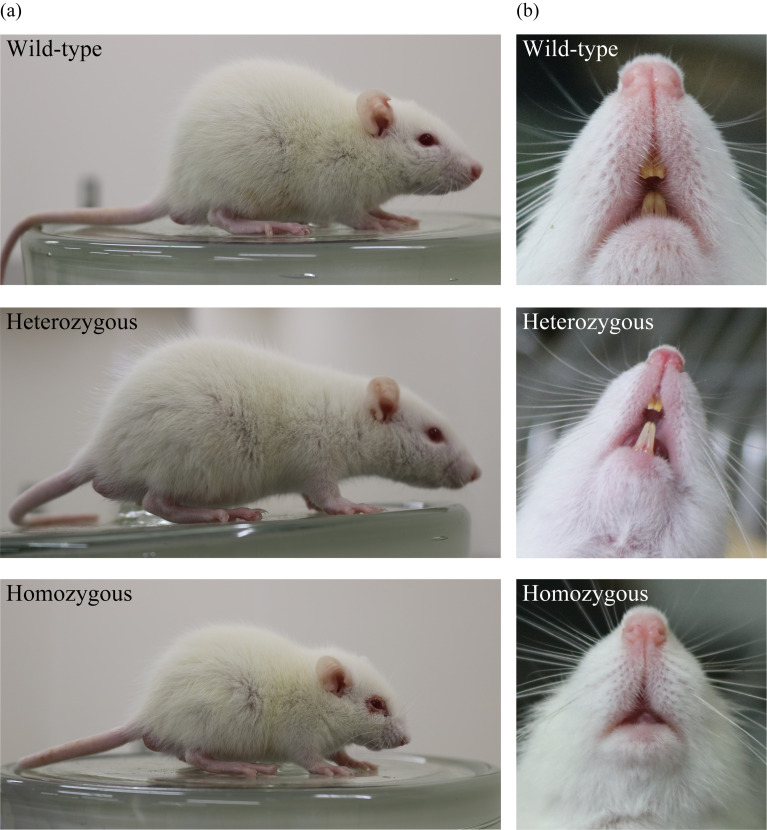
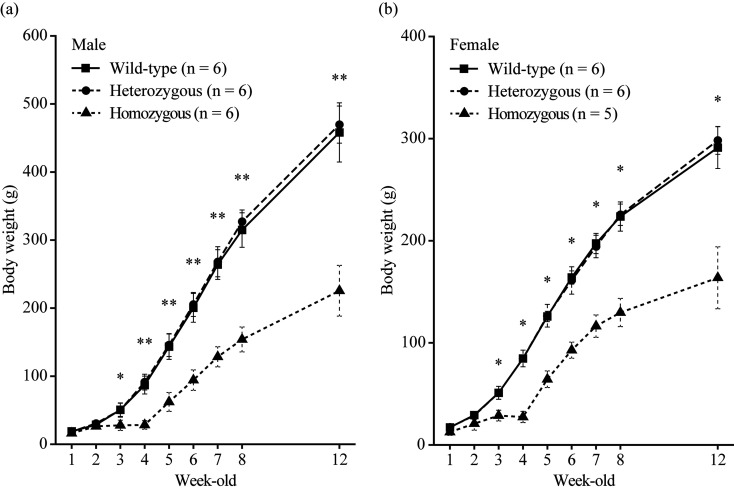
At birth, homozygous c-Fos KO rats could not be distinguished. However, after ~3 weeks, homozygous c-Fos KO rats were found to be smaller than other littermates and had shortened snouts and round heads (Fig. 2a). In contrast, heterozygous c-Fos KO rats seemed to be normal. Additionally, all homozygous c-Fos KO rats were toothless (Fig. 2b). The body weight of male homozygous c-Fos KO rats significantly decreased compared with that of WT rats after 3 weeks of age but that of heterozygous c-Fos KO rats did not decrease compared with that of WT rats (Fig. 3a). The body weight of female homozygous c-Fos KO rats was also similar (Fig. 3b). To confirm the fertility of homozygous c-Fos KO rats, one male and two female homozygous c-Fos KO rats were crossed. Offspring were obtained from all of 3 homozygous c-Fos KO rats, and an average of 8.2 pups were obtained.
Fig. 2.
Appearance of 4-week-old c-Fos KO rats. (a) Whole-body appearance of c-Fos KO rats. (b) Teeth of c-Fos KO rats.
Fig. 3.
Body weight of c-Fos KO rats. (a and b) Body weight of male (a) and female (b) c-Fos KO rats [female homozygous c-Fos KO rats (n=5) and others (n=6)]. The rats were weaned and fed powder diets at 4 weeks of age. The error bar shows the standard deviation, and the significant differences were tested using the Steel test. *P<0.05; **P<0.01.
Tooth and bone disorder in homozygous c-Fos KO rats
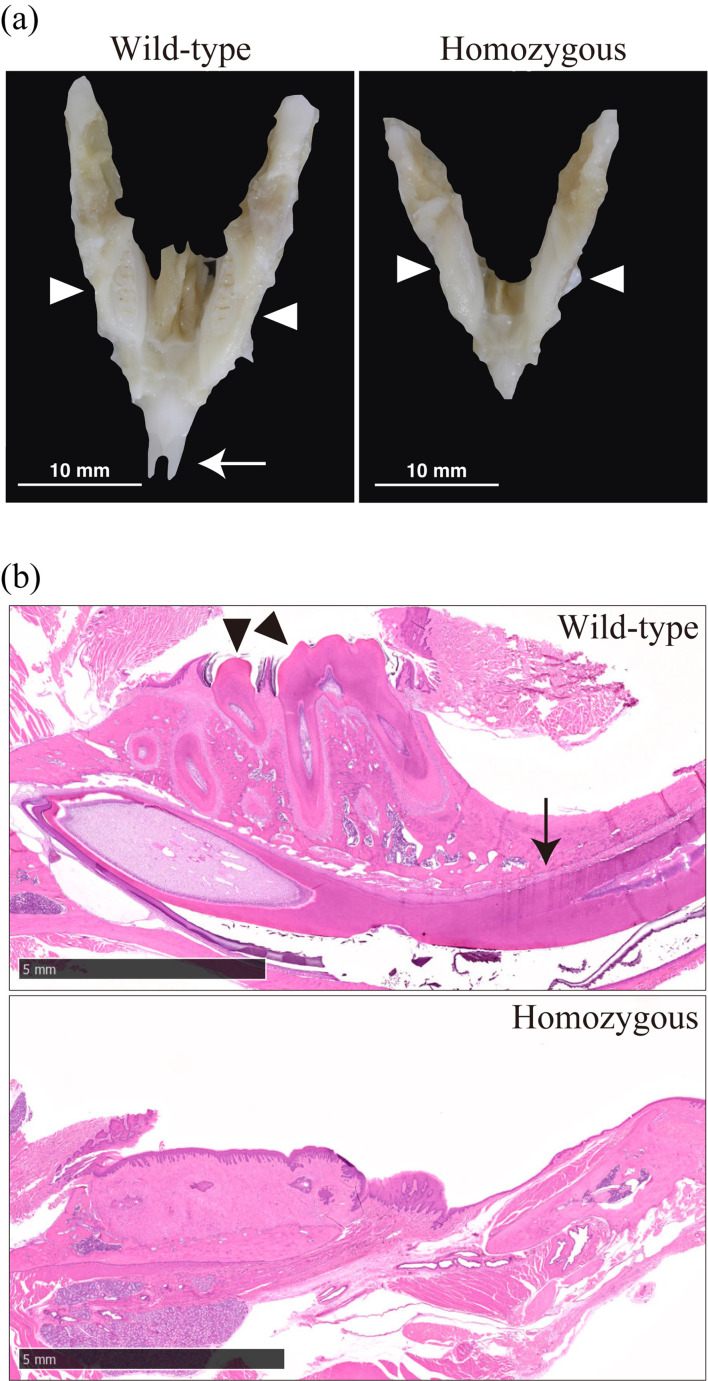
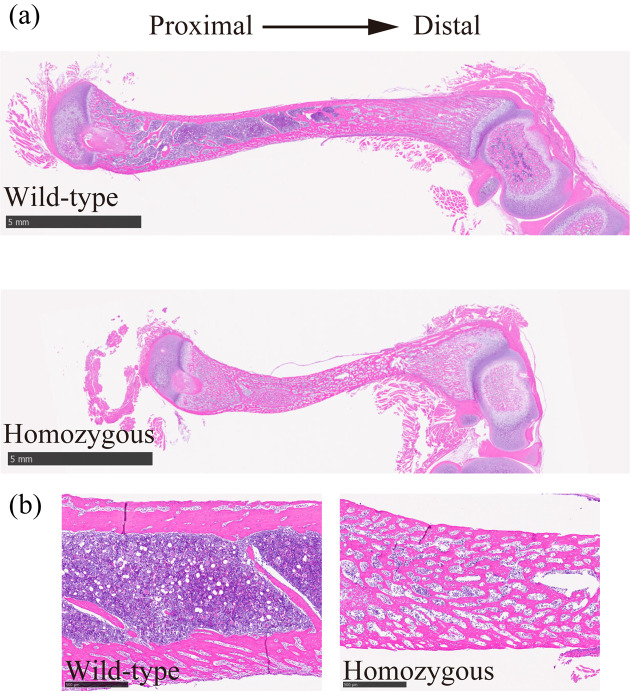
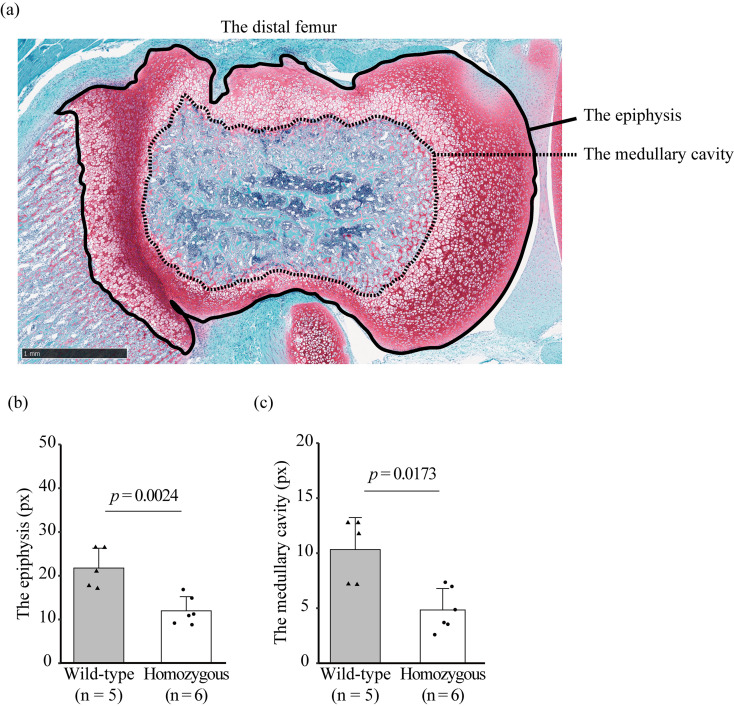
The jawbone of homozygous c-Fos KO rats was smaller than that of WT rats (Fig. 4a). H&E staining revealed that adult homozygous c-Fos KO rats had a tooth disorder (Fig. 4b). H&E staining of the femur showed that the diaphysis of homozygous c-Fos KO rats was short and had many small medullary cavities compared with that of WT rats (Figs. 5a and b). This study focused on the epiphysis of the distal femur (Fig. 6a). The distal epiphysis and medullary cavity of homozygous c-Fos KO rats were significantly smaller than those in WT rats (Figs. 6b and c).
Fig. 4.
c-Fos KO rats have a molar and incisor disorder. (a) Mandible of c-Fos KO rats. White arrows and white arrowheads show the incisor and molar, respectively. (b) H&E staining of the mandible of c-Fos KO rats. Black arrows and black arrowheads show the incisor and molar, respectively. The black bar indicates 5 mm.
Fig. 5.
Histological features of the femur of c-Fos KO rats at 4 weeks of age. (a) Overview of the femur stained with H&E. Scale bar, 5 mm. (b) Diaphysis of the femur stained with H&E. Scale bar, 1mm.
Fig. 6.
Disorder of the epiphysis of the distal femur in male c-Fos KO rats at 4 weeks of age. (a) Image of the area measured as the epiphysis and medullary cavity in the distal femur. The solid and dashed lines show the edge of the epiphysis and medullary cavity, respectively. Scale bar, 1 mm. (b and c) Area of the epiphysis (b) and medullary cavity (c) of the distal femur [WT (n=5) and homozygous c-Fos KO rats (n=6)]. The error bar shows the standard deviation, and the significant differences were tested using Student’s t-test (b) and Wilcoxon rank-sum test (c).
Discussion
The c-Fos KO rat strain was established using the CRISPR/Cas system, and the heterozygous c-Fos KO rat was normal in the appearance and growth. However, the homozygous c-Fos KO rat had a severe growth and bone development phenotype.
When using the CRISPR/Cas system, we needed to be aware off-target effects, of which unintended gene modifications could occur except for the target gene. However, founder rats were bred up to seven generations. We keep breeding c-Fos KO rats with Slc:SD rats. Unexpected off-target effects would fade away with each generation. Even if there was an off-target effect, c-Fos KO rat phenotypes were unlikely to have been affected by the off-target effect because the same phenotype was observed across all homozygous c-Fos KO rats.
The c-Fos gene was slightly expressed in homozygous c-Fos KO rats (Supplementary Fig. 1) despite of the fact that the deletion region extended to the promoter region and 1st ATG of c-Fos gene (Fig. 1b). Although some transcripts might be expressed from downstream of the deletion region, the c-Fos transcript in homozygous c-Fos KO rat wouldn’t be normal. Actually, the severe phenotype had emerged in homozygous c-Fos KO rat.
Homozygous c-Fos KO rat remarkably resembled c-Fos KO mice. Regarding the appearance, homozygous c-Fos KO rat had a shortened snout, round head, and lack of incisor, as shown in Fig. 2a. Johnson et al. described that c-Fos-deficient mice had a foreshortened snout, domed skull, and lack of incisor eruption [10]. The Mendelian ratio of homozygous c-Fos KO rat derived from crossbreeding of heterozygous c-Fos KO rats was significantly lower than that of the WT, which is similar to the c-Fos KO mice study (Supplementary Table 1). In addition, another c-Fos-deficient mouse strain that Wang et al. established had similar features [9]. The body weight of male homozygous c-Fos KO rat was approximately 40% decreased compared with that of WT and heterozygous c-Fos KO rat after 3 weeks of age (Fig. 3a). This rate was close to that of the c-Fos KO mice that Wang et al. generated [9]. The tooth defect of homozygous c-Fos KO rat is a serious problem for food intake for survival. Homozygous c-Fos KO rat were given a powder diet after weaning; then, these rats could survive beyond 12 weeks of age, and produce offspring.
Besides, homozygous c-Fos KO rat showed bone development abnormalities similar to those seen in c-Fos KO mice. c-Fos is an essential factor that induces osteoclast differentiation from progenitor cells [11, 12]. This suggests that homozygous c-Fos KO rat might have bone absorption abnormalities resulting from a deficiency of osteoclasts, similar to c-Fos KO mice. There is a positive correlation between body height and weight, and bone growth is essential for an increase in body height. Therefore, bone abnormalities often cause growth retardation. This possibility could explain why the body weight of homozygous c-Fos KO rats was lower than that of heterozygous and WT rats. c-Fos-deficient mice are used as model mice of osteopetrosis. Thereafter, c-Fos KO rat might be model rats of osteopetrosis.
To ensure proper expression of reporter genes, the KI technique is more rigorous than random integration of transgenes. In KI, the reporter gene is inserted into the endogenous target gene, and then target gene expression from the KI allele is often affected or disrupted. In some cases, the disruption of one allele might cause abnormal phenotypes, such as the disruption of methyl CpG binding protein 2 and fibroblast growth factor receptor 3 genes [23, 24]. The phenotype of disruption of target gene should be confirmed in detail to choose which gene modification style, e.g., conventional transgenic (TG), conditional KI, and other methods, is suitable for generating reporter animals. In present study, the c-Fos KO phenotypes were not shown in heterozygous c-Fos KO rat. Hence, the c-Fos locus is a permissible locus for KI.
We succeeded to establish c-Fos KO rats and observed their phenotype. The c-Fos KO rat phenotypes were not observed in the heterozygous c-Fos KO rat. However, KI rats targeted to the c-Fos locus must be maintained as heterozygous. Otherwise, when the c-Fos gene is null due to gene KI, it would not be suitable for neurophysiology experiments due to severe phenotypes. As a result, it might be inferred that the generation of c-Fos reporter KI rats is possible as in mice.
Funding
This study was supported by JSPS KAKENHI (grant no. JP20K21759, 21H03321 to S. Koba).
Supplementary
Acknowledgments
We thank Ms. Kaoru Shima for the support in animal care and Ms. Yui Yamane, Mr. Ryosuke Fujii, and Ms. Yumi Ono for the support in genotyping analysis. We thank Drs. Takeshi Hiyama, Miya Yoshino, Takao Mukuda, and Yuka Koyama for valuable discussion.
References
- 1.Curran T, Peters G, Van Beveren C, Teich NM, Verma IM. FBJ murine osteosarcoma virus: identification and molecular cloning of biologically active proviral DNA. J Virol. 1982; 44: 674–682. doi: 10.1128/jvi.44.2.674-682.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curran T, MacConnell WP, van Straaten F, Verma IM. Structure of the FBJ murine osteosarcoma virus genome: molecular cloning of its associated helper virus and the cellular homolog of the v-fos gene from mouse and human cells. Mol Cell Biol. 1983; 3: 914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen DR, Curran T. fra-1: a serum-inducible, cellular immediate-early gene that encodes a fos-related antigen. Mol Cell Biol. 1988; 8: 2063–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishina H, Sato H, Suzuki T, Sato M, Iba H. Isolation and characterization of fra-2, an additional member of the fos gene family. Proc Natl Acad Sci USA. 1990; 87: 3619–3623. doi: 10.1073/pnas.87.9.3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zerial M, Toschi L, Ryseck RP, Schuermann M, Müller R, Bravo R. The product of a novel growth factor activated gene, fos B, interacts with JUN proteins enhancing their DNA binding activity. EMBO J. 1989; 8: 805–813. doi: 10.1002/j.1460-2075.1989.tb03441.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu R, Boyle WJ, Meek J, Smeal T, Hunter T, Karin M. The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell. 1988; 54: 541–552. doi: 10.1016/0092-8674(88)90076-1 [DOI] [PubMed] [Google Scholar]
- 7.Sassone-Corsi P, Lamph WW, Kamps M, Verma IM. fos-associated cellular p39 is related to nuclear transcription factor AP-1. Cell. 1988; 54: 553–560. doi: 10.1016/0092-8674(88)90077-3 [DOI] [PubMed] [Google Scholar]
- 8.Rauscher FJ, 3rd, Cohen DR, Curran T, Bos TJ, Vogt PK, Bohmann D, et al. Fos-associated protein p39 is the product of the jun proto-oncogene. Science. 1988; 240: 1010–1016. doi: 10.1126/science.3130660 [DOI] [PubMed] [Google Scholar]
- 9.Wang ZQ, Ovitt C, Grigoriadis AE, Möhle-Steinlein U, Rüther U, Wagner EF. Bone and haematopoietic defects in mice lacking c-fos. Nature. 1992; 360: 741–745. doi: 10.1038/360741a0 [DOI] [PubMed] [Google Scholar]
- 10.Johnson RS, Spiegelman BM, Papaioannou V. Pleiotropic effects of a null mutation in the c-fos proto-oncogene. Cell. 1992; 71: 577–586. doi: 10.1016/0092-8674(92)90592-Z [DOI] [PubMed] [Google Scholar]
- 11.Grigoriadis AE, Wang ZQ, Cecchini MG, Hofstetter W, Felix R, Fleisch HA, et al. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science. 1994; 266: 443–448. doi: 10.1126/science.7939685 [DOI] [PubMed] [Google Scholar]
- 12.Tolar J, Teitelbaum SL, Orchard PJ. Osteopetrosis. N Engl J Med. 2004; 351: 2839–2849. doi: 10.1056/NEJMra040952 [DOI] [PubMed] [Google Scholar]
- 13.Morgan JI, Curran T. Role of ion flux in the control of c-fos expression. Nature. 1986; 322: 552–555. doi: 10.1038/322552a0 [DOI] [PubMed] [Google Scholar]
- 14.Morgan JI, Cohen DR, Hempstead JL, Curran T. Mapping patterns of c-fos expression in the central nervous system after seizure. Science. 1987; 237: 192–197. doi: 10.1126/science.3037702 [DOI] [PubMed] [Google Scholar]
- 15.Smeyne RJ, Schilling K, Robertson L, Luk D, Oberdick J, Curran T, et al. Fos-lacZ transgenic mice: mapping sites of gene induction in the central nervous system. Neuron. 1992; 8: 13–23. doi: 10.1016/0896-6273(92)90105-M [DOI] [PubMed] [Google Scholar]
- 16.Geusz ME, Fletcher C, Block GD, Straume M, Copeland NG, Jenkins NA, et al. Long-term monitoring of circadian rhythms in c-fos gene expression from suprachiasmatic nucleus cultures. Curr Biol. 1997; 7: 758–766. doi: 10.1016/S0960-9822(06)00334-4 [DOI] [PubMed] [Google Scholar]
- 17.Reijmers LG, Perkins BL, Matsuo N, Mayford M. Localization of a stable neural correlate of associative memory. Science. 2007; 317: 1230–1233. doi: 10.1126/science.1143839 [DOI] [PubMed] [Google Scholar]
- 18.Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron. 2013; 78: 773–784. doi: 10.1016/j.neuron.2013.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naito Y, Hino K, Bono H, Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015; 31: 1120–1123. doi: 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura K, Morimoto K, Shima K, Yoshimura Y, Kazuki Y, Suzuki O, et al. The effect of supplementation of amino acids and taurine to modified KSOM culture medium on rat embryo development. Theriogenology. 2016; 86: 2083–2090. doi: 10.1016/j.theriogenology.2016.07.001 [DOI] [PubMed] [Google Scholar]
- 21.Serikawa T, Mashimo T, Takizawa A, Okajima R, Maedomari N, Kumafuji K, et al. National BioResource Project-Rat and related activities. Exp Anim. 2009; 58: 333–341. doi: 10.1538/expanim.58.333 [DOI] [PubMed] [Google Scholar]
- 22.R Core Team. R: A language and environment for statistical computing, R Foundation for Statistical Computing. 2021; https://www.R-project.org/.
- 23.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001; 27: 322–326. doi: 10.1038/85899 [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Spatz MK, Kannan K, Hayk H, Avivi A, Gorivodsky M, et al. A mouse model for achondroplasia produced by targeting fibroblast growth factor receptor 3. Proc Natl Acad Sci USA. 1999; 96: 4455–4460. doi: 10.1073/pnas.96.8.4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.