Summary
Background
Given the importance of vaccination against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the prevention of severe coronavirus disease 2019 (COVID-19), detailed long-term analyses of neutralising antibody responses are required to inform immunisation strategies.
Methods
In this study, longitudinal neutralising antibody titres to an ancestral SARS-CoV-2 isolate and cross-neutralisation to delta and omicron isolates were analysed in individuals previously infected with SARS-CoV-2, vaccinated against COVID-19, or a complex mix thereof with up to two years of follow-up.
Findings
Both infection-induced and vaccine-induced neutralising responses against SARS-CoV-2 appeared to follow similar decay patterns. Following vaccination in previously infected individuals, neutralising antibody responses were more durable than prior to vaccination. Further, this study shows that vaccination after infection, as well as booster vaccination, increases the cross-neutralising potential to both delta and omicron SARS-CoV-2 variants.
Interpretation
Taken together, these results suggest that neither type of antigen exposure is superior for neutralising antibody durability. However, these results support vaccination to increase the durability and cross-neutralisation potential of neutralising responses, thereby enhancing protection against severe COVID-19.
Funding
This work was supported by grants from The Capital Region of Denmark’s Research Foundation, the Novo Nordisk Foundation, the Independent Research Fund Denmark, the Candys Foundation, and the Danish Agency for Science and Higher Education.
Keywords: Neutralising antibody, SARS-CoV-2, COVID-19, Longitudinal, Cross-neutralisation, Delta, Omicron, Virus isolate
Research in context.
Evidence before this study
Publications in relation to this study were searched in MEDLINE, PubMed and Embase using the search terms “severe acute respiratory syndrome coronavirus 2”, “SARS-CoV-2”, “COVID-19”, “Longitudinal”, “breadth/broad”, “neutralization/neutralisation”, “antibody”, “dynamics” and “kinetics”, which have been published up until 31 August 2022. Articles published in medRxiv and BioRxiv were not considered. A total of six major studies were found, which assessed longitudinal neutralising antibody responses as well as breadth of these responses after SARS-CoV-2 infection, COVID-19 vaccination, and/or a mix thereof. Common limitations amongst these studies included limited comparisons between the infected/vaccinated groups, limited follow up (up to one year), use of neutralisation to pseudo-typed models and lack of detection of possible reinfection.
Added value of this study
This study provides a thorough longitudinal analysis, with detection of possible infection and reinfection, as well as time-matched comparisons of neutralising antibody levels between infected, vaccinated, infected-vaccinated, vaccinated-boosted, and vaccinated-infected individuals with up to two years follow-up. The neutralising assays used in this study were of SARS-CoV-2 isolates, which clustered well with the sequences retrieved from infected individuals, and not pseudo-typed models. Breadth of neutralisation was also assessed using a select panel of each study group to understand the differences between each type of antigen exposure.
Implications of all the available evidence
Regardless of antigen exposure type, whether that be through vaccination or infection, the durability of neutralising titres in plasma are comparable, suggesting that neither type is superior. Additional antigen exposure, either through vaccination or infection, appears to improve the durability and breadth of neutralising responses when compared to that before vaccination or infection. If a boosting antigen is similar to that seen during a previous exposure, the breadth of neutralising responses is significantly increased to all other known variants, including the more recently discovered omicron variant.
Introduction
Since the beginning of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic ultimo 2019, coronavirus disease 2019 (COVID-19) has claimed the lives of millions worldwide.1 During late 2020, several COVID-19 vaccines were developed and given emergency use authorization worldwide. Initially, they showed high protection efficacy against development of severe COVID-19,2, 3, 4 but since then, the protection efficacy of these vaccines has dropped.5, 6, 7 This is most likely due to a combination of waning immunity and emerging SARS-CoV-2 variants of concern (VOCs), which, compared to ancestral variants, differ in their transmissibility levels and are more resistant to vaccine-induced immunity and immunity induced by previous infection.8, 9, 10, 11 To improve protective efficacies, booster vaccination programs have now been implemented as a standard of practice for many countries worldwide.
One of the main goals for vaccination against COVID-19 is the induction of neutralising antibodies (nAbs) that target the surface located spike (S) protein of the virus. Recent analyses have shown that nAbs are a strong correlate of protection from severe COVID-19,12,13 making them an ideal target for measuring protection efficacy. In the case of SARS-CoV-2, it is believed that most nAbs target epitopes are located within the S protein14,15. Analysis of emerging delta and omicron VOCs has shown that they carry specific amino acid changes in this protein, which likely account for the observed diminished neutralisation efficacy from prior infection or vaccination.16,17
In addition to anti-S antibodies, antibodies directed to the nucleocapsid (N) protein of the virus are also detected during SARS-CoV-2 infection.18 Although these antibodies have correlated with neutralising activity,19 their functionality remains putative. An advantage of eliciting antibodies to N is that they allow for discrimination of prior infection in those that have been vaccinated with Comirnaty®, Spikevax®, Vaxzevria® and Jcovden®, as these vaccines only target the S protein.
While there have been several studies on the dynamics of neutralising responses longitudinally,20, 21, 22, 23, 24, 25, 26 there is limited assessment of the durability of these responses following further antigen exposure, whether that be from infection or vaccination. Previously, our group studied virus neutralising antibody responses longitudinally in non-hospitalised individuals that had a SARS-CoV-2 infection and those vaccinated against COVID-19, using an ancestral SARS-COV-2 isolate.27, 28, 29 The present study assesses the durability of the longitudinal neutralisation responses of previously SARS-CoV-2 infected individuals who were vaccinated against COVID-19 in comparison to SARS-CoV-2 infection-naïve individuals who were vaccinated against COVID-19 with either a homologous or heterologous prime-boost vaccination regimen, and further boosted or exposed to SARS-CoV-2 infection. Further, a select panel of individuals had their cross-neutralisation potential to two major VOCs (delta and omicron) assessed. Overall, this study describes a highly detailed longitudinal analysis of antibody neutralisation responses against SARS-CoV-2.
Methods
Study cohort and SARS-CoV-2 screening by enzyme-linked immunosorbent assays
The Clinical, Virological and Immunological COVID-19 (CVIC) study is a prospective cohort of individuals either infected by SARS-CoV-2 or vaccinated against COVID-19. A total of 103 individuals with non-hospitalised COVID-19 and 55 individuals with hospitalised COVID-19 were recruited at Copenhagen University Hospital, Hvidovre, between 15 April 2020 to 1 February 2021. Additionally, 109 individuals vaccinated against COVID-19 with a homologous Comirnaty® prime-boost regimen (mRNA–mRNA group) and 21 individuals vaccinated against COVID-19 with a heterologous Vaxzevria® prime and either Comirnaty® or Spikevax® boost regimen (vector-mRNA group) were included between 27 December 2020 to 10 March 2021. For those infected, plasma was collected upon enrolment (baseline) and then at 1 week (1wk, hospitalised COVID-19 only), 1 month (1M, hospitalised COVID-19 only), 3M (hospitalised COVID-19 only), 6M, 12M and 24M (non-hospitalised COVID-19 only) post symptom onset (PSO). For those vaccinated, plasma was collected pre-vaccination, at 1M (completed 2-dose vaccine regimen), 5M, 9M (prior to booster vaccination [third vaccine dose]), 10M (1-month post booster vaccination [PBV]) and 14M post vaccination (PV).
All subjects were screened for the presence of anti-SARS-CoV-2 receptor binding domain (RBD) Abs via the WANTAI enzyme-linked immunosorbent assay (ELISA; Beijing Wantai, cat#: 256-WS-1096-96), according to the manufacturer's instructions. The mRNA–mRNA and vector-mRNA groups were screened at pre-vaccination to detect absence of prior infection and all groups were screened at baseline/1M PV to confirm seroconversion to infection/vaccination. Further, all individuals were required to report if they became SARS-CoV-2 PCR positive through routine diagnostic testing. Finally, to monitor for SARS-CoV-2 infection or reinfection events, all plasma in this study was tested for anti-SARS-CoV-2 nucleocapsid IgG using a EuroImmun semi-quantitative ELISA (PerkinElmer, cat#: EI 2606-9601-2 G). Assessment was performed according to the manufacturer's instructions with the exception that a 1/10 dilution of plasma was used instead of a 1/100 dilution. All tested plasma samples from vaccinated individuals were compared to their pre-vaccination time point, providing that this time point tested negative in the WANTAI ELISA. As there was a high degree of variability of the signal seen in healthy unexposed plasma, a signal/noise ratio of 3.0 or more was considered positive. Given that infected individuals did not have a pre-exposure time point in this study, all samples from these individuals were compared to the average signal from five healthy unexposed controls (WANTAI ELISA-negative). Demographic information regarding these 5 healthy controls can be found in Supplementary Table S1.1. Reinfection was defined as a 2-fold increase in the binding to the N protein compared to the time point prior (non-hospitalised and hospitalised groups).
Ethics
The study was approved by the Regional Ethical Committee (H-20025872) and Data Protection Agency (P-2020-357), respectively, and was conducted in compliance with the Declaration of Helsinki guidelines. All individuals included in this study were 18 years or older and able to read and speak adequate Danish to provide written informed consent. Participants were required to self-report their age, sex and ethnicity upon enrolment. Participants were included on a volunteer basis with ongoing inclusion throughout 2020 and 2021. Thus, all those that wanted to participate in this study regardless of age, sex or ethnicity were included. Study data was collected and managed using research electronic data capture (REDCap) tools hosted at Copenhagen University Hospital, Hvidovre.30
SARS-CoV-2 sequence acquisition from those infected
SARS-CoV-2 S protein sequences, where possible, were retrieved from next generation sequencing (NGS) data stored at the Department of Clinical Microbiology, Copenhagen University Hospital, Hvidovre. These sequences, along with the sequences of the isolates used in this study, were analysed using Mafft software (version 7.505) and a phylogenetic tree was constructed using Figtree software (version 1.4.4).
Neutralisation assay
All subjects were screened for neutralisation in Vero E6 cell-culture experiments using a D614G SARS-CoV-2 isolate (DK-AHH1, clade 20A, Genbank accession number MZ049597) obtained previously.29 Selected individuals were also screened against a delta SARS-CoV-2 isolate (DK-AHH3, clade 20J, accession number OP271297) and an omicron BA.1 SARS-CoV-2 isolate (DK-AHH4, clade 21K, accession number OP271296).
Neutralisation experiments were performed as previously described.28,29 In brief, virus was added to 2-fold serially diluted plasma at a 1:1 ratio and incubated at room temperature. Following 1h incubation, plasma/virus and antibody/virus complexes were then added to Vero E6 cells (RRID: CVCL_0574) seeded the day before (104 cells/well) in quadruplicate. After 48 h incubation, the cells were fixed and stained and spots representing virus infected cells were counted. Single outliers were removed as previously described.31 The percentage neutralisation was calculated as:
Statistics
50% inhibitory dilution neutralisation titres (ID50) of plasma were calculated in GraphPad Prism (version 9.4.1). Longitudinal figures were fitted with a coarse (5 points) LOWESS curve (GraphPad Prism). All statistical tests were performed in GraphPad Prism (version 9.4.1). Each specific statistical test performed is indicated in the figure legends. In brief, data was checked for normal distribution by using QQ-plots and assessed using the Shapiro–Wilk test and Kolmogorov–Smirnov test. Data that was not found to be normally distributed was analysed using the Kruskal–Wallis test and corrected for multiple comparisons using Dunn's test for multiple comparisons. For normally distributed paired datasets analysing one variable, a one-way ANOVA was used, which was corrected for multiple comparisons using Tukey's honest test. For normally distributed paired datasets analysing two different variables, a main effects 2way ANOVA was used, which was corrected for multiple comparisons using Tukey's honest test. Data that did not pass the normal distribution tests were plotted with the median and 95% confidence interval. Data that passed the normal distribution tests were plotted with the mean and standard deviation. Comparisons of sex between the groups was done using Fisher's exact test. Multivariate analyses comparing ID50 values between groups accounting for age and sex were conducted in RStudio (Rstudio Team (2022)). All analyses were two tailed and statistical significance was defined as a p value less than 0.05.
Role of funders
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Validation of cell lines, antibodies and reagents
All cell lines, antibodies and reagents included in this study were validated by the company or laboratory group from which they were purchased/gifted from, as previously described.28
Results
Inclusion of study participants
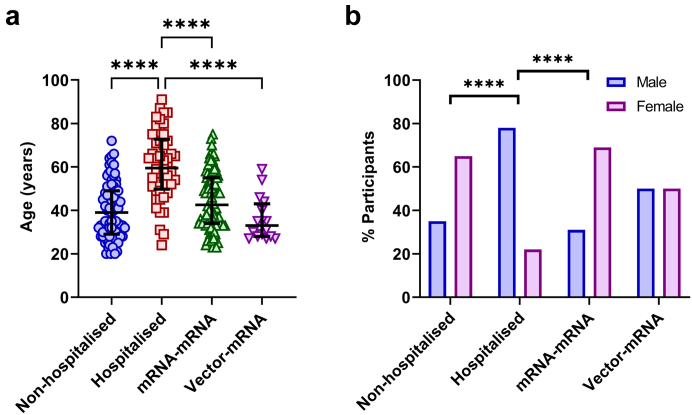
Of the 103 individuals from the non-hospitalised COVID-19 group, 89 were included in this study as three did not seroconvert to SARS-CoV-2 at their baseline sample and 11 dropped out. In the hospitalised COVID-19 group, 50 of the 55 individuals could be included in this study as five died. In the mRNA–mRNA group, 96 of 109 individuals were included in this study as two tested positive for SARS-CoV-2 S protein Abs at their pre-vaccination time point, three tested positive for N protein Abs at 1-month post-vaccination and eight dropped out. In the vector-mRNA group 16 out of 21 individuals were included as one tested positive for SARS-CoV-2 RBD Abs at the pre-vaccination time point and four dropped out. The median ages for the included participants in the non-hospitalised COVID-19, hospitalised COVID-19, mRNA–mRNA and vector-mRNA groups were 39 (interquartile range [IQR] = 29–49), 61 (IQR = 50–75), 43 (IQR = 34–55) and 35 (IQR = 28–43) years, respectively. The hospitalised group was found to be significantly older than the other groups (p < 0.0001, Fig. 1a). The percentage of females for each group were 65%, 22%, 69% and 50%, respectively. The hospitalised group had a significantly higher number of males compared to the non-hospitalised and mRNA–mRNA groups (p < 0.0001, Fig. 1b), but not the vector-mRNA group (p = 0.0543, Fig. 1b). No other significant differences were detected between the groups. Additional details on included individuals can be found in Supplementary Tables S1.2, S1.3, S1.4 and S1.5.
Fig. 1.
Demographic comparisons between the non-hospitalised (blue), hospitalised (red), mRNA–mRNA (green) and vector-mRNA (purple) groups. a) Comparison of age between each of the four groups. Statistical significance was determined by the Kruskal–Wallis test and corrected for multiple comparisons using Dunn's test (∗∗∗∗p < 0.0001). b) Comparison of sex between each of the four groups. Statistical significance was determined by Fischer's exact test (∗∗∗∗p < 0.0001).
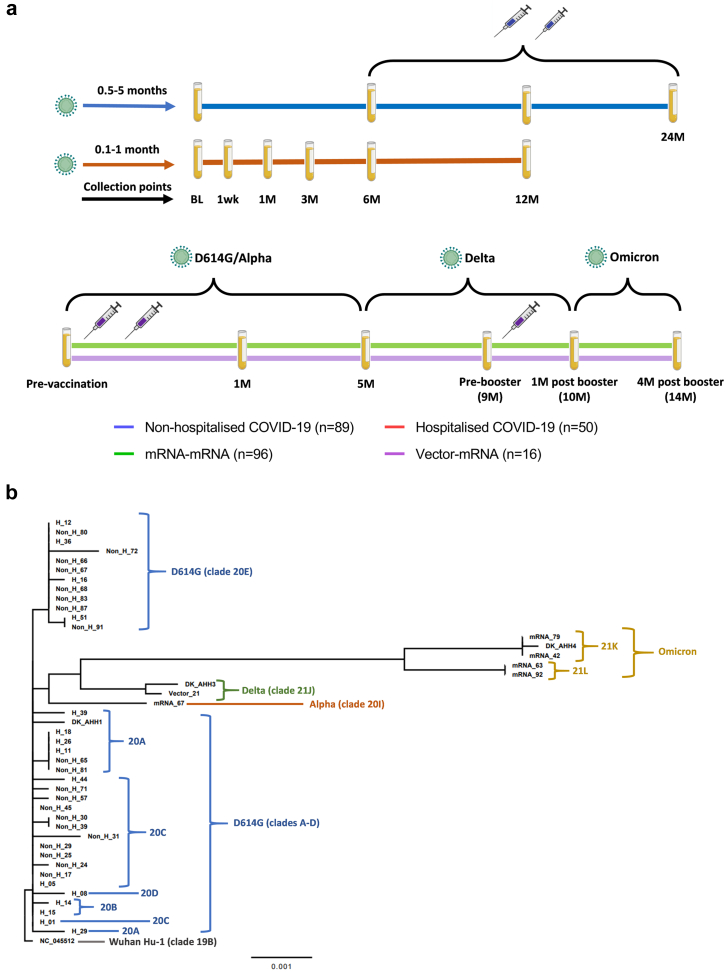
For both the non-hospitalised and hospitalised COVID-19 groups, vaccinations became available approximately six months PSO. Following completion of this study, a total of 60 (67%) and 34 (68%) individuals had been vaccinated in the non-hospitalised and hospitalised COVID-19 groups, respectively. For both the mRNA–mRNA and vector-mRNA groups, mRNA booster vaccinations became available approximately nine months post completed vaccination. Following completion of this study a total of 69 (72%) and 14 (87%) individuals had received a booster vaccination in the mRNA–mRNA and vector-mRNA groups, respectively. A summary of study follow up can be found in Fig. 2a.
Fig. 2.
a) Schematic depicting the time scale of infection and vaccination with approximate blood collection time points for all four groups included in this study. b) Phylogenetic analysis of retrieved SARS-CoV-2 spike protein sequences in comparison to the Wuhan-Hu-1 reference sequence (NC_045512). a) For non-hospitalized individuals (blue line), the baseline (BL) time point was collected between 0.5-5 months post symptom onset with follow-up time points at 6 months (6M), 12M and 24M post symptom onset. For hospitalized individuals (red line), the BL time point was collected between 0.1-1-month post symptom onset with follow-up time points at 1 week (1wk), 1M, 3M, 6M and 12M post symptom onset. The two syringes between 6M and 24M indicate the possible time in which the participants from both groups may have received their vaccination against COVID-19. For both vaccination groups (mRNA–mRNA group = green line, vector-mRNA group = purple line), blood was collected pre-vaccination, 1M post vaccination (2-dose regimen) and then at 5M and 9M (pre-booster vaccination), 10M (1M post booster vaccination) and 14M (4M post booster vaccination) post vaccination. The syringes indicate the time in which the participants received vaccine doses. The viruses above indicate the dominant circulating variants during the indicated time windows that could cause infection. b) Comparison of retrieved SARS-CoV-2 spike protein sequences from the non-hospitalised COVID-19 (Non-H), hospitalised COVID-19 (H) mRNA–mRNA (mRNA) and vector-mRNA (Vector) groups. Each colour is a clade shown in relation to its variant nomenclature (grey = Wuhan-Hu-1, blue = D614G, orange = alpha, green = delta and yellow = omicron). The scale represents the phylogenetic distance of 0.001 nucleotide substitutions per site.
Validation of study-specific SARS-CoV-2 isolates by analysis of retrieved S protein sequences
A summary of retrieved sequences can be found in Supplementary Tables S1.2, S1.3, S1.4 and S1.5. As shown in Fig. 2b, the majority of acquired sequences strongly associated with the DK-AHH1 ancestral isolate (clade 20A), indicating little genetic drift and positive selection within the S protein between infections. Although a large number of infected individuals did not have a sequence retrieved, given that the alpha SARS-CoV-2 variant (clade 20I) didn't start spreading until February 2021 in Denmark,32 it is most likely that infected individuals included before this time were also infected with D614G variants (clade 20) or preceding variants (clade 19). Similarly, those that were infected with delta or omicron variants were found to cluster well with the DK-AHH3 and DK-AHH4 isolates, respectively. This similarity between the retrieved S protein sequences and the isolates used in this study indicates that the 3 isolates used (DK-AHH1, DK-AHH3 and DK-AHH4) to perform virus neutralisation culture studies were good representative models for neutralisation analyses.
Detection of SARS-CoV-2 (re)infection
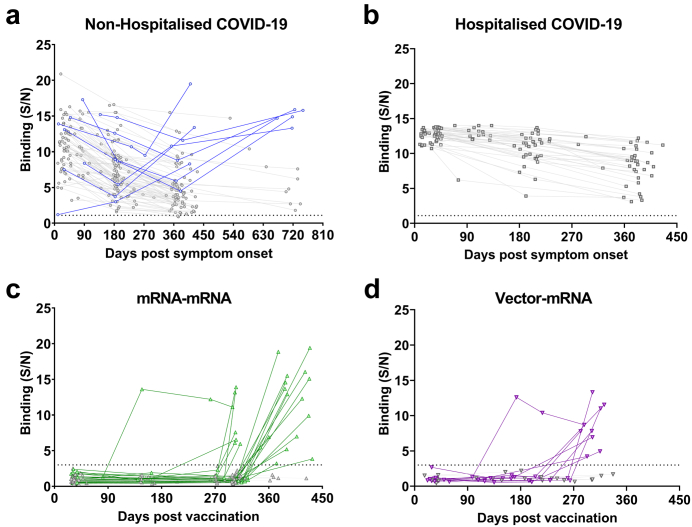
Of the 89 non-hospitalised individuals, 8 were found to have been reinfected with SARS-CoV-2, but no reinfections among the 50 hospitalised individuals were detected (Fig. 3a and b). Of the 96 individuals in the mRNA–mRNA group, 23 were found to have been infected with SARS-CoV-2 (Fig. 3c), of whom 22 were found to only be positive at their last time point and one subject became positive at 147 days post vaccination (DPV). Of the 16 individuals in the vector-mRNA group, nine individuals were found to have been infected with SARS-CoV-2 (Fig. 3d), of whom eight were found to be positive only at their last time point and one subject was found to be positive at 173 DPV.
Fig. 3.
Longitudinal anti-SARS-CoV-2 nucleocapsid antibody levels. The signal/noise (S/N) ratios of plasma antibody binding to the SARS-CoV-2 nucleocapsid (N) protein were determined for each of the 4 study groups. The S/N ratios for the non-hospitalised COVID-19 (a, blue, n = 89) and hospitalised COVID-19 (b, n = 50) groups were determined by the average binding of 5 unexposed healthy controls, with a S/N ratio over 1.1 determining positivity (dotted line). Reinfection was defined as a 2-fold S/N increase compared to the time point prior. The S/N ratio for the mRNA–mRNA (c, green, n = 96) and vector-mRNA (d, purple, n = 16) groups was determined by comparing the binding of each time point to the pre-vaccination time point. A S/N ratio above 3.0 was determined as positive (dotted line). Those highlighted in grey show individuals that did not get reinfected in the non-hospitalised COVID-19 and hospitalised COVID-19 groups and individuals that did not get infected in the mRNA–mRNA and vector-mRNA groups. Those in colour indicate individuals that became reinfected or infected.
Longitudinal neutralising titres against an ancestral SARS-CoV-2 isolate
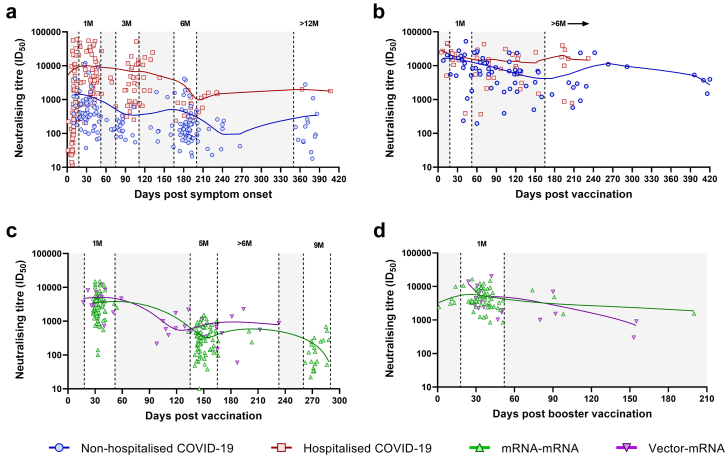
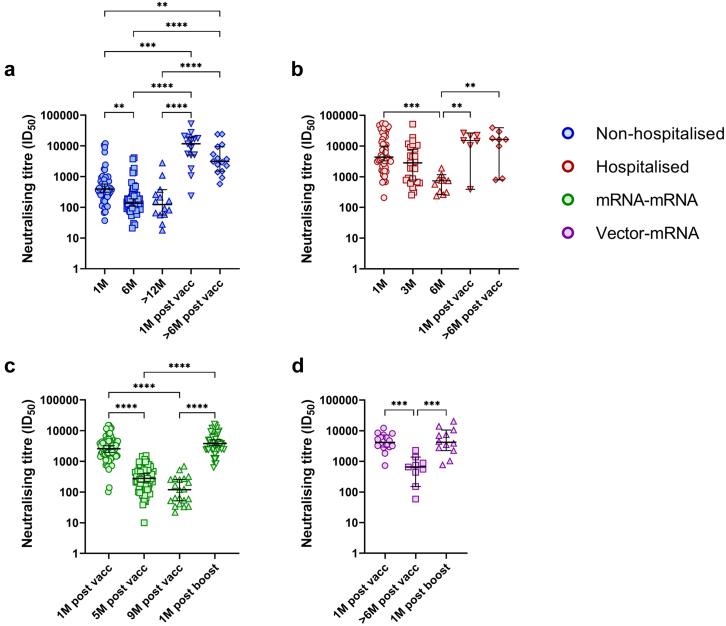
To understand the longitudinal dynamics of infection-induced and vaccine-induced nAbs, plasma neutralising titres against an ancestral SARS-CoV-2 isolate (DK-AHH1) were analysed over time in all subjects (Fig. 4). All time points with evidence of infection or reinfection, as determined by the N protein ELISA, were excluded from this analysis. Time points were stratified based on their days post symptom onset (DPO) or DPV. For the non-hospitalised and hospitalised COVID-19 groups (Fig. 4a), time points were stratified into 1M (18–52 days DPO), 3M (74–111 DPO, hospitalised COVID-19 group only), 6M (165–200 DPO) and >12M (350 or more DPO, non-hospitalised COVID-19 group only). Following vaccination in these groups (completed 2-dose regimen, Fig. 4b), time points were stratified into 1M (18–52 DPV) and >6M (165 or more DPV). For the mRNA–mRNA and vector-mRNA groups (Fig. 4c), time points were stratified into 1M (18–52 DPV), 5M (135–165 DPV, mRNA–mRNA group only), >6M (165–233 DPV, vector-mRNA group only) and 9M (260–290 DPV, mRNA–mRNA group only). Following mRNA booster vaccination (third dose, Fig. 4d), only a 1M (18–52 DPV) time point was assigned as many subjects acquired an infection thereafter. For all groups, there was a significant decline in neutralising titres from 1M to 5M (mRNA–mRNA group) or 6M (all other groups) (p < 0.01, Kruskal–Wallis test, Fig. 5). From 6M to >12M in the non-hospitalised COVID-19 group and from 5M to 9M in the mRNA–mRNA group, there was an additional drop in neutralising titres (Fig. 5a and c, respectively). Vaccination in the COVID-19 groups (Fig. 5a and b) and booster vaccination in the vaccinated groups (Fig. 5c and d) significantly increased neutralising titres compared to the time point prior (p < 0.01 Kruskal–Wallis test). For the non-hospitalised COVID-19 group, vaccination showed significantly higher neutralising titres than those observed at 1M PSO (p = 0.0005, Kruskal–Wallis test, Fig. 5a). For the other groups, vaccination (hospitalised COVID-19, Fig. 4b) or booster vaccination (Fig. 5c and d) showed comparable neutralising titres to those previously induced at 1M PSO/PV. For both COVID-19 groups, neutralising titres at >6M PV were significantly higher than those at the 6M PSO time point (p < 0.01, Kruskal–Wallis test, Fig. 5a and b).
Fig. 4.
Longitudinal analysis of neutralising titres (ID50) against DK-AHH1 (D614G variant) in the non-hospitalised (blue), hospitalised (red), mRNA–mRNA vaccinated (green) and vector-mRNA vaccinated (purple) groups. Overview of longitudinal neutralising titres with respect to days post symptom onset (non-hospitalised and hospitalised groups only, a), days post vaccination (completed 2-dose regimen, b and c) or days post booster vaccination (after a third vaccine dose, d). All graphs were fitted with a coarse (5 points) LOWESS curve. The top left shows the non-hospitalised COVID-19 (blue) and hospitalised COVID-19 (red) groups after infection (a) and the top right shows the same groups after vaccination (b). The bottom left shows the mRNA–mRNA (green) and vector-mRNA (purple) groups after vaccination (c) and the bottom right shows these same groups after booster vaccination (d). The white areas represent stratification of the time points. The grey shading represents areas where time points were not used for stratification analysis.
Fig. 5.
Within-group stratification analysis of neutralisation titres in non-hospitalized (blue [a]), hospitalized (red [b]), mRNA–mRNA (green [c]) and vector-mRNA (purple [d]) over time. Each symbol within each graph represents a different time point as shown on the X axis of the respective graph. Statistical significance was determined using the Kruskal–Wallis test and corrected for multiple comparisons using Dunn's test. (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001).
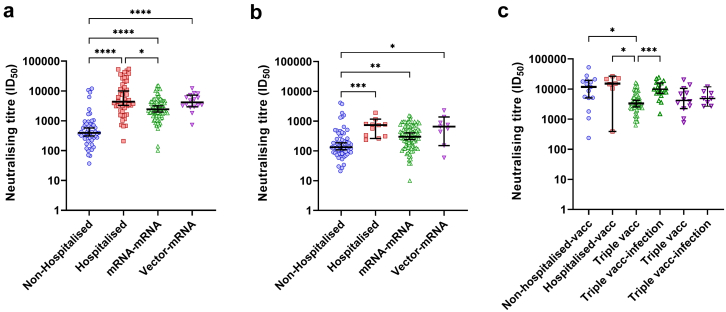
When the groups were compared at 1M PSO/PV, the hospitalised COVID-19 group exhibited the highest neutralising titres (Fig. 6a). The hospitalised COVID-19 group also had significantly higher neutralising titres at 6M PSO when compared to the non-hospitalised COVID-19 group (p = 0.0006, Kruskal–Wallis test, Fig. 6b). Both vaccine groups had significantly higher neutralising titres at 1M and 5/6M PV compared to 1M and 6M PSO in the non-hospitalised COVID-19 group, respectively (p < 0.0001 and p < 0.05, respectively, Kruskal–Wallis tests, Fig. 6a and b, respectively). However, at 1M PV, both COVID-19 groups had significantly higher neutralising titres when compared to the mRNA–mRNA group at 1M PBV (p < 0.05, Kruskal–Wallis test, Fig. 6c). Many in the mRNA–mRNA and vector-mRNA groups became infected after they had received their booster vaccination. Analysis of neutralising titres in these subjects showed that there was a significant boost in the mRNA–mRNA group when compared to the neutralising titres at 1M PBV (p = 0.0012, Kruskal–Wallis test, Fig. 6c). However, the same level of boosting was not observed in the vector-mRNA group.
Fig. 6.
Cross-group stratification analysis of neutralising titres. a) Comparison of neutralising titres after 1-month (1M) PSO for the non-hospitalised COVID-19 (blue) and hospitalised COVID-19 (red) groups and 1M post vaccination for the mRNA–mRNA (green) and vector-mRNA (purple) groups. b) Comparison of neutralising titres at 6M post symptom onset for the non-hospitalised and hospitalised groups, at 5M post vaccination for the mRNA–mRNA group and at the >6M time point for the vector-mRNA group. c) Comparison of neutralising titres at 1M post vaccination for the non-hospitalised COVID-19 and hospitalised COVID-19 groups, at 1M post mRNA booster vaccination for the mRNA–mRNA and vector-mRNA groups (triple vacc) and at the time point after infection for both the mRNA–mRNA and vector-mRNA groups (triple vacc-infection). Statistical significance was determined using the Kruskal–Wallis test and corrected for multiple comparisons using Dunn's test. (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001).
Given that the hospitalised COVID-19 group was significantly older and had a significantly higher proportion of males, these variables could confound the results above. To validate the findings above, multivariate analyses were done comparing the neutralising titres from each group to the hospitalised COVID-19 group, which took age and sex into account (Supplementary Table S2.1). At 1M PSO/PV, there was an increased significant difference in neutralising titres to the mRNA–mRNA group (p = 0.00013), but no further changes were observed against the non-hospitalised COVID-19 group (p < 0.0001) or vector-mRNA group (p > 0.05). At 6M PSO, when compared to the mRNA–mRNA group at 5M PV, there was now a significant difference (p = 0.013). Furthermore, when compared to the non-hospitalised COVID-19 group at 6M PSO, a slight increase in significance was observed (p = 0.00015). However, no changes in significance to the vector-mRNA group at 5M PV were found. Lastly, when the hospitalised COVID-19 group was compared to the other groups at 1M PV or 1M PBV, there was a gain in significance to the mRNA–mRNA group (triple vacc, p = 0.00028) while the other groups remained non-significant. Overall, the multivariate analyses conducted here validated the findings above.
Additional exposure to the spike protein through infection or vaccination increases cross-neutralising potency
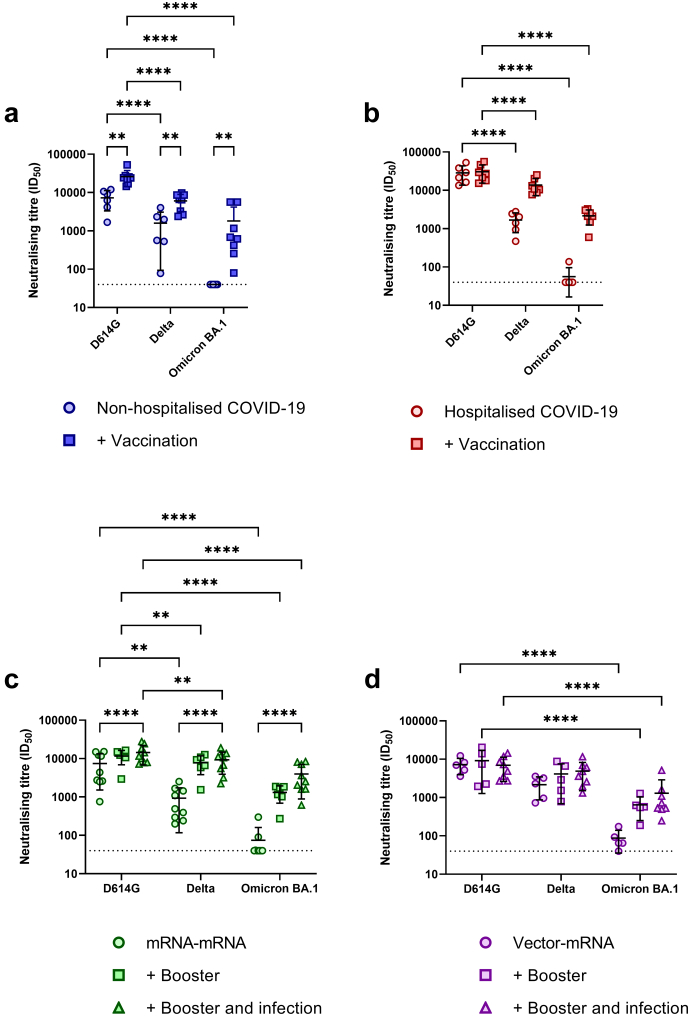
To test cross-neutralisation of variants, neutralisation against delta (DK-AHH3) and omicron (DK-AHH4) SARS-CoV-2 isolates was performed on a panel of subjects from each study group. A summary of this panel can be found in Supplementary Table S3.1. Selected subjects within the non-hospitalised and hospitalised COVID-19 groups were screened at their 1M PSO and 1M PV time points. For the mRNA–mRNA and vector-mRNA groups, subjects were screened at their 1M PV, 1M PBV (triple vacc) and post infection time points (triple vacc + infection). For all subject groups, infection or vaccination alone showed substantially lower neutralisation titres to both the delta and omicron isolates, with only some subjects showing detectable neutralisation above the limit of detection (LOD, ID50 of 40) to the omicron isolate (Fig. 7). Following vaccination in the non-hospitalised COVID-19 group, there was a significant boost in the neutralisation directed towards delta isolate (p = 0.0038, 2way ANOVA) and neutralisation was now detectable against the omicron isolate for all tested subjects (Fig. 7a). Although neutralisation titres were boosted following vaccination, neutralisation to the DK-AHH1 isolate remained significantly higher than the delta and omicron isolates (p < 0.0001, 2way ANOVA, 4.5-fold reduction and 18.5-fold reduction, respectively, Fig. 7a). Following vaccination in the hospitalised COVID-19 group, there were increases in cross-neutralisation to both the delta and omicron isolates, despite these increases not reaching significance (p > 0.05, 2way ANOVA, Fig. 7b). Compared to the DK-AHH1 isolate, neutralisation of the delta and omicron isolates remained significantly lower (p < 0.0001, 2way ANOVA, 2-fold and 13.6-fold reduction, respectively) after vaccination.
Fig. 7.
Analysis of cross-neutralisation to delta and omicron BA.1 SARS-CoV-2 isolates. Within-group analysis of neutralisation titres (ID50) to the DK-AHH1 (D614G), delta (DK-AHH3) and omicron (DK-AHH4) SARS-CoV-2 isolates at 1-month (1M) post symptom onset and 1M post vaccination (+vaccination) in the non-hospitalised COVID-19 (blue [a], n = 6 and n = 8, respectively) and hospitalised COVID-19 (red, [b], n = 6 and n = 7, respectively) groups and 1M post vaccination, 1M post booster vaccination (+booster) and post infection (+booster and infection) in the mRNA–mRNA (green [c], n = 9, n = 6 and n = 9, respectively) and vector-mRNA (purple [d], n = 5, n = 5 and n = 8, respectively) groups. Significance was determined by a main effects 2way ANOVA corrected for multiple comparisons using Tukey's Honest test.
Following booster vaccination in the mRNA–mRNA group, there were noticeable increases in neutralisation to all SARS-CoV-2 isolates (Fig. 7c). Furthermore, additional boosting was noticed against the omicron isolate following infection. Despite this boosting, compared to the DK-AHH1 isolate, neutralisation titres remained significantly lower against the delta isolate (p = 0.0048, 2way ANOVA, 1.5-fold reduction) and omicron isolate (p < 0.0001, 2way ANOVA, 8.8-fold reduction, Fig. 7c) at the 1M PBV time point. Although additional boosting against the omicron isolate was observed following infection, neutralising titres remained significantly lower when compared to the DK-AHH1 isolate (p < 0.0001, 2way ANOVA, 3.7-fold reduction, Fig. 7c).
Following booster vaccination and infection in the vector-mRNA group, no observable boosts in neutralisation titres against the DK-AHH1 and delta isolates were observed. While there were observable increases in the neutralisation titres against the omicron isolate following booster vaccination and infection, these remained non-significant when compared to post vaccination alone (p > 0.05, 2way ANOVA) and remained significantly lower than the neutralisation titres seen against the DK-AHH1 isolate (p < 0.0001, 2way ANOVA, Fig. 7d).
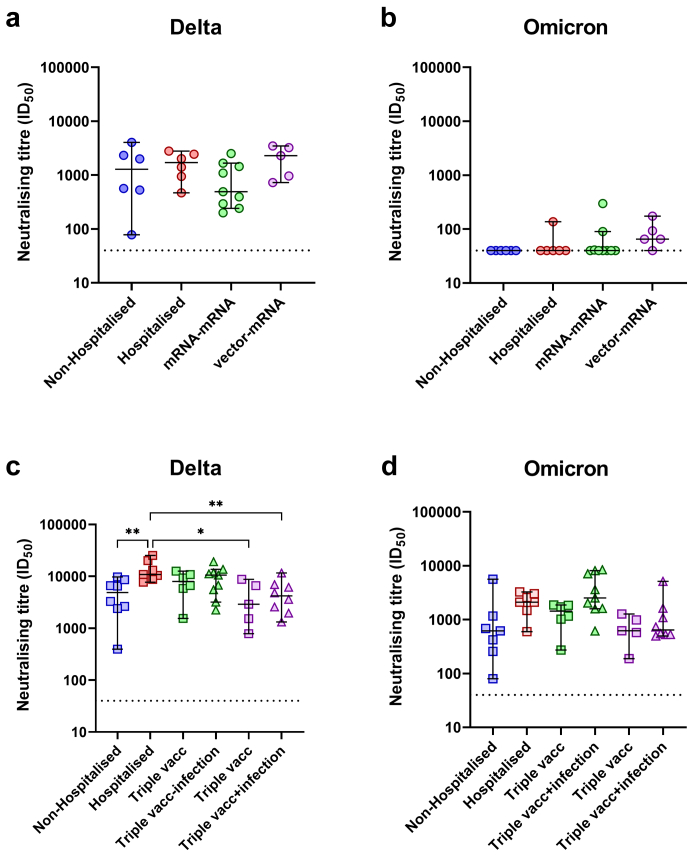
When the neutralisation titres between the groups were compared at 1M PSO/PV, no observable differences were detected against the delta isolate (Fig. 8a). However, the vector-mRNA group had the highest level of detectable neutralisation compared to any of the other groups against the omicron isolate (Fig. 8b). Following vaccination in the non-hospitalised and hospitalised COVID-19 groups and mRNA booster vaccination in the mRNA–mRNA and vector-mRNA groups, the hospitalised group had significantly higher neutralising titres when compared to the non-hospitalised group (p = 0.0086, one-way ANOVA) and to the vector-mRNA group both before and after infection (p < 0.05, one-way ANOVA, Fig. 8c). While infection in the boosted mRNA–mRNA group (3 vaccine doses) appeared to show the highest level of neutralisation against the omicron isolate, this was not significantly different when compared to any of the other groups (p > 0.05, one-way ANOVA, Fig. 8d).
Fig. 8.
Cross-group analysis of neutralisation titres to the delta and omicron SARS-CoV-2 isolates. Neutralising titres to the delta (a) and omicron (b) isolates at the 1M post symptom onset (non-hospitalised COVID-19 [n = 6] and hospitalised COVID-19 groups [n = 6]) and 1M post vaccination (mRNA–mRNA [n = 9] and vector-mRNA groups [n = 5]). Neutralising titres to the delta (c) and omicron (d) isolates at the 1M post vaccination (non-hospitalised [n = 8] and hospitalised groups [n = 7]) and 1M post mRNA booster (triple vacc) and post infection (triple vacc + infection) (mRNA–mRNA [n = 6 and n = 9, respectively] and vector-mRNA groups [n = 5 and n = 8, respectively]) (right) time points. Statistical significance was determined by the Kruskal–Wallis test and corrected for multiple comparisons using Dunn's test. (∗∗p < 0.01, ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001).
Discussion
Given that neutralising responses have been found to be a correlate of protection against severe COVID-19,12,13 longitudinal studies of the kinetics of neutralising responses provide valuable insights into the durability of protective immune responses and can help guide vaccine strategies. Herein, the results described provide several important insights. First, this study shows that while individuals in the hospitalised COVID-19 group displayed higher levels of neutralising responses, which is likely due to the relationship between disease severity and higher adaptive immune responses as reported by others,24,33,34 the level of neutralising responses declines in a similar manner regardless of disease severity or type of vaccination received. That is, after reaching a peak, neutralising responses wane until about 6 months PSO, from which they reach a plateau and decline at a slower rate. Interestingly, those that induced the highest neutralising responses initially also maintained the highest neutralising responses longitudinally, suggesting that the initial induced neutralising titre is reflective of the neutralising titre at plateau. The higher neutralising responses seen in the hospitalised COVID-19 group is likely due to the longer presence of viral antigen, which helps drive germinal centre activity and improve B cell maturation compared to shorter antigen exposure lengths.35, 36, 37 Second, although not significant, probably due to sample size, one noticeable feature was that those that received a heterologous prime-boost vaccine regimen (vector-mRNA group) appeared to have higher initial neutralising responses, which also appeared to be more durable, compared to those receiving a homologous prime-boost vaccine regimen (mRNA–mRNA group). This observation is also in agreement with other studies in which a heterologous prime-boost COVID-19 vaccine regimen was found to be superior at generating immune responses compared to a homologous one.21,38, 39, 40 One explanation for this could be that the heterologous prime-boost regimen reduces off-target immunity to non-S protein specific ingredients in the vaccines, such as the adenovirus vector in the Vaxzevria® vaccine and the lipids found in the Comirnaty® vaccine.
Compared to other recent longitudinal studies on neutralising antibody kinetics in SARS-CoV-2 infection and COVID-19 vaccination,20,22,26,41 this study was able to follow previously infected individuals for up to a year pre and post vaccination to map the durability of neutralising responses both with and without vaccination. In addition to this, because the N protein is not included in any of the vaccination formulas used here, SARS-CoV-2 (re)infection was accounted for by mapping serology to this protein. Interestingly, vaccination in those that were previously infected not only boosted neutralising titres but also appeared to increase the durability of these responses over time. Unfortunately, due to many participants becoming infected after their booster vaccination, the durability of neutralisation on booster vaccinated, infection-naïve individuals could not be determined. However, given that the vaccination and infection groups showed similar longitudinal neutralising kinetics, it may be possible that there is an increased durability of neutralising responses following booster vaccination. Interestingly, in another study, it was found that infection following vaccination or vaccination after infection induced more durable neutralising responses.25 Taken together, this would suggest that more durable neutralising responses can be induced through repeated antigen exposure and supports the use of booster vaccination.
In addition to the longitudinal data, analysis of cross-neutralisation to delta and omicron isolates was conducted to examine the cross-neutralising potential at different time points. Surprisingly, despite the hospitalised COVID-19 group having the highest neutralising titres prior to vaccination, there was limited cross-neutralisation seen to the delta isolate and almost no detectable neutralisation to the omicron isolate. By contrast, the vector-mRNA group displayed the highest level of cross-neutralisation at the 1M PV time point. This would suggest that the ability to cross-neutralise SARS-CoV-2 variants is not represented simply by the level of neutralising responses in the plasma. This is further shown in the hospitalised COVID-19 group where, following vaccination, no increase in neutralising titres was detected to the DK-AHH1 isolate but there was an, albeit non-significant, increase in cross-neutralisation to both the delta and omicron isolates. Similarly, although it didn't reach significance, there was also an observable increase in cross-neutralisation in the non-hospitalised group following vaccination and in the vaccine groups following both booster vaccination and infection. The lack of significance in these observations is likely a type II error driven by the small sample size. Given the likely level of similarity between the DK-AHH1 isolate, Comirnaty® and Vaxzevria® S protein sequences and the infecting viral variant during this study, it is likely that the B cell epitopes from any S protein re-exposure were conserved. Thus, this reduces the possibility for selection of B cells that recognize cross-reactive epitopes upon repeated S protein exposure. Although S-specific B cells were not analysed in this study, it has previously been shown that, in those that remained unexposed, memory B cells that recognize the RBD of alpha, beta, delta and omicron variants had the greatest boost compared to B cells targeting other areas of the S protein after booster vaccination.20 Given this, and that it is thought that only a fraction of memory B cells re-enter the germinal centre after antigen re-exposure,42 the observed boost in cross-neutralisation in this study is likely due to the expansion of RBD-specific B cells and not due to additional maturation. Regardless, this data supports vaccination to increase cross-neutralising responses in plasma and likely increase cross-protection from severe disease, as has been seen in the clinic.43 Interestingly, while early reports on SARS-CoV-2 omicron-specific S protein vaccine formulas have shown that it is suboptimal at inducing cross-neutralising responses to other variants of concern when compared to the vaccine formula containing the ancestral S protein sequence,44 a more recent report has shown that the newer bivalent mRNA booster can induce broader neutralising antibody responses to the current circulating variants when compared to the original mRNA vaccine formulas.45
This study is, however, not without limitations. Firstly, demographically speaking, the hospitalised COVID-19 group was not matched to the other groups. It has been previously shown that, following SARS-CoV-2 infection, older male individuals can develop higher neutralising responses compared to younger female individuals.46, 47, 48 However, hospitalisation from COVID-19 is predominantly among older individuals1 and therefore represents a difficulty in matching age between the groups. In contrast, neutralising responses following COVID-19 vaccination have been shown to be lower in older individuals.49,50 This may therefore reduce confoundment between the comparisons made between the hospitalised group and the vaccinated groups. In addition to this, multivariate analyses were conducted to account for the difference in age and sex between the hospitalised group and others and these analyses showed only minor differences, thus validating the differences observed. Secondly, symptomatic data was not reported in this study and has previously been shown to associate with neutralising antibody responses.51
Despite these limitations, this study has shown that the durability of neutralising responses for each study group follows similar patterns, whereby there is a decline after infection or vaccination until about 6 months, after which these responses plateau, and that the level of the plateau is reflected by the initial level of neutralising responses detected. In addition, vaccination in those previously infected generates an anamnestic neutralising response which, over time, appears to be more durable. Regarding cross-neutralisation, vaccination and booster vaccination greatly increases the level of cross-neutralisation. Taken together, this data supports the use of vaccination to increase the durability and cross-neutralising potential of neutralising responses.
Contributors
A.U., C.S., N.W. and J.B. conceived and designed the study; A.U., C.S., A.W., S.B., S.V., G.V., C.N.V., M.I.D., A-L.S. and N.W. collected and processed patient data and samples; A.U., C.F-A., L.S.M., A.B., U.F., S.R. and J.B. generated assays and experimental data; U.V.S. obtained the spike protein sequences for the study; A.U., C.S., N.W. and J.B. drafted the manuscript. Data was verified by C.F-A., S.B., U.F., S.R., N.W. and J.B. All authors critically revised the manuscript for important intellectual content and gave final approval for the submitted version.
Data sharing statement
Following publication, and in agreement with the Data Protection Agency, Denmark, the data generated in this study will be made available to those who provide a sound proposal. Proposals should be directed to jbukh@sund.ku.dk, and to gain access, data requestors will need to sign a data access agreement. All individual participant data will remain coded.
Declaration of interests
Nina Weis has been clinical investigator, lecturer or on advisory boards for Abbvie, Gilead, Glaxo Smith Kline and Merck Sharp Dohme and has received unrestricted grants for research from Abbvie and Gilead without relation to the presented work and for the remaining authors there are no conflicts of interest. Christina Sølund has received support from MSD Denmark to attend the Nordic HIV and Virology Conference in Stockholm 2022. Anni Assing Winckelmann has received support from Gilead Denmark to attend the Nordic HIV and Virology Conference in Stockholm 2022.
Acknowledgements
We would like to thank all the participants for their contribution to this study. We thank Bjarne Ørskov Lindhardt (Department of Infectious Diseases, Copenhagen University Hospital, Hvidovre) and Charlotte Menné Bonefeld (Department of Immunology and Microbiology, University of Copenhagen) for valuable support. We would also like to thank Jose Alfredo Samaniego Castruita for help with acquiring the SARS-CoV-2 sequences.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104475.
Appendix A. Supplementary data
References
- 1.Organization W.H. 2022. WHO Coronavirus Disease (COVID-19) situation reports.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ Available from. Accessed December 2022. [Google Scholar]
- 2.Voysey M., Clemens S.A.C., Madhi S.A., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397(10269):99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baden L.R., El Sahly H.M., Essink B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polack F.P., Thomas S.J., Kitchin N., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chemaitelly H., Tang P., Hasan M.R., et al. Waning of BNT162b2 vaccine protection against SARS-CoV-2 infection in Qatar. N Engl J Med. 2021;385(24):e83. doi: 10.1056/NEJMoa2114114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine-Tiefenbrun M., Yelin I., Alapi H., et al. Waning of SARS-CoV-2 booster viral-load reduction effectiveness. Nat Commun. 2022;13(1):1237. doi: 10.1038/s41467-022-28936-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall V., Foulkes S., Insalata F., et al. Protection against SARS-CoV-2 after Covid-19 vaccination and previous infection. N Engl J Med. 2022;386(13):1207–1220. doi: 10.1056/NEJMoa2118691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bates T.A., Leier H.C., Lyski Z.L., et al. Neutralization of SARS-CoV-2 variants by convalescent and BNT162b2 vaccinated serum. Nat Commun. 2021;12(1):5135. doi: 10.1038/s41467-021-25479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans J.P., Zeng C., Qu P., et al. Neutralization of SARS-CoV-2 omicron sub-lineages BA.1, BA.1.1, and BA.2. Cell Host Microbe. 2022:1093–1102.e3. doi: 10.1016/j.chom.2022.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Mulligan M.J., Landau N.R. Partial resistance of SARS-CoV-2 Delta variants to vaccine-elicited antibodies and convalescent sera. iScience. 2021;24(11) doi: 10.1016/j.isci.2021.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen P.R. Relative contagiousness of emerging virus variants: an analysis of the Alpha, Delta, and Omicron SARS-CoV-2 variants. Econom J. 2021 :25(3):739–761. [Google Scholar]
- 12.Khoury D.S., Cromer D., Reynaldi A., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27(7):1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 13.Cromer D., Steain M., Reynaldi A., et al. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: a meta-analysis. Lancet Microb. 2022;3(1):e52–e61. doi: 10.1016/S2666-5247(21)00267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerutti G., Guo Y., Zhou T., et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29(5):819–833 e7. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brouwer P.J.M., Caniels T.G., van der Straten K., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Y., Wang J., Jian F., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602(7898):657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Augusto G., Mohsen M.O., Zinkhan S., Liu X., Vogel M., Bachmann M.F. In vitro data suggest that Indian delta variant B. 1.617 of SARS-CoV-2 escapes neutralization by both receptor affinity and immune evasion. Allergy. 2022;77(1):111–117. doi: 10.1111/all.15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Elslande J., Oyaert M., Ailliet S., et al. Longitudinal follow-up of IgG anti-nucleocapsid antibodies in SARS-CoV-2 infected patients up to eight months after infection. J Clin Virol. 2021;136 doi: 10.1016/j.jcv.2021.104765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brochot E., Demey B., Touze A., et al. Anti-spike, anti-nucleocapsid and neutralizing antibodies in SARS-CoV-2 inpatients and asymptomatic individuals. Front Microbiol. 2020;11:584251. doi: 10.3389/fmicb.2020.584251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goel R.R., Painter M.M., Lundgreen K.A., et al. Efficient recall of Omicron-reactive B cell memory after a third dose of SARS-CoV-2 mRNA vaccine. Cell. 2022 doi: 10.1016/j.cell.2022.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marking U., Havervall S., Greilert-Norin N., et al. Duration of SARS-CoV-2 immune responses up to six months following homologous or heterologous primary immunization with ChAdOx1 nCoV-19 and BNT162b2 mRNA vaccines. Vaccines. 2022;10(3):359. doi: 10.3390/vaccines10030359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanshylla K., Di Cristanziano V., Kleipass F., et al. Kinetics and correlates of the neutralizing antibody response to SARS-CoV-2 infection in humans. Cell Host Microbe. 2021;29(6):917–929.e4. doi: 10.1016/j.chom.2021.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K., Long Q.X., Deng H.J., et al. Longitudinal dynamics of the neutralizing antibody response to SARS-CoV-2 infection. Clin Infect Dis. 2020:e531–e539. doi: 10.1093/cid/ciaa1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamayoshi S., Yasuhara A., Ito M., et al. Antibody titers against SARS-CoV-2 decline, but do not disappear for several months. eClinicalMedicine. 2021;32 doi: 10.1016/j.eclinm.2021.100734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walls A.C., Sprouse K.R., Bowen J.E., et al. SARS-CoV-2 breakthrough infections elicit potent, broad, and durable neutralizing antibody responses. Cell. 2022;185(5):872–880.e3. doi: 10.1016/j.cell.2022.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pradenas E., Trinité B., Urrea V., et al. Clinical course impacts early kinetics, magnitude, and amplitude of SARS-CoV-2 neutralizing antibodies beyond 1 year after infection. Cell Rep Med. 2022;3(2) doi: 10.1016/j.xcrm.2022.100523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solund C., Underwood A.P., Fernandez-Antunez C., et al. Analysis of neutralization titers against SARS-CoV-2 in health-care workers vaccinated with prime-boost mRNA-mRNA or vector-mRNA COVID-19 vaccines. Vaccines (Basel) 2022;10(1) doi: 10.3390/vaccines10010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Underwood A.P., Solund C., Fernandez-Antunez C., et al. Neutralisation titres against SARS-CoV-2 are sustained 6 months after onset of symptoms in individuals with mild COVID-19. EBioMedicine. 2021;71 doi: 10.1016/j.ebiom.2021.103519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez S., Fernandez-Antunez C., Galli A., et al. Overcoming culture restriction for SARS-CoV-2 in human cells facilitates the screening of compounds inhibiting viral replication. Antimicrob Agents Chemother. 2021;65(7):e0009721. doi: 10.1128/AAC.00097-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iglewicz B., Hoaglin D. In: The ASQC basic references in quality control: statistical techniques. Mykytka Edward F., editor. 1993. Volume 16: how to detect and handle outliers. [Google Scholar]
- 32.Michaelsen T.Y., Bennedbaek M., Christiansen L.E., et al. Introduction and transmission of SARS-CoV-2 lineage B.1.1.7, Alpha variant, in Denmark. Genome Med. 2022;14(1):47. doi: 10.1186/s13073-022-01045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crawford K.H.D., Dingens A.S., Eguia R., et al. Dynamics of neutralizing antibody titers in the months after severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis. 2021;223(2):197–205. doi: 10.1093/infdis/jiaa618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P., Liu L., Nair M.S., et al. SARS-CoV-2 neutralizing antibody responses are more robust in patients with severe disease. Emerg Microb Infect. 2020;9(1):2091–2093. doi: 10.1080/22221751.2020.1823890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu J.K., Crampton J.C., Cupo A., et al. Murine antibody responses to cleaved soluble HIV-1 envelope trimers are highly restricted in specificity. J Virol. 2015;89(20):10383–10398. doi: 10.1128/JVI.01653-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pauthner M., Havenar-Daughton C., Sok D., et al. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity. 2017;46(6):1073–1088.e6. doi: 10.1016/j.immuni.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tam H.H., Melo M.B., Kang M., et al. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc Natl Acad Sci USA. 2016;113(43):E6639–E6648. doi: 10.1073/pnas.1606050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groß R., Zanoni M., Seidel A., et al. Heterologous ChAdOx1 nCoV-19 and BNT162b2 prime-boost vaccination elicits potent neutralizing antibody responses and T cell reactivity against prevalent SARS-CoV-2 variants. EBioMedicine. 2022;75 doi: 10.1016/j.ebiom.2021.103761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaku C.I., Champney E.R., Normark J., et al. Broad anti–SARS-CoV-2 antibody immunity induced by heterologous ChAdOx1/mRNA-1273 vaccination. Science. 2022;375(6584):1041–1047. doi: 10.1126/science.abn2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atmar R.L., Lyke K.E., Deming M.E., et al. Homologous and heterologous Covid-19 booster vaccinations. N Engl J Med. 2022;386(11):1046–1057. doi: 10.1056/NEJMoa2116414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bates T.A., McBride S.K., Leier H.C., et al. Vaccination before or after SARS-CoV-2 infection leads to robust humoral response and antibodies that effectively neutralize variants. Sci Immunol. 2022 doi: 10.1126/sciimmunol.abn8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mesin L., Schiepers A., Ersching J., et al. Restricted clonality and limited germinal center reentry characterize memory B cell reactivation by boosting. Cell. 2020;180(1):92–106.e11. doi: 10.1016/j.cell.2019.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chenchula S., Karunakaran P., Sharma S., Chavan M. Current evidence on efficacy of COVID-19 booster dose vaccination against the Omicron variant: a systematic review. J Med Virol. 2022;94(7):2969–2976. doi: 10.1002/jmv.27697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He C., He X., Yang J., et al. Spike protein of SARS-CoV-2 Omicron (B.1.1.529) variant have a reduced ability to induce the immune response. Signal Transduct Target Ther. 2022;7(1):119. doi: 10.1038/s41392-022-00980-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurhade C., Zou J., Xia H., et al. Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1, and XBB.1 by parental mRNA vaccine or a BA.5-bivalent booster. Nat Med. 2022 doi: 10.1038/s41591-022-02162-x. :1. [DOI] [PubMed] [Google Scholar]
- 46.Schlickeiser S., Schwarz T., Steiner S., et al. Disease severity, fever, age, and sex correlate with SARS-CoV-2 neutralizing antibody responses. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.628971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Markmann A.J., Giallourou N., Bhowmik D.R., et al. Sex disparities and neutralizing-antibody durability to SARS-CoV-2 infection in convalescent individuals. mSphere. 2021;6(4) doi: 10.1128/mSphere.00275-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klein S.L., Pekosz A., Park H.S., et al. Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. J Clin Invest. 2020;130(11):6141–6150. doi: 10.1172/JCI142004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collier D.A., Ferreira I., Kotagiri P., et al. Age-related immune response heterogeneity to SARS-CoV-2 vaccine BNT162b2. Nature. 2021;596(7872):417–422. doi: 10.1038/s41586-021-03739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei J., Stoesser N., Matthews P.C., et al. Antibody responses to SARS-CoV-2 vaccines in 45,965 adults from the general population of the United Kingdom. Nat Microbiol. 2021;6(9):1140–1149. doi: 10.1038/s41564-021-00947-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aziz N.A., Corman V.M., Echterhoff A.K.C., et al. Seroprevalence and correlates of SARS-CoV-2 neutralizing antibodies from a population-based study in Bonn, Germany. Nat Commun. 2021;12(1):2117. doi: 10.1038/s41467-021-22351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.