Abstract
The phosphorylation of the RNA polymerase II (RNAP II) carboxy-terminal domain (CTD) plays a key role in mRNA metabolism. The relative ratio of hyperphosphorylated RNAP II to hypophosphorylated RNAP II is determined by a dynamic equilibrium between CTD kinases and CTD phosphatase(s). The CTD is heavily phosphorylated in meiotic Xenopus laevis oocytes. In this report we show that the CTD undergoes fast and massive dephosphorylation upon fertilization. A cDNA was cloned and shown to code for a full-length xFCP1, the Xenopus orthologue of the FCP1 CTD phosphatases in humans and Saccharomyces cerevisiae. Two critical residues in the catalytic site were identified. CTD phosphatase activity was observed in extracts prepared from Xenopus eggs and cells and was shown to be entirely attributable to xFCP1. The CTD dephosphorylation triggered by fertilization was reproduced upon calcium activation of cytostatic factor-arrested egg extracts. Using immunodepleted extracts, we showed that this dephosphorylation is due to xFCP1. Although transcription does not occur at this stage, phosphorylation appears as a highly dynamic process involving the antagonist action of Xp42 mitogen-activated protein kinase and FCP1 phosphatase. This is the first report that free RNAP II is a substrate for FCP1 in vivo, independent from a transcription cycle.
In many metazoans, early development is characterized by major changes in the global transcriptional activity of the zygote (13). After fertilization, development proceeds for a while in the absence of transcription and is dependent on the translation of maternal mRNAs stored during ovogenesis. Transcription resumes at a stage which is species specific and corresponds to the two-cell stage in mice. In Xenopus laevis, transcription resumes at the 12th cleavage division at a major developmental transition known as the midblastula transition. Important changes in RNA polymerase II (RNAP II) phosphorylation have been described to occur in worm (27), fly (20), mammalian (3), and amphibian (25) embryos during this period.
RNAP II phosphorylation is a key event in the transcription cycle (11, 12). The largest subunit of RNAP II (Rpb1) can be extensively phosphorylated on its carboxy-terminal domain (CTD), which is comprised of up to 52 repeats of the consensus heptapeptide Tyr-Ser-Pro-Thr-Ser-Pro-Ser. Thus, in somatic cells, two forms of RNAP II coexist in a dynamic equilibrium (15). The hypophosphorylated IIA form assembles into preinitiation complexes, whereas the hyperphosphorylated IIO form catalyzes transcript elongation and facilitates the recruitment of the pre-mRNA processing machinery. CTD dephosphorylation is required to recycle the polymerase at the end of each round of transcription.
Several CTD kinases have been identified (for reviews, see references 5, 21, and 28): the CDK8 subunit of the mediator complex in the RNAP II holoenzyme, the CDK7 subunit of the general transcription factor TFIIH which assembles in the preinitiation complex, the CDK9 subunit of P-TEFb (positive transcription elongation factor b) (26), and the extracellular signal-regulated kinase-type mitogen-activated protein kinases (MAPKs) (6). In contrast, only one CTD phosphatase has been characterized to date (1, 8, 9). Its activity is stimulated by the transcription factor TFIIF and repressed by TFIIB. Its only known substrate is the phosphorylated CTD within the RNAP II core enzyme. The corresponding FCP1 gene (for TFIIF-stimulated CTD phosphatase 1) is essential in Saccharomyces cerevisiae (18), and in vitro experiments have established that the phosphatase activity is necessary to recycle the RNAP II at the end of a transcription cycle (10).
Here we report the characterization of a CTD phosphatase activity from Xenopus cells and egg extracts. This activity was attributable to the frog orthologue of FCP1 and accounts for the RNAP II dephosphorylation that occurs shortly after fertilization. Furthermore, the finding that the dephosphorylation of RNAP IIO occurs prior to the onset of transcription indicates that free RNAP II is a natural substrate for FCP1.
MATERIALS AND METHODS
Cells.
A6 cells were propagated in Leibovitz medium supplemented with fetal calf serum (10%). Subconfluent cells, 24 h after plating, were lysed in PB buffer (50 mM Tris-HCl [pH 7.9], 10 mM MgCl2, 10 mM KCl, 0.1 mM EDTA, 10% glycerol, 0.5 mM dithiothreitol) supplemented with 0.5% Nonidet P-40; the lysate was centrifuged at 10,000 × g.
Transient transfections were performed in semiconfluent 50-mm-diameter dishes using the classical calcium phosphate procedure. Plasmid pSP64 was used as a carrier to bring the total amount of DNA to 10 μg per dish.
Eggs and embryo extracts.
Interphasic egg extract preparation was adapted from the method of Murray (24). Eggs were laid in high-salt Barth's medium (15 mM Tris-HCl [pH 7.4], 110 mM NaCl, 2 mM KCl, 1 mM MgSO4, 0.5 mM Na2HPO4, 2 mM NaHCO3), dejellied in high-salt Barth's medium supplemented with 2% cysteine (pH 7.8), and activated by 0.25 μg of calcium ionophore A23187/ml for 5 min. Activated eggs were rinsed in EB buffer (50 mM HEPES-KOH [pH 7.5], 50 mM KCl, 5 mM MgCl2, 2 mM dithiothreitol), packed by centrifugation at 100 × g, and crushed by centrifugation at 15,000 × g.
Meiotic cytostatic factor (CSF)-arrested egg extracts were prepared according to the method of Swedlow (29). Activation was performed by the addition of CaCl2 (1 mM) and incubation of the extracts at 22°C.
Alternatively, freshly laid eggs were fertilized using dilacerated testes. Embryos were allowed to develop at 22°C in development medium (1.5 mM Tris-HCl [pH 7.4], 8.8 mM NaCl, 0.2 mM KCl, 0.1 mM MgCl2, 100 U of penicillin G/ml, 100 μg of streptomycin/ml). Unfertilized eggs and embryos were dejellied in development medium supplemented with 2% cysteine (pH 7.8). Batches of 10 eggs or embryos were crushed in 500 μl of Laemmli buffer, and the resulting lysate was centrifuged at 15,000 × g for 15 min before being loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels for Western blot analysis.
Immunodepletions.
Twenty microliters of protein A-Sepharose beads (Amersham Pharmacia Biotech) equilibrated in PB buffer was incubated overnight at 4°C with 20 μl of anti-FCP1 serum or mock serum. The beads were washed with PB buffer and incubated with 40 μl of A6 cell extract or 10 μl of egg extract. After 30 min of shaking at 4°C, the supernatants were recovered.
CTD phosphatase assay.
Purified calf thymus RNAP IIA was labeled with casein kinase II (Boehringer-Mannheim) in the presence of [γ-32P]ATP. The labeled RNAP IIA was converted to RNAP IIO by incubation with CTDK1 and 2 mM cold ATP (8). 32P-labeled RNAP IIO (1,000 to 5,000 cpm, 50 fmol) was incubated with extracts at 30°C in PB buffer (20-μl final volume). When indicated, the reaction mixture contained 10 nM okadaic acid (Sigma) or 5 nM microcystin-LR (Sigma). Reactions were stopped by the addition of 20 μl of 2× Laemmli buffer. Samples were analyzed by SDS-PAGE and autoradiography.
CTD phosphate turnover assay.
Purified calf thymus RNAP IIA was uniformly labeled on the CTD repeats with CTDK1 in the presence of [γ-32P]ATP. The labeled RNAP II was fully phosphorylated into RNAP IIO by the subsequent addition of 2 mM cold ATP. 32P-labeled RNAP IIO was incubated with extracts at 22°C.
Cloning of Xenopus FCP1 and site-directed mutagenesis.
Total RNAs were extracted from A6 cells using the RNeasy kit (Qiagen). The primers ATA TGG ATC CAT GCA AAA TCG AGC TCG AGA and GCG GCC GCT AAT CTT CAA TTT ACC CTA ATA were used to amplify the full-length xFCP1 open reading frame by reverse transcription-PCR (ThermoscriptRT; Gibco BRL). The 2,943-bp fragment was cloned in the pcDNA-V5His-TOPO vector (Invitrogen) and sequenced. A BamHI/NotI restriction fragment was subcloned in the pGEX4T2 vector (Amersham Pharmacia Biotech) to generate plasmid pGEX4T2-xFCP1-wt.
Site-directed mutagenesis of pGEX4T2-xFCP1-wt was performed by successive PCRs. Primary PCRs were performed using the following primers: ATG GTT AAC TTA GAT CAG ACT and AGT CTG ATC TAA GTT AAC CAT for the D181N mutation; ATG GTT GAT TTA AAC CAG ACT and AGT CTG GTT TAA ATC AAC CAT for the D183N mutation; ATG GTT GAA TTA GAT CAG ACT and AGT CTG ATC TAA TTC AAC CAT for the D181E mutation; ATG GTT GAT TTA GAA CAG ACT and AGT CTG TTC TAA ATC AAC CAT for the D183E mutation; and upstream primers TCC CAC AAA TTG ATA AGT ACT and CAC ATA CTT TTT AAC TGT GAT C. Secondary PCR products were digested by BamHI and EcoRI and used to replace the BamHI/EcoRI fragment from pGEX4T2-xFCP1-wt, resulting in pGEX4T2-xFCP1-D181N, pGEX4T2-xFCP1-D183N, pGEX4T2-xFCP1-D181E, and pGEX4T2-xFCP1-D183E. Insertion of point mutations was checked by sequencing.
For eukaryotic expression plasmids, primers ATC TAT GCT AGC AAT GCA AAA TCG AGC TCG and CCG TCG CAT CGA TTC ATA TGA AAT CAT TCA were used to amplify the full-length xFCP1 cDNA from pGEX4T2-xFCP1-wt. An NheI-ClaI fragment of the PCR product (2,969 bp) was inserted in frame with the N-terminal FLAG epitope coding sequence in the pAdRSVFlag vector (a gift from François Giudiccelli). Correct insertion was confirmed by sequencing. pAdRSVFlag was derived from pAdRSVKrox20 by insertion of a FLAG epitope, a linker, and a splice donor-acceptor sequence (16).
Expression of recombinant proteins, electrophoresis, and Western blotting.
The pGEX4T2 expression plasmids were transformed in Escherichia coli BL21-DE3 cells, and fusion proteins were expressed by inducing an exponentially grown culture for 3 h at 37°C with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were harvested, resuspended in phosphate-buffered saline, and sonicated. The resulting lysates were bound to glutathione-Sepharose 4B columns, and fusion proteins were purified according to the manufacturer's instructions (Amersham Pharmacia Biotech).
Samples in Laemmli buffer were heated for 5 min at 95°C before being loaded onto 5% polyacrylamide gels. Western blotting was carried out with the following primary antibodies. Anti-Rpb1 recognizes a conserved epitope on Rpb1 outside the CTD and was kindly provided by E. K. Bautz (19). Anti-human FCP1 (anti-hFCP1) was kindly provided by J. Greenblatt (1). Anti-glutathione S-transferase (anti-GST) and anti-FLAG (M2) were purchased from Sigma. Anti-active MAPK (Promega) recognized the phosphorylated Xp42. Reactive bands were visualized using horseradish peroxidase-conjugated secondary antibodies (Promega) and chemiluminescence (Pierce).
Nucleotide sequence accession number.
The xFCP1 sequence data are available in GenBank under accession number AF348120.
RESULTS
CTD phosphatase activity in extracts from X. laevis cells and eggs.
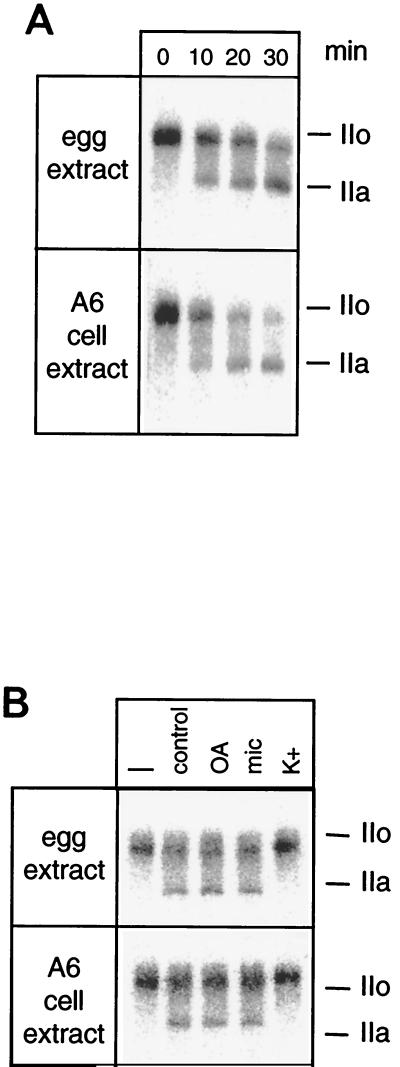
In X. laevis, fertilization is rapidly followed by a massive dephosphorylation of the RNAP II CTD (4, 25). To understand the basis of this phenomenon, the CTD phosphatase activity was investigated in interphasic egg extracts. Phosphatase activity was detected using 32P-labeled mammalian RNAP IIO and by taking advantage of the slower electrophoretic mobility of the Rpb1 subunit when hyperphosphorylated on its CTD (subunit IIo). Upon incubation in the frog egg extracts, the IIo subunit was gradually replaced by the hypophosphorylated IIa subunit (Fig. 1A). Similarly, a low-salt extract from A6 frog kidney cells dephosphorylated the CTD of mammalian RNAP II. The 32P end labeling did not significantly diminish during the incubations, indicating that the phosphatase activity did not remove the phosphate on the casein kinase II site that had been used to label the Rpb1 subunit. CTD phosphatase activity was processive in that the IIo subunit was transformed into the IIa subunit without observable intermediates. Furthermore, CTD phosphatase extracted in a low-salt buffer was insensitive to the usual phosphatase inhibitors, i.e., okadaic acid and microcystin. Activity was totally inhibited in the presence of moderate concentrations of K+ ions (Fig. 1B). These features matched those described for FCP1, the TFIIF-dependent CTD phosphatase that had been characterized in human and yeast cells (1, 8, 14). Thus, the CTD phosphatase activity detected in Xenopus cell extracts might be due to a frog orthologue of FCP1.
FIG. 1.
CTD phosphatase activity in Xenopus. (A) Interphasic egg extracts or low-salt extracts from Xenopus A6 cells were incubated with labeled RNAP IIO for the amount of time indicated. (B) Extracts were incubated for 15 min with labeled RNAP IIO in the absence (control) or presence of 10 nM okadaic acid (OA), 5 nM microcystin (mic), or 120 mM KCl (K+). The labeled IIO form was incubated in pure buffer (–). Dephosphorylation of the CTD was visualized by SDS-PAGE followed by autoradiography. The positions of subunits IIa and IIo are indicated in both panels.
Xenopus orthologue of the CTD phosphatase FCP1.
To clone the FCP1 frog orthologue, RNAs were isolated from A6 cells and primers were designed by taking advantage of a Xenopus ovary cDNA (GenBank accession number AJ132385) which had been reported to potentially code for a protein of 868 amino acids (aa), sharing homology with the hFCP1 sequence. A partial cDNA was subsequently amplified by reverse transcription-PCR and sequenced (GenBank accession number AF348120). It contains an open reading frame coding for a protein of 980 aa. The new cDNA was highly similar to the original ovary cDNA but differed by several point mutations, and more significantly, it coded for a protein with a 112-aa-long extension at its N terminus. The new amino acid sequence was more closely related to the hFCP1 sequence (961 aa), especially on its N-terminal extension (Fig. 2). Two highly conserved blocks of homology were separated by a stretch of about 200 aa with weaker similarities. These two blocks shared weak but still significant homologies with the yeast FCP1 sequence. On the basis of these homologies, the Xenopus protein was designated xFCP1 (for Xenopus FCP1 orthologue).
FIG. 2.
Alignment of FCP1 protein sequences. The human (Hs), Xenopus (Xl), and yeast (Sc) FCP1 sequences were deduced from their cDNA sequences (GenBank numbers AF154115, AF348120, and NC_001145). The homologies are in gray shaded boxes. The dark boxes enclose residues conserved in all three species. ◊, crucial aspartate residue.
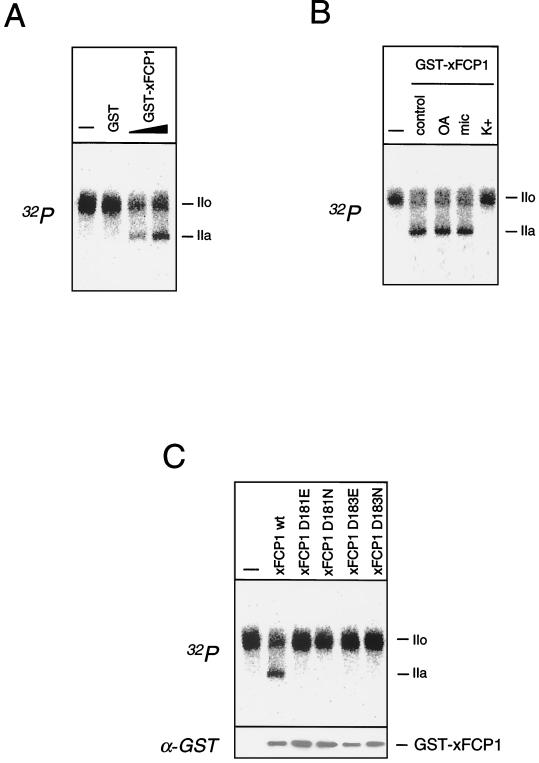
To test the activity of the putative xFCP1 protein, its cDNA was cloned adjacent to GST, and the fusion protein was expressed in bacteria. The resulting GST-xFCP1 protein efficiently dephosphorylated RNAP IIO, whereas GST alone did not (Fig. 3A). Dephosphorylation of RNAP IIO by GST-xFCP1 was insensitive to okadaic acid and microcystin but inhibited by mild concentrations of KCl (Fig. 3B). Kobor et al. have shown that the CTD phosphatase activity of the yeast FCP1 is knocked out by replacement of aspartate by either glutamate or asparagine in the LVVDLDQTII peptide motif (18). Both the hFCP1 and xFCP1 protein sequences contained the closely related motif LMVDLDQTLI. Replacement of either aspartate (D181 or D183) by glutamate or asparagine in the xFCP1 sequence suppressed the CTD phosphatase activity of the mutant recombinant GST fusion proteins, although the same amount of the proteins was used, as checked by anti-GST Western blot analysis (Fig. 3C). This finding highlights the similarity between the vertebrate and yeast FCP1 proteins.
FIG. 3.
X. laevis FCP1 exhibits CTD phosphatase activity. (A) Labeled RNAP IIO was incubated for 10 min with GST or increasing amounts of GST-xFCP1. (B) The experiment was carried out in the absence (control) or presence of 10 nM okadaic acid (OA), 5 nM microcystin (mic), or 120 mM KCl (K+). (C) Purified wild-type or mutant GST-xFCP1 fusion proteins were incubated for 10 min with labeled RNAP IIO. The amounts of the fusion proteins were checked by Western blot analysis with a GST-specific antibody. Dephosphorylation was visualized by SDS-PAGE and autoradiography. Unreacted RNAP IIO was loaded as a control (–) in each experiment. The positions of the subunits IIa and IIo are indicated in each panel.
CTD phosphatase activity in Xenopus extracts is attributable to xFCP1.
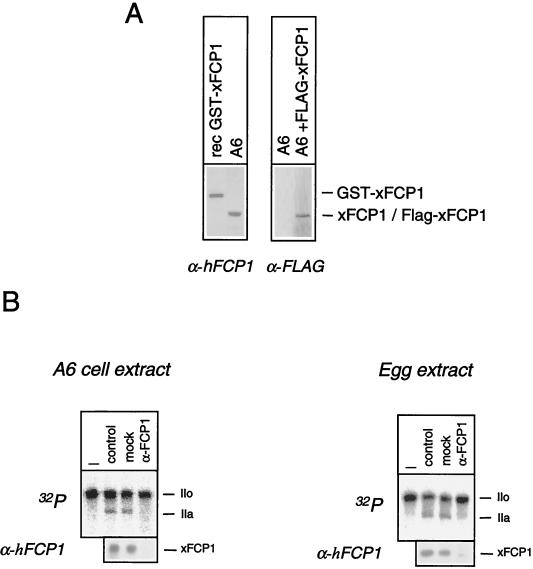
An immunochemical analysis was employed to establish whether or not the CTD phosphatase activity detected in Xenopus extracts corresponded to xFCP1. An antiserum raised against hFCP1 reacted with an ≈150-kDa polypeptide in human cell extracts (1, 14). The same antiserum reacted with a polypeptide of similar size in A6 cell extracts (Fig. 4A). The cDNA sequence is expected to encode a 110-kDa polypeptide. To make sure that the coding sequence was complete, a FLAG epitope was fused to the N terminus of xFCP1 within a eukaryotic expression vector. The resulting FLAG protein expressed in transfected A6 cells comigrated with the protein detected by the anti-FCP1 antiserum. The anti-hFCP1 reacted with the GST-xFCP1 fusion protein with an apparent molecular mass close to 180 kDa, in agreement with the addition of the GST moiety. Taken together, these experiments strongly suggest that the cloned xFCP1 cDNA encodes a full-length protein. In agreement with studies in both the yeast and mammalian systems, these results indicate that the xFCP1 protein migration is slower than expected from its theoretical molecular mass. As the anti-hFCP1 cross-reacted with the xFCP1 protein, it was used to immunodeplete A6 cell extract and egg extract (Fig. 4B). A mock-depleted extract largely retained CTD phosphatase activity as well as the xFCP1 protein, whereas the anti-hFCP1-depleted extract did not. These results indicate that xFCP1 accounts for the major CTD phosphatase activity in Xenopus extracts.
FIG. 4.
Xenopus CTD phosphatase activity is attributable to xFCP1. (A) Low-salt A6 cell extract or purified GST-xFCP1wt fusion protein was analyzed by SDS-PAGE and Western blotting with anti-hFCP1 (α-hFCP1). Total lysates from control A6 cells or cells transfected with 5 μg of the plasmid encoding FLAG-xFCP1 were analyzed with anti-FLAG (α-FLAG). The positions of recombinant GST-xFCP1, endogenous xFCP1, and transfected FLAG-xFCP1 are indicated. (B) Labeled RNAP IIO was incubated for 15 min with crude egg or low-salt A6 cell extracts (control) which had been either mock or xFCP1 (α-FCP1) depleted. CTD dephosphorylation was monitored as indicated above. Nonincubated labeled RNAP IIO was loaded as a control (–). The positions of the subunits IIa and IIo are indicated. The presence of xFCP1 was checked by Western blotting.
RNAP II dephosphorylation requires xFCP1.
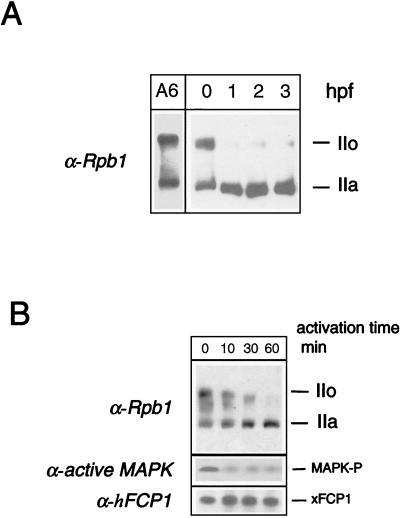
RNAP II phosphorylation undergoes major changes during the first hours of development (Fig. 5A). Fertilization triggers a rapid and massive dephosphorylation of the CTD. Xp42 MAPK deactivation has been proposed to be involved in this process (4). To evaluate the contribution of xFCP1, advantage was taken of the well-described preparation of Xenopus egg extracts capable of supporting elaborate cellular functions (24). The fertilization of metaphase II-arrested eggs can be mimicked by addition of calcium to a meiotic CSF-arrested egg extract. Interestingly, the dephosphorylation of RNAP IIO following calcium activation was paralleled by a deactivation of Xp42 MAPK (Fig. 5B, lower part). Dephosphorylation of the CTD was complete after 60 min (Fig. 5B, upper part), consistent with the pattern of CTD phosphorylation in parthenogenetically activated or fertilized eggs. As transcription resumes only at the midblastula transition, occurring 6 to 8 h after fertilization, the CTD dephosphorylation process takes place in the absence of transcription.
FIG. 5.
CTD phosphorylation state after fertilization in Xenopus. (A) CTD dephosphorylation after fertilization in Xenopus embryos. Batches of 10 unfertilized eggs or embryos were sampled; lysed at 0 (unfertilized eggs), 1, 2, or 3 h postfertilization (hpf); and analyzed by SDS-PAGE along with a whole-cell lysate from A6 cells. The phosphorylation state of the largest RNAP II subunit was analyzed by Western blotting using an anti-Rpb1 antibody. The positions of the subunits IIa and IIo are indicated. (B) CTD dephosphorylation after in vitro activation of a meiotic extract. Activation was performed at 22°C by adding Ca2+ to the CSF-arrested meiotic extract for the indicated time. The phosphorylation state of the CTD and the level of active MAPK were analyzed by Western blotting with anti-Rpb1 and anti-active-MAPK (MAPK-P) antibodies, respectively. The positions of the subunits IIa and IIo are indicated. The presence of xFCP1 was checked by Western blot analysis.
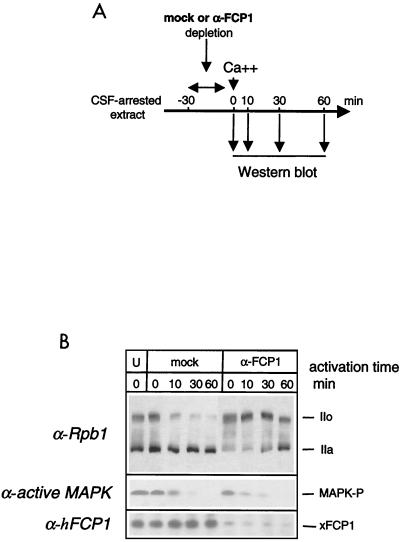
The consequence of xFCP1 depletion on the time course of CTD dephosphorylation was investigated (Fig. 6A). xFCP1 protein was removed by immunodepletion from a meiotic cell extract (Fig. 6B). Such immunodepletion resulted in a significant Rpb1 hyperphosphorylation during incubation with the antibody-coated beads at 4°C. As the FCP1-immunodepleted extract was activated with Ca2+, Rpb1 was slowly dephosphorylated and returned only after 1 h to the RNAP IIO and IIA distribution found in the initial nonactivated extract. In contrast, Rpb1 was rapidly dephosphorylated in the mock-depleted activated extract as it was in the untreated activated extract (compare Fig. 5B and 6B). These results indicated that xFCP1 mediates the dephosphorylation of RNAP II upon activation of Xenopus eggs. Note that the immunodepletion did not affect deactivation of Xp42 MAPK following the addition of calcium (Fig. 6B, lower part).
FIG. 6.
CTD dephosphorylation upon activation of a meiotic extract requires xFCP1. (A) Experimental scheme. The CSF-arrested extract was immunodepleted for 30 min at 4°C before activation by Ca2+ at 22°C for the indicated time. Samples were taken at the various time points as indicated (arrows) and analyzed by Western blotting with the appropriate antibodies. (B) CTD phosphorylation, active MAPK (MAPK-P), and FCP1 were analyzed in untreated (U), mock-depleted (mock), and anti-FCP1-depleted (α-FCP1) extracts. The positions of the subunits IIa and IIo are indicated.
Rapid CTD phosphate turnover in egg extracts.
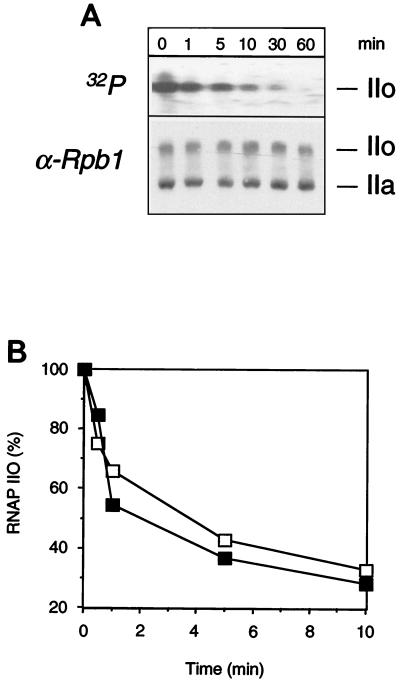
To test whether an increase in FCP1 activity would account for CTD dephosphorylation, the CTD phosphatase activity was monitored during the course of activation. However, a strong CTD kinase activity and the presence of ATP in the egg extracts interfered with the assay. Hence, in order to directly assay CTD phosphate turnover, RNAP IIO uniformly labeled on the CTD repeats was added to meiotic egg extract. The phosphate label was lost rapidly, within minutes, although the global IIo/IIa ratio revealed by Western blotting did not change (Fig. 7A). This result indicates a rapid phosphate turnover on the CTD. Indeed, the phosphorylation pattern of RNAP II results from the balance between CTD kinase and phosphatase activities. The egg extracts contain high levels of nonradioactive ATP that may be used to rephosphorylate the CTD. Xp42 kinase, the major CTD kinase in Xenopus egg extracts (4), is deactivated upon egg activation (Fig. 5B). When RNAP IIO labeled on the CTD repeats was added to activated egg extract, the phosphate label was lost as rapidly as in the meiotic extract, indicating that CTD phosphatase activity does not change upon egg activation (Fig. 7B). Thus, Xp42 kinase activity alone might determine the level of CTD phosphorylation.
FIG. 7.
Rapid CTD phosphate turnover in CSF-arrested and interphasic egg extracts. (A) 32P-labeled (on the CTD repeats) RNAP IIO was added to CSF-arrested egg extract. Samples were analyzed after increasing incubation times by autoradiography and Western blotting. The positions of the subunits IIa and IIo are indicated. (B) 32P-labeled RNAP IIO was added to an egg extract that was either unactivated (▪) or activated (□) by Ca2+ at 22°C for 60 min. The percentage of radioactivity associated with subunit IIo (arbitrary units) is plotted as a function of time.
DISCUSSION
These studies report the molecular cloning of the cDNA encoding the full-length Xenopus orthologue of the human and yeast FCP1 CTD phosphatases and examine the role of xFCP1 in early development. Fertilization triggers a rapid and massive CTD dephosphorylation. This pattern of dephosphorylation, which can be reproduced upon calcium activation of CSF-arrested egg extracts, is dependent on the activity of xFCP1. Finally, the rapid turnover of CTD phosphates that occurs during early development appears to be independent of transcription.
xFCP1, a nonconventional evolutionarily conserved protein phosphatase.
FCP1-related sequences are found in Schizosaccharomyces pombe, Aspergillus nidulans, Arabidopsis thaliana, Caenorhabditis elegans, and Drosophila melanogaster genomes. All of these putative FCP1 orthologues share the same two blocks of homology. The N-terminal domain, i.e., the FCP1 domain, contains a motif related to the consensus sequence ΨΨΨDXDX(T/V)ΨΨ (where Ψ designates hydrophobic amino acids) found in a family of phosphotransferases and phosphohydrolases using small phosphocompounds as substrates. Mutagenesis of the aspartate residues in this motif abolishes the CTD phosphatase activity of yeast FCP1 (18). We now show that replacement of either of the corresponding aspartate residues by glutamate or asparagine in vertebrate FCP1 has the same effect. Homology between members of the FCP1 family allows the definition of two functional domains in the protein. The N terminus contains the catalytic site with the double-aspartate motif, whereas the C terminus contains a putative TFIIF-interacting domain identified by biochemical or genetic studies of human and yeast cells (1, 9). A divergent zone is observed between the two domains. Despite the significant differences between the human and frog FCP1 sequences, mammalian RNAP IIO is an appropriate substrate for the frog enzyme.
CTD phosphorylation—a highly dynamic process even without transcription.
Previous studies have established that phosphorylation of the CTD is closely linked to the transcription cycle (12). Indeed, different kinases which utilize RNAP II as a substrate are present in the transcriptional apparatus. As a subunit of TFIIH, CDK7 or its yeast orthologue, KIN28, may phosphorylate the CTD during initiation, whereas CDK9, a subunit of P-TEFb, may act during the elongation process (21, 26, 28). Inhibition of KIN28 in S. cerevisiae (30) or inhibition of CDK9 in somatic vertebrate cells (7, 15) leads to a rapid dephosphorylation of the CTD. Conversely, inhibition of FCP1 in S. cerevisiae results in CTD hyperphosphorylation (18).
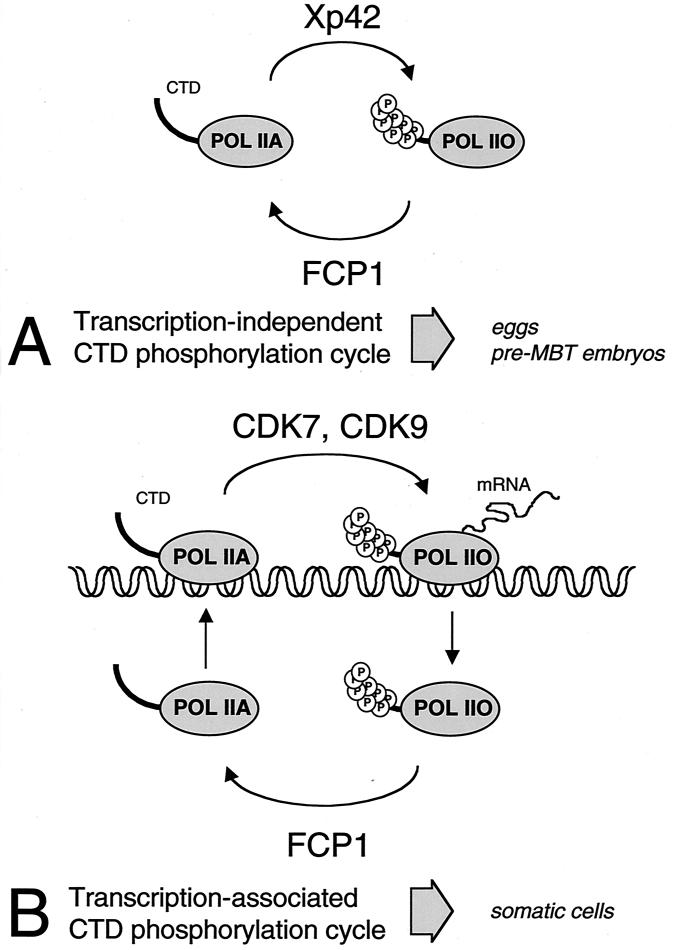
The CTD is phosphorylated when stage VI Xenopus oocytes enter meiosis, whereas egg activation or fertilization is followed by a rapid dephosphorylation of the CTD (4). Importantly, no transcription is detected at these stages. A minor CTD phosphorylation involving MAPK occurs independent of transcription in mammalian (6) and Xenopus cells (25). In the Xenopus oocyte and activated egg system, CTD phosphorylation correlates with the activity of Xp42 MAPK. When MAPK activation is prevented, meiotic CTD phosphorylation does not occur (4). As Xp42 is the major CTD kinase found in metaphase II egg extracts, it is likely to be a major player in CTD phosphorylation in this system in which no transcription is detected. In this report, xFCP1 is shown to account for the CTD phosphatase activity present in an egg extract. Removal of FCP1 from a meiotic extract leads to an enhanced CTD phosphorylation. Furthermore, CTD dephosphorylation is strongly attenuated in activated extracts depleted of FCP1. These findings indicate that phosphorylation of RNAP II in this system is tightly regulated and that Xp42 and FCP1 are the major players in this regulation. Since FCP1 CTD phosphatase activity does not change upon egg activation, Xp42 MAPK activity alone might determine the level of CTD phosphorylation. Since transcription does not occur in metaphase oocytes and early embryos, the MAPK- and FCP1-regulated phosphorylation of RNAP II proceeds independent of transcription (Fig. 8A).
FIG. 8.
A model of two distinct CTD phosphorylation cycles. The FCP1 CTD phosphatase may counteract distinct CTD kinases in early embryos (A) or somatic cells (B).
Dephosphorylation of RNAP II by FCP1 is uncoupled from transcription.
FCP1 has been shown to ensure the recycling of RNAP II (10). The CTD is phosphorylated upon entry into elongation and must be dephosphorylated to allow the formation of subsequent preinitiation complexes (12). CTD dephosphorylation after fertilization might thus contribute to the preparation of the transcriptional machinery for zygotic genome activation. It remains a challenge to establish at which point in the transcription cycle CTD dephosphorylation takes place. In this report, we show that xFCP1 efficiently dephosphorylates RNAP II during meiosis and during the first cell cycles that follow fertilization in X. laevis. As no transcription occurs at these stages, our data indicate that FCP1 dephosphorylates free RNAP II in vivo. In somatic cells, FCP1 is not tightly bound to chromatin because it is fully extracted in a low-salt buffer (14; this report). Furthermore, the CTD in elongation complexes is a poor substrate for FCP1 (22, 23). In transcribing cells, CTD rephosphorylation would occur on DNA-bound initiation complexes upon entry into elongation of transcription (Fig. 8B). A phosphorylated CTD might be required until pre-mRNA synthesis is completed, as it contributes to enhanced cleavage and polyadenylation of the transcript (2, 17). Although a change in the sensitivity of RNAP IIO in an elongation complex could indeed initiate the process of termination, it is tempting to speculate that CTD dephosphorylation could occur on free RNAP II released from DNA in the nucleoplasm following termination.
ACKNOWLEDGMENTS
This work was supported by grants from the Association pour la Recherche sur le Cancer (grant no. ARC 6250 to O.B.) and the Ligue Nationale Contre le Cancer (to O.B.).
We are much indebted to François Giudicelli, Jack Greenblatt, Olivier Jeanjean, Michael Kobor, and Daniela Marazziti for plasmids and reagents; to Olivier Hyrien and colleagues for Xenopus handling; and to Fréderic Gabriel and all members of the laboratory for help and discussions.
REFERENCES
- 1.Archambault J, Pan G, Dahmus G K, Cartier M, Marshall N F, Zhang S, Dahmus M E, Greenblatt J. FCP1, the RAP74-interacting subunit of a human protein phosphatase that dephosphorylates the carboxyl-terminal domain of RNA polymerase IIO. J Biol Chem. 1998;273:27593–27601. doi: 10.1074/jbc.273.42.27593. [DOI] [PubMed] [Google Scholar]
- 2.Barillà D, Lee B A, Proudfoot N J. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2001;98:445–450. doi: 10.1073/pnas.98.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellier S, Chastant S, Adenot P, Vincent M, Renard J-P, Bensaude O. Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J. 1997;16:6250–6262. doi: 10.1093/emboj/16.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellier S, Dubois M-F, Nishida E, Almouzni G, Bensaude O. Phosphorylation of the RNA polymerase II largest subunit during Xenopus laevis oocyte maturation. Mol Cell Biol. 1997;17:1434–1440. doi: 10.1128/mcb.17.3.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bensaude O, Bonnet F, Cassé C, Dubois M-F, Nguyen V-T, Palancade B. Regulated phosphorylation of the RNA polymerase II C-terminal domain (CTD) Biochem Cell Biol. 1999;77:1–7. [PubMed] [Google Scholar]
- 6.Bonnet F, Vigneron M, Bensaude O, Dubois M-F. Transcription-independent phosphorylation of the RNA polymerase II carboxy-terminal domain (CTD) requiring ERK kinase activity. Nucleic Acids Res. 1999;27:4399–4404. doi: 10.1093/nar/27.22.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassé C, Giannoni F, Nguyen V T, Dubois M-F, Bensaude O. The transcriptional inhibitors, actinomycin D and α-amanitin, activate the HIV-1 promoter and favor phosphorylation of the RNA polymerase II C-terminal domain. J Biol Chem. 1999;274:16097–16106. doi: 10.1074/jbc.274.23.16097. [DOI] [PubMed] [Google Scholar]
- 8.Chambers R S, Dahmus M E. Purification and characterization of a phosphatase from HeLa cells that dephosphorylates the C-terminal domain of RNA polymerase II. J Biol Chem. 1994;269:26243–26248. [PubMed] [Google Scholar]
- 9.Chambers R S, Wang B Q, Burton Z F, Dahmus M E. The activity of COOH-terminal domain phosphatase is regulated by a docking site on RNA polymerase II and by the general transcription factors IIF and IIB. J Biol Chem. 1995;270:14962–14969. doi: 10.1074/jbc.270.25.14962. [DOI] [PubMed] [Google Scholar]
- 10.Cho H, Kim T K, Mancebo H, Lane W S, Flores O, Reinberg D. A protein phosphatase functions to recycle RNA polymerase II. Genes Dev. 1999;13:1540–1552. doi: 10.1101/gad.13.12.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corden J L, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 12.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 13.Davidson E H. Gene activity in early development. 3rd ed. Orlando, Fla: Academic Press, Inc.; 1986. [Google Scholar]
- 14.Dubois M F, Marshall N F, Nguyen V T, Dahmus G K, Bonnet F, Dahmus M E, Bensaude O. Heat shock of HeLa cells inactivates a nuclear protein phosphatase specific for dephosphorylation of the C-terminal domain (CTD) of RNA polymerase II. Nucleic Acids Res. 1999;27:1338–1344. doi: 10.1093/nar/27.5.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubois M F, Nguyen V T, Bellier S, Bensaude O. Inhibitors of transcription such as 5,6-dichloro-1-β-d-ribofuranosyl benzimidazole (DRB) and isoquinoline sulfonamide derivatives (H-8 and H-7*) promote the dephosphorylation of the C-terminal domain (CTD) of RNA polymerase II largest subunit. J Biol Chem. 1994;269:13331–13336. [PubMed] [Google Scholar]
- 16.Giudicelli F, Taillebourg E, Charnay P, Gilardi-Hebenstreit P. Krox-20 patterns the hindbrain through both cell-autonomous and non cell-autonomous mechanisms. Genes Dev. 2001;15:567–580. doi: 10.1101/gad.189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirose Y, Manley J L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 18.Kobor M S, Archambault J, Lester W, Holstege F C, Gileadi O, Jansma D B, Jennings E G, Kouyoumdjian F, Davidson A R, Young R A, Greenblatt J. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol Cell. 1999;4:55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- 19.Kontermann R E, Liu Z, Schulze R A, Sommer K A, Queitsch I, Dubel S, Kipriyanov S M, Breitling F, Bautz E K. Characterization of the epitope recognized by a monoclonal antibody directed against the largest subunit of Drosophila RNA polymerase II. Biol Chem Hoppe-Seyler. 1995;376:473–481. doi: 10.1515/bchm3.1995.376.8.473. [DOI] [PubMed] [Google Scholar]
- 20.Leclerc V, Raisin S, Leopold P. Dominant-negative mutants reveal a role for the Cdk7 kinase at the mid-blastula transition in Drosophila embryos. EMBO J. 2000;19:1567–1575. doi: 10.1093/emboj/19.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee T I, Young R A. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 22.Lehman A L, Dahmus M E. The sensitivity of RNA polymerase II in elongation complexes to C-terminal domain phosphatase. J Biol Chem. 2000;275:14923–14932. doi: 10.1074/jbc.275.20.14923. [DOI] [PubMed] [Google Scholar]
- 23.Marshall N F, Dahmus M E. C-terminal domain phosphatase sensitivity of RNA polymerase II in early elongation complexes on the HIV-1 and adenovirus 2 major late templates. J Biol Chem. 2000;275:32430–32437. doi: 10.1074/jbc.M005898200. [DOI] [PubMed] [Google Scholar]
- 24.Murray A W. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 25.Palancade, B., S. Bellier, G. Almouzni, and O. Bensaude. Incomplete RNA polymerase II phosphorylation in Xenopus laevis early embryos. J. Cell Sci., in press. [DOI] [PubMed]
- 26.Price D H. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seydoux G, Dunn M A. Transcriptionally-repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of C. elegans and D. melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- 28.Svejstrup J Q, Vichi P, Egly J-M. The multiple roles of transcription/repair factor TFIIH. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 29.Swedlow J R. Chromosome assembly in vitro using Xenopus egg extracts. In: Bickmore W A, editor. Chromosome structural analysis: a practical approach. Oxford, United Kingdom: Oxford University Press; 1999. pp. 167–182. [Google Scholar]
- 30.Valay J-G, Simon M, Dubois M-F, Bensaude O, Facca C, Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]