Abstract
The avian homologue of the interferon regulatory factor 4 (IRF-4) and a novel splice variant lacking exon 6, IRF-4ΔE6, were isolated and characterized. Chicken IRF-4 is expressed in lymphoid organs, less in small intestine, and lungs. IRF-4ΔE6 mRNA, though less abundant than full-length IRF-4, was detected in lymphoid tissues, with the highest levels observed in thymic cells. IRF-4 is highly expressed in v-Rel-transformed lymphocytes, and the expression of IRF-4 is increased in v-Rel- and c-Rel-transformed fibroblasts relative to control cells. The expression of IRF-4 from retrovirus vectors morphologically transformed primary fibroblasts, increased their saturation density, proliferation, and life span, and promoted their growth in soft agar. IRF-4 and v-Rel cooperated synergistically to transform fibroblasts. The expression of IRF-4 antisense RNA eliminated formation of soft agar colonies by v-Rel and reduced the proliferation of v-Rel-transformed cells. v-Rel-transformed fibroblasts produced interferon 1 (IFN1), which inhibits fibroblast proliferation. Infection of fibroblasts with retroviruses expressing v-Rel resulted in an increase in the mRNA levels of IFN1, the IFN receptor, STAT1, JAK1, and 2′,5′-oligo(A) synthetase. The exogenous expression of IRF-4 in v-Rel-transformed fibroblasts decreased the production of IFN1 and suppressed the expression of several genes in the IFN transduction pathway. These results suggest that induction of IRF-4 expression by v-Rel likely facilitates transformation of fibroblasts by decreasing the induction of this antiproliferative pathway.
v-rel, which is derived from the c-rel proto-oncogene, is the acutely transforming oncogenic member of the Rel/NF-κB family (32, 42). The Rel/NF-κB family of transcription factors regulate gene expression from promoters or enhancers containing κB binding sites (GGGRNNYYCC, where R is a purine, Y is a pyrimidine, and N is any nucleotide) (7, 85). Rel/NF-κB proteins coordinate the expression of genes involved in natural and acquired immunity (56). In addition, the Rel/NF-κB proteins participate in the regulation of other processes such as apoptosis and embryonic development (8, 15, 29, 34, 36, 51). The function of Rel/NF-κB family members has been highly conserved during vertebrate evolution, and essentially the same five transcription factors—RelA, RelB, c-Rel, NF-κB1 (p50), and NF-κB2 (p52)—are present in birds and mammals (30).
Altered regulation of Rel/NF-κB activity has been linked to several pathological processes, including oncogenesis (25, 90). While rearrangements and amplifications of Rel/NF-κB genes have been found in human tumors, c-Rel is the only family member for which acute oncogenesis has been demonstrated (43, 59, 78). Mutations are necessary to activate c-Rel's oncogenic potential. The most highly oncogenic c-Rel mutant, v-Rel, transforms two different cell types, fibroblasts and cells of hematopoietic origin (26, 62). v-Rel-transformed fibroblasts acquire a distinct morphology, are capable of limited growth in soft agar, have a prolonged life span, and induce solid tumors (26, 59, 72, 74). Transformed hematopoietic cells become immortalized, and most of them form aggressive lymphomas in vivo. The ability of v-Rel to transform a particular cell type depends on its ability to induce or repress a specific set of genes. Approximately two dozen genes which exhibit elevated expression in v-Rel-transformed cells have been identified (32, 42). These genes encode cytokines, cell surface receptors, chaperones, genes involved in oxidative metabolism, apoptosis, and various transcription factors.
Interferon regulatory factors (IRFs) are transcription factors which bind to promoter elements that contain an AANNGAAA consensus sequence and either stimulate or repress transcription of these target genes (27). IRFs, originally identified as regulators of interferons (IFNs) and IFN-stimulated genes, are involved in the regulation of natural immunity against viruses, acquired immunity, apoptosis, and embryogenesis, functions also attributed to the Rel/NF-κB family (40, 66, 83, 88). Nine IRF family members have been described in mammals: IRF-1 through 7, IRF-8, also called IFN consensus sequence-binding protein (ICSBP), and IRF-9, originally described as IFN-stimulated gene factor 3-γ (ISGF3-γ). All IRF proteins possess a highly conserved DNA-binding domain with five regularly spaced tryptophans at their N termini. The C termini contain sequences involved in transcriptional activation and repression (66, 83). The IRF members except IRF-1 and IRF-2 have a conserved IRF association domain, which mediates interactions with both other IRF proteins and transcription factors of other families (83).
IRFs transactivate multiple genes that encode proteins with diverse effects on cell proliferation and differentiation, including genes of the IFN family. IFNs are multifunctional cytokines that play an important role in the induction of antiviral responses, cell growth, differentiation, and immunomodulation (40, 103). Type I IFNs (mammalian IFN-α, -β, -τ, and -ω as well as avian IFN1 and IFN2) are produced by a variety of cell types, while type II IFNs (IFN-γ) are secreted primarily by T cells and natural killer cells (101, 102). IFN type I cytokines are the major IFNs that respond to virus infection. Viral infection activates IRF, Rel/NF-κB, and bZIP transcription factors, leading to IFN synthesis (66). Secreted IFNs exert their effect by binding to cell surface receptors and activating the JAK/STAT signal transduction pathway, leading to DNA binding by a complex composed of STAT1, STAT2, and IRF-9 (103). This results in activation of the IFN-stimulated genes, which exert an antiviral and, in many cell types, antiproliferative effect (49).
Several IRF proteins have been associated with the development of cancer, and viral IRF homologues are present in oncogenic herpesviruses (28). IRF-1 and IRF-8 may function as antioncogenes, while IRF-2 and IRF-4 have some characteristics of oncogenes (39, 41, 47, 97). IRF-4 is activated by Tax in adult T-cell leukemia and is overexpressed in multiple myelomas as a result of a translocation of the IRF-4 gene into the immunoglobulin M (IgM) heavy-chain locus (47, 108, 110). In contrast to other IRF family members, IRF-4 is not induced by viral infection or IFNs but by proliferative stimuli (13, 68, 108). Furthermore, while most IRFs are expressed in a variety of cell types, IRF-4 is expressed predominantly in B cells and in activated T cells (66). Mice deficient in IRF-4 have defects principally in the development and function of B and T lymphocytes (71). However, IRF-4 is also detected in macrophages, lens cells, and melanocytes, suggesting that it may also be involved in the regulation of cells other than lymphocytes (35, 63, 64, 67, 94). IRF-4 inhibits the expression of some IFN-induced genes, presumably by interfering with the action of other IRFs (12, 68, 108).
In this work, we demonstrate that IRF-4 overexpression transforms primary chicken embryonic fibroblasts (CEFs). IRF-4 also synergistically cooperates with v-Rel in transformation of CEFs, while antisense IRF-4 expression inhibits v-Rel-mediated transformation. v-Rel transformation of CEFs is associated with the induction of expression of IFN1, the IFN1 receptor, STAT1, JAK1, and the IFN target gene 2′,5′-oligo(A) synthetase (OAS). IRF-4 expression, which is induced in CEFs following infection by v-Rel- and c-Rel-expressing viruses, alters the antiviral gene expression program and likely contributes to transformation of fibroblasts by decreasing the negative antiproliferative effect of the v-Rel-induced antiviral program.
MATERIALS AND METHODS
Cloning of chicken IRF-4 cDNA.
An 858-bp fragment of chicken IRF-4 was cloned by reverse transcription (RT)-PCR from chicken splenic mRNA. First-strand cDNA synthesis was carried out with avian myeloblastosis virus (AMV) reverse transcriptase (Promega, Madison, Wis.) and a locking-docking primer (LDP) specific for poly(A) RNA (5′-AATCTAGAATGTCGACATGCGCGCTTTTTTTTTTTTTTTTVN-3′, where V is an A, C, or G) at 42°C. Degenerate PCR primers I4-1 (5′-CACCTCGAGAAYGAYTTYGARGARYTNGT-3′) and I4-2 (5′-GCAGCGCGCGGRAAYTCYTCNCCRAARCA-3′) were designed based on the similarity between the amino acid sequences of human and mouse IRF-4 as well as differences in the sequence of the closest IRF family member, chicken IRF-8 (50). Two rounds of PCR were carried out with these primers using Taq DNA polymerase (Gibco-BRL Life Technologies, Grand Island, N.Y.). The second round used a 1-μl aliquot of the first-round PCR synthesis as a template. Each round consisted of 35 cycles of 1 min of denaturation at 94°C, 1 min of annealing, and 2 min of extension at 72°C. The annealing temperature was 55°C in the first round and 45°C in the second. The resulting 858-bp PCR fragment was cloned into the pGEM-T vector (Promega), creating pIRF858, and sequenced. The sequence of the 858-bp fragment, which corresponded to amino acids 105 to 392 of human IRF-4, was used to design an additional primer (5′-GGTACCAGGTTGCTCTGTGCTTCGGAGAG-3′) to amplify the 3′ region (1,920 bp) of the IRF-4 cDNA from chicken bursal mRNA by 3′SMART RACE PCR technology (Clontech Laboratories, Palo Alto, Calif.). The PCR products were cloned into the pGEM-T Easy vector (Promega), creating pIRF4CT, and sequenced.
In order to obtain the 5′ end of IRF-4 cDNA, the combined sequences of both PCR fragments and the N-terminal protein sequence of human IRF-4 were used to screen chicken expressed sequence tag (EST) databases (2, 106). Among the several positive ESTs, we identified one clone, pat.pk0027.c10.f (accession number AI980577), isolated from a concanavalin A (ConA)-activated chicken splenic T-cell library, which encoded the N terminus of chicken IRF-4. The complete sequencing of this cDNA clone determined that its 3′ region is 99.7% identical to the sequences of our PCR clones, with one notable exception. The pat.pk0027.c10.f clone was missing a region encoding 36 amino acids in the middle of the open reading frame (ORF) of IRF-4, suggesting that it may represent an alternatively spliced IRF-4 variant. To obtain the full ORF of IRF-4 including these 36 amino acids, primers which flanked the ORF on both sides (5′-GAGCTGGTCACAGAGGTGGTGCTCAG-3′ and 5′-TGCTATCCAGATCAGCTCCTCCACTC-3′) were used to PCR-amplify the cDNA from bursa as well as from ConA-activated splenic cells. Plasmids containing the cloned PCR fragments were designated pGE.IRF4-1 and pGE.IRF4-2 and sequenced.
Plasmids.
The pATH-I4#6 bacterial expression plasmid encodes a TrpE/IRF-4 fusion protein. This plasmid was created by cloning the PvuII-BssHII (828 nucleotides [nt]) fragment of pIRF858 between the SmaI and HindIII sites of pATH1 using a double-stranded adapter (5′-CGCGCGCTGAGTGACTGAGCTCA-3′ and 5′-AGCTTGAGCTCAGTCACTCAGCG-3′) containing a BssHII site, three stop codons in all reading frames, and a HindIII site (54).
pTZ-IRF-4ΔE6 was created by cloning the Eco72I-BglII fragment of pat.pk0027.c10 into pTZ-18R which was modified by the introduction of a double-stranded oligonucleotide adapter (5′-AATTGCTCGAGCACGTGGAATTCAGATCTATTCGCCATTCCTCTATTCAAGATAAGCGCGC-3′ and 5′-TCGAGCGCGCTTATTCTTGAATAGAGGAATGGCGAATAGATCTGAATT CCAGTGCTCGAGC-3′) between the EcoRI and HindIII sites. The adapter contained an XhoI site, an Eco72I site, the sequence encoding amino acids 436 to 445 of IRF-4 (containing a BglII site) followed by a stop codon, and a BssHII site. pTZ-IRF-4, which contains the full-length variant of IRF-4, was obtained by replacing the ApaI-PstI region of pTZ-IRF-4ΔE6 with an ApaI-PstI fragment from the pIRF858 clone.
The REV-T-based pREV-0 and pREV-TW (containing v-rel) reticuloendotheliosis virus (REV)-based retroviral vectors have been described previously (44, 81). The retroviral vector pRSN was created by cloning an adapter containing NotI, XhoI, MfeI, and SpeI sites (5′-TCGACGCGGCCGCCTCGAGCAATTGACTAGTG-3′ and 5′-CGCGCACTAGTCAATTGCTCGAGGCGGCCGCG-3′) between the XhoI and BssHII sites in pREV-0. The pCSV11S3 plasmid encodes an infectious genome of chicken syncytial virus (CSV) (23). The pREV-IRF-4 and pREV-IRF-4ΔE6 retroviral vectors were obtained by cloning the XhoI-BssHII fragments of pTZ-IRF-4 and pTZ-IRF-4ΔE6 into pREV-0. The retroviral vector pREV-antiIRF-4, which expresses IRF-4 in the antisense orientation, was obtained by cloning an XhoI-SpeI fragment containing nucleotides 73 to 1429 of the IRF-4 cDNA from pTZ-IRF-4 into the retroviral vector pRSN (Fig. 1A). The pRCAS plasmid encodes a replication-competent avian sarcoma-leukemia virus (46). pDS3 and pREP-A plasmids were used to prepare DS3 virus (84). Plasmids were cut with SalI and ligated to form concatemers before transfection of CEFs. pRCAS, pDS3, and pREP-A plasmids are derived from the same genomic clone of Schmidt-Ruppin Rous sarcoma virus (20). pRCASc-Rel was obtained by cloning of an HpaII-HpaII c-rel fragment from pBSc-rel#29 into a ClaI site in the RCAS vector (16). pDSv-Rel was constructed by insertion of v-rel into pDS3 as described previously (58). To prepare DSv-Rel virus, pDSv-Rel and pREP-A plasmids were cut with SalI and ligated to form concatemers before transfection of CEFs. Plasmid pcDNAchIFN1 was used for the production of IFN1 in COS-1 cells as described (99, 102).
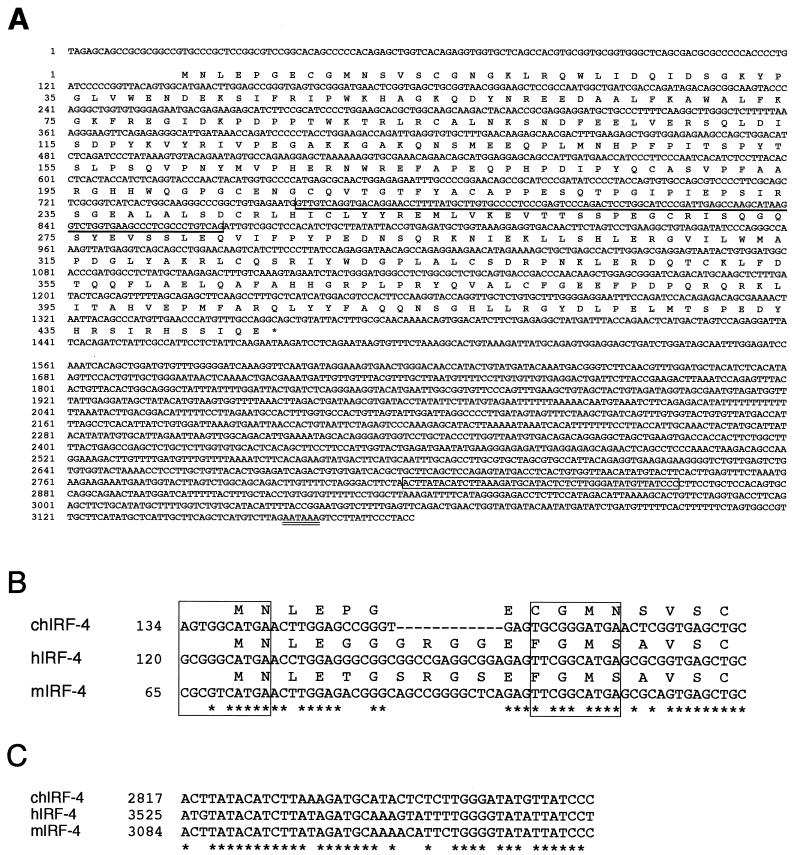
FIG. 1.
Sequence and selected features of chicken IRF-4 cDNA. (A) Nucleotide and predicted amino acid sequence. The position of exon 6, which is absent in the splice variant of IRF-4, is indicated by a single bold underline. The open box in the 3′ UTR identifies a 46-bp element that is conserved between the chicken, human, and mouse IRF-4 cDNAs. The polyadenylation signal is denoted by a double underline. The last C nucleotide at position 3178 was followed by tracts of multiple A nucleotides in two independently isolated clones (pIRF4CT and pat.pk0027.c10 clones). (B) Putative translation start sites in chicken (ch), human (h), and mouse (m) IRF-4 cDNA sequences. Asterisks mark positions that are identical in all three sequences. The boxes around each of the two possible ATG start codons circumscribe a 10-bp element homologous to the Kozak consensus sequence (57). Identical nucleotides are found in both potential start sites at the most critical positions: G at position −3 and A at position +1 relative to the ATG codon. The positions of the first nucleotide shown are indicated on the left side with respect to the full-length cDNA sequences. (C) The 46-bp conserved element in the 3′ UTR of chicken (ch), human (h), and mouse (m) IRF-4 cDNAs. This conserved motif is preceded by a stretch of 82 nt that also shows significant, albeit limited, similarity to the corresponding regions of the mammalian cDNAs (data not shown). For asterisks and position numbers, see legend to panel B.
The following plasmids were used for expression of various oncogenes in CEF cell cultures. pAPrP-C plasmid containing the genome of the Prague strain of Rous sarcoma virus was used for expression of v-src, and a pBUR2-II plasmid containing the genome of UR2 sarcoma virus was used for the expression of v-ros (70, 113). DS3 virus was used for the replication-defective UR2 as a helper. Additional oncogenes were expressed using pRCAS(A) or pRCAS-BP(A) retroviral vectors. These oncogenes included the activated chicken c-Ha-ras (tfch-ras), the oncogenic form of phosphatidylinositol-3 kinase (PI-3 kinase) (v-p3k), v-jun, v-qin, and v-ski (11, 17–19, 33).
Chickens, cell lines, and tissue culture.
Embryonated eggs of specific-pathogen-free White Leghorn chickens were obtained from Hy-Vac, Adel, Iowa (SPF-SC strain), and Charles River SPAFAS, North Franklin, Conn. (SPAFAS strain). Most of the work presented in this article was done with fibroblasts and animals of SPF-SC strain. Cells from SPAFAS birds were used only for the cloning of pGE.IRF4-2 cDNA clone, the detection of IRF-4 protein in DSv-Rel-transformed cells, and the experiment shown in Fig. 9B. CEFs were prepared from 10- or 11-day-old embryos. Cells were cultured with Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% newborn calf serum (Atlanta Biologicals, Norcross, Ga.), 5% chicken serum (Gibco-BRL Life Technologies, Grand Island, N.Y.), 100 U of penicillin, and 50 μg of streptomycin per ml. Secondary cultures of CEFs were used for transfection of plasmid DNA by a modified calcium phosphate method as described previously (44). pREV-0-based plasmids (30 μg) together with pCSV11S3 (1 μg), pRCAS-based plasmids (30 μg), pDS3- or pDSv-rel-pREP-A concatemers (4 μg), or pCSV11S3 (1 μg) alone were used for transfection per 100-mm-diameter tissue culture dish of CEFs. Viruses were harvested between 5 and 7 days after transfection, and the infectious titers of Rel-expressing viruses and CSV were determined by an immunochemical titration assay (44, 104).
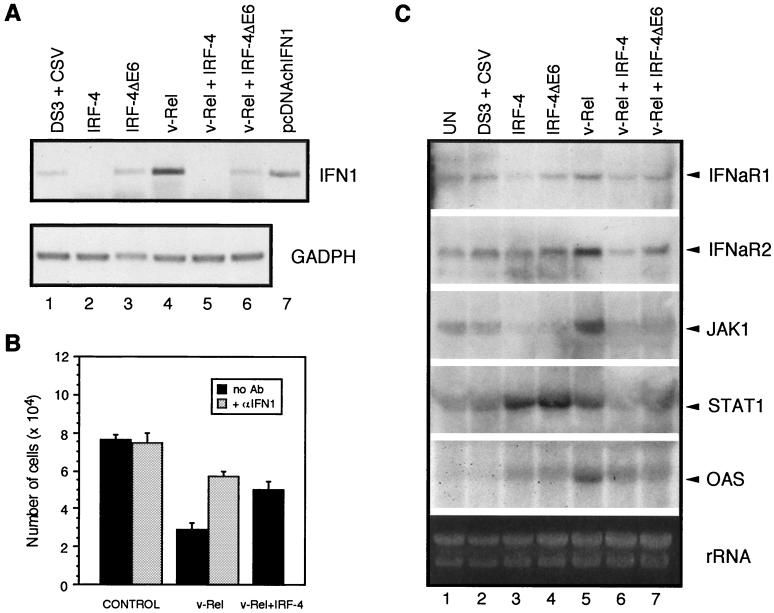
FIG. 9.
Modulation of the expression of genes of the IFN transduction pathway by IRF-4 and IRF-4ΔE6 in v-Rel-transformed fibroblasts. (A) The expression of IFN1 and the control gene GAPDH as determined by RT-PCR. Total RNA was prepared from the cultures described in the legend to Fig. 6. Products of PCR amplification from pcDNAchIFN1 (Table 1) using the same primers as in RT-PCR are shown (lane 7). Because IFN1 is an intronless gene, RNAs for cDNA synthesis were first treated with DNase I as described in Materials and Methods. PCR with pretreated RNA as a substrate confirmed that no detectable IFN1 DNA was present in the RNA samples (data not shown). (B) CEF cultures were infected with empty vector viruses DS3 and CSV for 1 week to induce resistance of CEF to subsequent viral infection. Cells were seeded to 24-well plates (104 cells per well), and medium was replaced with 0.5 ml of culture supernatant from v-Rel- or v-Rel- and IRF-4-transformed fibroblasts after 1 day. The supernatant fluids from DS3 virus-expressing CEFs were used as a control. The supernatant fluids from v-Rel-transformed and control cells were also incubated with monoclonal antibody 8A9 against IFN1 (purified immunoglobulin diluted 1:1,400) for 2 h on ice before application to CEF cultures. The number of cells in each culture was determined 2 days after exposure to the culture medium. The mean and standard errors (error bars) for three to six independent experiments are shown. (C) Expression of IFNaR1, IFNaR2, JAK1, STAT1, and OAS genes was determined by Northern blot analysis. RNA was prepared from the cultures shown in Fig. 6.
The simian COS-1 cell line was used for production of recombinant IFN1 (69). QT6 is a quail fibroblast cell line obtained by chemical mutagenesis (76). The 1757 duck cell line was derived from a duck sarcoma induced by avian leukosis virus (ALV) (82). All other cell lines used are of chicken origin. DT40 is a B-cell line established from an ALV-infected chicken bearing a bursa-derived lymphoid tumor (6). DT40 cells are transformed bursal stem cells which are continuously undergoing immunoglobulin gene diversification (53). The DT95 cell line is also derived from a chicken with an ALV-induced lymphoid leukosis and exhibits a more mature phenotype, including secretion of IgM (6). MSB-1 and RP-1 are T-cell lines established from a Marek's disease virus-induced lymphoma, 123/12 is a v-rel-transformed B-cell line, 160/2 is a v-rel-transformed T-cell line, and 123/6T is a macrophage-like v-rel-transformed cell line (3, 44, 80). AEV-1 is an avian erythroblastosis virus-transformed erythroid cell line (89). BM-2 is a macrophage-like cell line derived by transformation of yolk sac cells by AMV (77). C4-1 cells are a lymphoid non-virus-producing S2A3v-rel-transformed cell line (93, 111). The fibroblastoid cell line 26T6 was derived from a sarcoma induced by REV-Cgi, which expresses a c-rel mutant (43).
Transformation assay.
Transformed and control fibroblasts (105) were plated in 5 ml of DMEM containing 15% chicken serum and 0.35% Noble agar per 60-mm-diameter dish containing a bottom layer of 0.75% agar. Cells were refed with additional soft agar medium every 10 days. Plates were scored for the development of colonies 3 weeks after plating.
Preparation and treatment of white blood cells, lymphocytes, and bone marrow cells.
Peripheral white blood cells and lymphocyte-enriched fractions from bursa, thymus, and spleen used to prepare RNA for Northern and RT-PCR analyses were isolated using Histopaque (Sigma Chemical Co., St. Louis, Mo.). A single-cell suspension of bursal lymphocytes for the experiment shown in Fig. 3 was obtained by passing minced tissue through a nylon mesh followed by a nylon cell strainer (100 μm; Becton Dickinson Labware, Franklin Lakes, N.J.). Cells were stimulated with phorbol-12-myristate-13-acetate (PMA) (1 μg/μl) (Sigma). Bone marrow was isolated from femur and tibiotarsus bones. The bones were cut longitudinally with a razor blade, and the entire contents of the bone cavity were scraped out and used for RNA preparation.
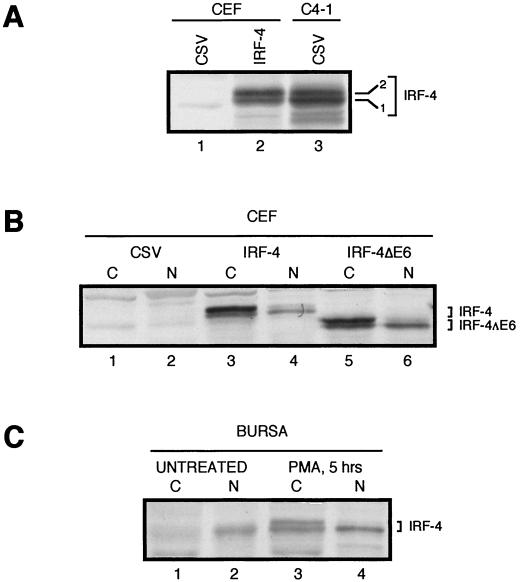
FIG. 3.
Comparison of chicken IRF-4 and IRF-4ΔE6 proteins expressed from retroviral vectors in CEFs with endogenously expressed IRF-4 in chicken B cells. Western blot analysis was performed using IRF-4 antiserum AI4-6. (A) Two isoforms of IRF-4. Lysate from control CSV-infected CEFs is shown in lane 1. Two isoforms of IRF-4 detected in whole-cell lysates of CEF infected with retroviruses expressing IRF-4 (lane 2) comigrate with two isoforms of IRF-4 from whole-cell lysates of the S2A3 v-rel-transformed splenic B-cell line C4-1 infected with CSV (lane 3). Whole-cell extracts from 4 × 105 cells were loaded in each lane. (B) Differential subcellular localization of the IRF-4 isoforms in CEF cultures. Cytoplasmic (C) and nuclear (N) lysates from 2 × 105 CEFs infected with CSV (lanes 1 and 2) or exogenously expressing IRF-4 or IRF-4ΔE6 from retroviral vectors (lanes 3 to 6) were loaded in each lane. (C) Differential subcellular localization of the IRF-4 isoforms in bursal cells. Cytoplasmic (C) and nuclear (N) lysates from normal untreated bursal cells (lanes 1 and 2) and bursal cells treated with PMA for 5 h (lanes 3 and 4). Extracts from 2.5 × 106 bursal lymphocytes were loaded in each lane.
Antisera.
The AI4-6 polyclonal antiserum was raised in a rabbit immunized with a TrpE/IRF-4 fusion protein purified from C600 bacteria transformed by the pATH-I4#6 expression plasmid. The TrpE/chIRF-4 fusion protein contains amino acids 112 to 386 of chicken IRF-4. Monoclonal antibody HY87 is specific for avian c-Rel and v-Rel (44). The goat anti-mouse IgG1 and goat anti-rabbit IgG biotinylated antibodies were purchased from Southern Biotechnology Associates, Inc., Birmingham, Ala. The monoclonal antibody 8A9, which recognizes chIFN1 but not chIFN2, was a generous gift from P. Staeheli (88a).
Western analysis.
Western analysis was performed as described previously (44). Briefly, harvested cells were washed, resuspended, and boiled in sodium dodecyl sulfate (SDS) sample buffer, and proteins were separated on an SDS-polyacrylamide gel using a Mini-Protean II apparatus (Bio-Rad Laboratories, Hercules, Calif.). Proteins were transferred to a PolyScreen polyvinylidene difluoride membrane (NEN Life Science Products, Inc., Boston, Mass.) and sequentially reacted with rabbit polyclonal antiserum AI4-6 or monoclonal antibody HY87, goat anti-mouse IgG1 or goat anti-rabbit IgG biotinylated antibodies, and streptavidin-linked alkaline phosphatase (Roche Molecular Biochemicals, Indianapolis, Ind.). Proteins were visualized by an enzymatic reaction using 5-bromo-4-chloro-3-indolylphosphate and 4-nitro blue tetrazolium chloride as substrates (Roche Molecular Biochemicals). Calibrated prestained molecular size markers from Bio-Rad Laboratories were used. Nuclear and cytoplasmic fractions of CEFs were prepared in hypotonic buffer (50 mM Tris-HCl [pH 8], 1.1 mM MgCl2, 0.5% Triton X-100) as previously described (75).
Northern analysis and probes.
Total RNA was isolated by RNAwiz (Ambion, Austin, Tex.). RNA was separated by electrophoresis in a 1% agarose gel in 20 mM MOPS (3-[N-morpholino]propanesulfonic acid) buffer and transferred to a HybondN+ membrane (Amersham Pharmacia Biotechnology, Piscataway, N.J.). Parallel samples with ethidium bromide were analyzed under identical conditions, and the gel was photographed. Filters were hybridized with [α-32P]dCTP-labeled DNA fragments using UltraHyb solution (Ambion) at 55°C. The probes used for hybridization were labeled by nick translation and are listed in Table 1.
TABLE 1.
DNA probes used for detection of mRNA expression on Northern blots
| Gene | GenBank accession no. | Fragment used (nt) | Reference |
|---|---|---|---|
| IRF-4 | AF320331 | 440–1296 or 473–1357 | This study |
| c-rela | X52193.1 | 16–1865 | 16 |
| IFNaR1 | AF082664.1 | 224–1271 | 91 |
| IFNaR2 | AF082665.1 | 445–1842 | 91 |
| JAK1 | AF096264.1 | 1500–3500 | 10 |
| STAT1 | AI981867.1 | pat.pk0067.e6.fbc | 106 |
| OAS | AW240080.1 | ptr1c.pk002.b9bd | Unpublished data |
The c-rel probe also detects the v-rel gene.
Whole insert of the indicated EST clone was excised and used to prepare the probe.
The EST clone was sequenced and determined to contain 3 kb of cDNA of the chicken homologue of the mammalian STAT1 gene. This cDNA encodes the C terminus of STAT1. Protein sequence is most similar to sequences of human and mouse STAT1 (84% identity), followed by other STAT family members (≤54%).
The EST clone was sequenced and determined to contain 1.6 kb of cDNA of the chicken OAS gene encoding full-length OAS protein of the A isoform (109).
Semiquantitative RT-PCR.
Total RNA (5 μg) together with 1 μl of 10 μM LDP (5′-AATCTAGAATGTCGACATGCGCGCTTTTTTTTTTTTTTTTVN-3′) and 1 μl of 10 μM Smart II primer (Clontech Laboratories, Inc.) was denatured for 10 min at 70°C in 11 μl of water. First-strand cDNA synthesis was carried out with 15 U of ThermoScript RT (Gibco-BRL Life Technologies), 2 μl of 10 μM deoxynucleoside triphosphate (dNTP) mix, and 2 μl of 100 μM dithiothreitol at 55°C for 1 h. For detection of IRF-4 and IRF-4ΔE6 by RT-PCR, 2.5 μl of the first-strand synthesis reaction was used together with 1 μl of Advantage cDNA polymerase mix (Clontech Laboratories, Inc.) and primers FP572 (5′-AGGAGCAGCCATTGATGAACC-3′) and BP878 (5′-CCTCTGGATAAGGGAAGATGACTTG-3′). PCRs were performed in a DNA thermal cycler 480 (Perkin-Elmer, Inc., Boston, Mass.) as follows: five cycles of 30 s at 94°C and 3 min at 72°C; five cycles of 30 s at 94°C, 30 s at 70°C, and 3 min at 72°C; and 25 to 30 cycles of 30 s at 94°C, 30 s at 65°C, and 3 min at 72°C.
The samples of RNA used for detection of IFN1 by RT-PCR were first treated with DNase I (“DNA-free” reagent; Ambion) to remove possible traces of DNA in the RNA preparations. First-strand synthesis was performed as above. The primers used in the PCR were either IFN13 and IFN15 (5′-CCTCCAGCTCCTCCGGGACATGGCTCC-3′ and 5′-GTGTTCCCAGGCGCAGGCGCTGTAATCG-3′), which are specific to chicken IFN1, or GAPDH1 and GAPDH2 (5′-TCATCTGAAGGGTGGTGCTAAGCGTG-3′ and 5′-TCTGGGCAGCACCTCTGTCATCTCTC-3′), which are specific for the chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (105). PCRs were performed as follows: five cycles of 30 s at 94°C and 3 min at 72°C; five cycles of 30 s at 94°C, 30 s at 70°C, and 3 min at 72°C; and 10 to 30 cycles of 30 s at 94°C, 30 s at 68°C, and 3 min at 72°C.
Nucleotide and protein sequence analysis.
The nucleotide sequence of the chicken IRF-4 clones was compared with the databases of GenBank, EMBL, DDBJ, and PDB utilizing a Blast 2.0 search engine (http://www.ncbi.nlm.nih.gov/BLAST) (4, 5). The same search engine was used to compare the protein encoded by the ORF of chicken IRF-4 with the protein sequence databases. The sequences of all IRFs similar to chicken IRF-4 were retrieved, and their amino acid homology was more precisely evaluated using the ClustalX program coupled with the MacBoxshade 2.15 analysis tool (48). The following protein sequence files from Swiss-Prot, translated GenBank, and DDJB databases were compared to the chicken IRF-4 sequence: sp Q90876, sp P10914, sp P15314, sp P23570, sp Q98925, sp P14316, sp P23906, sp Q14653, sp P70671, sp Q15306, sp Q64287, sp Q13568, sp P56477, sp O14896, gb AAB36714.1, dbj BAA24349.1, sp Q92985, sp P70434, sp Q90871, sp Q02556, sp P23611, sp Q00978, sp Q61179, and sp Q90643. Nucleotide sequences in the 5′ and 3′ untranslated regions (UTRs) of chicken IRF-4 cDNA were compared to sequences of mammalian IRF-4 (GenBank U52682 and U11692) using FASTA and Macaw 2.0.5 alignment tools to determine homology (61, 86, 98). Possible PEST regions were evaluated by the PEST-FIND program (available at http://www.at.embnet.org/embnet/tools/bio/PESTfind/) (92).
Nucleotide sequence accession numbers.
The sequence of chicken IRF-4 submitted to GenBank under accession number AF320331 is a composite of two cDNA clones, pat.pk0027.c10 and pGE.IRF4-2. The sequence of the IRF-4ΔE6 splice variant is based on the pat.pk0027.c10 clone and was submitted in a separate file with accession number AF320332. The four cDNA clones (pIRF858, pIRF4CT, and pGE.IRF4-1 and -2) that were obtained by RT-PCR differed in overlapping regions at a few nucleotide positions from pat.pk0027.c10 and from each other. Because at least some of these differences are probably caused by the low fidelity inherent to PCR, the sequence of pat.pk0027.c10 was taken to be the standard, as this plasmid was isolated directly from a plasmid-based cDNA library without a PCR amplification step. The fidelity of the sequence in the exon 6 region, which is not present in pat.pk0027.c10, is supported by the identity of this sequence in two independently isolated PCR clones, pGE.IRF4-2 and pIRF858.
RESULTS
Cloning and sequence analysis of chicken IRF-4.
Mammalian IRF-4 has been previously cloned and characterized (22, 35, 68, 108). Several of its properties suggest that IRF-4 expression may be important in v-Rel-mediated transformation. IRF-4 is expressed in lymphocytes, the hematopoietic target cells of v-Rel, and is induced by proliferative stimuli (62, 68). Furthermore, mice deficient in IRF-4, like those deficient in c-Rel, have defects in B-cell proliferation (55, 71). These similarities, together with the presence of κB sites in the IRF-4 promoter, suggest that IRF-4 expression may potentially be regulated by c-Rel and its oncogenic form, v-Rel (68).
To evaluate the role of IRF-4 in v-Rel-mediated transformation, we isolated the avian IRF-4 homologue. The avian IRF-4 cDNA is 3,178 nt long (Fig. 1A). The longest ORF encodes a protein of 445 amino acids with a predicted molecular mass of 51 kDa. The start codon of this ORF, at nt 140 to 142, is located downstream of an in-frame termination codon at nt 119 to 121. A second potential start codon is located at nt 164 to 166 (Fig. 1B). Both potential start codons are also found in mammalian IRF-4 genes (22, 108). The 5′ UTR of chicken IRF-4 is short and GC-rich, like the 5′ UTRs of mammalian IRF-4 homologues (Fig. 1A) (35, 68). The 1.7-kb 3′ UTR of avian IRF-4 is significantly shorter than the 3.5-kb 3′ UTRs of mammalian IRF-4 genes (35, 68). While chicken 3′ UTR sequences are generally divergent from their mammalian counterparts, a 46-bp-long conserved region of sequence homology was identified (Fig. 1A and C). The chicken 3′ UTR sequence contains a polyadenylation signal (AATAAA) at nt 3157 to 3162 located upstream of the poly(A) tract.
In addition to the 3,178-nt IRF-4 cDNA, we also identified a 3,070-nt cDNA isoform encoded by an EST clone isolated from a ConA-activated chicken splenic T-cell library (106). This isoform represents a novel, alternatively spliced form of IRF-4. The predicted exon structure of chicken IRF-4, based on the known exon-intron structure of mouse and human IRF-4, indicates that the alternative form of IRF-4 is missing exon 6 (Fig. 2A) (35, 68). This variant, IRF-4ΔE6, has not been described for mammalian IRF-4; however, a different type of alternative splicing has been detected in mouse and human IRF-4. About 50% of the mammalian IRF-4 clones contain an additional glutamine residue at the boundary between the fourth and fifth exons (35, 68). Four independently isolated clones of chicken IRF-4 did not contain this glutamine residue, suggesting that this form may not exist in the chicken.
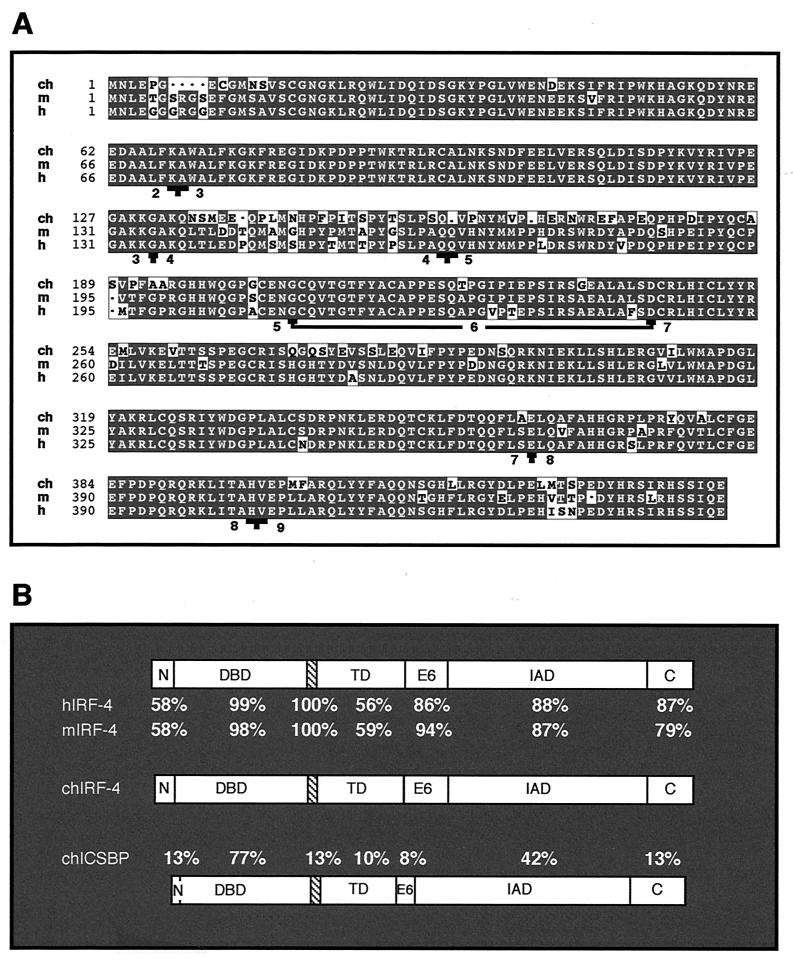
FIG. 2.
Comparison of predicted amino acid sequence of chicken IRF-4 with mouse and human IRF-4 and with chicken IRF-8 (chICSBP). (A) Sequence alignment of the chicken (ch), mouse (m), and human (h) IRF-4 protein sequences. Boundaries of individual exons of chicken IRF-4 were predicted based on the known exon-intron structure of the mouse and human IRF-4 genes. This prediction is supported by the highly conserved exon-intron structure of avian and mammalian IRF-8 genes (21, 52). Exons are numbered from 2 to 9, and the exon boundaries are indicated by a T sign. The underlined sequence encoded by exon 6 indicates the amino acids absent in IRF-4ΔE6. The codons for glycine 207 and aspartic acid 243 are spliced together in IRF4ΔE6 mRNA, forming a codon for aspartic acid. (B) Identical amino acids within predicted functional domains: N terminus (N), DBD, putative NLS (hatched box), transactivation domain (TD), exon 6 (E6), IAD, and the C terminus (C). These functional domains are located in chicken IRF-4 at the following amino acid positions: N terminus (1 to 15), DBD (16 to 126), putative NLS (127 to 134), transactivation domain (135 to 206), exon 6 (207 to 242), IAD (241 to 407), and C terminus (408 to 445). The region between amino acids 198 and 234 of chicken IRF-4 was determined by the program PEST-FIND to be a poor PEST sequence, with an assigned score of −3.95 (score ranges from −50 to +50, and only sequences with a score above +5 are considered likely to act as a PEST domain). Similar scores were obtained for homologous regions of human and mouse IRF-4.
Amino acid sequence analysis confirmed that cloned IRF-4 is the chicken homologue of mammalian IRF-4. The chicken IRF-4 sequence is 84.3 and 83.8% identical to human and mouse IRF-4 amino acid sequences, respectively (Fig. 2A). The identity drops off significantly when chicken IRF-4 is compared to chicken, human, and mouse IRF-8, with 38.4, 38.6, and 38.4% identity, respectively. Other vertebrate IRF family members show even less sequence identity (data not shown). The availability of the first nonmammalian IRF-4 enabled us to compare the evolutionary conservation of the different functional regions of IRF-4 (Fig. 2B). The DNA-binding domain (DBD) and IRF association domain (IAD), which are not only very similar between avian and mammalian IRF-4 but are also shared with other IRF family members, are highly conserved. In contrast, the N terminus and the middle proline-rich region of IRF-4 show the highest sequence variation between avian and mammalian IRF-4. The low evolutionary conservation of the middle proline-rich region is in agreement with its proposed function as a transactivation domain and a flexible connector of DBD and IAD (14, 79). The remaining three regions of IRF-4, the GAKKGAKQ motif, the region encoded by exon 6, and the C terminus, are not similar to the corresponding regions of other members of the IRF family, but they are highly conserved between avian and mammalian IRF-4, suggesting that they may provide important functions. The GAKKGAKQ motif, which immediately follows the DBD, has an amino acid composition similar to that of a nuclear localization sequence (NLS) and resides at the same location as the NLS of IRF-1 (96). The C-terminal sequences may be conserved because of their ability to regulate the DNA binding of IRF-4 (13).
Characterization of IRF-4 protein.
The protein product of the cloned IRF-4 cDNA was detected by Western analysis, confirming that the cDNA encodes a protein corresponding in size to the protein produced by the endogenous IRF-4 gene (Fig. 3). The coding region of IRF-4 was inserted into a retroviral vector and expressed in CEF cultures, in which no detectable IRF-4 protein is normally present. This protein was then compared with endogenous IRF-4 from the chicken C4-1 B-cell line transformed by the S2A3 v-rel oncogene (Fig. 3A). IRF-4 was expressed at comparable levels in both cell types and was detected as two closely migrating species with molecular weights corresponding to the theoretical molecular weight of IRF-4.
To further characterize the protein, its subcellular localization was established. In the cytoplasm of CEFs expressing IRF-4 from a retroviral vector, a slower-migrating IRF-4 species was more abundant than a faster-migrating form, while in the nucleus the faster-migrating form was prevalent (Fig. 3B, lanes 3 and 4). The exogenously expressed IRF-4ΔE6 protein variant had a similar subcellular localization as full-length IRF-4 (Fig. 3B, lanes 5 and 6). In bursal lymphocytes, very small amounts of the two IRF-4 isoforms were found, and most of the IRF-4 protein was located in the nucleus as the faster-migrating form. Both IRF-4 cytoplasmic isoforms dramatically increased in abundance, and the level of nuclear IRF-4 was elevated after stimulation with PMA (Fig. 3C, lanes 3 and 4). The presence of two IRF-4 species in fibroblasts and lymphoid cells is most likely the result of posttranslational modification, which may function to regulate nucleocytoplasmic transport. Regulation of subcellular localization by phosphorylation has been described for several other IRF family members (60, 66). Collectively, these experiments showed that exogenously expressed IRF-4 in CEF cultures has a similar migration pattern and subcellular distribution as the endogenous IRF-4 found in v-Rel-transformed or PMA-stimulated B cells.
Expression of IRF-4 in normal and transformed cells.
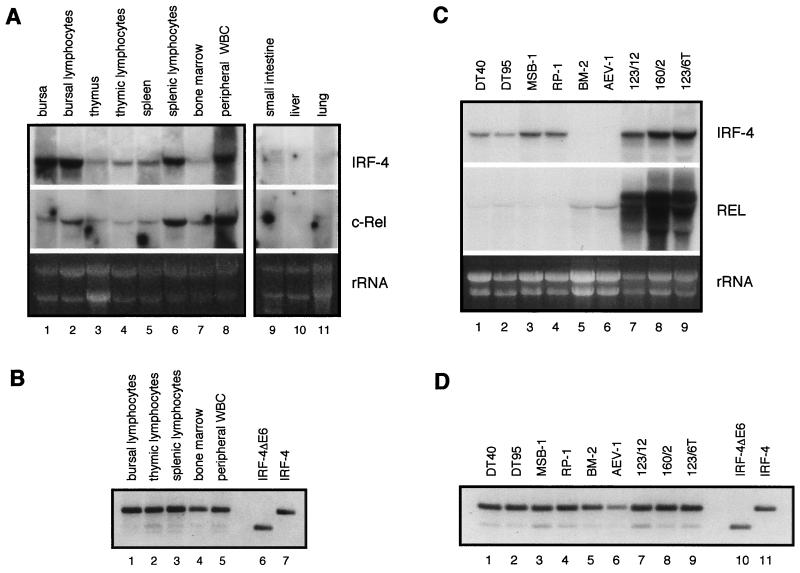
To determine whether chicken IRF-4 is expressed principally in lymphoid cells, as described for its mammalian counterpart, Northern analysis was performed using total RNA from a variety of organs of adult chickens, including spleen, bursa, thymus, bone marrow, peripheral blood, lungs, liver, intestine, gizzard, kidney, gonads, brain, muscles, heart, and skin (Fig. 4 and data not shown). High expression of chicken IRF-4 mRNA was detected exclusively in tissues of hematopoietic origin, with the highest level found in cells of the avian B-cell-specific organ, the bursa of Fabricius, followed by spleen, peripheral white blood cells, thymus, and bone marrow (Fig. 4A, upper panel). Higher expression was detected in purified lymphocyte populations than in the intact organ. Low levels of IRF-4 were also detected in the small intestine and lungs. Two mRNAs of different size were detected in the small intestine. In addition, Northern blot analysis revealed that the tissue-specific expression of c-rel correlates with that of IRF-4 (Fig. 4A, lower panel).
FIG. 4.
Expression patterns of chicken IRF-4, IRF-4ΔE6, and c-rel mRNA. Expression was analyzed by Northern blot analysis (panels A and C) and by RT-PCR (panels B and D). Total RNA (10 μg) was subjected to Northern blot analysis. The probes used to detect c-rel or v-rel and IRF-4 mRNA are described in Table 1. RT-PCR was performed on RNA samples used in Northern blot analysis. Forty PCR cycles were completed. (A) Expression of IRF-4 and c-rel mRNA in various tissues from a 1-month-old chicken. Peripheral white blood cells (WBC) and lymphocyte-enriched fractions from bursa, thymus, and spleen were obtained by Histopaque purification as described in Materials and Methods. The intensity of the rRNA stained with ethidium bromide is shown in the bottom panel (rRNA). (B) Expression of IRF-4 and IRF-4ΔE6 in bursal, splenic, and thymic lymphocytes, peripheral white blood cells, and bone marrow cells determined by RT-PCR. pREV-IRF-4ΔE6 and pREV-IRF-4 plasmids were PCR amplified with the same primers used for RT-PCR (lanes 6 and 7). (C) Expression of IRF-4 (upper panel), c-rel mRNA, and retrovirally expressed v-rel RNA (middle panel) in transformed cell lines as determined by Northern blotting. RNAs from B-cell lines DT40 and DT95, T-cell lines MSB-1 and RP-1, myeloblastoid cell line BM-2, erythroblastoid cell line AEV-1, v-rel-transformed B-cell line 123/12, T-cell line 160/2, and macrophage-like cell line 123/6T were analyzed. The intensity of the rRNA stained with ethidium bromide is shown in the bottom panel (rRNA). (D) Expression of IRF-4 and IRF-4ΔE6 in DT95, DT40, MSB-1, RP-1, BM-2, AEV-1, 123/12, 160/2, and 123/6T cell lines determined by RT-PCR. pREV-IRF-4ΔE6 and pREV-IRF-4 plasmids were PCR amplified with the same primers as used for RT-PCR (lanes 10 and 11).
In order to determine the expression of IRF-4ΔE6, RNA isolated from bursal, thymic, splenic, and peripheral blood lymphocytes and from bone marrow was analyzed by RT-PCR (Fig. 4B). IRF-4ΔE6 mRNA was detected at various levels in all of these tissues, with the highest expression detected in thymocytes, followed by lymphocytes from spleen and peripheral blood. In all tissues, the spliced form represented the minor fraction of the total IRF-4 mRNA.
Northern blot analysis of IRF-4 expression in immortalized avian cell lines demonstrated that its expression is highest in v-Rel-transformed cell lines of B, T, and non-T, non-B phenotypes (Fig. 4C, lanes 7 to 9). High expression was also detected in the Marek's disease virus-derived T-cell lines MSB-1 and RP-1 and in B-cell lines DT40 and DT95, derived from ALV-induced B-cell lymphomas. Low levels of IRF-4 expression were detected in the macrophage cell line BM-2 after longer exposure (data not shown). IRF-4 expression was not detected by Northern blot analysis in the AEV-1 cell line. However, IRF-4 was detected in all cell lines, including the one transformed by AEV-1, by RT-PCR (Fig. 4D). IRF-4ΔE6 was also detected as a minor component of total IRF-4 mRNA in all the cell lines. The highest expression of the spliced form was found in v-rel- and Marek's disease virus-transformed cell lines.
v-Rel and c-Rel induce expression of IRF-4 in transformed fibroblasts.
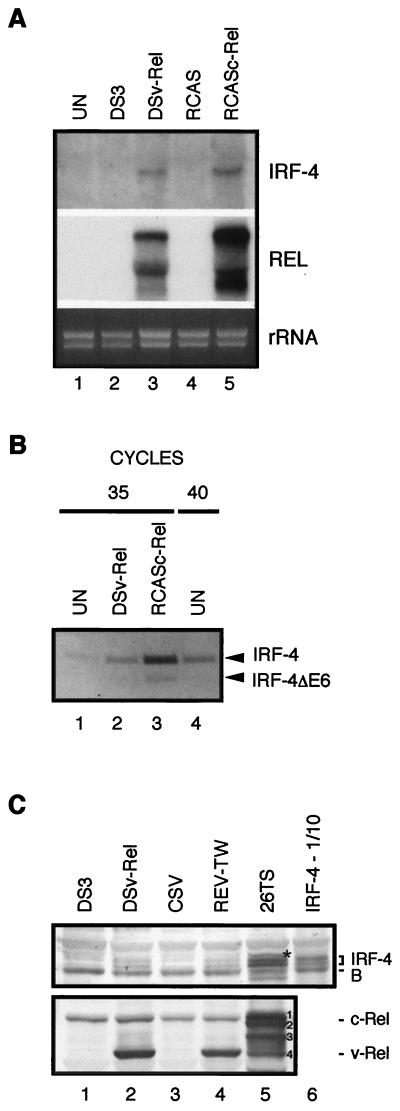
Both v-Rel and c-Rel transform CEFs (1, 26, 45, 59, 72, 74). To define whether IRF-4 may contribute to v-Rel- and c-Rel-mediated transformation, the steady-state level of IRF-4 mRNA in normal fibroblasts and transformed fibroblasts was determined by Northern blot analysis (Fig. 5). Though IRF-4 mRNA was not detected in normal CEFs, IRF-4 was expressed in both v-Rel- and c-Rel-transformed CEF cultures (Fig. 5A). RT-PCR revealed that expression of both the full-length and spliced IRF-4 variants was induced in transformed cells. As observed in the lymphoid cell lines, full-length IRF-4 was the predominant form (Fig. 5B, lanes 1 to 3). Although not detected by Northern analysis, a significant amount of IRF-4 mRNA was detected by RT-PCR in normal chicken fibroblasts (Fig. 5B, lane 4).
FIG. 5.
v-Rel and c-Rel induce the expression of IRF-4 in transformed fibroblasts. (A) Expression of IRF-4 mRNA in uninfected fibroblasts (UN) and fibroblasts either infected with empty retroviral vector (DS3), transformed by DSv-Rel, infected with empty retroviral vector RCAS, or transformed by RCASc-Rel. Total RNA (10 μg) was subjected to Northern blot analysis with the probes described in Table 1. The viral genomic and spliced RNAs in DSv-Rel- or RCASc-Rel-infected cells shown in the panel labeled Rel were detected with a c-rel probe. The endogenous c-rel mRNA was below the threshold of detection at this exposure. The membrane hybridized with a c-rel probe was exposed to film for 2 h, while the IRF-4 membrane was exposed for 48 h. The intensity of the rRNA stained with ethidium bromide is shown in the bottom panel (rRNA). (B) Expression levels of IRF-4 and IRF-4ΔE6 mRNA in uninfected fibroblasts (UN) and fibroblasts transformed by DSv-Rel or transformed by RCASc-Rel were determined by RT-PCR. Lanes 1 to 3 show the products from these cell types after 35 cycles, while lane 4 shows the product from uninfected fibroblasts after 40 cycles. The PCR products corresponding to IRF-4 and IRF-4ΔE6 are indicated on the right side of the panel. (C) Western blot analysis of IRF-4 and Rel expression in fibroblasts. Whole-cell extracts from fibroblasts either infected with empty retroviral vector DS3, transformed by DSv-Rel, infected with CSV helper virus, or transformed by REV-TW and from sarcoma-derived fibroblastoid cell line 26T6 were analyzed. Lane 6 contains a mixture of lysate from CEFs overexpressing IRF4 from a retroviral vector and lysate from CSV-infected control CEFs (1:9). Extracts from 3 × 105 to 4 × 105 CEFs and 26TS cells were loaded per lane, blotted, and stained with anti-IRF-4 AI4-6 serum (upper panel) or anti-Rel HY87 antibody (lower panel). Positions of the two IRF-4 bands, a prominent background band (B), v-Rel, and c-Rel are indicated on the right side. The bands marked with the asterisk and numbers 1 to 4 in lane 5 are discussed in the text.
In contrast to normal cells, the IRF-4 protein was detected in v-Rel- and c-Rel-transformed fibroblasts (Fig. 5C). As in v-Rel-transformed lymphoid cell lines, two closely migrating IRF-4 species were detected in CEFs transformed by v-Rel expressed from either ALV (DSv-Rel, lane 2) or REV-based retroviral vectors (REV-TW, lane 4). By comparing it to the standard shown in lane 6, we estimated that the amount of IRF-4 in v-Rel-transformed fibroblasts is 20 to 40 times lower than in v-Rel-transformed lymphoid cell lines or in fibroblasts overexpressing IRF-4 from retroviral vectors. However, in contrast to cells expressing large quantities of IRF-4, v-Rel-transformed fibroblasts had a faster-migrating form of IRF-4 localized almost exclusively in the nucleus (data not shown). Importantly, the IRF-4 protein was detected not only in fibroblasts transformed in vitro but also in the tumor-derived fibroblastoid cell line 26TS (lane 5). This cell line was established from a solid tumor induced by c-Relgi, a c-Rel mutant with two single amino acid substitutions in the middle region of the protein (81). The two proteins detected by staining with antibody specific to c-Rel and v-Rel (lane 5, bands 1 and 2) are the full-length and a C-terminal deletion mutant of c-Relgi mutant. Retrovirally expressed c-Rel is known to undergo frequent deletions of its C terminus in vivo that increase its tumorigenic potential (43). Two additional proteins (bands 3 and 4) are most likely degradation products of these abundantly expressed proteins. Apparently the high expression of mutant c-Rel proteins results in a high induction of IRF-4 expression, as the amount of IRF-4 in 26TS cells approaches almost one-quarter the amount detected in v-Rel-transformed lymphoid cells. In addition to the two IRF-4 species, a third protein is seen on the blot of 26TS cell lysates (marked by an asterisk), the identity of which is unknown. The induction of IRF-4 expression in Rel-transformed fibroblast cultures and the presence of IRF-4 protein in the fibroblastoid tumor cells suggest that IRF-4 may contribute to v-Rel- and c-Rel-mediated transformation of fibroblasts and that IRF-4 may also be physiologically regulated by c-Rel in fibroblasts.
To evaluate whether the induction of expression of IRF-4 is a general characteristic of transformed fibroblasts, the expression of IRF-4 in primary fibroblasts transformed by several oncogenes and in two sarcoma cell lines was analyzed (data not shown). In contrast to v-rel-transformed fibroblasts, fibroblasts transformed by v-src, v-ros, activated c-Ha-ras, v-ski, v-qin, v-jun, and v-PI-3 kinase and QT6 and 1757 sarcoma cell lines did not express IRF-4 mRNA, indicating that the induction of IRF-4 is not a general feature of fibroblast transformation.
IRF-4 transforms primary chicken fibroblasts and participates in transformation with v-Rel.
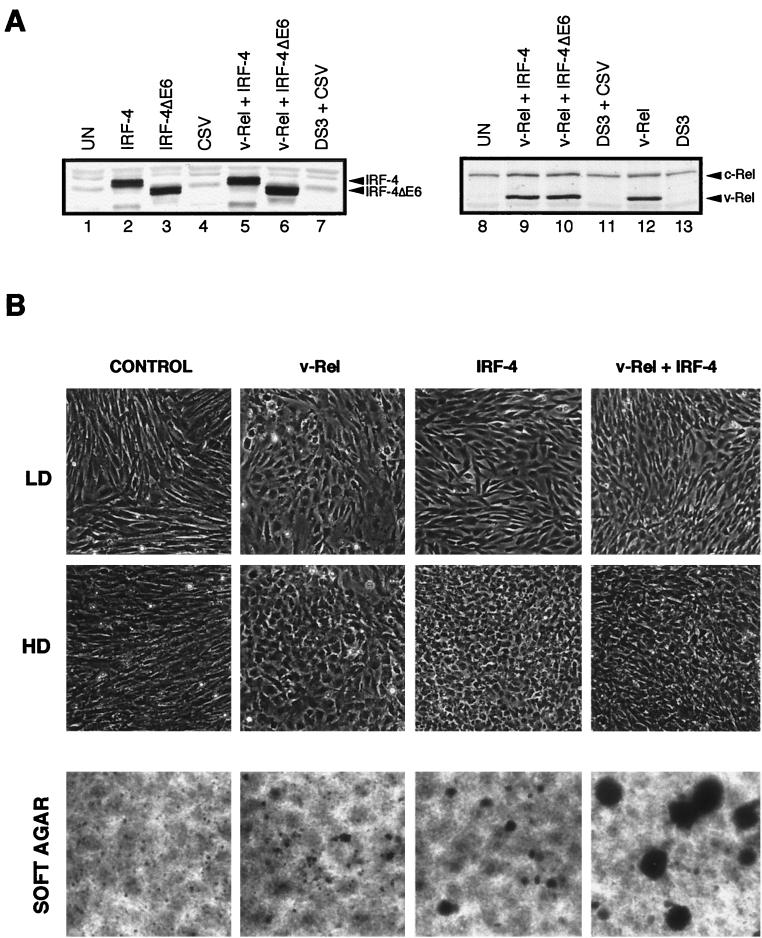
Preliminary experiments showed that fibroblasts which express IRF-4 or IRF-4ΔE6 from retroviral vectors become morphologically transformed. In order to characterize these transformed cells and analyze the contribution of IRF-4 and IRF-4ΔE6 to v-Rel-mediated transformation, CEF cultures were infected with retroviruses expressing v-Rel, IRF-4, or IRF-4ΔE6 or coinfected by viruses expressing v-Rel and IRF-4 or v-Rel and IRF-4ΔE6. The infected cells expressed high levels of the exogenously expressed proteins (Fig. 6A). Both v-Rel- and IRF-4-overexpressing cells became morphologically transformed 1 week after viral infection. The shape and orientation of cells expressing v-Rel were, however, distinct from those of cells expressing IRF-4 (Fig. 6B). v-Rel-transformed fibroblasts were rounded, with irregular edges, and disorganized. The size of these cells varied greatly, and the presence of giant cells was observed. IRF-4-transformed cells retained a more regular growth pattern and did not vary in size. At low density, they assumed a short cylindrical shape which at confluence changed to small, rounded, very densely packed cells. Cells overexpressing both genes had a morphology which was intermediate between those of v-Rel- and IRF-4-transformed cells: the cells had the disorganized growth pattern of v-Rel-transformed cells and, like IRF-4-transformed cells, grew to high density. Control cultures expressing the helper virus CSV or empty vector were morphologically indistinguishable from uninfected cells (data not shown). Cells expressing IRF-4ΔE6 or coexpressing v-Rel and IRF-4ΔE6 were morphologically similar to cells expressing IRF-4 or v-Rel and IRF-4, respectively. The morphological changes in these cells were, however, less pronounced than in those expressing full-length IRF-4 (data not shown).
FIG. 6.
Transformation of chicken fibroblasts by v-Rel and IRF-4. CEFs were infected by retroviruses expressing v-Rel, IRF-4, or IRF-4ΔE6 or coinfected by v-Rel-expressing viruses with either IRF-4- or IRF-4ΔE6-expressing viruses at a multiplicity of infection of 3. Control cells were left uninfected or infected with CSV and DS3 empty-vector retroviruses. The morphology of these cells, their ability to form soft agar colonies, and the level of v-Rel and retrovirally expressed IRF-4 or IRF-4ΔE6 expressed in them were analyzed between 2 and 3 weeks after infection. (A) Equal protein levels of exogenously expressed IRF-4, IRF-4ΔE6, and v-Rel in singly and doubly infected cultures. Whole-cell extracts from 2 × 105 cells from the cultures shown below were blotted and stained with anti-IRF-4 AI4-6 serum (left panel) or anti-Rel HY87 antibody (right panel). Positions of IRF-4 and Rel proteins are indicated on the right side of each panel. (B) Phase-contrast microphotography of low-density (LD) and high-density (HD) cultures. Original photographs were taken at 100× magnification; the printed images are decreased to 70% of the original. Since all three control groups had identical morphology, only the results for CSV-infected control cells are shown. The morphology of cells transformed by IRF-4ΔE6-infected or v-Rel- and IRF-4ΔE6-infected fibroblasts was less pronounced but otherwise similar to IRF-4- or v-Rel- and IRF-4-transformed cells, respectively. The growth of cells transformed by v-Rel, IRF-4, or v-Rel and IRF-4 in soft agar is shown in the bottom row of panels. Since none of the three control groups form colonies in soft agar, only the results for CSV-infected control cells are shown. CEF cultures infected with these viruses were plated in soft agar 4 weeks after infection, and the growth of colonies was scored 3 weeks after plating. Original photographs were taken at 40× magnification; the printed images are decreased to 56% of the original size.
The proliferation rate, saturation density, and ability to form soft agar colonies of cells infected with retroviruses expressing v-Rel, IRF-4, or IRF-4ΔE6 or coinfected by these viruses was analyzed 30 to 40 days after infection (Table 2). These transformation characteristics were compared with parameters determined for uninfected and DS3- and CSV-infected control cells. v-Rel-transformed cells did not proliferate more rapidly than control cells and grew to the same saturation density as control cells. However, in contrast to control cells, v-Rel-transformed cells formed colonies in soft agar. Cell cultures expressing IRF-4 or IRF-4ΔE6 proliferated approximately twice as rapidly as control cells. These cultures also reached a saturation density approximately two to three times higher than that of v-Rel-expressing and control cells. IRF-4 and IRF-4ΔE6, like v-Rel, induced the limited growth of colonies in soft agar. v-Rel and IRF-4 functioned synergistically to increase the proliferation rate, saturation density, and colony formation ability of the fibroblasts. Cells coexpressing v-Rel and either IRF-4 or IRF-4ΔE6 proliferated about four times more rapidly than v-Rel-infected cells and two times more rapidly than IRF-4- or IRF-4ΔE6-infected cells. Fibroblasts coexpressing v-Rel and IRF-4 (but not IRF-4ΔE6) grew to a saturation density four times higher than that of v-Rel-expressing cells and stopped growing when they reached 30 × 106 cells on 60-mm-diameter dishes (1.4 × 106/cm2). The coexpression of IRF-4 and v-Rel also significantly enhanced soft agar colony formation relative to fibroblast cells expressing either v-Rel or IRF-4. These cells produced about 12 times more colonies, which in turn grew to a diameter about three times greater than that of colonies formed by cells expressing v-Rel or IRF-4 alone (Table 2, Fig. 6B).
TABLE 2.
Characterization of CEF cultures transformed by v-Rel, IRF-4, IRF-4ΔE6, v-Rel plus IRF-4, and v-Rel plus IRF-4ΔE6a
| Virus | Morphological transformation | Mean proliferation (106 cells)b ± SD | Mean saturation density (106 cells/cm2)c ± SD | Mean no. of colonies in soft agard ± SD | Life spane (no. of doublings) |
|---|---|---|---|---|---|
| None | − | 2.53 ± 0.15 | 0.35 ± 0.03 | 0 | 40 |
| DS3 + CSV | − | 2.56 ± 0.25 | 0.37 ± 0.03 | 0 | 43 |
| DSv-Rel | + | 2.20 ± 0.20 | 0.31 ± 0.02 | 70.3 ± 14.3 | 36 |
| REV-IRF-4 | + | 4.63 ± 0.15 | 0.92 ± 0.04 | 61.6 ± 20.2 | 67 |
| REV-IRF-4ΔE6 | + | 4.50 ± 0.26 | 0.70 ± 0.06 | 61.3 ± 6.1 | 74 |
| REV-IRF-4 + DSv-Rel | + | 9.80 ± 0.36 | 1.40 ± 0.13 | 857.0 ± 49.2 | 84 |
| REV-IRF-4ΔE6 + DSv-Rel | + | 9.60 ± 0.85 | 0.71 ± 0.12 | 67.7 ± 19.1 | 81 |
CEF cultures were infected by retroviruses expressing v-Rel (DSv-Rel), IRF-4 (REV-IRF-4), and IRF-4ΔE6 (REV-IRF-4ΔE6) or coinfected by DSv-Rel and REV-IRF-4 or DSv-Rel and REV-IRF-4ΔE6 at a multiplicity of infection of 3. Control cells were infected with empty vector retroviruses (DS3 plus CSV) or left uninfected.
Fibroblasts from each culture were plated on 60-mm-diameter dishes (2 × 105 cells per dish), and the number of cells was determined after 48 h by counting with a hemacytometer. The mean ± standard deviation for three independent experiments is shown.
Fibroblasts from each culture were plated on six 60-mm-diameter dishes (2 × 105 cells per dish), and the medium was changed each day. The number of cells was determined each day by counting with a hemacytometer until maximum density was reached. The mean ± standard deviation for three independent experiments is shown.
CEF cultures (105 cells) were seeded in soft agar (0.37%) on top of a bed of hard agar (0.75%). The plates were scored for the development of colonies 3 weeks after seeding. The mean ± standard deviation for three independent experiments is shown.
Each culture was split 1:8 upon reaching confluence. The experiment was repeated twice with similar results.
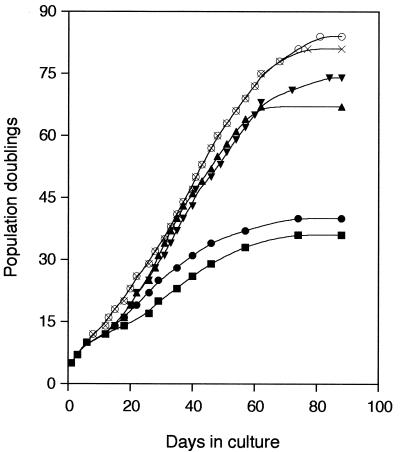
The life span of v-Rel- and IRF-4-transformed cells was also analyzed by monitoring the number of cell doublings in cell culture (Fig. 7). Cells coexpressing v-Rel and IRF-4 or v-Rel and IRF-4ΔE6 began to proliferate more rapidly than uninfected cultures after 1 week and continued to do so until 60 days after infection. Two weeks after infection, cells expressing IRF-4 or IRF-4ΔE6 also began to proliferate more rapidly than control cultures, while v-Rel-expressing cultures entered crisis at approximately 10 days, during which time they grew more slowly than control cells. When v-Rel-expressing cultures overcame this crisis, they began to proliferate at the same rate as uninfected cells or control cells infected with the empty vector. Finally, cells overexpressing both v-Rel and IRF-4 or IRF-4ΔE6 reached two times the number of generations of control cells or v-Rel-transformed cells (Table 2). Cells overexpressing IRF-4 or IRF-4ΔE6 alone doubled less frequently than these cells but still doubled approximately 30 more times than control or v-Rel-expressing cells. IRF-4 therefore cooperates synergistically with v-Rel in the transformation of CEFs.
FIG. 7.
Growth curves of cells expressing IRF-4, IRF-4ΔE6, v-Rel, and IRF-4 or IRF-4ΔE6, with v-Rel showing the cumulative increase in cell generations with time in culture. CEFs were infected with retroviruses expressing v-Rel (▪), IRF-4 (▴), or IRF-4ΔE6 (▾) or coinfected with retroviruses expressing v-Rel and IRF-4 (○) or v-Rel and IRF-4ΔE6 (×) at a multiplicity of infection of 3. Control cells were infected with empty vector retroviruses (•) or left uninfected. Uninfected fibroblasts had the same number of generations as fibroblasts infected by empty vector (data not shown). Each culture was split 1:8 when it reached confluence. This experiment was repeated twice with similar results.
Antisense IRF-4 RNA decreases soft agar colony formation and proliferation of v-Rel-transformed fibroblasts.
To determine if the induction of endogenous levels of IRF-4 in v-Rel-expressing fibroblasts contributes to their transformation, we used antisense RNA technology as a complementary approach to the coexpression studies described above. Western blot analysis of C4-1 cells demonstrated that the expression of IRF-4 in the antisense orientation reduced the levels of IRF-4 protein in v-Rel-transformed cells to less than 50% of wild-type levels (Fig. 8). v-Rel-transformed fibroblasts and lymphoid cells were infected with retroviruses expressing the IRF-4 gene in the antisense orientation. Both types of cells began to grow significantly more slowly (three to six times) than control cells infected with the CSV helper virus (Table 3). Interestingly, the growth of normal fibroblasts was also inhibited by the expression of the antisense IRF-4 construct. The ability of v-Rel-transformed fibroblasts expressing antisense IRF-4 to proliferate and to form colonies in soft agar was then determined (Table 3). In contrast to v-Rel-transformed fibroblasts, cells coexpressing v-Rel and antisense IRF-4 did not form colonies in soft agar. Collectively, the inability of v-Rel-transformed cells expressing antisense IRF-4 to form colonies in soft agar, the efficient transformation of fibroblasts by IRF-4 alone, and the cooperation of v-Rel and IRF-4 in fibroblast transformation suggest that increased levels of IRF-4 in v-Rel-transformed fibroblasts contribute to their transformation.
FIG. 8.
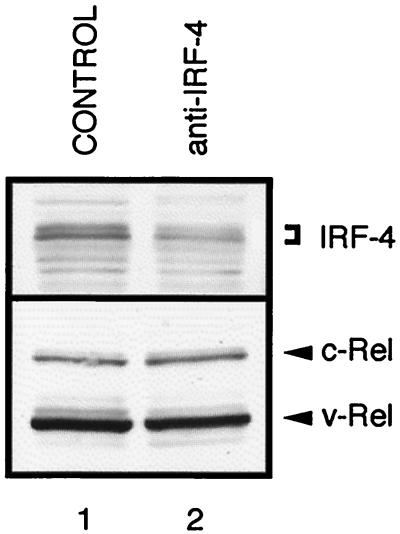
Expression of an antisense IRF-4 construct reduces IRF-4 expression in v-Rel-transformed cells. The S2A3 v-rel lymphoid cell line C4-1 was infected with REV–anti-IRF-4 (anti-IRF-4) or with CSV helper (control). Whole-cell extracts were prepared 48 h after infection and analyzed by Western blot using AI4-6 (anti-IRF-4) serum and HY87 (anti-Rel) antibody. The extracts from 105 cells were loaded in each lane. Positions of IRF-4 and Rel proteins are indicated on the right side of each panel.
TABLE 3.
Effect of antisense IRF-4 on proliferation and soft agar colony formation of v-Rel-transformed cellsa
| Virus | Cells | Mean proliferation (106 cells)b ± SD | Mean saturation density (106 cells/cm2)c ± SD | Mean no. of colonies in soft agard ± SD |
|---|---|---|---|---|
| None | CEF | 2.53 ± 0.15 | 0.35 ± 0.03 | 0 |
| DS3 + CSV | CEF | 2.56 ± 0.25 | 0.37 ± 0.03 | 0 |
| DS3 + REV-anti-IRF-4 | CEF | 0.63 ± 0.12 | 0.16 ± 0.02 | 0 |
| DSv-Rel + CSV | CEF | 2.40 ± 0.18 | 0.32 ± 0.03 | 65.4 ± 12.2 |
| DSv-Rel + REV-anti-IRF-4 | CEF | 0.83 ± 0.15 | 0.14 ± 0.04 | 0 |
| CSV | C4-1 | 90.3 ± 12.7 | NT | NT |
| REV-anti-IRF-4 | C4-1 | 15.6 ± 5.5 | NT | NT |
CEF cultures were infected with empty retroviral vector DS3 or a retrovirus expressing v-Rel (DSv-Rel) at a multiplicity of infection of 3. Control CEF cultures were left uninfected. Four weeks after infections, DS3- or DSv-Rel-infected fibroblasts were superinfected with REV–anti-IRF-4 or CSV 6 h after plating cells for proliferation and saturation density assays (see below). Since ALV-derived DS3 and DSv-Rel use different receptors to gain entry to the cell than reticuloendotheliosis virus-derived REV–anti-IRF-4 with CSV helper virus, the viruses did not interfere, and superinfection was highly efficient. For soft agar colony formation assays, DS3- or DSv-Rel-infected fibroblasts were superinfected 24 h before plating in soft agar medium. The lymphoid cell line C4-1, transformed by S2A3 v-rel, was infected with REV–anti-IRF-4 or CSV.
Fibroblasts from each culture were plated (2 × 105 cells per 60-mm-diameter dish) or C4-1 cells were seeded (2 × 106 cells in 5 ml of culture medium per bottle), and the number of cells was determined after 48 h by counting with a hemacytometer. The mean ± standard deviation for three independent experiments is shown.
Fibroblasts from each culture were plated on six 60-mm-diameter dishes (2 × 105 cells per dish), and the medium was changed each day. The number of cells was determined each day by counting with a hemacytometer until maximum density was reached. The mean ± standard deviation for three independent experiments is shown.
CEF cultures (105 cells) were seeded in soft agar (0.37%) on top of a bed of hard agar (0.75%). The plates were scored for the development of colonies 3 weeks after seeding. The mean ± standard deviation for three independent experiments is shown. NT, not tested.
IRF-4 represses v-Rel-induced expression of the negative cell proliferation regulator IFN1.
v-Rel-transformed fibroblasts do not proliferate more rapidly than control cells and enter crisis at 2 weeks after infection, at which time their proliferation rate decreases relative to control cells (Table 2). This is perplexing because v-Rel-transformed fibroblasts not only have increased levels of IRF-4 but also constitutively express elevated levels of c-Jun and c-Fos, which are known to induce cell proliferation (58). The expression of IRF-4 in v-Rel-transformed cells rescues v-Rel-transformed cells from their proliferation defect (Table 2). Since Rel/NF-κB, Jun/ATF-2, and IRF family members are known to cooperatively regulate the expression of type I IFNs, we determined whether v-Rel induces IFN, which in turn suppresses the proliferation of v-Rel-transformed cells (107). The expression of avian IFN1 mRNA in v-Rel-transformed cells was strongly increased relative to that in control cells (Fig. 9A, lanes 1 and 4).
Mammalian IFNs have antiproliferative effects on certain cell types, but the effect of avian IFNs on cell growth has not been reported (49). To determine whether IFN1 exerts an antiproliferative effect on avian fibroblasts, recombinant IFN1 was produced in COS-1 cells. Fibroblasts treated with recombinant IFN1 proliferated half as fast as control cells, suggesting that the induction of IFN1 by v-Rel may be responsible for the failure of these transformed cells to proliferate more rapidly than the control cells (data not shown). Therefore, the effect of IRF-4 on the expression of IFN1 in v-Rel-transformed and control fibroblasts was determined (Fig. 9A). IFN1 expression was repressed in control and in v-Rel-transformed cells by the expression of IRF-4 (and to a lesser degree by IRF-4ΔE6). Therefore, the increased levels of IRF-4 in v-Rel-transformed cells may partially eliminate the negative effects of IFN1 on cell proliferation and permit v-Rel-transformed cells to proliferate at rates equivalent to control cells. To test this hypothesis directly, culture fluids from v-Rel-transformed cells, cells transformed by v-Rel/IRF-4, and control cells were applied to CEF cultures, and their proliferation rate was evaluated (Fig. 9B). Supernatant fluids from v-Rel-transformed cells contained an inhibitor which strongly interfered with the proliferation of normal fibroblasts. This antiproliferative activity was partially reduced by preincubation with anti-IFN1 antibody. Cells exposed to supernatant fluids obtained from v-Rel/IRF-4-transformed cells proliferated significantly better than those exposed to culture fluids from v-Rel-transformed cells. These results indicate that v-Rel-transformed cells produce IFN1, which strongly inhibits cell proliferation, and that IRF-4 expression decreases the production of this inhibitor.
IRF-4 modulates expression of genes involved in IFN transduction pathway in v-Rel-transformed fibroblasts.
To determine whether the induction of IFN1 expression in v-Rel-transformed cells is coordinated with the expression of other components of the IFN pathway, the levels of mRNA encoding the IFN receptor (IFNaR1 and IFNaR2), the STAT1 transcription factor and its regulatory kinase, JAK1, and the IFN target gene OAS were analyzed (Fig. 9C, lanes 1, 2, and 5). The expression of all of these genes was elevated, indicating that the type I IFN signal transduction pathway is activated in v-Rel-transformed fibroblasts. In contrast, the expression of IFNaR1 and JAK1 was decreased in cells expressing IRF-4 alone, while the expression of STAT1 and OAS was increased in these cells (Fig. 9C, lane 3). The possibility that IRF-4 expression may alter the expression of these genes in v-Rel-transformed cells was explored (Fig. 9C, lanes 1 to 4, 6, and 7). As with IFN1, the expression of IFNaR1 and -2 and JAK1 was downregulated in cells coexpressing v-Rel and IRF-4s relative to cells expressing v-Rel alone. While the expression of STAT1 and OAS was also decreased, significant levels of OAS mRNA were still present in cells expressing both v-Rel and IRF-4. Collectively, these results suggest that the expression of IRF-4, and to a lesser extent IRF-4ΔE6, not only influences the expression of IFN1 but also modulates the expression of other genes in the IFN transduction pathway, likely decreasing the sensitivity of cells to the antiproliferative effects of IFN1. IRF-4, however, does not appear to be a general repressor of genes of the IFN transduction pathway, but is rather a modulator of their relative expression levels.
DISCUSSION
In this report we described the molecular cloning of chicken IRF-4 and the distribution of IRF-4 expression in tissues and cell lines and identified a novel splice variant of this gene. We further demonstrated that IRF-4 transforms primary fibroblast cells and that its expression is induced in fibroblasts by v-Rel and c-Rel and established a role for IRF-4 in the transformation of fibroblasts by v-Rel.
Alternative splice variant of IRF-4.
A differentially spliced variant of IRF-4 mRNA (IRF-4ΔE6) was isolated that does not contain exon 6 and consequently has a deletion of 36 amino acids immediately preceding the IAD. The amino acid sequence encoded by exon 6 is highly conserved between mammalian and avian IRF-4 family members, suggesting that this region may be functionally important. This region was described earlier as a potential PEST sequence (22). PEST sequences, which are rich in proline, glutamic acid, serine, and threonine amino acids, have been identified as destabilizing motifs that target proteins for rapid degradation (92). However, analysis of the IRF-4 sequence using the PEST motif prediction algorithm suggests that it is unlikely that this sequence serves as a PEST sequence (see legend to Fig. 2B). While the IRF-4ΔE6 protein could be overexpressed to slightly higher steady-state levels than IRF-4 in fibroblasts, this difference does not suggest a dramatically different half-life. Previously, a 96-amino-acid-long domain in mouse IRF-4 was identified that appears to mask IRF-4 transactivation activity (79). This domain includes all 36 residues encoded by exon 6, suggesting that the exon 6-encoded region may serve as a regulator of transactivation function. Various levels of IRF-4ΔE6 mRNA were detected by RT-PCR analysis in several lymphoid and hematopoietic tissues, albeit at lower abundance than full-length IRF-4. The levels of IRF-4ΔE6 varied in different tissues, with the highest levels detected in thymus cells. Collectively, these results suggest that IRF-4ΔE6 is differentially regulated and has overlapping but distinct functions compared with full-length IRF-4. Interestingly, a splice variant of human IRF-7, IRF-7B, was described that is lacking 29 amino acids encoding a region corresponding to exon 6 of IRF-4 (112). Since IRF-7 is not closely related to IRF-4, it is likely that alternative splicing of the region between the transactivation domain and IAD is used by additional members of the IRF family.
IRF-4 transforms primary fibroblast cells.
Several reports suggested that IRF-4 may function as an oncogene. IRF-4 was found to be translocated next to the immunoglobulin heavy-chain locus in 20% of multiple myeloma cell lines, and IRF-4 overexpression induced the formation of soft agar colonies in the permanent fibroblastoid cell line Rat-1 (47, 110). Furthermore, it was suggested that IRF-4 plays an important role in adult T-cell leukemia (108). Other reports, however, failed to show the ability of IRF-4 to induce leukemogenesis (95). The results presented in this article show that IRF-4 functions as a very potent oncogene in fibroblasts. The expression of IRF-4 in primary fibroblasts increases their saturation density and permits them to form colonies in soft agar. Moreover, our results demonstrated new characteristics associated with IRF-4 transformation: dramatic alteration of cell morphology, decrease of the doubling time to half that of normal fibroblasts, and significant increases in life span.
The fact that IRF-4 is involved in human oncogenesis is largely accepted, but how IRF-4 acts as an oncogene has not been explored. Low but significant levels of IFN1 mRNA, as well as mRNA for several components of the IFN signaling pathway, have been detected in our experiments in normal fibroblasts. It is possible that IRF-4 may, at least partly, regulate fibroblast proliferation by modulating expression of these genes. Our hypothesis is also consistent with the observation that overexpression of antisense IRF-4 reduces the proliferation rate of normal fibroblasts. Interestingly, the recent report of the isolation of a third viral IRF, vIRF-3, from human herpesvirus 8 (HHV-8) suggested a similar mechanism (65). HHV-8 is the etiological agent of Kaposi's sarcoma and also plays an important role in the pathogenesis of AIDS-associated body cavity-based lymphoma. vIRF-3, which is most homologous to IRF-4, has the ability to repress transactivation of the IFNA (IFN-α) gene normally caused by virus-mediated activation of IRF-3- and IRF-7.
v-Rel- and c-Rel-induced expression of IRF-4 in fibroblasts contributes to cell transformation.
The increased expression of IRF-4 found in v-Rel- and c-Rel-transformed fibroblasts suggests that IRF-4 is under the control of Rel transcription factors in CEFs. This is supported by a report by Grumont and Gerondakis, published while this study was in progress, which demonstrated that IRF-4 is transcriptionally regulated by c-Rel in murine B lymphocytes via κB sites present in the IRF-4 promoter (37). Therefore, it seems likely that IRF-4 is also a direct transcriptional target of v-Rel and c-Rel in chicken fibroblasts. The expression of about a dozen genes has been found to be changed in v-Rel-transformed cells (32, 42). However, only a few of these genes were connected with the transformation phenotype of cells. The AP-1 family of transcription factors and the chemokine mip-1β have been shown to play a functional role in v-Rel-mediated transformation of fibroblasts (58, 87). The ability of IRF-4 to transform fibroblasts suggested that an increased level of IRF-4 may also contribute to the transformed phenotype of these cells. To test this hypothesis, we performed experiments in which the level of IRF-4 in v-Rel-expressing fibroblasts was increased or decreased. Coexpression of v-Rel and IRF-4 synergistically increased transformation of fibroblasts, and expression of antisense IRF-4 RNA in v-Rel-transformed fibroblasts prevented the formation of soft agar colonies and reduced the proliferation of these cells. These results establish that IRF-4 contributes to transformation of fibroblasts by v-Rel.
v-Rel induces expression of genes with antiviral and antiproliferative functions in fibroblasts.
The expression of v-Rel results in the development of a cell type-specific cytopathic effect (81, 100). In most instances, v-Rel-expressing embryonic fibroblasts, despite their transformed morphology and ability to grow in soft agar, do not proliferate more rapidly than control cells (26, 74). REV-T-transformed fibroblast cultures produce less virus over time, suggesting that there may be a selective pressure against fibroblasts expressing high levels of v-Rel (12). Our studies provide a possible explanation for these observations. The expression of IFN1, which has an antiproliferative effect, is induced in v-Rel-transformed fibroblasts. Simultaneously with the expression of IFN1, the expression of IFNaR1, IFNaR2, STAT1, and JAK1 is elevated in v-Rel-transformed cells. The induction of expression of these genes of the IFN transduction pathway suggests that not only is the level of IFN1 increased, but the sensitivity of CEF cultures to its action is also increased. As a result, the increased expression of the IFN target gene OAS was also detected in v-Rel-transformed cells. The expression of IFN-induced genes has been shown in many instances to correlate with decreased cell proliferation (9).
v-Rel likely interacts directly with the promoters of at least some genes of the IFN/IRF signal transduction cascade. IFN1 itself may be a v-Rel target gene. It has been shown that the Rel/NF-κB family members cooperate with IRF-3, IRF-7, and ATF-2/Jun transcription factors in regulation of the mammalian IFN-β promoter (107). The lack of information concerning the regulation of the IFN type I receptors, STAT1, and JAK1 expression does not permit speculation about how v-Rel alters their expression.
IRF-4 modulates v-Rel-induced expression of members of the IFN signal transduction pathway.
Mammalian IRF-4 was suggested to be a repressor of IFN-induced gene expression (37, 94). Our results demonstrate that avian IRF-4 is also able to interfere with the induction of expression of IFN1 and several other components of IFN signaling in v-Rel-transformed cells. The downregulation of some components of the IFN signaling pathway by IRF-4 correlates with an increased transformation of fibroblasts coexpressing v-Rel and IRF-4, suggesting that alterations of the IFN pathway by IRF-4 may contribute to fibroblast transformation by v-Rel. However, we also found that IRF-4 induces the expression of some members of the IFN signal transduction pathway (STAT1 and OAS) in CEFs. It seems that IRF-4 acts as a modulator rather than a general repressor of IFN signaling, inhibiting certain genes while stimulating others. The final outcome of IFN signaling may depend on the absolute levels of IRF-4 activity in the cell as well as the relative ratio of the IRF-4 to Rel/NF-κB. Finally, we cannot exclude the possibility that some of the inhibitory effect of IRF-4 on the IFN signaling pathway in v-Rel-transformed cells may be the result of a direct influence of IRF-4 on the activity of v-Rel.
In summary, the increased expression of IFN1 and other genes of the IFN pathway in v-Rel-transformed cells suggests that the antiviral program can be switched on in fibroblast cells by Rel/NF-κB family members. This is in agreement with the crucial role of Rel/NF-κB proteins in cellular antiviral responses (31, 73). In contrast, the induction of IRF-4 expression by Rel/NF-κB members elicits a highly proliferative response (24, 38). The induction of IRF-4 modulates the v-Rel-induced IFN transduction pathway and gives cells a proliferative advantage. Our results suggest that two cell types transformed by v-Rel, lymphoid cells and fibroblasts, differ in the basal and v-Rel-inducible levels of IRF-4. IRF-4 levels are significantly higher in lymphoid cells than in fibroblasts; therefore, the ratio of IRF-4 to v-Rel in v-Rel-transformed lymphoid cells is higher than in v-Rel-transformed fibroblasts. This difference may explain why, in contrast to fibroblasts, v-Rel-transformed lymphoid cells proliferate better and can be immortalized. More generally, the difference in the level of both constitutive and Rel/NF-κB-inducible expression of IRF-4 in these types of cells may reflect the way fibroblasts and lymphoid cells respond to viral infection. The fibroblasts stop proliferation and are converted to antigen-presenting cells, while lymphoid cells actively proliferate to amplify cells which can specifically recognize and eliminate virus and virus-infected cells.
ACKNOWLEDGMENTS
We are grateful to many colleagues for providing chicken cDNA clones as well as clones of chicken oncogenes in retroviral vectors. We especially thank Joan Burnside and Robin Morgan (University of Delaware) for cDNA clones pat.pk0027.c10.f, pat.pk0067.e6.f, and ptr1c.pk002.b9, which were isolated as a part of the University of Delaware chicken EST project. We thank Thomas Gilmore (Boston University) for c-rel, Georges Lutfalla (CNRS, France) for IFNaR1 and IFNaR2, Christine Sick and Peter Staeheli (Universität Freiburg, Germany) for IFN1 and for monoclonal antibody specific to IFN1, Martin Zenke (Max-Delbrück-Center, Germany) for JAK1, Stephen H. Hughes (National Cancer Institute) for tfch-ras and v-ski, Peter K. Vogt (The Scripps Research Institute) for v-p3k, v-qin, and v-jun, and Lu-Hai Wang (Mount Sinai School of Medicine, New York) for v-ros. We thank Andrew Liss and Cullen Pendleton for careful reading of the manuscript and helpful comments. In addition, we are grateful to William Bargmann for providing DSv-Rel-transformed SPAFAS fibroblasts.
This work was supported by the Council for Tobacco Research 4163 and Public Health Service grant CA33192 from the National Cancer Institute.
REFERENCES
- 1.Abbadie C, Kabrun N, Bouali F, Smardova J, Stehelin D, Vandenbunder B, Enrietto P J. High levels of c-relexpression are associated with programmed cell death in the developing avian embryo and in bone marrow cells in vitro. Cell. 1993;75:899–912. doi: 10.1016/0092-8674(93)90534-w. [DOI] [PubMed] [Google Scholar]
- 2.Abdrakhmanov I, Lodygin D, Geroth P, Arakawa H, Law A, Plachy J, Korn B, Buerstedde J M. A large database of chicken bursal ESTs as a resource for the analysis of vertebrate function. Genome Res. 2000;10:2062–2069. doi: 10.1101/gr.10.12.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akiyama Y, Kato S. Two cell lines from lymphomas of Marek's disease. Biken J. 1974;17:105–116. [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baba T W, Giroir B P, Humphries E H. Cell lines derived from avian lymphomas exhibit two distinct phenotypes. Virology. 1985;144:139–151. doi: 10.1016/0042-6822(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 7.Baeuerle P A. The inducible transcription activator NF-κB: regulation by distinct protein subunits. Biochim Biophys Acta. 1991;1072:63–80. doi: 10.1016/0304-419x(91)90007-8. [DOI] [PubMed] [Google Scholar]
- 8.Barkett M, Gilmore T D. Control of apoptosis by Rel/NF-κB transcription factors. Oncogene. 1999;18:6910–6924. doi: 10.1038/sj.onc.1203238. [DOI] [PubMed] [Google Scholar]
- 9.Baron S, Tyring S K, Fleischmann W R, Jr, Coppenhaver D H, Niesel D W, Klimpel G R, Stanton G J, Hughes T K. The interferons. Mechanisms of action and clinical applications. JAMA. 1991;266:1375–1383. doi: 10.1001/jama.266.10.1375. [DOI] [PubMed] [Google Scholar]
- 10.Bartunek P, Koritschoner N P, Brett D, Zenke M. Molecular cloning, expression and evolutionary analysis of the avian tyrosine kinase JAK1. Gene. 1999;230:129–136. doi: 10.1016/s0378-1119(99)00080-3. [DOI] [PubMed] [Google Scholar]
- 11.Bos T J, Monteclaro F S, Mitsunobu F, Ball A R, Jr, Chang C H, Nishimura T, Vogt P K. Efficient transformation of chicken embryo fibroblasts by c-Jun requires structural modification in coding and noncoding sequences. Genes Dev. 1990;4:1677–1687. doi: 10.1101/gad.4.10.1677. [DOI] [PubMed] [Google Scholar]
- 12.Bose H R, Jr, Levine A S. Replication of the reticuloendotheliosis virus (strain T) in chicken embryo cell culture. J Virol. 1967;1:1117–1121. doi: 10.1128/jvi.1.6.1117-1121.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brass A L, Kehrli E, Eisenbeis C F, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 14.Brass A L, Zhu A Q, Singh H. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J. 1999;18:977–991. doi: 10.1093/emboj/18.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bushdid P B, Brantley D M, Yull F E, Blauerer G L, Hoffman L H, Nisswander L, Kerr L D. Inhibition of NF-κB activity results in disruption of the apical ectodermal ridge and aberrant limb morphogenesis. Nature. 1998;392:615–618. doi: 10.1038/33435. [DOI] [PubMed] [Google Scholar]
- 16.Capobianco A J, Simmons D L, Gilmore T D. Cloning and expression of a chicken c-rel cDNA: unlike p59v-rel, p68c-relis a cytoplasmic protein in chicken embryonic fibroblasts. Oncogene. 1990;5:257–265. [PubMed] [Google Scholar]
- 17.Chang H W, Aoki M, Fruman D, Auger K R, Bellacosa A, Tsichlis P N, Cantley L C, Roberts T M, Vogt P K. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 18.Chang H W, Li J, Vogt P K. Domains of the qin protein required for oncogenic transformation. Oncogene. 1996;13:441–444. [PubMed] [Google Scholar]
- 19.Colmenares C, Sutrave P, Hughes S H, Stavnezer E. Activation of the c-skioncogene by overexpression. J Virol. 1991;65:4929–4935. doi: 10.1128/jvi.65.9.4929-4935.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeLorbe W J, Luciw P A, Goodman H M, Varmus H E, Bishop J M. Molecular cloning and characterization of avian sarcoma virus circular DNA molecules. J Virol. 1980;36:50–61. doi: 10.1128/jvi.36.1.50-61.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dosch E, Zöller B, Redmann-Müller I, Nanda I, Schmid M, Viciano-Gofferge A, Jungwirth C. The genomic structure of the chicken ICSBP gene and its transcriptional regulation by chicken interferon. Gene. 1998;210:265–275. doi: 10.1016/s0378-1119(98)00063-8. [DOI] [PubMed] [Google Scholar]
- 22.Eisenbeis C F, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 23.Filardo E J, Lee M F, Humphries E H. Structural genes, not the LTRs, are the primary determinants of reticuloendotheliosis virus A-induced runting and bursal atrophy. Virology. 1994;202:116–128. doi: 10.1006/viro.1994.1328. [DOI] [PubMed] [Google Scholar]
- 24.Fini M E. Keratocyte and fibroblast phenotypes in the repairing cornea. Prog Retin Eye Res. 1999;18:529–551. doi: 10.1016/s1350-9462(98)00033-0. [DOI] [PubMed] [Google Scholar]
- 25.Foo S Y, Nolan G P. NF-κB to the rescue: RELs, apoptosis and cellular transformation. Trends Genet. 1999;15:229–235. doi: 10.1016/s0168-9525(99)01719-9. [DOI] [PubMed] [Google Scholar]
- 26.Franklin R B, Kang C-Y, Wan K M-M, Bose J H R. Transformation of chick embryo fibroblasts by reticuloendotheliosis virus. Virology. 1977;83:313–321. doi: 10.1016/0042-6822(77)90176-3. [DOI] [PubMed] [Google Scholar]
- 27.Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, Hakoshima T. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J. 1999;18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao S J, Boshoff C, Jayachandra S, Weiss R A, Chang Y, Moore P S. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene. 1997;15:1979–1985. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- 29.Gerondakis S, Grumont R, Rourke I, Grossmann M. The regulation and roles of Rel/NF-κB transcription factors during lymphocyte activation. Curr Opin Immunol. 1998;10:353–359. doi: 10.1016/s0952-7915(98)80175-1. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh S, May M J, Kopp E B. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 31.Gil J, Alcami J, Esteban M. Activation of NF-κB by the dsRNA-dependent protein kinase, PKR involves the IκB kinase complex. Oncogene. 2000;19:1369–1378. doi: 10.1038/sj.onc.1203448. [DOI] [PubMed] [Google Scholar]
- 32.Gilmore T D. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein v-Rel. Oncogene. 1999;18:6925–6937. doi: 10.1038/sj.onc.1203222. [DOI] [PubMed] [Google Scholar]
- 33.Givol I, Greenhouse J J, Hughes S H, Ewert D L. Retroviruses that express different ras mutants cause different types of tumors in chickens. Oncogene. 1992;7:141–146. [PubMed] [Google Scholar]
- 34.Grilli M, Memo M. Nuclear factor-κB/Rel proteins: a point of convergence of signalling pathways relevant in neuronal function and dysfunction. Biochem Pharmacol. 1999;57:1–7. doi: 10.1016/s0006-2952(98)00214-7. [DOI] [PubMed] [Google Scholar]
- 35.Grossman A, Mittrücker H-W, Nicholl J, Suzuki A, Chung S, Antonio L, Suggs S, Sutherland G R, Siderovski D P, Mak T W. Cloning of human lymphocyte-specific interferon regulatory factor (hLSIRF/hIRF4) and mapping of the gene to 6p23–p25. Genomics. 1996;37:229–233. doi: 10.1006/geno.1996.0547. [DOI] [PubMed] [Google Scholar]
- 36.Grossmann M, Nakamura Y, Grumont R, Gerondakis S. New insights into the roles of ReL/NF-κB transcription factors in immune function, hemopoiesis and human disease. Int J Biochem Cell Biol. 1999;31:1209–1219. doi: 10.1016/s1357-2725(99)00068-0. [DOI] [PubMed] [Google Scholar]
- 37.Grumont R J, Gerondakis S. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: modulation of interferon-regulated gene expression by Rel/nuclear factor κB. J Exp Med. 2000;191:1281–1291. doi: 10.1084/jem.191.8.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas A F, Wong J W, Iwahashi C K, Halliwell B, Cross C E, Davis P A. Redox regulation of wound healing? NF-κB activation in cultured human keratinocytes upon wounding and the effect of low energy HeNe irradiation. Free Radical Biol Med. 1998;25:998–1005. doi: 10.1016/s0891-5849(98)00135-x. [DOI] [PubMed] [Google Scholar]
- 39.Harada H, Kitagawa M, Tanaka N, Yamamoto H, Harada K, Ishihara M, Taniguchi T. Anti-oncogenic and oncogenic potentials of interferon regulatory factors-1 and -2. Science. 1993;259:971–974. doi: 10.1126/science.8438157. [DOI] [PubMed] [Google Scholar]
- 40.Harada H, Taniguchi T, Tanaka N. The role of interferon regulatory factors in the interferon system and cell growth control. Biochimie. 1998;80:641–650. doi: 10.1016/s0300-9084(99)80017-0. [DOI] [PubMed] [Google Scholar]
- 41.Holtschke T, Löhler J, Kanno Y, Fehr T, Giese N, Knobeloch K-P, Gabriele L, Waring J F, Bachmann M F, Zinkernagel R M, Morse III H C, Ozato K, Horak I. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 42.Hrdličková R, Nehyba J, Bose J H R. Reticuloendotheliosis viruses (Retroviridae) In: Granoff A, Webster R G, editors. Encyclopedia of virology. Vol. 3. London: Academic Press; 1999. pp. 1496–1503. [Google Scholar]
- 43.Hrdličková R, Nehyba J, Humphries E H. In vivo evolution of c-reloncogenic potential. J Virol. 1994;68:2371–2382. doi: 10.1128/jvi.68.4.2371-2382.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hrdličková R, Nehyba J, Humphries E H. v-rel induces expression of three avian immunoregulatory surface receptors more efficiently than c-rel. J Virol. 1994;68:308–319. doi: 10.1128/jvi.68.1.308-319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hrdličková R, Nehyba J, Roy A, Humphries E H, Bose H R., Jr The relocalization of v-Rel from the nucleus to the cytoplasm coincides with induction of expression of Ikba and nfkb1and stabilization of IκB-α. J Virol. 1995;69:403–413. doi: 10.1128/jvi.69.1.403-413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes S H, Greenhouse J J, Petropoulos C J, Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iida S, Rao P H, Butler M, Corradini P, Boccadoro M, Klein B, Chaganti R S K, Dalla-Favera R. Deregulation of MUM1/IRF4by chromosomal translocation in multiple myeloma. Nat Genet. 1997;17:226–230. doi: 10.1038/ng1097-226. [DOI] [PubMed] [Google Scholar]
- 48.Jeanmougin F, Thompson J D, Gouy M, Higgins D G, Gibson T J. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 49.Joklik W K. Interferons. In: Fields B N, Knipe D M, editors. Fundamental virology. 2nd ed. New York, N.Y: Raven Press; 1991. pp. 343–370. [Google Scholar]
- 50.Jungwirth C, Rebbert M, Ozato K, Degen H J, Schultz U, Dawid I B. Chicken interferon consensus sequence-binding protein (ICSBP) and interferon regulatory factor (IRF) 1 genes reveal evolutionary conservation in the IRF family. Proc Natl Acad Sci USA. 1995;92:3105–3109. doi: 10.1073/pnas.92.8.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanegae Y, Tavares A T, Belmonte J C I, Verma I M. Role of Rel/NF-κB transcription factors during the outgrowth of the vertebrate limb. Nature. 1998;392:611–614. doi: 10.1038/33429. [DOI] [PubMed] [Google Scholar]
- 52.Kanno Y, Kozak C A, Schindler C, Driggers P H, Ennist D L, Gleason S L, Darnell J J E, Ozato K. The genomic structure of the murine ICSBP gene reveals the presence of the gamma interferon-responsive element, to which an ISGFα subunit (or similar) molecule binds. Mol Cell Biol. 1993;13:3951–3963. doi: 10.1128/mcb.13.7.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim S, Humphries E H, Tjoelker L, Carlson L, Thompson C B. Ongoing diversification of the rearranged immunoglobulin light-chain gene in a bursal lymphoma cell line. Mol Cell Biol. 1990;10:3224–3231. doi: 10.1128/mcb.10.6.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koerner T J, Hill J E, Myers A M, Tzagoloff A. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- 55.Köntgen F, Grumont R J, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Mice lacking the c-relproto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 56.Kopp E B, Ghosh S. NF-κB and Rel proteins in innate immunity. Adv Immunol. 1995;58:1–27. doi: 10.1016/s0065-2776(08)60618-5. [DOI] [PubMed] [Google Scholar]
- 57.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kralova J, Liss A S, Bargmann W, Bose H R., Jr AP-1 factors play an important role in transformation induced by the v-reloncogene. Mol Cell Biol. 1998;18:2997–3009. doi: 10.1128/mcb.18.5.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kralova J, Schatzle J D, Bargmann W, Bose H R., Jr Transformation of avian fibroblasts overexpressing the c-rel proto-oncogene and a variant of c-rellacking forty C-terminal amino acids. J Virol. 1994;68:2073–2083. doi: 10.1128/jvi.68.4.2073-2083.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar K P, McBride K M, Weaver B K, Dingwall C, Reich N C. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol Cell Biol. 2000;20:4159–4168. doi: 10.1128/mcb.20.11.4159-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lawrence C E, Altschul S F, Boguski M S, Liu J S, Neuwald A F, Wootton J C. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 62.Lewis R B, McClure J, Rup B, Niesel D W, Garry R F, Hoelzer J D, Nazerian K, Bose H R., Jr Avian reticuloendotheliosis virus: identification of the hematopoietic target cell for transformation. Cell. 1981;25:421–431. doi: 10.1016/0092-8674(81)90060-x. [DOI] [PubMed] [Google Scholar]
- 63.Li W, Nagineni C N, Efiok B, Chepelinsky A B, Egwuagu C E. Interferon regulatory transcription factors are constitutively expressed and spatially regulated in the mouse lens. Dev Biol. 1999;210:44–55. doi: 10.1006/dbio.1999.9267. [DOI] [PubMed] [Google Scholar]
- 64.Li W, Nagineni C N, Hooks J J, Chepelinsky A B, Egwuagu C E. Interferon-γ signaling in human retinal pigment epithelial cells mediated by STAT1, ICSBP, and IRF-1 transcription factors. Investig Ophthalmol Vis Sci. 1999;40:976–982. [PubMed] [Google Scholar]
- 65.Lubyova B, Pitha P M. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J Virol. 2000;74:8194–8201. doi: 10.1128/jvi.74.17.8194-8201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mamane Y, Heylbroeck C, Genin P, Algarte M, Servant M J, LePage C, DeLuca C, Kwon H, Lin R, Hiscott J. Interferon regulatory factors: the next generation. Gene. 1999;237:1–14. doi: 10.1016/s0378-1119(99)00262-0. [DOI] [PubMed] [Google Scholar]
- 67.Marecki S, Atchison M L, Fenton M J. Differential expression and distinct functions of IFN regulatory factor 4 and IFN consensus sequence binding protein in macrophages. J Immunol. 1999;163:2713–2722. [PubMed] [Google Scholar]
- 68.Matsuyama T, Grossman A, Mittrucker H W, Siderovski D P, Kiefer F, Kawakami T, Richardson C D, Taniguchi T, Yoshinaga S K, Mak T W. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE) Nucleic Acids Res. 1995;23:2127–2136. doi: 10.1093/nar/23.12.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mellon P, Parker V, Gluzman Y, Maniatis T. Identification of DNA sequences required for transcription of the human alpha 1-globin gene in a new SV40 host-vector system. Cell. 1981;27:279–288. doi: 10.1016/0092-8674(81)90411-6. [DOI] [PubMed] [Google Scholar]
- 70.Meric C, Spahr P F. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J Virol. 1986;60:450–452. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mittrücker H-W, Matsuyama T, Grossman A, Kündig T M, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi P S, Mak T W. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- 72.Moore B E, Bose H R., Jr Expression of the v-reloncogene in reticuloendotheliosis virus-transformed fibroblasts. Virology. 1988;162:377–387. doi: 10.1016/0042-6822(88)90478-3. [DOI] [PubMed] [Google Scholar]
- 73.Mora A L, Chen D, Boothby M, Rubin D H. Lineage-specific differences among CD8+ T cells in their dependence of NF-κB/Rel signaling. Eur J Immunol. 1999;29:2968–2980. doi: 10.1002/(SICI)1521-4141(199909)29:09<2968::AID-IMMU2968>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 74.Morrison L E, Boehmelt G, Beug H, Enrietto P J. Expression of v-relin a replication competent virus: transformation and biochemical characterization. Oncogene. 1991;6:1657–1666. [PubMed] [Google Scholar]
- 75.Morrison L E, Boehmelt G, Enrietto P J. Mutations in the rel-homology domain alter the biochemical properties of v-reland render it transformation defective in chicken embryo fibroblasts. Oncogene. 1992;7:1137–1147. [PubMed] [Google Scholar]
- 76.Moscovici C, Moscovici M G, Jimenez H, Lai M M, Hayman M J, Vogt P K. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell. 1977;11:95–103. doi: 10.1016/0092-8674(77)90320-8. [DOI] [PubMed] [Google Scholar]
- 77.Moscovici C, Zeller N, Moscovici M G. Continuous lines of AMV-transformed nonproducer cells: growth and oncogenic potential in the chick embryo. In: Revoltella R F, et al., editors. Expression of differentiated functions in cancer cells. New York, N.Y: Raven Press; 1982. pp. 435–449. [Google Scholar]
- 78.Mosialos G, Hamer P, Capobianco A J, Laursen R A, Gilmore T D. A protein kinase A recognition sequence is structurally linked to transformation by p59v-rel and cytoplasmic retention of p68c-rel. Mol Cell Biol. 1991;11:5867–5877. doi: 10.1128/mcb.11.12.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nagulapalli S, Atchison M L. Transcription factor Pip can enhance DNA binding by E47, leading to transcriptional synergy involving multiple protein domains. Mol Cell Biol. 1998;18:4639–4650. doi: 10.1128/mcb.18.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nazerian K, Stephens E A, Sharma J M, Lee L M, Gailitis M, Witter R I. A nonproducer T lymphoblastoid cell line from Marek's disease transplantable tumor (JMV) Avian Dis. 1977;21:69–76. [PubMed] [Google Scholar]
- 81.Nehyba J, Hrdličková R, Humphries E H. Evolution of the oncogenic potential of v-rel: rel-induced expression of immunoregulatory receptors correlates with tumor development and in vitro transformation. J Virol. 1994;68:2039–2050. doi: 10.1128/jvi.68.4.2039-2050.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nehyba J, Svoboda J, Karakoz I, Geryk J, Hejnar J. Ducks: a new experimental host system for studying persistent infection with avian leukaemia retroviruses. J Gen Virol. 1990;71:1937–1945. doi: 10.1099/0022-1317-71-9-1937. [DOI] [PubMed] [Google Scholar]
- 83.Nguyen H, Hiscott J, Pitha P M. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 84.Okuno H, Suzuki T, Yoshida T, Hashimoto Y, Curran T, Iba H. Inhibition of jun transformation by a mutated fosgene: design of an anti-oncogene. Oncogene. 1991;6:1491–1497. [PubMed] [Google Scholar]
- 85.Pahl H L. Activators and target genes of Rel/NF-κB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 86.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Petrenko O, Ischenko I, Enrietto P J. Characterization of changes in gene expression associated with malignant transformation by the NF-κB family member, v-Rel. Oncogene. 1997;15:1671–1680. doi: 10.1038/sj.onc.1201334. [DOI] [PubMed] [Google Scholar]
- 88.Pitha P M, Au W C, Lowther W, Juang Y T, Schafer S L, Burysek L, Hiscott J, Moore P A. Role of the interferon regulatory factors (IRFs) in virus-mediated signaling and regulation of cell growth. Biochimie. 1998;80:651–658. doi: 10.1016/s0300-9084(99)80018-2. [DOI] [PubMed] [Google Scholar]
- 88a.Plachy J, Weining K C, Kremmer E, Puehler F, Hala K, Kaspers B, Staeheli P. Protective effects of type I and type II interferons toward Rous sarcoma virus-induced tumors in chickens. Virology. 1999;256:85–91. doi: 10.1006/viro.1999.9602. [DOI] [PubMed] [Google Scholar]
- 89.Rao A, Kline K, Sanders B G. Immune abnormalities in avian ertythroblastosis virus-infected chickens. Cancer Res. 1990;50:4764–4770. [PubMed] [Google Scholar]
- 90.Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 91.Reboul J, Gardiner K, Monneron D, Uze G, Lutfalla G. Comparative genomic analysis of the interferon/interleukin-10 receptor gene cluster. Genome Res. 1999;9:242–250. [PMC free article] [PubMed] [Google Scholar]
- 92.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 93.Romero P, Humphries E H. A mutant v-relwith increased ability to transform B lymphocytes. J Virol. 1995;69:301–307. doi: 10.1128/jvi.69.1.301-307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rosenbauer F, Waring J F, Foerster J, Wietstruk M, Philipp D, Horak I. Interferon consensus sequence binding protein and interferon regulatory factor-4/Pip form a complex that represses the expression of the interferon-stimulated gene-15 in macrophages. Blood. 1999;94:4274–4281. [PubMed] [Google Scholar]
- 95.Saito T, Yamagata T, Takahashi T, Honda H, Hirai H. ICSAT overexpression is not sufficient to cause adult T-cell leukemia or multiple myeloma. Biochem Biophys Res Commun. 1999;260:329–331. doi: 10.1006/bbrc.1999.0911. [DOI] [PubMed] [Google Scholar]
- 96.Schaper F, Kirchhoff S, Posern G, Köster M, Oumard A, Sharf R, Levi B Z, Hauser H. Functional domains of interferon regulatory factor I (IRF-1) Biochem J. 1998;335:147–157. doi: 10.1042/bj3350147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schmidt M, Nagel S, Proba J, Thiede C, Ritter M, Waring J F, Rosenbauer F, Huhn D, Wittig B, Horák I, Neubauer A. Lack of interferon consensus sequence binding protein (ICSBP) transcripts in human myeloid leukemia. Blood. 1998;91:22–29. [PubMed] [Google Scholar]
- 98.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 99.Schultz U, Rinderle C, Sekellick M J, Marcus P I, Staeheli P. Recombinant chicken interferon from Escherichia coliand transfected COS cells is biologically active. Eur J Biochem. 1995;229:73–76. doi: 10.1111/j.1432-1033.1995.0073l.x. [DOI] [PubMed] [Google Scholar]
- 100.Schwartz R C, Witte O N. A recombinant murine retrovirus expressing v-relis cytopathic. Virology. 1988;165:182–190. doi: 10.1016/0042-6822(88)90671-x. [DOI] [PubMed] [Google Scholar]
- 101.Sick C, Schultz U, Munster U, Meier J, Kaspers B, Staeheli P. Promoter structures and differential responses to viral and nonviral inducers of chicken type I interferon genes. J Biol Chem. 1998;273:9749–9754. doi: 10.1074/jbc.273.16.9749. [DOI] [PubMed] [Google Scholar]
- 102.Sick C, Schultz U, Staeheli P. A family of genes coding for two serologically distinct chicken interferons. J Biol Chem. 1996;271:7635–7639. doi: 10.1074/jbc.271.13.7635. [DOI] [PubMed] [Google Scholar]
- 103.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 104.Stoker A W, Bissell M J. Quantitative immunocytochemical assay for infectious avian retroviruses. J Gen Virol. 1987;68:2481–2485. doi: 10.1099/0022-1317-68-9-2481. [DOI] [PubMed] [Google Scholar]
- 105.Stone E M, Rothblum K N, Alevy M C, Kuo T M, Schwartz R J. Complete sequence of the chicken glyceraldehyde-3-phosphate dehydrogenase gene. Proc Natl Acad Sci USA. 1985;82:1628–1632. doi: 10.1073/pnas.82.6.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tirunagaru V G, Sofer L, Cui J, Burnside J. An expressed sequence tag database of T-cell-enriched activated chicken splenocytes: sequence analysis of 5251 clones. Genomics. 2000;66:144–151. doi: 10.1006/geno.2000.6189. [DOI] [PubMed] [Google Scholar]
- 107.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 108.Yamagata T, Nishida J, Tanaka T, Sakai R, Mitani K, Yoshida M, Taniguchi T, Yazaki Y, Hirai H. A novel interferon regulatory factor family transcription factor, ICSAT/Pip/LSIRF, that negatively regulates the activity of interferon-regulated genes. Mol Cell Biol. 1996;16:1283–1294. doi: 10.1128/MCB.16.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yamamoto A, Iwata A, Koh Y, Kawai S, Murayama S, Hamada K, Maekawa S, Ueda S, Sokawa Y. Two types of chicken 2′,5′-oligoadenylate synthetase mRNA derived from alleles at a single locus. Biochim Biophys Acta. 1998;1395:181–191. doi: 10.1016/s0167-4781(97)00148-6. [DOI] [PubMed] [Google Scholar]
- 110.Yoshida S, Nakazawa N, Iida S, Hayami Y, Sato S, Wakita A, Shimizu S, Taniwaki M, Ueda R. Detection of MUM1/IRF4-IgH fusion in multiple myeloma. Leukemia. 1999;13:1812–1816. doi: 10.1038/sj.leu.2401563. [DOI] [PubMed] [Google Scholar]
- 111.Zhang J Y, Bose H R., Jr Acquisition of new proviral copies in avian lymphoid cells transformed by reticuloendotheliosis virus. J Virol. 1989;63:1107–1115. doi: 10.1128/jvi.63.3.1107-1115.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang L, Pagano J S. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zong C S, Chan J L-K, Yang S-K, Wang L-H. Mutations of Ros differentially effecting signal transduction pathways leading to cell growth versustransformation. J Biol Chem. 1997;272:1500–1506. doi: 10.1074/jbc.272.3.1500. [DOI] [PubMed] [Google Scholar]